Abstract
Significance:
Thioredoxin interacting protein (TXNIP) is a member of the arrestin fold superfamily with important cellular functions, including cellular transport, mitochondrial energy generation, and protein cycling. It is the only arrestin-domain protein known to covalently bind to thioredoxin and plays roles in glucose metabolism, inflammation, apoptosis, and cancer.
Recent Advances:
The crystal structure of the TXNIP-thioredoxin complex provided details about this fascinating interaction. Recent studies showed that TXNIP is induced by endoplasmic reticulum (ER) stress, activates NLR family pyrin domain containing 3 (NLRP3) inflammasomes, and can regulate glucose transport into cells. The tumor suppressor role of TXNIP in various cancer types and the role of TXNIP in fructose absorption are now described.
Critical Issues:
The influence of TXNIP on redox state is more complex than its interaction with thioredoxin.
Future Directions:
It is incompletely understood which functions of TXNIP are thioredoxin-dependent. It is also unclear whether TXNIP binding can inhibit glucose transporters without endocytosis. TXNIP-regulated control of ER stress should also be investigated further. Antioxid. Redox Signal. 38, 442–460.
Keywords: TXNIP, arrestin, thioredoxin, redox state, inflammasome, glucose
The Arrestin Fold Protein Family
Thioredoxin interacting protein (TXNIP) is a member of a family of proteins believed to have structural similarities. Arrestin fold proteins are predicted to share a common arrestin domain structure and are widely present from single-celled organisms to metazoans, and many proteins in this superfamily play roles in metabolic regulation and intracellular signaling (Habourdin et al, 2013). The arrestin fold superfamily is composed of two main branches: the more ancient alpha-arrestin family and the well-studied beta-arrestin family (Patwari and Lee, 2012) (Fig. 1A).
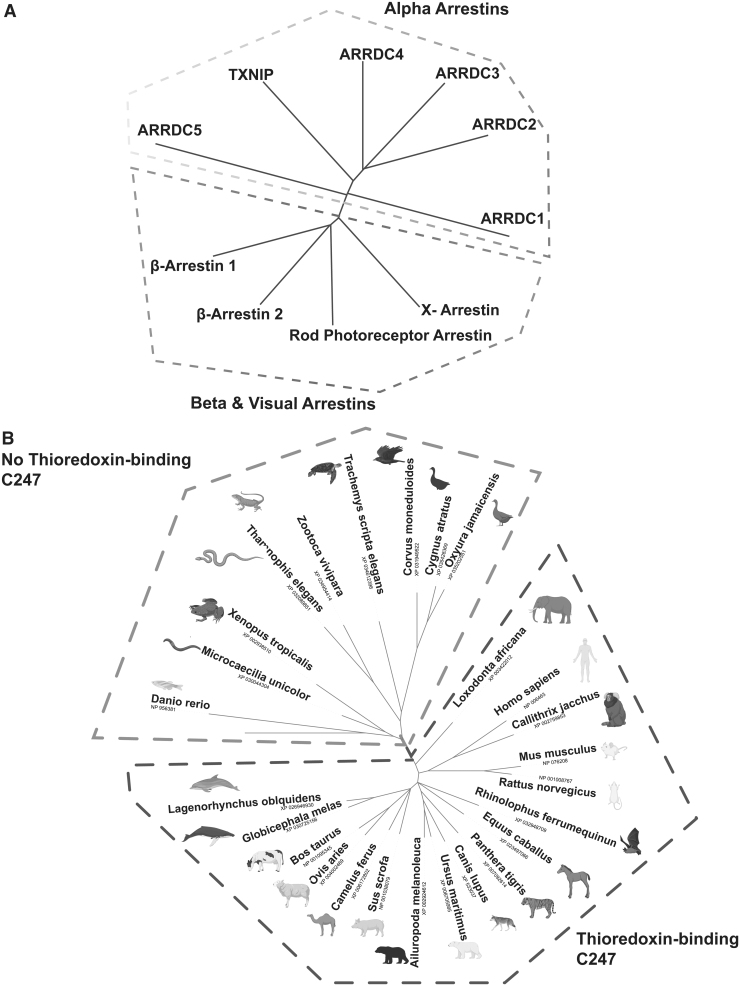
FIG. 1.
TXNIP as a member of the arrestin fold superfamily. (A) TXNIP is a member of the diverse arrestin fold superfamily that is composed of alpha (on top) and beta-arrestin (on bottom) branches. (B) The key cysteine in TXNIP (C247) for binding thioredoxin evolved in animals. Mammals bear a C247 cysteine residue, whereas nonmammalians such as fish and frogs do not (light gray: nonmammalians that do not have TXNIP C247 residue, dark gray: mammalians that have TXNIP C247 residue). TXNIP, thioredoxin interacting protein. (Drawn using iTOL, Letunic et al, 2019 Nuc. Acid. Res, PhytoL and NCBI COBALT, Created using Biorender.com)
Sequence-wise, alpha-arrestins, sometimes designated ARRDC for “Arrestin Domain-Containing,” have evolutionarily conserved PY motifs that bind to WW domains in their C-terminal tails, and beta-arrestins have an alpha-helix that is missing in alpha-arrestins in the arrestin N-terminal domain (Alvarez, 2008). Orthologues of the mammalian ARRDC family have been detected in lower organisms such as yeasts (Arrestin-related trafficking protein ARTs such as Smf1) (Nikko et al, 2008), archaea/bacteria (Spo0m) (Alvarez, 2008), and amoebae (Arrestin-domain containing proteins such as AdcA-F) (Guetta et al, 2010).
In primitive organisms such as Dictyostelium discoideum, ARRDC fold proteins respond to stimuli such as cyclic AMP (cAMP) and folate (Mas et al, 2018) resembling mammalian TXNIP, which responds to intracellular carbohydrate and cAMP levels (Patwari and Lee, 2012; Shalev, 2014).
The mammalian arrestin fold superfamily has diverse and essential intracellular functions for cellular homeostasis, including intracellular regulation of G-protein coupled receptors (GPCR) (Nabhan et al, 2010; Patwari and Lee, 2012; Shea et al, 2012; Shenoy et al, 2001). “Beta-arrestins” were extensively studied due to their roles in reducing rhodopsin and beta-adrenergic receptor signaling (Benovic et al, 1987; Dorey and Faure, 1977; Pfister et al, 1985). The “alpha-arrestins” also regulate several essential cellular processes in mammals, including glucose transport, fatty acid flux, mitochondrial citric acid cycle, carbohydrate response, extracellular vesicle generation, protein trafficking and ubiquitination, receptor recycling, thermogenesis, and adrenergic signaling (Farooq et al, 2022; Patwari and Lee, 2012; Wedegaertner et al, 2020).
The human alpha-arrestin family is composed of six members: ARRDC 1-5 and TXNIP (Fig. 1A). All alpha-arrestins have predicted structural homology with similar N- and C-terminal arrestin domains (Patwari et al, 2006). The alpha-arrestin protein subfamily members have potentially important tumor-suppressive and tumor growth regulatory roles in several cancers, including breast (Park et al, 2018), skin (Meylan et al, 2021), thyroid (Morrison et al, 2014), and lung cancers (Liang et al, 2021).
Several proteins in the alpha-arrestin family are known to control glucose metabolism in mammals. ARRDC3, which has been linked to human obesity in males, controls insulin receptor signaling in the liver to regulate glucose metabolism (Batista et al, 2020; Patwari et al, 2011). ARRDC4, a close homolog of TXNIP, can control glucose metabolism in vitro (Patwari et al, 2009). However, TXNIP is unique as it is the only arrestin fold protein to bind covalently to thioredoxin. Isolated from Vitamin D3-stimulated HL-60 cells, TXNIP was initially named “Vitamin-D-Upregulated-Protein 1” (VDUP1) (Chen and DeLuca, 1994). Following this, the protein was named “thioredoxin binding protein-2 (TBP-2)” from a yeast two-hybrid screen searching for thioredoxin binding proteins (Nishiyama et al, 1999).
The Yodoi lab showed that TBP-2 deficiency enhanced tumor growth factor (TGF)-β signaling (Masaki et al, 2012). TBP-2 was also found to improve insulin sensitivity and secretion without affecting obesity (Yoshihara et al, 2010). Later, TXNIP received its widely used name from Bodnar et al in 2002 when the absence of txnip was found through positional cloning to cause combined hyperlipidemia in mice (Bodnar et al, 2002).
This review will focus on TXNIP, its unique thioredoxin-binding property and biological functions that may depend on thioredoxin interaction; its regulation of carbohydrate and lipid metabolism; as well as the roles of TXNIP that are not yet well defined mechanistically. We will discuss the possibility that thioredoxin-binding serves as a cellular redox state sensor as a consequence of metabolic status rather than as a thioredoxin inhibitor, thus connecting the thioredoxin-interaction property with the regulation of metabolism by TXNIP.
Mechanism of the TXNIP-Thioredoxin Interaction
Thioredoxin is a thiol-oxidoreductase that protects cells from oxidative stress (Baba and Bhatnagar, 2018). It is conserved through all species, including bacteria, except for the bacterium Tropheryma whipplei because of its parasitic characteristics as obligate intracellular bacteria (Marth, 2015). Thioredoxin reduces protein disulfides through C32 and C35 at its active site (Eklund et al, 1991; Jeng et al, 1994; Patwari and Lee, 2012). The reduced thioredoxin reduces the disulfide bonds of target proteins and becomes oxidized. Nicotinamide adenine dinucleotide phosphate (NADPH)-dependent thioredoxin reductase restores the reduced thioredoxin to start a new cycle (Yamawaki et al, 2003).
Peroxiredoxin is oxidized by hydrogen peroxide and oxidizes a redox-sensitive target protein that is reduced by thioredoxin (Stancill and Corbett, 2021). Methionine sulfoxide reductases also require reduction by thioredoxin (Dobrovolska et al, 2012). The binding of transcription factor nuclear factor kappa B (NFkB) to DNA requires reduction by thioredoxin (Qin et al, 1995). Humans carry two thioredoxin (Txn) genes, Txn1 in the cytoplasm and Txn2 in mitochondria. Thioredoxin is critical for mammalian life, and mice with genetic loss of Txn1 and Txn2 die shortly after implantation (Matsui et al, 1996; Nonn et al, 2003).
Thioredoxin interaction with TXNIP requires reduced thioredoxin, and TXNIP will not bind to oxidized thioredoxin or catalytic mutants of thioredoxin. On TXNIP, the key cysteine C247 was shown to be an essential residue for thioredoxin interaction by mutational studies in cultured cells (Patwari et al, 2006). This residue appears in TXNIP in later metazoans and is conserved in mammals, and thus the probable thioredoxin-binding function of TXNIP may have evolved for specific purposes (Fig. 1B). In 2014, Hwang et al solved the crystal structure of TXNIP-thioredoxin complex by using a thioredoxin C35A mutant that could not dissociate from its target protein and TXNIP mutant (C120S/C170S/C205S/C267S) that behaved better in Escherichia coli expression (Fig. 2).
FIG. 2.
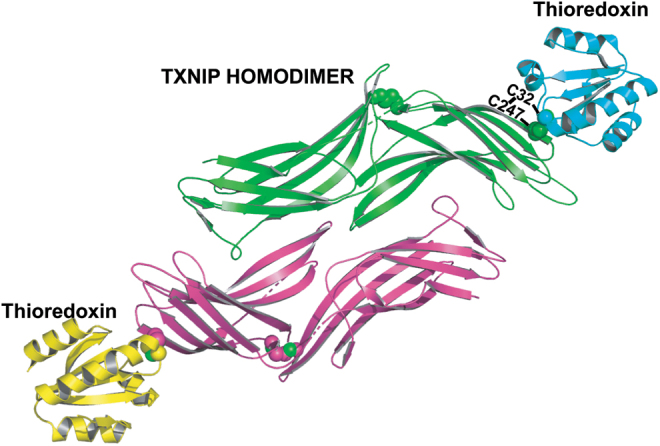
The TXNIP-thioredoxin interaction. Crystal structure of the TXNIP dimer-thioredoxin complex, resolution 2.7 Å, balls refer to bonding cysteine residues, green and pink are beta sheets and disoriented regions of TXNIP, and yellow and cyan are alpha helices and beta sheets of thioredoxin. Created using PyMOL Molecular Graphics System.
Because this model was built from in vitro studies of purified recombinant proteins where reducing agents were avoided, the second half of the thioredoxin reaction cycle could not be completed and TXNIP was left with a disulfide. Therefore, the structure itself did not support TXNIP as an inhibitor of thioredoxin function. However, it did point out how thioredoxin can reduce TXNIP if its C247 forms an internal disulfide with another cysteine residue. This interaction of TXNIP and thioredoxin may be important for TXNIP stability; thioredoxin may bind to TXNIP and render it more stable, avoiding insulin-induced degradation of TXNIP (Chutkow and Lee, 2011).
The interaction of TXNIP with cytoplasmic Txn1 has attracted attention, because TXNIP is mostly localized to the cytoplasm and the nucleus. In mitochondria, TXNIP forms a bond with thioredoxin2. This decreases thioredoxin2 binding to apoptosis signal-regulating kinase 1, inducing mitochondrial apoptosis via the release of cytochrome c and caspase-3 cleavage (Saxena et al, 2010).
TXNIP and Inflammasome Activation
A well-characterized biological process that depends on the TXNIP-thioredoxin1 interaction is the activation of inflammasomes. Inflammasomes are cytoplasmic multiprotein structures that are critical in the pathophysiology of multiple diseases and are formed on detection of warning signals for the cell (Broz and Dixit, 2016). The NLR family pyrin domain containing 3 (NLRP3) inflammasome is the most studied inflammasome complex and is composed of NLRP3, apoptosis-associated speck-like protein containing CARD (ASC), and caspase-1 (Zhao and Zhao, 2020). The NLRP3 inflammasome is reported to be associated with multiple infectious diseases, including acquired immune deficiency syndrome (AIDS) by human immunodeficiency virus (HIV)-1 (Pontillo et al, 2010) and tuberculosis (Souza de Lima et al, 2016), as well as metabolic conditions including type 2 diabetes (Lee et al, 2013) and obesity (Vandanmagsar et al, 2011).
Reactive oxygen species (ROS) and endoplasmic reticulum (ER) stress induce NLRP3 inflammasome complex activation, triggering an inflammatory response (Oslowski et al, 2012; Zhao et al, 2020; Zhou et al, 2010). In 2009, Zhou et al (2010) reported that TXNIP may have a pivotal role in ROS-mediated inflammasome activation using cultured cells. They built a model where in normoxic conditions, the TXNIP-thioredoxin interaction renders TXNIP unavailable for NLRP3 interaction (Zhou et al, 2010). On the activation of oxidative stress, TXNIP releases from thioredoxin and attaches to NLRP3 to activate it, and then caspase-1 cleaves interleukin (IL)-1β (Zhou et al, 2010).
This model requires most cellular TXNIP to exist as a covalent complex with thioredoxin in the cytoplasm without stimulation to keep NLRP3 inactive. However, this prerequisite is inconsistent with the reducing environment in the cytoplasm. Therefore, the detailed mechanism of TXNIP involvement in NLRP3 activation needs further investigation. On the other hand, thioredoxin itself may influence NLRP3 activation. In vitro, it was shown that thioredoxin1 suppressed the NLRP3 expression and subsequent IL-1β and caspase-1 secretion (Wang et al, 2020). The same article also demonstrated in vivo that thioredoxin1 reduced atherosclerosis development by inhibiting NLRP3 inflammasomes (Wang et al, 2020). On the other hand, thioredoxin was shown to enhance NFkB target DNA binding and NLRP3 inflammasome activation independent of TXNIP by another group (Muri et al, 2020).
Multiple inflammatory conditions are linked to the TXNIP-inflammasome axis. High glucose levels promote TXNIP expression and NLRP3 activation in a time‑ and dose‑dependent manner (Gu et al, 2019). In diabetes and atherosclerosis, the NLRP3 inflammasome mediates inflammation through caspase-1 activation and the secretion of pro-inflammatory cytokines IL-1β/IL-18 (Grebe et al, 2018; Schroder et al, 2010; Sun et al, 2017; Zhou et al, 2010). The ROS-TXNIP-NLRP3 biological axis may also participate in tubular injury of diabetic neuropathy (Han et al, 2018). Hepatic inflammation due to poor diet was linked to the TXNIP-NLRP3 axis (Mohamed et al, 2018).
TXNIP ablation improved high-fat diet-induced hepatosteatosis and inflammation by regulating the TLR2-NLRP3 inflammasome pathway (Mohamed et al, 2018). TXNIP was also found to play a role in fructose-induced inflammasome activation in the liver (Zhang et al, 2015); rats given fructose-containing drinking water for 8 weeks had elevated TXNIP and NLRP3 inflammasome levels in the liver. Interestingly, in txnip-silenced hepatocytes, fructose did not induce inflammasome activation (Zhang et al, 2015). The TXNIP-NLRP3 pathway also has effects on the colon.
For instance, the NLRP3 inflammasome may play a role in ulcerative colitis development (Zhen and Zhang, 2019), and TXNIP was found to be important for inflammation of colonic mucosa (Takahashi et al, 2007). In concordance with that concept, a flavonoid was shown to inhibit macrophage NLRP3 inflammasome activation recently in a model of ulcerative colitis by suppressing the TXNIP pathway (Liu et al, 2020).
The TXNIP-NLRP3 inflammasome plays roles in diverse organ systems, including the cardiovascular, respiratory, and nervous systems. For example, TXNIP activates the NLRP3 complex in myocardial ischemia/reperfusion injury in an ROS-dependent fashion (Liu et al, 2014). An essential part of the inflammatory response, macrophage activation, is also regulated by the TXNIP-NLRP3/inflammasome intracellular pathway (Lutz et al, 2016). The TXNIP-thioredoxin complex together with the nuclear factor erythroid 2–related factor 2 (NRF2) can also affect redox balance in a mouse model of Alzheimer's disease, and antioxidant treatment can suppress TXNIP-NLRP3 inflammasome activation by NRF2 induction (Wang et al, 2019).
The involvement of TXNIP in inflammasome complexes is not limited to the NLRP3 complex. A decrease in expression of the NLRP1 complex proteins was observed recently in human β cells treated with the small-molecule TXNIP inhibitor SRI-37330, which is a potential drug candidate for diabetes (Thielen et al, 2020).
TXNIP and Regulation of Glucose Metabolism by a Cellular Glucose Autoregulatory Circuit
Beyond its thioredoxin interacting ability, TXNIP regulation of metabolism in mouse models is clearly established. The first loss-of-function model arose when Bodnar et al (2002) identified a truncated txnip gene using positional cloning analysis of a combined hyperlipidemia mouse. Later, four other groups independently generated TXNIP knockout (KO) animals (Hui et al, 2008; Lee et al, 2005; Oka et al, 2006; Yoshioka et al, 2007), including two conditional deletion lines. Consistently, TXNIP global KO animals were reported to exhibit increased fasting plasma triglycerides, free fatty acids, ketone bodies, and hypoglycemia. These phenotypes were strong enough to be observed in fed mice in some cases (Oka et al, 2006; Sheth et al, 2005).
In 2007, Parikh et al made the discovery that TXNIP negatively regulates glucose uptake in peripheral tissues. Overexpressing TXNIP in cultured adipocytes reduced glucose uptake, whereas knocking down txnip enhanced glucose uptake. The lack of TXNIP also promoted adipogenesis in cultured cells (Chutkow and Lee, 2011; Chutkow et al, 2010), which was consistent with a higher adiposity in mice without txnip (Chen et al, 2008a; Chutkow et al, 2010; Sheth et al, 2005). TXNIP regulation of glucose uptake may explain the major metabolic phenotypes observed in mice.
The absence of TXNIP leads to more glucose taken up into peripheral tissues, and thus the TXNIP KO animal becomes hypoglycemic whereas hyperlipidemia follows due to excess intracellular glucose. Since excess glucose can be stored as fat (Chutkow et al, 2010) or secreted as lactate after glycolysis, this could result in a higher blood lactate level (Bodnar et al, 2002; Oka et al, 2006; Sheth et al, 2005). By allowing peripheral tissues to take up more glucose, txnip deletion can protect animals against high-fat diet-induced and genetically induced insulin resistance (Chen et al, 2008a; Chutkow et al, 2010; Hui et al, 2008). In 2009, Patwari et al demonstrated that TXNIP regulation of glucose uptake is independent of its interaction with thioredoxin using the TXNIP C247S mutant. As described earlier, a closely related alpha arrestin family member, ARRDC4, which does not possess C247 and does not interact with thioredoxin, also has the ability to inhibit glucose uptake (Patwari et al, 2009).
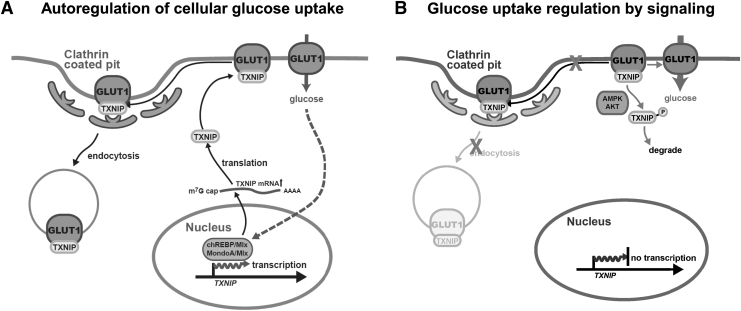
Further, TXNIP is one of the top genes regulated by glucose availability on the transcriptional level via carbohydrate response element binding protein (ChREBP)/Mlx and MondoA/Mlx (Cha-Molstad et al, 2009; Poungvarin et al, 2015; Stoltzman et al, 2008). These transcription factors bind to the carbohydrate response element on the TXNIP promoter to induce gene expression. This completes a cellular autoregulatory circuit for adjusting how much glucose should be allowed into the cell at a time. TXNIP protein is rapidly produced on glucose stimulation to reduce further glucose influx (Fig. 3A).
FIG. 3.
A TXNIP autoregulatory pathway of cellular glucose. Autoregulatory pathway of cellular glucose. (A) Glucose metabolites induce Txnip transcription via ChREBP/Mlx or MondoA/Mlx. TXNIP protein binds to GLUT1 on the plasma membrane and facilitates GLUT1 endocytosis via the clarthin pathway. (B) Activated AMPK from energy stress or activated Akt from growth factor stimulation phosphorylates TXNIP on Ser308, leading to its dissociation from GLUT1. This results in the immediate inhibition of GLUT1 endocytosis and acute increase in glucose uptake. AMPK, AMP-activated protein kinase; ChREBP, carbohydrate response element binding protein; GLUT1, glucose transporter 1.
Recently, TXNIP was shown to facilitate fructose absorption in the small intestine (Dotimas et al, 2016; Shah et al, 2020). A high-fructose diet induces both txnip, solute carrier family 2 member 2 (slc2a2), and slc2a5 gene expression in the intestine. Depending on how fructose was supplied (either in solid diet or in drinking water) and the type of TXNIP KO used (total KO or intestinal KO), upregulation of slc2a5 can be dependent on TXNIP. In intestinal-specific txnip deletion, TXNIP was found to be required for the apical localization of GLUT5.
In contrast to the glucose-ChREBP/MondoA/Mlx-TXNIP negative feedback loop, the absence of TXNIP reduces fructose uptake instead of promoting it, and the dependence of TXNIP induction by fructose on chREBP/Mlx has yet to be clarified (Kim et al, 2017). The studies also showed that energy-rich diets promote both txnip and slc2a5 expression and fructose absorption. Given the important role that fructose may play in metabolic disease, this finding indicates a potentially important interaction between glucose and fructose metabolism in the intestine epithelium.
Mechanism of TXNIP Regulation on Glucose Transport
In 2013, Wu et al discovered that TXNIP is an adaptor protein for clathrin-dependent endocytosis of the glucose transporter GLUT1. TXNIP contains a dileucine motif (L351, L352) in its flexible C-terminal tail that serves as either a clathrin or an AP2 binding site. A mutation of these two residues abolishes the clathrin-coated pit interaction, but not plasma membrane localization of TXNIP. However, the removal of the clathrin-interaction motif from TXNIP was sufficient to decrease the endocytosis rate of GLUT1. It was later observed that TXNIP also serves as an adaptor for GLUT2, GLUT3, and GLUT4 (Waldhart et al, 2017); GLUT4 is likely the physiological target of TXNIP within adipose tissue and muscle.
GLUT1-4 is a facilitator, thus glucose flows through these transporters in the direction of a high-to-low concentration with no other energy requirement. Cytoplasmic-free glucose concentration is usually kept lower than that of the blood by the rapid formation of glucose-6-phosphate. Therefore, how much glucose passes through these transporters depends on how many are present on the cell surface. Hence, higher expression of TXNIP will result in more transporter endocytosis and thus fewer available transporters on the plasma membrane for glucose. In the absence of TXNIP, endocytosis of transporters may be slowed, thus permitting greater glucose influx. It remains unclear whether the binding of TXNIP can inhibit glucose transporters even without endocytosis.
The interaction of TXNIP with both plasma membrane and glucose transporters requires phosphoinositides (Dykstra et al, 2021; Waldhart et al, 2017). TXNIP interaction with liposomes containing negatively charged phosphoinositides can be detected using a liposome-based floating assay. Using reconstituted GLUT1 in PI(4,5)P2-containing lipid nanodiscs and recombinant TXNIP, this interaction can be quantified with isothermal titration calorimetry. It was found that the bulk of binding energy comes from the ionic interaction between the acidic lipid and the basic residues K233 and R238 on TXNIP. However, there may still be some specificity coming from GLUT1-TXNIP interaction since there was no detectable interaction between rat GLUT5 and TXNIP using the same system. Together with the in vivo fructose uptake data, this suggests that the physical interaction between TXNIP and GLUT5 may be indirect (Dykstra et al, 2021; Waldhart et al, 2017).
These findings illustrate a major difference between beta-arrestins and TXNIP. First, beta-arrestins reside in the cytoplasm under basal conditions whereas TXNIP is already found on the plasma membrane. Second, beta-arrestin-GPCR interactions are initiated by beta-arrestins binding to the phosphorylated GPCR tails whereas the interaction with phospholipids comes later to stabilize the complex. On the other hand, this lipid interaction is crucial for TXNIP localization to the membrane to initiate interaction with GLUT1. Sequence-wise, R238 but not K233 is conserved in ARRDC3 and ARRDC4. It remains to be seen whether the basic pocket for lipids is conserved on the three-dimensional structural level for all alpha-arrestins.
In addition to the plasma membrane location, TXNIP is found in fast trafficking vesicles close to the plasma membrane, presumably in various stages of endosomes although their exact nature has not yet been confirmed. Mechanistically, this raises the possibility that TXNIP can bind to inositol lipids other than PI(4,5)P2. If so, whether a differential affinity toward the different types of head groups determines the binding status of TXNIP onto different vesicles and the role that TXNIP plays in glucose transporter recycling are areas in need of further study.
Signaling Intervention
Since cells constantly respond to intracellular metabolic needs and extracellular signals, both should be able to modify the TXNIP-enabled autoregulatory circuitry on glucose uptake (Fig. 3B). Given this, not surprisingly, it turns out that TXNIP also serves as a signaling hub. Both AMP-activated protein kinase (AMPK) and Akt phosphorylate TXNIP on S308, causing TXNIP to dissociate from glucose transporters, leading to an immediate increase in glucose uptake (Waldhart et al, 2017; Wu et al, 2013). Purified recombinant TXNIP from E. coli has its C-terminal tail extended away from the N-terminal arrestin-fold domains (Dykstra et al, 2021).
How phosphorylation triggers the release of TXNIP from glucose transporters remains currently unknown. Many phosphorylation sites have been detected on the C-terminal tail of TXNIP (phosphosite), and perhaps some of these sites have the same effect as pSer308 in terms of affecting glucose uptake.
On a physiological level, TXNIP regulation on GLUT4 downstream of insulin-phosphoinositide 3-kinases (PI3K)-Akt pathway exemplifies insulin control of GLUT4 endocytosis, a pathway that was previously thought to be passive since GLUT4 carries its own endocytosis signal (F5QQI and LL490) (Blot and McGraw, 2006). However, during insulin stimulation, transporter endocytosis must be stopped to maximize glucose flux. This is achieved through TXNIP phosphorylation, implying a major contribution of TXNIP as a signal-dependent endocytosis facilitator.
In diabetic patients, if the signaling strength through PI3K-Akt is weakened, insufficient TXNIP might be phosphorylated and the increase in glucose uptake would be dampened, resulting in the high blood glucose phenotype observed in insulin resistance. It is also possible that high blood glucose causes excess TXNIP expression to resist action of insulin-Akt on the single-cell level, contributing further to the insulin resistance phenotype.
TXNIP transcription is regulated by Akt. In the absence of insulin signaling (fasting), Akt is autoinhibited (Chu et al, 2020) and its substrate, forkhead box O (FOXO), stays within the nucleus to drive TXNIP expression (Noblet et al, 2021), thus inhibiting glucose uptake in insulin-responsive tissues. TXNIP has been reported to increase after fasting in muscles (Waldhart et al, 2017) and the liver (Oka et al, 2006). On insulin stimulation, activated AKT phosphorylates FOXO, which is then retained in the cytoplasm by 14-3-3 binding, and TXNIP expression is decreased, allowing more glucose influx (Hong et al, 2016; Tzivion et al, 2011). A similar action of AMPK on TXNIP gene and protein expression has also been reported (Shaked et al, 2011).
Glucose-Dependent Cellular Effects of TXNIP in Mitochondria
Genetic deletion of TXNIP results in both functional and structural mitochondrial defects in cardiac and skeletal muscle. Mechanistically, there may be multiple ways in which TXNIP affects mitochondria (Yoshioka et al, 2012; Yoshioka et al, 2007). One way glucose can affect mitochondria function is via rigidifying membranes by affecting their lipid composition (Waldhart et al, 2021). This change in lipid ratios can be a direct consequence of excess tissue glucose uptake in the absence of TXNIP, because it can be rescued by using a ketogenic diet (Elshaer et al, 2017; Waldhart et al, 2021).
De novo lipogenesis from glucose primarily generates C16:0, C18:0, C18:1, and other shorter and more saturated acyl-chains, which effectively reduces the percentage of dietary polyunsaturated long chain fatty acids (PUFAs) that are critical for optimal mitochondrial function. Functions of mitochondrial electron transport chain complexes, including the formation of the super complex as well as expression of other proteins (such as uncoupling protein 1 [UCP1]) (Hoang et al, 2013), require correct membrane lipid composition and fluidity.
This is illustrated by Barth syndrome in mutations of tafazzin, an enzyme involved in cardiolipin maturation (Gonzalez, 2005; Lu et al, 2016; Xu et al, 2006). In the case of brown adipose tissue (BAT), the functional defect becomes measurable under cold shock stress that requires a fast and dramatic increase in mitochondrial metabolic flux. Membrane lipid change is probably not the only cause of mitochondrial defects observed in TXNIP KO mice. In the BAT study, there are many gene expression changes in TXNIP KO not rescued by a ketogenic diet (Waldhart et al, 2021). This suggests there are non-glucose-dependent functions of TXNIP that require further investigation. Efforts also have been made in developing small molecules to inhibit TXNIP expression to manage insulin resistance (Li et al, 2017; Thielen et al, 2020).
Cell Cycle
Primary mouse embryonic fibroblasts (MEFs) isolated from TXNIP KO animals demonstrate Warburg-like metabolism: higher glucose uptake, more lactate production, less glucose oxidation, and faster cell doubling (Hui et al, 2008). Faster cell growth is made possible by having more glucose available to generate building blocks. In addition, in order for a cell to be divided into two daughter cells, mitochondria must be broken down and redistributed (Carlton et al, 2020). Faster cell division necessitates less tubular forms of mitochondria and thus less efficient oxidative phosphorylation (OXPHOS) and more reliance on glycolysis to provide ATP.
Using synchronized cells to profile proteins and metabolites during different stages of cell division, it was found that TXNIP expression was inhibited whereas GLUT1 expression increased at the beginning stages of the cell cycle to increase glucose uptake (Lee et al, 2017). Therefore, in the absence of oncogenic mutations, the availability of metabolites alone can potentiate cell growth in culture. In vivo, this may be relevant to tumor suppression by TXNIP. TXNIP KO on the C3 strain of mice developed hepatocellular carcinoma (Sheth et al, 2006), and TXNIP KO mice from another group were more susceptible to diethylnitrosamine-induced hepatocarcinogenesis (Kwon et al, 2011).
TXNIP absence also accelerates liver regeneration after a partial hepatectomy (Kwon et al, 2011). Together with the fact that mitochondrial oxidative capacity is decreased in the absence of TXNIP, glucose availability may directly influence cell replication. Therefore, it is not surprising that for rapidly dividing cells in some cancers, overexpressing TXNIP can be “tumor suppressive” (Chen et al, 2020). The earlier interpretation from the metabolite availability perspective is consistent with many reports that low TXNIP expression is associated with poor prognosis (Masutani, 2021) (also see later discussion). In addition, TXNIP protein itself can participate in cell cycle regulation (Jeon et al, 2005; Kamitori et al, 2018).
ER Stress
The ER stress partners with oxidative stress in the pathophysiology of several diseases (Chong et al, 2017). Overloading of ER stress can cause calcium release and oxidative stress, leading to inflammation (Zhang and Kaufman, 2008). The ER stress plays an essential role in type 1 and type 2 diabetes (Eizirik et al, 2008) due to its association with inflammation (Zhang and Kaufman, 2008). TXNIP was found to be the link that connects inflammation to ER stress (Oslowski et al, 2012). Peripheral blood mononuclear cells that were obtained from type 2 diabetes patients also showed increased TXNIP expression (Szpigel et al, 2018).
This finding also holds true in vitro; induction of ER stress in human monocyte-derived macrophages triggers TXNIP, NLRP3, and IL-1β production (Szpigel et al, 2018). TXNIP expression also regulates glucose metabolism in breast cancer cells; estrogen receptor activates the unfolded protein response (UPR) and represses TXNIP to divert glucose metabolism (Wang and Chen, 2021).
UPR is a signaling pathway that helps ER to retain its functionality. UPR is induced by stress factors such as inositol-requiring enzyme 1 (IRE1), the PRKR-like endoplasmic reticulum kinase (PERK), and activating transcription factor 6 (ATF6) (Hetz, 2012). The ER stress caused by chemicals such as thapsigargin and tunicamycin as part of UPR induces TXNIP expression in cultured INS-1 832/13 cells as well as primary human and mouse islets (Lerner et al, 2012; Oslowski et al, 2012). This induction is mostly via PERK and IRE1 pathways. TXNIP is required for the expression of IL-1β downstream of ER stress, leading to ER stress-mediated cell death.
Together with in vivo studies, it appears that the absence of TXNIP can protect β cells from apoptosis, increase β cell mass, and enhance insulin secretion (Chen et al, 2010; Chen et al, 2008a; Chen et al, 2008b; Hui et al, 2004; Oka et al, 2009; Yoshihara et al, 2010). The ER stress can reduce glucose uptake and lactate production (Wang et al, 2011); in this case, the authors used IL-3-dependent Bak−/−Bax−/− cells to avoid ER stress-induced cell death and explained the change in glucose uptake with reduced surface level of GLUT1, consistent with the increase in TXNIP previously reported. Therefore, ER stress-induced TXNIP expression results in decreased glucose uptake. However, the lack of glucose can lead to induction of glucose-regulated proteins that are involved in UPR (Ellgaard et al, 2018; Lee, 2014; Lee, 1992).
Folding proteins properly in the ER involves shuffling disulfide bridges. The sources of reducing power in ER to support protein folding are still unclear but most likely require glucose (further discussion below). Therefore, the glucose-TXNIP axis plays a role in the ER stress response, and further studies are needed to delineate the causal relationship under both mild and extreme ER stress conditions, particularly in vivo in metabolism and not in a chemical-induced manner.
TXNIP as Thioredoxin Inhibitor or Effector
Here, we shall discuss whether TXNIP acts as an inhibitor or effector of thioredoxin, and how its regulation of glucose metabolism can also affect cellular redox state. Glucose availability indirectly affects cellular redox state for the following reasons. The cytoplasm is generally thought to have a reduced environment with 1–10 mM glutathione (GSH) as the most abundant reducing agent. The cellular concentration of GSH is much higher than thioredoxin, but both have the same reducing potential (Schafer and Buettner, 2003). The regeneration of both GSH and thioredoxin uses the reducing potential in NADPH that can be generated via different pathways (Fig. 4).
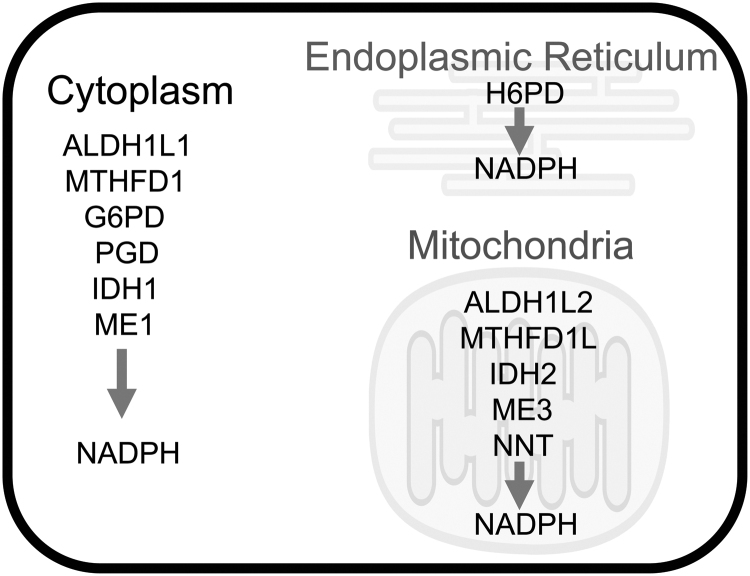
FIG. 4.
Enzymes that generate NADPH at different locations in cell. ALDH1L1/2, aldehyde dehydrogenase 1 family member L1/2; G6PD, glucose-6-phosphate dehydrogenase; H6PD, hexose-6-phosphate dehydrogenase/glucose 1-dehydrogenase; IDH1/2, isocitrate dehydrogenase [NADP(+)]; ME1/3, malic enzyme 1/3; MTHFD1/1L, methylenetetrahydrofolate dehydrogenase cyclohydrolase and formyltetrahydrofolate synthetase 1; NADPH, nicotinamide adenine dinucleotide phosphate; NNT, nicotinamide nucleotide transhydrogenase; PGD, phosphogluconate dehydrogenase.
There are separate pools of NADPH in the cytoplasm, ER, and mitochondria (Lewis et al, 2014; Maddocks et al, 2014; Mari et al, 2009; Rogoff et al, 2010). However, regardless of their location, the ultimate reducing power in NADPH comes from energy stored in the carbon–carbon bonds such as glucose, created by photosynthesis. As shown in Figure 4, aldehyde dehydrogenase 1 family member (ALDH1)L1/2, methylenetetrahydrofolate dehydrogenase cyclohydrolase and formyltetrahydrofolate synthetase 1 (MTHFD1/1L), function in 1-carbon metabolism; glucose-6-phosphate dehydrogenase (G6PD), phosphogluconate dehydrogenase (PGD), and hexose-6-phosphate dehydrogenase/glucose 1-dehydrogenase (H6PD) are directly downstream of glucose usage; isocitrate dehydrogenase [NADP(+)] (IDH1/2) and malic enzyme (ME)1/3 work in central carbon shuttling; and nicotinamide nucleotide transhydrogenase (NNT) uses the proton gradient to generate NADPH. Therefore, any process that affects cellular glucose availability can affect NADPH levels, thus affecting the available free thiol pool and ROS neutralization.
Given this reducing environment, TXNIP is most likely a thioredoxin effector, not inhibitor. That is, stress-induced oxidation of C247 in TXNIP is rescued by thioredoxin. To be a thioredoxin inhibitor, TXNIP needs to satisfy the following conditions in the cell: (1) maintain internal disulfide bonds so it can bind thioredoxin; (2) the cell contains at least equal amounts of oxidized TXNIP protein and reduced thioredoxin protein; (3) once the disulfide is transferred to thioredoxin, thioredoxin is not efficiently reduced by thioredoxin reductase; or (4) the reduced TXNIP has to become oxidized quickly to match the speed that thioredoxin is reduced by thioredoxin reductase to keep thioredoxin inhibited.
However, TXNIP is, undoubtedly, an important substrate of thioredoxin. Therefore, thioredoxin availability will affect TXNIP function, including its regulation of glucose transport and thus cellular NADPH concentration. Depending on the biological system, if TXNIP is the major effector of thioredoxin in the system, then it is possible that by having to rescue TXNIP, less free reduced thioredoxin is available for its other targets such as detoxifying H2O2 by peroxiredoxins (Buettner et al, 2013) or less NADPH is available for glutathione reductase to maintain the GSH pool (Fig. 5).
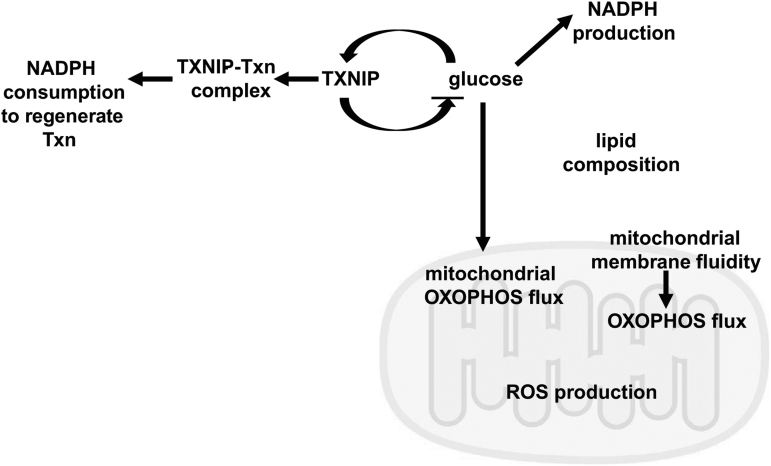
FIG. 5.
Potential mechanisms by which TXNIP can affect cellular redox state. Visual representation of our current knowledge regarding the potential ways in which TXNIP affects cellular redox state. TXNIP and glucose form an auto-feedback loop together, regulating each other. Glucose is needed to produce NADPH such as via the pentose phosphate pathway. Too much glucose can lead to increased ROS production by the mitochondria in two ways: (1) Increased metabolic flux through mitochondria increases ROS production; (2) excess glucose rigidifies mitochondrial membranes, leading to less efficient OXOPHOS flux and increased ROS. NADPH is consumed by intrinsic cellular defense mechanisms to counter ROS. NADPH is also consumed when oxidized Txn is regenerated. ROS, reactive oxygen species; Txn, thioredoxin.
We stress the point that the redox outcome will depend on the biological context. This is illustrated by the following evidence: The absence of TXNIP does not appear to affect liver thioredoxin level or activity (Sheth et al, 2005), even though there is a higher GSH:glutathione disulfide (GSSG) ratio (Hui et al, 2004). Thioredoxin activity is not different between TXNIP KO and wildtype mice in basal conditions (Sheth et al, 2005; Yoshioka et al, 2012) nor in human primary TXNIP−/− cells compared with controls (Katsu-Jiménez et al, 2019). However, TXNIP KO mice have a significant increase in the ratio of NADH to NAD+ in the fed state (Sheth et al, 2005). In cardiomyocytes, the TXNIP C247S mutation did not affect basal levels of either TXN1 or TXN2 activity or the GSH:GSSG ratio.
On the other hand, with streptozotocin-induced diabetes and acute myocardial infarction, TXNIP wildtype hearts have a lower GSH:GSSG ratio, lower TXN2 activity, but the same TXN1 activity as C247S hearts (Mukai et al, 2021; Nakayama et al, 2021). This illustrates the interconnection of cellular pools of reducing power. Depending on the biological context, TXNIP absence may be associated with reduced ROS such as in lung fibroblasts or higher ROS such as in transplanted bone marrow cells (Jung et al, 2013; Lee et al, 2005). Thus, the influence of TXNIP on cellular redox state is insufficiently explained by its interaction with thioredoxin. It may be a combination of glucose availability, NADPH production and usage, and ROS production that depends on the health of and flux through mitochondria and other organelles such as peroxisomes as well as the cell type (Fig. 5).
Other Functions of TXNIP
Earlier, we discussed TXNIP's interaction with thioredoxin and TXNIP's regulation of glucose in detail. Other functions of TXNIP such as its involvement in autophagy via binding to the regulated in development and DNA damage responses 1 (REDD1), a mammalian target of rapamycin (mTOR) regulator, will be discussed in the next section in the context of muscle function. Here, it is worth mentioning that in many cultured cells, TXNIP has a prominent nuclear localization (Nishinaka et al, 2004), though plasma membrane and vesicular TXNIP localization are also visible in cultured cells that grow in tight clusters. The exact function of nuclear TXNIP remains unclear.
A broad proteomics study identified proteins that bind to TXNIP using the proximity-based labeling method to understand the cellular functions of TXNIP and their redox-dependency using C247S mutation (Forred et al, 2016). Many protein interactions with TXNIP were nuclear, and some are redox-dependent since protein binding was disrupted by the C247S mutation. Nuclear TXNIP function may be a consequence of TXNIP stress response or redox sensing. Moreover, many interactions were glucose-dependent and triggered by hyperglycemia (Forred et al, 2016). However, not all interactions of TXNIP are glucose-triggered. The primary effect of TXNIP on the myocardium may arise from its oxidative actions (Domingues et al, 2021).
Given the evolution of TXNIP's ability to bind to thioredoxin, it is not surprising that TXNIP bears both thioredoxin-dependent and -independent regulatory roles (Cao et al, 2020) (Table 1). Studies suggest that induction of oxidative stress, inflammasomes, and cardiac hypertrophy are thioredoxin binding-dependent functions of TXNIP, whereas TXNIP's regulation of glucose uptake and insulin secretion are not thioredoxin binding-dependent (Spindel et al, 2012). Other alpha-arrestins in the family with a high sequence homology to TXNIP do not bind to thioredoxin, but they still have mammalian metabolic roles (Patwari and Lee, 2012; Patwari et al, 2006).
Table 1.
Thioredoxin-Dependent and -Independent Functions of Thioredoxin Interacting Protein
| Thioredoxin-dependent | Thioredoxin-independent |
|---|---|
| Hepatic gluconeogenesis (Chutkow et al, 2008) | Cellular glucose uptake (Patwari et al, 2009) |
| Inflammation/NLRP3 inflammasome activation (Zhou et al, 2010) | Angiogenesis (Dunn et al, 2014) |
| Mitochondrial energy metabolism (Richards et al, 2017; Yoshioka and Lee, 2014) | Hypoxia-inducible factor transcriptional activity (Farrell et al, 2010) |
| Cardiac function/hypertrophy (Yoshioka et al, 2004), myocardial infarction (Nakayama et al, 2021) | Induction by glucose (Richards et al, 2017) |
| Ventricular functional reserve (Mukai et al, 2021) | Survival of cone photoreceptors (Xue et al, 2021) |
NLRP3, NLR family pyrin domain containing 3.
TXNIP itself also has diverse metabolic roles, including regulation of hyperlipidemia, hypoglycemia, gluconeogenesis, glucotoxicity, glucose/fructose uptake, and feeding response (Bodnar et al, 2002; Chutkow et al, 2008; Dotimas et al, 2016; Parikh et al, 2007; Patwari et al, 2009; Shah et al, 2020; Shalev, 2008; Sheth et al, 2005). However, it is not yet understood whether all of these metabolic roles of TXNIP are regulated by thioredoxin binding. Numerous studies have mutated the key cysteine C247 to serine (C247S mutation) in the TXNIP protein in vitro to render it unable to bind thioredoxin.
Using this approach, it was shown that both TXNIP and the TXNIP-C247S mutation inhibit glucose uptake in 3T3-L1 adipocytes even though TXNIP-C247S samples had high thioredoxin activity (Patwari et al, 2009; Patwari et al, 2006). On the other hand, wildtype TXNIP protein rescued hepatocyte glucose production with a genetic TXNIP deletion, whereas the TXNIP-C247S mutant protein could not (Chutkow et al, 2008). These studies suggest that some metabolic functions of TXNIP may be redox-dependent, whereas others are not.
TXNIP and Its Actions on Diverse Tissues
Txnip expression is modulated by glucose (mediated by carbohydrate-response elements and associated transcription factors Mlx/MondoA), glutamine, fatty acids, and adenosine-containing molecules (Yu et al, 2010). The level of TXNIP is under tight regulation of physiological conditions. In HeLa cells, cycloheximide treatment causes rapid protein turnover (half-life <1 h) of the pre-existing endogenous TXNIP protein (Yu et al, 2010). TXNIP is expressed almost ubiquitously except in the central nervous system, where the expression is low (Tsubaki et al, 2020).
Because TXNIP is expressed in various tissues, including skeletal muscle, adipose, pancreas, and heart, it is not surprising that TXNIP appears to play roles in diverse disease processes (Fig. 6). Interesting insight on the role of TXNIP in human metabolism comes from new genetic investigations. Three members of a family were homozygous for a txnip mutation that ceases TXNIP protein expression; the loss of TXNIP resulted in lactic acidosis and low serum methionine levels (Katsu-Jiménez et al, 2019). TXNIP-deficient fibroblasts and myoblasts from these patients could not utilize pyruvate for OXPHOS.
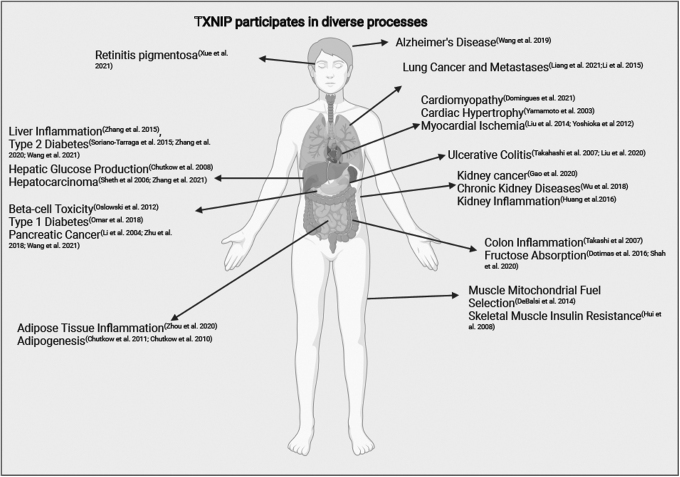
FIG. 6.
TXNIP participates in diverse processes. TXNIP has reported roles in many pathophysiologic processes. (Created in BioRender.com)
Consistent with this fascinating human finding, animal studies also showed that TXNIP KO mice are deficient in pyruvate utilization by the liver (Chutkow et al, 2008) and that lactate formation was higher in the hearts after ischemia–reperfusion injury (Yoshioka et al, 2012).
Here, we will review published data on TXNIP in specific tissues. These phenotypes are most likely a combination of known cellular functions of TXNIP discussed previously and yet to be discovered functions of TXNIP.
Heart
TXNIP's oxidative actions may determine its effect on the myocardium (Domingues et al, 2021). It is known that inhibiting endogenous thioredoxin in the heart increases oxidative stress and cardiac hypertrophy (Yamamoto et al, 2003). It was also recently shown that the C247S mutation of TXNIP in heart tissue protects mice from myocardial oxidative stress caused by streptozotocin-induced diabetes (Mukai et al, 2021). This single C247S amino acid mutation in the myocardium allowed the preservation of the antioxidative effects of thioredoxin in cardiomyocytes and reduced the adverse effects of myocardial infarctions (Nakayama et al, 2021).
TXNIP plays a fundamental role in myocardial energy homeostasis (Domingues et al, 2021; Yoshioka et al, 2012; Yoshioka et al, 2007). Single-nucleotide polymorphisms in the txnip gene were linked to arterial stiffness and coronary artery disease in a Brazilian population (Alvim et al, 2012). Also, differential expression of TXNIP variants in leukocytes is associated with acute myocardial infarction risk (Zhang et al, 2018). In clinical studies of coronary heart disease patients, TXNIP expression was detected to be higher in smokers than non-smokers (Mao et al, 2021); this could be related to ROS and inflammation-induced TXNIP expression.
TXNIP can control cardiac hypertrophy by regulating thioredoxin activity (Wang et al, 2002; Yoshioka et al, 2004), but it has dual roles on the heart. The absence of txnip was shown to not only protect the heart from ischemia injury in mice but also impair mitochondrial function (Yoshioka et al, 2012). The NLRP3 inflammasome-activating role of TXNIP may be particularly important for cardiac inflammation and cardiomyopathy (Liu et al, 2014; Peng et al, 2021; Wang et al, 2021a). TXNIP-NLRP3 activation in cardiac microvascular endothelial cells may participate in myocardial ischemia–reperfusion injury (Liu et al, 2014).
TXNIP was also recently discovered to participate in autophagy mechanisms. REDD1 is a negative regulator of mTOR, and TXNIP stabilizes REDD1 proteins, causing mTOR suppression (Jin et al, 2011). The REDD1-TXNIP complex was found to control stress-induced autophagy (Qiao et al, 2015). In recent studies, it was shown that TXNIP overexpression promoted cardiac autophagy and suppressed autophagosome clearance by increasing ROS in mice (Gao et al, 2020a).
Skeletal Muscle
The molecular roles of TXNIP in different organs are also apparent in the insulin signaling pathway. The absence of TXNIP activates Akt in skeletal muscle tissues and the heart, but not in liver and adipose tissue (Hui et al, 2008). TXNIP was found to control both insulin-dependent and insulin-independent pathways of glucose uptake in skeletal muscles in human studies (Parikh et al, 2007). Physical exercise and TXNIP have a reciprocal relationship. Exercise reduces the protein levels of the TXNIP-REDD1 complex in skeletal muscle (Chaves et al, 2022). Mitochondrial fuel selection of skeletal muscle fibers is also controlled by TXNIP (DeBalsi et al, 2014). The absence of TXNIP results in significant shortages in amino acid, ketone, and lactate-break down enzymes in mice (DeBalsi et al, 2014).
Liver
TXNIP is upregulated by both ChREBP and FOXO1 transcription factors in liver cells (Noblet et al, 2021). TXNIP KO mice were shown to be hypoglycemic and hypoinsulinemic, and they had diminished glucose production during a glucagon challenge, presenting a liver glucose homeostasis defect (Chutkow et al, 2008). Txnip null hepatocytes isolated from these mice could not produce sufficient glucose. Hepatic overexpression of TXNIP in wildtype mice caused increased serum glucose levels. Wildtype TXNIP protein rescued hepatocyte glucose production with a genetic txnip deletion, whereas the TXNIP-C247S mutant protein could not. TXNIP was also recently found to protect mice from fasting-induced liver steatosis through its effects on ER stress (Miyahara et al, 2022).
Adipose Tissue
TXNIP is abundantly expressed in adipose tissues. TXNIP controls the intake of glucose into adipocytes by acting as an adaptor for the glucose transporter. Adipose tissues of TXNIP KO mice absorb more glucose than wildtype controls under starved conditions (Waldhart et al, 2017). Therefore, the deletion of TXNIP promotes adiposity and adipogenesis (Chutkow et al, 2010) and TXNIP degradation is required for adipocyte differentiation (Chutkow and Lee, 2011). On the other hand, the TXNIP-C247S mutation does not impair adipocyte differentiation, but TXNIP-C247S is more rapidly degraded. Interestingly, although adiposity is increased in these mice, insulin sensitivity is still preserved. Adipokines also regulate the TXNIP-inflammasome complex. Omentin-1 can limit adipose tissue inflammation in obese mice by repressing TXNIP-NLRP3 activation (Zhou et al, 2020).
Pancreatic β-Islets
As mentioned earlier, the absence of TXNIP in muscle and adipose tissue allows more glucose influx into these insulin-responsive tissues without insulin signaling, and liver-specific deletion of txnip inhibits gluconeogenesis (Chutkow et al, 2008). These characteristics effectively keep blood glucose low in TXNIP KO mice and make them less susceptible to insulin resistance. A deficit in functional β cells is a common identifying factor in both type 1 and type 2 diabetes (Thielen and Shalev, 2018). We know that TXNIP levels are positively regulated by glucose in β cells as in other cells; however, there are incomplete data on which glucose transporter interacts with TXNIP in β cells.
Transcriptomic analyses identified txnip as the most upregulated gene in glucose-treated human β cells (Richards et al, 2017), and TXNIP expression is elevated in β cells during diabetes (Minn et al, 2005; Xu et al, 2013). TXNIP methylation in whole-blood DNA was associated with hemoglobin HbA1c levels, type 2 diabetes, and metabolic syndrome in human populations (Soriano-Tárraga et al, 2015; Wang et al, 2021c; Yamazaki et al, 2022; Zhang et al, 2020). In a recent study, type 1 diabetes patients had significantly lower serum vitamin D and serum TXNIP levels (Omar et al, 2018).
In addition, TXNIP is subject to O-GlcNAcylation in response to high glucose concentrations, and O-GlcNAcylation of TXNIP appeared to be elevated in pancreatic islets of diabetic rats (Filhoulaud et al, 2019). However, glucose-induced TXNIP expression is counteracted by the activation of insulin receptors through histone deacetylases and by palmitate, which reduces oxidative stress in pancreatic β cells (Panse et al, 2018).
Higher TXNIP levels induced by either high glucose or ER stress are associated with β cell death (Chen et al, 2008b). Culturing of INS-1 β cells and primary mouse and human islets with a glucose medium resulted in increased apoptosis levels (Shalev, 2008). TXNIP expression is induced by ER stress by PERK/IRE1 pathways and results in IL-1β transcription through NLRP3 inflammasome signaling to trigger β cell death (Oslowski et al, 2012). In addition, TXNIP was found to affect the function of β cells; TXNIP induces miR-204 by inhibiting the activity of signal transducer and activator of transcription 3 (STAT3) in β cells to inhibit mAF BZIP transcription factor A (MafA)-mediated insulin transcription (Xu et al, 2013). TXNIP KO mice show higher total pancreatic insulin content and β cell mass than wildtype mice (Masson et al, 2009). β cell-specific txnip deletion increased β cell mass through Akt/B cell lymphoma-extra large (Bcl-xL) signaling and protected against diabetes in mouse models (Chen et al, 2008a).
Kidney
TXNIP plays a significant role during nephropathy. High glucose levels increase TXNIP protein expression that triggers inflammatory and fibrotic responses in kidneys (Huang et al, 2016). In diabetic wildtype mice, renal injury and dysfunction indicators such as albuminuria and serum creatinine levels were increased, unlike in TXNIP KO mice (Shah et al, 2015). TXNIP also drives hyperglycemia-induced ROS generation by mitochondria in renal mesangial cells (Shah et al, 2013). Renal inflammation due to ureteral obstruction was suppressed in TXNIP KO mice, identifying TXNIP as a potential target to slow the development of chronic kidney diseases (Wu et al, 2018).
MicroRNAs can be triggering for the TXNIP-NLRP3 response. Exosomal microRNA miR-93-5p was found to target TXNIP-NLRP3 signaling to influence sepsis-induced acute kidney injury (Juan et al, 2021). Ablation of txnip was also shown to cause renal tubular lipid accumulation during diabetes by hindering the SREBP cleavage activating protein (SCAP) signaling pathway, yielding reduced cholesterol uptake and synthesis (Sun et al, 2021).
Lung
Hypoxia-Inducible factor (HIF)-1 is a protein complex that plays a crucial role in the hypoxia response (Ziello et al, 2007). TXNIP can inhibit HIF-1-mediated gene transcription in the murine lung (Farrell et al, 2010). Inflammation-associated lung diseases such as asthma are linked to TXNIP. Inflammatory nanoparticles that aggravate asthma symptoms were shown to induce inflammation via TXNIP upregulation in mice (Lim et al, 2021). The extracellular signal-regulated protein kinase (ERK) pathway can control the pro-inflammatory cytokine tumor necrosis factor (TNF)-alpha-directed TXNIP degradation in respiratory epithelial cells (Kelleher et al, 2019), and inhibition of this pathway could reduce ROS in lung epithelial cells. Ubiquitination and degradation of TXNIP by ERK were dependent on the Thr349-phosphorylation of TXNIP inside the C-terminal domain, identifying this specific amino acid as a potential target for E3 ubiquitin ligases (Kelleher et al, 2019).
Eye
TXNIP may play an important role in the retina. Viral gene delivery of txnip was found to ameliorate experimental Retinitis Pigmentosa in mice, which occurs due to the dysfunction or loss of cone-photoreceptor (Xue et al, 2021). Gene delivery of txnip prolonged the survival of cone photoreceptors and improved visual acuity in mice, and this saved the cones by improving their lactate metabolism. Interestingly, gene delivery of TXNIP C247S yielded even more improvement. The authors hypothesized that by losing the TXNIP-thioredoxin interaction, both TXNIP and thioredoxin might be more beneficial (Xue et al, 2021).
TXNIP and Cancer
TXNIP is associated with multiple cancer types and cancer cell malignancy (Meylan et al, 2021). TXNIP communicates with multiple fundamental intracellular pathways to act as a tumor suppressor. In a mouse strain that naturally had low TXNIP levels, hepatic carcinomas were common (Sheth et al, 2006). Lower TXNIP expression controlled by peroxisome proliferator-activated receptor gamma (PPARgamma) was found to be associated with advanced melanoma and metastases in patients with melanoma (Meylan et al, 2021). Further, in studies of malignant glioma, elevated TXNIP protein expression levels were associated with prolonged patient survival time (Zhang et al, 2017).
One of the cancer types where TXNIP plays a clear role through several molecular pathways is breast cancer. A decrease of TXNIP results in elevated proliferative activity and estrogen-induced cell growth in breast cancer (Park et al, 2018). A knockdown of the txnip gene in breast cancer cells was associated with glucose transporter 1 (GLUT1) induction and caused a substantial reduction in the cell-growth inhibitory effect of anti-estrogen agents. MicroRNAs were shown to be important in triggering metastasis via by TXNIP-HIF-1 signaling axis in breast cancer (Chen et al, 2015). TXNIP expression was discovered to be significantly correlated with HIF-1 expression in non-small cell lung cancer progression as well (Li et al, 2015).
Circular RNAs also suppressed tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression (Liang et al, 2021). A high expression of c-myelocytomatosis oncogene (C-MYC) and low expression of the txnip gene correlated with decreased survival in breast cancer (Shen et al, 2015). The MondoA-TXNIP axis is crucial for defining regulatory T cell identity and function in the progression of colorectal cancer (Lu et al, 2021). The effect of TXNIP on glucose uptake is also apparent in prostate cancer. Overexpression of TXNIP in prostate cancer cell lines inhibited glucose uptake, and TXNIP expression was lower in the human prostate cancer samples in a recent study (Xie et al, 2020).
TXNIP is also involved in kidney and liver cancers. Analysis of clinical data from The Cancer Genome Atlas Database identified TXNIP expression as a potential prognostic marker for patients with clear cell renal cell carcinoma (Gao et al, 2020b). Hepatitis B is known to cause hepatocellular carcinoma. Animal studies pointed out that TXNIP deficiency was sufficient to initiate hepatocellular cell carcinoma, and a truncated version of a Hepatitis B virus protein can cause hepatocarcinogenesis by downregulating TXNIP expression and reprogramming glucose metabolism (Sheth et al, 2006; Zhang et al, 2021).
In pancreatic cancer, TXNIP plays a different role. TXNIP protein and messenger RNA (mRNA) expression levels were higher in pancreatic cancer patients with diabetes, and TXNIP-Thioredoxin-ROS axis was found to be regulated via p38 MAPK and ERK pathways in these tumors (Li et al, 2014). Several MicroRNAs also control TXNIP-HIF-1a axis to control pancreatic cancer cell proliferation (Wang et al, 2021b; Zhu et al, 2018). Therefore, TXNIP has various significant roles in controlling tumor growth, and expression of TXNIP in cancer can sometimes be a predictor of cancer progression (Chen et al, 2020).
TXNIP and Vitamin D Related and Other Diseases
TXNIP was initially named as “Vitamin D3 upregulated protein 1” due to increased levels by vitamin D(3) administration (Chen and DeLuca, 1994). Therefore, TXNIP might play a role in vitamin D-associated diseases. For example, TXNIP expression in serum and bones was increased in rat models of glucocorticoid-induced osteoporosis (Mo et al, 2021). TXNIP was also suggested to mediate the negative effects of glucocorticoid on osteoblast function and osteoclastogenesis in glucocorticoid-treated Cushing Syndrome patients with secondary osteoporosis (Lekva et al, 2012).
TXNIP expression in Multiple Sclerosis patients was significantly lower and serum thioredoxin levels were higher than the controls in a recent study (Mahmoudian et al, 2017). Genetic variations in the TXNIP gene were also associated with hypertension in a Brazilian population (Alvim et al, 2012). TXNIP and Txn1 are differentially expressed in newly diagnosed multiple sclerosis patients or patients getting immunosuppressive treatments (Mahmoudian et al, 2017). miR-20a that plays a role in rheumatoid arthritis development may also play a part in the TXNIP-NLRP3-inflammasome pathway (Li et al, 2016).
Conclusions
Here, we described multiple roles of TXNIP, including important roles in metabolism. TXNIP directly facilitates glucose transporter GLUT1-4 endocytosis. The evolution of C247 in TXNIP suggests a unique capability of TXNIP as a redox sensor connecting metabolite flux and redox flux. Further investigation of TXNIP function should contain both metabolic and redox components, so we can have a more thorough interpretation of phenotypes. The nuclear function of TXNIP is one important area of future studies.
Abbreviations Used
- ALDH1
aldehyde dehydrogenase 1 family member 1
- AMPK
AMP-activated protein kinase
- ARRDC
arrestin domain-containing
- BAT
brown adipose tissue
- cAMP
cyclic AMP
- ChREBP
carbohydrate response element binding protein
- ER
endoplasmic reticulum
- ERK
extracellular signal-regulated protein kinase
- FOXO
forkhead box O
- G6PD
glucose-6-phosphate dehydrogenase
- GLUT
glucose transporter
- GPCR
G-protein coupled receptor
- GSH
glutathione
- GSSG
glutathione disulfide
- H6PD
hexose-6-phosphate dehydrogenase/glucose 1-dehydrogenase
- HIF
hypoxia-inducible factor
- IDH1/2
isocitrate dehydrogenase [NADP(+)]
- IL
interleukin
- IRE1
inositol-requiring enzyme 1
- KO
knockout
- ME
malic enzyme
- MTHFD1/1L
methylenetetrahydrofolate dehydrogenase cyclohydrolase and formyltetrahydrofolate synthetase 1
- mTOR
mammalian target of rapamycin
- NADPH
nicotinamide adenine dinucleotide phosphate
- NFkB
nuclear factor kappa B
- NLRP3
NLR family pyrin domain containing 3
- NNT
nicotinamide nucleotide transhydrogenase
- NRF2
nuclear factor erythroid 2–related factor 2
- OXPHOS
oxidative phosphorylation
- PERK
PRKR-like endoplasmic reticulum kinase
- PGD
phosphogluconate dehydrogenase
- PI3K
phosphoinositide 3-kinases
- REDD1
regulated in development and DNA damage responses 1
- ROS
reactive oxygen species
- slc2
solute carrier family 2
- TBP-2
thioredoxin binding protein-2
- Txn
thioredoxin
- TXNIP
thioredoxin interacting protein
- UPR
unfolded protein response
Authors' Contributions
All authors prepared and edited the article.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
S.D. was supported by NIH NIDDK Ruth L. Kirschstein NRSA Postdoctoral Fellow F32 Award (1F32DK126289). N.W. was funded by NIH R01 award (R01-GM120129). R.T.L. was funded by NIH R01 award (R01DK126688).
References
- Alvarez CE. On the origins of arrestin and rhodopsin. BMC Evol Biol 2008;8(1):222; doi: 10.1186/1471-2148-8-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvim RO, Santos PCJL, Ferreira NE, et al. Thioredoxin interacting protein (TXNIP) rs7212 polymorphism is associated with arterial stiffness in the Brazilian general population. J Hum Hypertens 2012;26(5):340–342; doi: 10.1038/jhh.2011.102 [DOI] [PubMed] [Google Scholar]
- Baba SP, Bhatnagar A. Role of thiols in oxidative stress. Curr Opin Toxicol 2018;7:133–139; doi: 10.1016/j.cotox.2018.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista TM, Dagdeviren S, Carroll SH, et al. Arrestin domain-containing 3 (Arrdc3) modulates insulin action and glucose metabolism in liver. Proc Natl Acad Sci U S A 2020;117(12):6733–6740; doi: 10.1073/pnas.1922370117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benovic JL, Kühn H, Weyand I, et al. Functional desensitization of the isolated beta-adrenergic receptor by the beta-adrenergic receptor kinase: Potential role of an analog of the retinal protein arrestin (48-kDa protein). Proc Natl Acad Sci U S A 1987;84(24):8879–8882; doi: 10.1073/pnas.84.24.8879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blot V, Mcgraw TE. GLUT4 is internalized by a cholesterol-dependent nystatin-sensitive mechanism inhibited by insulin. EMBO J 2006;25(24):5648–5658; doi: 10.1038/sj.emboj.7601462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar JS, Chatterjee A, Castellani LW, et al. Positional cloning of the combined hyperlipidemia gene Hyplip1. Nat Genet 2002;30(1):110–116; doi: 10.1038/ng811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Dixit VM. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat Rev Immunol 2016;16(7):407–420; doi: 10.1038/nri.2016.58 [DOI] [PubMed] [Google Scholar]
- Buettner GR, Wagner BA, Rodgers VG. Quantitative redox biology: An approach to understand the role of reactive species in defining the cellular redox environment. Cell Biochem Biophys 2013;67(2):477–483; doi: 10.1007/s12013-011-9320-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, He W, Pang Y, et al. Redox-dependent and independent effects of thioredoxin interacting protein. Biol Chem 2020;401(11):1215–1231; doi: 10.1515/hsz-2020-0181 [DOI] [PubMed] [Google Scholar]
- Carlton JG, Jones H, Eggert US. Membrane and organelle dynamics during cell division. Nat Rev Mol Cell Biol 2020;21(3):151–166; doi: 10.1038/s41580-019-0208-1 [DOI] [PubMed] [Google Scholar]
- Cha-Molstad H, Saxena G, Chen J, et al. Glucose-stimulated expression of Txnip is mediated by carbohydrate response element-binding protein, p300, and histone H4 acetylation in pancreatic beta cells. J Biol Chem 2009;284(25):16898–16905; doi: 10.1074/jbc.M109.010504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves AB, Miranda ER, Mey JT, et al. Exercise reduces the protein abundance of TXNIP and its interacting partner REDD1 in skeletal muscle: Potential role for a PKA-mediated mechanism. J Appl Physiol (1985) 2022;132(2):357–366; doi: 10.1152/japplphysiol.00229.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Dang BL, Huang JZ, et al. MiR-373 drives the epithelial-to-mesenchymal transition and metastasis via the miR-373-TXNIP-HIF1α-TWIST signaling axis in breast cancer. Oncotarget 2015;6(32):32701–32712; doi: 10.18632/oncotarget.4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Fontes G, Saxena G, et al. Lack of TXNIP protects against mitochondria-mediated apoptosis but not against fatty acid-induced ER stress-mediated beta-cell death. Diabetes 2010;59(2):440–447; doi: 10.2337/db09-0949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Hui ST, Couto FM, et al. Thioredoxin-interacting protein deficiency induces Akt/Bcl-xL signaling and pancreatic beta-cell mass and protects against diabetes. FASEB J 2008a;22(10):3581–3594; doi: 10.1096/fj.08-111690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Saxena G, Mungrue IN, et al. Thioredoxin-interacting protein: A critical link between glucose toxicity and beta-cell apoptosis. Diabetes 2008b;57(4):938–944; doi: 10.2337/db07-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KS, Deluca HF. Isolation and characterization of a novel cDNA from HL-60 cells treated with 1,25-dihydroxyvitamin D-3. Biochim Biophys Acta 1994;1219(1):26–32; doi: 10.1016/0167-4781(94)90242-9 [DOI] [PubMed] [Google Scholar]
- Chen Y, Ning J, Cao W, et al. Research progress of TXNIP as a tumor suppressor gene participating in the metabolic reprogramming and oxidative stress of cancer cells in various cancers. Front Oncol 2020;10:568574; doi: 10.3389/fonc.2020.568574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong WC, Shastri MD, Eri R. Endoplasmic reticulum stress and oxidative stress: A vicious nexus implicated in bowel disease pathophysiology. Int J Mol Sci 2017;18(4):771; doi: 10.3390/ijms18040771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu N, Viennet T, Bae H, et al. The structural determinants of PH domain-mediated regulation of Akt revealed by segmental labeling. Elife 2020;9; doi: 10.7554/eLife.59151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutkow WA, Birkenfeld AL, Brown JD, et al. Deletion of the alpha-arrestin protein Txnip in mice promotes adiposity and adipogenesis while preserving insulin sensitivity. Diabetes 2010;59(6):1424–1434; doi: 10.2337/db09-1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutkow WA, Lee RT. Thioredoxin regulates adipogenesis through thioredoxin-interacting protein (Txnip) protein stability. J Biol Chem 2011;286(33):29139–29145; doi: 10.1074/jbc.M111.267666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutkow WA, Patwari P, Yoshioka J, et al. Thioredoxin-interacting protein (Txnip) is a critical regulator of hepatic glucose production. J Biol Chem 2008;283(4):2397–2406; doi: 10.1074/jbc.M708169200 [DOI] [PubMed] [Google Scholar]
- Debalsi KL, Wong KE, Koves TR, et al. Targeted metabolomics connects thioredoxin-interacting protein (TXNIP) to mitochondrial fuel selection and regulation of specific oxidoreductase enzymes in skeletal muscle. J Biol Chem 2014;289(12):8106–8120; doi: 10.1074/jbc.M113.511535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolska O, Rychkov G, Shumilina E, et al. Structural insights into interaction between mammalian methionine sulfoxide reductase B1 and thioredoxin. J Biomed Biotechnol 2012;2012:586539; doi: 10.1155/2012/586539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues A, Jolibois J, Marquet De Rougé P, et al. The emerging role of TXNIP in ischemic and cardiovascular diseases; a novel marker and therapeutic target. Int J Mol Sci 2021;22(4):1693; doi: 10.3390/ijms22041693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorey C, Faure JP. Isolation and characterization of a retinal antigen inducing experimental autoimmune uveo-retinitis. Ann Immunol (Paris) 1977;128(1–2):229–232. [PubMed] [Google Scholar]
- Dotimas JR, Lee AW, Schmider AB, et al. Diabetes regulates fructose absorption through thioredoxin-interacting protein. eLife 2016;5:e18313; doi: 10.7554/eLife.18313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn LL, Simpson PJ, Prosser HC, et al. A critical role for thioredoxin-interacting protein in diabetes-related impairment of angiogenesis. Diabetes 2014;63(2):675–687; doi: 10.2337/db13-0417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra H, Larose C, Fisk C, et al. TXNIP interaction with GLUT1 depends on PI(4,5)P2. Biochim Biophys Acta Biomembr 2021;1863(12):183757; doi: 10.1016/j.bbamem.2021.183757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizirik DL, Cardozo AK, Cnop M. The role for endoplasmic reticulum stress in diabetes mellitus. Endocr Rev 2008;29(1):42–61; doi: 10.1210/er.2007-0015 [DOI] [PubMed] [Google Scholar]
- Eklund H, Gleason FK, Holmgren A. Structural and functional relations among thioredoxins of different species. Proteins 1991;11(1):13–28; doi: 10.1002/prot.340110103 [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Sevier CS, Bulleid NJ. How are proteins reduced in the endoplasmic reticulum? Trends Biochem Sci 2018;43(1):32–43; doi: 10.1016/j.tibs.2017.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshaer SL, Mohamed IN, Coucha M, et al. Deletion of TXNIP mitigates high-fat diet-impaired angiogenesis and prevents inflammation in a mouse model of critical limb ischemia. Antioxidants 2017;6(3):47; doi: 10.3390/antiox6030047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq AU, Gembus K, Sandow JJ et al. K-29 linked ubiquitination of Arrdc4 regulates its function in extracellular vesicle biogenesis. J Extracell Vesicles 2022;11(2):e12188; doi: 10.1002/jev2.12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell MR, Rogers LK, Liu Y, et al. Thioredoxin-interacting protein inhibits hypoxia-inducible factor transcriptional activity. Free Radic Biol Med 2010;49(9):1361–1367; doi: 10.1016/j.freeradbiomed.2010.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filhoulaud G, Benhamed F, Pagesy P, et al. O-GlcNacylation links TxNIP to inflammasome activation in pancreatic β cells. Front Endocrinol (Lausanne) 2019;10:291; doi: 10.3389/fendo.2019.00291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forred BJ, Neuharth S, Kim DI, et al. Identification of redox and glucose-dependent Txnip protein interactions. Oxid Med Cell Longev 2016;2016:5829063; doi: 10.1155/2016/5829063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Wang R, Li B, et al. TXNIP/Redd1 signalling and excessive autophagy: A novel mechanism of myocardial ischaemia/reperfusion injury in mice. Cardiovasc Res 2020a;116(3):645–657; doi: 10.1093/cvr/cvz152 [DOI] [PubMed] [Google Scholar]
- Gao Y, Qi JC, Li X, et al. Decreased expression of TXNIP predicts poor prognosis in patients with clear cell renal cell carcinoma. Oncol Lett 2020b;19(1):763–770; doi: 10.3892/ol.2019.11165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez IL. Barth syndrome: TAZ gene mutations, mRNAs, and evolution. Am J Med Genet A 2005;134(4):409–414; doi: 10.1002/ajmg.a.30661 [DOI] [PubMed] [Google Scholar]
- Grebe A, Hoss F, Latz E. NLRP3 inflammasome and the IL-1 pathway in atherosclerosis. Circ Res 2018;122(12):1722–1740; doi: 10.1161/circresaha.118.311362 [DOI] [PubMed] [Google Scholar]
- Gu C, Liu S, Wang H, et al. Role of the thioredoxin interacting protein in diabetic nephropathy and the mechanism of regulating NOD‑like receptor protein 3 inflammatory corpuscle. Int J Mol Med 2019;43(6):2440–2450; doi: 10.3892/ijmm.2019.4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetta D, Langou K, Grunwald D, et al. FYVE-dependent endosomal targeting of an arrestin-related protein in amoeba. PLoS One 2010;5(12):e15249; doi: 10.1371/journal.pone.0015249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habourdin C, Klein G, Araki T, et al. The arrestin-domain containing protein AdcA is a response element to stress. Cell Commun Signal 2013;11:91; doi: 10.1186/1478-1811x-11-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Xu X, Tang C, et al. Reactive oxygen species promote tubular injury in diabetic nephropathy: The role of the mitochondrial ros-txnip-nlrp3 biological axis. Redox Biol 2018;16:32–46; doi: 10.1016/j.redox.2018.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 2012;13(2):89–102; doi: 10.1038/nrm3270 [DOI] [PubMed] [Google Scholar]
- Hoang T, Smith MD, Jelokhani-Niaraki M. Expression, folding, and proton transport activity of human uncoupling protein-1 (UCP1) in lipid membranes: Evidence for associated functional forms. J Biol Chem 2013;288(51):36244–36258; doi: 10.1074/jbc.M113.509935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SY, Yu FX, Luo Y, et al. Oncogenic activation of the PI3K/Akt pathway promotes cellular glucose uptake by downregulating the expression of thioredoxin-interacting protein. Cell Signal 2016;28(5):377–383; doi: 10.1016/j.cellsig.2016.01.011 [DOI] [PubMed] [Google Scholar]
- Huang C, Zhang Y, Kelly DJ, et al. Thioredoxin interacting protein (TXNIP) regulates tubular autophagy and mitophagy in diabetic nephropathy through the mTOR signaling pathway. Sci Rep 2016;6(1):29196; doi: 10.1038/srep29196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui ST, Andres AM, Miller AK, et al. Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proc Natl Acad Sci U S A 2008;105(10):3921–3926; doi: 10.1073/pnas.0800293105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui TY, Sheth SS, Diffley JM, et al. Mice lacking thioredoxin-interacting protein provide evidence linking cellular redox state to appropriate response to nutritional signals. J Biol Chem 2004;279(23):24387–24393; doi: 10.1074/jbc.M401280200 [DOI] [PubMed] [Google Scholar]
- Hwang J, Suh HW, Jeon YH, et al. The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein. Nat Commun 2014;5:2958; doi: 10.1038/ncomms3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng M-F, Campbell AP, Begley T, et al. High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin. Structure 1994;2(9):853–868; doi: 10.1016/S0969-2126(94)00086-7 [DOI] [PubMed] [Google Scholar]
- Jeon JH, Lee KN, Hwang CY, et al. Tumor suppressor VDUP1 increases p27(kip1) stability by inhibiting JAB1. Cancer Res 2005;65(11):4485–4489; doi: 10.1158/0008-5472.CAN-04-2271 [DOI] [PubMed] [Google Scholar]
- Jin HO, Seo SK, Kim YS, et al. TXNIP potentiates Redd1-induced mTOR suppression through stabilization of Redd1. Oncogene 2011;30(35):3792–3801; doi: 10.1038/onc.2011.102 [DOI] [PubMed] [Google Scholar]
- Juan C-X, Mao Y, Cao Q, et al. Exosome-mediated pyroptosis of miR-93-TXNIP-NLRP3 leads to functional difference between M1 and M2 macrophages in sepsis-induced acute kidney injury. J Cell Mol Med 2021;25(10):4786–4799; doi: 10.1111/jcmm.16449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Kim MJ, Kim DO, et al. TXNIP maintains the hematopoietic cell pool by switching the function of p53 under oxidative stress. Cell Metab 2013;18(1):75–85; doi: 10.1016/j.cmet.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Kamitori K, Yamaguchi F, Dong Y, et al. Both Ser361 phosphorylation and the C-arrestin domain of thioredoxin interacting protein are important for cell cycle blockade at the G1/S checkpoint. FEBS Open Bio 2018;8(11):1804–1819; doi: 10.1002/2211-5463.12518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsu-Jiménez Y, Vázquez-Calvo C, Maffezzini C, et al. Absence of TXNIP in humans leads to lactic acidosis and low serum methionine linked to deficient respiration on pyruvate. Diabetes 2019;68(4):709–723; doi: 10.2337/db18-0557 [DOI] [PubMed] [Google Scholar]
- Kelleher ZT, Wang C, Forrester MT, et al. ERK-dependent proteasome degradation of Txnip regulates thioredoxin oxidoreductase activity. J Biol Chem 2019;294(36):13336–13343; doi: 10.1074/jbc.RA119.007733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Astapova I, Flier SN, et al. Intestinal, but not hepatic, ChREBP is required for fructose tolerance. JCI Insight 2017;2(24):e96703; doi: 10.1172/jci.insight.96703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HJ, Won YS, Yoon YD, et al. Vitamin D3 up-regulated protein 1 deficiency accelerates liver regeneration after partial hepatectomy in mice. J Hepatol 2011;54(6):1168–1176; doi: 10.1016/j.jhep.2010.09.025 [DOI] [PubMed] [Google Scholar]
- Lee AS. Mammalian stress response: Induction of the glucose-regulated protein family. Curr Opin Cell Biol 1992;4(2):267–273; doi: 10.1016/0955-0674(92)90042-b [DOI] [PubMed] [Google Scholar]
- Lee AS. Glucose-regulated proteins in cancer: Molecular mechanisms and therapeutic potential. Nat Rev Cancer 2014;14(4):263–276; doi: 10.1038/nrc3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Jedrychowski MP, Vinayagam A, et al. Proteomic and metabolomic characterization of a mammalian cellular transition from quiescence to proliferation. Cell Rep 2017;20(3):721–736; doi: 10.1016/j.celrep.2017.06.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HM, Kim JJ, Kim HJ, et al. Upregulated NLRP3 inflammasome activation in patients with type 2 diabetes. Diabetes 2013;62(1):194–204; doi: 10.2337/db12-0420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KN, Kang HS, Jeon JH, et al. VDUP1 is required for the development of natural killer cells. Immunity 2005;22(2):195–208; doi: 10.1016/j.immuni.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Lekva T, Ueland T, Bøyum H, et al. TXNIP is highly regulated in bone biopsies from patients with endogenous Cushing's syndrome and related to bone turnover. Eur J Endocrinol 2012;166(6):1039–1048; doi: 10.1530/eje-11-1082 [DOI] [PubMed] [Google Scholar]
- Lerner AG, Upton JP, Praveen PV, et al. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab 2012;16(2):250–264; doi: 10.1016/j.cmet.2012.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 2019;47(W1):W256–W259; doi.org/10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell 2014;55(2):253–263; doi: 10.1016/j.molcel.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Lin GY, Zhong L, et al. W2476 ameliorates beta-cell dysfunction and exerts therapeutic effects in mouse models of diabetes via modulation of the thioredoxin-interacting protein signaling pathway. Acta Pharmacol Sin 2017;38(7):1024–1037; doi: 10.1038/aps.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wu Z, Ma Q, et al. Hyperglycemia regulates TXNIP/TRX/ROS axis via p38 MAPK and ERK pathways in pancreatic cancer. Curr Cancer Drug Targets 2014;14(4):348–356; doi: 10.2174/1568009614666140331231658 [DOI] [PubMed] [Google Scholar]
- Li XF, Shen WW, Sun YY, et al. MicroRNA-20a negatively regulates expression of NLRP3-inflammasome by targeting TXNIP in adjuvant-induced arthritis fibroblast-like synoviocytes. Joint Bone Spine 2016;83(6):695–700; doi: 10.1016/j.jbspin.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Li Y, Miao LY, Xiao YL, et al. Hypoxia induced high expression of thioredoxin interacting protein (TXNIP) in non-small cell lung cancer and its prognostic effect. Asian Pac J Cancer Prev 2015;16(7):2953–2958; doi: 10.7314/apjcp.2015.16.7.2953 [DOI] [PubMed] [Google Scholar]
- Liang Y, Wang H, Chen B, et al. circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol Ther Nucleic Acids 2021;23:355–368; doi: 10.1016/j.omtn.2020.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JO, Lee SJ, Kim WI, et al. Titanium dioxide nanoparticles exacerbate allergic airway inflammation via TXNIP upregulation in a mouse model of asthma. Int J Mol Sci 2021;22(18):9924; doi: 10.3390/ijms22189924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zuo R, Wang K, et al. Oroxindin inhibits macrophage NLRP3 inflammasome activation in DSS-induced ulcerative colitis in mice via suppressing TXNIP-dependent NF-κB pathway. Acta Pharmacol Sin 2020;41(6):771–781; doi: 10.1038/s41401-019-0335-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lian K, Zhang L, et al. TXNIP mediates NLRP3 inflammasome activation in cardiac microvascular endothelial cells as a novel mechanism in myocardial ischemia/reperfusion injury. Basic Res Cardiol 2014;109(5):415; doi: 10.1007/s00395-014-0415-z [DOI] [PubMed] [Google Scholar]
- Lu Y, Li Y, Liu Q, et al. MondoA-thioredoxin-interacting protein axis maintains regulatory T-cell identity and function in colorectal cancer microenvironment. Gastroenterology 2021;161(2):575.e16–591.e16; doi: 10.1053/j.gastro.2021.04.041 [DOI] [PubMed] [Google Scholar]
- Lu YW, Galbraith L, Herndon JD, et al. Defining functional classes of Barth syndrome mutation in humans. Hum Mol Genet 2016;25(9):1754–1770; doi: 10.1093/hmg/ddw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz J, Blondonnet R, Belville C, et al. Roles of RAGE and TXNIP in NLRP3-inflammasome activation in human macrophages. Protectors of the lung: Innate immune responses in acute lung injury. Am J Respir Crit Care Med 2016;193:A4803; doi: 10.1164/ajrccm-conference.2016.193.1_MeetingAbstracts.A4803 [DOI] [Google Scholar]
- Maddocks OD, Labuschagne CF, Vousden KH. Localization of NADPH production: A wheel within a wheel. Mol Cell 2014;55(2):158–160; doi: 10.1016/j.molcel.2014.07.001 [DOI] [PubMed] [Google Scholar]
- Mahmoudian E, Khalilnezhad A, Gharagozli K, et al. Thioredoxin-1, redox factor-1 and thioredoxin-interacting protein, mRNAs are differentially expressed in multiple sclerosis patients exposed and non-exposed to interferon and immunosuppressive treatments. Gene 2017;634:29–36; doi: 10.1016/j.gene.2017.08.021 [DOI] [PubMed] [Google Scholar]
- Mao C, Li D, Zhou E, et al. Nicotine exacerbates atherosclerosis through a macrophage-mediated endothelial injury pathway. Aging (Albany NY) 2021;13(5):7627–7643; doi: 10.18632/aging.202660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari M, Morales A, Colell A, et al. Mitochondrial glutathione, a key survival antioxidant. Antioxid Redox Signal 2009;11(11):2685–2700; doi: 10.1089/ARS.2009.2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth T. Systematic review: Whipple's disease (Tropheryma whipplei infection) and its unmasking by tumour necrosis factor inhibitors. Aliment Pharmacol Ther 2015;41(8):709–724; doi: 10.1111/apt.13140 [DOI] [PubMed] [Google Scholar]
- Mas L, Cieren A, Delphin C, et al. Calcium influx mediates the chemoattractant-induced translocation of the arrestin-related protein AdcC in Dictyostelium. J Cell Sci 2018;131(19); doi: 10.1242/jcs.207951 [DOI] [PubMed] [Google Scholar]
- Masaki S, Masutani H, Yoshihara E, et al. Deficiency of thioredoxin binding protein-2 (TBP-2) enhances TGF-β signaling and promotes epithelial to mesenchymal transition. PLoS One 2012;7(6):e39900; doi: 10.1371/journal.pone.0039900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson E, Koren S, Razik F, et al. High beta-cell mass prevents streptozotocin-induced diabetes in thioredoxin-interacting protein-deficient mice. Am J Physiol Endocrinol Metab 2009;296(6):E1251–E1261; doi: 10.1152/ajpendo.90619.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani H. Thioredoxin-interacting protein in cancer and diabetes. Antioxid Redox Signal 2021;36(13–15):1001–1022; doi: 10.1089/ars.2021.0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, Oshima M, Oshima H, et al. Early embryonic lethality caused by targeted disruption of the mouse thioredoxin gene. Dev Biol 1996;178(1):179–185; doi: 10.1006/dbio.1996.0208 [DOI] [PubMed] [Google Scholar]
- Meylan P, Pich C, Winkler C, et al. Low expression of the PPARγ-regulated gene thioredoxin-interacting protein accompanies human melanoma progression and promotes experimental lung metastases. Sci Rep 2021;11(1):7847; doi: 10.1038/s41598-021-86329-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AH, Hafele C, Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology 2005;146(5):2397–2405; doi: 10.1210/en.2004-1378 [DOI] [PubMed] [Google Scholar]
- Miyahara H, Hasegawa K, Yashiro M, et al. Thioredoxin interacting protein protects mice from fasting induced liver steatosis by activating ER stress and its downstream signaling pathways. Sci Rep 2022;12(1):4819; doi: 10.1038/s41598-022-08791-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y, Lai W, Zhong Y, et al. TXNIP contributes to bone loss via promoting the mitochondrial oxidative phosphorylation during glucocorticoid-induced osteoporosis. Life Sci 2021;266:118938; doi: 10.1016/j.lfs.2020.118938 [DOI] [PubMed] [Google Scholar]
- Mohamed IN, Sarhan NR, Eladl MA, et al. Deletion of thioredoxin-interacting protein ameliorates high fat diet-induced non-alcoholic steatohepatitis through modulation of Toll-like receptor 2-NLRP3-inflammasome axis: Histological and immunohistochemical study. Acta Histochem 2018;120(3):242–254; doi: 10.1016/j.acthis.2018.02.006 [DOI] [PubMed] [Google Scholar]
- Morrison JA, Pike LA, Sams SB, et al. Thioredoxin interacting protein (TXNIP) is a novel tumor suppressor in thyroid cancer. Mol Cancer 2014;13(1):62; doi: 10.1186/1476-4598-13-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai N, Nakayama Y, Abdali SA, et al. Cardiomyocyte-specific Txnip C247S mutation improves left ventricular functional reserve in streptozotocin-induced diabetic mice. Am J Physiol Heart Circ Physiol 2021;321(2):H259–H274; doi: 10.1152/ajpheart.00174.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muri J, Thut H, Feng Q, et al. Thioredoxin-1 distinctly promotes NF-κB target DNA binding and NLRP3 inflammasome activation independently of Txnip. eLife 2020;9:e53627; doi: 10.7554/eLife.53627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Pan H, Lu Q. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep 2010;11(8):605–611; doi: 10.1038/embor.2010.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama Y, Mukai N, Wang BF, et al. Txnip C247S mutation protects the heart against acute myocardial infarction. J Mol Cell Cardiol 2021;155:36–49; doi: 10.1016/j.yjmcc.2021.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E, Sullivan JA, Pelham HRB. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep 2008;9(12):1216–1221; doi: 10.1038/embor.2008.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishinaka Y, Masutani H, Oka S, et al. Importin alpha1 (Rch1) mediates nuclear translocation of thioredoxin-binding protein-2/vitamin D(3)-up-regulated protein 1. J Biol Chem 2004;279(36):37559–37565; doi: 10.1074/jbc.M405473200 [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Matsui M, Iwata S, et al. Identification of thioredoxin-binding protein-2/vitamin D up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J Biol Chem 1999;274(31):21645–21650; doi: 10.1074/jbc.274.31.21645 [DOI] [PubMed] [Google Scholar]
- Noblet B, Benhamed F, O-Sullivan I, et al. Dual regulation of TxNIP by ChREBP and FoxO1 in liver. iScience 2021;24(3):102218; doi: 10.1016/j.isci.2021.102218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonn L, Williams RR, Erickson RP, et al. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol Cell Biol 2003;23(3):916–922; doi: 10.1128/mcb.23.3.916-922.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka S, Liu W, Masutani H, et al. Impaired fatty acid utilization in thioredoxin binding protein-2 (TBP-2)-deficient mice: A unique animal model of Reye syndrome. FASEB J 2006;20(1):121–123; doi: 10.1096/fj.05-4439fje [DOI] [PubMed] [Google Scholar]
- Oka S, Yoshihara E, Bizen-Abe A, et al. Thioredoxin binding protein-2/thioredoxin-interacting protein is a critical regulator of insulin secretion and peroxisome proliferator-activated receptor function. Endocrinology 2009;150(3):1225–1234; doi: 10.1210/en.2008-0646 [DOI] [PubMed] [Google Scholar]
- Omar DF, Kamal MM, El-Hefnawy MH, et al. Serum vitamin D and its upregulated protein, thioredoxin interacting protein, are associated with beta-cell dysfunction in adult patients with type 1 and type 2 diabetes. Can J Diabetes 2018;42(6):588–594; doi: 10.1016/j.jcjd.2018.02.012 [DOI] [PubMed] [Google Scholar]
- Oslowski CM, Hara T, O'Sullivan-Murphy B, et al. Thioredoxin-interacting protein mediates ER stress-induced β cell death through initiation of the inflammasome. Cell Metab 2012;16(2):265–273; doi: 10.1016/j.cmet.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse M, Kluth O, Lorza-Gil E, et al. Palmitate and insulin counteract glucose-induced thioredoxin interacting protein (TXNIP) expression in insulin secreting cells via distinct mechanisms. PLoS One 2018;13(5):e0198016; doi: 10.1371/journal.pone.0198016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh H, Carlsson E, Chutkow WA, et al. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med 2007;4(5):e158; doi: 10.1371/journal.pmed.0040158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Lee SH, Woo GH, et al. Downregulation of TXNIP leads to high proliferative activity and estrogen-dependent cell growth in breast cancer. Biochem Biophys Res Commun 2018;498(3):566–572; doi: 10.1016/j.bbrc.2018.03.020 [DOI] [PubMed] [Google Scholar]
- Patwari P, Chutkow WA, Cummings K, et al. Thioredoxin-independent regulation of metabolism by the alpha-arrestin proteins. J Biol Chem 2009;284(37):24996–25003; doi: 10.1074/jbc.M109.018093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwari P, Emilsson V, Schadt EE, et al. The arrestin domain-containing 3 protein regulates body mass and energy expenditure. Cell Metab 2011;14(5):671–683; doi: 10.1016/j.cmet.2011.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwari P, Higgins LJ, Chutkow WA, et al. The interaction of thioredoxin with Txnip. Evidence for formation of a mixed disulfide by disulfide exchange. J Biol Chem 2006;281(31):21884–21891; doi: 10.1074/jbc.M600427200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwari P, Lee RT. An expanded family of arrestins regulate metabolism. Trends Endocrinol Metab 2012;23(5):216–222; doi: 10.1016/j.tem.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Liu Y, Xu Y, et al. Cathelicidin-WA ameliorates diabetic cardiomyopathy by inhibiting the NLRP3 inflammasome. Cell Cycle 2021;20(21):2278–2290; doi: 10.1080/15384101.2021.1981631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister C, Chabre M, Plouet J, et al. Retinal S antigen identified as the 48K protein regulating light-dependent phosphodiesterase in rods. Science 1985;228(4701):891–893; doi: 10.1126/science.2988124 [DOI] [PubMed] [Google Scholar]
- Pontillo A, Brandão LA, Guimarães RL, et al. A 3′UTR SNP in NLRP3 gene is associated with susceptibility to HIV-1 infection. J Acquir Immune Defic Syndr 2010;54(3):236–240; doi: 10.1097/QAI.0b013e3181dd17d4 [DOI] [PubMed] [Google Scholar]
- Poungvarin N, Chang B, Imamura M, et al. Genome-wide analysis of ChREBP binding sites on male mouse liver and white adipose chromatin. Endocrinology 2015;156(6):1982–1994; doi: 10.1210/en.2014-1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao S, Dennis M, Song X, et al. A REDD1/TXNIP pro-oxidant complex regulates ATG4B activity to control stress-induced autophagy and sustain exercise capacity. Nat Commun 2015;6(1):7014; doi: 10.1038/ncomms8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Clore GM, Kennedy WM, et al. Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from the transcription factor NF kappa B. Structure 1995;3(3):289–297; doi: 10.1016/s0969-2126(01)00159-9 [DOI] [PubMed] [Google Scholar]
- Richards P, Rachdi L, Oshima M, et al. MondoA is an essential glucose-responsive transcription factor in human pancreatic β-cells. Diabetes 2017;67(3):461–472; doi: 10.2337/db17-0595 [DOI] [PubMed] [Google Scholar]
- Rogoff D, Black K, Mcmillan DR, et al. Contribution of hexose-6-phosphate dehydrogenase to NADPH content and redox environment in the endoplasmic reticulum. Redox Rep 2010;15(2):64–70; doi: 10.1179/174329210X12650506623249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena G, Chen J, Shalev A. Intracellular shuttling and mitochondrial function of thioredoxin-interacting protein. J Biol Chem 2010;285(6):3997–4005; doi: 10.1074/jbc.M109.034421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer FQ, Buettner GR. Redox State and Redox Environment in Biology. Springer: Dodrecht, 2003. [Google Scholar]
- Schroder K, Zhou R, Tschopp J. The NLRP3 inflammasome: A sensor for metabolic danger? Science 2010;327(5963):296–300; doi: 10.1126/science.1184003 [DOI] [PubMed] [Google Scholar]
- Shah A, Dagdeviren S, Lewandowski JP, et al. Thioredoxin interacting protein is required for a chronic energy-rich diet to promote intestinal fructose absorption. iScience 2020;23(9):101521; doi: 10.1016/j.isci.2020.101521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A, Xia L, Goldberg H, et al. Thioredoxin-interacting protein mediates high glucose-induced reactive oxygen species generation by mitochondria and the NADPH oxidase, Nox4, in mesangial cells. J Biol Chem 2013;288(14):6835–6848; doi: 10.1074/jbc.M112.419101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A, Xia L, Masson EA, et al. Thioredoxin-interacting protein deficiency protects against diabetic nephropathy. J Am Soc Nephrol 2015;26(12):2963–2977; doi: 10.1681/asn.2014050528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked M, Ketzinel-Gilad M, Cerasi E, et al. AMP-activated protein kinase (AMPK) mediates nutrient regulation of thioredoxin-interacting protein (TXNIP) in pancreatic beta-cells. PLoS One 2011;6(12):e28804; doi: 10.1371/journal.pone.0028804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev A. Lack of TXNIP protects beta-cells against glucotoxicity. Biochem Soc Trans 2008;36(Pt 5):963–965; doi: 10.1042/bst0360963 [DOI] [PubMed] [Google Scholar]
- Shalev A. Minireview: Thioredoxin-interacting protein: Regulation and function in the pancreatic β-cell. Mol Endocrinol 2014;28(8):1211–1220; doi: 10.1210/me.2014-1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea FF, Rowell JL, Li Y, et al. Mammalian alpha arrestins link activated seven transmembrane receptors to Nedd4 family e3 ubiquitin ligases and interact with beta arrestins. PLoS One 2012;7(12):e50557; doi: 10.1371/journal.pone.0050557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, O'Shea JM, Kaadige MR, et al. Metabolic reprogramming in triple-negative breast cancer through Myc suppression of TXNIP. Proc Natl Acad Sci U S A 2015;112(17):5425–5430; doi: 10.1073/pnas.1501555112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy SK, Mcdonald PH, Kohout TA, et al. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science 2001;294(5545):1307–1313; doi: 10.1126/science.1063866 [DOI] [PubMed] [Google Scholar]
- Sheth SS, Bodnar JS, Ghazalpour A, et al. Hepatocellular carcinoma in Txnip-deficient mice. Oncogene 2006;25:3528–3536; doi: 10.1038/sj.onc.1209394 [DOI] [PubMed] [Google Scholar]
- Sheth SS, Castellani LW, Chari S, et al. Thioredoxin-interacting protein deficiency disrupts the fasting-feeding metabolic transition. J Lipid Res 2005;46(1):123–134; doi: 10.1194/jlr.M400341-JLR200 [DOI] [PubMed] [Google Scholar]
- Soriano-Tárraga C, Jiménez-Conde J, Giralt-Steinhauer E, et al. Epigenome-wide association study identifies TXNIP gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Hum Mol Genet 2015;25(3):609–619; doi: 10.1093/hmg/ddv493 [DOI] [PubMed] [Google Scholar]
- Souza de Lima D, Ogusku MM, Sadahiro A, et al. Inflammasome genetics contributes to the development and control of active pulmonary tuberculosis. Infect Genet Evol 2016;41:240–244; doi: 10.1016/j.meegid.2016.04.015 [DOI] [PubMed] [Google Scholar]
- Spindel ON, World C, Berk BC. Thioredoxin interacting protein: Redox dependent and independent regulatory mechanisms. Antioxid Redox Signal 2012;16(6):587–596; doi: 10.1089/ars.2011.4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancill JS, Corbett JA. The role of thioredoxin/peroxiredoxin in the β-cell defense against oxidative damage. Front Endocrinol (Lausanne) 2021;12; doi: 10.3389/fendo.2021.718235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzman CA, Peterson CW, Breen KT, et al. Glucose sensing by MondoA:Mlx complexes: A role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc Natl Acad Sci U S A 2008;105(19):6912–6917; doi: 10.1073/pnas.0712199105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Chen J, Sun L, et al. Role of thioredoxin-interacting protein in diabetic fatty kidney induced by advanced glycation end-products. J Agric Food Chem 2021;69(40):11982–11991; doi: 10.1021/acs.jafc.1c03559 [DOI] [PubMed] [Google Scholar]
- Sun X, Hao H, Han Q, et al. Human umbilical cord-derived mesenchymal stem cells ameliorate insulin resistance by suppressing NLRP3 inflammasome-mediated inflammation in type 2 diabetes rats. Stem Cell Res Ther 2017;8(1):241; doi: 10.1186/s13287-017-0668-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpigel A, Hainault I, Carlier A, et al. Lipid environment induces ER stress, TXNIP expression and inflammation in immune cells of individuals with type 2 diabetes. Diabetologia 2018;61(2):399–412; doi: 10.1007/s00125-017-4462-5 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Masuda H, Ishii Y, et al. Decreased expression of thioredoxin interacting protein mRNA in inflamed colonic mucosa in patients with ulcerative colitis. Oncol Rep 2007;18(3):531–535; doi.org/10.3892/or.18.3.531 [PubMed] [Google Scholar]
- Thielen L, Shalev A. Diabetes pathogenic mechanisms and potential new therapies based upon a novel target called TXNIP. Curr Opin Endocrinol Diabetes Obes 2018;25(2):75–80; doi: 10.1097/med.0000000000000391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielen LA, Chen J, Jing G, et al. Identification of an anti-diabetic, orally available small molecule that regulates TXNIP expression and glucagon action. Cell Metab 2020;32(3):353.e8–365.e8; doi: 10.1016/j.cmet.2020.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubaki H, Tooyama I, Walker DG. Thioredoxin-interacting protein (TXNIP) with focus on brain and neurodegenerative diseases. Int J Mol Sci 2020;21(24):9357; doi: 10.3390/ijms21249357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta 2011;1813(11):1938–1945; doi: 10.1016/j.bbamcr.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Vandanmagsar B, Youm Y-H, Ravussin A, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med 2011;17(2):179–188; doi: 10.1038/nm.2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldhart AN, Dykstra H, Peck AS, et al. Phosphorylation of TXNIP by AKT mediates acute influx of glucose in response to insulin. Cell Rep 2017;19(10):2005–2013; doi: 10.1016/j.celrep.2017.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldhart AN, Muhire B, Johnson B, et al. Excess dietary carbohydrate affects mitochondrial integrity as observed in brown adipose tissue. Cell Rep 2021;36(5):109488; doi: 10.1016/j.celrep.2021.109488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Xu Y, Wang X, et al. Dl-3-n-butylphthalide inhibits NLRP3 inflammasome and mitigates Alzheimer's-like pathology via Nrf2-TXNIP-TrX axis. Antioxid Redox Signal 2019;30(11):1411–1431; doi: 10.1089/ars.2017.7440 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhao H, Xu H, et al. Targeting the TXNIP-NLRP3 interaction with PSSM1443 to suppress inflammation in sepsis-induced myocardial dysfunction. J Cell Physiol 2021a;236(7):4625–4639; doi: 10.1002/jcp.30186 [DOI] [PubMed] [Google Scholar]
- Wang P, Zheng D, Qi H, et al. Thioredoxin-interacting protein is a favored target of miR-125b, promoting metastasis and progression of pancreatic cancer via the HIF1α pathway. J Biochem Mol Toxicol 2021b;35(3):e22782; doi: 10.1002/jbt.22782 [DOI] [PubMed] [Google Scholar]
- Wang X, Eno CO, Altman BJ, et al. ER stress modulates cellular metabolism. Biochem J 2011;435(1):285–296; doi: 10.1042/BJ20101864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen S. TXNIP links anticipatory unfolded protein response to estrogen reprogramming glucose metabolism in breast cancer cells. Endocrinology 2021;163(1):bqab212; doi: 10.1210/endocr/bqab212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, De Keulenaer GW, Lee RT. Vitamin D(3)-up-regulated protein-1 is a stress-responsive gene that regulates cardiomyocyte viability through interaction with thioredoxin. J Biol Chem 2002;277(29):26496–26500; doi: 10.1074/jbc.M202133200 [DOI] [PubMed] [Google Scholar]
- Wang Y, Ji N, Gong X, et al. Thioredoxin-1 attenuates atherosclerosis development through inhibiting NLRP3 inflammasome. Endocrine 2020;70(1):65–70; doi: 10.1007/s12020-020-02389-z [DOI] [PubMed] [Google Scholar]
- Wang Z, Peng H, Gao W, et al. Blood DNA methylation markers associated with type 2 diabetes, fasting glucose, and HbA1c levels: An epigenome-wide association study in 316 adult twin pairs. Genomics 2021c;113(6):4206–4213; doi: 10.1016/j.ygeno.2021.11.005 [DOI] [PubMed] [Google Scholar]
- Wedegaertner H, Pan W-A, Trejo J. Regulation of alpha-arrestin function in G protein-coupled receptor signaling and trafficking. FASEB J 2020;34(S1):1–1; doi: 10.1096/fasebj.2020.34.s1.02800 [DOI] [Google Scholar]
- Wu M, Li R, Hou Y, et al. Thioredoxin-interacting protein deficiency ameliorates kidney inflammation and fibrosis in mice with unilateral ureteral obstruction. Lab Invest 2018;98(9):1211–1224; doi: 10.1038/s41374-018-0078-8 [DOI] [PubMed] [Google Scholar]
- Wu N, Zheng B, Shaywitz A, et al. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol Cell 2013;49(6):1167–1175; doi: 10.1016/j.molcel.2013.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Xie R, Xie S, et al. Thioredoxin interacting protein (TXNIP) acts as a tumor suppressor in human prostate cancer. Cell Biol Int 2020;44(10):2094–2106; doi: 10.1002/cbin.11418 [DOI] [PubMed] [Google Scholar]
- Xu G, Chen J, Jing G, et al. Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat Med 2013;19(9):1141–1146; doi: 10.1038/nm.3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Malhotra A, Ren M, et al. The enzymatic function of tafazzin. J Biol Chem 2006;281(51):39217–39224; doi: 10.1074/jbc.M606100200 [DOI] [PubMed] [Google Scholar]
- Xue Y, Wang SK, Rana P, et al. AAV-Txnip prolongs cone survival and vision in mouse models of retinitis pigmentosa. eLife 2021;10:e66240; doi: 10.7554/eLife.66240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Yang G, Hong C, et al. Inhibition of endogenous thioredoxin in the heart increases oxidative stress and cardiac hypertrophy. J Clin Invest 2003;112(9):1395–1406; doi: 10.1172/JCI17700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki H, Haendeler J, Berk BC. Thioredoxin. Circ Res 2003;93(11):1029–1033; doi: 10.1161/01.RES.0000102869.39150.23 [DOI] [PubMed] [Google Scholar]
- Yamazaki M, Yamada H, Munetsuna E, et al. DNA methylation level of the gene encoding thioredoxin-interacting protein in peripheral blood cells is associated with metabolic syndrome in the Japanese general population. Endocr J 2022;69(3):319–326; doi: 10.1507/endocrj.EJ21-0339 [DOI] [PubMed] [Google Scholar]
- Yoshihara E, Fujimoto S, Inagaki N, et al. Disruption of TBP-2 ameliorates insulin sensitivity and secretion without affecting obesity. Nat Commun 2010;1:127; doi: 10.1038/ncomms1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka J, Chutkow WA, Lee S, et al. Deletion of thioredoxin-interacting protein in mice impairs mitochondrial function but protects the myocardium from ischemia-reperfusion injury. J Clin Invest 2012;122(1):267–279; doi: 10.1172/JCI44927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka J, Imahashi K, Gabel SA, et al. Targeted deletion of thioredoxin-interacting protein regulates cardiac dysfunction in response to pressure overload. Circ Res 2007;101(12):1328–1338; doi: 10.1161/CIRCRESAHA.106.160515 [DOI] [PubMed] [Google Scholar]
- Yoshioka J, Lee RT. Thioredoxin-interacting protein and myocardial mitochondrial function in ischemia-reperfusion injury. Trends Cardiovasc Med 2014;24(2):75–80; doi: 10.1016/j.tcm.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka J, Schulze PC, Cupesi M, et al. Thioredoxin-interacting protein controls cardiac hypertrophy through regulation of thioredoxin activity. Circulation 2004;109(21):2581–2586; doi: 10.1161/01.Cir.0000129771.32215.44 [DOI] [PubMed] [Google Scholar]
- Yu F-X, Chai TF, He H, et al. Thioredoxin-interacting protein (Txnip) gene expression: Sensing oxidative phosphorylation status and glycolytic rate. J Biol Chem 2010;285(33):25822–25830; doi: 10.1074/jbc.M110.108290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Cheng C, Cao M, et al. TXNIP hypomethylation and its interaction with obesity and hypertriglyceridemia increase type 2 diabetes mellitus risk: A nested case-control study. J Diabetes 2020;12(7):512–520; doi: 10.1111/1753-0407.13021 [DOI] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature 2008;454(7203):455–462; doi: 10.1038/nature07203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Gao J, Wang X, et al. A novel indication of thioredoxin-interacting protein as a tumor suppressor gene in malignant glioma. Oncol Lett 2017;14(2):2053–2058; doi: 10.3892/ol.2017.6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang J-H, Chen X-Y, et al. Reactive oxygen species-induced TXNIP drives fructose-mediated hepatic inflammation and lipid accumulation through NLRP3 inflammasome activation. Antioxid Redox Signal 2015;22(10):848–870; doi: 10.1089/ars.2014.5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yan Q, Gong L, et al. C-terminal truncated HBx initiates hepatocarcinogenesis by downregulating TXNIP and reprogramming glucose metabolism. Oncogene 2021;40(6):1147–1161; doi: 10.1038/s41388-020-01593-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhong P, Xu Y, et al. Differential expression of TXNIP isoforms in the peripheral leukocytes of patients with acute myocardial infarction. Dis Markers 2018;2018(40):1–6; doi: 10.1155/2018/9051481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Zhao W. NLRP3 inflammasome—A key player in antiviral responses. Front Immunol 2020;11;211; doi: 10.3389/fimmu.2020.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Chen F, Yin Q, et al. Reactive oxygen species interact with NLRP3 inflammasomes and are involved in the inflammation of sepsis: From mechanism to treatment of progression. Front Physiol 2020;11:571810; doi: 10.3389/fphys.2020.571810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Zhang H. NLRP3 inflammasome and inflammatory bowel disease. Front Immunol 2019;10:276; doi: 10.3389/fimmu.2019.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhang Z, Qian G, et al. Omentin-1 attenuates adipose tissue inflammation via restoration of TXNIP/NLRP3 signaling in high-fat diet-induced obese mice. Fundam Clin Pharmacol 2020;34(6):721–735; doi: 10.1111/fcp.12575 [DOI] [PubMed] [Google Scholar]
- Zhou R, Tardivel A, Thorens B, et al. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol 2010;11(2):136–140; doi: 10.1038/ni.1831 [DOI] [PubMed] [Google Scholar]
- Zhu G, Zhou L, Liu H, et al. MicroRNA-224 promotes pancreatic cancer cell proliferation and migration by targeting the TXNIP-mediated HIF1α pathway. Cell Physiol Biochem 2018;48(4):1735–1746; doi: 10.1159/000492309 [DOI] [PubMed] [Google Scholar]
- Ziello JE, Jovin IS, Huang Y. Hypoxia-inducible factor (HIF)-1 regulatory pathway and its potential for therapeutic intervention in malignancy and ischemia. Yale J Biol Med 2007;80(2):51–60. [PMC free article] [PubMed] [Google Scholar]