Abstract
The RimM protein in Escherichia coli is associated with free 30S ribosomal subunits but not with 70S ribosomes. A ΔrimM mutant shows a sevenfold-reduced growth rate and a reduced translational efficiency, probably as a result of aberrant assembly of the ribosomal 30S subunits. The slow growth and translational deficiency can be partially suppressed by increased synthesis of the ribosome binding factor RbfA. Here, we have identified 14 chromosomal suppressor mutations that increase the growth rate of a ΔrimM mutant by increasing the expression of rbfA. Nine of these mutations were in the nusA gene, which is located upstream from rbfA in the metY-nusA-infB operon; three mutations deleted the transcriptional terminator between infB and rbfA; one was an insertion of IS2 in infB, creating a new promoter for rbfA; and one was a duplication, placing a second copy of rbfA downstream from a promoter for the yhbM gene. Two of the nusA mutations were identical, while another mutation (nusA98) was identical to a previously isolated mutation, nusA11, shown to decrease termination of transcription. The different nusA mutations were found to increase the expression of rbfA by increasing the read-through of two internal transcriptional terminators located just downstream from the metY gene and that of the internal terminator preceding rbfA. Induced expression of the nusA+ gene from a plasmid in a nusA+ strain decreased the read-through of the two terminators just downstream from metY, demonstrating that one target for a previously proposed NusA-mediated feedback regulation of the metY-nusA-infB operon expression is these terminators. All of the nusA mutations produced temperature-sensitive phenotypes of rimM+ strains. The nusA gene has previously been shown to be essential at 42°C and below 32°C. Here, we show that nusA is also essential at 37°C.
The Escherichia coli RimM protein shows affinity for free 30S ribosomal subunits but not for 70S ribosomes (3) and is important for the maturation of the 30S subunits (4). Mutants lacking RimM show a severalfold-decreased growth rate and a reduced translational efficiency (3). These defects can be partially suppressed by increased expression of the ribosome binding factor RbfA (4) encoded by the metY-nusA-infB operon. RbfA is a cold shock protein that is essential for the resumption of growth after a downshift in temperature (19); however, it also has an important function at higher temperatures, since an rbfA mutant shows a twofold-lower growth rate than an rbfA+ strain at 42°C (9). RbfA is important for the maturation of the 30S ribosomal subunits, possibly by stabilizing the 5′-terminal helix of 16S rRNA (4, 9). Overexpression of rbfA suppresses a cold-sensitive mutation in this 16S rRNA helix (9). Previously, we isolated 23 mutations that increase the growth rate of a ΔrimM mutant and that were shown to be tightly linked to the rbfA gene (4) of the metY-nusA-infB operon (Fig. 1A). This operon contains, in the direction of transcription, the metY gene encoding a minor form of the initiator tRNA, the p15a open reading frame, the nusA gene for the transcriptional elongation factor NusA (8, 15, 16, 45), the infB gene encoding the translation initiation factor IF2 (38, 43), the rbfA gene (9, 47), and the truB gene for the tRNA(Ψ55) synthase (34, 47). The operon is transcribed from two promoters located upstream from metY, P−1 (12) and P1 (16, 27), and a minor promoter between metY and p15a, P2 (40). The rpsO and pnp genes located downstream from truB and encoding the ribosomal protein S15 and polynucleotide phosphorylase, respectively (39, 41), are also transcribed from these promoters (47); however, the major promoter for these two genes is that just upstream from rpsO (42). The cleavage by RNase III at sites between metY and p15a on the polycistronic mRNA initiates the rapid degradation of the downstream RNA (40). Internal transcriptional terminators are found between metY and p15a (16, 40) and between infB and rbfA (47). NusA negatively feedback regulates the expression of the metY-nusA-infB operon, and the two terminators between metY and p15a were suggested to be the regulatory target (30, 37, 40). In this paper, we describe the identification of 14 suppressor mutations in the metY-nusA-infB operon, which increase the growth rate of a rimM mutant by increasing the expression of the rbfA gene. Nine of the mutations were localized to the nusA gene and found to result in a deficiency in feedback regulation at the two terminators between metY and p15a and also at the terminator just upstream from rbfA. Of the other mutations, three had the transcriptional terminator between infB and rbfA deleted; one was an insertion of IS2 in infB, creating a new promoter for rbfA; and one was a duplication, placing a second copy of rbfA downstream from a putative promoter for yhbM.
FIG. 1.
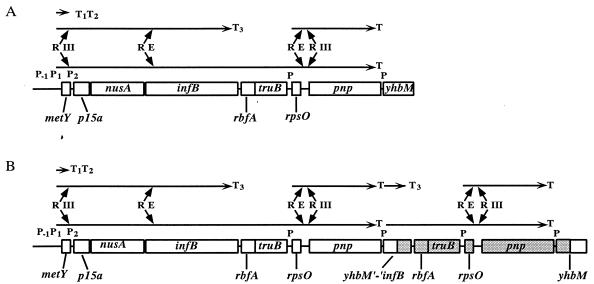
Genetic organization of the metY-nusA-infB operon region on the chromosomes of wild-type E. coli (A) and strains that contain a duplication covering the 3′ part of infB to the 5′ half of yhbM (B; shaded region). P−1, P1, P2, and P indicate the locations of promoters; T, T1, T2, and T3 indicate different transcriptional terminators; while R E and R III show sites for the RNA-processing enzymes RNase E and RNase III, respectively. The horizontal arrows represent transcriptional products. For references and explanations of gene symbols, see the introduction.
MATERIALS AND METHODS
Bacterial strains and plasmid constructions.
The strains and plasmids used are listed in Table 1. The low-copy-number lacZ fusion vector pGOB100 was constructed in the following way. First, a 300-bp fragment containing the rrnBt1 and rrnBt2 transcriptional terminators was amplified by PCR using the oligonucleotides 5′-TTTTGGTACCGATGGTAGTGTGG-3′ and 5′-TTTTGGATCCGTAGATATGACGACAGG-3′, trimmed with KpnI and BamHI and inserted into the low-copy-number vector pCL1921. To facilitate later constructions of lacZ fusions, the unique BamHI site of the plasmid obtained was removed by inserting the oligonucleotide 5′-GATCGTCGAC-3′. This plasmid was named pMW348. Next, the unique EagI site downstream from lacZ in the fusion vector pTL61T was converted to a KpnI site by inserting the oligonucleotide 5′-GGCCAGGTACCT-3′. Finally, the EcoRI-KpnI lacZ fragment of the resulting plasmid was cloned into plasmid pMW348.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype | Origin or referencea |
|---|---|---|
| Strains | ||
| GOB375 | MW100 argG2424::miniTn10Cm | |
| GOB383 | PW100 argG2424::miniTn10Cm | |
| GOB388 | MW100 nusA98 argG2424::miniTn10Cm | |
| GOB390 | GOB388 argG+ | |
| GOB417 | GOB375 nusA97 argG+ | |
| GOB421 | GOB375 nusA93 argG+ | |
| GOB423 | GOB375 nusA95 argG+ | |
| GOB425 | GOB375 nusA92 argG+ | |
| GOB426 | GOB375 nusA94 argG+ | |
| GOB427 | GOB375 nusA91 argG+ | |
| GOB428 | GOB375 nusA96 argG+ | |
| GOB434 | MW100 lacI′-rplSt-PmetY-metY-metYt1t2-lacZ | |
| GOB435 | MW100 lacI′-rplSt-PmetY-metY-lacZ | |
| GOB438 | MW100 lacI′-rplSt-lacZ | |
| GOB440 | MW100 lacI′-rplSt-metYt1t2-lacZ | |
| GOB492 | GOB434 nusA98 argG2424::miniTn10Cm | |
| GOB494 | GOB434 argG2424::miniTn10Cm | |
| GOB496 | GOB435 nusA98 argG2424::miniTn10Cm | |
| GOB498 | GOB435 argG2424::miniTn10Cm | |
| GOB600 | GOB492 ΔrimM102 yfiB::nptI | |
| GOB604 | GOB494 ΔrimM102 yfiB::nptI | |
| GOB608 | GOB496 ΔrimM102 yfiB::nptI | |
| GOB612 | GOB498 ΔrimM102 yfiB::nptI | |
| GOB699 | GOB434 nusA92 argG2424::miniTn10Cm | |
| GOB702 | GOB435 nusA92 argG2424::miniTn10Cm | |
| GOB734 | GOB434 nusA96 argG2424::miniTn10Cm | |
| GOB736 | GOB435 nusA96 argG2424::miniTn10Cm | |
| GOB742 | GOB434 nusA93 argG2424::miniTn10Cm | |
| GOB744 | GOB435 nusA93 argG2424::miniTn10Cm | |
| GOB750 | GOB434 nusA94 argG2424::miniTn10Cm | |
| GOB752 | GOB435 nusA94 argG2424::miniTn10Cm | |
| GOB758 | GOB434 nusA95 argG2424::miniTn10Cm | |
| GOB760 | GOB435 nusA95 argG2424::miniTn10Cm | |
| GOB766 | GOB434 nusA91 argG2424::miniTn10Cm | |
| GOB768 | GOB435 nusA91 argG2424::miniTn10Cm | |
| GOB788 | GOB792 nusA98 | |
| GOB790 | GOB794 nusA98 | |
| GOB792 | MW100 nusA+ DUP[ffh+ to yfiB′]nptI[ffh+ (P+A)rpsP-lacZ Δ(rpsP-trmD) ′rplS yfiB′] | |
| GOB794 | MW100 nusA+ DUP[ffh+ to yfiB′]nptI[ffh+ PrpsP-lacZ Δ(rpsP-trmD) ′rplS yfiB′] | |
| GOB796 | GOB792 nusA93 | |
| GOB798 | GOB794 nusA93 | |
| GOB800 | GOB792 nusA95 | |
| GOB802 | GOB794 nusA95 | |
| GOB804 | GOB792 nusA94 | |
| GOB806 | GOB794 nusA94 | |
| GOB808 | GOB792 nusA91 | |
| GOB810 | GOB794 nusA91 | |
| GOB812 | GOB792 nusA96 | |
| GOB814 | GOB794 nusA96 | |
| GOB816 | GOB792 nusA92 | |
| GOB818 | GOB794 nusA92 | |
| GOB821 | GOB434 nusA97 argG2424::miniTn10Cm | |
| GOB823 | GOB435 nusA97 argG2424::miniTn10Cm | |
| GOB838 | MW100 lacI′-rplSt-PmetY-metY-TrplS-lacZ | |
| GOB840 | MW100 lacI′-rplSt-Ptet-metYt1t2-lacZ | |
| GOB842 | MW100 lacI′-rplSt-Ptet-lacZ | |
| GOB868 | GOB838 nusA94 argG2424::miniTn10Cm | |
| GOB870 | GOB840 nusA94 argG2424::miniTn10Cm | |
| GOB872 | GOB842 nusA94 argG2424::miniTn10Cm | |
| JML012 | MW100 ΔnusA/pmetY-pl5a-nusA-infB′ | |
| JML087 | MW100 ΔnusA/pJML007 | |
| JML125 | MW100 sdr-53 (infB::IS2) argG2424::miniTn10Cm | |
| JML126 | MW100 sdr-40 (ΔinfBt3) argG2424::miniTn10Cm | |
| JML127 | MW100 sdr-44 (ΔinfBt3) argG2424::miniTn10Cm | |
| JML128 | MW100 sdr-54 (ΔinfBt3) argG2424::miniTn10Cm | |
| MW37 | Hfr P4X ΔrimM102 yfiB::nptI | 36 |
| MW38 | Hfr P4X rimM+yfiB::nptI | 36 |
| MW100 | Hfr P4X | 54 |
| PW093 | MW37 sdr-27 = nusA97 (ΔA in codon 412) | 4 |
| PW100 | MW37 sdr-34 = nusA98 (GGC to GAC in codon 181) | 4 |
| PW101 | MW37 sdr-35 = nusA93 (ATC to AAC in codon 114) | 4 |
| PW104 | MW37 sdr-38 = nusA92 (ATC to AAC in codon 49) | 4 |
| PW105 | MW37 sdr-39 = nusA94 (GTG to GAG in codon 142) | 4 |
| PW106 | MW37 sdr-40 = ΔinfBt3 | 4 |
| PW107 | MW37 sdr-41 = nusA92 (ATC to AAC in codon 49) | 4 |
| PW109 | MW37 sdr-43 = DUP (′infB to yhbM′) | 4 |
| PW110 | MW37 sdr-44 = ΔinfBt3 | 4 |
| PW114 | MW37 sdr-48 = nusA96 (ACT to CCT in codon 198) | 4 |
| PW115 | MW37 sdr-49 = nusA95 (GTC to GAC in codon 197) | 4 |
| PW116 | MW37 sdr-50 = nusA91 (ATT to AAT in codon 23) | 4 |
| PW119 | MW37 sdr-53 = infB::IS2 | 4 |
| PW120 | MW37 sdr-54 = ΔinfBt3 | 4 |
| Plasmids | ||
| pBAD30 | bla cat′ araC | 13 |
| pCL1921 | Strr Spcr | 21 |
| pGOB117 | rep(Ts) cat ′lacI′-rplSt-lacZ-rrnBt1t2 | |
| pGOB118 | rep(Ts) cat ′lacI′-rplSt-metYt1t2-lacZ-rrnBt1t2 | |
| pGOB121 | rep(Ts) cat ′lacI′-rplSt-PmetY-metY-metYt1t2-lacZ-rrnBt1t2 | |
| pGOB122 | rep(Ts) cat ′lacI′-rplSt-PmetY-metY-lacZ-rrnBt1t2 | |
| pGOB126 | rep(Ts) cat ′lacI′-rplSt-PmetY-metY-TrplSt-lacZ-rrnBt1t2 | |
| pGOB131 | rep(Ts) cat ′lacI′-rplSt-Ptet-metYt1t2-lacZ-rrnBt1t2 | |
| pGOB134 | rep(Ts) cat ′lacI′-rplSt-Ptet-lacZ-rrnBt1t2 | |
| pJML001 | rep(Ts) cat PmetY-metY-p15a-ΔnusA-infB′ | |
| pJML007 | bla cat′ araC PBAD-nusA | |
| pMAK700 | rep(Ts) cat | 14 |
| pMAK705 | rep(Ts) cat | 14 |
| pTL61T | bla lacZ | 22 |
Unless otherwise noted, the origin was this study.
Two different transcriptional fusions between metY and lacZ were constructed in pGOB100: DNA fragments containing either PmetY-metY and the terminators metYt1 and metYt2 between metY and p15a or PmetY-metY without the terminators were amplified by PCR using the upstream oligonucleotide 5′-TTTTGAATTCAACAAATGAAAGTGAAC-3′ and the downstream oligonucleotides 5′-TTTTGGATCCGCAGTGTGGATGTGCGACC-3′ and 5-TTTTGGATCCGAACCCTATAACCGCAACTG-3′, respectively, trimmed with EcoRI-BamHI and cloned into pGOB100. The EcoRI site of the two resulting plasmids was converted to an SphI site using the oligonucleotide 5′-AATTGCATGC-3′, yielding plasmids pGOB115 and pGOB116, respectively. To enable the introduction of these two lacZ fusions into the lac operon region of the chromosome, an 880-bp fragment containing most of lacI was amplified by PCR using the oligonucleotides 5′-TTTTAAGCTTCTCTTATCAGACCGTTTCC-3′ and 5′-TTTTGGATCCAGTTGCAGCAAGCGGTCC-3′, trimmed with HindIII and BamHI, and inserted into the temperature-sensitive vector pMAK700. Into the resulting plasmid, the trmD operon transcriptional terminator, rplSt, was cloned on a BamHI-SphI fragment consisting of two complementary oligonucleotides: 5′-GATCGGGCTGGCCAATTGGCTGGCCCTTTTTTGCATG-3′ and 5′-CAAAAAAGGGCCAGCCAATTGGCCAGCCC-3′. Finally, into the SphI site of this plasmid, pGOB119, the two transcriptional fusions on plasmids pGOB115 and pGOB116 were inserted, yielding plasmids pGOB121 and pGOB122, respectively. In a similar way, two control constructions were made: a derivative (pGOB117) of pGOB122 which did not contain any DNA from the metY region and a derivative (pGOB118) of pGOB121 which lacked the metY promoter fragment and only contained the 245-bp metYt1-metYt2 terminator fragment.
To combine the metY promoter fragment with the trmD operon transcriptional terminator, rplSt, a 307-bp fragment containing rplSt was amplified by PCR using the oligonucleotides 5′-TTTTGGATCCGTGAGCGTACTGGTAAGG- 3′ and 5′-TTTTGGATCCAAACGGGCGAATGTCGTGG-3′, trimmed with BamHI, and inserted into plasmid pGOB122, yielding plasmid pGOB126.
To study the transcriptional read-through of the metYt1 and metYt2 terminators when transcription was initiated from Ptet of pBR322, two different fusions of Ptet and lacZ were constructed. First, a fragment carrying Ptet was amplified by PCR using the oligonucleotides 5′-AGATCCAGTTCGATGTAACC-3′ and 5′-TTTTGGATCCAATTTAACTGTGATAAACTACC-3′, cleaved with EcoRI and BamHI, and inserted into plasmid pGOB100, yielding plasmid pMW368. A 262-bp metYt1-metYt2 terminator fragment was amplified by PCR using the oligonucleotides 5′-TTTTGGATCCTTCTGGAAAGTGCTCC-3′ and 5′-TTTTGGATCCGCAGTGTGGATGTGCGACC-3′, trimmed with BamHI, and inserted into pMW368, yielding plasmid pMW381. The EcoRI sites of plasmids pMW368 and pMW381 were converted to an SphI site using the oligonucleotide 5′-AATTGCATGC-3′, yielding plasmids pGOB129 and pGOB127, respectively. The transcriptional fusions on these two plasmids were cloned into the SphI site of plasmid pGOB119, yielding plasmids pGOB134 and pGOB131, respectively.
The constructions on the temperature-sensitive plasmids pGOB117, pGOB118, pGOB121, pGOB122, pGOB126, pGOB131, and pGOB134 were integrated into the lacI-lacZ region of the chromosome of strain MW100 following the allelic replacement procedure described by Hamilton et al. (14), yielding strains GOB438, GOB440, GOB434, GOB435, GOB838, GOB840, and GOB842, respectively. That the constructions had replaced the wild-type lacI-lacZ region was confirmed by PCR.
Strains MW208 and MW210 were used as donors for the transfer of transcriptional fusions of PrpsP and lacZ to different nusA mutants (Table 1). The construction of strains MW208 and MW210 will be published elsewhere. The two strains carry two copies each of the chromosomal region normally containing the ffh+ rpsP+-rimM+-trmD+-rplS+ yfiB′ genes. Between the two copies is the nptI gene, conferring resistance to kanamycin and neomycin. In the right-hand copy of the duplicated region, fusions of PrpsP and lacZ have been integrated, replacing the segment containing rpsP+-rimM+-trmD+-rplS′. The wild-type rpsP+-rimM+-trmD+-rplS operon contains, between PrpsP and rpsP, a terminatorlike structure which is active in vitro (6) and probably functions as a transcriptional attenuator. In strain MW208, the fusion point is downstream from the transcriptional attenuator, while in strain MW210, it is upstream from the attenuator.
In order to examine the effect of induced synthesis of the wild-type NusA protein on the read-through of the two terminators between metY and p15a, plasmid pJML007 was constructed. A DNA fragment containing the nusA gene was amplified by PCR using the oligonucleotides 5′-TTTTGAATTCCCCACTTTTAATAGTCTGG-3′ and 5′-TTTTGGTACCTGTTCCTTCCTGCTACAG-3′, trimmed with EcoRI and KpnI, and inserted into the expression vector pBAD30.
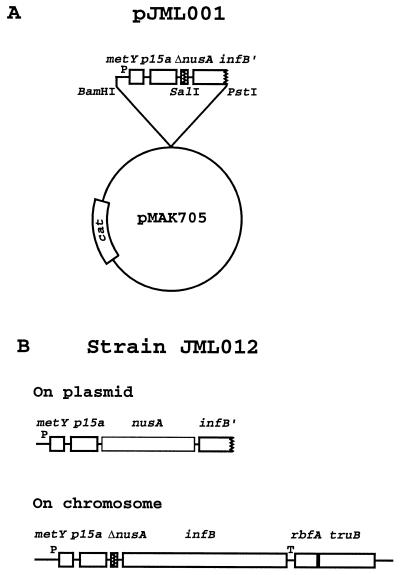
To investigate whether the nusA gene was essential at 37°C, an in-frame deletion of nusA was constructed (see Fig. 7). The region upstream from nusA was amplified by PCR using the oligonucleotides 5′-TTTTGGATCCATTCAACAAATGAAAGTGAAC-3′ and 5′-TTTTGTCGACGGCTTCAACTACAGC-3′, while the region downstream from nusA was amplified with the oligonucleotides 5′-TTTTGTCGACCGTAATATTTGCTGGTTCGG-3′ and 5′-GCATCACACCGTCGTCGG-3′. The two DNA fragments were cleaved with BamHI-SalI and SalI-PstI, respectively, and ligated to the BamHI-PstI-digested plasmid vector pMAK705. The nusA deletion on the resulting plasmid, pJML001, was substituted for the wild-type nusA gene on the chromosme of strain MW100 following the allelic replacement procedure described by Hamilton et al. (14). The resulting strain, JML012, contained the nusA deletion on the chromosome and the wild-type nusA gene on the temperature-sensitive plasmid (see Fig. 7B), as confirmed by PCR analyses.
FIG. 7.
Deletion of the chromosomal nusA gene. (A) Plasmid pJML001 carries a metY-nusA-infB operon fragment containing an in-frame deletion of nusA cloned into the temperature-sensitive allelic replacement vector pMAK705. (B) Genetic organization of the chromosomal metY-nusA-infB operon containing the nusA deletion and that of the complementing nusA+ plasmid after resolution of cointegrates formed between plasmid pJML001 and the chromosome of the wild-type strain, MW100. P represents the P−1 and P1 promoters, and T represents the infBt3 terminator.
Media and growth conditions.
Rich medium was either rich morpholinepropanesulfonic acid (MOPS) (33) or Luria-Bertani (LB) (1) medium supplemented with medium E plus vitamin B1 and 0.4% glucose (52). Cultures were grown at 37°C, and growth was monitored at 600 nm using a Shimadzu UV-1601 spectrophotometer.
Assay of β-galactosidase.
The β-galactosidase activity was measured after permeabilization of whole cells with toluene as described previously (25).
PCR amplification of chromosomal DNA and DNA sequencing.
Regions of the E. coli chromosome were amplified by PCR from colonies resuspended in H2O (26, 44). Pfu DNA polymerase from Stratagene cloning systems, La Jolla, Calif., was used if the obtained fragments were to be cloned into plasmids, and Taq DNA polymerase from Roche Diagnostics Scandinavia AB, Bromma, Sweden, was used in all other cases. The obtained fragments were separated on agarose gels, cut out, and purified using GENE-CLEAN from Bio 101 Inc., La Jolla, Calif. DNA sequencing of PCR fragments and plasmid DNA was done with a Thermo Sequenase II dye terminator cycle-sequencing premix kit from Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England, using an ABI 377 XL DNA Sequencer from PE Applied Biosystems, Stockholm, Sweden.
Northern blot analysis.
Total RNA was prepared according to the method of von Gabain et al. (53) and subjected to Northern blot analysis essentially as described by Sambrook et al. (46). That equal amounts of the RNAs from the different strains grown in LB medium were used was determined both by spectrophotometric measurements at 260 nm and by ethidium bromide staining of aliquots of the RNA electrophoresed on agarose gels. DNA fragments used as probes were purified as described above and labeled with [α-32P]dATP using the Megaprime DNA labeling system from Amersham Pharmacia Biotech.
Primer extension analysis.
Total RNA was prepared as described by von Gabain et al. (53). Primer extension was performed using 3.75 μg of RNA, 32P end-labeled primer (5′-CAGGTGGAAGGGCTGTTCAC-3′), and AMV reverse transcriptase from Roche Diagnostics Scandinavia AB.
Analysis of proteins by 2-D gel electrophoresis.
Steady-state cultures of bacterial cells were grown in rich MOPS medium at 37°C to an optical density at 600 nm of 0.5, labeled for 15 min with 250 μCi of [35S]methionine each (>1,000 Ci/mmol), and chased with 0.167 ml of 0.2 M methionine for 3 min. Extracts were prepared essentially as described by VanBogelen and Neidhardt (51). O'Farrell two-dimensional (2-D) polyacrylamide gels (35) were used to analyze the protein expression pattern. One million counts per minute was loaded onto each first-dimension isoelectric focusing gel (Millipore Intertech, Bedford, Mass.) containing ampholines 3 to 10 and Duracryl acrylamide from Oxford Glycosystems. The first dimension was run as described by the manufacturer, whereas the second dimension was 10 to 17.5% gradient polyacrylamide slab gels containing sodium dodecyl sulfate. The gels were dried and exposed to a PhosphorImager screen and analyzed using ImageQuant software from Molecular Dynamics, Inc.
RESULTS
Identification of 14 suppressor mutations linked to rbfA.
Previously, we isolated several suppressor mutations (sdr-27 to sdr-55; for suppressor to deletion of rimM) that increased the growth rate and translational efficiency of a ΔrimM102 mutant at 37°C (4). Twenty-three of the mutations were shown to be tightly linked to the metY-nusA-infB operon. For one of these strains, PW109 (ΔrimM102 sdr-43), we showed that an increased synthesis of the cold shock protein RbfA encoded by the fifth gene of the metY-nusA-infB operon (Fig. 1A) was responsible for the suppression at 37°C. The results from Northern blot experiments with probes corresponding to different parts of the metY-nusA-infB operon (4; G. O. Bylund and P. M. Wikström, unpublished results) prompted us to examine, by Southern hybridization, whether sdr-43 was a duplication that covered rbfA and truB. The results from this experiment (data not shown) demonstrated that the sdr-43 strain PW109 contained two copies of the rbfA gene and that the 5′ half of the yhbM gene downstream from pnp had been joined to the 3′ part of infB (Fig. 1B). The proposed hybrid region was successfully PCR amplified with a downward-facing yhbM primer and an upward-facing infB primer. The DNA sequence of the obtained PCR product showed that position 369 of yhbM had been fused to position 2295 of infB (data not shown). A putative promoter for yhbM that would explain two very abundant mRNA species (2.6 and 0.9 kb in length) that are present in sdr-43 strains but not in sdr+ strains (4) was identified 21 bp downstream from the transcriptional terminator for pnp. The shorter mRNA probably results from termination at the transcriptional terminator just upstream from rbfA, since it hybridizes to an infB probe but not to an rbfA probe, whereas the longer mRNA also contains the rbfA and truB genes and therefore seems to be responsible for the suppression (4).
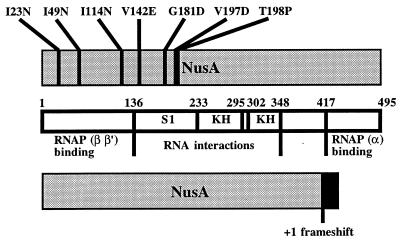
Since the suppressor mutation in strain PW109 increased the synthesis of RbfA (4), we found it conceivable that the other suppressor mutations linked to the metY-nusA-infB operon would also do so. We reasoned that three regions were likely to contain suppressor mutations which could increase expression of rbfA: (i) the region immediately upstream from rbfA, including the transcriptional terminator infBt3 between infB and rbfA; (ii) the region between metY and p15a, containing an internal promoter, an RNase III site the processing at which has been shown to decrease the stability of the downstream part of the mRNA (40), and two internal transcriptional terminators, metYt1 and metYt2; and (iii) the nusA gene, since the NusA protein feedback regulates the expression of the metY-nusA-infB operon (7, 30, 37, 40). Therefore, we sequenced one or more of these candidate regions in 13 of the suppressor strains. In three of the suppressor strains, the major part of the infBt3 terminator between infB and rbfA had been deleted (strain PW106, nucleotides [nt] −41 to −25; strain PW110, nt −40 to −30; and strain PW120, nt −39 to −30, relative to the rbfA start codon). Strain PW119, which was cold sensitive for growth, contained an insertion of IS2, just before the penultimate codon of infB. Nine of the suppressor strains were found to have mutations in nusA (Table 1 and Fig. 2). Two of these strains contained the same mutation (nusA92) in codon 49, substituting asparagine for isoleucine. Of the other seven mutations, six also resulted in single-amino-acid substitutions in the N-terminal half of NusA, while one was a deletion of a single base pair, resulting in replacement of the last 84 amino acids of NusA with 23 amino acids encoded by the +1 reading frame. One of the mutations (nusA98) was identical to a previously isolated conditional-lethal mutation, nusA11 (28, 31), substituting aspartate for glycine in position 181 (8, 17).
FIG. 2.
Locations and natures of different alterations in NusA that suppress the slow growth of a ΔrimM102 mutant. The linear functional map of NusA shown has been modified from reference 23, integrating information from reference 24. S1 and KH represent different motifs found to be important for binding to RNA (2, 11, 49).
The nusA mutations increase the amounts of two metY-nusA-infB operon mRNAs.
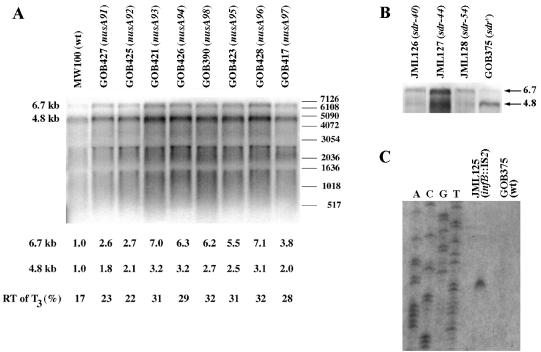
Previously, we found that the suppressor strain PW100 (ΔrimM102 sdr-34), here shown to contain a mutation in nusA (nusA98), expressed dramatically increased levels of two metY-nusA-infB operon mRNA species, 4.8 and 6.7 kb in size, compared to those in the ΔrimM102 mutant MW37 (4). The nusA98 mutation is identical to nusA11, which has been shown to reduce transcription termination (18, 28, 29). Therefore, we wanted to examine whether the different nusA mutations isolated here increased the read-through of transcriptional terminators internal to the metY-nusA-infB operon, leading to an increased synthesis of RbfA, which would then explain the suppression of the slow growth of the ΔrimM102 mutant MW37. The difference in expression of the 4.8- and 6.7-kb transcripts between strains PW100 (ΔrimM102 nusA98) and MW37 (ΔrimM102) might be partly a secondary effect resulting from the 2.5-fold growth rate difference between the two strains (the specific growth rates, k = ln2/g, where g is the mass doubling time in hours, were 0.92 and 0.37, respectively, in LB medium). Therefore, to assess any direct effects of the different nusA mutations on the read-through of metY-nusA-infB operon transcriptional terminators, the amounts of the 4.8- and 6.7-kb mRNAs were determined for rimM+ strains containing the different nusA mutations and showing only minor growth rate differences at 37°C. The levels of the 4.8- and 6.7-kb mRNAs as determined by using the region corresponding to the p15a gene (Fig. 1A) as a probe in a Northern blot experiment were higher in all of the nusA mutants than in the nusA+ strain MW100 (Fig. 3A). The shorter of the two mRNAs probably corresponds to an RNase III-processed form of the initial transcript of 5.0 kb, which starts upstream from metY and terminates at the infBt3 terminator just before rbfA (4, 40, 47). The longer transcript results from read-through of the infBt3 terminator (4), and its size suggests that the 3′ end is between rpsO and pnp. The larger amounts of the 4.8-kb transcript in the nusA mutants relative to the nusA+ strain suggest that the nusA mutations increased the read-through of the metYt1 and metYt2 terminators between metY and p15a, although other explanations, such as increased promoter activities, could not be excluded. However, the relative differences between the amounts of the 6.7-kb transcript for each of the nusA mutants and that in the nusA+ strain were higher (2.6- to 7.1-fold) than those for the 4.8-kb transcript (1.8- to 3.2-fold), clearly indicating that all of the nusA mutations increased the read-through of the infBt3 terminator preceding rbfA. In fact, the calculated read-through of that terminator increased 1.3- to 1.9-fold due to the nusA mutations (Fig. 3A). Thus, NusA is important for transcription termination at least at the infBt3 terminator, but likely also at the metYt1 and metYt2 terminators between metY and p15a. Further, we note that there is a correlation between the degree of suppression and mRNA expression levels, since the nusA mutations of the slowest-growing suppressor strains, PW107 (nusA92) and PW116 (nusA91) (k = 0.53 and 0.66, respectively) increased the read-through of the infBt3 terminator and the amount of the 6.7-kb transcript to a lesser extent than did the suppressor mutations in the faster-growing strains, PW101 (nusA93), PW105 (nusA94), PW100 (nusA98), PW115 (nusA95), PW114 (nusA96), and PW093 (nusA97), which had specific growth rates, k, between 0.79 and 0.92.
FIG. 3.
Transcriptional analyses of the metY-nusA-infB operon in different mutants. (A) Quantitation of metY-nusA-infB operon mRNAs in wild-type (wt) and nusA mutant strains by Northern blot analysis. Five micrograms of total RNA was subjected to electrophoresis in an agarose gel containing formaldehyde, transferred to a Hybond N filter, and probed with a radiolabeled PCR fragment corresponding to the p15a gene. The strains used (with the relevant genetic markers in parentheses) are indicated above the respective lanes. The sizes of the 32P-end-labeled fragments of the 1-kb DNA ladder (GIBCO BRL Life Technologies Inc., Gaithersburg, Md.) are indicated. The 6.7-kb transcript results from read-through of the metYt1 and metYt2 terminators between metY and p15a and the infBt3 terminator just upstream from rbfA, while the 4.8-kb transcript terminates at the infBt3 terminator (Fig. 1). The amounts of these transcripts (determined by quantitation of the radioactivity using a PhosphorImager from Molecular Dynamics, Inc.) in the different nusA mutants were normalized to those for the nusA+ strain MW100. The read-through (RT) of the infBt3 terminator was calculated as the amount of radioactivity in the 6.7-kb band divided by the sum of the radioactivity in the 4.8- and 6.7-kb bands. (B) Northern blot analysis showing the effect of deletions in the infBt3 transcriptional terminator on the amount of the 6.7-kb transcript relative to that of the 4.8-kb transcript. (C) Identification of the 5′ end of the mRNA resulting from transcription initiation at a new promoter created by the insertion of IS2 in infB. Primer extension analyses of mRNA and DNA sequencing of a PCR fragment covering the 3′ part of infB and the 5′ part of rbfA from a wild-type strain were performed using a 32P-end-labeled primer binding to positions −54 to −73 relative to the start codon of rbfA. The primer extension product obtained for strain JML125 (infB::IS2) corresponds to an mRNA 5′ end at the A 6 nt downstream from the −10 region of the proposed promoter.
The suppressor mutations increase the amount of the RbfA protein.
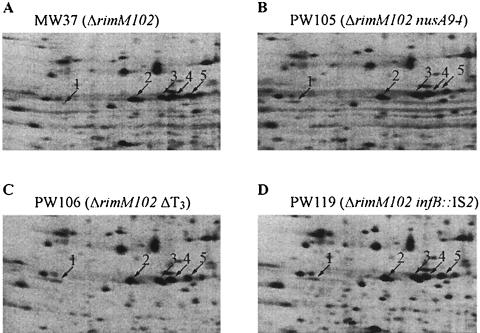
Conceivably, the three suppressor mutations deleting the major part of the terminator infBt3 between infB and rbfA increased the amount of the 6.7-kb read-through transcript relative to that of the 4.8-kb transcript resulting from termination at this terminator (Fig. 3B). As mentioned above, one of the suppressor strains contained an insertion of IS2 in infB. There are a number of examples where IS2 activates the transcription of genes located downstream from the insertion point, probably by creating hybrid promoters (10). We note that in the infB sequence there is a four-out-of-six match (underlined) (TACCAT) to the consensus sequence for a −10 promoter region 17 bp downstream from a postulated −35 region in the left end of the IS2 insertion (10). To examine whether the proposed promoter could initiate transcription, mRNA from strain JML125 containing the infB::IS2 mutation was subjected to primer extension analysis using a primer binding upstream from the infBt3 terminator. A primer extension product corresponding to an mRNA 5′ end 6 nt downstream from the −10 hexamer of the proposed promoter was obtained for strain JML125, whereas no primer extension product was seen for the control strain, GOB375 (Fig. 3C), indicating that the IS2 insertion in infB had created a new promoter for rbfA. To examine whether the suppressor mutations increased the synthesis of the RbfA protein, total protein extracts from suppressor mutants were analyzed by 2-D protein gel electrophoresis. The amounts of RbfA in the suppressor mutants PW105 (ΔrimM102 nusA94), PW106 (ΔrimM102 ΔinfBt3), and PW119 (ΔrimM102 infB::IS2) were severalfold higher than in the suppressor-free ΔrimM102 mutant MW37 (Fig. 4). Thus, these findings indicate that the suppressor mutations increasing the growth rate of the ΔrimM102 mutant MW37 were obtained because they increase the synthesis of RbfA.
FIG. 4.
Synthesis of individual proteins at 37°C in the ΔrimM102 mutant and three different suppressor strains. Total cell extracts of the indicated strains labeled with [35S]methionine were separated on 2-D gels. Only the relevant part of each gel is shown. The indicated proteins are as follows: 1, RbfA; 2, H-NS; 3, 4, and 5, ribosomal protein S6. The position of RbfA on the gels was determined previously (4), and the identities of the other proteins were obtained by comparing the gels with those of VanBogelen et al. (50).
The nusA98 mutant is deficient in NusA-mediated transcriptional feedback regulation at the terminators between metY and p15a.
Previously, the metYt1 and metYt2 terminators between metY and p15a were suggested to be the target for the NusA-mediated negative-feedback regulation of transcription of the metY-nusA-infB operon (30, 37). However, formally the possibility that the region upstream from the two terminators, which contains promoters and the metY gene, was the site of regulation could not be excluded. Furthermore, other results indicated that the regulatory site is located further downstream (7). To investigate whether the metYt1 and metYt2 terminators were a target for the NusA-mediated regulation, the effect of increased synthesis of NusA from an expression vector on the read-through of the terminators was examined. Transcriptional fusions between metY with or without the terminators and lacZ were constructed and integrated into the lacI-lacZ region of the chromosome of a rimM+ strain (Fig. 5; see Materials and Methods), and the effect of arabinose-induced synthesis of wild-type NusA from the PBAD promoter in plasmid pJML007 on the activity of β-galactosidase was measured. The read-through of the two terminators was approximately 25% when no arabinose was added or when the expression vector did not contain the nusA gene, as judged from a comparison of the β-galactosidase activity of the terminator-containing lacZ fusion with that of the fusion lacking the two terminators (Table 2). The expression of the terminator-containing lacZ fusion of strain GOB492 dropped almost twofold, whereas that of the lacZ fusion lacking the two terminators (strain GOB496) was not significantly affected when NusA synthesis was induced with 0.2% arabinose. This suggests that the target for the NusA-mediated feedback regulation of the metY-nusA-infB operon expression indeed is the terminators between metY and p15a. Further, the read-through was 1.7-fold higher in the nusA98 mutant than in the nusA+ strain at 37°C when synthesis of the wild-type NusA protein from plasmid pJML007 was not induced, indicating that the mutant NusA protein is deficient in feedback regulation at the terminators (Table 2). However, induction of wild-type NusA protein synthesis with 0.2% arabinose in the nusA98 mutant restored termination to wild-type levels. The higher β-galactosidase activity (i.e., enzyme activity per unit of optical density of the culture) of the fusion lacking the terminator in the nusA98 strains relative to that in nusA+ strains seemed to result from spontaneous lysis of the nusA98 strains (leading to an underestimation of the cell culture density). However, an increased lysis was also observed for the nusA98 strains that carried the lacZ fusion containing the metYt1 and metYt2 terminators, and thus, the calculated transcriptional read-through was not affected by this lysis.
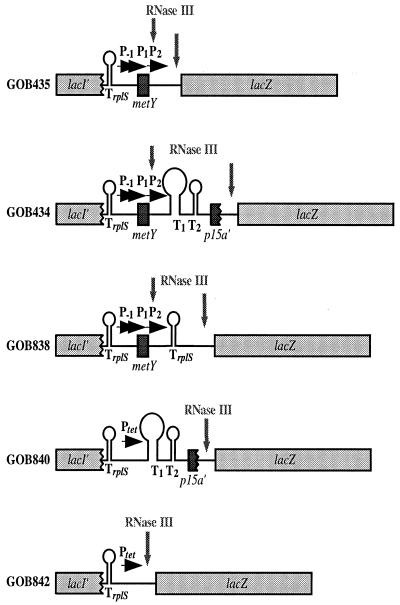
FIG. 5.
Transcriptional fusions integrated into the lacI-lacZ region of the chromosome. The fusion point in strain GOB434 is 245 bp downstream from that in strain GOB435. P−1, P1, and P2 indicate promoters of the metY operon, and Ptet is the promoter for the tetracycline resistance gene of plasmid pBR322; T1 and T2 indicate the terminators between metY and p15a, and TrplS is the terminator of the trmD operon. Two different RNase III-processing sites are indicated, one (left-hand arrow) native to the region between metY and p15a and the other (right-hand arrow) present in the lacZ fusion vector used (22). For a description of the construction of the different fusions, see Materials and Methods.
TABLE 2.
NusA-mediated transcriptional feedback regulation at the terminators between metY and p15aa
| Relevant genotype | β-Galactosidase activity
|
Read-throughb
|
||||
|---|---|---|---|---|---|---|
| P
|
P + T
|
|||||
| − | + | − | + | − | + | |
| nusA+/pJML007(nusA+) | 5,782 (5,804, 5,760) | 6,050 (6,461, 5,638) | 1,472 (1,490, 1,453) | 860 (874, 846) | 0.25 | 0.14 |
| nusA+/pBAD30 | 5,952 (5,938, 5,966) | 5,599 (5,720, 5,318) | 1,498 (1,537, 1,460) | 1,410 (1,454, 1,367) | 0.25 | 0.25 |
| nusA98/pJML007(nusA+) | 7,408 (8,070, 6747) | 6,363 (6,352, 6,374) | 3,112 (3,164, 3060) | 1,476 (1,527, 1,425) | 0.42 | 0.23 |
| nusA98/pBAD30 | 7,782 (8,148, 7,417) | 7,521 (7,860, 7,182) | 3,298 (3,434, 3,161) | 2,991 (3,070, 2,912) | 0.42 | 0.40 |
Expression of the PmetY-metY-lacZ (P) and PmetY-metY-metYt1t2-lacZ (P + T) fusions is shown in Miller units. The average expression from two independent experiments is shown for each strain, with the actual values from the respective experiment within parentheses. pBAD30 is the plasmid vector control for pJML007 (nusA+). + indicates that a concentration of 0.2% arabinose was used for induction of the pBAD promoter on the two plasmids; − indicates no arabinose.
Calculated as (P + T)/P.
To investigate whether the effect of the nusA98 mutation on the read-through of the terminators between metY and p15a was different in a ΔrimM102 and in a rimM+ background, the expression of the lacZ fusions was also measured in strains containing the ΔrimM102 mutation. The expression of the lacZ fusions in the ΔrimM102 strains was slightly lower than that in the rimM+ strains; however, the read-through of the terminators was not dependent on the allelic state of rimM either in nusA+ or in nusA98 strains (Table 3).
TABLE 3.
Effect of the nusA98 mutation on the read-through of transcriptional terminators between metY and p15a in a ΔrimM mutanta
| Relevant genotype | β-galactosidase activity
|
Read-throughb | |
|---|---|---|---|
| P | P + T | ||
| ΔrimM102 nusA+ | 4,204 (3,712–4,681) | 1,024 (998–1,070) | 0.24 |
| ΔrimM102 nusA98 | 4,455 (3,324–5,346) | 1,898 (1,658–2,338) | 0.43 |
| rimM+nusA+ | 5,063 (4,726–5,584) | 1,226 (1,123–1,293) | 0.24 |
| rimM+nusA98 | 6,491 (5,698–7,282) | 2,638 (2,446–2,767) | 0.41 |
Expression of the PmetY-metY-lacZ (P) and PmetY-metY-metYt1t2-lacZ (P + T) fusions is shown in Miller units. The average expression shown is from four independent experiments for the ΔrimM102 strains and from five independent experiments for the rimM+ strains. The variation in expression is shown within parentheses. The expression of a control construction (strain GOB438) lacking the metY insert and the cloning cassette was 17 Miller units, while a similar construction (strain GOB440) containing the metYt1 and metYt2 terminators expressed 3 Miller units of β-galactosidase activity in a ΔrimM102 nusA+ strain.
Calculated as (P + T)/P.
The new nusA mutations increase the read-through of the terminators between metY and p15a.
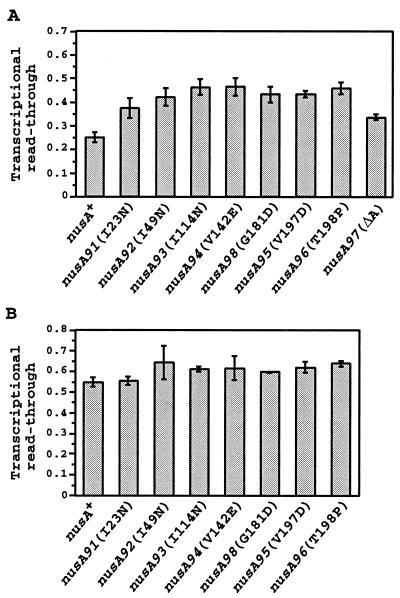
To examine whether the new nusA mutations isolated here also increased the read-through of the metYt1 and metYt2 terminators between metY and p15a, the different nusA mutations were introduced into the lacZ fusion strains and the β-galactosidase activity was measured. The eight different nusA mutations had only minor effects on the expression of the fusion containing the metY promoter fragment (data not shown); however, they increased the read-through of the terminators between metY and p15a 1.3- to 1.9-fold (Fig. 6A). This is in agreement with the higher levels of the 4.8-kb mRNA in the different nusA mutants relative to that in the nusA+ strain (Fig. 3A). To investigate whether the different nusA mutations also affected the termination at a completely different internal terminator, the read-through of the trmD operon attenuator (5, 6) was measured. The nusA mutations were introduced into strains containing either a fusion of the trmD operon promoter PrpsP and lacZ or PrpsP, the attenuator, and lacZ. The read-through of the attenuator was calculated as the ratio of the β-galactosidase activity of the fusion containing the attenuator to that of the fusion lacking the attenuator. Evidently, the nusA mutations had little or no effect on the read-through of the trmD operon attenuator (Fig. 6B). This suggests either that the different nusA mutations were specific for the terminators within the metY-nusA-infB operon or, perhaps more likely, that NusA does not enhance termination at the trmD operon attenuator.
FIG. 6.
Transcriptional read-through of terminators in different nusA mutants. The read-through of the terminators T1 and T2 between metY and p15a (A) and that of the attenuator upstream from rpsP (B) were calculated as the ratio of the β-galactosidase activity from a lacZ fusion containing the respective terminator(s) to the activity from a fusion lacking the terminator(s) in the genetic backgrounds indicated. The standard deviations are shown as error bars.
NusA-mediated feedback regulation is not promoter or terminator specific.
To investigate whether the observed NusA-mediated feedback regulation of transcription termination was promoter or terminator specific, we made two different chimeric promoter-terminator constructions fused to lacZ and quantified the read-through in nusA+ and nusA94 strains. First, the terminator, rplSt, of the trmD operon was substituted for the fragment containing the metYt1 and metYt2 terminators in the fusion described earlier (Fig. 5). From measurements of the β-galactosidase activities of the different constructions, the transcriptional read-through of metYt1 and metYt2 was found to be more than twofold higher than that of rplSt (Table 4), demonstrating that the rplSt terminator is more efficient than the metYt1 and metYt2 terminators. However, the effect of the nusA94 mutation on the read-through of the rplSt terminator was as pronounced as that on the read-through of the metYt1-metYt2 terminators (Table 4), showing that the NusA-mediated transcriptional termination was not absolutely dependent on the metYt1 and metYt2 terminators. To examine whether the feedback regulation at the metYt1 and metYt2 terminators requires the presence of the native promoters for the metY-nusA-infB operon, Ptet from pBR322 was substituted for the fragment containing the P−1, P1, and P2 promoters in the fusions between metY, with or without the metYt1 and metYt2 terminator fragment, and lacZ described above (Fig. 5). The effect of the nusA94 mutation on the read-through of the metYt1 and metYt2 terminators was comparable for the fusions containing either the Ptet promoter or the fragment with the metY promoters (Table 4). Thus, the NusA-mediated feedback regulation at the metYt1 and metYt2 terminators is not dependent on the presence of the metY operon promoters. Interestingly, the read-through of the metYt1 and metYt2 terminators was severalfold higher in both nusA+ and nusA94 strains when the native metY promoter fragment was present than when the Ptet promoter fragment was used.
TABLE 4.
NusA-mediated feedback regulation of transcriptional termination in different chimeric promoter-terminator constructions
| Promoter | Terminator | Transcriptional read-througha
|
||
|---|---|---|---|---|
| nusA+ | nusA94 | nusA94/nusA+ | ||
| PmetY | metYt1t2 | 0.25 (0.22–0.27) | 0.42 (0.40–0.43) | 1.7 |
| PmetY | rplSt | 0.089 (0.070–0.12) | 0.20 (0.20–0.21) | 2.2 |
| Ptet | metYt1t2 | 0.035 (0.033–0.037) | 0.073 (0.063–0.081) | 2.1 |
Transcriptional read-through was calculated as the ratio of the β-galactosidase activity of PmetY-metYt1t2-lacZ to that of PmetY lacZ, that of PmetY-TrplSt-lacZ to that of PmetY-lacZ, and that of Ptet-metYt1t2-lacZ to that of Ptet-lacZ in nusA as well as nusA94 strains. The values presented are the averages from three independent experiments, with the variation shown within parentheses.
The nusA gene is essential at 37°C.
A deletion of the nusA gene is lethal at 42°C in a rho+ strain but not in rho mutants (55). Also, the nusA(Am113) amber mutation is lethal at 42°C in a strain with a temperature-sensitive supF6 tRNA suppressor (32). Further, rho+ strains with the nusA11 missense mutation cannot grow at 42°C (28, 31). All of the nusA mutations isolated here were also found to confer a temperature-sensitive phenotype (data not shown). Since the nusA mutations were selected as fast-growing derivatives of a ΔrimM102 mutant at 37°C, it was surprising that all were temperature sensitive. This made us consider the possibility that the NusA protein, or at least its function in transcription termination, was essential at 42°C but not at 37°C. Therefore, we constructed an in-frame deletion of nusA in the plasmid vector pMAK705 (Cmr) containing a temperature-sensitive replicon (Fig. 7). Clones carrying cointegrates between the recombinant plasmid pJML001 and the chromosome were selected for at 44°C on chloramphenicol plates. The obtained clones were grown at 30°C to select for resolution of the cointegrates. (Cells containing cointegrates grow slowly at 30°C due to replication of the chromosome from the plasmid replicon.) One of these segregants (JML012) was shown by PCR to carry the nusA deletion on the chromosome and the wild-type nusA gene on the plasmid (Fig. 7). The plating efficiency of JML012 at 37°C was only 10% of that at 30°C in a viable-count experiment (data not shown). The surviving colonies were Cmr, suggesting that they still contained the complementing plasmid, either free in the cytoplasm or as a cointegrate. Upon restreaking of these colonies, most continued to show a low plating efficiency and exhibited a heterogeneous growth phenotype, indicating a further loss of the plasmid with a concomitant death of the cells. Colonies that grew well at 37°C were Cmr and grew poorly at 30°C, typical for clones with the plasmid integrated into the chromosome. These findings suggested that the NusA protein is also essential at 37°C. This was also corroborated by an experiment in which the expression of nusA+ was from the PBAD promoter in plasmid pJML007; the temperature-sensitive plasmid (Cmr) that carries nusA+ in strain JML012 with nusA deleted on the chromosome (Fig. 7) was replaced by pJML007 (Cbr) containing nusA+ under the control of the arabinose-inducible PBAD promoter by transformation and selection for Cbr at 44°C in the presence of 0.2% arabinose. One Cms transformant, JML087, was tested for the ability to grow at 30, 37, and 44°C on plates lacking or containing arabinose. At all three temperatures, strain JML087 grew only in the presence of arabinose, demonstrating that nusA is essential at these temperatures.
DISCUSSION
In this report, we describe the identification of 14 mutations in the metY-nusA-infB operon that suppress the slow growth of a ΔrimM102 mutant. These suppressor mutations increase the expression of rbfA, encoded by the fifth gene of the metY-nusA-infB operon, in accordance with the previous finding that overexpression of RbfA suppresses the slow growth and translational deficiency of the ΔrimM102 mutant (4). RimM and RbfA are both crucial for efficient maturation of the 30S ribosomal subunits (4). The plethora of suppressor mutations that increase the expression of rbfA emphasizes the importance of elevated levels of RbfA in strains lacking RimM and seems to connect the function of RbfA to that of RimM in ribosome maturation.
Nine of the isolated suppressor mutations were localized to the nusA gene encoding the transcriptional elongation factor NusA, important for termination and antitermination of transcription. Transcription of the metY-nusA-infB operon can be repressed by overexpression of nusA from plasmids and derepressed by nusA mutations (30, 37). It was suggested that the two transcriptional terminators (metYt1 and metYt2) between metY and p15a were the target for the autoregulation (30, 37). The addition of NusA to an in vitro transcription system completely prevented the read-through of the metYt1 terminator by the RNA polymerase (40). Similarly, in vitro transcription of the nusA gene was inhibited by a plasmid expressing nusA; however, in this case it was concluded that the two terminators were not the target for the regulation (7). Here we demonstrate that one target for NusA-mediated autoregulation in vivo resides within a 245-nt region that contains the metYt1 and metYt2 terminators. Previously, a 7- to 10-fold plasmid-mediated overexpression of nusA was found to reduce the expression of chromosomal metY-nusA-infB operon lacZ fusions by 50% (37). A similar reduction in the read-through (from 0.25 to 0.14) of the terminators was obtained when we induced the expression of a plasmid-carried copy of nusA from a PBAD promoter with 0.2% arabinose in a nusA+ strain. This amount of arabinose also reduced the read-through to the same extent (from 0.42 to 0.23) in the nusA98 mutant (and restored termination to wild-type levels), indicating that the amount of NusA produced from the plasmid under these conditions was similar to that expressed from the nusA+ gene on the chromosome. Thus, as little as an approximately twofold overexpression of nusA is sufficient to repress efficiently the read-through of the terminators.
The effect of the different nusA mutations on the transcriptional read-through at the metYt1 and metYt2 terminators varied from 1.3- to 1.9-fold (Fig. 6A), as calculated from the results obtained with lacZ transcriptional fusions. Similarly, the effect of the mutations on the read-through at the infBt3 terminator just upstream from rbfA was 1.3- to 1.9-fold, as judged from the quantification of the amounts of the 4.8- and 6.7-kb transcripts detected in the Northern blotting experiment (Fig. 3A). Interestingly, the relative derepression (1.8- to 3.2-fold) in the nusA mutants of the 4.8-kb transcript, which requires the read-through of the metYt1 and metYt2 terminators, was higher than the 1.3- to 1.9-fold increase in read-through of these terminators. Possibly, this difference could be accommodated by invoking other sites on the mRNA at which NusA could promote transcriptional termination in accordance with in vitro results that suggest that NusA acts downstream from the metYt1 and metYt2 terminators (7). Alternatively, the difference could result from an increased metY promoter activity in the nusA mutants; however, we find this explanation unlikely, since the difference in expression of the metY-lacZ fusion lacking the metYt1 and metYt2 terminators between the nusA+ strain and the nusA mutants was less than 10% in the experiments shown in Fig. 6A (data not shown).
The effect of the nusA94 mutation on the read-through of different terminators in some chimeric promoter-terminator constructions was dependent neither on the native promoters nor on the native terminators of the metY-nusA-infB operon, suggesting that this mutation has a general effect on transcription termination. Similarly, the nusA11 mutation, identical to nusA98 described here, has been shown to decrease termination efficiency at different terminators (18, 28, 29). However, neither of the nusA mutations studied here seemed to affect the read-through of the trmD operon attenuator. Conceivably, NusA might not be involved in termination at this terminator structure because of the short distance between the promoter and the attenuator (5), which could decrease the possibility for NusA to bind to the RNA polymerase before it reaches the terminator structure. Interestingly, the read-through of the metYt1 and metYt2 terminators was severalfold higher when transcription initiation was from the native metY promoter fragment than when it was from the Ptet promoter in both a wild-type and a nusA94 mutant strain. Thus, the metY promoter fragment (including the metY gene) seems to have an NusA-independent antitermination function.
Sequence and structural alignments have suggested that NusA contains similarities to the proposed RNA binding domains S1 (2) and KH (11), which seem important for interactions with RNA during termination and antitermination (23). Four of the amino acid substitutions in NusA studied here, V142E (nusA94), G181D (nusA98), V197D (nusA95), and T198P (nusA96), are in the region of homology to the S1 RNA binding domain, supposedly explaining their negative effect on the NusA-mediated feedback regulation at the terminators between metY and p15a and at that just upstream from rbfA. We note that the V142E substitution is in one of the most conserved positions of the S1 region whereas the G181D, V197D, and T198P substitutions are in less conserved positions (2). However, since all four substitutions result in dramatic changes of the amino acid side chain in the respective position, they might have altered the structure of the entire S1 homology region. Two other substitutions in the same region, L183R and R199A, cause defects in the interaction between NusA and the nut site RNA (24), corroborating the importance of this region of NusA in termination and antitermination of transcription.
The 79 C-terminal amino acids of NusA prevent the RNA binding regions of NusA from interacting with RNA. However, interactions between the C-terminal domain of the α subunit of the RNA polymerase and the 79 C-terminal amino acids of NusA seem to allow the RNA binding regions of NusA to bind to RNA (24). Since NusA lacking the 79 C-terminal amino acids binds RNA alone and is proficient in transcription termination (23, 24), it is surprising that the nusA97 mutation, which results in the substitution of 23 amino acids encoded by the +1 reading frame for the C-terminal 84 amino acids of NusA (due to a deletion of 1 nt in codon 412), confers a reduced termination at the internal terminators of the metY-nusA-infB operon. We note that the truncated NusA protein seems to be more affected in its ability to promote termination at the infBt3 terminator just upstream from rbfA than at the metYt1 and metYt2 terminators between metY and p15a (cf. Fig. 3A and 6A). Possibly, the 23 new amino acids added to the C-terminally truncated NusA interfere with the ability of NusA to interact with some terminators.
NusA is essential for bacterial growth at temperatures above 42 and below 32°C (28, 31, 32, 48, 55). Since all of the nusA mutations isolated here conferred temperature-sensitive phenotypes, we considered it possible that NusA was not essential at 37°C. However, here we show that NusA is also essential at this temperature by controlling the expression of nusA from an inducible promoter. Further, we discovered that the temperature sensitivity of the nusA mutants studied here could be partially suppressed by increasing the sodium chloride concentration in the medium from 0.5 to 1 to 2% (data not shown). However, the termination function of NusA at 37°C was not restored by increased levels of sodium chloride, as demonstrated by efficient suppression of the slow growth of a ΔrimM102 mutant at increased levels of sodium chloride (data not shown). We suggest that the temperature sensitivity conferred by the nusA mutations is the result of increased degradation of the mutant NusA proteins at high temperature and that this proteolysis can be inhibited by exogenous salt, as suggested for many other temperature-sensitive mutants (20). However, we cannot exclude the possibility that the nusA mutations isolated here reduce termination at a terminator(s) that is essential at high temperatures and that the effect of this reduced termination can be suppressed by increased concentration of salt by some unknown mechanism.
The high frequency of nusA mutations among the suppressors of the ΔrimM102 mutation together with the straightforward complementation of obtained mutations by arabinose-induced expression of wild-type NusA from plasmid pJML007 make this an excellent system for isolating several new termination-deficient nusA mutants.
ACKNOWLEDGMENTS
This work was supported by the Swedish Natural Science Research Council (B-BU 9911), the Carl Trygger Foundation, the Magnus Bergvall Foundation, and the Kempe Foundations.
G. R. Björk, T. G. Hagervall, J. Johansson, and O. P. Persson are acknowledged for their helpful comments on the manuscript.
REFERENCES
- 1.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bycroft M, Hubbard T J P, Proctor M, Freund S M V, Murzin A G. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 3.Bylund G O, Persson B C, Lundberg L A C, Wikström P M. A novel ribosome-associated protein is important for efficient translation in Escherichia coli. J Bacteriol. 1997;179:4567–4574. doi: 10.1128/jb.179.14.4567-4574.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bylund G O, Wipemo L C, Lundberg L A C, Wikström P M. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J Bacteriol. 1998;180:73–82. doi: 10.1128/jb.180.1.73-82.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byström A S, Hjalmarsson K J, Wikström P M, Björk G R. The nucleotide sequence of an Escherichia coli operon containing genes for the tRNA(m1G)methyltransferase, the ribosomal proteins S16 and L19 and a 21-K polypeptide. EMBO J. 1983;2:899–905. doi: 10.1002/j.1460-2075.1983.tb01519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byström A S, von Gabain A, Björk G R. Differentially expressed trmD ribosomal protein operon of Escherichia coli is transcribed as a single polycistronic mRNA species. J Mol Biol. 1989;208:575–586. doi: 10.1016/0022-2836(89)90149-6. [DOI] [PubMed] [Google Scholar]
- 7.Cenatiempo Y, Deville F, Brot N, Weissbach H. In vitro expression of the Escherichia coli nusA-infB operon. J Biol Chem. 1987;262:152–157. [PubMed] [Google Scholar]
- 8.Craven M G, Friedman D I. Analysis of the Escherichia coli nusA10(Cs) allele: relating nucleotide changes to phenotypes. J Bacteriol. 1991;173:1485–1491. doi: 10.1128/jb.173.4.1485-1491.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dammel C S, Noller H F. Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev. 1995;9:626–637. doi: 10.1101/gad.9.5.626. [DOI] [PubMed] [Google Scholar]
- 10.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 11.Gibson T J, Thompson J D, Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 1993;324:361–366. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- 12.Granston A E, Thompson D L, Friedman D I. Identification of a second promoter for the metY-nusA-infB operon of Escherichia coli. J Bacteriol. 1990;172:2336–2342. doi: 10.1128/jb.172.5.2336-2342.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishii S, Ihara M, Maekawa T, Nakamura Y, Uchida H, Imamoto F. The nucleotide sequence of the cloned nusA gene and its flanking region of Escherichia coli. Nucleic Acids Res. 1984;12:3333–3342. doi: 10.1093/nar/12.7.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishii S, Kuroki K, Imamoto F. tRNAMetf2 gene in the leader region of the nusA operon in Escherichia coli. Proc Natl Acad Sci USA. 1984;81:409–413. doi: 10.1073/pnas.81.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito K, Egawa K, Nakamura Y. Genetic interaction between the β′ subunit of RNA polymerase and the arginine-rich domain of Escherichia coli nusA protein. J Bacteriol. 1991;173:1492–1501. doi: 10.1128/jb.173.4.1492-1501.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin D J, Cashel M, Friedman D I, Nakamura Y, Walter W A, Gross C A. Effects of rifampicin resistant rpoB mutations on antitermination and interaction with nusA in Escherichia coli. J Mol Biol. 1988;204:247–261. doi: 10.1016/0022-2836(88)90573-6. [DOI] [PubMed] [Google Scholar]
- 19.Jones P G, Inouye M. RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol Microbiol. 1996;21:1207–1218. doi: 10.1111/j.1365-2958.1996.tb02582.x. [DOI] [PubMed] [Google Scholar]
- 20.Kohno T, Schmid M, Roth J. Effect of electrolytes on growth of mutant bacteria. In: Rains D W, Valentine R C, Hollaender A, editors. Genetic engineering of osmoregulation. New York, N.Y: Plenum Publishing Corp.; 1980. pp. 53–57. [Google Scholar]
- 21.Lerner C G, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linn T, St. Pierre R. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol. 1990;172:1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mah T-F, Li J, Davidson A R, Greenblatt J. Functional importance of regions in Escherichia coli elongation factor NusA that interact with RNA polymerase, the bacteriophage lambda N protein and RNA. Mol Microbiol. 1999;34:523–537. doi: 10.1046/j.1365-2958.1999.01618.x. [DOI] [PubMed] [Google Scholar]
- 24.Mah T F, Kuznedelov K, Mushegian A, Severinov K, Greenblatt J. The α subunit of E. coli RNA polymerase activates RNA binding by NusA. Genes Dev. 2000;14:2664–2675. doi: 10.1101/gad.822900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura Y, Mizusawa S. In vivo evidence that the nusA and infB genes of E. coli are part of the same multi-gene operon which encodes at least four proteins. EMBO J. 1985;4:527–532. doi: 10.1002/j.1460-2075.1985.tb03660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura Y, Mizusawa S, Court D L, Tsugawa A. Regulatory defects of a conditionally lethal nusAts mutant of Escherichia coli. Positive and negative modulator roles of NusA protein in vivo. J Mol Biol. 1986;189:103–111. doi: 10.1016/0022-2836(86)90384-0. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura Y, Mizusawa S, Tsugawa A, Imai M. Conditionally lethal nusAts mutation of Escherichia coli reduces transcription termination but does not affect antitermination of bacteriophage lambda. Mol Gen Genet. 1986;204:24–28. doi: 10.1007/BF00330182. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura Y, Plumbridge J, Dondon J, Grunberg-Manago M. Evidence for autoregulation of the nusA-infB operon of Escherichia coli. Gene. 1985;36:189–193. doi: 10.1016/0378-1119(85)90085-x. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura Y, Tsugawa A. Role of Escherichia coli NusA protein in transcription termination: ts mutant and RNA recognition. In: Calendar R, Gold L, editors. Sequence specificity in transcription and translation. 30. Alan R. New York, N.Y: Liss, Inc.; 1985. pp. 185–196. [Google Scholar]
- 32.Nakamura Y, Uchida H. Isolation of conditionally lethal amber mutations affecting synthesis of the nusA protein of Escherichia coli. Mol Gen Genet. 1983;190:196–203. doi: 10.1007/BF00330640. [DOI] [PubMed] [Google Scholar]
- 33.Neidhardt F C, Bloch P L, Pedersen S, Reeh S. Chemical measurement of steady-state levels of ten aminoacyl-transfer ribonucleic acid synthetases in Escherichia coli. J Bacteriol. 1977;129:378–387. doi: 10.1128/jb.129.1.378-387.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nurse K, Wrzesinski J, Bakin A, Lane B G, Ofengand J. Purification, cloning, and properties of the tRNA Ψ55 synthase from Escherichia coli. RNA. 1995;1:101–112. [PMC free article] [PubMed] [Google Scholar]
- 35.O'Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 36.Persson B C, Bylund G O, Berg D E, Wikström P M. Functional analysis of the ffh-trmD region of the Escherichia coli chromosome by using reverse genetics. J Bacteriol. 1995;177:5554–5560. doi: 10.1128/jb.177.19.5554-5560.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plumbridge J A, Dondon J, Nakamura Y, Grunberg-Manago M. Effect of NusA protein on expression of the nusA, infB operon in E. coli. Nucleic Acids Res. 1985;13:3371–3388. doi: 10.1093/nar/13.9.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plumbridge J A, Howe J G, Springer M, Touati-Schwartz D, Hershey J W B, Grunberg-Manago M. Cloning and mapping of a gene for translational initiation factor IF2 in Escherichia coli. Proc Natl Acad Sci USA. 1982;79:5033–5037. doi: 10.1073/pnas.79.16.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Portier C, Régnier P. Expression of the rpsO and pnp genes: structural analysis of a DNA fragment carrying their control regions. Nucleic Acids Res. 1984;12:6091–6102. doi: 10.1093/nar/12.15.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Régnier P, Grunberg-Manago M. Cleavage by RNase III in the transcripts of the metY-nusA-infB operon of Escherichia coli releases the tRNA and initiates the decay of the downstream mRNA. J Mol Biol. 1989;210:293–302. doi: 10.1016/0022-2836(89)90331-8. [DOI] [PubMed] [Google Scholar]
- 41.Régnier P, Grunberg-Manago M, Portier C. Nucleotide sequence of the pnp gene of Escherichia coli encoding polynucleotide phosphorylase. Homology of the primary structure of the protein with the RNA-binding domain of ribosomal protein S1. J Biol Chem. 1987;262:63–68. [PubMed] [Google Scholar]
- 42.Régnier P, Portier C. Initiation, attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J Mol Biol. 1986;187:23–32. doi: 10.1016/0022-2836(86)90403-1. [DOI] [PubMed] [Google Scholar]
- 43.Sacerdot C, Dessen P, Hershey J W B, Plumbridge J A, Grunberg-Manago M. Sequence of the initiation factor IF2 gene: unusual protein features and homologies with elongation factors. Proc Natl Acad Sci USA. 1984;81:7787–7791. doi: 10.1073/pnas.81.24.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 45.Saito M, Tsugawa A, Egawa K, Nakamura Y. Revised sequence of the nusA gene of Escherichia coli and identification of nusA11 (ts) and nusA1 mutations which cause changes in a hydrophobic amino acid cluster. Mol Gen Genet. 1986;205:380–382. doi: 10.1007/BF00430455. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Sands J F, Régnier P, Cummings H S, Grunberg-Manago M, Hershey J W. The existence of two genes between infB and rpsO in the Escherichia coli genome: DNA sequencing and S1 nuclease mapping. Nucleic Acids Res. 1988;16:10803–10816. doi: 10.1093/nar/16.22.10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schauer A T, Carver D L, Bigelow B, Baron L S, Friedman D I. λ N antitermination system: functional analysis of phage interactions with the host NusA protein. J Mol Biol. 1987;194:679–690. doi: 10.1016/0022-2836(87)90245-2. [DOI] [PubMed] [Google Scholar]
- 49.Siomi H, Matunis M J, Michael W M, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.VanBogelen R A, Abshire K Z, Pertsemlidis A, Clark R L, Neidhardt F C. Gene-protein database of Escherichia coli K-12, edition 6. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2067–2117. [Google Scholar]
- 51.VanBogelen R A, Neidhardt F C. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5589–5593. doi: 10.1073/pnas.87.15.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 53.von Gabain A, Belasco J G, Schottel J L, Chang A C, Cohen S N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci USA. 1983;80:653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wikström P M, Byström A S, Björk G R. Nonautogenous control of ribosomal protein synthesis from the trmD operon in Escherichia coli. J Mol Biol. 1988;203:141–152. doi: 10.1016/0022-2836(88)90098-8. [DOI] [PubMed] [Google Scholar]
- 55.Zheng C, Friedman D I. Reduced Rho-dependent transcription termination permits NusA-independent growth of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:7543–7547. doi: 10.1073/pnas.91.16.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]