Summary
A rare sub-population of mouse embryonic stem cells (mESCs), the 2-cell-like cell, is defined by the expression of MERVL and 2-cell-stage-specific transcript (2C transcript). Here, we report that the ribosomal proteins (RPs) RPL14, RPL18, and RPL23 maintain the identity of mESCs and regulate the expression of 2C transcripts. Disregulation of the RPs induces DUX-dependent expression of 2C transcripts and alters the chromatin landscape. Mechanically, knockdown (KD) of RPs triggers the binding of RPL11 to MDM2, an interaction known to prevent P53 protein degradation. Increased P53 protein upon RP KD further activates its downstream pathways, including DUX. Our study delineates the critical roles of RPs in 2C transcript activation, ascribing a novel function to these essential proteins.
Keywords: 2C-like cells, MERVL, RPs, P53
Graphical abstract
Highlights
-
•
Disregulation of ribosomal proteins (RPs) activates MERVL and 2-cell-like state
-
•
RP loss leads to interaction of RPL11 and MDM2
-
•
RP KD activates P53 and its downstream pathways including DUX
-
•
RP regulates 2C activation through RPL11-MDM2-P53-DUX axis
In this article, Loh and colleagues show that ribosomal proteins (RPs) RPL14, RPL18, and RPL23 maintain the identity of mouse embryonic stem cells (mESCs) and regulate the expression of 2C transcripts through a unique RP-RPL11-MDM2-P53-DUX cascade.
Introduction
In the course of mammalian development, a zygote undergoes multiple cleavage events to form a blastocyst, transitioning via 2-cell and 4-cell intermediate stages (O’connor, 1939). During this process, blastomeres shift from a totipotent to a pluripotent state with increased lineage specificity and restrictions on potential cell fates. Derived from the inner cell mass of blastocytes, mESCs are widely regarded as pluripotent due to their capacity to give rise to all embryonic lineages except the extraembryonic lineages (Beddington and Robertson, 1989; Kaufman and Evans, 1981; Martin, 1981). Interestingly, a rare (∼1%–2%) population of mESCs, known as the 2-cell-like cells (2CLCs), was characterized by traits of expanded pluripotency and a transcriptional profile similar to the totipotent 2-cell stage in terms of the activation of endogenous retrovirus MERVL and other 2-cell-stage-specific genes (2C genes) such as the Zscan4 family (Eckersley-Maslin et al., 2016; Macfarlan et al., 2012; Wu et al., 2016; Zhang et al., 2019). Activation of MERVL and other 2C genes were shown to catalyze the transition from mESCs to an early embryonic expanded potential state (Yang et al., 2020), accentuating the significance of studying their upstream regulatory mechanisms. Recent studies have demonstrated critical roles for various epigenetic and transcriptional factors in 2C activation (Eckersley-Maslin et al., 2019; Fang et al., 2018; Gautam et al., 2017; Hendrickson et al., 2017; Hu et al., 2020; De Iaco et al., 2017; Yan et al., 2019; Yang et al., 2015). However, the importance of molecular players in cellular metabolism in regulating 2C transcripts has yet to be fully understood.
Aside from their basic role in translation as a component of ribosomes, ribosomal proteins (RPs) are also involved in multiple cellular physiological and pathological processes (Kim et al., 2014; Zhou et al., 2015). Multiple studies have revealed the accumulation of individual RPs in the nuclear compartment via transportation from the cytoplasm for the assembly of ribosome precursors with rRNA. Subsequently, it was discovered that RPs were able to regulate gene expression through other transcription factors or pathways, such as with c-Myc (Challagundla et al., 2011; Dai et al., 2007; Zhou et al., 2013) and the MDM2-P53 cascade (Bhat et al., 2004; Dai et al., 2004). Processes such as rRNA synthesis and ribosomal biogenesis were reported to be essential for self-renewal of ESCs and the maintenance of pluripotency (Atlasi et al., 2020; Zhang et al., 2020). Meanwhile, reduction of rRNA synthesis along with 2C transcript activation was observed to be induced by the knockdown of LINE1, Ncl (Percharde et al., 2018), and Ythdc1 (Chen et al., 2021). A recent study demonstrated that 2C transcript activation could be achieved by inhibiting rRNA synthesis (Yu et al., 2021). However, the roles of RPs and their related molecular players in regulating pluripotency have yet to be uncovered.
Result
siRNA screen of RPs reveals their 2C regulatory functions
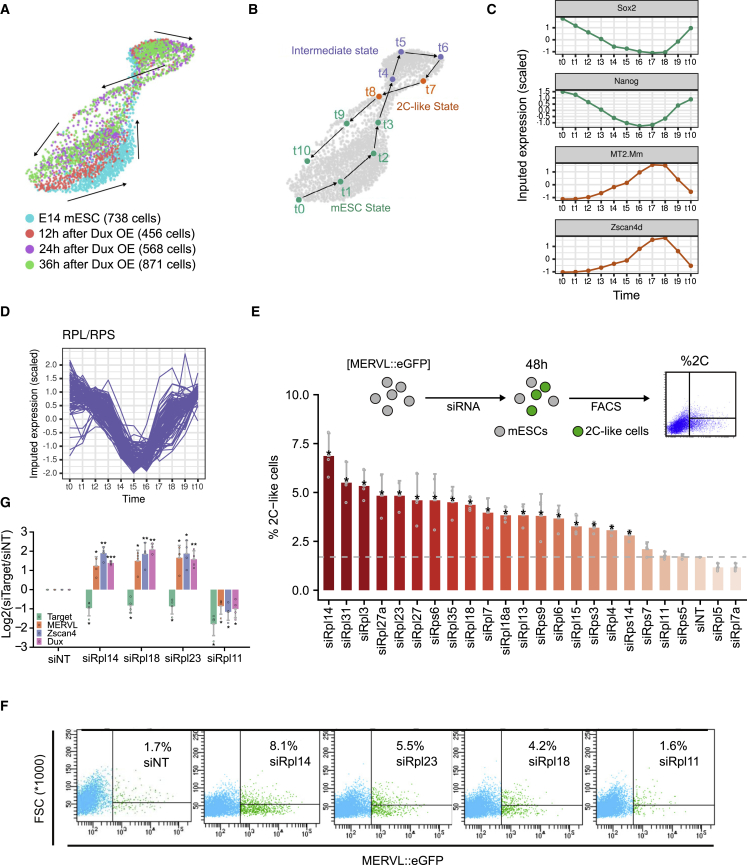
We reanalyzed the publicly available single-cell RNA sequencing (RNA-seq) data of DUX overexpressed mESCs in which cells were annotated as mESC state, intermediate state, or 2C-like state based on clustering analysis (Fu et al., 2019) (Figure S1A). With trajectory analysis (Gautam et al., 2021), we classified the annotated cells into 11 different stages (Figures 1A and 1B). During the mESC to 2CLC transition, it was observed that 2C transcripts, like MT2-Mm and Zscan4, were activated, while the expression of pluripotency factors including Sox2 and Nanog was decreased (Figure 1C). Surprisingly, RPs were generally lowly expressed in the state prior to the entry into the 2CLCs from mESCs, indicating their possible role as barriers in 2CLC transition (Figure 1D). RNAi has been used as a powerful tool to systemically identify key regulators in pluripotent stem cells (Loh et al., 2006; Toh et al., 2016). Here, we performed gene perturbation via small interfering RNA (siRNA) screening across 23 selected Rpl and Rps candidates, defined by their lower expression in the 2-cell stage, compared with other developmental stages or in mESCs (Wu et al., 2016). Out of the 23 candidates, knockdowns (KDs) of 18 targets significantly increase transition to 2CLCs (Figures 1E, 1F, and S1B). We selected and validated the positive targets, Rpl14, Rpl18, and Rpl23, and the negative target, Rpl11, with qRT-PCR and confirmed the induction of MERVL and key 2C genes upon KD (Figure 1G). These findings were further echoed in experiments that performed KD using the short hairpin RNA (shRNA) system, thereby indicating the regulatory impact of RPs exerted on 2C transcripts. (Figures S1C and S1D).
Figure 1.
siRNA screen reveals 2C regulatory functions of ribosomal proteins
(A) Single-cell predicted trajectory illustrates the dynamic of mESCs upon Dux overexpression at 12, 24, and 36 h. Reanalysis of data from Fu et al. (2019).
(B) 11 stages of the predicted trajectory in (A), stages 7 and 8 were annotated as 2C-like state, stages 4–6 were intermediate states, and the rest of the stages were normal mESCs.
(C and D) Dynamics of the expression of selected genes for (C) pluripotency, 2 cell stage, and (D) ribosomal proteins (RPs) throughout the 11 stages as defined in (B).
(E) Quantification of the percentage of 2C-like cells upon knockdown of the RPs utilizing siRNA screening. Shown are mean ±standard deviation (SD). Data from three independent experiments were combined and shown.
(F) Plots of flow cytometry show the percentage of 2C-like cells upon knockdown of Rpl11, Rpl14, Rpl18, and Rpl23.
(G) qRT-PCR validates the expression of key 2C genes upon the knockdown of Rpl11, Rpl14, Rpl18, and Rpl23 utilizing the siRNA system. Two-tailed Student’s t test. Shown are mean ± SD. Data from four independent experiments were combined and shown. Zscan4: common sequences of Zscan4 family.
Rpl14, Rpl18, and Rpl23 regulate MERVL and 2C genes
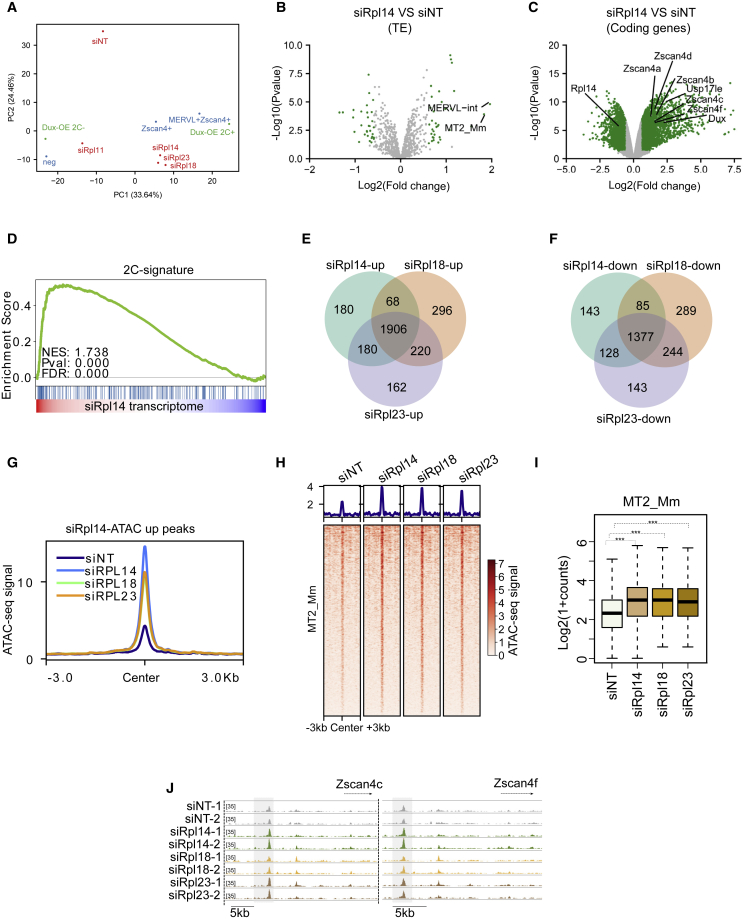
To explore the functionality of RPs as 2C regulators, we performed transcriptomic analysis of mESCs transfected with siRNA targeting Rpl14, Rpl18, Rpl23, and Rpl11 (Figure S1A; Table S2). Principal-component analysis (PCA) compared with published datasets of DUX-induced 2CLCs (Hendrickson et al., 2017) and MuERVL+_ Zscan4+/Zscan4+ cells (Eckersley-Maslin et al., 2016) revealed that the KD of Rpl14, Rpl18, and Rpl23 induced transcriptome profiles that were highly similar to the 2CLCs (Figure 2A), while KD of Rpl11 maintained a cellular identity proximal to 2C marker-negative cells. The expression of MERVL and its long terminal repeat (LTR) (MT2_Mm) were observed to be significantly elevated (Figures 2B and S2B), alongside 2-cell genes such as the Zscan4 family (Figures 2C and S2C), in siRpl14, siRpl18, and siRpl23 KD but not in Rpl11 KD. Gene set enrichment analysis (GSEA) illustrated that 2C genes were significantly up-regulated in siRpl14, siRpl18, and siRpl23 (Figures 2D and S2D). KD of Rpl14, Rpl18, and Rpl23 induced a large amount of common regulated genes (Figures 2E and 2F), implying their similar regulatory roles. Thereafter, we performed assay for transposase-accessible chromatin sequencing (ATAC-seq) on siRpl14, siRpl18, and siRpl23 KDs (Figure S2E). We identified 2,650 differently enriched peaks (1,418 peaks enriched in siRpl14 and 1,232 peaks enriched in siNT) for Rpl14 KD (Figure S2F) and similar numbers of peaks for other RPs. Consistent with the transcriptomic expression, KD of RPs shared a considerable amount of gained ATAC peaks (Figure 2G). Importantly, upon KD of RPs, increased chromatin accessibility was observed at MT2_Mm loci (Figures 2H and 2I). As expected, some key 2C genes, such as Zscan4c and Zscan4f, were enriched of ATAC-seq signal upon RP KD (Figure 2J). Collectively, these data revealed that the disturbance of Rpl14, Rpl18, and Rpl23, but not Rpl11, activates the expression of MERVL and 2C genes.
Figure 2.
Loss of RPs elicits changes in the expression and chromatin accessibility of MERVL and 2C genes in mESCs
(A) PCA plot indicates the transcriptomic profile upon knockdown of selected candidates in mESCs are high in resemblance to the cells in 2C-like state with MERVL+Zscan4+, Zscan4+, and the 2C-positive populations induced by Dux overexpression for RP knockdown (KD) RNA-seq samples. Data from three independent experiments were combined and shown.
(B and C) Volcano plot showing the (B) the differentially expressed transposable elements (green data points represent transposable elements with p < 0.05 and |fold change| > 1.5) and (C) the differentially expressed genes (green data points present genes with p < 0.05 and |fold change|> 1.5)upon Rpl14 KD.
(D) GSEA reveals the enrichment of 2C-signature genes (Wu et al., 2016) in the transcriptome of Rpl14 KD at 48 h.
(E and F) Venn diagram representing the common (E) up- and (F) down-regulated gene in all the performed KDs of RPs.
(G) Average plot shows the ATAC-seq signal at differentially up-regulated accessible peaks upon KD of respective RP. For RP KD ATAC-seq samples, data from two independent experiments were combined and shown.
(H) Heatmap shows the chromatin accessibility of MT2_Mm (MERVL-LTR) upon KD of respective RP.
(I) Bar plot shows the ATAC-seq signal of MT2_Mm upon KD of RPs. One-sided Wilcoxon test.
(J) Genome browser visualization illustrates the ATAC-seq signal increased at the upstream regions of the key 2C genes upon RP KDs.
Rpl14 KD activates MERVL and 2C genes through the RPL11-MDM2-P53-Dux axis
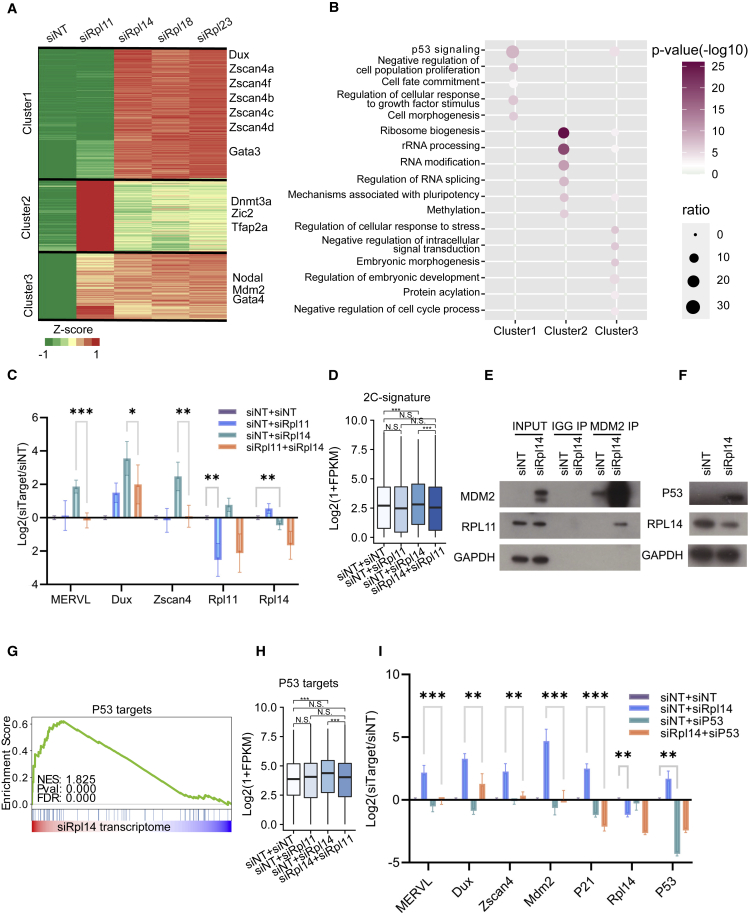
Hierarchical clustering analysis of RNA-seq data revealed that the KD of Rpl14, Rpl18, and Rpl23 largely up-regulated cluster 1 genes, which comprised genes involved in cell fate commitment and the key 2C genes (Figures 3A and 3B; Table S3), such as Dux and the Zscan4 family. Meanwhile, Rpl11 KD was shown to have distinct functions as it extensively activated cluster 2 genes, represented by ribosome biogenesis and RNA-splicing pathways.
Figure 3.
Rpl14 KD activates MERVL and 2C programs through the RPL11-MDM2-P53-Dux axis
(A) Heatmap represents the expression of pluripotent-related factors, key 2C genes, and 2C-related genes in clusters 1, 2, and 3, respectively, upon KD of Rpl11, Rpl14, Rpl18, and Rpl23.
(B) Representative enriched GO terms of biological process of the three clusters as defined in (A). For each GO term, the ratio represents the percentage of its contained genes that overlaps with the genes in different clusters.
(C) qRT-PCR reveals the expression of key 2C genes and respective Rpls, upon single or double KD of Rpl11 and Rpl14. Data from four independent experiments were combined and shown. Two-tailed Student’s t test.
(D) Barplot illustrates the expression of 2C-signature genes upon single and combinatorial KD of Rpl11 and Rpl14. One-sided Wilcoxon test. Data from three independent experiments were combined and shown.
(E) Native Co-IP analysis in mESCs reveals the robust interaction between MDM2 and RPL11 upon siRpl14 KD.
(F) Western blot analysis indicates protein expression of P53 upon siRpl14 KD.
(G) GSEA shows the enrichment of the targets for P53 in the transcriptome of Rpl14 KD.
(H) Barplot describes the expression of P53 targets upon single and combinatorial KD of Rpl11 and Rpl14. One-sided Wilcoxon test. Data from three independent experiments were combined and shown.
(I) qRT-PCR represents the expression of key 2C genes and P53 targets upon single or double KD of Rpl14 and p53. Two-tailed Student’s t test. Data from three independent experiments were combined and shown.
We then explored in depth the mechanism of Rpl14, our top target, in regulating the 2C-like state in mESCs. In human cancer cell lines, KD of RPs, including Rpl14, might cause nucleolar stress and activate P53 pathways (Fujiyama et al., 2020; Fumagalli et al., 2012; Horn and Vousden, 2008; Russo and Russo, 2017; Watanabe et al., 2018). More specifically, failure of pre-ribosome assembly due to a lack of sub-units led to increased free-floating RPL11 and RPL5, which further bound to and suppressed the P53 inhibitor MDM2 (Russo and Russo, 2017). Hence, we performed simultaneous KDs of both Rpl11 and Rpl14 to decipher their functional relationship. Intriguingly, up-regulation of MERVL and 2C genes by Rpl14 KD was counteracted by Rpl11 KD (Figures 3C, 3D, and S3A; Table S4), confirming RPL11 as an essential downstream effector of RPL14 in 2C regulation. Moreover, RPL11 was shown to interact with MDM2 upon RPL14 KD by native coimmunoprecipitation (Co-IP) (Figure 3E), an interaction previously reported to inhibit MDM2-mediated P53 degradation (Fumagalli et al., 2012; Horn and Vousden, 2008; Sasaki et al., 2011). Western blot and transcriptional analysis further confirmed the downstream increase in P53 protein expression (Figure 3F) and activation of P53 targeting pathways (Figure 3G). Consistently, ATAC-seq analysis indicated enrichment of the P53 motif in differential enriched peaks upon RP KD (Figure S2G). Of note, the activated P53 pathway showed a synchronous expression pattern with 2C genes, as their expressions induced by RPL14 KD were both rescued upon RPL11 KD (Figures 3D, 3H, and S3B), further indicating the involvement of MDM2-P53 in RPL11-mediated RP-induced 2C activation. This possibility was validated as the KD of p53 rescued the Rpl14-KD-induced increase in MERVL and 2C gene expression, including Dux (Figure 3I). Moreover, we sorted the Rpl14 KD population into 2C-positive and 2C-negative cells using the fluorescence-activated cell sorting (FACS) machine to investigate their gene expression profile (Figure S3C). Together with the higher expression of MERVL and 2C genes in the siRPL14-2C-positive cells compared with the siRPL14-2C-negative cells, we also observed a significantly more robust activation of P53, confirming the critical role of P53 in mediating 2C transition (Figure S3D). Intriguingly, activated P53 was recently shown to directly bind and promote Dux expression, thus enhancing 2C transition (Grow et al., 2021). Therefore, the 2C transition induced by RP KD was mediated at least in part through the activated RPL11-MDM2-P53 pathways, which consequently promoted the expression of Dux and its downstream 2C genes.
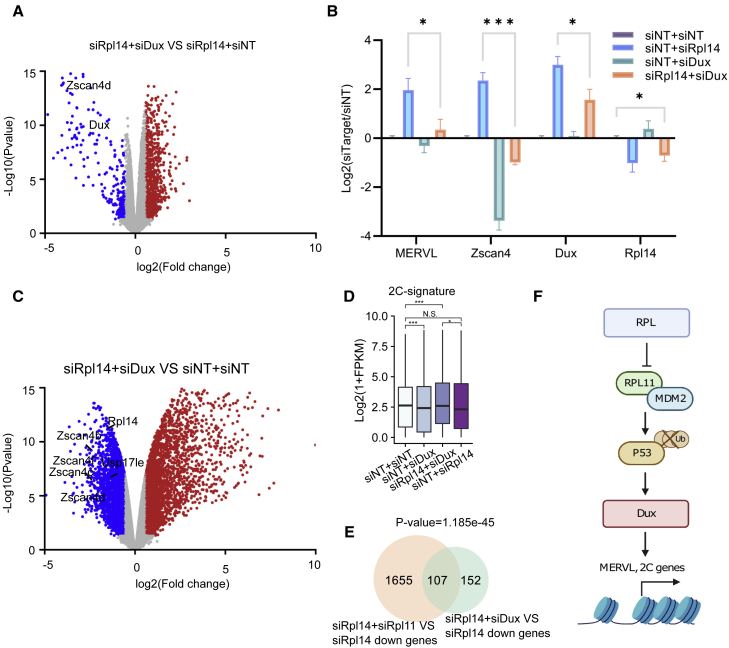
Rpl14-based induction of 2C transcripts is Dux dependent
DUX was reported to bind to and activate MERVL and multiple 2C genes (Hendrickson et al., 2017; De Iaco et al., 2017) and function as an indispensable mediator of multiple 2C regulators, including P53 (Chen et al., 2021; Grow et al., 2021; Liu et al., 2021; Percharde et al., 2018). We therefore performed a concurrent KD of Dux and Rpl14. Both RNA-seq and qRT-PCR showed that the up-regulation of MERVL, Zscan4, and other 2C genes in Rpl14 KD was rescued by the KD of Dux (Figures 4A and 4B; Table S4). Expression of 2C genes showed statistically insignificant differences between siNT and double KD of Rpl14 and Dux, indicating that Dux plays a dominant downstream role in complementing the action of Rpl14 in the induction of the 2C program (Figures 4C and 4D). A large portion (41%) of the siDux-rescued genes, including most of the 2C genes, were rescued by siRpl11 upon siRpl14 KD (Figure 4E), further confirming Dux as the downstream factor of RPL11. TRIM28, the reported direct upstream regulator of Dux (Percharde et al., 2018), emerged as another key player when we further examined the functional mediators between RPL14 and DUX. We compared our transcriptome data with a publicly available RNA-seq dataset of TRIM28 (De Iaco et al., 2017) with ZMYM2 (Yang et al., 2020), a reported DUX downstream 2C regulator, as a control. A strong correlation was shown between siRpl14-up-regulated and shTrim28-up-regulated genes, which was expected since both factors are Dux upstream regulators (Figure S4A). Notably, a significant number of commonly regulated genes was also observed between Trim28 KD in Dux knockout (KO) and double KD of Rpl14 and Dux, pointing to their possible involvement in the same regulatory cascade (Figure S4B). Therefore, we examined TRIM28 binding on genome upon Rpl14 KD. We observed a decrease in enrichment at Dux loci and rRNA loci following Rpl14 KD (Figures S4C and S4D), revealing TRIM28 as one of the downstream effectors of RPL14 induced 2C activation.
Figure 4.
Rpl14-based induction of 2C programs is Dux dependent
(A) Volcano plot of differentially expressed genes between double KD of Dux and Rpl14 compared with siRpl14. The known 2C marker expressions were labeled out. Data from three independent experiments were combined and shown.
(B) qRT-PCR represents the gene expression of key 2C genes, MERVL, and respective ribosomal proteins upon single or combinatorial KD of Dux and Rpl14. Two-tailed Student’s t test. Shown are mean ± SD. Data from three independent experiments were combined and shown.
(C) Volcano plot of differentially expressed genes between double KD of Dux and Rpl14 compared with siNT.
(D) Bar plots reveal the expression of 2C-signature genes upon single or combinatorial KD of Rpl14 and Dux. One-sided Wilcoxon test. Data from three independent experiments were combined and shown.
(E) Venn diagram representing the down-regulated genes in siRpl11 + siRpl14 and siDux + siRpl14 compared with siRpl14. Fisher’s exact test.
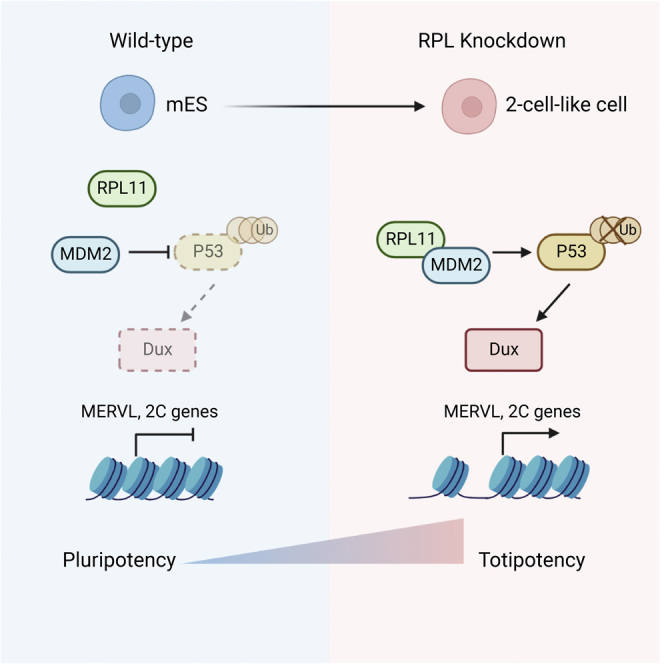
(F) Proposed model of RPs acting as regulators for 2C transcriptome. RP KD led to increased free-floating RPL11, which binds to MDM2, thus inhibiting P53 ubiquitination and degradation. This was followed by DUX expression and 2C activation. Created with biorender.com.
Discussion
We uncovered RPs—namely RPL14, RPL18, and RPL23—as key players involved in 2C transcripts. Our investigations confirmed their regulatory effects on MERVL and 2C genes at the gene expression level as well as in the maintenance of chromatin accessibility. Moreover, our results indicated that RP-dependent 2C regulation is primarily based on the MDM2-P53-DUX axis mediated by RPL11 (Figure 4F). In parallel, impaired binding of TRIM28 on Dux upon RP KD further promoted 2C activation. From these observations, we have elucidated a novel pathway in mESCs that connects RPs and 2C transcript activation, shedding light on research for RP functions and expanded pluripotency domains.
While disruption of rRNA synthesis was recently shown to be associated with 2C activation (Chen et al., 2021; Percharde et al., 2018; Yu et al., 2021), our study further establishes individual RPs as crucial triggers for 2C activation. These findings position the RPs, one of the most essential cellular components, as upstream 2C regulators, operating through their functional interactions with transcriptional factors and epigenetic modifiers. Multiple stresses, including DNA damage (Atashpaz et al., 2020), DNA replication stress (Grow et al., 2021), and splicing inhibition (Shen et al., 2021), were reported to trigger 2C activation. Given the sensitivity of embryos to environmental stress and the high level of RP synthesis in early embryogenesis, we speculate that embryos might use the described mechanism here as a stress response to ensure their development.
Meanwhile, our study also illustrates the distinct role of different RPs and their interplay in the regulatory cascade, deepening the understanding of this fundamental protein family. Of late, a growing volume of studies has been focusing on the extraribosomal function of RPs and have thus demonstrated their distinct regulatory roles in several molecular mechanisms (Wang et al., 2015), including proliferation (Kim et al., 2004; Lindström and Zhang, 2008; Volarević et al., 2000); apoptosis (He and Sun, 2007; Jang et al., 2004); DNA repair (Hegde et al., 2004; Kim et al., 1995); cellular development (Anderson et al., 2007; Flygare et al., 2005); and differentiation (Da Costa et al., 2003; Zhan et al., 2010). Due to the abundance of regulatory interactions between RPs, it is worth exploring the possible involvement of RPs via multiple pathways in the regulation of 2C activation and expanded pluripotency.
In particular, we show the involvement of MDM2-P53 and its downstream cascades in RP-dependent regulation of 2C transcripts and identify RPL11 as one of the direct mediators of this cascade. In human and mouse cells, RPL11 was reported as one of the critical mediators between RP imbalance and stress response pathways, along with c-Myc (Challagundla et al., 2011; Dai et al., 2007; Zhou et al., 2013) and MDM2-P53 (Bhat et al., 2004; Dai et al., 2004; Sasaki et al., 2011). Interestingly, three out of the five RPL/RPS proteins whose KD showed no effect on 2CLC transition, namely RPL11, RPL5, and RPL7, were reported to bind and inhibit MDM2 as a complex (Bhat et al., 2004; Nicolas et al., 2016; Sasaki et al., 2011; Takafuji et al., 2017). Therefore, it is highly likely that the other two “negative” RPs we screened out, RPS5 and RPL7A, are also a part of this essential mediating complex.
Since the mode of RPL-based 2C regulation is Dux dependent, other Dux-related 2C regulators are highly likely involved in this mode of regulation. Transcriptional factors were shown to be critical in cell fate transition of ESCs (Loh et al., 2006; Mzoughi et al., 2017; Ng et al., 2008; Maury et al., 2015; Wang et al., 2020). Specifically, the TRIM28-NCL complex was identified as the top target as it is an effector regulating both rRNA synthesis as well as 2C transcript expression (Percharde et al., 2018; Sun et al., 2021; Yu et al., 2021). The confirmation of TRIM28 as a downstream factor of RPs expands this stress response cascade. However, further mechanistic and biochemical investigations would be required to delineate other downstream partners and histone modifications that might be involved in this mode of regulation via RPs and related TRIM28 machinery. Interestingly, decreased expression of rRNA was observed upon Rpl14 KD (Figure S4E), indicating that dissociation of TRIM28 could be potentially caused by the disturbance of liquid-liquid phase separation (Yu et al., 2021). Meanwhile, LINE1 was recently shown to directly repress Dux expression, thus also repressing MERVL, by recruiting the TRIM28-NCL complex to establish the inhibitory histone marker H3K9me2 (De Iaco et al., 2017). Intriguingly, LINE1 was significantly up-regulated in RP KDs, indicating a possible synergistic effect of LINE1 and RPL on the inhibitory function of TRIM28. Thus, our studies on the unique roles of RPs are strongly indicative of their potential roles as significantly powerful upstream regulators of the 2C state.
The discovery of RPs as 2C transcript regulators adds a whole new layer of depth to the investigation of the 2C fate. With the growing relevance of the fields of expanded pluripotency and totipotency, these findings lay the crucial groundwork for further investigation and delineation of 2C fate determinants while also unraveling alternative roles for RPs—currently a relatively unexplored domain. Future studies could benefit from advanced platforms like single-cell sequencing, which couples both transcriptome and chromatin accessibility to provide additional insights after RP KD, on top of bulk RNA-seq (Xing et al., 2020a, 2020b).
Experimental procedures
Resource availability
Corresponding author
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Loh Yuin Han (yhloh@imcb.a-star.edu.sg).
Materials availability
Not applicable.
Cells and cell culture
mESCs (embryonic day 14 [E14]) were cultured on a 0.1% gelatin-coated plate with DMEM medium supplemented with 15% fetal bovine serum (FBS); 1,000 units/mL recombinant leukaemia inhibitory factor (LIF); 0.1 mM 2-mercaptoethanol; 2 mM L-glutamine; 0.1 mM MEM non-essential amino acids (NEAAs); and 50 U/mL penicillin/streptomycin. The 2C-reporter cell line was established by transfecting 2C-EGFP reporter (Addgene plasmid #69071) into E14 cells with Lipofectamin 2000, followed by 72 h of selection (500 mg/mL of G418), and single cells derived clone picking. Clones with reporter expressing were picked and cultured. All cell lines with plasmid transfected were maintained in the medium with the respectable antibiotic. All cultures were incubated at 37°C in 5% CO2.
siRNA KD
siRNAs against the potential targets were diluted to a working condition of 12.5 nM with the culture medium and added to E14 cell line with Dharmafect (Horizon Discovery). For double KD of siRpl14 with siRpl11 or siDux, an equal amount of siRNA (12.5 nM for each) was used. RNA was harvested at 48 h post-transfection.
shRNA KD
shRNA sequence-targeting Rpls were cloned into pSUPER.puro plasmid. Two μg shRNA plasmid was transfected with Lipofectamin 2000 (Invitrogen) to 2 × 106 cells seeded for 24 h in a 6-well plate. Puromycin selection (1.5 μg/mL) was applied 24 h post-transfection. Cells were harvested 72 h post-transfection and extracted for total RNA. The KD efficiency of the shRNAs were quantified by qRT-PCR. The shRNA sequences used in this study are listed in Table S1.
Chromatin IP (ChIP)
Cells knocked down with siRpl14 for 48 h were crosslinked by 1% formaldehyde for 10 min at room temperature. ChIP was performed as previously described (Warrier et al., 2022) using the following antibody: Trim28 (Bethyl A300-274A). Inputs and IP samples were analyzed by qPCR. Primers for this study are listed in Table S1.
Co-IP
ESCs knocked down with siRl14 for 48 h were lysed with IPH lysis buffer (25 mM Tris-HCl [pH 7.4], 150 mM Nacl, 1 mM EDTA, 1% NP-40, and 5% glycerol) on ice for 30 min, followed by repeated resuspension with a 21G needle for 50 times. The lysate was centrifuged at 16,000 × g at 4°C for 25 min. The supernatant was collected. Dynabeads protein G (Invitrogen; 50 μL per sample) was washed twice with IPH lysis buffer and incubated with antibody of MDM2 (Santa Cruz #sc965) and immunoglobulin G (IgG; Santa Cruz #sc2025) at room temperature for 3 h. The beads were isolated with magnetic stand and washed three times. Each time, the beads were resuspended in IPH lysis buffer, rotated for 5 min, and placed on the stand for 30 s. The beads were then incubated with the lysate at 4°C overnight. After incubation, the beads were again washed three times with IPH buffer and resuspended in elution buffer (4% SDS, 40 nM DTT, 100 mM Tris). Eluted protein was then sent for western blot analysis.
Single-cell RNA-seq data analysis
For the analysis of gene expression trends during the transition of 2CLCs, we used the public ESC and 2CLC single-cell data available under GEO: GSM3436751, GSM3436752, GSM3436753, and GSM3436754 (Fu et al., 2019). We directly used the mm10 processed data, metadata, and cell state annotation deposited by the authors. Trajectories and pseudotimes were computed using Palantir (Setty et al., 2019). Briefly, diffusion components were determined using the run_diffusion_maps function with the parameter “n_components = 7”, and t-stochastic neighbor embedding (tSNE) projection was used to visualize the data. The imputed expression values were further scaled, and a total of 11 cells were manually selected for data visualization.
Data resource
Previously published sequencing data that were reanalyzed here are available in the GEO under GEO: GSE66582 (Wu et al., 2016) (mouse embryo pre-implantation development stage RNA-seq), GSE75751 (Eckersley-Maslin et al., 2016) (Zscan+, and Zscan+_MuERVL+ RNA-seq), GSE85627 (Hendrickson et al., 2017) (Dux overexpression RNA-seq), GSE119819 (Yang et al., 2020) (Zmym2 KO RNA-seq), and GSE94324 (De Iaco et al., 2017) (Trim28 KD and Dux KO plus Trim28 KD RNA-seq).
Statistic tests
All statistical analysis was performed with R (www.r-project.org/) or with Prism 9.0. The statistical test used in the boxplot analysis was by one-sided Wilcoxon test or paired-end t test. The p values for the Venn diagram were calculated by Fisher’s exact test. Details of the statistical test and numbers of repeat are illustrated in each figure legend; p values <0.05 were considered statistically significant: ∗p <0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Author contributions
Y.Y. designed and performed research, analyzed data, and wrote the paper. Y.Z. designed research, performed most of the bioinformatics analyses, and revised the paper. T.W.S. and T.W. conducted research, analyzed data, and revised the paper. K.H. performed part of the bioinformatics analyses and revised the paper. R.J.R.T. and J.X.P. conducted research. R.T., Y.-C.L., H.L., and J.X. analyzed data. Y.-H.L. designed research, analyzed data, and wrote the paper.
Acknowledgments
We thank Qiu Ye Bao, Hao Fei Wang, Tao Yu, Hong Nguyen, and Xin Tian for technical support. We are grateful to Hui Li Guo, Wee Wei Tee, and Shi Feng Xue for helpful discussions. H.L. is supported by National Institutes of Health (AG056318, AG61796, and CA208517); the Glenn Foundation for Medical Research; Mayo Clinic Center for Biomedical Discovery; Center for Individualized Medicine, Mayo Clinic; Mayo Clinic Cancer Center; and the David F. and Margaret T. Grohne Cancer Immunology and Immunotherapy Program. J.X. is supported by the Institute for Water and Wetland Research, Radboud University; by the Department of Biological Sciences, National University of Singapore; and by the Joint Center for Single Cell Biology, Radboud University-Shanghai Jiao Tong University-Shandong Agricultural University. Y.-H.L. is supported by the NRF Investigatorship award - NRFI2018-02; IAF-PP grant - H1801a0021; NRF grant - NRF2019-THE002-0001; NMRC grant - OFIRG21nov-0088; A∗STAR grants - W22W3D0007 and C211318012; and A∗STAR BMRC - Use-Inspired Basic Research award. We are grateful to the Biomedical Research Council, Agency for Science, Technology and Research, Singapore, for research funding.
Conflict of interest
The authors declare no competing interests.
Published: January 12, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.12.007.
Contributor Information
Jian Xu, Email: j.xu@science.ru.nl.
Yuin-Han Loh, Email: yhloh@imcb.a-star.edu.sg.
Supplemental information
Data availability
The RNA-seq and ATAC-seq data reported in this paper have been depostied to NCBI Gene Expression Omnibus (GEO) with the following accession number: GSE179124, GSE185232, and GSE167177. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
- Anderson S.J., Lauritsen J.P.H., Hartman M.G., Foushee A.M.D.G., Lefebvre J.M., Shinton S.A., Gerhardt B., Hardy R.R., Oravecz T., Wiest D.L. Ablation of ribosomal protein L22 selectively impairs αβ T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–772. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Atashpaz S., Shams S.S., Gonzalez J.M., Sebestyén E., Arghavanifard N., Gnocchi A., Albers E., Minardi S., Faga G., Soffientini P., et al. ATR expands embryonic stem cell fate potential in response to replication stress. Elife. 2020;9:1–30. doi: 10.7554/eLife.54756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlasi Y., Jafarnejad S.M., Gkogkas C.G., Vermeulen M., Sonenberg N., Stunnenberg H.G. The translational landscape of ground state pluripotency. Nat. Commun. 2020;11:1–13. doi: 10.1038/s41467-020-15449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddington R.S., Robertson E.J. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. doi: 10.1242/dev.105.4.733. [DOI] [PubMed] [Google Scholar]
- Bhat K.P., Itahana K., Jin A., Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challagundla K.B., Sun X.X., Zhang X., Devine T., Zhang Q., Sears R.C., Dai M.S. Ribosomal protein L11 recruits miR-24/miRISC to repress c-myc expression in response to ribosomal stress. Mol. Cell Biol. 2011;31:4007–4021. doi: 10.1128/MCB.05810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Liu W., Guo J., Liu Y., Liu X., Liu J., Dou X., Le R., Huang Y., Li C., et al. Nuclear m6A reader YTHDC1 regulates the scaffold function of LINE1 RNA in mouse ESCs and early embryos. Protein Cell. 2021;12:455–474. doi: 10.1007/s13238-021-00837-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa L., Narla G., Willig T.N., Peters L.L., Parra M., Fixler J., Tchernia G., Mohandas N. Ribosomal protein S19 expression during erythroid differentiation. Blood. 2003;101:318–324. doi: 10.1182/blood-2002-04-1131. [DOI] [PubMed] [Google Scholar]
- Dai M.S., Sears R., Lu H., Dai M., Sears R., Lu H. Feedback regulation of c-myc by ribosomal protein L11 feedback regulation of c-myc by ribosomal protein L11. Cell Cycle. 2007;6:2735–2741. doi: 10.4161/cc.6.22.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M.-S., Zeng S.X., Jin Y., Sun X.-X., David L., Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A., Planet E., Coluccio A., Verp S., Duc J., Trono D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017;49:941–945. doi: 10.1038/ng.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin M., Alda-Catalinas C., Blotenburg M., Kreibich E., Krueger C., Reik W. Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev. 2019;33:194–208. doi: 10.1101/gad.321174.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin M.A., Svensson V., Krueger C., Stubbs T.M., Giehr P., Krueger F., Miragaia R.J., Kyriakopoulos C., Berrens R.V., Milagre I., et al. MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs. Cell Rep. 2016;17:179–192. doi: 10.1016/j.celrep.2016.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H.T., El Farran C.A., Xing Q.R., Zhang L.F., Li H., Lim B., Loh Y.H. Global H3.3 dynamic deposition defines its bimodal role in cell fate transition. Nat. Commun. 2018;9:1537. doi: 10.1038/s41467-018-03904-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flygare J., Kiefer T., Miyake K., Utsugisawa T., Hamaguchi I., Da Costa L., Richter J., Davey E.J., Matsson H., Dahl N., et al. Deficiency of ribosomal protein S19 in CD34+ cells generated by siRNA blocks erythroid development and mimics defects seen in Diamond-Blackfan anemia. Blood. 2005;105:4627–4634. doi: 10.1182/blood-2004-08-3115. [DOI] [PubMed] [Google Scholar]
- Fu X., Wu X., Djekidel M.N., Zhang Y. Myc and Dnmt1 impede the pluripotent to totipotent state transition in embryonic stem cells. Nat. Cell Biol. 2019;21:835–844. doi: 10.1038/s41556-019-0343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama H., Tsuji T., Hironaka K., Yoshida K., Sugimoto N., Fujita M. GRWD1 directly interacts with p53 and negatively regulates p53 transcriptional activity. J. Biochem. 2020;167:15–24. doi: 10.1093/jb/mvz075. [DOI] [PubMed] [Google Scholar]
- Fumagalli S., Ivanenkov V.V., Teng T., Thomas G. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev. 2012;26:1028–1040. doi: 10.1101/gad.189951.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam P., Hamashima K., Chen Y., Zeng Y., Makovoz B., Parikh B.H., Lee H.Y., Lau K.A., Su X., Wong R.C.B., et al. Multi-species single-cell transcriptomic analysis of ocular compartment regulons. Nat. Commun. 2021;12:5675–5714. doi: 10.1038/s41467-021-25968-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam P., Yu T., Loh Y.H. Regulation of ERVs in pluripotent stem cells and reprogramming. Curr. Opin. Genet. Dev. 2017;46:194–201. doi: 10.1016/j.gde.2017.07.012. [DOI] [PubMed] [Google Scholar]
- Grow E.J., Weaver B.D., Smith C.M., Guo J., Stein P., Shadle S.C., Hendrickson P.G., Johnson N.E., Butterfield R.J., Menafra R., et al. p53 convergently activates Dux/DUX4 in embryonic stem cells and in facioscapulohumeral muscular dystrophy cell models. Nat. Genet. 2021;53:1207–1220. doi: 10.1038/s41588-021-00893-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Sun Y. Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene. 2007;26:2707–2716. doi: 10.1038/sj.onc.1210073. [DOI] [PubMed] [Google Scholar]
- Hegde V., Wang M., Deutsch W.A. Human ribosomal protein S3 interacts with DNA base excision repair proteins hAPE/Ref-1 and hOGG1. Biochemistry. 2004;43:14211–14217. doi: 10.1021/bi049234b. [DOI] [PubMed] [Google Scholar]
- Hendrickson P.G., Doráis J.A., Grow E.J., Whiddon J.L., Lim J.W., Wike C.L., Weaver B.D., Pflueger C., Emery B.R., Wilcox A.L., et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017;49:925–934. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn H.F., Vousden K.H. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene. 2008;27:5774–5784. doi: 10.1038/onc.2008.189. [DOI] [PubMed] [Google Scholar]
- Hu Z., Tan D.E.K., Chia G., Tan H., Leong H.F., Chen B.J., Lau M.S., Tan K.Y.S., Bi X., Yang D., et al. Maternal factor NELFA drives a 2C-like state in mouse embryonic stem cells. Nat. Cell Biol. 2020;22:175–186. doi: 10.1038/s41556-019-0453-8. [DOI] [PubMed] [Google Scholar]
- Jang C.Y., Lee J.Y., Kim J. RpS3, a DNA repair endonuclease and ribosomal protein, is involved in apoptosis. FEBS Lett. 2004;560:81–85. doi: 10.1016/S0014-5793(04)00074-2. [DOI] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Kim J., Chubatsu L.S., Admon A., Stahl J., Fellous R., Linn S. Implication of mammalian ribosomal protein S3 in the processing of DNA damage. J. Biol. Chem. 1995;270:13620–13629. doi: 10.1074/jbc.270.23.13620. [DOI] [PubMed] [Google Scholar]
- Kim J.H., You K.R., Kim I.H., Cho B.H., Kim C.Y., Kim D.G. Over-expression of the ribosomal protein L36a gene is associated with cellular proliferation in hepatocellular carcinoma. Hepatology. 2004;39:129–138. doi: 10.1002/hep.20017. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Leslie P., Zhang Y. Ribosomal proteins as unrevealed caretakers for cellular stress and genomic instability. Oncotarget. 2014;5:860–871. doi: 10.18632/oncotarget.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström M.S., Zhang Y. Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J. Biol. Chem. 2008;283:15568–15576. doi: 10.1074/jbc.M801151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Gao M., He J., Wu K., Lin S., Jin L., Chen Y., Liu H., Shi J., Wang X., et al. The RNA m6A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature. 2021;591:322–326. doi: 10.1038/s41586-021-03313-9. [DOI] [PubMed] [Google Scholar]
- Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury J.J.P., El Farran C.A., Ng D., Loh Y.H., Bi X., Bardor M., Choo A.B.H. RING1B O-GlcNAcylation regulates gene targeting of polycomb repressive complex 1 in human embryonic stem cells. Stem Cell Res. 2015 Jul;15:182–189. doi: 10.1016/j.scr.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Mzoughi S., Zhang J., Hequet D., Teo S.X., Fang H., Xing Q.R., Bezzi M., Seah M.K.Y., Ong S.L.M., Shin E.M., et al. PRDM15 safeguards naive pluripotency by transcriptionally regulating WNT and MAPK-ERK signaling. Nat. Genet. 2017;49:1354–1363. doi: 10.1038/ng.3922. [DOI] [PubMed] [Google Scholar]
- Ng J.H., Heng J.C.D., Loh Y.H., Ng H.H. Transcriptional and epigenetic regulations of embryonic stem cells. Mutat. Res. 2008;647:52–58. doi: 10.1016/j.mrfmmm.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Nicolas E., Parisot P., Pinto-monteiro C., de Walque R., Lafontaine D.L.J., De Vleeschouwer C. Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat. Commun. 2016;7:11390. doi: 10.1038/ncomms11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’connor R.J. Experiments on the development of isolated blastomers of mouse eggs. J. Anat. 1939;74:34–44.5. [Google Scholar]
- Biechele S., Lin C.J., Shen X., Guan J., Peixoto G.A., Bulut-Karslioglu A., Percharde M., Huang B., Yin Y., Ramalho-Santos M., et al. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell. 2018;174:391–405.e19. doi: 10.1016/j.cell.2018.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A., Russo G. Ribosomal proteins control or bypass p53 during nucleolar stress. Int. J. Mol. Sci. 2017;18:140–216. doi: 10.3390/ijms18010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Kawahara K., Nishio M., Mimori K., Kogo R., Hamada K., Itoh B., Wang J., Komatsu Y., Yang Y.R., et al. Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat. Med. 2011;17:944–951. doi: 10.1038/nm.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M., Kiseliovas V., Levine J., Gayoso A., Mazutis L., Pe’er D. Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 2019;37:451–460. doi: 10.1038/s41587-019-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Yang M., Li S., Zhang J., Peng B., Wang C., Chang Z., Ong J., Du P. Mouse totipotent stem cells captured and maintained through spliceosomal repression. Cell. 2021:1–17. doi: 10.1016/j.cell.2021.04.020. [DOI] [PubMed] [Google Scholar]
- Sun Z., Yu H., Zhao J., Tan T., Pan H., Zhu Y., Chen L., Zhang C., Zhang L., Lei A., et al. LIN28 coordinately promotes nucleolar/ribosomal functions and represses the 2C-like transcriptional program in pluripotent stem cells. Protein Cell. 2022;13:490–512. doi: 10.1007/s13238-021-00864-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takafuji T., Kayama K., Sugimoto N., Fujita M. GRWD1, a new player among oncogenesis-related ribosomal/nucleolar proteins. Cell Cycle. 2017;16:1397–1403. doi: 10.1080/15384101.2017.1338987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh C.X.D., Chan J.W., Chong Z.S., Wang H.F., Guo H.C., Satapathy S., Ma D., Goh G.Y.L., Khattar E., Yang L., et al. RNAi reveals phase-specific global regulators of human somatic cell reprogramming. Cell Rep. 2016;15:2597–2607. doi: 10.1016/j.celrep.2016.05.049. [DOI] [PubMed] [Google Scholar]
- Volarević S., Stewart M.J., Ledermann B., Zilberman F., Terracciano L., Montini E., Grompe M., Kozma S.C., Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- Wang H.F., Warrier T., Farran C.A., Zheng Z.H., Xing Q.R., Fullwood M.J., Zhang L.F., Li H., Xu J., Lim T.M., Loh Y.H. Defining essential enhancers for pluripotent stem cells using a features-oriented CRISPR-cas9 screen. Cell Rep. 2020;33:108309. doi: 10.1016/j.celrep.2020.108309. [DOI] [PubMed] [Google Scholar]
- Wang W., Nag S., Zhang X., Wang M.H., Wang H., Zhou J., Zhang R. Ribosomal proteins andHuman diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Med. Res. Rev. 2015;35:225–285. doi: 10.1002/med.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrier T., El Farran C., Zeng Y., Ho B.S.Q., Bao Q., Zheng Z.H., Bi X., Ng H.H., Ong D.S.T., Chu J.J.H., et al. SETDB1 acts as a topological accessory to Cohesin via an H3K9me3-independent , genomic shunt for regulating cell fates. Nucleic Acids Res. 2022;50:7326–7349. doi: 10.1093/nar/gkac531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Fujiyama H., Takafuji T., Kayama K., Matsumoto M., Nakayama K.I., Yoshida K., Sugimoto N., Fujita M. GRWD1 regulates ribosomal protein L23 levels via the ubiquitin-proteasome system. J. Cell Sci. 2018;131:jcs213009. doi: 10.1242/jcs.213009. [DOI] [PubMed] [Google Scholar]
- Wu J., Huang B., Chen H., Yin Q., Liu Y., Xiang Y., Zhang B., Liu B., Wang Q., Xia W., et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–657. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- Xing Q.R., El Farran C.A., Gautam P., Chuah Y.S., Warrier T., Toh C.X.D., Kang N.Y., Sugii S., Chang Y.T., Xu J., et al. Diversification of reprogramming trajectories revealed by parallel single-cell transcriptome and chromatin accessibility sequencing. Sci. Adv. 2020;6:1–19. doi: 10.1126/sciadv.aba1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Q.R., Farran C.A.E., Zeng Y.Y., Yi Y., Warrier T., Gautam P., Collins J.J., Xu J., Dröge P., Koh C.G., et al. Parallel bimodal single-cell sequencing of transcriptome and chromatin accessibility. Genome Res. 2020;30:1027–1039. doi: 10.1101/gr.257840.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.L., Zhang C., Hao J., Wang X.L., Ming J., Mi L., Na J., Hu X., Wang Y. 2019. DPPA2/4 and SUMO E3 Ligase PIAS4 Opposingly Regulate Zygotic Transcriptional Program. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B.X., El Farran C.A., Guo H.C., Yu T., Fang H.T., Wang H.F., Schlesinger S., Seah Y.F.S., Goh G.Y.L., Neo S.P., et al. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell. 2015;163:230–245. doi: 10.1016/j.cell.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Huang X., Zang R., Chen J., Fidalgo M., Sanchez-Priego C., Yang J., Caichen A., Ma F., Macfarlan T., et al. DUX-miR-344-ZMYM2-Mediated activation of MERVL LTRs induces a totipotent 2C-like state. Cell Stem Cell. 2020;26:234–250.e7. doi: 10.1016/j.stem.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Sun Z., Tan T., Pan H., Zhao J., Zhang L., Chen J., Lei A., Zhu Y., Chen L., et al. rRNA biogenesis regulates mouse 2C-like state by 3D structure reorganization of peri-nucleolar heterochromatin. Nat. Commun. 2021;12:6365–6421. doi: 10.1038/s41467-021-26576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y., Melian N.Y., Pantoja M., Haines N., Ruohola-Baker H., Bourque C.W., Rao Y., Carbonetto S. Dystroglycan and mitochondrial ribosomal protein L34 regulate differentiation in the drosophila eye. PLoS One. 2010;5:e10488. doi: 10.1371/journal.pone.0010488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Wang J., Shen X., Huang B., Zhou H., Zhang H., Wu Z., Lu J.Y., Huang B., Zhou H., et al. DEAD-box helicase 18 counteracts PRC2 to safeguard ribosomal DNA in pluripotency regulation. Cell Rep. 2020;30:81–97.e7. doi: 10.1016/j.celrep.2019.12.021. [DOI] [PubMed] [Google Scholar]
- Zhang W., Chen F., Chen R., Xie D., Yang J., Zhao X., Guo R., Zhang Y., Shen Y., Göke J., et al. Zscan4c activates endogenous retrovirus MERVL and cleavage embryo genes. Nucleic Acids Res. 2019;47:8485–8501. doi: 10.1093/nar/gkz594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Hao Q., Liao J.M., Liao P., Lu H. Ribosomal protein S14 negatively regulates c-myc activity. J. Biol. Chem. 2013;288:21793–21801. doi: 10.1074/jbc.M112.445122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Liao W.J., Liao J.M., Liao P., Lu H. Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol. 2015;7:92–104. doi: 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and ATAC-seq data reported in this paper have been depostied to NCBI Gene Expression Omnibus (GEO) with the following accession number: GSE179124, GSE185232, and GSE167177. All other data supporting the findings of this study are available from the corresponding author on reasonable request.