Abstract
Many photosynthetic bacteria use inorganic sulfur compounds as electron donors for carbon dioxide fixation. A thiosulfate-induced cytochrome c has been purified from the photosynthetic α-proteobacterium Rhodovulum sulfidophilum. This cytochrome c551 is a heterodimer of a diheme 30-kDa SoxA subunit and a monoheme 15-kDa SoxX subunit. The cytochrome c551 structural genes are part of an 11-gene sox locus. Sequence analysis suggests that the ligands to the heme iron in SoxX are a methionine and a histidine, while both SoxA hemes are predicted to have unusual cysteine-plus-histidine coordination. A soxA mutant strain is unable to grow photoautotrophically on or oxidize either thiosulfate or sulfide. Cytochrome c551 is thus essential for the metabolism of both these sulfur species. Periplasmic extracts of wild-type R. sulfidophilum exhibit thiosulfate:cytochrome c oxidoreductase activity. However, such activity can only be measured for a soxA mutant strain if the periplasmic extract is supplemented with purified cytochrome c551. Gene clusters similar to the R. sulfidophilum sox locus can be found in the genome of a green sulfur bacterium and in phylogenetically diverse nonphotosynthetic autotrophs.
The environmentally most abundant reduced inorganic sulfur species are sulfide and thiosulfate. These species are converted to sulfate in the oxidative half of the sulfur cycle, primarily by bacterial action. Photosynthetic sulfur-oxidizing bacteria use sulfur compounds as the electron donor for reductive carbon dioxide fixation during photolithotrophic growth (4, 5). In these organisms electrons obtained from the sulfur compounds are initially fed into the photosynthetic electron transfer chain. Light energy is then used to drive movement of the electrons onto the more reducing electron carriers NAD(P)+ and ferredoxin. Photosynthetic sulfur oxidation is an ancient metabolism, and most anoxygenic photosynthetic bacteria have at least a limited ability to use such compounds (4). In nonphotosynthetic (colorless) sulfur bacteria, sulfur compounds are oxidized to support chemolithotrophic growth (12, 21). In these bacteria the sulfur compounds function primarily as respiratory electron donors, providing energy for cellular metabolism via oxidative phosphorylation. The electron acceptor is either oxygen or inorganic nitrogen compounds. A minority of the electrons derived from the sulfur species are used for carbon dioxide fixation.
In our laboratory we seek to understand photolithotrophic sulfur metabolism using the genetically accessible α-proteobacterium Rhodovulum sulfidophilum (formerly [f.] Rhodobacter sulfidophilus, formerly Rhodopseudomonas sulfidophila) (17, 18) as our model organism. R. sulfidophilum is able to carry out the complete eight-electron oxidation of sulfide or thiosulfate to sulfate. The thiosulfate oxidation pathway in R. sulfidophilum is thiosulfate inducible, and sulfite is a free intermediate in the process (36).
Photosynthetic α-proteobacteria operate a cyclical light-driven electron transport chain. Excitation of the photosynthetic reaction center by light of the appropriate wavelength results in transfer of electrons from the reaction center to ubiquinone. The resultant ubiquinol is then oxidized by the cytochrome bc1 complex with a periplasmic c-type cytochrome, normally cytochrome c2, acting as the electron acceptor. The cycle is completed by transfer of electrons from the ferrocytochrome c to the oxidized reaction center. During photolithotrophic growth the reductant for carbon dioxide fixation is provided in the form of NADH. This is produced by reverse electron transfer from ubiquinol through the NADH:ubiquinone oxidoreductase (complex I). The electrons removed from the photosynthetic electron transfer chain in this way are replaced by oxidation of an inorganic electron donor. It is therefore anticipated that the electron transfer pathways associated with thiosulfate and sulfide oxidation in R. sulfidophilum will utilize cytochrome c2 and/or ubiquinone as the terminal oxidant.
In an earlier study it was observed that R. sulfidophilum grown autotrophically with thiosulfate as electron donor expresses a c-type cytochrome that is not found in heterotrophically grown cells (36). The cytochrome is therefore a good candidate to be a component of the thiosulfate oxidation pathway. Taking this observation as our starting point, we have purified the thiosulfate-induced cytochrome and shown it to be a heterodimeric protein containing three covalently bound heme groups. The structural genes encoding the cytochrome are part of a large genetic locus involved in inorganic sulfur metabolism. Biochemical and genetic evidence is presented that confirms a function for the cytochrome in thiosulfate oxidation. Unexpectedly, the cytochrome is also essential for sulfide oxidation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
R. sulfidophilum DSM1374T (strain W4 in reference 17) or a spontaneous rifampin-resistant derivative, strain 3.1, was used throughout this study. R. sulfidophilum was normally cultured at 30°C in sealed glass vessels placed in front of tungsten lamps. R. sulfidophilum was routinely grown on a basal salts medium, RCV-N, adapted from the RCV medium of Weaver et al. (16, 52). The constituents of this medium are 5 mM KH2PO4, 5 mM K2HPO4, 7.5 mM (NH4)2SO4, 425 mM NaCl, 0.5 mM MgSO4, 0.5 mM CaCl2, 85 μM FeSO4, 50 μM disodium EDTA, 11 μM H2BO3, 1.8 μM MnSO4, 0.9 μM NaMoO4, 0.2 μM ZnSO4, and 40 nM Cu(NO3)2, and the pH is adjusted to 6.5. After autoclaving, 1 mg of thiamine-HCl, 1 mg of nicotinic acid, 0.1 mg of biotin. and 0.2 mg of para-aminobenzoic acid per liter are added from a 1,000-fold-concentrated filter-sterilized stock. For photoheterotrophic growth, RCV-N was supplemented with 30 mM d,l-disodium malate to give medium RCV-NM. For photolithotrophic growth, RCV-N was modified by replacing the KH2PO4 and K2HPO4 components with 20 mM Tricine and 1 mM K2HPO4 and adjusting the pH of the medium to 7.8 with NaOH. After autoclaving, 40 mM NaHCO3 was added from a filter-sterilized 0.6 M stock or, for large-scale cultures, directly as a powder. This basal medium constitutes RCV-A. Control experiments demonstrate that R. sulfidophilum does not use the Tricine in RCV-A as a carbon source. For photolithotrophic growth with thiosulfate as electron donor, RCV-A was additionally supplemented after autoclaving with 100 mM Na2S2O3 from a separately autoclaved 1 M stock solution to give medium RCV-AT. The pH of the culture medium was monitored every 12 h during photolithotrophic growth, and an additional 40 mM NaHCO3 was added if the pH fell to 7.5. For photomixotrophic growth with thiosulfate as electron donor and malate as carbon source, RCV-AT medium was supplemented with 20 mM d,l-disodium malate to give medium RCV-ATM. For photolithotrophic growth of R. sulfidophilum strains with sulfide as electron donor, RCV-A was supplemented with 3.5 mM Na2S to produce medium RCV-AS. For photomixotrophic growth on sulfide plus malate, RCV-AS was supplemented with 20 mM d,l-disodium malate to produce medium RCV-ASM. For photolithotrophic growth with formate as electron donor, RCV-A medium was supplemented with 50 mM sodium formate.
For single-colony isolation, R. sulfidophilum was routinely cultured phototrophically at 30°C on LB-agar medium (32) in a Don Whitley Mark 3 anaerobic cabinet (atmosphere of 10% H2, 10%CO2, and 80%N2) custom modified with tungsten bulb strip lighting in the roof. For photolithotrophic growth on solid media with sulfide as electron donor, RCV-A agar plates were incubated in an illuminated anaerobic jar (Oxoid) containing an Anaerogen (Oxoid) hydrogen-free anoxia-generating sachet. A sulfidic atmosphere was generated over the plates by taping an open Eppendorf tube containing a mixture of 0.1 g of thioacetamide in 1 ml of 0.2 M HCl (19, 46) to the inside surface of the jar.
Escherichia coli strains DH5α [supE44 ΔlacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 λ−], used in routine cloning experiments, and S17-1 [endA1 recA1 gyrA96 hsdR17supE44 λ− Δ(lac-proAB)/F′(traD36 proAB+ lacZΔM15)] (47) were grown on LB medium. Antibiotics, when required, were added to E. coli or R. sulfidophilum cultures at the following final concentrations: ampicillin, 50 μg/ml; kanamycin, 25 μg/ml; spectinomycin, 25 μg/ml; streptomycin, 50 μg/ml; rifampin, 50 μg/ml; and gentamicin, 5 μg/ml.
Preparation of periplasmic extract from R. sulfidophilum
Cells were harvested by centrifugation at 12,000 × g for 20 min at 4°C and then resuspended in 20 ml of spheroplasting buffer (50 mM Tris-HCl, pH 8.0, 0.5 M sucrose, 1.5 mM disodium EDTA) per g (wet weight) of cells. The resuspended cells were incubated with 600 μg of lysozyme (Sigma L2879) ml−1 for 30 min at 30°C. Spheroplasts were removed by centrifugation at 12,000 × g for 20 min at 4°C. The supernatant was further centrifuged at 200,000 × g and 4°C for 1 h to remove fine membrane fragments.
Periplasmic extracts for use in the thiosulfate:cytochrome c oxidoreductase activity assays were prepared by the same protocol except that prior to fractionation the cells were washed three times with growth medium lacking electron donor, the cells were then resuspended in 4 ml of spheroplasting buffer per g (wet weight) of cells, and lysozyme was added to a final concentration of 3 mg ml−1.
Purification of R. sulfidophilum cytochrome c551
A 50-liter total culture volume of R. sulfidophilum grown photolithotrophically with thiosulfate as electron donor was harvested at early stationary phase (A650 = 1 to 1.3) by crossflow ultrafiltration, and a periplasmic extract prepared as described above. The first two chromatography steps in the purification procedure were carried out at 4°C, with the remaining column separations performed at room temperature. The periplasmic extract was loaded onto a Q Sepharose Fast Flow (Amersham Pharmacia Biotech) anion-exchange column (2.6-cm diameter by 55-cm height) that had previously been equilibrated with 50 mM Tris-HCl, pH 7.8. The column was then washed with 300 ml of the equilibration buffer and developed with a linear gradient of 0 to 1 M NaCl in 1.5 liters of equilibration buffer. Fractions in the hemoprotein peak (as judged by absorbance at 410 nm) eluting at approximately 0.45 M NaCl were pooled. Solid (NH4)2SO4 was added to a concentration of 25% saturation, and the sample was applied to a Phenyl Sepharose 6 Fast Flow high sub (Amersham Pharmacia Biotech) hydrophobic interaction column (1.6-cm diameter by 20-cm height) that had been preequilibrated with 50 mM Tris-HCl, pH 7.8, and 25% saturation (NH4)2SO4. The column was developed with a linear gradient of 25 to 0% saturation (NH4)2SO4 in 50 mM Tris-HCl, pH 7.8, and the hemoprotein fractions [eluting around 13% saturation (NH4)2SO4] were pooled. Ultrafiltration with an Amicon Diaflo YM3 membrane was used to exchange the buffer of the pooled fractions for 20 mM HEPES-NaOH (pH 7.8)–150 mM NaCl and then to concentrate the fractions to 1 ml. The concentrated sample was subject to fast protein liquid chromatography size exclusion chromatography on a Superdex 75 HiLoad (Amersham Pharmacia Biotech) column (1.6-cm diameter by 60-cm height) equilibrated with 20 mM HEPES-NaOH (pH 7.8)–150 mM NaCl. The highest purity cytochrome c551-containing fractions were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, pooled, and subjected to separation by anion-exchange perfusion chromatography on a 1.6-ml SP12 VAB POROS 20HQ column run on a BioCAD Sprint system. The column running buffer was 20 mM sodium acetate, pH 5.0, and the column was developed with a 110-ml linear gradient of 0 to 0.75 M NaCl. Immediately following chromatography, 0.2 volume of 100 mM HEPES-NaOH, pH 7.8, was added to each collected fraction. Fractions containing pure cytochrome c551 were identified by SDS-PAGE analysis. Routinely these fractions were exchanged into 10 mM HEPES-NaOH, pH 7.0, and concentrated by ultrafiltration, and 50-μl aliquots were flash frozen in liquid N2 for long-term storage.
Analytical methods.
SDS-PAGE analysis employed the buffer system of Laemmli (26). Following electrophoresis, c-type cytochromes were detected by a heme-linked peroxidase stain (50). For peptide sequencing, purified SoxAX complex was subjected to SDS-PAGE with 2 mM thioglycolate added to the cathode buffer. The gel was blotted on a polyvinylidene difluoride (Bio-Rad Sequi-Blot PVDF) membrane and stained with Ponceau S, and the SoxA and SoxX bands were excised. N-terminal peptide sequences were determined directly from the membrane-immobilized subunits. Tryptic peptides of the SoxA subunit were obtained by digesting the membrane-bound protein and separating the resultant fragments by microbore reverse-phase high-pressure liquid chromatography.
Electrospray mass spectra were acquired on a Micromass Platform instrument, using 1:1:0.001 CH3CN/H2O/HCOOH as sample solvent and horse heart myoglobin as calibration standard. Approximately 40 pmol of sample was used per run. Data were analyzed with the supplier's Masslynx software.
Analytical ultracentrifugation experiments were carried out in a Beckman Optima XL/I centrifuge with absorbance optics using an An50Ti rotor. Aliquots (110 μl) of SoxAX dialyzed against 10 mM HEPES (pH 7.0)–100 mM NaCl buffer were placed in the sample sectors of charcoal-filled Epon two-channel centerpieces. The reference sectors were loaded with 120 μl of dialysis buffer. Centrifugation was performed at 15,000 rpm and 20°C. Equilibrium data were collected at 410 nm, with five readings averaged for each scan and data from five scans combined. Equilibrium was confirmed by the absence of any difference between data collected 4 h or more apart. Partial specific volumes were calculated from the SoxA and SoxX amino acid sequences using the program SEDNTERP, neglecting the hemes. Data were analyzed using software supplied by Beckman with the centrifuge.
Thiosulfate concentrations in growth media were measured by an iodometric method (20) after removal of cells by centrifugation. Sulfide concentrations were measured by the method of Hallenbeck et al. (15).
Cloning and sequence analysis of R. sulfidophilum sox locus.
Standard molecular genetic techniques were carried out as described (32, 45). R. sulfidophilum genomic DNA was prepared using the Wizard Genomic DNA purification kit from Promega. Degenerate oligonucleotides 5′-GA(C/T)CC(C/G)CT(C/G)GT(C/G)ATCAA(C/T)GG-3′ and 5′-C(G/T)(C/G)AC(C/G)(C/G)(A/T)(C/G)GG(C/G)CC(C/T)TC(C/G)AC-3′ were designed on the basis of, respectively, the SoxA internal peptide sequence RGNGLSVEGPSVR and the SoxA amino-terminal sequence GPDDPLVINGEIEIVTRAPT and used to amplify an 800-bp internal soxA fragment using R. sulfidophilum DSM1374T chromosomal DNA as the template. The amplified soxA fragment was cloned into plasmid Bluescript SK(+) (Stratagene) and then used as a hybridization probe to screen an R. sulfidophilum DSM1374T genomic library (31) constructed in the vector SuperCos (Stratagene). Cosmid 207 containing the entire sox locus was identified by this procedure and used by MWG Biotech and Genome Express to determine the DNA sequence of the sox region. The DNA sequence was analyzed by the GCG v10.1 (11) and Lasergene (DNAstar Inc.) software packages together with Web facilities. Predicted proteins were compared with the protein databases using the BlastP program (1) with the BLOSUM62 scoring matrix. The possible presence of signal peptides on the sox gene products was assessed using the program SignalP (37), while proteins were tested for the presence of potential transmembrane helices using the program TMHMM (48).
Construction of soxA mutant strains.
Suicide plasmids for the construction of the soxA mutant strains were prepared as follows. A 1,082-bp fragment starting 508 bp upstream of soxA was amplified from the R. sulfidophilum chromosome by PCR with primers 5′-AAAATCTAGACCAATACCGTGAAAGTCACCATCGGCGGCT-3′ and 5′-AAAAGGATCCAGATCTCGCGGCCCTTCTCCCAGGTCGACT-3′. The product was digested with XbaI and BamHI and cloned into the same sites in the polylinker of suicide vector pARO181 (38), producing plasmid psoxA-N. A second fragment of the R. sulfidophilum chromosome covering the 1,716-bp region ending 1,502 bp after the soxA stop codon was amplified using primers 5′-AAAAGGATCCGACCATCTGAGCCAGGGCCAGATCAACGGC-3′ and 5′-AAAAGGTAC CGAGGATGGTGTGCAGCATCTCGCCCG TCAT-3′, digested with BamHI and KpnI, and cloned into BamHI- and KpnI-cut psoxA-N to produce plasmid psoxA. The 2-kb BamHI fragment of pUX-Ω (40) bearing the Ω (Specr, Strepr) cartridge was then cloned into the BamHI site of plasmid psoxA to produce the plasmid psoxA::Ω. Plasmid psoxA::Gmr was constructed by cloning the BamHI fragment of pWKR189I (34) into the BamHI site of psoxA and then selecting a construct in which the gentamicin resistance gene was transcribed in the same direction as soxA. The final constructs were verified by DNA sequencing.
Interposon-containing plasmids were mobilized from E. coli S17.1 into R. sulfidophilum 3.1 (Rifr) by mixing exponential-phase donor and recipient cells in a 1 to 4 ratio and spotting the mixture onto nitrocellulose filters placed on LB agar plates. Following overnight aerobic incubation at 30°C, recombinant donor cells were selected by plating dilutions of the mating mixture on LB agar containing the appropriate interposon-selective antibiotic together with rifampin and incubating the plates in the illuminated anaerobic cabinet for 4 days. Recombinant cells were then screened for the loss of vector-encoded kanamycin resistance by replica plating. Southern hybridization was used to confirm the presence in the mutant strains of an interposon in the soxA gene and loss of the vector.
RNA purification and analysis.
Cultures (50 ml) of R. sulfidophilum strains were harvested in exponential growth phase, resuspended in 200 μl of 10 mM Tris-HCl (pH 8.0)–1 mM disodium EDTA and 0.4 mg of lysozyme per ml. RNA was then prepared from the cells using the Sigma total mammalian RNA miniprep kit. The RNA was further purified by RQ1 DNase I (Promega) treatment followed by extraction with phenol and then with phenol-chloroform (5:1). Each reverse transcriptase PCR (RT-PCR) reaction contained 2 μg of the purified RNA. The reactions were carried out using the Promega Access RT-PCR kit. The soxC-specific primers were 5′-AAGGAAGATTACCGGCTGATG-3′ and 5′-CGTATCGGTGTATTTCGAGGTC-3′. The soxF-specific primers were 5′-GGCAAGACCTATTACACCTGCT-3′ and 5′-TGTTTCGAGAAGTTTTCCTTGG-3′. Each set of primers amplifies an approximately 500-bp fragment.
Measurement of thiosulfate:cytochrome c oxidoreductase activity in periplasmic extracts.
Reactions were carried out at 30°C in 0.5-ml capacity glass cuvettes stoppered with a butyl rubber septum and rendered anoxic by bubbling with oxygen-free nitrogen for 5 min. The reaction mixture comprised 0.4 ml of freshly prepared periplasmic extract together with 65 μl of spheroplasting buffer and 10 μl of a 100 mM Na2S2O3 solution to give a final concentration of electron donor in the assay of 2 mM. The reaction was initiated by the addition of 25 μl of equine heart ferricytochrome c (Sigma) from a 1 mM stock. Cytochrome c reduction was monitored at 550 nm, and initial rates were calculated using ΔΕ550 nm(ferrocytochrome c − ferricytochrome c) = 20 mM−1 cm−1. All other donor-cytochrome c oxidoreductase activity assays were carried out in a Belle Technology anaerobic glove box (<10 ppm O2) fitted with a World Precision Instruments modular diode array spectrophotometer.
RESULTS
Purification and characterization of a thiosulfate-induced cytochrome c
Neutzling and coworkers have previously observed that R. sulfidophilum grown photolithotrophically with thiosulfate as electron donor contains a c-type cytochrome that is not present in photoheterotrophically grown cells (36). This cytochrome has an α-band visible absorption maximum at 551 nm in the reduced state and is therefore designated cytochrome c551. We determined that cells grown photomixotrophically on thiosulfate plus malate also produced cytochrome c551, confirming a correlation between the occurrence of cytochrome c551 and the presence of thiosulfate in the growth medium. The thiosulfate induction of cytochrome c551 expression suggests that this hemoprotein is likely to have a function in thiosulfate oxidation.
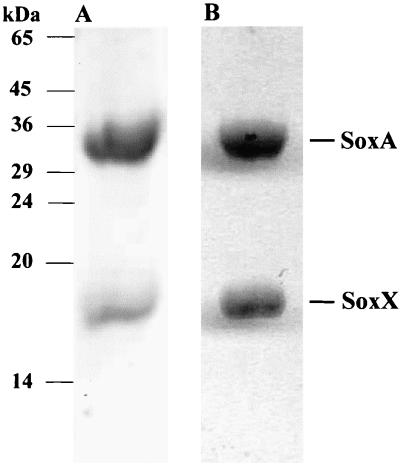
Cytochrome c551 was purified to homogeneity from a periplasmic extract of R. sulfidophilum cells cultured photolithotrophically with thiosulfate as electron donor. The purified preparation contains two polypeptides with apparent molecular masses of 18 kDa and 30 kDa under SDS-PAGE (Fig. 1) and 15,357.3 ± 0.8 Da and 30,177 ± 4 Da by electrospray mass spectrometery. We designated the larger polypeptide SoxA and the smaller protein SoxX. The alkaline pyridine ferrohemochrome spectrum of cytochrome c551 has a single α-band absorption maximum at 550 nm. This is characteristic of c-type cytochromes in which the heme group is covalently linked to the protein by two thioether linkages. SoxA and SoxX both stained for heme-linked peroxidase activity following denaturing electrophoresis (Fig. 1), indicating that both subunits of cytochrome c551 possess covalently bound heme. Solvent extraction experiments gave no evidence for additional non-covalently bound hemes or other chromophores.
FIG. 1.
Purified cytochrome c551 is composed of two heme-binding subunits. Purified cytochrome c551 was subjected to nonreducing SDS-PAGE in a 15% polyacrylamide gel. Lane A was stained with Coomassie brilliant blue R250 to detect proteins. Lane B was treated with a heme-linked peroxidase stain to identify polypeptides possessing covalently bound heme groups. The electrophoretic mobility of marker proteins is indicated to the side of the gel.
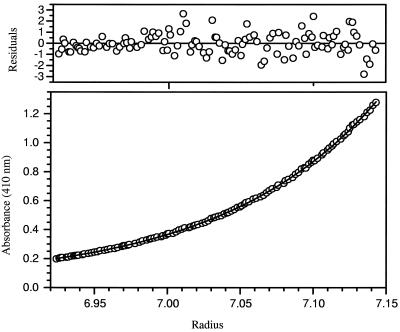
The copurification of SoxA and SoxX is presumptive evidence that the two proteins form a complex. To rigorously assess the oligomeric state of cytochrome c551, the hydrodynamic properties of the purified material were analyzed by analytical ultracentrifugation. Sedimentation velocity experiments carried out in a 10 mM sodium HEPES–100 mM NaCl, pH 7.0, buffer at 20°C gave no indication that multiple components were present (data not shown). Thus, SoxA and SoxX form a single tight complex under these conditions. Sedimentation equilibrium studies in the same buffer system at three different protein concentrations (0.14, 0.07, and 0.035 mg ml−1) gave three essentially identical sedimentation curves and allowed the calculation of a mass of 48,742 Da for native cytochrome c551 (Fig. 2). This value corresponds closely to the mass of a 1 to 1 complex of the SoxA and SoxX polypeptides (45,534 Da), and so we conclude that cytochrome c551 is a heterodimeric SoxA1-SoxX1 complex.
FIG. 2.
Determination of the native molecular mass of cytochrome c551 by sedimentation equilibrium studies. The sample contained 0.14 mg of cytochrome c551 per ml in 10 mM sodium HEPES–100 mM NaCl, pH 7.0. The sample was sedimented at 18,000 × g for 72 h at 20°C. Sedimentation of the cytochrome was monitored by visible spectroscopy using the heme absorption maximum at 410 nm. The lower panel shows the sedimentation curve for cytochrome c551 together with the simulated single-component sedimentation curve for a species with a weight average molecular mass of 48,742 Da. Residuals between experimental data and the fitted curve are shown in the top panel.
Purified cytochrome c551 was not reduced by thiosulfate (2 mM), did not exhibit thiosulfate:cytochrome c oxidoreductase activity with either R. sulfidophilum ferricytochrome c2 or equine heart ferricytochrome c as electron acceptor, and did not possess thiosulfate:ferricyanate oxidoreductase activity. The ability of cytochrome c551 to oxidize other sulfur compounds with equine heart ferricytochrome c as electron acceptor was also tested. However, no oxidation of sulfite (1 mM) or enhancement of the rate of chemical reduction of cytochrome c by 50 μM sulfide was observed.
Cloning of R. sulfidophilum genomic locus encoding cytochrome c551.
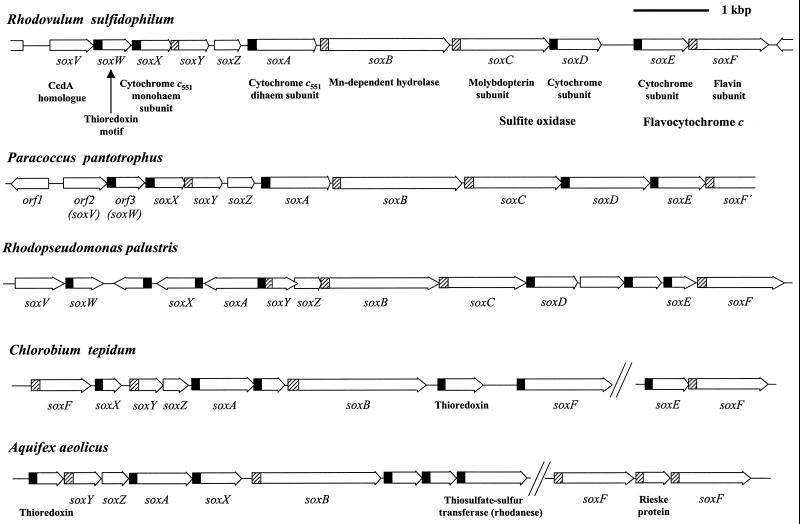
Amino-terminal and five internal peptide sequences were obtained from the SoxA subunit of purified R. sulfidophilum cytochrome c551. Degenerate primers designed on the basis of the amino-terminal and one of the internal peptide sequences were used to amplify an 800-bp fragment of the SoxA-encoding gene. This fragment was then used to isolate a cosmid containing the full soxA gene together with approximately 45 kbp of the surrounding chromosomal DNA. Sequence analysis of the cosmid shows soxA to be one of a set of 11 closely spaced and identically orientated open reading frames that potentially form a transcriptional unit (Fig. 3 and 4). Two of the open reading frames at the soxA locus (soxC and soxD) overlap, while the remaining genes are separated by noncoding regions of between 27 and 430 bp. Amino-terminal sequence analysis of the small subunit of cytochrome c551 allows identification of one of the soxA-linked open reading frames as the SoxX structural gene (Fig. 5). The genes in the soxA cluster show sequence similarity to a chromosomal region involved in thiosulfate oxidation in the nonphotosynthetic α-proteobacterium Paracoccus pantotrophus (formerly Paracoccus denitrificans GB17, formerly Thiosphaera pantotropha) (13, 53, 54) (Fig. 3). We have therefore adopted the P. pantotrophus sox designations for the corresponding open reading frames in R. sulfidophilum. A soxA-like gene has also recently been identified in another nonphotosynthetic α-proteobacterium, Thiobacillus sp. strain KCT001, where it is the site of transposon insertion in a thiosulfate oxidation-defective mutant (35) (Fig. 4). Homologues of many of the genes in the R. sulfidophilum soxA gene cluster can be found grouped in the genome sequences of another thiosulfate-oxidizing photosynthetic α-proteobacterium, Rhodopseudomonas palustris, the nonphotosynthetic hyperthermophilic bacterium Aquifex aeolicus, and the green sulfur bacterium Chlorobium tepidum, supporting the proposed functional relationship of the R. sulfidophilum open reading frames (Fig. 3). These genes have been assigned the same designations as their R. sulfidophilum homologues in Fig. 3. Sequence comparisons show that the SoxA protein of C. tepidum is equivalent to a cytochrome c551 of Chlorobium limicola strain Tassajara (23) (Fig. 4). Synthesis of this cytochrome c551 in Chlorobium species is correlated with the ability of the strain to use thiosulfate as a photosynthetic electron donor (23), and there is biochemical evidence for an ill-defined role of this cytochrome in thiosulfate oxidation (25).
FIG. 3.
Schematic overview of the soxA locus of R. sulfidophilum and related gene clusters in other bacteria. The predicted identities of the gene products are indicated under the gene designations where appropriate. Signal peptide coding regions are indicated in black for Sec signal peptides and are hatched for Tat signal peptides. Both C. tepidum and A. aeolicus produce multiple proteins with homology to the R. sulfidophilum SoxF flavoprotein. The sequence of the R. sulfidophilum soxA locus was determined in the present work and has been deposited in the GenBank database with accession number AY005800. Preliminary sequence data for C. tepidum were obtained from the Institute for Genomic Research website (http://www.tigr.org) and for R. palustris from http://www.jgi.doe.gov/JGI_microbial/html/rhodo_homepage.html. The P. pantotrophus sequence data are from references 13, 53, and 54, while the A. aeolicus sequence is from reference 9.
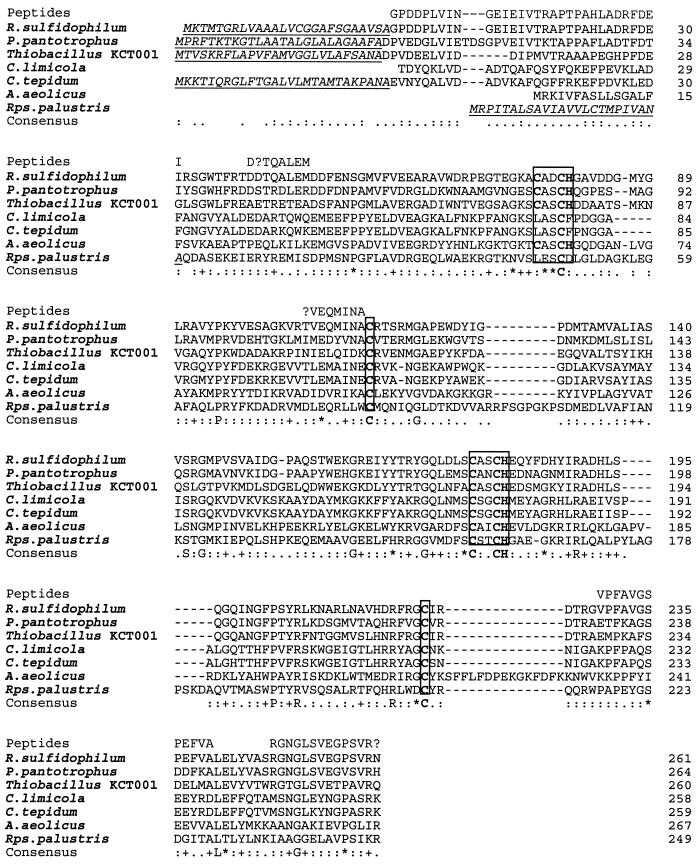
FIG. 4.
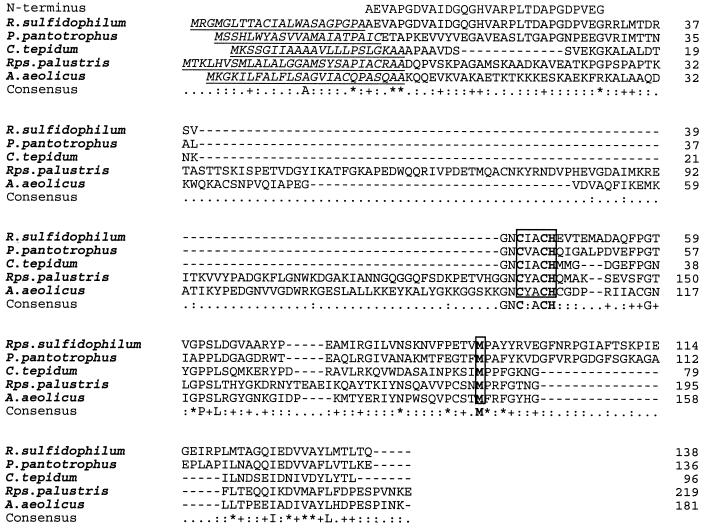
Multiple sequence analysis of SoxA proteins. The amino acid sequences of the amino terminus of the mature R. sulfidophilum SoxA protein and of internal SoxA peptides determined by protein sequencing in this work are shown on the line marked “Peptides.” Experimentally determined (R. sulfidophilum and P. pantotrophus) or predicted signal peptides are underlined. No signal peptide is shown for the A. aeolicus protein due to uncertainty in the identity of the precursor start codon. The consensus c-type cytochrome Cys-Xaa-Xaa-Cys-His heme attachment sites and conserved cysteines that are the proposed heme iron ligands in R. sulfidophilum SoxA are boxed. The sources of the sequence data are the same as in Fig. 3 with the addition of C. limicola cytochrome c551 from reference 23 and Thiobacillus sp. strain KCT001 SoxA from reference 35. The numbering on the individual sequences refers to the mature protein.
FIG. 5.
Multiple sequence analysis of SoxX proteins. The amino terminus of the mature R. sulfidophilum SoxX protein determined by protein sequencing is given in the line labeled “N-terminus.” Experimentally determined (R. sulfidophilum and P. pantotrophus) or predicted SoxX signal peptides are underlined. The consensus c-type cytochrome Cys-Xaa-Xaa-Cys-His heme attachment site and the proposed methionine distal heme iron ligand are boxed. The sources of the sequence data are as for Fig. 3. The numbering on the individual sequences refers to the mature protein.
With the exception of SoxV, all 11 R. sulfidophilum Sox gene products are predicted to be water-soluble proteins. Nine of these water-soluble proteins are predicted to be synthesized as precursor proteins, with N-terminal signal sequences directing export of the protein to the periplasm (Fig. 3). Five of these proteins have standard signal peptides that target the unfolded precursor by the Sec pathway (41), while the other four proteins have twin arginine signal peptides that mediate export of the prefolded, often cofactor-containing, precursor via the Tat apparatus (2, 3) (Fig. 3).
Sequence analysis of SoxA and SoxX.
Each subunit of cytochrome c551 contains covalently bound heme (Fig. 1), and consistent with this observation, the SoxA sequence contains two, and the SoxX sequence contains one, Cys-Xaa-Xaa-Cys-His c-type cytochrome heme attachment motif (Fig. 4 and 5). The assignment of two c-type heme groups (616.5 Da of additional mass for each heme, assuming protonated propionate groups under the assay conditions) to the large subunit is supported by the similarity of the calculated mass of diheme-modified mature SoxA protein (30,142.3 Da) to the experimentally determined mass of the large subunit (30,177 ± 4 Da). However, having taken the two heme groups into account, the experimentally determined mass exceeds the calculated mass by approximately 35 Da, suggesting that SoxA is subject to a posttranslational modification. This extra mass might arise from an additional sulfur atom (32 Da) or two additional oxygen atoms (32 Da). The calculated mass of the mature SoxX protein with one covalently bound heme (15,360.2 Da) is in good agreement with the mass of the cytochrome c551 small subunit determined by mass spectrometry (15,357.3 ± 0.8 Da).
In hemoproteins the heme iron is coordinated by either one or two axial protein ligands. In structurally characterized c-type cytochromes, the histidines of the two Cys-Xaa-Xaa-Cys-His heme attachment sites are invariably found to coordinate the iron of the heme attached to the adjacent cysteine residue. Thus, the three heme groups of R. sulfidophilum cytochrome c551 should each have at least one histidine iron ligand. Detailed spectroscopic analysis of R. sulfidophilum cytochrome c551 suggests the presence of one methionine-plus-histidine-coordinated heme and two further heme groups with spectroscopic properties dominated by thiolate, and thus presumably cysteine, coordination (7a). Due to their functional importance, amino acids that act as heme ligands are almost invariably conserved between related hemoproteins. A multisequence comparison of R. sulfidophilum SoxA and SoxX sequences with their homologues is therefore useful in attempting to assign specific amino acid residues as sixth ligands to the three spectroscopically defined hemes. In making this sequence comparison, it is necessary to take into account the fact that the Chlorobium and R. palustris SoxA sequences lack the more amino-terminal heme binding motif found in the R. sulfidophilum protein and may thus retain the ligands for only one of the two hemes present in the R. sulfidophilum protein (Fig. 4). Only SoxX subunits contain a conserved methionine residue (Met92 in the mature R. sulfidophilum protein), suggesting that SoxX contains the His/Met-coordinated heme (Fig. 4 and 5). This inference is supported by the observation that, while the SoxX proteins do not show significant overall sequence similarity to other proteins, they do contain the key highly conserved amino acids of class I c-type cytochromes (33) and are likely to have a similar overall structure. In class I cytochromes the heme is attached close to the N terminus of the polypeptide and the heme iron is bound by the histidine from the heme attachment motif and a more carboxy-terminal methionine residue that corresponds to the conserved Met92 of R. sulfidophilum SoxX. In addition to the cysteines of the heme attachment motifs, SoxA, but not SoxX, contains two conserved cysteine residues, Cys114 and Cys222 (R. sulfidophilum mature SoxA protein numbering). On the basis of this sequence conservation, these cysteine residues are predicted to be the thiolate ligands of the remaining two heme groups. Remarkably, the monoheme Chlorobium and R. palustris SoxA sequences conserve both Cys114 and Cys222 equivalents. The Cys114 equivalent in Chlorobium limicola cytochrome c551 has been shown to form a disulfide bond with a cysteine located at the same position as the amino-terminal heme attachment motif in R. sulfidophilum SoxA (23). Given that to form a disulfide bond, both cysteine residues in the C. limicola protein must be in close proximity, we infer that Cys114 is likely to provide a ligand to the heme attached to the amino-terminal heme binding motif in the homologous R. sulfidophilum SoxA structure. It is possible that the disulfide bridge in Chlorobium cytochrome c551 and the amino-terminal heme in R. sulfidophilum SoxA have similar functions since both moieties are redox active and would cross-link the protein structure. If Cys114 coordinates the amino-terminal heme group, then Cys222 is predicted to act as a ligand to the more carboxy-terminal heme.
Sequence analysis of other R. sulfidophilum soxA locus gene products.
Friedrich and coworkers have previously presented analyses of sequence features in the sox gene products of P. pantotrophus (13, 53, 54). Here we present additional insights into Sox protein structure derived from multiple sequence alignments of the R. sulfidophilus proteins with those of P. pantotrophus and the other bacteria analyzed in Fig. 3. In addition we identify significant structural differences between the R. sulfidophilus and P. pantotrophus SoxD proteins.
The SoxY and SoxZ proteins of P. pantotrophus form a complex corresponding to the thiosulfate-binding enzyme A of the Paracoccus versutus (formerly Thiobacillus versutus, formerly Thiobacillus A2) thiosulfate-oxidizing system (13, 27, 30). Multiple sequence analysis reveals a remarkable degree of sequence conservation in the decapeptide Val-Lys-Val-Thr-Ile-Gly-Gly-Cys-Gly-Gly at the carboxyl terminus of SoxY. This peptide is invariant between the SoxY proteins of R. sulfidophilum, P. pantotrophus, R. palustris, and C. tepidum, and there are only two changes (Val for Ile and loss of the carboxyl-terminal Gly) in the A. aeolicus protein. This is by far the highest level of sequence conservation between the sox gene products of these phylogenetically widespread organisms and suggests a critical role for the decapeptide in thiosulfate oxidation. P. pantotrophus SoxY is covalently modified by a species of approximately 117 Da in mass (13). Given the earlier demonstration of thiosulfate binding by P. versutus enzyme A (30), we tentatively infer this modifying group to be thiosulfate (112 Da). The conserved cysteine residue in the SoxY carboxyl-terminal decapeptide is an obvious candidate to form covalent adducts with thiosulfate-derived species since no other conserved cysteine residues are found in either SoxY or SoxZ. The carboxyl-terminal context and adjacent glycine residues potentially render this conserved cysteine highly accessible and mobile. An alternative substrate binding site is suggested by the observation that members of the ubiquitin superfamily also contain a carboxyl-terminal Gly-Gly sequence that they use to form thioether adducts (44, 51). Genome comparisons suggest an intimate functional connection between cytochrome c551 and the SoxYZ complex since the soxY and soxZ genes are interpolated between the two cytochrome c551 structural genes not only in R. sulfidophilum but also in P. pantotrophus and C. tepidum (Fig. 3). Plausibly the substrate for cytochrome c551 is a thiosulfate-derived species covalently bound to the SoxYZ complex.
The SoxB protein is thought to have a hydrolytic function. The N-terminal domain of the SoxB protein has sequence similarity to 5′-nucleotidases and related enzymes (24) and is the probable location of a dinuclear Mn2+ site (6). Intriguingly, while the SoxB proteins have Tat signal peptides, the periplasmically located members of the homologous 5′-nucleotidase family are synthesized as precursors with Sec signal peptides.
The products of the overlapping soxC and soxD genes are homologous to the molybdopterin and diheme cytochrome c subunits, respectively, of the sulfite:cytochrome c oxidoreductase (sulfite dehydrogenase) of P. pantotrophus (42, 54). However, the 191-residue mature R. sulfidophilum SoxD protein lacks the final 169 amino acids of the corresponding P. pantotrophus polypeptide. Since this region contains the second consensus heme attachment motif, the R. sulfidophilum SoxD is a monoheme (class I) c-type cytochrome. It is possible that this deletion relative to the P. pantotrophus protein renders the R. sulfidophilum sulfite:cytochrome c oxidoreductase defective since R. sulfidophilum, uniquely among lithotrophic bacteria, liberates substantial quantities of sulfite as a free intermediate in the oxidation of thiosulfate to sulfate (36). In addition, and in agreement with an earlier report (36), we have been unable to detect substantive enzymatic sulfite:cytochrome c oxidoreductase activity in periplasmic extracts of R. sulfidophilum. Intriguingly, R. sulfidophilum, R. palustris, and P. pantotrophus SoxD proteins conserve a potentially redox-active Cys-(Xaa)3-Cys motif.
SoxF is homologous to the catalytic flavin subunit of Allochromatium vinosum flavocytochrome c (7), an enzyme that is generally held to have the physiological function of oxidizing sulfide to elemental sulfur or polysulfides (4, 43). SoxE contains two c-type heme attachment motifs. The amino-terminal half of the protein bearing the first of these motifs shows high sequence similarity to the c2 family of class I c-type cytochromes and to the carboxy-terminal domain of the P. pantotrophus SoxD protein. SoxE may be the redox partner of the SoxF flavoprotein, since genes coding for SoxE-like proteins also lie adjacent to soxF genes in P. pantotrophus, R. palustris, and C. tepidum (Fig. 3) and since experiments reported below suggest that soxE and soxF form a separate transcription unit in R. sulfidophilum.
SoxV is homologous to proteins of the CcdA protein family. These are integral membrane proteins containing six transmembrane helices (10). Proteins of the CcdA and related DsbD families function in disulfide bond isomerization and heme attachment to c-type cytochromes. These proteins catalyze movement of electrons from cytoplasmic thioredoxins to periplasmic thioredoxin-like proteins using a pair of cysteine residues that are also conserved in the SoxV protein (8, 10, 14, 49). The periplasmic SoxW protein exhibits weak sequence similarity to thioredoxins, including the presence of an active-site Cys-Xaa-Xaa-Cys motif, and is therefore likely to be the periplasmic redox partner of SoxV. Involvement of the SoxVW system in thiosulfate oxidation in R. sulfidophilum is suggested by the observation that all currently characterized soxA loci contain a gene coding for a periplasmically located thioredoxin (Fig. 3).
Insertional mutagenesis of the R. sulfidophilum soxA gene.
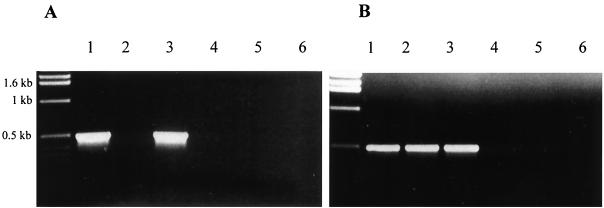
To test the involvement of R. sulfidophilum cytochrome c551 in photolithotrophic thiosulfate oxidation, the soxA gene was disrupted by interposon mutagenesis and the phenotype of the resultant mutant strains was assessed. Two types of mutants were constructed. In both cases, interposon insertion was combined with deletion of a major part of the soxA coding region. In strain soxA::Ω, the interposon contains transcriptional and translational terminators. Thus, in this strain not only is soxA inactivated but also any downstream genes that rely on transcriptional readthrough from soxA for expression. In the second mutant, soxA::Gmr, the promoter of the antibiotic resistance gene carried on the interposon is expected to drive transcription of the genes downstream from soxA. Thus, although the level of transcriptional readthrough from soxA is likely to differ from the parental strain, soxA::Gmr was anticipated to be a null mutant for soxA alone. The RT-PCR experiments shown in Fig. 6 were undertaken to test transcription of genes downstream of soxA in the mutant strains. Transcription of soxC is blocked in mutant soxA::Ω (Fig. 6A, lane 2). This shows that soxC is transcribed exclusively from promoters upstream of (or in) soxA. In contrast, soxC-containing mRNA is detected in mutant soxA::Gmr (Fig. 6A, lane 3). This confirms that the gentamicin resistance gene of the interposon drives transcription of the genes lying downstream of soxA. Intriguingly, soxF transcripts could be detected in both soxA mutant strains (Fig. 6B, lanes 2 and 3). Since the interposon insertion in strain soxA::Ω prevents soxC transcription, it follows that soxF can be transcribed from a promoter lying between soxC and soxF. This promoter is presumably located in the 430-bp soxD-soxE intergenic region. Given the implied function of soxEF in sulfide oxidation, this internal sox promoter could explain the reported differential expression of thiosulfate- and sulfide-oxidizing capabilities in R. sulfidophilum (36).
FIG. 6.
Assessment of transcriptional readthrough in soxA mutant strains using RT-PCR. RT-PCR experiments using primers in soxC (A) or soxF (B) employed total RNA isolated from strains cultured photomixotrophically with malate as carbon source and thiosulfate as electron donor. Lanes 1 to 3 in each panel are the complete RT-PCRs, while lanes 4 to 6 are control experiments in which reverse transcriptase was omitted. The sources of the RNA were R. sulfidophilum strains 3.1 (lanes 1 and 4), soxA::Ω (lanes 2 and 5), and soxA::Gmr (lanes 3 and 6).
Periplasmic extracts were prepared from cultures of each of the two soxA mutant strains grown photomixotrophically on thiosulfate plus malate and fractionated by anion-exchange chromatography. The cytochrome c551-containing hemoprotein peak exhibited by parental strain 3.1 cultured under these conditions was absent from the elution profiles of the mutant periplasm separations. This analysis confirms that the soxA strains have a defect in the production of native cytochrome c551 complexes.
Neither soxA mutant was capable of photolithotrophic growth with thiosulfate as electron donor. However, both strains grew photolithotrophically when the electron donor was either formate or hydrogen, indicating that the soxA mutants were specifically defective in thiosulfate metabolism. Washed cells of photomixotrophic (thiosulfate-malate) cultures of parental strain 3.1 catalyzed the light- and carbon dioxide-dependent oxidation of thiosulfate at a mean rate of 6 nmol of S2O32− oxidized min−1 (g of cells [wet weight])−1. In contrast, photomixotrophically grown cultures of the two soxA mutants did not consume thiosulfate.
Periplasmic extracts prepared from the parental strain cultured in thiosulfate-containing media reduce equine heart ferricytochrome c with thiosulfate as electron donor (Table 1), while periplasmic extracts from heterotrophically grown cells exhibit substantially lower specific activity in the same assay (Table 1). Thiosulfate:cytochrome c oxidoreductase activity can also be measured using R. sulfidophilum ferricytochrome c2 as electron acceptor, but for experimental convenience the commercially available equine heart protein was used. Thiosulfate:cytochrome c oxidoreductase activity was zero in periplasmic extracts prepared from photomixotrophically (thiosulfate plus malate) grown cells of the two soxA mutant strains (Table 1). However, supplementation of the mutant periplasmic extracts with purified cytochrome c551 complex led to the reappearance of substantial thiosulfate:cytochrome c oxidoreductase activity (Table 1) even though the purified cytochrome itself does not exhibit such activity. For mutant soxA::Gmr maximal activity was obtained with 20 μg of cytochrome c551 per ml of periplasmic extract.
TABLE 1.
Thiosulfate:cytochrome c oxidoreductase activities in periplasmic extracts of R. sulfidophilum strains
| Strain | Culture conditions (electron donor, carbon source)a | Assay supplementb | Mean activity [nmol of cytochrome c reduced min−1 (ml of periplasmic extract)−1] |
|---|---|---|---|
| Parental 3.1 | S2O32−, HCO3− | 12 ± 2 | |
| S2O32−, malate | 27 ± 3 | ||
| None, malate | 3.5 ± 0.5 | ||
| soxA::Gmr | S2O32−, malate | NDc | |
| S2O32−, malate | SoxAX | 18 ± 3 | |
| soxA::Ω | S2O32−, malate | ND | |
| S2O32−, malate | SoxAX | 9 ± 1 |
Cells were harvested at an optical density at 650 nm in the range of 0.8 to 1.3.
Assays were supplemented with 20 μg of cytochrome c551 complex per ml where indicated.
ND, nondetectable.
Taken together, these various data indicate that SoxA is required for thiosulfate oxidation in R. sulfidophilum. In addition, although we have been unable to establish the precise reaction catalyzed by cytochrome c551, the biochemical reconstitution data demonstrate that our purified protein is enzymatically active.
R. sulfidophilum is capable of photolithotrophic growth with sulfide as electron donor (17). However, neither soxA mutant grew photoautotrophically in medium containing 3.5 mM Na2S. In mixotrophic sulfide-plus-malate culture the soxA mutant strains did not consume detectable sulfide over 24 h, while over the same time period parallel cultures of parental strain 3.1 completely oxidized 3.5 mM sulfide to sulfate. Washed cells of photomixotrophic (thiosulfate plus malate) cultures of parental strain 3.1 catalyzed the light- and carbon dioxide-dependent oxidation of Na2S at a mean rate of 0.8 nmol of H2S oxidized min−1 (g of cells [wet weight])−1. The soxA mutants did not oxidize sulfide in this assay. It was not possible to analyze sulfide:cytochrome c oxidoreductase activity in cell extracts because even in the parental strain we could not detect activity significantly above the background rate of chemical reduction of the cytochrome.
Sulfide is a toxic compound, and it was conceivable that the apparent defect in sulfide oxidation exhibited by the soxA mutant strains was due to a change in level of sulfide tolerance of the cells rather than a defect in sulfide metabolism. In an attempt to distinguish between these two possibilities, we tested autotrophic growth of R. sulfidophilum on agar plates in a sulfidic atmosphere using conditions that allow growth of the closely related but more sulfide-sensitive bacterium Rhodobacter capsulatus (46). After a 3-week incubation, single colonies of the parental strain 3.1 had formed but only trace growth was evident for either soxA mutant strain. However, if malate was included in the plates, strong growth of the soxA mutants was observed, suggesting that the mutant strains were not sensitive to the prevailing sulfide concentrations. R. capsulatus grown in the sulfidic atmosphere in the presence of malate deposits elemental sulfur around the colonies as the product of sulfide oxidation (46). This behavior was not observed for any of the three R. sulfidophilum strains. We conclude that SoxA is essential for photolithotrophic sulfide oxidation in R. sulfidophilum and that the pathways of thiosulfate and sulfide oxidation in this organism have common components.
DISCUSSION
A thiosulfate-induced hemoprotein, SoxAX, has been purified from the marine photosynthetic bacterium R. sulfidophilum. Analysis of a soxA-specific null mutant demonstrates that SoxAX is an obligate component of the photolithotrophic thiosulfate oxidation pathway. More unexpectedly, SoxAX was also shown to be essential for photosynthetic oxidation of sulfide. The biochemical, biophysical, and sequence data presented here, together with spectroscopic data (7a), allow assignment of cysteine residues as heme iron ligands in the SoxA protein, making SoxA the first c-type cytochrome for which cysteine heme ligation has been described. The R. sulfidophilum soxA gene is part of a large cluster of genes coding for proteins with homology to components of sulfur oxidation pathways in other thiosulfate-oxidizing organisms (Fig. 3). The bacteria possessing these gene clusters span a wide range of phylogenetic and physiological groupings, indicating that the mechanism of lithotrophic thiosulfate oxidation is conserved between at least some photosynthetic and facultatively chemolithotrophic bacteria. This conclusion is supported by a recent analysis of the phylogenetic distribution of soxB genes (39). The five bacterial species analyzed in Fig. 3 conserve a core set of Sox components, namely, SoxAX, SoxB, SoxYZ, a flavocytochrome c (not always at the soxA locus), and a periplasmic thioredoxin, suggesting that these are the minimal components of the pathway. Additional enzymatic activities coded at the soxA loci in some of the organisms, for example, sulfite:cytochrome c oxidoreductase, may not be required for sulfur metabolism in all these bacteria.
With sequence data suggesting an identical mechanism of thiosulfate oxidation in the bacterial species detailed in Fig. 3, it is of some interest to reexamine the available biochemical data on these processes in each organism to see how each might provide insight into the operation of their common sulfur oxidation pathway. Characterization of thiosulfate oxidation in the two Paracoccus species, P. versutus and P. pantotrophus, has led to a model in which a periplasmic thiosulfate-oxidizing multienzyme system (TOMES) fully oxidizes thiosulfate to sulfate and feeds electrons into the respiratory chain at the level of cytochrome c (reviewed in reference 21). The components of the TOMES system are a thiosulfate-binding enzyme A (SoxYZ), enzyme B (SoxB), and cytochrome c552.5 (29), which spectroscopic data identify as analogous to the cytochrome c551 (SoxAX) complex of R. sulfidophilum (7a, 21). A sulfite dehydrogenase (SoxCD) has been reported either to be an additional essential component of this process (54) or to facilitate the reaction (13, 28). Since there are no readily detectable free intermediates in the TOMES system, it has been proposed that the sulfur species remain protein bound during the oxidation process. This could explain our failure to detect sulfur-oxidizing activities with R. sulfidophilum cytochrome c551 unless other periplasmic proteins are also present (Table 1). However, the observation that R. sulfidophilum produces sulfite as a free intermediate in the oxidation of either thiosulfate or sulfide (36) supports the idea that sulfite at least is produced during operation of the Sox pathway. Since only some of the sox gene products are required for complete thiosulfate oxidation in the TOMES system, this model would imply that the additional conserved sox genes are involved specifically either in sulfide oxidation (perhaps SoxEF) or in the biosynthesis or maintenance of the TOMES system (perhaps SoxVW).
The cytochrome c551 component of the Sox pathway is required for the oxidation of sulfide as well as thiosulfate in R. sulfidophilum. A crucial question is then whether sulfide is the product of thiosulfate metabolism or vice versa, or whether the metabolic pathways for the two compounds are initially distinct and later converge. The observation that the ability to oxidize sulfide but not thiosulfate is constitutive in R. sulfidophilum (36) argues against thiosulfate being formed from sulfide oxidation. In addition, experiments in which C. vibrioforme f. thiosulfatophilum was fed thiosulfate differentially labeled at the sulfane and sulfone sulfur positions have been interpreted in terms of an initial reductive cleavage of thiosulfate to sulfide plus sulfite (22). At possible variance with these conclusions, both C. vibrioforme f. thiosulfatophilum and C. limicola f. thiosulfatophilum have also been observed to produce thiosulfate during oxidation of sulfide (4). However, there may be more than one mechanism for sulfide oxidation in the green sulfur bacteria, and caution is therefore required in the interpretation of these experiments. Since our soxA mutagenesis data suggest that there is only one pathway of sulfide oxidation in R. sulfidophilum, this bacterium may be a tractable system in which to study the photosynthetic oxidation of sulfide by the Sox pathway.
It is noteworthy that the soxA::Gmr mutant is unable to grow on or oxidize sulfide even though the soxEF genes, encoding a probable sulfide dehydrogenase, are still transcribed in the mutant (Fig. 6). Similarly, while the purified SoxAX complex does not initiate thiosulfate oxidation, the soxA::Gmr mutant is incapable of utilizing thiosulfate. It is also pertinent to note that a P. pantotrophus ΔsoxC mutant is unable to grow on thiosulfate (54). These observations suggest strong cooperativity between the individual steps of the Sox pathway. This behavior would be expected of a system in which the pathway intermediates remain protein bound.
ACKNOWLEDGMENTS
This work was supported by U.K. Biotechnology and Biological Research Council (BBSRC) grant 83/P09311 to B.C.B. and by the BBSRC and the Engineering and Physical Sciences Research Council through core funding to the Center for Metalloprotein Spectroscopy and Biology. P.J.L. was the recipient of a BBSRC studentship. B.C.B. is R. J. P. Williams Senior Research Fellow at Wadham College, Oxford.
We thank W. Klipp for supplying plasmid pWKR189I and A. Willis for performing the peptide sequencing. We acknowledge S. G. Haigh and P. Barrell for their input into the preliminary stages of this project and J. Mayne, J. Thornton, and D. Clarke for assistance in protein purification and cell culture.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 3.Berks B C, Sargent F, Palmer T. The Tat protein export pathway. Mol Microbiol. 2000;35:260–274. doi: 10.1046/j.1365-2958.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 4.Brune D C. Sulfur oxidation by phototrophic bacteria. Biochim Biophys Acta. 1989;975:189–221. doi: 10.1016/s0005-2728(89)80251-8. [DOI] [PubMed] [Google Scholar]
- 5.Brune D C. Sulfur compounds as photosynthetic electron donors. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Amsterdam, The Netherlands: Kluwer; 1995. pp. 847–870. [Google Scholar]
- 6.Cammack R, Chapman A, Lu W-P, Karagouni A, Kelly D P. Evidence that protein B of the thiosulfate-oxidizing system of Thiobacillus versutus contains a binuclear manganese cluster. FEBS Lett. 1989;253:239–243. [Google Scholar]
- 7.Chen Z -W, Koh M, van Driessche G, van Beeumen J J, Bartsch R G, Meyer T E, Cusanovich M A, Mathews F S. The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium. Science. 1994;266:430–432. doi: 10.1126/science.7939681. [DOI] [PubMed] [Google Scholar]
- 7a.Cheesman, M. R., P. J. Little, and B. C. Berks. Heme ligation in a c-type cytochrome involved in thiosulfate oxidation: EPR and MCD of SoxAX from Rhodovulum sulfidophilum. Biochemistry, in press. [DOI] [PubMed]
- 8.Chung J, Chen T, Missiakas D. Transfer of electrons across the cytoplasmic membrane by DsbD, a membrane protein involved in thiol-disulphide exchange and protein folding in the bacterial periplasm. Mol Microbiol. 2000;35:1099–1109. doi: 10.1046/j.1365-2958.2000.01778.x. [DOI] [PubMed] [Google Scholar]
- 9.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olson G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 10.Deshmukh M, Brasseur G, Daldal F. Novel Rhodobacter capsulatus genes required for the biogenesis of various c-type cytochromes. Mol Microbiol. 2000;35:123–138. doi: 10.1046/j.1365-2958.2000.01683.x. [DOI] [PubMed] [Google Scholar]
- 11.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich C G. Physiology and genetics of sulfur-oxidizing bacteria. Adv Microbiol Physiol. 1998;39:235–289. doi: 10.1016/s0065-2911(08)60018-1. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich C G, Quentmeier A, Bardischewsky F, Rother D, Kraft R, Kostka S, Prinz H. Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J Bacteriol. 2000;182:4677–4687. doi: 10.1128/jb.182.17.4677-4687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon E H J, Page M D, Willis A C, Ferguson S J. Escherchia coli DipZ: anatomy of a transmembrane protein disulphide reductase in which three pairs of cysteine residues, one in each of three domains, contribute differentially to function. Mol Microbiol. 2000;35:1360–1374. doi: 10.1046/j.1365-2958.2000.01796.x. [DOI] [PubMed] [Google Scholar]
- 15.Hallenbeck P C, Clark M A, Barrett E L. Characterization of anaerobic reduction by Salmonella typhimurium and purification of the anaerobically induced sulfite reductase. J Bacteriol. 1989;171:3008–3015. doi: 10.1128/jb.171.6.3008-3015.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanlon S P, Holt R A, Moore G R, McEwan A G. Isolation and characterization of a strain of Rhodobacter sulfidophilus: a bacterium which grows autotrophically with dimethylsulphide as electron donor. Microbiology. 1994;140:1953–1958. [Google Scholar]
- 17.Hansen T A, Veldkamp H. Rhodopseudomonas sulfidophila, nov. spec., a new species of the purple nonsulfur bacteria. Arch Mikrobiol. 1973;92:45–58. doi: 10.1007/BF00409510. [DOI] [PubMed] [Google Scholar]
- 18.Hiraishi A, Ueda Y. Intrageneric structure of the genus Rhodobacter: transfer of Rhodobacter sulfidophilus and related marine species to the genus Rhodovulum gen. nov. Int J Syst Bacteriol. 1994;44:15–23. [Google Scholar]
- 19.Irgens R L. Thioacetamide as a source of hydrogen sulfide for colony growth of purple sulfur bacteria. Curr Microbiol. 1983;8:183–186. [Google Scholar]
- 20.Kelly D P, Wood A. Synthesis and determination of thiosulfate and polythionates. Methods Enzymol. 1994;243:475–501. [Google Scholar]
- 21.Kelly D P, Shergill J K, Lu W-P, Wood A P. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Leeuwenhoek. 1997;71:95–107. doi: 10.1023/a:1000135707181. [DOI] [PubMed] [Google Scholar]
- 22.Khanna S, Nicholas D J D. Utilization of tetrathionate and 35S-labelled thiosulfate by washed cells of Chlorobium vibrioforme f. sp. thiosulfatophilum. J Gen Microbiol. 1982;128:1027–1034. [Google Scholar]
- 23.Klarskov K, Verté F, van Driessche G, Meyer T E, Cusanovich M A, van Beeumen J. The primary structure of soluble cytochrome c-551 from the phototrophic green sulfur bacterium Chlorobium limicola, strain Tassajara, reveals a novel c-type cytochrome. Biochemistry. 1998;37:10555–10562. doi: 10.1021/bi9806706. [DOI] [PubMed] [Google Scholar]
- 24.Knöfel T, Sträter N. X-ray structure of the Escherichia coli periplasmic 5′-nucleotidase containing a dimetal catalytic site. Nat Struct Biol. 1999;6:448–453. doi: 10.1038/8253. [DOI] [PubMed] [Google Scholar]
- 25.Kusai K, Yamanaka T. The oxidation mechanism of thiosulfate and sulfide in Chlorobium thiosulfatophilum: roles of cytochrome c-551 and cytochrome c-553. Biochim Biophys Acta. 1973;325:304–314. doi: 10.1016/0005-2728(73)90106-0. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:280–285. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Lu W-P, Kelly D P. Purification and some properties of two principal enzymes of the thiosulfate-oxidizing multi-enzyme system from Thiobacillus A2. J Gen Microbiol. 1983;129:3549–3564. [Google Scholar]
- 28.Lu W-P, Kelly D P. Properties and role of sulphite cytochrome c oxido-reductase purified from Thiobacillus versutus (A2) J Gen Microbiol. 1984;130:1683–1692. [Google Scholar]
- 29.Lu W-P, Kelly D P. Purification and characterization of two essential cytochromes of the thiosulfate-oxidizing multi-enzyme system from Thiobacillus A2 (Thiobacillus versutus) Biochim Biophys Acta. 1984;765:106–117. [Google Scholar]
- 30.Lu W-P, Swoboda B E P, Kelly D P. Properties of the thiosulfate-oxidizing multi-enzyme system from Thiobacillus versutus. Biochim Biophys Acta. 1985;828:116–122. [Google Scholar]
- 31.Masuda S, Matsumoto Y, Nagashima K V P, Shimada K, Inoue K, Bauer C E, Matsuura K. Structural and functional analyses of photosynthetic regulatory genes regA and regB from Rhodovulum sulfidophilum, Roseobacter denitrificans, and Rhodobacter capsulatus. J Bacteriol. 1999;181:4205–4215. doi: 10.1128/jb.181.14.4205-4215.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 33.Moore G R, Pettigrew G W. Cytochromes c: evolutionary, structural and physicochemical aspects. Berlin, Germany: Springer-Verlag; 1990. [Google Scholar]
- 34.Moreno-Vivien C, Schmehl M, Masepohl B, Arnold W, Klipp W. DNA sequence and genetic analysis of the Rhodobacter capsulatus nifENX region: homology between NifX and NifB suggests involvement of NifX in processing of the iron-molybdenum cofactor. Mol Gen Genet. 1989;216:353–363. doi: 10.1007/BF00334376. [DOI] [PubMed] [Google Scholar]
- 35.Mukhopadhyaya P N, Deb C, Lahiri C, Roy P. A soxA gene, encoding a diheme cytochrome c, and a sox locus, essential for sulfur oxidation in a new sulfur lithotrophic bacterium. J Bacteriol. 2000;182:4278–4287. doi: 10.1128/jb.182.15.4278-4287.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neutzling O, Pfleiderer C, Trüper H G. Dissimilatory sulphur metabolism in phototrophic ‘non-sulphur’ bacteria. J Gen Microbiol. 1985;131:791–798. [Google Scholar]
- 37.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 38.Parke D. Construction of mobilizable vectors derived from plasmids RP4, pUC18, and pUC19. Gene. 1990;93:135–137. doi: 10.1016/0378-1119(90)90147-j. [DOI] [PubMed] [Google Scholar]
- 39.Petri R, Podgorsek L, Imhoff J F. Phylogeny and distribution of the soxB gene among thiosulfate-oxidizing bacteria. FEMS Microbiol Lett. 2001;197:171–178. doi: 10.1111/j.1574-6968.2001.tb10600.x. [DOI] [PubMed] [Google Scholar]
- 40.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 41.Pugsley A P. The complete general-secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quentmeier A, Kraft R, Kostka S, Klockenkämper R, Friedrich C G. Characterization of a new type of sulfite dehydrogenase from Paracoccus pantotrophus GB17. Arch Microbiol. 2000;173:117–125. doi: 10.1007/s002039900118. [DOI] [PubMed] [Google Scholar]
- 43.Reinartz M, Tschäpe J, Brüser T, Trüper H G, Dahl C. Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch Microbiol. 1998;170:59–68. doi: 10.1007/s002030050615. [DOI] [PubMed] [Google Scholar]
- 44.Rudolph M J, Wuebbens M M, Rajagopalan K V, Schindelin H. Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation. Nat Struct Biol. 2001;8:42–46. doi: 10.1038/83034. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.Schütz M, Maldener I, Griesbeck C, Hauska G. Sulfide-quinone reductase from Rhodobacter capsulatus: requirement for growth, periplasmic localization, and extension of gene sequence analysis. J Bacteriol. 1999;181:6516–6523. doi: 10.1128/jb.181.20.6516-6523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon R U, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 48.Sonnhammer E L L, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. In: Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editors. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. Menlo Park, Calif: AAAI Press; 1998. pp. 175–182. [PubMed] [Google Scholar]
- 49.Stewart E J, Katzen F, Beckwith J J. Six conserved cysteines of the membrane protein DsbD are required for the transfer of electrons from the cytoplasm to the periplasm of Escherichia coli. EMBO J. 1999;18:5963–5971. doi: 10.1093/emboj/18.21.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas P E, Ryan D, Levin W. An improved staining procedure for the detection of peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 51.Wang C, Xi J, Begley T P, Nicholson L K. Solution structure of ThiS and implications for the evolutionary roots of ubiquitin. Nat Struct Biol. 2001;8:47–51. doi: 10.1038/83041. [DOI] [PubMed] [Google Scholar]
- 52.Weaver P T, Wall J D, Gest H. Characterization of Rhodopseudomonas capsulata. Arch Microbiol. 1975;105:207–216. doi: 10.1007/BF00447139. [DOI] [PubMed] [Google Scholar]
- 53.Wodara C, Kostka S, Egert M, Kelly D P, Friedrich C G. Identification and sequence analysis of the soxB gene essential for sulfur oxidation of Paracoccus denitrificans GB17. J Bacteriol. 1994;176:6188–6191. doi: 10.1128/jb.176.20.6188-6191.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wodara C, Bardischewsky F, Friedrich C G. Cloning and characterization of sulfite dehydrogenase, two c-type cytochromes, and a flavoprotein of Paracoccus denitrificans GB17: essential role of sulfite dehydrogenase in lithotrophic sulfur oxidation. J Bacteriol. 1997;179:5014–5023. doi: 10.1128/jb.179.16.5014-5023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]