Abstract
Rhizobium etli modifies lipopolysaccharide (LPS) structure in response to environmental signals, such as low pH and anthocyanins. These LPS modifications result in the loss of reactivity with certain monoclonal antibodies. The same antibodies fail to recognize previously isolated R. etli mutant strain CE367, even in the absence of such environmental cues. Chemical analysis of the LPS in strain CE367 demonstrated that it lacked the terminal sugar of the wild-type O antigen, 2,3,4-tri-O-methylfucose. A 3-kb stretch of DNA, designated as lpe3, restored wild-type antigenicity when transferred into CE367. From the sequence of this DNA, five open reading frames were postulated. Site-directed mutagenesis and complementation analysis suggested that the genes were organized in at least two transcriptional units, both of which were required for the production of LPS reactive with the diagnostic antibodies. Growth in anthocyanins or at low pH did not alter the specific expression of gusA from the transposon insertion of mutant CE367, nor did the presence of multiple copies of lpe3 situated behind a strong, constitutive promoter prevent epitope changes induced by these environmental cues. Mutations of the lpe genes did not prevent normal nodule development on Phaseolus vulgaris and had very little effect on the occupation of nodules in competition with the wild-type strain.
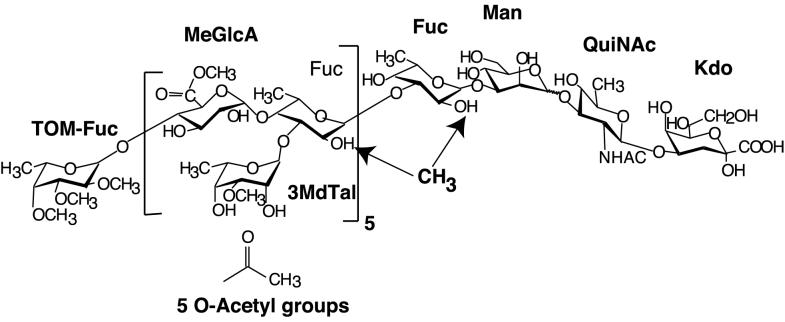
The carbohydrate backbones of lipopolysaccharides (LPSs) of gram-negative bacteria often are highly decorated with substituent chemical groups. Striking examples can be found among Rhizobium spp. and related bacteria (28). For instance, the LPS O chain of Rhizobium etli CE3 (18) is heavily substituted with moieties that should confer hydrophobic character: O-methylations, O- and N-acetylations, and esterification of a repeating carboxyl group (Fig. 1). The hypothetical hydrophobicity is most pronounced at the nonreducing end, where the O-chain repeating units are capped by a terminal deoxysugar in which all of the hydroxyl groups are methylated.
FIG. 1.
Structure of O chain of R. etli CE3. The methylation of fucose residues, particularly the internal fucoses, is variable, whereas the 3-O-methylation of 6-deoxytalose and the methyl esterification of glucuronic acid is invariant. The QuiNAc-Kdo disaccharide at the reducing end is partially and variably degraded during mild acid hydrolysis. The locations of the O-acetyl groups are unknown. Abbreviations: TOM-Fuc, 2,3,4-tri-O-methylfucose; MeGlcA, glucuronyl methyl ester; Fuc, fucose; 3MdTal, 3-O-methyl-6-deoxytalose; Man, mannose; QuiNAc, 2-N-acetamido-2,6-dideoxyglucose (2-N-acetylquinovosamine); Kdo, 3-deoxy-d-manno-2-octulonic acid.
A number of interesting questions arise from considering these substituents. One is the mechanism of synthesis; for instance, whether the O-methyl, methyl ester, and O-acetyl groups are added during synthesis of the nucleotide diphosphosugars or after polymerization of the sugar residues. Another issue is the functions of these substituents. With bacteria such as rhizobia, which are known to interact intimately with multicellular hosts, one question is whether such decorations influence these cell-cell interactions.
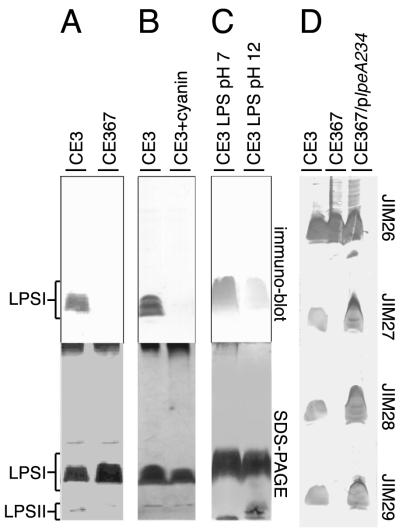
The LPS structures of R. etli and Rhizobium leguminosarum change during the course of infection of their legume hosts and in response to environmental cues, such as plant-released anthocyanins, low pH, and low oxygen concentrations (28, 33). Whether these changes are required for successful bacterial-host interaction remains to be determined. In the case of R. etli CE3, detergent gel electrophoresis and sugar composition analyses indicate that the LPS structure has been altered only slightly after growth in these conditions (16, 34, 40), leading to speculation that the changes involve the chemical substituents that decorate the main carbohydrate backbone. The main tools in tracking these induced LPS changes have been three monoclonal antibodies (MAbs). Depending on the particular LPS alteration, one or more of these antibodies exhibit greatly decreased affinity or do not bind at all to the altered LPS (16, 34, 40) (e.g., Fig. 2B).
FIG. 2.
LPS antigenicity of mutant CE367 and wild-type CE3 after various treatments. Purified LPS or LPS in cell lysates was subjected to SDS-PAGE, electroblotted onto nitrocellulose, and probed with MAbs. (A to C) The blot was probed with MAb JIM28 (immunoblot), and the lower images show meta-periodate-silver staining of the residual components in the SDS-PAGE gel after blotting. Bands are labeled LPS I (containing O chain) or LPS II (lacking the O chain) as established in previous work (8). (A) R. etli CE3 and Tn5gus-mutant CE367 cells were grown in TY medium. Approximately equal numbers of cells of each strain were lysed in SDS-containing buffer, and the lysates were separated by SDS-PAGE. (B) R. etli CE3 cells were grown in TY medium or TY supplemented with 50 μM cyanin (CE3+cyanin). The cultured bacteria were then processed for SDS-PAGE as described above. (C) The LPS of strain CE3 purified by Sepharose 4B chromatography was incubated in SDS-PAGE buffer titrated to pH 7 or 12 with NaOH at room temperature for 1 h before analysis by SDS-PAGE and immunoblotting. (D) The LPS I regions of four blots are shown after being probed with MAb JIM26, JIM27, JIM28, or JIM29. Strains CE3, CE367, and CE367, carrying plpeA234, were grown in TY medium, and SDS lysates of the cells were processed as described above.
By screening for mutants of R. etli CE3 that are not recognized by one of these MAbs in the absence of such environmental cues, mutant strain CE367 was isolated in a previous study (40). The LPS of this mutant appeared to migrate normally on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, suggesting that the overall structure had suffered very little truncation, but it was not recognized by any of the three antibodies after growth under any condition (Lpe− [lipopolysaccharide epitopes] phenotype) (Fig. 2). Studying the defects of CE367, therefore, should provide insight into the process of the maturation of the R. etli LPS into the fully realized O antigen that is presented on the bacterial surface and insight into the function of the structural feature that the mutant lacks. When analysis of this strain began, it also seemed plausible that its deficiency might correlate with one of the environmentally induced changes in LPS structure.
The present report describes the cloning and genetic analyses of a gene cluster (lpe3) in which the mutation of strain CE367 is located. The LPS structure of mutant CE367 was analyzed in detail and found to lack the terminal tri-O-methylated fucose of the wild type. The effect of this structural deficiency on symbiosis with Phaseolus vulgaris also was assessed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. Rhizobium cultures were grown at 30°C in a rotating shaker at 150 rpm in TY liquid medium (tryptone, yeast extract, and CaCl2), in defined YGM medium at pH 5 as described previously (16, 40), or in YGM medium titrated to pH 7 with NaOH. Cells of Escherichia coli strains were grown in Luria-Bertani medium at 37°C supplemented with the appropriate antibiotic (kanamycin [50 μg/ml], gentamicin [20 μg/ml], ampicillin [100 μg/ml], chloramphenicol [30 μg/ml]). Cyanin, delphinidin, and seed exudate preparations for induction of the LPS modification were performed as described previously (16, 34).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 23 |
| MT616 | pro thi endA hsdR supE44 recA56, pRK2013Km::Tn9 | 17 |
| Rhizobium etli | ||
| CE3 | str-1 Lpe+ Ndv+ Fix+ | 36 |
| CE367 | str-1 lpeA367::Tn5-gusA1 Ndv+ Fix+ | 40 |
| CE426 | str-1 mTn5SsgusA11 (inserted at unknown site) Lpe+ Ndv+ Fix+ | This work |
| CE430 | str-1 ΔlpeA::Km Ndv+ Fix+ | This work |
| CE431 | str-1 ORF2::Km Ndv+ Fix+ | This work |
| CE432 | str-1 Ω(pJQ200::Km::ΔlpeA insert at XbaI site of lpe3) Lpe+ Ndv+ Fix+ | This work |
| CE445 | str-1 lpeA::Km Ndv+ Fix+ | This work |
| CE462 | str-1 ORF3::Km Ndv+ Fix+ | This work |
| Plasmids | ||
| pBluescript | Cloning vector | Stratagene |
| pRK404E1 | incP, Tcr, with second EcoRI site of pRK404 (15) eliminated | G. Roberts |
| pJQ200SK− | Suicide plasmid, Gmr, sacB traJ lacZα | 37 |
| pJQ200uCi | Suicide plasmid, Gmr, sacB traJ lacZα | 37 |
| pUC4K | Contains Kmr cassette used for generation of lpe mutants | Pharmacia |
| PCAM111 | Smr Spr mTn5SsgusA11 (carried in pUT vector) | 45 |
| pSB504 | pRK600Ω::Tn5-gusA7 | 39 |
::Km, insertion of Kanamycin resistance cassette from pUC4K; ORF2 and ORF3 are ORFs within lpe3 not shown to be required individually for Lpe+; Ndv+ Fix+, elicits normally developed nitrogen-fixing nodules on P. vulgaris.
PAGE and analysis of LPS antigenicity.
MAbs JIM26, JIM27, JIM28, and JIM29 have been described previously (16, 34, 40) and were a generous gift from N. J. Brewin (Norwich, United Kingdom). The routine analyses of the LPS antigenicity of SDS-extracted LPS or rhizobial colonies, SDS-PAGE, electrotransfer, and probing of electroblotted nitrocelluose or colony lifts with MAbs were performed as described previously (16, 34). While the data presented here are from assays of LPS separated by PAGE in the presence of detergents (e.g., see Fig. 2), the lack of antibody binding was also confirmed by performing colony blottings and dot blottings of intact cells from liquid cultures. For analysis by taurodeoxycholate (TDOC) PAGE, SDS extracts of the R. etli strains were separated on an 18% polyacrylamide gel in which TDOC was substituted for SDS at the same concentration (wt/vol) (27). Electrophoresis was at 75 V for 60 min, followed by 110 V for 105 min, and staining was performed as described previously (8).
Analysis of LPS structure.
Strains CE3 and CE367 were grown in TY medium in 60- and 300-liter fermenter batches, washed bacterial pellets were extracted by the hot-phenol method, and the LPS was purified by Sepharose 4B chromatography as described previously (8, 9). Purified LPS was mild acid hydrolyzed in 1% acetic acid at 100°C for 2 h, and lipid A was removed by centrifugation. The O-chain polysaccharide was separated from the core oligosaccharides by size exclusion chromatography of the supernatant by using Bio-Gel P2 equilibrated with deionized water.
Glycosyl composition was analyzed by the combined gas chromatography-mass spectrometry (GC-MS) of alditol acetates and trimethylsilyl methyl glycosides (46). Glycosyl linkage analysis was performed by permethylation (Hakomori method), conversion to partially methylated alditol acetates (PMAAs), and GC-MS analysis (46). The uronic acid linkages were identified by sequential permethylation reduction of the carboxymethyl groups with lithium triethylborodeuteride and conversion to the PMAAs (19). The location of endogenous O-methyl ether groups was determined by methylation analysis using trideuteromethyliodide.
Matrix-assisted laser desorption ionization (mass spectrometry) (MALDI [MS])) was performed using a Kratos Analytical Kompact SEQ MALDI-time-of-flight (TOF) spectrometer system in the positive mode. Approximately 1 μl of a 1-mg/ml LPS solution was mixed with 1 μl of the dihydroxybenzoic acid matrix and applied to the probe for analysis. Nuclear magnetic resonance (NMR) analysis was performed using either a Varian 300 or 600 MHz spectrometer. The sample was dissolved in D2O and lyophilized. This step was repeated, and then the sample was dissolved in D2O and analyzed at 29°C. Chemical shifts were measured relative to that of the HOD signal (* 4.78) which was assigned relative to sodium trimethylsilylpropionate (TSP).
Recombinant DNA techniques.
Genomic DNA was isolated from R. etli strains by a method employing cetyltrimethyl ammonium bromide (CTAB) (4) for use in cloning (38) or Southern blot analyses (4). E. coli cells were transformed (23) and plasmids were isolated from E. coli (14) as previously described. DNA was recovered from agarose gels (6) and amplified or modified with enzymes purchased from New England Biolabs (Beverly, Mass.). Custom primers were synthesized by Operon Technologies (Alameda, Calif.).
Cloning of lpe3.
An inverse PCR strategy was used to isolate wild-type DNA regions adjacent to the Tn5-gus insertion in CE367. R. etli DNA flanking one side of the Tn5gusA1 (39) of CE367 (pE1-Tn5; Fig. 3) was cloned from an EcoRI digest of CE367 genomic DNA by virtue of conferring kanamycin resistance after ligation into pBluescript KS(−) and transformation of E. coli DH5α cells. The R. etli genomic portion was sequenced by use of T7 and GUS sequencing primers (United States Biochemical, Cleveland, Ohio). By Southern blotting, the DNA sequence hybridized to a single 3-kb fragment of CE3 genomic DNA that had been digested with HindIII.
FIG. 3.
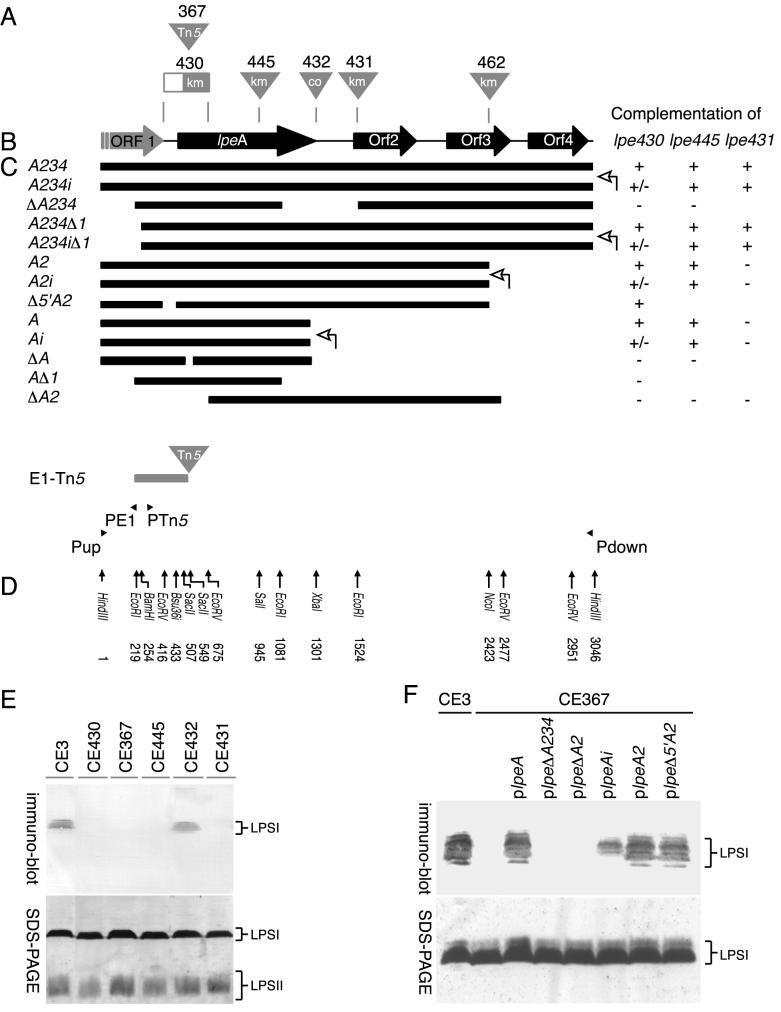
lpe3 genetic locus. (A) Sites of mutations. Triangles represent insertions. The rectangle depicting the mutation in strain CE430 indicates deleted lpe DNA (nt 416 to 675) as well as the site of the kanamycin resistance cassette insertion. Tn5, insertion of the Tn5gus transposon into lpeA in strain CE367; km, insertion of a kanamycin resistance cassette at the indicated position; co, the cointegration of plasmid pJQ200SK− at the indicated site. (B) Representation of the ORFs (large arrows) inferred from the DNA sequence of lpe3. (C) The solid bars represent the extents of R. etli DNA cloned into broad-host-range plasmid pRK404E1 and used for complementation analyses. In the text, the name of each construct has the prefix plpe, which was omitted in this figure. The lac promoter of pRK404E1 is on the left unless indicated otherwise with an open arrow on the right (on the plasmids with suffix “i”). To the right of each construct are tabulated the phenotypes conferred when this DNA was transferred into CE430, CE445, or CE431. (+, +/−, and − indicate strong, weak, and no binding, respectively, with MAb JIM28. See panel F for examples. Where there is no entry, complementation was not tested.) The solid arrowheads in the lower portion of the panel indicate the relative genomic position of the PCR primers used to clone the genomic sequence from R. etli CE3. E1-Tn5 depicts the EcoRI fragment cloned from strain CE367 in the first step of cloning the lpe3 DNA. (D) Positions within the lpe3 sequence of restriction sites referred to in the text. (E) The effects of mutations in lpeA (CE430, CE367, and CE445), the region between lpeA and ORF2 (CE432), or ORF2 (CE431). After growth of the strains in TY medium, the LPS was extracted, separated by SDS-PAGE (below), blotted, and reacted with MAb JIM28 (above) as described in Fig 2. (F) Examples of the complementation results tabulated in panel C. The LPS of strain CE3, strain CE367 (lane 2), or transconjugants of CE367 carrying the indicated lpe3 deletion constructs was analyzed as described for panel E.
Total DNA from a HindIII digest of CE3 genomic DNA that had been religated in a large volume was linearized with BamHI. The fragment of interest (DNA locus lpe3) was isolated by an inverse PCR approach using Vent DNA polymerase. The diverging primers used were PE1 (5′-GTGGTACCTCGAGGAACAAACCGTAAGGCCA-3′) and PTn5 (5′-ACCTGCAGGATCCTACCTGCTGGGTGGATTC-3′), which each anneal to sequences flanking opposite sides of a unique BamHI site of lpe3 at nucleotide (nt) 254, with their 5′ ends proximal to the BamHI-site (Fig. 3). Sequence analysis of the resulting PCR product cloned into pJQ200SK− (37) with a T3 sequencing primer (Promega, Madison, Wis.) gave the nucleotide sequence proximal to both of the lpe3 HindIII sites (nt 1 and 3046; Fig. 3).
The primers Pup (5′-TCACTCGAGAAGCTTGATGCATTCATTGAACTGCG-3′) and Pdown (5′-TCTCTAGAGCTCGAACGAACTGAATGTCGACC-3′) were designed to these sequences to amplify the lpe3 region from HindIII-digested CE3 genomic DNA (Fig. 3). This PCR product was ligated into pBluescript KS(−) for analysis and manipulations.
Sequence analysis of lpe3.
Sequence data was obtained by sequencing overlapping EcoRI-, EcoRV-, NcoI-, and XbaI-digested DNA fragments subcloned into pBluescript KS(−) as double-stranded templates with T7 and T3 primers. The sequence was obtained by a combination of the manual Fidelity DNA Sequencing System (Oncor, Md.) using α-35S-labeled dATP or automated sequencing at the University of Wisconsin—Milwaukee (ABI Prism; Perkin-Elmer, Norwalk, Conn.). Open reading frames (ORFs) were identified with Orf Finder (http://www.ncbi.nlm.nih.gov/gOrf/gOrf.html). Various BLAST searches of the nonredundant sequence database and the database of unfinished microbial genomes at the National Center for Biotechnology Information (NCBI) were performed with nucleotide and deduced amino acid sequences as query sequences (http://www.ncbi.nlm.nih.gov [1, 2]). Analysis of the inferred protein structures based on the deduced amino acid sequence were performed through the Baylor College of Medicine Search Launcher (http://www.hgsc.bcm.tmc.edu /SearchLauncher/) and the ExPASy Molecular Biology Server (http://www.expasy.ch/). Coil predictions were performed (http://www.isrec.isb-sib.ch) according to methods described by Lupas et al. (30). ORFs were compared to each other (http://bibiserv.techfak.uni-bielefeld.de/dialign/), consensus analysis was performed (http://www.toulouse.inra.fr/multalin.html), pairwise alignments of the identified ORFs were performed (http://www.ch.embnet.org/software/LALIGN_form.html), and the consensus was calculated (http://pbil.univ-lyon1.fr/). Transmembrane prediction analyses were performed online (http://psort.nibb.ac.jp:8800/) (32) and elsewhere (7, 24, 25, 29).
Site-directed mutagenesis and complementation analyses.
Site-directed mutagenesis was performed by insertional ligation of the Km cassette of pUC4K (Pharmacia) (42) at the deletion between nt 416 and 675 (EcoRV) to obtain mutation ΔlpeA430::Km, at nt 945 (SalI) for mutation lpeA445::Km, nt 1524 (EcoRI) for mutation of ORF2, lpe-431::Km, and nt 2423 (NcoI) for mutation of ORF3, lpe-462::Km (Fig. 3). lpe3 DNA fragments containing these insertions were subcloned into suicide plasmid pJQ200uci or pJQ200SK− (37). E. coli DH5α was transformed with these constructs. The constructs were transferred into R. etli CE3 cells by triparental mating (22) with mobilizing strain MT616 (17). CE3 transconjugants containing these constructs were selected on TY agar plates supplemented with 50 μg of kanamycin/ml, 20 μg of nalidixic acid/ml, 100 μg of streptomycin/ml, and 20 μg of gentamicin/ml. Double recombinants were selected on TY agar plates supplemented with 50 μg of kanamycin/ml, 20 μg of nalidixic acid/ml, 100 μg of streptomycin/ml, and 8% sucrose. At least 50 putative double recombinants were tested for their antigenicity to the MAbs by colony blottings, and 10 were tested by probing the antigenicity of blotted LPS after separation of SDS extracts of TY-cultured cells on SDS-PAGE, as described above. In each case, >95% of the colonies tested had the same antigenic phenotype.
Strain CE432 was the result of a single recombination event that inserted a complex cointegrate at nt 1301 (XbaI) of the lpe3 locus (Fig. 3). Included within this insertion was pJQ200SK−, ligated to the first 30 nt of lpe3, followed by the Km cassette and then a fragment of lpe3 from nt 945 to 1301.
For complementation analyses of these mutants, deletion fragments of lpe3 derived from various restriction digests (Fig. 3) were cloned into vector pRK404E1. Recovery of the Lpe+ phenotype was tested after introduction of the cloned deletion fragment into an Lpe− mutant by triparental mating. At least 50 tetracycline-resistant transconjugant colonies were tested for their Lpe phenotype by using colony blottings. In each case, the electroblotted LPSs of three or more purified transconjugant clones were also probed for reaction with MAb JIM28 after SDS-PAGE separation of SDS extracts of cells cultured in liquid TY.
Gus activity.
Strains carrying gusA fusions were grown in TY liquid medium to stationary phase. The cultures were diluted to 2% (vol/vol) in 50 ml of fresh YGM medium (pH 5.0 or 7.0) in 150-ml Erlenmeyer flasks and shaken at 200 rpm for 0 to 5 h at 30°C. At indicated time points, the cultures were collected by centrifugation at 12,000 × g for 10 min and snap frozen. Samples containing equal amounts of protein were separated by SDS-PAGE (50 μg of total protein/lane) for assessment of JIM28 antigenicity. Other aliquots of the same sample were washed twice in 50 mM sodium phosphate, pH 7.0, resuspended in phosphate buffer, and lysed in SDS and chloroform. The aqueous supernatants were resuspended at equal protein concentrations, and β-glucuronidase (GUS) activities of aliquots of the lysed cells were assessed by colorimetric assay as described previously (20). GUS activity was standardized to protein concentration as measured with the bicinchoninic acid assay (Pierce). Both the GUS and protein colorimetric assays were measured on an EL340 microtiter reader (Bio-Tek Instruments).
Nodulation assay.
For competition assays, CE3 and CE367 cells were grown to 8.5 × 108 and 1.0 × 109 CFU/ml, respectively, in TY, and 102 to 104 CFU were resuspended in 150 μl of plant nutrient solution (RBN) (36) and applied onto seeds that were surface sterilized with ethanol and planted in RBN agar (36) in 250-ml Erlenmeyer flasks. Nodules were harvested 20 days after inoculation, and the released bacteria were counted using the Miles and Misra drop-plate method (44) by spotting extracts onto TY plates supplemented with streptomycin, nalidixic acid, and kanamycin to assess the relative abundance of mutant strain CE367. The colonies were also tested with JIM28 to verify the Lpe− phenotype of the recovered bacteria.
Nucleotide sequence accession number.
The sequencing data has been submitted to GenBank as “lpe” under accession number AF333486.
RESULTS
SDS-PAGE, immunochemical, and structural analyses of the LPS of strain CE367.
The LPS of mutant strain CE367 did not react with MAb JIM27, JIM28, or JIM29 after growth under any conditions, including those that lead wild-type strain CE3 to produce LPS that is a strong antigen for these antibodies (Fig. 2). The LPS of this mutant also reacted weakly to total rabbit antisera generated against the LPS of CE3 cells grown in TY, indicating that the major epitopes of CE3 LPS are missing from CE367 (data not shown). On the other hand, the mutation had little effect on LPS migration in SDS-PAGE gels (Fig. 2A) or binding to antibody JIM26 (Fig. 2D). Nor was the proportion of LPS I and LPS II altered in the mutant, indicating that the LPS population carried near-normal amounts of O antigen. In terms of decreased reaction with MAb JIM28, the effect of the mutation in strain CE367 is similar to various treatments of wild-type strain CE3 and its LPS, including growth at low pH or in anthocyanins (Fig. 2B) and incubation of the isolated LPS at pH 12 (Fig. 2C). The antigenic properties of the mutant LPS were stable to phenol-water extraction, nuclease and protease treatment, and subsequent purification by Sepharose 4B chromatography.
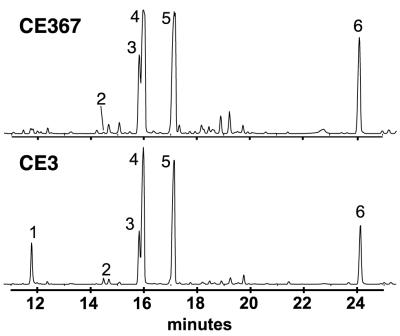
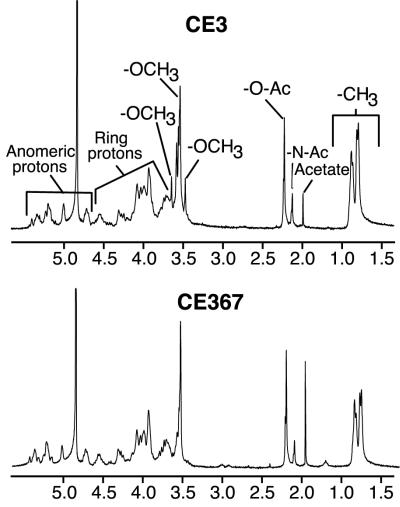
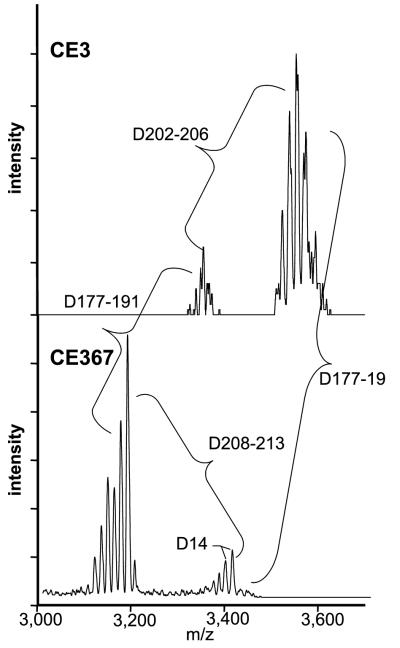
The O chain of the purified CE367 LPS completely lacked 2,3,4-tri-O-methylfucose (TOM-Fuc) and its level of 2,3-di-O-methylfucose (DOM-Fuc) was greatly decreased (Fig. 4). Otherwise, the sugar composition of the mutant LPS was very similar to that of CE3 LPS (Table 2). In the NMR spectra for the CE3 and CE367 O chains (Fig. 5), the only obvious difference was the absence of several of the minor -OCH3 proton resonances in the mutant spectrum. This result is consistent with the absence of the TOM-Fuc and DOM-Fuc residues in the mutant. Figure 6 shows the MALDI-TOF (MS) spectra for CE3 and CE367 O-chain polysaccharides. In each spectrum, there were two sets of peaks, corresponding to the intact O chain and the O chain truncated by the loss of the Kdo residue (18). Clearly, the mutant O chain was smaller and the mass difference was consistent with the size of the TOM-Fuc residue.
FIG. 4.
GC-MS total ion chromatographs of the alditol acetates derived from the O-chain polysaccharides from strains CE367 and CE3. Peak 1, 2,3,4-tri-O-methylfucose; peak 2, 2,3-di-O-methylfucose; peak 3, 2-O-methylfucose; peak 4, 3-O-methyl-6-deoxytalose; peak 5, fucose; peak 6, mannose. The rest of the two chromatographs were very similar, including the QuiNAc peak, which emerged at a much later retention time (not shown). The very small signal at the position of peak 1 in the case of CE367 did not have the MS of TOM-Fuc, and by ion scanning of the chromatogram, TOM-Fuc was undetectable.
TABLE 2.
Sugar composition of CE367 lipopolysaccharide
| Sugarb | Mole ratios found in straina
|
|
|---|---|---|
| CE367 | CE3 | |
| TOM-Fuc | NDc | 0.7d |
| DOM-Fuc | Trace | 0.1d |
| QuiN | 0.8 | 0.8 |
| 2OMeFuc | 0.9 | 0.9 |
| 3OMe6dTal | 3.3 | 2.9 |
| Fuc | 3.4 | 3.1 |
| GlcA | 3.2 | 3.1 |
| Man | 1.8 | 1.7 |
| Gal | 1.0 | 1.0 |
| GalA | 2.8 | 2.8 |
| Kdo | 1.8 | 1.7 |
| GlcN | 0.9 | 0.9 |
Entries are the molar ratios of the sugars normalized to 1.0 galactose.
Underlined sugars are found only in the portion of the LPS operationally designated as the O antigen (Fig. 1). QuiN, quinovosamine; 2OMeFuc, 2-O-methylfucose; 3OMe6dTal, 3-O-methyl-6-deoxytalose; Fuc, fucose; GlcA, glucuronic acid; Man, mannose; Gal, galactose; GalA, galacturonic acid; Kdo, 3-deoxy-d-manno-2-octulonic acid; GlcN, glucosamine.
ND, none detected (<0.01).
FIG. 5.
Proton NMR spectra of O chains from strains CE3 and CE367. The resonances were assigned as indicated.
FIG. 6.
Positive-mode MALDI-TOF (MS) spectra of O chains from strains CE3 and CE367. The masses are as indicated. The expected mass difference conferred by the absence of the TOM-Fuc residue is 188. The spacing between the two sets of peaks in each spectrum (202 to 206 and 208 to 213) is consistent with the loss of a Kdo residue (anhydro Kdo, 202 amu; Kdo, 220 amu). The range in mass differences is likely due to the heterogeneity introduced due to the presence of a mixture of molecules with Kdo, anhydro-Kdo, and no Kdo. The spacing indicated by Δ14 may represent heterogeneity in O-methylation (the difference between a hydroxyl and -OCH3 being 14 amu).
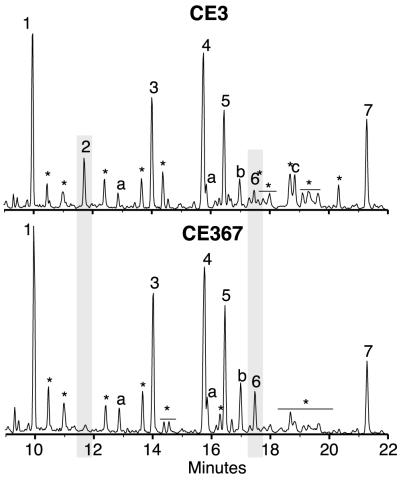
Figure 1 shows the structure of the CE3 O-chain polysaccharide as reported by Forsberg et al. (18). In that structure, the nonreducing end of the polysaccharide is capped with the TOM-Fuc residue, which is linked directly to the glucuronyl (GlcA) residue in the O-chain repeat unit. All of the above data were consistent with the possibility that the CE367 O chain lacks not only the methyl groups of TOM-Fuc but the entire residue, in which case the O chain should end in a GlcA residue. As a further test of this possibility, the CE367 and CE3 O chains were methylated using trideuteromethyliodide. Figure 7 shows a GC profile of the PMAAs obtained for both CE3 and CE367 O-chain polysaccharides. The glycosyl linkages observed for the CE3 O chain were consistent with those previously reported (18). As expected, the CE3 O chain contained endogenously methylated TOM-Fuc, as evidenced by the presence of 1,5-di-O-acetyl-2,3,4-tri-O-methylfucitol (Fig. 7, peak 2, with signature ions of m/z 118, 131, 162, and 175 in its mass spectrum). Scanning for these ions confirmed that this PMAA was completely missing in the O chain from CE367. Instead, there was a concomitant increase in terminally linked GlcA, as evidenced by the presence of 1,5,6-tri-O-acetyl-2,3,4-tri-O-deuteromethyl-6,6-dideuterioglucitol (m/z 121, 168, 194, and 241; Fig. 7, peak 6). These data suggest that the CE367 O-chain polysaccharide ends with a terminal GlcA residue at the nonreducing end instead of the TOM-Fuc residue present in the CE3 O chain.
FIG. 7.
GC-MS total ion chromatographs of the PMAAs derived from the trideuteriomethyliodide methylation of the CE3 and CE367 O-chain polysaccharides. The PMAAs of the glycosyl residues are as follows. Peak 1, terminally linked 3-O-methyl-6-deoxytalose; peak 2, 2,3,4-tri-O-methylfucose; peak 3, 3-linked fucose (and 3-linked 2-O-methylfucose); peak 4, 3,4-linked fucose (and 3,4-linked 2-O-methylfucose); peak 5, 3-linked mannose; peak 6, terminally linked glucuronic acid; peak 6*, a small amount of terminally linked glucuronic acid mixed in with some non-carbohydrate material; peak 7, 4-linked glucuronic acid; peaks a, undermethylated 3-O-methyl-6-deoxytalosyl residues; peaks b, undermethylated fucosyl residues; peak c, a small amount of galacturonic acid presumably due to slight contamination of the O antigen with core oligosaccharides; peaks ∗, non-carbohydrate contaminants from the derivatization process.
The methylation data show that both O chains have the same degree of variable endogenous methylation of the 3,4- and 3-linked Fuc residues. For both O chains, the PMAA derivatives from the 3-linked Fuc residue consisted of 92% 1,3,5-tri-O-acetyl-2,4-di-O-trideuteriomethylfucitol (m/z 121, 134, 240, and 253), and 8% 1,3,5-tri-O-acetyl-4-O-trideuteriomethyl-2-O-methylfucitol (m/z 118, 134, 237, and 250). These percentages were estimated from the m/z 121-to-118 ion intensity ratio. In both O chains, approximately 37% of the 3,4-linked Fuc residue was found to be endogenously methylated at O-2.
Locus of lpe.
A 3,046-bp stretch of DNA designated lpe3, in which the Tn5gus insertion of CE367 is situated, was cloned from parent wild-type strain R. etli CE3 by using an inverse PCR approach. Southern hybridization using lpe3 as a probe against wild-type genomic DNA digested with HindIII, PstI, or KpnI revealed a single 3-, 9-, or 14-kb DNA fragment, respectively, indicating that lpe3 is probably present as a single-copy locus. The 3-kb fragment, lpe3, was sufficient to restore the reactivity of the LPS to antibodies JIM27, JIM28, and JIM29 when expressed from broad-host-range plasmid pRK404E1 in CE367 (Fig. 2D, CE367/plpeA234).
Five hypothetical ORFs on the same DNA strand—ORF1, lpeA, ORF2, ORF3, and ORF4—were inferred from the nucleotide sequence of lpe3 (Fig. 3B). The Tn5gus insertion of mutant CE367 was mapped to ORF lpeA by analyzing the EcoRI fragment cloned from this mutant (Fig. 3C, fragment E1-Tn5). Separately, two other mutations in lpeA (Fig. 3, ΔlpeA430::Km and lpeA445::Km) were generated in the lpe3 DNA. Exchanging this mutated DNA with the wild-type locus in CE3 resulted in mutants CE430 and CE445. Like CE367 LPS, CE430 and CE445 LPSs did not bind detectably to MAbs JIM27, JIM28 and JIM29 (Fig. 3E, JIM28). Insertion of a kanamycin resistance cassette in downstream ORF2 (lpe-431::Km, resulting in strain CE431) also led to an LPS that was not recognized by MAb JIM27, JIM28 (Fig. 3E), or JIM29. Mutation lpe-462::Km in ORF3 (Fig. 3A) also effected loss in antigenicity (tested only with JIM28; data not shown).
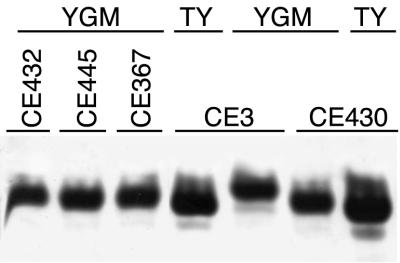
After chemical analysis indicated that the LPS of Lpe− mutant strain CE367 was one residue smaller than the wild-type LPS, the migration of the mutant LPSs on gel electrophoresis was examined more closely. Occasionally, SDS-PAGE revealed a slight difference between Lpe− mutants and wild-type CE3 in LPS I migration after growth on TY (e.g., Fig. 3F). On TDOC gels, also, a slight difference was noted, and after growth at low pH, the difference in migration between mutant and wild-type LPS I was more obvious (Fig. 8). Nevertheless, as for the wild type (40), growth at low pH results in the mutant LPS I migrating substantially slower, compared with its migration after growth in TY and other media at neutral pH (Fig. 8).
FIG. 8.
lpe locus and LPS I migration on TDOC PAGE at low pH. Wild-type CE3, lpeA mutants CE367, CE430, and CE445, and Lpe mutant CE431 (mutated in ORF2) cells were grown in YGM at pH 5. CE3 and lpeA mutant CE430 cells also were grown in TY medium at neutral pH for comparison. The cells were lysed in SDS buffer and separated by TDOC PAGE. The portion of the stained gel containing the LPS I bands is shown.
The sugar compositions of the phenol-water extracts (crude LPS) of strain CE445 (lpeA::Km) and CE431 (ORF2::Km) were essentially the same as that of strain CE367. In particular, the crude LPS of all three mutant strains lacked both TOM-Fuc and DOM-Fuc as revealed by alditol acetate analysis.
Complementation analysis to define genes within lpe locus.
To assess the minimal DNA sequence required for restoration of the wild-type antigenicity in the lpe mutants, various extents of the lpe3 DNA were cloned into expression vector pRK404E1 and transferred into the mutants. The restoration of wild-type antigenicity was then tested by immunoblottings of colonies, and LPS was separated by SDS-PAGE. The results of these experiments are tabulated on the right of Fig. 3C, with a few examples documented in Fig. 3F.
Complementation of lpeA mutants supports the extent of the lpeA ORF depicted in Fig. 3B. Deletion constructs that removed the first 109 nt 3′ of the first possible methionine start codon for this ORF (e.g., Fig. 3, plpeΔ5′A2) were sufficient to restore wild-type antigenicity to lpeA mutants CE367 and CE430, but an in-frame deletion construct further downstream (in plpeΔA) did not (Fig. 3C and F). Thus, translation of lpeA likely commences downstream of ORF1 from an inferred Shine-Dalgarno site as indicated in Fig. 3, rather than within ORF1 as predicted by the first possible start codon. The 3′ terminus of the lpeA ORF as shown in Fig. 3 also is consistent with the complementation by these and the other deletion constructs. Restoration of antigenicity required DNA with this complete ORF, but not beyond the XbaI restriction site, when the recipient carried any of the lpeA mutations (e.g., Fig. 3C and F, compare the effects of plpeΔA234, plpeA234Δ1, plpeA, plpeΔA, plpeAΔ1, and plpeΔA2).
The restoration of the wild-type antigenicity in Lpe− mutant CE431 (ORF2::Km) required not only ORF2 but also DNA that was 3′ of ORF2. For example, plpeA234 complemented lpe-431, whereas plpeA2 did not (Fig. 3C). As all complementing sequences included ORF4 as well as ORF3, one or both of these ORFs apparently were required for the production of the wild-type antigenicity. This inference was supported by the subsequent isolation of mutation lpe-462::Km in ORF3 (Fig. 3A) and the Lpe− phenotype conferred by this mutation. Moreover, the complementation of CE431 by the larger XbaI-HindIII fragment of lpe3 DNA was diminished by the presence of mutation lpe-462::Km (data not shown).
Complete complementation of strains CE445 (mutated in the 3′ third of lpeA) or CE431 (mutated in ORF2) did not depend on the orientation of the lpe3 genes relative to Plac of pRK404E1. However, only sense-constructs with respect to Plac yielded complete complementation of lpeA mutants CE367 and CE430, which are mutated in the 5′ third of lpeA. This inference follows from comparing the extents of MAb binding in lpeA mutant recipients after transfer of inverse constructs, such as plpeAi, with the results yielded by corresponding sense constructs, such as plpeA (Fig. 3C and F).
Previously cloned R. etli CE3 lps regions α (pCOS109.11) (10, 11), β (pCOS126) (10, 21), and γ (pCOS309.1, pCOS309.2, pCOS309.3, pCOS309.7, pCOS309.8) (10) did not complement the lpe mutations of CE367 or CE431. Nor did lpe3 (plpeA234) complement LPS mutant CE166, which carries a mutation defining another lps region (35).
Sequence comparisons with previously identified ORFs.
The predicted polypeptide sequence of LpeA exhibits greatest similarity to hypothetical, undefined proteins from Streptomyces coelicolor, Mesorhizobium loti, Mycobacterium tuberculosis, and Caulobacter crescentus, with the best match yielding 25% identity and 45% similarity over a stretch of 229 amino acids. The closest matches to proteins of known function were with FkbM, a 31-O-methyltransferase in Streptomyces spp. (31), and NoeI (23% identity and 39% similarity over a stretch of 143 amino acids), which is required for 2-O-methylation of the fucose residue of a lipooligosaccharide Nod factor of Rhizobium sp. strain NGR234 (26). The N-terminal 13 amino acids of LpeA are predicted to be periplasmic, followed by one α-helical span through the inner membrane, and the remainder are predicted to be cytoplasmic.
ORF2, ORF3, and ORF4 have only very weak matches with short stretches of proteins in the database. ORF2 may code for an inner membrane protein with weak similarity to a rat carboxyl methyltransferase. In a stretch of 39 amino acids, 13 are identical and 23 are similar. ORF3 most resembles a protein of Xylella fastidiosa that is described as a member of a family of S-adenosyl methionine-dependent methyltransferases involved in cell division. In a stretch of 75 amino acids, 23 are identical and 39 are similar. ORF3 and LpeA group to clusters of putative and demonstrated orthologous S-adenosyl methionine-dependent methyltransferases by similarity analysis of ORFs from all of the presently known complete genome sequences (41).
Expression of lpeA::gusA fusion in CE367 after environmental cues that induce LPS modifications.
Various growth conditions affect the formation of the epitopes of antibodies JIM27, JIM28, and JIM29. These include low pH (Fig. 9A), the presence of anthocyanins (Fig. 2B), and the bacteroid state (34, 40). Unlike an lpeA mutation, these conditions do not affect the binding of all three antibodies equally. However, it was conceivable that one or more environmental cues lead to a weaker LPS antigen by reducing the transcriptional activity across lpeA. One test of this hypothesis made use of the gusA fusion of mutant CE367 (lpeA::Tn5gusA). Growth of CE367 under conditions that led to greatly decreased wild-type LPS antigenicity to MAbs JIM28 and JIM29—including low pH (YGM medium) or the presence of seed exudate or delphinidin—did not affect transcription across lpeA, as monitored by GUS activity (Fig. 9B and data not shown). The GUS activity of CE367 was high in nodules, whereas binding of R. etli CE3 bacteroid LPS to MAb JIM28 and JIM29 is relatively low (40). Thus, these data provide no support for the idea that the loss of antigenicity triggered by the above environmental cues occurs by transcriptional repression of lpeA. This conclusion is supported by other experiments, in which the strong, constitutive promoter Paph was cloned in a sense orientation at the 5′ end of lpe3 in plpeA234. When CE3 carried this construct and was grown in the presence of seed exudate, delphinidin, or YGM at pH 5, the LPS modifications still occurred (data not shown).
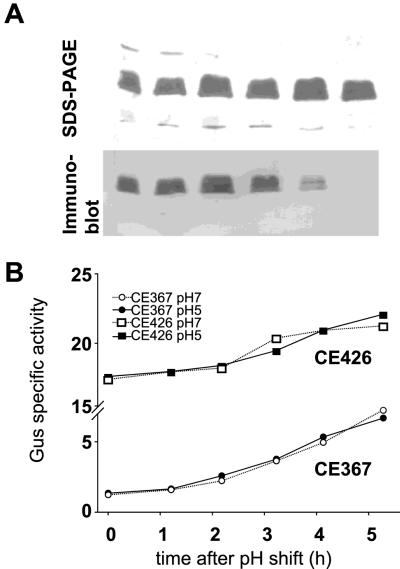
FIG. 9.
Lack of effect of growth medium pH on expression of the lpeA::Tn5gusA fusion of CE367. Cells of Tn5gus-mutant CE367 and CE426, an Lpe+ derivative of CE3 that constitutively expresses GUS, were grown in YGM buffered at pH 5 or pH 7. (A) CE426 cells growing at pH 5 were lysed in SDS at hourly intervals, and the extracted LPS was probed with JIM28 as described for Fig. 2. The time at which each sample was harvested corresponds to the scale shown on the x axis of panel B. (B) Chloroform extracts of the SDS lysates were assayed for specific GUS activity. The unit on the y axis is nanomoles of p-nitrophenol (min)−1 (milligrams of total protein)−1.
Although these experiments demonstrated no repression of lpeA under conditions known to induce loss of the JIM28 epitope, the data of Fig. 9B suggest that this gene may be regulated in response to other factors. At both pH 7 and pH 5, as the bacteria emerged from lag phase, the lpeA::Tn5gusA was differentially induced relative to total protein and the constitutive gusA of control strain CE426.
Minimal effect of lpeA on symbiosis with P. vulgaris.
No obvious difference in the abundance, location, and time of emergence of nodules or bacteroid abundance was observed when P. vulgaris was inoculated with strain CE3, CE367, CE430, CE431, or CE432 alone. In experiments to test the ability of CE367 to compete for nodule occupancy with the wild type, no significant difference in the ratio of CFU inoculated per CFU recovered from nodules was observed when roots were inoculated with total CFU that varied from 102 to greater than 104 and CE367-to-CE3 CFU ratios of 1:5, 1:1, and 5:1. When seeds were inoculated directly, only when the total inoculum was below 103 CFU was any reproducible difference observed. In a representative experiment, the CFU ratios of CE3 to CE367 in the inocula were 20/80, 47/53, and 76/24, and the CFU ratios of CE3 to CE367 recovered from the nodules were 24/76, 63/37, and 84/16, respectively. The mean enrichment in CE3 from inoculum to nodule occupant was 9% of the total population. If the lpe genes had a beneficial effect on nodulation efficiency of P. vulgaris, it was barely perceptible in these experiments.
DISCUSSION
Sugar composition, methylation, NMR, and MALDI-TOF (MS) analysis support the conclusion that the CE367 O-chain polysaccharide differs from that of CE3 in one primary feature. It is missing the TOM-Fuc residue and instead is terminated by a GlcA residue. All other known structural features of the wild-type LPS—including the sugars, their linkages, the variable methylation at O-2 of the 3- and 3,4-linked Fuc residues, the methyl esterification of GlcA, and the O-acetyl content—appear to be identical in mutant CE367. It has been speculated that the addition of the terminal TOM-Fuc residue might be an essential part of the regulation of O-chain length because it necessarily prevents the addition of further repeating units (18). However, this appears not to be the case. The mutant has the same remarkable uniformity in the number of repeating units as that exhibited by the wild type. With regard to LPS synthesis, it is also interesting that the absence of TOM-Fuc and DOM-Fuc does not seem to decrease the relative number of LPS molecules carrying the O chain (LPS I), whereas the absence of 2-O-methylation of the internal Fuc residues in strain CE395 leads to a 50% decrease in the relative amount of LPS I (35).
One possible function of this residue might be as a structural feature involved in liganding to another biological molecule. An artificial illustration is in the binding of antibodies to the LPS. The lpe3 locus, which is required for the presence of TOM-Fuc in the O chain, is required also for maturation of the R. etli CE3 O chain into a structure that is recognized by three rat MAbs and the majority of the antibodies of polyclonal sera developed in rabbits against the wild-type LPS. Previous studies have indicated that the epitopes recognized by MAbs JIM27, JIM28, and JIM29 probably overlap but are not identical (40). Mutations in the lpe3 locus are unique in eliminating the binding of all three of these antibodies. The TOM-Fuc residue may be shared spatially among the epitopes, or it may affect the structure of one or more of the epitopes by a conformation effect at a distance.
Of these three antibodies, JIM28 and its requirements for binding have been studied the most extensively. The absence of TOM-Fuc in CE367 is the third documented change in the O chain that is correlated with decreased binding of this antibody. Loss of reactivity after growth in anthocyanins and at low pH is correlated with increased 2-O-methylation of the internal fucose residues (5; J. Box and K. D. Noel, unpublished data), and conversely, the absence of this 2-O-methylation of internal fucoses in mutant strain CE395 (35) is correlated with increased binding of the antibody (J. Box, V. J. Neumann, and K. D. Noel, unpublished data). Treatment of the LPS at pH 12, which should have eliminated both O-acetylation and the methyl esterified to GlcA in the O chain (18), also eliminated JIM28 binding (Fig. 2C). If this base-sensitive feature, the TOM-Fuc residue, and the 2-hydroxyl of an internal fucose residue were all in contact with the antibody, the binding site would have spanned at least the last three residues of the O chain (Fig. 1). X-ray crystallographic studies have documented two types of structural interactions of antibodies with LPS oligosaccharides (13, 43), burying of the terminal sugar residue in a hydrophobic cavity at the antibody binding site (43), and an extended “groove” in the antibody that interacts by hydrophobic stacking and hydrogen bonding with at least three sugar residues in a branched repeating unit (12, 13). The hypothetical interaction of the R. etli CE3 O chain might have features in common with both of these models, with TOM-Fuc fitting into a hydrophobic pocket of an extended groove on the antibody.
Although the sequences of the putative lpe3 ORFs did not provide strong matches with sequences of known function in the database, the weak matches were consistent with hypothetical roles in the methylation of fucose. In concert with this role, preliminary sequencing of DNA upstream of lpe3 has revealed that ORF1 has greater sequence similarity than LpeA to NoeI (the 2-O-methylase of fucose in Nod factors [26]). It also has uncovered an ORF with great similarity to fucose synthetases and another with sequence similarity to putative glycosyltransferases (J. Box, D. M. Duelli, and K. D. Noel, unpublished data). Hence, lpe3 may be part of a larger locus that specifies both the methylation of fucose and its addition to the end of the O antigen. In regard to the specific biochemistry involved, two findings from the LPS structural analysis should be noted. One, not only the methyl groups of TOM-Fuc but also the fucose itself are missing from CE367 LPS. Two, other fucose residues and their variable 2-O-methylations are not affected. If this locus is only involved in the 2,3,4-tri-O-methylation, the absence of unmethylated terminal fucose would favor the idea that methylation occurs at the level of GDP-Fuc, before transfer to the O chain.
Genetic analysis suggests that the lpe3 genes are expressed in at least two separate transcriptional units, one ending in lpeA and the other including ORF2. At least one downstream gene was required for complementation of the insertion mutation in ORF2, perhaps because this insertion has polar effects on expression of ORF3 or ORF4. Indeed, a kanamycin resistance cassette that is believed to be very similar to the one used in this study has strongly polar effects, regardless of the orientation of the insertion (3). Similarly, the insertion in ORF3 (lpe-462::Km) may be affecting the expression of ORF4. In any case, it also remains to be seen whether the lpe genes are expressed in R. etli as the polypeptides inferred from ORF analysis. Hence, a conservative interpretation of the genetic data is that at least two lpe genes are required for epitope synthesis: lpeA and a gene downstream of ORF2.
The results of this study provide no support for the possibility that CE3 responds to low pH or the presence of anthocyanins by repressing lpeA transcription. Such results do not rule out the regulation of the lpe gene products in response to such conditions, but structural analysis of the LPS produced during growth at low pH lends no support to that idea either. Although TOM-Fuc is absent, DOM-Fuc is present (5). On the other hand, recent results indicate that one effect of growth in seed exudate is the absence of TOM-Fuc and DOM-Fuc from the synthesized O antigen (J. Box and K. D. Noel, unpublished data). In that case, another gene responsible for adding the terminal residue, such as the gene for the glycosyltransferase, may be repressed or the effect may be posttranscriptional.
The biological functions of the TOM-Fuc residue and the lpe3 locus remain obscure. Recent work in progress indicates that another decoration of the R. etli CE3 LPS, the variable 2-O-methylation of the internal fucose residues of the O chain, is relatively unimportant when the bacteria are inoculated on roots but becomes important when the bacteria are inoculated directly onto P. vulgaris seeds (J. Box and K. D. Noel, unpublished data). The benefit in this latter case may derive from coping with toxic compounds released from the seed. In this connection, it may be significant that all of the known antigenic modifications of the LPS in R. leguminosarum and R. etli are induced by what could be considered stress conditions. The results of this study indicate that the terminal residue of the O antigen and its three O-methyl groups have little effect, if any, on the symbiotic proficiency of this bacterium, even when the bacteria are inoculated directly on the host seed. Indeed, if loss of this structure is one of the LPS changes induced by the plant, this result is as expected. The benefit of having the TOM-Fuc residue under other conditions may be related to surviving or thriving under conditions that have been common during the evolution of this species but which thus far have not been tested in the laboratory.
ACKNOWLEDGMENTS
We acknowledge the services of the DNA sequencing facilities at the University of Wisconsin—Milwaukee. We thank Kevin Barleben and Stacie Lambert for research assistance and S. Sharma, Kate Wilson, and Mark McBride for plasmids.
This work was funded by a grant from the Marquette University Committee on Research, Department of Energy grant DE-FG02-98ER20307 (to K.D.N.), National Institutes of Health grant GM39583 (to R.W.C.), as well as DOE grant DE-FG09-93ER20097 to the CCRC. D.M.D. was supported in part by a predoctoral fellowship from the A.J. Schmitt Foundation.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arps P J, Winkler M E. Structural analysis of the Escherichia coli K-12 hisT operon by using a kanamycin resistance cassette. J Bacteriol. 1987;169:1061–1070. doi: 10.1128/jb.169.3.1061-1070.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 5.Bhat U R, Carlson R W. Chemical characterization of pH-dependent structural epitopes of lipopolysaccharides from Rhizobium leguminosarum biovar phaseoli. J Bacteriol. 1992;174:2230–2235. doi: 10.1128/jb.174.7.2230-2235.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle J S, Lew A M. An inexpensive alternative to glassmilk for DNA purification. Trends Genet. 1995;11:8. doi: 10.1016/s0168-9525(00)88977-5. [DOI] [PubMed] [Google Scholar]
- 7.Brendel V, Bucher P, Nourbakhsh I R, Blaisdell B E, Karlin S. Methods and algorithms for statistical analysis of protein sequences. Proc Natl Acad Sci USA. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson R W, Kalembassa S, Turowski D, Pachori P, Noel K D. Characterization of the lipopolysaccharide from a Rhizobium phaseoli mutant that is defective in infection thread development. J Bacteriol. 1987;169:4923–4928. doi: 10.1128/jb.169.11.4923-4928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson R W, Sanders R E, Napoli C, Albersheim P. Host-symbiont interactions. III. Purification and characterization of Rhizobium lipopolysaccharides. Plant Physiol. 1978;62:912–917. doi: 10.1104/pp.62.6.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cava J R, Elias P M, Turowski D A, Noel K D. Rhizobium leguminosarum CFN42 genetic regions encoding lipopolysaccharide structures essential for complete nodule development on bean plants. J Bacteriol. 1989;171:8–15. doi: 10.1128/jb.171.1.8-15.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cava J R, Tao H, Noel K D. Mapping of complementation groups within a Rhizobium leguminosarum CFN42 chromosomal region required for lipopolysaccharide synthesis. Mol Gen Genet. 1990;221:125–128. [Google Scholar]
- 12.Cygler M. Recognition of carbohydrates by antibodies. Res Immunol. 1994;145:36–40. doi: 10.1016/s0923-2494(94)80040-5. [DOI] [PubMed] [Google Scholar]
- 13.Cygler M, Rose D R, Bundle D R. Recognition of a cell-surface oligosaccharide of pathogenic Salmonella by an antibody Fab fragment. Science. 1991;253:442–445. doi: 10.1126/science.1713710. [DOI] [PubMed] [Google Scholar]
- 14.Del Sal G, Manfioletti G, Schneider C. The CTAB-DNA precipitation method: a common mini-scale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. BioTechniques. 1989;7:514–520. [PubMed] [Google Scholar]
- 15.Ditta G, Schmidhauser T, Yakobson E, Lu P, Liang X W, Finlay D R, Guiney D, Helinski D R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985;13:149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- 16.Duelli D M, Noel K D. Compounds exuded by Phaseolus vulgaris that induce a modification of a Rhizobium etli lipopolysaccharide. Mol Plant-Microbe Interact. 1997;10:903–910. [Google Scholar]
- 17.Finan T M, Kunkel B, De Vos G F, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsberg L S, Bhat U R, Carlson R W. Structural characterization of the O-antigenic polysaccharide of the lipopolysaccharide from Rhizobium etli strain CE3. A unique O-acetylated glycan of discrete size, containing 3-O-methyl-6-deoxy-L-talose and 2,3,4-tri-O-methyl-L-fucose. J Biol Chem. 2000;275:18851–18863. doi: 10.1074/jbc.M001090200. [DOI] [PubMed] [Google Scholar]
- 19.Forsberg L S, Carlson R W. The structures of the lipopolysaccharides from Rhizobium etli strains CE358 and CE359. The complete structure of the core region of R. etli lipopolysaccharides. J Biol Chem. 1998;273:2747–2757. doi: 10.1074/jbc.273.5.2747. [DOI] [PubMed] [Google Scholar]
- 20.Gallagher S R, editor. GUS protocols. Using the GUS gene as a reporter of gene expression. New York, N.Y: Academic Press, Inc.; 1992. [Google Scholar]
- 21.Garcia de los Santos A, Brom S. Characterization of two plasmid-borne lps loci of Rhizobium etli required for lipopolysaccharide synthesis and for optimal interaction with plants. Mol Plant-Microbe Interact. 1997;7:891–902. doi: 10.1094/MPMI.1997.10.7.891. [DOI] [PubMed] [Google Scholar]
- 22.Glazebrook J, Walker G C. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 1991;204:398–418. doi: 10.1016/0076-6879(91)04021-f. [DOI] [PubMed] [Google Scholar]
- 23.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 24.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann K, Stoffel W. TMBASE—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 26.Jabbouri S, Relic B, Hanin M, Kamalaprija P, Burger U, Prome D, Prome J C, Broughton W J. nolO and noeI (HsnIII) of Rhizobium sp. NGR234 are involved in 3-O-carbamoylation and 2-O-methylation of Nod factors. J Biol Chem. 1998;273:12047–12055. doi: 10.1074/jbc.273.20.12047. [DOI] [PubMed] [Google Scholar]
- 27.Kannenberg E L, Rathbun E A, Brewin N J. Molecular dissection of structure and function in the lipopolysaccharide of Rhizobium leguminosarum strain 3841 using monoclonal antibodies and genetic analysis. Mol Microbiol. 1992;6:2477–2487. doi: 10.1111/j.1365-2958.1992.tb01424.x. [DOI] [PubMed] [Google Scholar]
- 28.Kannenberg E L, Reuhs B L, Forsberg L S, Carlson R W. Lipopolysaccharides and K-antigens: their structures, biosynthesis, and functions. In: Spaink H, Kondorosi A, Hooykaas P, editors. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 119–154. [Google Scholar]
- 29.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 31.Motamedi H, Shafiee A, Cai S J, Streicher S L, Arison B H, Miller R R. Characterization of methyltransferase and hydroxylase genes involved in the biosynthesis of the immunosuppressants FK506 and FK520. J Bacteriol. 1996;178:5243–5248. doi: 10.1128/jb.178.17.5243-5248.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 33.Noel K D, Duelli D M. Rhizobium lipopolysaccharide and its role in symbiosis. In: Triplett E W, editor. Prokaryotic nitrogen fixation: a model system for analysis of a biological process. Wymondham, United Kingdom: Horizon Scientific Press; 2000. pp. 415–431. [Google Scholar]
- 34.Noel K D, Duelli D M, Tao H, Brewin N J. Antigenic change in the lipopolysaccharide of Rhizobium etli CFN42 induced by exudates of Phaseolus vulgaris. Mol Plant-Microbe Interact. 1996;9:180–186. [Google Scholar]
- 35.Noel K D, Forsberg L S, Carlson R W. Varying the abundance of O antigen in Rhizobium etli and its effect on symbiosis with Phaseolus vulgaris. J Bacteriol. 2000;182:5317–5324. doi: 10.1128/jb.182.19.5317-5324.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noel K D, Sanchez A, Fernandez L, Leemans J, Cevallos M A. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J Bacteriol. 1984;158:148–155. doi: 10.1128/jb.158.1.148-155.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sharma S B, Signer E R. Temporal and spatial regulation of the symbiotic genes of Rhizobium meliloti in planta revealed by transposon Tn5-gusA. Genes Dev. 1990;4:344–356. doi: 10.1101/gad.4.3.344. [DOI] [PubMed] [Google Scholar]
- 40.Tao H, Brewin N J, Noel K D. Rhizobium leguminosarum CFN42 lipopolysaccharide antigenic changes induced by environmental conditions. J Bacteriol. 1992;174:2222–2229. doi: 10.1128/jb.174.7.2222-2229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tatusov R L, Galperin M Y, Natale D A, Koonin E V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor L A, Rose R E. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 1988;16:358. doi: 10.1093/nar/16.1.358. . (Erratum, 16:7762.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villeneuve S, Souchon H, Riottot M-M, Mazie J-C, Lei P-S, Glaudemans C P J, Kovac P, Fournier J-M, Alzari P M. Crystal structure of an anti-carbohydrate antibody directed against Vibrio cholerae O1 in complex with antigen: molecular basis for serotype specificity. Proc Natl Acad Sci USA. 2000;97:8433–8438. doi: 10.1073/pnas.060022997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vincent J M. A manual for the practical study of root nodule bacteria. Vol. 15. Oxford, United Kingdom: Blackwell Scientific Publications; 1970. [Google Scholar]
- 45.Wilson K J, Sessitsch A, Corbo J C, Giller K E, Akkermans A D, Jefferson R A. beta-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other gram-negative bacteria. Microbiology. 1995;141:1691–1705. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]
- 46.York W S, Darvill A G, McNeil M, Stevenson T T, Albersheim P. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 1985;118:3–40. [Google Scholar]