Summary
Sex differences exist for many lung pathologies, including COVID-19 and pulmonary fibrosis, but the mechanistic basis for this remains unclear. Alveolar type 2 cells (AT2s), which play a key role in alveolar lung regeneration, express the X-linked Ace2 gene that has roles in lung repair and SARS-CoV-2 pathogenesis, suggesting that X chromosome inactivation (XCI) in AT2s might impact sex-biased lung pathology. Here we investigate XCI maintenance and sex-specific gene expression profiles using male and female AT2s. Remarkably, the inactive X chromosome (Xi) lacks robust canonical Xist RNA “clouds” and less enrichment of heterochromatic modifications in human and mouse AT2s. We demonstrate that about 68% of expressed X-linked genes in mouse AT2s, including Ace2, escape XCI. There are genome-wide expression differences between male and female AT2s, likely influencing both lung physiology and pathophysiologic responses. These studies support a renewed focus on AT2s as a potential contributor to sex-biased differences in lung disease.
Keywords: X chromosome inactivation, Xist RNA, heterochromatic modifications, XCI escape genes, Ace2, alveolar type 2 cells, sex differences, sex-biased gene expression, lung progenitors
Highlights
-
•
XCI maintenance and sex-specific gene expression profiles of alveolar type 2 cells
-
•
The Xi lacks Xist RNA “clouds” and less heterochromatic mark enrichment
-
•
68% of expressed X-linked genes in mouse AT2s, including Ace2, escape XCI
-
•
Genome-wide sex-specific gene expression profile differences in AT2s
Anguera, Vaughan, and colleagues show that lung alveolar type 2 (AT2s) cells maintain X chromosome inactivation with fewer epigenetic modifications on the inactive X, with very high numbers of X-linked genes that “escape” transcriptional silencing. AT2 cells also exhibit significant genome-wide sex-biased gene expression, which likely influences lung physiology and pathophysiologic responses.
Introduction
COVID-19, the disease caused by SARS-CoV-2, commonly presents as pneumonia, with those most severely affected progressing to acute respiratory distress syndrome (ARDS) (CDC COVID-19 Response Team, 2020), itself bearing a mortality rate as high as 45%. As previously observed with related coronaviruses SARS-CoV and MERS (Channappanavar et al., 2017), older males are at significantly higher risk for severe or fatal outcomes from COVID-19 (Chen et al., 2020; Galbadage et al., 2020; Lau et al., 2021; Peckham et al., 2020; Takahashi et al., 2020; Wenham et al., 2020). Since the onset of the pandemic, this male sex bias is incredibly consistent across the globe, with increases in mortality among older genetic (XY) males (Galbadage et al., 2020; Lau et al., 2021; Peckham et al., 2020; Takahashi et al., 2020). A greater understanding of the underlying mechanisms contributing to this male bias may reveal novel insights for the treatment or reduction of disease severity.
Severe COVID-19/ARDS is characterized by diffuse alveolar damage, wherein large regions of alveolar epithelium are ablated either directly by SARS-CoV-2 infection or indirectly by virus-induced inflammatory stresses (e.g., “cytokine storms”). Within the alveolar parenchyma, alveolar type 2 cells (AT2s), responsible for pulmonary surfactant production, are also extremely well characterized as alveolar progenitor cells. Upon injury, AT2s can proliferate, restore the loss of surfactant-producing cells, and differentiate to re-establish gas-exchanging alveolar type I cells (AT1s) (Barkauskas et al., 2013; Evans et al., 1975). Whether any of the sex differences in COVID-19 might reflect sex differences within alveolar progenitor cells themselves has not been explored. However, there have been a few developmental studies that report sex differences in cell number and functionality of fetal lung epithelial cells, surfactant phospholipid content in fetal rat lungs, and expression of apolipoprotein genes in fetal lung RNA (Kaltofen et al., 2015; McCoy et al., 1999; Simard et al., 2006). To date, there are no unbiased investigations of genome-wide transcriptional profiling of male and female AT2s.
Female mammals use X chromosome inactivation (XCI) to equalize expression of X-linked genes between the sexes. XCI is initiated during female embryonic development by the expression of the long non-coding RNA Xist (Marahrens et al., 1997; Penny et al., 1996), which recruits chromatin modifiers that deposit heterochromatic histone modifications across the entire X chromosome, resulting in allele-specific gene silencing (Kohlmaier et al., 2004; Yildirim et al., 2013; Zhao et al., 2008). Xist deletion or loss in hematopoietic stem cells and human pluripotent stem cells results respectively in partial X-reactivation with increased expression of certain X-linked genes, dependent on cell type (Yildirim et al., 2013) (Anguera et al., 2012; Mekhoubad et al., 2012). While XCI has been considered a stable attribute of female somatic cells, we have found that XCI in lymphocytes is unusually dynamic. Our research found that mature lymphocytes from female mice and humans lack Xist RNA “clouds” and enrichment of heterochromatic marks on the inactive X chromosome (Xi) and that these epigenetic modifications return in stimulated lymphocytes (Syrett et al., 2017, 2019; Wang et al., 2016). We also discovered that other immune cells have dispersed Xist RNA localization patterns (Syrett et al., 2018). Whether atypical XCI maintenance extends to additional cell types that could contribute to sex-biased differences in COVID severity is unknown.
Interestingly, the X chromosome encodes angiotensin-converting enzyme 2 (ACE2), an enzyme centrally involved in multiple aspects of SARS-CoV-2 pathogenesis, including acting as the major viral receptor (Huang et al., 2020; Katsura et al., 2020). However, because ACE2 also plays an important role in degradation of bradykinin and angiotensin II (which contributes to lung injury), it is difficult to predict how increased or decreased levels of ACE2 contribute to disease severity beyond the minimal expression level required for initial infection (Imai et al., 2005). Indeed, Ace2 knockout mice exhibit much more severe lung injury after acid aspiration and H5N1 influenza infection, while administration of recombinant Ace2 is protective (Imai et al., 2005; Zou et al., 2014). Thus, if ACE2 “escapes” XCI in AT2s, it is unclear how this might affect pathophysiology upon SARS-CoV-2 infection.
Here we assessed whether AT2s might, like lymphocytes, exhibit non-canonical XCI maintenance, and whether this might contribute toward sex-biased expression of both X-linked and autosomal genes. We find that AT2s from mouse and human lungs lack the canonical robust Xist “clouds” typical of somatic cells and exhibit very low enrichment of two heterochromatic modifications, H3K27me3 and H2AK119-ubiquitin, on the Xi. Allele-specific quantification of the Xi revealed that about 50% of the X-linked genes expressed in AT2 cells “escape” XCI in adult mouse AT2s. Finally, we performed whole-transcriptome differential expression analysis, observing a strikingly large number of gene expression differences between male and female AT2s, which likely contribute to sex differences in lung injury and repair in multiple settings including COVID-19.
Results
Female AT2 cells have faint and dispersed Xist RNA pinpoints
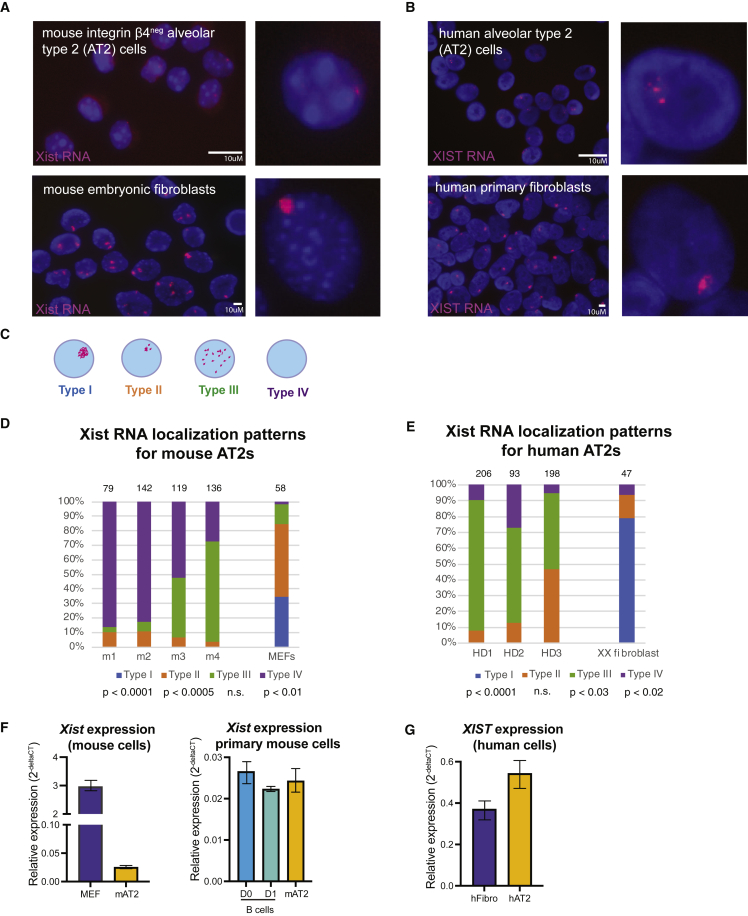
To isolate AT2 cells from adult female mouse lungs, we gated on CD45neg Epcampos Integrin β4pos cells, as we previously demonstrated that this sort scheme results in >96% enrichment for AT2s (Chapman et al., 2011; Vaughan et al., 2015; Weiner et al., 2019). Consistent with previous results, sorted cells were approximately 99% pure (Figure S1). AT2 cells were immediately cytospun and fixed, and used for Xist RNA fluorescence in situ hybridization (RNA FISH) using oligo probes specific for repetitive regions within Xist RNA (Syrett et al., 2017). Surprisingly, unlike transformed mouse embryonic fibroblasts (MEFs) which have robust Xist RNA “clouds,” female mouse AT2 cells had very faint pinpoints of Xist RNA or completely lacked Xist RNA signals (Figure 1A). Mouse AT2 cells contained two Xs using X-paints for DNA FISH, and Xist pinpoints could identify the Xi in some cells (Figure S1D). To determine whether human female AT2 cells also lacked robust XIST RNA localization at the Xi, we isolated human AT2 cells from explanted lungs of healthy donors as previously described, utilizing the AT2-specific HTII-280 antibody (Xi et al., 2017). Sorted human AT2 cells were used for XIST RNA FISH using human-specific oligo probes (Wang et al., 2016), and these cells also exhibited pinpoints of XIST RNA, instead of robust XIST RNA signals typical of human fibroblasts (Figure 1B). Human AT2 cells also contained two X chromosomes, and sequential DNA-RNA FISH could identify the Xi in some nuclei (Figure S1E). We classified the Xist/XIST RNA FISH localization patterns into four groups, as done previously for lymphocytes: type I nuclei have the canonical robust Xist RNA “clouds”; type II nuclei exhibit diffuse Xist RNA signals within a nuclear area encompassing the X chromosome; type III nuclei have dispersed pinpoints of Xist RNA across the nucleus; type IV nuclei lack Xist RNA signals (Figure 1C) (Pyfrom et al., 2021; Syrett et al., 2019; Wang et al., 2016). We found that Xist RNA patterns in mouse AT2s were predominantly types III and IV, and had significantly fewer type I and II patterns compared with MEFs (Figure 1D). Interestingly, human AT2 cells had more detectable XIST RNA transcripts than mouse AT2s, and consisted of mostly type II and type III patterns (Figure 1E). Human AT2 cells, similar to mouse AT2s, also had significantly fewer type I patterns compared with human fibroblasts. Xist was abundantly expressed in both mouse and human AT2 cells, at similar levels to primary mouse B cells and also fibroblasts (Figures 1F and 1G). Taken together, female AT2 cells from both mouse and human lack robust Xist/XIST RNA localization at the Xi.
Figure 1.
Xist RNA is not robustly localized at the Xi in mouse and human AT2 cells
(A) Representative field images of Xist RNA FISH in female mouse AT2 cells isolated from lung. Scale bar is 10 micron.
(B) Representative field images of XIST RNA FISH in human female AT2 cells. Scale bar is 10 micron.
(C) Cartoon showing each category of Xist RNA localization pattern observed in AT2 cells.
(D and E) Quantification of XIST RNA localization patterns for (D) mouse AT2 cells and (E) human AT2 cells. Number of nuclei counted is above each column. Statistical significance determined using t test with unequal variance comparing AT2 with fibroblasts for each category of Xist localization pattern (types I, II, III, IV).
(F) qRT-PCR for Xist expression in mouse AT2 cells, transformed female MEFs (with multiple X chromosomes), and primary B cells from female mice. Error bars denote standard error of mean.
(G) qRT-PCR for XIST expression in human AT2 cells and primary female fibroblasts (IMR-90). Error bars denote standard error of mean.
The Xi in female AT2 cells has reduced levels of heterochromatic foci
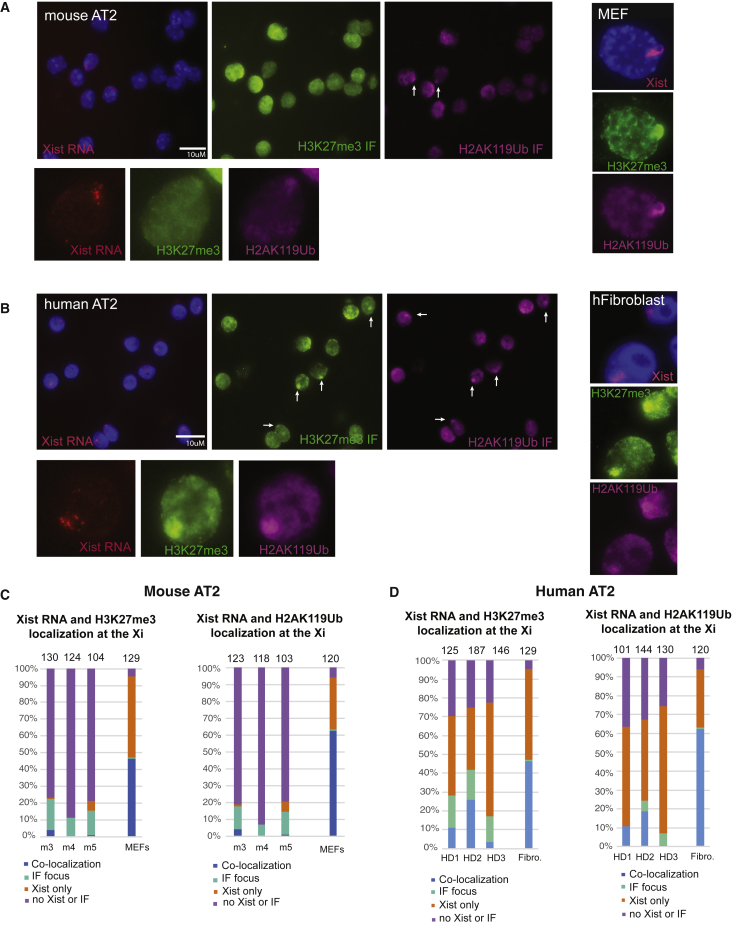
In most somatic cells, the Xi is enriched for heterochromatic modifications that can be visualized cytologically by immunofluorescence (IF) (Payer and Lee, 2008; Plath et al., 2003). Because AT2 cells lacked robust Xist/XIST RNA clouds, we next performed sequential Xist RNA FISH and IF for two histone modifications, H3K27me3 and H2AK119-ubiquitin (Ub), typically associated with XCI. Mouse AT2 cells had low levels of detectable foci of H3K27me3 and H2AK119Ub (white arrows in Figure 2A), and foci were present in cells that had more Xist RNA signals (type II). Human AT2 cells had more H3K27me3 and H2AK119Ub foci compared with mouse cells (white arrows in Figure 2B), and IF foci were present in cells with type II XIST RNA. We quantified the number of nuclei with co-localization of Xist RNA and IF focus, nuclei containing just an IF focus, nuclei with just Xist RNA signal, and nuclei lacking both Xist RNA and IF focus, for both mouse and human AT2 cells. We found that less than 5% of mouse AT2 nuclei exhibited co-localization of Xist RNA and heterochromatic focus (H3K27me3 or H2AK119Ub), and about 15%–20% of nuclei contained a focus of either mark (Figure 2C). The majority of mouse AT2 nuclei lacked IF foci and Xist RNA signals. Human AT2 cells had higher percentages of nuclei with co-localization of XIST RNA and a heterochromatic focus (blue bars; 5%–25%) (Figure 2D), in keeping with human AT2s exhibiting more detectable Xist RNA patterns. Similar to mouse cells, human AT2s exhibited 5%–15% of nuclei with a heterochromatic focus. Fibroblasts from mouse and human samples contained high levels of co-localization of Xist/XIST RNA signal with heterochromatic foci. In sum, the Xi in mouse and human AT2 cells has reduced enrichment of two heterochromatic histone modifications, H3K27me3 and H2AK119Ub, a finding more prominent in mouse than in human cells.
Figure 2.
Heterochromatic modifications H3K27me3 and H2AK119-ubiquitin (Ub) are not enriched at the Xi in mouse and human AT2 cells
(A) Sequential Xist RNA FISH followed by IF detection for H3K27me3 (FITC) and H2AK119Ub (Cy5) for mouse AT2 cells; a representative field image (same left to right) is shown. Zoomed-in images for a representative nucleus are shown below the field images. White arrows denote IF foci. Control IF staining using MEFs is shown. Scale bar is 10 micron.
(B) Sequential Xist RNA FISH followed by IF detection for H3K27me3 (FITC) and H2AK119Ub (Cy5) for human AT2 cells; representative field image is shown. Zoomed-in images for a representative nucleus are shown below the field images. White arrows denote IF foci. Control IF staining using human fibroblasts (293T) is shown. Scale bar is 10 micron.
(C) Quantification of Xist RNA and IF co-localization patterns for AT2 cells isolated from three distinct female mice (m3, m4, m5) and MEFs from n = 2 female mice.
(D) Quantification of Xist RNA and IF co-localization patterns for AT2 cells isolated from three distinct healthy human donors (HD1, HD2, HD3) and 293T cells (two replicate experiments). Number of nuclei counted is above each column.
Over half of the expressed X-linked genes escape XCI in female AT2 cells, including Ace2
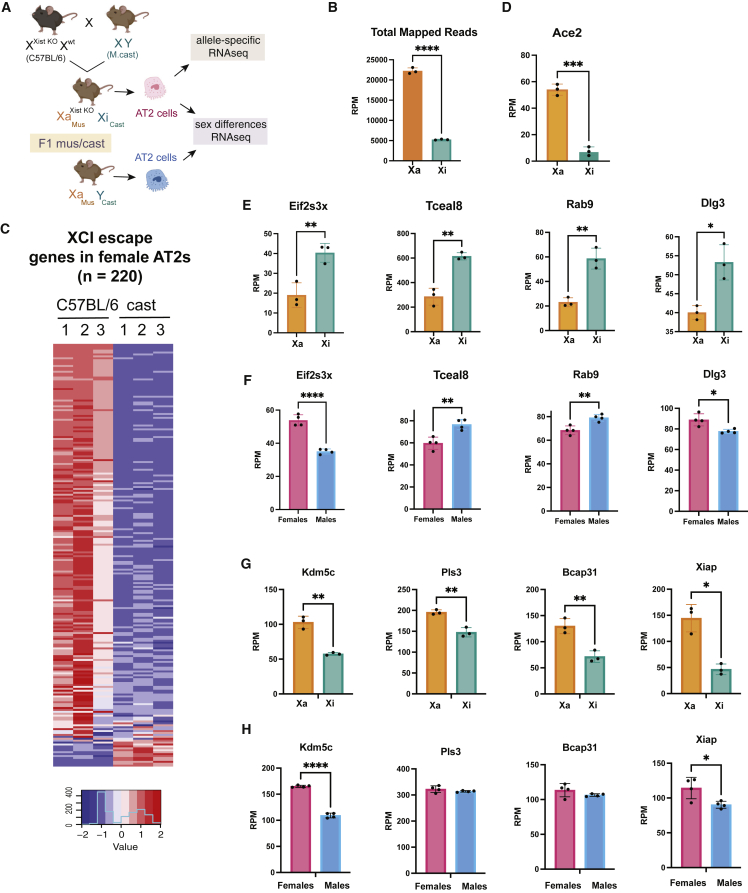
The atypical localization of Xist RNA and heterochromatic modifications H3K27me3 and H2AK119Ub at the Xi in AT2 cells suggests that there could be increased transcription from the Xi. XCI silences most genes across the X in female cells, but there are genes that “escape” XCI and are transcribed from both the Xi and active X chromosome (Xa) (Berletch et al., 2010, 2015). About 3%–7% of the mouse X chromosome escapes XCI in brain, ovary, and splenic tissues (Berletch et al., 2015), and human tissues exhibit much higher levels of XCI escape that ranges from 25% to 35% (Tukiainen et al., 2017). To determine the allele-specific expression of X-linked genes in mouse AT2 cells, we mated C57BL/6 females heterozygous for a deletion of exons 1–3 of Xist (Csankovszki et al., 1999) (XXist KO Xwt) to Mus castaneus (cast) male mice to generate F1 mus/cast progeny where female mice (XaXist KO Xicast) have completely skewed XCI across all tissues, as all Xi are derived from the cast allele, which can be distinguished from the Xa (C57BL/6) allele by multiple SNPs (Figure 3A). We isolated AT2 cells from four female F1 mus/cast mice for RNA sequencing (RNA-seq) to determine allele-specific reads based on SNPs. We also isolated AT2 cells from four male F1 mus/cast mice for RNA-seq to determine sex-specific gene expression profiles, confirming extremely high sort purity in all samples (Figures S1A–S1C).
Figure 3.
Female AT2 cells have over 200 X-linked genes that escape XCI
(A) Schematic diagram showing mouse mating strategy to generate F1 mus/cast female mice with skewed XCI. In these mice, the maternal X (C57BL/6) carries the Xist deletion and is the active X (Xa), and the paternal X is always inactivated (Xi) and expresses Xist. We also isolated AT2 cells from male F1 mus/cast mice to determine sex-specific gene expression profiling.
(B) Allelic specific reads (RPM) for each expressed X-linked gene which map to the Xa (orange) or Xi (teal). Error bars are standard error of the mean.
(C) Heatmap showing the allele-specific expression for XCI escape genes in AT2 cells. Each replicate sample (1–3) is shown for reads derived from the C57BL/6 or cast allele.
(D) Ace2 is an XCI escape gene in AT2 cells.
(E) Normalized haploid mapped reads from the Xa (orange) and Xi (teal) for four XCI escape genes where there is more expression from the Xi compared with the Xa (Xi > Xa). Error bars are standard error of the mean.
(F) Normalized diploid mapped reads from female (pink) and male (blue) AT2 samples for four XCI escape genes with Xi > Xa. Error bars are standard error of the mean.
(G) Normalized haploid mapped reads from the Xa (orange) and Xi (teal) for the top three XCI escape genes with the highest expression in female AT2 cells. Error bars are standard error of the mean.
(H) Normalized diploid mapped reads from female (pink) and male (blue) samples for the top three XCI escape genes. Statistical significance determined using unpaired t tests for panels B–H (excluding C). Error bars are standard error of the mean.
To determine the reads that mapped to each parental genome, we separately aligned RNA-seq reads to the C57BL/6 and castaneus EiJ genomes. Reads that did not map were then aligned to the other genome to generate uniquely mapped reads in two categories: (1) C57BL/6 reads that contain C57BL/6 specific SNPs; and (2) castaneus reads that contain specific castaneus SNPs (Figure S1F). We calculated diploid gene expression for all mapped reads to determine reads per million (RPM), and defined the SNP-based haploid gene expression for Xi (Xi-SRPM) or Xa (Xa-SRPM) to be allele-specific SNP-containing exonic reads per 10 million uniquely mapped reads (Berletch et al., 2015). To validate our allele-specific mapping, we examined autosomal imprinted gene expression in female and male AT2 cell samples (Figures S2A and S2B). Meg3 is a maternally expressed gene and was expressed exclusively from the maternal C57BL/6 allele in both male and female AT2 cells (Figure S2A). Peg3 is paternally expressed and was predominantly expressed from the cast allele in both male and female samples (Figure S2B). We also validated that reads from the Xist/Tsix region were predominantly expressed from the cast allele in female AT2 cells, and detected some expression (∼40 RPM) from Xa in this region (Figures S2C and S2D). The allelic distribution of mapped reads across autosomal chromosomes was similar for C57BL/6 and cast alleles and the geometric mean of ratios was nearly equal to 1, yet we observed a significant skew for allelic X-linked gene expression in AT2 cells (Figure S2E). We observed significantly higher reads that mapped to the Xa compared with the Xi, yet we also found that about 25% of total X-linked mapped reads were expressed from the Xi, unlike the total mapped reads for chromosome 1, which were similar for both alleles (Figure S2F). Indeed, the geometric mean of ratios for autosomal allelic expression were similar for C57BL/6 and cast reads, yet we observed an expected X-linked skew for Xa reads (C57BL/6) relative to Xi reads (cast) (Figure S2G). Taken together, while the Xi in female AT2 cells is still dosage compensated, there is a very significant amount of transcription specifically from the Xi.
For female F1 mus/cast samples (n = 3), we used a published binomial model to identify escape genes and estimated a statistical confidence of escape probability (Berletch et al., 2015). We found that there are 465 X-linked genes with detectable expression in AT2 cells (RPM >1), and 455 of these genes are expressed in both male and female AT2 cells (Figure S3A). Approximately 327 X-linked genes are expressed allele-specifically (with detectable SNPs) in female AT2 cells (Figure S3B). Using the binomial model to identify XCI escape genes in AT2s, we determined that 223 (68%) genes escape XCI at a 99% confidence with a p-value cutoff of 0.01 (Figure 3C and Table S1). The majority of AT2 XCI escape genes are novel, and just nine of these genes have been previously reported to escape XCI in mouse brain, ovary, and spleen tissue (Berletch et al., 2015). We also found that Ace2 escapes XCI in AT2 cells (Figure 3D) and has higher expression in AT2 cells compared with fibroblasts (Figure S3C). XCI escape genes were distributed across the X and did not cluster in specific regions (Figure S3D), perhaps reflecting the paucity of Xist RNA and heterochromatic modifications across the Xi (Figures 1 and 2). Surprisingly, we observed ∼20 X-linked genes which have higher expression from the Xi compared with the Xi (Figures 3C and S3B; Table S1). Some XCI escape genes with Xi/Xa ratios >1 include Gla, Bex1, Pin4, Eif2s3x, Tceal8, Rab 9, Dlg3, and Xist (Figure 3E and Table S1). We observed that XCI escape genes with Xi/Xa ratios >1 were expressed in a sex-biased fashion, but the direction of the bias varied (Figure 3F). For example, Eif2s3x was expressed at higher levels in female AT2 cells whereas Tceal8 exhibited higher expression in male AT2 cells (Figure 3F). The majority of XCI escape genes expressed in female AT2 cells had higher expression from the Xa (Figure 3G; complete list in Table S1), and some also exhibited sex-biased expression (Figure 3H), suggesting that expression of many escape genes may be otherwise compensated at the transcriptional level to normalize expression between sexes. Gene ontology (GO) analysis for the 223 XCI escape genes revealed significant enrichment of pathways for regulation of dosage compensation, nucleotide metabolism, chromatin organization, and regulation of cellular component organization (Table S3C). Thus 68% of all X-linked genes expressed in female AT2 cells escape XCI, and some of these X-linked genes exhibit significant sex-biased expression.
Next we examined allelic expression for autosomal genes that are typical markers for AT2 cells. In keeping with their homeostatic physiological function as the primary producer of pulmonary surfactant, we found that the surfactant protein gene Sftpc was expressed at significantly higher levels from the maternal allele compared with the paternal allele (Figures S3E and S3F) in both male and female AT2s (Figure S3G). We also observed that Lamp3, Muc1, and Abac3 exhibited allelic differences in expression in female AT2s (Figure S3F). However, these allelic expression differences did not significantly impact overall expression on comparing female with male samples (Figure S3G).
Sex-biased gene expression is a prominent feature of mouse AT2s
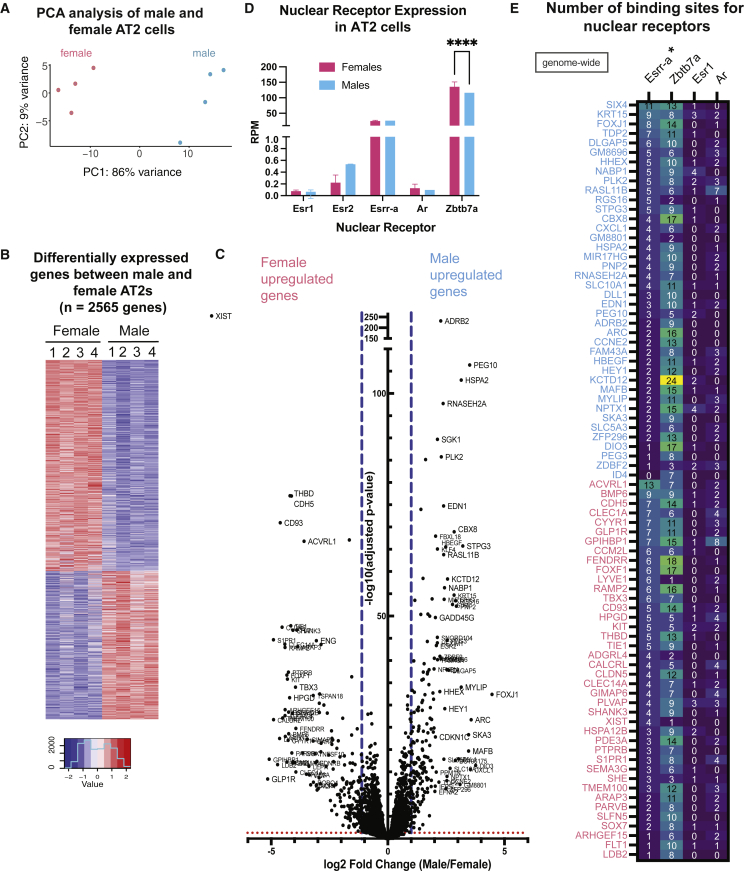
We compared the global gene expression profiles in male and female AT2s using principal component analysis (PCA) for both autosomal and sex-linked genes. We observed that female AT2 samples grouped apart from male samples in the PC1 component (with 86% variance; Figure 4A). Indeed, we found that there were 2,565 genes that were differentially expressed between male and female AT2s (RPM in all four female samples >1; adjusted p < 0.05 across all four replicates; Figures 4B and 4C). The majority of the differentially expressed genes (DEGs) were autosomal, yet we observed some X-linked genes that were significantly overexpressed greater than log2 fold change in male AT2s (GM6568, Rbm3, Rtl5, Zfp711) and female AT2s (Tspan7, Tex11, Slc25A43, Mtcp1) (Figure S4A). While the relatively high number of DEGs was somewhat surprising, we note that only 510 genes show greater than 2.0 fold change between male and females, highlighting genes whose expression differences are most likely to result in phenotypic manifestations. We found that the β2-adrenergic receptor (Adrb2) was the most upregulated gene in male AT2 cells, along with various imprinted genes including Peg10 and Gnas (Figure S4B), which is part of the G-protein-coupled receptor-regulated adenylyl cyclase signal transduction pathway downstream of Adrb2. As expected, Xist was the most significantly upregulated gene in female AT2 cells (Figures 4C and S4A). GO analyses revealed that the female-specific enrichment included pathways of autophagy, protein modification, catabolic processes, and phosphatidylinositol biosynthetic processes (Figure S4C and Table S3A). The top five pathways enriched in male samples included nonsense-mediated decay, mRNA processing, Parkinson’s disease, cholesterol biosynthesis, and nucleobase-containing biosynthetic pathways (Figure S4C and Table S3B). While we did observe some differential expression of genes typically not associated with the AT2 lineage (e.g., Foxj1), equivalent expression of essentially all major AT2 marker genes strongly indicates that these unexpected expression differences are biologically valid.
Figure 4.
AT2 cells exhibit sex-biased expression of autosomal and sex chromosome genes
(A) Principal component analyses (PCA) for the four replicate male (blue) and female (pink) AT2 samples.
(B) Heatmap showing DEGs (a total of 2,565 genes) between replicate male and female AT2 samples.
(C) Volcano plot of female upregulated genes (left) and male upregulated genes (right) expressed in AT2 cells. Blue dashed lines indicate the cutoff for log2 fold change of expression (x axis), and the y axis is the adjusted p value for significance of differential expression.
(D) Normalized diploid mapped reads for male (blue) and female (pink) AT2 samples of nuclear receptor transcription factors. Statistical significance determined using Student’s unpaired t test. Error bars are standard error of the mean.
(E) Numbers of binding sites for hormone (Ar, Esr1) and nuclear receptor (Esrr-α and Zbtb7a) transcription factors in promoter regions (2 kb of TSS) of the top 40 DEGs (genome-wide) upregulated in male (blue) or female (pink) AT2 cells.
Because of the large number of sex-specific DEGs, we asked whether AT2 cells exhibited expression differences in hormone or nuclear receptor transcription factors and whether genes expressed in AT2 cells were enriched for hormone receptor binding sites. We examined the expression levels for known hormone and nuclear receptors in male and female AT2s and found that the nuclear orphan receptor Esrr-α and transcription factor Zbtb7a, which mediates AR activity, were significantly expressed at RPM values greater than 1, unlike estrogen receptors Esr1 and Esr2, and the androgen receptor Ar (Figure 4D). We found a significant difference in expression of Zbtb7a between male and female AT2s, yet expression levels for Esrr-α were similar for both sexes. Next, we determined the number of binding sites for hormone (Esr1, Ar) and nuclear receptors (Esrr-α, Zbtb7a) present within 2 kb of the transcriptional start site (TSS) for the top 40 DEGs (genome-wide) in male and female AT2s (Figure 4E). We found that there were more nuclear receptor binding sites (Esrr-α, Zbtb7a) compared with hormone receptor sites (Esr1, Ar) present in these genes, which exhibited sex-biased expression in AT2s. Comparing binding sites between male and female DEGs, we found that Esrr-α sites were significantly higher (p = 0.048) in male upregulated DEGs, yet the number of binding sites for Zbtb7a, Esr1, and Ar were similar for both male and female upregulated genes (Figure 4E). Next, we determined the hormone and nuclear receptor binding sites for the XCI escape genes expressed in AT2 cells. We observed that there were higher numbers of nuclear receptor binding sites compared with hormone receptor sites for XCI escape genes (Figure S4D). There were similar numbers of Esrr-α and Zbtb7a binding sites across XCI escape genes, irrespective of whether there was more (top) or less (bottom) expression from the Xi (Figure S4D). This suggests that differential binding of these nuclear receptors may not account for the differences in expression between the two groups of XCI escape genes. One exception could be the XCI escape gene Gla, which has the most Esr1 binding sites in this list and exhibits higher expression from the Xi compared with the Xa. We observed some Ar binding sites across both groups of XCI escape genes, which suggests that Ar could increase transcription from some genes on the Xa in male cells. In conclusion, while AT2 cells have low expression of the estrogen receptors (Esr1, Esr2) and the androgen receptor (Ar), there is higher expression of the nuclear orphan receptor Esrr-α and transcription factor Zbtb7a, which may influence sex-biased expression genome-wide.
Discussion
The prevailing model of XCI maintenance indicates that all somatic cells utilize the same epigenetic mechanisms to maintain transcriptional silencing of the Xi for dosage compensation of X-linked genes between males and females. Here we show that female AT2 cells from mouse and human lung contain an Xi that lacks canonical enrichment of Xist RNA and heterochromatic modifications H3K27me3 and H2AK119Ub, suggesting that there could be more expression or “XCI escape” in this cell type. Our studies demonstrate that AT2 cells have the highest levels of XCI escape for mouse cells reported to date, where approximately 68% of expressed X-linked genes are transcribed from the Xi and Xa, and XCI escape genes are distributed across the Xi chromosome. We propose that the reductions with Xist RNA, H3K27me3, and H2AK119Ub enrichment contribute toward increased XCI escape across the Xi in AT2 cells. Our experiments demonstrate that AT2 cells, similar to immune cells (Pyfrom et al., 2021; Syrett et al., 2018, 2019; Wang et al., 2016), do not strictly follow the canonical “rules” of XCI maintenance, which likely reflects a necessity for specific increased expression from the Xi for biological functions.
One surprising discovery is that about 20 XCI escape genes exhibited higher expression from the Xi compared with the Xa in AT2 cells, which is another unique feature of XCI maintenance specific to these cells. XCI escape genes are typically expressed at lower levels from the Xi compared with expression from the Xa (Berletch et al., 2010, 2015). We predicted that increased Xi expression would perhaps result in higher female expression for these genes; however, we observed that the sex-biased expression did not follow a trend that consistently favored either male or female. Our results are in agreement with previous work suggesting that XCI escape is a significant source of sex-biased expression (Balaton and Brown, 2016; Johnston et al., 2008; Tukiainen et al., 2017), although in this study we did not observe consistently higher female expression for XCI escape genes. Possible mechanisms that might contribute toward allele-specific expression differences of XCI escape genes could involve epigenetic modification enrichment differences between the X chromosomes (such as DNA methylation or histone modifications) or transcription factors that would preferentially activate one allele more than the other. Our observation that XCI escape genes expressed in AT2 cells exhibited high numbers of Esrr-α binding sites makes it intriguing to consider that there may be differential binding of the Esrr-α receptor and co-activator (or co-repressor) proteins that results in allelic expression differences.
The SARS-CoV-2 receptor, Ace2, is located on the X chromosome, and an important unanswered question has been whether this gene escapes XCI in cell types important in COVID-19. Previous work using hybrid fibroblast cell lines indicated that ACE2 exhibits variable escape (Balaton et al., 2015; Tukiainen et al., 2017); however, some of these cells are immortalized and thus do not reflect a physiological context. We hypothesized that if ACE2 escaped XCI in AT2 cells, women might produce more ACE2 protein and be subject to higher viral infection/titers yet also might exhibit enhanced lung repair compared with men. Even though Ace2 is biallelically expressed in mouse AT2s, we did not observe sex differences in the steady-state levels of Ace2 mRNA, although there still could be differences in Ace2 protein levels in these cells. Our studies demonstrate that Ace2 is an XCI escape gene in mouse AT2 cells and may also likely escape XCI in human AT2 cells. We were unable to detect biallelic expression of ACE2 in human AT2 cells because of poor signal resolution using RNA FISH (data not shown), so it remains an open question whether ACE2 escapes XCI in human AT2 cells. However, because there are significantly higher numbers of XCI escape genes in human cells compared with mouse (Berletch et al., 2010, 2011), it is very likely that ACE2 would also escape XCI in human AT2s. While Ace2 escapes XCI in AT2 cells, we found that there are similar levels of Ace2 expression in male and female AT2s. This result could be explained by observations that XCI escape genes sometimes, but not always, contribute toward sex-biased expression (Tukiainen et al., 2017).
While fetal lungs and fetal lung epithelial cells have been shown to exhibit some sex-specific differences in cell number, gene expression, and cellular function (Kaltofen et al., 2015; McCoy et al., 1999; Simard et al., 2006), this study is the first to directly assess genome-wide transcriptional differences between male and female adult AT2s. Over 2,500 genes exhibited significant sex-related differences of expression in mouse AT2 cells. Our findings suggest that these expression differences may underlie the observed sex differences in pulmonary disease prevalence and severity. Human males are at higher risk for severe COVID-19 disease (Scully et al., 2020; Takahashi et al., 2020), and males also experience greater prevalence and severity of interstitial lung disease and pulmonary fibrosis (Kawano-Dourado et al., 2021). Intriguingly, these pathologies are increasingly recognized as being driven by defects in the lung epithelium, primarily AT2 cells (Camelo et al., 2014; Katzen and Beers, 2020; Parimon et al., 2020), reinforcing the relevance of the AT2 transcriptomic differences between the sexes. However, females are more likely to develop ARDS after traumatic injury (Heffernan et al., 2011), and once ARDS has developed women are at higher risk of death (McNicholas et al., 2019), which may reflect the fact that females have stronger immune responses compared with males (Fischer et al., 2015; Klein and Flanagan, 2016). How biallelic ACE2 expression, and whether there are sex differences in ACE2 protein levels in AT2 cells, contributes to ARDS-induced disease warrants further investigation.
The underlying drivers of the sex-specific transcriptional differences observed here are almost certainly multifactorial. We found that some genes exhibiting greater expression in females also demonstrate escape from XCI, and that there are many DEGs genome-wide. Likely candidates for these sex-biased gene signatures include hormone receptors, which are expressed at low levels (below 1 RPM) in AT2 cells. However, we cannot exclude the possibility that these proteins are present, albeit at low concentrations, in AT2 cells, and likely influence gene expression. We found that there are both Esr1 and Ar binding sites in nearly all of the top 40 male and female DEGs (Figure 4E). The DEGs contained higher numbers of Esrr-α and Zbtb7a binding sites compared with Esr1 and Ar across both male and female upregulated DEGs. While we did not detect significant gene expression differences for Esrr-α, we found that that there were significantly fewer Zbtb7a reads in male AT2 samples (Figure 4D), which might result in less Zbtb7a protein and transcriptional repression, contributing to increased expression of male upregulated DEGs (Figure 4E, genes in blue).
In summary, our studies have revealed unexpected differences in XCI maintenance in both mouse and human AT2s, and we propose that reduced Xist RNA and heterochromatic histone modifications could contribute to the high levels of XCI escape across the Xi in mouse AT2 cells. Additionally, we observed a surprisingly large number of genes exhibiting sex-specific expression, which suggests that there could be both genetic and epigenetic contributions for sex biases in lung injury and repair, including both acute (ARDS) and progressive (interstitial lung) disease. Our work is an important first step for understanding the role of sex in lung injury and chronic pathology, and could ultimately contribute to the design of novel sex-specific therapeutic approaches to improve lung repair.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Montserrat C. Anguera (anguera@vet.upenn.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The RNA-seq data are available in the Gene Expression Omnibus database under accession GEO: GSE203166.
AT2 cell isolation
Lungs were inflated with 5 U/mL dispase II in Hanks’ balanced salt solution (Thermo Fisher Scientific), tying off the trachea and cutting lobes away from the main stem bronchi. Lungs were incubated in dispase for 45 min shaking at room temperature, and mechanically dissociated by pipetting in sort buffer (SB; Dulbecco’s modified Eagle’s medium [DMEM; Thermo Fisher] + 2% cosmic calf serum [CC; Thermo Fisher] + 1% penicillin/streptomycin [P/S]). After pelleting at 550 × g for 5 min at 4°C, the whole-lung suspension was treated with Red Blood Cell Lysis buffer (Millipore Sigma) for 5 min, pelleted, and resuspended in SB + 1:1,000 DNase I (Millipore Sigma) for a 45-min recovery period while shaking at 37°C. Whole-lung suspension was then resuspended in SB + 1:50 TruStain FcX (anti-mouse CD16/32) antibody (BioLegend) for a 10-min blocking period at 37°C. For sorts from C57BL6 mice, AT2 cells were collected using allophycocyanin (APC)/Cy7-conjugated rat anti-mouse CD45 antibody (1:200; BioLegend, 30-F11), Alexa Fluor 488-conjugated rat anti-mouse CD326 (EpCAM) antibody (1:200; BioLegend, G8.8), and phycoerythrin (PE)-conjugated rat anti-mouse CD104 (integrin β4) antibody (1:100; BioLegend, 346-11A). Staining was performed for 45 min at 4°C, followed by a final spin-down at 550 × g for 5 min at 4°C. Stained cells and fluorescence-minus-one controls were then resuspended in SB + 1:1,000 DNase + 1:1,000 Draq7 (Beckman Coulter) as a live/dead stain. All fluorescence-activated cell sorting (FACS) was done on a BD FACS Aria Fusion (BD Biosciences), and cells were collected in Falcon round-bottom polystyrene tubes (Thermo Fisher) in 300 μL of DMEM + 20% CC + 2% P/S. Purity of sorted AT2s was routinely performed for each sample as demonstrated in Figure S1.
RNA FISH
For Xist RNA FISH, AT2 cells (mouse and human) were immediately cytospun onto glass slides after FACS, then incubated in ice-cold cytoskeletal (CSK) buffer containing 0.5% Triton X-100 for 3 min, fixed in 4% paraformaldehyde for 10 min, and dehydrated using an ethanol series. For Xist RNA FISH we used two Cy3-labeled oligonucleotide probes, which target repetitive regions within Xist exon 1 (Wang et al., 2016). Images were obtained using a Nikon Eclipse microscope and were categorized by the type of Xist RNA localization patterns as shown in Figure 1C and as described previously (Syrett et al., 2017, 2018; Wang et al., 2016).
XCI gene escape from allele-specific RNA sequencing
Genes that escape XCI were identified using three thresholds of escape, as previously described (Berletch et al., 2015). In brief, diploid gene expression was first calculated in RPKM (reads per kilobase of exon length per million mapped reads), and genes were called expressed if their diploid RPKM was >1. For every X-linked gene that passed this threshold, haploid gene expression was calculated in SRPM, and genes which had an Xi-SRPM >2 were considered to be expressed from the Xi. Finally, a binomial model estimating the statistical confidence of escape probability was applied to the genes passing the first two thresholds. This model compares the proportion of Xi-specific reads with the total X-linked reads (Xi + Xa) and calculates a 95% confidence interval. If the 95% lower confidence limit of a gene’s escape probability was greater than 0, it was called an escape gene.
RNA-seq methods
Using the same fastq files generated for the allele-specific alignment, a separate pipeline was used to quantify standard, diploid gene expression. Reads were aligned to the mm10 genome build using the ultrafast STAR aligner (Dobin et al., 2013) with default parameters and “—quantmode GeneCounts” to quantify gene expression. Read counts generated by STAR were used as input for the DeSeq2 R package to identify DEGs between male and female samples. RPM were also calculated, and a gene was considered expressed if it had an RPM >1 in all female samples. GO analyses were determined using the Metascape platform (Zhou et al., 2019). RNA-seq data are available in the GEO database under accession GEO: GSE203166.
Author contributions
M.C.A. and A.E.V. wrote and edited the manuscript. M.C.A. made the figures from data collected and generated by I.S. and S.P. The AT2 cells were isolated by A.W., G.Z., and A.E.V. The Xist/XIST RNA FISH and IF of human and mouse AT2 cells was performed by I.S. All bioinformatic analyses were performed by S.P. The allele-specific RNA-seq pipeline was developed by X.Y. while training in the lab of B.D.G. A.D. performed genotyping, F1 mus × cast mice organization, and mouse preparation for sorting.
Acknowledgments
We would like to thank L. King for assistance with editing of the manuscript; Z. Beethem for modifications to the code for calculating the confidence intervals for XCI escape genes; D. Beiting of PennVet CHMI for RNA-seq library preparation; N. Jiwrajka for imaging assistance; and all members of the Anguera and Vaughan labs for helpful discussions. We also greatly appreciate the efforts of L. Carrel, H. Markus, and D. Liu for RNA-seq analyses of hAT2 cells. We also thank BioRender for using some of their stock images for figures in this paper. This research was supported by an Administrative Supplement to NIH R01 AI134834 (to M.C.A.), NIH R21 AI164006 (to M.C.A.), Penn IRM COVID pilot grant (to M.C.A. and A.E.V.), Penn Vet COVID/CHMI pilot grant (to M.C.A. and A.E.V.), Lupus Research Foundation Insight award (to M.C.A.), NIH 1F31GM136073-01 to I.S., NIH 1F32AI154797-01 to S.P., and NIH R01HL153539 and Lisa Dean Moseley Research grant to A.V.
Conflict of interests
The authors declare no competing interests.
Published: January 12, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.12.005.
Contributor Information
Andrew E. Vaughan, Email: vaughan@vet.upenn.edu.
Montserrat C. Anguera, Email: anguera@vet.upenn.edu.
Supplemental information
Gene ID, gene name, and chromosome are also shown.
References
- Anguera M.C., Sadreyev R., Zhang Z., Szanto A., Payer B., Sheridan S.D., Kwok S., Haggarty S.J., Sur M., Alvarez J., et al. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell. 2012;11:75–90. doi: 10.1016/j.stem.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaton B.P., Brown C.J. Escape artists of the X chromosome. Trends Genet. 2016;32:348–359. doi: 10.1016/j.tig.2016.03.007. [DOI] [PubMed] [Google Scholar]
- Balaton B.P., Cotton A.M., Brown C.J. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol. Sex Differ. 2015;6:35. doi: 10.1186/s13293-015-0053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas C.E., Cronce M.J., Rackley C.R., Bowie E.J., Keene D.R., Stripp B.R., Randell S.H., Noble P.W., Hogan B.L.M. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch J.B., Yang F., Disteche C.M. Escape from X inactivation in mice and humans. Genome Biol. 2010;11:213. doi: 10.1186/gb-2010-11-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch J.B., Yang F., Xu J., Carrel L., Disteche C.M. Genes that escape from X inactivation. Hum. Genet. 2011;130:237–245. doi: 10.1007/s00439-011-1011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch J.B., Ma W., Yang F., Shendure J., Noble W.S., Disteche C.M., Deng X. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015;11:e1005079. doi: 10.1371/journal.pgen.1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camelo A., Dunmore R., Sleeman M.A., Clarke D.L. The epithelium in idiopathic pulmonary fibrosis: breaking the barrier. Front. Pharmacol. 2014;4:173. doi: 10.3389/fphar.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC COVID-19 Response Team Severe outcomes among patients with coronavirus disease 2019 (COVID-19) — United States, February 12–March 16, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:343–346. doi: 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fett C., Mack M., Ten Eyck P.P., Meyerholz D.K., Perlman S. Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J. Immunol. 2017;198:4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman H.A., Li X., Alexander J.P., Brumwell A., Lorizio W., Tan K., Sonnenberg A., Wei Y., Vu T.H. Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. J. Clin. Invest. 2011;121:2855–2862. doi: 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Jiang Q., Xia X., Liu K., Yu Z., Tao W., Gong W., Han J.J. Individual variation of the SARS-CoV2 receptor ACE2 gene expression and regulation. Aging Cell. 2020;19:e13168. doi: 10.1111/acel.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G., Panning B., Bates B., Pehrson J.R., Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat. Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Cabral L.J., Stephens R.J., Freeman G. Transformation of alveolar type 2 cells to type 1 cells following exposure to NO2. Exp. Mol. Pathol. 1975;22:142–150. doi: 10.1016/0014-4800(75)90059-3. [DOI] [PubMed] [Google Scholar]
- Fischer J., Jung N., Robinson N., Lehmann C. Sex differences in immune responses to infectious diseases. Infection. 2015;43:399–403. doi: 10.1007/s15010-015-0791-9. [DOI] [PubMed] [Google Scholar]
- Galbadage T., Peterson B.M., Awada J., Buck A.S., Ramirez D.A., Wilson J., Gunasekera R.S. Systematic review and meta-analysis of sex-specific COVID-19 clinical outcomes. Front. Med. 2020;7:348. doi: 10.3389/fmed.2020.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan D.S., Dossett L.A., Lightfoot M.A., Fremont R.D., Ware L.B., Sawyer R.G., May A.K. Gender and acute respiratory distress syndrome in critically injured adults: a prospective study. J. Trauma. 2011;71:878–883. doi: 10.1097/TA.0b013e31822c0d31. discussion 883-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Pearce R., Zhang Y. De novo design of protein peptides to block association of the SARS-CoV-2 spike protein with human ACE2. Aging (Albany NY) 2020;12:11263–11276. doi: 10.18632/aging.103416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C.M., Lovell F.L., Leongamornlert D.A., Stranger B.E., Dermitzakis E.T., Ross M.T. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet. 2008;4:e9. doi: 10.1371/journal.pgen.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltofen T., Haase M., Thome U.H., Laube M. Male sex is associated with a reduced alveolar epithelial sodium transport. PLoS One. 2015;10:e0136178. doi: 10.1371/journal.pone.0136178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura H., Sontake V., Tata A., Kobayashi Y., Edwards C.E., Heaton B.E., Konkimalla A., Asakura T., Mikami Y., Fritch E.J., et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell. 2020;27:890–904.e8. doi: 10.1016/j.stem.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen J., Beers M.F. Contributions of alveolar epithelial cell quality control to pulmonary fibrosis. J. Clin. Invest. 2020;130:5088–5099. doi: 10.1172/JCI139519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano-Dourado L., Glassberg M.K., Assayag D., Borie R., Johannson K.A. Sex and gender in interstitial lung diseases. Eur. Respir. Rev. 2021;30:210105. doi: 10.1183/16000617.0105-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S.L., Flanagan K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- Kohlmaier A., Savarese F., Lachner M., Martens J., Jenuwein T., Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau E.S., McNeill J.N., Paniagua S.M., Liu E.E., Wang J.K., Bassett I.V., Selvaggi C.A., Lubitz S.A., Foulkes A.S., Ho J.E. Sex differences in inflammatory markers in patients hospitalized with COVID-19 infection: insights from the MGH COVID-19 patient registry. PLoS One. 2021;16:e0250774. doi: 10.1371/journal.pone.0250774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y., Panning B., Dausman J., Strauss W., Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- McCoy D.M., Salome R.G., Kusner D.J., Iyar S.S., Mallampalli R.K. Identification of sex-specific differences in surfactant synthesis in rat lung. Pediatr. Res. 1999;46:722–730. doi: 10.1203/00006450-199912000-00003. [DOI] [PubMed] [Google Scholar]
- McNicholas B.A., Madotto F., Pham T., Rezoagli E., Masterson C.H., Horie S., Bellani G., Brochard L., Laffey J.G., LUNG SAFE Investigators and the ESICM Trials Group Demographics, management and outcome of females and males with acute respiratory distress syndrome in the LUNG SAFE prospective cohort study. Eur. Respir. J. 2019;54:1900609. doi: 10.1183/13993003.00609-2019. [DOI] [PubMed] [Google Scholar]
- Mekhoubad S., Bock C., de Boer A.S., Kiskinis E., Meissner A., Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parimon T., Yao C., Stripp B.R., Noble P.W., Chen P. Alveolar epithelial type II cells as drivers of lung fibrosis in idiopathic pulmonary fibrosis. Int. J. Mol. Sci. 2020;21:2269. doi: 10.3390/ijms21072269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B., Lee J.T. X chromosome dosage compensation: how mammals keep the balance. Annu. Rev. Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Peckham H., de Gruijter N.M., Raine C., Radziszewska A., Ciurtin C., Wedderburn L.R., Rosser E.C., Webb K., Deakin C.T. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020;11:6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny G.D., Kay G.F., Sheardown S.A., Rastan S., Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Plath K., Fang J., Mlynarczyk-Evans S.K., Cao R., Worringer K.A., Wang H., de la Cruz C.C., Otte A.P., Panning B., Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Pyfrom S., Paneru B., Knox J.J., Cancro M.P., Posso S., Buckner J.H., Anguera M.C. The dynamic epigenetic regulation of the inactive X chromosome in healthy human B cells is dysregulated in lupus patients. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2024624118. e2024624118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully E.P., Haverfield J., Ursin R.L., Tannenbaum C., Klein S.L. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol. 2020;20:442–447. doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard M., Provost P.R., Tremblay Y. Sexually dimorphic gene expression that overlaps maturation of type II pneumonocytes in fetal mouse lungs. Reprod. Biol. Endocrinol. 2006;4:25. doi: 10.1186/1477-7827-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrett C.M., Sindhava V., Hodawadekar S., Myles A., Liang G., Zhang Y., Nandi S., Cancro M., Atchison M., Anguera M.C. Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS Genet. 2017;13:e1007050. doi: 10.1371/journal.pgen.1007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrett C.M., Sindhava V., Sierra I., Dubin A.H., Atchison M., Anguera M.C. Diversity of epigenetic features of the inactive X-chromosome in NK cells, dendritic cells, and macrophages. Front. Immunol. 2018;9:3087. doi: 10.3389/fimmu.2018.03087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrett C.M., Paneru B., Sandoval-Heglund D., Wang J., Banerjee S., Sindhava V., Behrens E.M., Atchison M., Anguera M.C. Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight. 2019;4:e126751. doi: 10.1172/jci.insight.126751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Ellingson M.K., Wong P., Israelow B., Lucas C., Klein J., Silva J., Mao T., Oh J.E., Tokuyama M., et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588:315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukiainen T., Villani A.C., Yen A., Rivas M.A., Marshall J.L., Satija R., Aguirre M., Gauthier L., Fleharty M., Kirby A., et al. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan A.E., Brumwell A.N., Xi Y., Gotts J.E., Brownfield D.G., Treutlein B., Tan K., Tan V., Liu F.C., Looney M.R., et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517:621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Syrett C.M., Kramer M.C., Basu A., Atchison M.L., Anguera M.C. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc. Natl. Acad. Sci. USA. 2016;113:E2029–E2038. doi: 10.1073/pnas.1520113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A.I., Jackson S.R., Zhao G., Quansah K.K., Farshchian J.N., Neupauer K.M., Littauer E.Q., Paris A.J., Liberti D.C., Scott Worthen G., et al. Mesenchyme-free expansion and transplantation of adult alveolar progenitor cells: steps toward cell-based regenerative therapies. NPJ Regen. Med. 2019;4:17. doi: 10.1038/s41536-019-0080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenham C., Smith J., Morgan R., Gender and COVID-19 Working Group COVID-19: the gendered impacts of the outbreak. Lancet. 2020;395:846–848. doi: 10.1016/S0140-6736(20)30526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y., Kim T., Brumwell A.N., Driver I.H., Wei Y., Tan V., Jackson J.R., Xu J., Lee D.K., Gotts J.E., et al. Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat. Cell Biol. 2017;19:904–914. doi: 10.1038/ncb3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim E., Kirby J.E., Brown D.E., Mercier F.E., Sadreyev R.I., Scadden D.T., Lee J.T. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Yan Y., Shu Y., Gao R., Sun Y., Li X., Ju X., Liang Z., Liu Q., Zhao Y., et al. Angiotensin-converting enzyme 2 protects from lethal avian influenza A H5N1 infections. Nat. Commun. 2014;5:3594. doi: 10.1038/ncomms4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene ID, gene name, and chromosome are also shown.
Data Availability Statement
The RNA-seq data are available in the Gene Expression Omnibus database under accession GEO: GSE203166.