Abstract
Alzheimer’s disease (AD) is an age-related multifactorial neurodegenerative disorder. Advances in genome technology, including next generation sequencing have uncovered complex genetic effects in AD by analyzing both common and rare functional variants. Multiple lines of evidence suggest that the pathogenesis of AD is influenced by multiple genetic components rather than single genetic factor. Previous genetic studies on AD have predominantly included European ancestry cohorts; hence, the non-European population may be underrepresented, potentially leading to reduced diversity in AD genetic research. Additionally, ethnic diversity may result in dissimilar effects of genetic determinants in AD. APOE genotypes are a well-established genetic risk factor in AD, with the East Asian population having a higher risk of AD associated with the APOE ε4 allele. To date, seven genome-wide association studies (GWAS) have been conducted in East Asians, which report a total of 26 AD-associated loci. Several rare variants, including the p.H157Y variant in TREM2, and the p.G186R and p.R274W variants in SHARPIN are associated with risk of AD in East Asians. Extending genetic studies to diverse populations, including East Asians is necessary, which could yield more comprehensive insights into AD, and here we review the recent findings regarding the genetic determinants of AD from an East Asian perspective.
Subject terms: Alzheimer's disease, Medical genetics
Introduction
Alzheimer’s disease (AD) is an age-related neurodegenerative disorder and a leading cause of dementia. Although aging is the largest risk factor, it is not sufficient for the development of AD. The etiology of AD is complex as it involves a combination of genetic and environmental factors [1]. Studies in biological twin have estimated that the heritability of AD ranged from 58 to 79% [2]. Heritability of AD calculated based on common single nucleotide polymorphisms (SNP) was estimated to be 33% [3]. Precise knowledge of the genetic determinants of AD is essential to understand the neurobiological pathogenesis of AD.
Previous genetic studies have identified many disease-associated genes and risk variants in AD [4]. In particular, APOE locus is a well-established genetic risk factor for AD [5]. Genome-wide association studies (GWAS) have identified 38 different loci associated with AD [6, 7], and recent utilization of whole exome/genome sequencing (WES/WGS) and next generation sequencing (NGS) have revealed that rare coding variants not only play an important role but also have significant effects in the pathogenesis of AD [8]. Furthermore, accumulating evidence suggests that pathogenesis of AD is influenced by multiple genetic components rather than a single genetic factor [4].
Diverse genetic architectures among different ethnic groups may differentially influence how these genetic factors contribute to the pathogenesis of AD. Previous genetic studies of AD have been largely conducted in European ancestry cohorts with potential underrepresentation of non-European populations, leading to a lack of ethnic diversity in genetic research on AD. This can impede our ability to fully understand the contribution of the genetic component in the pathogenesis of AD from the viewpoint of global healthcare policy. As extending genetic studies to other populations including East Asians, could yield more comprehensive genetic insights into AD pathogenesis, this review article summarizes the recent findings on the genetic contribution to AD from an East Asian perspective.
APOE Genotypes
APOE as risk factor for AD
Apolipoprotein E, encoded by APOE, is a secreted multifunctional protein that plays central roles in lipid metabolism and the pathogenesis of neurodegenerative disorders, including AD. In the 1970s and 1980s, genetic research on APOE was mainly conducted from the viewpoint of dyslipidemia, and it was in the 1990s that it was reported that APOE genotypes confer major risk of AD [9]. Since then, genetic risk of AD associated with APOE ε4 and the protective role of ε2 have been confirmed worldwide [5]. APOE is now recognized as the strongest susceptibility gene for late-onset sporadic AD. This should be taken into account when evaluating clinical and pathological features of AD.
Three kinds of APOE alleles including ε2 (rs429358-rs7412, T-T [Cys-Cys]), ε3 (rs429358-rs7412, T-C [Cys-Arg]), and ε4 (rs429358-rs7412, C-C [Arg-Arg]), have been extensively evaluated as determinants of disease susceptibility. The APOE ε4 allele is associated with an approximately 4-fold higher risk in clinically diagnosed subjects, and this risk rises to 6-fold in patients with neuropathological confirmation [10]. Notably, the presence of such a susceptibility gene with relatively large effect size appears to be a rare phenomenon in common diseases with a sporadic occurrence. Meta-analyses of the effects of APOE genotypes on AD have been reported in Caucasians [11–13], Chinese [14], Indians [15], and Iranians [16]. However, there have been no report of such meta-analyses in the Japanese population.
Larger effects of APOE ε4 in East Asians
An interaction between ethnicity and the effect of APOE genotype on AD risk has gained much attention. Specifically, while the effect of APOE ε4 is weaker in African American and Hispanic populations, its effect is higher in East Asian populations, including the Japanese (Table 1) [11]. Variable effects of the ε4 allele across populations can be partly explained by differences in the frequency of the ε4 allele in general population of each ethnic group. We have previously reported that the odds ratio for AD with the ε4 allele is higher in East Asians than in Europeans [17]. The frequency of the rs405509 genotypes in the promoter region of APOE are different between East Asian and European populations with the frequency of the T/T genotype being significantly higher in East Asians. Functional experiments using a reporter assay have demonstrated that the T genotype at rs405509 resulted in lower expression of APOE [17]. Thus, the modifying effect of rs405509 may explain the ethnic variability in the effects of the APOE ε4 allele.
Table 1.
Genetic risk and protective effects of APOE genotypes on AD in different population
| Population | Genotype: OR (95% CI) | Reference | |||||
|---|---|---|---|---|---|---|---|
| ε2*2 | ε2*3 | ε2*4 | ε3*3 | ε3*4 | ε4*4 | ||
| Japanese | 1.1 (0.1–17.2) | 0.9 (0.4–2.5) | 2.4 (0.4–15.4) | 1.0 (Ref) | 5.6 (3.9–8.0) | 33.1 (13.6–80.5) | [11] |
| Japanese | NA | 0.7 (0.3–1.6) | NA | 1.0 (Ref) | 3.9 (1.9–8.0) | 21.8 (8.6–55.3) | [13] |
| Caucasians: clinic/autopsy | 0.6 (0.2–2.0) | 0.6 (0.5–0.8) | 2.6 (1.6–4.0) | 1.0 (Ref) | 3.2 (2.8–3.8) | 14.9 (10.8–20.6) | [11] |
| Caucasians: clinic/autopsy | NA | 0.6 (0.3–1.2 | NA | 1.0 (Ref) | 4.3 (3.3–5.5) | 15.6 (10.9–22.5) | [13] |
| Caucasians: autopsy | 0.1 (0.1–0.4) | 0.4 (0.3–0.5) | 2.7 (1.7–4.4) | 1.0 (Ref) | 6.1 (5.–7.4) | 31.2 (16.6–58.8) | [10] |
| Caucasians: population-based | 0.9 (0.3–2.8) | 0.6 (0.5–0.9) | 1.2 (0.8–2.0) | 1.0 (Ref) | 2.7 (2.2–3.2) | 12.5 (8.8–17.7) | [11] |
| Caucasians: population-based | NA | 0.3 (0.2–0.6) | NA | 1.0 (Ref) | 2.8 (2.3–3.5) | 11.8 (7.0–19.8) | [13] |
| African Americans | 2.4 (0.3–22.7) | 0.6 (0.4–1.7) | 1.8 (0.4–8.1) | 1.0 (Ref) | 1.1 (0.7–1.8) | 5.7 (2.3–14.1) | [11] |
| Hispanics | 2.6 (0.2–33.3) | 0.6 (0.3–1.3) | 3.2 (0.9–11.6) | 1.0 (Ref) | 2.2 (1.3–3.4) | 2.2 (0.7–6.7) | [11] |
OR odds ratio, CI confidence interval, NA not available, Ref referece
Rare missense variants of APOE
Recent research in APOE has focused on the identification of the rare missense variants (MAF < 1%) and their functional significance [18]. The Christchurch variant rs121918393 (APOEChc: CGC > aGC, p.Arg[R]136Ser[S]) [19–21] and the Jacksonville variant rs199768005 (APOEJax: GTG > GaG, p.Val[V]236Glu[E]) [22, 23] have been identified as protective variants against AD in Caucasians, with the APOEChc variant apparently reducing the effects of the pathogenic PSEN1 variant (GAA > GcA, p.Glu[E]280Ala[A]), which is a highly penetrant and causative mutation for dominantly inherited AD [20, 21]. Individuals carrying the PSEN1 p.E280A mutation typically develop mild cognitive impairment at a median age of 44 years (95% confidence interval [CI]: 43‒45 years) and dementia at a median age of 49 years (95% CI: 49‒50 years). Surprisingly, a woman with homozygous APOEChc variant and carrying the PSEN1 p.E280A mutation did not exhibit mild cognitive impairment until her 70 s even though abundant accumulation of amyloid-β (Aβ) was seen in the brain. However, tau accumulation in the brain, which is a major component of neurofibrillary tangles, was clearly limited, and the degree of hippocampal atrophy was also mild, suggesting that APOEChc may exhibit an anti-tau effect.
On the other hand, APOEJac was found to show an anti-Aβ effect [23] as amount of Aβ and senile plaques in the brain of APOEJax carriers was found to be significantly lower than that of control subjects. Additionally, biochemical analysis showed that APOEJax variant inhibited self-aggregation of ApoE, which may in turn inhibit the accumulation of Aβ. Genetic analysis demonstrated that APOEJax was equally or more protective against AD than the ε2 allele [22]. Further work is warranted to elucidate the molecular networks affected by the APOEChc and APOEJax variants. Importantly, these variants are not listed in the Japanese database of the Tohoku Medical Megabank, and it is possible that these are seen only in Caucasian. Hence, additional rare variants of APOE in AD patients of East Asian origin must be explored.
Among the missense variants of APOE identified so far, those evaluated for pathogenicity in the human genome variant database ClinVar have been summarized in Table 2. Currently, 37 variations are listed, including rs429358 and rs7412, which determine the three alleles ε2, ε3, and ε4, as well as APOEChc (rs121918393) and APOEJax (rs199768005). Many of listed variants are associated with dyslipidemia and atherosclerosis, and only three are relevant to AD (Variation ID: 242765 [rs769452], 17864 [rs429358], 694585 [rs429358 - rs121918393]). Six variants are found only in East Asians including in the Japanese (rs121918392, rs587778876, rs121918397, rs267606663, rs140808909, and rs190853081); however, none of these have been described in relation to AD. As AD can be influenced by vascular disorders that may be caused by disruption of lipid metabolism, it is important to assign biological significance to missense variants of APOE.
Table 2.
Missense variants of APOE listed at the human genomic variant database ClinVar
| Variation ID | Variant type | dbSNP ID | Locationa | Amino acid changeb | CADD (GRCh38-v1.6) | Anotation of the variant | Related disease | |
|---|---|---|---|---|---|---|---|---|
| 17849 | SNV | rs121918392 | c.61 G > A | p.Glu21Lys | 20.20 | Pathogenic | Hyperlipoproteinemia, type III; and atherosclerosis (APOE ε5) | |
| 440842 | SNV | rs201672011 | c.91 G > A | p.Glu31Lys | 15.87 | Pathogenic | NA | |
| 441264 | Haplotype | rs201672011 - rs769455 | c.[91 G > A;487 C > T] | p.Glu31Lys - p.Arg163Cys | 15.87–28.40 | Pathogenic | Familial hyperlipoproteinemia, type III | |
| 17880 | SNV | rs121918399 | c.127 C > T | p.Arg43Cys | 23.30 | Likely pathogenic | Lipoprotein glomerulopathy | |
| 242765 | SNV | rs769452 | c.137 T > C | p.Leu46Pro | 0.72 | Conflicting interpretations: benign, likely benign, uncertain significance | Alzheimer’s disease | |
| 441268 | Haplotype | rs769452 - rs429358 | c.[137 T > C;388 T > C] | p.Leu46Pro - p.Cys130Arg | 0.72–16.65 | Pathogenic/likely pathogenic | Familial hypercholesterolemia | |
| 17871 | SNV | rs28931576 | c.178 A > G | p.Thr60Ala | 15.46 | Pathogenic | NA | |
| 441269 | Haplotype | rs11083750 - rs429358 | c.[305 C > G;388 T > C] | p.Pro102Arg - p.Cys130Arg | 23.20–16.65 | Association | NA | |
| 441270 | Haplotype | rs28931577 - rs267606662 | c.[349 G > A;508 G > C] | p.Ala117Thr - p.Ala170Pro | 27.00–17.41 | Pathogenic | NA | |
| 88639 | SNV | rs587778876 | c.364 C > A | p.Leu122Met | 24.00 | Not provided | Major depressive disorder | |
| 17864 | SNV | rs429358 | c.388 T > C | p.Cys130Arg | 16.65 | Conflicting interpretations: pathogenic, likely pathogenic, risk factor, drug response, uncertain significance | Alzheimer’s disease; lipoprotein glomerulopathy; and warfarin response | |
| 694585 | Haplotype | rs429358 - rs121918393 | c.[388 = ;460 C > A526 = ] | p.Cys130Arg - p.Arg154Ser | 16.65–25.30 | Protective | Alzheimer’s disease (APOEε3_Christchurch) | |
| 440870 | Haplotype | rs429358 - rs387906567 | c.[388 T > C;478 C > T] | p.Cys130Arg - p.Arg160Cys | 16.65–28.60 | Pathogenic | Familial hyperlipoproteinemia, type III | |
| 441267 | Haplotype | rs429358 - rs267606661 | c.[388 T > C;805 C > G] | p.Cys130Arg - p.Arg269Gly | 16.65–23.30 | Pathogenic | Familial hyperlipoproteinemia, type III | |
| 917851 | SNV | rs1969863273 | c.422 A > G | p.Gln141Arg | 19.85 | Uncertain significance | Familial hypercholesterolemia; familial hyperlipoproteinemia, type III; and hyperlipoproteinemia | |
| 478904 | SNV | rs267606664 | c.434 G > A | p.Gly145Asp | 24.50 | Uncertain significance | Hypercholesterolemia | |
| 441262 | Haplotype | rs267606664 - rs7412 | c.[434 G > A;526 C > T] | p.Gly145Asp - p.Arg176Cys | 24.50–24.60 | Pathogenic | Apolipoproteinemia E1 | |
| 88640 | SNV | rs587778877 | c.451 C > A | p.Leu151Met | 20.70 | Not provided | Major depressive disorder | |
| 17874 | SNV | rs28931578 | c.455 G > A | p.Arg152Gln | 24.90 | Pathogenic | NA | |
| 17850 | SNV | rs121918393 | c.460 C > A | p.Arg154Ser | 25.30 | Pathogenic | Familial hyperlipoproteinemia, type III | |
| 375636 | SNV | rs200703101 | c.461 G > T | p.Arg154Leu | 27.80 | Likely pathogenic | Abnormal circulating lipid concentration | |
| 17851 | SNV | rs769455 | c.487 C > T | p.Arg163Cys | 28.40 | Benign | Familial hyperlipoproteinemia, type III | |
| 17865 | SNV | rs121918397 | c.488 G > A | p.Arg163His | 22.70 | Pathogenic | Familial hyperlipoproteinemia, type III | |
| 17879 | SNV | rs121918397 | c.488 G > C | p.Arg163Pro | 25.60 | Pathogenic | Lipoprotein glomerulopathy | |
| 17858 | SNV | rs121918394 | c.490 A > C | p.Lys164Gln | 25.50 | Pathogenic | Hyperlipoproteinemia, type III | |
| 17857 | SNV | rs121918394 | c.490 A > G | p.Lys164Glu | 26.10 | Pathogenic | Familial hyperlipoproteinemia, type III | |
| 1077013 | SNV | NA | c.494 G > C | p.Arg165Pro | 27.50 | Likely pathogenic | Lipoprotein glomerulopathy | |
| 126456 | 3 bp microsatellite | rs515726148 | c.497TCC[1] | p.Leu167del | – | Pathogenic | Sea-blue histiocyte syndrome | |
| 17848 | SNV | rs7412 | c.526 C > T | p.Arg176Cys | 24.60 | Drug response | Hypercholesterolemia; familial hyperlipoproteinemia, type III; warfarin response; atorvastatin response efficacy | |
| 441265 | Haplotype | rs7412 - rs267606663 | c.[526 C > T;725 G > A] | p.Arg176Cys - p.Arg242Gln | 24.60–8.57 | Pathogenic | Familial hyperlipoproteinemia, type III | |
| 441266 | Haplotype | rs7412 - rs199768005 | c.[526 C > T;761 T > A] | p.Arg176Cys - p.Val254Glu | 24.60–25.20 | Pathogenic | Familial hyperlipoproteinemia, type III | |
| 17862 | SNV | rs121918396 | c.683 G > A | p.Trp228Ter | 35.00 | Pathogenic | Hyperlipoproteinemia, type III (APOE ε3_Washington); and familial hyperlipoproteinemia, type III | |
| 1315806 | SNV | rs567353589 | c.688 G > A | p.Glu230Lys | 11.14 | Uncertain significance | Lipoprotein glomerulopathy | |
| 17859 | SNV | rs121918395 | c.736 C > T | p.Arg246Cys | 24.20 | Pathogenic | NA | |
| 441263 | Haplotype | rs140808909 - rs190853081 | c.[784 G > A;787 G > A] | p.Glu262Lys - p.Glu263Lys | 23.20–24.80 | Pathogenic | Hyperlipoproteinemia, type III; and atherosclerosis (APOE ε7) | |
| 17875 | SNV | rs121918398 | c.875 G > A | p.Arg292His | 25.20 | Pathogenic | NA | |
| 17876 | SNV | rs28931579 | c.940 A > C | p.Ser314Arg | 13.28 | Pathogenic | NA | |
SNV rs429358 and rs7412, which determine the three common APOE alleles (ε2, ε3, and ε4) are shown in bold. Genomic variants detected only in East Asians are underlined: database searched, gnomAD v2.1.1 and v3.1.2
[Web site] CADD https://cadd.gs.washington.edu; ClinVar https://www.ncbi.nlm.nih.gov/clinvar; gnomAD https://gnomad.broadinstitute.org.
CADD combined annotation dependent depletion, NA not assigned, SNV single nucleotide variant
aPosition on the APOE-encoding sequence, 1–954 bp (ATG [Met] - TGA [Ter]: 317 aa).
bPosition on the immature APOE protein consisting of 317 amino acids, including the N-terminal signal peptide region with 18 amino acids (position, 1–18)
GWAS
East Asian populations
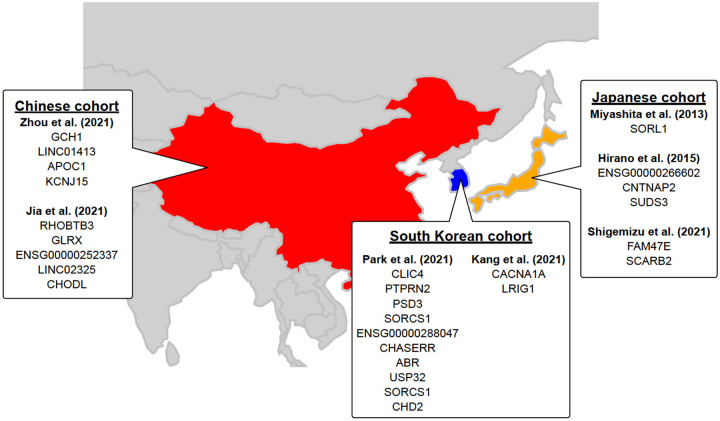
GWASs have been performed worldwide to identify common genetic factors that can explain clinical phenotypes, wherein the association between all autosomal SNPs, which are mainly genotyped by SNP arrays, and phenotypes are evaluated. The most recent GWAS for AD was performed in a European population, including 1,126,563 individuals and identified 38 susceptibility loci [7]. It is essential to perform GWASs using samples from each ethnic population to identify race-specific AD susceptibility loci. To date, 7 GWASs have been conducted in East Asians with samples from Japan, China and South Korea, and they have identified 26 AD associated loci (Fig. 1).
Fig. 1.
Overview of the genetic loci reported by seven GWAS in East Asian population. Note that the neighbor genes of each SNP shown below are mapped to GENCODE Release 39 (GRCh38.p13) based on rs numbers and may differ from the neighbor genes listed in the original paper
Japanese cohorts
The first GWAS for AD in East Asia was reported from Japan in 2013. This study included a discovery cohort of 1008 AD patients and 1016 healthy subjects, and identified 6 SNPs outside the APOE region [24]. Among these, SNP rs4598682 in SORL1 was confirmed in a replication cohort that included 885 AD patients and 985 healthy controls. Importantly, SNPs in SORL1 have also been identified as susceptibility loci in European populations [7, 25], and in a transethnic meta-analysis that included South Korean and Caucasian cohorts.
The second GWAS for AD in East Asians was published in 2015 [26], which was a meta-analysis of a discovery cohort (816 AD patients and 7992 healthy subjects) and a replication cohort (1011 AD patients and 7212 healthy subjects). This GWAS identified rs1992269 located at 18p11.32, and meta-analysis after stratification of the discovery and replication cohorts by APOE ε4 carrier and non-carrier status identified rs802571 in the intron of CNTNAP2 and rs11613092 in the intergenic region between SUDS3 and SRRM4. However, a meta-analysis of APOE ε4 carriers did not yield any significant SNPs associated with AD.
Shigemizu et al. investigated a discovery cohort of 8036 individuals, including approximately 2000 individuals who had participated in a previous study [24, 27]. They identified 134 markers located in nine genes that satisfied the significance level in the discovery cohort, and their evaluation in the replication cohort revealed the presence of rs920608 on FAM47E and SCARB2.
Chinese cohorts
Two GWASs have been conducted in the Chinese population since 2018. Zhou et al. obtained WGS data from 477 AD patients and 2187 healthy subjects [28], and association analysis, which excluded the APOE region, identified four SNPs located in GCH1, APOC1, KCNJ15, and LINC01413. Additionally, a transethnic meta-analysis of three European cohorts (ADNI, ADC, and LOAD) also identified rs72713460, which was located 11.7 kb downstream from GCH1 and rs928771, located in the intron of KCNJ15. Jia et al. analyzed 1595 AD patients and 2474 healthy subjects, and identified 34 candidate SNPs [29], that were validated in a replication cohort of 2234 AD patients and 7319 healthy subjects. Four novel SNPs were present in the 34 candidate SNPs, and among these novel SNPs, rs3777215 was located in the intron regions of RHOBTB3 and GLRX, while rs6859823 was located in the intergenic region of ENSG00000251574 and ENSG00000252337, both of which are RNA genes. Further, rs234434 was located between RNA gene ENSG00000285584 and noncoding RNA LINC02325, and rs2255835 was located in the intron region of CHODL.
South Korean cohorts
Two GWAS have been recently reported from South Korea. Park et al. focused on APOE ε4 carriers and individuals regardless of ε4 status [30]. In the GWAS focusing on APOE ε4 carriers, a discovery cohort including 331 AD patients and 169 healthy subjects and a replication cohort of 190 AD patients and 97 healthy subjects, whose samples were analyzed by WGS and a custom array. Two SNPs were identified in this analysis: rs1890078, located 54 kb upstream of SORCS1, and rs12594991, located in the intron of CHD2. The authors also analyzed samples from 874 AD patients and 1063 healthy subjects, including the APOE ε4 carriers described above, and identified nine suggestive variants. These included two SNPs located around SORCS1, which were present only in ε4 carriers. Kang et al. performed a GWAS using their own South Korean cohort and Japanese samples used previously [24, 31]. The discovery cohort included 1172 South Korean AD patients and 1119 South Korean healthy subjects, while the replication cohort used samples from 976 Japanese AD patients and 980 Japanese healthy subjects. At a significance level of P < 5 × 1e-5, only APOE regions were associated in both cohorts. Next, a stratified analysis of APOE ε4 carriers and noncarriers yielded no significant SNPs in ε4 carriers, but rs189753894, located upstream of 7 kb from CACNA1A, and rs2280575, present in the intron of LRIG1, were found in ε4 noncarriers. Interestingly, these two SNPs had the same directionality of effect in both South Korean and Japanese cohorts and satisfied a significance level of P < 5e-8 during a meta-analysis.
Intriguingly, no significant SNPs were found in APOE ε4 carriers in two GWAS populations [26, 31], suggesting that APOE genotypes in ε4 carriers may account for almost all genetic determinants in AD. In contrast, several SNPs been identified in ε4 noncarriers, but they were not common, and they had a much smaller effect size than SNPs in the APOE region, suggesting that polygenic effects may play a role in the pathogenesis of AD in ε4 noncarriers.
East Asian specific loci
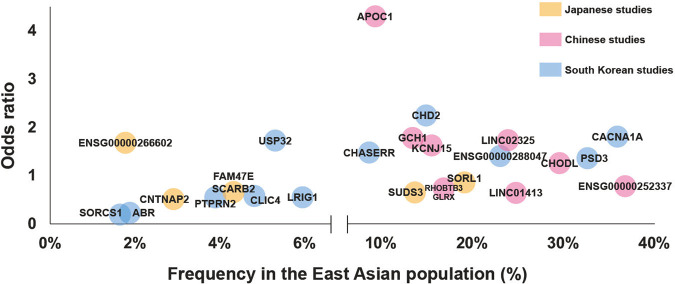
We have summarized the statistics for AD-susceptibility loci found in the three countries in Table 3 and Fig. 2, and show large differences in frequency between East Asian and European populations for some variants. For example, rs189753894 near CACNA1A, found in APOE ε4 noncarriers in the South Korean population, had an MAF of 0.3598 in East Asian populations, while the MAF in the European population was 0.02503. On the other hand, rs12594991, which is located in the intron of CHD2 and was found in another South Korean cohort, was less frequent in East Asians compared to Europeans. Thus, these observations explain ethnicity specific AD-susceptibility loci in East Asians.
Table 3.
Statistics of AD susceptibility loci found in Japanese, Chinese and South Korean populations
| SNPa | CHR:POS (GRCh38)b | Nearest genec | Location | The number of subjects in East Asiand (Discovery cohort / Replication cohort) | Healthy controls | Stratification | ORe | MAF (gnomAD v3.1.2)f | Genotyping platform | Reference | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AD patients | East Asian | non-Finnish European | MAF differenceg | |||||||||
| Japanese studies | ||||||||||||
| rs4598682 | 11:121,505,242 | SORL1 | intron | 891/885 | 844/985 | All samples | 0.75 | 0.192 | 0.01884 | 0.173 | SNP array, PCR assay | [24] |
| rs1992269 | 18:1,872,316 | ENSG00000266602 | intron | 816/1011 | 7992/7212 | All samples | 1.66 | 0.01777 | 0.1819 | −0.164 | SNP array, PCR assay | [26] |
| rs802571 | 7:146,265,094 | CNTNAP2 | intron | 489/528 | 6463/5824 | APOEε4 non-carrier | 0.52 | 0.02928 | 0.03317 | −0.004 | SNP array, PCR assay | [26] |
| rs11613092 | 12:118,455,443 | SUDS3 | intergenic | 323/480 | 1484/1364 | APOEε4 carrier | 0.61 | 0.1367 | 0.06899 | 0.068 | SNP array, PCR assay | [26] |
| rs920608 | 4:76,217,307 | FAM47E, SCARB2 | intron | 3962/1216 | 4074/2446 | All samples | 0.65 | 0.0433 | 0.009 | 0.034 | SNP array, PCR assay | [27] |
| Chinese studies | ||||||||||||
| rs72713460 | 14:54,830,325 | GCH1 | intergenic | 477 | 2187 | All samples | 1.74 | 0.1339 | 0.2188 | −0.085 | WGS | [28] |
| rs2591054 | 15:57,320,212 | LINC01413 | intron | 477 | 2187 | All samples | 0.61 | 0.2489 | 0.4126 | −0.164 | WGS | [28] |
| rs73052335 | 19:44,916,825 | APOC1 | intron | 477 | 2187 | All samples | 4.27 | 0.09286 | 0.1247 | −0.032 | WGS | [28] |
| rs928771 | 21:38,291,838 | KCNJ15 | intron | 477 | 2187 | All samples | 1.59 | 0.1549 | 0.4975 | −0.343 | WGS | [28] |
| rs3777215 | 5:95,786,296 | RHOBTB3, GLRX | intron | 1595/2234 | 2474/5085 | All samples | 0.69 | 0.1683 | 0.2307 | −0.062 | SNP array, MALDI-TOFMS | [29] |
| rs6859823 | 5:106,218,683 | ENSG00000252337 | intergenic | 1595/2234 | 2474/5085 | All samples | 0.74 | 0.3688 | 0.425 | −0.056 | SNP array, MALDI-TOFMS | [29] |
| rs234434 | 14:97,354,683 | LINC02325 | intergenic | 1595/2234 | 2474/5085 | All samples | 1.71 | 0.2398 | 0.3077 | −0.068 | SNP array, MALDI-TOFMS | [29] |
| rs2255835 | 21:18,119,346 | CHODL | intron | 1595/2234 | 2474/5085 | All samples | 1.23 | 0.296 | 0.6536 | −0.358 | SNP array, MALDI-TOFMS | [29] |
| South Korean studies | ||||||||||||
| rs12063304 | 1:24,745,177 | CLIC4 | intergenic | 543/331 | 169/894 | All samples | 0.57 | 0.04822 | 0.03179 | 0.016 | WGS, SNP array | [30] |
| rs80020083 | 7:158,491,056 | PTPRN2 | intron | 543/331 | 169/894 | All samples | 0.56 | 0.03888 | 0.0003527 | 0.039 | WGS, SNP array | [30] |
| rs967326 | 8:18,689,751 | PSD3 | intron | 543/331 | 169/894 | All samples | 1.33 | 0.3275 | 0.0523 | 0.275 | WGS, SNP array | [30] |
| rs144835823 | 10:107,242,334 | SORCS1 | intergenic | 543/331 | 169/894 | All samples | 0.32 | 0.01719 | 0.0000294 | 0.017 | WGS, SNP array | [30] |
| rs78442236 | 10:107,273,420 | SORCS1 | intergenic | 543/331 | 169/894 | All samples | 0.21 | 0.01676 | 0.0000294 | 0.017 | WGS, SNP array | [30] |
| rs74352072 | 11:119,921,300 | ENSG00000288047 | intergenic | 543/331 | 169/894 | All samples | 1.39 | 0.2301 | 0.0004703 | 0.230 | WGS, SNP array | [30] |
| rs79919241 | 15:92,891,403 | CHASERR | intron | 543/331 | 169/894 | All samples | 1.45 | 0.08674 | 0.046 | 0.041 | WGS, SNP array | [30] |
| rs201351606 | 17:1,179,646 | ABR | intron | 543/331 | 169/894 | All samples | 0.23 | 0.01897 | 0.00004415 | 0.019 | WGS, SNP array | [30] |
| rs117665140 | 17:60,203,564 | USP32 | intron | 543/331 | 169/894 | All samples | 1.74 | 0.05327 | 0.00001471 | 0.053 | WGS, SNP array | [30] |
| rs1890078 | 10:107,218,478 | SORCS1 | intergenic | 331/190 | 169/97 | APOEε4 carrier | 0.43 | 0.0792 | 0.0689 | 0.010 | WGS, SNP array | [30] |
| rs12594991 | 15:92,973,197 | CHD2 | intron | 331/190 | 169/97 | APOEε4 carrier | 2.21 | 0.1496 | 0.5297 | −0.380 | WGS, SNP array | [30] |
| rs189753894 | 19:13,513,675 | CACNA1A | intron | 976/815 | 621/435 | APOEε4 non-carrier | 1.787 | 0.3598 | 0.02503 | 0.335 | SNP array | [31] |
| rs2280575 | 3:66,492,439 | LRIG1 | intron | 976/815 | 621/435 | APOEε4 non-carrier | 0.544 | 0.05939 | 0.2958 | −0.236 | SNP array | [31] |
SNP single nucleotide polymorphism, CHR chromosome, POS genomic postion, MAF minor allele frequency, OR odds ratios calculated according to the minor allele
aEach SNP is a representative SNP identified from the East Asian cohort
bGenomic position was based on GRCh38
cThe nearest genes were based on GENCODE V39
dThe number of subjects shows QC-passed East Asian subjects used in each study
eOdds ratio indicates a value calculated in the final East Asian cohort, not including non-East Asian cohort(s)
fMAF was provided by gnomAD v3.1.2.
gMAF difference was calculated by subtracting MAF in East Asian from that in non-Finnish European
Fig. 2.
Effects and frequencies of the AD susceptible loci in East Asian population
Notably, none of the GWASs mentioned above identified the same loci, excluding the APOE region, even in the same country. One reason for these inconsistent results may be differences in the genetic background among East Asian populations. Although Japanese, Chinese, and South Koreans share genetic extensions, genetic clusters in each population are clearly distinct [32]. Even within the same country, there are several subpopulations with slightly different genetic backgrounds [33, 34]. Furthermore, there are concerns that because these GWAS are relatively smaller compared to the large GWAS in Caucasians, there may be insufficient statistical power. Thus, in the future, integrated analysis of multiple cohorts from multiple neighboring countries can help to resolve these limitations.
Rare variants
The advent of NGS has facilitated genetic analysis at the resolution of a single nucleotide, thereby shifting the focus from common variants to the identification of rare variants. Much attention has been paid to low-frequency functional variants involving amino acid alterations, because functional rare variants may be directly linked to disease pathogenesis due to their biological consequences. Thus, rare variants with functional relevance are likely to provide a better understanding of disease etiology than common variants with small effect size that are located in the noncoding regions and are the focus of GWAS. Indeed, many rare functional variants have been successfully identified in AD in recent years, which have shed new light on the pathogenesis of AD.
The first well-known rare variant for AD is the p.R47H variant (rs75932628) in TREM2, which was independently identified by two research groups in 2013 [35, 36]. Since then, multiple studies have attempted to validate its genetic association with AD, and a recent GWAS of nearly 100,000 individuals has estimated an odds ratio of 2.08 with a P value of 2.7 × 10−15 for this variant [37]. Although the allele frequency of p.R47H is as low as 0.8% [37], it confers a high risk for AD, which is comparable to that of APOE ε4. Crucially, such a large effect size is characteristic of functional rare variants, which is in contrast to common variants with a small effect size.
However, genetic studies in East Asian populations have been unable to replicate the significance of the p.R47H variant in TREM2, because it is rarely found in this population. To date, thousands of Chinese and Japanese have been screened for p.R47H variant, and only three Japanese carriers of this variant have been reported (Table 4) [38–42]. This observation is also true for the rare variant p.A673T (rs63750847) in APP, which was identified in Icelanders and was shown to have a strong protective effect against age-related cognitive decline as well as AD [43]. The p.A673T variant was observed in control subjects aged over 85 years at a frequency of 0.45%, which is higher than that seen in AD patients [43]. However, this variant has never been reported in East Asian populations (Table 3) [44, 45].
Table 4.
Rare variants associated with AD in East Asian populations
| Gene | dbSNP | Coding DNA | Protein | Minor allele frequency | Association study in East Asians | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| European (non-Finnish) | South Asian | East Asian | Ethnicity | P value | OR | 95% CI | ||||
| APP | rs63750847a | c.2017G > A | p.Ala673Thr | 0.0349% | 0.0000% | 0.0000% | Chinese | No carriers found | ||
| Chinese | No carriers found | |||||||||
| MLKL | rs763812068 | c.142 C > T | p.Gln48Ter | 0.0008% | 0.0000% | 0.1003% | Chinese | 0.006 | NA | NA |
| TREM2 | rs75932628a | c.140 G > A | p.Arg47His | 0.2466% | 0.2321% | 0.0000% | Japanese | 1.00 | 0.57 | 0.05–6.30 |
| Chinese | No carriers found | |||||||||
| Chinese | No carriers found | |||||||||
| rs201280312 | c.389 C > T | p.Ala130Val | 0.0000% | 0.0000% | 0.0109% | Chinese | 0.13 | NA | NA | |
| rs2234255 | c.469 C > T | p.His157Tyr | 0.0294% | 0.0719% | 0.1805% | Japanese | 0.53 | 1.73 | 0.49–6.13 | |
| Chinese | 0.005 | 11.01 | 1.38–88.05 | |||||||
| rs200820365 | c.547 A > T | p.Ser183Cys | 0.0000% | 0.0000% | 0.8525% | Chinese | 0.035 | 2.75 | 1.03–7.34 | |
| rs150277350 | c.574 G > A | p.Ala192Thr | 0.0080% | 0.0294% | 0.0401% | Chinese | 0.18 | NA | NA | |
| rs1391283629 | c.600 G > C | p.Trp200Cys | 0.0000% | 0.0000% | 0.0056% | Chinese | 1.00 | NA | NA | |
| rs2234256 | c.632 T > C | p.Leu211Pro | 0.1115% | 5.8400% | 0.1604% | Japanese | 0.30 | 0.71 | 0.39–1.28 | |
| PLD3 | rs145999145a | c.694 G > A | p.Val232Met | 0.5325% | 0.0851% | 0.0150% | Chinese | No carriers found | ||
| SHARPIN | rs572750141 | c.556 G > A | p.Gly186Arg | 0.0018% | 0.0065% | 0.0558% | Japanese | 0.000012 | 6.1 | 2.4–15.5 |
| rs77359862 | c.820 C > T | p.Arg274Trp | 0.0086% | 0.0818% | 3.7530% | Japanese | 0.0016 | 1.43 | 1.15–1.78 | |
| UNC5C | rs137875858a | c.2504 C > T | p.Thr835Met | 0.0496% | 0.0033% | 0.0000% | Chinese | No carriers found | ||
| NA | c.2508 C > G | p.Val836Val | NA | NA | NA | Chinese | 0.36 | NA | NA | |
| rs368284839 | c.2510 C > A | p.Thr837Lys | 0.0000% | 0.0000% | 0.1203% | Chinese | 0.13 | NA | NA | |
| rs779272234 | c.2527 A > G | p.Ser843Gly | 0.0008% | 0.0000% | 0.1203% | Chinese | 0.36 | NA | NA | |
| rs372767649 | c.2580 G > C | p.Gln860His | 0.0000% | 0.0294% | 0.1253% | Chinese | 0.017 | NA | NA | |
Minor allele frequencies were obtained from gnomAD v2.1.1
CI confidence interval, NA not available, OR odds ratio
arare variants proven in Caucasians
Nevertheless, several other rare variants that are significantly associated with AD have been reported in East Asians. For example, TREM2 p.H157Y (rs2234255) has been detected not only in Caucasians [35] but also in Chinese [40] and Japanese [24], and the significance of this variant has been confirmed in the Chinese population (Table 4) [40]. Moreover, two rare variants, p.G186R (rs572750141) and p.R274W (rs77359862), identified in the coding regions of SHARPIN, have been reported to be associated with late-onset AD in the Japanese (Table 3) [46, 47]. Similarly, the p.R274W variant in SHARPIN has been associated with brain atrophy in Korean patients with AD [48]. Thus, these two are examples of rare variants that are relatively frequent in East Asians (Table 4), but have yet to be verified in other ethnic groups.
Notably, these findings raise the notion that rare variants may exist in an ethnicity dependent manner, and that they seem to exhibit a mutually exclusive behavior, i.e., wherein one rare variant seen in an ethnic group may not be found in other ethnic groups. This is probably because not enough time has passed since these rare variants arose and they are yet to spread to other populations. Alternatively, rare variants might be subjected to a selection pressure that could be detrimental to human survival, making it harder for them to spread from one population to another. Hence, to explore the significance of rare variants, it would be advantageous to analyze their impact in a genetically homogeneous population. Nonetheless, further genetic research will uncover additional rare variants associated with AD among diverse populations, and such identification may pose difficulties in validating inter-racial reproducibility. It may not be surprising even if the significance of these rare variants is not replicated in another population, and it is possible that another rare variant(s) within the same gene may be found in ethnically divergent populations. Hence, it is important to evaluate pathogenicity of each of these rare variants and utilize gene-based approaches, while also taking into account other variants observed in the same gene. Crucially, due to their rarity, genetic analysis of thousands of samples will be required to confirm significant differences.
Future directions
During the last 20 years, numerous relevant susceptibility loci, genes and pathways associated with AD have been identified, and they have provided robust clues that have helped further our understanding of the complex pathogenesis of AD. It has also become apparent that genetic diversity among the various ethnic groups can affect disease risk, treatment efficacy, and safety. An important goal of genetic research in AD is the identification of medically actionable information that can help in the management of AD patients. Polygenic risk score, which is constructed as a weighted sum of allele counts, has been used to predict the development of AD [49]. Recent work suggests that genetic contributions to AD may be oligogenic, i.e., influenced by a limited set of common genetic variants [50]. Additional research is needed to better understand the genetic mechanisms underlying AD pathogenesis among different ethnic groups, and this could be achieved by facilitating data sharing and international collaboration. These efforts will lead to testable working hypothesis for the development of therapeutics, which would ultimately accelerate the use of precision medicine in the management of AD.
Acknowledgements
The authors are grateful to Drs. Koichi Ozaki and Shumpei Niida (National Center for Gerontology and Geriatrics), Dr. Akihiro Nakaya (University of Tokyo), Profs. Jungsoo Gim and Kun Ho Lee (Chosun University), and Prof. Jianping Jia (Capital Medical University) for their collaborative support. The authors would like to thank all lab members at Department of Molecular Genetics, Brain Research Institute, Niigata University and at Department of Genome Informatics, Graduate School of Medicine, Osaka University. This work was supported in part by AMED JP21dk0207045 and JP22dk0207060 to AM, MK, and TI, AMED JP22wm0525019 to MK and TI, and MEXT KAKENHI Grant Numbers 21K07271 and 21H03537 to AM.
Author contributions
TI designed the study. AM, MK, NH and TI wrote the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Akinori Miyashita, Masataka Kikuchi, Norikazu Hara.
References
- 1.Sims R, Hill M, Williams J. The multiplex model of the genetics of Alzheimer’s disease. Nat Neurosci. 2020;23:311–22. doi: 10.1038/s41593-020-0599-5. [DOI] [PubMed] [Google Scholar]
- 2.Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–74. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 3.Ridge PG, Mukherjee S, Crane PK, Kauwe JS. Alzheimer’s Disease Genetics Consortium. Alzheimer’s disease: analyzing the missing heritability. PLoS One. 2013;8:e79771. doi: 10.1371/journal.pone.0079771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellenguez C, Grenier-Boley B, Lambert JC. Genetics of Alzheimer’s disease: where we are, and where we are going. Curr Opin Neurobiol. 2020;61:40–48. doi: 10.1016/j.conb.2019.11.024. [DOI] [PubMed] [Google Scholar]
- 5.Belloy ME, Napolioni V, Greicius MD. A quarter century of APOE and Alzheimer’s disease: progress to date and the path forward. Neuron. 2019;101:820–38. doi: 10.1016/j.neuron.2019.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuyvers E, Sleegers K. Genetic variations underlying Alzheimer’s disease: evidence from genome-wide association studies and beyond. Lancet Neurol. 2016;15:857–68. doi: 10.1016/S1474-4422(16)00127-7. [DOI] [PubMed] [Google Scholar]
- 7.Wightman DP, Jansen IE, Savage JE, Shadrin AA, Bahrami S, Holland D, et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer's disease. Nat Genet. 2021;53:1276–82. doi: 10.1038/s41588-021-00921-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoogmartens J, Cacace R, Van, Broeckhoven C. Insight into the genetic etiology of Alzheimer’s disease: A comprehensive review of the role of rare variants. Alzheimers Dement (Amst) 2021;13:e12155. doi: 10.1002/dad2.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 10.Reiman EM, Arboleda-Velasquez JF, Quiroz YT, Huentelman MJ, Beach TG, Caselli RJ, et al. Exceptionally low likelihood of Alzheimer's dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat Commun. 2020;11:667. doi: 10.1038/s41467-019-14279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–56. doi: 10.1001/jama.1997.03550160069041. [DOI] [PubMed] [Google Scholar]
- 12.Rubinsztein DC, Easton DF. Apolipoprotein E genetic variation and Alzheimer’s disease. a meta-analysis. Dement Geriatr Cogn Disord. 1999;10:199–209. doi: 10.1159/000017120. [DOI] [PubMed] [Google Scholar]
- 13.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 14.Liu M, Bian C, Zhang J, Wen F. Apolipoprotein E gene polymorphism and Alzheimer’s disease in Chinese population: a meta-analysis. Sci Rep. 2014;4:4383. doi: 10.1038/srep04383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agarwal R, Tripathi CB. Association of apolipoprotein E genetic variation in Alzheimer’s disease in Indian population: a meta-analysis. Am J Alzheimers Dis Other Demen. 2014;29:575–82. doi: 10.1177/1533317514531443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abyadeh M, Djafarian K, Heydarinejad F, Alizadeh S, Shab-Bidar S. Association between apolipoprotein E gene polymorphism and alzheimer’s disease in an Iranian population: a meta-analysis. J Mol Neurosci. 2019;69:557–62. doi: 10.1007/s12031-019-01381-1. [DOI] [PubMed] [Google Scholar]
- 17.Choi KY, Lee JJ, Gunasekaran TI, Kang S, Lee W, Jeong J, et al. APOE Promoter Polymorphism-219T/G is an Effect Modifier of the Influence of APOE ε4 on Alzheimer's Disease Risk in a Multiracial Sample. J Clin Med. 2019;8:1236. doi: 10.3390/jcm8081236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khani M, Gibbons E, Bras J, Guerreiro R. Challenge accepted: uncovering the role of rare genetic variants in Alzheimer’s disease. Mol Neurodegener. 2022;17:3. doi: 10.1186/s13024-021-00505-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wardell MR, Brennan SO, Janus ED, Fraser R, Carrell RW. Apolipoprotein E2-Christchurch (136 Arg–Ser). New variant of human apolipoprotein E in a patient with type III hyperlipoproteinemia. J Clin Invest. 1987;80:483–90. doi: 10.1172/JCI113096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arboleda-Velasquez JF, Lopera F, O’Hare M, Delgado-Tirado S, Marino C, Chmielewska N, et al. Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote: a case report. Nat Med. 2019;25:1680–3. doi: 10.1038/s41591-019-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zalocusky KA, Nelson MR, Huang Y. An Alzheimer’s-disease-protective APOE mutation. Nat Med. 2019;25:1648–9. doi: 10.1038/s41591-019-0634-9. [DOI] [PubMed] [Google Scholar]
- 22.Medway CW, Abdul-Hay S, Mims T, Ma L, Bisceglio G, Zou F, et al. ApoE variant p.V236E is associated with markedly reduced risk of Alzheimer’s disease. Mol Neurodegener. 2014;9:11. doi: 10.1186/1750-1326-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu CC, Murray ME, Li X, Zhao N, Wang N, Heckman MG, et al. APOE3-Jacksonville (V236E) variant reduces self-aggregation and risk of dementia. Sci Transl Med. 2021;13:eabc9375. doi: 10.1126/scitranslmed.abc9375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, et al. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS One. 2013;8:e58618. doi: 10.1371/journal.pone.0058618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reitz C, Rogaeva E, Lee JH, Tokuhiro S, Bettens K, Zou F, et al. Association of SORL1 gene variants with Alzheimer’s disease: A meta-analysis. Arch Neurol. 2011;68:99–106. doi: 10.1001/archneurol.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirano A, Ohara T, Takahashi A, Aoki M, Fuyuno Y, et al. A genome-wide association study of late-onset Alzheimer’s disease in a Japanese population. Psychiatr Genet. 2015;25:139–46. doi: 10.1097/YPG.0000000000000090. [DOI] [PubMed] [Google Scholar]
- 27.Shigemizu D, Mitsumori R, Akiyama S, Miyashita A, Morizono T, Higaki S, et al. Ethnic and trans-ethnic genome-wide association studies identify new loci influencing Japanese Alzheimer’s disease risk. Transl Psychiatry. 2021;11:151. doi: 10.1038/s41398-021-01272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou X, Chen Y, Mok KY, Zhao Q, Chen K, Chen Y, et al. Identification of genetic risk factors in the Chinese population implicates a role of immune system in Alzheimer’s disease pathogenesis. Proc Natl Acad Sci U S A. 2018;115:1697–1706. doi: 10.1073/pnas.1715554115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia L, Li F, Wei C, Zhu M, Qu Q, Qin W, et al. Prediction of Alzheimer’s disease using multi-variants from a Chinese genome-wide association study. Brain. 2021;144:924–37. doi: 10.1093/brain/awaa364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park JH, Park I, Youm EM, Lee S, Park JH, Lee J, et al. Novel Alzheimer’s disease risk variants identified based on whole-genome sequencing of APOE e4 carriers. Transl Psychiatry. 2021;11:296. doi: 10.1038/s41398-021-01412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang S, Gim J, Lee J, Gunasekaran TI, Choi KY, Lee JJ, et al. Potential novel genes for late-onset Alzheimer’s disease in East-Asian descent identified by APOE-stratified genome-wide association study. J Alzheimers Dis. 2021;82:1451–60. doi: 10.3233/JAD-210145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Lu D, Chung YJ, Xu S. Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations. Hereditas. 2018;155:19. doi: 10.1186/s41065-018-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Zheng H, Bei JX, Sun L, Jia WH, Li T, et al. Genetic structure of the Han Chinese population revealed by genome-wide SNP variation. Am J Hum Genet. 2009;85:775–85. doi: 10.1016/j.ajhg.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeuchi F, Katsuya T,Kimura R, Nabika T, Isomura M, Ohkubo T, et al. The fine-scale genetic structure and evolution of the Japanese population. PLoS One. 2017;12:e0185487. doi: 10.1371/journal.pone.0185487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, et al. TREM2 variants in Alzheimer’s disease. N. Engl J Med. 2013;368:117–27. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl J Med. 2013;368:107–16. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51:414. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu J-T, Jiang T, Wang Y-L, Wang H-F, Zhang W, Hu N, et al. Triggering receptor expressed on myeloid cells 2 variant is rare in late-onset Alzheimer’s disease in Han Chinese individuals. Neurobiol Aging. 2014;35:937.e1–3. doi: 10.1016/j.neurobiolaging.2013.10.075. [DOI] [PubMed] [Google Scholar]
- 39.Jiao B, Liu X, Tang B, Hou L, Zhou L, Zhang F, et al. Investigation of TREM2, PLD3, and UNC5C variants in patients with Alzheimer’s disease from mainland China. Neurobiol Aging. 2014;35:2422.e9–2422.e11. doi: 10.1016/j.neurobiolaging.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 40.Jiang T, Tan L, Chen Q, Tan M-S, Zhou J-S, Zhu X-C, et al. A rare coding variant in TREM2 increases risk for Alzheimer’s disease in Han Chinese. Neurobiol Aging. 2016;42:e1–3. doi: 10.1016/j.neurobiolaging.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 41.Bonham LW, Sirkis DW, Fan J, Aparicio RE, Tse M, Ramos EM, et al. Identification of a rare coding variant in TREM2 in a Chinese individual with Alzheimer’s disease. Neurocase. 2017;23:65–9. doi: 10.1080/13554794.2017.1294182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyashita A, Wen Y, Kitamura N, Matsubara E, Kawarabayashi T, Shoji M, et al. Lack of genetic association between TREM2 and late-onset Alzheimer’s disease in a Japanese population. J Alzheimers Dis. 2014;41:1031–8. doi: 10.3233/JAD-140225. [DOI] [PubMed] [Google Scholar]
- 43.Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488:96–9. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 44.Ting SKS, Chong M-S, Kandiah N, Hameed S, Tan L, Au W-L, et al. Absence of A673T amyloid-β precursor protein variant in Alzheimer’s disease and other neurological diseases. Neurobiol Aging. 2013;34:2441.e7–8. doi: 10.1016/j.neurobiolaging.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 45.Liu YW, He YH, Zhang YX, Cai WW, Yang LQ, Xu LY, et al. Absence of A673T variant in APP gene indicates an alternative protective mechanism contributing to longevity in Chinese individuals. Neurobiol Aging. 2014;35:935.e11–935.e12. doi: 10.1016/j.neurobiolaging.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 46.Asanomi Y, Shigemizu D, Miyashita A, Mitsumori R, Mori T, Hara N, et al. A rare functional variant of SHARPIN attenuates the inflammatory response and associates with increased risk of late-onset Alzheimer’s disease. Mol Med. 2019;25:20. doi: 10.1186/s10020-019-0090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asanomi Y, Shigemizu D, Akiyama S, Miyashita A, Mitsumori R, Hara N, et al. A functional variant of SHARPIN confers increased risk of late-onset Alzheimer’s disease. J Hum Genet. 2022;67:203–8. doi: 10.1038/s10038-021-00987-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park JY, Lee D, Lee JJ, Gim J, Gunasekaran TI, Choi KY, et al. A missense variant in SHARPIN mediates Alzheimer’s disease-specific brain damages. Transl Psychiatry. 2021;11:1–9. doi: 10.1038/s41398-021-01680-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker E, Escott-Price V. Polygenic risk scores in Alzheimer’s disease: current applications and future directions. Front Digit Health. 2020;2:14. doi: 10.3389/fdgth.2020.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q, Sidorenko J, Couvy-Duchesne B, Marioni RE, Wright MJ, Goate AM, et al. Risk prediction of late-onset Alzheimer's disease implies an oligogenic architecture. Nat Commun. 2020;11:4799. doi: 10.1038/s41467-020-18534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]