Abstract
Background
Portable, low-field-strength (0.064-T) MRI has the potential to transform neuroimaging but is limited by low spatial resolution and low signal-to-noise ratio.
Purpose
To implement a machine learning super-resolution algorithm that synthesizes higher spatial resolution images (1-mm isotropic) from lower resolution T1-weighted and T2-weighted portable brain MRI scans, making them amenable to automated quantitative morphometry.
Materials and Methods
An external high-field-strength MRI data set (1-mm isotropic scans from the Open Access Series of Imaging Studies data set) and segmentations for 39 regions of interest (ROIs) in the brain were used to train a super-resolution convolutional neural network (CNN). Secondary analysis of an internal test set of 24 paired low- and high-field-strength clinical MRI scans in participants with neurologic symptoms was performed. These were part of a prospective observational study (August 2020 to December 2021) at Massachusetts General Hospital (exclusion criteria: inability to lay flat, body habitus preventing low-field-strength MRI, presence of MRI contraindications). Three well-established automated segmentation tools were applied to three sets of scans: high-field-strength (1.5–3 T, reference standard), low-field-strength (0.064 T), and synthetic high-field-strength images generated from the low-field-strength data with the CNN. Statistical significance of correlations was assessed with Student t tests. Correlation coefficients were compared with Steiger Z tests.
Results
Eleven participants (mean age, 50 years ± 14; seven men) had full cerebrum coverage in the images without motion artifacts or large stroke lesion with distortion from mass effect. Direct segmentation of low-field-strength MRI yielded nonsignificant correlations with volumetric measurements from high field strength for most ROIs (P > .05). Correlations largely improved when segmenting the synthetic images: P values were less than .05 for all ROIs (eg, for the hippocampus [r = 0.85; P < .001], thalamus [r = 0.84; P = .001], and whole cerebrum [r = 0.92; P < .001]). Deviations from the model (z score maps) visually correlated with pathologic abnormalities.
Conclusion
This work demonstrated proof-of-principle augmentation of portable MRI with a machine learning super-resolution algorithm, which yielded highly correlated brain morphometric measurements to real higher resolution images.
© RSNA, 2022
Online supplemental material is available for this article.
See also the editorial by Ertl-Wagner amd Wagner in this issue.
An earlier incorrect version appeared online. This article was corrected on February 1, 2023.
Summary
Synthetic super-resolved images generated by a machine learning algorithm from portable low-field-strength (0.064-T) brain MRI had good agreement with real images at high field strength (1.5–3 T).
Key Results
■ In 24 patients who presented with neurologic symptoms, a machine learning super-resolution algorithm generated 1-mm isotropic synthetic magnetization-prepared rapid gradient-echo high-spatial-resolution images from low-field-strength (0.064-T) brain MRI sequences.
■ Synthetic images had highly correlated region of interest volumes compared with real 1-mm isotropic high-field-strength (1.5–3-T) MRI scans when segmented with automated methods for individual regions (eg, hippocampal volume [r = 0.85], thalamus [r = 0.84], and whole cerebrum [r = 0.92]).
Introduction
A portable, low-field-strength (0.064-T) MRI system has been recently studied (1,2). However, portable MRI has limitations that include a reduced signal-to-noise ratio and limited spatial image resolution (1.6 × 1.6 × 5 mm3) compared with high-field-strength MRI at 1.5–3 T. Therefore, a key goal is to maximize the extraction of available information by transforming lower spatial resolution low-field-strength MRI data into higher spatial resolution images (1 × 1 × 1 mm3 isotropic) while maintaining accuracy.
Quantitative morphometry is central to many neuroimaging studies (3,4). Morphometry of low-field-strength MRI would substantially extend its use for research and clinical neuroimaging. Existing MRI segmentation tools (eg, FreeSurfer, https://surfer.nmr.mgh.harvard.edu [5]; FSL, https://fsl.fmrib.ox.ac.uk/fsl [6]; and SPM, https://www.fil.ion.ucl.ac.uk/spm [7]) have prerequisites in terms of image resolution (typically 1 mm3 isotropic), MRI pulse sequence (often T1 weighted), and signal-to-noise ratio. These prerequisites are often satisfied with a magnetization-prepared rapid gradient-echo acquisition or one of its variants, acquired with high-field-strength MRI. Unfortunately, such requirements are not met by low-field-strength MRI, therefore precluding application of segmentation tools to these data sets.
Modern super-resolution methods use convolutional neural networks (CNNs) to generate a high-spatial-resolution output from low-spatial-resolution input or inputs (8,9). However, application to low-field-strength MRI would require compiling a large data set of paired and accurately aligned low-resolution and high-resolution images, which is difficult because of nonlinear spatial distortions. One common alternative is to down-sample high-spatial-resolution scans to obtain paired images (8). However, this approach may fail when processing low-field-strength MRI because down-sampled high-field-strength MRI does not resemble low-field-strength MRI closely enough. This problem is known as domain shift (10).
Our study presents low-field SynthSR (hereafter, referred to as LF-SynthSR), a method to train a CNN that uses low-field-strength (0.064-T) T1-and T2-weighted brain MRI sequences to generate an image with 1-mm isotropic spatial resolution and the appearence of a magnetization-prepared rapid gradient-echo acquisition. We demonstrate the potential of LF-SynthSR in quantitative neuroradiology by correlating brain morphometry measurements between the synthetic and ground truth high-field-strength images.
Materials and Methods
MRI Data
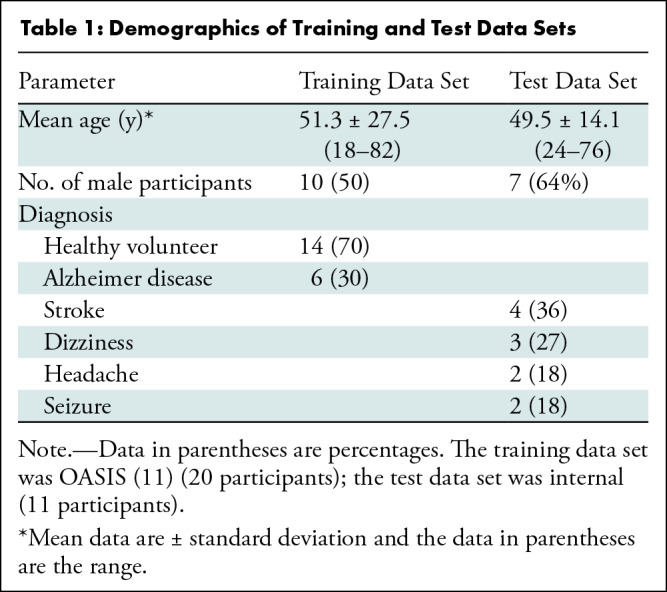
Training data set.—To train the super-resolution CNN, we used a high-field-strength MRI data set composed of 1-mm isotropic magnetization-prepared rapid gradient-echo acquisition scans in 20 participants (10 men; mean age, 51.3 years ± 27.5 [SD]; Table 1) from the Open Access Series of Imaging Studies (11), a neuroimaging, clinical, cognitive, and biomarker data set for normal aging and Alzheimer disease. In addition to the raw images, we used corresponding segmentations of 39 regions of interest (ROIs), as follows: 36 brain ROIs, segmented manually; and three extracerebral ROIs, segmented automatically with a publicly available Bayesian algorithm that is resilient to changes in MRI pulse sequence and segments extracerebral tissue (12). The training set was also artificially augmented to better model pathologic tissue like stroke or hemorrhage (Appendix S1).
Table 1:
Demographics of Training and Test Data Sets
Test data set.—Evaluation relied on secondary analysis of a data set of paired low- and high-field-strength scans from the same participants. The low-field-strength data included T1- and T2-weighted scans acquired with a 0.064-T portable MRI scanner (Swoop; Hyperfine). Reference standard segmentations and associated ROI volumes were obtained by processing high-field-strength MRI scans (1.5–3 T) from the same participants with a publicly available machine learning approach (SynthSeg; https://surfer.nmr.mgh.harvard.edu/fswiki/SynthSeg [13,14]). Details regarding the scanning protocols, pulse sequences, and parameters are in Appendix S1.
Synthesis, super-resolution, and segmentation of low-field-strength MRI.—The training data set was used to train a CNN (a three-dimensional U-net [15,16]) to predict 1-mm isotropic magnetization-prepared rapid gradient-echo acquisition scans from the T1- and T2-weighted low-field-strength MRI examinations in each participant. The proposed method, LF-SynthSR, builds on our previous method, SynthSR (17). Like its predecessor, LF-SynthSR uses a synthetic data generator (Fig 1) to expose the CNN to images of highly varying appearance during training, which yields a CNN that is robust against domain shift.
Figure 1:
Architecture of the proposed convolutional neural network (CNN). The synthetic data generator was based on our previously published technique, SynthSR (17). It used the training set to produce minibatches, which consisted of synthetic low-field-strength MRI and corresponding high-field-strength MRI and high-spatial-resolution segmentations. The super-resolution CNN (a U-net [16], the most widespread segmentation CNN in medical imaging) predicted high-spatial-resolution intensities that were compared with the ground truth high-field-strength MRI to update the CNN weights. In addition, the predicted intensities were fed to a segmentation U-net pretrained to segment 1-mm3 isotropic magnetization-prepared rapid gradient-echo (MPRAGE) scans (with frozen weights), and the output was compared with the ground truth segmentation to inform training. The upper half of the figure (inside the red dotted line) corresponds to the architecture in the original SynthSR publication. The segmentation head (inside the purple dotted line) was added for improving the accuracy of the synthesis.
CNN training minimized an intensity loss equal to the sum of absolute differences between the ground truth and predicted intensities. Compared with the original SynthSR, our method also includes a semantic segmentation loss in the architecture (Fig 1), which uses a pretrained segmentation CNN with so-called frozen weights (ie, not updated during training). This loss ensures accurate synthesis in regions such as the globus pallidum, where subtle intensity differences may lead to large segmentation mistakes. Further details can be found in Appendix S1.
At test time, the low-field-strength MRI data were processed by co-registering the T1- and T2-weighted low-field-strength MRI scans with NiftyReg (18) and feeding them to the trained CNN to obtain the synthetic 1-mm magnetization-prepared rapid gradient-echo acquisition output. These synthetic magnetization-prepared rapid gradient-echo acquisition scans were subsequently segmented with SynthSeg (13,14).
The code to train and apply the CNN is available (github.com/BBillot/SynthSR and github.com/freesurfer/freesurfer/tree/dev/mri_synthsr). A ready-to-use implementation is also available as part of FreeSurfer (surfer.nmr.mgh.harvard.edu/fswiki/SynthSR).
Experimental setup.— We compared the ability of several segmentation tools to generate accurate ROI volumes from the low-field-strength scans, relative to the reference standard volumes obtained from the high-field-strength MRI scans. We considered four publicly available segmentation tools (FreeSurfer [5], SynthSeg [13,14], FSL-FIRST [6], and SAMSEG [12]) directly applied to the low-field-strength MRI and to LF-SynthSR. We note that SAMSEG can jointly exploit T1- and T2-weighted information, whereas FreeSurfer and FSL-FIRST can only use the T1-weighted scans and SynthSeg can only use one scan at the time (either T1- or T2-weighted scans).
We also computed so-called abnormality maps using the technique described in Appendix S1. In short, these are z scores that quantify the statistical deviation of every low-field-strength MRI voxel from the expected image intensities while accounting for partial-volume effects. They provide an additional data layer that quantitatively highlights potential abnormalities on native low-field-strength scans.
Statistical Analysis
The agreement between ROI volumes was measured with the Pearson correlation coefficient and its statistical significance was measured with the Student t test. This agreement was further analyzed with Bland-Altman plots (19), including Kolmogorov-Smirnov tests of normality of the differences. Correlations obtained with different methods were compared with the Steiger Z test for dependent correlations (20). All statistical analyses were performed by using software (Matlab R2020a; MathWorks).
Because the proposed method LF-SynthSR produces images that look similar to real magnetization-prepared rapid gradient-echo acquisition scans, we expect the correlations to be strong (r > 0.70). A sample size calculation with an α value of .05 (one-tailed test) and a β value of .20 yielded the following: 11 images, r value of 0.70; eight images, r value of 0.80; and six images, r value of 0.90.
Results
Data Set Characteristics
The test set consists of imaging data from 24 participants from a prospective observational study performed from August 2020 to December 2021 at Massachusetts General Hospital. Participants who presented with neurologic symptoms and who were scheduled for standard-of-care neuroimaging (ie, high-field-strength MRI) were approached for participation in low-field-strength scanning, therefore yielding paired low-field-strength (0.04-T) and high-field-strength (1.5–3-T) data. Exclusion criteria consisted of an inability to lie flat, a body habitus preventing low-field-strength MRI, and the presence of MRI contraindications. From this 24-person sample, we excluded individuals with incomplete coverage of the cerebrum in the low-field-strength images (five participants), severe motion artifacts (three participants), and large stroke lesion with distortion from mass effect (five participants), which yielded a final sample of 11 participants (mean age, 49.5 years ± 14.1; seven men; two participants with stroke lesions; Table 1). Written consent was obtained from all participants with the approval of the local institutional review board.
Qualitative Results
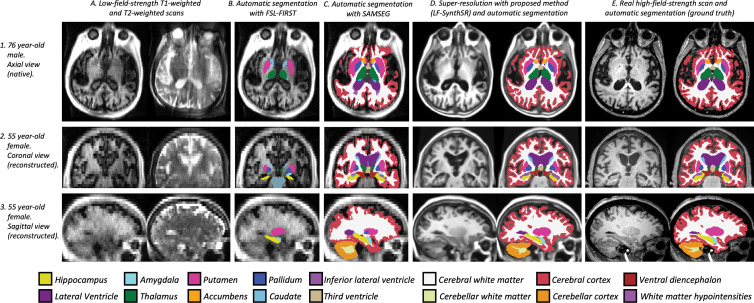
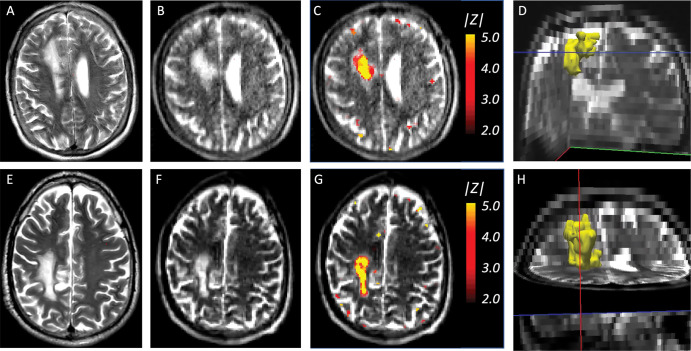
Examples of super-resolution and segmentation of three representative participants are shown in Figure 2. Direct segmentation of the low-field-strength MRI scans with FSL-FIRST and SAMSEG led to mistakes (eg, FSL-FIRST misplaced the basal ganglia and undersegmented the hippocampus). Likewise, SAMSEG could not cope with the low resolution of the scans and segmented most ROIs poorly, especially the convoluted cerebral cortex. LF-SynthSR, however, successfully recovered the high-frequency information that was missing in the low-field-strength scan (especially between axial sections, as highlighted at the coronal and sagittal views) and yielded an automated segmentation that was much closer to the ground truth from the high-field-strength scan.
Figure 2:
Qualitative comparison of the proposed method (LF-SynthSR) with automated segmentation tools in three different participants; image sections are shown in both native (axial) and orthogonal reconstructed projections (coronal, sagittal). Row 1 shows an axial section (native), row 2 shows a coronal section (reconstructed), and row 3 shows a sagittal section (also reconstructed). The images show (A) low-field-strength T1-weighted and T2-weighted MRI scans, automated segmentations produced by (B) FSL-FIRST (6) and (C) SAMSEG (12), (D) the output of LF-SynthSR and its automated segmentation with SynthSeg (13,14), and (E) registered real high-field-strength scan and its automated segmentation with SynthSeg.
Quantitative Results
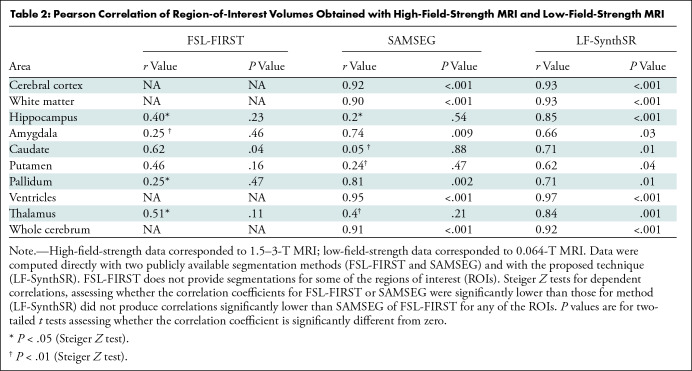
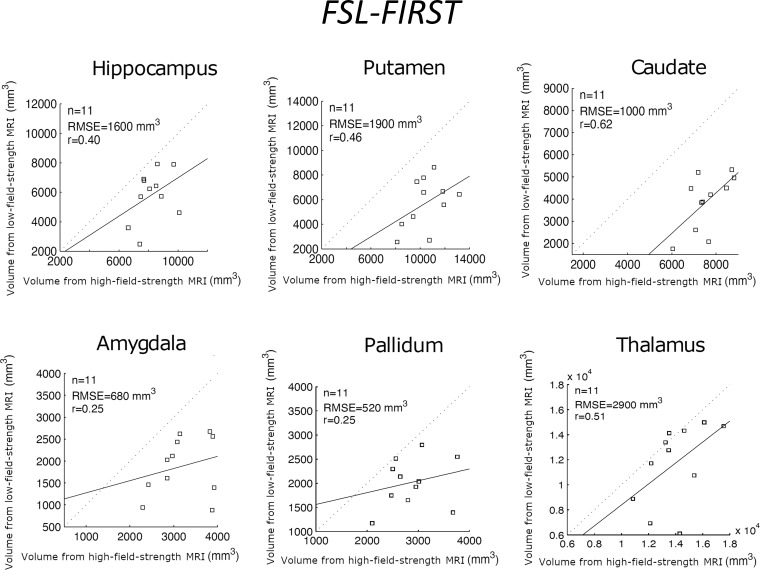
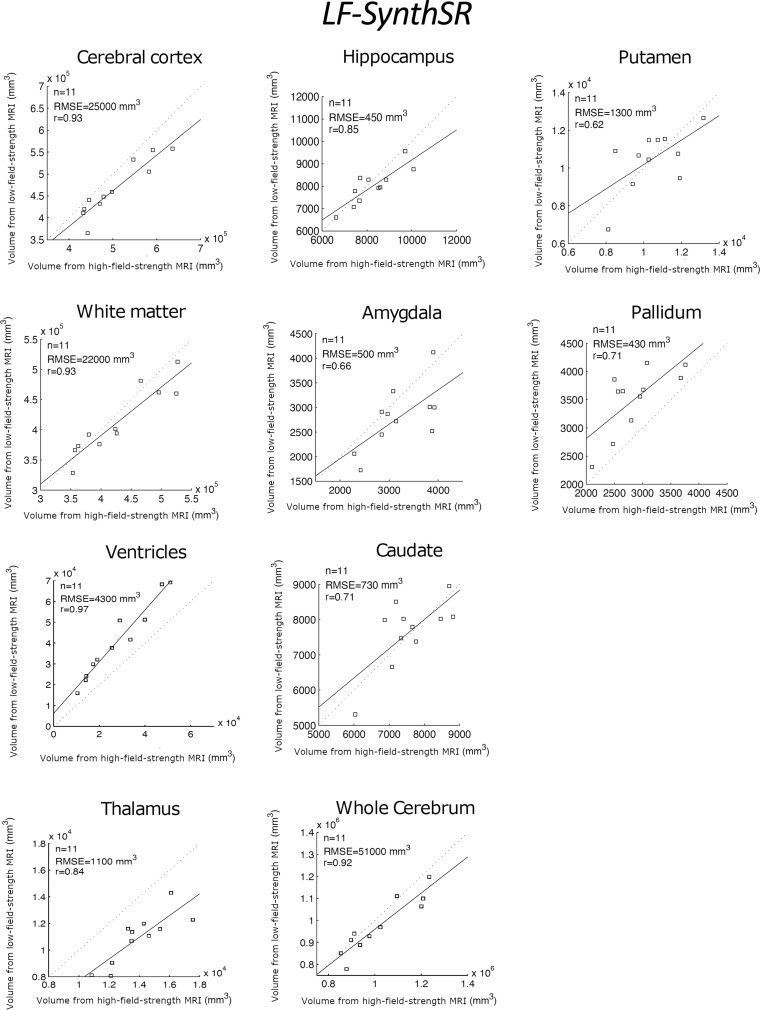
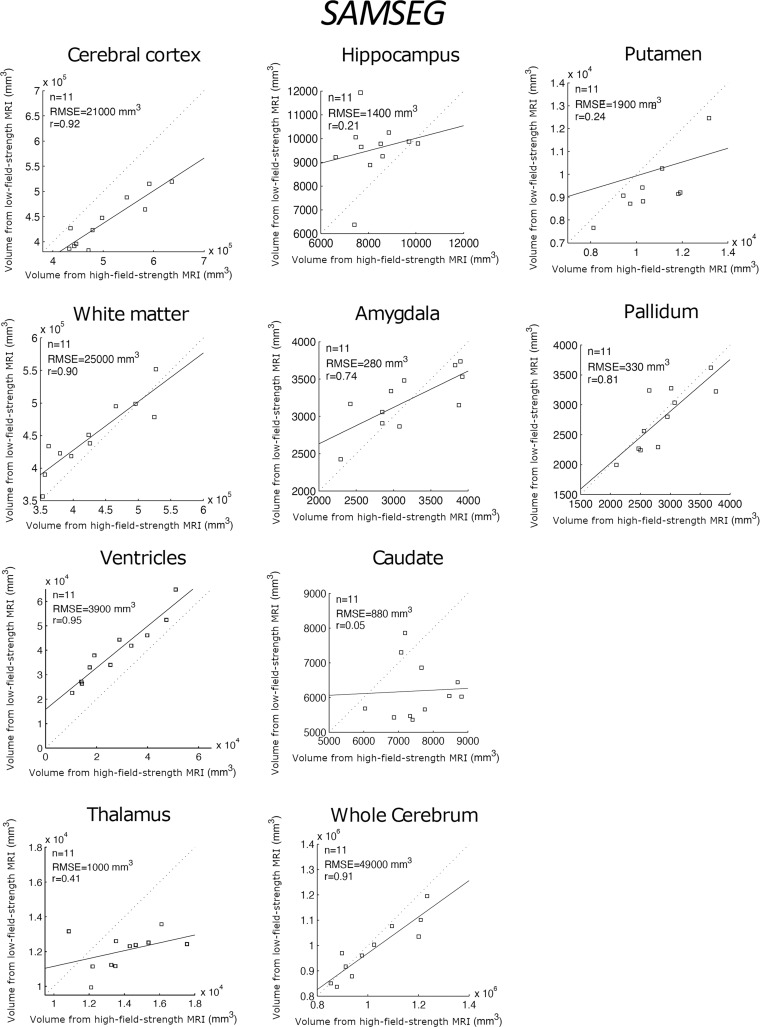
Table 2 shows the correlation between the reference standard volumes (derived from the high-field-strength MRI scans) and the volumes estimated by the different competing methods from the low-field-strength MRI data. Scatterplots for each brain ROI, including the root mean square errors, can be found in Figures 3–5; Bland-Altman plots are shown in Figures S1–S3. FreeSurfer and SynthSeg were unable to produce usable segmentations from the low-field-strength MRI scans and did not yield correlated ROI volumes compared with the reference standard. FSL-FIRST and SAMSEG were able to generate usable segmentations in some ROIs, with volumes that were moderately correlated with the reference standard (Table 2; Figs 3, 4). However, LF-SynthSR was able to generate volumes that were more highly correlated with the ground truth: The correlation was very strong (r > 0.8) for hippocampus, thalamus, ventricles, white matter, and cortex; and moderately strong (r > 0.6) for amygdala, caudate, putamen, and pallidum (Table 2, Fig 5). The Bland-Altman plots also showed that LF-SynthSR exhibited reduced bias: LF-SynthSR showed minor biases in the pallidum, ventricles, and thalamus, whereas FSL-FIRST and SAMSEG showed biases for nearly every ROI.
Table 2:
Pearson Correlation of Region-of-Interest Volumes Obtained with High-Field-Strength MRI and Low-Field-Strength MRI
Figure 3:
Scatterplots for FSL-FIRST, comparing the volumes of regions of interest derived from the high-field-strength MRI scans (ground truth) and from the low-field-strength MRI scans using the publicly available automated segmentation method FSL-FIRST (6). The root mean square error (RMSE) and correlation coefficient (r value) are shown.
Figure 5:
Scatterplots for the proposed technique (LF-SynthSR), comparing the volumes of regions of interest derived from the high-field-strength MRI scans (ground truth) and from the low-field-strength MRI scans using LF-SynthSR. The root mean square error (RMSE) and correlation coefficient (r value) are shown.
Figure 4:
Scatterplots for SAMSEG, comparing the volumes of regions of interest derived from the high-field-strength MRI scans (ground truth) and from the low-field-strength MRI scans using the publicly available automated segmentation method SAMSEG (12). The root mean square error (RMSE) and correlation coefficient (r value) are shown.
Correlation coefficients compared by using a Steiger Z test (P values in Table 2) showed that our method produced significantly higher correlations for the amygdala and pallidum (compared with FSL-FIRST), caudate and putamen (compared with SAMSEG), and hippocampus and thalamus (compared with both). LF-SynthSR did not produce correlations significantly lower than SAMSEG of FSL-FIRST for any of the ROIs.
Finally, Figure 6 shows the abnormality maps for the two participants with stroke lesions. These data layers visually correlated well with the lesions and described them voxel by voxel in a quantitative fashion.
Figure 6:
Stroke lesions and abnormality maps for the two participants with stroke in the test set that satisfied the inclusion criteria. (A) Axial section of a T2-weighted high-field-strength (3-T) MRI scan in a 51-year-old male participant, without contrast ehnancement, shows a stroke lesion. (B) Approximately corresponding section in a T2-weighted low-field-strength (0.064-T) MRI scan in the same participant. (C) Abnormality map (ie, absolute value of Z scores) for the same section, thresholded at |z| = 1.96 (ie, the 95% CI), overlaid on the low-field-strength scan. (D) Three-dimensional rendering of the main cluster of the abnormality map, shows the whole stroke lesion. (E) Coronal section in a 63-year-old female participant, acquired with the same imaging protocol as A, also with a stroke lesion. (F) Approximately corresponding section in low-field-strength scan. (G) Abnormality map thresholded at the 95% CI. (H) Three-dimensional rendering.
Discussion
In this study, we present LF-SynthSR, a technique to train a convolutional neural network, which uses low-field-strength (0.064-T) T1- and T2-weighted brain MRI sequences to generate synthetic images with 1-mm isotropic resolution and the appearance of a magnetization-prepared rapid gradient-echo scan. In a secondary analysis of 11 participants who presented with neurologic symptoms, automated segmentation of the synthetic images yielded region-of-interest volumes that were highly correlated with those derived from real high-field-strength (1.5–3-T) MRI scans, as follows: r value for hippocampal volume, 0.85 (P < .001); r value for thalamus, 0.84 (P = .001); and r value for the whole cerebrum, 0.92 (P < .001). Z score maps of signal intensities visually correlated well with pathologic findings.
Compared with LF-SynthSR, well-established segmentation techniques such as FSL-FIRST and SAMSEG struggled to find the boundaries of ROIs because of the poor spatial resolution of the scans, rendering their output unusable. FSL-FIRST produced P values greater than .05 for the correlation of every ROI other than the caudate, and SAMSEG produced significant correlations (P < .05) for some structures (amygdala and pallidum, and larger structures like the cortex, white matter, and ventricles), but the correlations for the remaining ROIs (hippocampus, caudate, putamen, and thalamus) were weak and not significant (P < .05).
LF-SynthSR may improve the image quality of low-field-strength MRI scans to the point that they are usable not only by automated segmentation methods but potentially also with registration and classification algorithms. The quantitative results demonstrated that the volume correlations were significant (P < .05) for all ROIs, with strong correlations for the large ROIs (cortex, white matter, ventricles, and whole brain: r > 0.9; hippocampus: r = 0.85; and thalamus: r = 0.84) and strong correlations for all other structures (r > 0.6 in all cases). We further leveraged LF-SynthSR to highlight areas of pathologic findings by generating an additional data layer that quantifies the statistical deviation from the expected image intensities. This approach could be used to augment the detection of abnormal lesions.
Our study had three main limitations. The sample size was small because of the limited number of participants with paired low-field-strength or high-field-strength MRI data. Second, the validation was limited to a single downstream task (segmentation), which is a quantitative proxy for the goal of detecting clinically relevant findings. Third, our method did not estimate the uncertainty in the output.
In conclusion, LF-SynthSR overcomes the limitations of previous super-resolution tools with low-field-strength MRI and enables quantitative morphometry of low-field-strength MRI. Future work will need to include the incorporation of uncertainty estimates to improve the reliability of our method and its extension to enhance the detection of normal and abnormal findings on images.
Acknowledgments
Acknowledgments
M.S.R. acknowledges the gracious support of the Kiyomi and Ed Baird MGH Research Scholar Award.
M.S.R. and W.T.K. are co-senior authors.
K.N.S., M.S.R., and W.T.K. supported by the American Heart Association (Collaborative Science Award 17CSA3355004). K.N.S. supported by the National Institutes of Health (U24NS107136, U24NS107215, R01NR018335, U01NS106513), the American Heart Association (18TPA34170180, 17CSA33550004), and a Hyperfine Research grant. W.T.K. supported by the National Institutes of Health (R01 NS099209 and R21 NS120002) and American Heart Association (17CSA33550004 and 20SRG35540018). J.E.I. supported by the National Institutes of Health (1R01AG070988), the BRAIN Initiative (1RF1MH123195), the European Research Council (starting grant 677697, project BUNGE-TOOLS) and Alzheimer’s Research UK (interdisciplinary grant ARUK-IRG2019A-003). The Hyperfine device was provided to Massachusetts General Hospital as part of a research agreement.
Disclosures of conflicts of interest: J.E.I. No relevant relationships. R.S. No relevant relationships. S.L. No relevant relationships. B.B. No relevant relationships. P.S. No relevant relationships. B.M. Support for travel from MGH Department of Emergency Medicine. J.N.G. Grants from Pfizer, Takeda, Octapharma; consulting fees from CSL Behring, Alexion, NControl; stock or stock options in NControl, Cayuga. K.N.S. No relevant relationships. M.S.R. Grant from GE Healthcare; patent royalties from MGH; consulting fees from Synex Medical, Foqus, Nanalysis, O2M Technologies; patents at MGH; serves on the executive committee of the Experimental NMR Conference; founder and equity holder in Hyperfine, Vizma Life Sciences, Intact Data Service, Q4ML. W.T.K. Consulting fees from NControl Therapeutics; patent no. 16/486,687 pending; associate editor, Neurotherapeutics Journal; stock or stock options in Woolsey Pharmaceuticals; receipt of equipment, materials, drugs, medical writing, gifts, or other services from NControl Therapeutics.
Abbreviations:
- CNN
- convolutional neural network
- ROI
- region of interest
References
- 1. Sheth KN , Mazurek MH , Yuen MM , et al . Assessment of brain injury using portable, low-field magnetic resonance imaging at the bedside of critically ill patients . JAMA Neurol 2020. ; 78 ( 1 ): 41 – 47 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mazurek MH , Cahn BA , Yuen MM , et al . Portable, bedside, low-field magnetic resonance imaging for evaluation of intracerebral hemorrhage . Nat Commun 2021. ; 12 ( 1 ): 5119 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Frangou S , Modabbernia A , Williams SCR , et al . Cortical thickness across the lifespan: Data from 17,075 healthy individuals aged 3-90 years . Hum Brain Mapp 2022. ; 43 ( 1 ): 431 – 451 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dima D , Modabbernia A , Papachristou E , et al . Subcortical volumes across the lifespan: Data from 18,605 healthy individuals aged 3-90 years . Hum Brain Mapp 2022. ; 43 ( 1 ): 452 – 469 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fischl B , Salat DH , Busa E , et al . Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain . Neuron 2002. ; 33 ( 3 ): 341 – 355 . [DOI] [PubMed] [Google Scholar]
- 6. Patenaude B , Smith SM , Kennedy DN , Jenkinson M . A Bayesian model of shape and appearance for subcortical brain segmentation . Neuroimage 2011. ; 56 ( 3 ): 907 – 922 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ashburner J , Friston KJ . Unified segmentation . Neuroimage 2005. ; 26 ( 3 ): 839 – 851 . [DOI] [PubMed] [Google Scholar]
- 8. Wang Z , Chen J , Hoi SC . Deep Learning for Image Super-Resolution: A Survey . IEEE Trans Pattern Anal Mach Intell 2021. ; 43 ( 10 ): 3365 – 3387 . [DOI] [PubMed] [Google Scholar]
- 9. Yang W , Zhang X , Tian Y , Wang W , Xue JH , Liao Q . Deep learning for single image super-resolution: A brief review . IEEE Trans Multimed 2019. ; 21 ( 12 ): 3106 – 3121 . [Google Scholar]
- 10. Wang M , Deng W . Deep visual domain adaptation: A survey . Neurocomputing 2018. ; 312 : 135 – 153 . [Google Scholar]
- 11. Marcus DS , Wang TH , Parker J , Csernansky JG , Morris JC , Buckner RL . Open Access Series of Imaging Studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults . J Cogn Neurosci 2007. ; 19 ( 9 ): 1498 – 1507 . [DOI] [PubMed] [Google Scholar]
- 12. Puonti O , Iglesias JE , Van Leemput K . Fast and sequence-adaptive whole-brain segmentation using parametric Bayesian modeling . Neuroimage 2016. ; 143 : 235 – 249 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Billot B , Greve DN , Van Leemput K , Fischl B , Iglesias JE , Dalca A . A Learning Strategy for Contrast-agnostic MRI Segmentation . Proc Mach Learn Res 2020. ; 121 : 75 – 93 . https://proceedings.mlr.press/v121/billot20a.html . [Google Scholar]
- 14. Billot B , Robinson E , Dalca AV , Iglesias JE . Partial Volume Segmentation of Brain MRI Scans of Any Resolution and Contrast . In: Martel AL , Abolmaesumi P , Stoyanov D , et al . eds . Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science, vol 12267 . Cham, Switzerland: : Springer; , 2020. ; 177 – 187 . [Google Scholar]
- 15. Çiçek Ö , Abdulkadir A , Lienkamp SS , Brox T , Ronneberger O . 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation . In: Ourselin S , Joskowicz L , Sabuncu M , Unal G , Wells W , eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science, vol 9901 . Cham, Switzerland: : Springer; , 2016. ; 424 – 432 . [Google Scholar]
- 16. Ronneberger O , Fischer P , Brox T . U-Net: Convolutional Networks for Biomedical Image Segmentation . In: Navab N , Hornegger J , Wells WM , Frangi AF , eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, vol 9351 . Cham, Switzerland: : Springer; , 2015. ; 234 – 241 . [Google Scholar]
- 17. Iglesias JE , Billot B , Balbastre Y , et al . Joint super-resolution and synthesis of 1 mm isotropic MP-RAGE volumes from clinical MRI exams with scans of different orientation, resolution and contrast . Neuroimage 2021. ; 237 : 118206 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Modat M , Cash DM , Daga P , Winston GP , Duncan JS , Ourselin S . Global image registration using a symmetric block-matching approach . J Med Imaging (Bellingham) 2014. ; 1 ( 2 ): 024003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Altman DG , Bland JM . Measurement in medicine: the analysis of method comparison studies . Statistician 1983. ; 32 ( 3 ): 307 – 317 . [Google Scholar]
- 20. Steiger JH . Tests for comparing elements of a correlation matrix . Psychol Bull 1980. ; 87 ( 2 ): 245 – 251 . [Google Scholar]
- 21. Milletari F , Navab N , Ahmado SA . V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation . In: 2016 Fourth International Conference on 3D Vision (3DV) , Stanford, CA , October 25–28, 2016 . Piscataway, NJ: : IEEE; , 2016. ; 565 – 571 . [Google Scholar]
- 22. Clevert DA , Unterthiner T , Hochreiter S . Fast and Accurate Deep Network Learning by Exponential Linear Units (ELUs) . arXiv preprint arXiv:1511.07289. https://arxiv.org/abs/1511.07289. Posted November 23, 2015. Accessed October 13, 2022 . [Google Scholar]
- 23. Abadi M , Barham P , Chen J , et al . Tensorflow: A system for large-scale machine learning . In: OSDI’16: Proceedings of the 12th USENIX conference on Operating Systems Design and Implementation , November 2016 . New York, NY: : Association for Computing Machinery; , 2016. ; 265 – 283 . https://dl.acm.org/doi/10.5555/3026877.3026899 . [Google Scholar]
- 24. Leys C , Ley C , Klein O , Bernard P , Licata L . Detecting outliers: Do not use standard deviation around the mean, use absolute deviation around the median . J Exp Soc Psychol 2013. ; 49 ( 4 ): 764 – 766 . [Google Scholar]
- 25. Dempster AP , Laird NM , Rubin DB . Maximum likelihood from incomplete data via the EM algorithm . J R Stat Soc Ser B Methodol 1977. ; 39 ( 1 ): 1 – 22 . https://www.jstor.org/stable/2984875 . [Google Scholar]
- 26. Prados F , Cardoso MJ , Kanber B , et al . A multi-time-point modality-agnostic patch-based method for lesion filling in multiple sclerosis . Neuroimage 2016. ; 139 : 376 – 384 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chard DT , Jackson JS , Miller DH , Wheeler-Kingshott CA . Reducing the impact of white matter lesions on automated measures of brain gray and white matter volumes . J Magn Reson Imaging 2010. ; 32 ( 1 ): 223 – 228 . [DOI] [PubMed] [Google Scholar]
- 28. Valverde S , Oliver A , Lladó X . A white matter lesion-filling approach to improve brain tissue volume measurements . Neuroimage Clin 2014. ; 6 : 86 – 92 . [DOI] [PMC free article] [PubMed] [Google Scholar]



![Architecture of the proposed convolutional neural network (CNN). The synthetic data generator was based on our previously published technique, SynthSR (17). It used the training set to produce minibatches, which consisted of synthetic low-field-strength MRI and corresponding high-field-strength MRI and high-spatial-resolution segmentations. The super-resolution CNN (a U-net [16], the most widespread segmentation CNN in medical imaging) predicted high-spatial-resolution intensities that were compared with the ground truth high-field-strength MRI to update the CNN weights. In addition, the predicted intensities were fed to a segmentation U-net pretrained to segment 1-mm3 isotropic magnetization-prepared rapid gradient-echo (MPRAGE) scans (with frozen weights), and the output was compared with the ground truth segmentation to inform training. The upper half of the figure (inside the red dotted line) corresponds to the architecture in the original SynthSR publication. The segmentation head (inside the purple dotted line) was added for improving the accuracy of the synthesis.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/4bf8/9968773/92285f020c58/radiol.220522.fig1.jpg)