Abstract
A gene cluster containing the mevalonate pathway genes (open reading frame 2 [ORF2] to ORF7) for the formation of isopentenyl diphosphate and a geranylgeranyl diphosphate (GGDP) synthase gene (ORF1) had previously been cloned from Streptomyces griseolosporeus strain MF730-N6, a diterpenoid antibiotic, terpentecin (TP) producer (Y. Hamano, T. Dairi, M. Yamamoto, T. Kawasaki, K Kaneda, T. Kuzuyama, N. Itoh, and H. Seto, Biosci. Biotech. Biochem. 65:1627–1635, 2001). Sequence analysis in the upstream region of the cluster revealed seven new ORFs, ORF8 to ORF14, which were suggested to encode TP biosynthetic genes. We constructed two mutants, in which ORF11 and ORF12, which encode a protein showing similarities to eukaryotic diterpene cyclases (DCs) and a eubacterial pentalenene synthase, respectively, were inactivated by gene disruptions. The mutants produced no TP, confirming that these cyclase genes are essential for the production of TP. The two cyclase genes were also expressed in Streptomyces lividans together with the GGDP synthase gene under the control of the ermE* constitutive promoter. The transformant produced a novel cyclic diterpenoid, ent-clerod-3,13(16),14-triene (terpentetriene), which has the same basic skeleton as TP. The two enzymes, each of which was overproduced in Escherichia coli and purified to homogeneity, converted GGDP into terpentetriene. To the best of our knowledge, this is the first report of a eubacterial DC.
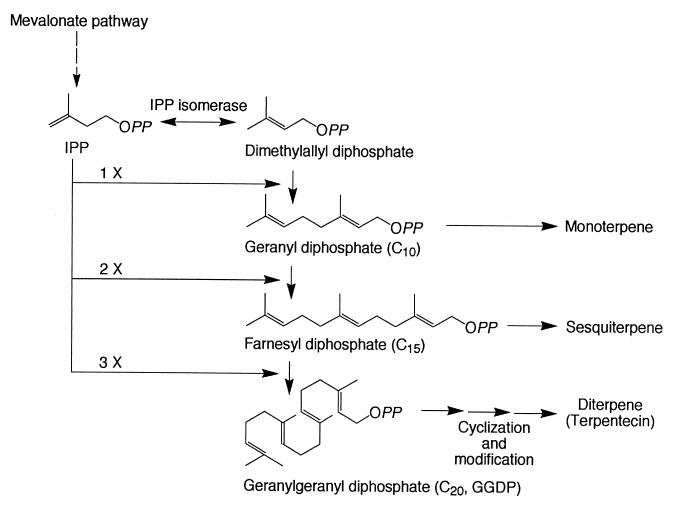
Isoprenoids are the largest single family of compounds found in nature, with over 22,000 known examples (12), and can be classified into several groups based on the number of C5 units derived from isopentenyl diphosphate (IPP), such as monoterpenes (C10), sesquiterpenes (C15), diterpenes (C20), and triterpenes (C30), etc. (12). These compounds are biosynthesized from the corresponding prenyl diphosphate. Geranyl diphosphate gives rise to monoterpenes, farnesyl diphosphate gives rise to sesquiterpenes, and geranylgeranyl diphosphate (GGDP) gives rise to diterpenes (9, 29) (Fig. 1). In many cases, these prenyl diphosphates undergo a range of cyclizations to produce the parent skeletons of each class, followed by a variety of modifications to give many thousands of different isoprenoid metabolites (9, 29).
FIG. 1.
Formation of different isoprenoid metabolites. OPP, diphosphate.
A variety of isoprenoid synthases (cyclases), most of which are from plants and fungi, have been purified and extensively studied (12). As for the genes encoding isoprenoid cyclases, more than 30 eukaryotic isoprenoid synthases have been cloned as cDNAs (9, 29). On the other hand, there have been few reports about eubacterial isoprenoid cyclases and genes because the vast majority of isoprenoids are produced by eukaryotes. Pentalenene synthase, a sesquiterpene cyclase from a Streptomyces strain (10), and squalene-hopene cyclases, triterpene cyclases from bacteria, are the only examples (22, 32, 34, 35, 42, 46). There are no reports, to the best of our knowledge, about eubacterial monoterpene cyclases and diterpene cyclases (DCs).
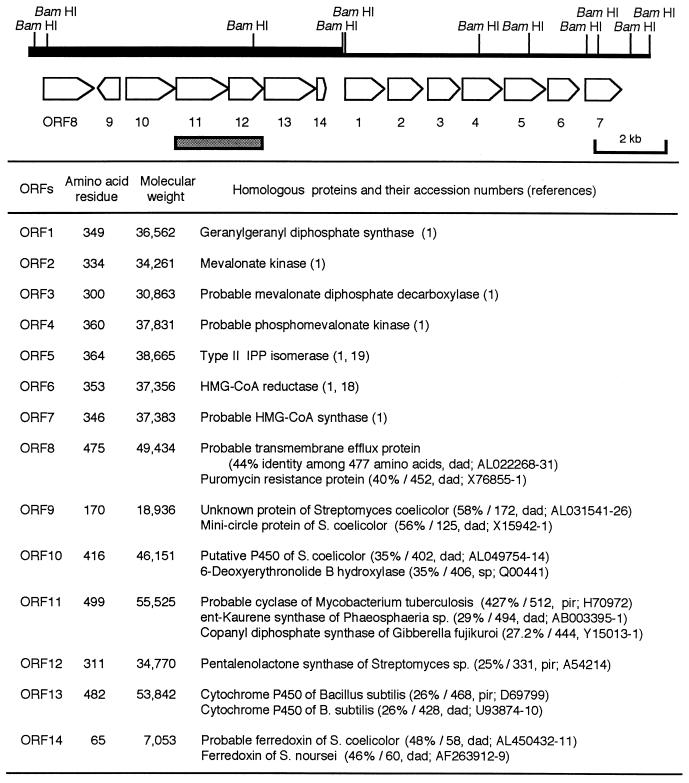
We have been studying the biosynthesis of isoprenoid antibiotics produced by actinomycetes. Although actinomycetes produce approximately 70% of all natural compounds, a very limited number of isoprenoid compounds are known to be produced by them (30). The gene cluster containing the mevalonate pathway genes used to synthesize IPP had previously been cloned from Streptomyces griseolosporeus strain MF730-N6, a diterpene antibiotic terpentecin (TP) producer (15). The GGDP synthase gene encoding the enzyme catalyzing the formation of GGDP, which is the direct precursor of TP, was also identified in the upstream region of the mevalonate pathway gene cluster (15) (open reading frame 1 [ORF1] in Fig. 2). Considering that the biosynthetic genes for almost all of the antibiotics produced by actinomycetes are known to be clustered in the genomic DNA region (11, 27), the TP biosynthetic genes are also expected to exist in the flanking region of the GGDP synthase gene.
FIG. 2.
ORFs deduced by sequencing analysis. The thick and thin bars represent the DNA fragments used for sequencing analysis in this study and in the previous study, respectively. A gray box shows the DNA fragment used for Northern blot analysis. The arrows indicate the extents and directions of the ORFs. CoA, coenzyme A; HMG, 3-hydroxy-3-methylglutaryl; dad, DNA Data Bank of Japan; sp, Swissprot; pir, Protein Identification Resource.
In this paper, we describe the cloning and sequencing analysis of seven genes that were newly found in the flanking regions of the mevalonate pathway gene cluster of strain MF730-N6. In particular, we focused on the two cyclase genes, which encode proteins showing similarities to eukaryotic DCs (ORF11) and a eubacterial pentalenene synthase (ORF12). Mutant constructions to examine if these genes would encode the TP biosynthetic enzymes and heterologous expression of the cyclase genes in Streptomyces lividans and Escherichia coli are mainly described.
MATERIALS AND METHODS
Chemicals.
[α-32P]dCTP was obtained from Amersham. GGDP was purchased from Sigma. The other chemicals used were all analytical grade.
Bacterial strains.
S. griseolosporeus strain MF730-N6 (a TP producer), which was formerly classified in the genus Kitasatosporia (41), was used for the cloning experiment. The media and growth conditions used for strain MF730-N6 were those described by Tamamura et al. (41). S. lividans TK23 (17) and pWHM860 (a gift from C. R. Hutchinson, Kosan Biosciences Inc., Hayward, Calif.), a derivative of plasmid pWHM3 (44), were used for heterologous expression of the TP biosynthetic genes. E. coli M15/pREP4 and plasmid pQE30 (Qiagen) were used for the expression of His-tagged proteins. E. coli JM110 (rpsL thr leu thi lacY galK ara tonA tsx dam dcm supE44/F′ [traD proAB lacIqΔM15]) (Toyobo, Osaka, Japan) and plasmids pUC118 and pUC119 were used for sequencing analysis. If necessary, ampicillin was added to the medium to a final concentration of 100 μg/ml.
DNA isolation and manipulation.
Plasmids from E. coli were prepared by using a Qiagen Plasmid Kit (QIAGEN, Inc., Chatsworth, Calif.). All restriction enzymes, T4 ligase, and calf intestinal alkaline phosphatase were obtained from Toyobo and used in accordance with the manufacturer's protocols. Transformation of E. coli with plasmid DNA by electroporation was performed under standard conditions by using a BTX ECM 600 electroporation system (Biotechnologies and Experimental Research, Inc., San Diego, Calif.). The transformation protocols used for S. lividans and S. griseolosporeus were essentially the same as those described by Hopwood et al. (17) and Dairi et al. (13), respectively. Other general procedures were performed as described by Maniatis et al. (26).
Sequence analysis.
A cosmid clone, pSG003 (15), which carried the mevalonate pathway gene cluster and its flanking region, was used for sequencing analysis. Several restriction enzyme-digested fragments were subcloned to pUC118 or pUC119. After construction of a series of plasmids, sequencing was done by the dideoxy-chain termination method of Sanger et al. (37) with an automated DNA sequencer (Li-cor model 4000L). Finally, the nucleotide sequence of an 8.5-kb BamHI fragment was determined.
Disruption of ORF11 and ORF12.
A 5.9-kb SacI fragment carrying the ORF11 and ORF12 genes was subcloned into pGEM5Zf(+) (Promega, Madison, Wis.) to give pCYC1. The plasmid was digested with SmaI, which existed in the inserted DNA, and the site was changed into a HindIII site by a linker to introduce a frameshift mutation into the ORF11 gene. The plasmid was digested with SacI, and the resultant fragment was subcloned into the same sites of pEN101 (encodes thiostrepton resistance) (13) to give pCYC1FS. Plasmid pCYC1 was digested with BstEII, treated with the Klenow fragment of DNA polymerase I to fill in the sticky ends of this site, and self-ligated, in which process ORF12 was inactivated by a frameshift mutation. The plasmid was digested with SacI, and the resultant fragment was subcloned into the same sites of pEN101 to give pCYC2FS. Plasmids pCYC1FS and pCYC2FS were independently introduced into strain MF730-N6, and thiostrepton-resistant colonies were selected. After protoplasting and regeneration of the transformant, thiostrepton-sensitive colonies were collected. Among them, a frameshifted mutation in ORF11 or ORF12 was confirmed by Southern blot analysis.
Complementation of the cyclase gene-inactivated mutant.
To complement the ORF11- and ORF12-inactivated mutants, two plasmids, pEN-ORF11 and pEN-ORF12, which carried the ORF11 gene and the ORF12 gene, respectively, were constructed. To obtain the entire ORF11 gene without the excessive flanking region, PCR amplification was carried out under standard conditions. The 5′ and 3′ primers with an additional restriction site (underlined) had the respective sequences 5′-ATCAGCCGACGCTTCTAGACGCTCCCCGTC-3′ (ORF11-N) and 5′-AAGCCGCGAGCAAGCTTTCGGCATCC-3′ (ORF11-C). After sequence confirmation, the XbaI-HindIII fragment was inserted into the same sites of pWHM860 to give pWHM-ORF11, in which the ORF11 gene was expressed under the control of the ermE* promoter. The plasmid was digested with HindIII, and the site was changed into a BglII site by a linker. The plasmid, which had two BglII sites (one is the site newly created by the linker, and the other existed in the upstream region of the ermE* promoter), was digested with BglII, and the resultant fragment was subcloned into the same site of pEN101 (13) to give pEN-ORF11. Plasmid pEN-ORF12 was constructed by the same method as pEN-ORF11 by using the primers 5′-ATGGGCAAGGACCTCTAGACGCCTTTCCGG-3′ (ORF12-N) and 5′-GTAACGGACGGCAAGCTTCTCGGATCGAGC-3′ (ORF12-C). Plasmids pEN-ORF11 and pEN-ORF12 were introduced into the ORF11-inactivated mutant and the ORF12-inactivated mutant, respectively, and TP productivity was examined.
Expression of the cyclase genes in S. lividans.
A 1.2-kb BamHI-HincII fragment carrying the GGDP synthase gene was inserted into the BamHI-SmaI sites of pGEM7Z (Promega). The BamHI-XbaI fragment (the latter site existed in multilinker sites) was subcloned into the same site of pWHM860 to construct pWHM-GGDP. The entire cyclase genes (ORF11 and ORF12) was amplified by PCR with the ORF11-N primer and the ORF12-C primer. After sequence confirmation, the XbaI-HindIII fragment was inserted into the same sites of pWHM-GGDP to give pWHM-TER1, in which the GGDP synthase gene and the two cyclase genes were expressed under the control of the ermE* promoter.
Isolation of a compound produced by an S. lividans transformant.
S. lividans harboring pWHM-TER1 was grown in many 300-ml Erlenmeyer flasks containing SK-no. 2 medium (30 ml) (14) and thiostrepton (10 μg/ml). Fermentation was carried out for 7 days at 30°C with agitation (200 rpm). The culture broth (15 liters) was centrifuged, and the precipitated mycelial cake was suspended in 15 liters of acetone. After vigorous shaking, the suspension was filtered and the acetone filtrate was concentrated to dryness in vacuo. The dried material was dissolved in 200 ml of chloroform and water (1:1). After centrifugation to separate the emulsion, the organic layer was recovered. The aqueous layer was extracted twice with chloroform. The combined organic layer was evaporated to dryness under reduced pressure. The dried material was dissolved in a small volume of chloroform and acetone (1:1) and subjected to chromatography on thin-layer chromatography plates (silica gel 60F254; Merck) developed in hexane. The material (Rf, 0.47 to 0.68) was subjected to extraction with acetone, followed by filtration and concentrated to dryness. The dried material was dissolved in a small volume of acetonitrile and then fractionated by preparative high-performance liquid chromatography (HPLC; Merck Mightisil RP-8 column, 250 by 20 mm; mobile phase, 100% acetonitrile; flow rate, 5 ml/min; detection wavelength, 210 nm).
NMR spectroscopy.
1H and 13C nuclear magnetic resonance (NMR) spectra were recorded at 500 and 125 MHz, respectively, by using a JEOL A500 spectrometer. One- and two-dimensional experiments (correlation spectroscopy [COSY], nuclear Overhauser and exchange spectroscopy [NOESY], heteronuclear single quantum coherence [HSQC], heteronuclear multi quantum coherence [HMQC], and a difference decoupling experiment) were performed at ambient temperature. The sample (4.4 mg) was dissolved in 0.35 ml of CDCl3.
Mass spectroscopy.
High-resolution electron ionization (EI) mass spectra were obtained by using a JEOL HX-110 mass spectrometer.
Structure determination.
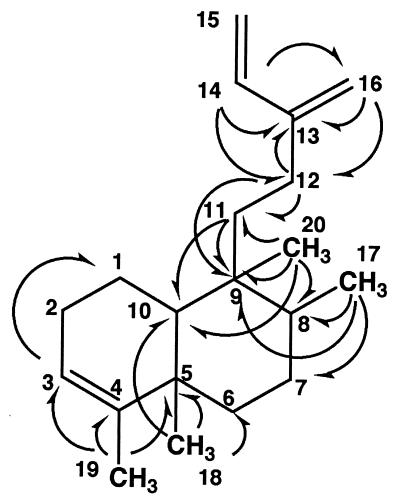
The molecular formula of terpentetriene was determined to be C20H32 by high-resolution EI mass spectral data (observed Mr, 272.2547; calculated Mr, 272.2504), which indicated the degree of unsaturation of terpentetriene to be 5. The 13C-NMR spectral data of terpentetriene (Table 1) showed the presence of four CH3, six CH2, two CH, two C, two CH2⩵, two CH⩵, and two C⩵ groups. These data, together with its molecular formula, indicated that the hydrocarbon terpentetriene is a diterpenoid with two rings. The 1H- and 13C-NMR spectral data obtained by phase-sensitive COSY, HMQC, and heteronuclear multiple-bond connectivity (HMBC) experiments (Fig. 3 and Table 1) disclosed partial structures compatible with the planar structure shown in Fig. 3, although conclusive evidence was not available due to the overlapping of proton signals at around 1.44 and 1.65 δ.
TABLE 1.
1H- and 13C-NMR data of terpentetriene and ent-clerod-3,13(16),14-triene
| Group no.a, group | Terpentetriene
|
ent-clerod- 3,13(16), 14-triene
|
|
|---|---|---|---|
| C-13 | H-1b | C-13 | |
| 1, CH2 | 17.85 | ca. 1.42 (m), ca. 1.64 (m) | 18.3 |
| 2, CH2 | 26.85 | ca. 1.97 (m), ca. 2.04 (m) | 26.9 |
| 3, CH⩵ | 120.24 | 5.143 (br. s) | 120.4 |
| 4, C⩵ | 144.55 | 144.5 | |
| 5, C | 38.31 | 38.2 | |
| 6, CH2 | 30.20 | ca. 1.435 (13, 13, 6), ca. 1.46 (m) | 38.8 |
| 7, CH2 | 25.59 | 1.299 (14, 3, 3, 3), ca. 1.95 (13, 4, 4) | 27.5 |
| 8, CH | 35.16 | ca. 1.65 (m) | 38.3 |
| 9, C | 37.69 | 38.8 | |
| 10, CH | 45.13 | ca. 1.43 (m) | 46.5 |
| 11, CH2 | 38.31 | 1.179 (13, 13, 5), ca. 1.63 (m) | 37.5 |
| 12, CH2 | 24.57 | 2.136 (14, 14, 5), 2.185 (14, 14, 5) | 24.7 |
| 13, C⩵ | 147.75 | 147.6 | |
| 14, CH⩵ | 139.24 | 6.372 (18, 11) | 139.0 |
| 15, CH2⩵ | 113.02 | 5.242 (18, br), 5.043 (11, br) | 112.9 |
| 16, CH2⩵ | 115.24 | 4.99 (s) | 115.5 |
| 17, CH3 | 14.88 | 0.96 (7) | 16.1 |
| 18, CH3 | 20.63 | 1.051 (s) | 20.0 |
| 19, CH3 | 18.01 | 1.582 (br. s) | 16.0 |
| 20, CH3 | 20.38 | 0.968 (s) | 18.4 |
FIG. 3.
Structure of terpentetriene revealed by NMR spectral analysis. Arrows indicate 1H-13C long-range couplings observed by HMBC experiments.
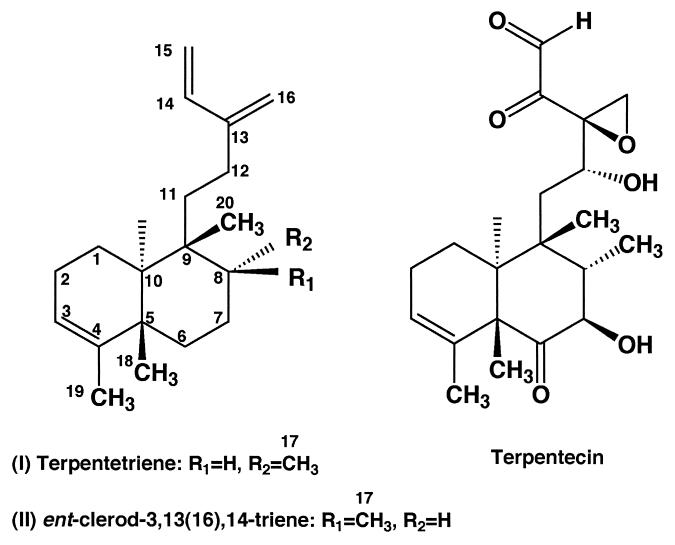
In view of the carbon skeleton of the diterpenoid TP and its biosynthetic formation mechanism, we assumed that terpentetriene might be a diterpenoid possessing the same carbon skeleton as TP. Thus, we compared the 13C-NMR spectral data of terpentetriene with those of ent-clerod-3,13(16),14-triene (31), which was found by a database search to have the same planar structure. As summarized in Table 1, the carbon chemical shifts of ring A (C-1 to -5), its appended methyls (C-18 and -19), and the side chain (C-12 to -16) were completely identical between terpentetriene and ent-clerod-3,13(16),14-triene. The chemical shifts of the remaining carbons of ring B (C-6 to -10) and its appended methyls (C-17 and -20), however, are different, suggesting that they are stereoisomers of each other at C-8 and/or C-9. A strong nuclear Overhauser effect (NOE) observed between the two methyls C-18 and -20 but not between the methyl C-18 and methylene C-11 of terpentetriene confirmed that these two methyl groups are both in an axial orientation. Thus, the only difference between terpentetriene and ent-clerod-3,13(16),14-triene was the stereochemistry at C-8, with the methyl C-17 of terpentetriene being in the axial orientation, as shown in Fig. 4. This conclusion was corroborated by a large upfield shift of C-6 caused by a strong γ effect of the axial methyl group (C-17). Another conclusive evidence was obtained by a difference-decoupled spectrum prepared by irradiation of C-17 methyl protons. H-8 was observed as a triplet with J = 3.5 Hz, showing that this proton was in an equatorial orientation. Thus, the structure of terpentetriene was determined to be a stereoisomer at C-8 of ent-clerod-3,13(16),14-triene, as shown in Fig. 4. The identical stereostructures of terpentetriene and TP strongly suggest that terpentetriene is a biosynthetic intermediate of TP and its absolute stereochemistry is reasonably assumed as shown in Fig. 4 based on the established absolute stereochemistry of TP (43).
FIG. 4.
Structures of terpentetriene (I), ent-clerod-3,13(16),14-triene (II), and TP.
Overproduction of the cyclases in E. coli.
Two sets of primers were used to amplify ORF11 and ORF12. The primers were designed as follows (restriction sites are underlined): ORF11, 5′-CGCGGATCCAAGGACCGCGCTGCCGACCCG-3′ (forward) and 5′-CGCGGATCCTCAGTACCTGCCCACGACGGC-3′ (reverse); ORF12, 5′-CGGGGTACCCCCGACGCGATCGAGTTCGAG-3′ (forward) and 5′-CGGGGTACCTCAGCGGTAGCGGTTCGTCTC-3′ (reverse). PCR was carried out under standard conditions. After sequence confirmation, a 1.5-kb BamHI fragment (ORF11) and a 940-bp KpnI fragment (ORF12) were each inserted into the same site of pQE30. Plasmids pQE30-ORF11 and pQE30-ORF12, in which recombinant proteins were expressed as N-terminal six-His-tagged fusion proteins, were selected.
E. coli M15(pREP4) harboring pQE30-ORF11 or pQE30-ORF12 was grown at 37°C in Luria broth with appropriate antibiotics. Expression of the recombinant protein was induced by adding 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when the optical density at 600 nm reached about 0.8. Cultivation was continued for additional 12 h at 18°C. Purification of His-tagged recombinant proteins was done in accordance with the manufacturer's (Qiagen) protocols. Purified proteins were analyzed by sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis (PAGE).
Enzymatic synthesis of terpentetriene in vitro.
The reaction mixture contained 50 mM potassium phosphate (pH 6.5), 10 mM MgCl2, 5 mM 2-mercaptoethanol, 0.1% Tween 80, 40 μM GGDP, and 150 μg each of the purified ORF11 and ORF12 proteins per ml. The reaction was carried out at 30°C for 3 h. The product was extracted with chloroform and analyzed by reversed-phase HPLC. The analytical conditions were essentially the same as those described above, except for the column (Mightisil RP-18 GP, 250 by 4.6 mm) and the flow rate (1 ml/min).
Hybridization.
Northern blot hybridization with a 32P-labeled DNA fragment made by nick translation (5 × 108 cpm/mg) was done as described previously (14). The filter was washed twice with a buffer containing 0.3× SSC (1× SSC is 0.15 M NaCl plus 0.015 sodium citrate) and 0.2% SDS for 30 min at 68°C. Total RNA was isolated from S. griseolosporeus strain MF730-N6 grown at 30°C for 2 days in production medium as described previously (14).
Nucleotide sequence accession number.
The DNA sequence of an 8.5-kb BamHI fragment containing the upstream region of the GGDP synthase has been deposited in the DDBJ, EMBL, and GenBank databases under accession number AB048795.
RESULTS
Nucleotide sequences of the flanking region of the mevalonate pathway gene cluster.
We have recently cloned the gene cluster containing the mevalonate pathway genes for the formation of IPP and the gene encoding the enzyme catalyzing the formation of GGDP, which is the direct precursor of TP (15). In many cases, antibiotic biosynthetic genes cloned from actinomycetes are clustered in the genomic DNA region (11, 27). Therefore, the TP biosynthetic genes are also expected to exist in the flanking region of the GGDP synthase gene. To examine this possibility, the DNA sequence of an 8.5-kb BamHI fragment containing the upstream region of the GGDP synthase gene was determined (Fig. 2). Computer analysis of the DNA sequence by Frame Analysis (6) showed seven ORFs (ORF8 to ORF14) in the same direction (Fig. 2), except for ORF9. ORF11 and ORF12 appeared to be translationally coupled.
To understand the functions of individual ORFs deduced by DNA sequencing, we searched the databases with their translated products by means of the sequence similarity search programs FASTA (33) and BLAST (3). The results are summarized in Fig. 2. In brief, each of the ORFs had significant similarity to efflux proteins responsible for antibiotic resistance (ORF8), an unknown protein found in the genomic DNA of Streptomyces coelicolor (ORF9), P450-like hydroxylation proteins (ORF10), DCs from eukaryotes (ORF11), a pentalenene synthase of Streptomyces sp. (ORF12), P450-like hydroxylation proteins (ORF13), and ferredoxin (ORF14), respectively. TP was previously reported to be synthesized from GGDP after successive cyclization, hydroxylation, and epoxidation (18), suggesting that the ORFs found in this study would encode TP biosynthetic genes.
Features of ORF11 and ORF12 products deduced by primary structures.
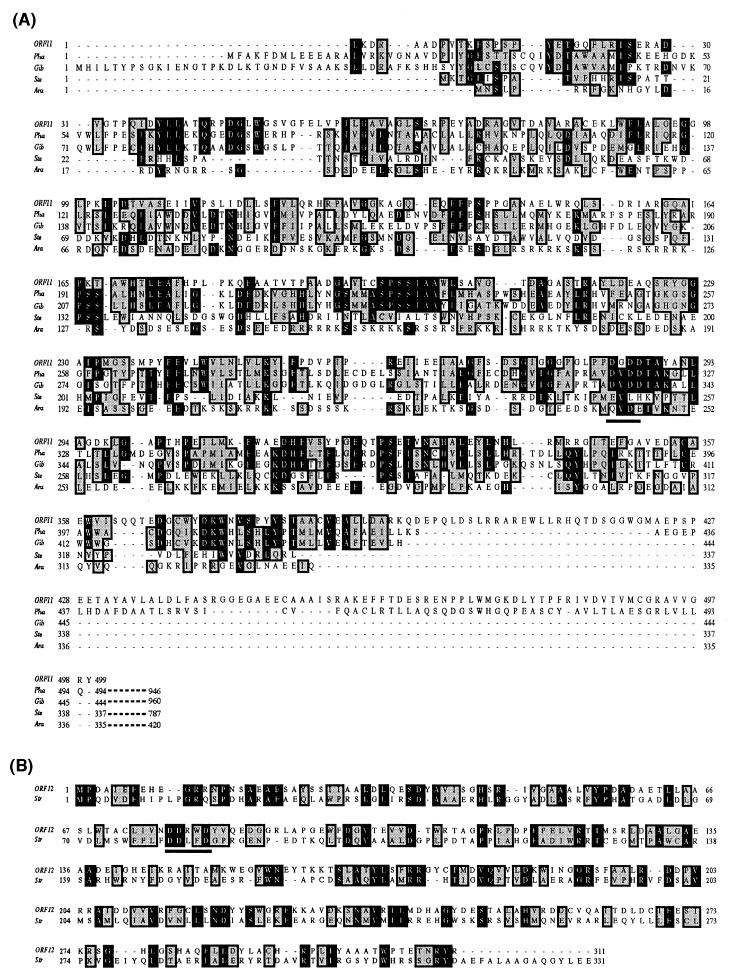
The predicted amino acid sequence of the ORF11 product has significant homology with those of the N-terminal halves of DCs from plants and fungi: 29% identity over 494 amino acids with the ent-kaurene synthase (KS) from Phaeosphaeria sp., 27% identity over 444 amino acids with the (−)-copalyl diphosphate synthase (CPS) from Gibberella fujikuroi, 22% identity over 337 amino acids with the CPS from Stevia rebaudiana, 23% identity over 335 amino acids with the KS from Arabidopsis thaliana, 24% identity over 346 amino acids with the taxadiene synthase from Taxus brevifolia, and 21% identity over 383 amino acids with the KS from Pisum sativum. Sequence alignment with eukaryotic DCs revealed that the motifs QXXDGSW and DXDDTA, which were proposed to stabilize an intermediate cation during the cyclization process and to mediate substrate binding by chelation of divalent metal ions, respectively (9, 29), are conserved in the ORF11 product (43QRPDGLW, 284DGDDTA) (Fig. 5).
FIG. 5.
Alignment of the ORF11 and ORF12 products with homologous proteins. Multiple alignments of the ORF11 product with eukaryotic DCs (A) and an alignment of the ORF12 product with pentalenene synthase (B) are shown. Identical (black) and similar (gray) amino acid residues are boxed. Aspartate-rich motifs (DXDD and DDXXD) are underlined. The numbers on the left and right are the residue numbers for each protein. Abbreviations and accession numbers: Pha, KS of Phaeosphaeria sp., GenBank accession no. AB003395-1; Gib, CPS of G. fujikuroi, GenBank accession no. Y15013-1; Ste, CPS of S. rebaudiana, GenBank accession no. AF034545-1; Ara, KS of A. thaliana, GenBank accession no. AC004044-7; Str, pentalenolactone synthase of Streptomyces sp., GenBank accession no. A54214.
The sole protein homologous to the ORF12 product was the pentalenene synthase cloned from Streptomyces sp. strain UC5319 (5) (Fig. 5). These proteins share 25% sequence identity over 331 amino acids. The ORF12 product has the DDXXD motif (9, 29), which was also suggested to be involved in the coordination of divalent metal ions for substrate binding (77DDRWD).
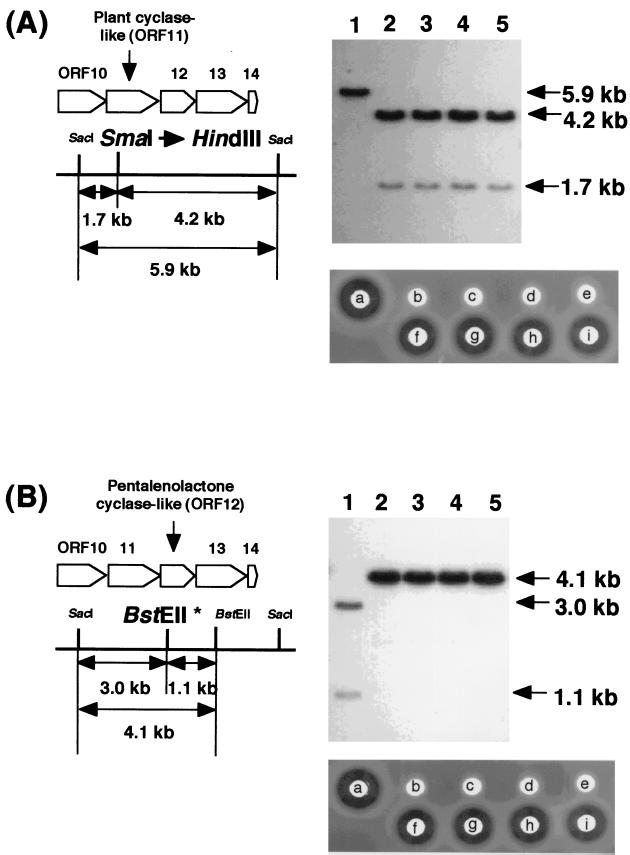
Construction of ORF11- and ORF12-inactivated strains.
To explore the possibility that the ORF11 and ORF12 genes code for cyclases responsible for TP biosynthesis, we constructed two mutant strains in which ORF11 or ORF12 was inactivated by the gene replacement technique originally developed for Streptomyces strains by Anzai et al. (4). Two plasmids, pCYC1FS and pCYC2FS, in which ORF11 and ORF12 were inactivated by a frameshift mutation, respectively, were constructed. After introduction of each plasmid into the TP producer, protoplasts from thiostrepton-resistant colonies were prepared and regenerated on RSG medium (13). Genomic DNAs of the regenerated thiostrepton-sensitive (plasmid-free) colonies were prepared, and double digested with SacI-HindIII (ORF11-inactivated strain) or SacI-BstEII (ORF12-inactivated strain) and hybridized by Southern blotting using a 5.9-kb SacI fragment carrying ORF11 and ORF12 as a probe (Fig. 6). In our experiments, about half of the regenerated colonies lost plasmids and the ORF11- and ORF12-inactivated strains emerged from plasmid-cured colonies at a frequency of about 10%. Four strains were randomly isolated from each of the ORF11- and ORF12-inactivated strains to examine TP productivity. Both types of mutants produced no TP, as judged by bioassay (Fig. 6) and HPLC analysis (data not shown). As described below, the ORF11 and ORF12 genes were suggested to be polycistronically transcribed with the downstream genes by Northern blot analysis. Therefore, to confirm that the loss of TP productivity of the ORF11- or ORF12-inactivated strain is not due to polar effects on the downstream genes, complementation experiments were carried out. Two plasmids, pEN-ORF11 and pEN-ORF12, which carried the ORF11 and ORF12 genes, respectively, were constructed and introduced into the corresponding mutants. As shown in Fig. 6, each of the mutants recovered TP productivity, confirming that the loss of TP productivity was due not to a polar effect of the frameshift mutation but to disruption of the ORF11 or ORF12 gene.
FIG. 6.
Southern blot analyses of the genomic DNAs of the ORF11- and ORF12-inactivated strains and bioassay for the TP productivity of these mutants and their complemented transformants. SacI-HindIII digests of DNAs from ORF11-inactivated strains (A, lanes 2 to 5) and SacI-BstEII digests of DNAs from ORF12-inactivated strains (B, lanes 2 to 5) were, respectively, subjected to Southern blot analyses with a 32P-labeled 5.9-kb SacI fragment as a probe. SacI-HindIII (A) and SacI-BstEII digests (B) of DNAs from S. griseolosporeus strain MF730-N6 (wild type) are shown in lane 1. Arrows indicate the sizes of the hybridized fragments. The hybridized fragments are schematically shown. Bioassays for the TP production of the wild-type strain (A, a), the ORF11-inactivated (A, b to e) and ORF12-inactivated (B, b to e) strains, the ORF11-inactivated strain harboring pEN-ORF11 (A, f to i), and the ORF12-inactivated strain harboring pEN-ORF12 (B, f to i) are shown.
Expression of the cyclase genes in S. lividans.
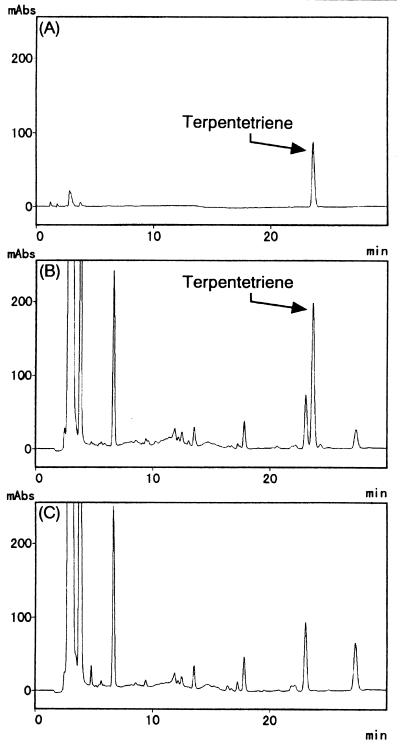
Considering that both the ORF11 and ORF12 products were essential for TP production and had significant similarities to isoprenoid cyclases, these proteins were expected to catalyze the cyclization of GGDP. To confirm this possibility and to elucidate the structure of the reaction product formed by these enzymes, heterologous expression of the ORF11 and ORF12 genes was employed. We constructed plasmid pWHM-TER1, in which the GGDP synthase, ORF11, and ORF12 genes were expressed under the control of the constitutive ermE* promoter (7, 8, 38). We transformed S. lividans TK23 with the plasmid and investigated the production of new compounds in the transformant. S. lividans TK23 harboring pWHM-TER1 was cultivated in liquid medium, and the metabolites were analyzed by HPLC. A new compound, which was eluted with a retention time longer than that of geranylgeraniol, was specifically detected in the culture broth of the transformant harboring pWHM-TER1. We propose to name this new compound terpentetriene based on structural similarities to TP and the presence of three double bonds. The product was purified, and its structure was determined (see Materials and Methods and Fig. 4).
Enzymatic synthesis of terpentetriene in vitro.
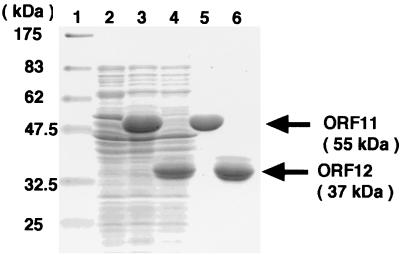
To verify the enzymatic function of ORF11 and ORF12, these ORF products were overproduced in E. coli. Soluble protein extracts from E. coli harboring pQE30-ORF11, pQE30-ORF12, or pQE30 (no insert) were analyzed by SDS-PAGE. His-tagged ORF11, with a molecular mass of about 55 kDa, and His-tagged ORF12, with a molecular mass of about 37 kDa, which were in good agreement with the values calculated from the deduced amino acid sequences of the enzymes, were expressed at a high level (Fig. 7, lanes 3 and 4). The expressed proteins were then purified (Fig. 7, lanes 5 and 6) and used for an enzyme assay. Incubation of both of the purified recombinant proteins with GGDP resulted in the formation of terpentetriene (Fig. 8), confirming that the ORF11 and ORF12 products were the DCs.
FIG. 7.
Electrophoresis of overproduced and purified cyclases. Molecular mass markers (lane 1); total soluble proteins from the E. coli harboring pQE30 (lane 2), pQE30-ORF11 (lane 3), or pQE30-ORF12 (lane 4); purified, His-tagged ORF11 (lane 5); and purified, His-tagged ORF12 (lane 6) were analyzed by SDS–10% PAGE. Proteins were stained with Coomassie brilliant blue R-250.
FIG. 8.
HPLC analysis of the product generated by recombinant cyclases. An authentic terpentetriene standard (A) and chloroform extracts of the reaction products generated by the purified, His-tagged ORF11 and ORF12 products in the presence of GGDP (B) or the absence of GGDP (C) were analyzed by reversed-phase HPLC. mAbs, milliabsorbance.
Northern blot analysis of DC genes.
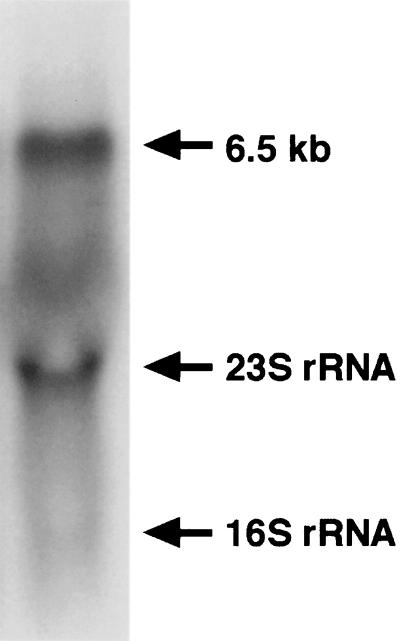
The ORF11 and ORF12 genes were shown to be essential for TP production by gene disruption experiments (Fig. 6). In many cases, antibiotic biosynthetic genes cloned from actinomycetes are clustered in the genomic DNA region (11, 27). Therefore, other ORFs found in this study were also expected to code for TP biosynthetic enzymes. To study this possibility, Northern blot analysis was performed with the XbaI-HindIII fragment carrying the two cyclase genes as a probe. As shown in Fig. 9, a message of 6.5 kb was specifically detected. Considering that the mevalonate pathway gene cluster (ORF1 to ORF7) seemed to be polycistronically transcribed (15), we surmised that the genes encoding ORF10 to ORF14 were also polycistronically transcribed and that these gene products would participate in the biosynthesis of TP.
FIG. 9.
Northern blot analysis of the gene cluster essential for TP production. Total RNA was isolated from S. griseolosporeus strain MF730-N6 grown at 30°C for 2 days and analyzed by Northern blotting using the XbaI-HindIII fragment carrying the two cyclase genes as a probe (Fig. 1). The arrowheads indicate the positions of 23S and 16S rRNAs and the sizes of hybridized fragments. A background signal due to hybridization to degraded RNAs occurred.
DISCUSSION
By sequencing analysis of the flanking region of the mevalonate pathway gene cluster cloned from a diterpenoid antibiotic (TP) producer, S. griseolosporeus strain MF730-N6, we have found seven additional ORFs, which were suggested to encode the TP biosynthetic gene cluster. Among the ORFs identified by the sequencing analysis, the two genes encoding proteins homologous to isoprenoid cyclases were extensively analyzed and confirmed to be DCs by heterologous expression and in vitro experiments. The ORF11 and ORF12 products catalyzed the cyclization of GGDP to form terpentetriene, a compound with the same planar structure as ent-clerod-3,13(16),14-triene, which was previously isolated from Jungermannia infusca (31). However, the compound isolated in this study had a structure with stereochemistry different from that of J. infusca, showing that our compound is a novel diterpenoid. The genes for the eubacterial isoprenoid cyclases, the pentalenolactone synthase from Streptomyces sp. strain UC5319 (10) and the squalene-hopene cyclases from Bacillus acidocaldarius (32), Rhodopseudomonas palustris (22), Zymomonas mobilis (35), Bradyrhizobium japonicum (34), Methylococcus capsulatus (42), and Alicyclobacillus acidocaldarius (46), have been characterized. However, none of the genes encoding DCs have been isolated from a prokaryote. Thus, this is the first report of the cloning and identification of DCs of eubacterial origin.
Although we do not know the detailed reaction mechanism catalyzed by the ORF11 and ORF12 products, both of the proteins were essential for terpentetriene production because both the ORF11- and ORF12-inactivated strains produced no TP and neither the ORF11 product nor the ORF12 product alone could catalyze the cyclization of GGDP to produce terpentetriene (data not shown). DCs are classified into two major types with respect to their modes of cyclization (25). One type of reaction is initiated by ionization of GGDP to an allylic carbocation, followed by cyclization and deprotonation to the olefin (9, 29). Casbene synthase (28) and taxadiene synthase (23, 24) are representatives of this class. The other type of reaction is initiated by protonation at the 14, 15 double bond of GGDP (2, 16, 40) in a manner similar to that of the squalene-hopene cyclase from Alicyclobacillus acidocaldarius (1, 32). Croteau and coworkers suggested that the presence of the DXDD or DDXXD motif in DCs might be used to determine which mode of cyclization would be involved in the reaction catalyzed by uncharacterized DCs (45). They found that the N-terminal region of the abietadiene synthase, which contains the DXDD motif, resembles those of enzymes catalyzing protonation-initiated isoprenoid cyclization, whereas the C-terminal region of the cyclase, which has the DDXXD motif, resembles those of enzymes catalyzing ionization-dependent isoprenoid cyclization. The following fact supports their assumption. In higher plants, two different DCs, CPS and KS, participate in the biosynthesis of (−)-kaurene, the precursor of the gibberellin plant hormone (16). CPS, which contains the DXDD motif and lacks the DDXXD motif (2, 5, 36, 39, 40), is known to catalyze the protonation-initiated cyclization of GGDP to (−)-copalyl diphosphate. A separate enzyme, kaurene synthase B, which has the DDXXD motif and no DXDD motif (36, 47, 48), was shown to transform this intermediate to (−)-kaurene via ionization-dependent cyclization. The predicted amino acid sequences of ORF11 and ORF12 have the DXDD and DDXXD motifs, respectively. Therefore, the ORF11 product catalyzes cyclization by protonation of the terminal double bond of GGDP, resulting in the formation of an intermediate with a phosphodiester, as does CPS; the ORF12 product completes the reaction sequence by ionization-dependent cyclization, as does KS. Kawaide et al. also reported that the DVDD motif of the KS from the fungus Phaeosphaeria sp. strain L487, which is a bifunctional enzyme catalyzing the two-step cyclization reaction from GGDP to ent-kaurene via (−)-copalyl diphosphate, is essential for CPS activity (20, 21). However, they also suggested that the DDVLD motif is involved in both the CPS and KS reactions (20, 21). Therefore, to elucidate the detailed reaction mechanisms catalyzed by the ORF11 and ORF12 products, more studies are required to clarify the function of each the ORF products. These studies are now in progress, and the results will be reported in the near future.
ACKNOWLEDGMENTS
We thank Y. Asakawa of Tokushima Bunri University for 1H- and 13C-NMR spectra of ent-clerod-3,13(16),14-triene, H. Kobayashi of the University of Tokyo for collecting MS spectra, M. Hamada of the Institute of Microbial Chemistry for providing the TP producer, and Y. Ono for excellent technical assistance.
This work was supported in part by RFTF (JSPS-RFTF96I00301) from JSPS and by a Grant-in-Aid for Scientific Research (C) to T.D. from JSPS.
REFERENCES
- 1.Abe I, Rohmer M, Prestwich G D. Enzymatic cyclization of squalene and oxidosqualene to sterol and triterpenes. Chem Rev. 1993;93:2189–2206. [Google Scholar]
- 2.Ait-Ali T, Swain S M, Reid J B, Sun T P, Kamiya Y. The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J. 1997;11:443–454. doi: 10.1046/j.1365-313x.1997.11030443.x. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Anzai H, Kumada Y, Hara O, Murakami T, Ito T, Takano E, Imai S, Satoh A, Nagaoka K. Replacement of Streptomyces hygroscopicus genomic segments with in vitro altered DNA sequences. J Antibiot. 1988;41:226–233. doi: 10.7164/antibiotics.41.226. [DOI] [PubMed] [Google Scholar]
- 5.Bensen R J, Johal G S, Crane V C, Tossberg J T, Schnable P S, Meeley R B, Briggs S P. Cloning and characterization of the maize An1 gene. Plant Cell. 1995;7:75–84. doi: 10.1105/tpc.7.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 7.Bibb M J, Janssen G R, Ward J M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38:215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- 8.Bibb M J, White J, Ward J M, Janssen G R. The mRNA for the 23S rRNA methylase encoded by the ermE gene of Saccharopolyspora erythraea is translated in the absence of a conventional ribosome-binding site. Mol Microbiol. 1994;14:533–545. doi: 10.1111/j.1365-2958.1994.tb02187.x. [DOI] [PubMed] [Google Scholar]
- 9.Bohlmann J, Meyer-Gauen G, Croteau R. Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA. 1998;95:4126–4133. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cane D E, Sohng J K, Lamberson C R, Rudnicki S M, Wu Z, Lloyd M D, Oliver J S, Hubbard B R. Pentalenene synthase. Purification, molecular cloning, sequencing, and high-level expression in Escherichia coli of a terpenoid cyclase from Streptomyces UC5319. Biochemistry. 1994;33:5846–5857. doi: 10.1021/bi00185a024. [DOI] [PubMed] [Google Scholar]
- 11.Chater K F. The improving prospects for yield increase by genetic engineering in antibiotic-producing streptomycetes. Bio/Technology. 1990;8:115–121. doi: 10.1038/nbt0290-115. [DOI] [PubMed] [Google Scholar]
- 12.Connolly J D, Hill R A. Dictionary of terpenoids. New York, N.Y: Chapman & Hall; 1992. [Google Scholar]
- 13.Dairi T, Motohira Y, Kuzuyama T, Takahashi S, Itoh N, Seto H. Cloning of the gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from terpenoid antibiotic-producing Streptomyces strains. Mol Gen Genet. 2000;262:957–964. doi: 10.1007/pl00008664. [DOI] [PubMed] [Google Scholar]
- 14.Dairi T, Nakano T, Aisaka K, Katsumata R, Hasegawa M. Cloning and nucleotide sequence of the gene responsible for chlorination of tetracycline. Biosci Biotech Biochem. 1995;59:1099–1106. doi: 10.1271/bbb.59.1099. [DOI] [PubMed] [Google Scholar]
- 15.Hamano Y, Dairi T, Yamamoto M, Kawasaki T, Kaneda K, Kuzuyama T, Itoh N, Seto H. Cloning of a gene cluster encoding enzymes responsible for the mevalonate pathway from a terpenoid-antibiotic-producing Streptomyces strain. Biosci Biotech Biochem. 2001;65:1627–1635. doi: 10.1271/bbb.65.1627. [DOI] [PubMed] [Google Scholar]
- 16.Hedden P, Kamiya Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- 17.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schremp H. Gene manipulation of Streptomyces, a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 18.Isshiki K, Tamamura T, Sawa T, Naganawa H, Takeuchi T, Umezawa H. Biosynthetic studies of terpentecin. J Antibiot. 1986;34:1634–1635. doi: 10.7164/antibiotics.39.1634. [DOI] [PubMed] [Google Scholar]
- 19.Kaneda K, Kuzuyama T, Takagi M, Hayakawa Y, Seto H. An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc Natl Acad Sci USA. 2001;98:932–937. doi: 10.1073/pnas.020472198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawaide H, Imai R, Sassa T, Kamiya Y. Ent-kaurene synthase from the fungus Phaeosphaeria sp. L487. cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase in fungal gibberellin biosynthesis. J Biol Chem. 1997;272:21706–21712. doi: 10.1074/jbc.272.35.21706. [DOI] [PubMed] [Google Scholar]
- 21.Kawaide H, Sassa T, Kamiya Y. Functional analysis of the two interacting cyclase domains in ent-kaurene synthase from the fungus Phaeosphaeria sp. L487 and a comparison with cyclases from higher plants. J Biol Chem. 2000;275:2276–2280. doi: 10.1074/jbc.275.4.2276. [DOI] [PubMed] [Google Scholar]
- 22.Kleemann G, Kellner R, Poralla K. Purification and properties of the squalene-hopene cyclase from Rhodopseudomonas palustris, a purple non-sulfur bacterium producing hopanoids and tetrahymanol. Biochim Biophys Acta. 1994;1210:317–320. doi: 10.1016/0005-2760(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 23.Koepp A E, Hezari M, Zajicek J, Vogel B S, LaFever R E, Lewis N G, Croteau R. Cyclization of geranylgeranyl diphosphate to taxa-4(5),11(12)-diene is the committed step of taxol biosynthesis in Pacific yew. J Biol Chem. 1995;270:8686–8690. doi: 10.1074/jbc.270.15.8686. [DOI] [PubMed] [Google Scholar]
- 24.Lin X, Hezari M, Koepp A E, Floss H G, Croteau R. Mechanism of taxadiene synthase, a diterpene cyclase that catalyzes the first step of taxol biosynthesis in Pacific yew. Biochemistry. 1996;35:2968–2977. doi: 10.1021/bi9526239. [DOI] [PubMed] [Google Scholar]
- 25.MacMillan J, Beale M H. In: Comprehensive natural products chemistry. Barton D, Nakanishi K, Meth-Cohn O, editors. 2. Diterpene biosynthesis. Oxford, United Kingdom: Elsevier; 1999. pp. 217–243. [Google Scholar]
- 26.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 27.Martin J F, Liras P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu Rev Microbiol. 1989;43:173–206. doi: 10.1146/annurev.mi.43.100189.001133. [DOI] [PubMed] [Google Scholar]
- 28.Mau C J D, West C A. Cloning of casbene synthase cDNA: evidence for conserved structural features among terpenoid cyclases in plants. Proc Natl Acad Sci USA. 1994;91:8497–8501. doi: 10.1073/pnas.91.18.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCaskill D, Croteau R. Prospects for the bioengineering of isoprenoid biosynthesis. Adv Biochem Eng Bio/Technology. 1997;55:107–146. doi: 10.1007/BFb0102064. [DOI] [PubMed] [Google Scholar]
- 30.Miyado S. Research on antibiotic screening in Japan over the last decade: a producing microorganism approach. Actinomycetologica. 1993;7:100–106. [Google Scholar]
- 31.Nagashima F, Takaoka S, Asakawa Y. Diterpenoids from the Japanese liverwort Jungermannia infusca. Phytochemistry. 1998;49:601–608. [Google Scholar]
- 32.Ochs D, Kaletta C, Entian K D, Beck-Sickinger A, Poralla K. Cloning, expression, and sequencing of squalene-hopene cyclase, a key enzyme in triterpenoid metabolism. J Bacteriol. 1992;174:298–302. doi: 10.1128/jb.174.1.298-302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perzl M, Muller P, Poralla K, Kannenberg E L. Squalene-hopene cyclase from Bradyrhizobium japonicum: cloning, expression, sequence analysis and comparison to other triterpenoid cyclases. Microbiology. 1997;143:1235–1242. doi: 10.1099/00221287-143-4-1235. [DOI] [PubMed] [Google Scholar]
- 35.Reipen I G, Poralla K, Sahm H, Sprenger G A. Zymomonas mobilis squalene-hopene cyclase gene (shc): cloning, DNA sequence analysis, and expression in Escherichia coli. Microbiology. 1995;141:155–161. doi: 10.1099/00221287-141-1-155. [DOI] [PubMed] [Google Scholar]
- 36.Richman A S, Gijzen M, Starratt A N, Yang Z, Brandle J E. Diterpene synthesis in Stevia rebaudiana: recruitment and up-regulation of key enzymes from the gibberellin biosynthetic pathway. Plant J. 1999;19:411–421. doi: 10.1046/j.1365-313x.1999.00531.x. [DOI] [PubMed] [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt-John T, Engels J W. Promoter constructions for efficient secretion expression in Streptomyces lividans. Appl Microbiol Biotechnol. 1992;36:493–498. doi: 10.1007/BF00170190. [DOI] [PubMed] [Google Scholar]
- 39.Smith M W, Yamaguchi S, Ait-Ali T, Kamiya Y. The first step of gibberellin biosynthesis in pumpkin is catalyzed by at least two copalyl diphosphate synthases encoded by differentially regulated genes. Plant Physiol. 1998;118:1411–1419. doi: 10.1104/pp.118.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun T P, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamamura T, Sawa T, Isshiki K, Masuda T, Homma Y, Iinuma H, Naganawa H, Hamada M, Takeuchi T, Umezawa H. Isolation and characterization of terpentecin, a new antitumor antibiotic. J Antibiot. 1985;38:1664–1669. doi: 10.7164/antibiotics.38.1664. [DOI] [PubMed] [Google Scholar]
- 42.Tippelt A, Jahnke L, Poralla K. Squalene-hopene cyclase from Methylococcus capsulatus (Bath): a bacterium producing hopanoids and steroids. Biochim Biophys Acta. 1998;1391:223–232. doi: 10.1016/s0005-2760(97)00212-9. [DOI] [PubMed] [Google Scholar]
- 43.Uosaki Y, Kawada S, Nakano H, Saitoh Y, Sano H. UCT4B, a new antitumor antibiotic with topoisomerase II mediated DNA cleavage activity. Structure determination. J Antibiot. 1993;46:235–240. doi: 10.7164/antibiotics.46.235. [DOI] [PubMed] [Google Scholar]
- 44.Vara J, Lewandowska-Skarbek M, Wang Y-G, Donadio S, Hutchinson C R. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus) J Bacteriol. 1989;171:5872–5881. doi: 10.1128/jb.171.11.5872-5881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogel B S, Wildung M R, Vogel G, Croteau R. Abietadiene synthase from grand fir (Abies grandis). cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase involved in resin acid biosynthesis. J Biol Chem. 1996;271:23262–23268. doi: 10.1074/jbc.271.38.23262. [DOI] [PubMed] [Google Scholar]
- 46.Wendt K, Poralla K, Schulz G E. Structure and function of a squalene cyclase. Science. 1997;277:1811–1815. doi: 10.1126/science.277.5333.1811. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi S, Saito T, Abe H, Yamane H, Murofushi N, Kamiya Y. Molecular cloning and characterization of a cDNA encoding the gibberellin biosynthetic enzyme ent-kaurene synthase B from pumpkin (Cucurbita maxima L.) Plant J. 1996;10:203–213. doi: 10.1046/j.1365-313x.1996.10020203.x. [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi S, Sun T P, Kawaide H, Kamiya Y. The GA2 locus of Arabidopsis thaliana encodes ent-kaurene synthase of gibberellin biosynthesis. Plant Physiol. 1998;116:1271–1278. doi: 10.1104/pp.116.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]