Abstract
Parkinson’s disease (PD) is a neurodegenerative disorder primarily characterized by motor dysfunction. Aging is the greatest risk factor for developing PD. Recent molecular genetic studies have revealed that genetic factors, in addition to aging and environmental factors, play an important role in the development of the disorder. Studies of familial PD have identified approximately 20 different causative genes. PRKN is the most frequently detected causative gene in Japan. The PRKN gene is located at a common fragile site, and both copy number variants as well as single nucleotide variants are frequently detected. The location and variety of variant types makes an accurate genetic diagnosis difficult with conventional genetic testing. In sporadic PD, genome-wide association studies have revealed more than 200 genes that are potential drivers for the development of PD. Many of these studies have been conducted in Caucasian populations alone, which has limited the identification of all genetic risk factors for sporadic PD, particularly as genetic backgrounds vary widely by race. The Global Parkinson’s Genetics Program is a global undertaking meant to address the issue of regional differences in genetic studies of PD.
Subject terms: Parkinson's disease, Genome-wide association studies, Medical genomics
Introduction
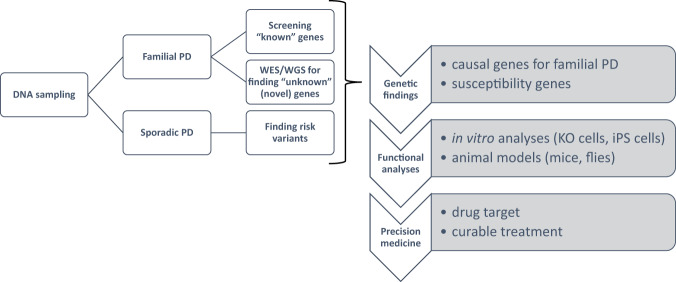
Parkinson’s disease (PD) is a progressive neurodegenerative disease whose main symptoms are motor dysfunctions, such as tremor, rigidity, bradykinesia, and postural instability. Selective degeneration of dopaminergic neurons in the substantia nigra of the midbrain underlies these symptoms. PD patients often have comorbid non-motor symptoms that include autonomic dysfunctions, such as constipation and orthostatic hypotension, as well as psychiatric symptoms, such as anxiety and depression. Thus, PD is not a disease of the central nervous system alone, but rather a systemic disease. Because the prevalence of PD increases with age, it is clear that aging is a risk factor for its development. Environmental factors are also thought to be important, as exposure to pesticides and other chemicals can cause PD symptoms. Advances in genetic research methods have provided insights into how genetic backgrounds contribute to the development of PD. Accordingly, PD is a complex genetic disease that is caused by a combination of aging, environmental factors, and genetic factors. There are two major ways to identify genetic factors in PD (Fig. 1). One is to investigate rare Mendelian forms of PD and identify the causative genes. The other is to identify risk variants from genetic statistical analyses of large groups of subjects. While there are advantages and disadvantages in each approach, both are necessary to provide a more complete picture of this disease. Genetic studies have identified over 200 PD-related genes [1]. However, regional differences, associated with variations in ethno-social factors, have complicated molecular genetic studies of PD. Here, we discuss the current status and challenges of molecular genetics research on PD.
Fig. 1.
Strategies to identify genetic factors in PD and applications of the findings. The search for genes associated with PD relies on the molecular genetic characteristics of PD patients and the classification of samples into familial PD and sporadic PD groups. After PD-associated genes are discovered, cellular and animal PD models are used to reveal pathophysiological functions and for precision medicine applications
Molecular genetics of Mendelian forms of PD
It has been a quarter of a century since the alpha-synuclein gene (SNCA) was reported as the causative gene for the autosomal dominant form of PD [2]. In the early days of Mendelian PD research, the prevailing approach used to discover disease-related genes and variants included identifying loci from positional cloning of large families, using artificial chromosome libraries, such as yeast artificial chromosomes (YACs) and bacterial artificial chromosomes (BACs) to detect the genes and identify patient-specific variants by sequence analysis. This approach led to the discovery of SNCA and PRKN [2, 3]. With the completion of the Human Genome Project and the advent of next-generation sequencers, the throughput has been dramatically improved, and the resulting gene maps and sequences have become more readily available [4]. Together, these molecular genetic Mendelian PD studies have identified 24 genes or loci that have been registered in the Online Mendelian Inheritance in Man (OMIM) database as being involved in the development of PD (Table 1).
Table 1.
PD-related genes registered in OMIM
| Loci | Chromosomal position | Gene |
|---|---|---|
| PARK1/4 | 4q21.3-q22 | SNCA |
| PARK2 | 6q25.2-27 | PRKN |
| PARK3 | 2p13 | unknown |
| PARK5 | 4p13 | UCH-L1 |
| PARK6 | 1p36.12 | PINK1 |
| PARK7 | 1p36.23 | DJ-1 |
| PARK8 | 12q12 | LRRK2 |
| PARK9 | 1p36 | ATP13A2 |
| PARK10 | 1p32 | unknown |
| PARK11 | 2q36-q37 | GIGYF2 |
| PARK12 | Xq21-q25 | unknown |
| PARK13 | 2p13.1 | HTRA2 |
| PARK14 | 22q13.1 | PLA2G6 |
| PARK15 | 22q11.2-qter | FBXO7 |
| PARK16 | 1q32 | unknown |
| PARK17 | 16q12 | VPS35 |
| PARK18 | 3q27-qter | EIF4G1 |
| PARK19 | 1p31.3 | DNAJC6 |
| PARK20 | 21q22.11 | SYNJ1 |
| PARK21 | 3q22.1 | DNAJC13 |
| PARK22 | 7p11.2 | CHCHD2 |
| PARK23 | 15q22.2 | VPS13C |
| PARK24 | 10q22.1 | PSAP |
PRKN
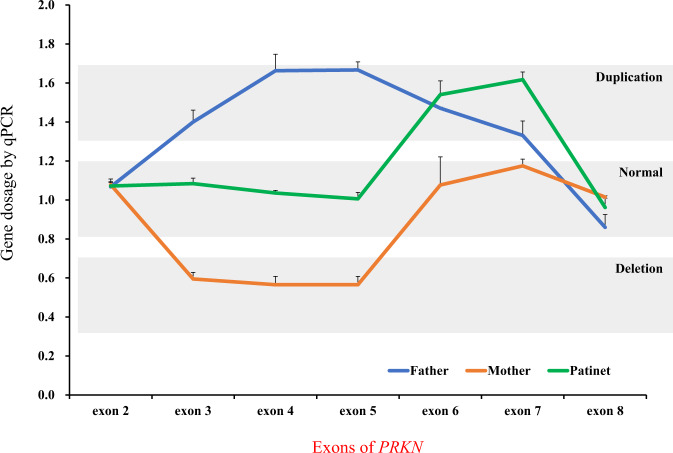
The most frequent causative gene of PD in Japan is the PRKN gene, which is the causative gene of autosomal recessive juvenile PD, as reported by Kitada et al. in 1998 [3]. We recently published the results of a study of the PRKN gene in over 2000 cases, and identified biallelic variants in PRKN that are observed in 8.1% (98/1204) of familial PD and 5.8% (65/1118) of sporadic PD cases [5]. It is important to note that this population included many patients who were clinically diagnosed as likely to have PRKN variants and were actively selected for genetic analysis. Therefore, the sampling bias is likely to have been strong, and the frequency of PRKN variants is estimated to be a little lower than that observed in the study. We also found PD patients with a putative pathogenic monoallelic rare variant in the PRKN gene, with a frequency of 2.5% (57/2322). A comparison of clinical symptoms between the biallelic and monoallelic variants showed that the age at onset was 29.6 ± 9.8 years for the biallelic variants and 45.2 ± 15.9 years for the monoallelic variant, which indicates an earlier age of onset for the biallelic variants [5]. Additionally, more cases of Hoehn and Yahr stage IV or V were observed in the biallelic variants in patients who had the disease for at least 15 years [5]. The PRKN gene is the causative gene of autosomal recessive juvenile PD, and therefore, a person with a monoallelic variant is presumed to be an unaffected carrier. However, the frequency of the PRKN monoallelic variant is slightly higher in Japanese PD patients than the Leucine rich-repeat kinase 2 (LRRK2) variant, a cause of autosomal dominant PD (discussed further below). Thus, the PRKN monoallelic variant may have some influence on the development of PD. Because the PRKN gene is located at a common fragile site, it is prone to site-specific gaps and breaks due to chromosomal fragility [6]. Indeed, the PRKN gene is a cluster of deletions and duplications in exons 2 through 7. Quantitative PCR (qPCR) and Multiplex Ligation-dependent Probe Amplification (MLPA) are simple and widely used tests for copy number variations such as deletions and duplications. However, these methods only target a part of a gene, and the results therefore need to be interpreted carefully. For example, we reported a pseudo-heterozygous variant in a PD patient whose father had a duplication in exons 3–7 and whose mother had a deletion in exons 3–5. The patient inherited the mutant allele from both parents and developed juvenile PD, but was incorrectly identified as having a heterozygous duplication in exons 6–7 due to the gene dosages of the parental mutant alleles canceling each other out in qPCR testing (Fig. 2) [7]. Thus, genetic testing of only the patient may miss the biallelic variants, and therefore, it is advisable to test the patient as well as close relatives, including those who have not yet developed the disease. However, some variants, such as variants in the promoter region and small inversions involving only a partial region of the PRKN gene, may be difficult to identify even with trio analysis. When small inversions are present, the abnormality cannot be detected by sequence analysis using the Sanger method or by copy number variation analysis using the qPCR or MLPA methods. Therefore, even if one genetic test is negative, diligent testing should be continued if there is clinical suspicion of gene mutation [8]. The long-read sequencing method is a useful genetic analysis method that should help identify the underlying genetic variants.
Fig. 2.
Pseudo-heterozygotes in the PRKN gene. Copy number variations tested by qPCR showed that the PD patient (green) had a heterozygous duplication in exons 6–7. The PD patient inherited a duplication of exons 3–7 from the father (blue) and a deletion of exons 3–5 from the mother (orange), resulting in compound heterozygous variants in the PRKN gene
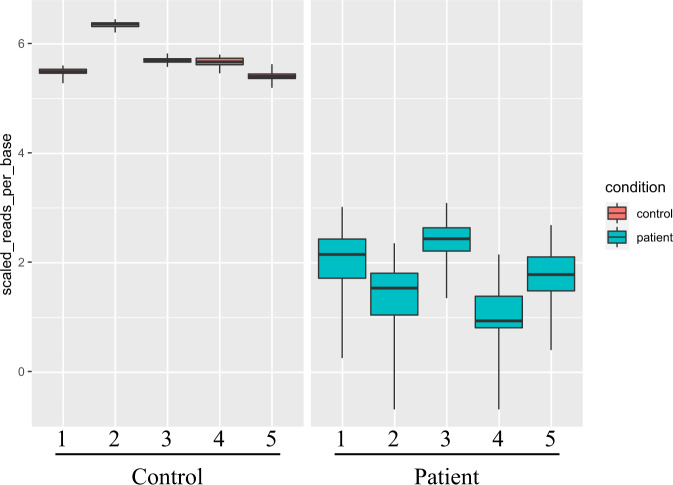
For a more complete analysis, it is necessary to consider genes other than PRKN that are involved in the pathogenesis of the disease. We have conducted RNAseq analysis of patients with both a monoallelic variant in the PRKN gene and the juvenile PD phenotype, and found patient-specific gene expression changes (Fig. 3). Although this downregulated gene has a different chromosomal location from the PRKN gene, it has been reported to be expressed in the nervous system and may be functionally related to PRKN. Thus, for autosomal recessive diseases with monoallelic variants in genes such as PRKN, other loci should be considered. These strategies include (1) seeking deletions, duplications, and often-missed biallelic variants, such as those in transcriptional regulatory regions, (2) seeking biallelic or polygenic variants, (3) investigating patient susceptibility to the disease, and (4) considering other unrelated causes. Therefore, it is necessary to make careful assessments based on family history and clinical considerations in the diagnosis of PD.
Fig. 3.
RNAseq analysis of autosomal recessive juvenile parkinsonism (ARJP) patients with PRKN single heterozygous variants. RNAseq analysis of five ARJP patients (right half) and five controls (left half) shows one example of a gene whose expression is prominently and significantly downregulated only in ARJP patients
PTEN-induced kinase 1 (PINK1)
The second most frequent gene after PRKN as a causative gene for autosomal recessive PD is PINK1. PINK1 is a PD gene discovered in 2004 from a genetic analysis of families in Sicily, Italy [9]. Subsequent follow-up studies revealed many PD-related variants. PINK1 is a mitochondrial kinase. Single nucleotide variants (SNVs) in PINK1 are more common than copy number variants (CNVs) in PRKN. We performed genetic analysis of PINK1 in 1700 PD patients and identified PINK1 variants in 1.8% of young-onset familial PD and 0.009% of young-onset sporadic PD [10]. PINK1 and parkin are known to cooperate and are involved in mitochondrial quality control [11]. We found a total of four PD patients with digenic PRKN-PINK1 variants [10, 12]. These digenic variants seem to have younger age of onset than biallelic variants in PINK1 or PRKN [12]. However, the small number of cases makes it unclear whether these differences are significant.
Leucine rich-repeat kinase 2 (LRRK2)
The most frequent PD causative gene in Europe and the United States is LRRK2, which was first reported in 2004 as the causative gene of the autosomal forms of PD [13, 14]. Originally, the LRRK2 gene was mapped to chromosome 12 in a study of Japanese families with an autosomal form of PD [15]. The causative locus, PARK8, was mapped to chromosome 12 and linked to LRRK2. Although this gene is historically associated with Japan, the frequency of the LRRK2 gene in PD in Japan is very low compared with Caucasians [16]. Conversely, the LRRK2 gene p.G2019S variant is highly prevalent in Europe, the United States and Arab countries, including those in North Africa [17]. We analyzed approximately 1400 PD patients and estimated that the frequency of LRRK2 variants in Japan is 1.7% in familial PD, 0.3% in sporadic PD, and 1.0% in all PD patients [16]. Most PD patients with LRRK2 variants had families in which the mode of inheritance was thought to be autosomal dominant. However, some PD patients appeared to have sporadic PD with no known cases in the family, indicating incomplete penetrance. Clinical features such as tremors (78.3%), postural instability (73.9%), gait with small steps (55.6%), constipation (40.0%) and olfactory disturbances (41.7%) were also frequently observed, and there was no significant difference in clinical symptoms among pathogenic variants [16].
Other genes in familial PD
In 2011, Vacuolar protein sorter-35 (VPS35) was reported as the first causative gene for autosomal dominant PD based on whole-exome analysis using next-generation sequencing [18]. Since the report of VPS35, one or two genes a year have been reported as causative PD genes by studies using the same methodology. In a report from Japan, coiled-coil-helix-coiled-coil-helix domain containing 2 (CHCHD2) and prosaposin (PSAP) were reported as novel genes for familial PD, though they have a very low frequency in Mendelian PD [19, 20]. CHCHD2 is the first PD-causing gene involved in the mitochondrial respiratory chain complex, and PSAP is also involved in a lysosomal disease, similar to glucocerebrosidase (GBA1) [19, 20]. Despite their very low frequencies, these genes are important for understanding the pathogenesis of PD and as potential therapeutic targets. In particular, age-dependent decline in motor function and loss of nigrostriatal dopaminergic neurons have been observed in mouse models of PSAP, suggesting that PSAP may be useful as an animal model of PD [20, 21]. Other familial PD-causing genes include DJ-1, ATP13A2, GIGYF2, HTRA2, PLA2G6, FBXO7, EIF4G1, DNAJC6, SYNJ1, and DNAJC13, all of which are infrequent in Japanese PD. However, the functions of these gene products, including intracellular trafficking, oxidative stress, mitochondria, phospholipid membranes and ubiquitin-proteasome system, are all predicted to be involved in the pathogenesis of PD [22–31].
Difficulties in discovering new PD-causing genes
One of the problems with Mendelian PD research is the lack of research targets. It has been reported that the percentage of Mendelian-inherited diseases for which the cause could be identified by whole-exome analysis is about 20% [32]. Because PD is a complex genetic disease with a late onset, the rate of cause-identification is expected to be even lower. One approach to overcome this limitation is to collect as many subjects as possible in the same family with Mendelian PD. In the case of manifest PD, collecting more genetic samples over multiple generations yields a higher probability of identifying the cause of the disease, but in many cases, the parents or grandparents with PD had died by the time the proband developed the disease. For example, in the PARK8 family mentioned above, the causative gene locus was successfully identified after more than 20 years of intra-family research and DNA sampling, including from those who did not develop the disease [33–35]. Even though genetic analysis technology has evolved, and high-throughput techniques have improved dramatically, new discoveries will likely remain difficult to make. For example, when whole-exome sequencing is performed using small PD families, hundreds of candidate causative variants will be identified; however, with few exceptions, there is only one causal variant per family, regardless of its size. Investigators must therefore implement various techniques to narrow down the number of candidate variants from hundreds to one. In familial PD studies, it is expected that for at least 80% of the families, a single candidate variant and the causative gene will remain unidentified.
PD risk genes and variants
Genome-wide association studies (GWAS)
GWAS have been conducted to identify genetic factors that contribute to the pathogenesis of sporadic PD. To identify these factors, GWAS and meta-analysis on GWAS (Meta-GWAS) have been employed. In GWAS, microarray technology is used to genotype relatively frequent single nucleotide polymorphisms (SNPs). SNPs with large differences in frequency between the patient and control groups can be identified. Genes located near hit-SNPs, which are significantly more or less frequent in the patient group, are presumed to be more likely to influence the development of sporadic PD.
As a result of multiple GWAS, several important genes have been identified as being risk factors for PD. Among them, SNCA and LRRK2 were found to be causative genes for the Mendelian forms of PD, but GWAS revealed that they are also involved in the development of sporadic PD [36, 37]. Two GWAS studies in Europeans and Japanese populations have shown that LRRK2 is a common risk factor for PD regardless of race. However, Europeans share a unique variant, p.G2019S, while p.R1628P and p.G2385R are variants unique to Asians [38, 39]. According to the GWAS catalog, about 200 genes related to PD have been reported so far [1]. Furthermore, a recent meta-analysis of GWAS identified 90 independent variants in 78 genomic regions associated with PD; however, how these variants affect pathogenesis remains largely unknown [40]. Therefore, it is likely that more genes will be reported to be involved in PD pathogenesis in future Meta-GWAS. Clarifying how these many genes may be related to or affect PD onset as well as other clinical manifestations may require new approaches that combine artificial intelligence, deep learning, gene expression data, metabolomics, and other technologies. The findings from these combined approaches are expected to contribute to the prediction of PD onset and prognosis as well as to the implementation of targeted medicine strategies and therapeutics.
GBA1
GBA1 is the causative gene for Gaucher disease (GD), a lysosomal disorder [41]. GD is a recessively inherited disease caused by variants in the GBA1 gene. Notably, in GD families, many GD-naive GBA1 variant carriers have family members who develop PD [42, 43]. This suggests that a monoallelic GBA1 variant may be a risk factor for developing PD, and in fact, GBA1 was found to be a common and frequent risk factor for the development of PD in different populations, including Japan [44, 45]. Variants of GBA1 known to be associated with PD development include p.E326K, p.T369M, p.N370S and p.L444P, which decrease the activity of glucocerebrosidase, the enzyme encoded by GBA1, and consequently reduce the ability to degrade alpha-synuclein in lysosomes [46]. Variants of GBA1 have different frequencies in different races. For example, p.E326K has a frequency of 1–5% in the general European population, but is very rare in Asia.
Polygenic risk score (PRS)
PD is thought to be caused by a complex interaction of multiple genetic risk factors with environmental factors and aging. The identification of novel risk genes and variants for PD by GWAS is a promising approach for discovering a small number of common causative genes or risk variants. The PRS is calculated for each individual based on the weighted sum of the risk variants, assuming that the patient has many risk variants. PRS has been shown to correlate with the actual risk of developing the disease, and by examining the distribution of scores within a population, it is possible to identify individuals at particularly high risk for the disease. Foo et al. identified 11 PD-risk SNVs in a meta-GWAS of Asian samples [47]. Two of these (SV2C and WBSCR17) were novel PD risk genes. The PRS was calculated using these SNVs, and it was shown that the age of onset decreased with each additional copy of the risk allele [47]. Similar PRS calculations using the previously reported 90 risk variants in a European sample showed no significant difference in risk prediction in spite of the 8-fold greater number of SNVs [47]. Thus, these findings suggest that the PRS calculation based on GWAS for each race is significant for disease prediction, and that the weights of individual risk SNVs differ by race.
Efforts to address regional and racial differences in PD
Genetic regional and racial differences in PD are observed for many genes. For example, the MAPT gene, encoding the microtubule-associated protein tau, was detected as a risk factor for PD in a GWAS of a Caucasian population, but not in a GWAS of a Japanese population [36, 37]. However, several Japanese families have been affected by variants in MAPT [48, 49]. Most molecular genetic studies of disease have been conducted primarily in Caucasians for various geopolitical, cultural, linguistic, and economic reasons [50]. However, as the examples of GBA1 and MAPT show, the lack of molecular genetic studies in multiracial populations is an important issue that must be resolved in future PD research.
The Global Parkinson’s Genetics Program (GP2) is a worldwide international consortium established under the auspices of Aligning Science Across Parkinson’s (ASAP) to actively promote research in regions where genomic research has not been actively conducted before and where English is not the native language [51]. For this reason, the GP2 web page (https://gp2.org) is available in Arabic, Chinese, French, German, Japanese and Spanish, in addition to English. GP2 has hubs in various locations, such as the East Asian Parkinson Disease Genomics Consortium (EAPDGC) in East Asia, which includes Japan, the Genomics Consortium Africa (IPDGC Africa) in Africa, and the Latin American Research Consortium on the Genetics of PD (LARGE-PD) [52–54]. Each hub collects thousands of patient and control samples to determine racial genetic background, polygenic risk scores, and other metrics. GP2 makes its data and analysis scripts available to the public and runs educational programs on how to use them to perform various analyses in the cloud. Through such open science, GP2 is expanding its reach beyond Caucasians to include both patients who have not previously been included in studies and researchers who have not previously had the opportunity to conduct large-scale genomic analysis studies.
Conclusion
In this review, we described the current status of molecular genetics research on PD, its challenges, and the efforts to address them. Studying the molecular genetics of PD is a crucial first step in understanding the disease, because it advances our knowledge of the relationship between phenotype and genotype. Whether by revealing the full etiopathogenesis of the disease, uncovering novel pathogenetic mechanisms, or leading to other exciting findings, future molecular genetic studies are likely to foster major advances in our understanding of this debilitating neurodegenerative disorder in the near future.
Acknowledgements
We thank Barry Patel, PhD, from Edanz (https://jp.edanz.com/ac), for editing a draft of this manuscript.
Author contributions
Writing original draft: MF. Review and revise manuscript: MF, KN, YL, NH.
Funding
This work was supported by JSPS KAKENHI [Grant Numbers; 19K08003 to MF, 21K07283 to YL, and 18H04043 and 21H04820 to NH]. This study was in part supported by the following: High Technology Research Center Grant from the Ministry of Education, Culture, Sports, Science and Technology, Japan; Subsidies for Current Expenditures to Private Institutions of Higher Education from the Promotion and Mutual Aid Corporation for Private Schools of Japan; and GAPFREE from AMED (21ak0101125h0002). This work was performed, in part, at the Intractable Disease Research Center, Juntendo University Graduate School of Medicine.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D12.. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–7. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 3.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 4.International Human Genome Sequencing C. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 5.Yoshino H, Li Y, Nishioka K, Daida K, Hayashida A, Ishiguro Y, et al. Genotype-phenotype correlation of Parkinson’s disease with PRKN variants. Neurobiol Aging. 2022;114:117–28. doi: 10.1016/j.neurobiolaging.2021.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012;28:22–32. doi: 10.1016/j.tig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Funayama M, Yoshino H, Li Y, Kusaka H, Tomiyama H, Hattori N. Pseudo-heterozygous rearrangement mutation of parkin. Mov Disord. 2012;27:552–5. doi: 10.1002/mds.24906. [DOI] [PubMed] [Google Scholar]
- 8.Rajagopalan R, Gilbert MA, McEldrew DA, Nassur JA, Loomes KM, Piccoli DA, et al. Genome sequencing increases diagnostic yield in clinically diagnosed Alagille syndrome patients with previously negative test results. Genet Med. 2021;23:323–30. doi: 10.1038/s41436-020-00989-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–60. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 10.Hayashida A, Li Y, Yoshino H, Daida K, Ikeda A, Ogaki K, et al. The identified clinical features of Parkinson’s disease in homo-, heterozygous and digenic variants of PINK1. Neurobiol Aging. 2021;97:146.e1–46.e13. doi: 10.1016/j.neurobiolaging.2020.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–21. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funayama M, Li Y, Tsoi TH, Lam CW, Ohi T, Yazawa S, et al. Familial Parkinsonism with digenic parkin and PINK1 mutations. Mov Disord. 2008;23:1461–5. doi: 10.1002/mds.22143. [DOI] [PubMed] [Google Scholar]
- 13.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 14.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–7. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Funayama M, Hasegawa K, Kowa H, Saito M, Tsuji S, Obata F. A new locus for Parkinson’s disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann Neurol. 2002;51:296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Ikeda A, Yoshino H, Oyama G, Kitani M, Daida K, et al. Clinical characterization of patients with leucine-rich repeat kinase 2 genetic variants in Japan. J Hum Genet. 2020;65:771–81. doi: 10.1038/s10038-020-0772-4. [DOI] [PubMed] [Google Scholar]
- 17.Correia Guedes L, Ferreira JJ, Rosa MM, Coelho M, Bonifati V, Sampaio C. Worldwide frequency of G2019S LRRK2 mutation in Parkinson’s disease: a systematic review. Parkinsonism Relat Disord. 2010;16:237–42. doi: 10.1016/j.parkreldis.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Vilarino-Guell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, et al. VPS35 mutations in Parkinson disease. Am J Hum Genet. 2011;89:162–7. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funayama M, Ohe K, Amo T, Furuya N, Yamaguchi J, Saiki S, et al. CHCHD2 mutations in autosomal dominant late-onset Parkinson’s disease: a genome-wide linkage and sequencing study. Lancet Neurol. 2015;14:274–82. doi: 10.1016/S1474-4422(14)70266-2. [DOI] [PubMed] [Google Scholar]
- 20.Oji Y, Hatano T, Ueno SI, Funayama M, Ishikawa KI, Okuzumi A, et al. Variants in saposin D domain of prosaposin gene linked to Parkinson’s disease. Brain. 2020;143:1190–205. doi: 10.1093/brain/awaa064. [DOI] [PubMed] [Google Scholar]
- 21.Matsuda J, Kido M, Tadano-Aritomi K, Ishizuka I, Tominaga K, Toida K, et al. Mutation in saposin D domain of sphingolipid activator protein gene causes urinary system defects and cerebellar Purkinje cell degeneration with accumulation of hydroxy fatty acid-containing ceramide in mouse. Hum Mol Genet. 2004;13:2709–23. doi: 10.1093/hmg/ddh281. [DOI] [PubMed] [Google Scholar]
- 22.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–9. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 23.Ramirez A, Heimbach A, Grundemann J, Stiller B, Hampshire D, Cid LP, et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet. 2006;38:1184–91. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 24.Lautier C, Goldwurm S, Durr A, Giovannone B, Tsiaras WG, Pezzoli G, et al. Mutations in the GIGYF2 (TNRC15) gene at the PARK11 locus in familial Parkinson disease. Am J Hum Genet. 2008;82:822–33. doi: 10.1016/j.ajhg.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strauss KM, Martins LM, Plun-Favreau H, Marx FP, Kautzmann S, Berg D, et al. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson’s disease. Hum Mol Genet. 2005;14:2099–111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- 26.Paisan-Ruiz C, Bhatia KP, Li A, Hernandez D, Davis M, Wood NW, et al. Characterization of PLA2G6 as a locus for dystonia-parkinsonism. Ann Neurol. 2009;65:19–23. doi: 10.1002/ana.21415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Fonzo A, Dekker MC, Montagna P, Baruzzi A, Yonova EH, Correia Guedes L, et al. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology. 2009;72:240–5. doi: 10.1212/01.wnl.0000338144.10967.2b. [DOI] [PubMed] [Google Scholar]
- 28.Chartier-Harlin MC, Dachsel JC, Vilarino-Guell C, Lincoln SJ, Lepretre F, Hulihan MM, et al. Translation initiator EIF4G1 mutations in familial Parkinson disease. Am J Hum Genet. 2011;89:398–406. doi: 10.1016/j.ajhg.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edvardson S, Cinnamon Y, Ta-Shma A, Shaag A, Yim YI, Zenvirt S, et al. A deleterious mutation in DNAJC6 encoding the neuronal-specific clathrin-uncoating co-chaperone auxilin, is associated with juvenile parkinsonism. PLoS One. 2012;7:e36458. doi: 10.1371/journal.pone.0036458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krebs CE, Karkheiran S, Powell JC, Cao M, Makarov V, Darvish H, et al. The Sac1 domain of SYNJ1 identified mutated in a family with early-onset progressive Parkinsonism with generalized seizures. Hum Mutat. 2013;34:1200–7. doi: 10.1002/humu.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quadri M, Fang M, Picillo M, Olgiati S, Breedveld GJ, Graafland J, et al. Mutation in the SYNJ1 gene associated with autosomal recessive, early-onset Parkinsonism. Hum Mutat. 2013;34:1208–15. doi: 10.1002/humu.22373. [DOI] [PubMed] [Google Scholar]
- 32.Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–7. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nukada H, Kowa H, Saitoh T, Tazaki Y, Miura S. [A big family of paralysis agitans (author’s transl)] Rinsho Shinkeigaku. 1978;18:627–34. [PubMed] [Google Scholar]
- 34.Hasegawa K, Kowa H. Autosomal dominant familial Parkinson disease: older onset of age, and good response to levodopa therapy. Eur Neurol. 1997;38:39–43. doi: 10.1159/000113460. [DOI] [PubMed] [Google Scholar]
- 35.Hasegawa K, Funayama M, Matsuura N, Furusawa H, Sakai F, Kowa H, et al. Analysis of alpha-synuclein, parkin, tau, and UCH-L1 in a Japanese family with autosomal dominant parkinsonism. Eur Neurol. 2001;46:20–4. doi: 10.1159/000050751. [DOI] [PubMed] [Google Scholar]
- 36.Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41:1303–7. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 37.Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–12. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tolosa E, Vila M, Klein C, Rascol O. LRRK2 in Parkinson disease: challenges of clinical trials. Nat Rev Neurol. 2020;16:97–107. doi: 10.1038/s41582-019-0301-2. [DOI] [PubMed] [Google Scholar]
- 39.Ross OA, Soto-Ortolaza AI, Heckman MG, Aasly JO, Abahuni N, Annesi G, et al. Association of LRRK2 exonic variants with susceptibility to Parkinson’s disease: a case-control study. Lancet Neurol. 2011;10:898–908. doi: 10.1016/S1474-4422(11)70175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuji S, Choudary PV, Martin BM, Stubblefield BK, Mayor JA, Barranger JA, et al. A mutation in the human glucocerebrosidase gene in neuronopathic Gaucher’s disease. N. Engl J Med. 1987;316:570–5. doi: 10.1056/NEJM198703053161002. [DOI] [PubMed] [Google Scholar]
- 42.Lwin A, Orvisky E, Goker-Alpan O, LaMarca ME, Sidransky E. Glucocerebrosidase mutations in subjects with parkinsonism. Mol Genet Metab. 2004;81:70–3. doi: 10.1016/j.ymgme.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 43.Aharon-Peretz J, Rosenbaum H, Gershoni-Baruch R. Mutations in the glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N. Engl J Med. 2004;351:1972–7. doi: 10.1056/NEJMoa033277. [DOI] [PubMed] [Google Scholar]
- 44.Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N. Engl J Med. 2009;361:1651–61. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitsui J, Mizuta I, Toyoda A, Ashida R, Takahashi Y, Goto J, et al. Mutations for Gaucher disease confer high susceptibility to Parkinson disease. Arch Neurol. 2009;66:571–6. doi: 10.1001/archneurol.2009.72. [DOI] [PubMed] [Google Scholar]
- 46.Blauwendraat C, Reed X, Krohn L, Heilbron K, Bandres-Ciga S, Tan M, et al. Genetic modifiers of risk and age at onset in GBA associated Parkinson’s disease and Lewy body dementia. Brain. 2020;143:234–48. doi: 10.1093/brain/awz350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foo JN, Chew EGY, Chung SJ, Peng R, Blauwendraat C, Nalls MA, et al. Identification of Risk Loci for Parkinson Disease in Asians and Comparison of Risk Between Asians and Europeans: A Genome-Wide Association Study. JAMA Neurol. 2020;77:746–54. doi: 10.1001/jamaneurol.2020.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakayama S, Shimonaka S, Elahi M, Nishioka K, Oji Y, Matsumoto SE, et al. Tau aggregation and seeding analyses of two novel MAPT variants found in patients with motor neuron disease and progressive parkinsonism. Neurobiol Aging. 2019;84:240.e13–40.e22. doi: 10.1016/j.neurobiolaging.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 49.Ikeda A, Shimada H, Nishioka K, Takanashi M, Hayashida A, Li Y, et al. Clinical heterogeneity of frontotemporal dementia and Parkinsonism linked to chromosome 17 caused by MAPT N279K mutation in relation to tau positron emission tomography features. Mov Disord. 2019;34:568–74. doi: 10.1002/mds.27623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sirugo G, Williams SM, Tishkoff SA. The Missing Diversity in Human Genetic Studies. Cell. 2019;177:1080. doi: 10.1016/j.cell.2019.04.032. [DOI] [PubMed] [Google Scholar]
- 51.Global Parkinson’s Genetics P. GP2: The Global Parkinson’s Genetics Program. Mov Disord. 2021;36:842–51. doi: 10.1002/mds.28494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mok KY, East Asian Parkinson Disease Genomics C. The East Asian Parkinson Disease Genomics Consortium. Lancet Neurol. 2021;20:982. doi: 10.1016/S1474-4422(21)00373-2. [DOI] [PubMed] [Google Scholar]
- 53.Rizig M, Okubadejo N, Salama M, Thomas O, Akpalu A, Gouider R, et al. The International Parkinson Disease Genomics Consortium Africa. Lancet Neurol. 2021;20:335. doi: 10.1016/S1474-4422(21)00100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zabetian CP, Mata IF, Latin American Research Consortium on the Genetics of PD. LARGE-PD: Examining the genetics of Parkinson’s disease in Latin America. Mov Disord. 2017;32:1330–31. doi: 10.1002/mds.27081. [DOI] [PubMed] [Google Scholar]