Abstract
The epidemiological situation of SARS‐CoV‐2 in humans and animals is continually evolving. To date, animal species known to transmit SARS‐CoV‐2 are American mink, raccoon dog, cat, ferret, hamster, house mouse, Egyptian fruit bat, deer mouse and white‐tailed deer. Among farmed animals, American mink have the highest likelihood to become infected from humans or animals and further transmit SARS‐CoV‐2. In the EU, 44 outbreaks were reported in 2021 in mink farms in seven MSs, while only six in 2022 in two MSs, thus representing a decreasing trend. The introduction of SARS‐CoV‐2 into mink farms is usually via infected humans; this can be controlled by systematically testing people entering farms and adequate biosecurity. The current most appropriate monitoring approach for mink is the outbreak confirmation based on suspicion, testing dead or clinically sick animals in case of increased mortality or positive farm personnel and the genomic surveillance of virus variants. The genomic analysis of SARS‐CoV‐2 showed mink‐specific clusters with a potential to spill back into the human population. Among companion animals, cats, ferrets and hamsters are those at highest risk of SARS‐CoV‐2 infection, which most likely originates from an infected human, and which has no or very low impact on virus circulation in the human population. Among wild animals (including zoo animals), mostly carnivores, great apes and white‐tailed deer have been reported to be naturally infected by SARS‐CoV‐2. In the EU, no cases of infected wildlife have been reported so far. Proper disposal of human waste is advised to reduce the risks of spill‐over of SARS‐CoV‐2 to wildlife. Furthermore, contact with wildlife, especially if sick or dead, should be minimised. No specific monitoring for wildlife is recommended apart from testing hunter‐harvested animals with clinical signs or found‐dead. Bats should be monitored as a natural host of many coronaviruses.
Keywords: SARS‐CoV‐2, mink, wildlife, public health, monitoring, prevention, control
Summary
Since the entry into force of the Commission Implementing Decision (EU) 2021/788 laying down the monitoring measures in the EU for mink, other animals of the family Mustelidae and raccoon dog, the epidemiological situation and scientific knowledge of COVID‐19 in the EU has evolved and improved both in humans and animals. Therefore, the risks for animals and humans need to be reassessed based on new scientific findings and on the availability of control measures. In this opinion, EFSA was asked to review the scientific literature related to animal species susceptible to SARS‐CoV‐2 infection that play a role in its epidemiology. An assessment of the current epidemiological situation and of the risk for human and animal health posed by SARS‐CoV‐2 infection in animal species of concern was also conducted, which should serve to recommend options for reviewing the monitoring strategies for SARS‐CoV‐2 infection in animal species of concern. Finally, the main possible options for the prevention and control of COVID‐19 in both humans and susceptible animals were explored, highlighting their strengths and drawbacks.
The criterion used to classify the animal species of concern for SARS‐CoV‐2 epidemiology was the ability to shed infectious virus and to transmit SARS‐CoV‐2 to other individuals. The species assessed were American mink (Neogale vison), raccoon dog (Nyctereutes procyonoides), cat (Felis catus), Syrian hamster (Mesocricetus auratus), ferret (Mustela furo), house mouse (Mus musculus, for some virus variants only), Egyptian fruit bat (Rousettus aegyptiacus), deer mouse species (Peromyscus spp., not present in Europe) and white‐tailed deer (Odocoileus virginianus). Since SARS‐CoV‐2 variants continue to arise, new animal species fulfilling the above‐mentioned criterion may be detected over time with the continuous potential emergence of new host species.
Since the current assessment covers a range of points that deserve specific considerations in different contexts – i.e. susceptibility to the virus, risk for animal and public health, monitoring approach and preventive and control measures – the animal species considered were grouped according to the categories reflecting those contexts: farmed animals, companion animals, wildlife (referring to free‐ranging wildlife, thus excluding for example captive wild animals such as in zoo), and animals kept in zoos (from now on referred to as zoo animals).
Among farmed animals, American mink farmed for fur production have the highest likelihood to become infected from humans or animals and transmit SARS‐CoV‐2 within animal populations and to in‐contact humans. This is due to both the inherent susceptibility to SARS‐CoV‐2 infection of this species and the characteristics of the mink farming system, with a high density of animals kept in contiguous cages. During the still ongoing COVID‐19 pandemic, a vast majority of the reported outbreaks of SARS‐CoV‐2 in animals globally were from farmed mink. In the period, the present document refers to (February 2021 to November 2022), 50 outbreaks of SARS‐CoV‐2 were reported, of those 44 were reported in 2021 in seven MSs, while only six were reported in 2022 in two MSs, thus representing a decreasing trend. The genomic analysis of SARS‐CoV‐2 sequences showed major mink‐specific clusters, high rates of virus evolution within the mink population and emergence of mink‐specific variants with a potential to spill back into the human population.
The introduction of SARS‐CoV‐2 into mink farms is usually from infected humans; thus, this probability is associated with the SARS‐CoV‐2 level of circulation in the surrounding human population. Continuous and proper implementation of biosecurity measures in mink farms including the use of non‐pharmaceutical interventions (NPI) for all humans accessing mink farms can reduce the probability of introduction. Once introduced into a mink farm, SARS‐CoV‐2 spreads efficiently within the animal population, resulting in extensive virus circulation and risk of spill‐over to humans in contact with the mink, as well as to other susceptible animals with access to mink and their local environment.
The public health impact of the possible spill‐over of SARS‐CoV‐2 from mink farms to humans depends on the respective virus variant, effectiveness of the vaccine for this variant in vaccinated people including the time period after the vaccination, previous exposure to other SARS‐CoV‐2 variants and health status of the individual person. While the risk, determined by the probability of infection and the impact of the disease, for an occupationally exposed person to a SARS‐CoV‐2 infected mink has been assessed as low to moderate, in persons without or with limited exposure to farmed mink is estimated to be negligible to very low.
Concerning the monitoring of SARS‐CoV‐2 in mink farms, given the current epidemiological situation in the EU, where a substantial decrease of outbreaks in mink farms was reported in 2022 compared to 2020 and 2021, and where the majority of the human population has acquired some level of immunity to SARS‐CoV‐2, the most appropriate monitoring approach on animals would be the one based on testing dead animals with a suspicion of SARS‐CoV‐2 infection or animals showing clinical signs compatible with SARS‐CoV‐2 infection, with sampling triggered by increased mortality (compared to the baseline mortality rate) or morbidity in mink, or farm personnel testing positive. In fact, the primary purposes of monitoring of mink farms are to confirm outbreaks based on suspicion in order to apply preventive measures. In addition, genomic surveillance of circulating variants in mink and other species is also considered relevant to monitor virus evolution.
Regarding prevention and control of SARS‐CoV‐2 introduction into mink farms, since the most important source of introduction of SARS‐CoV‐2 into mink farms is via infected humans, systematic and frequent testing of people entering mink farms for SARS‐CoV‐2 infection using rapid antigen test and/or PCR is a prerequisite for the early detection of infected humans that may come into contact with mink. Also, the ban of non‐essential visits to farms and the use of personal protective equipment will reduce the exposure of animals to potentially infected people and reduce the probability of introduction of the virus. Biosecurity measures such as cleaning, disinfection, pest control (e.g. rodents) and restricted access to other animals on the farm (such as cats, dogs, bats, etc.) can also reduce the risk of virus introduction into the farm, and its further spread within and from the farm. The risk of further virus spread and secondary outbreaks to other farms can be reduced by restriction of mink movement and/or by testing for SARS CoV‐2 in mink prior to movement, especially in mink farms located in areas with known infected farms. Vaccines against COVID‐19 are protective against severe disease, hospitalisation and death, however, do not fully prevent virus transmission to and from humans as well as between humans and mink. In general, preventive and control measures applied to reduce the risk of transmission between mink and humans will only be effective if implemented consistently.
Among companion animal species, cats, ferrets and several hamster species are those most at risk of SARS‐CoV‐2 infection, which most likely originates from an infected human; in such situations, there is a very low risk of spillback infection to humans, and little or no animal‐to‐animal transmission, as indicated by genomic analysis. Categories of people with high contact rates to companion animals from different households (e.g. veterinarians) may have a higher risk of infection from companion animals. Under field conditions, cats and hamsters have been associated with mild to moderate respiratory, gastrointestinal or systemic signs of disease and they can shed virus. In general, SARS‐CoV‐2 transmitted by companion animals to humans are considered to have no or very low probability of having impact on virus circulation in the general population, and there is a low frequency of species‐adapted mutations.
Therefore, there is no need for specific monitoring programmes of SARS‐CoV‐2 infection in companion animals; some testing activity can be generally limited to owners, zoo workers or veterinarians in contact with these animals. In case of clinical signs compatible with SARS‐CoV‐2 disease, animal testing may be important for possible quarantine measures or application of proper therapies. Moreover, testing of individuals in stray communities (especially cats) could be justified, apart from research objectives, in case of suspected SARS‐CoV‐2 clinical cases or abnormal mortality rates in these communities.
The number of wildlife species that are globally reported to be naturally infected by SARS‐CoV‐2 grows steadily, also due to the active research in this field, which should be promoted. These include several wild carnivores and the white‐tailed deer in North America. Only the latter has been demonstrated, both free living or captive in game reserves, to maintain and possibly spill back the virus to humans. Nevertheless, in the EU, no cases of infected wildlife (with viral or RNA isolation) have been reported so far.
The situation of white‐tailed deer in the EU is very different from that in North America: The abundance of this deer species in the EU is very limited (less than 1% of the total deer population) and it is present only in two countries (Czechia and Finland); thus, it is unknown whether these animals may be able to support the persistence of SARS‐CoV‐2 infection in the European context. Moreover, white‐tailed deer are also kept farmed in some places in North America, which may increase the risk of transmission to and from any susceptible species, but this practice is not seen in the EU. Therefore, the risk of transmission of SARS CoV‐2 infection from humans to white‐tailed deer and backward, causing a severe disease, is considered very low.
Regarding wild carnivores, due to their elusive and solitary behaviour, to their low density and to the low numbers hunted, there is a very low probability for these species of maintaining the infection or representing a risk for other animal species or for public health. The latter is also due to limited human exposure, even for occupationally exposed people (rangers, hunters, researchers, etc.).
In any case, as a preventive measure, humans dealing with wildlife should follow biosecurity measures to minimise direct contact with wild animals, especially sick and dead animals. Furthermore, safe disposal of garbage and waste from human communities in both urban and rural settings is advised to reduce the risks of SARS‐CoV‐2 spillover to wildlife.
The probability of transmission from bats to humans or the emergence of SARS‐CoV‐2‐related or new coronaviruses has been assessed as none to very low, since transmission of SARS‐CoV‐2 or other coronaviruses from bats to humans and backwards has not been observed and there is a limited human population having direct contact with these animals in Europe. However, since bats are a natural host of many coronaviruses, the monitoring of these species remains important.
As a result of the above‐mentioned arguments, for wild species that may be considered as possible targets for SARS‐CoV‐2 monitoring (such as white‐tailed deer, wild carnivores, bats, rodents such as wild synanthropic mice and rats), no specific regulated monitoring activities would be needed, apart from testing of hunter‐harvested animals showing clinical signs or dead‐found individuals and sequencing the virus isolates to monitor its evolution.
Regarding animal species kept in zoos, there are reports of both experimental and natural infection with SARS CoV‐2, mainly felids and non‐human great apes. In zoos, susceptible species can acquire the infection mainly from in‐contact infected zoo workers; however, this is still at very low risk and there is no report of spillback transmission from animals to humans. Transmission between susceptible animals in the same enclosure could occur at moderate probability once an animal is infected, although transmission between animals kept in zoos is difficult to prove, because they are usually exposed to the same infectious source (e.g. infectious caretaker). Overall, animals kept in zoos do not represent a major public health risk in relation to SARS‐CoV‐2, the risk being considered very low for occupationally or activity‐related exposed people and negligible to very low for the general population.
No specific regulated monitoring activities on animals are needed in this animal category, apart from suspicion‐based testing and isolation of animals with clinical signs or testing in the frame of other veterinary checks. The main prevention is based on regular testing of zoo workers, self‐isolation when positive, use of PPE and good hygiene practice (e.g. avoiding close contact, tool disinfection, etc.) is expected to significantly reduce the risk of transmission from humans to animals.
1. Introduction
1.1. Background and terms of reference as provided by the EC
The scientific report of January 2021 produced by EFSA, in collaboration with ECDC, on ‘Monitoring of SARS‐CoV‐2 infection in mustelids’ (EFSA, 2021), along with the rapid risk assessment of November 2020 by ECDC have been the basis for the current monitoring measures in the EU for mink, other animals of the family Mustelidae and raccoon dogs as provided by Commission Implementing Decision (EU) 2021/7881.
The epidemiological situation has evolved in the EU since the adoption of these measures in May 2021 along with the scientific knowledge on the spread of SARS‐CoV‐2 in both humans and animals and what role individual epidemiological units play for the disease spread. This includes availability of new measures such as vaccination (for both animals and humans), refined diagnostic techniques, better understanding of biosecurity requirements and the risks related to genetic mutations of SARS‐CoV‐2. The risks for humans need to be reassessed based on new scientific findings and on the availability of control measures.
SARS‐CoV‐2 infections in mink have been reported by Member States to the Commission in line with the monitoring requirements. Further data has been collected by the OIE via the notification by its Member Countries.
The adequacy of the current monitoring system in the EU should be reviewed in light of the above to ensure proportionate measures are put in place to address the significant risks which could exist.
Terms of Reference
There is a need to revise and update the measures put in place by Member States to face the challenges posed by the epidemiological situation of SARS‐CoV‐2 in mink, other animals of the family Mustelidae and raccoon dogs.
The coordinated monitoring provided by Implementing Decision (EU) 2021/788 may need to be reviewed in view of the control measures available in response to the different epidemiological scenarios and possible evolution of the disease agent.
In view of the above, and in accordance with Article 29 of Regulation (EC) No 178/2002, the Commission asks EFSA for scientific opinion about:
Reviewing updated relevant scientific literature available globally related to SARS‐CoV‐2 infection in animal species of concern in the epidemiology of SARS‐CoV2.
Assess the current epidemiological situation in the EU and elsewhere as regards the risk for human and animal health posed by SARS‐CoV‐2 infection in animal species of concern with a view to review the design of the existing monitoring performed by the Member States for minks, other animals of the family Mustelidae and raccoon dogs.
In different epidemiological scenarios, recommend options for reviewing the monitoring strategies indicating possible objectives and suitable methodologies, in particular as regards scope, sampling, frequencies and testing methods taking into account existing risk mitigating measures.
Explore the main possible options for disease prevention and control measures suitable to address the risks under different plausible scenarios indicating the strengths and drawbacks of each set of measures.
1.2. Interpretation of the terms of reference
Regarding term of reference (ToR) 1, i.e. reviewing scientific literature related to SARS‐CoV‐2 infection in animal species of concern in the epidemiology of SARS‐CoV2, an extensive literature review has been conducted, focusing on susceptibility of wild and domestic animal species reported under field and laboratory conditions, considering different diagnostic tests (e.g. virus isolation, RNA and antibody detection), infection dynamic, pathogenesis, immunity and further transmission of the virus. The results are presented in Sections 5.1 and 5.2, where the animal species are grouped in the following categories: (1) farmed animals, (2) companion animals, (3) wildlife (including feral animals) and (4) animals kept in zoos (from now one referred as ‘zoo animals’). The phylogenetic analysis of sequence data from animal isolates deposited in GISAID until October 2022 is reported in Section 5.3, and this has been conducted by ECDC. Given the high rate and continuous publication of studies about potential new susceptible species along 2022, the evidence collected by the literature review has been complemented by that from new studies indicated by the experts of the established EFSA ad‐hoc working group, up to November 2022.
Concerning ToR 2, an updated epidemiological situation in the above‐mentioned animal categories is presented in Section 3.4. In particular, for fur farmed animals, mostly mink, the situation of reported outbreaks and related monitoring in MSs where mink are still bred is discussed. The risk for human and animal health posed by SARS‐CoV‐2 infection in animal species, which is also a part of ToR2, is addressed in Section 3.4, where, for the animal species considered as susceptible and relevant in the epidemiology of SARS‐CoV‐2, the probability of transmission in different pathways between animals and between animals and humans is assessed. A dedicated section about public health aspects is Section 3.5, which has been drafted by ECDC. Here, besides an overview of the virus variants circulating in the human population, the measures to prevent and control infection or spread of SARS‐CoV‐2 at the animal–human interface are discussed as well as the risk assessment for public health represented by animals infected by SARS‐CoV‐2.
The options for reviewing the monitoring strategies for SARS Cov‐2 (ToR 3) are discussed in Section 3.6. Here, keeping the current legislative requirements of the EC Decision 2021/788 as a reference, the monitoring approaches for SARS‐CoV‐2 in the different categories of animals and in different scenarios are discussed. An update about diagnostic tools that could be used for monitoring purposes, compared to what was already assessed in a previous EFSA report (EFSA, 2021), is reported.
The main disease prevention and control measures in humans and in the different animal categories are discussed in Sections 3.5 and 3.7, respectively, based on information provided by MSs and based on expert knowledge (ToR 4). The measures include, among others, biosecurity, movement controls, vaccination and non‐pharmaceutical interventions; the strengths and drawbacks of each of those are discussed.
Further details about problem formulation are provided in Annex A.1.
2. Data and methodologies
A systematic literature review was performed by an external contractor with stringent inclusion criteria (literature search protocol available at this link: https://doi.org/10.5281/zenodo.7559990) to retrieve a list of wild and domestic mammals that are susceptible to SARS‐CoV‐2 under laboratory conditions. Due to the stringent inclusion criteria and to the constantly and rapidly evolving results from the research about SARS‐CoV‐2 in animals, the presented lists are not exhaustive, and therefore, only publications published between 1 January 2020 and 15 February 2022 were included. Therefore, in order to provide the most up‐to‐date information, additional publications on various animal species issued up to December 2022 were added by the expert panel, to ensure that all relevant animal species were listed. Literature protocol and data extraction tables are available at this link: https://doi.org/10.5281/zenodo.7559990.
Data about the current epidemiological situation in the EU were retrieved from the literature as well as from outbreak reports submitted by MSs to the European Commission, from the World Organisation for Animal Health (WOAH) database2 and from ProMedmail3 notifications and they were analysed and presented by descriptive epidemiology.
The probability of transmission of SARS‐CoV‐2 from the different animal categories considered in the present opinion (i.e. farmed animals, companion animals, wildlife, zoo animals) to humans or other animals and vice versa and scenarios were assessed by consensus agreement among experts. In order to make a proper assessment of the likelihood of an event, it is necessary to come up with a well‐defined question or quantity of interest (QoI), such that the true answer or value could be determined, at least in principle. By doing so, ambiguities that could contribute to uncertainty are minimised. In addition, it helps to ensure that the range of probabilities provided in the assessment are reflecting only uncertainty and not also variability (that could be interpreted differently by the different experts if not made explicit). This assessment does not imply making ‘additional judgements’ (for which insufficient knowledge exists), but rather to translate the judgements already done into transparent and clear statements that can be unambiguously understood by the readers (and risk managers). With this approach, once a range of probabilities is agreed, uncertainty becomes evident as will be reflected by the width of the interval (EFSA, 2014). Proposed probability ranges used alone or in combination were:
Very high: > 90%
High: 66–90%
Moderate: 33–< 66%
Low: 10–< 33%
Very low < 10%
The risk assessment for human health posed by SARS‐CoV‐2 infection in animals is based on evidence available to ECDC at the time of publication. It follows the ECDC rapid risk assessment methodology, where the overall risk is determined by a combination of the probability of infection (taking into consideration the assessment of the probability of transmission of SARS‐CoV‐2 for the different animal categories, as explained above) and the level of impact of the disease on the affected individuals or general population and it is assessed on a qualitative scale as in ECDC (2019), as displayed below in Figure 1.
Figure 1.

Risk ranking matrix as in ECDC (2019)
The revision of monitoring approaches and the assessment on strengths and drawbacks of preventive and control measures were done based on data retrieved from MSs and literature and addressed based on expert knowledge.
3. Assessment
3.1. Animal species susceptible to SARS‐CoV‐2 under experimental conditions
Susceptibility to SARS‐CoV‐2 infection can be determined by detection of indicators of productive infection such as isolation of infectious virus or viral RNA from host's secretions/excretions (ante‐mortem) or organs (post‐mortem) and/or seroconversion. Based on these indicators, animal species that were described in the literature (see literature protocol and data extraction at this link https://doi.org/10.5281/zenodo.7559990) as susceptible to SARS‐CoV‐2 experimental infection (see also Table A.1 in Annex A.2) were grouped in three categories: (1) companion animals, (2) farmed animals, (3) wildlife and (4) zoo animals (Figure 2).
Figure 2.

- Detection of viral RNA refers to either detection in at least two consecutive ante‐mortem respiratory samples, or any post‐mortem sample. A black cross indicates the absence of a feature, while a red check mark indicates the presence of a feature. Both signs mean disagreement among studies considered; ‘nd’ means that this feature was not determined. Please note that for some species for which extensive literature is available (e.g. ferrets and Syrian hamsters), only a selection of a few representative references were included.
3.1.1. Companion animals (cat, dog, ferret, rabbit, mice and several hamster species)
Chinese, Djungarian, Roborovski and Syrian hamsters displayed clinical signs post infection, while Campbell's dwarf hamsters remained without clinical signs (Chan et al., 2020; Imai et al., 2020; Sia et al., 2020; Trimpert et al., 2020; Bertzbach et al., 2021; Gerhards et al., 2021). In cats and ferrets, both the presence and absence of clinical signs were reported (such as fever, diarrhoea, sneezing, arching of back) (Bosco‐Lauth et al., 2020; Gaudreault et al., 2020; Halfmann et al., 2020; Kim et al., 2020; Richard et al., 2020; Schlottau et al., 2020; Shi et al., 2020; Bao et al., 2021; Chiba et al., 2021; Gaudreault et al., 2021; Kutter et al., 2021; Marsh et al., 2021; Ryan et al., 2021; Ciurkiewicz et al., 2022). No clinical signs were reported for other companion animals such as dogs and rabbits (Bosco‐Lauth et al., 2020; Shi et al., 2020; Montagutelli et al., 2021; Pan et al., 2021), neither for laboratory animals such as mice (laboratory mouse strains C54BL/6 and BALB/c). Nevertheless, animals usually recover from the experimental infection with the exception of Roborovski hamsters, for which infection with SARS‐CoV‐2 is severe, requiring euthanasia.
Viral RNA was detected in all investigated companion animal species except in dogs, and virus could be isolated from hamsters, cats, ferrets, rabbits and laboratory mice. Noteworthy, for laboratory mice, this was only the case for B.1.1.7, B.1.351 and P.1 isolates.
Cats, dogs, ferrets, laboratory mice, rabbits and Syrian hamsters developed antibodies against SARS‐CoV‐2. For ferrets, one report did not detect antibodies post infection while Campbell's, Chinese, Djungarian and Roborovski hamsters were not tested for the presence of antibodies.
Transmission of SARS‐CoV‐2 from a donor animal to a recipient by direct contact has been demonstrated for cats, ferrets, laboratory mice and Syrian hamsters. There was no evidence of virus transmission between dogs. Rabbits, Campbell's dwarf, Chinese, Djungarian and Roborovski hamsters were not investigated for virus transmission.
A large number of publications are available describing the susceptibility of ferrets and hamsters. In another literature review, performed within the project COVRIN of the One Health European Joint program (Grant agreement 773830), efficient direct contact as well as indirect contact transmission between ferrets and between hamsters was widely reported (de Vries et al., 2021; Dowall et al., 2021; Kutter et al., 2021; Mok et al., 2021; Neary et al., 2021; Page et al., 2021; Patel et al., 2021; Peacock et al., 2021; Cox et al., 2021a,b; Kim et al., 2022).
3.1.2. Farmed animals
For cattle, mink and pigs, reports disagree concerning the presence of clinical signs post SARS‐CoV‐2 infection (Schlottau et al., 2020; Shi et al., 2020; Meekins et al., 2020; Ulrich et al., 2020; Buckley et al., 2021; Falkenberg et al., 2021; Pickering et al., 2021; Shuai et al., 2021; Sikkema et al., 2022; Virtanen et al., 2022). One out of two references describe fever in cattle; one out of two references describe anorexia, diarrhoea, lethargy and respiratory signs in mink; and fever and ocular discharge was described for pigs in two independent references out of six in total. No clinical signs were observed in raccoon dogs (Freuling et al., 2020).
Viral RNA was detected in ante‐mortem and/or post‐mortem samples of cattle, mink and raccoon dogs, while for pigs, both detection as well as the absence of viral RNA post infection were reported. Virus could be isolated from raccoon dogs and mink, while for pigs and cattle, virus isolation was not undertaken.
Seroconversion was observed in racoon dogs, mink and cattle, while for pigs, antibodies were detected only in three out of six references.
Transmission of SARS‐CoV‐2 was observed for mink and raccoon dogs, and no transmission was observed for cattle and pigs.
Goats, sheep, alpacas and horses are not included in the figure, because the available publications did not pass the inclusion criteria of the systematic literature review (time of publication and use of animals to model human infections). No viral RNA could be detected in ante‐mortem respiratory samples from sheep, alpacas and horses and no virus could be isolated and no neutralising antibodies could be detected on day 14 post infection. For goats, viral RNA could be detected in swabs from the respiratory tract in some animals, but no virus could be isolated and no antibodies were detected post infection (Bosco‐Lauth et al., 2021b). In a study published in September 2022, SARS‐CoV‐2 was detected in experimentally infected goats in nasal swabs and tissues by PCR, and seroneutralisation was confirmed via ELISA. However, the viral amount and tissue distribution suggest a low susceptibility of goats, thus no relevant role of goats in the epidemiology of SARS‐Cov‐2 (Fernández‐Bastit et al., 2022).
3.1.3. Zoo and wild animals
In this category, we refer to African green monkey (Chlorocebus sabaeus), bank vole (Myodes glareolus), bushy‐tailed woodrat (Neotoma cinerea), Chinese tree shrew (Tupaia belangeri chinensis), cynomolgus macaques (Macaca fascicularis), deer mouse (Peromyscus maniculatus), Egyptian fruit bat (Rousettus aegyptiacus), raccoon (Procyon lotor), red fox (Vulpes vulpes), rhesus macaque (Macaca mulatta), skunk (Mephitis mephitis) and white‐tailed deer (Odocoileus virginianus). Clinical signs were observed in African green monkeys (fever and reduced appetite or hypothermia and respiratory distress; (Blair et al., 2021; Woolsey et al., 2021)), cynomolgus macaques (nasal discharge; (Rockx et al., 2020)) and white‐tailed deer (fever or ocular discharge; (Palmer et al., 2021; Cool et al., 2022)) post experimental infection with SARS‐CoV‐2. Reports disagree regarding clinical signs in Chinese tree shrews (fever in one out of two references (Xu et al., 2020; Zhao et al., 2020)) and rhesus macaques (no signs in two references; fever, respiratory distress, weight loss, hunched posture and nasal discharge or reduced appetite and weight loss in two out of four references (Munster et al., 2020; Shan et al., 2020; Blair et al., 2021; Yadav et al., 2021)). No clinical signs were observed in bank voles (Ulrich et al., 2021), bushy‐tailed woodrats (Bosco‐Lauth et al., 2021a), deer mice (Fagre et al., 2021; Griffin et al., 2021), Egyptian fruit bats (Schlottau et al., 2020), raccoons (Bosco‐Lauth et al., 2021a; Francisco et al., 2022) and skunks (Bosco‐Lauth et al., 2021a; Francisco et al., 2022).
Viral RNA was detected and virus could be isolated in all investigated zoo and wild animal species, except raccoons. For Chinese tree shrews, virus isolations were not performed.
Seroconversion was observed in all animals. One out of four references did not observe antibodies in rhesus macaques post‐infection, and antibodies were not assessed for Chinese tree shrews.
Transmission of SARS‐CoV‐2 was successfully demonstrated in deer mice, Egyptian fruit bats and white‐tailed deer. No transmission took place in bank voles, raccoons and skunks. For all other animal species, transmission was not investigated.
Common marmosets (Callithrix jacchus) and hamadryas baboons (Papio hamadryas) are listed in Figure 2, although the publications did not pass the inclusion criteria of the systematic literature review. In both species, viral RNA was detected post‐infection (Singh et al., 2021).
In a study published in September 2022, two red foxes (Vulpes vulpes) and coyotes (Canis latrans)4 were tested for susceptibility to SARS‐CoV‐2 at experimental inoculation. Only red foxes became infected and shed infectious virus. The authors concluded that the role of red foxes in SARS CoV‐2 transmission should be carefully evaluated, given the wide distribution of this species, its frequent proximity to humans, and that it preys, scavenges upon or otherwise interacts with species demonstrated to be susceptible to SARS‐CoV‐2, including felids, skunks, rodents and white‐tailed deer (Porter et al., 2022).
3.1.4. Time to detection of infection following experimental infection
The period from inoculation until viral RNA detection varied between 1 and 3 days post‐inoculation for most species. Longer incubation periods were observed for animals infected by direct contact (up to 8 days), for Chinese tree shrews (6–8 days), African green monkeys and rhesus macaques (up to 7 days).
Seroconversion took place within the following days post‐inoculation: 3–11 (pigs), 5–11 (cats), 6–14 (deer mice), 7–12 (cattle), 7 (white‐tailed deer), 7–14 (African green monkey), 7–17 (ferrets), 8 (bank voles, bats, raccoon dogs), 8–28 (skunks), 9–28 (raccoons), 10–14 (rhesus macaques), 12–14 (mice), 7–16 (Syrian hamsters), 14 (dogs, cynomolgus macaques), 18 (mink), 21 (rabbits).
Noteworthy, the above‐mentioned time from infection to seroconversion depends on the individual study designs and sampling schemes, leaving the possibility that, e.g. antibodies would have been detectable at earlier time points post‐infection, if an earlier blood sample would have been taken.
Sequence or structure‐based predictions of animal susceptibility due to homology of human ACE2 receptor and their agreement with in vivo data has been systematically assessed by Fischhoff et al. (2021). Sequence‐based predictions revealed a low affinity of SARS‐CoV‐2 to mink and ferret ACE2, for instance, while, in fact, these species were highly susceptible. The agreement between structure‐based predictions of species susceptibility and factual susceptibility is high, although not always accurate.
Furthermore, the following animal species were experimentally infected with SARS‐CoV‐2, but did not seroconvert, and neither viral RNA nor infectious virus could be isolated post‐infection: big brown bats, black‐tailed prairie dogs, cottontail rabbits, coyotes, fox squirrels and Wyoming ground squirrels. These species are therefore considered not susceptible to SARS‐CoV‐2.
3.2. Animal species detected infected with SARS CoV‐2 under field conditions
In the present section, the information obtained by literature review,5 from WOAH database and from ProMEDmail about animal species that have been detected as positive for SARS‐CoV‐2 worldwide, either as PCR positive or serologically positive, are presented. The animal species are grouped as farmed animals, companion animals and zoo and wild animals.
Figures 3 and 4 show animal species that were found to be susceptible to SARS‐CoV‐2 under field (natural) conditions, based on the literature review done on 15 February 2022. Moreover, in Annex A.3 (Table A.2), the results obtained from literature review about field infection of different animal species to SARS CoV‐2 are shown, as proportion of positive animals in each epidemiological unit, tested by PCR/virus isolation or serological test.
Figure 3.
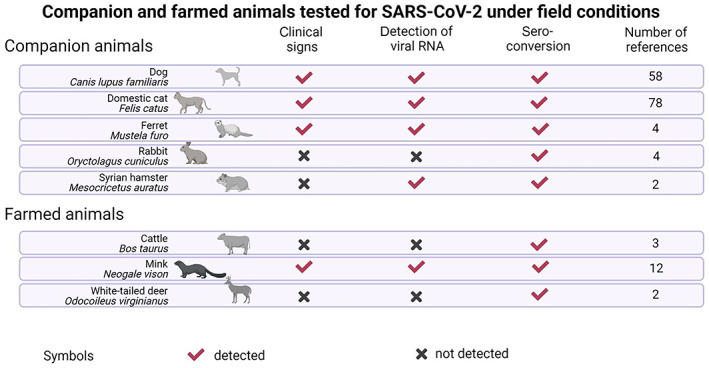
Companion and farm animal species that have been reported as being susceptible to SARS‐CoV‐2 infection under field conditions based on seroconversion and/or detection of viral RNA and able to further transmit (based on the literature search carried out on 15 February 2022)
Figure 4.
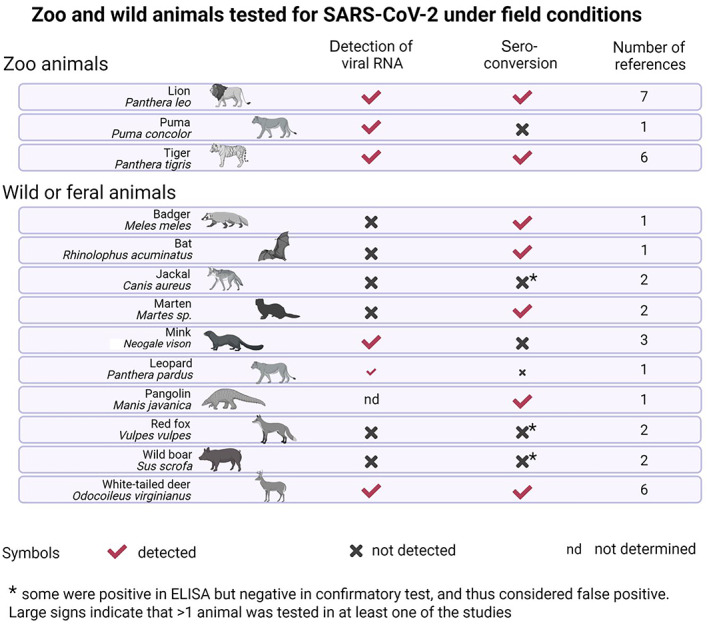
Zoo and wild animal species that are susceptible to SARS‐CoV‐2 infection under field conditions based on seroconversion and/or detection of viral RNA (based on literature search carried out on 15 February 2022)
Table A.2.
Proportion of animals positive (as median and inter‐quartile range (IQR)) to SARS CoV‐2 infection in the field, per species and epidemiological unit following passive or active monitoring and according to virus detection/isolation or serological tests. Based on literature screening, only species found positive are included in the table, and studies including only one animal of this species are excluded from the calculations of proportions of positives
| Passive monitoring (suspicion‐based) | Active monitoring (random sampling) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Species | No. of references | Total no. of animal | % positive tested animals in each reference (virus isolation or PCR‐positive) (median; min–max) | % serologically positive animals in each reference (Median; min–max) | No. of references | Total no. of animal | % positive tested animals in each reference (virus isolation or PCR‐positive) (median; 25th–75th) | % serologically positive animals in each reference (Median; min–max | ||
| Companion animals/farmed animals | Cat | Felis catus | 51 | 725 | 100%; 0–100%) | (33.33%; 0–100%) | 27 | 12,531 | (0%; 0–2,48%) | (1.79%; 0–22.4%) |
| Cattle | Bos taurus | 1 | 16 | (0%) | 2 | 1,144 | (1.1%; 0–36.36%) | |||
| White‐tailed deer (farmed) | Odocoileus virginianus | 2 | 36 | (0%) | (95.24%; 90.47–100%) | |||||
| Dog | Canis familiaris | 34 | 823 | (1.95%; 0–100%) | (19.16%; 0–100%) | 24 | 10,971 | 7/3542 (0%; 0–1.32%) | (1.47%; 0–33.33%) | |
| Ferret | Mustela furo | 2 | 30 | (50.0%; 0–100%) | (87.93%; 80.0–100%) | 2 | 198 | (7.04%) | (1.57%) | |
| American mink | Neogale vison | 8 | 22,611 | (61.26%; 0.26–100%) | (78.34%; 2–100%) | 4 | 1,121 | (16.48%; 0.68–33.44%) | 98.7%; 7.7%–100.0%) | |
| Syrian Hamster | Mesocricetus auratus | 1 | 97 | 16.55% | 12.4% | |||||
| Rabbit | Oryctolagus cuniculus | 2 | 9 | (0%) | 2 | 173 | (0.69%; 0–1.38%) | |||
| zoo animals | Lion | Panthera leo | 7 | 30 | (100%; 1166.6–100%) | (100%; 11%–100%) | ||||
| Tiger | Panthera tigris | 7 | 22 | (100%; 62.5–100%) | (100%; 100%–100%) | |||||
| Gorilla | Gorilla gorilla | 2 | 16 | (50.0%; 37.5–62.5%) | ||||||
| Amur Leopard cat | Prionailurus bengalensis | 1 | 2 | 50% | ||||||
| Puma | Puma concolor | 1 | 1 | 100% | ||||||
| wild animals | Badger | Meles meles | 1 | 10 | (20%) | |||||
| Bat | Rhinolophus acuminatus | 1 | 208 | (13%) | (0.46%) | |||||
| White‐tailed deer | Odocoileus virginianus | 6 | 1,464 | 62 (24%; 0%–100% | (24%; 0%–100%) | |||||
| Jackal | Canis aureus | 2 | 66 | (0%) | (2.3%; 0–4.6%) | |||||
| Leopard | Panthera pardus | 1 | 1 | (100%) | ||||||
| Marten | Martes spp. | 1 | 14 | (21.42%) | ||||||
| American mink | Neogale vison | 3 | 108 | (0%; 0–25.0%) | (0%) | |||||
| Pangolin | Manis javanica | 1 | 10 | (10%) | ||||||
| Wild boar | Sus scrofa | 2 | 247 | (0%) | (1.95%; 0%–3.9%) | |||||
| Stray animals | cat | Felis catus | 4 | 332 | (64,29%) | (0.95%; 0–3.51%) | ||||
| dog | Canis familiaris | 1 | 8 | (62.5%) | ||||||
Based on the extensive literature review, the following animal species were tested with PCR test and/or serological test but results were negative (Table 1).
Table 1.
Animal species under field conditions tested with PCR test and/or serological test with negative results
| Species | PCR | Reference | Serological test | Reference |
|---|---|---|---|---|
| Alpaca (Vicugna pacos) | na | neg | Deng et al. (2020) | |
| Bamboo rat (Rhizomys spp.) | na | neg | Deng et al. (2020) | |
| Bear (Ursus spp.) | na | neg | Deng et al. (2020) | |
| Beech marten (Martes foina) | neg | Davoust et al. (2022) | neg | Davoust et al. (2022) |
| Buffalo (Bubalus bubalis) | neg | Cerino et al. (2021) | na | |
| Camel (Camelus dromedarius) | na | neg | Deng et al. (2020) | |
| Deer (red deer, roe deer, fallow deer) (Cervus elaphus, Capreolus capreolus, Dama dama) | na | neg | Moreira‐Soto (2022) | |
| Giant panda (Ailuropoda melanoleuca) | na | neg | Deng et al. (2020) | |
| Goat (Capra hircus) | neg | Cerino et al. (2021) | na | |
| Guinea pig (Cavia porcellus) | neg | Ruiz‐Arrondo et al. (2021) | na | |
| Horse (Equus ferus caballus) | neg | Cerino et al. (2021) | neg | Deng et al. (2020) |
| Leopard cat (Prionailurus bengalensis) | na | neg | Deng et al. (2020) | |
| Masked civet (Paguma larvata) | na | neg | Deng et al. (2020) | |
| Mouse (Mus musculus) | neg | Ip et al. (2021) | na | |
| Non‐human primates of genera Callithrix, Callicebus and Alouatta spp. | neg | Sacchetto et al. (2021) | na | |
| Pig (Sus scrofa domesticus) | neg | Cerino et al. (2021) | neg | Deng et al. (2020) |
| Polecat (Mustela putorius) | neg | Davoust et al. (2022) | neg | Davoust et al. (2022) |
| Porcupine (Hystrix spp.) | na | neg | Deng et al. (2020) | |
| Raccoon (Procyon lotor) | neg | Ip et al. (2021) | ||
| Rat (Rattus rattus) | neg | Colombo et al. (2021) | neg | Colombo et al. (2021) |
| Red panda (Ailurus fulgens) | na | neg | Deng et al. (2020) | |
| Rhinoceros (Rhinoceros spp.) | na | neg | Deng et al. (2020) | |
| Sheep (Ovis aries) | neg | Cerino et al. (2021) | neg | Deng et al. (2020); Villanueva‐Saz et al. (2021a) |
| Skunk (Mephitis mephitis) | neg | Ip et al. (2021) | na | |
| Tamarin (Saguinus spp.) | neg | Sacchetto et al. (2021) | na | |
| Weasel (Mustela nivalis) | na | neg | Deng et al. (2020) |
neg: tested negative; na: not available.
3.2.1. Farmed animals detected infected with SARS CoV‐2 under field conditions
To date, the only farmed animals that were tested positive following natural infection (detection of SARS‐CoV‐2 RNA by PCR) are mink (Molenaar et al., 2020; Oreshkova et al., 2020; Domańska‐Blicharz et al., 2021; EFSA, 2021; Hammer et al., 2021; Rabalski et al., 2021; Rasmussen et al., 2021), raccoon dogs (one outbreak reported in Poland in 2021), and to a certain extent also ferrets (Shi et al., 2020; Giner et al., 2021; Gortázar et al., 2021; Račnik et al., 2021). The former two species are bred mainly in the fur industry, while ferrets are bred mainly as companion, research or as work animals for rabbit hunting and rabbit control. The size of ferret breeding centres may be between 10 and 100 ferrets per facility.
In the following section, the epidemiological situation in farmed mink is reported in MSs where mink farming is still practiced, also considering molecular epidemiology and monitoring scheme applied.
3.2.1.1. Fur animals
EU data
From the beginning of the pandemics up to 31 January 2021 (the reporting period which the first EFSA report refers to (EFSA, 2021)), 401 outbreaks of SARS‐CoV‐2 in mink farms were reported in Europe (Table 3), mostly in Denmark (290) and the Netherlands (69), where around 60% of the total mink farms in EU were until the end of 2020, then the mink farming was stopped in those two MSs. From 1 February 2021 until 30 November 2022, period in which the number of mink farms was approximately stable in EU (around 700 farms), 50 SARS‐CoV‐2 outbreaks were detected in farmed mink and raccoon dogs in the EU, of those 44 were reported in 2021 in seven MSs, while only six were reported in 2022 in two MSs (Figure 6). In all the affected establishments, farmed mink were raised, apart from one Polish farm where both raccoon dogs (300 animals) and mink (5,000 animals) were raised (detection in December 2021).
Table 3.
Data on outbreaks of SARS‐CoV‐2 in mink farms in the EU in 2020 until January 2021 (EFSA, 2021)
| Country | Number of mink farms in the country at the time of SARS‐Cov‐2 virus first detection | Number of infected mink farms | Date of first and last SARS‐CoV‐2 virus detection in mink farms | Number of farms where clinical signs of SARS‐CoV‐2 were observed | Likely source of virus origin |
|---|---|---|---|---|---|
| Denmark | 1,147 | 290 | 15 June to 7 December | 145 | Human‐to‐animal transmission suspected or confirmed in some outbreaks. Unclear in most outbreaks |
| France | 4 | 1 | 20 November | 0 | Undetermined, but most probably humans on the farm |
| Greece | 91 | 21 | 13 November to 8 January | 8 | Human‐to‐animal transmission suspected in most outbreaks |
| Italy | 9 | 1 | 10 October | 0 | Human‐to‐animal transmission |
| Lithuania | 86 | 2 | 24 November | 2 | Human‐to‐animal transmission |
| Netherlands | 126 | 69 | 24 April to 4 November | 62 | Partly human to mink transmission and partly unclear, but with a strong spatial component |
| Poland | 272 | 1 | 27 January 2021 | 0 | Unknown |
| Spain | 29 | 3 | 22 June to 22 January | 0 | Human‐to‐animal transmission suspected |
| Sweden | 35 | 13 | 23 October to 11 November | No information | Human‐to‐animal transmission |
Figure 6.

- Source: EC, EFSAThe vertical grey line indicates when the Commission Implementing Decision (EU) 2021/788 of 12 May 2021 laying down rules for the monitoring and reporting of infections with SARS‐CoV‐2 in certain animal species (OJ L 173, 17.5.2021, p. 6) entered into force.
The geographical distribution of the affected mink establishments is presented in Figure 5 and the monthly distribution of outbreaks is presented in Figure 6. Clinical signs of SARS‐CoV‐2 infections were detected in 4 out of 50 affected mink establishments, and humans were identified as the possible source of virus introduction in 12 mink establishments, whereas the source of the infection was not identified in the other 38 SARS‐CoV‐2 outbreaks. No clinical signs of SARS‐CoV‐2 infections were detected in the affected raccoon dog establishment and the source of the virus introduction was not identified.6
Figure 5.

- *: This designation is without prejudice to positions on status and is in line with United Nations Security Council Resolution 1,244 and the International Court of Justice Opinion on the Kosovo Declaration of Independence.
In Tables 2 and 3, data on number of mink farms in the MS, number of infected farms, with clinical signs and where likely human source was confirmed, in the two reporting periods (February 2021 until November 2022, and 2020, until 31 January 2021, respectively) are reported.
Table 2.
Data on outbreaks of SARS‐CoV‐2 in mink farms in the EU from February 2021 to November 2022
| Country | Number of mink farms in the MS as of January 2022 | Number of infected mink farms in the reporting period | Date of first and last SARS‐Cov‐2 virus detection in mink farms in the reporting period | Start of systematic monitoring | Number of outbreaks where clinical signs of SARS‐CoV‐2 were observed | Number of outbreaks with likely human source of virus origin in the reporting period (if known) |
|---|---|---|---|---|---|---|
| Finland | 133 | 0 | – | January 2021 | – | |
| Greece | 91 | 4 | February 2021 to August 2021 | November 2020 | 3 | 2 |
| Italy | 5 | 1 | – | February 2021 | 1 | |
| Latvia | 5 | 1 | April 2021 | January 2021 | 0 | 1 |
| Lithuania | 71 | 13 | November–December 2021 | November 2021 | 0 | Unknown |
| Poland | 261 | 15 | June 2021–July 2022 | December 2020 | 0 | 1 |
| Spain | 27 | 15 | March–October 2021 | May 2021 | 1 | 6 |
| Sweden | 22 | 1 | August 2021 | October 2020 | 0 | 1 |
In the following section, we present the epidemiological situation and monitoring activities (reporting period February 2021–March 2022) carried out in MSs where mink farming is still present.
3.2.1.1.1. Finland
Fur‐farmed animals
In Finland, the farmed species targeted by SARS‐CoV‐2 monitoring were mink, raccoon dogs and sable (Martes zibellina). Up to January 2022, 133 mink farms were registered, including a total of 190,500 breeding animals and around 500,000 production stocks as well as 50 farms with raccoon dogs, including a total of 13,700 breeding animals, and one sable farm with 310 breeding animals.
Epidemiological situation
Finland has not reported any outbreaks of SARS‐CoV‐2 in mink or other fur animal farms. A voluntary preventive vaccination scheme in mink (implemented in November 2021) was in place on January–February 2022 covering approximately 95% of breeding females. The vaccination campaign is currently not active.
Monitoring scheme
In December 2020, monitoring of SARS‐CoV‐2 infection started in all fur farms (mink and other animals of the family Mustelidae), and at the beginning of 2021, raccoon dog farms were included in the monitoring, although the sampling scheme was not completely harmonised among farms. Since June 2021, a harmonised active monitoring scheme foresees that all fur farms submit five dead animals every 2 weeks for SARS‐CoV‐2 testing to the laboratory of the Finnish Food Authority. Throat swab samples from dead animals are taken by laboratory staff and samples are analysed by RT‐PCR. If the number of dead animals is insufficient to reach the threshold of five animals per 2 weeks, further live animals are sampled by municipal veterinarians. So far, 14,165 samples from 229 farms have been tested, all negative.
Furthermore, passive monitoring is carried out in all farms. Suspicion is triggered if farmed fur animals show clinical signs, in which case 30 samples are taken from the animals of the farm, or if a person tested positive at SARS‐CoV‐2 has been in contact with fur animals, in which case 60 samples are taken from the animals of the farm. Until March 2022, a total of 568 samples from 11 farms were tested, all negative.
Other species
Wild mustelids or wild raccoon dog: Those animals found dead or hunted animals with clinical signs associated with SARS‐CoV‐2 are sampled for SARS‐CoV‐2 tests. From wild animals, mostly oral swabs or organ samples are used and tested by PCR (E gene). In total, 278 raccoon dogs, three otters and three badgers have been tested for SARS‐CoV‐2, all negative.
Wild white‐tailed deer: In total, 36 samples from oral swab/lymph node have been tested by PCR (E gene), all negative for SARS‐CoV‐2.
Companion animals: Six cats and three dogs have been sampled in case of a suspicion of SARS‐CoV‐2 (e.g. a pet had SARS‐CoV‐2 signs and had been in contact with a person having COVID‐19). Oral swabs have been tested by PCR (E gene). One cat and one dog were found positive in 2021 (Delta variant) and 2022 (Omicron variant), respectively.
3.2.1.1.2. France
In France, there is currently only one mink farm left, where passive monitoring is carried out. No outbreaks have been reported in the period from February 2021 to September 2022.
3.2.1.1.3. Greece
By 2022, in Greece, the only farmed species targeted by SARS‐CoV‐2 monitoring are mink. There are 91 mink farms, 89 of which are located in the region of Western Macedonia (Regional Units Kastoria, Kozani, Grevena). In March 2022, the total population was 470,000 breeding animals and approximately 1.6 million production stock that are pelted annually. As 90% of Greece's fur production is exported to Russia, following the sanctions, the above‐mentioned numbers can decrease. Raccoon dogs are not bred in Greece.
Epidemiological situation
Until 29 January 2021, the reporting period to which the previous EFSA report refers (EFSA, 2021), SARS‐CoV‐2 was detected on 21 out of the 91 mink farms currently present in Greece. Until November 2022, four more outbreaks were reported, two in February 2021 in the regional Units of Kastoria and Grevena, and two more in August 2021 in the regional Unit of Kozani.
The first one in Kastoria was suspected based on the first observation of reduced feed intake on 26 January 2021, while respiratory symptoms and increased mortality were first detected on 29 January 2021. It was estimated that around 70% of all 716 farmed animals showed respiratory symptoms, and 55 minks died since the estimated date of virus introduction. The outbreak was confirmed on 5 February by RT‐PCR, but it was not possible to relate it to human infection, as none of the farm personnel or owners was found positive in either molecular or antibody testing.
The second outbreak reported in February 2021 was in Grevena. It was detected in minks, which were tested because two positive workers were positive in the framework of the early warning system (weekly testing) for farm personnel in place since November 2020. Animals showed no clinical signs and mortality appeared in line with baseline mortality. There was no conclusive evidence on the possible source of the virus, although transmission from infected workers is suspected.
The third outbreak was reported in Kozani, in August 2021, on a farm with 30,000 animals, of which 25,600 were juveniles, and it was detected because an increase in daily deaths (80 juvenile) followed by respiratory symptoms in 50 juvenile animals was noticed. Based on that, the owner notified veterinary authorities on 8 November 2021, and samples from dead animals were collected and confirmed positive by RT‐PCR. Farm personnel were vaccinated and tested all negative.
Similarly, the fourth outbreak, reported in the same regional unit as the third at the end of August 2021, was suspected due to death (180 animals) and respiratory signs in animals, approx. 50% of them (total 14,200 animals, out of which 2,700 breeders). One worker, who was vaccinated 2 weeks before, tested positive by PCR.
In Greece, the control measures applied included movement control inside the country, zoning, traceability, quarantine, monitoring biosecurity measures including mandatory use of PPE. Affected animals are not culled.
Genomic analysis
The analysis of genomes isolated in 20 infected farms showed that B.1.1.305 in the prefecture of Kastoria and B.1.1.218 in the prefectures of Kozani and Grevena are the two main lineages detected in mink and farm personnel, as it is the case in the general population. In each distinct farm, the same lineage was identified in mink and farm personnel.
The most frequent mutations in the S protein are the D614G, N501T and P812L, both for the general human population and minks. Preliminary data indicate that mink were infected by humans in most of the cases. Mink‐related mutations in the S protein have been detected (Y453F) in six human cases directly related to farms and in animals from four establishments, but it has not been detected in the small sample of the general population. None of the other mutations described on by ECDC (ECDC, 2020a) has been found so far, neither in humans (farm personnel/owners and community) nor animals. Although spillback into the local community has not been observed, further investigation in the general population by extending the number of sequenced genomes is ongoing.
Monitoring scheme
Since the first case of SARS‐CoV‐2 in mink in Greece in November 2020, a monitoring plan has been implemented as follows:
Mink farm personnel were regularly tested with PCR and rapid antigen tests during the reporting period (February 2001–March 2022), by PCR weekly until May 2021, PCR every 14 days from June to December 2021 and rapid antigen tests weekly from January 2022 to March 2022; and results are notified to veterinary authorities.
As regards mink, passive monitoring was implemented by clinical investigations and laboratory testing (oropharyngeal swabs tested by RT‐PCR) upon notification of clinical signs related to SARS‐CoV‐2 or increased mortality in mink, or in case humans in the establishment tested positive. Under this scheme, 24 of 29 samples from three mink farms and 14 out of 40 samples from farm personnel from two farms tested positive. From February 2021 to May 2021, when the 2021/788 Decision entered into force, 10 samples were collected from farms with increased mortality and 20 from establishments, where SARS‐CoV‐2 cases in workers or their families were notified. The sampling scheme has been adjusted to the Decision since it was adopted in May 2021.
Mink farms were subjected also to active monitoring whenever animals were moved. In these cases, 20 samples were taken for testing (oropharyngeal swabs‐real‐time PCR) from the establishment of origin prior to each movement (design prevalence: 15%); no positive cases were detected.
Sequencing was conducted on samples from both human (100% samples) and animals (30% samples) that were tested positive at farm level during the reporting period (February 2021–March 2022). Moreover, sequencing was performed on positive samples from the community in the workers' places of residence. All sequencing results were compared and no mink variants were detected in samples collected from the community in the areas where mink farms are located, and no spillover infection to the community was detected.
Since the 2021/788 Decision entered into force, also other kept and wild animals belonging to mustelids have been monitored according to the section 2 of Annex III of Decision 2021/788.
3.2.1.1.4. Italy
As of January 2022, in Italy, there were five mink farms with 6,055 breeding stock, mainly concentrated in northern Italy (two farms in Lombardy and two in Emilia Romagna region) and one farm in central Italy (Abruzzo). No other animal species is object of systematic monitoring for SARS‐CoV‐2 infection.
Epidemiological situation
So far, only one positive animal in one farm was detected by PCR in Italy, in January 2021, out of 10,823 samples tested from six farms, up to March 2022. No clinical signs were observed in the farm. No other animals in the farm were tested positive at PCR, while seropositive animals were detected. A further outbreak was reported in November 2022 in a mink farm composed of breeding 1,523 animals in Emilia Romagna region; this was detected within the surveillance plan, which provides controls on live and dead animals. In this regard two animals tested positive at RT‐PCR, one out of 55 live animals tested and one out of four dead animals, both asymptomatic.
Monitoring scheme
There is a weekly control of all dead animals in mink farms, coupled with 60 oropharyngeal swab (target prevalence 5%) every 15 days (in 2022, it is on weekly frequency) as random sampling to be tested by PCR.
3.2.1.1.5. Latvia
In Latvia, the only farmed species targeted by SARS‐CoV‐2 monitoring are mink. As of January 2022, there are four mink farms with a total of 117,954 breeding animals and an estimated production stock of 250,000 animals to be pelted by the end of 2022.
Epidemiological situation
The only outbreak in mink in Latvia was reported in April 2021, confirmed on 10 April 2021 in Iecava county, approximately 50 km south from Riga, in a farm with 64,000 female breeders. No clinical signs nor unusual mortality was reported in the farm in the period around detection until the pelting season in November. The disease was detected in the frame of monitoring programme, i.e. weekly sampling of at least one dead mink from the each farm. For the confirmation, 10 additional samples (dead mink) were taken, and the presence of SARS‐CoV‐2 virus was confirmed in nine dead minks by RT‐PCR. After confirmation of the outbreak, weekly sampling of five dead minks was established to monitor the epidemiological situation in the affected farm. The epidemiological investigation showed that SARS‐CoV‐2 was introduced in the mink farm by an infected worker.
From July 2021, the number of dead minks to be tested weekly was increased to 10 to get more isolates for sequencing, as from July all PCR‐positive mink samples were sequenced. Apart from dead mink testing, also 20 live mink were sampled every month (for PCR and serological testing). After pelting, up to 19 dead minks were sampled every week. In total, more than 500 dead minks and around 200 live minks were tested from April 2021 to the beginning of March 2022. In the second week of March 2022, the weekly testing was stopped due to the absence of PCR‐positive results.
Interestingly, the continuous surveillance in the affected farm showed fluctuating virus circulation in animals: the seroprevalence in samples taken from live minks was around 70%, and three infection waves were observed during the period of April–November 2021, when animals were pelted. After pelting, a significant reduction in virus prevalence in the affected mink farm was observed. The interpretation by Latvian authorities was that the reduction of animal numbers and density also might have an impact on virus circulation.
Since outbreak detection, weekly screening tests were performed also on farm personnel, who did not experience any serious clinical signs, nor were they hospitalised. Until the beginning of pelting season in November, all farm personnel were fully vaccinated against SARS‐Cov‐2, and, among those, no positive cases were reported in 2022 (32 workers were tested positive in 2021).
Genomic analysis
The results of the phylogenetic analysis in mink showed that the genome sequences belong to the Pangolin lineages B.1, B.1.177, B.1.177.60 (SARS‐CoV‐2 Alpha variant), which were also the dominant lineages in the Latvian human population at the beginning of 2021. Some mutations in spike protein gene (e.g. Y453F, F486I, N501T, P681R, T478K, L452R) in virus isolates obtained from mink were also identified.
The phylogenetic analysis of the virus isolated from farm personnel showed eight cases in which virus mutations were related to those from infected minks, indicating backward transmission of the virus from animals to humans in the affected holding. However, the circulating type of SARS‐CoV‐2 virus detected in the mink farm was not detected in human population living close to the farm, nor elsewhere in Latvia.
Monitoring scheme
From January until October 2021, weekly sampling and testing of one dead mink from every mink farm to SARS‐CoV‐2, oropharyngeal swabs by RT‐PCR.
From July 2021, weekly screening (saliva laboratory testing by RT‐PCR) of mink farm personnel.
From September 2021, following the risk assessment done by the Food and Veterinary Service (FVS), the active monitoring based on weekly testing in all mink farms was changed to passive surveillance based on suspicion in mink and confirmed SARS‐CoV‐2 cases in farm personnel.
The risk assessment included:
-
–
the results of the laboratory tests performed so far on the mink regarding the presence of SARS‐CoV‐2 in the farm;
-
–
the active monitoring in all mink farms based on oropharyngeal swabs to be tested by RT‐PCR targeting 25% prevalence with 95% confidence. Samples were chosen from dead mink, but if they were not sufficiently available to achieve the required sample size, additional samples were taken from live mink.
-
–
the vaccination status of the persons working in the holding, the procedures for laboratory examinations (screening) and the results of the examinations regarding the SARS‐CoV‐2;
-
–
compliance of the biosecurity measures implemented in the farm as set by national legislation.
The owner or keeper of the mink farm continue to send to FVS electronically data:
-
–
number of minks in the holding.
-
–
number of minks that have died and been killed in the farm weekly.
In accordance with national legislation, the owner or keeper of minks must immediately notify the FVS about suspicion of SARS‐CoV‐2 in minks, e.g. if the animals show symptoms of acute respiratory infection, digestive system dysfunction, depression, immobility, withdrawal from food or water or increased animal mortality.
Other animals
SARS‐CoV‐2 monitoring was implemented also on wild animals of the family Mustelidae, as well as raccoon dogs. For this purpose, dead animal carcasses were submitted for laboratory testing in the frame of rabies passive surveillance; no positive results were found.
3.2.1.1.6. Lithuania
In Lithuania, the only animal species targeted by SARS‐CoV‐2 monitoring are mink. As of January 2022, there are 120 registered mink farms, of which 71 (62 in January 2021) were active (animals were present) with a total of 277,043 breeding animals, and an estimated production stock of 600,000 animals to be pelted by end of 2022.
Epidemiological situation
In Lithuania, the first two outbreaks were reported in mink farms in November and December 2020, and, under an intensified active monitoring, 13 further outbreaks were detected in November and December 2021. As it was the pelting season, 10 of the infected herds with more than 78,000 animals in total only pelted young animals and kept breeding stock. In the other three infected mink herds, all animals were killed and pelted. However, the mink mortality and morbidity did not increase and remained within the norm or even below; thus, no clinical signs were observed on farms. The disease was very mild, and it was detected only due to intensified targeted sampling.
Monitoring scheme
Passive monitoring is conducted based on suspicion due to increased morbidity or mortality. During the pelting season in November and December 2021, the veterinary authority decided to implement also an active monitoring campaign on mink farms over the country, 1,219 samples from 57 out of 62 farms were tested and 41 samples from 13 farms were found as positive. Blood samples were also taken from 19 farms for antibody detection, by targeting 10% prevalence with 95% of confidence. Out of 19 tested farms, 16 were found with antibodies (300/570 samples, 52%).
From 2021 until March 2022, 621 samples from 14 farms were tested and 24 samples from two farms were positive by PCR (oral swabs). In addition to passive surveillance, the veterinary authority is notified in case of positive farm personnel (currently in 2022 most of the farm personnel are vaccinated, and vaccinated workers are not tested anymore on a regular basis) or increased mink mortality, and following an epidemiological investigation is conducted. Furthermore, not less than the last five dead animals per week are sampled for testing. Samples in case of suspicion are collected from dead animals, pelted animals and clinically affected animals.
Genomic analysis
For all virus positive samples, sequencing was performed, and the results indicated that for almost all samples, SARS‐CoV‐2 virus Delta mutation was dominant.
3.2.1.1.7. Poland
In Poland, the farmed species targeted by SARS‐CoV‐2 monitoring are mink and raccoon dogs. As of January 2022, there are 261 mink farms (272 in January 2021), with a total of 1,988,272 breeding animals, and an estimated production stock of 344,958 animals to be pelted by the end of 2022. Besides, there are 28 raccoon dog farms, with 4,701 animals in total.
Epidemiological situation
The first outbreak in farmed mink in Poland was confirmed in January 2021, followed by another one in June 2021. During the months of November and December 2021, through active surveillance, further nine outbreaks were reported in mink farms, without clinical signs. In 2022, four additional outbreaks have been reported, in January, July, September and October, respectively.
Genomic analysis
Molecular tests are conducted on E gene fragments of Sarbecoviruses for virus monitoring; positive samples are confirmed in tests aimed at N and RdRp genes fragments by using in‐house PCR based on Corman et al. (Euro surveillance, 2019), and NGS sequencing is done on positive samples.
Monitoring scheme
In 2021, monitoring was carried out in accordance with the monitoring rules in the Polish legislation, i.e. throat or nasopharyngeal swabs were taken from at least 10 dead mink or mink with clinical signs. In the absence of clinical symptoms, the tests were carried out twice a year (20 live mink) with a minimum of 8 weeks sampling interval on the farm.
In 2022, in accordance with the provisions of Commission Implementing Decision (EU) 2021/788, passive surveillance is carried out, in all mink and raccoon dog farms in Poland. In addition, active monitoring is carried out on farms with over 500 adult livestock at the beginning of the production cycle. Between February 2021 until March 2022, 11,853 samples from 594 farms were tested by PCR, and 104 positive samples from 11 farms were detected.
Farm personnel monitoring is carried out when the second alternative sampling scheme is in place as of (EU) 2021/788.
Other animals
Other kept (other than farmed) or wild animals are subject to passive surveillance in accordance with Annex III, Section 2, of Commission Implementing Decision (EU) 2021/788. Samples are taken from all dead animals or animals with clinical signs related to SARS‐CoV‐2. In 2022, six badgers, one ferret and one marten (road kills) were tested for SARS‐CoV‐2, all with negative results.
3.2.1.1.8. Spain
As of January 2022, there are 27 mink farms in Spain with 104,000 breeding animals and around 340,000 animals for pelting.
Epidemiological situation
In 2021 and up to November 2022, there have been 17 outbreaks reported in Spain in mink farms; 14 were reported in summer 2021, between June and October 2021, most linked to the implementation of a new monitoring scheme (see below).
Monitoring scheme
In accordance with Commission Implementing Decision (EU) 2021/788 of 12 May 2021, a National Program for Prevention, Surveillance and Control of SARS‐CoV‐2 in American mink farms was developed by the Spanish Ministry of Agriculture, Fisheries and Food (MAPA) in collaboration with the Coordination Centre for Health Alerts and Emergencies (CCAES) and the Autonomous Communities with mink farms.
The programme comprises (i) the prevention measures for SARS‐CoV‐2 infection, (ii) the surveillance and early detection of SARS‐CoV‐2 infection and (iii) SARS‐CoV‐2 control activities.
The surveillance on animals is based on two components:
A passive surveillance component focused on the detection and communication to the Official Veterinary Services (OVS) of any clinical sign compatible with SARS‐CoV‐2 infection, followed by sampling of sick animals and PCR test carried out in the NRL;
A targeted active surveillance component including the PCR testing of oropharyngeal swabs from 8 found dead animals on the farm every 2 weeks.
Through passive surveillance, 2,043 samples from 21 farms were tested from February 2021 to March 2022, 21 samples from five farms were tested positive at PCR; in these the infection was detected after the detection of positive workers, but without signs or abnormal mortalities in mink. So far, no positive farms have been detected in 2022.
By the active surveillance, tests on 740 samples from 27 farms identified 63 positive animals from 10 farms.
Moreover, a monitoring is conducted on positive farms as follow‐up. Every 2 weeks, oropharyngeal swabs are taken from 30 adult animals (older than 13 months) and from 60 offspring. This monitoring is maintained until a negative result is obtained by RT‐PCR in two consecutive samplings.
Monitoring of infection in farm personnel is also in place with two components:
A passive surveillance component for the early detection of cases, consisting of communication of any compatible clinical symptom to the human health authorities.
An active surveillance where farm personnel are subject to regular random screening tests.
In case of detection of virus in a farm, positive samples are subjected to sequencing processes to investigate genetic variants and possible mutations.
Passive surveillance for detection of cases compatible with SARS‐CoV‐2 is conducted in other domestic and wild mustelids and raccoon dogs as foreseen in Annex I of Commission Implementing Decision (EU) 2021/788 with no detection of cases of infection.
Independently from the governmental monitoring scheme, wildlife research groups and rescue centres have performed some research activities of SARS‐CoV‐2 infection in susceptible species, mainly wild mink. Two infected wild mink were initially suspected to be positive (Aguilo‐Gisbert et al., 2021), which they turned out to be negative according to confirmatory analysis in official accredited laboratory.7 Infection was detected in dogs and cats from positive households (Barroso‐Arévalo et al., 2021a, 2021b; Miró et al., 2021).
Genomic analysis
The sequencing conducted in the isolates from Spain revealed that the most frequent lineage detected was B.1.1.7 (Alpha variant). The mutations identified were D614G, N501T (a site related to an adaptation to the host and to antigenic drift), A222V (characteristic of the human cases), Y453F (very rarely, detected in Galicia in March 2021 – described in samples from several Danish mink farms (Hammer et al., 2021; Rasmussen et al., 2021)), and F486V and D796H (the two changes that always appear together, detected in 2020).
3.2.1.1.9. Sweden
During the current reporting period (February 2021–November 2022), mink is the only farmed species targeted by SARS‐CoV‐2 monitoring in Sweden. In February 2022, there were 22 mink farms with approximately 60,000 breeding animals. Breeding was banned during the season 2021 as a preventive measure after the outbreaks of SARS‐CoV‐2 that affected large parts of the Swedish mink industry in 2020 but is again allowed during the season 2022. There is no racoon dog breeding in the country.
Epidemiological situation
During the previous reporting period (until January 2021), SARS‐CoV‐2 was detected in mink in 13 farms. Moreover, a serological screening conducted during fall 2020, which covered the majority of mink fur farms active at the time, suggested that most of them had been exposed to the virus. During the current reporting period, movement restrictions and strict biosecurity measures have been in place for all mink farms in Sweden.
During the present reporting period, one outbreak of SARS‐CoV‐2 in mink has been confirmed and reported in August 2021. The farm in question, with 11,000 breeding animals at that time, was located in Skara municipality in the southwest of Sweden. No increased morbidity or mortality had been observed on the farm, and samples were taken for analysis within the surveillance programme covering all mink farms in Sweden in accordance with Commission Implementing Decision (EU) 2021/788. At the time of the outbreak, all people associated with the farm had either had the infection (COVID‐19 confirmed in farm personnel in November 2020) or been vaccinated, or both. Moreover, a serological screening carried out in December 2020 demonstrated that also the mink on the farm had been exposed to SARS‐CoV‐2 although virus could not be detected at that time. In spite of this, SARS‐CoV‐2 might have been introduced again to the farm, most likely through one of the farm personnel, although this was never confirmed.
Genomic analysis
Whole genome sequencing of the virus demonstrated that the virus belonged to sublineage B.1.1.464 (clade 20B) of SARS‐CoV‐2. None of the amino acid mutations described on the spike protein and considered associated with adaptation to mink was present in the sequence. At the time, matching sequences from sublineage B.1.1.464 had previously been described from at least 14 countries globally in samples originating from people. Moreover, this sublineage had also been detected in mink in two other countries. B1.1.464 had not been detected previously in Sweden.
Monitoring scheme
A monitoring scheme has been in place in Sweden since fall 2020, which foresees that all fur farms submit animals found dead, or throat swabs from animals found dead, to the National Veterinary Institute for SARS‐CoV‐2 testing using RT‐PCR. The scheme was initially run on a voluntary basis but has been compulsory since 2021‐05‐12 in accordance with Implementing Decision (EU) 2021/788. Based on a risk assessment with positive outcome, the monitoring has been based on the alternative sampling scheme for the monitoring of animals provided by Annex III in the decision.
During this reporting period, 1,143 samples from 28 farms have been tested, with positive results only in one out of six samples submitted from the outbreak farm described above.
To have a better overview of the situation, the monitoring scheme described above has been complemented with two serological screenings during 2020 and 2021, respectively. During this reporting period, 30 mink serum samples from each of 25 farms (out of the 28 farms active at that time) were submitted to the National Veterinary Institute between June and November 2021, and analysed by indirect ELISA (IDvet, ID Screen SARS‐CoV‐2 Double Antigen Multi‐species ELISA). Samples from 12 farms tested positive in the screening, 11 of which were positive also during the screening conducted during the previous reporting period. In these 11 farms, the proportion of positive samples were lower (in 10 farms) or equal in this screening compared to the previous screening. In one farm, which was negative in the screening in 2020, 16 of 30 samples tested positive suggesting that the animals on the farm had been exposed to the virus during this reporting period. No increased morbidity or mortality had been observed on the farm.
3.2.1.2. Natural infection in other livestock species
By screening the scientific literature and WOAH outbreak dashboard,8 camel, cattle, horse, chicken, swine, goat, sheep and rabbit have never been detected positive for SARS CoV‐2 infection at PCR under field conditions (Deng et al., 2020; Cerino et al., 2021; Pomorska‐Mól et al., 2021; Ruiz‐Arrondo et al., 2021; Villanueva‐Saz et al., 2021a). Serological evidence was detected in rabbit and cattle in a limited number of studies (Fritz et al., 2022; Wernike et al., 2022).
3.2.2. Companion animals detected as infected with SARS‐CoV‐2 under field conditions
3.2.2.1. Cat
In general, cases in cats have been associated with mild to moderate respiratory (e.g. coughing, sneezing, shortness of breath, increased respiratory rate, congestion and eye discharge) or gastrointestinal (e.g. vomiting, mouth ulcers, diarrhoea) or general non‐specific syndromes (e.g. lethargy, fever, lack of appetite, cardiac or neural signs). The analysis of available global data shows 51 references reporting 725 suspected cases of cats based on clinical signs or epidemiological links with SARS‐CoV‐2 infections. Several of these studies were case reports of detected infected cats associated with infected households and showing clinical signs. A systematic review on clinical presentation of infection in cats identified 70 cases (out of a total 124 reviewed cases) with clear data on clinical presentation. Out of these reviewed cases, 38 (54%) were asymptomatic and 32 (46%) were symptomatic. From all cases reviewed, there were six severe cases, where cats died or were euthanised due to disease complications (Giraldo‐Ramirez et al., 2021).
For cases where infection was confirmed by virus isolation or identification of virus genome in respiratory and/or anal samples, high levels of virus loads, indirectly measured by detection of virus genome (10^6.8 RNA copies/ml sample or RT‐PCR cycle threshold, (CT) values < 21) were observed, particularly in some symptomatic cases (Gonzales et al., 2021; Piewbang et al., 2022; Sila et al., 2022).
From all articles identified in the SLR, a subset (N = 28) was selected where observational studies were reported. These studies were either random or opportunistic surveys, which reported serological results. Data on the proportion of infected cats grouped by their habitat conditions (household pets, feral/stray (free roaming cats) or cats kept in shelters) were collected from each selected study. A summary of the distribution of the reported percentage of seropositive cats among the studies is presented in Table 4. While there is large heterogeneity in the study approach (study design, diagnostic methods and timing of sampling) and the reported findings, it could be seen that the observed percent of seropositive cats among the studies was higher (two to three times) in those cats – household pets or feral – which were exposed to a known infected source (e.g. infected household member(s), infected mink farms (Boklund et al., 2021; van Aart et al., 2021; Zhao et al., 2021; Amman et al., 2022)),than in those cats whose close environment reported no infection (e.g. household members not infected) or this information (e.g. infection situation in the household) was unknown (Fritz et al., 2020; Patterson et al., 2020; Bessière et al., 2022; Kannekens‐Jager et al., 2022; Oliveira et al., 2022). Observed levels of seropositivity in shelters with an unknown source of infection (median 10.4% [interquartile range (IQR): 1.7–13.0]) appear to be higher than seropositive levels observed in feral/stray cats with unknown source of exposure (median 2.5% [IQR: 0–9]) (Table 4), which could reflect exposure of shelter cats to infection via contact with infected humans in the shelter or contact with infected cats (Piewbang et al., 2022).
Table 4.
Summary of apparent seroprevalence (%) observed in observational studies reported in the literature
| Species | Epidemiological unit (a) | Source of exposure (b) | Number of Studies | Distribution of number of animals sampled | Distribution of apparent prevalence | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | Q1 | Q2 | Median | Min | Q1 | Q2 | Max | ||||
| Cats | Feral/stray | Infected (c) | 5 | 26 | 15 | 44 | 50.0 | 17.7 | 22.7 | 53.3 | 64.3 |
| Feral/stray | unknown | 6 | 50 | 27 | 92 | 2.5 | 0.0 | 0.4 | 9.2 | 19.2 | |
| Household | Infected | 12 | 21 | 15 | 38 | 20.0 | 0.0 | 7.9 | 23.5 | 52.1 | |
| Household | Not_infected | 1 | 38 | 2.6 | |||||||
| Household | Unknown | 3 | 16 | 16 | 385 | 6.3 | 0.1 | 3.2 | 6.3 | 6.3 | |
| Shelter | Unknown | 4 | 67 | 46 | 102 | 10.4 | 0.0 | 1.7 | 13.0 | 22.5 | |
| Vet clinic | Infected | 2 | 11 | 9 | 12 | 7.7 | 0.0 | 3.8 | 11.5 | 15.4 | |
| Vet clinic | Not_infected | 2 | 21 | 15 | 56 | 0.0 | 0.0 | 0.0 | 3.3 | 6.7 | |
| Vet clinic | Unknown | 7 | 48 | 24 | 127 | 0.0 | 0.0 | 0.0 | 1.2 | 22.9 | |
| Dogs | Feral/stray | Infected (c) | 1 | 8 | 62.5 | ||||||
| Household | Infected | 11 | 16 | 13 | 53 | 12.8 | 5.2 | 11.4 | 17.1 | 40.7 | |
| Household | Not_infected | 1 | 133 | 1.5 | |||||||
| Household | Unknown | 4 | 238 | 22 | 454 | 0.4 | 0.0 | 0.0 | 1.9 | 4.9 | |
| shelter | Unknown | 2 | 20 | 14 | 25 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Vet clinic | Infected | 2 | 14 | 11 | 17 | 12.5 | 0.0 | 6.3 | 18.8 | 25.0 | |
| Vet clinic | Not_infected | 1 | 17 | 13 | 22 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
| Vet clinic | Unknown | 7 | 251 | 55 | 500 | 0.8 | 0.0 | 0.2 | 1.8 | 9.1 | |
Within each selected study, one or multiple epidemiological units (households, veterinary clinics and/or feral cats from different geographical locations) were sampled. Only epidemiological units where equal or more than 10 animals were sampled were selected for data extraction. Data on number of animals sampled and number of positives were collected per epidemiological unit. Percentage of seropositive animals (apparent prevalence) was estimated per epidemiological unit.
The source of exposure to infection is known: ‘infected’ source (e.g. infected humans in the household), ‘not_infected’ source (e.g. animals coming from households where no infection in humans was confirmed) or ‘unknown’ (no information was available or not provided about the situation regarding infection in humans or other pets in the epidemiological unit).
Overall, it is possible to infer that cats are highly susceptible to the infection and that they could shed virus in comparable levels to those reported for people, which may lead to efficient cat‐to‐cat direct‐contact transmission (R0 > 1) (Gonzales et al., 2021; Gerhards et al., 2022; Piewbang et al., 2022) and potential cat‐to‐human transmission events (Piewbang et al., 2022; Sila et al., 2022). Cats from households with infected people have the highest risk of infection, and if these cats are allowed outdoor roaming, they may represent a risk for transmission to other cats (domestic or stray/feral cats) they contact outdoors. Similarly, feral cats with access to infected sources, such as infected mink farms, may represent a risk for transmission to both the feral cat population and household cats they would encounter when roaming around residential neighbourhoods in the vicinity of mink farms (Amman et al., 2022). The low levels of infection observed in stray/feral cats with unknown source of exposure may be a result of limited contact with potential sources of infection, such as contact with household cats, and even between feral cat colonies. The latter would limit spread of infection within feral cats. However, feral colonies and household cats nearby mink farms may appear to represent the cat population with the highest risk of becoming infected.
3.2.2.2. Dog
The SLR identified 34 studies evaluating the presence of infection in dogs. As done for cats, most studies were opportunistic, which followed infected households, reported individual case reports or random surveys, mostly involving clients from veterinary clinics. A crude overview of the reports from the studies identified in the SLR shows that the frequency of reported virus (genome) detections in dogs is lower than that in cats (data not shown) and that infections in dogs were mainly asymptomatic. Observational studies where data on detection of infection (assessed by serological tests) could be extracted are summarised in Table 4. As observed for cats, the percentage of positives detected was higher in dogs exposed to an infected source (mainly a household with infected people). Studies where both dogs and cats were sampled, reported higher percent of positivity in cats than in dogs, with one study in particular reporting the odds for infection in cats being 7.6 (95% CI: 1.9–44.4) higher than in dogs (Colitti et al., 2021).
In summary, dogs are susceptible to infection; if infected, they are mostly asymptomatic and may shed low levels of virus, which may limit between‐dog transmission.
3.2.2.3. Hamster
There is evidence that hamsters can be naturally infected with SARS‐CoV‐2 and cause human infections (Kok et al., 2022; Yen et al., 2022). Kok et al. (2022) reported that following a case of an infected worker in a pet shop in Hong Kong, 7 out of 69 swab specimens collected from hamsters were confirmed positive for SARS‐CoV‐2 infection by RT‐PCR. The warehouse supplying this pet‐shop chain was investigated on 17 January 2022, with 511 swabs randomly collected from hamsters (n = 137), rabbits (n = 204), guinea pigs (n = 52), chinchillas (n = 116) and mice (n = 2). One Syrian hamster swab was positive for SARS‐CoV‐2 infection by RT‐PCR.
As follow‐up investigation, swabs and serum samples were collected from the Syrian and dwarf hamsters in the pet shop and the warehouse. Eight (50%) of 16 Syrian hamsters had evidence of infection, with four animals positive by both serology and RT‐PCR, three animals tested positive by RT‐PCR alone and one animal tested positive by serology alone. In the warehouse, 7 (58%) out of 12 Syrian hamsters had evidence of infection by serology. Of those, two were also positive by RT‐PCR. None of the dwarf hamsters were confirmed positive for SARS‐CoV‐2. Genetic and phylogenetic analysis of the viruses suggested that importation of SARS‐CoV‐2‐infected hamsters to Hong Kong was a likely source of this outbreak. Eighty‐two human cases with an epidemiological link to hamsters were detected leading to multiple zoonotic transmission events to humans. SARS‐CoV‐2 viral genomes from human and hamster cases in this cluster belong to the Delta variant of concern (AY.127) that had not been circulating locally before the outbreak.
Further investigations were then performed on 100 randomly selected euthanised dwarf hamsters housed in the warehouse, as the small number of rabbits and chinchillas had already been removed from the warehouse prior to the warehouse visit. The lung tissues of three of these 100 (3.0%) hamsters tested positive for the SARS‐CoV‐2 by RT‐PCR (Kok et al., 2022).
3.2.3. Wild animals detected as infected with SARS CoV‐2 under field conditions
Among wildlife, white‐tailed deer in North America, where it is very abundant with over 30 million animals (EAZWV, 2022), as free living or kept in game reserves, is one of the species raising most concern in SARS‐CoV‐2 epidemiology. Detection of SARS‐CoV‐2 in white‐tailed deer has been described in seven publications, all describing surveys in different areas of USA or Canada (Chandler et al., 2021; Hale et al., 2021; Cool et al., 2022; Kotwa et al., 2022; Palermo et al., 2022; Vandegrift et al., 2022; Pickering et al., 2022a). The surveys cover in total 72 areas (7 states in USA and a high and low density region in Canada) from which in total 962 animals have been sampled for PCR‐test, leading to a median of 16 animals (IQR: 9–32%) per sampled area, and 821 (median 27 animals, IQR: 5–21%) have been sampled for serology. The median percentage of PCR positives was 27% (IQR: 9–62%), while the median percentage of serologically positives was 16% (IQR: 0–67%). From the Canadian Animal Health Surveillance System's (CAHSS) dashboard, it appears that infection with SARS‐CoV‐2 has been detected in 47 white‐tailed deer across Canada (seven states). From USDA's dashboard,9 it appears that SARS‐CoV‐2 has been detected in white‐tailed deer in 24 states. Furthermore, detections of SARS‐CoV‐2 in mule deer have been reported in USA as well as in Canada.3, 10 From Germany and Austria, 232 samples from hunted red deer and roe deer were collected for SARS‐CoV‐2 surrogate virus neutralisation test (sVNT), with none of them found positive (Moreira‐Soto et al., 2022). White‐tailed deer are not native in Europe; some individuals were imported from North America in the last century, but the abundance in Europe is very limited; currently few individuals are present in Czechia (700 animals) and a more consistent population in Finland, approx. 100,000 individuals (EFSA, 2021). White‐tailed deer are also kept farmed in some places in North America, while this practice is not seen in the EU. Farming may increase the contact between animals and humans, so it may increase the risk of transmission to and from any susceptible species.
SARS‐CoV‐2 has been detected in wild American mink (or mink escaped from farms) in USA (Shriner et al., 2021). In a study from Spain, 162 European or American mink were tested by ELISA and/or RT‐qPCR, and none were found positive (Villanueva‐Saz et al., 2022). Furthermore, five seropositive mustelids (three martens and two badgers) have been detected in France (Davoust et al., 2022). One PCR‐positive juvenile wild leopard (Panthera pardus) was reported in India (Mahajan et al., 2022).
Large numbers of red foxes, jackals and wild boar have been tested in Croatia, all with negative results at PCR. Few results were serologically positive (6/153 wild boar, 3/65 jackals and 6/204 red foxes); however, this was considered as false‐positive results, due to negative or weak positive results in VNT (Jemeršić et al., 2021).
In August 2022, the detection of RNA and antibodies against the SARS‐CoV‐2 Gamma variant infection was reported in four specimens of Chaetophractus villosus (big hairy armadillo) in Argentina (Arteaga et al., 2022).
From a study performed in Eastern USA, 18 different wildlife species were sampled, with SARS‐CoV‐2 detected by PCR in the Virginia opossum (Didelphis virginiana) and equivocal detections in additional species red fox (Vulpes vulpes), white‐tailed deer (Odocoileus virginianus) Eastern grey squirrel (Sciurus carolinensis), Eastern cottontail rabbit (Sylvilagus floridanus) and bobcat (Lynx rufus). Species considered human commensals like squirrels and raccoons had seroprevalence ranging between 62 and71%, and sites with high human use had three times higher seroprevalence than low human‐use areas (Goldberg et al., 2022).
3.2.4. Zoo animals detected as infected with SARS‐CoV‐2 under field conditions
Detection of SARS‐CoV‐2 in lions, tigers and pumas in zoos was described in 10 publications, of which four described multispecies outbreaks. In total, SARS‐CoV‐2 was detected by PCR in 25 lions, 15 tigers and 1 puma. The median percentage of PCR positives was 100% in all three species; however, for lions, the 25‐percentile was 22%, while 73% for tigers. And pumas were only reported in one reference, with two pumas included, of which one puma was tested and found positive. The median group size for lion and tiger was 2–4, with a maximum of 18, and 8, respectively.
In reports by WOAH,11 detections of SARS‐CoV‐2 by PCR have been described in numerous zoo or captive wild species, most often felines (tiger, lion, puma, leopard, fishing cat, Eurasian lynx and, Canada lynx), but also in other families (South American coati (Nasua nasua), spotted hyena (Crocuta crocuta), hippopotamus, giant anteater (Myrmecophaga tridactyla), West Indian manatee (Trichechus manatus), black‐tailed marmoset (Mico melanurus), common squirrel monkey (Saimiri spp.), mandrill (Mandrillus sphinx), otter (Lutra lutra) and gorilla (Gorilla gorilla)).
3.3. Sequence data of animal species infected with SARS‐CoV‐2
As of 13 October 2022 (week 39/2022), there are 2093 sequences collected from animal hosts deposited in the GISAID EpiCoV database (Figure 7, Table 5). Inclusion criteria were non‐human and non‐environmental host sources, collection dates from January 2020 and excluding non‐SARS‐CoV‐2 sequences from bats. The sequences related to the Variants of Concern (VOC), belonging to the Ancestral (pre‐Alpha), Alpha, Beta, Delta, Gamma and Omicron clades, are indicated specifically. The microreact site, a web application to visualise data and sharing genomic epidemiology of the sequences is available. 12
Figure 7.
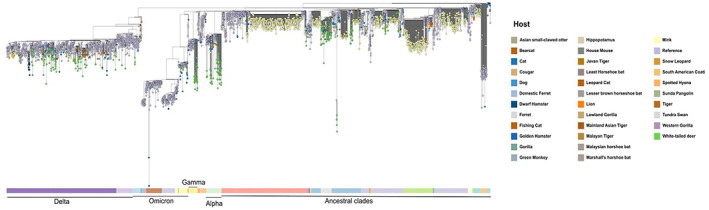
- The tree is constructed using Animal sequences from GISAID EpiCoV, using Nextclade tool by including clade specific reference genomes from Nextclade (link). Human reference genomes are coloured Lavender blue in the tree, whereas the colouring patterns for Animal sequences are based on their hosts. Mainly, mink sequences (yellow), companion animals – cats, dogs and hamsters (shades of blue), wild cats – lions, tigers, mainland Asian tiger, leopard cats, fishing cat and bear cats (shades of orange) and white‐tailed deer (green).
Table 5.
Number of deposited SARS‐CoV‐2 sequences from animal sources by animal host from GISAID database, 13 January 2020–13 October 2022 (week 39/2022)
| Host | Common name | Number of sequences |
|---|---|---|
| Neogale vison | Mink/American Mink | 1,323 |
| Odocoileus virginianus | White‐tailed deer | 323 |
| Felis catus | Cat | 133 |
| Canis lupus familiaris | Dog | 93 |
| Panthera leo | Lion | 72 |
| Mus musculus | House mouse | 23 |
| Panthera tigris | Tiger | 22 |
| Mesocricetus auratus | Golden (syn. Syrian) Hamster | 18 |
| Panthera tigris jacksoni | Malayan Tiger | 13 |
| Gorilla gorilla | Western Gorilla | 13 |
| Panthera uncia | Snow Leopard | 12 |
| Aonyx cinereus | Asian small‐clawed otter | 8 |
| Panthera tigris | Mainland Asian Tiger | 6 |
| Chlorocebus sabaeus | Green Monkey | 4 |
| Rhinolophus malayanus | Malayan horseshoe bat | 4 |
| Gorilla gorilla gorilla | Lowland Gorilla | 3 |
| Phodopus roborovskii | Dwarf Hamster | 3 |
| Rhinolophus pusillus | Least horseshoe bat | 3 |
| Cygnus columbianus | Tundra Swan | 2 |
| Mustela putorius furo | Domestic Ferret | 2 |
| Panthera tigris sondaica | Sumatran Tiger/Javan Tiger | 2 |
| Arctictis binturong | Bearcat | 1 |
| Crocuta crocuta | Spotted Hyena | 1 |
| Hippopotamus amphibius | Hippopotamus | 1 |
| Manis javanica | Sunda Pangolin | 1 |
| Mustela furo | Ferret | 1 |
| Nasua nasua | South American Coati | 1 |
| Prionailurus bengalensis euptilurus | Leopard Cat | 1 |
| Prionailurus viverrinus | Fishing Cat | 1 |
| Puma concolor | Cougar | 1 |
| Rhinolophus marshalli | Marshall's horseshoe bat | 1 |
| Rhinolophus stheno | Lesser brown horseshoe bat | 1 |
3.3.1. Sequence data of farmed animals infected with SARS CoV‐2 (mink)
More than half of the total animal sequence depositions in GISAID EpiCoV database are derived from mink (1,323 sequences). Mink sequences cluster into distinct major mink‐specific clusters and most of them are located in ancestral clades (pre‐alpha clades, Figure 8). These clusters show high intra‐cluster variability, indicating mink‐to‐mink transmission, high rates of virus evolution within the mink population and emergence of mink‐specific variants with potential to spill back into the human population, which has previously been described (ECDC, 2020d; Oude Munnink et al., 2021). Sequence data within clusters exhibit high intra‐cluster variability, which indicates high level of transmission and accelerated evolution within the cluster. The latest pre‐alpha virus sample collected from mink belongs to clade 20C in Lithuania, collected on 26 November 2021, and there are no pre‐alpha viruses detected in 2022 and only six mink‐related sequences were reported in 2022. Fewer smaller mink clusters have been detected in VOC clades, but mostly sporadic mink sequences are reported in these VOC clades – belonging to Alpha, Delta and Omicron. Only two mink‐related Omicron sequences have been reported indicating that human‐to‐mink transmission has been probably mitigated over time if testing has been continued. No sequences of SARS‐CoV‐2 originating from raccoon dogs have been uploaded to GISAID.
Figure 8.
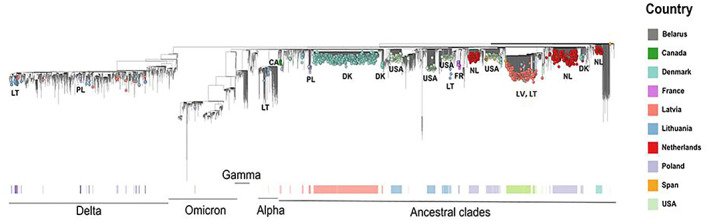
- The tree is constructed using Animal sequences from GISAID EpiCoV, using Nextclade tool by including clade specific reference genomes from Nextclade (link).
Mutations identified in outbreaks of SARS‐CoV‐2 in mink and related human cases:
SARS‐CoV‐2 variants detected in mink outbreaks and related human cases were part of at least five closely related clusters; each cluster was characterised by a specific mink‐related variant, identified in humans and animals from infected mink farms. Some mutations, such as Y453F in the receptor‐binding domain (RBD) of the spike protein, have been suggested as an adaptation of the virus to mink and has been observed in many of the SARS‐CoV‐2 strains in different countries, independently of the clustering (ECDC, 2020d). To note, the Y453F mutation has also been observed in human cases not related to mink. One of the clusters (Cluster 5 variant strains), which was reported as circulating in August and September 2020, carried a deletion of two amino acids (69–70) and two additional mutations in the S protein (I692V, M1229I) in addition to the Y453F mutation in the RBD.
Spillover of SARS‐CoV‐2 from mink to humans
The majority of spill‐over events from animals is related to mink and in some occasions, this spill‐over caused circulation of mink‐related viruses in the general population, as observed in Denmark (ECDC, 2020d; EFSA, 2021). Also other countries (e.g. The Netherlands) reported transmission of SARS‐CoV‐2 from mink to people (Larsen and Paludan, 2020; Oreshkova et al., 2020; Devaux et al., 2021; Hammer et al., 2021; Larsen et al., 2021; Lassaunière et al., 2021; Lu et al., 2021; Oude Munnink et al., 2021; Sharun et al., 2021; Wang et al., 2021; Rabalski et al., 2022).
3.3.2. Sequence data of companion animals infected with SARS CoV‐2
In total, 247 sequences have been deposited for companion animals, divided into cats (133), dogs (93) and hamsters (21) (Figures 9, 10, 11 ). These sequences are spread all over the SARS‐CoV‐2 clades and have much less tendency of clustering together. There is one Syrian hamster cluster (12 sequences) in the Delta clade mixed with human sequences reported from Hong Kong and previously described (Kok et al., 2022). Otherwise, companion animal sequences mostly spread within human sequences. However, some sequences cluster within mink clusters, indicating some spillover from the minks. Little to no tendency in terms of clustering indicates sporadic transmission from humans, with little or no animal‐to‐animal transmission among companion animals. Furthermore, unlike mink, which are farmed and kept in large numbers, accelerated evolution and emergence of species‐specific variants was not observed in companion animal sequences. Some cat sequences cluster within mink clusters pointing to sporadic transmission events either during outbreaks or from infected humans, possibly from cats visiting mink farms, or from households of mink workers.
Figure 9.
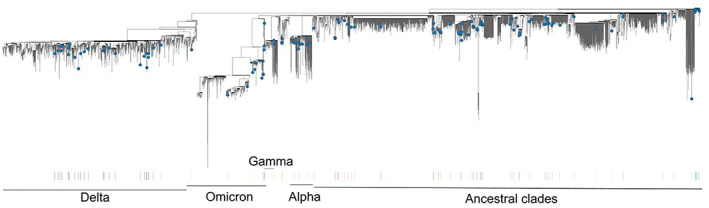
- The tree is constructed using Animal sequences from GISAID EpiCoV, using Nextclade tool by including clade‐specific reference genomes from Nextclade (link).
Figure 10.
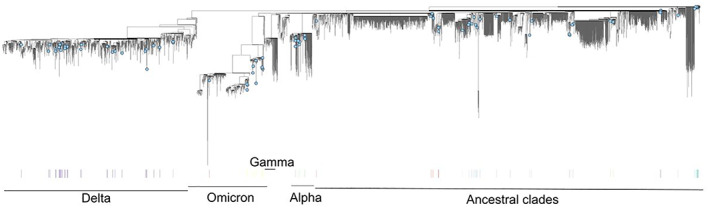
- The tree is constructed using Animal sequences from GISAID EpiCoV, using Nextclade tool by including clade‐specific reference genomes from Nextclade (link).
Figure 11.
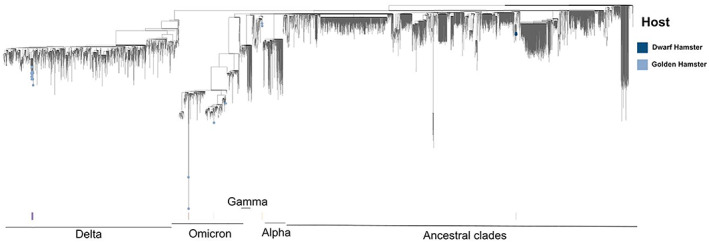
- The tree is constructed using Animal sequences from GISAID EpiCoV, using Nextclade tool by including clade‐specific reference genomes from Nextclade (link).
3.3.3. Sequence data of wild animals infected with SARS CoV‐2
Around 130 sequences have been deposited in the GISAID EpiCoV database for wild cats (lions, tigers, Mainland Asian Tiger, Leopard Cats, Fishing Cat and Bear Cats, Figure 12). These wild felids sequences have a higher tendency of clustering than companion animals. There is one lion‐associated cluster from Spain from November 2020 (19 sequences). This cluster also contains one sequence from dog and two human sequences and has previously been described (Fernández‐Bellon et al., 2021). Another lion‐related cluster was identified in sequences deposited from the USA in October 2021, also containing one spotted Hyena‐related sequence. Another cluster containing several different wild cats, collected in September–November 2021, uploaded from the USA, also containing human sequences. These clustering patterns indicates animal‐to‐animal transmission for wild cats in zoo settings, but there is no clear sign of species‐specific evolution suggesting viral adaptation in these cats in these settings. These wild‐cat‐related sequences are spread all over the clades except the Omicron clade.
Figure 12.

- The tree is constructed using Animal sequences from GISAID EpiCoV, using Nextclade tool by including clade‐specific reference genomes from Nextclade (link).
White‐tailed deer
In total, 323 white‐tailed deer‐related sequences have been deposited in GISAID EpiCoV database, originating from studies performed in the USA and Canada (Figure 13) (Hale et al., 2021; Caserta et al., 2022). Similar to mink, these white‐tailed deer sequences form a significant number of animal‐specific clusters indicating intra‐species transmission and there are signs of accelerated evolution within these clusters. Some of the clusters include human sequences, but not all of them. Sequences are spread across all clades including Omicron, except the index virus clade (Lineage A, clade 19). There were two sampling periods for white‐tailed deer sequences, the first in autumn–winter 2020 (shown in light green colour dots, Figure 12) and the second in autumn 2021 (shown in dark green colour dots, Figure 12). For the first season, mostly all the sequences belong to the ancestral clades, except one sequence in the Alpha clade. One small cluster (four sequences) previously described (Caserta et al., 2022) was highly divergent, indicating adaptation of the virus to new host species, and there is one reference human sequence in this cluster which indicates limited spill‐back to the human population for this variant. During the second season, the sequences are spread over all the VOC clades, mainly Alpha and Delta clades, with few sequences in the Omicron clade. Some ancestral divergent clade sequences were observed in this second season. This indicates new SARS‐CoV‐2 introductions to deer population in 2022, with remaining ancestral variants circulating for a longer period and at least one novel deer‐adapted variant still circulating. It also indicates a continuous transmission from humans to deer populations with deer‐related intra‐host evolution resulting in divergent clusters. The findings of sequence data analysis have been also described in a recent publication (Pickering et al., 2022b).
Figure 13.
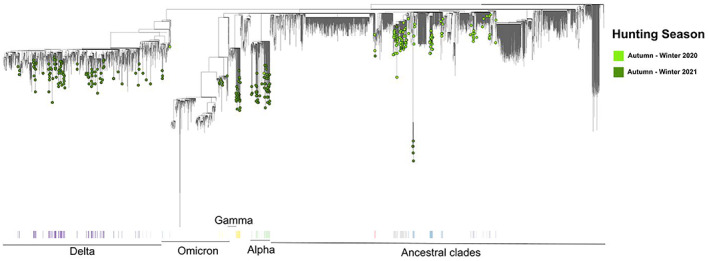
- The tree is constructed using Animal sequences from GISAID EpiCoV, using Nextclade tool by including clade‐specific reference genomes from Nextclade (link).
As only very few sequences from other wild animals have been deposited in GISAID EpiCoV compared to white‐tailed deer, it is difficult to ascertain whether the spread of SARS‐CoV‐2 within the white‐tailed deer population in North America is unique, or if other similar events are ongoing in other animal species and geographic regions.
3.4. Probability of transmission between animals, and between animals and humans, posed by SARS‐CoV‐2 infection in animal species of concern
In this section, for the animal species considered as particularly susceptible (Section 3.1) and thus potentially important in the epidemiology of SARS‐CoV‐2, the probability of SARS CoV‐2 transmission between animals and between animals and humans is addressed, as requested in ToR 2.
The possible epidemiological scenarios and related transmission pathways in farmed animals, companion animals, zoo animals and wildlife are presented in diagrams, tables and supported by explanatory text. The probability of transmission of the virus in each single pathway is qualitatively assessed by consensus of the experts based on the risk and exposure factors identified and supported by the evidence provided in Section 3.1. The reasoning is further explained either in the tables or in the text below the tables in Section 3.3.1. The probability of transmission is estimated assuming no preventive and control measures specific to SARS‐CoV‐2 in the animal species and in the human exposed groups are applied (e.g. no use of non ‐pharmaceutical interventions (NPI), nor infection prevention and control (see Section 3.5.3)). An additional characterisation of the risk assessment is composed by the amount or quality of evidence available (categorised as low, moderate, high) as explained in Table 6. The possible preventive and control measures and monitoring approaches are also indicated for each transmission pathway, and next to each, it is indicated in brackets, whether the measure may affect the susceptibility or the exposure.
Table 6.
Ratings used to describe the amount or quality of the evidence available (EFSA AHAW Panel, 2015)
| Name | Explanation |
|---|---|
| High | No or limited information or data are lacking, incomplete, inconsistent or conflicting. No subjective judgement is introduced. No unpublished data are used. |
| Moderate | Some information or data are lacking, incomplete, inconsistent or conflicting. Subjective judgement is introduced with supporting evidence. Unpublished data are sometimes used. |
| Low |
The majority of information or data are lacking, incomplete, inconsistent or conflicting. Subjective judgement may be introduced without supporting evidence. Unpublished data are frequently used. |
3.4.1. Farmed animals
This section refers to probability of transmission between farmed animals susceptible to SARS‐CoV‐2, and from them to humans and vice versa. Farmed animals include fur‐farmed animals such as mink, raccoon dogs, sable, ferrets, foxes, but from now on we refer to mink, which is used as the type animal in the category, given that it is considered the worst case. In Figure 14, the pathways of transmission (both direct and indirect transmission, the former as by direct contact with infected animals or humans, the latter as transmission, e.g. by contaminated environment and fomites, etc.) within and between mink farms are presented, while in Table 7, the probability of transmission (P), possible control measures and monitoring approach are presented. The reasoning is described more extensively in the text below the table.
Figure 14.
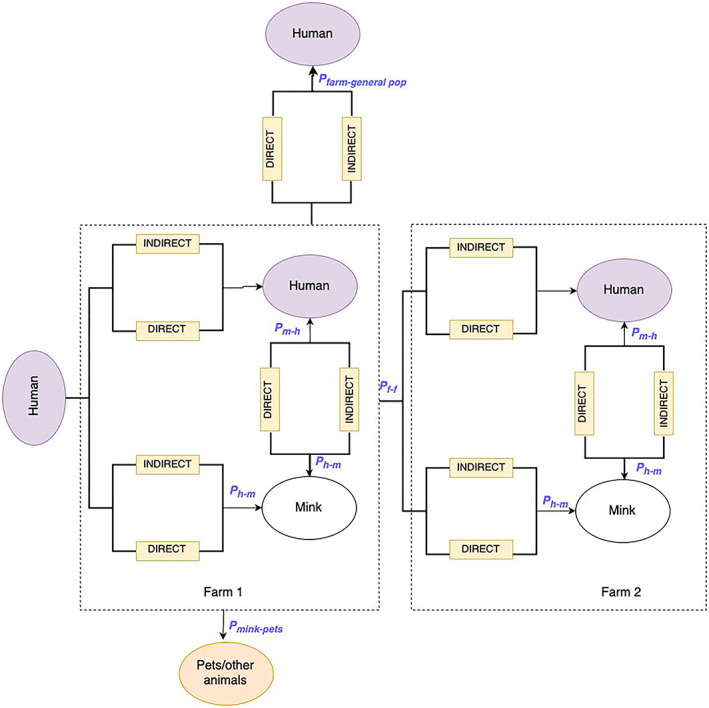
Diagram of transmission pathways of SARS‐CoV‐2 in mink
Table 7.
Transmission pathways of SARS‐CoV‐2 for different scenarios in mink farms, related preventive and control measures and probability of transmission. Mink is used as a worst‐case example of farmed animals
| Transmission pathways (quantity of interest) | Risk factors/exposure factors | Probability of transmission (in the absence of control measures specific to animals in question) | Potential preventive and control measures in animals/farm (in brackets is indicated influence on probability of exposure or susceptibility) | Potential preventive and control measures in humans | Possible monitoring approaches to reduce the probability of transmission15 |
|---|---|---|---|---|---|
|
Human to mink (Ph‐m) ‘What proportion of mink farms in the EU will be infected in the next 12 months due to contact with an infectious human?’ |
|
|
|
|
|
|
Mink to mink (intra‐farm) (Pm‐m) What is the proportion of mink that will become infected in infected mink farms in the EU in the next 12 months? |
|
|
|
|
|
|
Mink to humans (Pm‐h) What is the proportion of people entering/present in infected mink farms in the EU next 12 months that will become infected from mink? |
|
|
|
|
|
|
Farm to farm (Pf‐f) What proportion of infected mink farms in the EU will generate at least one secondary case through direct or indirect farm‐to‐farm transmission of SARS‐CoV‐2 in the next 12 months? |
|
|
|
|
|
|
Spread from farm to general population (Pfarm‐ general pop) What proportion of infected mink farms in the EU will generate circulation (i.e. not limited to sporadic detections) of mink adapted SARS‐CoV‐2 strains in general population to people not associated with mink farms in the next 12 months? |
|
|
|
|
|
|
Mink to companion animals /other animals at farm In the next 12 months, out of the mink farms becoming infected in the EU what is the proportion in which at least one pet or other animal of susceptible species (including wild animals) in the farm will become infected (due to transmission from the mink), given such animals are present on the farms? |
|
|
– | – |
NPI: non‐pharmaceutical interventions; PPE: personal protective equipment.
Human to mink (Ph‐m):
The epidemiological situation, and in particular the level of virus activity and the respective virus variant circulating in the general population in the area is expected to be a driving force in the probability of transmission from humans to mink. However, despite a relatively low incidence in the general population in 2020, the first outbreaks in mink occurred, most likely caused by a mixture of low awareness, limited testing and control measures in the early phases of the pandemic, and a prolonged infectious pressure from infected humans to mink. In addition, introduction likely occurred via people with asymptomatic infection during times when testing was limited or not performed.
Lower testing volumes in the general population will also add limitation to the available data about the level of SARS‐CoV‐2 circulation in the population locally and beyond. This will limit the assessment of when more stringent measures are needed on a farm to prevent virus introduction.
An increase in numbers of workers and visitors will lead to an increased probability of transmission between humans and mink, for example as result of seasonal increase of workload, e.g. during pelting operations.
Non‐pharmaceutical intervention (NPI) measures (Section 3.5.3) such as maintaining distance, hand and respiratory hygiene, wearing face masks, testing and isolating positive tested people and their contacts have been shown to effectively reduce the transmission rate between humans, and most likely from humans to mink. However, experience shows that it is difficult to ensure compliance to all measures during full working days. Limitation of access for visitors and testing for workers and visitors can effectively reduce the risk of virus introduction, if the time from testing to test results is short and the sensitivity of tests is high. However, in many countries, at the time of writing this opinion, SARS‐CoV‐2 is no longer considered such a threat to the population, and such that health care is overwhelmed and would require testing, and public health measures such as wearing facemasks in public places or indoor areas. Indeed, vaccination against SARS‐CoV‐2 reduced hospitalisations, intensive care units (ICU) admissions and deaths due to SARS‐CoV‐2.
Vaccination in humans has shown to be a powerful tool to prevent severe and fatal disease during the SARS‐CoV‐2 pandemic, reduce disease overall, lower virus load and reduce the time of infectivity in infected people; however, vaccination cannot be considered a control measure of the viral spread e.g. to prevent the introduction into a farm.
Explanation about estimation of probability of transmission (low to moderate, from 10 to 66% of all farms): the assessment by the experts indicated a high uncertainty regarding the potential number of adequate contacts between infected humans in the next 12 months, mostly due to possible variation in the incidence of infection in humans (which will influence that of workers/visitors of mink farms). This is expected to be the case even if human transmission to mink (i.e. an infected human entering a mink farm) is very likely, given adequate contact (evidence is available for this as observed in past epidemics in DK and NL).
Mink to mink (intra‐farm) (Pm‐m)
Experience from outbreaks in mink shows that the virus spreads rapidly throughout the farm (Boklund et al., 2021; Hammer et al., 2021), and from two farms, an R0 of 2.9 and a growth rate of 0.293 during a within‐farm epidemic have been estimated (Chaintoutis et al., 2021). In periods with kits (young mink), the density in farms increases, which may lead to even higher transmission rates. However, as virus has been often detected in air within 1 m from mink and may be present up to 3 m away, even reduced density can result in high probability of spread between mink, although likely at a lower speed. Experimental studies on ferrets have shown a higher probability of transmission between animals by direct contact than by airborne transmission, although airborne transmission up to 1 metre has been described (Richard et al., 2020; Kutter et al., 2021).
Previous exposure or vaccination of mink is likely to reduce the transmission rate, but experience is scarce and data lacking. One vaccine study with mink showed that one of three vaccinated mink became infectious, and that vaccinated mink were PCR‐positive for a shorter time period (Shuai et al., 2021). Vaccination has been used in mink in USA and Finland. No farms have been tested positive in Finland, neither before nor after vaccination.
Within‐farm biosecurity may reduce the probability of spread between sections of a farm. However, it is likely that introductions from workers will occur into several sections of the farm (Chaintoutis et al., 2021), leading to a limited effect of within‐farm biosecurity.
Explanation about estimation of probability of transmission (high to very high, from 66 to > 90%): While there is a large uncertainty on the proportion of mink farms becoming infected, mink can readily transmit the virus (R0 > 1) and thus, given the close contact between animals in mink farms, once the infection is present in at least one animal, it is very likely that it will reach a large proportion of the exposed mink population within farm.
Mink to humans (Pm‐h)
Evidence from previous outbreaks shows an increased incidence among mink workers (Larsen and Paludan, 2020; Lu et al., 2021; Oude Munnink et al., 2021). Moreover, characterisation of virus circulating in mink and farm personnel has demonstrated transmission from humans to mink as well as from mink to humans (Hammer et al., 2021; Lu et al., 2021). The probability of transmission from mink to humans will vary with the number of persons exposed on the farm (farm personnel, visitors including family members) as well as applied personal protective measures. Farm size is likely to have an effect on the virus load in the environment, leading to increased risk of transmission in large farms depending on number of animals and in seasons with increased animal density. Testing does not reduce the risk of infection of the individuals tested, but reduces the risk of further spread by early detection of infection/contamination. NPIs instead reduce the risk of individual infection and of further spread to others (depending on the NPI measure). Early detection will in theory increase the effectiveness of PPE by reducing the exposure before detection if PPE is not worn routinely. In practise, it can be difficult to detect outbreaks early enough to have the optimal effect of additional PPE. However, the effect will be influenced by several factors, such as animal density, transmission rate and frequency of contact between workers/visitor and animals. Similar effects can be obtained by measures reducing the spread between mink (Pm‐m). Monitoring infections with the purpose of early detection is described (EFSA, 2021).
Explanation about estimation of probability of transmission (very low to very high, from < 10 to > 90%): in infected mink farms, there is a large uncertainty regarding the risk for workers due to future circulating strains, biosecurity measures, etc. Furthermore, the average proportion of people infected from mink on an infected farm, varies with farm size and thereby with the numbers of workers/visitors of the farm.
Farm to farm (Pf‐f)
Experience from previous outbreaks indicates that spread between farms occur and that farms size and distance between farms is associated with the risk of occurrence (Boklund et al., 2021; Lu et al., 2021). Direct or indirect transmission by humans is believed to be the main route of transmission (Lu et al., 2021). However, from other contagious husbandry diseases, it is well known that the extent of movement between farms of animals, people and equipment influence the risk of spread of disease. Therefore, movement restrictions, limited access to farms for visitors and quarantine time between visits for workers and visitors can reduce the probability of transmission.
Explanation about estimation of probability of transmission (Moderate, from 33 to 66%): multiple factors may play a role here (particularly biosecurity), but assuming possible exposure (e.g. close distance or high farm density and/or movement of animals) the probability of transmission is considered moderate. In some situations, where only antibodies are detected, phylogenetic analyses are not possible, and therefore, the relations between farms can be difficult to describe and, as such, it is a source of uncertainty.
Spread to general population (Pf‐society)
Experience from Denmark showed that the SARS‐CoV‐2 variant related to mink spread from farm personnel to the general population. After the first three outbreaks on mink farms in June 2020, its spread led to at least 90 people infected with this mink‐related variant (Larsen et al., 2021). However, after all mink were culled in the country, this variant was no longer found in the sequenced samples from Denmark. In the Netherlands, three human community cases infected with a mink strain were found (Lu et al., 2021). Furthermore, SARS‐CoV‐2 was not detected in air samples outside farms in the Netherlands (de Rooij et al., 2021) or in Denmark (Boklund et al., 2021).
Explanation about estimation of probability of transmission: (very low < 10%) The probability that mink‐related strains establish in the general population can be assumed to depend on how well adapted these viruses are for transmission between humans, and on the level and viral fitness of other circulating strains at the time. The risk of spread to the general population can be reduced by minimising the risk of mink outbreaks and the risk of transmitting the mink‐adapted viruses to workers at mink farms and thereby lowering the risk of transmission to the general population in the local area of the affected mink farm and beyond. Also, interventions on the animal side, if feasible (e.g. compartmentalisation, etc.), to reduce the spread among the animal population will reduce the risk of workers to be exposed to infected animals (and consequently to the general population) as well as to aerosol or contaminated environment such as surfaces or dust particles in the air. Vaccination and acquired immunity in human population from previous SARS‐CoV‐2 infection have shown to lower the risk of getting infected and also reduce the period of infectiousness, which contributes to reduce the spread to the general population, but this effect is decreasing over time and with any new variant. Frequent testing of farm personnel and visitors on farms could be a useful monitoring approach, the latter with the purpose of early detection.
Mink to other animals at farm (companion animals, wild, feral animals)
The probability of transmission from mink to companion animals (e.g. cats, dogs) will vary with the number of the latter at risk on the farm and the viral load. Companion animals are considered at risk if they have access to the farm area. Similarly, animals of susceptible species living in the surroundings of the farm can be at risk, if they have access to the farm area (e.g. feral cats). Wild mink in surroundings of farms in USA have tested positive, and virus was found in two escaped mink in the Netherlands, 450 and 650 m from culled farms17 (Lu et al., 2021). Furthermore, several cats (many feral) and dogs living on infected Danish, Dutch and Utah (USA) farms have been tested positive, for either virus or antibodies (van Aart et al., 2021; Amman et al., 2022) (Table 4).
High levels of biosecurity, efficient fencing and closed farms, including automated closure of gates and doors, can reduce the risk of companion animals as well as wildlife entering the farms, and thereby the probability of transmission. Monitoring of wildlife and feral cats cannot reduce the probability of transmission; however, it can help supporting information on potential spread between species.
Explanation about estimation of probability of transmission (Low to moderate, from 10 to 66%): Mink are likely highly infectious and effective contact with other animals in the farm is considered likely if the latter are present. However, as few animals of other species have been tested on infected mink farms, there is uncertainty on the probability of each susceptible animal getting infected.
3.4.2. Companion animals
This section refers to probability of transmission between companion animals and humans. Examples are given for cats and hamsters, as these are considered the species at highest risk, and include cats living in households having access to outdoor, stray (feral) cats (cats without owners that may often live in colonies and roam freely), dogs and hamsters in pet shops or at breeding centre. In Figures 15 and 16, the pathways of transmission are presented, while in Tables 8 and 9, the probability of transmission (P), possible control measures and monitoring approach are shown, respectively, for cats and hamsters.
Figure 15.

Diagram of transmission pathways of SARS‐CoV‐2 between dogs or cats and humans
Figure 16.

Diagram of transmission pathways of SARS‐CoV‐2 between hamsters19 and humans
Table 8.
Transmission pathways of SARS CoV‐2 for different scenarios in cats (referring to EU), probability of transmission and related preventive and control measures
| Pathways | Risk factors | Probability of transmission (P) | Potential preventive and control measures in animals | Potential preventive and control measures in humans | Possible monitoring approach |
|---|---|---|---|---|---|
|
Human > cat In the next 12 months, out of the households with at least one SARS‐CoV‐2 infected person living with one or more cats in the EU, what is the proportion in which at least one of the cats in the household will become infected? |
|
|
|
|
|
|
Cat > Human In the next 12 months, out of the cats that will become infected with SARS‐CoV‐2 in the EU, what is the proportion that will transmit the infection to one or more humans in the same household? |
|
Quality of evidence: low |
|
|
|
|
Cat > cat (in same household, shelter) In the next 12 months, out of SARS‐CoV‐2 infected cats in the EU, what is the proportion that will transmit the infection to at least one other cat in the same household or shelter? |
|
Quality of evidence: moderate |
|
|
|
|
Cat ‐> other household cats, stray cats 18 In the next 12 months, out of SARS‐CoV‐2 infected household cats in the EU, what is the proportion that will transmit the infection to one or more cats that do not live in the same household (including stray cats)? |
|
Very Low to low (< 10–33%) Reasoning: Transmission between cats can be sustained (Gonzales et al., 2021; Gerhards et al., 2022), however, the duration and intensity of the contact between cats influences the transmission. Quality of evidence: low |
|
|
|
|
Stray cat > other animals In the next 12 months, out of SARS‐CoV‐2 infected stray cats in the EU, what is the proportion that will transmit the infection to one or more ‘non‐stray’ cats or other animals? |
|
Very low to low (< 10–33%) Reasoning: The duration and intensity of the contact between a cat and another animal influences the transmission probability. Quality of evidence: low |
|
|
Table 9.
Transmission pathways of SARS CoV‐2 for different scenarios in hamsters (referring to EU), probability of transmission and related preventive and control measures
| Pathways | Risk factors/periods/areas | Probability of transmission (P) | Potential preventive and control measures in animals | Potential preventive and control measures in humans | Possible monitoring approach |
|---|---|---|---|---|---|
|
Human > hamster In the next 12 months, what proportion of hamsters in pet shops in the EU will be infected with SARS‐CoV‐2 due to contact with infected people? |
|
Reasoning:
Quality of evidence: low to moderate, there is only one publication describing natural infection of hamsters (Yen et al.). Hamsters however are used frequently as animal models for SARS‐CoV‐2 and high susceptibility to infection has been demonstrated by numerous publications. |
|
|
|
|
Hamster > hamster In the next 12 months, out of SARS‐CoV‐2 infected hamsters in the EU, what is the proportion that will transmit the infection to at least one other hamster? |
|
Reasoning:
|
|
|
|
|
Hamster > human In the next 12 months, out of SARS‐CoV‐2 infected hamsters in the EU, what is the proportion that will transmit the infection to one or more humans? |
|
Low (10–33%) Reasoning: Hamsters shed high levels of virus. Quality of evidence: low to moderate, because there is only one publication describing the transmission of SARS‐CoV‐2 from hamsters to humans (Yen et al.). Considering that hamsters shed high levels of virus, which can be detected in the air, and that virus transmits rapidly among hamsters, high amounts of infectious virus can be reached in situations where several hamsters are kept. |
|
|
|
Cats and dogs
Hamsters
3.4.3. Wild animals
This section refers to probability of transmission between wild animals and humans. In Figures 17 and 18, the pathways of transmission are presented. The white‐tailed deer is until now the only wildlife species in North America (either as free ranging or as captive in game reserves) found positive at a significant prevalence. Therefore, only white‐tailed deer is indicated in the figure (indicated as deer) and the related table. However, the assessment and considerations done for white‐tailed deer might be extrapolated to other wildlife species with a high susceptibility to infection and potential for sustained infectivity if identified in the future (Table 10).
Figure 17.

Diagram of transmission pathways of SARS‐CoV‐2 between white‐tailed deer (or other wildlife species with high susceptibility to infection and potential for sustained infectivity) and humans
Figure 18.

Diagram of transmission pathways of SARS‐CoV‐2 between zoo animals and humans
Table 10.
Transmission pathways of SARS CoV‐2 for different scenarios in white‐tailed deer (WTD), probability of transmission and related preventive and control measures
| Possible transmission pathways | Risk factors/periods/areas | P of transmission | Possible monitoring approach | Potential preventive and control measures in/for animals | Potential preventive and control measures in humans |
|---|---|---|---|---|---|
|
Human > deer In the next 12 months, what proportion of the WTD in the EU will get infected with SARS‐CoV‐2 due to contact with infected people? |
|
Very low to low, < 10%–33% quality of evidence: moderate (Palmer et al., 2021; Vandegrift et al., 2022) |
To set up integrated monitoring of relevant species (Cardoso et al., 2021) |
|
|
| Reasoning to support estimation of P of transmission: several variants circulating among humans spilled over to deer, although no case in EU, the majority of white‐tailed deer WTD in the EU are in Finland, where the circulation of SARS‐CoV‐2 has been among the lowest in the EU all through the pandemic. Nevertheless, this could be linked to the diversity of hosts and the limited and generally non‐systematic sampling. | |||||
|
Deer > deer In the next 12 months, out of SARS‐CoV‐2 infected WTD in the EU, what is the proportion that will transmit the infection to one or more WTD? |
|
|
|
Not applicable | |
|
Deer > humans In the next 12 months, out of SARS‐CoV‐2 infected WTD in the EU, what is the proportion that will transmit the infection to one or more humans? |
|
|
|
|
|
3.4.4. Zoo animals
This section refers to probability of transmission between zoo animals and humans. In Figure 18, the pathways of transmission are presented (Table 11).
Table 11.
Transmission pathways of SARS‐CoV‐2 for different scenarios in the EU in zoo animals (e.g. great apes, felines), probability of transmission and related preventive and control measures
| Pathways | Risk factors | Probability of transmission (P) | Potential preventive and control measures in animals | Potential preventive and control measures in humans | Possible monitoring approach |
|---|---|---|---|---|---|
|
Human > animals In the next 12 months, what is the proportion of zoos in the EU with susceptible animal species in which at least one animal will become infected with SARS‐CoV‐2 due to contact with infected people? |
|
Very low to low, 10–33% Quality of evidence: moderate Reasoning: few case reports available, although susceptible species present in zoos has not been regularly tested for contact with SARS‐CoV‐2. |
|
|
|
|
Animal > animal (same species, same enclosure) In the next 12 months, SARS‐CoV‐2 infected zoo animals in the EU, what is the proportion that transmit the virus to at least one more zoo animal in the same enclosure? |
|
Moderate, 33–66% Quality of evidence: moderate Reasoning: case reports available, close contact between animals, although difficult to be proven, because the animals belonging to the same outbreak were usually exposed to the same infectious source (e.g. positive caretaker (EAZWV, 2022)). |
|
|
|
|
Animal > animal (in other enclosures) In the next 12 months, out of SARS‐CoV‐2 infected zoo animals in the EU, what is the proportion that will transmit the infection to at least one other zoo animal in another enclosure in the same zoo? |
|
Very low < 10% Quality of evidence: moderate Reasoning: no confirmed cases, low chance of transmission, difficult to confirm, whether spread between enclosures, or multiple introduction. For infection maintenance, zoo populations are usually too small |
|
|
|
|
Animal > human In the next 12 months, out of SARS‐CoV‐2 infected zoo animals in the EU, what is the proportion that will transmit the infection to one or more humans? |
|
Very low < 10% Quality of evidence: high Reasoning: no reported cases |
|
|
|
3.5. Risk for human health posed by SARS‐CoV‐2 infection in animal species and preventive measures
3.5.1. Overview of variant viruses in humans by cases over time
On 30 January 2020, the World Health Organization (WHO) declared that the outbreak of COVID‐19 constitutes a Public Health Emergency of International Concern (PHEIC). On 11 March 2020, the Director‐General of WHO declared the COVID‐19 outbreak a pandemic. As of 4 December 2022, 179 million COVID‐19 cases and 1.18 million COVID‐19‐related deaths have been reported from EU/EEA countries. The last Rapid Risk Assessment by ECDC was published 27 January 2022 (ECDC, 2022a).
Different SARS‐CoV‐2 variants have emerged over the course of the pandemic, and variants of concern (VOC) which have been dominant in the EU/EEA countries are shown in Figure 19 below by number of human cases. The emergence and circulation of VOC clades of the Ancestral (pre‐Alpha), Alpha, Beta, Delta, Gamma and Omicron variants with sublineage (BA.1–BA.5) clades are indicated in Figure 19. Information on variants that ECDC is currently monitoring is available on a dedicated webpage,21 together with a timeline about the Omicron variant emergence.22
Figure 19.
- The proportion of the circulating variant viruses was estimated from the SARS‐CoV‐2 sequences submitted to GISAID EpiCov database based on the collection date of the submissions. The average proportions of variants were plotted for the EU/EEA countries. Omicron variants are depicted based on its sublineages (BA.1–BA.5) in the figure, ‘other’ indicates the lineages that are other than Alpha, Beta, Gamma, Delta and Omicron. In the pre‐Alpha period, ‘other’ includes ancestral lineages belonging to nextclade A and B clades whereas now during Omicron circulation, ‘other’ includes recombinants and unassigned lineages.
3.5.2. Measures to prevent and control infection or spread of SARS‐CoV‐2 at the animal–human interface: monitoring
3.5.2.1. SARS‐CoV‐2 surveillance in people
Information from comprehensive testing of SARS‐CoV‐2 was used to monitor the circulation in the general population during the first 2 years of the COVID‐19 pandemic. With countries moving into more routine monitoring approaches of SARS‐CoV‐2, also surveillance systems are being adapted and combined with other respiratory viruses.
These integrated respiratory virus surveillance systems should be able to monitor the spread and intensity of SARS‐CoV‐2 transmission to guide control measures and mitigate the impact of COVID‐19. Integrated surveillance systems for respiratory viruses should also monitor the possible emergence of new variants as well as other relevant events. Countries are encouraged to integrate SARS‐CoV‐2 into existing surveillance frameworks, such as those for influenza, where representative sentinel surveillance systems in primary and secondary care are already established and remain the central surveillance method for acute respiratory infections. These sentinel systems rely on common syndromic case definitions that combine epidemiological data and virological testing that can cover multiple respiratory viruses such as influenza, COVID‐19, and potentially other respiratory virus infections. Indicators of severity such as hospitalisations, admissions to ICU and mortality are key parameters for risk assessments and public health decision as well as assess the impact of vaccination in the population over time (ECDC/WHO, 2022b).
With the shift from comprehensive testing to sentinel surveillance, data on SARS‐CoV‐2 circulation in the population, particularly at local or regional level, might be limited, impacting also the assessment about the risk for animal settings through available data. Over the course of the COVID‐19 pandemic, different other parameters such as hospital or ICU admissions, outbreaks in long‐term care or health‐care facilities might be other parameters that have been used for situational assessments instead of the reported incidence in the populations. These parameters focus on severity and burden on health care and not on the overall circulation in the population. These parameters, however, are delayed indicators of the actual virus circulation in the population.
3.5.2.2. Genomic surveillance
Genomic monitoring is a key part of SARS‐CoV‐2 surveillance with the objective to monitor circulation, evolution and dominance of known and emerging variant viruses in the population, describe key mutations and relevant sites during evolutionary processes, inform vaccine composition decisions or outbreak analyses, and identify new emerging SARS‐CoV‐2 variant viruses early. This should also include the analysis of viruses from animal sources and their relation to viruses circulating in humans in a One Health approach. Genetic surveillance needs to be integrated into the national surveillance strategies for respiratory viruses.
Whole genome sequencing (WGS), or at least complete or partial spike (S)‐gene sequencing, is the best method for characterising a specific variant. Alternative methods, such as diagnostic screening nucleic acid amplification technique (NAAT)‐based assays, have been developed for early detection and pre‐screening to allow prevalence calculation of variants of concern (VOC), variants of interest (VOI) and variants under monitoring (VUM). Many of these methods can accurately identify the different variants, while others will require confirmation by sequencing of at least the complete or partial S‐gene genomic region in a subset of samples.
Timely sharing of SARS‐CoV‐2 consensus sequences is crucial. Sequences should be deposited in the Global Initiative on Sharing All Influenza Data (GISAID) database, or other public databases. Related sequencing raw data should be deposited in the European Nucleotide Archive (ENA) and raw data, if available (ECDC/WHO, 2022a).
3.5.2.3. Wastewater surveillance
Wastewater monitoring is a tool to monitor the overall situation of SARS‐CoV‐2 in the population without specific testing of individuals and has been useful to identify upsurges of infections. Viruses from wastewater surveillance can be sequenced to identify the currently circulating variant but also to identify new or emerging viruses. The European Commission adopted a recommendation asking EU MSs to establish wastewater monitoring to track COVID‐19 and its variants.23 The Joint Research Centre (JRC) coordinates this network on wastewater surveillance with the aim to build on the EU capacities to detect future threats and trends arising from emerging pathogens and pollutants of emerging concern for public health. At the time of writing this document, wastewater or sewage testing for SARS‐CoV‐2 has been implemented in 1370 wastewater plants across the EU for the monitoring of the activity levels in a community and has shown promising results with timely data as well as the identification of new and emerging variants in the population.
Variant monitoring through WGS can also be applied in wastewater, sewage or environmental samples but requires specialised bioinformatic analyses. The value of this surveillance covers also the One Health area (ECDC/WHO, 2022a).
3.5.2.4. Testing
For the testing, different commercial detection assays for SARS‐CoV‐2 RNA or antigen, and serological assays for SARS‐CoV‐2 specific antibodies are available on the market with CE‐IVD (in vitro diagnostic device) marking. More information on CE‐IVD‐marked COVID‐19 rapid antigen tests can be found in ECDC's technical report (ECDC, 2021). Information on these assays can be found in the test directory of the Foundation for Innovative New Diagnostics (FIND) and in the JRC COVID‐19 In Vitro Diagnostic Devices and Test Methods Database24 of the European Commission. The characteristic amino acid substitutions of variants can be found in ECDC's updated list of variants of concern (VOC), variants of Interest (VOI) and variants under monitoring (VUM).21
The specific tests currently recommended by the WHO for the diagnosis and confirmation of SARS‐CoV‐2 are described at WHO webpages.25
Testing strategies of symptomatic or people exposed to infected contacts have been successfully applied over the course of the pandemic to identify SARS‐CoV‐2‐infected people and prevent further transmission through the implementation of public health measures like isolation at home. Population‐based testing strategies have been recently revised in several EU/EEA countries with a focused approach on targeted testing of patients with respiratory illness in secondary health care or selected other groups e.g. risk groups in long‐term care facilities or health‐care workers. Different occupational settings are also part of a more risk based or targeted testing approach.
Testing at the workplace needs to be embedded in the occupational safety and health management approach. There are different legal frameworks and requirements in the different EU/EEA countries concerning testing in the workplace and many countries have lifted the requirements for testing at occupational settings. When testing strategies are designed and implemented at enterprise level, workers (or their representatives) should be consulted and clearly informed, in a language, they can understand, about the procedures set out in the enterprise.
The use of rapid antigen detection tests (RADTs) and/or self‐test RADTs in occupational settings can complement, but not replace, occupational safety and health measures including existing non‐pharmaceutical interventions aimed at preventing the introduction and spread of SARS‐CoV‐2 (ECDC/EUOSHA, 2021).
Measures to prevent and control infection or spread of SARS‐CoV‐2 at the animal–human interface – non‐pharmaceutical interventions (NPI)
Non‐pharmaceutical interventions (NPI) are public health measures that aim to prevent and/or control transmission of communicable diseases such as respiratory virus infections e.g. SARS‐CoV‐2 in the community. NPIs are more effective to reduce transmission than vaccination, while COVID‐19 vaccination is clearly the most effective measure to reduce the health impact. NPIs have played a critical role in reducing transmission rates and the impact of COVID‐19 in the EU/EEA. NPIs will continue to be one public health tool against COVID‐19, however, based on the national situation, countries have lifted requirements following the SARS‐CoV‐2 vaccine roll‐out that prevents those vaccinated from severe disease and avert deaths in the population.26
Most important NPIs relevant for the animal–human interface are detailed below.
3.5.2.5. Hygiene measure
Coughing and sneezing hygiene, frequent washing of hands with soap and water for at least 20 s, or applying hand hygiene solutions, such as alcohol‐based hand rub or gels are recommended in all settings and are simple measures to reduce the spread and exposure.
3.5.2.6. Keeping distance
Keeping physical distance (e.g. 1–2 m) to other people or animals likely infected with SARS‐CoV‐2 as well as avoiding physical contact reduce the risk to spread the infection or get infected. This is also applicable for occupational environments and contact between farm/shop/zoo personnel and visitors as well as animals takes place. However, some working duties might not be possible without contact to animals.
3.5.2.7. Personal protective equipment (PPE)
Personal protective equipment (PPE) prevents from contact, droplet and airborne transmission of pathogens but also provides protection from physical harm and other hazards. PPE includes respiratory protection e.g. filtering facepiece (FFP) respirators, goggles or face shields for eye, gowns for body and gloves for hand protection. PPE need to be available and worn appropriately according to the situation and risk. PPE might be different based on situation and needs, e.g. for a mink farm worker, a ranger or a veterinarian. Also, the recommended PPE may differ in regular working situations, compared to situations where SARS‐CoV‐2 has been confirmed at an establishment and its personnel has to continue working on the premises to feed and take care of the animals (ECDC, 2020b).
3.5.2.8. Face masks
Coronaviruses are transmitted primarily from person to person via respiratory droplets, either by being inhaled or deposited on mucosal surfaces, including aerosols produced when coughing and speaking. A public health policy for wearing a face mask in public spaces should be considered in areas with community transmission when the public health objective is to limit community transmission (ECDC, 2022b).
FFP respirators should be worn by all people in contact to SARS‐CoV‐2 positive tested animals at the workplace.
People tested positive for SARS‐CoV‐2 could also consider limiting the contact and wearing a face mask when in close contact to companion animals in the household to limit the spread of the infection to the respective animal.
3.5.2.9. Ventilation
Based on evidence from several SARS‐CoV‐2 outbreak investigations that transmission also occurs in closed, poorly ventilated spaces, even without close proximity to the source, improved ventilation minimises the role of aerosols in transmission of SARS‐CoV‐2. This could be considered in occupational settings, especially in closed animal facilities such as pet shops and other indoor facilities where animals are kept in close distance and where SARS‐CoV‐2 outbreaks would lead to high virus presence in the environment including air (ECDC, 2020c, 2022b).
3.5.2.10. Stay‐at‐home/isolation
In general, people are advised to stay at home when feeling sick or having COVID‐19‐like respiratory symptoms. Countries have lifted the requirement of COVID‐19 positive tested people to isolate at home for a particular period of time until the infectivity is considered low. However, in special circumstances, it might be requested for people to self‐isolate after a positive test to avoid either the introduction into a farm, e.g. when a new SARS‐CoV‐2 variant circulates in the population, or to avoid spread of new variants to the general population, if the infection was likely acquired at the work place through close contact with animals.
3.5.3. Measures to prevent and control infection or spread of SARS‐CoV‐2 at the animal–human interface – vaccination of humans as protective and control measure
National authorities in the EU make final decisions on the roll‐out of vaccines, including booster doses and type of vaccines, considering factors such as the spread of infection, the impact of COVID‐19 in different populations and the emergence of new variants. These elements will determine which vaccines people receive and when, based on their level of risk and the epidemiological situation.
The latest ECDC report on vaccination strategies and deployment plans in the EU/EEA countries is available.27 Some countries have particular recommendations for occupational settings related to nursing home staff, healthcare and social–health personnel but not related to groups working at the animal–human interface. The report also details the respective vaccines and timing of vaccinations. Currently recommendations for booster doses are usually targeted to risk group and the elderly population to prevent severe disease and deaths in a population at risk, see also ECDC and EMA joint statement.28 Detailed and up‐to‐date information on the vaccine rollout and country‐specific disclaimers on the data may be found in the ECDC Vaccine Tracker.29
As of December 2022, eight COVID‐19 vaccines have received conditional marketing authorisation in the EU/EEA, following evaluation by the European Medicines Agency (EMA), and are part of the EU Coronavirus Vaccines Strategy Portfolio including most recently authorised adapted COVID‐19 vaccines including the bivalent original/Omicron BA.1 and the bivalent original/BA.4–5.30
Safe and effective COVID‐19 vaccines are a powerful tool for ensuring public health and controlling the pandemic. Results from observational studies carried out to date have shown that the vaccines authorised in the EU/EEA are highly protective against severe COVID‐19, hospitalisation and death.
Overall, vaccines against COVID‐19 have shown to substantially lower the risk of severe disease and death and to lower the risk of infection and symptomatic illness. In addition, the risk of exposure is lowered due to a shorter infectious period and lower virus titres in vaccinated people. Vaccination and natural immunity from exposure to the circulating virus has increased the level of protection across the population in many countries across the EU/EEA. However, the emergence of different SARS‐CoV‐2 variants with different antigenic properties over the course of the pandemic has impacted the effectiveness of available vaccines. In addition, waning immunity has also contributed to declining protection over time.
With newly available adapted vaccines and newly emerging immune‐evasive variants, vaccines are still the measure of choice to prevent severe disease and reduce somewhat risk of infection but are not a measure that alone can grant full control of the spread of SARS‐CoV‐2. This is also true for possibly antigenically modified SARS‐CoV‐2 viruses that originate from animal sources.
Published literature indicates that vaccine effectiveness (VE) against severe outcomes caused by Omicron remains high, including among older age groups, with continuous strong protection generally at around 80–90% about 2–3 months after receiving the first booster. Results of studies with a follow‐up period of 3–6 months after the first booster dose are heterogenous, but generally show a gradual decrease in effectiveness against severe COVID‐19 outcomes (VE estimates in the range of 53–100%). The available studies indicate that a first booster dose provides strong protection against severe disease in all the investigated age groups, and there are no clear signs of a more rapid waning in elderly groups.
VE following a second booster dose against severe disease remains high during the short follow‐up period covered in the studies available so far and appears to restore the slightly reduced protection seen 4 months after the first booster dose. Depending on the specific outcome and study, protection is in the range of 40–77% when compared to the third dose (incremental or relative VE) and in the range of 66–86%, when compared to the unvaccinated. Recent analysis of VE from the UK shows some waning of protection following a fourth dose.31 This analysis estimated VE against the Omicron variant for several sub‐lineages (BA.1, BA.2, BA.4 and BA.5) and found that VE against hospitalisation was enhanced by a fourth dose and the incremental VE after 2–4 weeks was 58.6%. This incremental VE decreased to 19.2% at 15 or more weeks after receiving the fourth dose.
In summary, studies conducted during the period when the Omicron subvariants BA.1 and BA.2 were dominant have found that VE against infection with the Omicron variant wanes over time, starting from around 2–3 months after completing the primary series. Similarly, the effectiveness against documented infection wanes after the administration of a first mRNA vaccine booster dose, from estimates within the range of 45–66% in the first 3 months to around 25–45% between 3 and 6 months after the booster dose. A second booster improves VE against infection, but this seems to wane rapidly, as seen within the short follow‐up period available so far after the second booster dose.
Studies suggest that booster doses in general have a modest effect and limited duration in preventing Omicron transmission in the population.
Duration of immunity is a complex issue and, to date, the correlation between measured immunity (e.g. levels of antibodies) and clinical protection from SARS‐CoV‐2 infection has yet to be established. The presence of memory T cells could prevent severe disease in infected individuals for a long period of time, although the durability of these cells and role in protecting from infection (and onward transmission) remains unclear.
Recent studies from the UK, Denmark and South Africa have not found a big difference in VE against different outcomes, including against severe disease, between sublineages BA.1/BA.2 compared with BA.4/BA.5. However, some other studies have found that protection following third or fourth doses against severe outcomes was lower for BA.5 compared to BA.1/BA.2 (Collie et al., 2022; Davies et al., 2022; Hansen et al., 2022; Kirsebom et al., 2022; Kislaya et al., 2022).
A recent analysis from the UK found that, overall, there was no evidence of reduced VE against hospitalisation for the Omicron sublineage BA.4.6 as compared to other BA.4 or BA.5 sublineages.32
Real‐world evidence is essential to measure the impact that the new Omicron‐adapted bivalent vaccines have in preventing infection and disease, since the approval of these adapted vaccines was based on studies collecting data related to safety and immunogenicity. The first VE estimates following vaccination with mRNA bivalent BA.4/BA.5 vaccines against infection have recently been published from the USA (Link‐Gelles, 2022). This study shows that bivalent boosters provided additional protection against symptomatic infection in individuals who had previously received 2, 3 or 4 monovalent vaccine doses. They found that for 18‐ to 49‐year‐olds, the bivalent vaccines were 42% effective against infection, for 50‐ to 64‐year‐olds, 28% effective against infection and for those over 65 years, the bivalent vaccine was 22% effective against infection (see also other topic page on COVID‐19 vaccination by ECDC).33,34,35
Vaccine effectiveness against transmission of the Omicron variant
One study from the UK, that compared VE against transmission of Omicron and Delta variants after vaccination, found a protective effect in contacts (adjusted risk ratio (aRR) 0.88, 95% CI: 0.79–0.97, p = 0.0129) or index cases (aRR 0.78 (95% CI: 0.69–0.88)) having received three doses (compared to two doses) in household settings (Allen et al., 2022). In non‐household settings, a protective effect was observed for contacts having received three doses compared to two doses (0.76 (95% CI: 0.61–0.94)), but there was no evidence of differences in protection based on the number of doses received by those exposing the contacts (Allen et al., 2022). This protective effect of a first booster dose was less pronounced for Omicron compared to Delta. Data from a Danish household study showed a secondary attack rate (SAR) of 31% related to Omicron and 21% for the Delta variant (Lyngse et al., 2021). For unvaccinated household members SAR secondary attack rates of 29% and 28% were observed for Omicron and Delta, respectively, while the SAR were 32% (Omicron) and 19% (Delta) for fully vaccinated, respectively. For booster‐vaccinated individuals, Omicron was associated with a SAR was 25% for Omicron and 11% for Delta, while the corresponding estimate for Delta was only 11%, thus indicating that Omicron is generally 2.7–3.7 times more infectious than the Delta among vaccinated individuals (Lyngse et al., 2021). The secondary attack rate in a household study conducted in Norway for Omicron was estimated at 51% (95% CI: 48–54) compared to 36% (95% CI: 33–40) with Delta giving a significantly higher risk of infection in households with Omicron relative to Delta (Jalali et al., 2022). Generally, the SAR in households with booster vaccinated primary cases and contacts was lower than in households with unvaccinated primary cases and contacts, however primary cases who were booster vaccinated were found to have a considerably higher risk (RR: 4.34; 95% CI: 1.52–25.16) of transmitting SARS‐CoV‐2 to their household contacts with Omicron compared to Delta (Jalali et al., 2022). Moreover, booster vaccinated primary cases with Delta have 80% lower risk of Delta transmission (RR: 0.18; 95% CI: 0.01–0.70) relative to the unvaccinated primary cases (Jalali et al., 2022).
A study using the epidemiologic data from the SARS‐CoV‐2 surveillance within the California state prison system found that vaccination and prior infection were each associated with comparable reductions in infectiousness during SARS‐CoV‐2 infection and additional doses of vaccination against SARS‐CoV‐2 and more recent vaccination led to greater reductions in infectiousness (Tan et al., 2023).
Protection against infections in vaccinated people by the different variants (data for the different variants)
The data from a systematic review and meta‐analysis of 18 peer‐reviewed studies has shown that the pooled mean vaccine effectiveness for all vaccines and ages against symptomatic COVID‐19 was 87% (95% CI: 86–87%) with variation based on vaccine type; for mRNA‐1,273 and BNT162b2, VE were 92% (95% CI: 88–96%) and 85% (95% CI: 85–86%), respectively (Ssentongo et al., 2022). Mean vaccine effectiveness (VE) declined over time and reached 94% (95% CI: 93–94%), 78% (95% CI, 55–100%) and 64% (95% CI: 24–100%) 1, 3 and 4 months after vaccination (Ssentongo et al., 2022). Pooled results from another meta‐analysis demonstrated a 71% (OR = 0.3, 95% CI: 0.2–0.5) reduction in SARS‐CoV‐2 infection rates among subjects who received a booster shot compared with those who did not receive a booster shot of COVID‐19 vaccine (Zhu et al., 2022).
A large meta‐analysis has shown that the VE of full vaccination against any infection and symptomatic infection with the Alpha variant was 88.0% (95% CI: 83.0–91.5) (Zeng et al., 2022). In a multicentre cohort study, VE against SARS‐CoV‐2 infection with the Alpha variant was estimated at 70% 21 days or more following the first dose and 85% 7 days or more after the second dose of BNT162b2 vaccination, respectively (Hall et al., 2021). In another study during the predominance of both Alpha (B.1.1.7) and Beta (B.1.351), vaccine effectiveness (VE) of BNT162b2 was estimated at 90% for Alpha infection and 75% for Beta infection (Abu‐Raddad et al., 2021).
The effectiveness of vaccination against symptomatic disease after vaccination with one dose was notably lower among persons with the Delta variant (30.7%; 95% CI: 25.2–35.7) than among those with the Alpha variant (48.7%; 95% CI: 45.5–51.7) (Bernal et al., 2021). In a case–control study, the effectiveness of two doses of BNT162b2 was 94% against B.1.1.7 and 88% against Delta (B.1.617.2) (Bernal et al., 2021). A meta‐analysis, involving ~17.2 million people of whom 61.1% fully vaccinated with two doses of COVID‐19 vaccines demonstrated that vaccines significantly lower the risk of exposure (RR = 0.2, 95% CI: 0.1–0.5) against the Delta variant among the fully vaccinated population by 80% compared to the unvaccinated population (Mahumud et al., 2022). Overall, the effectiveness of COVID‐19 vaccines against the Delta variant was 86% (RR = 0.1, 95% CI: 0.1–0.5) (Mahumud et al., 2022). During the dominance period of the Delta variant, the booster‐vaccinated subjects demonstrated a significant reduction in infection rates compared with non‐booster‐vaccinated subjects (Zhu et al., 2022).
During the period of dominance of the Omicron variant, the pooled results of studies with a total sample of around one hundred million participants showed that full vaccination significantly lowered the risk of infection (odds ratio (OR) = 0.6, 95% CI: 0.6–0.7) against the Omicron variant (Zou et al., 2022). Additionally, the results indicated that a two‐dose vaccination plus booster significantly lowered the risk of infection (OR = 0.4, 95% CI: 0.4–0.5) against the Omicron variant compared to the unvaccinated group (Zou et al., 2022). The pooling of these same studies also showed that the standard two‐dose vaccination plus a booster significantly lowered the risk of infection (OR = 0.6, 95% CI: 0.5–0.7) against the Omicron variant compared to the two‐dose vaccination without booster (Zou et al., 2022). Another meta‐analysis demonstrated that booster‐vaccinated subjects displayed a 47% reduction in infection rates compared with those who did not receive the booster vaccine (OR = 0.5, 95% CI: 0.4–0.8).
Björk et al. (2022) showed that the VE for the Omicron subvariants BA.1 and BA.2, after at least three doses remained above 80%, however, the VE after two doses declined substantially from 90% (95% CI: 78–95) during Omicron BA.1 dominance to 54% (95% CI: 13–75) during BA.2 dominance. Furthermore, there was a marked decline in protection against severe COVID‐19 during the Omicron BA.2 dominance among persons with two vaccine doses only (Björk et al., 2022). A vaccine effectiveness study has shown that among those who received two doses of any vaccine, VE against symptomatic disease was 64% (95% CI: 59–68%) and 67% (95% CI: 54–76%) for BA.1 and BA.2, respectively, within the first 2 weeks of receiving the second dose (Kirsebom et al., 2022). The numbers drop to 17% (95% CI: 15–19%) and 24% (95% CI: 20–28%) after 25 or more weeks for BA.1 and BA.2, respectively. Additionally, among those who received any booster dose following immunisation with a primary course of any vaccine, VE increased to 71% (70–73%) and 72% (67–77%) for BA.1 and BA.2 respectively, after a week. Over time, this waned to 46% (95% CI: 44–47%) and 48% (95% CI: 45–51%), respectively, at 15 or more weeks after receiving the booster dose (Kirsebom et al., 2022). A different VE study demonstrated that the effectiveness of a three‐dose (booster) vaccination with BNT162b2 for BA.1 was 60% (95% CI: 53–65%), while the VE for BA.2 was 52% (95% CI: 48–56%) (Altarawneh et al., 2022). A recently published study, investigating the VE of four vs. three doses of mRNA‐based vaccines reported a VE estimate of 47% (95% CI: 44–47) against Omicron infection at 12 or more days after the fourth dose (Bar‐On et al., 2022).
Full vaccination with the currently available vaccines provides a statistically significant protection against the Omicron variant (OR = 0.6, 95% CI: 0.6–0.7) within a period of time, but it is not as effective as it was against the Delta variant (RR = 0.15, 95% CI: 0.1–0.3) (Mahumud et al., 2022). The booster provides additional protection against Omicron (OR = 0.4, 95% CI: 0.4–0.5); however, it is also less effective compared to the reported protection effectiveness against Delta (OR = 0.065, 95% CI: 0.06–0.07) (Accorsi et al., 2022). Additionally, those who received the two‐dose vaccination plus booster were also reported to have fewer hospitalisations and lower disease severity compared to the unvaccinated population (Lauring et al., 2022). The new bivalent vaccines have been shown to induce an increase in neutralisation of the B.1.1.529 Omicron variant (Regev‐Yochay et al., 2022).
A living systematic review is being carried out on the efficacy, effectiveness and safety of COVID‐19 vaccines that are currently, or soon to be, authorised in the EU/EEA.36 The aim of the review is to provide a living, continuously updated overview on the evidence by specific vaccine product, age groups and SARS‐CoV‐2 variants. Overall, COVID‐19 vaccines continue to provide a high level of protection against severe outcomes of SARS‐CoV‐2 but lower and relatively fast‐waning protection against infection by the virus. Effectiveness of newer vaccines against infection is still under investigation. The virus continuously evolves and new variants or sub‐lineages with immunity‐evading properties may emerge at any time (Külper‐Schiek et al., 2022).
If vaccinated people are infected, can they still transmit the virus? How does this differ between vaccinated and non‐vaccinated? How is this related to the different variants?
Overall, COVID‐19 vaccines, due to the new immune escape variants, have not shown to be anymore an effective tool at individual level to prevent infection and onward transmission of the virus although the COVID‐19 vaccines, still maintain high effectiveness against severe disease.
The results from a cohort study have demonstrated that even though the initial genomic viral load between vaccinated and unvaccinated individuals was similar, fully vaccinated individuals had a shorter duration of viable viral shedding and a lower secondary attack rate than partially vaccinated or unvaccinated individuals (Jung et al., 2022). This implies that COVID‐19 vaccinations lead to a more rapid virus clearance, thus shortening the infectious period.
Household studies have shown that vaccination reduced onward transmission of the Alpha variant from infected and previously vaccinated people (Harris et al., 2021; Layan et al., 2022; Prunas et al., 2022; Salo et al., 2022). Another study showed transmission of the Alpha variant was 68% (95% CI: 52–79) lower from SARS‐CoV‐2 infected index person 2 weeks after the second vaccination with BNT162b2 than from unvaccinated index patients (Eyre et al., 2022).
Although viral loads were similar in vaccinated and unvaccinated people infected with the Delta variant (Brown et al., 2021), the duration of viral shedding may have been shorter for vaccinated (Chia et al., 2022). This was also shown in another study where 2 weeks after the second BNT162b2 vaccination, transmission of the Delta variant was reduced by 50% (95% Cl: 35–61) and by 24% (95% CI: 20–28) after 12 weeks (Eyre et al., 2022).
In a study conducted in Switzerland from April 2020 to February 2022, it was shown that full vaccination significantly reduced infectious viral load in Delta breakthrough infection cases compared to unvaccinated individuals (Puhach et al., 2022). Fully vaccinated individuals with Delta variant infection had a faster mean rate of viral load decline than did unvaccinated individuals with pre‐Alpha, Alpha or Delta variant infections (Singanayagam et al., 2022).
Vaccine‐associated reductions in onward transmission of the Alpha and Delta variants declined over time after the second vaccination in index patients (Eyre et al., 2022).
For Omicron, an increased transmission rate for unvaccinated individuals and a reduced transmission for booster‐vaccinated individuals was observed, compared to fully vaccinated individuals. These findings show that vaccinated individuals, particularly those recently having received a booster dose, do not transmit the virus to the same extent as unvaccinated individuals (Lyngse et al., 2021).
For Omicron BA.1 breakthrough infections in patients with completed primary course vaccination resulted in significantly lower infectious viral loads (Puhach et al., 2022). Moreover, a significantly lower infectious viral load was observed for booster‐vaccinated individuals compared to fully vaccinated subjects (Puhach et al., 2022). Similarly, for infections occurring 7–30 days after the booster vaccination, a more than sixfold reduction in viral load was noted; however, this booster‐associated viral‐load reducing effectiveness rapidly declined for breakthrough infections occurring 31–60 and 61–120 days after the booster shot, respectively (Levine‐Tiefenbrun et al., 2022).
How long are vaccinated vs. non‐vaccinated able to transmit the virus or at least have detectable virus (period of infectiousness) also by variant?
Duration of infectiousness and peak viral load may differ among virus variants. Even though viral load is a common proxy for infectiousness, a correlation between viral loads and infectiousness is not fully established (Owusu et al., 2021).
One study found that the duration of virus shedding (median) was shorter in vaccinated compared to unvaccinated participants, but this was only effective from day six of the infection (Garcia‐Knight et al., 2022). Overall, the study identified a more rapid decrease of the viral RNA level and a shorter detection of infectious virus in fully vaccinated compared to unvaccinated (Garcia‐Knight et al., 2022). Even though full vaccination may be inefficient at reducing infectiousness during the early phase of the infection, it likely leads to a reduction in the duration of infectiousness (Accorsi et al., 2022; Garcia‐Knight et al., 2022; Singanayagam et al., 2022).
For people infected with the Delta variant similar levels of genome copy numbers were detected in vaccinated compared to unvaccinated during the first 3 days of infection but after this period, the detected genome copy numbers declined faster in vaccinated patients (Puhach et al., 2022) In contrast, infectious virus levels for Delta were substantially lower in vaccinated patients at all days after symptom onset with the biggest effect at days 3–5 and 5 days after onset of symptoms infectious virus was detectable in 54% vaccinated and 85% unvaccinated patients, indicating a shorter period of infectiousness for vaccinated individuals (Puhach et al., 2022).
Bouton et al. (2022) found that the median time for Omicron culture conversion was 2 days for boosted participants with Omicron, 3 days for vaccinated, unboosted participants with Omicron, and 3 days for participants with Delta. Moreover, only 17% of their study cohort failed to culture‐convert by day six and no major differences in culture conversion or viral load decay between the Delta and Omicron variants were found. These results are consistent with previous data that showed no major differences in Omicron infection duration when compared with Delta (Hay et al., 2022). A different study observed that the median time from initial positive PCR assay to culture conversion was 4 days in the Delta virus group and 5 days in the Omicron group, whereas the median time from symptom onset to culture conversion was 6 and 8 days, respectively, for Delta and Omicron (Boucau et al., 2022).
Immunity in the population acquired over time through vaccination and natural infection with SARS‐CoV‐2 is described at ECDC pages.37
3.5.4. Treatment and pharmaceutical prophylaxis of COVID‐19
Different medicinal products have been studied to assess their safety and efficacy as potential agents for pharmaceutical prophylaxis or treatment of COVID‐19. These include corticosteroids, immunomodulatory agents, monoclonal antibodies, antivirals, COVID‐19 convalescent plasma and other therapeutic agents.38 Some antiviral monoclonal antibodies (bamlanivimab/etesevimab, casirivimab/imdevimab) have been studied for post‐exposure prophylaxis (PEP) but none has been recommended so far due to low effectiveness against Omicron.
3.5.5. Risk assessment
This assessment is based on information available to ECDC at the time of publication and, unless otherwise stated, the assessment of risk refers to the risk that existed at the time of writing. It follows the ECDC rapid risk assessment methodology, with relevant adaptations (ECDC, 2019). The overall risk for public health was determined by a combination of the probability of transmission, taking into consideration the assessment presented in Section 3.4, and its consequences (impact of the disease) for individuals or the population (ECDC, 2019). Limitations have been described in the previous rapid risk assessment and EFSA document on mink (EFSA, 2021).
Which detailed threats to humans exist related to SARS‐CoV‐2 in animals?
SARS‐CoV‐2 has the ability to transmit between humans, but also transmit from humans to animals, between animals and from animals back to humans. Not all animal species are susceptible to be infected or can spread the virus back to humans. The relevant animal species that may play a role in SARS‐CoV‐2 epidemiology are discussed in Sections 3.1–3.4 and in the following section.
SARS‐CoV‐2 can adapt in animals and undergo evolutionary processes that result in viral characteristics that may have a public health impact; however, this has not been observed so far (Tan et al., 2022). These animal species‐specific mutations might be located in similar but also different genome sites compared to humans. Mink‐specific mutations have been described with characteristic amino acid changes not observed in humans before, establishing a ‘finger‐print’ in SARS‐CoV‐2 viruses, which could be traced in humans following animal exposure and infection with mink‐specific viruses.
- Another threat is related to the creation of a reservoir in animals where for instance slightly different animal‐adapted SARS‐CoV‐2 viruses could co‐circulate while being related to the SARS‐CoV‐2 viruses circulating in the human population at the same time.
- However, a SARS‐CoV‐2 variant virus could also circulate among the animal population over a longer period that does not or very little relate to the respective virus variant in the human population. Such variants in animals would also undergo evolutionary mechanisms in the animal host, which could lead to infecting a more naïve human population with the evolved variants and reintroduction into the general population. This could equally have an impact on vaccine effectiveness, disease severity and viral transmission.
Spill‐over of SARS‐CoV‐2 from one animal species to another species could introduce different evolutionary and species‐related mutations and processes that could lead to viruses with altered genetic and antigenic profile, as described in point 2. However, to consider continuous animal‐to‐animal transmission, a large susceptible population might be needed to maintain the infection over a longer time period time.
A specific susceptible species may be an intermediate host/vector in the transmission process but not represent a reservoir.
How does SARS‐CoV‐2 in animals represents a threat for individual people/specific exposed groups and the general public?
SARS‐CoV‐2 infected animals do pose a threat to those people in direct unprotected contact with them. This threat could be related to single individuals of the populations, e.g. in a household, or to specific occupationally exposed groups in the animal sector (e.g. mink farm personnel, rangers, veterinarians, zoo or pet shop workers, etc.). Not only a single person but also a larger group of people could be considered occupationally exposed to the animal species identified as a source of potential transmission (see below and in Section 3.5.5).
A wider spread of animal‐related viruses can occur when viruses transmitted to those primarily exposed are further transmitted to local contacts, and then they are transferred to a wider group reaching the general population causing the replacement of the circulating variant with an animal‐derived SARS‐CoV‐2 virus.
Therefore, the risk for the individual and general population will be discussed separately in the risk assessment part.
How is the probability of transmitting SARS‐CoV‐2 from infected animals to individual people/specific exposed groups and the general public evaluated?
The probability of humans to get infected with animal‐derived SARS‐CoV‐2 is dependent of the specific animal species to which the person is exposed, the level and intensity of exposure, as well as the likelihood of the animal to get infected and to become infectious. As outlined in Section 3.4, not every animal species is susceptible and can transmit the virus to the same or another species. In addition, the level of infectiousness (correlated with virus load) as well its duration in the animal species determines the probability of infection.
The likelihood of exposure is dependent of the setting (e.g. workplace, household, zoo, wildlife, etc.), level, and quality of exposure, which is determined by the kind and level of protection of the human, the number and frequency of exposure events over time, as well as the duration of exposure. Long repeated unprotected exposure of people (e.g. mink farm personnel activity over longer periods of time) increases the probability to be exposed to infected animals and acquire the infection.
How is SARS‐CoV‐2 in animals transmitted to humans?
Transmission pathways from animals to humans and vice versa as well as between animals likely occur similarly to the identified routes between people, e.g. through droplets or aerosol in close proximity to infected animals as well as through contaminated hands, surfaces or other materials and the environment. Aerosol sampling around cages of infected mink showed that virus particles were detectable up to 3 m away.
Which animal species are more likely to pose a threat for a human and for public health in relation to SARS‐CoV‐2?
Farmed animals:
Among farmed animals in the EU, farmed mink are the most likely to get infected and transmit SARS‐CoV‐2 among the animal population and onwards to humans. Transmission events within these populations as well as between farm personnel and the animals and back have been observed over the course of the COVID‐19 pandemic. In addition, species‐related evolution of ‘older’ SARS‐CoV‐2 variants that are not anymore circulating in the general human population has been observed, e.g. in Latvia.
No sequence data from SARS‐CoV‐2 infected raccoon dogs has been provided to GISAID EpiCoV database indicating possibly very limited infections in this animal species and raccoon dogs would not be considered a risk species based on these data.
For other farmed animals screened by the literature review (Sections 3.1 and 3.2) such as cattle and pigs, the evidence that these species can be infected is limited, no further transmission has been described including to humans and very few sequences have been reported. Therefore, they are not an animal species that represents a public health risk.
Risk assessment for individual people/groups and general public:
Mink farms have restricted access and are confined in specific areas or regions that minimise the possibility of exposure to potentially infected mink for individuals not associated with the mink farm. The risk for a person without or with limited exposure to farmed mink based on the probability to get infected and develop severe disease from farmed animals is estimated to be none to very low.
Transmission events from farmed mink to humans as well as humans to farmed mink concern directly exposed individuals with unprotected close contact with the animal or with the environment within a farm in the presence of ongoing virus circulation. Exposed individuals are part of different occupational groups working at the premises such as farm owners, veterinarians, workers or seasonal workers involved in, e.g. culling or pelting activities. The risk for an occupationally exposed individual of this defined group to get infected when in unprotected contact to an infected farmed mink (highest probability to be infected) and impact of this infection (probability to develop severe disease) is estimated to be low‐to‐moderate dependent and subjected to uncertainty in relation to the respective virus variant, the effectiveness of the vaccine for this variant in vaccinated people including the time period after the vaccination, previous exposure to other SARS‐CoV‐2 variants and the health status of the individual (i.e. presence of comorbid conditions). The use of PPE and other measures can reduce the risk.
Mink‐related variants transmission from farmed mink to occupationally exposed groups and further spread to the local and even general population have been observed in 2020 and 2021 before the implementation of measures in mink farms. Transmission events from humans to mink and vice versa have been reported only sporadically in 2022. The risk of the spread of a virus variant with mink‐specific mutations or of the re‐introduction of an older variant virus that circulated in minks into the general population causing severe disease is estimated to be very low‐to‐low. This is, however, dependent on the implemented measures at farm level, the follow‐up of exposed people, the respective virus variant, the effectiveness of the vaccine for this variant (in those vaccinated) and the previous exposure to other SARS‐CoV‐2 variants.
Companion animals:
Among companion animals, hamsters, cats and ferrets are considered the most probable to get infected as well as transmit the virus to the same species, to other animal species and to humans. Since the emergence of the Omicron variant, also mice and rats have been identified to get infected and possibly be able to further spread the virus. Dogs are able to get infected but do not transmit the virus further very consistently indicating a lower risk associated with this species. An outbreak involving humans and forming a larger cluster among companion animals has only been observed in a pet shop setting with hamsters in Hong Kong (Kok et al., 2022). Limited clustering and sporadic transmission to humans has been observed for companion animals overall has been reported with viruses lacking species‐specific separate evolution.
The assessment is based on experimental evidence, observational studies as well as available sequence data.
Risk assessment for individual people/groups and general public:
Cats represent the species with the highest number of animals living in close contact to humans across all EU/EEA MSs and have the highest contact frequency with humans. As displayed in Section 3.4.2, the contact between different cats can occur between those living in the same household, with other family cats in the neighbourhood as well as to stray cats when having outdoor access and to other cats in holiday shelters. This contact pattern, including to humans, is unlikely to be sufficient to result in a long or even permanent circulation of the virus or to establish a virus reservoir.
The probability of cats to get infected is dependent on humans in the same household being infected and transmitting the virus to the cat (see Section 3.4.2).
In a household, an infected person is the main source of infection both for other persons as well as for companion animals, due to similar and more timely exposure.
The probability for a human to get infected from a cat with outdoor access that got infected from another cat outside the household is very low. This probability is higher (very low‐to‐low) for occupationally exposed groups with higher number of close contacts to different cats from different households such as veterinarians. The related disease severity is estimated to be equal to the situation in human‐to‐human transmission.
Similarly, the risk is estimated to be very low for non‐occupationally exposed individuals in close contact with hamsters and higher (very low‐to‐low) for groups occupationally exposed to hamsters.
The probability of companion animals to have an impact on the virus circulation in the general population is none to‐very low although smaller outbreaks related to hamsters have been observed outside the EU; the related disease severity is estimated to be equal to the situation in human‐to‐human transmission and therefore the overall risk (determined by the probability and severity) is considered very low.
Zoo and wild animals:
Of animals kept in zoos, primates (apes) as well as feline species have been identified to be susceptible and to transmit the virus to other animals.
In wildlife, carnivores such as foxes, skunks, raccoon dogs, etc., have been found infected but so far there is no evidence that the virus was transmitted among the same species, to other species or to humans. Wild carnivores are mostly solitary living animals, which limits the circulation and spread of SARS‐CoV‐2 among a wild carnivore population of, e.g. wild mink, ferrets or foxes (EFSA, 2021).
A different situation has been reported for white‐tailed deer in the USA and Canada, which has been shown to have become a possible reservoir species with longer circulation among deer herds and is also able to potentially transmit the virus back to humans (Pickering et al., 2022b). Species‐specific virus evolution has been observed in white‐tailed deer. However, white‐tailed deer might not play a major role in establishing a reservoir in Europe as there is a much smaller local population compared to North America.
Rodents, such as wild synanthropic mice and rats in urban settings, are widespread susceptible species, often living in colonies close or even inside human settlements. The virus could spread in these populations over a longer period, thus species‐specific mutations could emerge and potentially represent a public health risk. However, such a scenario has not been observed so far, also with limited sequence data reported from those populations, and the evidence for this is limited at the moment.
Bats are a classical animal reservoir for coronaviruses and are also susceptible to SARS‐CoV‐2. However, very little is known about the possible role of European bats in the emergence of potential zoonotic viruses. Bats of the family Rhinolophidae were identified in the previous EFSA report as of possible concern (EFSA, 2021) since SARS‐CoV‐related Betacoronaviruses were identified in Rhinolophus ferrumequinum. Bats of the Rhinolophidae family, from which SARS‐CoV‐2 (betacoronavirus) is thought to have originated, are present in central and southern Europe.
Risk assessment for individual people/specific groups and general public:
The probability for an individual person to get infected from an animal in a zoo or from an animal in the wilderness is considered to be none to very low and the disease severity associated with such an infection estimated to be similar to the one observed among the human population. The risk is therefore estimated to be none to very low.
The probability for occupationally or activity‐related exposed people or groups such as zoo workers, hunters, rangers or forest workers to be in contact with infected animals or their droppings and get infected might be slightly higher with a similar level of disease severity as for viruses circulating in the general human population. The risk is therefore estimated to be very low.
The probability to have an emerging virus from zoo or wild animals circulating in the general population in the EU/EEA is considered none to very low. The associated disease severity is expected to be comparable to viruses circulating among the general human population. The overall risk is estimated to be none to very low.
Wild carnivores do not represent a public health risk also due to limited human exposure and lack of continuous circulation in such wildlife populations.
Transmission of SARS‐CoV‐2 or other coronaviruses from bats to humans and backwards has not been observed in Europe. The probability of transmission from bats to humans or the emergence of SARS‐CoV‐2‐related or new coronaviruses has been assessed as none to very low for the time period of the next 12 months according to the description in previous sections. This assessment is based also on the limited human population having direct contact with these animals. However, since bats are natural host of many coronaviruses, the monitoring of these species is still important.
Table 12 shows the assessment of public health risk for different animal categories and species, for individuals, occupationally exposed and general population.
Table 12.
Overview of assessment of public health risk for different animal categories and species, for individuals, occupationally exposed and general population
| Category and animal species | Risk for an individual | Risk for occupationally exposed | Risk for general population |
|---|---|---|---|
| Farmed animals (mink) | None to very low | Low to moderate | Very low to low |
| Companion animals (Cat, hamster, mouse, rat and ferret) | Very low | very low to low | none to very low |
| Wildlife (White‐tailed deer, bats) | None to very low | Very low | None to very low |
| Zoo animals | None to very low | Very low | None to very low |
3.6. Revision of monitoring strategies
In this section, the monitoring approaches for SARS‐Cov‐2 in animals are discussed for different categories of animals to be targeted, in the light of the changing and evolving epidemiological situation and new control measures for both animal and public health. As baseline, the current legislative requirements are presented, as well as the information about monitoring plans of SARS‐CoV‐2 in farmed mink in place in MSs, which is reported Section 3.2.1.1.
3.6.1. Current legislative requirements of monitoring SARS‐CoV‐2 in mustelids and raccoon dogs
The current legislation in force for the monitoring and reporting of infections with SARS‐CoV‐2 in animals is the Commission Decision 788/2021. The target species are mink (Neogale vison and other animals belonging to the family Mustelidae) and raccoon dogs, because these species are susceptible to SARS‐CoV‐2 infection and often farmed in large numbers, thus potentially supporting the transmission of infection in the farms at high rates and probability of variant emergence. In addition, although the introduction of the infection into the farms is usually caused by infected farm personnel, the transmission of the SARS‐CoV‐2 virus back to humans (by American mink) has been observed (Oude Munnink et al., 2021). The sampling scheme is applied in establishments with more than 500 adult breeders at the beginning of the cycle and it is summarised in Table 13.
Table 13.
Sampling scheme alternatives as foreseen by EC Decision 788/2021
| Default scheme | 1st alternative scheme | 2nd alternative scheme | ||
|---|---|---|---|---|
| Prerequisites | – | Favourable outcome of a risk assessment + risk mitigating measures | Risk assessment + risk mitigating measures + testing employees | |
| Events triggering sampling | – | – | Increased mortality compared to the baseline mortality rate or animals with clinical signs related to SARS‐CoV‐2 | Detection of SARS‐CoV‐2 in employees |
| Sampling population | Dead and sick animals from each epidemiological unit. If no dead or sick animals > > randomly from live animals to reach the expected sample size. | Dead and sick animals from each epidemiological unit. If no dead or sick animals > > randomly from live animals to reach the expected sample size. | Only dead and sick animals from each epidemiological unit | |
|
Sampling frequency |
Weekly | Every 2 weeks | Following ‘events’ as above | |
| Sample matrix | Oropharyngeal swabs from live or dead animals | Oropharyngeal swabs or expiration air | Oropharyngeal swabs from live or dead animals | |
| Diagnostic test | Detection of SARS‐CoV‐2 virus genome. | Detection of SARS‐CoV‐2 virus genome. | Detection of SARS‐CoV‐2 virus genome. | |
| Design prevalence | 5% prevalence with 95% confidence. | 20% prevalence with 95% confidence. | 50% prevalence with 95% confidence. | 5% prevalence with 95% confidence. |
| Estimated amount of samples required (e.g. in a 5,000 mink farm) | 60 | 15 | 5 | 60 |
3.6.2. Animal categories and monitoring approach
The general aim of SARS‐CoV‐2 monitoring is to provide relevant information for planning and implementing appropriate preventive and control measures for preserving public and animal health.
However, the changes of the epidemiological situation of SARS‐CoV‐2 at global and EU levels have led the countries to modify the objectives and the approaches followed for the monitoring of SARS‐CoV‐2 infection in humans. The reduction of mortality and severe disease in the human population due to COVID‐19 following the implementation of the COVID‐19 vaccination led to the relaxation of public health and social control measures as well as a modified overall testing strategy in the countries with decreasing testing intensity for the early detection of the infection in the general population.39 Nevertheless, genomic surveillance of the emergence of new variants of the virus remains a relevant objective, especially in relation to the possible emergence of variants more capable of escaping the immune response induced by the vaccines. The World Health Organization (WHO) together with the Food and Agriculture Organization of the United Nations (FAO) and the World Organisation for Animal Health (WOAH) have recently raised concern about the risk of the establishment of animal reservoirs and virus evolution in novel hosts, potentially leading to the emergence of new SARS‐CoV‐2 variants (WHO‐FAO‐WOAH, 2022).
In this context, the information coming from monitoring programs in animal populations can support the assessment of risks of SARS‐CoV‐2 transmission from animals to humans, with particular regard to the possible selection and emergence of new variants in animal populations, for which humans might be more susceptible or available vaccines less effective.
However, it must be highlighted that humans currently represent the main population maintaining the circulation of SARS‐CoV‐2 virus, and that the introduction of the virus into animal populations is caused in almost all cases by infected people. Spillback of infection, from animals to humans, has been identified only in farmed minks, white‐tailed deer and hamsters (Oude Munnink et al., 2020; Yen et al., 2022). Therefore, considering the different purposes and opportunities of human exposure, the four animal categories considered in this opinion (farmed animals, companion animals, wild animals and zoo animals) will be considered separately.
In particular, based on the results of the assessment of susceptible species and their ability of transmitting SARS‐CoV‐2 (Sections 3.1 and 3.2), the following species are considered in each animal category:
farmed animals: minks, raccoon dogs;
companion animals: cats, hamsters, ferrets;
wild animals: white‐tailed deer and other susceptible wildlife including carnivores and bats;
zoo animals: wild Felidae, great apes.
Concerning the main possible objectives of monitoring programmes of SARS‐CoV‐2 infection in animals, the four objectives already reported in a previous EFSA report (EFSA, 2021) have been considered (early detection of SARS‐CoV‐2, measuring exposure to SARS‐CoV‐2, confirmation of SARS‐CoV‐2 infection in suspected animals, monitoring virus evolution). The monitoring scenarios are reported in Table 14. As explained in Section 3.6.2.1 below, the objective of early detection being no longer considered relevant according to the current epidemiological situation, but kept in the table for comparison.
Table 14.
Monitoring scenarios for animal categories and monitoring objective
| Early detection of SARS‐CoV‐2 | Measuring exposure to SARS‐CoV‐2 | Confirmation of SARS‐CoV‐2 infection in suspected animals | Monitoring virus evolution | |
|---|---|---|---|---|
| Farmed animals (minks, raccoon dogs) | X | X | X | |
| Companion animals (household cats, hamsters, ferrets) | X | X | ||
| Stray cats | X | X | X | |
| Wild animals | X | X | X | |
| Zoo animals | X | X |
3.6.2.1. Farmed animals
In the EU context, this category includes mainly mink and other Mustelidae and carnivores (e.g. sable, raccoon dogs, farmed for fur production). The current legislation is aiming at the early detection of the infection in farmed animals, especially in establishments with a high number of individuals, to quickly apply all measures needed to halt the transmission and prevent potential risk for humans in the establishments. Given the current epidemiological situation of SARS‐CoV‐2 in the EU, this objective has lost its original importance. In fact, from 44 outbreaks reported in 2021 in the whole EU, in 2022 only six were reported (Section 3.2.1). This improvement may be linked to several factors, among those the general reduced population of mink in the EU, and the measures in place to prevent the virus introduction into farms through farm personnel, like the application of a repeated testing regime of workers, use of PPE and possibly vaccination of farm personnel. On the other hand, the genomic surveillance of viruses circulating in this category of animals is still important, given the possibility of selection and emergence of new genetic variants of the virus, especially in farms with a large number of animals.
Likewise, the confirmation of SARS‐CoV‐2 infection in suspected animals remains a potentially relevant objective, in order to allow the farmers to apply some preventive measures (quarantine, targeted culling, vaccination, etc.) in the effort to reduce the virus circulation within the farm and prevent health consequences for the animals kept.
For the above‐reported reasons, a surveillance approach following the 2nd alternative scheme foreseen in the Commission Decision 788/2021, based on the investigation of dead animals or animals showing clinical signs compatible with SARS‐CoV‐2 infection, with sampling triggered by increased mortality (compared to the baseline mortality rate) or morbidity in mink, or farm personnel testing positive, would be the most appropriate for confirming the infection in the farms and monitoring the virus evolution.
In case of confirmation of infection in farm personnel or other persons in close contact with the animals, and in the absence of observable clinical signs in the animals, a random sample of individuals should be tested with the purpose to detect the infection, assuming a 20% prevalence (with 95% confidence). Considering the time needed to establish the infection in 20% of subjects, it could be preferable to wait for or to repeat sampling after 8–10 days (according to the epidemic model reported in the previous EFSA report (EFSA et al., 2021)), according to the presumable time of exposure of the worker, in order to have a higher chance to detect the infection in the animals, if present. However, in case the veterinary authorities intend to reduce the time for the verification of the health status of the animals and, therefore, the impact of the disease, a larger sample of individuals should be tested, considering the expected prevalence in the farm in relation to the most probable time of virus introduction, in line with the epidemic model reported in the previous EFSA report (EFSA, 2021). Simultaneous testing of workers (rapid test or PCR) would improve the sensitivity and detect infected workers, especially if no PPE, in particular face masks or FFP respirators, are worn in the premises.
National veterinary authorities might also voluntarily consider to periodically assess the situation in the farms, following a more active monitoring scheme. In this situation, sampling during pelting can be a reasonable approach to reduce logistic difficulties and sampling costs. Serological assays and PCR tests can be used for assessing the level of exposure and infection, respectively, of the farmed mink population.
In any case, regardless of the type of objective, a representative sample of PCR‐positive samples should be subjected to genomic characterisation through genome sequencing in order to ensure a proper surveillance on the circulating genetic variants of the virus. If many positive animals are detected and the sequencing capacity is limited, a subset should be selected, at least to represent each positive farm or epidemiological unit. Samples metadata (e.g. epidemiological link, spatial data, temporal information) should be collected, also to properly select the samples to be sequenced and allow comparative analysis.
3.6.2.2. Companion animals
Companion animals (dogs, cats and ferrets) may be infected by SARS‐CoV‐2 virus when in close contact with infected people. Cats may also develop clinical signs (see Section 3.1). Concerning the possibility of back transmission to humans, two reports from Thailand singled out this possibility (Piewbang et al., 2022; Sila et al., 2022).
It must be assumed, therefore, that infected companion animals in households may coexist with infected owners. In this circumstance, testing the animals in the household, especially those showing severe clinical signs, may be relevant to confirm the SARS‐CoV‐2 infection and, if needed, to apply proper therapy.
In any case, when cats coexist with infected persons in the same household, the outdoor access of these animals should be limited and possibly avoided to reduce any possible risk of further transmission. In addition, genome sequencing on positive samples taken from these animals may be useful, especially when the sequences were not obtained from the owner's samples or in case other epidemiological circumstances (e.g. unusual clinical picture observed) may suggest a more in‐depth characterisation of the virus involved.
Concerning the possible circulation and persistence of the SARS‐CoV‐2 virus in stray cat and stray dog communities, available literature reports contrasting results. According to some published surveys, serological evidence of SARS‐CoV‐2 infection was detected in stray cat and stray dog populations in Spain, and northern Italy (Farnia et al., 2020; Spada et al., 2021; Villanueva‐Saz et al., 2021b), whereas other surveys in Italy in stray cats failed to find any evidence of virus circulation (Stranieri et al., 2022). Surveys in feral cats frequenting infected mink farms in Denmark, the Netherlands and Utah (Boklund et al., 2021; van Aart et al., 2021, Amman et al., 2022) or with access to hospital's waste in Iran (Farnia et al., 2020) have reported a seroprevalence ranging from 17.7% to 64.3%. Amman et al. (2022) used trackers to follow the roaming activity of cats frequenting infected mink farms in Utah and observed that these cats would freely move between farms and the surrounding residential properties. To date, it is rather difficult to evaluate the importance of these studies in relation to the possibility of circulation of virus and its persistence in stray cat communities. More research studies on the possible persistence of the infection in stray animal communities should be performed to better clarify the possible role of these populations in the maintenance and evolution of the virus. However, apart from research objectives, testing of individual animals in stray communities could be justified when suspected SARS‐CoV‐2 clinical cases or abnormal mortality rates possibly due to infectious diseases are observed.
3.6.2.3. Wild animals
The wild animal species that may be considered as possible targets for SARS‐CoV‐2 monitoring are white‐tailed deer, as well as other susceptible wildlife, including carnivores, bats and rodents such as wild synanthropic mice and rats (those living in or close to human settlements).
Given the low number of white‐tailed deer in the EU (Finland and Czech Republic), compared to USA and Canada, it is unknown whether white‐tailed deer under the European conditions may be able to sustain the persistence of SARS‐CoV‐2 infection and circulation (in terms of ecology, population density, likelihood of exposure by contacts with humans, etc.), as it seems possible in North America. Therefore, no specific regulated monitoring activities, apart from testing hunter‐harvested showing signs related to SARS CoV2 infection or dead‐found individual animals, would be needed for white‐tailed deer populations.
Monitoring based on suspicion can be conducted in other wildlife species, including found dead individuals (especially carnivores) and animals showing respiratory signs suggesting possible SARS‐CoV‐2 infection. Further research studies might be relevant to monitor the possible introduction and persistence of the virus into the white‐tailed deer population in Europe, as well as in other potential susceptible European deer species, in particular roe deer and red deer. A similar approach can be followed for wild Mustelidae and wild canids (e.g. fox that has been found susceptible in one experimental study (Porter et al., 2022)), for which no evidence of a role in the circulation and maintenance of the SARS‐CoV‐2 infection currently exists. In case of increased mortality in these wildlife populations, SARS‐CoV‐2 infection should be included as a differential diagnosis. In addition, research studies should investigate the possible role of bats in the European context, especially those belonging to the family Rhinolophidae (Delahay et al., 2021; EFSA, 2021).
From all types of surveillance of wildlife for SARS‐CoV‐2, positive samples should be subjected to genomic characterisation to monitor the circulating virus genetic characteristics.
3.6.2.4. Zoo animals
The infection of zoo animals with SARS‐CoV‐2 can be considered an accidental and sporadic event from infected workers, with little relevance from the epidemiological point of view. Therefore, no specific systematic monitoring activities are considered necessary in these animals; only testing sporadic mortality cases and animals showing clinical signs or in contact with positive tested workers or animals may be relevant to confirm the SARS‐CoV‐2 infection and, in case needed, to apply a proper therapy, especially for high‐value individuals. Usual preventive measures (quarantine, isolation, etc.) should be then foreseen for infected animals, especially if kept in enclosure with other susceptible species or individuals. A repeated testing scheme could be justified for zoo workers in order to prevent the transmission to these animals.
3.6.3. New development in diagnostics/sample matrices for SARS‐CoV‐2 in animals
Depending on the various objectives of the surveillance, different testing method approaches have to be considered: viral nucleic acid detection tests for acute infection and serological tests for population studies. Relevant aspects regarding diagnostic tests are described in a previous EFSA report (EFSA, 2021) and are still applicable. In particular, caution needs to be taken if new variants arise that may not be detected by previously validated tests.
The limitations of diagnostic tests must be taken into account. Very few diagnostic tests have been validated in animals, especially in wild populations. Direct diagnostic assays (e.g. RT‐PCR) may be affected by a low sensitivity due to the limited knowledge of the viral loads and routes of virus shedding as well as the temporal window during which the virus can be detected on animal samples. Antigen tests, as used for humans, are not recommended for use in animals due to the unknown specificity and sensitivity in animal samples.
Serological tests (e.g. neutralisation tests or ELISAs), so relevant for retrospective or prospective population studies, may be seriously affected by a low specificity, due to potential cross reaction with antibodies against other coronaviruses frequently present in animal populations. Noteworthy, some animals can also have antibodies against coronaviruses to which they are not susceptible.
Samples from the environment of animals can also be collected for surveillance studies. This could include water, air or surface sampling for environmental SARS‐CoV‐2 RNA.
3.7. Options for disease prevention and control measures
In this chapter, the main possible measure for prevention and control of SARS‐CoV‐2 infection in farmed animals are presented and their advantages and drawbacks discussed based on feedback gathered from MSs, assessment by ECDC (for the measures to be applied on humans) and on expert knowledge. The control measures to be applied in the scenarios for companion animals in the household, wild and feral animals and zoo animals are indicated in Section 3.4; because these are driven by general principles of good hygiene practice, an assessment about their advantages and limitations is not conducted.
3.7.1. Farmed animals
This section refers to main control measures applied in MSs, where farmed animals susceptible to SARS‐CoV‐2 are bred (i.e. mink, raccoon dogs, sable, ferrets, foxes). Humans are considered to be the most important source of introduction of SARS‐CoV‐2 into the farm, with 12 reported occurrences of human introductions to fur farms in the current reporting period, up to November 2022 (Section 3.2.1.1).
3.7.1.1. Farm personnel and visitors
3.7.1.1.1. Health self‐assessment (‘stay‐at‐home/isolation’)
In general, the risk of transmission from persons (workers/visitors) to mink can be reduced by staying at home when feeling sick or testing positive for SARS‐CoV‐2 or other pathogens that could be transmitted to animals, e.g. influenza. For details, see Section 3.5.3.
3.7.1.1.2. Systematic testing of personnel/visitors at predetermined frequency
To prevent introduction of infection from humans, systematic frequent testing using rapid antigen test and/or PCR of personnel and visitors for SARS‐CoV‐2 infection has been recommended as an option. As already indicated in the previous report by EFSA (2021), this is a prerequisite for the early detection of infection in people that may enter the farm and come into contact with animals, and therefore considered a key measure to prevent introduction of SARS‐CoV‐2 into the farm. Early detection of infection in people is only efficient if followed by restricted access to the farm for people tested positive. More details are provided in Section 3.5.3.
- Advantages:
-
○Theoretically good effectiveness
-
○Limited costs
-
○Easy application and verification (a record of test results can be kept)
-
○Relatively early detection of infected workers including the asymptomatic ones.
-
○
- Challenges and drawbacks
-
○Limited (and depending on the type of test) sensitivity of the test during early stages of infection and potentially for emerging variants
-
○High frequency of testing necessary to ensure early detection
-
○Additional cost for farm or workers might decrease compliance
-
○Possible organisational difficulties (e.g. to train people for properly making nasopharyngeal swabs or to recruit sanitary personnel)
-
○Some personnel/visitors may ‘escape’ testing at sampling locations (farmers may not have the authority to impose testing) or skip mandatory self‐tests.
-
○
3.7.1.1.3. Temperature screening
The systematic testing has sometimes been coupled with temperature screening and health check for clinical signs for whoever enters the farm at any time (personnel, visitors, etc.), in combination with a SARS‐CoV‐2 rapid antigen test.
Nevertheless, based on the experience with different diseases over the past outbreaks and pandemics, temperature screening is an ineffective, unspecific and resource‐intensive measure that adds little benefit. Fever is not a specific symptom that clearly identifies a SARS‐CoV‐2 infection. A proportion of COVID‐19 cases will not be identified through temperature measurement because of being asymptomatic‐ or pre‐symptomatic or under antipyretic medication. Furthermore, a proportion of transmission happens before symptom onset. In addition, the technical implementation of this measure is quite complex (equipment, calibration, thresholds, performance, sensitivity and specificity, etc.).40
Compared to the application of screening by COVID‐19 rapid test, this measure does not provide any advantage in such settings nor prevents introduction of SARS‐CoV‐2 into the facilities.
3.7.1.1.4. Use of personal protective equipment (PPE) for farm personnel and visitors
Personal protective equipment (PPE) impacts the risk of transmission from infected persons to animals or vice versa. PPE will help to limit the exposure to and spread of pathogens but is dependent on the level of applied PPE. To reduce the risk of transmission, people in contact with SARS‐CoV‐2‐infected animals should wear FFP masks or respirators as well as goggles.
The wearing of specific clothing, aprons, rubber boots and additional protective equipment needs to be considered according to the work area in consultation with the respective occupational safety and health authorities. Limitations in the preventive effect could be due to difficulties in maintaining compliance for a long period, working conditions could be difficult especially when temperatures are high or in case of chronic respiratory disorders of farm personnel. It may also require long‐term awareness from farm personnel and work environment rules/regulations might require special types of masks or limited time working with masks. See also ECDC's scientific evidence basis assessment for the considerations for the use of face masks in the community in the context of the SARS‐CoV‐2 Omicron variant of concern (ECDC, 2022b).
3.7.1.1.5. Vaccination of personnel
Vaccines continue to provide high levels of protection against severe COVID‐19, hospitalisation and death. Vaccination of personnel may influence the risk of spread from personnel and visitors to mink and vice versa, however, with waning immunity against infection and the possible emergence of new immune‐escape variants, protection against infection and onward transmission could be limited in time and magnitude. Vaccination recommendations need to be in line with the national recommendations and discussed with occupational health and safety authorities. For details, please see Section 3.5.4.
3.7.1.2. General on‐farm biosecurity measures
Biosecurity measures aim at reducing the risk of introduction of pathogens into the farm and its further spread in and from the farm. The measures described in this section are focused on mechanical transmission, or transmission between farmed animals and other species, while transmission from infected persons to mink and vice versa is described in Section 3.7.1.1.
Application of biosecurity measures can be verified by official controls and record keeping, although the level of on‐farm biosecurity and the compliance may vary between farms and may change over time in the individual farm, according to what has been reported by MSs.
3.7.1.2.1. Restricted access for animals and visitors to farm, including tracing of visitors
To reduce the risk of introduction of pathogens, non‐essential visits of people to the farm should not be allowed. Farm personnel or visitors with symptoms compatible with SARS‐CoV‐2 or having tested positive should not be permitted to enter fur farm premises (see Section 3.7.1.1).
To allow tracing of introduction of SARS‐CoV‐2 from persons entering farms, records (electronic or physical) of all workers and visitors entering should be kept and be up to date. To reduce the risk of spread between farms, rotation of farm workers between farms should be limited and farm personnel should be discouraged to breed or keep other mink at home.
In closed farms, openings (doors, windows, holes, enclosures) should be fixed to prevent animals from escaping and entering the farm. Ventilation would represent an issue only for indoor farms, although mink farms are generally structures covered with a roof but open on the sides, so natural air circulation is sufficient. In all farms, but especially in open farms, efficient fencing can reduce the access of other animals to the farm area. Automatic closure of gates and doors can help keeping the openings closed.
- Advantages
-
○Basic biosecurity measures reduce the risk of introduction of pathogens in general. Each individual measure might not be highly effective by itself, while in combination, they can reduce the overall risk.
-
○Fences and locked doors can reduce the risk of uninvited persons to enter the farm and reduce the risk of pets and wildlife to enter the farm area.
-
○Reducing the numbers of events (e.g. visits by persons or animals) will reduce the overall risk of disease introduction.
-
○
- Challenges and drawbacks
-
○Availability of trained personnel might be limited in certain periods of the year with high workloads in mink farms.
-
○It might be difficult to control whether visitors have visited other farms previously.
-
○Fences and doors require proper facilities (e.g. fences, which, in some countries, such as Finland, are mandatory in raccoon dog farms as a rule for invasive alien species).
-
○Building and maintenance costs.
-
○Difficult to ensure daily compliance of closed doors/fences.
-
○Requires long‐term awareness from farm personnel.
-
○In case of restricted/delayed access of workers, this may delay operations at farm.
-
○
3.7.1.2.2. Changing work clothes for farm personnel
Changing clothes will reduce the risk of mechanical transmission of infectious pathogens from and to the farm. In a situation, where the worker or person entering the farm is infected, the effect of changing clothes will be limited. However, if for example the personnel entering a farm have had contact with an infected family or other persons, the risk of introduction can be reduced by changing of clothes, boots, washing hands, etc. In some farms, changing room or areas are available, where clean clothes and boots, a sink for hand wash, disinfectants, etc. are available.
- Advantages
-
○Basic biosecurity measures reduce the risk of introduction of pathogens in general. Each individual measure might not be highly effective by itself, while in combination, they can reduce the overall risk.
-
○Relatively limited costs, as some level of biosecurity is often already in place in farms.
-
○
- Challenges and drawbacks
-
○Supply costs (if not already implemented).
-
○Proper facilities are required (e.g. locker rooms, washing machine on farm).
-
○Building and maintenance costs (if not in place).
-
○Requires long‐term awareness from farm personnel.
-
○
3.7.1.2.3. Cleaning and disinfection equipment/vehicles
For other pathogens, transport vehicles are often considered a risk, and cleaning disinfection and tracing is used as general biosecurity measures. For SARS‐CoV‐2, the transmission of SARS‐CoV‐2 through transport vehicles has not been described. Furthermore, sharing equipment between farms might pose a risk of mechanical transmission of pathogens. Avoiding sharing equipment and vehicles, and proper cleaning and disinfection, if sharing is needed, can reduce the risk of potential mechanical transmission.
- Advantages
-
○Basic biosecurity measures reduce the risk of introduction of pathogens in general. Each individual measure might not be highly effective by itself, while in combination, they can reduce the overall risk.
-
○
- Challenges and drawbacks
-
○Requires proper facilities, and efficient disinfectants.
-
○Especially during winter months, temperatures can be a challenge in relation to cleaning, and disinfection of equipment and vehicles.
-
○Difficult to ensure daily and sufficient compliance.
-
○Requires long‐term awareness from farm personnel.
-
○
3.7.1.2.4. Rodent control
The use of rodent control is a basic biosecurity measure often used in farms to reduce the risk of spread of pathogens, although in the case of SARS‐CoV‐2, spread of the virus by rodents has not been reported.
- Advantages
-
○Basic biosecurity measures reduce the risk of introduction of pathogens in general. Each individual measure might not be highly effective by itself, while in combination, they can reduce the overall risk.
-
○Prevention of entry and presence of rodents can be based also on keeping the area where mink are as clean as possible, reducing feed rests and having a clear, open zone between the fence and the animal cages.
-
○
- Challenges and drawbacks
-
○Requires suitable equipment and rodenticides.
-
○Supply costs.
-
○Risk for animals and humans if misused, due to rodenticides toxicity.
-
○
3.7.1.3. Animal movement control, including pre‐movement testing and tracing
In known infected farms and surrounding areas, restriction of movements can prevent or reduce the risk of further virus spread and secondary outbreaks. By testing prior to movements, the risk of moving infected animals can be reduced. Furthermore, keeping records of movements can make easy the tracing after detections of infected farms.
- Advantages
-
○Potentially effective in preventing spread to other holdings.
-
○Ease of implantation, since not many movements are performed in farmed mink, and movements most often occur in restricted time periods.
-
○
- Challenges and drawbacks
-
○Only efficient if early detection occurs in infected farms.
-
○Negative impact on trade and potential financial losses can limit compliance.
-
○Negative impact on farm management and productivity (e.g. inability to replace breeding animals or transfer animals in order to decrease density following increased litter sizes).
-
○If testing is required prior to movements, additional costs are incurred.
-
○Records (electronic or physical) must be kept and be up to date, which might be a challenge for small farms.
-
○At certain times (pelting), movement restrictions may cause problems in the removal of skins or transfer of carcasses for pelting.
-
○
3.7.1.4. Awareness raising
This is to increase awareness about how to prevent and control SARS‐CoV‐2 infection, and it is considered a very important measure. Especially among farm personnel, it is important to remind about how SARS‐CoV‐2 in animals spreads and how to prevent animal and human infection and routinely remind them about biosafety and biosecurity measures against SARS‐CoV‐2 on the farm. Authorities and fur industry usually spread information and operating instructions through different channels to farm personnel. Workers from other countries should be provided information in their own language for regular work arrangements as well as during outbreak situations with specifically targeted information leaflets.
- Advantages
-
○Potentially enhances and improves application of biosecurity measures.
-
○Potentially increases reporting of abnormal morbidity and mortality (already foreseen by the legislation).
-
○
- Challenges and drawbacks
-
○May cause disquiet due to possible economic losses and public health concerns.
-
○Translation in different languages may be needed, especially in case of farm personnel from abroad.
-
○
3.7.1.5. Culling and disposal of animal in an infected farm
In the case of an infected farm, the veterinary authorities may order the immediate killing of all farmed animals under official surveillance with the aim of preventing the spread of the disease.
- Advantages
-
○It can reduce the virus load thus reducing the risk of spread, if the infection is detected early.
-
○If infection is detected early, culling all animals on the farm will prevent extensive virus circulation and consequently the risk of mutations that might pose a risk to public health.
-
○
- Challenges and drawbacks
-
○Only efficient, if early detection can be ensured.
-
○Extensive secondary spread from infected farms have only been reported from areas with very dense population, meaning that the probability is not always high.
-
○High costs, large need of resources, human resources and equipment (official vets, dedicated companies).
-
○Costs for infected farms (entire loss of breeding and production animals) as well as for the administrations for the compensation to farmers and for the destruction of skins and other possibly contaminated by products.
-
○Emotional impairment for farmers and other involved people.
-
○Public concerns about culling animals.
-
○Increased exposure to humans involved in culling activities, however, no or very limited number of infections have been observed in people related to culling activities likely due to the wearing of PPE.
-
○
3.7.1.6. Zoning around infected farms
Implementation of zones around infected farms is a general preventive measure used for outbreaks of other pathogens. In the zones, increased surveillance and movement restrictions are typically implemented. The effect of movement restrictions is described in Section 3.7.1.3, while the effect of surveillance is described in Section 3.6.
For SARS‐CoV‐2, distance to infected farms was described as a risk factor (EFSA, 2021). However, as introductions from infected persons is considered the highest risk of transmission, the effect of zoning on the risk of SARS‐CoV‐2 transmission is considered to be limited. While airborne transmission is one of several pathways that in general might increase the risk in zones around infected farms, which might be managed by zoning restrictions, SARS‐CoV‐2 has not been detected in air samples in distances > 3 m from infected mink. Moreover, zoning is a way to delimitate and differentiate geographical zones with a different health status. Here, since SARS‐CoV‐2 is present everywhere outside the outbreak, zoning would not be a meaningful measure.
3.7.1.7. Vaccination of animals
In the EU, vaccination of animals against SARS‐CoV‐2 was only applied in mink in Finland with a product developed by the Finnish Fur Breeders' Association (FIFUR), whose usage permit was granted by the Finnish Food Authority according to national procedures that foresee a provisional authorisation of a vaccine in the event of a serious animal disease epidemic. Approximately 95% of breeding females in all farms were vaccinated early in 2022 when only breeding animals were present at farm.
The vaccine product contains subunits of the SARS‐CoV‐2 bivalent RBD‐mFc fusion protein produced in mammalian cells as antigen and aluminium hydroxide as adjuvant.
Data provided by the producer showed that the vaccine was well tolerated in mink and it was able to induce a humoral immune response characterised, inter alia, by virus‐neutralising antibodies, tested 5 weeks after vaccination.
Regarding the protection induced by vaccination, the experimental studies performed by the producer suggested that the symptoms (sneezing, anorexia and diarrhoea) were less common and severe in vaccinated than in control animals. Data regarding other possible impact of the vaccination, such as the spread of the virus, the efficacy versus infection and the onset or duration of immunity were not complete to allow a reliable assessment. Overall, it is possible to conclude that the vaccine was able to provide a certain degree of protection against severe disease caused by Delta‐variant of SARS‐CoV‐2 in mink, but it did not prevent infection. Vaccination of mink is no longer in use in Finland, due to its limited effectiveness against Omicron variant, as reported by the producer.
Another vaccine product was submitted to the European Medicines Agency (EMA) for a centralised procedure of authorisation, but they have been withdrawn during the process.41
3.7.2. Options for public health response
A close collaboration and communication between animal and public health sectors as well as occupational safety and health authorities in a One Health approach is crucial to identify transmission events at the human–animal interface and prevent further spread.
Testing approaches together with genetic monitoring of virus samples from people exposed to animals with confirmed SARS‐Cov‐2 infection as well as from animal sources are key to understand factors related to transmission as well as evolutionary host‐driven mechanisms. It is important to collect representative samples for further characterisation of viruses from the general population as well as from animals infected with SARS‐CoV‐2.
PPE is a crucial measure for workers to prevent exposure to SARS‐CoV‐2‐infected animals or contaminated workplaces. Information about outbreaks should be immediately shared with public and occupational safety and health authorities to enable appropriate follow‐up of exposed people and the implementation of control measures.
4. Conclusions and recommendations
Based on the aspects to be assessed as indicated in the ToRs (susceptibility, risk for animal and public health, monitoring approach and preventive and control measures), the current assessment is structured according to different animal categories: farmed animals (mink), companion animals, wildlife and animals kept in zoos, and the conclusions are presented accordingly.
General conclusions about animal species of concern in the epidemiology of SARS‐CoV2
This scientific opinion classified animal species of potential epidemiological concern to be those that shed infectious virus and are able to transmit SARS‐CoV‐2 to other animals or humans. Such species of epidemiological concern assessed here are American mink (Neogale vison), raccoon dog (Nyctereutes procyonoides), cat (Felis catus), Syrian hamster (Mesocricetus auratus), ferret (Mustela furo), house mouse (Mus musculus, for some virus variants only), Egyptian fruit bat (Rousettus aegyptiacus), deer mouse species (Peromyscus spp., not present in Europe) and white‐tailed deer (Odocoileus virginianus).
Of note, there is uncertainty on the list provided in the previous bullet: most experimental infections to determine species susceptibility were performed with ancestral (pre‐Alpha clades) virus isolates. Furthermore, as variants continue to arise, and new host species are being detected over time, species susceptibility and virus transmission capacity may change, with the continuous potential emergence of new host species.
Farmed animals (mink)
Susceptibility, epidemiological situation
American mink is a highly susceptible species, and outbreaks have occurred in MSs from April 2020 until the end of the reporting period considered in this document, although with a decreasing number of outbreaks in 2022 compared to 2021. Species‐specific viral evolution of SARS‐CoV‐2 has been observed in this species.
Among animals in the EU, mink farmed for fur production have the highest likelihood to become infected and transmit SARS‐CoV‐2 within animal populations and to in‐contact humans e.g. farm personnel, and subsequently to the general population. This can be explained by the inherent susceptibility to SARS‐CoV‐2 infection of the species, in combination with the characteristics of the systems in which farmed mink are kept, with large numbers of animals in a limited area.
During the still ongoing SARS‐CoV‐2 pandemic, a vast majority of the outbreaks of SARS‐CoV‐2 reported globally in animals have occurred in mink for fur production. In the reporting period (February 2021–November 2022), 50 outbreaks of SARS‐CoV‐2 were reported, of those 44 were reported in 2021 in 7 MSs, while only 6 were reported in 2022 in 2 MSs, thus representing a decreasing trend.
Most sequenced mink isolates grouped into distinct major mink‐specific clusters, were geographically clustered and showed high intra‐cluster variability, indicating mink‐to‐mink transmission, high rates of virus evolution within the mink population and emergence of mink‐specific variants with a potential to spill back into the human population.
Risk for animal and public health
SARS‐CoV‐2 can spread from humans to mink and from mink to humans during exposure.
The probability of introduction of SARS‐CoV‐2 from humans to mink farms is associated with the SARS‐CoV‐2 level of circulation in the surrounding general human population. This probability can be reduced through continuous and proper implementation of biosecurity measures in mink farms including the use of non‐pharmaceutical interventions (NPI) for all humans accessing mink farms (Section 3.5.3).
Once introduced into a mink farm, SARS‐CoV‐2 spreads efficiently within the farm from animal to animal, resulting in extensive virus circulation and risk of spill‐over to humans in contact with the mink, as well as to other susceptible animals with access to mink and their local environment.
The extensive circulation of SARS‐CoV‐2 in an infected mink farm drives virus adaptation, resulting in the potential generation of mink‐adapted virus variants.
- In general and for all animal species, the public health impact of the possible spill‐over of SARS‐CoV‐2 from mink farms to humans depends on several factors: the respective virus variant, effectiveness of the vaccine for this variant in vaccinated people including the time period after the vaccination, previous exposure to other SARS‐CoV‐2 variants and health status of the individual person:
-
○The risk (determined by probability of infection and impact of the disease), for an occupationally exposed person to a SARS‐CoV‐2‐infected mink is assessed as low to moderate. This assessment is subjected to uncertainty related to the impact of variants potentially emerging in mink.
-
○The risk based on the probability to get infected and develop severe disease for a person without or with limited exposure to farmed mink is estimated to be none to very low.
-
○The risk of the spread of a SARS‐CoV‐2 variant with mink‐specific mutations, or re‐introduction of an older variant virus that circulated in mink, into the general human population causing severe COVID‐19 is estimated to be very low‐to‐low.
-
○
Monitoring approach at farm level
- In the current epidemiological situation in the EU, where a substantial decrease of outbreaks in mink farms has been reported in 2022 compared to 2020 and 2021, and where the majority of the human population has acquired some level of immunity to SARS‐CoV‐2, the risk for the general population represented by infected mink is considered very low‐to‐low; therefore:
-
○The primary purposes of monitoring of mink farms are to confirm outbreaks based on suspicion and monitor virus evolution (sequencing isolates).
-
○The confirmation of SARS‐CoV‐2 infection in suspected animals remains a relevant objective, in order to allow the farmers to apply preventive measures in terms of reducing the risk of secondary outbreaks.
-
○A monitoring approach based on testing of dead animals or with clinical signs suggesting possible SARS‐CoV‐2 infection is considered appropriate, with sampling triggered by increased mortality or morbidity in mink, or farm personnel testing positive.
-
○
The genomic surveillance of viruses circulating in mink and in general in all animal species is considered relevant to monitor the circulating genetic variants of the virus and comparing the genetic type from mink to currently circulating variants in humans. Virus isolates from positive samples representing at least every epidemiological unit should be subjected to genomic characterisation and genome sequences shared with the scientific community.
As an additional monitoring strategy, sampling at pelting and use of serological tests can help in assessing the number of mink farms that have been infected to monitor possible changes in prevalence.
Decreasing testing for SARS‐CoV‐2 in the human population and testing with sequencing of specimens from severely ill COVID‐19 patients in hospital may delay the early detection of animal‐associated SARS‐CoV‐2 variant viruses. It is therefore important to expand and increase the national sentinel surveillance for SARS‐CoV‐2 in primary (as well as secondary) health care units to achieve higher representativeness across the population and higher number of available specimens for genomic analysis. This will also enable the detection of new virus variants in the population.
Options for preventive and control measures
- Humans are considered the most important source of introduction of SARS‐CoV‐2 into a farm.
-
○Systematic frequent (e.g. at least weekly) testing for SARS‐CoV‐2 infection using rapid antigen test and/or PCR of personnel and visitors is a prerequisite for the early detection of infection in humans that may enter the farm and come into contact with mink and is therefore a key measure to prevent introduction of SARS‐CoV‐2 into the farm.
-
○Similarly, the limitation or ban of non‐essential visits of humans to farms will reduce the frequency of contacts and thereby the risk of transmission from visitors.
-
○
The risk of transmission from persons (e.g. workers, visitors) to farmed mink can be reduced by staying at home when feeling sick or testing positive for SARS‐CoV‐2.
Measures applied to reduce the risk of transmission between mink and humans will only have effect if used consistently. The level of on‐farm biosecurity and the compliance can be verified by official controls and record keeping, although may vary between farms and may change over time in an individual farm.
Wearing of personal protective equipment for persons in contact with mink is a useful measure to reduce the probability of introduction and transmission of SARS‐CoV‐2 virus into farms from people. These measures also may reduce the risk for people of being infected by mink.
Biosecurity measures applied at farm (cleaning, disinfection, pest control and restricted access to other animals than mink that may be present at farm) aim at reducing the risk of virus introduction into the farm, and its further spread in and from the farm (e.g. transmission between farmed animals and other susceptible or possibly susceptible species such as cats, dogs, bats, etc.).
The risk of further virus spread and secondary outbreaks to other farms can be reduced by restriction of mink movement and/or by testing for SARS CoV‐2 prior to movement, especially in farms located in areas with known infected farms.
Current vaccines against SARS‐CoV‐2 do not fully prevent virus transmission to and from vaccinated humans as well as between humans and mink. Vaccines are protective against severe disease, hospitalisation and death. Experimental vaccines for mink conferred a certain degree of protection against severe disease caused by Delta‐variant of SARS‐CoV‐2 but they did not prevent infection, and they not effective against Omicron variants.
Recommendation: Equally important in relation to farm personnel is the provision of information material and training to all farmworkers, including guest workers, in mink farms about biosafety and biosecurity measures against SARS‐CoV‐2 on the farm in their own language, as well as providing them access to health care.
Companion animals
Susceptibility, epidemiological situation
Among companion animal species, cats, ferrets and hamsters are the species most at risk of infection. In these species, serious illness from SARS‐CoV‐2 has sporadically been observed.
In experimental infections, hamsters, ferrets and cats can display clinical signs. Usually, they recover spontaneously. No clinical signs have been reported for dogs, rabbits, rats and mice.
Transmission of SARS‐CoV‐2 from a donor animal to a recipient of the same species by direct contact has been demonstrated for cats, ferrets, hamsters and mice, while no evidence exists for virus transmission between dogs.
Under field conditions, cats and hamsters have been associated with mild to moderate respiratory, gastrointestinal or systemic signs of disease and they can shed virus.
Deposited sequences of SARS‐CoV‐2 obtained from infected companion animals are spread all over the SARS‐CoV‐2 clades and have limited tendency of clustering together, indicating sporadic transmission from humans, with little or no animal‐to‐animal transmission among companion animals.
Viral sequence analysis indicates low frequency of species‐adapted mutations.
Risk for animal and public health and related control measures
SARS‐CoV‐2 infection of companion animals is most likely originating from an infected human.
The risk for cats and hamsters of spreading SARS‐CoV‐2 back to humans (probability of infection and severity of disease of a pet owner as well as in the general population) is assessed as very low.
Humans with high contact rates to companion animals from different households (e.g. veterinarians) have a higher risk (very low to low) to get infected from a companion animal.
The probability of companion animals to have an impact on the virus circulation in the general population is none to very low.
Monitoring approach
-
–
For companion animals, there is no need for specific monitoring programmes, taking into consideration that sporadic cases of transmission to humans may occur, but generally limited to owners, zoo workers or veterinarians in contact with these animals.
-
○
In some companion animal species, in case of clinical signs compatible with SARS‐CoV‐2 disease, animal testing may be important for possible quarantine measures or application of proper therapies.
-
○
In addition, apart from research objectives, testing of individuals in stray communities (especially cats) could be justified, in case of suspected SARS‐CoV‐2 clinical cases or abnormal mortality rates in these communities.
-
○
Wildlife
Susceptibility, epidemiological situation
The number of wildlife species that have been reported naturally SARS‐CoV‐2 infected grows steadily, also due to the active research in this field, including several wild carnivores and the white‐tailed deer in North America.
The epidemiological role of susceptible wildlife in the EU context for the maintenance of virus circulation, and as a possible public health risk, depends on their abundance and the level of exposure to human population and to other wild or domesticated animals.
In the EU, no cases of infected wildlife (with viral isolation or RNA detection) have been reported so far.
So far, only North American white‐tailed deer, both free living or captive in game reserves, have been demonstrated to maintain and possibly spill back the infection to humans.
Recommendation: Further epidemiological research should be promoted in a broad range of wildlife species, including feral species, and geographical regions.
Risk for animal and public health
Differently from the situation in North America, very low numbers of white‐tailed deer are present in the EU (less than 1% of the total EU deer population) only in two countries. It is unknown whether these animals may be able to support the persistence of SARS‐CoV‐2 infection in the European context. Population density, aggregation and mating season can facilitate this possible occurrence.
- In the EU, the risk of transmission of SARS CoV‐2 infection from humans to white‐tailed deer and backward causing severe disease is considered very low. Risk factors are events that increase exposure of white‐tailed deer to humans, such as hunting. In general, any activities that bring animals close to humans or even proximity of animals to urban settings and likelihood of exposure to contaminated wastewater or garbage.
-
○Recommendation: Action should be taken to avoid overabundance or aggregation of white‐tailed deer, and in general all game species (such as by avoiding feeding sites, monitoring group size) to control this transmission pathway.
-
○Recommendations: Further research studies might be relevant to monitor the possible role of white‐tailed deer and other deer or other wildlife populations in Europe for the presence and eventual persistence of SARS‐CoV‐2 infection.
-
○Recommendation: Good hunting practices (avoiding feeding or baiting) are advised. Humans dealing with wildlife should follow biosecurity measures in general minimising direct contact with wild animals, especially sick and dead animals. Furthermore, safe disposal of garbage and waste from human communities in both urban and rural settings is advised to reduce the risks of SARS‐CoV‐2 spill‐over to wildlife.
-
○
Regarding wild carnivores, due to their elusive and solitary behaviour, to their low density and to the low numbers hunted, there is a very low probability of maintaining the infection or representing a risk for other animal species or for public health also due to limited human exposure, even for occupationally exposed people (rangers, hunters, researchers, etc.).
The probability of transmission of viruses from bats to humans or the emergence of SARS‐CoV‐2‐related or new coronaviruses has been assessed as none to very low for the next 12 months, since transmission of SARS‐CoV‐2 or other coronaviruses from bats to humans and backwards has not been observed in Europe and on the limited human population having direct contact with these animals. However, since bats are natural hosts of many coronaviruses, the monitoring of these species is still important.
Monitoring approach
-
–
Based on the current knowledge, the main wild animal species that may be considered as possible targets for SARS‐CoV‐2 monitoring are white‐tailed deer, wild carnivores (e.g. wild mustelids, felids and canids), bats and rodents such as wild synanthropic mice and rats (those living in or close to human settlements).
-
○
No specific regulated monitoring activities would be needed for wildlife in the EU, apart from monitoring based on suspicion, i.e. testing of animals showing clinical signs suggesting possible SARS‐CoV‐2 infection or dead‐found animals. Positive samples should be subjected to genomic analysis to monitor virus evolution and genome sequences shared with the scientific community.
-
○
-
–
Insectivorous bats, especially those belonging to the genus Rhinolophus, are hosts for a range of betacoronaviruses and thus also of potential relevance for SARS‐CoV‐2 monitoring. Recommendations: Research studies should investigate the possible role of bats in the European context, especially those belonging to the family Rhinolophidae.
Zoo animals
Susceptibility and epidemiological situation
There are reports of both experimental and natural infection of animal species kept in zoos with SARS CoV‐2, mainly felids and great apes.
Risk for animal and public health and related control measures
Zoo animals, such as felids and great apes, can acquire the infection mainly from infected zoo workers in contact with them, still at very low risk. There is no report of spillback transmission from animals to humans in zoos.
Regular testing of workers, self‐isolation when positive, use of PPE and good hygiene practice (e.g. avoiding close contact, disinfection of tools), as well as good ventilation in closed enclosures can significantly reduce the risk of transmission from humans to animals.
Transmission between susceptible animals in the same enclosure could occur once an animal is infected, although transmission between animals kept in zoos is difficult to be proven, because if they belong to the same outbreak they are usually exposed to the same infectious source (e.g. positive caretaker). The probability of transmission to animals in other enclosures is considered very low.
Testing and isolation of animals with clinical signs, even by non‐invasive monitoring by air sampling or rope‐based oral fluid sampling, or testing in the frame of other veterinary checks, as well as reducing animal density and avoiding sharing same air in ventilation systems, might reduce the risk of further transmission.
- Animals kept in zoos overall do not represent a major public health risk in relation to SARS‐CoV‐2.
-
○The risk for occupationally or activity‐related exposed people or groups such as zoo workers, rangers or forest workers to be in contact with infected animals or their droppings and get infected and develop severe is estimated to be very low.
-
○The risk (probability to be infected and develop severe disease) for an individual person to get infected from an animal in a zoo as well as the probability to have an emerging virus from zoo or wild animals circulating in the general population is considered to be none to very low.
-
○
Monitoring approach
The same conclusions as those indicated for companion animals are valid for zoo animals.
Abbreviations
- COVID‐19
Coronavirus disease 2019
- ECDC
European Centre for Disease Prevention and Control
- ELISA
Enzyme‐linked immunosorbent assay
- EMA
European Medicines Agency
- EWS
Early Warning System
- FP
Fusion peptide
- MS
Member State
- PPE
Personal protective equipment
- RADT
rapid antigen detection tests
- RT‐PCR
Reverse transcription polymerase chain reaction
- RBD
Receptor‐binding domain
- SARS‐CoV‐2
Severe acute respiratory syndrome coronavirus 2
- ToR
Term of Reference
- VNT
Virus neutralisation test
- WHO
World Health Organization
Annexes
A.1 Protocol for the assessment of the SARS‐CoV‐2 in animals: susceptibility of animal species, monitoring, prevention and control
A.1.1 Background and ToRs as provided by the requestor
See Section 1.1.
A.1.2 Problem formulation
Here, a summary of the initial considerations are described. With this mandate, the European Commission requests EFSA's support in assessing the adequacy of the current monitoring system for SARS‐CoV‐2 in mustelids and raccoon dogs in the EU, which should be reviewed in light of the evolvement of the epidemiological situation and along with the new scientific knowledge on the spread of SARS‐CoV‐2 in both humans and animals. For the assessment of the following problems are formulated:
- susceptibility of animals
- susceptibility in natural and laboratory conditions, species where SARS‐CoV‐2 or its RNA or antibodies to SARS‐CoV‐2 been detected in experimental studies or in the field, and their geographical distribution;
- dynamic of infection and pathogenesis;
- role in inter and intra‐species transmission;
- genetic analysis of the virus variants in each species where it is isolated;
- The group animal species to be considered.
- Risk for animals posed by SARS‐CoV‐2 infection in animals species of concern
- Once susceptible species or group of species susceptible to SARS ‐CoV‐2 are identified, the possible exposure and transmission pathways and related risk between these species and other species are assessed.
- Risk for humans posed by SARS‐CoV‐2 infection in animal species of concern
- Which species identified in ToR 1 to whom humans may be exposed and the following:
- Exposure assessment: social groups exposed to which relevant animal species;
- Genetic variants and assessment of the possibility that virus can persist in animal population with the emergence of new variants potentially able to escape vaccine efficacy;
- Circulation, severity;
- Diagnostics, immunity, vaccination, treatment;
- Options for public health response for each social groups.
-
Revision of the monitoring system
The main problem is to assess which monitoring approach in mink/raccoon dogs should be applied and whether it should be extended to other species. The monitoring scenarios is depicted by animal category and the related monitoring objective.
-
Possible options for disease prevention and control measures
Identification and assessment of effectiveness of main preventive and control options in place in EU, highlighting strengths and drawbacks.
A.1.3 Clarifications of the scope of the request: framework, population and geographical area of concern, definitions
Scope: to assess health risk in animals and humans and to provide recommendations for revision of the current monitoring system for SARS‐CoV‐2 in mustelids and raccoon dogs in the EU.
Geographical area: EU for the monitoring approach; worldwide for screening the literature.
Population: mustelids and raccoon dogs as population target for the current monitoring system and assessment of any other species considered susceptible to SARS‐CoV‐2 that may represent a risk for animals and humans and should be considered in the monitoring plans.
A.1.4 Translation of ToRs into assessment questions and subquestions
ToR 1 ‘Reviewing updated relevant scientific literature available globally related to SARS‐CoV‐2 infection in animals species of concern in the epidemiology of SARS‐CoV2’ has been translated in the following assessment questions:
Which mammal species have been reported test positive to SARS‐CoV‐2 in the field (either at RNA, isolated virus, antibodies), and their geographical distribution?
Which wild and domestic mammal species are susceptible to SARS‐CoV‐2 in laboratory conditions?
Dynamic of infection and pathogenesis:
What are confirmed infection routes for these species?
What is the length of the incubation period in these species?
What are the clinical signs, if any, their severity, duration, etc.?
Which species are able to shed the virus (even in the absence of clinical signs)?
Do these species develop protective immunity?
Which of these species can further transmit the disease to same/other species?
What are the genetic virus variants in each species where it is isolated?
In which animals vaccines against SARS‐CoV‐2 have been tested, and what are the results in terms of immunogenicity, protection to challenge or safety?
Which diagnostic tests are used in animals to detect SARS CoV‐2?
Type of test
Test performance (Se, Sp)
Sample matrix used
ToR2 ‘Assess the current epidemiological situation in the EU and elsewhere as regards the risk for human and animal health posed by SARS‐CoV‐2 infection in animals species of concern with a view to review the design of the existing monitoring performed by the Member States for minks, other animals of the family Mustelidae and raccoon dogs’ is addressed the first part as descriptive epidemiology of SARS‐CoV‐2 outbreaks in animals in EU and worldwide, while the second part is translated into the question what is the possible exposure and transmission pathways and related risk between the species assessed as susceptible and other species, mainly due to their capacity of being infected and further transmit the virus?
ToR 3 about recommending options for reviewing the monitoring strategies in different epidemiological scenarios, first the scenarios are defined by the question:
Which monitoring objectives should be set for each susceptible animal category?
For each defined scenario, the following subquestions should be answered:
-
○
What type of surveillance would be needed (active/passive)?
-
○
What type of Diagnostic test?
-
○
What type of Sample matrix?
-
○
What type of Target population?
-
○
What is the sampling frequency?
-
○
What is the duration of active monitoring plan?
-
○
where applicable, what would be the design prevalence and sample size?
The ToR 4 about disease prevention and control measures can be translated into the following questions:
What are the preventive measures for SARS‐CoV‐2 in animals, and their pros and cons?
What are the control measures for SARS‐CoV‐2 in animals, and their pros and cons?
A.1.5 Assessing and synthesising evidence (including uncertainty analysis)
Details of the methodology used for the analysis of the data retrieved by the literature review as well as data from other sources will be provided in the methodology section of the opinion.
All sources of uncertainty identified during the assessment will be recorded, and their impact on the scientific assessment will be assessed collectively.
A.2 Animal species considered susceptible to SARS‐CoV‐2 based on detection of viral RNA in ante‐mortem or post‐mortem samples or based on seroconversion
Table A.1.
Animal species considered susceptible to SARS‐CoV‐2 based on detection of viral RNA in ante‐mortem or post‐mortem samples, or based on seroconversion (please note that some references contain multiple studies)
| Animal species | Specification animal species and age | Route of inoculation [IN = intranasal; IT = intratracheal; PO = per oral; IV = intravenous] | SARS‐CoV‐2 isolate | Clinical signs | Detection of viral RNA in ante‐mortem samples of airways (at least 2 consecutive days) | Detection of viral RNA or antigen in any post mortem samples | Isolation of infectious virus | Seroconversion (ELISA or Neutralisation assay) | Direct transmission | References |
|---|---|---|---|---|---|---|---|---|---|---|
|
African Green Monkey Chlorocebus aethiops |
Adult | IN and IT | SARS‐CoV‐2/INMI1‐Isolate/2020/Italy | Fever and reduced appetite | Yes | Yes | Yes | Yes | Not investigated | Woolsey et al. (2021) |
| 16 years | Aerosol | 2019‐nCoV/USA‐WA 1/2020 | Hypothermia and respiratory distress | Not investigated | Not investigated | Blair et al. (2020) | ||||
| IN, PO, IT and conjunctival | ||||||||||
|
Bank vole Myodes glareolus |
7–9 weeks | IN | 2019_nCoV Muc‐IMB‐1 | No | Yes | Yes | Yes | Yes | No evidence | Ulrich et al. (2021) |
|
Bushy‐tailed woodrat Neotoma cinerea |
Not reported | IN | WA1/2020WY96 | No | Yes | Yes | Yes | Yes | Not investigated | Bosco‐Lauth et al. (2021) |
| Campbell's dwarf hamster Phodopus campbelli | 5–7 weeks | IN | BetaCoV/Germany/BavPat1/2020 | No | Yes | Yes | Yes | Not investigated | Not investigated | Trimpert et al. (2020) |
|
Cat Felis catus |
15–18 weeks | IN, IT, PO and ocular | UT‐NCGM02/Human/2020/Tokyo | No | Yes | Yes |
Not investigated |
Yes |
Yes | Halfmann et al. (2020) |
| 5–7 months | IN and PO | USA‐WA1/2020 | Gaudreault et al. (2021) | |||||||
| 6–9 months | IN | SARS‐CoV‐2/Ctan/human/2020/Wuhan | Yes | Shi et al. (2020) | ||||||
| 4.5–5 months | IN and PO | SARS‐CoV‐2 USA‐WA1/2020 | Mild fever | Not investigated | Gaudreault et al. (2020) | |||||
| 5–8 years | IN | WA1/2020WY96 | No | Yes | Bosco‐Lauth et al. (2020) | |||||
| 19 weeks | IN, IT, PO and ocular | UT‐NCGM02/Human/2020/Tokyo | Not investigated | Chiba et al. (2021) | ||||||
| 8 months – 1.5 years | IN | SARS‐CoV‐2/WH‐09/human/2020/CHN/ MT093631.2 | Arching of back, diarrhoea | Not investigated | Yes | Bao et al. (2021) | ||||
|
Cattle Bos taurus |
6 weeks | IT | TGR1/NY/20 | Fever | Yes | Yes | Not investigated | Yes | Not investigated | Falkenberg et al. (2021) |
| IV | No | |||||||||
| 4–6 months | IN | 2019 nCoV Muc‐IBM‐1 | No | Not investigated | No evidence | Ulrich et al. (2020) | ||||
|
Chinese hamster Cricetulus griseus |
5–7 weeks | IN | BetaCoV/Germany/BavPat1/2020 | Weight loss | Yes | Yes | Yes | Not investigated | Not investigated | Bertzbach et al. (2020) |
|
Chinese tree shrew Tupaia belangeri chinensis |
1 year and 5–6 years | IN, PO and ocular | SARS‐CoV‐2 strain 107 | No | No | Yes | Not investigated | Not investigated | Not investigated | Xu et al. (2020) |
| 6–12 months and 2–4 years and 5–7 years | IN | Not reported | Fever | Yes | Zhao et al. (2020) | |||||
|
Cynomolgus macaque Macaca fascicularis |
4–20 years | IT and IN | BetaCoV/Munich/BavPat1/2020 | Nasal discharge | Yes | Yes | Yes | Yes | Not investigated | Rockx et al. (2020) |
|
Deer mouse Peromyscus maniculatus nebrascensis |
6 months | IN | 2019‐nCoV/USA‐WA1 | No | Yes | Yes | Yes | Yes | Yes | Fagre et al. (2021) |
| 8–32 weeks | GISAID # ID_EPI_ISL_425177 | Griffin et al. (2021) | ||||||||
| Djungarian hamster Phodopus sungorus | 5–7 weeks | IN | BetaCoV/Germany/BavPat1/2020 | Weight loss | Yes | Yes | Yes | Not investigated | Not investigated | Trimpert et al. (2020) |
|
Dog Canis lupus familiaris |
3 months | IN | SARS‐CoV‐2/Ctan/human/2020/Wuhan | No | No | No | No | Yes | No evidence | Shi et al. (2020) |
| 5–6 years | WA1/2020WY96 | Not investigated | Bosco‐Lauth et al. (2020) | |||||||
|
Egyptian fruit bat Rousettus aegyptiacus |
1–5 years old | IN | 2019‐nCoV Muc‐IMB‐1 | No | Yes | Yes | Yes | Yes | Yes | Schlottau et al. (2020) |
|
Ferret Mustela furo |
6–9 months | IN | 2019‐nCoV Muc‐IMB‐1 | No |
Yes |
Yes |
Yes | Yes | Yes | Schlottau et al. (2020) |
| 3–4 months | SARS‐CoV‐2/F13/environment/2020/Wuhan | Anorexia |
Not investigated |
Shi et al. (2020) | ||||||
| 8 mtonhs | IT | BavPat1/2020 |
No |
Not investigated |
Not investigated | Ciurkiewicz et al. (2022) | ||||
| 4 months | IN | hCoV‐19/Australia/VIC01/2020 | No | Marsh et al. (2021) | ||||||
| 6 months | BetaCoV/Munich/BavPat1/2020 | Yes | Yes | Kutter et al. (2021) | ||||||
| 7 months | Victoria/01/2020 | Reduced activity, sneezing | Not investigated | Ryan et al. (2021) | ||||||
| 7 months | BetaCoV/Munich/BavPat1/2020 | No | Not investigated | Yes | Yes | Yes | Richard et al. (2020) | |||
| 12–20 months | MNC‐nCoV02 | Fever | Yes | Kim et al. (2020) | ||||||
|
Mink Neogale vison |
– | IN | SARS‐CoV‐2 HRB25 | No signs | Yes | Yes | Yes | Yes | Not investigated | Shuai et al. (2021) |
| Not reported | hCoV‐19/Finland/THL‐202126660/2021 | Anorexia, diarrhoea, lethargy, respiratory signs | Not investigated | Yes | Virtanen et al. (2022) | |||||
|
Mouse Mus musculus |
Strain C54BL/6: 8 weeks |
IN | 19 nCoV‐CDC‐Tan‐GDPCC | No | Not investigated | Yes | Not investigated | Yes | Yes | Pan et al. (2021) |
|
Strain BALB/c: 8 weeks |
Not investigated | Not investigated | ||||||||
|
Strain C54BL/6: 8 weeks |
France/GES‐1973/2020 | No | Montagutelli et al. (2021) | |||||||
| France/IDF‐IPP11324/2020 |
Yes |
|||||||||
| France/IDF‐IPP05174/2021 | ||||||||||
| France/IDF‐IPP00078/2021 | ||||||||||
| FrenchGuiana/IPP03772/2021 | ||||||||||
| France/IDF‐IPP02260/2021 | ||||||||||
| France/HDF‐IPP11602/2021 |
No |
|||||||||
|
Strain BALB/c: 8 weeks |
France/GES‐1973/2020 | |||||||||
| France/IDF‐IPP11324/2020 |
Yes |
|||||||||
| France/IDF‐IPP05174/2021 | ||||||||||
| France/IDF‐IPP00078/2021 | Yes | Yes | ||||||||
| FrenchGuiana/IPP03772/2021 | Not investigated | Not investigated | ||||||||
| France/IDF‐IPP02260/2021 | ||||||||||
| France/HDF‐IPP11602/2021 | No | |||||||||
|
Pig Sus scrofa domesticus |
6 weeks | IN | SARS‐CoV‐2/Ctan/human/2020/Wuhan | No | No | Not investigated | Not investigated | No | No evidence | Shi et al. (2020) |
| 3 months | IT and aerosol | SARS‐CoV‐2/human/NL/Lelystad/2020 | Yes | Yes | Yes | Not investigated | Sikkema et al. (2022) | |||
| 3 weeks | IV | TGR/NY/20 | Fever | No | No evidence | Buckley et al. (2021) | ||||
| IT | Yes | |||||||||
| IN | No | |||||||||
| 8 weeks | IN and PO | hCoV‐19/Canada/ON‐VIDO‐01/2020 | Ocular discharge | Pickering et al. (2021) | ||||||
| 9 weeks | IN | 2019‐nCoV Muc‐IMB‐1 | No | No | Not investigated | No | Schlottau et al. (2020) | |||
| 5 weeks | PO, IN and IT | USA‐WA1/2020 | No | Meekins et al. (2020) | ||||||
|
Rabbit Oryctolagus cuniculus |
3 months | IN | BetaCoV/Munich/BavPatl/2020 | No | Yes | Yes | No (on DPI 1 only) | Yes | Not investigated | Mykytyn et al. (2021) |
|
Raccoon Procyon lotor |
10 weeks | IN | USA‐WA1/2020 | No | No | No | No | Yes | No evidence | Francisco et al. (2022) |
| Not stated | WA1/2020WY96 | Not investigated | Bosco‐Lauth et al. (2021) | |||||||
|
Raccoon dog Nyctereutes procyonoides |
Adult | IN | 2019_nCoV Muc‐IMB‐1 | No | Yes | Yes | Yes | Yes | Yes | Freuling et al. (2020) |
|
Red fox Vulpes vulpes |
3–5 months | IN | WA1/2020WY96 | Lethargy | Yes | Yes | Yes | Yes | Not investigated | Porter et al. (2022) |
|
Rhesus macaque Macaca mulatta |
7–12 years | IN and IT | Not reported | No | Yes | Yes | Yes | No | Not investigated | Yadav et al. (2021) |
| 4–6 years | IN, IT, PO and ocular | nCoV‐WA1‐2020 | Fever, respiratory distress, weight loss, hunched posture, nasal discharge | Yes | Munster et al. (2020) | |||||
| 6–11 years | IT | SARS‐CoV‐2 IVCAS 6.7512 | Reduced appetite, weight loss | Not investigated | Shan et al. (2020) | |||||
| 13–15 years | Aerosol | 2019‐nCoV/USA‐WA 1/2020 | No | Not investigated | Blair et al. (2020) | |||||
| PO, IN, IT and conjunctival | ||||||||||
|
Roborovski dwarf hamster Phodopus roborovskii |
5–7 weeks | IN | BetaCoV/Germany/BavPat1/2020 | Humane euthanasia 3–5 DPI due to severe clinical symptoms (weight loss, respiratory distress, hypothermia, ruffled fur) | Yes | Yes | Yes | Not investigated | Not investigated | Trimpert et al. (2020) |
|
Skunk Mephitis mephitis |
Not reported | IN | WA1/2020WY96 | No | Yes | Yes | Yes | Yes | Not investigated | Bosco‐Lauth et al. (2021) |
| 10 weeks | USA‐WA1/2020 | No evidence | Francisco et al. (2022) | |||||||
|
Syrian hamster Mesocricetus auratus |
4–5 weeks | IN | BetaCoV/Hong Kong/VM20001061/2020 | Weight loss | Yes | Yes | Yes | Yes | Yes | Sia et al. (2020) |
| 1 months and 7–8 months | IN and ocular | SARS‐CoV‐2/UT‐NCGM02/Human/2020/Tokyo | Not investigated | Not investigated | Imai et al. (2020) | |||||
| 6–10 weeks | IN | SARS‐CoV‐2 isolated from human patient | Not investigated | Not investigated | Yes | Chan et al. (2020) | ||||
|
White‐tailed deer Odocoileus virginianus |
6 weeks | IN | TGR/NY/2 | Fever | Yes | Yes | Yes | Yes | Yes | Palmer et al. (2021) |
| 2 years | IN and PO | SARS‐CoV‐2/human/USA/WA1/2020 lineage A and SARS‐CoV‐2/human/USA/CA_CDC_5574/2020 lineage B.1.1.7 | Ocular discharge | Cool et al. (2022) |
A.3 Proportion of animals positive (as median and inter‐quartile range (IQR)) to SARS CoV‐2 infection in the field
Table A.2 summarises all mammal species in which SARS‐CoV‐2 positivity either at virus detection, PCR or serologically was reported in peer‐reviewed literature (based on the SLR), or reported to the World Animal Health Organisation (OIE), by country. OIE reports were queried in the WAHIS system on 8 April 2022.
Based on passive (following evidence of clinical signs or epidemiological links to human or animal cases of SARS CoV‐2 infections, suspicion‐based) or active (e.g. random sampling) monitoring, in Table 17, the results obtained from SLR about field infection of different species to SARS CoV‐2 are shown, as proportion of positive animals in each epidemiological unit, tested by PCR/virus isolation or serological test.
Suggested citation: EFSA AHAW Panel (EFSA Panel on Animal Health and Animal Welfare) , Nielsen SS, Alvarez J, Bicout DJ, Calistri P, Canali E, Drewe JA, Garin‐Bastuji B, Gonzales Rojas JL, Gortázar C, Herskin M, Michel M, Miranda Chueca MA, Padalino B, Pasquali P, Roberts HC, Spoolder H, Velarde A, Viltrop A, Winckler C, Adlhoch C, Aznar I, Baldinelli F, Boklund A, Broglia A, Gerhards N, Mur L, Nannapaneni P and Ståhl K, 2023. SARS‐CoV‐2 in animals: susceptibility of animal species, risk for animal and public health, monitoring, prevention and control. EFSA Journal 2023;21(2):7822, 108 pp. 10.2903/j.efsa.2023.7822
Requestor: European Commission
Question number: EFSA‐Q‐2022‐00139
Panel members: Saxmose Nielsen, Julio Alvarez, Dominique Joseph Bicout, Paolo Calistri, Elisabetta Canali, Julian Ashley Drewe, Bruno Garin‐Bastuji, José Luis Gonzales Rojas, Christian Gortázar, Mette Herskin, Virginie Michel, Miguel Ángel Miranda Chueca, Barbara Padalino, Paolo Pasquali, Helen Clare Roberts, Hans Spoolder, Karl Ståhl, Antonio Velarde, Arvo Viltrop and Christoph Winckler.
Declarations of interest: If you wish to access the declaration of interests of any expert contributing to an EFSA scientific assessment, please contact interestmanagement@efsa.europa.eu.
- Verena Oswaldi, Anna Karagiannis, Marzia Gnocchi, trainees at BIOHAW Unit of EFSA; Erik Alm, Maja Vukovikj, Diamantis Plachouras, Kate Olsson (European Center for Disease Prevention and Control, ECDC); Member State representatives who provided data about SARS‐CoV‐2 outbreaks and monitoring: Finland: Riikka‐Elina Lahdenperä, Terhi Laaksonen and Ari Kauppinen (Finnish Food Authority); Greece: Sokratis Perdikaris (Ministry of Rural Development and Food); Italy: Andrea Maroni Ponti, Luigi Ruocco (General Directorate of animal health, Ministry of Health); Latvia: Edvins Olsevskis (Food and Veterinary Service); Lithuania:‐ Marius Masiulis, Paulius Bušauskas and Vilija Grigaliūnienė (Emergency Response Division, State Food and Veterinary Service); Poland: Agnieszka Warda and Anna Hoffman (General Veterinary Inspectorate) and Katarzyna Domańska‐Blicharz (National Veterinary Research Institute); Spain: Luis José Romero González, Germán Cáceres Garrido, Sergio Bonilla García (Ministry of Agriculture, Fisheries and Food); Sweden: Emelie Pettersson and Siamak Zohari (National Veterinary Institute, SVA). Noemi Garcia del Blanco, Ewa Shaw, Alberto Contreras Fuentetaja, Jordi Torren (EMA, European Medicine Agency, Amsterdam); One Health European Joint Programme, project COVRIN. European Union's Horizon 2020 Research and Innovation programme, grant agreement No 773830; Joanna Korpela and Jussi Peura from FiFur – Fur Farming in Finland.
EFSA may include images or other content for which it does not hold copyright. In such cases, EFSA indicates the copyright holder and users should seek permission to reproduce the content from the original source.
Adopted: 19 January 2023
Notes
Commission Implementing Decision (EU) 2021/788 of 12 May 2021 laying down rules for the monitoring and reporting of infections with SARS‐CoV‐2 in certain animal species (OJ L 173, 17.5.2021, p. 6).
Red foxes are not reported in Figure 1 since the investigation was carried out after the date of literature review (until Feb 2022).
Disclaimer: The designations employed and the presentation of material on this map do not imply the expression of any opinion whatsoever on the part of the European Food Safety Authority concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries.
Source of figure at https://microreact.org/project/7SSAQ1zRZ9CaJNJDYKm4xD-20221026-animal-sc2-publication-country
The monitoring approaches listed here are examples of which approach can be taken, but are not necessarily linked with what is discussed in the Section 3.5 on monitoring revision.
https://food.ec.europa.eu/horizontal-topics/committees/paff-committees/animal-health-and-welfare_en
Other cats here means cats living in households having access to outdoor, and stray cats are those without owners that may live in colonies.
It could apply also for cat or dog or other susceptible animals in a pet shop or breeding center.
A non‐invasive sampling method consisting of a cotton rope, often with a bait, that is hung and let chewed by animals, out of which oral fluid can be collected and tested.
References
- Abu‐Raddad LJ, Chemaitelly H and Butt AA, 2021. Effectiveness of the BNT162b2 Covid‐19 Vaccine against the B.1.1.7 and B.1.351 Variants. The New England Journal of Medicine, 385, 187–189. 10.1056/NEJMc2104974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accorsi EK, Britton A, Fleming‐Dutra KE, Smith ZR, Shang N, Derado G, Miller J, Schrag SJ and Verani JR, 2022. Association between 3 doses of mRNA COVID‐19 vaccine and symptomatic infection caused by the SARS‐CoV‐2 Omicron and Delta variants. JAMA, 327, 639–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilo‐Gisbert J, Padilla‐Blanco M, Lizana V, Maiques E, Munoz‐Baquero M, Chillida‐Martinez E, Cardells J and Rubio‐Guerri C, 2021. First description of SARS‐CoV‐2 infection in two feral American Mink (Neovison vison) caught in the wild. Animals (Basel), 11, 1–13. 10.3390/ani11051422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen H, Tessier E, Turner C, Anderson C, Blomquist P, Simons D, Lochen A, Jarvis CI, Groves N and Capelastegui F, 2022. Comparative transmission of SARS‐CoV‐2 Omicron (B. 1.1. 529) and Delta (B. 1.617. 2) variants and the impact of vaccination: national cohort study, England. medRxiv, 2022‐02. 10.1101/2022.02.15.22271001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarawneh HN, Chemaitelly H, Ayoub H, Tang P, Hasan MR, Yassine HM, Al‐Khatib HA, Smatti MK, Coyle P and Al‐Kanaani Z, 2022. Effects of previous infection and vaccination on symptomatic omicron infections. The New England Journal of Medicine, 387, 21–34. 10.1056/NEJMoa2203965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman BR, Cossaboom CM, Wendling NM, Harvey RR, Rettler H, Taylor D, Kainulainen MH, Ahmad A, Bunkley P, Godino C, Tong S, Li Y, Uehara A, Kelleher A, Zhang J, Lynch B, Behravesh CB and Towner JS, 2022. GPS tracking of free‐roaming cats (Felis catus) on SARS‐CoV‐2‐Infected Mink farms in Utah. Viruses, 14, 1–14. 10.3390/v14102131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga FL, Jodar MN, Mondino M, Portu A, Boeris M, Joly A, Jar A, Mundo S, Castro E, Alvarez D, Torres C, Viegas M and Bratanich A, 2022. An outbreak of SARS‐CoV‐2 in big hairy armadillos (Chaetophractus villosus) associated with Gamma variant in Argentina three months after being undetectable in humans. bioRxiv, 2022‐08. 10.1101/2022.08.23.503528 [DOI] [Google Scholar]
- Bao L, Song Z, Xue J, Gao H, Liu J, Wang J, Guo Q, Zhao B, Qu Y, Qi F, Gong S, Liu M, Lv Q, Li D, Han Y, Zhao W, Deng S, Liu Y, Xiang Z, Yang B, Deng W, Yu H, Cong Z, Wei Q, Xu J, Gao GF and Qin C, 2021. Susceptibility and attenuated transmissibility of SARS‐CoV‐2 in domestic cats. The Journal of Infectious Diseases, 223, 1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar‐On YM, Goldberg Y, Mandel M, Bodenheimer O, Amir O, Freedman L, Alroy‐Preis S, Ash N, Huppert A and Milo R, 2022. Protection by a fourth dose of BNT162b2 against omicron in Israel. New England Journal of Medicine, 386, 1712–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso‐Arévalo S, Barneto A, Ramos ÁM, Rivera B, Sánchez R, Sánchez‐Morales L, Pérez‐Sancho M, Buendía A, Ferreras E, Ortiz‐Menéndez JC, Moreno I, Serres C, Vela C, Risalde MÁ, Domínguez L and Sánchez‐Vizcaíno JM, 2021a. Large‐scale study on virological and serological prevalence of SARS‐CoV‐2 in cats and dogs in Spain. Transboundary and Emerging Diseases, 6, e759–e774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso‐Arévalo S, Rivera B, Domínguez L and Sánchez‐Vizcaíno JM, 2021b. First detection of sars‐cov‐2 b.1.1.7 variant of concern in an asymptomatic dog in Spain. Viruses, 13, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal JL, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, Stowe J, Tessier E, Groves N and Dabrera G, 2021. Effectiveness of Covid‐19 vaccines against the B. 1.617.2 (Delta) variant. New England Journal of Medicine, 385, 585–594. 10.1056/NEJMoa2108891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertzbach LD, Vladimirova D, Dietert K, Abdelgawad A, Gruber AD, Osterrieder N and Trimpert J, 2021. SARS‐CoV‐2 infection of Chinese hamsters (Cricetulus griseus) reproduces COVID‐19 pneumonia in a well‐established small animal model. Transboundary and Emerging Diseases, 68, 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessière P, Vergne T, Battini M, Brun J, Averso J, Joly E, Guérin J‐L and Cadiergues M‐C, 2022. SARS‐CoV‐2 infection in companion animals: prospective serological survey and risk factor analysis in France. Viruses, 14, 1–6. 10.3390/v14061178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk J, Bonander C, Moghaddassi M, Rasmussen M, Malmqvist U, Inghammar M and Kahn F, 2022. COVID‐19 vaccine effectiveness against severe disease from SARS‐CoV‐2 Omicron BA.1 and BA.2 subvariants–surveillance results from southern Sweden, December 2021 to March 2022. Eurosurveillance, 27, 2200322. 10.2807/1560-7917.ES.2022.27.18.220032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair RV, Vaccari M, Doyle‐Meyers LA, Roy CJ, Russell‐Lodrigue K, Fahlberg M, Monjure CJ, Beddingfield B, Plante KS and Plante JA, 2021. Acute respiratory distress in aged, SARS‐CoV‐2–infected African green monkeys but not rhesus macaques. The American Journal of Pathology, 191, 274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boklund A, Hammer AS, Quaade ML, Rasmussen TBL, Strandbygaard B, Jørgensen CS, Olesen AS, Hjerpe FB, Petersen HH, Jensen TK, Mortensen S, Calvo‐Artavia FF, Lefèvre SK, Nielsen SS, Halasa T, Belsham GJ and Bøtner A, 2021. SARS‐CoV‐2 in Danish mink farms: course of the epidemic and a descriptive analysis of the outbreaks in 2020. Animals, 11, 1–16. 10.3390/ani11010164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco‐Lauth AM, Hartwig AE, Porter SM, Gordy PW, Nehring M, Byas AD, VandeWoude S, Ragan IK, Maison RM and Bowen RA, 2020. Experimental infection of domestic dogs and cats with SARS‐CoV‐2: pathogenesis, transmission, and response to reexposure in cats. Proceedings of the National Academy of Sciences of the United States of America, 117, 26382–26388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco‐Lauth AM, Root JJ, Porter SM, Walker AE, Guilbert L, Hawvermale D, Pepper A, Maison RM, Hartwig AE, Gordy P, Bielefeldt‐Ohmann H and Bowen RA, 2021a. Peridomestic mammal susceptibility to severe acute respiratory syndrome Coronavirus 2 infection. Emerging Infectious Diseases, 27, 2073–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco‐Lauth AM, Walker A, Guilbert L, Porter S, Hartwig A, McVicker E, Bielefeldt‐Ohmann H and Bowen RA, 2021b. Susceptibility of livestock to SARS‐CoV‐2 infection. Emerging Microbes & Infections, 10, 2199–2201. 10.1080/22221751.2021.2003724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucau J, Marino C, Regan J, Uddin R, Choudhary MC, Flynn JP, Chen G, Stuckwisch AM, Mathews J and Liew MY, 2022. Duration of shedding of culturable virus in SARS‐CoV‐2 Omicron (BA.1) infection. New England Journal of Medicine, 387, 275–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton TC, Atarere J, Turcinovic J, Seitz S, Sher‐Jan C, Gilbert M, White L, Zhou Z, Hossain MM and Overbeck V, 2022. Viral dynamics of Omicron and delta severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) variants with implications for timing of release from isolation: a longitudinal Cohort Study. Clinical Infectious Diseases, 76(3), e227–e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CM, Vostok J, Johnson H, Burns M, Gharpure R, Sami S, Sabo RT, Hall N, Foreman A, Schubert PL, Gallagher GR, Fink T, Madoff LC, Gabriel SB, MacInnis B, Park DJ, Siddle KJ, Harik V, Arvidson D, Brock‐Fisher T, Dunn M, Kearns A and Laney AS, 2021. Outbreak of SARS‐CoV‐2 infections, including COVID‐19 vaccine breakthrough infections, associated with large public gatherings ‐ Barnstable County, Massachusetts, July 2021. MMWR. Morbidity and Mortality Weekly Report, 70, 1059–1062. 10.15585/mmwr.mm7031e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley A, Falkenberg S, Martins M, Laverack M, Palmer MV, Lager K and Diel DG, 2021. Intravenous, intratracheal, and intranasal inoculation of swine with sars‐cov‐2. Viruses, 13, 1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso B, García‐Bocanegra I, Acevedo P, Cáceres G, Alves PC and Gortázar C, 2021. Stepping up from wildlife disease surveillance to integrated wildlife monitoring in Europe. Research in Veterinary Science, 144, 149–156. 10.1016/j.rvsc.2021.11.003 [DOI] [PubMed] [Google Scholar]
- Caserta LC, Martins M, Butt SL, Hollingshead NA, Covaleda LM, Ahmed S, Everts M, Schuler KL and Diel DG, 2022. White‐tailed deer (Odocoileus virginianus) may serve as a wildlife reservoir for nearly extinct SARS‐CoV‐2 variants of concern. bioRxiv, 2022‐09. 10.1101/2022.09.02.506368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerino P, Buonerba C, Brambilla G, Atripaldi L, Tafuro M, Concilio DD, Vassallo L, Conte GL, Cuomo MC, Maiello I, D'Auria J, Cardinale D, Viscardi M, Rofrano G, Gallo A, Brusco P, Pizzolante A, Cicalese V, Galdi P, Galdi L, Vita SD, Volzone P, Vuolo GD, Coppola A, Pierri B and Fusco G, 2021. No detection of SARS‐CoV‐2 in animals exposed to infected keepers: results of a COVID‐19 surveillance program. Future Science OA, 7, FSO711. 10.2144/fsoa-2021-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaintoutis SC, Thomou Z, Mouchtaropoulou E, Tsiolas G, Chassalevris T, Stylianaki I, Lagou M, Michailidou S, Moutou E, Koenen JJH, Dijkshoorn JW, Paraskevis D, Poutahidis T, Siarkou VI, Sypsa V, Argiriou A, Fortomaris P and Dovas CI, 2021. Outbreaks of SARS‐CoV‐2 in naturally infected mink farms: impact, transmission dynamics, genetic patterns, and environmental contamination. PLoS Pathogens, 17, e1009883. 10.1371/journal.ppat.1009883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JF‐W, Zhang AJ, Yuan S, Poon VK‐M, Chan CC‐S, Lee AC‐Y, Chan W‐M, Fan Z, Tsoi H‐W, Wen L, Liang R, Cao J, Chen Y, Tang K, Luo C, Cai J‐P, Kok K‐H, Chu H, Chan K‐H, Sridhar S, Chen Z, Chen H, To KK‐W and Yuen K‐Y, 2020. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID‐19) in a Golden Syrian Hamster model: implications for disease pathogenesis and transmissibility. Clinical Infectious Diseases, 71, 2428–2446. 10.1093/cid/ciaa325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JC, Bevins SN, Ellis JW, Linder TJ, Tell RM, Jenkins‐Moore M, Root JJ, Lenoch JB, Robbe‐Austerman S, DeLiberto TJ, Gidlewski T, Torchetti MK and Shriner SA, 2021. SARS‐CoV‐2 exposure in wild white‐tailed deer (Odocoileus virginianus). Proceedings of the National Academy of Sciences of the United States of America, 118, e2114828118. 10.1073/pnas.2114828118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia PY, Ong SWX, Chiew CJ, Ang LW, Chavatte J‐M, Mak T‐M, Cui L, Kalimuddin S, Chia WN and Tan CW, 2022. Virological and serological kinetics of SARS‐CoV‐2 Delta variant vaccine breakthrough infections: a multicentre cohort study. Clinical Microbiology and Infection, 28, 612.e611–612.e617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Halfmann PJ, Hatta M, Maemura T, Fan S, Armbrust T, Swartley OM, Crawford LK and Kawaoka Y, 2021. Protective immunity and persistent lung sequelae in domestic cats after SARS‐CoV‐2 infection. Emerging Infectious Diseases, 27, 660–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciurkiewicz M, Armando F, Schreiner T, de Buhr N, Pilchová V, Krupp‐Buzimikic V, Gabriel G, von Köckritz‐Blickwede M, Baumgärtner W, Schulz C and Gerhauser I, 2022. Ferrets are valuable models for SARS‐CoV‐2 research. Veterinary Pathology, 59, 661–672. 10.1177/03009858211071012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colitti B, Bertolotti L, Mannelli A, Ferrara G, Vercelli A, Grassi A, Trentin C, Paltrinieri S, Nogarol C, Decaro N, Brocchi E and Rosati S, 2021. Cross‐sectional serosurvey of companion animals housed with sars‐cov‐2‐infected owners, Italy. Emerging Infectious Diseases, 27, 1919–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collie S, Nayager J, Bamford L, Bekker L‐G, Zylstra M and Gray G, 2022. Effectiveness and durability of the BNT162b2 vaccine against omicron sublineages in South Africa. New England Journal of Medicine, 387, 1332–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo VC, Sluydts V, Mariën J, Vanden Broecke B, Van Houtte N, Leirs W, Jacobs L, Iserbyt A, Hubert M, Heyndrickx L, Goris H, Delputte P, De Roeck N, Elst J, Ariën KK, Leirs H and Gryseels S, 2021. SARS‐CoV‐2 surveillance in Norway rats (Rattus norvegicus) from Antwerp sewer system, Belgium. Transboundary and Emerging Diseases, 69, 3016–3021. 10.1111/tbed.14219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cool K, Gaudreault NN, Morozov I, Trujillo JD, Meekins DA, McDowell C, Carossino M, Bold D, Mitzel D, Kwon T, Balaraman V, Madden DW, Artiaga BL, Pogranichniy RM, Roman‐Sosa G, Henningson J, Wilson WC, Balasuriya UBR, García‐Sastre A and Richt JA, 2022. Infection and transmission of ancestral SARS‐CoV‐2 and its alpha variant in pregnant white‐tailed deer. Emerging Microbes and Infections, 11, 95–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J and Schmidt ML, 2020. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Eurosurveillance, 25, 2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RM, Wolf JD, Lieber CM, Sourimant J, Lin MJ, Babusis D, DuPont V, Chan J, Barrett KT and Lye D, 2021a. Oral prodrug of remdesivir parent GS‐441524 is efficacious against SARS‐CoV‐2 in ferrets. Nature Communications, 12, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RM, Wolf JD and Plemper RK, 2021b. Therapeutically administered ribonucleoside analogue MK‐4482/EIDD‐2801 blocks SARS‐CoV‐2 transmission in ferrets. Nature Microbiology, 6, 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M‐A, Morden E, Rosseau P, Arendse J, Bam J‐L, Boloko L, Cloete K, Cohen C, Chetty N and Dane P, 2022. Outcomes of laboratory‐confirmed SARS‐CoV‐2 infection during resurgence driven by Omicron lineages BA.4 and BA.5 compared with previous waves in the Western Cape Province, South Africa. International Journal of Infectious Diseases, 127, 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoust B, Guérin P, Orain N, Fligny C, Flirden F, Fenollar F, Mediannikov O and Edouard S, 2022. Evidence of antibodies against SARS‐CoV‐2 in wild mustelids from Brittany (France). Transboundary and Emerging Diseases, 69, e3400–e3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RD, Schmitz KS, Bovier FT, Predella C, Khao J, Noack D, Haagmans BL, Herfst S, Stearns KN and Drew‐Bear J, 2021. Intranasal fusion inhibitory lipopeptide prevents direct‐contact SARS‐CoV‐2 transmission in ferrets. Science, 371, 1379–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahay RJ, de la Fuente J, Smith GC, Sharun K, Snary EL, Flores Girón L, Nziza J, Fooks AR, Brookes SM, Lean FZX, Breed AC and Gortazar C, 2021. Assessing the risks of SARS‐CoV‐2 in wildlife. One Health Outlook, 3, 1–14. 10.1186/s42522-021-00039-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Jin Y, Liu Y, Sun J, Hao L, Bai J, Huang T, Lin D, Jin Y and Tian K, 2020. Serological survey of SARS‐CoV‐2 for experimental, domestic, companion and wild animals excludes intermediate hosts of 35 different species of animals. Transboundary and Emerging Diseases, 67, 1745–1749. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Devaux CA, Pinault L, Delerce J, Raoult D, Levasseur A and Frutos R, 2021. Spread of mink SARS‐CoV‐2 variants in humans: a model of sarbecovirus interspecies evolution. Frontiers in Microbiology, 12, 2848. 10.3389/fmicb.2021.675528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domańska‐Blicharz K, Orłowska A, Smreczak M, Niemczuk K, Iwan E, Bomba A, Lisowska A, Opolska J, Trȩbas P, Potyrało P, Kawiak‐Sadurska M and Rola J, 2021. Mink SARS‐CoV‐2 infection in Poland ‐ short communication. Journal of Veterinary Research (Poland), 65, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowall S, Salguero FJ, Wiblin N, Fotheringham S, Hatch G, Parks S, Gowan K, Harris D, Carnell O and Fell R, 2021. Development of a hamster natural transmission model of SARS‐CoV‐2 infection. Viruses, 13, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EAZWV (European Association of Zoo and Wildlife Veterinarians) , 2022. Science‐based facts and knowledge about wild animals, zoos and Sars‐CoV‐2 virus. Available online: https://cdn.ymaws.com/www.eazwv.org/resource/resmgr/files/transmissible_diseases_handbook/5th_ed_transmissible_diseases_handbook/chapters/2_covid19_faq_v9_17_january_.pdf
- ECDC (European Centre for Disease Prevention and Control) , 2019. Operational tool on rapid risk assessment methodology. Stockholm. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/operational-tool-rapid-risk-assessment-methodolgy-ecdc-2019.pdf
- ECDC (European Centre for Disease Prevention and Control) , 2020a. Detection of new SARS‐CoV‐2 variants related to mink ‐ 12 November 2020. ECDC: Stockholm; 2020. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/RRA-SARS-CoV-2-in-mink-12-nov-2020.pdf
- ECDC (European Centre for Disease Prevention and Control) , 2020b. Guidance for wearing and removing personal protective equipment in healthcare settings for the care of patients with suspected or confirmed COVID‐19. Stockholm. Available online: https://www.ecdc.europa.eu/en/publications-data/guidance-wearing-and-removing-personal-protective-equipment-healthcare-settings
- ECDC (European Centre for Disease Prevention and Control) , 2020c. Heating, ventilation and air‐conditioning systems in the context of COVID‐19. Stockholm. Available online: https://www.ecdc.europa.eu/en/publications-data/heating-ventilation-air-conditioning-systems-covid-19
- ECDC (European Centre for Disease Prevention and Control) , 2020d. Rapid Risk Assessment: Detection of new SARS‐CoV‐2 variants related to mink. Stockholm. Available online: https://www.ecdc.europa.eu/en/publications-data/detection-new-sars-cov-2-variants-mink
- ECDC (European Centre for Disease Prevention and Control) , 2021. Options for the use of rapid antigen detection tests for COVID‐19 in the EU/EEA – first update, 26 October 2021. Stockholm. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/Options-for-the-use-of-rapid-antigen-tests-for-COVID-19-first-update.pdf
- ECDC (European Centre for Disease Prevention and Control) , 2022a. Assessment of the further spread and potential impact of the SARS‐CoV‐2 Omicron variant of concern in the EU/EEA, 19th update. Stockholm Available online: https://www.ecdc.europa.eu/en/publications-data/covid-19-omicron-risk-assessment-further-emergence-and-potential-impact
- ECDC (European Centre for Disease Prevention and Control) , 2022b. Considerations for the use of face masks in the community in the context of the SARS‐CoV‐2 Omicron variant of concern. Stockholm. Available online: https://www.ecdc.europa.eu/en/publications-data/using-face-masks-community-reducing-covid-19-transmission
- ECDC/EUOSHA (European Centre for Disease Prevention and Control/European Agency for Safety and Health at Work) , 2021. Considerations on the use of rapid antigen detection (including self‐) tests for SARS‐CoV‐2 in occupational settings. Stockholm/Bilbao. Available online: https://www.ecdc.europa.eu/en/publications-data/considerations-use-rapid-antigen-detection-including-self-tests-sars-cov-2
- ECDC/WHO (European Centre for Disease Prevention and Control/World Health Organization Regional Office for Europe) , 2022a. Methods for the detection and identification of SARS‐CoV‐2 variants: second update. ECDC and WHO European Region: Stockholm and Copenhagen. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/Methods-for-the-detection-char-SARS-CoV-2-variants_2nd%20update_final.pdf
- ECDC/WHO (European Centre for Disease Prevention and Control/World Health Organization Regional Office for Europe) , 2022b. Operational considerations for respiratory virus surveillance in Europe. Stockholm and Copenhagen. Available online: https://www.ecdc.europa.eu/en/publications-data/operational-considerations-respiratory-virus-surveillance-europe
- EFSA (European Food Safety Authority) , 2014. Guidance on expert knowledge elicitation in food and feed safety risk assessment. EFSA Journal 2014;12(6):3734, 278 pp. 10.2903/j.efsa.2014.3734 [DOI] [Google Scholar]
- EFSA AHAW Panel (EFSA Panel on Animal Health and Welfare) , 2015. Scientific opinion on the survival, spread and establishment of the small hive beetle (Aethina tumida). EFSA Journal 2015;13(12):4328, 77 pp. 10.2903/j.efsa.2015.4328 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority) , ECDC (European Centre for Disease Prevention and Control) , Boklund A, Gortazar C, Pasquali P, Roberts H, Nielsen SS, Stahl K, Stegeman A, Baldinelli F, Broglia A, Van Der Stede Y, Adlhoch C, Alm E, Melidou A and Mirinaviciute G, 2021. Monitoring of SARS‐CoV‐2 infection in mustelids. EFSA Journal 2021;19(3):6459, 68 pp 10.2903/j.efsa.2021.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre DW, Taylor D, Purver M, Chapman D, Fowler T, Pouwels KB, Walker AS and Peto TEA, 2022. Effect of Covid‐19 vaccination on transmission of alpha and delta variants. The New England Journal of Medicine, 386, 744–756. 10.1056/NEJMoa2116597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagre A, Lewis J, Eckley M, Zhan S, Rocha SM, Sexton NR, Burke B, Geiss B, Peersen O, Bass T, Kading R, Rovnak J, Ebel GD, Tjalkens RB, Aboellail T and Schountz T, 2021. SARS‐CoV‐2 infection, neuropathogenesis and transmission among deer mice: Implications for spillback to New World rodents. PLoS Pathogens, 17, e1009585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg S, Buckley A, Laverack M, Martins M, Palmer MV, Lager K and Diel DG, 2021. Experimental inoculation of young calves with sars‐cov‐2. Viruses, 13, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnia P, Aghajani J, Farnia P, Ayoubi S, Ghanavi J, Nadji SA, Hoffner S and Velayati AA, 2020. Evidence for SARS‐CoV‐2 circulating among stray dogs and cats: should we worry about our pets during the COVID‐19 Pandemic? Biomedical and Biotechnology Research Journal (BBRJ), 4, 49–55. [Google Scholar]
- Fernández‐Bastit L, Roca N, Romero‐Durana M, Rodon J, Cantero G, García Ó, López C, Pérez M, López R, Carrillo J, Izquierdo‐Useros N, Blanco J, Clotet B, Pujols J, Vergara‐Alert J, Segalés J and Lorca‐Oró C, 2022. Susceptibility of domestic goat (Capra aegagrus hircus) to experimental infection with severe acute respiratory syndrome Coronavirus 2 (SARS‐CoV‐2) B.1.351/Beta Variant. Viruses, 14, 1–13. 10.3390/v14092002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Bellon H, Rodon J, Fernández‐Bastit L, Almagro V, Padilla‐Solé P, Lorca‐Oró C, Valle R, Roca N, Grazioli S, Trogu T, Bensaid A, Carrillo J, Izquierdo‐Useros N, Blanco J, Parera M, Noguera‐Julián M, Clotet B, Moreno A, Segalés J and Vergara‐Alert J, 2021. Monitoring natural SARS‐CoV‐2 infection in lions (Panthera leo) at the Barcelona zoo: viral dynamics and host responses. Viruses, 13, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischhoff IR, Castellanos AA, Rodrigues JP, Varsani A and Han BA, 2021. Predicting the zoonotic capacity of mammals to transmit SARS‐CoV‐2. Proceedings of the Royal Society B, 288, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco R, Hernandez SM, Mead DG, Adcock KG, Burke SC, Nemeth NM and Yabsley MJ, 2022. Experimental susceptibility of North American Raccoons (Procyon lotor) and striped Skunks (Mephitis mephitis) to SARS‐CoV‐2. Frontiers in Veterinary Science, 8, 1654. 10.3389/fvets.2021.715307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freuling CM, Breithaupt A, Müller T, Sehl J, Balkema‐Buschmann A, Rissmann M, Klein A, Wylezich C, Höper D, Wernike K, Aebischer A, Hoffmann D, Friedrichs V, Dorhoi A, Groschup MH, Beer M and Mettenleiter TC, 2020. Susceptibility of Raccoon dogs for experimental SARS‐CoV‐2 infection. Emerging Infectious Diseases, 26, 2982–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz M, Rosolen B, Krafft E, Becquart P, Elguero E, Vratskikh O, Denolly S, Boson B, Vanhomwegen J, Gouilh MA, Kodjo A, Chirouze C, Rosolen SG, Legros V and Leroy EM, 2020. High prevalence of SARS‐CoV‐2 antibodies in pets from COVID‐19+ households. One Health, 11, 100192. 10.1016/j.onehlt.2020.100192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz M, de Riols de Fonclare D, Garcia D, Beurlet S, Becquart P, Rosolen SG, Briend‐Marchal A and Leroy EM, 2022. First evidence of natural SARS‐CoV‐2 infection in domestic rabbits. Veterinary Sciences, 9, 1–4. 10.3390/vetsci9020049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Knight M, Anglin K, Tassetto M, Lu S, Zhang A, Goldberg SA, Catching A, Davidson MC, Shak JR and Romero M, 2022. Infectious viral shedding of SARS‐CoV‐2 Delta following vaccination: a longitudinal cohort study. PLoS Pathogens, 18, e1010802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreault NN, Trujillo JD, Carossino M, Meekins DA, Morozov I, Madden DW, Indran SV, Bold D, Balaraman V, Kwon T, Artiaga BL, Cool K, García‐Sastre A, Ma W, Wilson WC, Henningson J, Balasuriya UBR and Richt JA, 2020. SARS‐CoV‐2 infection, disease and transmission in domestic cats. Emerging Microbes and Infections, 9, 2322–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreault NN, Carossino M, Morozov I, Trujillo JD, Meekins DA, Madden DW, Cool K, Artiaga BL, McDowell C, Bold D, Balaraman V, Kwon T, Ma W, Henningson J, Wilson DW, Wilson WC, Balasuriya UBR, García‐Sastre A and Richt JA, 2021. Experimental re‐infected cats do not transmit SARS‐CoV‐2. Emerging Microbes and Infections, 10, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhards NM, Cornelissen JBWJ, van Keulen LJM, Harders‐Westerveen J, Vloet R, Smid B, Vastenhouw S, van Oort S, Hakze‐van der Honing RW, Gonzales JL, Stockhofe‐Zurwieden N, de Jong R, van der Poel WHM, Vreman S, Kortekaas J, Wichgers Schreur PJ and Oreshkova N, 2021. Predictive value of precision‐cut lung slices for the susceptibility of three animal species for SARS‐CoV‐2 and validation in a refined hamster model. Pathogens, 10, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhards NM, Gonzales JL, Vreman S, Ravesloot L, van den Brand JM, Doekes HP, Egberink HF, Stegeman A, Oreshkova N and van der Poel WH, 2022. Efficient direct and limited environmental transmission of SARS‐CoV‐2 lineage B.1.22 in domestic cats. bioRxiv, 2022‐2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giner J, Villanueva‐saz S, Tobajas AP, Pérez MD, González A, Verde M, Yzuel A, García‐garcía A, Taleb V, Lira‐navarrete E, Hurtado‐guerrero R, Pardo J, Santiago L, Paño JR, Ruíz H, Lacasta D and Fernández A, 2021. Sars‐cov‐2 seroprevalence in household domestic ferrets (Mustela putorius furo). Animals, 11, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AR, Langwig KE, Marano J, Sharp AK, Brown KL, Ceci A, Kailing MJ, Briggs R, Roby C, Brown AM, Weger‐Lucarelli J, Finkielstein CV and Hoyt JR, 2022. Wildlife exposure to SARS‐CoV‐2 across a human use gradient. bioRxiv, 2022‐11. 10.1101/2022.11.04.515237 [DOI] [Google Scholar]
- Gonzales JL, De Jong MC, Gerhards NM and Van der Poel WH, 2021. The SARS‐CoV‐2 reproduction number R0 in cats. Viruses, 13, 1–11. 10.3390/v13122480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gortazar C, Diez‐Delgado I, Barasona JA, Vicente J, De La Fuente J and Boadella M, 2015. The wild side of disease control at the wildlife‐livestock‐human interface: a review. Frontiers in Veterinary Science, 1, 1–12. 10.3389/fvets.2014.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gortázar C, Barroso‐Arévalo S, Ferreras‐Colino E, Isla J, de la Fuente G, Rivera B, Domínguez L, de la Fuente J and Sánchez‐Vizcaíno JM, 2021. Natural SARS‐CoV‐2 infection in kept ferrets, Spain. Emerging Infectious Diseases, 27, 1994–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin BD, Chan M, Tailor N, Mendoza EJ, Leung A, Warner BM, Duggan AT, Moffat E, He S, Garnett L, Tran KN, Banadyga L, Albietz A, Tierney K, Audet J, Bello A, Vendramelli R, Boese AS, Fernando L, Lindsay LR, Jardine CM, Wood H, Poliquin G, Strong JE, Drebot M, Safronetz D, Embury‐Hyatt C and Kobasa D, 2021. SARS‐CoV‐2 infection and transmission in the North American deer mouse. Nature Communications, 12, 1–10. 10.1038/s41467-021-23848-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale VL, Dennis PM, McBride DS, Nolting JM, Madden C, Huey D, Ehrlich M, Grieser J, Winston J, Lombardi D, Gibson S, Saif L, Killian ML, Lantz K, Tell R, Torchetti M, Robbe‐Austerman S, Nelson MI, Faith SA and Bowman AS, 2021. SARS‐CoV‐2 infection in free‐ranging white‐tailed deer. Nature, 602, 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann PJ, Hatta M, Chiba S, Maemura T, Fan S, Takeda M, Kinoshita N, Hattori SI, Sakai‐Tagawa Y, Iwatsuki‐Horimoto K, Imai M and Kawaoka Y, 2020. Transmission of SARS‐CoV‐2 in domestic cats. The New England Journal of Medicine, 383, 592–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall VJ, Foulkes S, Saei A, Andrews N, Oguti B, Charlett A, Wellington E, Stowe J, Gillson N and Atti A, 2021. COVID‐19 vaccine coverage in health‐care workers in England and effectiveness of BNT162b2 mRNA vaccine against infection (SIREN): a prospective, multicentre, cohort study. The Lancet, 397, 1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer AS, Quaade ML, Rasmussen TB, Fonager J, Rasmussen M, Mundbjerg K, Lohse L, Strandbygaard B, Jørgensen CS, Alfaro‐Núñez A, Rosenstierne MW, Boklund A, Halasa T, Fomsgaard A, Belsham GJ and Bøtner A, 2021. SARS‐CoV‐2 transmission between mink (Neovison vison) and humans, Denmark. Emerging Infectious Diseases, 27, 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CH, Friis NU, Bager P, Stegger M, Fonager J, Fomsgaard A, Gram MA, Engbo Christiansen L, Ethelberg S and Legarth R, 2022. Risk of reinfection, vaccine protection, and severity of infection with the BA.5 omicron subvariant: a Danish nation‐wide population‐based study. The Lancet. Infectious Diseases, 23, 167–176. 10.1016/S1473-3099(22)00595-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RJ, Hall JA, Zaidi A, Andrews NJ, Dunbar JK and Dabrera G, 2021. Effect of vaccination on household transmission of SARS‐CoV‐2 in England. New England Journal of Medicine, 385, 759–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JA, Kissler SM, Fauver JR, Mack C, Tai CG, Samant RM, Connolly S, Anderson DJ, Khullar G, MacKay M, Patel M, Kelly S, Manhertz A, Eiter I, Salgado D, Baker T, Howard B, Dudley JT, Mason CE, Nair M, Huang Y, DiFiori J, Ho DD, Grubaugh ND and Grad YH, 2022. Quantifying the impact of immune history and variant on SARS‐CoV‐2 viral kinetics and infection rebound: a retrospective cohort study. eLife, 11, e81849. 10.7554/eLife.81849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Iwatsuki‐Horimoto K, Hatta M, Loeber S, Halfmann PJ, Nakajima N, Watanabe T, Ujie M, Takahashi K, Ito M, Yamada S, Fan S, Chiba S, Kuroda M, Guan L, Takada K, Armbrust T, Balogh A, Furusawa Y, Okuda M, Ueki H, Yasuhara A, Sakai‐Tagawa Y, Lopes TJS, Kiso M, Yamayoshi S, Kinoshita N, Ohmagari N, Hattori S‐i, Takeda M, Mitsuya H, Krammer F, Suzuki T and Kawaoka Y, 2020. Syrian hamsters as a small animal model for SARS‐CoV‐2 infection and countermeasure development. Proceedings of the National Academy of Sciences of the United States of America, 117, 16587–16595. 10.1073/pnas.2009799117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip HS, Griffin KM, Messer JD, Winzeler ME, Shriner SA, Killian ML, Torchetti MK, Deliberto TJ, Amman BR, Cossaboom CM, Harvey RR, Wendling NM, Rettler H, Taylor D, Towner JS, Behravesh CB and Blehert DS, 2021. An opportunistic survey reveals an unexpected coronavirus diversity hotspot in North America. Viruses, 13, 1–12. 10.3390/v13102016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalali N, Brustad HK, Frigessi A, MacDonald EA, Meijerink H, Feruglio SL, Nygård KM, Rø GØI, Madslien EH and De Blasio BF, 2022. Increased household transmission and immune escape of the SARS‐CoV‐2 Omicron compared to Delta variants. Nature Communications, 13, 1–5. 10.1038/s41467-022-33233-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemeršić L, Lojkić I, Krešić N, Keros T, Zelenika TA, Jurinović L, Skok D, Bata I, Boras J, Habrun B and Brnić D, 2021. Investigating the presence of SARS CoV‐2 in free‐living and captive animals. Pathogens, 10, 1–12. 10.3390/pathogens10060635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Kim JY, Park H, Park S, Lim JS, Lim SY, Bae S, Lim Y‐J, Kim EO and Kim J, 2022. Transmission and infectious SARS‐CoV‐2 shedding kinetics in vaccinated and unvaccinated individuals. JAMA Network Open, 5, e2213606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannekens‐Jager MM, de Rooij MM, de Groot Y, Biesbroeck E, de Jong MK, Pijnacker T, Smit LA, Schuurman N, Broekhuizen‐Stins MJ and Zhao S, 2022. SARS‐CoV‐2 infection in dogs and cats is associated with contact to COVID‐19‐positive household members. Transboundary and Emerging Diseases, 69, 4034–4040. 10.1111/tbed.14713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Seiler P, Jones JC, Ridout G, Camp KP, Fabrizio TP, Jeevan T, Miller LA, Throm RE, Ferrara F, Fredrickson RL, Lowe JF, Wang L, Odemuyiwa SO, Wan XF and Webby RJ, 2020. Antibody responses to SARS‐CoV‐2 antigens in humans and animals. Vaccines, 8, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y‐I, Yu K‐M, Koh J‐Y, Kim E‐H, Kim S‐M, Kim EJ, Casel MAB, Rollon R, Jang S‐G and Song M‐S, 2022. Age‐dependent pathogenic characteristics of SARS‐CoV‐2 infection in ferrets. Nature Communications, 13, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom FC, Andrews N, Stowe J, Toffa S, Sachdeva R, Gallagher E, Groves N, O'Connell A‐M, Chand M and Ramsay M, 2022. COVID‐19 vaccine effectiveness against the omicron (BA.2) variant in England. The Lancet Infectious Diseases, 22, 931–933. 10.1016/S1473-3099(22)00309-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislaya I, Casaca P, Borges V, Sousa C, Ferreira BI, Fonte A, Fernandes E, Matias Dias C, Duarte S and Almeida JP, 2022. Comparative COVID‐19 vaccines effectiveness in preventing infections, hospitalizations, and deaths with SARS‐CoV‐2 BA.5 and BA.2 omicron lineages: a case‐case and cohort study using electronic health records in Portugal. SSRN Electronic Journal. 10.2139/ssrn.4180482 [DOI] [Google Scholar]
- Kok K‐H, Wong S‐C, Chan W‐M, Wen L, Chu AW‐H, Ip JD, Lee L‐K, Wong IT‐F, Lo HW‐H and Cheng VC‐C, 2022. Co‐circulation of two SARS‐CoV‐2 variant strains within imported pet hamsters in Hong Kong. Emerging Microbes and Infections, 11, 689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwa JD, Massé A, Gagnier M, Aftanas P, Blais‐Savoie J, Bowman J, Buchanan T, Chee H‐Y, Dibernardo A, Kruczkiewicz P, Nirmalarajah K, Soos C, Yip L, Lindsay LR, Lung O, Pickering B and Mubareka S, 2022. First detection of SARS‐CoV‐2 infection in Canadian wildlife identified in free‐ranging white‐tailed deer (Odocoileus virginianus) from southern Québec, Canada. bioRxiv, 022‐2001. 10.1101/2022.01.20.476458 [DOI] [Google Scholar]
- Külper‐Schiek W, Piechotta V, Pilic A, Batke M, Dreveton L‐S, Geurts B, Koch J, Köppe S, Treskova M and Vygen‐Bonnet S, 2022. Facing the Omicron variant—how well do vaccines protect against mild and severe COVID‐19? Third interim analysis of a living systematic review. Frontiers in Immunology, 13, 1–13. 10.3389/fimmu.2022.940562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter JS, de Meulder D, Bestebroer TM, Lexmond P, Mulders A, Richard M, Fouchier RA and Herfst S, 2021. SARS‐CoV and SARS‐CoV‐2 are transmitted through the air between ferrets over more than one meter distance. Nature Communications, 12, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen CS and Paludan SR, 2020. Corona's new coat: SARS‐CoV‐2 in Danish minks and implications for travel medicine. Travel Medicine and Infectious Disease, 38, 101922. 10.1016/j.tmaid.2020.101922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen HD, Fonager J, Lomholt FK, Dalby T, Benedetti G, Kristensen B, Urth TR, Rasmussen M, Lassaunière R and Rasmussen TB, 2021. Preliminary report of an outbreak of SARS‐CoV‐2 in mink and mink farmers associated with community spread, Denmark, June to November 2020. Eurosurveillance, 26, 2100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassaunière R, Fonager J, Rasmussen M, Frische A, Polacek C, Rasmussen TB, Lohse L, Belsham GJ, Underwood A and Winckelmann AA, 2021. In vitro characterization of fitness and convalescent antibody neutralization of SARS‐CoV‐2 Cluster 5 variant emerging in mink at Danish farms. Frontiers in Microbiology, 12, 1679. 10.3389/fmicb.2021.698944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring AS, Tenforde MW, Chappell JD, Gaglani M, Ginde AA, McNeal T, Ghamande S, Douin DJ, Talbot HK and Casey JD, 2022. Clinical severity and mRNA vaccine effectiveness for omicron, delta, and alpha SARS‐CoV‐2 variants in the United States: a prospective observational study. medRxiv, 2022‐02. 10.1101/2022.02.06.22270558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layan M, Gilboa M, Gonen T, Goldenfeld M, Meltzer L, Andronico A, Hozé N, Cauchemez S and Regev‐Yochay G, 2022. Impact of BNT162b2 vaccination and isolation on SARS‐CoV‐2 transmission in Israeli households: an observational study. American Journal of Epidemiology, 191, 1224–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine‐Tiefenbrun M, Yelin I, Alapi H, Herzel E, Kuint J, Chodick G, Gazit S, Patalon T and Kishony R, 2022. Waning of SARS‐CoV‐2 booster viral‐load reduction effectiveness. Nature Communications, 13, 1237. 10.1038/s41467-022-28936-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link‐Gelles R, 2022. Effectiveness of bivalent mRNA vaccines in preventing symptomatic SARS‐CoV‐2 infection—increasing community access to testing program, United States, September–November 2022. Morbidity and Mortality Weekly Report, 71, 1526–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Sikkema RS, Velkers FC, Nieuwenhuijse DF, Fischer EA, Meijer PA, Bouwmeester‐Vincken N, Rietveld A, Wegdam‐Blans MC and Tolsma P, 2021. Adaptation, spread and transmission of SARS‐CoV‐2 in farmed minks and associated humans in the Netherlands. Nature Communications, 12, 6802. 10.1038/s41467-021-27096-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyngse FP, Mortensen LH, Denwood MJ, Christiansen LE, Møller CH, Skov RL, Spiess K, Fomsgaard A, Lassauniere R and Rasmussen M, 2021. Household transmission of the SARS‐CoV‐2 Omicron variant in Denmark. Nature Communications, 13, 5573. 10.1038/s41467-022-33328-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Karikalan M, Chander V, Pawde AM, Saikumar G, Semmaran M, Lakshmi PS, Sharma M, Nandi S and Singh KP, 2022. Detection of SARS‐CoV‐2 in a free ranging leopard (Panthera pardus fusca) in India. European Journal of Wildlife Research, 68, 1–5. 10.1007/s10344-022-01608-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahumud RA, Ali MA, Kundu S, Rahman MA, Kamara JK and Renzaho AM, 2022. Effectiveness of COVID‐19 vaccines against delta variant (B.1.617.2): a meta‐analysis. Vaccines, 10, 277. 10.3390/vaccines10020277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh GA, McAuley AJ, Brown S, Pharo EA, Crameri S, Au GG, Baker ML, Barr JA, Bergfeld J, Bruce MP, Burkett K, Durr PA, Holmes C, Izzard L, Layton R, Lowther S, Neave MJ, Poole T, Riddell SJ, Rowe B, Soldani E, Stevens V, Suen WW, Sundaramoorthy V, Tachedjian M, Todd S, Trinidad L, Williams SM, Druce JD, Drew TW and Vasan SS, 2021. In vitro characterisation of SARS‐CoV‐2 and susceptibility of domestic ferrets (Mustela putorius furo). Transboundary and Emerging Diseases, 69, 297–307. 10.1111/tbed.13978 [DOI] [PubMed] [Google Scholar]
- Meekins DA, Morozov I, Trujillo JD, Gaudreault NN, Bold D, Carossino M, Artiaga BL, Indran SV, Kwon T, Balaraman V, Madden DW, Feldmann H, Henningson J, Ma W, Balasuriya UBR and Richt JA, 2020. Susceptibility of swine cells and domestic pigs to SARS‐CoV‐2. Emerging Microbes and Infections, 9, 2278–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miró G, Regidor‐Cerrillo J, Checa R, Diezma‐Díaz C, Montoya A, García‐Cantalejo J, Botías P, Arroyo J and Ortega‐Mora LM, 2021. SARS‐CoV‐2 infection in one cat and three dogs living in COVID‐19‐positive households in Madrid, Spain. Frontiers in Veterinary Science, 8, 779341. 10.3389/fvets.2021.779341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok BW‐Y, Liu H, Deng S, Liu J, Zhang AJ, Lau S‐Y, Liu S, Tam RC‐Y, Cremin CJ and Ng TT‐L, 2021. Low dose inocula of SARS‐CoV‐2 alpha variant transmits more efficiently than earlier variants in hamsters. Communications Biology, 4, 1–8. 10.1038/s42003-021-02640-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar RJ, Vreman S, Hakze‐van der Honing RW, Zwart R, de Rond J, Weesendorp E, Smit LAM, Koopmans M, Bouwstra R, Stegeman A and van der Poel WHM, 2020. Clinical and pathological findings in SARS‐CoV‐2 disease outbreaks in farmed mink (Neovison vison). Veterinary Pathology, 57, 653–657. [DOI] [PubMed] [Google Scholar]
- Montagutelli X, Prot M, Levillayer L, Salazar EB, Jouvion G, Conquet L, Beretta M, Donati F, Albert M and Gambaro F, 2021. Variants with the N501Y mutation extend SARS‐CoV‐2 host range to mice, with contact transmission. bioRxiv, 2021‐03. 10.1101/2021.03.18.436013 [DOI] [Google Scholar]
- Moreira‐Soto A, Walzer C, Czirják GÁ, Richter MH, Marino SF, Posautz A, De Yebra RP, McEwen GK, Drexler JF and Greenwood AD, 2022. Serological evidence that SARS‐CoV‐2 has not emerged in deer in Germany or Austria during the COVID‐19 pandemic. Microorganisms, 10, 748. 10.3390/microorganisms10040748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez‐Pérez L, Schulz J, Meade‐White K, Okumura A, Callison J, Brumbaugh B, Avanzato VA, Rosenke R, Hanley PW, Saturday G, Scott D, Fischer ER and de Wit E, 2020. Respiratory disease in rhesus macaques inoculated with SARS‐CoV‐2. Nature, 585, 268–272. 10.1038/s41586-020-2324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neary M, Box H, Sharp J, Tatham L, Curley P, Herriott J, Kijak E, Arshad U, Hobson JJ and Rajoli RK, 2021. Evaluation of intranasal nafamostat or camostat for SARS‐CoV‐2 chemoprophylaxis in Syrian golden hamsters. bioRxiv, 2021–2007. 10.1101/2021.07.08.451654 [DOI] [Google Scholar]
- Oliveira A, Pereira MA, Mateus TL, Mesquita JR and Vala H, 2022. Seroprevalence of SARS‐CoV‐2 in client‐owned cats from Portugal. Veterinary Sciences, 9, 1–18. 10.3390/vetsci9070363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreshkova N, Molenaar RJ, Vreman S, Harders F, Oude Munnink BB, Van Der Honing RWH, Gerhards N, Tolsma P, Bouwstra R, Sikkema RS, Tacken MGJ, De Rooij MMT, Weesendorp E, Engelsma MY, Bruschke CJM, Smit LAM, Koopmans M, Van Der Poel WHM and Stegeman A, 2020. SARS‐CoV‐2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance, 25, 2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, van der Spek A, Tolsma P, Rietveld A, Brouwer M, Bouwmeester‐Vincken N, Harders F, Hakze‐van der Honing R, Wegdam‐Blans MCA, Bouwstra RJ, GeurtsvanKessel C, van der Eijk AA, Velkers FC, Smit LAM, Stegeman A, van der Poel WHM and Koopmans MPG, 2020. Transmission of SARS‐CoV‐2 on mink farms between humans and mink and back to humans. Science, 371, 172–177. 10.1126/science.abe5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, van der Spek A, Tolsma P, Rietveld A, Brouwer M, Bouwmeester‐Vincken N, Harders F, Hakze‐van der Honing R, Wegdam‐Blans MCA, Bouwstra RJ, GeurtsvanKessel C, van der Eijk AA, Velkers FC, Smit LAM, Stegeman A, van der Poel WHM and Koopmans MPG, 2021. Transmission of SARS‐CoV‐2 on mink farms between humans and mink and back to humans. Science, 371, 172–177. 10.1126/science.abe5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu D, Pomeroy MA, Lewis NM, Wadhwa A, Yousaf AR, Whitaker B, Dietrich E, Hall AJ, Chu V and Thornburg N, 2021. Persistent SARS‐CoV‐2 RNA shedding without evidence of infectiousness: a cohort study of individuals with COVID‐19. The Journal of Infectious Diseases, 224, 1362–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA and Brennan SE, 2021. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Systematic Reviews, 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo PM, Orbegozo J, Watts DM and Morrill JC, 2022. SARS‐CoV‐2 neutralizing antibodies in white‐tailed deer from Texas. Vector Borne and Zoonotic Diseases, 22, 62–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer MV, Martins M, Falkenberg S, Buckley A, Caserta LC, Mitchell PK, Cassmann ED, Rollins A, Zylich NC, Renshaw RW, Guarino C, Wagner B, Lager K and Diel DG, 2021. Susceptibility of white‐tailed deer (odocoileus virginianus) to SARS‐CoV‐2. Journal of Virology, 95, e00083–e00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T, Chen R, He X, Yuan Y, Deng X, Li R, Yan H, Yan S, Liu J, Zhang Y, Zhang X, Yu F, Zhou M, Ke C, Ma X and Zhang H, 2021. Infection of wild‐type mice by SARS‐CoV‐2 B.1.351 variant indicates a possible novel cross‐species transmission route. Signal Transduction and Targeted Therapy, 6, 420. 10.1038/s41392-021-00848-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel DR, Field CJ, Septer KM, Sim DG, Jones MJ, Heinly TA, Vanderford TH, McGraw EA and Sutton TC, 2021. Transmission and protection against reinfection in the ferret model with the SARS‐CoV‐2 USA‐WA1/2020 reference isolate. Journal of Virology, 95, e02232‐02220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson EI, Elia G, Grassi A, Giordano A, Desario C, Medardo M, Smith SL, Anderson ER, Prince T, Patterson GT, Lorusso E, Lucente MS, Lanave G, Lauzi S, Bonfanti U, Stranieri A, Martella V, Solari Basano F, Barrs VR, Radford AD, Agrimi U, Hughes GL, Paltrinieri S and Decaro N, 2020. Evidence of exposure to SARS‐CoV‐2 in cats and dogs from households in Italy. Nature Communications, 11, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, Kugathasan R, Penn R, Brown JC and Sanchez‐David RY, 2021. The furin cleavage site in the SARS‐CoV‐2 spike protein is required for transmission in ferrets. Nature Microbiology, 6, 899–909. [DOI] [PubMed] [Google Scholar]
- Pickering B, Lung O, Maguire F, Kruczkiewicz P, Kotwa JD, Buchanan T, Gagnier M, Guthrie JL, Jardine CM and Marchand‐Austin A, 2022a. Highly divergent white‐tailed deer SARS‐CoV‐2 with potential deer‐to‐human transmission. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering B, Lung O, Maguire F, Kruczkiewicz P, Kotwa JD, Buchanan T, Gagnier M, Guthrie JL, Jardine CM, Marchand‐Austin A, Massé A, McClinchey H, Nirmalarajah K, Aftanas P, Blais‐Savoie J, Chee H‐Y, Chien E, Yim W, Banete A, Griffin BD, Yip L, Goolia M, Suderman M, Pinette M, Smith G, Sullivan D, Rudar J, Vernygora O, Adey E, Nebroski M, Goyette G, Finzi A, Laroche G, Ariana A, Vahkal B, Côté M, McGeer AJ, Nituch L, Mubareka S and Bowman J, 2022b. Divergent SARS‐CoV‐2 variant emerges in white‐tailed deer with deer‐to‐human transmission. Nature Microbiology, 7, 2011–2024. 10.1038/s41564-022-01268-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering BS, Smith G, Pinette MM, Embury‐Hyatt C, Moffat E, Marszal P and Lewis CE, 2021. Susceptibility of domestic swine to experimental infection with severe acute respiratory syndrome coronavirus 2. Emerging Infectious Diseases, 27, 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piewbang C, Poonsin P, Lohavicharn P, Wardhani SW, Dankaona W, Puenpa J, Poovorawan Y and Techangamsuwan S, 2022. SARS‐CoV‐2 transmission from human to pet and suspected transmission from pet to human, Thailand. Journal of Clinical Microbiology, 60, e0105822. 10.1128/jcm.01058-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomorska‐Mól M, Turlewicz‐Podbielska H, Gogulski M, Ruszkowski JJ, Kubiak M, Kuriga A, Barket P and Postrzech M, 2021. A cross‐sectional retrospective study of SARS‐CoV‐2 seroprevalence in domestic cats, dogs and rabbits in Poland. BMC Veterinary Research, 17, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port JR, Yinda CK, Owusu IO, Holbrook M, Fischer R, Bushmaker T, Avanzato VA, Schulz JE, Martens C and van Doremalen N, 2021. SARS‐CoV‐2 disease severity and transmission efficiency is increased for airborne compared to fomite exposure in Syrian hamsters. Nature Communications, 12, 1–15. 10.1038/s41467-021-25156-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter S, Hartwig A, Bielefeldt‐Ohmann H, Bosco‐Lauth A and Root JJ, 2022. Susceptibility of wild canids to SARS‐CoV‐2. Emerging Infectious Disease, 28, 1852–1855. 10.3201/eid2809.220223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunas O, Warren JL, Crawford FW, Gazit S, Patalon T, Weinberger DM and Pitzer VE, 2022. Vaccination with BNT162b2 reduces transmission of SARS‐CoV‐2 to household contacts in Israel. Science, 375, 1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhach O, Adea K, Hulo N, Sattonnet P, Genecand C, Iten A, Jacquérioz F, Kaiser L, Vetter P, Eckerle I and Meyer B, 2022. Infectious viral load in unvaccinated and vaccinated individuals infected with ancestral, delta or omicron SARS‐CoV‐2. Nature Medicine, 28, 1491–1500. 10.1038/s41591-022-01816-0 [DOI] [PubMed] [Google Scholar]
- Rabalski L, Kosinski M, Smura T, Aaltonen K, Kant R, Sironen T, Szewczyk B and Grzybek M, 2021. Severe acute respiratory syndrome coronavirus 2 in farmed mink (neovison vison), Poland. Emerging Infectious Diseases, 27, 2333–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabalski L, Kosinski M, Mazur‐Panasiuk N, Szewczyk B, Bienkowska‐Szewczyk K, Kant R, Sironen T, Pyrc K and Grzybek M, 2022. Zoonotic spill‐over of SARS‐CoV‐2: mink‐adapted virus in humans. Clinical Microbiology and Infection, 28, 451.e451–451.e454. 10.1016/j.cmi.2021.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Račnik J, Kočevar A, Slavec B, Korva M, Rus KR, Zakotnik S, Zorec TM, Poljak M, Matko M, Rojs OZ and Županc TA, 2021. Transmission of SARS‐CoV‐2 from human to domestic ferret. Emerging Infectious Diseases, 27, 2450–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TB, Fonager J, Jørgensen CS, Lassaunière R, Hammer AS, Quaade ML, Boklund A, Lohse L, Strandbygaard B, Rasmussen M, Michaelsen TY, Mortensen S, Fomsgaard A, Belsham GJ and Bøtner A, 2021. Infection, recovery and re‐infection of farmed mink with SARS‐CoV‐2. PLoS Pathogens, 17, e1010068. 10.1371/journal.ppat.1010068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev‐Yochay G, Gonen T, Gilboa M, Mandelboim M, Indenbaum V, Amit S, Meltzer L, Asraf K, Cohen C and Fluss R, 2022. Efficacy of a fourth dose of COVID‐19 mRNA vaccine against omicron. New England Journal of Medicine, 386, 1377–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M, Kok A, de Meulder D, Bestebroer TM, Lamers MM, Okba NMA, Fentener van Vlissingen M, Rockx B, Haagmans BL, Koopmans MPG, Fouchier RAM and Herfst S, 2020. SARS‐CoV‐2 is transmitted via contact and via the air between ferrets. Nature Communications, 11, 1–6. 10.1038/s41467-020-17367-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Oude Munnink BB, de Meulder D, van Amerongen G, van den Brand J, Okba NMA, Schipper D, van Run P, Leijten L, Sikkema R, Verschoor E, Verstrepen B, Bogers W, Langermans J, Drosten C, Fentener van Vlissingen M, Fouchier R, de Swart R, Koopmans M and Haagmans BL, 2020. Comparative pathogenesis of COVID‐19, MERS, and SARS in a nonhuman primate model. Science, 368, 1012–1015. 10.1126/science.abb7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Arrondo I, Portillo A, Palomar AM, Santibáñez S, Santibáñez P, Cervera C and Oteo JA, 2021. Detection of SARS‐CoV‐2 in pets living with COVID‐19 owners diagnosed during the COVID‐19 lockdown in Spain: a case of an asymptomatic cat with SARS‐CoV‐2 in Europe. Transboundary and Emerging Diseases, 68, 973–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KA, Bewley KR, Fotheringham SA, Slack GS, Brown P, Hall Y, Wand NI, Marriott AC, Cavell BE and Tree JA, 2021. Dose‐dependent response to infection with SARS‐CoV‐2 in the ferret model and evidence of protective immunity. Nature Communications, 12, 1–13. 10.1038/s41467-020-20439-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetto L, Chaves BA, Costa ER, de Menezes Medeiros AS, Gordo M, Araújo DB, Oliveira DBL, da Silva APB, Negri AF, Durigon EL, Hanley KA, Vasilakis N, de Lacerda MVG and Nogueira ML, 2021. Lack of evidence of severe acute respiratory syndrome coronavirus 2 (Sars‐cov‐2) spillover in free‐living neotropical non‐human primates, brazil. Viruses, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo J, Hägg M, Kortelainen M, Leino T, Saxell T, Siikanen M and Sääksvuori L, 2022. The indirect effect of mRNA‐based COVID‐19 vaccination on healthcare workers' unvaccinated household members. Nature Communications, 13, 1–7. 10.1038/s41467-022-28825-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, Höper D, Mettenleiter TC, Balkema‐Buschmann A, Harder T, Grund C, Hoffmann D, Breithaupt A and Beer M, 2020. SARS‐CoV‐2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. The Lancet Microbe, 1, e218–e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, Höper D, Mettenleiter TC, Balkema‐Buschmann A, Harder T, Grund C, Hoffmann D, Breithaupt A and Beer M, 2020. SARS‐CoV‐2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. The Lancet Microbe, 1, e218–e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C, Yao Y‐F, Yang X‐L, Zhou Y‐W, Gao G, Peng Y, Yang L, Hu X, Xiong J, Jiang R‐D, Zhang H‐J, Gao X‐X, Peng C, Min J, Chen Y, Si H‐R, Wu J, Zhou P, Wang Y‐Y, Wei H‐P, Pang W, Hu Z‐F, Lv L‐B, Zheng Y‐T, Shi Z‐L and Yuan Z‐M, 2020. Infection with novel coronavirus (SARS‐CoV‐2) causes pneumonia in Rhesus macaques. Cell Research, 30, 670–677. 10.1038/s41422-020-0364-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharun K, Tiwari R, Natesan S and Dhama K, 2021. SARS‐CoV‐2 infection in farmed minks, associated zoonotic concerns, and importance of the One Health approach during the ongoing COVID‐19 pandemic. Veterinary Quarterly, 41, 50–60. 10.1080/01652176.2020.1867776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liu P, Liang L, Cui P, Wang J, Zhang X, Guan Y, Tan W, Wu G, Chen H, Bu Z and Bu Z, 2020. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS‐coronavirus 2. Science, 368, 1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner SA, Ellis JW, Root JJ, Roug A, Stopak SR, Wiscomb GW, Zierenberg JR, Ip HS, Torchetti MK and DeLiberto TJ, 2021. SARS‐CoV‐2 exposure in escaped mink, Utah, USA. Emerging Infectious Diseases, 27, 988–990. 10.3201/eid2703.204444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai L, Zhong G, Yuan Q, Wen Z, Wang C, He X, Liu R, Wang J, Zhao Q, Liu Y, Huo N, Deng J, Bai J, Wu H, Guan Y, Shi J, Tian K, Xia N, Chen H and Bu Z, 2021. Replication, pathogenicity, and transmission of SARS‐CoV‐2 in minks. National Science Review, 8, 1–8. 10.1093/nsr/nwaa291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia SF, Yan L‐M, Chin AWH, Fung K, Choy K‐T, Wong AYL, Kaewpreedee P, Perera RAPM, Poon LLM, Nicholls JM, Peiris M and Yen H‐L, 2020. Pathogenesis and transmission of SARS‐CoV‐2 in golden hamsters. Nature, 583, 834–838. 10.1038/s41586-020-2342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikkema RS, Tobias T, Oreshkova N, de Bruin E, Okba N, Chandler F, Hulst MM, Rodon J, Houben M, van Maanen K, Bultman H, Meester M, Gerhards NM, Bouwknegt M, Urlings B, Haagmans B, Kluytmans J, GeurtsvanKessel CH, van der Poel WHM, Koopmans MPG and Stegeman A, 2022. Experimental and field investigations of exposure, replication and transmission of SARS‐CoV‐2 in pigs in the Netherlands. Emerging Microbes and Infections, 11, 91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sila T, Sunghan J, Laochareonsuk W, Surasombatpattana S, Kongkamol C, Ingviya T, Siripaitoon P, Kositpantawong N, Kanchanasuwan S and Hortiwakul T, 2022. Suspected cat‐to‐human transmission of SARS‐CoV‐2, Thailand, July–September 2021. Emerging Infectious Diseases, 28, 1485–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singanayagam A, Hakki S, Dunning J, Madon KJ, Crone MA, Koycheva A, Derqui‐Fernandez N, Barnett JL, Whitfield MG and Varro R, 2022. Community transmission and viral load kinetics of the SARS‐CoV‐2 delta (B. 1.617. 2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. The Lancet Infectious Diseases, 22, 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Singh B, Ganatra SR, Gazi M, Cole J, Thippeshappa R, Alfson KJ, Clemmons E, Gonzalez O, Escobedo R, Lee T‐H, Chatterjee A, Goez‐Gazi Y, Sharan R, Gough M, Alvarez C, Blakley A, Ferdin J, Bartley C, Staples H, Parodi L, Callery J, Mannino A, Klaffke B, Escareno P, Platt RN, Hodara V, Scordo J, Gautam S, Vilanova AG, Olmo‐Fontanez A, Schami A, Oyejide A, Ajithdoss DK, Copin R, Baum A, Kyratsous C, Alvarez X, Ahmed M, Rosa B, Goodroe A, Dutton J, Hall‐Ursone S, Frost PA, Voges AK, Ross CN, Sayers K, Chen C, Hallam C, Khader SA, Mitreva M, Anderson TJC, Martinez‐Sobrido L, Patterson JL, Turner J, Torrelles JB, Dick EJ, Brasky K, Schlesinger LS, Giavedoni LD, Carrion R and Kaushal D, 2021. Responses to acute infection with SARS‐CoV‐2 in the lungs of rhesus macaques, baboons and marmosets. Nature Microbiology, 6, 73–86. 10.1038/s41564-020-00841-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spada E, Vitale F, Bruno F, Castelli G, Reale S, Perego R, Baggiani L and Proverbio D, 2021. A pre‐and during pandemic survey of sars‐cov‐2 infection in stray colony and shelter cats from a high endemic area of Northern Italy. Viruses, 13, 1–11. 10.3390/v13040618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ssentongo P, Ssentongo AE, Voleti N, Groff D, Sun A, Ba DM, Nunez J, Parent LJ, Chinchilli VM and Paules CI, 2022. SARS‐CoV‐2 vaccine effectiveness against infection, symptomatic and severe COVID‐19: a systematic review and meta‐analysis. BMC Infectious Diseases, 22, 1–12. 10.1186/s12879-022-07418-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranieri A, Lauzi S, Giordano A, Galimberti L, Ratti G, Decaro N, Brioschi F, Lelli D, Gabba S and Amarachi NL, 2022. Absence of SARS‐CoV‐2 RNA and anti‐SARS‐CoV‐2 antibodies in stray cats. Transboundary and Emerging Diseases, 69, 2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CCS, Lam SD, Richard D, Owen CJ, Berchtold D, Orengo C, Nair MS, Kuchipudi SV, Kapur V, van Dorp L and Balloux F, 2022. Transmission of SARS‐CoV‐2 from humans to animals and potential host adaptation. Nature Communications, 13, 2988. 10.1038/s41467-022-30698-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan ST, Kwan AT, Rodríguez‐Barraquer I, Singer BJ, Park HJ, Lewnard JA, Sears D and Lo NC, 2023. Infectiousness of SARS‐CoV‐2 breakthrough infections and reinfections during the Omicron wave. Nature Medicine, 1–8. 10.1038/s41591-022-02138-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimpert J, Vladimirova D, Dietert K, Abdelgawad A, Kunec D, Dökel S, Voss A, Gruber AD, Bertzbach LD and Osterrieder N, 2020. The Roborovski dwarf hamster is a highly susceptible model for a rapid and fatal course of SARS‐CoV‐2 infection. Cell Reports, 33, 108488. 10.1016/j.celrep.2020.108488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich L, Wernike K, Hoffmann D, Mettenleiter TC and Beer M, 2020. Experimental Infection of Cattle with SARS‐CoV‐2. Emerging Infectious Diseases, 26, 2979–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich L, Michelitsch A, Halwe N, Wernike K, Hoffmann D and Beer M, 2021. Experimental SARS‐CoV‐2 infection of bank voles. Emerging Infectious Diseases, 27, 1193–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Aart AE, Velkers FC, Fischer EAJ, Broens EM, Egberink H, Zhao S, Engelsma M, Hakze‐van der Honing RW, Harders F, de Rooij MMT, Radstake C, Meijer PA, Oude Munnink BB, de Rond J, Sikkema RS, van der Spek AN, Spierenburg M, Wolters WJ, Molenaar RJ, Koopmans MPG, van der Poel WHM, Stegeman A and Smit LAM, 2021. SARS‐CoV‐2 infection in cats and dogs in infected mink farms. Transboundary and Emerging Diseases, 69, 3001–3007. 10.1111/tbed.14173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandegrift KJ, Yon M, Surendran‐Nair M, Gontu A, Amirthalingam S, Nissly RH, Levine N, Stuber T, DeNicola AJ, Boulanger JR, Kotschwar N, Aucoin SG, Simon R, Toal K, Olsen RJ, Davis JJ, Bold D, Gaudreault NN, Richt JA, Musser JM, Hudson PJ, Kapur V and Kuchipudi SV, 2022. Detection of SARS‐CoV‐2 Omicron variant (B.1.1.529) infection of white‐tailed deer. bioRxiv, 2022.2002.2004.479189. 10.1101/2022.02.04.479189 [DOI] [Google Scholar]
- Villanueva‐Saz S, Giner J, Fernández A, Lacasta D, Ortín A, Ramos JJ, Ferrer LM, de Arcaute MR, Tobajas AP, Pérez MD, Verde M, Marteles D, Hurtado‐Guerrero R, Pardo J, Santiago L, González‐Ramírez AM, Macías‐León J, García‐García A, Taleb V, Lira‐Navarrete E, Paño‐Pardo JR and Ruíz H, 2021a. Absence of sars‐cov‐2 antibodies in natural environment exposure in sheep in close contact with humans. Animals, 11, 1984. 10.3390/ani11071984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva‐Saz S, Giner J, Tobajas AP, Pérez MD, González‐Ramírez AM, Macías‐León J, González A, Verde M, Yzuel A, Hurtado‐Guerrero R, Pardo J, Santiago L, Paño‐Pardo JR, Ruíz H, Lacasta DM, Sánchez L, Marteles D, Gracia AP and Fernández A, 2021b. Serological evidence of SARS‐CoV‐2 and co‐infections in stray cats in Spain. Transboundary and Emerging Diseases, 69, 1056–1064. 10.1111/tbed.14062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva‐Saz S, Giner J, Palomar AM, Gómez MA, Põdra M, Aranda MC, Jiménez MÁ, Lizarraga P, Hernández R and Portillo A, 2022. No Evidence of SARS‐CoV‐2 Infection in Wild Mink (Mustela lutreola and Neogale vison) from Northern Spain during the First Two Years of Pandemic. Animals, 12, 1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen J, Aaltonen K, Kegler K, Venkat V, Niamsap T, Kareinen L, Malmgren R, Kivelä O, Atanasova N, Österlund P, Smura T, Sukura A, Strandin T, Dutra L, Vapalahti O, Nordgren H, Kant R and Sironen T, 2022. Experimental infection of mink with SARS‐COV‐2 Omicron variant and subsequent clinical disease. Emerging Infectious Disease journal, 28, 1286. 10.3201/eid2806.220328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Didelot X, Bi Y and Gao GF, 2021. Assessing the extent of community spread caused by mink‐derived SARS‐CoV‐2 variants. The Innovation, 2, 100128. 10.1016/j.xinn.2021.100128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernike K, Böttcher J, Amelung S, Albrecht K, Gärtner T, Donat K and Beer M, 2022. Serological screening suggests single SARS‐CoV‐2 spillover events to cattle. bioRxiv, 2022.2001.2017.476608. 10.1101/2022.01.17.476608 [DOI] [Google Scholar]
- WHO‐FAO‐WOAH (Food and Agriculture Organization (FAO), World Organisation for Animal Health (WOAH) and World Health Organization (WHO)) , 2022. Joint statement on the prioritization of monitoring SARS‐CoV‐2 infection in wildlife and preventing the formation of animal reservoirs. Available online: https://www.who.int/news/item/07-03-2022-joint-statement-on-the-prioritization-of-monitoring-sars-cov-2-infection-in-wildlife-and-preventing-the-formation-of-animal-reservoirs
- Woolsey C, Borisevich V, Prasad AN, Agans KN, Deer DJ, Dobias NS, Heymann JC, Foster SL, Levine CB, Medina L, Melody K, Geisbert JB, Fenton KA, Geisbert TW and Cross RW, 2021. Establishment of an African green monkey model for COVID‐19 and protection against re‐infection. Nature Immunology, 22, 86–98. 10.1038/s41590-020-00835-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Yu D‐D, Ma Y‐H, Yao Y‐L, Luo R‐H, Feng X‐L, Cai H‐R, Han J‐B, Wang X‐H and Li M‐H, 2020. COVID‐19‐like symptoms observed in Chinese tree shrews infected with SARS‐CoV‐2. Zoological Research, 41, 517–526. 10.24272/j.issn.2095-8137.2020.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav PD, Kumar S, Agarwal K, Jain M, Patil DR, Maithal K, Mathapati B, Giri S, Mohandas S, Shete A, Sapkal G, Patil DY, Dey A, Chandra H, Deshpande G, Gupta N, Nyayanit D, Kaushal H, Sahay R, Tripathy A, Jain R, Kumar A, Sarkale P, Baradkar S, Rajanathan C, Raju HP, Patel S, Shah N, Dwivedi P, Singh D and Abraham P, 2021. Assessment of immunogenicity and protective efficacy of ZyCoV‐D DNA vaccine candidates in Rhesus macaques against SARS‐CoV‐2 infection. bioRxiv, 2021.2002.2002.429480. 10.1101/2021.02.02.429480 [DOI] [Google Scholar]
- Yen H‐L, Sit TH, Brackman CJ, Chuk SS, Gu H, Tam KW, Law PY, Leung GM, Peiris M and Poon LL, 2022. Transmission of SARS‐CoV‐2 delta variant (AY. 127) from pet hamsters to humans, leading to onward human‐to‐human transmission: a case study. The Lancet, 399, 1070–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng B, Gao L, Zhou Q, Yu K and Sun F, 2022. Effectiveness of COVID‐19 vaccines against SARS‐CoV‐2 variants of concern: a systematic review and meta‐analysis. BMC Medicine, 20, 1–15. 10.1186/s12916-022-02397-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang J, Kuang D, Xu J, Yang M, Ma C, Zhao S, Li J, Long H, Ding K, Gao J, Liu J, Wang H, Li H, Yang Y, Yu W, Yang J, Zheng Y, Wu D, Lu S, Liu H and Peng X, 2020. Susceptibility of tree shrew to SARS‐CoV‐2 infection. Scientific Reports, 10, 16007. 10.1038/s41598-020-72563-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Schuurman N, Li W, Wang C, Smit LAM, Broens EM, Wagenaar JA, van Kuppeveld FJM, Bosch BJ and Egberink H, 2021. Serologic screening of severe acute respiratory syndrome coronavirus 2 infection in cats and dogs during first coronavirus disease wave, the Netherlands. Emerging Infectious Diseases, 27, 1362–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Liu S and Zhang D, 2022. Effectiveness of COVID‐19 vaccine booster shot compared with non‐booster: a meta‐analysis. Vaccines, 10, 1–9. 10.3390/vaccines10091396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Huang D, Jiang Q, Guo Y and Chen C, 2022. The vaccine efficacy against the SARS‐CoV‐2 omicron: a systemic review and meta‐analysis. Frontiers in Public Health, 10, 940956. 10.3389/fpubh.2022.940956 [DOI] [PMC free article] [PubMed] [Google Scholar]


