Abstract
The recombinational rescue of chromosome replication was investigated in Escherichia coli strains with the unidirectional origin oriR1, from the plasmid R1, integrated within oriC in clockwise (intR1CW) or counterclockwise (intR1CC) orientations. Only the intR1CC strain, with replication forks arrested at the terminus, required RecA for survival. Unlike the strains with RecA-dependent replication known so far, the intR1CC strain did not require RecBCD, RecF, RecG, RecJ, RuvAB, or SOS activation for viability. The overall levels of degradation of replicating chromosomes caused by inactivation of RecA were similar in oriC and intR1CC strains. In the intR1CC strain, RecA was also needed to maintain the integrity of the chromosome when the unidirectional replication forks were blocked at the terminus. This was consistent with suppression of the RecA dependence of the intR1CC strain by inactivating Tus, the protein needed to block replication forks at Ter sites. Thus, RecA is essential during asymmetric chromosome replication for the stable maintenance of the forks arrested at the terminus and for their eventual passage across the termination barrier(s) independently of the SOS and some of the major recombination pathways.
Recombination and replication are intimately coupled processes with a large number of shared gene functions. The involvement of the replication mechanism in recombination and repair is well established (11, 43, 46). The extensive and vital role of recombination in chromosome replication was not evident until recently, although its key role in the replication of several bacteriophages has been recognized (25, 29, 42). Early evidence for the involvement of recombination in Escherichia coli chromosome replication came from special situations in which the initiation factor-dependent, oriC-specific assembly of replisomes could be bypassed through homologous-recombination-dependent replication processes called SDRs (for stable DNA replication) (21, 22, 30). SDR is constitutive (cSDR) in rnh mutants when initiation from oriC is inhibited and stable R loops can act as cryptic initiation sites. SDR can also be induced (iSDR) in response to DNA damage in UV-irradiated, thymidine-starved, or drug-treated cells in which recombinogenic invasion at double-strand-break (DSB) sites leads to the formation of branched structures and/or D loops, which act as the assembly sites for a new replisome. RecA is believed to be critical for replication initiation for both SDRs (51, 52), but each has its own distinctive recombination-repair pathway (20). DSBs have also been shown to act as hot spots for recombination-mediated initiation of replication (1, 12, 25).
Even normal replication, initiated from oriC in wild-type E. coli, relies on recombination-repair pathways for the apparently unhindered progression of replication forks over the full length of the bacterial chromosome (40, 47). The importance of the recombination-repair pathways in maintaining genome stability during replication in E. coli and other organisms is now generally recognized, and various models have been suggested for the reactivation of stalled or collapsed replication forks through homologous recombination (reviewed in references 8, 13, 24, 27, 29, 37, 39, 45, and 47). However, the recombination pathways involved in assisting the replication forks during their normal course of chromosome replication are not necessarily RecA dependent, since recA mutations are not lethal by themselves. The requirement for RecA in E. coli viability has been seen in SDR strains (see above) and in strains with large chromosomal inversions or under integrative suppression, depending on the position of the inversion or of the integrated replicon on the chromosome (33, 36). However, the detailed mechanism by which RecA sustains ongoing replication in such cases is not yet clearly understood.
We have constructed intR1 strains in which the basic replicon of plasmid R1 (oriR1) is integrated into the minimal oriC by a small deletion (4, 23). Plasmid R1 replicates unidirectionally, and interestingly, intR1 strains with different orientations of the integrated oriR1 exhibit drastically different growth and division phenotypes. The intR1 strain with clockwise initiation from the integrated oriR1 (intR1CW) replicates the chromosome bidirectionally and exhibits near-normal growth and division. Its homologue, intR1CC, with counterclockwise initiation, shows slower growth, filamentation, and poor nucleoid segregation (23, 34). The cell cycle defects of the intR1CC strain were partially suppressible by inactivation of the tus gene, coding for the Ter site binding protein Tus, needed for the polar block of the replication forks at the terminus (9). In the present work, we have further characterized the chromosomal replication in the intR1CC strain as predominantly unidirectional and have shown that active RecA is essential for its viability. The recombination function of RecA is required to prevent extensive degradation of chromosomes with replication forks blocked at the terminus, allowing the completion of a round of replication for the whole chromosome.
MATERIALS AND METHODS
Bacterial strains.
The E. coli strains used in this work are listed in Table 1. Introduction of rec and tus mutations into the intR1 strains or of the intR1 origin into other backgrounds, when necessary, was performed by generalized phage P1 transduction following standard procedures (41). SOS inactivation and Rec− and Ruv− phenotypes were confirmed by testing the sensitivity of the strains to exposure to UV radiation from a germicidal lamp.
TABLE 1.
Escherichia coli K-12 and intR1 derivatives
| Designation | Relevant genotypea | Origin |
|---|---|---|
| BR3674 | recA::Tn10 | D. K. Chattoraj |
| CC3775 | recD1901::Tn10 | J. Sawitzke |
| EC1005 | metB nalA relA1 spoT1 Ir F− | 14 |
| EC1005 tus::Kanr | EC1005 × P1 (PK2517) | This work |
| GY6781 | Δ(pro-lac) mal::Tn9 lexA1(Ind−) rpsL sfiA::lacZ | R. Devoret |
| JAS34 | recF332::Tn3 | J. A. Sawitzke |
| JAS73 | recJ2001::Kanr | J. A. Sawitzke |
| KHG962 | tus::Tn3 (W3110 background) | T. Horiuchi |
| MG1655 | λ− F− | 19 |
| MG1655 tus::Kanr | MG1655 × P1 (PK2517) | This work |
| N2057 | ruvA60::Tn10 | R. G. Lloyd |
| N2101 | recB268::Tn10 | 31 |
| N3793 | recG263::Kanr | R. G. Lloyd |
| N8954 | recA::Cmr (JM103 background) | J. L. Rosner |
| PK1012 | P2sig5; prophage inserted at 47 min; dnaA(Ts) | 15 |
| PK2517 | tus::Kanr | P. Kuempel |
| SK129 | recB270 recC271 (AB1157 background) | 26 |
| TH765 | srl300::Tn10 recA200(Ts) λ−argA21 spoT thi-1 deoB13 | T. Hill, 32 |
| intR1 strains | ||
| EC::71CW | EC1005 ΔoriC::pOU71; rep-Td; Ampr; initiating clockwise | 4 |
| EC::71CC | EC1005 ΔoriC::pOU71; rep-Td; Ampr; initiating counterclockwise | 4 |
| EC::71CW tus::Kanr | EC::71CW-L × P1 (PK2517) | 9 |
| EC::71CC tus::Kanr | EC::71CC-L × P1 (PK2517) | 9 |
| LK211 | ΔoriC::pKN1562; Kanr; initiating clockwise | 23 |
| LK348 | ΔoriC::pKN1562; Kanr; initiating counterclockwise | 23 |
| MG::71CW-L | MG1655 ΔoriC::pOU71; rep-Td; Ampr; initiating clockwise | 34 |
| MG::71CC-L | MG1655 ΔoriC::pOU71; rep-Td; Ampr; initiating counterclockwise | 34 |
| MG::71CW-L tus::Kanr | MG::71CW-L × P1 (PK2517) | This work |
| MG::71CC-L tus::Kanr | MG::71CC-L × P1 (PK2517) | This work |
| MG::71CC-L tus::Kanr-pBADtus | MG::71CC-L tus::Kanr transformed with pBADtus (Cmr) (pBRI) | B. Peter, this work |
| MG::71CC-L tus::KanrrecA::Tn10 | MG::71CC-L tus::Kanr × P1 (BR3674) | This work |
| MG::71CC-L tus::KanrrecA::Tn10-pBADtus | MG::71CC-L tus::KanrrecA::Tn10 transformed with pBADtus (Cmr) | B. Peter, this work |
| TH765::CC | TH765 × P1 (LK348) | This work |
| TH765::CC tus::Tn3 | TH765::CC × P1 (KHG962) | This work |
Symbols according to Berlyn et al. (2). Amp, ampicillin; Cm, chloramphenicol; Kan, kanamycin; IN, chromosomal inversion; rep-Td, replication temperature dependent. Tn3 confers ampicillin resistance, and Tn10 confers tetracyclin resistance.
Culture media and growth conditions.
Bacterial cultures were grown at 30°C in minimal medium M9 (35) with glucose (0.2%), Casamino Acids (0.2%), the necessary supplements (10 to 50 μg of thymine/ml and 10 μg of thiamine/ml), and the appropriate antibiotics (50 μg of ampicillin/ml, 30 μg of chloramphenicol/ml, 50 μg of kanamycin/ml, and 15 μg of tetracycline/ml). Growth was monitored by measurement of optical density at 550 nm (OD550). Samples for flow cytometry and extraction of genomic DNA were collected at an OD550 of 0.1 to 0.15 after at least 5 to 10 generations of exponential growth. Cells harvested for extraction of replicating chromosomes were killed rapidly with 0.1 M NaN3 at 0°C.
Thymidine incorporation.
Incorporation of [3H]thymidine during exponential growth and retention of prelabeled DNA in replicating and nonreplicating cells were estimated by measurement of trichloroacetic acid (TCA)-insoluble radioactivity in samples of the cultures under different growth conditions. For continuous labeling for two or three generations, 2.5 μCi of [3H]thymidine/ml was added along with 50 μg of deoxyadenosine/ml to a culture grown in minimal medium (M9-glucose supplemented with 0.2% Casamino Acids) containing 10 μg of nonradioactive thymine/ml. One-milliliter samples were taken at different times for measurement of the cumulative incorporation. For pulse labeling, 10 μCi of [3H]thymidine was added to 1-ml samples of the culture. Labeling was stopped after 2 min by the addition of 5 ml of ice-cold 10% TCA containing 10 μg of cold thymidine/ml. To measure the degradation of prelabeled DNA, the cells labeled for two or three generations were filter washed (0.22-μm pore size; Millipore) two or three times with equal volumes of prewarmed medium free of radioactive thymidine. They were then resuspended in the same medium without radioactivity and split into two equal parts, which were incubated at 30 and 42°C. Samples (1 ml) were taken at different times and transferred to 5 ml of ice-cold 10% TCA. Alternatively, the culture labeled with [3H]thymidine for two or three generations was incubated for 3 h in the presence of 200 μg of rifampin/ml to allow ongoing replication to run out, filter washed as described above, and resuspended in rifampin-containing medium at 30 and 42°C. Samples (1 ml) were treated with TCA as described above. The TCA-insoluble material was trapped on GF/A filters (Whatman), which were washed with 5% TCA, 95% ethanol, and diethyl ether and dried under a lamp. The dried filters were placed in plastic vials with 4 ml of scintillation fluid (Quicksafe N; Zinsser Analytic) and counted in a Beckman scintillation counter (LS-3801) at the standard setting for tritium.
Flow cytometry.
Exponentially growing bacteria (OD550 = 0.1) and rifampin (200 μg/ml)-treated cells were fixed in 70% ethanol and stored at 4°C. The fixed cells were washed, stained, and run in a Bio-Rad Bryte HS flow cytometer as described earlier (34).
Southern blots.
Chromosomal DNA was purified with the Qiagen kit for total genomic DNA preparation that allows preservation of replication intermediates (53). Purified DNA samples were digested with BglI, electrophoresed, transferred to nylon membranes, and hybridized as described by Maisnier-Patin et al. (34). The probe used for hybridization was generated by PCR using the primers 5′-TTCTCCTGTGGTTGTTGC-3′ and 5′-CCTGTTCTGGCGTCATAA-3′.
RESULTS
Asymmetric chromosome replication and prolonged arrest of replication fork(s) in intR1CC strain.
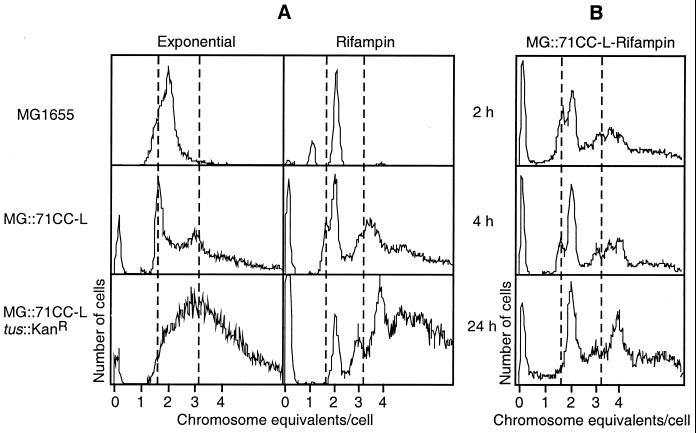
As reported earlier, chromosome replication patterns in the two genetically identical intR1 strains were very different. Despite unidirectional initiation from the integrated oriR1, the clockwise replication underwent a postinitiation conversion to bidirectional symmetry, while the counterclockwise replication failed to do so. Marker frequency analysis of the intR1CC strain was ineffective in detecting any gene dosage gradient across the chromosome suggesting the possibility of reinitiation from random sites during a single round of replication (34). However, a comparison of the flow cytometric profiles of the wild-type strain, MG1655, and its intR1CC derivative was more revealing and showed a predominance of unidirectionally replicating chromosomes. Figure 1 shows the DNA content distributions in the exponentially growing and the rifampin-treated cells. The fluorescence intensities were calibrated in terms of chromosome equivalents, using wild-type bacteria either at stationary phase or after completion of runout replication in the presence of 200 μg of rifampin/ml. The exponentially growing MG1655 cells showed a continuum of DNA content ranging from one to four chromosomes, indicating ongoing replication without any detectable pause. In contrast, the exponential culture of the intR1CC derivative showed sharp peaks representing cells with 1.6 and 3.2 chromosomes rising above the population of cells with ongoing replication. The appearance of distinct subpopulations of cells with fixed DNA contents during steady-state growth implied prolonged arrest in the progress of the replication forks at sites over halfway from the start site. As replication in intR1CC starts in the vicinity of oriC, 1.6 chromosomes was consistent with replication of one arm of the chromosome being blocked in the terminus region. Replication runout in the presence of rifampin was complete for the bidirectionally replicating chromosomes within 3 h (sharp peaks corresponding to cells with 1, 2, and 4 complete chromosome equivalents of DNA [Fig. 1A, top right]). Similar treatment of the intR1CC strain led to a gradual conversion of the 1.6- and 3.2-chromosome peaks into 2- and 4-chromosome-equivalent peaks, respectively. Figure 1B shows that longer runout times are needed for the completion of the round of replication producing integral numbers of chromosomes per cell, indicating that the replication arrest was not permanent and the forks overcame the terminus barriers. Runout experiments with chloramphenicol (250 μg/ml) showed similar profiles (data not shown). The flow cytometry profiles for intR1CC, after the addition of rifampin or chloramphenicol, never showed any one chromosome-equivalent peak, and the total fluorescence remained about the same after 2, 3, 4, and 24 h in the presence of the drugs, indicating that there was no significant degradation or loss of chromosomal DNA. The intR1CW derivative with bidirectional chromosome replication shows flow cytometric profiles similar to those of the MG1655 parent (data not shown), suggesting that the distinctive replication pattern of the intR1CC chromosome is not a consequence of replication initiation from the integrated R1 origin per se but of its failure to convert to bidirectionality.
FIG. 1.
Flow cytometric analysis of the DNA content distribution in E. coli MG1655 and its intR1 derivatives grown exponentially in M9-glucose medium at 30°C. (A) DNA content in cells harvested from the exponential phase of growth (left) and after exposure to 200 μg of rifampin/ml for at least two generations (right). The fixed cells were stained with ethidium bromide and mithramycin A as described in Materials and Methods prior to examination by flow cytometry. The x axis shows the average amount of DNA per cell expressed as chromosome equivalents (calibrated from stationary-phase cells), and the y axis shows the number of cells with a specific DNA content. (B) DNA content distribution in MG::71CC-L exposed to rifampin for different times.
Inactivation of the gene coding for Tus, the termination protein, eliminated the cell population with fractional chromosome contents both in the exponential and in the rifampin runout cultures of intR1 strains (Fig. 1A, row 3). This indicated that in the absence of Tus the replication forks were no longer arrested and that the block observed in the Tus+ strain must be predominantly at one or more Ter sites. There were cells with 2, 3, 4, etc., chromosome equivalents represented by sharp peaks, but a large number of cells with higher DNA content and replicative intermediates were also present.
Location of the blocked replication forks on the chromosome.
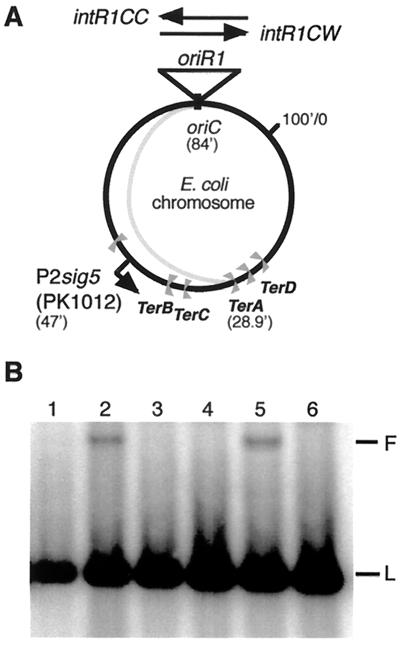
The sharp peak in the flow cytogram corresponding to 1.6 chromosomes in exponentially growing cells (Fig. 1A, row 2, left column) suggested that most of the half-replicated chromosomes were of similar size. Elimination of the half chromosomes by inactivation of the tus gene indicated that all forks were arrested at a Ter site. As they all started from within oriC (see above), replication block at TerA, the first Ter site encountered by the counterclockwise replication forks, was consistent with the length of the half-replicated chromosome detected by flow cytometry (Fig. 2A). We extracted the total DNA from an exponentially growing culture of the intR1CC strain, cleaved the DNA with restriction enzymes, and analyzed it by agarose gel electrophoresis. Southern blots from such gels were probed with a 32P-labeled PCR fragment containing the pyrF gene sequence located near the TerA site; branched structures corresponding to replication forks arrested at TerA were observed as slowly migrating bands (Fig. 2B, lane 5). No such band was visible for replicating chromosomes from the MG::71CW-L, MG::71CW-L tus::Kanr, or MG::71CC-L tus::Kanr strains (Fig. 2B, lanes 3, 4, and 6, respectively). As a quantitative control, we analyzed the DNA from replicating chromosomes of the integratively suppressed strain PK1012 dnaA(Ts) (15), which, under nonpermissive conditions, initiates replication from the origin of bacteriophage P2 integrated at 47 min on the E. coli map (Fig. 2A). Arrested forks at TerA were seen only during integrative suppression at 42°C, when the P2 origin was active, but not when the chromosome replicated bidirectionally from oriC at the permissive temperature (Fig. 2B, lanes 2 and 1, respectively). The band intensities of the slowly migrating fragments representing the forks arrested at TerA were similar for PK1012 and MG::71CC-L, suggesting that these bands were representative of replication from their respective origins.
FIG. 2.
Demonstration of blocked forks at the Ter sites in the replication terminus region by Southern blot analysis. (A) Circular map of the E. coli chromosome showing the sites of integration of the mini R1 plasmid and of the P2 origin in the P2 sig5 strain PK1012 and the location of the Ter sites. The extra arm (shaded) shows the half chromosome as the product of unidirectional replication from oriR1 in intR1CC strains. (B) Southern blot analysis at the TerA site. Lanes 1 and 2, DNA from PK1012 grown at 30 and 42°C, respectively; lane 3, MG::71CW-L; lane 4, MG::71CW-L tus::Kanr; lane 5, MG::71CC-L; lane 6, MG::71CC-L tus::Kanr. L indicates the linear fragment, and F indicates the branched structure.
intR1CC, but not intR1CW, strain requires RecA activity for viability.
Since replication arrests can induce DSBs requiring recombination-repair mechanisms for replication restart (28, 40), we tested the dependence of the intR1 strains on RecA for viability. As shown in Table 2, null mutations in the recA gene could not be transduced into intR1CC derivatives in two different genetic backgrounds (MG1655 and EC1005). intR1CW derivatives of both strains carrying the same recA allele were viable. The distinct responses to recA mutation elicited in intR1CW and intR1CC strains could not be at the level of initiation, since RepA oriR1 constitutes the sole replicon for both of the intR1 strains. This was tested by suppression of initiation from oriR1 by overproduction of the antisense RNA inhibitor of RepA (the rate-limiting initiator protein) that led to cell death for the intR1CW as well as the intR1CC strain (reference 4 and data not shown). Hence, the RecA function must be required at one or more stages downstream of initiation of replication.
TABLE 2.
RecA dependence for viability tested by P1 transduction of recA::Tn10 into intR1 strains
| Host strain | Relative transduction frequency |
|---|---|
| MG1655 | 1.0a |
| MG::71CW-L | 0.16a |
| MG::71CC-L | 0ac |
| MG::71CW-L tus::Kanr | 0.30a |
| MG::71CC-L tus::Kanr | 0.38a |
| EC1005 | 0.63b |
| EC::CW-L | 0.61b |
| EC::CC-L | 0bc |
| EC1005 tus::Kanr | 0.80b |
| EC::71CW-L tus::Kanr | 0.63b |
| EC::71CC-L tus::Kanr | 0.60b |
Relative to the number of transductants obtained with the wild-type parent, MG1655.
Ratio between the number of recA::Tn10 transductants and the number of met+ transductants.
Tiny colonies emerged on selective plates after 3 to 4 days of incubation, but none were temperature sensitive. Temperature sensitivity is a characteristic feature of the intR171CW and intR171CC strains.
Only the recombinase activity of RecA was required for the survival of the intR1CC strains, since the recA430 allele, coding for the protease-deficient mutant RecA, could be sustained in intR1CC (data not shown). Thus, RecA-activated cleavage of the SOS repressor did not seem to be required for the viability of the intR1CC strain (see below).
Transduction with phage P1 was used to replace the oriC region in the recA200(Ts) strain TH765 with the intR1CW (from the LK211 strain) and intR1CC (from the LK348 strain) replicons. The transductants selected at the permissive temperature for the recipient were then tested for growth at 30 and 42°C. Only the intR1CC recA200(Ts) strain was temperature sensitive for viability (Table 3), confirming that the RecA activity was essential for the viability of the intR1CC but not for the intR1CW strain. When these replicons were introduced into the recBC(Ts) strain, the resulting strains did not show any temperature sensitivity (Table 3), suggesting that RecBC was not essential for the maintenance of the intR1CC strain. In order to identify the specific recombination pathway involved in the RecA-mediated rescue of intR1CC chromosome replication, we tested the sustainability of the loss of several recombination-repair functions (RecB, RecD, RecF, RecG, RecJ, and RuvAB) by transducing the mutant alleles (from some of the strains listed in Table 1) into the intR1CC background. We found them all to be dispensable (Table 4); comparable numbers of transductants (100 to 400) were obtained for the intR1CW and intR1CC strains, as well as for the oriC strains (data not shown). The intR1CC strain also survived the introduction of the lexA(Ind−) mutation, which renders the strain incapable of SOS induction (Table 4). Bernander et al. (3) had previously demonstrated that the intR1CW strain shows a mild constitutive SOS activity (through the reporter gene din::lacZ); a similar or higher level of SOS activity could be detected in the intR1CC strain as well (data not shown). However, survival with the lexA(Ind−) mutation ruled out dependence on the SOS pathway for the viability of the intR1CC strain.
TABLE 3.
RecA and RecBC dependence for viability and temperature sensitivity for growth of the intR1 strains carrying temperature-sensitive recA200 and recBC
| Host strain | Donor lysate | Viability of transductantsa
|
|
|---|---|---|---|
| 30°C | 42°C | ||
| TH765 recA200(Ts) | P1(LK211) | + | + |
| P1(LK348) | + | − | |
| SK129 recBC(Ts) | P1(LK211) | + | + |
| P1(LK348) | + | + | |
Total number of Kanr transductants varied between 400 and 995. +, viable; −, not viable.
TABLE 4.
Recombination mutations and inactivation of SOS pathway sustained by the MG::71CC-L strain
| Donor lysate | Mutation transduced | Growth of transductants |
|---|---|---|
| P1(N2101) | recB268::Tn10 | +a |
| P1(CC3375) | recD1901::Tn10 | + |
| P1(JAS34) | recF332::Tn3 | + |
| P1(N3793) | recG263::Kanr | + |
| P1(JAS73) | recJ2001::Kanr | + |
| P1(N2057) | ΔruvA60::Tn10 | + |
| P1(GY6781) | lexA1(Ind−) | + |
The total number of transductants was above 100 on selective plates.
Table 2 shows the consequences of inactivation of the tus gene for the lethality caused by the recA mutation in the intR1CC strains. Transduction of recA::Tn10 into the intR1 tus::Kanr strains was equally efficient for both orientations of the integrated R1 replicon (compare the intR1CW and intR1CC strains, respectively, of intR1 tus::Kanr in Table 2). Thus, the vital function performed by RecA in maintaining the intR1CC strain could be dispensed with when the Ter site(s) was rendered ineffective in blocking the replication forks.
Comparison of chromosome stability in the absence of RecA activity in oriC and intR1CC strains.
RecA was shown to protect replicating DNA from UV-induced degradation (16). Chromosome stability is also seriously compromised during normal growth, in the absence of active RecA, as evidenced by extensive degradation of the bacterial DNA in E. coli recA strains (7, 50). Furthermore, chromosome partitioning is perturbed in the RecA-deficient strains, which give rise to the formation of anucleate cells at high frequency (54). However, RecA deficiency does not confer lethality upon either oriC or intR1CW strains in which chromosome replication is bidirectional (Table 2). To examine whether the unidirectional chromosome replication in the intR1CC strain causes excessive DNA degradation or formation of anucleate cells leading to cell death in the absence of RecA, we compared the rates of DNA synthesis and degradation of the intR1CC strain with that of its oriC parent under RecA+ and RecA− conditions.
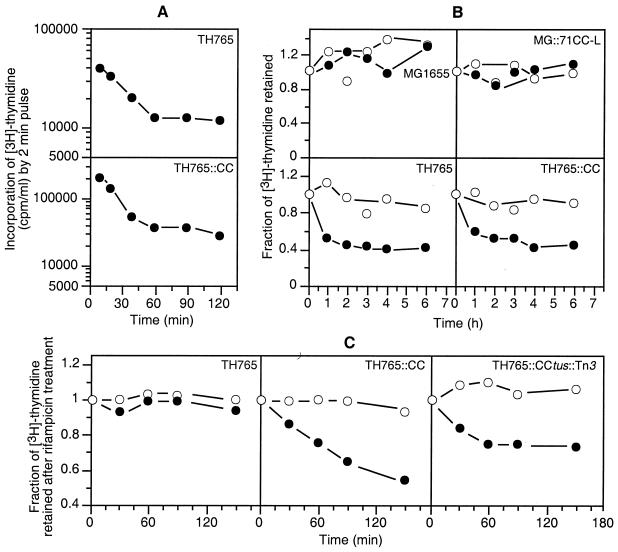
The recA200(Ts) strain TH765 (32) and its TH765::CC derivative were used to compare the rates of DNA synthesis and degradation at low (30°C) and high (42°C) temperatures. Figure 3A shows the DNA synthesis rates in the two strains as measured by [3H]thymidine incorporation (2-min pulse) during exponential growth in minimal medium, as described in Materials and Methods. At 42°C, when RecA was inactivated, the rate of DNA synthesis in both strains fell four- to fivefold within 1 h before it reached a steady lower rate. Thus, the effects of the recA mutation on the DNA synthesis rates were similar for chromosome replication from oriC and oriR1.
FIG. 3.
Role of RecA in DNA synthesis and degradation in strains MG1655, MG::71CC-L, TH765, TH765::CC, and TH765::CC tus::Tn3 grown exponentially in M9-glucose medium. (A) Change in the rates of DNA synthesis, as measured from 2-min pulse incorporation of [3H]thymidine in 1 ml of culture after a shift of temperature from 30 to 42°C. At different times, [3H]thymidine (10 μCi) was added to 1 ml of culture and incubated for 2 min before incorporation was stopped with 5 ml of ice-cold 10% TCA. After 30 min in ice, the cell suspensions were filter washed on Whatman GF/A filters, and TCA-insoluble counts were measured as described in Materials and Methods. (B) DNA degradation as measured from [3H]thymidine-labeled DNA retained in replicating cells. The cells were continuously labeled with 2.5 μCi of [3H]thymidine/ml for two to three generations. The [3H]thymidine was removed at time zero, and the cultures were incubated at 30 and 42°C. The total count per milliliter of culture was normalized to 1.0 at time zero. The degradation at different times was estimated relative to the total count at time zero, and the ratio of counts per minute was plotted against time. (C) DNA degradation in nonreplicating cells was measured as for panel B, except that replication was allowed to run out by incubating the culture with 200 μg of rifampin/ml for 3 h. The [3H]thymidine was removed at time zero before the shift to 42°C, as the cells were filter washed with prewarmed medium containing rifampin. Degradation of the labeled DNA was measured, and the data were plotted as for panel B. Open symbols, 30°C; closed symbols, 42°C.
Chromosome stability, in the presence and absence of RecA, was tested by measuring the loss of radioactivity from prelabeled bacterial DNA in oriC and intR1CC strains. As shown in Fig. 3B, the prelabeled chromosomal DNA in RecA+ strains remained intact for up to 6 h at both 30 and 42°C, irrespective of whether the bacterial chromosome was replicated from oriC or from the integrated oriR1. However, in the recA(Ts) strains, 60% of the label was degraded within 1 h of the temperature upshift, confirming the vital role of RecA in the maintenance of genome stability. The extents of and the time courses for degradation were similar for both oriC- and oriR1-driven chromosomes. Next, we wanted to test whether the RecA deficiency caused degradation of nonreplicating chromosomes. This was achieved by exposing an exponentially growing TH765 recA200(Ts) culture to rifampin (200 μg/ml) for more than one generation (2 h), allowing the ongoing replication to complete but preventing any new initiation. The tritium-labeled thymidine was washed off rapidly by filtration, and the culture was resuspended in prewarmed medium without [3H]thymidine and allowed to grow at 30 and 42°C in the presence of rifampin. Measurements of TCA-soluble counts in samples taken at different times for about 2 to 3 h showed no significant DNA degradation, even in the absence of RecA activity (Fig. 3C, 42°C). However, similar treatment of the TH765::CC derivative showed extensive degradation at 42°C without any new initiation of replication, suggesting that these cells might still retain replication intermediates (Fig. 3C). Evidently, the half-replicated chromosomes arrested at the TerA site in the intR1CC strain were targets for degradation in the absence of RecA. In the TH765::CC tus::Tn3 strain, the DNA degradation was slightly reduced but still quite extensive (Fig. 3C). Somewhat lesser but measurable degradation in the Tus-deficient cells possibly manifested an abundance of incompletely replicated chromosomes, due possibly to the slower movement of the replication forks in the “wrong” direction.
The recA(Ts) derivatives of both oriC and intR1CC strains exhibited similar increases in the frequency of anucleate-cell production upon upshift from a permissive (30°C) to a nonpermissive (42°C) temperature (data not shown). Formation of anucleate cells in the RecA-deficient intR1CC strain compared to that in the oriC strain was not sufficiently excessive to justify cell death.
Cell death in the absence of RecA requires active Tus protein.
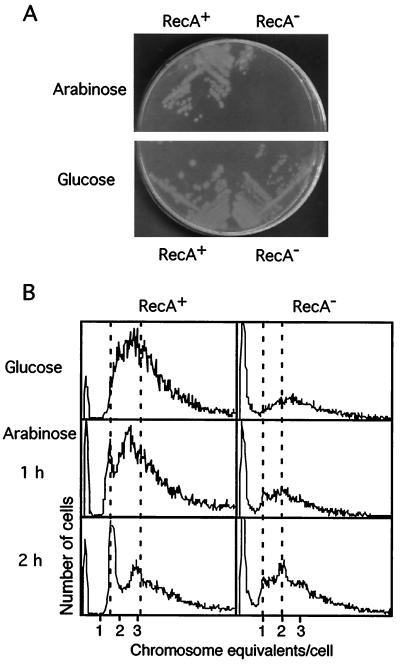
To demonstrate further the role of the Ter-Tus block of the replication forks in cell death in the absence of RecA activity, we introduced the null mutation recA::Tn10 into an intR1CC strain after knocking out the tus gene (tus::Kanr). A pBAD18 plasmid derivative with the tus gene under the control of the arabinose-inducible promoter was then introduced into the MG::CC tus::Kanr recA::Tn10 strain. Figure 4A shows the effect of induction of Tus production in this background; the intR1CC strain was able to form colonies in the absence of RecA as long as the production of the Tus protein was not induced by a change of carbon source from glucose to arabinose.
FIG. 4.
Viability of the MG::71CC-L tus::Kanr (RecA+) and MG::71CC-L tus::Kanr recA::Tn10 (RecA−) strains of E. coli harboring the plasmid pBADtus and analysis of their DNA content distributions by flow cytometry. (A) Growth on minimal-medium plates containing either glucose or arabinose plus glycerol as carbon sources. (B) DNA content in cells harvested from the exponential phase of growth in M9-glucose medium and after a shift to M9 medium containing 0.2% arabinose and glycerol as carbon sources to induce tus gene expression.
Fate of arrested replication forks at TerA in the intR1CC strain with or without active RecA.
We examined the fate of the half-replicated chromosomes in the presence and absence of RecA using the plasmid with inducible Tus production. Figure 4B shows the flow cytograms of exponentially growing cells of the MG::CC tus::Kanr(pBADtus) and MG::CC tus::Kanr recA::Tn10(pBADtus) strains at different times after induction of active Tus protein. In the absence of Tus, both RecA+ and RecA− strains showed similar flow cytometric profiles during exponential growth, with broad peaks characteristic of ongoing replication and the absence of any sharp peak representing cells with half-replicated chromosomes, as none of the Ter-Tus blocks were active. After Tus production was induced by a switch from glucose- to arabinose-containing medium, the RecA+ culture started to show the presence of cells with 1.6 chromosomes as a sharp peak that increased in size with time, indicative of blockage at the TerA site. There were also peaks corresponding to 3.2 and higher numbers of chromosome equivalents. For the RecA− strain, the half-replicated chromosomes were not visible; instead, new peaks corresponding to cells with 1 and 2 chromosomes emerged after some time. This suggested that the half-replicated chromosomes arrested at the terminus were rapidly degraded in the absence of RecA. The large population of anucleate cells produced in all RecA-deficient cells is seen as a sharp peak in the right-hand graphs of Fig. 4B.
DISCUSSION
The results presented in this work describe a RecA-dependent rescue of replication forks arrested at the Ter-Tus barriers on the chromosome. Since 5 to 20% of the forks in a normal bidirectionally replicating system fail to arrive simultaneously at the TerC region (reference 44 and data not shown), such rescue mechanisms are of some physiological significance. The intR1 derivative with asymmetric chromosome replication used in this study accentuates the value of the rescue operations, without which the round of chromosome replication could not be completed.
Similar dependence on RecA has been reported for E. coli strains with large chromosomal inversions (33) or ectopic termination blocks (18, 49) or in cases of integrative suppression of oriC (36). Among all these RecA-dependent strains with diverse replication mechanisms from different origins (oriC, oripBR322, oriP2sig5, and oriR1), the only shared characteristic seems to be the prolonged arrest of replication forks at the Ter sites due to asymmetry in the overall chromosome replication. Thus, RecA-mediated rescue seems to assume growing importance with increasing deviation from bidirectional symmetry of chromosome replication.
The rescue pathway(s) involved in assisting the arrested forks to overcome the Ter-Tus block in the intR1CC strain did not require RecBCD, RecF, RecG, RecJ, or RuvAB. It differs from the RuvAB-, RecBCD-, RecF-, or RecG-mediated mechanisms activated in E. coli when the replication forks are blocked because cells are deficient for the nonessential helicase Rep (47) or due to DNA lesions (6, 38). It is also different from the recombinational rescue of strains with replication forks blocked at a Ter-Tus complex (17, 18, 49), which were described in terms of iSDR-type replication restart from a RecBCD-mediated D-loop structure (20). However, the Ter-Tus systems used in these studies were not the natural arrest sites but rather a Ter site within the lac gene and an inverted Ter duplex cassette in the terminus region. These might not faithfully mimic the events at the natural arrest site(s). Furthermore, Luria-Bertani medium was used as the growth medium in both studies reported earlier (16, 48). E. coli strains in which completion of chromosome replication is prevented due to arrested replication forks have often been found to display rich-medium sensitivity (Rms phenotype) (33). Fast growth, with multiple replication forks (four to eight at least) traversing the chromosome at the same time possibly caused DSBs due to colliding replication forks, resulting in activation of SOS and possibly a need for the RecBCD repair pathway to survive. This might mask the RecBCD-independent pathway activated for survival of the strain with asymmetric chromosome replication. All bacterial cultures in the present work were grown in minimal medium at a growth rate such that there was never more than one round of replication ongoing at the same time, even in the wild-type cells (see the rifampin runout for the MG1655 strain in Fig. 1A).
The RecA-mediated rescue of the intR1CC strains is also different from both cSDR and iSDR pathways. Unlike iSDR, intR1CC is RecBCD independent, and unlike cSDR, it does not require the absence of RNase H, which would allow the formation of a stable R loop (9, 20). Furthermore, cSDR initiation is known to be rifampin sensitive, which would be inconsistent with the conversion of the cells with 1.6 chromosomes into ones with 2.0 chromosomes during rifampin runout (Fig. 1). The function of RecA in intR1CC strains may bear comparison with the recently discovered mechanism demonstrated to act at blocked replication forks in E. coli cells deficient for the essential replicative helicase DnaB (48). In such cells, RecA is believed to promote homologous recombination and formation of Holliday junctions without the requirement for the RecBCD, RecFOR, or RecG pathway or SOS induction. Further investigation into the genetics and biochemistry of replication restart after the forks are arrested at the Ter-Tus barriers seems necessary for clearer understanding of the mechanism(s) invoked for rescuing blocked replication forks during replication.
Comparison of synthesis and degradation of DNA in the absence of RecA in cells with oriC- or oriR1-driven chromosome replication showed that the DNA synthesis rates were reduced almost equally (three- to fivefold) in both (Fig. 2B). Thus, a deficiency of RecA seemed to affect oriC and oriR1 cells similarly, and the nonspecific DNA degradation could not be the reason for intR1CC cell death. However, measurement of DNA degradation in the absence of DNA synthesis (rifampin-treated cells) showed a major distinction between intR1CC and oriC strains. In the absence of replication, the genomic DNA replicated from oriC was stable, while chromosomes from rifampin-treated intR1CC cells showed continuing degradation for 3 h in the absence of RecA activity, suggesting that replication was not complete (no runout yet). This was consistent with the flow cytometric profiles showing the presence of a considerable fraction of cells with 1.6 chromosomes and with higher-order replication intermediates in the intR1CC cells after rifampin treatment. These half-replicated chromosomes from the counterclockwise arm (counterclockwise from the origin to TerA) were susceptible to rapid degradation when not protected by RecA, as visualized in the experiment with inducible Tus expression in RecA− cells with the intR1CC origin: a 1-chromosome-equivalent peak replaced the 1.6-chromosome peak.
Combining the DNA synthesis and degradation data with flow cytometry and Southern blot analysis, we showed that RecA prevents the rapid and complete degradation of a chromosome segment arrested at the polar block of a Ter site. Despite the prolonged block of the unidirectional replication fork at TerA, the intR1CC strains survive and complete replication, albeit taking much longer than would be warranted by uniform progression of replisomes. RecA protects the arrested forks at Ter-Tus blocks by possibly binding to the exposed single-stranded DNA. In addition, RecA may play a vital role in the recombination-mediated regeneration of the forks at the Ter sites or at other sites on the chromosome, along with replication and/or recombination proteins yet to be identified. In RecA-deficient cells, the chromosomal arm with the arrested fork may be degraded too rapidly for the slower kinetics of replisome reassembly, resulting in cell death. The process of RecA-mediated pairing and promotion of strand exchange to set up a new replication fork structure may also be responsible for dislodging the Tus protein from the Ter site.
It could be argued that replication in the intR1CC strain might rely on RecA-dependent initiations from alternate cryptic origins, as for cSDR strains with inactive oriC (20); elimination of blockage in the intR1CC recA tus strain would render it viable. However, nonviability of the intR1 strains (both intR1CW and intR1CC [see Results]) in the absence of RepA activity argues strongly against the presence of alternative initiation activity. Furthermore, cSDR-type replication requires stable R loops, which are highly unlikely in the presence of RNase H.
The progress of replication forks in the “forbidden” direction in the intR1 tus strain might be slower due to pauses arising from collisions with transcriptional complexes (5, 10). Evidently, rescue from these replication pauses can be performed without RecA, as the intR1CC tus recA strain is viable, but the cells would be filled with replication intermediates for a long time after the addition of rifampin. This is supported by the long runout time (data not shown) and the DNA degradation (less than in Tus+) seen for the intR1CC strain in the absence of Tus protein (Fig. 3C).
Accidental and frequent arrests of the replication forks shown from accumulation of DSBs during replication (reviewed in references 8, 39, and 45), might be rather short-lived compared to the blocks of the replication fork(s) at the Ter-Tus complex in the terminus region. If RecA were essential for the recombinogenic rescue of these replication forks arrested anywhere on the chromosome, then null mutation in the recA gene would be fatal. It is not. But RecA recombinase deficiency in E. coli strains with a high degree of positional or directional asymmetry in chromosome replication is lethal and needs to be suppressed by removal of the termination blocks.
The Ter-Tus blocks are used during normal bidirectional replication in wild-type cells growing exponentially in minimal medium (reference 44 and data not shown) when coordination between the replication forks is lost. In such situations, one fork arrives earlier and is blocked at the first Ter site encountered while the other is arrested upstream on the other arm of the chromosome due to DNA lesions or steric hindrances, etc. An understanding of the RecA-mediated mechanism that makes prolonged arrests of the replication forks at the terminus tolerable might provide valuable insights into the physiology of the E. coli replication terminus and the evolution of its role in the cell cycle. Moreover, investigation into the genetics and biochemistry of replication restart might help in better understanding the mechanism(s) invoked for rescuing altered cell cycles, in addition to furthering our appreciation of the intimate link between recombination and replication.
ACKNOWLEDGMENTS
We thank D. K. Chattoraj, T. Hill, T. Horiuchi, P. Kuempel, B. Michel, B. Peters, J. Sawitzke, and J. L. Rosner for kindly providing the plasmids and strains.
S.M.-P. was supported by a Marie Curie fellowship from the EU (Biotech2; contract no. BIO4CT985017). This research was funded by grants from the Swedish Cancer Society and the Swedish Natural Science Research Council.
REFERENCES
- 1.Asai T, Bates D B, Kogoma T. DNA replication triggered by double-stranded breaks in E. coli: dependence on homologous recombination functions. Cell. 1994;78:1051–1061. doi: 10.1016/0092-8674(94)90279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berlyn M K B, Low K B, Rudd K E. Linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 1715–1902. [Google Scholar]
- 3.Bernander R, Åkerlund T, Nordström K. Inhibition and restart of initiation of chromosome replication: effects on exponentially growing Escherichia coli cells. J Bacteriol. 1995;177:1670–1682. doi: 10.1128/jb.177.7.1670-1682.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernander R, Merryweather A, Nordström K. Overinitiation of replication of the Escherichia coli chromosome from an integrated runaway-replication derivative of plasmid R1. J Bacteriol. 1989;171:674–683. doi: 10.1128/jb.171.2.674-683.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewer B J. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53:679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- 6.Courcelle J, Carswell-Crumpton C, Hanawalt P C. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox M M. Recombinational DNA repair in bacteria and the RecA protein. Prog Nucleic Acids Res Mol Biol. 1999;63:311–366. doi: 10.1016/s0079-6603(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 8.Cox M M, Goodman M F, Kreuzer K N, Sherratt D J, Sandler S J, Marians K J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 9.Dasgupta S, Bernander R, Nordström K. In vivo effect of the tus mutation on cell division in an Escherichia coli strain where chromosome replication is under the control of plasmid R1. Res Microbiol. 1991;142:177–180. doi: 10.1016/0923-2508(91)90027-8. [DOI] [PubMed] [Google Scholar]
- 10.French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992;258:1362–1365. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- 11.Friedman D I. Interaction between bacteriophage lambda and its Escherichia coli host. Curr Opin Genet Dev. 1992;2:727–738. doi: 10.1016/s0959-437x(05)80133-9. [DOI] [PubMed] [Google Scholar]
- 12.George J W, Kreuzer K N. Repair of double-strand breaks in bacteriophage T4 by a mechanism that involves extensive DNA replication. Genetics. 1996;143:1507–1520. doi: 10.1093/genetics/143.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodman M F. Coping with replication ‘train wrecks’ in Escherichia coli using Pol V, Pol II and RecA proteins. Trends Biochem Sci. 2000;25:189–195. doi: 10.1016/s0968-0004(00)01564-4. [DOI] [PubMed] [Google Scholar]
- 14.Grinsted J, Saunders J R, Ingram L C, Sykes R B, Richmond M H. Properties of an R factor which originated in Pseudomonas aeruginosa 1822. J Bacteriol. 1972;110:529–537. doi: 10.1128/jb.110.2.529-537.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill T M, Henson J M, Kuempel P L. The terminus region of the Escherichia coli chromosome contains two separate loci that exhibit polar inhibition of replication. Proc Natl Acad Sci USA. 1987;84:1754–1758. doi: 10.1073/pnas.84.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horii Z, Suzuki K. Degradation of the DNA of Escherichia coli K12 rec-(JC1569b) after irradiation with ultraviolet light. Photochem Photobiol. 1968;8:93–105. doi: 10.1111/j.1751-1097.1970.tb05976.x. [DOI] [PubMed] [Google Scholar]
- 17.Horiuchi T, Fujimura Y. Recombinational rescue of the stalled DNA replication fork: a model based on analysis of an Escherichia coli strain with a chromosome region difficult to replicate. J Bacteriol. 1995;177:783–791. doi: 10.1128/jb.177.3.783-791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horiuchi T, Fujimura Y, Nishitani H, Kobayashi T, Hidaka M. The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J Bacteriol. 1994;176:4656–4663. doi: 10.1128/jb.176.15.4656-4663.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen K F. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kogoma T, Lark K G. Characterization of the replication of Escherichia coli DNA in the absence of protein synthesis: stable DNA replication. J Mol Biol. 1975;94:243–256. doi: 10.1016/0022-2836(75)90081-9. [DOI] [PubMed] [Google Scholar]
- 22.Kogoma T, Skarstad K, Boye E, von Meyenburg K, Steen H B. RecA protein acts at the initiation of stable DNA replication in rnh mutants of Escherichia coli K-12. J Bacteriol. 1985;163:439–444. doi: 10.1128/jb.163.2.439-444.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koppes L, Nordström K. Insertion of an R1 plasmid into the origin of replication of the E. coli chromosome: random timing of replication of the hybrid chromosome. Cell. 1986;44:117–124. doi: 10.1016/0092-8674(86)90490-3. [DOI] [PubMed] [Google Scholar]
- 24.Kowalczykowski S C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 25.Kreuzer K N, Saunders M, Weislo L J, Kreuzer H W. Recombination-dependent DNA replication stimulated by double-strand breaks in bacteriophage T4. J Bacteriol. 1995;177:6844–6853. doi: 10.1128/jb.177.23.6844-6853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kushner S R. Differential thermolability of exonuclease and endonuclease activities of the recBC nuclease isolated from thermosensitive recB and recC mutants. J Bacteriol. 1974;120:1219–1222. doi: 10.1128/jb.120.3.1219-1222.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 28.Kuzminov A. Instability of inhibited replication forks in E. coli. Bioessays. 1995;17:733–741. doi: 10.1002/bies.950170810. [DOI] [PubMed] [Google Scholar]
- 29.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lark K G, Lark C A. recA-dependent DNA replication in the absence of protein synthesis: characteristics of a dominant lethal replication mutation, dnaT, and requirement for recA+ function. Cold Spring Harbor Symp Quant Biol. 1979;43:537–549. doi: 10.1101/sqb.1979.043.01.059. [DOI] [PubMed] [Google Scholar]
- 31.Lloyd R G, Buckman C, Benson F E. Genetic analysis of conjugational recombination in Escherichia coli K12 strains deficient in RecBCD enzyme. J Gen Microbiol. 1987;133:2531–2538. doi: 10.1099/00221287-133-9-2531. [DOI] [PubMed] [Google Scholar]
- 32.Lloyd R G, Low B, Godson G N, Birge E A. Isolation and characterization of an Escherichia coli K-12 mutant with a temperature-sensitive recA− phenotype. J Bacteriol. 1974;120:407–415. doi: 10.1128/jb.120.1.407-415.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louarn J M, Bouché J P, Legendre F, Louarn J, Patte J. Characterisation and properties of very large inversions of the E. coli chromosome along the origin-to-terminus axis. Mol Gen Genet. 1985;201:467–476. doi: 10.1007/BF00331341. [DOI] [PubMed] [Google Scholar]
- 34.Maisnier-Patin S, Dasgupta S, Krabbe M, Nordström K. Conversion to bidirectional replication after unidirectional initiation from R1 plasmid origin integrated at oriC in Escherichia coli. Mol Microbiol. 1998;30:1067–1079. doi: 10.1046/j.1365-2958.1998.01136.x. [DOI] [PubMed] [Google Scholar]
- 35.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 36.Mao Y M, Shi Q, Li Q G, Sheng Z J. recA gene dependence of replication of the Escherichia coli chromosome initiated by plasmid pUC13 integrated at predetermined sites. Mol Gen Genet. 1991;225:234–240. doi: 10.1007/BF00269854. [DOI] [PubMed] [Google Scholar]
- 37.Marians K J. Replication and recombination intersect. Curr Opin Genet Dev. 2000;10:151–156. doi: 10.1016/s0959-437x(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 38.McGlynn P, Lloyd R G. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 39.Michel B. Replication fork arrest and DNA recombination. Trends Biochem Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- 40.Michel B, Ehrlich S D, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller J. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 42.Mosig G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 43.Motamedi M R, Szigety S K, Rosenberg S M. Double-strand-break repair recombination in Escherichia coli: physical evidence for a DNA replication mechanism in vivo. Genes Dev. 1999;13:2889–2903. doi: 10.1101/gad.13.21.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelletier A J, Hill T M, Kuempel P L. Location of sites that inhibit progression of replication forks in the terminus region of Escherichia coli. J Bacteriol. 1988;170:4293–4298. doi: 10.1128/jb.170.9.4293-4298.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination or “gimme a break.”. Genes Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- 46.Rupp W D. DNA repair mechanisms. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 2277–2294. [Google Scholar]
- 47.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 48.Seigneur M, Ehrlich S D, Michel B. RuvABC-dependent double-strand breaks in dnaBts mutants require RecA. Mol Microbiol. 2000;38:565–574. doi: 10.1046/j.1365-2958.2000.02152.x. [DOI] [PubMed] [Google Scholar]
- 49.Sharma B, Hill T M. Insertion of inverted Ter sites into the terminus region of the Escherichia coli chromosome delays completion of DNA replication and disrupts the cell cycle. Mol Microbiol. 1995;18:45–61. doi: 10.1111/j.1365-2958.1995.mmi_18010045.x. [DOI] [PubMed] [Google Scholar]
- 50.Skarstad K, Boye E. Degradation of individual chromosomes in recA mutants of Escherichia coli. J Bacteriol. 1993;175:5505–5509. doi: 10.1128/jb.175.17.5505-5509.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torrey T A, Kogoma T. Genetic analysis of constitutive stable DNA replication in rnh mutants of Escherichia coli K12. Mol Gen Genet. 1987;208:420–427. doi: 10.1007/BF00328133. [DOI] [PubMed] [Google Scholar]
- 52.von Meyenburg K, Boye E, Skarstad K, Koppes L, Kogoma T. Mode of initiation of constitutive stable DNA replication in RNase H-defective mutants of Escherichia coli K-12. J Bacteriol. 1987;169:2650–2658. doi: 10.1128/jb.169.6.2650-2658.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu J R, Gilbert D M. Rapid DNA preparation for 2D gel analysis of replication intermediates. Nucleic Acids Res. 1995;23:3997–3998. doi: 10.1093/nar/23.19.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zyskind J W, Svitil A L, Stine W B, Biery M C, Smith D W. RecA protein of Escherichia coli and chromosome partitioning. Mol Microbiol. 1992;6:2525–2537. doi: 10.1111/j.1365-2958.1992.tb01429.x. [DOI] [PubMed] [Google Scholar]