Abstract
Our working hypothesis is that the major molecular chaperones DnaK and GroE play central roles in the ability of oral bacteria to cope with the rapid and frequent stresses encountered in oral biofilms, such as acidification and nutrient limitation. Previously, our laboratory partially characterized the dnaK operon of Streptococcus mutans (hrcA-grpE-dnaK) and demonstrated that dnaK is up-regulated in response to acid shock and sustained acidification (G. C. Jayaraman, J. E. Penders, and R. A. Burne, Mol. Microbiol. 25:329–341, 1997). Here, we show that the groESL genes of S. mutans constitute an operon that is expressed from a stress-inducible ςA-type promoter located immediately upstream of a CIRCE element. GroEL protein and mRNA levels were elevated in cells exposed to a variety of stresses, including acid shock. A nonpolar insertion into hrcA was created and used to demonstrate that HrcA negatively regulates the expression of the groEL and dnaK operons. The SM11 mutant, which had constitutively high levels of GroESL and roughly 50% of the DnaK protein found in the wild-type strain, was more sensitive to acid killing and could not lower the pH as effectively as the parent. The acid-sensitive phenotype of SM11 was, at least in part, attributable to lower F1F0-ATPase activity. A minimum of 10 proteins, in addition to GroES-EL, were found to be up-regulated in SM11. The data clearly indicate that HrcA plays a key role in the regulation of chaperone expression in S. mutans and that changes in the levels of the chaperones profoundly influence acid tolerance.
The ability to adapt to changes in environmental conditions is essential for the survival of microorganisms. Bacteria in oral biofilms are constantly subjected to acid shock and sustained acidification as a result of the rapid accumulation of acids generated from glycolysis by the acidogenic oral microflora. Central to the tolerance of environmental insults by microorganisms is the production of a variety of stress proteins, including the molecular chaperones GroEL and DnaK (12, 26). These proteins perform essential roles in cellular metabolism by assisting in the folding of newly synthesized or denatured proteins, as well as in the assembly, transport, and degradation of cellular proteins (9).
In bacteria, transcriptional regulators and alternative ς factors have been shown to be components of complex regulatory pathways that tightly control the transcription of heat shock genes under various conditions (28). In Escherichia coli, the primary mechanism for induction of the general stress response involves the modulation by DnaK of the activity of the alternative ς factor RpoH (ς32), which controls transcription of a variety of stress genes, including groEL and dnaK (8, 15). In the gram-positive soil bacterium Bacillus subtilis, four classes of stress-regulated genes have been identified (14, 20). Class I genes, encoding GroEL and DnaK, constitute the CIRCE regulon and are negatively controlled by HrcA (37, 40). Class II genes are controlled by the alternative ς factor ςB, and class III genes were recently shown to be negatively regulated by CtsR (14). Finally, class IV stress genes comprise those genes that are not regulated by one of the systems described above.
Control of class I stress gene expression in B. subtilis is governed by the widespread HrcA-CIRCE system. HrcA, a repressor protein, negatively regulates transcription of class I stress genes by binding to a DNA element called CIRCE (for controlling inverted repeat of chaperone expression) located in the regulatory regions of stress genes (20, 40). It also has been found that GroEL modulates the activity of the HrcA repressor (27), stabilizing and preventing aggregation of HrcA, which allows HrcA to function.
Streptococcus mutans, a bacterial pathogen associated with human dental caries, is well adapted to tolerate rapid changes in dental plaque pH, as well as fluctuations in carbohydrate availability and multiple environmental stresses. The ability of this organism to survive large and rapid changes in its environment, and in particular to grow and metabolize carbohydrates at low pH, is considered to be an important virulence factor. S. mutans is inherently more acid resistant than many other oral bacteria and is able to mount an acid tolerance response (ATR). The ATR is characterized by increased acid resistance, enhanced glycolytic capacities, and increased activity of the proton-translocating enzyme F1F0-ATPase (5, 18, 35). The ATR has also been found to confer cross-protection against multiple environmental insults, including UV radiation and oxidative stress (34). The basis for the ATR and cross-protection has been partially defined by the demonstration that acid-adapted cells have higher ATPase activities (5), as well as the identification of a low-pH-inducible DNA repair pathway (17) and an H+-glucose symporter that operates at pH 5.0 (13). Additionally, several proteins are induced in response to growth at low pH, although the roles of these induced gene products in acid tolerance have not been evaluated (19, 39). Finally, separate studies have shown that two genes, ffh (16) and sgp (2), are required for acid tolerance. In spite of progress made on acid tolerance in oral streptococci, many of the gene products necessary for the inherent acid resistance of S. mutans and for the induction and phenotypic manifestation of the ATR by oral streptococci are yet to be defined.
The production of stress proteins, including the molecular chaperones GroEL and DnaK, is a central feature of bacterial stress responses (12). Unlike for E. coli and B. subtilis, there is very little information on modulation of stress gene expression in lactic acid bacteria. The first detailed description of the transcriptional organization and regulation of a stress response in lactic acid bacteria was for the dnaK operon of S. mutans, containing the hrcA, grpE, and dnaK genes, which are transcribed from a ςA-type promoter (22). Additionally, by using a continuous chemostat culture, it was demonstrated that levels of dnaK mRNA and DnaK were increased in response to acid shock and remained elevated in acid-adapted cells (i.e., cells that had induced the ATR), suggesting that DnaK is intimately involved in responses to environmental acidification.
In this report, we identified and characterized the groE operon of S. mutans (groES-groEL) and explored the regulation of groEL expression in response to various stresses, including heat, acidic pH, ethanol, and H2O2. To evaluate the role of HrcA as a repressor protein in stress gene regulation, an HrcA-deficient strain was constructed, and the effects of inactivation of hrcA on groE and dnaK expression were assessed. The mutant, which produced elevated levels of GroEL and diminished levels of DnaK, was used to establish a molecular link between chaperone gene expression and acid tolerance.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains DH10B and M15 were grown in Luria broth, and S. mutans UA159 was grown in brain heart infusion broth (BHI). When needed, kanamycin (40 μg ml−1 for E. coli or 500 μg ml−1 for S. mutans) or ampicillin (100 μg ml−1) was added to the medium. To investigate the response of S. mutans to different stress conditions, cells were grown in BHI at 37°C to mid-exponential phase (optical density at 600 nm [OD600] ≅ 0.6), and aliquots of the cultures were then heat shocked at 42°C or incubated in the presence of ethanol (20 mM) or H2O2 (2 mM) for different periods of time. For studies involving acid shock and acid adaptation, S. mutans was grown in a continuous chemostat culture as previously described (22).
DNA manipulations.
Chromosomal DNA was prepared from S. mutans as previously described (10). Plasmid DNA was isolated from E. coli by using QIAgen columns (Qiagen, Chatsworth, Calif.), and restriction and DNA-modifying enzymes were obtained from Life Technologies, Inc. (LTI; Gaithersburg, Md.), New England Biolabs (Beverly, Mass.), or MBI Fermentas (Amherst, N.Y.). PCRs were carried out with 100 ng of S. mutans chromosomal DNA by using Vent DNA polymerase, and PCR products were purified with the QIAquick kit (Qiagen). DNA was introduced into S. mutans by natural transformation (33) and into E. coli by electroporation (36). Southern blot analyses were carried out at high stringency as described by Sambrook et al. (36).
Overexpression and purification of HrcA and antibody production.
The hrcA gene was amplified from S. mutans UA159 by PCR with primers containing restriction sites (BamHI and PstI) to facilitate cloning into the plasmid expression vector pQE30 (Qiagen). The resulting plasmid (pJL1), which produced a recombinant protein with an in-frame fusion of six consecutive histidine residues to the N terminus of HrcA, was used to transform E. coli M15. His-tagged HrcA was purified from isopropyl-β-d-thiogalactopyranoside (IPTG)-treated cells under denaturing conditions by following the protocols recommended by the supplier (Qiagen). The recombinant protein (0.4 mg) was further purified by excision from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and used to elicit a polyclonal antiserum in rabbits (Lampire, Pipersville, Pa).
Construction of an hrcA insertion mutant.
A 2.1-kb fragment containing the hrcA gene was amplified by PCR and cloned onto pGEM-5Zf(+) (Promega, Madison, Wis.) to generate plasmid pJL2. The hrcA gene was then inactivated by replacing a region containing the ςA-type promoter located upstream of hrcA (P1) and a 5′ portion of the structural gene with a kanamycin resistance gene (ΩKm element) that is flanked by strong transcription and translation terminators (32). To allow for expression of genes downstream of hrcA in the dnaK operon, which appear to be essential for cell viability (see Results), the Streptococcus salivarius urease promoter (PureI) (11) was also cloned immediately downstream of the insertion site of ΩKm into the hrcA gene. Specifically, a 0.31-kb fragment of hrcA in pJL2 was replaced by a 3.3-kb ΩKm-PureI fragment to give plasmid pJL6. Circular plasmid pJL6 was used to transform S. mutans, and transformants were selected on BHI agar with kanamycin. Chromosomal DNAs from the mutant and wild-type strains were digested with BglII and probed with the hrcA gene and the ΩKm element. The absence of HrcA in the recombinant strains was further confirmed by Western blot analysis. One such isolate, designated SM11, was used throughout the studies.
Primer extension and RNA analysis.
Total RNA was extracted from cells of S. mutans UA159 and the hrcA mutant strain (SM11) growing in chemostat cultures or from mid-exponential-phase batch cultures by using a protocol described by Chen et al. (11). Primer extensions were carried out with the oligonucleotide designated EXTGROE (5′-CTAGCACCAGCAAGGAC-3′) by a protocol described previously (1) with primer annealing and reverse transcription performed at 42°C. For quantitative slot blot analysis, equivalent amounts of denatured RNA were transferred to nylon membranes (GeneScreen Plus; NEN Life Science products, Inc., Boston, Mass.) by a slot blot apparatus (LTI) as described by Sambrook et al. (36). For Northern blot analyses, total RNA was separated on a 1.0% formaldehyde gel following the protocol described elsewhere (1). RNAs were UV cross-linked to the membranes, and filter membranes were probed with S. mutans groEL or dnaK probes labeled with [α-32P]dATP (NEN Life Science). All hybridizations and washes were carried out under high-stringency conditions. Signals obtained on autoradiographs were quantified with an IS1000 digital imaging system (Alpha Innotech Corp, San Leandro, Calif.). Reverse transcriptase PCR (RT-PCR) was performed with the ThermoScript RT-PCR System from LTI.
Protein electrophoresis and Western blotting.
Cells grown as described above were centrifuged, washed, and homogenized in the presence of glass beads by using a Bead Beater (Biospec, Bartlesville, Okla.), as described previously (11). Whole-cell lysates were separated by SDS-PAGE, blotted onto Immobilon-P membranes (Millipore, Bedford, Mass.), and subjected to Western blot analysis by standard techniques (1). Two-dimensional (2-D) electrophoresis was performed according to the method of O'Farrell (29) at Kendrick Labs, Inc. (Madison, Wis.). Isoeletrofocusing was carried out with 2.0% pH 4 to 8 ampholines (Gallard-Schlesinger, Garden City, N.Y.), and 2-D electrophoresis was carried out in a SDS–10% polyacrylamide gel. Gels were stained with Coomassie blue and dried between sheets of cellophane.
pH drop and acid killing.
The glycolytic capacities of the HrcA-deficient (SM11) and wild-type (UA159) strains were compared by pH drop experiments according to the protocol described by Belli and Marquis (5). Briefly, cells from steady-state chemostat cultures of UA159 or SM11 were harvested, washed with 1 culture volume of cold distilled water, and resuspended in a solution of 50 mM KCl and 1 mM MgCl2 in 1/10 of the original culture volume. The suspension was titrated with 0.1 M KOH to a pH of 7.2, and pH drops were initiated by addition of 55.6 mM glucose. In order to determine the ability of SM11 to resist acid killing, steady-state cultures of the wild-type and mutant strains grown under the conditions described above were washed once with 1% peptone (pH 7.0) and resuspended in 1/10 of the original volume in 1% peptone (pH 3.0). Samples were stirred continuously at room temperature, aliquots of cells were removed at predetermined intervals, and the viable counts of each culture were determined by plating on BHI agar.
F1F0-ATPase assays.
The F1F0-ATPase activity of permeabilized cells prepared from chemostat-grown cells was determined according to the method of Belli and Marquis (5). Briefly, cells were collected, washed once with membrane buffer (75 mM Tris [pH 7.0], 10 mM MgSO4), and concentrated 25-fold in the same buffer. Cells were then permeabilized in the presence of toluene, collected by centrifugation, and resuspended in the same volume of membrane buffer. Permeabilized samples were mixed with 50 mM Tris-maleate buffer (pH 6.0) containing 10 mM MgSO4, and the reactions were initiated by adding 0.5 M ATP (pH 6.0) to a final concentration of 5 mM at 37°C. Samples were removed at 0, 15, 30, and 45 min and assayed for inorganic phosphate released from ATP with the Fiske-Subbarow reagent (Sigma, St. Louis, Mo).
RESULTS
Genetic organization of the dnaK operon.
Previous studies in our laboratory indicated that the dnaK operon of S. mutans contained the hrcA, grpE, and dnaK genes. BLAST search analysis of databases maintained as part of the S. mutans Genome Sequencing Project at the University of Oklahoma's Advanced Center for Genome Technology identified the dnaK operon (starting at position 83510) and revealed the presence of an open reading frame (ORF4) of 1,131 bp located 3′ to dnaK and starting 531 bp from the dnaK stop codon (Fig. 1A). The deduced amino acid sequence of ORF4 had a predicted molecular mass of 40.8 kDa and showed a high degree of similarity (up to 83%) to DnaJ proteins of other bacteria. Analysis of the region 3′ to dnaJ revealed an ORF (ORF5) of 747 bp starting 305 bp from the dnaJ stop codon. ORF5 was predicted to encode a 249-amino-acid polypeptide with a predicted molecular mass of 28 kDa that exhibited high levels of similarity (86%) to a putative tRNA pseudouridine synthase A (TruA) protein from Streptococcus pyogenes. Immediately downstream of truA is an ORF similar to that coding for phosphomethylpyrimidine kinases, which are generally involved in thiamine biosynthesis, followed by two ORFs with the highest degree of similarity to those coding for hypothetical proteins of S. pyogenes. None of the ORFs 3′ to dnaJ share homology to genes that have been proven to be transcribed as part of eubacterial dnaK operons, nor do they share homology with proteins required for stress tolerance.
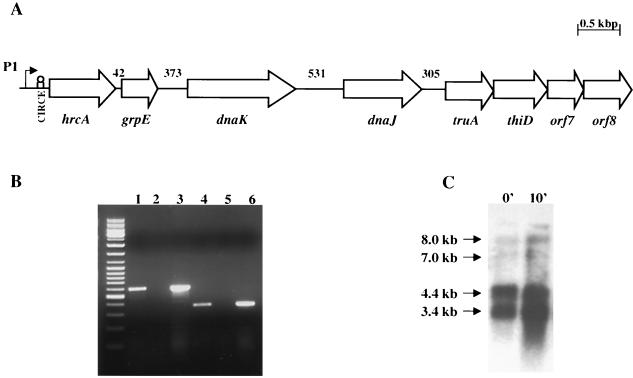
FIG. 1.
(A) Transcriptional organization of the S. mutans dnaK operon. (A) Schematic diagram of the S. mutans dnaK operon and genes downstream of the operon. Upstream of hrcA is the fruAB operon involved in the utilization of extracellular polysaccharides (22). truA is predicted to encode a tRNA-modifying enzyme and thiD phosphomethyl pyrimidine kinase, which is possibly involved in thiamine biosynthesis. The numbers above the schematic indicate the size of the intergenic regions in base pairs. (B) PCR products generated by RT-PCR. Shown are ethidium bromide-stained 1% agarose gels of products obtained with primers spanning the intergenic regions between dnaK-dnaJ (lanes 1 to 3) and dnaJ-truA (lanes 4 to 6). Products from the PCR were derived with the following: cDNA prepared from S. mutans RNA (lanes 1 and 4); a negative control using S. mutans RNA, but omitting RT (lanes 2 and 5); and S. mutans chromosomal DNA (lanes 3 and 6). A DNA ladder mix from Fermentas was used as the molecular weight markers. (C) Northern blot analysis of S. mutans UA159 dnaK. Cells were grown at 37°C to mid-exponential phase and then subjected to heat shock at 42°C. RNA was isolated from cells before (0′) and after (10′) heat shock for 10 min. Total RNA (10 μg per lane) was separated in a 1.0% gel, transferred to a nylon membrane, and hybridized to a dnaK-specific probe.
The use of RT-PCR revealed evidence for a transcript containing the dnaK-dnaJ sequences, indicating that dnaJ is probably cotranscribed with hrcA, grpE, and dnaK from the ςA-type promoter (P1) (22) located 5′ to hrcA (Fig. 1B). Surprisingly, with a set of primers containing internal sequences of the dnaJ and truA genes, a band corresponding to the correct molecular size was observed, suggesting that transcription of the dnaK operon may proceed beyond the dnaJ gene (Fig. 1B). Northern analysis (Fig. 1C), revealed the presence of two major mRNA species migrating at 3.4 and 4.4 kb, consistent with the sizes of the dnaKJ and grpE-dnaKJ transcripts, and these transcripts were increased in cells that had undergone heat shock for 10 min. These mRNAs represented over 99% of the total dnaK-containing mRNA as assessed by densitometry. Of note, the two major transcripts found to hybridize with a dnaK probe were present in Northern blots as revealed by a dnaJ probe (data not shown), further confirming the RT-PCR results that showed that dnaJ is cotranscribed with dnaK. Also consistent with the RT-PCR results, it was found that a small fraction (<1%) of the total mRNA hybridizing to the dnaK and dnaJ probes was present as transcripts of approximately 7.0 and 8.0 kb (Fig. 1C). The most reasonable interpretation of these data is that termination of the highly expressed dnaK operon is not 100% efficient, and in some cases, the downstream genes can be cotranscribed as part of the operon.
Genetic organization and transcriptional mapping of the groE operon.
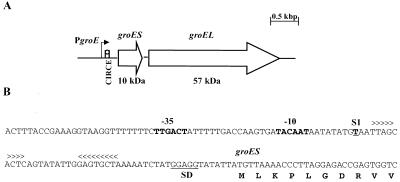
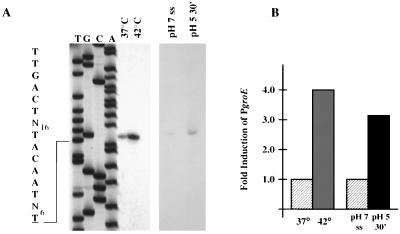
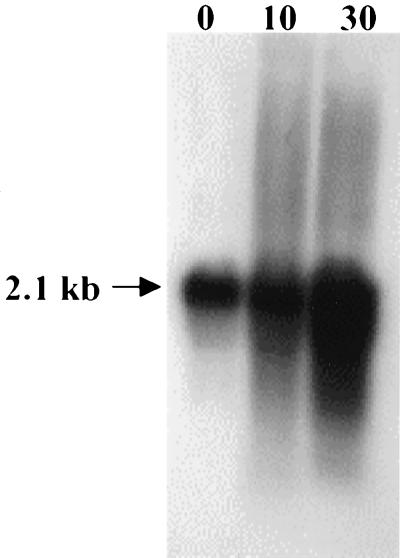
The groES and groEL genes from S. mutans were identified in the S. mutans genome by BLAST searches (Fig. 2). The groE operon is located from position 1834692 through position 1832649 and is transcribed in the opposite orientation to the dnaK operon. The groES gene (285 bp) encoded a polypeptide of 95 amino acids with a predicted molecular mass of 10 kDa. The groEL gene (1,626 bp) encoded a 542-amino-acid protein with a predicted molecular mass of 57 kDa. As expected, the proteins encoded by groES and groEL showed high levels of similarity to known GroES and GroEL proteins found in other gram-positive bacteria, including S. pyogenes (71% GroES and 91% GroEL), Streptococcus pneumoniae (81% GroES and 89% GroEL), and Streptococcus agalactiae (80% GroES and 90% GroEL). To locate the transcriptional initiation site(s) of the groE operon and to determine whether transcription was induced in response to heat shock and acidic conditions, primer extensions were performed with total RNA isolated from mid-log cultures subjected to heat shock or from chemostat cultures growing at steady state at pH 7.0 that had been subjected to an acid shock at pH 5.0. A single transcription initiation site was identified 48 nucleotides 5′ to the translational initiation site. A ςA-type promoter (TTGACT-N16-TACAAT), located 57 bp 5′ to the groES start codon, was identified. Densitometric analysis of this single band demonstrated that transcription from this promoter, designated PgroE, was increased fourfold in heat-shocked cells and threefold in acid-shocked cells (Fig. 3). A perfect CIRCE element (TTAGCACTC-N9-GAGTGCTAA) was identified starting 11 bp 3′ to the −10 element and 19 bp from the groES start codon. Northern analyses with a fragment of the groEL gene as a probe revealed the presence of a single transcript of 2.1 kb, indicating that groEL can be cotranscribed with groES from PgroE (Fig. 4).
FIG. 2.
(A) Schematic diagram of the groE operon of S. mutans. (B) Sequence of the PgroE region. The ςA-type −35 and −10 regions are in boldface. A transcriptional initiation site, S1, mapped by primer extension, is shown in boldface and underlined. The CIRCE element is marked with arrowheads. The predicted Shine-Dalgarno (SD) sequence is underlined.
FIG. 3.
Mapping of the S. mutans groE promoter (PgroE) by primer extension. (A) Total RNA was isolated from mid-log-phase cultures grown at 37°C or 30 min after heat shock at 42°C or from chemostat-grown cells that have reached steady state at pH 7.0 (pH 7 ss) or 30 min after acid shock at pH 5.0. Adjacent to the primer extension is a sequencing reaction performed with plasmid pJL14 with the same primer used in the primer extension reaction (EXTGROE). Indicated on the left are the sequences of the −35 and −10 regions of the ςA-type promoter (boldface) and the corresponding transcriptional initiation site (boldface and underlined). Images of primer extension products under each stress condition were obtained from different exposures of X-ray film with a digital imaging system. (B) Bar graph showing PgroE induction as determined by densitometry with an IS1000 digital imaging system.
FIG. 4.
Northern blot analysis of S. mutans UA159 groEL. Cells were grown at 37°C to the mid-log phase and then subjected to heat shock at 42°C. RNA was isolated from cells before (time zero) and after heat shock for 10 and 30 min. Total RNA (10 μg per lane) was separated in a 1.0% gel, blotted to nylon membrane, and hybridized to a groEL-specific probe.
Induction of groEL mRNA and GroEL protein in response to environmental stresses.
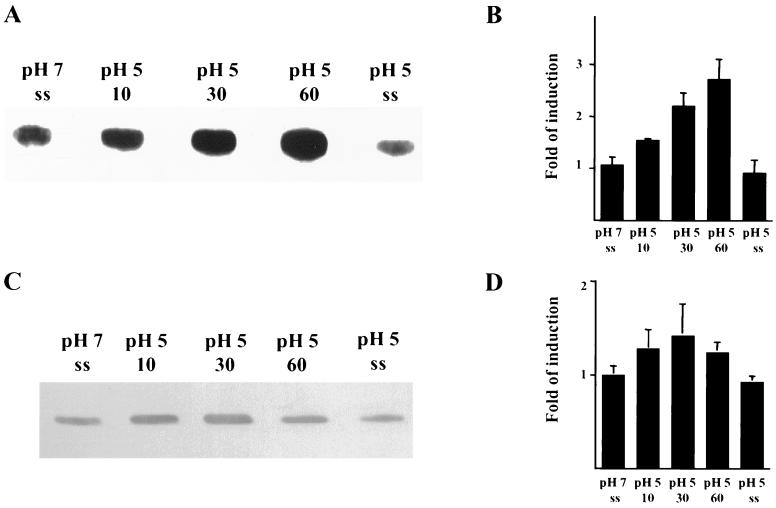
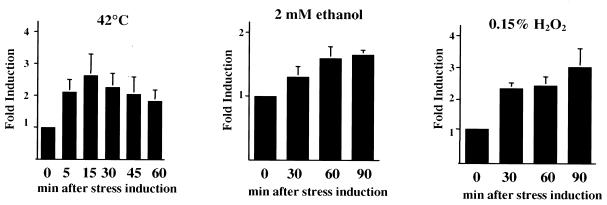
To determine whether groEL expression is modulated by environmental conditions, slot blots of total RNA and Western blot analyses were performed. Steady-state continuous culture or batch cultures of UA159 grown under the conditions described in Materials and Methods were submitted to different stresses. Slot blot analysis revealed that the levels of groEL mRNA were induced approximately 2.5-fold when pH 7.0 steady-state cultures were submitted to acid shock (pH 5.0) for up to 1 h. In contrast to what was previously observed for dnaK (22), no significant differences in the groEL mRNA levels in steady-state cells grown at either pH 7.0 or 5.0 were observed (Fig. 5A and B). The results of Western blot analyses also indicated that the levels of S. mutans GroEL were elevated following acid shock, but were not altered in steady-state pH 5.0 cells compared with steady-state pH 7.0 cells (Fig. 5C and D). As seen with the induction of dnaK mRNA and DnaK (22), the magnitude of the increase in groEL RNA is much greater than for the gene product. This is most likely attributable to the fact that both DnaK and GroEL are highly abundant proteins and that large increases in the amount of mRNA are needed to effect significant changes in the absolute quantity of these proteins in the cell. Although acid is believed to be the most prominent stress factor with which S. mutans is confronted, organisms in dental biofilms are subjected to many other types of stresses. In addition to acid shock, it was found that the levels of groEL mRNA and GroEL were elevated when S. mutans was subjected to heat shock, oxidative stress, and exposure to ethanol (Fig. 6).
FIG. 5.
Induction of groEL mRNA and GroEL in reponse to changes in environmental pH. (A) Slot blot of total RNA isolated from samples grown in the chemostat under the conditions indicated in the figure (ss, steady-state). RNase-treated samples were used as controls (data not shown). Hybridization was performed with an internal fragment of groEL. (B) Bar graph depicting induction of groEL mRNA as measured by densitometry. The graph shows the averages and standard deviations of three independent experiments. (C) Western blot analysis of GroEL levels with a polyclonal antibody against S. pyogenes GroEL (1:1,000). (D) Bar graph showing induction of GroEL as measured by densitometry.
FIG. 6.
Quantitative slot blot analysis of groEL mRNA in reponse to environmental stresses. Total RNA was isolated from mid-log-phase cultures that were submitted to the conditions indicated in the figure. The calculated mRNA induction profile of each stress condition was determined by densitometry (n = 2).
Inactivation of the S. mutans hrcA gene leads to constitutive expression of groES and groEL.
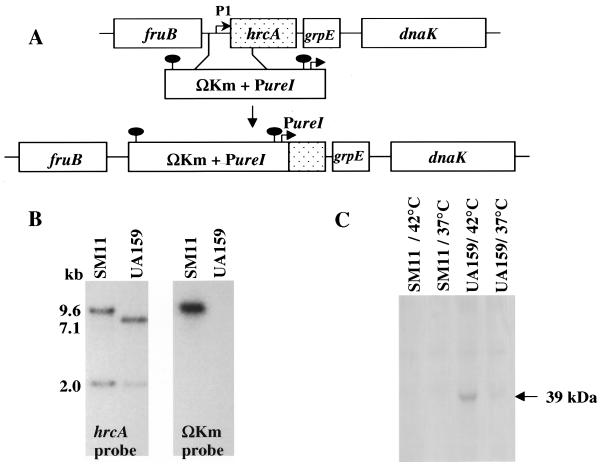
To determine the role of HrcA in stress gene regulation of S. mutans, the hrcA gene was disrupted by double crossover insertion of a kanamycin resistance gene flanked by strong transcription and translation terminators (ΩKm) (32) and followed by the S. salivarius urease promoter (PureI), (11) (Fig. 7). Initially, attempts to inactivate hrcA by inserting the strongly polar ΩKm cassette alone resulted in isolation of only single crossover insertions. Similar results were obtained when attempts were made to isolate strains with insertions of ΩKm into the dnaK gene (data not shown). By using PureI to drive the expression of the genes 3′ to hrcA in the dnaK operon, a strain with a double crossover insertion into the hrcA gene was then isolable. Correct integration of ΩKm was confirmed by Southern blotting and by Western blotting with an anti-S. mutans HrcA antibody (Fig. 7). It is interesting that, as previously observed at the mRNA level (22), the levels of HrcA protein were elevated after heat shock (Fig. 7C), consistent with the fact that hrcA is transcribed from a stress-inducible promoter. Also, similar to other organisms, HrcA proved to be highly insoluble. The protein could only be purified from recombinant E. coli strains and was only detectable in S. mutans when the cell lysates were obtained under denaturing conditions.
FIG. 7.
Construction of an hrcA mutant strain (SM11). (A) Schematic diagram of the insertional inactivation of the hrcA gene in S. mutans UA159. The gene was inactivated by replacing a region containing the ςA-type promoter (P1) upstream of hrcA and a 5′ portion of the structural gene by a cassete containing the ΩKm element and the PureI promoter. The solid oval markings indicate the transcriptional and translational terminators of the ΩKm element. (B) Southern hybridization to S. mutans chromosomal DNA by using the hrcA gene and the ΩKm element as probes. Indicated are the sizes of hybridizing bands as determined by comparison with DNA standards. (C) Western blot analysis of HrcA protein in response to heat shock (42°C). Total protein lysates of UA159 and SM11 were obtained under denaturing conditions and probed with anti-S. mutans HrcA antibody (diluted 1:500).
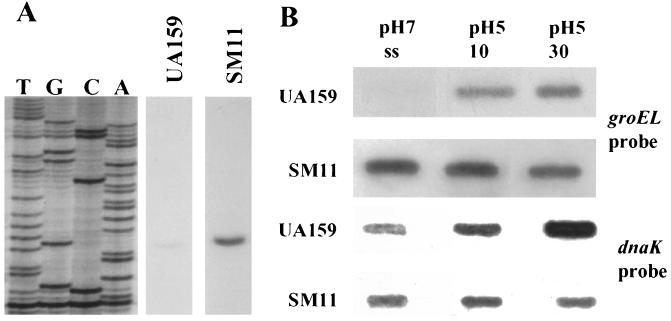
Primer extension analysis with RNA from wild-type and mutant strains demonstrated that inactivation of hrcA led to derepression of PgroE under both homeostatic and acidic conditions, with 5- and 3.4-fold inductions, respectively, when compared to the wild-type strain growing under the same conditions (Fig. 8A). It was not possible to obtain primer extension data from the ςA-type promoter (P1) of the dnaK operon in strain SM11, because the hrcA mutant was constructed such that P1 and the 5′ portion of hrcA were replaced by the ΩKm/PureI cassette. Instead, the expression from P1 in an hrcA mutant strain was analyzed by primer extension in a strain in which the hrcA locus was inactivated by single crossover insertion of a suicide plasmid (HRCERM) (22). The results indicated that HrcA also represses transcription of P1 in the dnaK operon (data not shown).
FIG. 8.
Primer extension and slot blot analysis of an hrcA mutant strain (SM11). (A) Total RNA was isolated from pH 7.0, steady-state, chemostat-grown cells of S. mutans UA159 or SM11. Equal amounts of RNA were hybridized to the primer EXTGROE (Fig. 2) and subjected to primer extension analysis. Images of primer extension products from UA159 and SM11 RNAs were obtained from the same piece of X-ray film with a digital imaging system. Intervening lanes were omitted to present the relevant data. (B) Slot blot of total RNA from UA159 and SM11 probed with internal fragments of the groEL or dnaK genes.
The levels of groEL and dnaK mRNA in the mutant and wild-type strains were compared under different stress conditions. Under acid stress and all other conditions tested, the levels of groEL mRNA were significantly elevated in the mutant strain, demonstrating that HrcA represses groEL transcription (Fig. 8B). Similar results were obtained when blots were probed with groES (data not shown). Diminished levels of dnaK mRNA were observed (Fig. 8B), probably because transcription of dnaK from PureI was not as efficient or the mRNA was not as stable as when transcription arises from the cognate dnaK promoter (P1). Western blot analysis with anti-S. pyogenes GroEL and DnaK polyclonal antibodies (25) also confirmed the results obtained by RNA analysis indicating that GroEL was constitutively derepressed in the hrcA mutant strain, whereas the levels of DnaK were diminished relative to UA159 (data not shown).
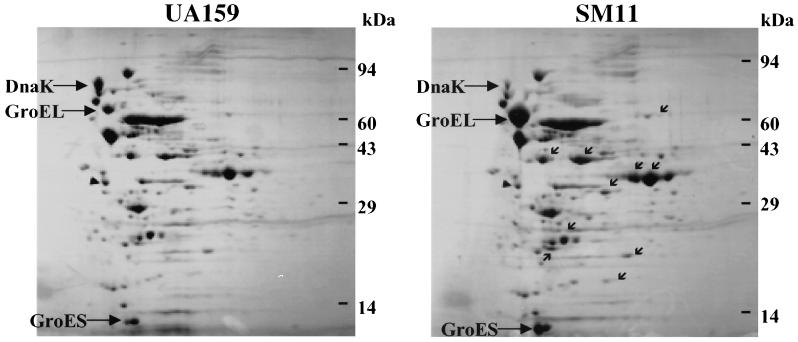
A 2-D PAGE approach was used to assess differences in protein expression of exponential phase cells of SM11 and UA159. Strong induction of the GroEL (4.2-fold) and GroES (4.5-fold) proteins was observed in SM11, whereas DnaK expression was approximately 45% lower in SM11 (Fig. 9), consistent with the Western blot data. Interestingly, in addition to alterations in molecular chaperone expression, at least 10 other proteins appeared to be up-regulated in the mutant strain.
FIG. 9.
2-D protein pattern of S. mutans strains UA159 and SM11. Samples were grown in BHI and harvested in the exponential growth phase (OD600 ≅ 0.6). GroEL, GroES, and DnaK are identified. Small arrows indicate other protein spots with increased expression in the SM11 strain.
Acid tolerance of the hrcA mutant strain.
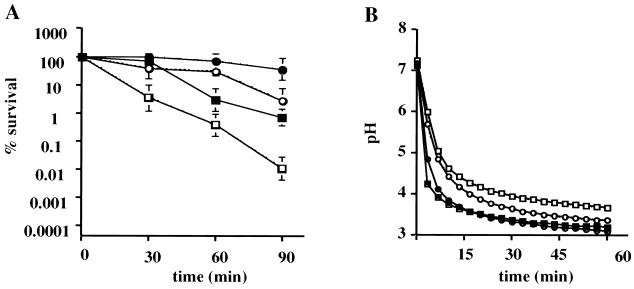
At 37°C, SM11 had a generation time of about 70 min, slower than the 50-min doubling time of the wild-type strain. To determine if SM11 had altered resistance to low pH, acid killing and pH drop experiments were performed with cells obtained from steady-state continuous culture. Steady-state pH 7.0 (unadapted) or pH 5.0 (acid-adapted) cultures of SM11 and UA159 were subjected to acidification at pH 3.0. Regardless of the growth conditions, the mutant strain was killed more rapidly at pH 3.0 than the wild-type strain growing under the same conditions (Fig. 10A), indicating an acid-sensitive phenotype for SM11. Consistent with the acid-sensitive phenotype, pH drop experiments revealed that UA159 was consistently able to lower the pH through glycolysis to values lower than that which could be achieved by strain SM11 (Fig. 10B and Table 1). In spite of the overall acid-sensitive phenotype and changes in the final pH values achieved, acid-adapted SM11 cells (grown at pH 5) were able to reduce the pH to values lower than those of the unadapted SM11 cells (grown at pH 7) and became more resistant to acid killing, indicating that the hrcA mutant strain is able to mount an ATR.
FIG. 10.
Acid tolerance response of S. mutans UA159 (solid symbols) and SM11 (open symbols). Cells were collected from a steady-state chemostat held at pH 7.0 (squares) or pH 5.0 (circles) and subjected to acid killing or pH drop experiments. (A) Survival of UA159 and SM11 during acid challenge at pH 3.0. Cell viability at each time point is expressed as the percentage of viable cells (CFU per milliliter of culture) at time zero. (B) Glycolytic pH profile of UA159 and SM11. Glucose was added to the cell suspensions to initiate glycolysis, and pH drops were continuously monitored for 1 h. Data points were collected every 10 s, and data for every 2 min and 20 s are presented. The graph shows the averages and standard deviations of three independent experiments.
TABLE 1.
Acid-adaptive response of S. mutans UA159 and SM11
| Strain | Growth pH | 1-h glycolytic pH (final pH)a | ATPase activity (nmol of PO4/mg [dry wt])b |
|---|---|---|---|
| UA159 | 7.0 | 3.303 (±0.08) | 0.0578 (±0.0062) |
| 5.0 | 3.208 (±0.111) | 0.2429 (±0.0658) | |
| SM11 | 7.0 | 3.562 (±0.107) | 0.0689 (±0.0103) |
| 5.0 | 3.33 (±0.075) | 0.1862 (±0.062) |
Differences in the final pH reached from pH 7-grown cells were statistically significant (P < 0.05). Although the differences in the final glycolytic pH values achieved by pH 5-grown cells of UA159 and SM11 were not statistically significant, because of day-to-day variation in the final pH values achieved by a given strain (about 0.1 pH unit), in every instance, the mutant was never able to lower the pH as low as the wild-type strain.
ATPase activity is expressed as the amount of PO4 liberated 30 min after the addition of ATP. The results presented here are the means and standard deviations of three independent experiments.
It has been demonstrated previously that acid adaptation of oral streptococci is correlated with an increase in proton-extruding ATPase activity (5). The F1F0-ATPase activity of both UA159 and SM11 was elevated in acid-adapted cells when compared to that in unadapted cells (4.2- and 2.7-fold increases, respectively). When comparing the ATPase activity of the two strains, the levels obtained from unadapted cells were very similar for UA159 and SM11. However, acid-adapted cells of SM11 showed substantially lower levels of ATPase than acid-adapted cells of UA159 (Table 1).
DISCUSSION
The dnaK operon (hrcA-grpE-dnaK) has been partially characterized in S. mutans (22). In the present study, we identified an ORF with high homology to dnaJ that was located 531 bp 3′ to dnaK. Frequently, in other gram-positive bacteria, dnaK operons consist of hrcA, grpE, and dnaKJ as well as other genes (4, 21, 30). Our results clearly indicate that the S. mutans dnaK operon consists of at least four genes: hrcA, grpE, and dnaKJ. In B. subtilis, it has been demonstrated that the dnaK operon is heptacistronic (hrcA grpE dnaK dnaJ orf35 orf28 orf50) (21). A putative protein showing strong homology to the B. subtilis ORF35 protein (63% similarity) was found 3′ to dnaJ in Clostridium acetobutylicum and Staphylococcus aureus (4, 30). However, BLAST searches against the S. mutans genome did not indicate the presence of an ORF35 homologue near the dnaK locus. Instead, there is an ORF 3′ to dnaJ that has high levels of similarity to a putative tRNA pseudouridine synthase A gene (truA) from S. pyogenes, followed by a gene that may participate in pyrimidine metabolism or thiamine biosynthesis, and then by two genes with products of unknown function. Although we were able to detect transcriptional readthrough from the dnaK operon into truA by using the very sensitive RT-PCR technique, we question the biological significance of this finding, based on the fact that the genes downstream of dnaJ seem unlikely to participate in stress tolerance, and the overwhelming majority of the transcripts we observed by Northern blotting terminate at dnaJ. Thus, we believe that the dnaK operon is a four-gene operon that terminates at dnaJ, and any transcription into the truA gene probably arises because the terminator 3′ to dnaJ is not 100% efficient.
It is also noteworthy that the organization of the S. mutans dnaK operon is unique when compared with the same operon of other eubacteria and gram-positive cocci. In particular, there are two unusually large intergenic regions between grpE and dnaK and dnaK and dnaJ in the dnaK operon of S. mutans. Using reporter gene fusions and primer extension analysis, no functional promoter was found within the 373-bp grpE-dnaK intergenic region (22). In B subtilis, dnaJ is transcribed from a ςA-dependent promoter preceding the entire dnaK operon, as well as from a vegetative promoter located downstream of dnaK (21). Preliminary results obtained by primer extension analysis and reporter gene fusions have not revealed a functional promoter in the dnaKJ intergenic region of S. mutans (data not shown). Thus, there do not appear to be internal promoters driving dnaJ expression in the S. mutans dnaK operon, and transcription of dnaJ is probably dependent on P1. We propose that the intergenic regions play a role in posttranscriptional regulation of chaperone expression, perhaps in the processing or stability of the dnaKJ mRNAs. Along these lines, the Northern blots indicate that hrcA may not be a part of the most abundant transcripts, which most likely contain grpE-dnaKJ and dnaKJ. Although this is consistent with the fact that the DnaK chaperone complex is far more abundant than HrcA, it appears that mRNA processing may be an important component of regulation of expression of the genes in the dnaK operon, as has been shown in B. subtilis (21).
A previous report from our laboratory (22) and the present study demonstrated that groEL and dnaK are part of the general stress response of S. mutans and that the hcrA gene product is a major component controlling dnaK and groE expression. We are primarily interested in the role of the DnaK and GroE chaperone machines in acid tolerance and regulation of adaptation to acidic conditions. In particular, it has been demonstrated that the glycolytic activity of oral bacteria in biofilms on the tooth and other surfaces of the mouth can lower the pH from 7 to 4 in just a few minutes (23). Bacteria present in the biofilms on teeth must be able to withstand these rapid and substantial fluctuations in the environmental pH. Previously, it was shown that rapid acidification of steady-state cultures of S. mutans growing at neutral pH resulted in increases in the levels of dnaK mRNA and DnaK protein, and the levels remained elevated (approximately 1.8-fold) in steady-state cultures grown at pH 5.0 (22). As was observed for dnaK, the levels of groEL mRNA and GroEL were elevated during acid shock. However, after cells had achieved steady state at pH 5.0, which is known to result in full induction of the ATR, the levels of groEL mRNA and GroEL protein were indistinguishable from those seen in steady-state cultures at pH 7.0. Thus, both GroEL and DnaK are induced during the acid shock response, whereas acid adaptation involves maintenance of elevated levels of DnaK, but not of GroEL.
Despite the fact that DnaK is important for survival under extreme conditions, the role of this protein in acid tolerance and acid adaptation has not been defined. In order to evaluate the role of S. mutans DnaK in stress responses, we have tried to construct a DnaK-deficient strain by inserting a variety of different antibiotic resistance cassettes in the 5′ portion of the dnaK gene. Taking into consideration that a dnaK mutant strain would probably exhibit a temperature- and acid-sensitive phenotype, in addition to using standard conditions to isolate a dnaK mutant strain (e.g., nonbuffered BHI and incubation at 37°C), we used buffered media and a lower temperature (25°C) to try to allow growth of putative mutants. However, all attempts to produce a dnaK mutant failed. Also, as indicated above, we were unable to use the ΩKm element to isolate an hrcA mutant, likely due to the polar effects of this insertion on the expression of the downstream genes. These results indicate that, in contrast to E. coli and B. subtilis, a dnaK mutant of S. mutans may not be isolable, at least under the conditions tested. Similar findings have been reported for Streptomyces coelicolor, in which a dnaK mutation was apparently lethal (7). Such observations clearly point to a central role of the DnaK protein in physiologic homeostasis in S. mutans.
The presence of a gene with high homology to hrcA in the dnaK operon and the identification of the CIRCE element in the promoter regions of the S. mutans groE and dnaK operons suggested that HrcA could negatively regulate these two operons. To circumvent problems of DnaK deficiency, yet still be able to investigate the role of HrcA, an hrcA mutant strain (SM11) was isolated by inserting the ΩKm fragment containing an outward-reading promoter (PureI), such that the genes downstream of hrcA could still be expressed. The data obtained here strongly indicated that the groE and dnaK operons of S. mutans were negatively controlled by HrcA in a manner similar to that of class I heat shock genes in B. subtilis. Although there was about a 50% reduction of dnaK expression in the hrcA mutant strain, the cells were viable and grew normally, albeit more slowly than the wild-type strain.
Compared to the parental strain, S. mutans SM11 was more sensitive to acid killing and showed a reduced capacity to produce acid at low pH values. The F1F0-ATPase is essential for pH homeostasis in S. mutans, and it is thought that an increase in the activity of this enzyme is an important contributor to the mounting of the ATR by this organism (35). Consistent with these ideas, acid-adapted cells of SM11 showed diminished levels of F1F0-ATPase compared to those in UA159 cells, whereas unadapted cells from both strains had comparable levels of the enzyme. This suggests that under conditions of acid stress, S. mutans is unable to produce sufficient ATPase or cannot maintain the integrity of the ATPase in the face of cytoplasmic or membrane acidification. The basis for lower ATPase activity is not entirely clear at this point, but it is well established that the DnaK chaperone and its cochaperones, GrpE and DnaJ, play an important role in essential cellular processes, including assisting in the folding of nascent or denatured proteins, as well as in translocation of proteins. Therefore, it is possible that the DnaK-DnaJ-GrpE complex is involved in the biogenesis of the ATPase complex or in stabilizing this complex at low pH. It is reasonable to speculate that under normal conditions, the reduced amount of DnaK that is present in SM11 is still sufficient to assist F1F0-ATPase folding, assembly, or membrane insertion, explaining why comparable levels of ATPase activity were observed in both the wild-type and mutant strain growing at pH 7.0. Under acid stress conditions, there is an apparent need for increased ATPase activity to maintain the internal pH near neutrality, but DnaK may be titrated by denatured proteins, resulting in insufficient levels of DnaK to chaperone the ATPase complex. Interestingly, overexpression of the GroESL chaperonin was not sufficient to compensate for the decrease in DnaK in terms of biogenesis of the F-ATPase. In many cases, the GroE chaperonin seems to be more important in processing oligomeric complexes than the DnaK machinery. However, it has been suggested that the folding of proteins larger than 60 to 70 kDa, as is the case for the ATPase complex, cannot be properly assisted by GroESL (9). From the data obtained here, it can be hypothesized that DnaK and not GroESL is responsible for assisting the biogenesis of the ATPase complex. If this hypothesis is correct, involvement of DnaK with F-ATPases could be at the level of deaggregation, facilitation of proper folding and assembly, or participation of DnaK in insertion of the F0 portion into the membrane. The concept of an interaction between DnaK and F-ATPase is supported by studies that have demonstrated that depletion of hsp70, a eukaryotic DnaK homologue, affected the translocation of the β subunit of the F1F0-ATPase of Saccharomyces cerevisae (3).
It is also important to note that the acid-sensitive phenotype of SM11 could be unrelated to a direct effect of changes in the levels of DnaK or GroESL. It has been reported that GroEL positively modulates the HrcA repressor activity (27). One possibility is that GroEL regulates other stress response repressors as it does HrcA. In this case, the mutant strain, which had high levels of GroEL, could display a stress-sensitive phenotype as a result of a failure to derepress other pathways needed for stress tolerance. In fact, the hrcA mutant strain does have enhanced sensitivity to killing by heat (50°C) and H2O2 (0.15%) (data not shown). Additionally, 10 other protein spots were elevated in the mutant strain. Whether HrcA negatively regulates these proteins or if the altered expression of these polypeptides occurs as a consequence of changes in expression of GroEL or DnaK is not known. However, after searching the available sequence of the S. mutans genome database, we have found no evidence of the presence of a CIRCE element in regions that are not linked to the groE and dnaK operons, suggesting that HrcA may not directly impact genes other than those in the dnaK and groE operons.
Other studies have demonstrated that altered expression of GroEL and DnaK can lead to enhanced susceptibility to stresses and reduced cell viability (6, 24, 31, 38). For example, in E. coli and Haemophilus ducreyi, the overexpression of DnaK and DnaJ, which are known to negatively regulate the stress response in these organisms, leads to decreased expression of GroEL and GroES, resulting in diminished cell survival (6, 31). In Lactococcus lactis, a C-terminal deletion of DnaK resulted in increased expression of all CIRCE-containing heat shock operons, including the dnaK operon itself, and led to a pronounced temperature-sensitive phenotype (24).
The results presented here support that the repressor of class I heat shock gene expression, HrcA, and the major molecular chaperones of S. mutans play central roles in growth and stress tolerance by these important pathogens. In addition, we have shown that DnaK may play a key role in acid tolerance, perhaps by participating in the biogenesis or stabilization of the ATPase complex. Studies are under way to dissect the molecular genetics and biochemistry of the genes in the dnaK and groE operons in tolerance of environmental acidification.
ACKNOWLEDGMENTS
We thank B. A. Roe, R. Y. Tian, H. G. Jia, Y. D. Qian, S. P. Linn, L. Song, R. E. McLaughlin, M. McShan, and J. Ferreti for their efforts in the Streptococcus mutans Genome Sequencing Project, which is supported by a grant from the National Institute of Dental and Craniofacial Research.
This study was supported by grants DE11549 and DE12236 from the National Institute for Dental and Craniofacial Research.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley; 1987. [Google Scholar]
- 2.Baev D, England R, Kuramitsu H K. Stress-induced membrane association of the Streptococcus mutans GTP-binding protein, an essential G protein, and investigation of its physiological role by utilizing an antisense RNA strategy. Infect Immun. 1999;67:4510–4516. doi: 10.1128/iai.67.9.4510-4516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker J, Walter W, Yan W, Craig E A. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens S F, Narberhaus F, Bahl H. Cloning, nucleotide sequence and structural analysis of the Clostridium acetobutylicum dnaJ gene. FEMS Microbiol Lett. 1993;114:53–60. doi: 10.1016/0378-1097(93)90141-n. [DOI] [PubMed] [Google Scholar]
- 5.Belli W A, Marquis R E. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl Environ Microbiol. 1991;57:1134–1138. doi: 10.1128/aem.57.4.1134-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum P, Ory J, Bauernfeind J, Krska J. Physiological consequences of DnaK and DnaJ overproduction in Escherichia coli. J Bacteriol. 1992;174:7436–7444. doi: 10.1128/jb.174.22.7436-7444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bucca G, Brassington A M E, Schonfeld H-J, Smith C P. The HspR regulon of Streptomyces coelicolor: a role for the DnaK chaperone as a transcriptional co-repressor. Mol Microbiol. 2000;38:1093–1103. doi: 10.1046/j.1365-2958.2000.02194.x. [DOI] [PubMed] [Google Scholar]
- 8.Bukau B. Regulation of the Escherichia coli heat shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 9.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 10.Burne R A, Schilling K, Bowen W H, Yasbin R E. Expression, purification, and characterization of an exo-β-d-fructosidase of Streptococcus mutans. J Bacteriol. 1987;169:4507–4517. doi: 10.1128/jb.169.10.4507-4517.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y-Y M, Weaver C A, Mendelsohn D R, Burne R A. Transcriptional regulation of the Streptococcus salivarius 57.1 urease operon. J Bacteriol. 1998;180:5769–5775. doi: 10.1128/jb.180.21.5769-5775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig E A, Gambill B D, Nelson R J. Heat shock proteins: molecular chaperones of protein biogenesis. Microbiol Rev. 1993;57:402–414. doi: 10.1128/mr.57.2.402-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cvitkovitch D G, Boyd D A, Thevenot T, Hamilton I R. Glucose transport by a mutant of Streptococcus mutans unable to accumulate sugars via the phosphoenolpyruvate phosphotransferase system. J Bacteriol. 1995;177:2251–2258. doi: 10.1128/jb.177.9.2251-2258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derre I, Rapoport G, Msadek T. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol Microbiol. 1999;31:117–131. doi: 10.1046/j.1365-2958.1999.01152.x. [DOI] [PubMed] [Google Scholar]
- 15.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 1382–1399. [Google Scholar]
- 16.Gutierrez J A, Crowley P J, Cvitkovitch D G, Brady L J, Hamilton I R, Hillman J D, Bleiweis A S. Streptococcus mutans ffh, a gene encoding a homologue of the 54 kDa subunit of the signal recognition particle, is involved in resistance to acid stress. Microbiology. 1999;145:357–366. doi: 10.1099/13500872-145-2-357. [DOI] [PubMed] [Google Scholar]
- 17.Hahn K, Faustoferri R C, Quivey R G J. Induction of an AP endonuclease activity in Streptococcus mutans during growth at low pH. Mol Microbiol. 1999;31:1489–1498. doi: 10.1046/j.1365-2958.1999.01292.x. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton I R, Buckley N D. Adaptation by Streptococcus mutans to acid tolerance. Oral Microbiol Immunol. 1991;6:65–71. doi: 10.1111/j.1399-302x.1991.tb00453.x. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton I R, Svensater G. Acid-regulated proteins induced by Streptococcus mutans and other oral bacteria during acid shock. Oral Microbiol Immunol. 1998;13:292–300. doi: 10.1111/j.1399-302x.1998.tb00710.x. [DOI] [PubMed] [Google Scholar]
- 20.Hecker M, Schumann W, Volker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 21.Homuth G, Masuda S, Mogk A, Kobayashi Y, Schumann W. The dnaK operon of Bacillus subtilis is heptacistronic. J Bacteriol. 1997;179:1153–1164. doi: 10.1128/jb.179.4.1153-1164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jayaraman G C, Penders J E, Burne R A. Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol Microbiol. 1997;25:329–341. doi: 10.1046/j.1365-2958.1997.4671835.x. [DOI] [PubMed] [Google Scholar]
- 23.Jensen M E, Polansky P J, Schachtele C F. Plaque sampling and telemetry for monitoring acid production on human buccal tooth surfaces. Arch Oral Biol. 1982;27:21–31. doi: 10.1016/0003-9969(82)90172-8. [DOI] [PubMed] [Google Scholar]
- 24.Koch B, Kilstrup M, Vogensen F K, Hammer K. Induced levels of heat shock proteins in a dnaK mutant of Lactococcus lactis. J Bacteriol. 1998;180:3873–3881. doi: 10.1128/jb.180.15.3873-3881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemos J A C, Burne R A, Castro A C D. Molecular cloning, purification and immunological responses of recombinants GroEL and DnaK from Streptococcus pyogenes. FEMS Immunol Med Microbiol. 2000;28:121–128. doi: 10.1111/j.1574-695X.2000.tb01465.x. [DOI] [PubMed] [Google Scholar]
- 26.Lindquist S, Craig E A. The heat shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 27.Mogk A, Homuth G, Scholz C, Kim L, Schmid F X, Schumann W. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997;16:4579–4590. doi: 10.1093/emboj/16.15.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narberhaus F. Negative regulation of bacterial heat shock genes. Mol Microbiol. 1999;3:1–8. doi: 10.1046/j.1365-2958.1999.01166.x. [DOI] [PubMed] [Google Scholar]
- 29.O'Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 30.Ohta T, Saito K, Kuroda M, Honda K, Hirata H, Hayashi H. Molecular cloning of two new heat shock genes related to the hsp70 genes in Staphylococcus aureus. J Bacteriol. 1994;176:4779–4783. doi: 10.1128/jb.176.15.4779-4783.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons L M, Limberger R J, Shayegani M. Alterations in levels of DnaK and GroEL result in diminished survival and adherence of stressed Haemophilus ducreyi. Infect Immun. 1997;65:2413–2419. doi: 10.1128/iai.65.6.2413-2419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Casal J, Caparon M G, Scott J R. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry D, Kuramitsu H K. Genetic transformation of Streptococcus mutans. Infect Immun. 1981;32:1295–1297. doi: 10.1128/iai.32.3.1295-1297.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quivey R G, Jr, Faustoferri R C, Clancy K A, Marquis R E. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol Lett. 1995;126:257–262. doi: 10.1111/j.1574-6968.1995.tb07427.x. [DOI] [PubMed] [Google Scholar]
- 35.Quivey R G, Jr, Kuhnert W L, Hahn K. Adaptation of oral streptococci to low pH. Adv Microbiol Physiol. 2000;42:239–274. doi: 10.1016/s0065-2911(00)42004-7. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schulz A, Schumann W. hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J Bacteriol. 1996;178:1088–1093. doi: 10.1128/jb.178.4.1088-1093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulz A, Tzschaschel B, Schumann W. Isolation and analysis of mutants of the dnaK operon of Bacillus subtilis. Mol Microbiol. 1995;15:421–429. doi: 10.1111/j.1365-2958.1995.tb02256.x. [DOI] [PubMed] [Google Scholar]
- 39.Svensater G, Sjogreen B, Hamilton I R. Multiple stress responses in Streptococcus mutans and the induction of general and stress-specific proteins. Microbiology. 2000;146:107–117. doi: 10.1099/00221287-146-1-107. [DOI] [PubMed] [Google Scholar]
- 40.Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]