Summary
Multiple chromatin modifiers associated with H3K9me3 play important roles in the transition from embryonic stem cells to 2-cell (2C)-like cells. However, it remains elusive how H3K9me3 is remodeled and its association with totipotency. Here, we integrated transcriptome and H3K9me3 profiles to conduct a detailed comparison of 2C embryos and 2C-like cells. Globally, H3K9me3 is highly preserved and H3K9me3 dynamics within the gene locus is not associated with gene expression change during 2C-like transition. Promoter-deposited H3K9me3 plays non-repressive roles in the activation of genes during 2C-like transition. In contrast, transposable elements, residing in the nearby regions of up-regulated genes, undergo extensive elimination of H3K9me3 and are tended to be induced in 2C-like transitions. Furthermore, a large fraction of trophoblast stem cell-specific enhancers undergo loss of H3K9me3 exclusively in MERVL+/Zscan4+ cells. Our study therefore reveals the unique H3K9me3 profiles of 2C-like cells, facilitating the further exploration of totipotency.
Keywords: pluripotency, totipotency, H3K9me3, transposable elements (TEs)
Graphical abstract

Highlights
-
•
H3K9me3 is highly preserved during 2C-like transition
-
•
Transcriptional burst of TEs occurs predominantly in MERVL+/Zscan4+ cells
-
•
H3K9me3 barrier of cell fate segregation is erased in MERVL+/Zscan4+ cells
In this article, Zhang and colleagues show that removal of H3K9me3 is accompanied with the transcriptional burst of TEs residing in the nearby regions of up-regulated genes, whereas H3K9me3 remains constant in these regions in Zscan4+ cells. Furthermore, a large fraction of trophoblast stem cell-specific enhancers undergo loss of H3K9me3 exclusively in MERVL+/Zscan4+ cells.
Introduction
Histone 3 lysine 9 trimethylation (H3K9me3) is crucial for cell fate control (Becker et al., 2016; Nicetto and Zaret, 2019). H3K9me3 participates in the formation of heterochromatin to prevent the inappropriate expression of cell-type incompatible genes and repeats (Becker et al., 2016; Burton et al., 2020; Hawkins et al., 2010; Martens et al., 2005; Nakayam et al., 2001; Rea et al., 2000; Zhu et al., 2013). Following fertilization, H3K9me3 undergoes extensive reprogramming (Burton et al., 2020; Li, 2002; Nicetto and Zaret, 2019; Wang et al., 2018). Disruption in the reprogramming of H3K9me3-dependent heterochromatin is detrimental to early embryogenesis (Burton et al., 2020; Wang et al., 2018). Meanwhile, different classes of H3K9 methyltransferases control distinct genomic regions and display highly embryonic-stage-specific expression patterns, indicating orchestrated and complex regulation of H3K9me3 in early embryos (Burton et al., 2020; Wang et al., 2018). H3K9me3 is a key barrier of the reprogramming process toward totipotency (Chung et al., 2015; Liu et al., 2016; Matoba et al., 2014). In addition to the natural reprogramming process through fertilization, totipotency can also be acquired through somatic cell nuclear transfer (SCNT). The low efficiency of SCNT is closely linked with the failure to remove H3K9me3 of the repetitive elements in the reprogramming-resistant regions (Matoba et al., 2014).
Recently, it has been reported that a rare group of ESCs are capable of transiently entering a state resembling 2-cell (2C) stage embryos (Macfarlan et al., 2012; Zalzman et al., 2010). In addition to showing 2C-like transcriptome feature, the striking hallmark of these so-called 2C-like cells is their expanded developmental potential to form both embryonic and extraembryonic tissues (Macfarlan et al., 2012). The scarcity of early embryos strongly hinders the study of the underlying principle of totipotency, whereas the identification of 2C-like cells provides a unique in vitro model for understanding the molecular basis of totipotency. By using 2C-like cells, the essential regulators and their functions in the establishment of totipotency have been identified (Chen and Zhang, 2019; De Iaco et al., 2017, 2019; Eckersley-Maslin et al., 2019; Guo et al., 2019; Hendrickson et al., 2017; Percharde et al., 2018; Tian et al., 2019; Whiddon et al., 2017; Yan et al., 2019; Zhang et al., 2019).
MERVL−/Zscan4+ cells (termed Zscan4+ cells hereafter) and MERVL+/Zscan4+ cells are the main subpopulations during 2C-like transition, sharing similar transcriptome features (Eckersley-Maslin et al., 2016). MERVL+/Zscan4+ cells arise primarily from Zscan4+ cells (Rodriguez-Terrones et al., 2018). However, MERVL+/Zscan4+ cells are capable of contributing to embryonic and extraembryonic tissues, while Zscan4+ cells show lower competency than both ESCs and MERVL+/Zscan4+ cells (Amano et al., 2013; Macfarlan et al., 2012; Zhang et al., 2021). Multiple chromatin modifiers associated with H3K9me3 have been implicated in the mediation of the 2C-like transition (Maksakova et al., 2013; Wu et al., 2020). However, the role of H3K9me3 in the pluripotency-to-totipotency transition remains largely unknown.
To further explore the epigenetic signatures underlying totipotency and the different competency between MERVL+/Zscan4+ cells and Zscan4+ cells, we mapped the genome-wide profiles of H3K9me3 in these cells and conducted detailed comparisons among 2C embryos, MERVL+/Zscan4+ cells, Zscan4+ cells, and ESCs. We find that removal of H3K9me3 is accompanied with the transcriptional burst in a large fraction of transposable elements (TEs) in MERVL+/Zscan4+ cells, whereas H3K9me3 is highly preserved or newly acquired in the TE regions with mild transcriptional activation in Zscan4+ cells. Furthermore, we also find that a large fraction of trophoblast stem cell-specific enhancers undergo loss of H3K9me3 exclusively in MERVL+/Zscan4+ cells during 2C-like transitions.
Results
Genome-wide profiling of H3K9me3 in 2C-like ESCs
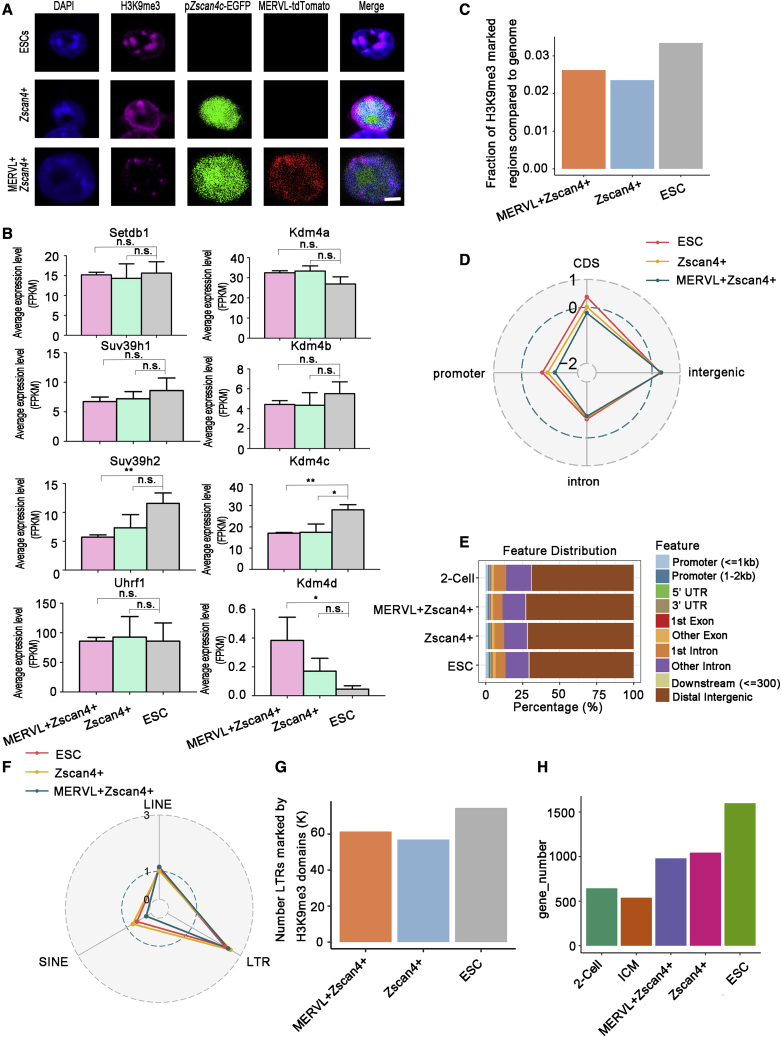
ESC lines containing MERVL-tdTomato and pZscan4c-EGFP fluorescent reporters were constructed and validated as we previously described (Zhang et al., 2021). We first analyzed the distribution of H3K9me3 by immunofluorescence (Figure 1A). Consistent with a previous study (Akiyama et al., 2015), there were larger and fewer DAPI-dense domains in Zscan4+ cells (Figure 1A). Besides, Zscan4+ cells exhibited co-localization of H3K9me3 clusters and DAPI-rich regions akin to the pattern in ESCs (Figure 1A). In contrast, MERVL+/Zscan4+ cells showed small and faint H3K9me3 spots in nuclei and significantly reduced H3K9me3 deposition in DAPI-rich regions compared with ESCs and Zscan4+ cells (Figure 1A). This observation was consistent with prior findings that MERVL+/Zscan4+ cells display molecular features of chromatin decondensation and activation of pericentromeric heterochromatin (Ishiuchi et al., 2015).
Figure 1.
Genome-wide profiling of H3K9me3 in MERVL+/Zscan4+ cells and Zscan4+ cells
(A) Immunostaining analysis with a H3K9me3 antibody. Zscan4 expression was visualized with EGFP (green), and MERVL expression was visualized with tdTomato (red). Cell nuclei were visualized with DAPI. Scale bar, 20 μm. Two independent experiments were performed.
(B) Mean RNA expression level (FPKM, fragments per kilobase of transcript per million) of H3K9me3 writers and erasers determined by RNA-seq data (Eckersley-Maslin et al., 2016). n = 3; mean ± SD; n.s., not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001; unpaired Student’s t test.
(C) Bar plot showing the fraction of genomic regions covered by H3K9me3 peaks in the indicated cells. The value of the whole fraction (the whole genome) is equal to “1.”
(D) Radar chart showing the enrichment score (log ratio of observed/random) of H3K9me3 peaks in promoters, the coding sequence (CDS), intergenic and intron regions. The enrichment score >0 indicates that H3K9me3 domain is enriched in the indicated regions.
(E) The genome-wide distribution of H3K9me3 peaks in 2-cell embryos, and the indicated cells.
(F) Radar chart showing the enrichment score (log ratio of observed/random) of H3K9me3 peaks in TEs. The enrichment score >0 indicates that H3K9me3 domain is enriched in the indicated regions.
(G) Bar plot showing the number of H3K9me3-marked LTR regions in the indicated cells.
(H) Bar plot showing the number of H3K9me3-marked promoter regions in 2-cell embryos, ICM, and the indicated cells.
See also Figure S1.
Next, we sorted Zscan4+ cells and MERVL+/Zscan4+ cells by fluorescence-activated cell sorting and performed chromatin immunoprecipitation sequencing (ChIP-seq) to generate the genome-wide H3K9me3 profiles. The two replicates of H3K9me3 for each cell type had Pearson’s correlation coefficients >0.9, indicating the high reproducibility of the H3K9me3 data (Figure S1A). Principal-component analysis based on H3K9me3 signals revealed that Zscan4+ cells were similar to ESCs, while MERVL+/Zscan4+ cells exhibited an intermediate state between ESCs and 2-cell embryos along the PC1 axis (constituting 99% of the variation) (Figure S1B). Notably, MERVL+/Zscan4+ cells were much closer to ESCs than to 2C embryos on the PC1 axis. Next, we performed peak calling analysis using model-based analysis for ChIP-seq (MACS2) to identify H3K9me3-enriched domains in ESCs, Zscan4+ cells, and MERVL+/Zscan4+ cells (Figure S1C). MERVL+/Zscan4+ cells, Zscan4+ cells, and ESCs shared similar H3K9me3 peak length density patterns (Figure S1D). In addition, MERVL+/Zscan4+ cells exhibited decreased H3K9me3 signal for peaks compared with that of ESCs and Zscan4+ cells (Figure S1E). Next, we investigated the dynamics of H3K9me3 domains during 2C-like transition. Extensive loss and gain of H3K9me3 domains were observed during 2C-like transition, especially in the transition from Zscan4+ cells to MERVL+/Zscan4+ cells (Figures S1F–S1K). Accompanied with the major change (Figures S1F–S1K), MERVL+/Zscan4+ cells exhibited significantly decreased expression levels of H3K9 methyltransferases Suv39h2 and H3K9me3 eraser Kdm4c compared with ESCs (Figure 1B). In addition, expression levels of Setdb1 remained constant in both Zscan4+ cells and MERVL+/Zscan4+ cells compared with ESCs (Figure 1B). Although Setdb1 is responsible for the inhibition of 2C-like transition (Wu et al., 2020), the above data suggest that depletion of Setdb1 may not be required in the spontaneous 2C-state entry.
Next, we examined the genome-wide distribution of the H3K9me3. As illustrated in Figure 1C, the fraction of the genome covered by H3K9me3 peaks was reduced in Zscan4+ cells and MERVL+/Zscan4+ cells compared with that in ESCs. In addition, MERVL+/Zscan4+ cells displayed slightly higher fraction of the genome covered by H3K9me3 peaks than that in Zscan4+ cells (Figure 1C). This result seems inconsistent with the immunofluorescent staining in Figure 1A. One possible explanation is that uniquely assigned reads were used in this genomic analysis (Figure 1C) and the status of low mappability repeat regions, typically enriched for H3K9me3, may be underrepresented in this result. In addition, H3K9me3 peak signal in MERVL+/Zscan4+ cells was lower than Zscan4+ cells and ESCs (Figure S1E). We then assigned H3K9me3 peaks to genome features. All three cell types and 2C embryos shared similar H3K9me3 distribution patterns with preferential enrichment in the intergenic regions (Figures 1D and 1E). Globally, H3K9me3 was not enriched in the promoter regions in all three cell types (Figure 1D). In addition, H3K9me3 domains were highly enriched in LTRs in Zscan4+ cells and MERVL+/Zscan4+ cells (Figure 1F). The number of LTRs marked by H3K9me3 was decreased in Zscan4+ cells and MERVL+/Zscan4+ cells compared with ESCs (Figure 1G). A similar trend was also observed in the promoter regions (Figure 1H).
H3K9me3 dynamics at gene loci during 2C-like transition
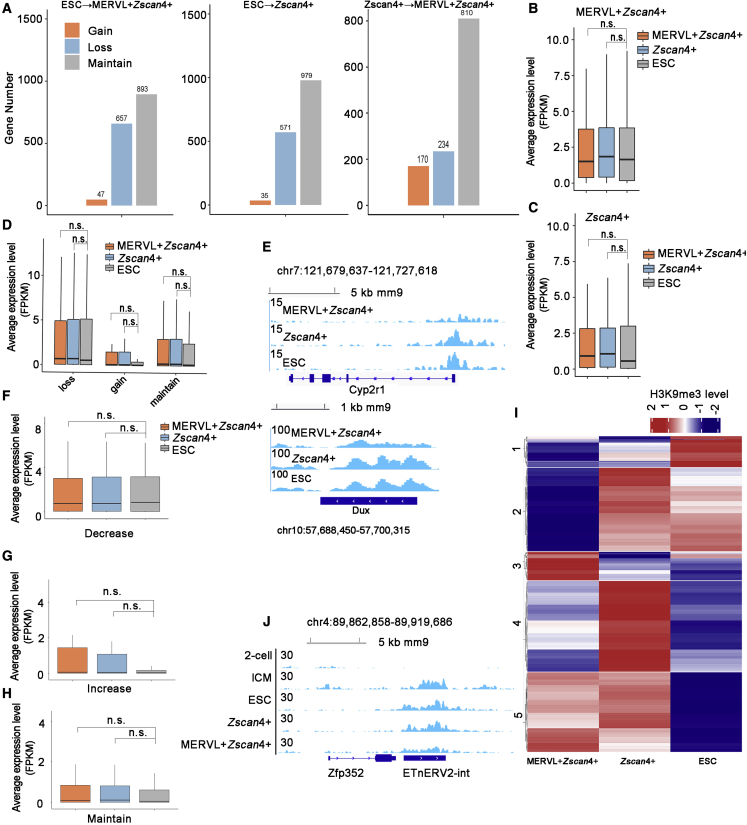
Next, we investigated the regulatory role of H3K9me3 in gene expression during 2C-like transition. The PCA plot of promoter H3K9me3 signals showed that ESCs and Zscan4+ cells were clustered together (Figure S2A). Similar to Figure S1B, MERVL+/Zscan4+ cells were much closer to ESCs and Zscan4+ cells than 2C embryos along the PC1 axis (constituting 95% of the variation) (Figure S2A). Among the 1,598 genes decorated with H3K9me3 peaks at the promoter regions in ESCs, 657 and 571 genes showed loss of promoter H3K9me3 in MERVL+/Zscan4+ cells and Zscan4+ cells, respectively (Figure 2A). The newly formed H3K9me3-marked promoters were rare in MERVL+/Zscan4+ cells and Zscan4+ cells (Figures 2A and S2B). Only a small fraction of genes (47 genes in MERVL+/Zscan4+ cells and 35 genes in Zscan4+ cells compared with ESCs; 170 genes during the transition from Zscan4+ cells to MERVL+/Zscan4+ cells) newly acquired promoter H3K9me3 during 2C-like transition (Figure 2A). Of note, Zscan4+ cells and MERVL+/Zscan4+ cells possessed far more H3K9me3 marked promoters than early embryos (Figure 1H). Next, we investigated whether the dynamics of promoter H3K9me3 during 2C-like transition could reflect the feature of preimplantation development. By using the genome-wide map of H3K9me3 in preimplantation embryos (Wang et al., 2018), we identified 381 genes displaying ICM-specific H3K9me3 in promoter regions compared with 2-cell embryos (Figure S2C). Among the genes that lost promoter H3K9me3 during 2C-like transition, only a small fraction carried ICM-specific-H3K9me3 marks (36 genes in MERVL+/Zscan4+ cells and 49 genes in Zscan4+ cells) (Figure S2D), suggesting that reprogramming of promoter H3K9me3 during 2C-like transition does not recapitulate embryonic development in reverse. Although showing enhanced expression in 2C embryos during preimplantation development (Figure S2E), genes lost ICM-specific-H3K9me3 marks during 2C-like transition were failed to be up-regulated in MERVL+/Zscan4+ cells and Zscan4+ cells (Figures 2B and 2C), indicating a lack of other regulatory controllers.
Figure 2.
H3K9me3 dynamics at gene loci during 2C-like transitions
(A) Bar plot showing the number of genes with gain, loss, or maintenance of H3K9me3 marks in promoters during 2C-like transition.
(B and C) Boxplot showing the mean RNA expression level (FPKM) of genes loss ICM-specific-H3K9me3 mark during the transition from ESCs to MERVL+/Zscan4+ cells (B) or from ESCs to Zscan4+ cells (C) determined by RNA-seq data (Eckersley-Maslin et al., 2016). n = 3 biological replicates.
(D) Boxplot for the mean RNA expression level (FPKM) of genes showing loss, gain or maintenance of promoter H3K9me3 during the transition from ESCs to MERVL+/Zscan4+ cells. n = 3 biological replicates.
(E) Integrative genomics viewer of H3K9me3 signals at the Cyp2r1 (top) and Dux (bottom) range in the indicated cells.
(F–H) Mean RNA expression level of genes showing decreased (F), increased (G), or maintained (H) intronic H3K9me3 signals in MERVL+/Zscan4+ cells compared with ESCs determined by RNA-seq (Eckersley-Maslin et al., 2016). n = 3 biological replicates.
(I) Heatmap showing the dynamics of H3K9me3 domains in the nearby regions ( 20 kb around TSS) of up-regulated genes during 2C-like transition. Each row represents the Z score of H3K9me3 signal subtract input.
(J) Integrative genomics viewer of H3K9me3 signals at the Zfp352 region in the indicated cells. The center of the boxplots in Figure 2 represents the median value and the lower and upper lines represent the 25% and 75% quantiles, respectively. Statistical analysis of Figure 2, p value, Wilcoxon rank-sum test, n.s. represents not significant, ∗ represents p < 0.05, ∗∗ represents p < 0.01, and ∗∗∗ represents p < 0.001.
See also Figure S2.
Next, we investigated the correlation between H3K9me3 dynamics and gene expression change during 2C-like transition. Globally, neither gain nor loss of H3K9me3 at promoter regions was associated with transcriptional change in MERVL+/Zscan4+ cells compared with ESCs (Figure 2D). Generally, genes activated during 2C-like transition displayed low fold enrichment of H3K9me3 at promoter regions in all three cell types (Figure S2F). In addition, a small subset of up-regulated genes retained high levels of H3K9me3 during 2C-like transition, suggesting that promoter H3K9me3 is not the major controller to silence 2C-like transcriptome in ESCs. For instance, the up-regulated gene Cyp2r1 remained high H3K9me3 enrichment at the promoter region in Zscan4+ cells (Figures 2E and S2G), indicating a non-repressive role of promoter H3K9me3 during 2C-like transition. Interestingly, the expression pattern of Cyp2r1 was associated with totipotency in early preimplantation development (Figure S2H). Another representative example was Dux (Figures 2E and S2I), an essential driver governing 2C-like transition (De Iaco et al., 2017; Hendrickson et al., 2017; Whiddon et al., 2017).
We observed that a large portion of H3K9me3 peaks were located in introns in all three cell types (Figure 1E). Although H3K9me3 was not enriched in the intronic regions in general (Figure 1D), strong H3K9me3 enrichment in the intronic regions of 2C-specific genes was found in all three cell types (Figure S2J). Intronic heterochromatin plays vital roles in transcriptional controls (Espinas et al., 2020; Karimi et al., 2011; Peaston et al., 2004). We found that 345 genes exhibited reduced H3K9me3 levels at intronic regions in MERVL+/Zscan4+ cells, among which, 102 genes showed 2C-specific expression pattern in preimplantation development while failed to be up-regulated in 2C-like cells (Table S1, Figure S2K). In addition, neither decreased nor increased intronic H3K9me3 levels were associated with gene expression change during 2C-like transition (Figures 2F–2H and S2L–S2N). To summarize, the above evidence suggests that H3K9me3 dynamics within the gene locus is not globally associated with gene expression change in 2C-like cells.
H3K9me3 dynamics in the vicinity of up-regulated genes during 2C-like transition
H3K9me3 is capable of mediating gene repression via modulation of the activity of promoters or enhancers (Vahedi et al., 2015). Since few genes are regulated by promoter-deposited H3K9me3, we next analyzed dynamics of H3K9me3 domains in the vicinity ( 20 kb around TSS) of up-regulated genes during 2C-like transition (Figure 2I). The dynamics of H3K9me3 domains displayed different patterns (Figure 2I). We focused on the cluster 1 and cluster 2, which included 237 domains marked by high levels of H3K9me3 in ESCs (Figure 2I). A significant portion of these domains maintained high H3K9me3 levels during the transition from ESCs to Zscan4+ cells (Figure 2I). By contrast, H3K9me3 levels were severely reduced in most of these domains in MERVL+/Zscan4+ cells. In addition, the corresponding genes in the vicinity displayed 2C-specific expression pattern during early embryogenesis (Table S2). Interestingly, concomitant with the reduced H3K9me3 levels in the nearby regions, the corresponding genes displayed gradually up-regulated expression levels during 2C-like transition (Figure S2O). It has been previously reported that ERVs can act as regulatory elements to stimulate the transcription of the nearby genes (Fueyo et al., 2022; Macfarlan et al., 2012; Rowe et al., 2013). Next, we focused on ERVs and identified 2,073 ERV elements residing in these regions (Table S2). A representative case was ETnERV2-int/ERVK, located ∼7 kb downstream of the 2C-specific gene Zfp352 (Figure 2J), which remained a high H3K9me3 level in Zscan4+ cells while displaying severely reduced H3K9me3 level in MERVL+/Zscan4+ cells, accompanied with gradually enhanced expression levels in 2C-like transition (Figures 2J and S2P). In summary, these results suggest that diminished H3K9me3 in the vicinity of up-regulated genes may act as enhancers to further promote the 2C-like transcriptome.
TEs transcription burst occurs predominantly in MERVL+/Zscan4+ cells
Although sharing similar gene expression pattern, we and others have reported that MERVL expression is significantly different between MERVL+/Zscan4+ cells and Zscan4+ cells (Eckersley-Maslin et al., 2016; Zhang et al., 2021). To expand this observation to other TEs, we generated comprehensive profiles of repeat transcriptome in all three cell types. MERVL+/Zscan4+ cells displayed a more extensive change in repeats transcription than Zscan4+ cells (Figure 3A, Table S3). Among 785 repeat subfamilies, 210 differentially expressed TE subfamilies were identified in MERVL+/Zscan4+ cells while only 42 differentially expressed TE subfamilies were found in Zscan4+ cells (Table S3). Down-regulated TEs were rare in 2C-like transition (four subfamilies in MERVL+/Zscan4+ cells and one subfamily in Zscan4+ cells) (Table S3). Nearly all the highly up-regulated TEs belonged to ERVs (Table S3). Interestingly, TEs located in reprogramming-resistant regions (RRR) enriched of ERVs, previously identified as barriers for SCNT (Matoba et al., 2014), were also significantly up-regulated during 2C-like transition (Figure 3B).
Figure 3.
Expression change and H3K9me3 dynamics in TEs during 2C-like transitions
(A) Volcano plots showing differentially expressed TEs between MERVL+/Zscan4+ cells and ESCs (left panel) or Zscan4+ cells and ESCs (right panel). Red dots represent upregulated TEs and blue dots represent downregulated TEs (TE families with RPKM change greater than 2-fold or less than 0.5-fold). See methods in supplemental experimental procedures for details. n = 3 biological replicates.
(B) Boxplot showing the average expression levels of TEs (RPKM) located in the RRR regions as defined in (Matoba et al., 2014) determined by RNA-seq (Eckersley-Maslin et al., 2016). n = 3 biological replicates.
(C) Bar plots showing the average expression levels (RPKM) of indicated ERVs in the indicated cells determined by RNA-seq (Eckersley-Maslin et al., 2016). n = 3 biological replicates.
(D) Bar plots showing the average expression levels of indicated ERVs in preimplantation development determined by RNA-seq (Deng et al., 2014). n = 4 biological replicates.
(E) Heatmaps showing H3K9me3 signals (left) and expression levels (right) of TEs in the indicated cells. Left panel, each row represents the Z score of H3K9me3 signal subtract input. Right panel, each row represents the Z score of expression level (RPKM). The heatmap in the right panel was generated using the same order of H3K9me3 clusters.
(F) Integrative genomic viewer of H3K9me3 signals at the indicated loci in the indicated cells.
(G) Boxplot showing the average expression levels (RPKM) of TEs included in Figure S3L during early embryo development determined by RNA-seq (Deng et al., 2014). n = 4 biological replicates.
(H) Bar plot showing the average expression levels of elements belonging to the ERVK subfamilies in the nearby regions of up-regulated genes (included in Figure S3L) or across the whole genome in the indicated cells. n = 3 biological replicates. The center of the boxplots in Figure 3 represents the median value and the lower and upper lines represent the 25% and 75% quantiles, respectively. Statistical analysis of Figure 3, p value, Wilcoxon rank-sum test, n.s. represents not significant, ∗ represents p < 0.05, ∗∗ represents p < 0.01, and ∗∗∗ represents p < 0.001.
See also Figure S3.
Next, we mainly focused on ERVs that display highly stage-specific expression pattern in preimplantation embryos (Fadloun et al., 2013; Zhang et al., 2019). First, we analyzed the expression of the four major ERV families (ERV1, ERVK, ERVL, and MalR). Overall, the global transcription of ERVs displayed graded expression pattern, ranging from modest (in Zscan4+ cells) to intense increase (in MERVL+/Zscan4+ cells) (Figure S3A). ERVL was the most highly up-regulated ERV family in 2C-like cells and 2C embryos (Figures S3A and S3B). Expression levels of ERV families varied between 2C-like cells and 2C embryos (Figures S3A and S3B). For example, MalR, the second highly expressed ERV family in 2C embryos (Figure S3B), exhibited a moderate expression level similar to the ERV1 and ERVK family in 2C-like cells (Figure S3A).
Next, we conducted detailed analysis within each ERV family. In the ERVL family, MERVL and MT2 were the most highly up-regulated elements in both 2C-like cells and 2C embryos (Figure S3C, Table S3), consistent with previous reports (Macfarlan et al., 2012). We also observed that other ERV elements (ORR1A3-int/MaLR, ORR1A0-int/MaLR, ORR1A1/MaLR, RLTR19/ERVK), which were highly expressed in 2-cell and 4-cell embryos, were significantly reactivated in MERVL+/Zscan4+ cells (Figures 3C and 3D; Table S3). Similarly, these repeats displayed mild magnitude of activation or remained unchanged in Zscan4+ cells (Figures 3C and 3D; Table S3). In contrast, we also observed that ERVs (such as MMETn-int/ERVK), highly expressed in ESCs, were significantly down-regulated in 2C-like cells, similar to the pattern in early embryos (Figures 3C and 3D; Table S3).
Collectively, these results suggest that burst of LTR transcription associated with totipotency in early embryos predominantly occurs in MERVL+/Zscan4+ cells other than in Zscan4+ cells.
Low H3K9me3 enrichment is associated with TE transcriptional burst in MERVL+/Zscan4+ cells
H3K9me3 is the major mechanism for the transcriptional silencing of TEs in ESCs (Bulut-Karslioglu et al., 2014; Matsui et al., 2010; Rowe et al., 2010). H3K9me3 was highly enriched in LTRs and LINEs in ESCs (Figure 1F). Clustering analysis of H3K9me3 occupancy on these regions indicated that MERVL+/Zscan4+ cells and Zscan4+ cells displayed a pattern intermediate to those of ESCs and 2C embryos along the PC1 axis (describes the greatest variance in the datasets) (Figure S3D). Compared with the distance to 2C embryos, MERVL+/Zscan4+ cells and Zscan4+ cells were much closer to ESCs along the PC1 axis (Figure S3D). In general, LTRs with highest H3K9me3 enrichment (IAP/ERVK and MMERVK10C-int/ERVK) in ESCs belonged to the ERVK family and displayed gradually decreased H3K9me3 levels during 2C-like transition (Figures S3E–S3G). ERVL and MalR, which highly contribute to the ERV transcriptome in the oocytes (Franke et al., 2017; Peaston et al., 2004; Waterston et al., 2002), displayed low H3K9me3 enrichment in all three cell types (Figures S3E and S3G).
Next, we investigated H3K9me3 dynamics of individual TE elements. Six clusters of TEs were identified (Figure 3E). Generally, TEs maintained high or further acquired H3K9me3 deposition in the transition from ESCs to Zscan4+ cells (Figure 3E). One representative example was RLTR8/ERVK, highly expressed in 2-cell stage embryos, showing extensively increased H3K9me3 enrichment in Zscan4+ cells (Figures 3E and S3H; Table S3). Surprisingly, we observed a modest increased expression level of TEs in Zscan4+ cells (Figures 3E and S3I). Thus, H3K9me3 seems to play a non-repressive role in TE regulation for Zscan4+ cells (Figure S3I). However, due to the highly repetitive nature of TEs, one alternative possibility is that the average H3K9me3 level may not reflect the status of the transcriptional active elements inside a given family.
Either increase or decrease in H3K9me3 level at TE regions was observed in MERVL+/Zscan4+ cells compared with ESCs (Figure 3E). A large portion of TEs displayed decreased H3K9me3 enrichment or maintained low levels in MERVL+/Zscan4+ cells, correlated with the transcriptional burst (cluster 1–3, Figure 3E, Table S3). Although 2C-specific retrotransposon MERVL displayed increased H3K9me3 signals in MERVL+/Zscan4+ cells compared with ESCs (Figure 3E, cluster 5), MERVL displayed extremely low fold enrichment of H3K9me3 in all three cell types (Figures S3J and S3K), in line with previous findings (Chen et al., 2021; Cossec et al., 2018; Maksakova et al., 2013).
Cluster 1 represented repeats with gradually reduced H3K9me3 level during 2C-like transition (Figure 3E). Seven out of 23 elements in cluster 1 belonged to the ERVK family (Figure 3E, Table S3), in line with the previous findings that H3K9me3 is the major suppressor of ERVK transposons (Karimi et al., 2011). TEs associated with totipotent cells in early embryogenesis were also found in cluster 1 (Figure 3E, Table S3). A representative example was LINE1 elements (L1MD_ Gf) with critical role in open chromatin status at the beginning of development (Figures 3E and 3F) (Jachowicz et al., 2017). In addition, RLTR10C/ERVK, especially expressed in 4-cell embryos (Table S3), was also included in cluster 1 (Figures 3E and 3F).
Since a group of up-regulated genes showed reduced H3K9me3 enrichment in the nearby regions during 2C-like transitions (Figure 2I, cluster 1 and cluster 2), we next investigated the behavior of TEs within these regions. We focused on TEs displaying extensive H3K9me3 loss in these regions (Figure S3L). In contrast to the general trend of H3K9me3 maintenance or acquisition, TEs belonging to this subtype showed mild H3K9me3 reduction in Zscan4+ cells compared with ESCs (Figure S3L). Furthermore, TEs of this subtype showed markedly increased expression in MERVL+/Zscan4+ cells compared with ESCs, resembling the pattern of their in vivo counterparts (Figures S3M and 3G). We chose ERVK as a representative example. Compared with the whole subfamilies, individual elements defined in Figure S3L showed more dramatic reactivation, ranging from a modest increase in Zscan4+ cells to a robust increase in MERVL+/Zscan4+ cells (Figure 3H). MMETn−int/ERVK was an interesting example. MMETn−int elements in the vicinity of up-regulated genes showed enhanced expression during 2C-like transition (Figure 3H). In contrast, the average expression of the whole MMETn−int family displayed an opposite pattern (Figure 3H). Collectively, these results indicate that TEs, residing in the regions undergoing H3K9me3 loss near the up-regulated genes, display a strong tendency of reactivation during 2C-like transition.
Lineage-specific H3K9me3 is removed in MERVL+/Zscan4+ cells
MERVL+/Zscan4+ cells are superior to Zscan4+ cells with the potential to contribute to both embryonic and extraembryonic lineages (Zhang et al., 2021). H3K9me3 plays vital role in restricting the conversion of ESCs toward trophoblast stem cells (TSCs) (Lohmann et al., 2010; Yuan et al., 2009). Next, we aimed to study whether H3K9me3 barrier associated with cell fate segregation is erased during 2C-like transition.
The total number of H3K9me3 peaks was markedly reduced in TSCs compared with ESCs (Figure S4A). Next, we analyzed regions decorated with ESC-specific H3K9me3 peaks during 2C-like transition. First, we focused on H3K9me3 occupancy at promoter regions. We identified 933 genes carrying ESC-specific H3K9me3 and 120 genes carrying TSC-specific H3K9me3. We defined these genes as ESC-specific H3K9me3 genes or TSC-specific H3K9me3 genes, respectively. Globally, TSC-specific H3K9me3 genes maintained low H3K9me3 level in all three cell types (Figure S4B). Next, we focused on ESC-specific H3K9me3 genes. In general, ESC-specific H3K9me3 genes displayed progressively decreased H3K9me3 levels, ranging from a moderate reduced level in Zscan4+ cells to massive loss in MERVL+/Zscan4+ cells (Figure 4A). We then investigated dynamics of ESC-specific H3K9me3 genes (Figure 4B). Most of these genes showed decreased H3K9me3 signals in MERVL+/Zscan4+ cells while retained at relatively high H3K9me3 level in Zscan4+ cells (Figure 4B), as exemplified by Gpa33 and Gm6578 (Figure 4C). In addition, genes displaying loss of ESC-specific H3K9me3 were not induced in Zscan4+ cells and MERVL+/Zscan4+ cells (Figure S4C), suggesting that TSC-specific transcriptional network is not activated during 2C-like transition.
Figure 4.
Lineage-specific H3K9me3 is removed in MERVL+/Zscan4+ cells
(A) Boxplot showing the H3K9me3 signals (RPKM) at promoters marked with ESC-specific H3K9me3 in the indicated cells. n = 2 biological replicates.
(B) Heatmap generated from the cluster analysis of H3K9me3 dynamics in ESC-specific H3K9me3-marked promoters in the indicated cells. Each row represents the Z score of H3K9me3 signal subtract input.
(C) Integrative Genomics viewer of H3K9me3 signals around Gpa33 and Gm6578 in the indicated cells.
(D) Boxplot showing the H3K9me3 signals of TSC-specific enhancers in the indicated cells. n = 2 biological replicates.
(E) Integrative Genomics viewer of H3K9me3 signals in the regions of TSC-specific enhancer in the indicated cells.
(F) Boxplot showing the H3K9me3 signals in the region of TSC-specific super-enhancers in the indicated cells. N = 2 biological replicates.
(G) Integrative Genomics viewer of H3K9me3 signals in the region of TSC-specific super enhancer in the indicated cells.
(H) Radar chart showing the enrichment score (log ratio of observed/random) of H3K9me3 peaks in TEs.
(I) Integrative Genomics viewer of H3K9me3 signals in the region of TSC-specific LTR enhancer in the indicated cells. The center of the boxplots in Figure 4 represents the median value and the lower and upper lines represent the 25% and 75% quantiles, respectively. Statistical analysis of Figure 4, p value, Wilcoxon rank-sum test, n.s. represents not significant, ∗ represents p < 0.05, ∗∗ represents p < 0.01, and ∗∗∗ represents p < 0.001.
See also Figure S4.
A prior study has highlighted the importance of enhancers in the segregation of TSCs and ESCs and identified 29,568 TSC-specific enhancers based on p300 occupancy (Lee et al., 2019). Generally, MERVL+/Zscan4+ cells displayed significantly reduced H3K9me3 levels in TSC-specific enhancers while Zscan4+ cells still possessed high levels similar to ESCs (Figure 4D). A total of 1,642 TSC-specific enhancers showed dynamic features of H3K9me3 during 2C-like transition (Figure S4D, Table S4); 1,053 TSC-specific enhancers remained high H3K9me3 levels or even acquired H3K9me3 in Zscan4+ cells (cluster 3, cluster 5, and cluster 6 in Figure S4D, Table S4). In MERVL+/Zscan4+ cells, 1,148 TSC-specific enhancers remained low or showed decreased H3K9me3 levels (Figures 4E and S4D; Table S4). Super-enhancers (SEs) regulate key genes linked with cell identity (Hnisz et al., 2013; Vahedi et al., 2015). Globally, TSC-specific SEs showed markedly decreased H3K9me3 levels in MERVL+/Zscan4+ cells, while TSC-specific SEs in Zscan4+ cells remained highly enriched H3K9me3 similar to ESCs (Figures 4F and 4G). A total of 196 TSC-specific SEs showed dynamic features of H3K9me3 during 2C-like transition, among which a large fraction (82%, 160 of 196) displayed decreased level in MERVL+/Zscan4+ cells compared with ESCs (Figures S4E and 4G; Table S4). The opposite pattern was observed in Zscan4+ cells (Figures S4E and 4G; Table S4). Targeted genes of TSC-specific SEs with decreased H3K9me3 level were not activated during 2C-like transition (Figure S4F), supporting the notion that TSC-specific transcriptional network is silenced in 2C-like cells.
LTRs are capable of functioning as enhancers to maintain cell-type-specific transcriptional program and segregate embryonic and trophoblast lineages (Chuong et al., 2013; Lee et al., 2019; Todd et al., 2019). Interestingly, LTRs decorated with ESC-specific H3K9me3 showed substantially reduced H3K9me3 signals in MERVL+/Zscan4+ cells (Figure S4G). Furthermore, LTRs were highly enriched in TSC-specific enhancers (Figure 4H). Next, we reanalyzed a previously published dataset to select LTRs overlapped with TSC-specific enhancers (Lee et al., 2019) and termed these regions as “TSC-specific LTR enhancer.” We examined H3K9me3 dynamics of TSC-specific LTR enhancers during 2C-like transition. Compared with ESCs, 304 TSC-specific LTR enhancers exhibited significantly decreased H3K9me3 levels in MERVL+/Zscan4+ cells (Table S4). In contrast, H3K9me3 levels of TSC-specific LTR enhancers remained constant in Zscan4+ cells (Table S4). Representative example was RLTR13D5 elements (Table S4), critical LTR enhancers of TSC identity (Chuong et al., 2013). Next, we investigated the contribution of specific LTR subfamilies to the identified TSC-specific LTR enhancers. Interestingly, LTRIS2/ERV1 was significantly overrepresented in the TSC-specific LTR enhancers undergoing H3K9me3 loss in MERVL+/Zscan4+ cells (Figure 4I, Table S4). This observation suggests a potential role of LTRIS2 element in the expanded potential of 2C-like cells.
Discussion
In the current study, we find that removal of H3K9me3 in TEs and TSC-specific enhancers predominantly occurred in MERVL+/Zscan4+ cells while H3K9me3 remains constant in Zscan4+ cells.
Unlike the global loss of DNA methylation in 2C-like cells (Eckersley-Maslin et al., 2016), H3K9me3 pattern is highly retained and only a small fraction of the genome undergoes H3K9me3 reprogramming during 2C-like transition. H3K9me3 signature of 2C-like cells and ESCs are generally quite different from their vivo counterparts (2C embryos and ICM). Thus, 2C-like transition does not recapitulate embryonic development in reverse. During 2C-like transition, loss of promoter-deposited H3K9me3 is not the major driver for transcriptional activation. Genes activated in this process show low fold enrichment of H3K9me3 at promoters in the three cell types analyzed. With regard to TEs, maintenance or gain of high H3K9me3 was observed in Zscan4+ cells compared with ESCs, accompanied with a modest increased expression level. DNA methylation and H3K9 methylation are major controllers for silencing TEs in ESCs (Karimi et al., 2011). H3K9me3 may be a compensate mechanism for the extensive loss of DNA methylation to restrict the activation of TEs in Zscan4+ cells. Although a large portion of activated TEs display decreased H3K9me3 enrichment in MERVL+/Zscan4+ cells, we still observed de novo H3K9me3 accumulation at some TE families. One possible explanation is that H3K9me3 may not confer transcriptional repression in these regions. Alternatively, it is also possible that transcriptionally active elements inside these subfamilies may undergo removal of H3K9me3.
TEs may be closely linked with higher competency of MERVL+/Zscan4+ cells. Indeed, we have previously demonstrated that MERVL activation is capable of conferring expanded potential in ESCs (Yang et al., 2020). MERVL is able to serve as alternative promoters or enhancers to drive the expression of genes restricted to totipotent cells (Macfarlan et al., 2012; Zhang et al., 2019). In addition to MERVL, other TEs, especially expressed in totipotent cells during early development, are also induced in MERVL+/Zscan4+ cells, which may be critical to shape the transcription network during 2C-like transition.
It has been shown that the current strategy of the ESC to TSC conversion is incomplete (Cambuli et al., 2014). A subset of TSC-specific enhancers decorated with H3K9me3 in ESCs undergoes elimination of H3K9me3 during 2C-like transition. Interestingly, LTRIS2/ERV1, highly enriched in TSC-specific LTR enhancers, displays loss of H3K9me3 in 2C-like cells. Thus, further investigation of LTRIS2/ERV1 as potential ERV-based enhancers in the specification of extraembryonic cell lineage will deepen our understanding of the first cell lineage separation.
In summary, our study provides a detailed comparison of H3K9me3 profiles among 2-cell embryos, 2C-like cells, and ESCs. The exploration of epigenetic factors to further reprogram the epigenome of 2C-like cells in the future may help us to understand the molecular mechanisms of totipotency.
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shaorong Gao (gaoshaorong@tongji.edu.cn).
Materials availability
All plasmids or mouse lines generated in this study are available from the lead contact with a completed Materials Transfer Agreement/without restriction.
Mice
The specific pathogen-free grade mice (SPF) grade mice were housed in the animal facility at Tongji University, Shanghai, China. All the mice had free access to food and water. All experiments were performed in accordance with the University of Health Guide for the Care and Use of Laboratory Animals and were approved by the Biological Research Ethics Committee of Tongji University.
Peak enrichment analysis
Enrichment of H3K9me3 peaks in repeat elements was calculated as observed versus random counts. The observed counts were calculated as the probability of H3K9me3 peaks overlapping with related genomic regions using the “intersectBed” function of bedtools with parameter -f 0.5. The random peaks were generated using the “shuffleBed” function. The random counts were calculated the same as the observed counts.
Clustering analysis
The K-means clustering of H3K9me3 levels was conducted using ComplexHeatmap (Gu et al., 2016).
Statistical analysis
R (version 3.6.1) was used for statistics analysis. Pearson’s correlation coefficient was calculated using the cor.test function with default parameters to evaluate reproducibility of replicates. Wilcoxon rank-sum test was used for RNA expression comparisons using the wilcox.test function in R. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Author contributions
R.L. and S.G. conceived and designed the experiments. H.L. and J.S. performed most of the experiments. Y.Z. performed the computational analysis. R.L., Y.Z., H.L., and J.S. designed and performed the data analysis. Y.D., Z.L., M.G., X.K., Y.Z., A.Z., J.Y., J.S., Z.S., K.S., Y.G., J.C., L.K., Y.W., C.L., and H.W. assisted with the experiments. R.L., Y.Z., and S.G. wrote the manuscript.
Acknowledgments
We are grateful to our colleagues in the laboratory for their assistance with the experiments and in the preparation of this manuscript. This work was primarily supported by the National Natural Science Foundation of China (32122030, 31721003, 32000562, 32170589, 31970758). This work was also supported by the Shanghai Rising-Star Program (A type) of the Science and Technology Commission of Shanghai Municipality (21QA1409500), and the Science and Technology Innovation Action Plan of the Science and Technology Commission of Shanghai Municipality (21JC1405500).
Conflict of interests
The authors declare no competing interests.
Published: January 12, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.12.006.
Contributor Information
Rongrong Le, Email: lerongrong@tongji.edu.cn.
Shaorong Gao, Email: gaoshaorong@tongji.edu.cn.
Yanping Zhang, Email: yanpingzhang@tongji.edu.cn.
Supplemental information
Data and code availability
The accession number in GEO for the RNA-seq and ChIP-seq data reported in this paper is GEO: GSE199042. The accession number for the published H3K9me3 ChIP-seq data of early embryos is GEO: GSE97778 (Wang et al., 2018). The accession number for the published RNA-seq data of 2C-like cells is GEO: GSE75751 (Eckersley-Maslin et al., 2016). The accession number for the published RNA-seq data of early embryos is GEO: GSE45719 (Deng et al., 2014).
References
- Akiyama T., Xin L., Oda M., Sharov A.A., Amano M., Piao Y., Cadet J.S., Dudekula D.B., Qian Y., Wang W.D., et al. Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res. 2015;22:307–318. doi: 10.1093/dnares/dsv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano T., Hirata T., Falco G., Monti M., Sharova L.V., Amano M., Sheer S., Hoang H.G., Piao Y., Stagg C.A., et al. Zscan4 restores the developmental potency of embryonic stem cells. Nat. Commun. 2013;4:1966. doi: 10.1038/ncomms2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J.S., Nicetto D., Zaret K.S. H3K9me3-Dependent heterochromatin: barrier to cell fate changes. Trends Genet. 2016;32:29–41. doi: 10.1016/j.tig.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut-Karslioglu A., De La Rosa-Velázquez I.A., Ramirez F., Barenboim M., Onishi-Seebacher M., Arand J., Galán C., Winter G.E., Engist B., Gerle B., et al. Suv39h-Dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell. 2014;55:277–290. doi: 10.1016/j.molcel.2014.05.029. [DOI] [PubMed] [Google Scholar]
- Burton A., Brochard V., Galan C., Ruiz-Morales E.R., Rovira Q., Rodriguez-Terrones D., Kruse K., Le Gras S., Udayakumar V.S., Chin H.G., et al. Heterochromatin establishment during early mammalian development is regulated by pericentromeric RNA and characterized by non-repressive H3K9me3. Nat. Cell Biol. 2020;22:767–778. doi: 10.1038/s41556-020-0536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambuli F., Murray A., Dean W., Dudzinska D., Krueger F., Andrews S., Senner C.E., Cook S.J., Hemberger M. Epigenetic memory of the first cell fate decision prevents complete ES cell reprogramming into trophoblast. Nat. Commun. 2014;5:5538. doi: 10.1038/ncomms6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Liu W.Q., Guo J.Y., Liu Y.Y., Liu X.L., Liu J., Dou X.Y., Le R.R., Huang Y.X., Li C., et al. Nuclear m6A reader YTHDC1 regulates the scaffold function of LINE1 RNA in mouse ESCs and early embryos. Protein Cell. 2021;12:455. doi: 10.1007/s13238-021-00837-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhang Y. Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat. Genet. 2019;51:947–951. doi: 10.1038/s41588-019-0418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y.G., Matoba S., Liu Y., Eum J.H., Lu F., Jiang W., Lee J.E., Sepilian V., Cha K.Y., Lee D.R., Zhang Y. Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells. Cell Stem Cell. 2015;17:758–766. doi: 10.1016/j.stem.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Chuong E.B., Rumi M.A.K., Soares M.J., Baker J.C. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 2013;45:325–329. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossec J.C., Theurillat I., Chica C., Búa Aguín S., Gaume X., Andrieux A., Iturbide A., Jouvion G., Li H., Bossis G., et al. SUMO safeguards somatic and pluripotent cell identities by enforcing distinct chromatin states. Cell Stem Cell. 2018;23:742–757.e8. doi: 10.1016/j.stem.2018.10.001. [DOI] [PubMed] [Google Scholar]
- De Iaco A., Planet E., Coluccio A., Verp S., Duc J., Trono D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017;49:941–945. doi: 10.1038/ng.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A., Coudray A., Duc J., Trono D. DPPA2 and DPPA4 are necessary to establish a 2C-like state in mouse embryonic stem cells. EMBO Rep. 2019;20:e47382. doi: 10.15252/embr.201847382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q., Ramsköld D., Reinius B., Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin M.A., Svensson V., Krueger C., Stubbs T.M., Giehr P., Krueger F., Miragaia R.J., Kyriakopoulos C., Berrens R.V., Milagre I., et al. MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs. Cell Rep. 2016;17:179–192. doi: 10.1016/j.celrep.2016.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin M., Alda-Catalinas C., Blotenburg M., Kreibich E., Krueger C., Reik W. Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev. 2019;33:194–208. doi: 10.1101/gad.321174.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinas N.A., Tu L.N., Furci L., Shimajiri Y., Harukawa Y., Miura S., Takuno S., Saze H. Transcriptional regulation of genes bearing intronic heterochromatin in the rice genome. PLoS Genet. 2020;16:e1008637. doi: 10.1371/journal.pgen.1008637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadloun A., Le Gras S., Jost B., Ziegler-Birling C., Takahashi H., Gorab E., Carninci P., Torres-Padilla M.E. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013;20:332–338. doi: 10.1038/nsmb.2495. [DOI] [PubMed] [Google Scholar]
- Franke V., Ganesh S., Karlic R., Malik R., Pasulka J., Horvat F., Kuzman M., Fulka H., Cernohorska M., Urbanova J., et al. Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome Res. 2017;27:1384–1394. doi: 10.1101/gr.216150.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fueyo R., Judd J., Feschotte C., Wysocka J. Roles of transposable elements in the regulation of mammalian transcription. Nat. Rev. Mol. Cell Biol. 2022 doi: 10.1038/s41580-022-00457-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Guo M., Zhang Y., Zhou J., Bi Y., Xu J., Xu C., Kou X., Zhao Y., Li Y., Tu Z., et al. Precise temporal regulation of Dux is important for embryo development. Cell Res. 2019;29:956–959. doi: 10.1038/s41422-019-0238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins R.D., Hon G.C., Lee L.K., Ngo Q., Lister R., Pelizzola M., Edsall L.E., Kuan S., Luu Y., Klugman S., et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson P.G., Doráis J.A., Grow E.J., Whiddon J.L., Lim J.W., Wike C.L., Weaver B.D., Pflueger C., Emery B.R., Wilcox A.L., et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017;49:925–934. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T., Enriquez-Gasca R., Mizutani E., Bošković A., Ziegler-Birling C., Rodriguez-Terrones D., Wakayama T., Vaquerizas J.M., Torres-Padilla M.E. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 2015;22:662–671. doi: 10.1038/nsmb.3066. [DOI] [PubMed] [Google Scholar]
- Jachowicz J.W., Bing X., Pontabry J., Bošković A., Rando O.J., Torres-Padilla M.E. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 2017;49:1502–1510. doi: 10.1038/ng.3945. [DOI] [PubMed] [Google Scholar]
- Karimi M.M., Goyal P., Maksakova I.A., Bilenky M., Leung D., Tang J.X., Shinkai Y., Mager D.L., Jones S., Hirst M., Lorincz M.C. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.K., Jang Y.J., Kim M., LeBlanc L., Rhee C., Lee J., Beck S., Shen W., Kim J. Super-enhancer-guided mapping of regulatory networks controlling mouse trophoblast stem cells. Nat. Commun. 2019;10:4749. doi: 10.1038/s41467-019-12720-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Liu W., Liu X., Wang C., Gao Y., Gao R., Kou X., Zhao Y., Li J., Wu Y., Xiu W., et al. Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell Discov. 2016;2:16010. doi: 10.1038/celldisc.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann F., Loureiro J., Su H., Fang Q., Lei H., Lewis T., Yang Y., Labow M., Li E., Chen T., Kadam S. KMT1E mediated H3K9 methylation is required for the maintenance of embryonic stem cells by repressing trophectoderm differentiation. Stem Cell. 2010;28:201–212. doi: 10.1002/stem.278. [DOI] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova I.A., Thompson P.J., Goyal P., Jones S.J., Singh P.B., Karimi M.M., Lorincz M.C. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenet. Chromatin. 2013;6:15. doi: 10.1186/1756-8935-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J.H.A., O'Sullivan R.J., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S., Liu Y., Lu F., Iwabuchi K.A., Shen L., Inoue A., Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159:884–895. doi: 10.1016/j.cell.2014.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Leung D., Miyashita H., Maksakova I.A., Miyachi H., Kimura H., Tachibana M., Lorincz M.C., Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Nakayama J., Rice J.C., Strahl B.D., Allis C.D., Grewal S.I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nicetto D., Zaret K.S. Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr. Opin. Genet. Dev. 2019;55:1–10. doi: 10.1016/j.gde.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaston A.E., Evsikov A.V., Graber J.H., de Vries W.N., Holbrook A.E., Solter D., Knowles B.B. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Percharde M., Lin C.J., Yin Y., Guan J., Peixoto G.A., Bulut-Karslioglu A., Biechele S., Huang B., Shen X., Ramalho-Santos M. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell. 2018;174:391–405.e19. doi: 10.1016/j.cell.2018.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O'Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Terrones D., Gaume X., Ishiuchi T., Weiss A., Kopp A., Kruse K., Penning A., Vaquerizas J.M., Brino L., Torres-Padilla M.E. A molecular roadmap for the emergence of early-embryonic-like cells in culture. Nat. Genet. 2018;50:106–119. doi: 10.1038/s41588-017-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H.M., Jakobsson J., Mesnard D., Rougemont J., Reynard S., Aktas T., Maillard P.V., Layard-Liesching H., Verp S., Marquis J., et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Rowe H.M., Kapopoulou A., Corsinotti A., Fasching L., Macfarlan T.S., Tarabay Y., Viville S., Jakobsson J., Pfaff S.L., Trono D. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013;23:452–461. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q., Wang X.F., Xie S.M., Yin Y., Zhou L.Q. H3.3 impedes zygotic transcriptional program activated by Dux. Biochem. Biophys. Res. Commun. 2019;522:422–427. doi: 10.1016/j.bbrc.2019.11.114. [DOI] [PubMed] [Google Scholar]
- Todd C.D., Deniz Ö., Taylor D., Branco M.R. Functional evaluation of transposable elements as enhancers in mouse embryonic and trophoblast stem cells. Elife. 2019;8:e44344. doi: 10.7554/eLife.44344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G., Kanno Y., Furumoto Y., Jiang K., Parker S.C.J., Erdos M.R., Davis S.R., Roychoudhuri R., Restifo N.P., Gadina M., et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558–562. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu X., Gao Y., Yang L., Li C., Liu W., Chen C., Kou X., Zhao Y., Chen J., et al. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 2018;20:620–631. doi: 10.1038/s41556-018-0093-4. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Whiddon J.L., Langford A.T., Wong C.J., Zhong J.W., Tapscott S.J. Conservation and innovation in the DUX4-family gene network. Nat. Genet. 2017;49:935–940. doi: 10.1038/ng.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Liu H., Wang Y., He J., Xu S., Chen Y., Kuang J., Liu J., Guo L., Li D., et al. SETDB1-Mediated cell fate transition between 2C-like and pluripotent states. Cell Rep. 2020;30:25–36.e6. doi: 10.1016/j.celrep.2019.12.010. [DOI] [PubMed] [Google Scholar]
- Yan Y.L., Zhang C., Hao J., Wang X.L., Ming J., Mi L., Na J., Hu X., Wang Y. DPPA2/4 and SUMO E3 ligase PIAS4 opposingly regulate zygotic transcriptional program. PLoS Biol. 2019;17:e3000324. doi: 10.1371/journal.pbio.3000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Huang X., Zang R., Chen J., Fidalgo M., Sanchez-Priego C., Yang J., Caichen A., Ma F., Macfarlan T., et al. DUX-miR-344-ZMYM2-Mediated activation of MERVL LTRs induces a totipotent 2C-like state. Cell Stem Cell. 2020;26:234–250.e7. doi: 10.1016/j.stem.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P., Han J., Guo G., Orlov Y.L., Huss M., Loh Y.H., Yaw L.P., Robson P., Lim B., Ng H.H. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 2009;23:2507–2520. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalzman M., Falco G., Sharova L.V., Nishiyama A., Thomas M., Lee S.L., Stagg C.A., Hoang H.G., Yang H.T., Indig F.E., et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Chen F., Chen R., Xie D., Yang J., Zhao X., Guo R., Zhang Y., Shen Y., Göke J., et al. Zscan4c activates endogenous retrovirus MERVL and cleavage embryo genes. Nucleic Acids Res. 2019;47:8485–8501. doi: 10.1093/nar/gkz594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huang Y., Dong Y., Liu X., Kou X., Zhao Y., Zhao A., Sun J., Su Z., Li Z., et al. Unique patterns of H3K4me3 and H3K27me3 in 2-Cell-like embryonic stem cells. Stem Cell Rep. 2021;16:458–469. doi: 10.1016/j.stemcr.2021.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Adli M., Zou J.Y., Verstappen G., Coyne M., Zhang X., Durham T., Miri M., Deshpande V., De Jager P.L., et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number in GEO for the RNA-seq and ChIP-seq data reported in this paper is GEO: GSE199042. The accession number for the published H3K9me3 ChIP-seq data of early embryos is GEO: GSE97778 (Wang et al., 2018). The accession number for the published RNA-seq data of 2C-like cells is GEO: GSE75751 (Eckersley-Maslin et al., 2016). The accession number for the published RNA-seq data of early embryos is GEO: GSE45719 (Deng et al., 2014).