Abstract
The endoplasmic reticulum (ER) and microtubule (MT) network form extensive contact with each other and their interconnection plays a pivotal role in ER maintenance and distribution as well as MT stability. The ER participates in a variety of biological processes including protein folding and processing, lipid biosynthesis, and Ca2+ storage. MTs specifically regulate cellular architecture, provide routes for transport of molecules or organelles, and mediate signaling events. The ER morphology and dynamics are regulated by a class of ER shaping proteins, which also provide the physical contact structure for linking of ER and MT. In addition to these ER-localized and MT-binding proteins, specific motor proteins and adaptor-linking proteins also mediate bidirectional communication between the two structures. In this review, we summarize the current understanding of the structure and function of ER-MT interconnection. We further highlight the morphologic factors which coordinate the ER-MT network and maintain the normal physiological function of neurons, with their defect causing neurodegenerative diseases such as Hereditary Spastic Paraplegia (HSP). These findings promote our understanding of the pathogenesis of HSP and provide important therapeutic targets for treatment of these diseases.
Keywords: Endoplasmic Reticulum, ER-MT, ER membrane protein, ER shaping proteins, Hereditary Spastic Paraplegia, HSP, Microtubule dynamics
1. Introduction
Eukaryotic cells are compartmentalized into different organelles in order to perform complex cellular functions. The endoplasmic reticulum (ER) is one of the largest and most complicated organelles, and is composed of several subdomains including perinuclear membrane sheets as well as tubule-like structures that extend from the nuclear envelope all the way to the cell membrane [1]. This ER tubule network is linked to several other organelles and participates in numerous biological processes. Studies have consistently found that the ER forms contacts with other organelles, including mitochondria, Golgi apparatus, and endosomes [2]. Along with many other organelles, the ER is closely associated with the microtubule (MT) network [3], [4]. ER tubules transport along MTs bidirectionally by binding to the positive end of an MT or sliding along the MT using molecular motor proteins [5]. Nocodazole-induced MT depolymerization results in the retraction of peripheral ER tubules and their interconversion to perinuclear ER cisternae [3]. MTs therefore play a pivotal role in ER morphogenesis. Numerous studies have identified a number of ER-localized proteins directly interacting with MTs, including the ER membrane protein CLIMP-63 [4], and p180 [6], receptor expression-enhancing protein 1 (REEP1) [7], and Sec61β [8], all of which contain MT-binding domains. These findings highlight the physiological importance of the ER-MT association.
Hereditary spastic paraplegias (HSPs) are a diverse group of heritable neurological disorders, which are clinically characterized by spastic and weakness in lower-extremities, and pathologically by retrograde axonal degeneration. To date, no etiological treatments for prevention or reversal of motor neuron degeneration in HSP have been documented [9]. Symptomatic treatments via physical or occupational therapy and drugs have improved muscle spasticity only temporarily and improved balance, strength, and agility. Over 80 genetic forms of HSP have been defined by genetic linkage analyses [10]. Gene functional studies have led to several hypotheses regarding the pathogenesis of HSP including ER morphology defect, axonal transport dysfunction, and axonal myelin abnormality [11]. More than half of HSP cases result from autosomal dominant mutations in ATL1, REEP1, SPAST, and RTNs, all of which are either ER-localized or MT-binding proteins. These results indicate that dysfunctional ER-MT interaction may be a major pathogenesis factor of HSP [10], [12].
In this review, we focus on the interconnection between the ER and MTs as well as the linking molecules responsible for ER distribution and integrity and their role in maintenance of ER position and MT stability. We provide an overview of three kinds of molecules which mediate ER-MT connections, which work together to sustain a normal ER distribution, particularly for axon development and maintenance. We further highlight how impaired maintenance of axonal ER-MT interaction, local ER stress, MT instability, and protein synthesis failure act as prominent contributors to the pathogenesis of HSP. In conclusion, we provide perspectives on potential therapeutic strategies targeting ER-MT interaction.
2. ER and MT contacts
2.1. Structure and function of the ER and MTs
The ER network, including the nuclear envelope, as well as a connected peripheral network of tubules and interspersed sheets is highly dynamic, with tubules continuously forming and retracting [13]. Network formation and maintenance involves curvature stabilizing proteins such as RTNs, and proteins participating in ER remodeling, including atlastins and Lunapark (LNP) [14]. RTNs and REEPs generate curves of membrane [15], [16]. Atlastins tether and fuse the ER tubules together to form three-way junctions, which are thereafter stabilized by LNP [17]. Atlastins and RTNs counterbalance one another dynamically to regulate ER fragmentation and fusion [18]. The integrity of ER is essential for synthesis of transmembrane and secreted protein, as well as folding and processing of proteins, and finally plays a role in lipid synthesis and Ca2+ homeostasis [19], [20].
MTs cooperate with actin filaments and intermediate filaments to construct the cytoskeletal system, provide routes for transport of molecules or organelles, and maintain cell shape, division, and polarity [21], [22], [23], [24]. MTs are composed of α- and β-tubulin, which are heterodimeric and noncovalently join to form a stable hollow tubular structure [25]. MTs are initially nucleated from MT organizing centers that contain γ-tubulin, which serves as a structural template for high-efficiency nucleation [26], [27]. Tubulin dimers hydrolyze GTP to GDP, facilitating addition to the growing MT lattice, which weakens the affinity for adjacent dimers and favors disassembly. The GDP attached to β-tubulin which is incorporated into an MT can only be replaced by GTP when tubulin returns to its heterodimeric form during the continuous cycling of MT polymerization and depolymerization [28]. Both extrinsic [MT-associated proteins (MAPs) and physical barriers] and intrinsic [tubulin concentration, isoforms, and posttranslational modifications (PTMs)] factors control MT dynamics, enabling both global and local tuning of MT growth and shrinkage. MTs are modified with distinct PTMs, including acetylation, tyrosination, and glutamylation, and some of these PTMs influence the binding affinity and/or activity of several MAPs [29], [30].
2.2. Molecules involved in ER-MT connections
The ER network forms abundant membrane contacts with other organelles, such as mitochondria, Golgi apparatus, and endosomes [2]. Electron microscopy in the 1970 s and 1980 s revealed close connections between the ER and MT. Further, live-cell fluorescence microscopy has been used to visualize the dynamic changes taking place across ER and MT interactions. In the lamellipodia of thinly spread cells, Mark Terasaki and his colleagues observed that MTs and ER tubules have similar distributions, however, this is not observed with respect to intermediate filaments [3]. Other evidence of ER-MT connections are evident in the tip of ER tubule being linked to the tip of a MT, and the fact that ER distribution and retraction occurs concurrently with MTs [31], [32]. During this activity, the interactions mediating movement of ER along MT are linked through motor proteins or other linking proteins. In recent years, a growing body of evidence has suggested that the ER is closely associated with MTs [33], [34], [35]. It was found in 1998 that p63, an integral membrane protein within the reticular subdomain of the rough ER, directly binds MTs both in vivo and in vitro, providing evidence of ER-MT connections at the molecular level. Functional domain analysis of p63 revealed an ER rearrangement domain near the N-terminus and a central MT-binding domain. Due to its similar functioning to cytoskeleton-linking membrane proteins (CLIMPs), it was renamed CLIMP-63 [4]. Thereafter, more proteins with similar features were identified (this will be explored in more detail in the following section).
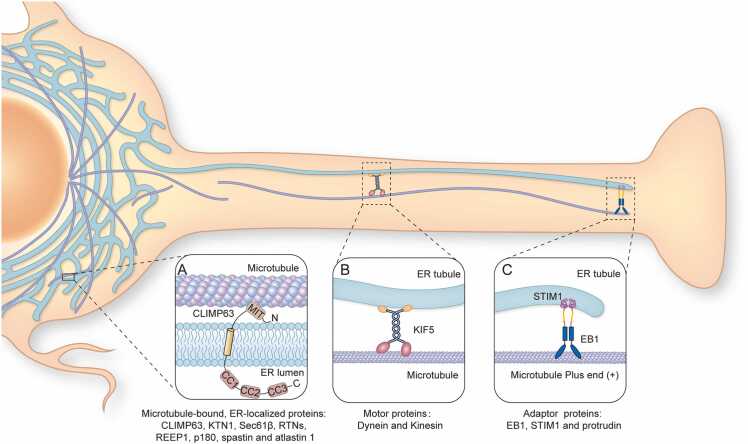
Current investigation has found that three kinds of molecules mediate ER-MT connections: microtubule-bound ER-localized proteins, motor proteins, and adaptor proteins (Fig. 1). The microtubule-bound, ER-localized proteins include CLIMP-63, p180, REEP1, and Sec61β. Mutation and functional-domain truncation experiments have demonstrated that CLIMP-63 has an ER-rearrangement domain near the N-terminus and a central MT-binding region [4] (Fig. 1A). A recent study also demonstrated that CLIMP63 is able to bind the centrosomal MT in the perinuclear region [36]. Another integral ER membrane protein, p180, contains a short luminal and single-transmembrane domain followed by a long cytosolic tail featuring a basic decapeptide repeat at the N-terminus and an acidic coiled-coil (CC) domain at the C terminus. The highly basic N-terminal region of p180 is responsible for binding to MTs [6]. REEP1 remodels the ER network by directly interacting with MTs through its C-terminal cytoplasmic domain [7].
Fig. 1.
Three kinds of molecules mediate ER-MT connections: ER-localized MT-binding proteins, motor proteins, and adaptor linking proteins. Three enlarged detail diagrams are presented separately: A) ER-localized MT-binding protein CLIMP-63 has a MT-interacting and trafficking (MIT) domain that binds MT; B) KIF5 (a Kinesin) mediates ER movement along a microtubule; C) ‘TAC’ (tip attachment complex) mediates ER moving with the elongating plus end of a microtubule, through interaction between STIM1 on the ER and EB1 on the microtubule.
The tubular ER network is dynamic and constantly rearranges its structure along the MT cytoskeleton [13]. In animal cells, these rearrangements, referred to as ER sliding, occur bidirectionally and are mediated by motor proteins [37]. The MT-associated protein families of kinesins and dyneins serve to distribute intracellular cargo along MTs, with kinesins working in the anterograde direction and dyneins working in the retrograde direction [38]. The typical protein used is Kinesin-1 (also known as KIF5) [39] (Fig. 1B). Immunofluorescence experiments using anti-Kinesin-1 antibodies illustrate punctate staining patterns which are often strongly associated with ER membranes. Suppression of kinesin heavy chain expression within cultured rat hippocampal neurons by short hairpin RNA (shRNA) results in the retraction of the ER network from the periphery into the center of the cell, without impacting the distribution of MTs [40]. Furthermore, a variety of tubulin isoforms, posttranslational modifications, and MT-associated proteins create a tubulin code and provide a precisely regulated mechanism of organelles and molecules transported in an orderly fashion along MTs [36], [41]. For example, KIF5 binds and moves along MTs marked with acetylated tubulin. Acetylated microtubules are predominantly bundled, which allows for this movement. This bundling enhances kinesin run lengths and provides a greater number of available kinesin binding sites [42].
The tip attachment complex (TAC) is important for the linkage of the ER and MTs; the tip of the ER tubule is bound to the tip of a dynamic MT, and the ER tubule grows and shrinks in concert with the dynamics of the plus-end of the MT [43], [44]. The integral ER membrane protein STIM1 mediates TAC formation, however it lacks a MT-binding domain. In contrast, STIM1 interacts with MT indirectly through the STIM1-MT end binding protein 1 (EB1) complex [45] (Fig. 1C). STIM-EB1 regulates ER tubule localization at the cell periphery, and the KIF5 complex transports ER tubules to the cell periphery. Additionally, a complex structure can be formed between three proteins: protrudin, a vesicle-associated membrane protein-associated protein (VAP), and the kinesin protein KIF5. VAP is present in ER where it can bind to protrudin and protrudin binds to KIF5 thereafter [46], [47]. Protrudin then facilitates the interaction of KIF5 with VAP family proteins including Rab11, Surf4, and RTNs. This suggests that protrudin serves as an adaptor protein and that the protrudin-KIF5 complex contributes greatly to the transport of proteins in neurons [47].
2.3. ER and MTs are interdependent
In vitro studies have demonstrated that the de novo formation of the ER is possible using small membrane vesicles prepared from Xenopus laevis eggs independent of MTs [48]. However, some studies have demonstrated that the interaction between the ER and MTs regulates the eventual shape of the ER. For example, nocodazole-induced MT depolymerization leads to the retraction of peripheral ER tubules and their interconversion to perinuclear ER cisternae [3]. The MT cytoskeleton helps position and support ER membranes, as well as actively participating in remodeling the ER. During nutrient starvation, cells increase CLIMP63 protein levels to shuttle ER towards the perinuclear region, thereby clustering lysosomes for efficient autophagic degradation. Thereafter, cells harness enhanced p180–MT binding to redistribute ER for a proper reset [36]. Conversely, a recent study demonstrated that MT dynamics and numbers are affected by the ER. Researchers used the Streptavidin-SBP system to produce immediate and sustained retention of ER tubules within the soma. They found that retraction of ER tubules to the perinuclear area results in a drastic increase in the number of the characteristic comets (marking MT plus-end tips) in regions of the soma lacking ER tubules, highlighting a decrease in MT stability [40].
MT diversity can be achieved via numerous post-translational modifications including acetylation, tyrosination, and glutamylation, all of which together constitute key elements of the tubulin code [41]. Modifications are dynamic and rapidly reversible. ER distribution is mediated via specific membrane-bound proteins, which bind to distinct levels and types of glutamylated MTs. For example, Kinectin 1 (KTN1) preferentially binds to perinuclear, polyglutamylated MTs with long glutamate chains, whereas p180 binds glutamylated MTs with either short or long chains. Therefore, MTs have key roles in ER distribution [36]. Cells dynamically tune ER distribution through differential MT modifications, which is certainly important functionally. Dysregulation of ER shaping and MT polyglutamylation may lead to different neurodegenerative diseases [49], [50].
Within a polarized neuron ER tubules can access the axon, while ER cisternae are excluded from the axon through retrograde transport back to the soma [40]. Tubular ER is able to recognize the tubulin code and move along MTs to establish and maintain a proper distribution and function; altered ER distribution is mediated by ER tubule-shaping proteins such as p180 alongside the MT. p180 induces MT acetylation through MTB-1, which further stabilizes p180 MTs [6]. Therefore, tubular ER selectively moves along acetylated MT, further demonstrating that ER distribution is broadly sensitive to MT modifications. Under nutrient starvation conditions, cells modulate CLIMP63 protein levels and p180–MT binding to glutamylated MT to move ER and lysosomes bidirectionally for adequate autophagic responses [36]. Nonetheless, the mechanisms underlying the dynamic regulation of ER and MT across different physiological and pathological conditions are topics of future study.
3. Physical functions of ER-MT interactions in axon development and maintenance
A neuron consists of a single axon and multiple dendrites, forming a highly polarized structure which establishes a complex network. During neuronal development, axon formation is critical for neuronal polarity establishment [51]. The neurites are indistinguishable until one of them develops into the axon, while the remaining ends develop into dendrites. Local cytoskeleton rearrangements determine the neuronal symmetry, in which a stable and uniform pool of parallel MT bundles is reorganized with their plus ends towards the axon terminus [52], [53]. The specialized axon and dendrites form separate compartments which depend on a unique set of motors and cargos to transport organelles and molecules. The preference of KIF5 for modified MTs is also important for axon specification in developing neurons, which extend multiple MT-rich neurites outward from the cell body [54]. Outside of motor proteins, MT-associated proteins have been proposed to be the major players in regulating MT remodeling within axons [55]. Nevertheless, numerous studies have indicated that specific ER membrane proteins can directly bind MTs and regulate their reorganization and stability [56]. We further summarize the mechanism of ER-MT connections regulating axon specification and development below.
The ER is organized into two distinct structural and functional interconnected domains determined by their membrane shape: ER cisternae and ER tubules [1]. In mature neurons, the two different ER shapes are asymmetrically distributed within the axonal, dendritic, and soma compartments. The ER distributes along the entire axon mainly in its tubular form [57]. ER tubules localize to the axon at very early stages of neuronal development. Alongside axon growth, the localization of the axonal ER depends on the cooperation of the MT plus-end motor protein KIF5 and various ER tubule-shaping proteins (Fig. 1B). Accordingly, different ER-shaping proteins are distributed throughout dendrites and axons. For example, Sec61β, RTN4A, DP1, and atlastin 1 are all distributed in both the axon and dendrites, whereas the ER cisternae proteins CLIMP63 and KTN1 are distributed only in the soma and dendrites [40]. Specific ER-resident transmembrane proteins control the shape of the ER. For example, CLIMP63, p180, and KTN1 induce a flattened structural organization of ER cisternae, whereas proteins such as RTNs and REEP1 promote highly curved ER [15], [58]. Therefore, ER-shaping proteins control ER cisternae and tubule-level balance and determine the ER distribution across the axon. A recent study reported a specialized ER structure called an ER ladder in axons. ER ladders are composed of rungs that wrap tightly around MT bundles and dynamic rails for sliding across MT [34].
Recent studies suggest that neuronal polarity establishment depends upon axonal ER-MT connections. Disrupting the MT cytoskeleton alters ER tubule organization, and these alterations in the ER disrupt proper MT organization. Throughout unpolarized neuron stages, recruitment of ER tubules to one neurite initiates primary axon formation, whereas ER retention in the perinuclear area or disruption of ER tubules inhibits neuronal polarization. When ER tubules reach the axon initiation site, those containing proteins, such as p180, further induce MT stabilization which fully supports axon formation [40]. Consistently, KIF5 is found more frequently in developing axons than dendrites [59]. This is likely induced by increased posttranslational modifications of axonal MTs and the preference of KIF5 to bind to these modified tracks [42]. These studies suggest that axon developmental initiation requires proper ER-MT connections.
ER-MT connections are essential for achieving neuron polarization, and importantly, for maintaining it. MTs are required for stabilization of ER tubules and their transport along the axon. Additionally, ER tubules are critically important for organizing axonal MTs [60]. A balance between ER cisterna and ER tubule-shaping proteins as well as fusion between ER membranes regulates ER organization within the mature neuron [14], [58]. Investigating the precise role of ER-MT connections in axon formation and maintenance in response to extracellular signals will be of interest to future studies.
4. The implication of ER-MT contact defects in HSP
HSP is an inherited neurodegenerative disease defined by a length-dependent axonopathy of corticospinal motor neurons, resulting in prominent lower extremity spasticity and gait difficulties [11]. Because of the remarkable genetic heterogeneity across HSPs, a genetic classification scheme has emerged and HSPs are commonly identified by their affected genes and spastic gait (SPG) loci, SPG1–80, assigned in order of locus identification [10], [61]. Over half of HSP genes result from autosomal dominant mutations in SPAST (SPG4), ATL1 (SPG3A), or REEP1 (SPG31). More importantly, mutations in SPAST cause 40% of the autosomal dominant cases of HSP. Overall, the common feature of these gene products is that they are ER-localized and MT-associated, leading us to therefore speculate that these gene mutations may disrupt ER-MT interaction [7], [62].
4.1. Disassociation of ER-MT contact is implicated in HSP pathogenesis
As ER-MT interaction is essential to axon development and maintenance, observance of HSP-associated proteins indicates developmental defects or retrograde degeneration of the long corticospinal tract axons, thus implying pathogenesis of HSP [62]. A recent study demonstrated that the HSP proteins atlastin 1 and REEP1 interact within the tubular ER membrane in corticospinal neurons to coordinate ER shaping and MT dynamics [56]. Indeed, the same ER-resident protein mutations have been reported to cause neurological disorders through significant MT disorganization mediated by spastin [63], ER stress mediated by REEP1 [64], and severe axonal degeneration mediated by RTNs, spastin, and atlastin 1 [7], [65], similar to spastic paraplegia syndrome in humans. Thus, defects in tubular ER shaping and network interactions with the MT cytoskeleton appear to be the predominant pathogenic mechanism of HSP.
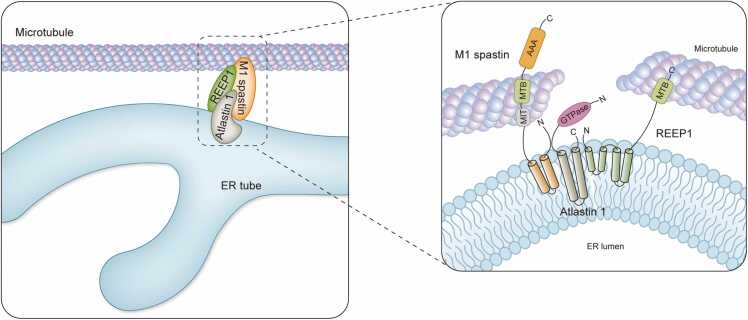
Spastin is an ATPase protein which severs MTs and plays a critical role in cytoskeleton regulation [66]. Spastin contains four functional domains: N-terminal sequence (only in M1 isoform), an ATPase domain, a MT-binding domain, and a MT-interacting and trafficking (MIT) domain [67], [68] (Fig. 2). M1 Spastin inserts itself into the ER membrane and interacts with RTNs, atlastin 1, and REEP1 to regulate ER morphogenesis [7], [69]. Functionally, spastin breaks longer MTs into shorter ones, which is critical for efficient MT transport in a rapid and concerted fashion within the axon [70]. Functional studies have demonstrated that spastin is an MT-severing enzyme. Most of its missense gene mutations are clustered within the AAA domain. In vivo these mutations lead to at least a partial loss of microtubule-severing activity [67], [71]. Outside of its function with respect to MTs, numerous studies have shown that the long M1 spastin isoform physically interacts with REEP1 and atlastin 1 to coordinate ER morphogenesis and localization along MTs, which plays a critical role in axon branching, elongation, and regeneration [7], [72]. M1 spastin is especially enriched within the spinal cord [73] and the three most prevalent SPG mutations consistently lead to HSP disease, highlighting the critical role of the integrity of ER-MT in disease pathogenesis [7].
Fig. 2.
HSP proteins spastin, atlastin 1, and REEP1 interact within the tubular ER membrane to coordinate ER shaping and MT dynamics (modified from Park et al. [7]). The three proteins (M1 spastin, atlastin 1, and REEP1) form protein complexes within the tubular ER, interacting with each other through hydrophobic hairpin domains in each of these proteins. The enlarge image shows the membrane topology of spastin, atlastin 1, and REEP1. The M1 isoform of the spastin ATPase binds to microtubules through its MIT domain and MTB domain, and is involved in microtubule severing, coupling changes in ER morphology with microtubule dynamics. Atlastin 1 interacts with spastin in their N-termini. REEP1 makes direct contact with the microtubular cytoskeleton through its C-terminal cytoplasmic domain.
ATL1 is identified as the second-most abundant mutant gene that causes an HSP [12]. ATL1 encodes the dynamin-family GTPase. Atlastin 1 interacts with spastin in their N-termini [66] (Fig. 2). A mutation of ATL1 outside of the GTPase domain causing HSP has been demonstrated functionally to prevent interaction with spastin [74], suggesting that HSP could result from a lack of normal interaction between these two proteins. The ER tubule shaping protein REEP1 also binds to MTs (Fig. 2) and interacts robustly with atlastin 1 [7]. Therefore, atlastin 1 may also disrupt the balance of ER tubules by influencing their distribution, shape, and function. Importantly, treatment with MT-targeting drugs has a demonstrated beneficial effect on axon development of human SPG3A neurons [75]. Through knockdown of SPAST or overexpression of disease mutant SPAST, animal models mimicking human HSP phenotypes in Drosophila, zebrafish, C. elegans, and mouse, our understanding of disease pathogenesis and the development of effective therapeutic drugs has been deepened. Recently, vinblastine, a MT-destabilizing drug, consistently alleviates synapse and muscle defects in the Drosophila model of ATL1 mutants [60]. It has also been shown that either SPAST knockdown or mutation significantly relieves the loss of synapses and ameliorates eclosion ratios in the Drosophila model [76]. Moreover, similar results are obtained using a mouse model. Vinblastine treatment rescues axonal swelling pathology of iPSCs from SPAST mutant patients and SPAST-knockdown hESCs derived neurons [77], [78], [79]. Studies from other authors have suggested that only mild stabilization may be beneficial [80]. However, this result is disputed [81]. Vinblastine dosage and multi-target response may contribute to the different outcomes seen in different prior publications. Taken together, these results suggest that integrity of the ER-MT interaction may be one of the predominant pathogenic mechanisms of HSP, and targeting ER-MT is a potential therapy for HSP.
4.2. ER-MT contact deficiency contributes to ER morphology and distribution
Shaping and positioning of organelles, signaling complexes, and other molecules properly within highly polarized neurons all depend on motor proteins [82]. An autosomal dominant forms, SPG10, is caused by mutations in KIF5A [83], one of the three KIF5 heavy chain proteins in mammals which act as a plus end–directed MT motor protein involved in anterograde transport of membranous organelles in nerve axons including ER [38] (Fig. 1B). Most mutations are missense changes in the motor domain with sites in the vicinity of the MT and nucleotide binding regions. For example, the N256S mutation results in substitution of a highly conserved asparagine residue in the switch II loop/helix motif of the MT binding site with a serine residue, preventing stimulation of motor ATPase activity by MT-binding [83]. In vitro studies have shown that N256S-KIF5A is unable to generate single-motor processive motion in MT gliding and bead motility assays, resulting in decreased gliding and transport velocities [84], [85]. Mutations in the KIF5A gene with SPG10 have given direct evidence for motor-based transport impairments in HSP.
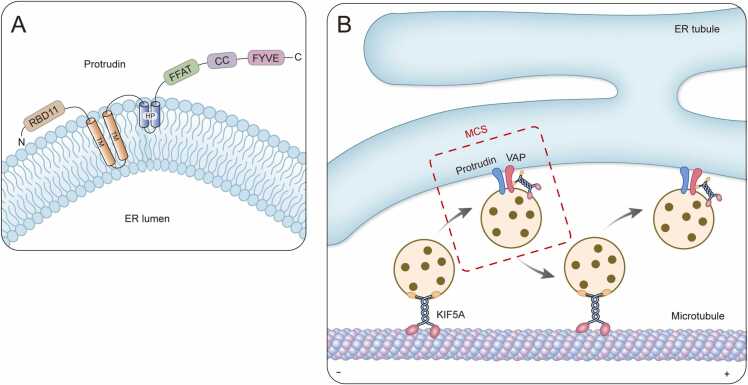
Protrudin is an ER resident membrane protein, which regulates polarized vesicular trafficking in neurons. Mutations within the protrudin gene (ZFYVE27, also known as SPG33) are found in a subset of individuals [86]. However, the sequence variant has also been identified in several normal control populations across different ethnic backgrounds. The p.G191V mutation was a benign polymorphism or variant in different ethnic backgrounds and still requires further investigation and validation [87]. Protrudin has a FFAT motif responsible for binding the ER protein VAP, a coiled-coil domain, and a FYVE domain with Zn2+-binding motifs at its C-terminal end [88] (Fig. 3A). Protrudin has a hydrophobic HP domain that shapes high-curvature ER tubules. Mutations in this domain give rise to abnormal ER morphology and increased susceptibility to ER stress [89]. This may be a major contributor to HSP pathogenesis. Animal and cell culture studies have demonstrated that protrudin plays a central role in directional endosomal trafficking through its function as a tethering factor at membrane contact sites (MCSs). At MCSs formed between the ER and endosomes, protrudin promotes the transfer of endosomes from the ER to MTs for their polarized transport [46] (Fig. 3B). VAP residing in the ER binds to the consistent FFAT (two phenylalanines in an acidic tract) motif of protrudin. Protrudin is then able to bind to KIF5 through its FFAT motif and coiled-coil domain, serving as an adaptor protein, which links the motor protein KIF5 and its cargo molecules including Rab11, VAP family members, and Surf4. The protrudin-KIF5 complex contributes to the transport of these proteins in neurons and is essential for neurite elongation [47]. Research has demonstrated that protrudin contributes to the regulation of ER morphology and function through interactions with other HSP-related proteins including spastin, PLP1, atlastin 1, REEP1, REEP5, KIF5A, KIF5B, KIF5C, and RTNs [89], [90]. VAP is an important determinant of the subcellular localization of protrudin within the ER. Given that mutations in genes for ZFYVE27, KIF5A, and RTNs give rise to HSP, protrudin-containing complexes appear to be fundamental to neuronal function associated with HSP pathogenesis [47], [89].
Fig. 3.
Protrudin regulates KIF5-dependent endosome trafficking along microtubules at MCSs. A) Membrane topology of protrudin. Mutations in the HP domain of protrudin give rise to ER stress in neurons and eventual axonopathy. B) At MCSs formed between the ER and endosomes, protrudin promotes the transfer of endosomes from the ER to MTs for their polarized transport. Protrudin and VAP tether the endosome to the ER, and then charge with KIF5. The KIF5 binding endosome is released to the microtubule for KIF5-mediated transport toward the plus-end of the MT. Protrudin plays a vital role in directional endosomal trafficking through its function as a tethering factor at MCSs.
4.3. ER-MT contact deficiency contributes to ER dysfunction
Aside from the role of MT-ER interacting proteins in regulating ER structure and distribution, these proteins are also involved ER functioning, including promoting ER stress resistance, Ca2+ homeostasis, and autophagic degradation of ER, which may also implicate in the pathogenesis of HSP. Knockdown of REEP1 aggravates Tau-mediated neurodegeneration. Conversely, the overexpression of REEP1 protein rescues these phenotypes through resistance to ER stress [64]. These results suggest that improving ER stress is a promising treatment strategy for HSP disease. Liang and his colleagues demonstrate that atlastin deficiency inhibits ER-autophagy and that atlastins participate downstream of the FAM134B ER-autophagy receptor. The similarities between FAM134B and ATL1 mutations in the context of clinical manifestation, suggest that mutations of MT-ER interacting proteins impair ER-autophagy, thus contributing to HSP [91]. In addition, the expression of spastin K467R, lack of spastin [92], and mutant atlastin 1 or depletion of atlastin 1 [93] reduce calcium entry capacity and thereby result in enlarged ER. Importantly, both enlarged ER and improper Ca2+ homeostasis can be rescued by the MT-destabilizing drug vinblastine [92]. It remains to be seen whether HSP is caused by deficits in MT-ER interacting proteins inducing ER dysfunction. Several mechanistic studies will hopefully shed light on the molecular basis of the disease and provide an effective therapeutic strategy.
5. Summary and outlook
Recent improvements in the ability to visualize the dynamic interaction of ER-MT within live cells have revealed ER-MT interaction domains and functions that were not fully understood even a few years ago. While we have come a long way in our understanding of factors that influence the ER-MT shape and function, it is clear that the responsibilities of ER-MT extend well beyond traditionally studied functions. The ER-MT interaction platform is extraordinarily dynamic, but information about the physical cues and molecular pathways that regulate these connections is lacking. Outside of ER morphology imbalance and intracellular trafficking defects, other molecular pathogenesis, such as mitochondrial dysfunction, abnormalities in axonal pathfinding or myelination and lipid metabolism, also play critical roles in HSP. As we know, all membrane-bound organelles in the cytoplasm (cell membranes, Golgi apparatus, vacuoles, endosomes, peroxisomes, and mitochondria) are a continuous structure. Previous studies have also demonstrated that ER-microtubule interactions specify the cellular distribution of many other organelles [36], however, it is still unclear whether the formation of ER/MT connections also requires the participation of other membrane-bound organelles. MT code diversity may provide the regulatory basis for these complex biological processes. Future work addressing these questions will greatly improve our understanding of ER-MT functions in the cell.
CRediT authorship contribution statement
Conceptualization, X.W., Y.L. and Y.Z.; Writing - original draft, X.W. and C.F.; Writing - review & editing, X.W., Y.L. and Y.Z.; Funding acquisition, Y.Z.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the Natural Science Foundation of Shanghai (No. 22ZR1442400) and the National Natural Science Foundation of China (No. 32170974) to Yan Zou.
Contributor Information
Yanfen Liu, Email: liuyf@shanghaitech.edu.cn.
Yan Zou, Email: zouyan@shanghaitech.edu.cn.
References
- 1.Friedman J.R., Voeltz G.K. The ER in 3D: a multifunctional dynamic membrane network. Trends Cell Biol. 2011;21(12):709–717. doi: 10.1016/j.tcb.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.English A.R., Voeltz G.K. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb Perspect Biol. 2013;5(4) doi: 10.1101/cshperspect.a013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terasaki M., Chen L.B., Fujiwara K. Microtubules and the endoplasmic reticulum are highly interdependent structures. J Cell Biol. 1986;103(4):1557–1568. doi: 10.1083/jcb.103.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klopfenstein D.R., Kappeler F., Hauri H.P. A novel direct interaction of endoplasmic reticulum with microtubules. EMBO J. 1998;17(21):6168–6177. doi: 10.1093/emboj/17.21.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lane J.D., Allan V.J. Microtubule-based endoplasmic reticulum motility in Xenopus laevis: activation of membrane-associated kinesin during development. Mol Biol Cell. 1999;10(6):1909–1922. doi: 10.1091/mbc.10.6.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogawa-Goto K., Tanaka K., Ueno T., Tanaka K., Kurata T., Sata T., Irie S., Munro S. p180 Is Involved in the Interaction between the Endoplasmic Reticulum and Microtubules through a Novel Microtubule-binding and Bundling Domain. Mol Biol Cell. 2007;18(10):3741–3751. doi: 10.1091/mbc.E06-12-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park S.H., Zhu P.P., Parker R.L., Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120(4):1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Y., Zhang G., Lin S., Shi J., Zhang H., Hu J. Sec61beta facilitates the maintenance of endoplasmic reticulum homeostasis by associating microtubules. Protein Cell. 2018;9(7):616–628. doi: 10.1007/s13238-017-0492-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyyazhagan A., Kuchi Bhotla H., Pappuswamy M., Orlacchio A. The puzzle of hereditary spastic paraplegia: from epidemiology to treatment. Int J Mol Sci. 2022;23(14):7665. doi: 10.3390/ijms23147665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hedera P.: Hereditary Spastic Paraplegia Overview. In: GeneReviews((R)). Edited by Adam M.P., Everman D.B., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., Amemiya A. Seattle (W.A.); 1993. [PubMed]
- 11.Fink J.K. Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol. 2013;126(3):307–328. doi: 10.1007/s00401-013-1115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCorquodale D.S., 3rd, Ozomaro U., Huang J., Montenegro G., Kushman A., Citrigno L., Price J., Speziani F., Pericak-Vance M.A., Zuchner S. Mutation screening of spastin, atlastin, and REEP1 in hereditary spastic paraplegia. Clin Genet. 2011;79(6):523–530. doi: 10.1111/j.1399-0004.2010.01501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawes C., Kiviniemi P., Kriechbaumer V. The endoplasmic reticulum: a dynamic and well-connected organelle. J Integr Plant Biol. 2015;57(1):50–62. doi: 10.1111/jipb.12297. [DOI] [PubMed] [Google Scholar]
- 14.Wang S., Tukachinsky H., Romano F.B., Rapoport T.A. Cooperation of the ER-shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. Elife. 2016;5 doi: 10.7554/eLife.18605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., Rapoport T.A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124(3):573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 16.Wang N., Clark L.D., Gao Y., Kozlov M.M., Shemesh T., Rapoport T.A. Mechanism of membrane-curvature generation by ER-tubule shaping proteins. Nat Commun. 2021;12(1):568. doi: 10.1038/s41467-020-20625-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X., He Y., Huang X., Guo Y., Li D., Hu J. Reciprocal regulation between lunapark and atlastin facilitates ER three-way junction formation. Protein Cell. 2019;10(7):510–525. doi: 10.1007/s13238-018-0595-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espadas J., Pendin D., Bocanegra R., Escalada A., Misticoni G., Trevisan T., Velasco Del Olmo A., Montagna A., Bova S., Ibarra B., et al. Dynamic constriction and fission of endoplasmic reticulum membranes by reticulon. Nat Commun. 2019;10(1):5327. doi: 10.1038/s41467-019-13327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson N., Powis K., High S. Post-translational translocation into the endoplasmic reticulum. Biochim Biophys Acta. 2013;1833(11):2403–2409. doi: 10.1016/j.bbamcr.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Schwarz D.S., Blower M.D. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci. 2015;73(1):79–94. doi: 10.1007/s00018-015-2052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yogev S., Cooper R., Fetter R., Horowitz M., Shen K. Microtubule organization determines axonal transport dynamics. Neuron. 2016;92(2):449–460. doi: 10.1016/j.neuron.2016.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terenna C.R., Makushok T., Velve-Casquillas G., Baigl D., Chen Y., Bornens M., Paoletti A., Piel M., Tran P.T. Physical mechanisms redirecting cell polarity and cell shape in fission yeast. Curr Biol. 2008;18(22):1748–1753. doi: 10.1016/j.cub.2008.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schatten G., Simerly C., Schatten H. Microtubule configurations during fertilization, mitosis, and early development in the mouse and the requirement for egg microtubule-mediated motility during mammalian fertilization. Proc Natl Acad Sci USA. 1985;82(12):4152–4156. doi: 10.1073/pnas.82.12.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallenfang M.R., Seydoux G. Polarization of the anterior-posterior axis of C. elegans is a microtubule-directed process. Nature. 2000;408(6808):89–92. doi: 10.1038/35040562. [DOI] [PubMed] [Google Scholar]
- 25.Nogales E., Wolf S.G., Downing K.H. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391(6663):199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 26.Kollman J.M., Polka J.K., Zelter A., Davis T.N., Agard D.A. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466(7308):879–882. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y., Wong M.L., Alberts B., Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378(6557):578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- 28.Vemu A., Szczesna E., Zehr E.A., Spector J.O., Grigorieff N., Deaconescu A.M., Roll-Mecak A. Severing enzymes amplify microtubule arrays through lattice GTP-tubulin incorporation. Science. 2018;361(6404):eaau1504. doi: 10.1126/science.aau1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valenstein M.L., Roll-Mecak A. Graded control of microtubule severing by tubulin glutamylation. Cell. 2016;164(5):911–921. doi: 10.1016/j.cell.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janke C., Magiera M.M. The tubulin code and its role in controlling microtubule properties and functions. Nat Rev Mol Cell Biol. 2020;21(6):307–326. doi: 10.1038/s41580-020-0214-3. [DOI] [PubMed] [Google Scholar]
- 31.Lee C., Chen L.B. Dynamic behavior of endoplasmic reticulum in living cells. Cell. 1988;54(1):37–46. doi: 10.1016/0092-8674(88)90177-8. [DOI] [PubMed] [Google Scholar]
- 32.Waterman-Storer C.M., Salmon E.D. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr Biol. 1998;8(14):798–806. doi: 10.1016/s0960-9822(98)70321-5. [DOI] [PubMed] [Google Scholar]
- 33.Tikhomirova M.S., Kadosh A., Saukko-Paavola A.J., Shemesh T., Klemm R.W. A role for endoplasmic reticulum dynamics in the cellular distribution of microtubules. Proc Natl Acad Sci USA. 2022;119(15) doi: 10.1073/pnas.2104309119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zamponi E., Meehl J.B., Voeltz G.K. The ER ladder is a unique morphological feature of developing mammalian axons. Dev Cell. 2022;57(11) doi: 10.1016/j.devcel.2022.05.002. 1369-1382 e1366. [DOI] [PubMed] [Google Scholar]
- 35.Pavez M., Thompson A.C., Arnott H.J., Mitchell C.B., D'Atri I., Don E.K., Chilton J.K., Scott E.K., Lin J.Y., Young K.M., et al. STIM1 is required for remodeling of the endoplasmic reticulum and microtubule cytoskeleton in steering growth cones. J Neurosci. 2019;39(26):5095–5114. doi: 10.1523/JNEUROSCI.2496-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng P., Obara C.J., Szczesna E., Nixon-Abell J., Mahalingan K.K., Roll-Mecak A., Lippincott-Schwartz J., Blackstone C. ER proteins decipher the tubulin code to regulate organelle distribution. Nature. 2022;601(7891):132–138. doi: 10.1038/s41586-021-04204-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedman J.R., Webster B.M., Mastronarde D.N., Verhey K.J., Voeltz G.K. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J Cell Biol. 2010;190(3):363–375. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein L.S., Yang Z. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu Rev Neurosci. 2000;23:39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- 39.DeBoer S.R., You Y., Szodorai A., Kaminska A., Pigino G., Nwabuisi E., Wang B., Estrada-Hernandez T., Kins S., Brady S.T., et al. Conventional kinesin holoenzymes are composed of heavy and light chain homodimers. Biochemistry. 2008;47(15):4535–4543. doi: 10.1021/bi702445j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farías G.G., Fréal A., Tortosa E., Stucchi R., Pan X., Portegies S., Will L., Altelaar M., Hoogenraad C.C. Feedback-driven mechanisms between microtubules and the endoplasmic reticulum instruct neuronal polarity. Neuron. 2019;102(1) doi: 10.1016/j.neuron.2019.01.030. 184-201.e188. [DOI] [PubMed] [Google Scholar]
- 41.Roll-Mecak A. The tubulin code in microtubule dynamics and information encoding. Dev Cell. 2020;54(1):7–20. doi: 10.1016/j.devcel.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balabanian L., Berger C.L., Hendricks A.G. Acetylated microtubules are preferentially bundled leading to enhanced kinesin-1 motility. Biophys J. 2017;113(7):1551–1560. doi: 10.1016/j.bpj.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterman-Storer C.M., Gregory J., Parsons S.F., Salmon E.D. Membrane/microtubule tip attachment complexes (TACs) allow the assembly dynamics of plus ends to push and pull membranes into tubulovesicular networks in interphase Xenopus egg extracts. J Cell Biol. 1995;130(5):1161–1169. doi: 10.1083/jcb.130.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Garcia R., Volkov V.A., Chen C.Y., Katrukha E.A., Olieric N., Aher A., Grigoriev I., Lopez M.P., Steinmetz M.O., Kapitein L.C., et al. Mechanisms of motor-independent membrane remodeling driven by dynamic microtubules. Curr Biol. 2020;30(6) doi: 10.1016/j.cub.2020.01.036. 972-987 e912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grigoriev I., Gouveia S.M., van der Vaart B., Demmers J., Smyth J.T., Honnappa S., Splinter D., Steinmetz M.O., Putney J.W., Hoogenraad C.C., et al. STIM1 Is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr Biol. 2008;18(3):177–182. doi: 10.1016/j.cub.2007.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shirane M. Roles of protrudin at interorganelle membrane contact sites. Proc Jpn Acad Ser B Phys Biol Sci. 2019;95(7):312–320. doi: 10.2183/pjab.95.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsuzaki F., Shirane M., Matsumoto M., Nakayama K.I. Protrudin serves as an adaptor molecule that connects KIF5 and its cargoes in vesicular transport during process formation. Mol Biol Cell. 2011;22(23):4602–4620. doi: 10.1091/mbc.E11-01-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dreier L., Rapoport T.A. In vitro formation of the endoplasmic reticulum occurs independently of microtubules by a controlled fusion reaction. J Cell Biol. 2000;148(5):883–898. doi: 10.1083/jcb.148.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sree S., Parkkinen I., Their A., Airavaara M., Jokitalo E. Morphological heterogeneity of the endoplasmic reticulum within neurons and its implications in neurodegeneration. Cells. 2021;10(5):970. doi: 10.3390/cells10050970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baird F.J., Bennett C.L. Microtubule defects & neurodegeneration. J Genet Syndr Gene Ther. 2013;4:203. doi: 10.4172/2157-7412.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deiters V.S., Guillery R.W. Otto Friedrich Karl Deiters (1834-1863) J Comp Neurol. 2013;521(9):1929–1953. doi: 10.1002/cne.23316. [DOI] [PubMed] [Google Scholar]
- 52.Schelski M., Bradke F. Neuronal polarization: from spatiotemporal signaling to cytoskeletal dynamics. Mol Cell Neurosci. 2017;84:11–28. doi: 10.1016/j.mcn.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 53.Witte H., Neukirchen D., Bradke F. Microtubule stabilization specifies initial neuronal polarization. J Cell Biol. 2008;180(3):619–632. doi: 10.1083/jcb.200707042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konishi Y., Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat Neurosci. 2009;12(5):559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- 55.Cassimeris L., Spittle C. Regulation of microtubule-associated proteins. Int Rev Cytol. 2001;210:163–226. doi: 10.1016/s0074-7696(01)10006-9. [DOI] [PubMed] [Google Scholar]
- 56.Zhu P.P., Hung H.F., Batchenkova N., Nixon-Abell J., Henderson J., Zheng P., Renvoise B., Pang S., Xu C.S., Saalfeld S., et al. Transverse endoplasmic reticulum expansion in hereditary spastic paraplegia corticospinal axons. Hum Mol Genet. 2022;31(16):2779–2795. doi: 10.1093/hmg/ddac072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramirez O.A., Couve A. The endoplasmic reticulum and protein trafficking in dendrites and axons. Trends Cell Biol. 2011;21(4):219–227. doi: 10.1016/j.tcb.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 58.Shibata Y., Shemesh T., Prinz W.A., Palazzo A.F., Kozlov M.M., Rapoport T.A. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143(5):774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacobson C., Schnapp B., Banker G.A. A change in the selective translocation of the Kinesin-1 motor domain marks the initial specification of the axon. Neuron. 2006;49(6):797–804. doi: 10.1016/j.neuron.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 60.Lee M., Paik S.K., Lee M.J., Kim Y.J., Kim S., Nahm M., Oh S.J., Kim H.M., Yim J., Lee C.J., et al. Drosophila Atlastin regulates the stability of muscle microtubules and is required for synapse development. Dev Biol. 2009;330(2):250–262. doi: 10.1016/j.ydbio.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 61.Elert-Dobkowska E., Stepniak I., Krysa W., Ziora-Jakutowicz K., Rakowicz M., Sobanska A., Pilch J., Antczak-Marach D., Zaremba J., Sulek A. Next-generation sequencing study reveals the broader variant spectrum of hereditary spastic paraplegia and related phenotypes. Neurogenetics. 2019;20(1):27–38. doi: 10.1007/s10048-019-00565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sonda S., Pendin D., Daga A. ER morphology in the pathogenesis of hereditary spastic paraplegia. Cells. 2021;10(11):2870. doi: 10.3390/cells10112870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jeong B., Kim T.H., Kim D.S., Shin W.H., Lee J.R., Kim N.S., Lee D.Y. Spastin contributes to neural development through the regulation of microtubule dynamics in the primary cilia of neural stem cells. Neuroscience. 2019;411:76–85. doi: 10.1016/j.neuroscience.2019.05.024. [DOI] [PubMed] [Google Scholar]
- 64.Appocher C., Klima R., Feiguin F. Functional screening in Drosophila reveals the conserved role of REEP1 in promoting stress resistance and preventing the formation of Tau aggregates. Hum Mol Genet. 2014;23(25):6762–6772. doi: 10.1093/hmg/ddu393. [DOI] [PubMed] [Google Scholar]
- 65.Montenegro G., Rebelo A.P., Connell J., Allison R., Babalini C., D'Aloia M., Montieri P., Schule R., Ishiura H., Price J., et al. Mutations in the ER-shaping protein reticulon 2 cause the axon-degenerative disorder hereditary spastic paraplegia type 12. J Clin Invest. 2012;122(2):538–544. doi: 10.1172/JCI60560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanderson C.M., Connell J.W., Edwards T.L., Bright N.A., Duley S., Thompson A., Luzio J.P., Reid E. Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum Mol Genet. 2006;15(2):307–318. doi: 10.1093/hmg/ddi447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shoukier M., Neesen J., Sauter S.M., Argyriou L., Doerwald N., Pantakani D.V., Mannan A.U. Expansion of mutation spectrum, determination of mutation cluster regions and predictive structural classification of SPAST mutations in hereditary spastic paraplegia. Eur J Hum Genet: EJHG. 2009;17(2):187–194. doi: 10.1038/ejhg.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.White S.R., Evans K.J., Lary J., Cole J.L., Lauring B. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J Cell Biol. 2007;176(7):995–1005. doi: 10.1083/jcb.200610072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mannan A.U., Boehm J., Sauter S.M., Rauber A., Byrne P.C., Neesen J., Engel W. Spastin, the most commonly mutated protein in hereditary spastic paraplegia interacts with Reticulon 1 an endoplasmic reticulum protein. Neurogenetics. 2006;7(2):93–103. doi: 10.1007/s10048-006-0034-4. [DOI] [PubMed] [Google Scholar]
- 70.Baas P.W., Vidya Nadar C., Myers K.A. Axonal transport of microtubules: the long and short of it. Traffic. 2006;7(5):490–498. doi: 10.1111/j.1600-0854.2006.00392.x. [DOI] [PubMed] [Google Scholar]
- 71.Sauter S., Miterski B., Klimpe S., Bonsch D., Schols L., Visbeck A., Papke T., Hopf H.C., Engel W., Deufel T., et al. Mutation analysis of the spastin gene (SPG4) in patients in Germany with autosomal dominant hereditary spastic paraplegia. Hum Mutat. 2002;20(2):127–132. doi: 10.1002/humu.10105. [DOI] [PubMed] [Google Scholar]
- 72.Rao K., Stone M.C., Weiner A.T., Gheres K.W., Zhou C., Deitcher D.L., Levitan E.S., Rolls M.M. Spastin, atlastin, and ER relocalization are involved in axon but not dendrite regeneration. Mol Biol Cell. 2016;27(21):3245–3256. doi: 10.1091/mbc.E16-05-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Solowska J.M., Morfini G., Falnikar A., Himes B.T., Brady S.T., Huang D., Baas P.W. Quantitative and functional analyses of spastin in the nervous system: implications for hereditary spastic paraplegia. J Neurosci. 2008;28(9):2147–2157. doi: 10.1523/JNEUROSCI.3159-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Evans K., Keller C., Pavur K., Glasgow K., Conn B., Lauring B. Interaction of two hereditary spastic paraplegia gene products, spastin and atlastin, suggests a common pathway for axonal maintenance. Proc Natl Acad Sci USA. 2006;103(28):10666–10671. doi: 10.1073/pnas.0510863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu P.-P., Denton K.R., Pierson T.M., Li X.-J., Blackstone C. Pharmacologic rescue of axon growth defects in a human iPSC model of hereditary spastic paraplegia SPG3A. Hum Mol Genet. 2014;23(21):5638–5648. doi: 10.1093/hmg/ddu280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Orso G., Martinuzzi A., Rossetto M.G., Sartori E., Feany M., Daga A. Disease-related phenotypes in a Drosophila model of hereditary spastic paraplegia are ameliorated by treatment with vinblastine. J Clin Invest. 2005;115(11):3026–3034. doi: 10.1172/JCI24694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Denton K.R., Lei L., Grenier J., Rodionov V., Blackstone C., Li X.J. Loss of spastin function results in disease-specific axonal defects in human pluripotent stem cell-based models of hereditary spastic paraplegia. Stem Cells. 2014;32(2):414–423. doi: 10.1002/stem.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abrahamsen G., Fan Y., Matigian N., Wali G., Bellette B., Sutharsan R., Raju J., Wood S.A., Veivers D., Sue C.M., et al. A patient-derived stem cell model of hereditary spastic paraplegia with SPAST mutations. Dis Model Mech. 2013;6(2):489–502. doi: 10.1242/dmm.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fassier C., Tarrade A., Peris L., Courageot S., Mailly P., Dalard C., Delga S., Roblot N., Lefevre J., Job D., et al. Microtubule-targeting drugs rescue axonal swellings in cortical neurons from spastin knockout mice. Dis Model Mech. 2013;6(1):72–83. doi: 10.1242/dmm.008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fan Y., Wali G., Sutharsan R., Bellette B., Crane D.I., Sue C.M., Mackay-Sim A. Low dose tubulin-binding drugs rescue peroxisome trafficking deficit in patient-derived stem cells in Hereditary Spastic Paraplegia. Biol Open. 2014;3(6):494–502. doi: 10.1242/bio.20147641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rehbach K., Kesavan J., Hauser S., Ritzenhofen S., Jungverdorben J., Schule R., Schols L., Peitz M., Brustle O. Multiparametric rapid screening of neuronal process pathology for drug target identification in HSP patient-specific neurons. Sci Rep. 2019;9(1):9615. doi: 10.1038/s41598-019-45246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279(5350):519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 83.Reid E., Kloos M., Ashley-Koch A., Hughes L., Bevan S., Svenson I.K., Graham F.L., Gaskell P.C., Dearlove A., Pericak-Vance M.A., et al. A Kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10) Am J Hum Genet. 2002;71(5):1189–1194. doi: 10.1086/344210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ebbing B., Mann K., Starosta A., Jaud J., Schols L., Schule R., Woehlke G. Effect of spastic paraplegia mutations in KIF5A kinesin on transport activity. Hum Mol Genet. 2008;17(9):1245–1252. doi: 10.1093/hmg/ddn014. [DOI] [PubMed] [Google Scholar]
- 85.Wang L., Brown A. A hereditary spastic paraplegia mutation in kinesin-1A/KIF5A disrupts neurofilament transport. Mol Neurodegener. 2010;5:52. doi: 10.1186/1750-1326-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mannan A.U., Krawen P., Sauter S.M., Boehm J., Chronowska A., Paulus W., Neesen J., Engel W. ZFYVE27 (SPG33), a novel spastin-binding protein, is mutated in hereditary spastic paraplegia. Am J Hum Genet. 2006;79(2):351–357. doi: 10.1086/504927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martignoni M., Riano E., Rugarli E.I. The role of ZFYVE27/protrudin in hereditary spastic paraplegia. Am J Hum Genet. 2008;83(1):127–128. doi: 10.1016/j.ajhg.2008.05.014. ; author reply 128-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saita S., Shirane M., Natume T., Iemura S.I., Nakayama K.I. Promotion of neurite extension by protrudin requires its interaction with vesicle-associated membrane protein-associated protein. J Biol Chem. 2009;284(20):13766–13777. doi: 10.1074/jbc.M807938200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hashimoto Y., Shirane M., Matsuzaki F., Saita S., Ohnishi T., Nakayama K.I. Protrudin regulates endoplasmic reticulum morphology and function associated with the pathogenesis of hereditary spastic paraplegia. J Biol Chem. 2014;289(19):12946–12961. doi: 10.1074/jbc.M113.528687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chang J., Lee S., Blackstone C. Protrudin binds atlastins and endoplasmic reticulum-shaping proteins and regulates network formation. Proc Natl Acad Sci USA. 2013;110(37):14954–14959. doi: 10.1073/pnas.1307391110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liang J.R., Lingeman E., Ahmed S., Corn J.E. Atlastins remodel the endoplasmic reticulum for selective autophagy. J Cell Biol. 2018;217(10):3354–3367. doi: 10.1083/jcb.201804185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vajente N., Norante R., Redolfi N., Daga A., Pizzo P., Pendin D. Microtubules stabilization by mutant spastin affects ER morphology and Ca(2+) handling. Front Physiol. 2019;10:1544. doi: 10.3389/fphys.2019.01544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li J., Yan B., Si H., Peng X., Zhang S.L., Hu J. Atlastin regulates store-operated calcium entry for nerve growth factor-induced neurite outgrowth. Sci Rep. 2017;7:43490. doi: 10.1038/srep43490. [DOI] [PMC free article] [PubMed] [Google Scholar]