Abstract
Colonies formed by bacteria, archaea, fungi, and viral groups and their genomes, metabolites, and expressed proteins constitute complex human microbiomes. An increasing evidences showed that carcinogenesis and disease progression were link to microbiomes. Different organ sources, their microbial species, and their metabolites are different; the mechanisms of carcinogenic or procancerous are also different. Here, we summarize how microbiomes contribute to carcinogenesis and disease progression in cancers of the skin, mouth, esophagus, lung, gastrointestinal, genital, blood, and lymph malignancy. We also insight into the molecular mechanisms of triggering, promoting, or inhibiting carcinogenesis and disease progress induced by microbiomes or/and their secretions of bioactive metabolites. And then, the strategies of application of microorganisms in cancer treatment were discussed in detail. However, the mechanisms by which human microbiomes function are still poorly understood. The bidirectional interactions between microbiotas and endocrine systems need to be clarified. Probiotics and prebiotics are believed to benefit human health via a variety of mechanisms, in particular, in tumor inhibition. It is largely unknown how microbial agents cause cancer or how cancer progresses. We expect this review may open new perspectives on possible therapeutic approaches of patients with cancer.
Keywords: cancer therapy, carcinogenesis, microbiome, microorganism, tumor
Several microbes, such as Fn, Helicobacter pylori, Fusobacterium, Staphylococcus aureus, Peptostreptococcus anaerobius, and Porphyromonas gingivalis, promote cancer progression. Other microbes, such as Lactobacillus, Bifidobacterium, Clostridium butyricum, Bacillus subtilis, Bifidobacteria, Lactobacoilli, Streptococcus thermophilus, Leptotrichia, Bacteroides thetaiotaomicron, and Faecalibacterium prausnitzii, can inhibit cancer progression. Possible therapeutic approaches for cancer include fecal microbiome transplantation (FMT), probiotics, synbiotics, antibiotics, oncolytic virus, and nanospheres.
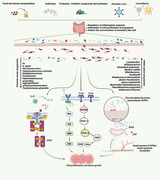
1. INTRODUCTION
Approximately 1 × 1012 types of microbiomes are known to exist on Earth. Numerous microorganisms live in the gut, the skin, the lungs, and the oral cavity. Additionally, the human microbiome, often called the “hidden organ,” contributes more genetic information than the whole human genome combined. 1 , 2 , 3 Advances in recent years have uncovered a close relationship between the human microbiome and its ability to extract nutrients, process carbohydrates, and create antibodies. There are many possible mechanisms for the influence of the microbiomes on biological processes. Microbiomes play an important role in converting food into energy, nutrition, and bioactive molecules, such as vitamins, lipids, and amino acids. Furthermore, the human microbiomes contribute to mucosal and immune system development, as well as to protection from external pathogens. 4 , 5 , 6 However, the system consists of trillions of microbiomes.
Using bioinformatics and advanced sequencing technologies, scientists are discovering the relationship between the microbiome and disease. Changing environmental conditions can disrupt the balance of microbiomes, resulting in dysregulation of bodily functions and disease. 7 , 8 , 9
There are more cancer deaths worldwide than any other disease. 10 , 11 , 12 The risk factors of cancer are very complex, and environmental factors account for a high proportion of common risk factors. The symbiotic microbiome is a major regulator of carcinogenesis. 10 , 13 A comparison of cancer growth in germ‐free mouse models or models where antibiotics are used to deplete the gut microbiome with mouse models maintained on normal feed suggests that there is a strong correlation between cancer growth and the microbiome. 14 , 15 Overall, it has been demonstrated that dysbiosis of the microbiome can initiate and progress cancer .
In the 1970s, a pathologist from the Royal Perth Hospital in Australia (J. Robin Warren) found that Helicobacter pylori (H. pylori) bacteria were frequently observed in the gastric tissue of chronic gastritis patients. Warren and Barry J. Marshall speculated that H. pylori may therefore cause gastritis. 16 To confirm the relationship between this bacteria and chronic gastritis, Marshall underwent an endoscopy to confirm that he was H. pylori negative and then infected himself with the bacteria. He then developed gastritis and recorded the development of his disease. 17 H. pylori is considered a common gut microorganism that causes chronic gastritis, gastric ulcers, and gastric atrophy. Additionally, it contributes to gastric cancer development. Approximately half of the population worldwide are infected with H. pylori. 18 , 19 , 20
In the early 2000s, microbiomes and cancer have been studied systematically by researchers. A total of seven viruses, two parasites, and one bacteria are human carcinogens, according to the International Agency for Research on Cancer. These include Epstein–Barr virus (EBV), hepatitis B virus, hepatitis C virus, Kaposi sarcoma herpesvirus, human immunodeficiency virus‐1, human papillomaviruses (HPV), human T‐cell lymphotropic virus Type 1, Opisthorchis viverrini and Clonorchis sinensis, Schistosoma haematobium, and H. pylori 21 , 22 (Table 1). Other fungi also are human carcinogens, such as Aspergillus flavus. An increasing number of microbiomes have been identified as promoters but not inducers of cancer. The gut microbiomes is a collection of microorganisms in the human gastrointestinal tract, which makes up 70% of all microbiomes in the human system. Bacteria are also found in the skin, oral and nasal cavity, lungs, esophagus, and vaginal area. However, the number of bacteria in the extraintestinal organs constitutes 3% or less of all bodily microbiomes. Gut microbiomes secretions pass into the local tissues and circulatory system, interacting with cancer cells. 21 , 23 Other organs such as the lungs, 24 breasts, 24 , 25 skin, 25 and urogenital system 26 are also host to microorganisms, indicating that the organ‐specific microbiome may play a role in carcinogenesis through alteration of the local microenvironment via microbiomes and their metabolites. Importantly, secreted microbial products from microbiomes in the gastrointestinal tract, lungs, breasts, and female reproductive tract can enter the circulation, activate immune signals, and participate in the immune response and immune surveillance. 26 , 27 , 28
TABLE 1.
Human carcinogenic or cancer‐promoting microbiomes in cancer.
| Microbes | Cancer |
|---|---|
| Carcinogenic microbes | |
| Viruses and parasites | |
| Epstein‐Barr virus | Burkitt lymphoma, 41 virus‐associated T‐cell and NK‐cell lymphomas, 42 cancer of the nasopharynx, 43 lymphoepithelioma‐like carcinomas, 44 cancer of the stomach 45 |
| Hepatitis B virus | Hepatocellular carcinoma, 46 cholangiocarcinoma, 47 non‐Hodgkin lymphoma, 48 , 49 cancer of the pancreas, 49 Hodgkin disease 50 |
| Hepatitis C virus | Hepatocellular carcinoma, 51 gallbladder cancer, 52 lymphoid malignancies, 53 leukemias, 53 cancer of the thyroid 54 |
| Kaposi sarcoma herpesvirus | Kaposi sarcoma, 55 primary effusion lymphoma, 56 multiple myeloma 56 |
| Human immunodeficiency virus‐1 | Kaposi sarcoma, 57 non‐Hodgkin lymphoma, 58 Hodgkin lymphoma, 58 cervical and anogenital cancers, 59 the skin cancer, 60 the conjunctiva cancer, 61 the lung cancer, 62 the liver cancer, 63 the lip cancer, 64 the head and neck cancer 65 |
| Human papillomaviruses | The cervix cancer, 66 the vulva cancer, 67 the vagina cancer, 67 the penis cancer, 68 the anus cancer, 68 the oral cavity cancer, 69 the oropharynx and tonsil cancer, 70 the esophagus cancer, 68 the larynx cancer, 68 the skin cancer, 71 the nose and nasal sinuses cancer, 68 the lung cancer, 72 the colon and rectum cancer, 68 the breast cancer, 73 the ovary cancer, 74 the prostate cancer, 75 the urinary bladder and urethra cancer 76 |
| Human T‐cell lymphotropic virus Type 1 | T‐cell malignancies, 77 cutaneous T‐cell lymphoma, 77 B‐ and T‐cell lymphomas, 78 nonlymphomatous tumors 79 |
| Opisthorchis viverrini and Clonorchis sinensis | Cholangiocarcinoma, 80 hepatocellular carcinoma 80 |
| Schistosoma haematobium | The urinary bladder cancer, 81 , 82 the female genital tract cancer, 82 the prostate cancer 83 |
| Bacteria | |
| Helicobacter pylori | The stomach cancer, 84 Gastric mucosa‐associated lymphoid tissue (MALT) lymphoma, 85 the esophagus cancer, 86 the liver cancer, 87 the colorectum cancer, 88 the pancreas cancer, 89 the lung cancer, 90 the head and neck cancer, 91 Childhood leukemia 92 |
| Cancer‐promoting microbes | Cancer |
|---|---|
| Bacteria | |
|
Enterotoxigenic Bacteroides fragili Peptostreptococcus anaerobius Fusobacteriumnuleatum |
The colorectum cancer 93 , 94 |
|
Hemophilus, Porphyromonas gingivalis, Prevotella |
The esophagus cancer 95 |
|
Micromonospra Pporphyromonas gingivalis Neisseria, Fusobacteria Escherichia Coli, Streptococcus faecalis Helicobacter hepaticus |
The liver cancer 96 |
| Fermentation bacillus, lactobacillus | The esophagus cancer 97 |
| Streptococcus pneumoniae | The esophagus cancer 97 |
| Staphylococcus aureus | The skin cancer 98 |
| Tuberculous bacillus (TB) | The lung cancer 99 |
| Actinomyces, Falclella, Campylobacter | The urinary bladder cancer 100 |
| Fungi | |
| Aspergillus, Malassezia, Malasus, Pseudospecies, Asppersporus, Machyus | The colorectum cancer 101 |
| Microbe‐induced metabolite | |
| Deoxycholic acid | The colorectum cancer 102 |
Fungi have been used to treat human diseases since ancient times. Many biologically active compounds extracted from fungi are currently the foci of extensive research. Cohort studies conducted independently in China and Europe found that the amount of malasella increased in colorectal cancer (CRC) patients, while the amount of yeast decreased. 29 The distribution of Aspergillus, Malassezia, Malasus, Pseudospecies, Asppersporus, and Machyus was altered in CRC patients.
Overall, a number of mechanisms remain to be explored to understand how microbiomes influence the development of cancer, including:
Influence on host cell proliferation and death: Some microbiomes bind to E‐cadherins, which trigger β‐Catenin activation through polarity changes of barrier disruption. Other microbiomes can activate the β‐Catenin signaling pathway through cytotoxin‐associated gene A (CagA) or avirulence gene (AvrA), leading to cell proliferation dysregulation, stem cell‐like growth capability, and loss of polarity.
Alteration of immune system activity: Microbiomes and microbial metabolism can cause cancer by driving inflammatory responses and promoting cancer immune evasion by binding to human natural killers (NKs) and T cells via the inhibitory receptor TIGIT. 30 , 31 , 32 There is evidence that some invasive Campylobacter species induce an IL‐18‐driven proinflammatory response that leads to tumor progression. 33 , 34
Mechanistic effects on host metabolism: Microbiomes produce short‐chain fatty acids (SCFAs), mainly butyrate, acetate, and propionate, through fiber fermentation. 35 , 36 , 37 This indirectly promotes carcinogenesis through host metabolism and direct destruction of host DNA or the utilization of host superoxide to degrade DNA. Apart from SCFAs, other bacteria metabolites also were involved in carcinogenesis. 38 A high‐fat diet increases secondary bile acids such as deoxycholic acid (DCA) and lithocholic acid (LCA), which can cause colonic inflammation and cancer 39 , 40 (Figure 1).
FIGURE 1.
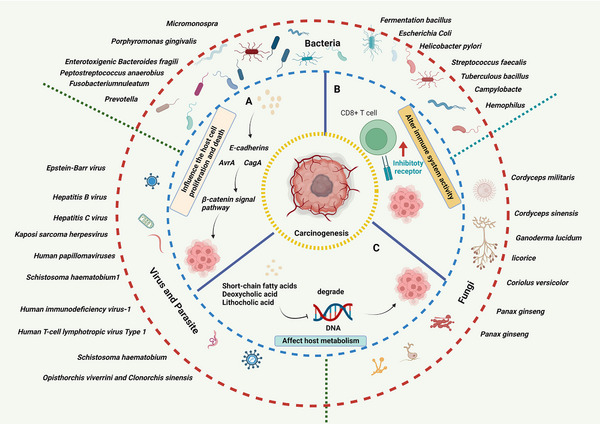
Human carcinogenic microbiomes and cancer. (A) Influence host cell proliferation and death. Microbiomes bind to E‐cadherins, which trigger β‐Catenin activation through polarity changes of barrier disruption or activate the β‐Catenin signaling pathway through CagA or AvrA, leading to dysregulation of cell proliferation. (B) Altering immune system activity. Microbiomes and microbial metabolism bind to inhibitory receptor TIGIT on human immune cells, drive inflammatory responses, and promote cancer immune evasion. (C) Effects on host metabolism. SCFAs, DCA, and LCA, which produced by microbiomes, promote carcinogenesis through host metabolism.
2. HUMAN CARCINOGENIC OR CANCER‐PROMOTING MICROBIOMES
According to the different pathogenic of human microbiomes, we divided the tumor‐causing microorganisms into carcinogenic microorganisms and cancer‐promoting microorganisms. Oncogenic microorganisms can directly induce somatic mutations, promote tumor cell proliferation and tumorigenesis. Cancer‐promoting microorganisms cannot cause tumorigenesis by themselves, but they can secrete cytokines to affect the tumor microenvironment and promote tumorigenesis. In this section, we review the oncogenic microorganisms and cancer‐promoting microorganisms in human physiological systems.
2.1. Gut microbiomes and CRC
Increasing evidence has revealed that gastrointestinal microbiome play a critical role in colon cancer carcinogenesis and progression. Kado et al. reported that adenocarcinomas of the ileocecum and cecum were detected in 70% of TCR‐β (−/−)/p53(−/−) mice in the conventionally fed group but not in the germ‐free group, suggesting that intestinal microbiomes play a major role in the development of adenocarcinoma of the colon in this animal model. 103
Further, an Massachusetts Institute of Technology (MIT) team found that mice infected with citrobacter murine were also more likely to develop colon cancer. 104 In 2003, Susan Erdman's team at MIT showed that H. pylori caused colon cancer in immune‐compromised mice. 105 In 2006, Erdman's team infected a particular mouse model with H. hepaticus and continued to observe subsequent changes in the mice. Unexpectedly, many of the mice developed breast cancer, showing that mice infected with H. hepaticus are likely to develop breast cancer. 106 In 2013, Schloss’ team compared tumor‐bearing mice treated with antibiotics prior to the induction of cancer or proinflammatory treatments to tumor‐bearing mice that received only cancer and inflammatory treatments. They found that the size and number of tumors in the antibiotic‐treated mice were significantly smaller than in the control group. In addition, when the researchers transferred microbes from the healthy mice to the antibiotic‐treated or germ‐free mice, the latter's exposure to carcinogens led to more malignant tumor tissue size. 107 In 2014, Patrick Schloss’ team from the University of Michigan sequenced the 16S ribosomal RNA(16S rRNA) gene from 90 samples of human feces. The samples were provided by patients with colon cancer, patients with precancerous adenoma, and healthy individuals. 108 The results showed that fecal samples from cancer patients were significantly different from samples from healthy individuals in that an abnormal increase in common oral bacteria clostridium difficile or porphyria was observed. A similar study was subsequently carried out by Professor Peer Bork's team at the European Molecular Biology Laboratory. They sequenced the microbiomes of 156 fecal samples, some of which were from colon cancer patients, and found that the relative concentration of 22 types of bacteria predicted cancer. 109 Bork's team found that microbial composition predicted CRC risk with 50% accuracy. When combined with blood tests, the method improved diagnostic accuracy to 70%. However, it remains unclear whether changes in colon cancer patients’ microbiomes predict disease progression. 110
Several factors have been identified as significant and distinctive in CRC's development, including the presence of specific microbiome strains. Bacteria that are abundant in the early stages through the metastatic stages of CRC include Fusobacterium nucleatum and Solobacterium moorei. Several bacteria, such as Atopobium parvulum and Actinomyces odontolyticus, have been found to be in abundance only in adenomas and intramucosal carcinomas. 111 Researchers examined the DNA of samples taken from CRC patients and found that Atopobium, Porphyromonas Bacteroides, and Fusobacterium were abundant in the samples. There are four bacteria that are associated with CRC: Bacteroides fragilis, Escherichia coli, Enterococcus faecalis, and Streptococcus gallolyticus. CRC patients’ fecal and tumor samples contained higher number of F. nucleatum, Parvimonas, Peptostreptococcus, Porphyromonas, and Prevotella strains. 112
There are different ways in which bacteria can cause cancer. F. nucleatum stimulates CRC growth by provoking signaling by Toll‐like receptor 2 (TLR2)/Toll‐like receptor 4 (TLR4) and inhibiting apoptosiss. 113 Peptostreptococcus contributes to the development of cancer by producing metabolites that produce an acidic and hypoxic tumor microenvironment and increase microbiome colonization. Several types of bacteria, such as E. coli, are genotoxic. 112 Oncometabolites are a collection of metabolites produced by the gut microbiome and by human cancers. Several metabolites, including L‐2‐hydroxyglutarate, succinate, fumarate, D‐2‐hydroxyglutarate, and lactate, accumulate in cancer cells. Interestingly, lactic acid serves as fuel for cancer cells and contributes to cancer progression, whereas butyrate suppresses proinflammatory genes and tumor growth. 112 , 114 , 115
This suggests that microbes in the body contribute to cancer development. At the very least, these studies suggest that microbial composition does influence CRC. Schloss speculated that inflammation caused by gut microbiomes creates a favorable environment for tumor formation and development. Upsetting the balance of the microbiome can result in the release of reactive oxygen species (ROS) that damage cells and their genetic material. 116 In addition, inflammation increases the release of growth factors and angiogenic factors, which may accelerate the spread of cancer.
2.2. The skin microbiomes and skin cancer
Thousands of microorganisms live on the skin including bacteria, fungi, and viruses. 117 , 118 Like microbiomes in the gut, skin microbiomes also play important roles in resisting the invasion of harmful substances. As the largest organ in the body, the skin hosts many beneficial microorganisms, forming a physical barrier against pathogens. Recent studies have found that skin microbial dysbiosis leads to chronic inflammation, immune escape, and skin cancer. 119 , 120
In a tumor biopsy study, researchers used swab samples and 16S rRNA gene sequencing and found that the growth rate of S. aureus in the skin of squamous cell carcinoma patients was higher than in healthy individuals, and the increase in S. aureus was closely related to skin keratosis (precancerous lesions of squamous cell carcinoma). 121 Malignant melanoma (MM) is the most malignant skin tumor, accounting for about 75% of all skin cancer‐related deaths. 122 The skin microbiome in animal models of melanoma showed differences in significance between microbe composition and microbial diversity when compared with normal skin. 123 Fusobacterium and Eucoccus (Trueperella) were found to be increased in melanoma skin samples. 123 However, the regulatory effect of the skin microbiome on the oncogenic pathways that induce mutations remains unclear, and there is still much to learn about the role of microbiota in skin cancer.
2.3. Oral microbiomes and oral cancer
The association between oral microbiomes and distant cancers falls into two categories. First, microbiomes do not directly participate in the pathogenesis of cancer. However, microbial changes and carcinogenesis are correlated, and therefore microbial changes can be used as markers of cancer. 124 Second, microorganisms are directly connected to tumorigenesis . 125 Significantly higher levels of Peptostreptococcus, Peptococcus, Fusobacterium, Prevotella (especially P. melaninogenica), Porphyromonas, Veillonella (mainly Veillonella parvula), Haemophilus, Capnocytophaga, Porphyromonas gingivalis, Rothia, and Streptococcus have been detected in oral squamous cell carcinoma (OSCC) samples. 126 , 127
2.4. The esophagus microbiomes and esophagus cancer
The microbial composition of the esophagus is relatively stable, consisting mainly of Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Clostridium, and Saccharibacteria. 128 Esophageal microorganisms are mainly classified into two types. The type I microbiome is composed of Gram‐positive bacteria and is closely associated with the normal esophagus. Type I is dominated by Firmicutes, and Streptococcus is the most dominant genus with a high relative abundance. The type II community is enriched with Gram‐negative bacteria and is mainly associated with esophageal abnormalities. Many of the bacteria in the type II microbiome are associated with the onset of esophageal cancer such as Chaveamella, Prevotella, Haemophilus, Neisseria, Granule bacteria, and Fusobacterium. 129
Studies have shown that when esophagitis or esophageal cancer occurs, the microbial diversity of the patient's esophagus changes significantly, and the abundance of streptococcal species decreases. Gram‐negative anaerobic bacteria or microaerophiles such as Peperamella, Prevotella, Fusobacterium, and Neisseria are increased in esophageal cancer. 130 This suggests that the Gram‐negative anaerobic microbiome may be associated with abnormal disease status and that adjusting the proportion of esophageal Gram‐positive and Gram‐negative bacteria may reduce the incidence of esophageal disease. During esophageal epithelial injury, the esophageal‐mucosal barrier is disrupted, and the bacteria translocate, affecting the esophageal microenvironment and immune homeostasis. Compared with Barrett's esophagus (BE), patients with esophageal adenocarcinoma exhibited a reduced intraesophageal pH and increases in fermented Bacillus and Lactobacillus. 131 The abundance of Streptococcus pneumoniae was shown to be high in rat animal models of BE and esophageal adenocarcinoma. 132 There is a strong association between obesity and BE and esophageal adenocarcinoma. Abdominal adipose tissue and its secretion of proinflammatory cytokines are recognized risk factors for BE and esophageal adenocarcinoma. Obese patients are also known to experience gut microbiome changes. These changes induce inflammation and change the proportion of Streptococcus and Prevotella in the upper digestive tract, which may also contribute to the development of BE and esophageal adenocarcinoma. 132 , 133
2.5. The lungs microbiomes and lung cancer
Lung cancer is a complex disease caused by interactions between the host and environmental factors. 134 Treatment for lung cancer includes surgery, chemotherapy, radiotherapy, targeted therapy, and immunotherapy. 135 The early diagnosis rate of lung cancer is low; and most lung cancer patients are diagnosed in the advanced stages, which have a high mortality rate. Microorganisms can maintain a microecological balance and regulate host immune responses, both of which are crucial for responding to different environmental factors. Although healthy lung tissue is considered a sterile environment, the development of high‐throughput technology has identified microbes in the microenvironment of healthy lung tissue including Bacteroides, Firmicutes, Proteobacteria, Streptococcus, Pseudomonas, Periconella, and Prevotella, where they maintain a balance between health and morbidity. 136
Interestingly, 16s RNA sequencing of lung tissue showed that the abundance of Bacteriaceae, Trichospiraceae, and Ruminococcaceae in lung tissue was closely related to lung cancer risk, recurrence‐free survival, and disease‐free survival in lung cancer patients. However, the molecular mechanisms of saliva, sputum, and fecal microbes underlying the occurrence and prevention of lung cancer are not yet understood. 137 Currently, the understanding of the lung microbiome and its impact on lung cancer and treatment is still in its infancy. Epidemiological survey data indicate that Mycobacterium tuberculosis (TB) is associated with lung cancer development. 138 Compared with healthy subjects or other lung diseases, lung cancer patients have a lower alpha diversity than beta diversity. 139 Studies have found that many gram‐negative bacteria such as H. influenzae, Enterobacter, and E. coli are colonized in the airways of lung cancer patients. 140 Peericonella, Neisseria, and Selenomonas have been found in the sputum of patients with lung squamous cell carcinoma and lung adenocarcinoma carcinoma. 141
2.6. The urogenital tract microbiomes and urogenital tract cancer
Although most bacteria are concentrated in the gastrointestinal tract, other parts of the body such as the genitourinary tract contain unique microbiomes. 142 Molecular biology and cell and organ culture techniques have shown that many tissues traditionally considered sterile (including the bladder, prostate, uterus, fallopian tubes, and ovaries) harbor microbial communities when in a disease state. 143 Anatomically, the female genital tract can be separated into the lower reproductive tract (vagina and cervix) and the upper reproductive tract (uterus, fallopian tubes, and ovaries), each with its site‐specific microenvironment. Most of the bacteria in the female reproductive tract are in the vagina, with low microbial diversity in healthy women. Lactobacillus (L. Chris, L. garnei, L. James, and L. vaginalis) is the dominant flora in the female lower reproductive tract (mainly in women in sub‐Saharan Africa). 144 The increased consumption of lactic acid bacteria was found to be involved in a variety of female diseases such as sexually transmitted diseases, preterm birth, spontaneous abortion, and pelvic inflammation. 145 Owing to the presence of lactic acid, the microenvironment of the vagina remains at pH < 4.5, which is the main mechanism for the self‐cleaning of the vagina. Studies have shown that vaginal flora disorders are closely associated with gynecological tumors. For example, it is well established that papillomaviruses (HPV) infection and cervical cancer occurrence are closely related. 144 A recent study found that during HPV infection, the microenvironment of the cervix changes, and HPV cooperatively promotes tumorigenesis. 146 Clinical studies showed that vaginal lactobacillus depletion and overgrowth of anaerobic bacteria were positively associated with an increased risk of cervical cancer. Lactobacillus predominance and a significant increase in vaginal microbiome diversity were found in patients with cervical precancerous lesions and cancer, compared with healthy women. 147 In patients at high risk of cervical cancer, local cervicovaginal conditions such as increased proinflammatory cytokines and cancer‐specific biomarkers, vaginal pH abnormalities, and excessive depletion of lactic acid bacteria can directly or indirectly lead to cervical cancer.
Endometrial cancer is the most common gynecological cancer in developed countries. Several factors including obesity, inflammation, metabolic imbalance, and postmenopausal estrogen therapy are major risk factors for the type I endometrial cancer. 148 These factors are associated with changes in the gut and vaginal microbiome. 117 , 149 The close association between the gut microbiome, estrogen metabolism, and obesity suggests that the microbiome may contribute in the etiology of endometrial cancer. 117 , 119 Estrogen compounds can influence the vaginal microbial community, indicating that regulating estrogen can affect the vaginal microbiome and therefore influence endometrial hyperplasia and cancer development. 149 A study using a 3D human vaginal epithelial model showed that A. vaginae induced the production of proinflammatory cytokines and antimicrobial peptides. Antibiotics, porphyrin, dialysis bacteria, Ruminococcus, anaerobic bacteria, Treponema, Bacteroides, and Spirospira were found to be significantly enriched in tumors. Proteobacteria, Porphyromonas, and an abnormal vaginal pH (>4.5) also occur in patients with endometrial cancer. 150
In addition, cancer of the ovary is one of the deadliest malignancies in women. Dysbiosis of the genital microbiome has been implicated in ovarian cancer development and has been suggested as a potential biomarker of the disease. Clinical data suggest a unique, potentially pathogenic profile in the ovarian microbiome among ovarian cancer patients, with organisms such as Brucella, Mycoplasma, and Chlamydia. 151 Moreover, Proteobacteria, especially Acinetobacter are increased in the ovarian microbiome. Although preliminary studies have demonstrated that the presence of various bacteria in ovarian cancer tissues is associated with ovarian inflammation and that the highly hypoxic tumor microenvironment favors the recruitment and growth of anaerobic microorganisms, the causal relationship between the microbiome and ovarian cancer remains unclear. 152 Studying tissue specificity of the ovarian cancer microbiome and determining its role in tumors could aid in understanding the association between the female reproductive tract microbiome and ovarian cancer.
Traditionally, urine is thought to be sterile. However, recent studies have found that microorganisms are present in the urinary tract and bladder, and that the urinary microbiome may change with age. The female urinary microbiome contains mainly Lactobacillus and Gardnerella, while the male microbiome is primarily Corynebacterium, Staphylococcus, and Streptococcus. Microorganisms in the urethra are strongly associated with cancers in the genitourinary system. 153 Most current studies focus only on bacteria in the urogenital tract, with less attention to other microbes such as fungi and viruses. Many infectious‐related cytokines lead to chronic inflammation, which, in turn, drives cellular carcinogenesis. Microorganisms in the urinary tract can control the pathogenic bacteria growth and inhibit tumorigenesis, but the role of the urogenital microbiome in regulating pathogenic infections and mediating cancer development requires further clarification.
It is not yet clear whether the urinary microbiome affects bladder cancer progression, or whether the composition, diversity, or abundance of microorganisms is associated with bladder cancer. A high abundance of Fusobacterium, Actinomyces, Facracter, and Campylobacter was found in the urine of patients with bladder cancer, while Streptococcus and Corynebacterium were higher in the urine of healthy people. The abundance of bacteria was increased in patients with bladder cancer compared with urine from nontumor patients, but there was no difference in diversity. 154 Studies of prostate cancer suggest that the urinary microbiome in patients with prostate cancer may contain proinflammatory bacteria, and the chronic proliferative inflammation caused by these proinflammatory bacteria may be a risk factor for the development of prostate cancer. However, the association between prostatitis and prostate cancer and the urinary microbiome remains unclear. 155 , 156
As research on genitourinary microorganisms and genitourinary cancers is in the early stages, data from large populations are needed to reveal the relationship between genitourinary microorganisms and cancer. Analysis of the role of genitourinary microorganisms in tumorigenesis is promising for the treatment of cancer.
2.7. The blood and lymphatic system microbiomes and cancer
Currently, studies on leukemia and microbiomes have primarily focused on the relationship between childhood leukemia and gut microbes. Acute leukemia is the most common cancer in children and the most common cause of cancer‐related deaths in childhood. It was found that patients without infectious complications had an increased relative abundance of Bacteroidetes and Proteus faecacterium, while patients with concurrent infections had an increased abundance of aerobic bacteria. 157 Because chemotherapy damages the intestinal mucosal barrier, bacteria translocate into the blood, which can lead to diarrhea, abdominal pain, low diversity of enterococcus and porphyraceae, and increased abundance of C reactive protein. 158 Microorganisms mainly secrete SCFAs such as butyrate. Butyrate has a key nutritional effect on the intestinal epithelial barrier, and when chemotherapeutic drugs destroy the intestinal epithelial barrier, the microbial abundance of the Trichospiraceae family decreases, aggravating microbial infection. 159 The genetic predisposition for leukemia is also closely related to commensal microorganisms. 160 Bone marrow suppression and immunosuppression are common in children with leukemia. One study found that after the induction and reinduction of patients, microbiome diversity in children's feces with leukemia was decreased significant, and the proteobacteria including Enterobacteriaceae and Pseudomonas were also reduced. Analysis of the changes in intestinal microbial diversity and composition before and after treatment can be used to monitor the occurrence of adverse chemotherapy reactions. 161
Although recent studies have explored the relationship between the gut microbiome and acute childhood leukemia, the relationship between human T cell leukemia virus type 1 (HTLV‐1)‐related adult T cell leukemia (ATL) and intestinal microbes is rarely reported. The prognosis of ATL is very poor; the median survival of acute ATL is 8 months, the 4‐year overall survival is 11%, and the treatment options are very limited. The FBXW7 proteins in the F‐box protein family were found to play a role in tumor protein phosphorylation‐dependent ubiquitination and proteasomal degradation. 162 The absence of FBXW7 could serve as an independent prognostic marker and was found to be significantly associated with tumor cell resistance to chemotherapeutic agents and poor disease prognosis. 163 , 164 , 165 It is a gut commensal anaerobic bacterium that produces large amounts of thioredoxin and nitroreductase, reduces oxidative stress associated with CYB5RL mutations, and inhibits cell death. However, it is unclear whether FBXW7 is involved in leukemogenesis. 166 , 167 Notch1 signaling activation promotes proliferation in adult acute leukemia cells and regulates hepatic glucose production and lipid synthesis. It has been shown that inhibiting Notch1 signaling in HepG2 cells increased the expression of fatty acid oxidation genes and fatty acid oxidation rate. 168 In adult acute leukemia, JAG1 overexpression contributed to the Notch1 signaling pathway and the migration of HTLV‐1 ‐transformed ATL cells. 169
This suggests that gut microbes may be involved in ATL development through the SCFA pathway. Because the gut microbiome develops with age, further confirmation of whether the fecal microbiome can provide information on the entire gut microbiome is needed. Fasting and breastfeeding can both modulate the gut microbiome to reverse the progression of childhood leukemia. However, the specific microorganisms that promote or reverse leukemogenesis and progression remain poorly defined. Therefore, the identification of specific microorganisms closely related to leukemogenesis and progression, and elucidation of gut microbiome mechanisms promotes or reverses leukemia is imperative in providing new strategies for the treatment of leukemia.
Very few studies have focused on lymphoid tissue and microorganisms. In a study on the relationship between changes in human lymphatic structure/intestinal function and gut microbes, researchers determined that the abundance of Prevotella and Bacteroidetes‐Prevotella‐porphyromona (BPP) increased in the gut microbiome. This was the first human study to link changes in lymphatic structure/intestinal function and the gut microbiome. 170 Subsequent studies found that the incidence of H. pylori‐negative mucosa‐associated lymphatic tissue (MALT) lymphoma was related to the presence of H. pylori in some patients. Interestingly, alpha diversity was significantly reduced in H. pylori‐negative MALT lymphoma patients compared to controls (p = 0.04). In addition, Burkholderia and Sphingomonas were significantly increased in MALT lymphoma patients, but Prevotella and Pellona were lower, suggesting that an altered microbiome may be involved in the pathogenesis of H. pylori‐negative MALT lymphoma. 171 Patients with gastrointestinal follicular lymphoma (GI‐FL) had significantly less alpha diversity compared to controls. The microbial composition between the two groups was significantly different, and the content of sporophyte, Rhodella, Prevotella, and Doupecaceae was significantly lower in GI‐FL patients, suggesting that this microbiome may be play a role in the pathogenesis of GI‐FL. 172
2.8. Others
2.8.1. The hepatitis viruses and liver cancer
Hepatitis B and C viruses play a role in the pathogenesis of primary hepatocellular carcinoma (HCC). 173 By integrating viral genes and mutating viral proteins (mostly HBx and PreS), HBV can induce unstable insertion and deletion of host genes. 174 Genotype C and D of chronic hepatitis B patients with a promoter A1762T/G1764A mutation are at higher risk of HCC than those with genotype A and B. 175 DNA integration of HBV occurs more frequently in tumor tissues compared with adjacent normal liver tissues. Approximately 40% of HBV gene break points located in the core area of virus enhancer. 176 HBV DNA integrated into the host genome and caused genetic damage and chromosomal instability. It has a selective advantage for tumor progression. Viral proteins (HBx, HBc, and PreS) mutants can affect cell function, activate oncogenic pathways and increase the sensitivity of hepatocytes to these mutants. HBx viral protein play a role in carcinogenesis by inducing the cytoplasmic isolation of chromosomal maintenance protein 1 (Crm1) and promoting nuclear factor kappa‐B (NF‐κB) enter into the nucleus. On the other hand, HBx viral protein can also promote carcinogenesis by maintaining centrosome integrity though Crm 1 and affecting mitosis checkpoint. Telomerase activation is observed in over 90% of HCC patients and is closely related to HCC. 177 C‐terminally truncated middle surface protein of hepatitis B virus (MHBst) and wild‐type/truncated HBx proteins activate the transcripts promoter of TERT in HBV‐associated HCC. 178 Many signaling pathways were activated in HCC such as Wnt/β‐Catenin, cell cycle, oxidative stress metabolic pathway, Ras/ MAPK, and so on. 179
Whether HCV can induce HCC is unknown, but it is a primary risk factor for the disease. It depends on the interaction among virus, host and environmental factors when HCV carriers developed into HCC. The HCV genome cannot be integrated into the host genome, but it can damage the host liver cells DNA through viral proteins and ROS producing by some certain carcinogenesis pathways, causing gene mutation and HCC occurrence. 180 It is possible for HBV carriers or HCV carriers to develop cancer due to some certain factors, such as sex, age, family history, viral load, infection time, coinfection with other virus (HDV, HIV, HBV/HDV), environmental factors, and so on. 181 The intestinal microbiomes also might be involved in the development of HCC.
2.8.2. The HPV and cervical cancer
Human papillomavirus is the most common sexually transmitted virus in the world. 182 In addition, more than 99% of cervical cancer cases are caused by infection with high‐risk HPVs (hrHPVs). 183 Since 85–90% of hrHPVs infections are transient and can be spontaneously cleared by the host's immune system, HPVs infection alone cannot cause the disease. There is only a 10–15% chance that cervical intraepithelial neoplasia will progress to invasive cervical cancer with persistent infections. 184
Additionally to their transformational properties, HPV‐infected cells actively influence the local microenvironment. As a result of creating a supportive postinfection microenvironment (PIM), virus‐infected cells play an important role in cervical cancer development. 185 , 186 A PIM is formed by interactions between virus‐infected cells, immune cells, and host stroma, as well as their derived components (chemokines, cytokines, extracellular vesicles, and metabolites). 187 As a result, PIM actively alters the local microenvironment, thereby promoting the persistence and spread of the virus and the development and progression of cervical cancer. Understanding PIM properties and its role in HPV infection and disease progression can help identify potential biomarkers to improve diagnosis and prognosis.
2.8.3. The EBV and nasopharyngeal carcinoma
In humans, EBV is the first virus that causes cancer. There are several diseases that can be caused by or associated with EBV infection, including infectious mononucleosis, burkitt lymphoma, and nasopharyngeal carcinoma (NPC). 188 A NPC is a rare epithelial cancer of the nose and throat, divided into three pathological subtypes (keratinized squamous carcinoma, nonkeratinized squamous carcinoma, and basic squamous carcinoma). 189 NPC is almost always caused by EBV infection, which is the most common pathogenic factor. 190 When EBV is infected into B lymphocytes, it will remain latent at the cytoplasm for a long time. As a result, EBV latent infection is a characteristic of precancerous nasopharyngeal epithelium. Infected epithelial cells express viral genes such as EBER, EBNA1, LMP1, and LMP2A, which can initiate nasopharyngeal epithelial tumor growth. 191 Additionally, it is not fully understood how EBV infection contributes to NPC development and progression. According to recent genomic analyses, the NF‐κB signaling pathway, caused by overexpression of the EBV viral oncoprotein, may play a role in NPC. 192
3. MECHANISMS BY WHICH HUMAN CARCINOGENIC MICROBIOMES PROMOTE CANCER
Carcinogenic microbiomes can suppress human immune system directly, active immunosuppressive factors in tumor microenvironment, inhibit immune cells activity in tumor microenvironment, induce cell mutation, and promote cell proliferation. We reviewed the mechanism of human carcinogenic microbiomes promoting cancer in this section.
3.1. Alteration in immune system activity
Gut microbiomes not only participate in the pathogenesis of CRC but also secrete inflammatory factors and destroy the integrity of the intestinal barrier, 193 activating cellular metabolic pathways and promoting breast cancer, 131 , 194 and HCC. 132 , 195 One study using a tumor‐bearing mouse model showed that H. pylori and interleukin‐22 (IL‐22) upregulated the expression of matrix metalloproteinase 10 (MMP10) in gastric epithelial cells through the extracellular signal‐regulated kinase (ERK) pathway, producing chemokine ligand 16 (CXCL16). 196 , 197 This recruited CD8+ T cells and induced an inflammatory response, which damaged the gastric mucosa and promoted gastric carcinogenesis . 198 In a mouse model of CRC, enterotoxin Bacteroides fragilis (ETBF) stimulated the immune response through T helper 17 cells (Th17) and promoted colon tumor growth. 137 , 138 , 199 Similarly, E. coli expressing the genomic island polyketide synthase (pks+) enhanced tumorigenesis in preclinical CRC models and were found to be enriched in human CRC tissues . 200 , 201 Furthermore, pks + E. coli produces the genotoxin colibactin, which alkylates DNA, resulting in the formation of DNA adducts (a form of potentially mutagenic DNA damage) in colonic epithelial cells. The recent biochemical resolution of this process illustrates how host‐microbe interactions may lead to cancer. 202 Streptococcus gastrop (Peptostreptococcus anaerobius) interacts with TLR2 and TLR4 in colon cells to regulate a variety of immune cell types including myeloid‐derived suppressor cells (MDSCs), tumor‐associated macrophages (TAMs), and granulocyte tumor‐related neutrophils, thus promoting colon carcinogenesis. 203 F. nucleatum is associated with various types of cancer including CRC. 204 F. nucleatum was shown to bind to epithelial cadherin (E‐cadherin) through FadA and Fap2, inducing β‐Catenin signaling, regulating the inflammatory and carcinogenic response, and promoting tumorigenesis. 123 , 205 P. anaerobius has been shown to bind to tumor cell surface integrins via a cell surface protein, putative cell wall binding repeat 2 (PCWBR2), activating the PI3K–AKT pathway and the NF‐κB cascade and inducing an inflammatory response that promoted tumor cell proliferation. 203 P. anaerobius recruited tumor‐infiltrating, promoted cell proliferation, and triggered an inflammatory response in the tumor microenvironment through the PI3K/AKT/NF‐kB signaling pathway. 129
Various studies have demonstrated several different mechanisms in OSCC such as the promotion of the production of inflammatory cytokines as well as cellular proliferation and invasion by P. gingivalis and F. nucleatum. 206 F. nucleatum interacted with T‐cell immunoglobulin and ITIM domain (TIGIT) to inhibit NK cell cytotoxicity, which induced lymphocyte death through the TLR4/MYD88 pathway and promoted tumor development and induced resistance to chemotherapy 207 (Figure 2A). F. nucleatum stimulated cell proliferation and induced increased levels of IL‐2 and MMPs necessary for tumor invasion and metastasis. 208 Oncogene transcription activity increased greatly due to these changes, as well as levels of proinflammatory cytokines. 208 Fusobacterium may promote tumor proliferation through Fap2, T cell immunoglobulin, and TIGIT, and inhibit the cytotoxicity of NK cells. 209 , 210 Skin swabs from patients with acral melanin showed a high abundance of the Corynebacterium genus in stage III/IV MM patients compared with stage I/II patients. Corynebacterium induced IL‐17 production, upregulated IL‐6, and activated STAT3 signaling, which induced melanoma growth (Figure 2B). This indicates that Corynebacterium can promote the development of MM through an IL‐17‐dependent pathway. Intratumoral injection of Corynebacterium acnes induced Th1 cytokines such as IL‐12, tumor necrosis factor (TNF), and interferon (IFN), and inhibited the growth of melanoma cells. 211 , 212
FIGURE 2.
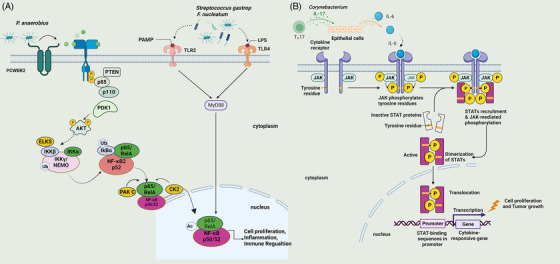
Alteration of immune system activity. (A) P. anaerobius recruited tumor‐infiltrating, promoted cell proliferation, and triggered the inflammatory response in the tumor microenvironment through the PI3K/AKT/NF‐kB signaling pathway. F. nucleatum induced lymphocyte death through the TLR4/MYD88 pathway, promoting tumor development. (B) Corynebacterium induced IL‐17 production, upregulated IL‐6, and activated the JAK/STAT3 signaling pathway to induce cell proliferation and tumor growth.
Studies have shown that Fusobacterium upregulated cervical IL‐4 and transforming growth factor 1 expression and inhibited the vaginal immune microenvironment. 133 In the vaginal microenvironment of cervical cancer patients, proinflammatory factors such as IL‐36, chemotaxis factors such as IFN‐induced protein 10 (IP10), macrophage inflammatory protein 1 (MIP‐1β), and RANTES, hematopoiesis factors such as FLT3 ligand, and adaptive immune response cytokines such as IL‐2, IL‐4, and soluble CD40 ligand were associated with increased nonlactobacilli microbial growth. 133 S. gastrop (P. anaerobius) has been shown to interact with TLR2 and TLR4 on colon cells to regulate a variety of immune cells including MDSCs, TAMs, and granulocyte tumor‐related neutrophils, thus promoting colon carcinogenesis. 203 Additionally, the specific chemokine genes that are upregulated in ESCC tissues with F. nucleatum suggest that F. nucleatum might contribute to the growth of esophageal tumors by activating chemokines such as CCL20. 213
Studies have confirmed that the secretion of S. epidermidis activated regulatory T cells and inhibited inflammatory responses in the skin. Both animal and cell experiments have confirmed that S. epidermidis produces 6‐n‐hydroxyaminophosphorus (6HAP), which inhibits DNA synthesis and UV‐induced tumor growth. 214 , 215 Furthermore, it was also reported that S. epidermidis and its derived LTA upregulated TRAF1, CASP14, and CASP5 and improved the survival of melanoma cells. 216 C. acnes promoted apoptosis, enhanced fecal porphyrin secretion, upregulated TNFα, and promoted the death of residual melanoma cells after radiotherapy. 217 Inhibition of S. aureus infection with topical mupirocin and the oral antibiotic dicloxacillin also improved clinical symptoms of CTCL, which was likely linked to an increase in IL‐2 and STAT3 signaling activation. 218
Skin microbial dysbiosis, activation of the skin immune system, production of microbial metabolites and toxins, destruction of barriers and UV radiation may all play a role in skin cancer occurrence and development. 219 However, the regulatory effect of the skin microbiome on the oncogenic pathways that induce mutations is not clear, and further studies are needed to elucidate the role of the skin microbiome in skin cancer.
Cancer‐causing bacteria are largely induced by inflammation‐associated cytokines. 208 Leakage of microbial products from the intestinal lumen to the peripheral circulation increases the levels of lipopolysaccharide (LPS) in the peripheral circulation and activates chronic inflammation, leading to the development of non‐Hodgkin's lymphoma (AIDS‐NHL) associated with acquired immunodeficiency syndrome (AIDS). 220 Microorganisms can regulate the immune response, alter epigenetic markers of cells, modify target cells, activate B lymphocytes, and cause patients with Sjogren's syndrome (SS) to develop B cell lymphoma. 221 , 222 , 223 S. aureus is closely associated with cutaneous T‐cell lymphoma and promotes the progression of this disease. 224 Staphylococcal‐toxin (a‐hemolysin) inhibits cytotoxic responses by T cells and lead to immune escape. 225 Staphylococcal enterotoxin produced by S. aureus can activate the STAT5 protein, causing an increase in Th2 cells. 226 Further, Staphylococcal enterotoxin interferes with the removal of malignant tumor cells by cytotoxic T cells, inhibiting autoimmunity and increasing tumor immune tolerance. 225
Epithelial biopsy specimens of BE are enriched in proinflammatory cytokines, especially IL‐1b. 24 Moreover, a number of inflammatory factors are released under inflammatory conditions, such as IL‐6 and IL‐23, which promote the immune response between microorganisms and hosts by activating the inflammatory response, which may ultimately lead to cancer. 227 The esophageal type II microflora can produce abundant LPS due to the presence of Gram‐negative bacteria. 207 , 228 In addition to affecting cyclooxygenase 1/2, LPS can directly affect the lower esophageal sphincter, resulting in an increase in intragastric pressure and promoting the GERD occurrence, which leads to EAC development. 207 , 229 There is interesting evidence that in patients with BE and EAC, TLR4 is being expressed more strongly as a natural ligand of LPS. 207 It is thought that TLR4 receptor activation triggers the NF‐B pathway, which contributes to inflammation‐related carcinogenesis, 230 , 231 and is implicated in early BE. 232 Thus, inflammation and malignant transformation of the esophagus can be triggered by activating LPS‐TLR4‐NF‐kB. 207 , 233 As well, After BE cells were treated with LPS, Nadatani et al. observed increased expression of NOD‐like receptor protein 3, caspase‐1 activity, and secretion of IL‐1b and IL‐18. They hypothesized that LPS activates ROS, thereby promoting cancer development. 121 , 234 Recent studies have found that the host‐lung microbiome interaction is closely related to lung cancer development. An animal model of lung cancer using K‐ras/p53‐mutant (KP) mice showed a decreased incidence of lung cancer in germ‐free mice compared with SPF mice. Reduced airway microbial microbe diversity was found in SPF tumor‐bearing mice. In germ‐free mice, expression of IL‐1, IL‐23, IL‐17, and ROR‐γt were decreased in γδ T cells from peripheral lymph nodes and the spleen. 235 Treatment with aerosolized antibiotics in tumor‐bearing mice revealed a reduced number of regulatory T cells (Tregs), enhanced activation of immune effector cells, increased immune surveillance function in the lungs, and suppressed tumor metastasis, which was correlated with decreased enrichment of Streptococcus, Proteobacteria, and Actinobacteria in lung tissue. 149 , 235 These studies suggest that the lung microbiome has a direct link to immune regulation, but exactly how this microbiome affects immune cells in the tumor microenvironment is currently unknown.
3.2. Influence on host cell proliferation and death
In a mouse CRC model, ETBF stimulated the immune response via T helper 17 cells (Th17) and promoted colon tumor growth. 123 , 236 Additionally, a preclinical study found that E. coli expressing genetically altered polyketide synthase (pks+) enhanced tumor formation in mice. pks+ expression was found to be significantly enriched in human cancer tissues. 200 DNA adducts (potentially mutagenic DNA damage) are created when pks+ E. coli produce the genotoxin colibactin. This process was demonstrated via the recent biochemical resolution of this interaction as a possible cause of cancer. 202 F. nucleatum is associated with various types of cancer, including CRC. F. nucleatum was shown to bind to epithelial cadherin (E‐cadherin) through FadA and Fap2, inducing β‐Catenin signaling and regulating the inflammatory and carcinogenic responses, ultimately promoting tumorigenesis. 123 P. gingivalis and F. nucleatum have been shown to induce the production of inflammatory cytokines, cell proliferation, and cellular invasion in OSCC through a variety of different mechanisms. 206 P. gingivalis stimulated the formation of IL, TNF‐α, and MMP while inhibiting apoptosis. 206 , 237
There is no doubt that chemokines, along with their receptors, play an important role in the development and progression of cancer. 238 , 239 , 240 Cancer cells were empowered by CCL20 stimulation in vitro, according to Wang et al. 241 EC may be caused by genetic toxins or cancer‐promoting metabolites produced from bacteria, as well as microbiome components. To illustrate, by secreting swelling toxins, Gram‐negative bacteria may damage host DNA, 242 , 243 , 244 and subsequent repair of that damage may cause EC. 245 Gabriel et al. found that mammalian epithelial cells exposed to E. coli exhibited DNA damage responses, followed by cell division with signs of incomplete DNA repair, resulting in anaphase bridges and chromosome aberrations. A significant increase in DNA mutation frequency was observed among cells exposed to E. coli, as well as the formation of anchorage‐independent colonies, demonstrating that E. coli infection has the potential to be mutagenic and transformative. 243
Moreover, H. pylori produce cytotoxin‐associated gene A (CagA) or vacuolating cytotoxin A and can cause inflammation and cancer. 246 By increasing the level of ROS in the body, CagA can induce DNA damage by upregulating host‐mediated ROS production. 214 , 247 It is also known that vacuolating cytotoxin A has a tendency to alter membrane permeability and cause an increase in apoptosis rates. 209 The study by Li et al. demonstrated that a CagA1‐positive H. pylori can break the DNA in squamous epithelial cells in the esophagus, leading to atypical hyperplasia and causing ESCC carcinogenesis. Further mechanistic research found that H. pylori infection induces ROS production in the cytoplasm, which promotes the DNA damage response (Figure 3A). 248
FIGURE 3.
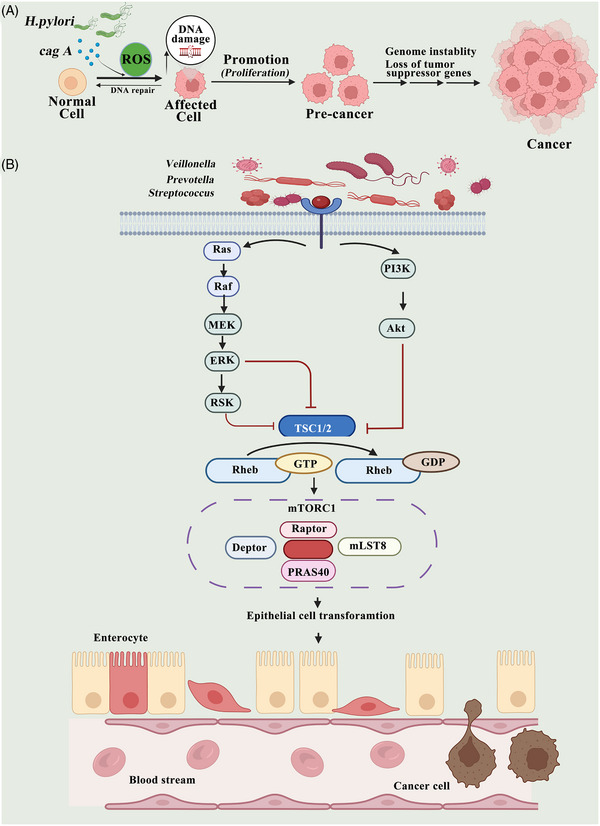
Influence on host cell proliferation and death. (A) Cytotoxin‐associated gene A (CagA) produced by H. pylori can stimulate inflammation and lead to the development of cancer. CagA induces DNA damage through host‐mediated upregulation of the production of reactive oxygen species in the cytoplasm and promotion of the DNA damage response. (B) The connection between microbial components and lung cancer can be explained by the production of metabolites. Veillonella, Prevotella, and Streptococcus bacteria induce epithelial cell transformation and promote epithelial cell transformation through activation of the PI3K and ERK signaling pathway.
Dysbiosis of the lung microbiome may influence cancer progression by causing dysregulated immune responses, characterized by the overactivity of inflammatory cells such as M1 macrophages, Th1 cells, or γδ T cells. 249 , 250 Currently, it is unclear exactly how lung cancer is related to the airway microbiome and immune regulation. 251 The connection between microbial components and lung cancer can be explained by the production of metabolites; however, this has been insufficiently researched compared to the relationship between dysbiosis and chronic inflammation. Researchers have discovered that as a result of exposure to Veillonella, Prevotella, and Streptococcus bacteria, epithelial cells are transformed both in vitro and in vivo by activating ERK and PI3K (Figure 3B). 131 , 252 , 253
4. MICROBIOME TREATMENT
The gut microbiome can modulate the efficacy of anticancer drugs. 254 Alterations in the gut microbiome are associated with tumor resistance to chemotherapeutic agents or immune checkpoint inhibitors (ICIs), 255 , 256 and modulation of the gut microbiome by antibiotics, probiotics, fecal microbiome transplantation (FMT), and nanotechnology may enhance the antitumor effects of chemotherapeutic agents and ICIs. 257
In recent years, the application of precision medicine in cancer treatment has greatly improved the disease‐free survival and quality of life of cancer patients. However, treatment failure due to patient hyporesponsiveness to immunotherapy is a great challenge in cancer treatment. Owing to the complexity of the host immune response, the mechanisms of immunotherapy are not well defined. 136 , 137 Microorganisms can promote tumor regression by enhancing the host immune response. There are new possibilities for effective cancer treatment with the microbiome. 139 The microbial treatment of cancer primarily includes fecal transplantation, targeted use of antibiotics, probiotics, bacteria, prebiotics, and phages. 140 , 141 FMT is the process of introducing feces from healthy donors (or the cryopreserved microbial content) into a patient's colon. 258 Interestingly, the fecal enema was used to treat pseudomembranous colitis clinically and achieved better results than FMT. FMT has been approved for the treatment of recurrent C. difficile infections, based on C. difficile treatment guidelines (2013). Increasing evidence has also demonstrated the beneficial effects of FMT in the treatment of other diseases such as intractable functional constipation, inflammatory bowel disease, and hematological malignancies. 22 In recent years, clinical trials of FMT have been carried out (NCT03812705, NCT04729322, NCT04163289, NCT03819296). For example, patients with PD‐1 refractory melanoma are being treated with FMT alone or in combination with pembrolizumab (NCT03353402). Some of the patients treated with the combination therapy had increased T lymphocyte infiltration into the tumor tissue and achieved remission of clinical symptoms. In clinical studies using FMT treatment for acute myeloid cell leukemia, multiple myeloma, myelodysplastic syndrome, and lung cancer, 40% of patients initially developed neutropenia. However, the final study results indicated that FMT therapy was safe and effective. 259 FMT is a relatively new approach for altering gut microbiome composition, so a long‐term safety assessment is still needed (Table 2).
TABLE 2.
Clinical trials in microbiome treatment (https://www.clinicaltrials.gov/).
| NCT number | Title | Status | Conditions | Interventions | Phase | |
|---|---|---|---|---|---|---|
| 1 | NCT04857697 | Effects of Probiotics on the Gut Microbiome and Immune System in Operable Stage I‐III Breast or Lung Cancer | Recruiting |
Anatomic Stage I Breast Cancer AJCC v8 Anatomic Stage II Breast Cancer AJCC v8 Anatomic Stage III Breast Cancer AJCC v8 Breast Adenocarcinoma Stage I Lung Cancer AJCC v8 Stage II Lung Cancer AJCC v8 Stage III Lung Cancer AJCC v8 |
Procedure: Biopsy Procedure: Biospecimen Collection Drug: Probiotic Procedure: Therapeutic Conventional Surgery |
Early Phase 1 |
| 2 | NCT03819296 | Role of Gut Microbiome and Fecal Transplant on Medication‐Induced GI Complications in Patients With Cancer | Recruiting |
Clinical Stage 0 Cutaneous Melanoma AJCC v8 Clinical Stage I Cutaneous Melanoma AJCC v8 Clinical Stage IA Cutaneous Melanoma AJCC v8 Clinical Stage IB Cutaneous Melanoma AJCC v8 Clinical Stage II Cutaneous Melanoma AJCC v8 Clinical Stage IIA Cutaneous Melanoma AJCC v8 Clinical Stage IIB Cutaneous Melanoma AJCC v8 Clinical Stage IIC Cutaneous Melanoma AJCC v8 Clinical Stage III Cutaneous Melanoma AJCC v8 Clinical Stage IV Cutaneous Melanoma AJCC v8 |
Other: Best Practice Other: Biospecimen Collection Procedure: Endoscopic Procedure Procedure: Fecal microbiome Transplantation Biological: Infliximab Other: Laboratory Biomarker Analysis Drug: Prednisone Biological: Vedolizumab |
Phase 1 Phase 2 |
| 3 | NCT05020574 | Microbiome and Association With Implant Infections | Recruiting |
Breast Cancer Breast Cancer Female Genetic Predisposition to Disease |
Drug: Cephalexin | Phase 2 |
| 4 | NCT04552418 | Intestinal Microbiome Modification With Resistant Starch in Patients Treated With Dual Immune Checkpoint Inhibitors | Recruiting | Solid Tumor | Drug: Potato starch | Early Phase 1 |
| 5 | NCT04875728 | The Impact of an Antibiotic (Cefazolin) Before Surgery on the Microbiome in Patients With Stage I‐II Melanoma | Recruiting |
Clinical Stage I Cutaneous Melanoma AJCC v8 Clinical Stage IA Cutaneous Melanoma AJCC v8 Clinical Stage IB Cutaneous Melanoma AJCC v8 Clinical Stage II Cutaneous Melanoma AJCC v8 Clinical Stage IIA Cutaneous Melanoma AJCC v8 Clinical Stage IIB Cutaneous Melanoma AJCC v8 Clinical Stage IIC Cutaneous Melanoma AJCC v8 Pathologic Stage I Cutaneous Melanoma AJCC v8 Pathologic Stage IA Cutaneous Melanoma AJCC v8 Pathologic Stage IB Cutaneous Melanoma AJCC v8 |
Drug: Cefazolin Procedure: Resection |
Phase 1 |
| 6 | NCT03686202 | Feasibility Study of Microbial Ecosystem Therapeutics (MET‐4) to Evaluate Effects of Fecal Microbiome in Patients on Immunotherapy | Recruiting | All Solid Tumors | Biological: MET‐4 | Early Phase 1 |
| 7 | NCT03959241 | TAC/MTX vs. TAC/MMF/PTCY for Prevention of Graft‐ versus‐Host Disease and Microbiome and Immune Reconstitution Study (BMT CTN 1703/1801) | Active, not recruiting |
Acute Leukemia Chronic Myelogenous Leukemia (CML) Myelodysplasia Lymphoma |
Procedure: Mobilized Peripheral Blood Stem Cell graft with Tacrolimus/Methotrexate Drug: Tacrolimus Drug: Methotrexate Procedure: Mobilized Peripheral Blood Stem Cell graft with Tacrolimus/Mycophenolate Mofetil/ Post‐Transplant Cyclophosphamide Drug: Mycophenolate Mofetil Drug: Cyclophosphamide |
Phase 3 |
| 8 | NCT03817125 | Melanoma Checkpoint and Gut Microbiome Alteration With Microbiome Intervention | Active, not recruiting | Metastatic Melanoma |
Drug: Placebo for antibiotic Drug: Vancomycin pretreatment Drug: Nivolumab Drug: Matching Placebo for SER‐401 Drug: SER‐401 |
Phase 1 |
| 9 | NCT03812705 | Fecal microbiome Transplantation for Steroid Resistant/Dependent Acute GI GVHD | Recruiting | Hematopoietic and Lymphoid Cell Neoplasm | Biological: fecal microbiome transplantation | Phase 2 |
| 10 | NCT04729322 | Fecal microbiome Transplant and Re‐introduction of Anti‐PD‐1 Therapy (Pembrolizumab or Nivolumab) for the Treatment of Metastatic Colorectal Cancer in Anti‐PD‐1 Non‐Responders | Recruiting |
Metastatic Colorectal Adenocarcinoma Metastatic Small Intestinal Adenocarcinoma Stage IV Colorectal Cancer AJCC v8 |
Procedure: Biopsy Procedure: Fecal microbiome Transplantation Drug: Fecal microbiome Transplantation Capsule |
Early Phase 1 |
| 11 | NCT04163289 | Preventing Toxicity in Renal Cancer Patients Treated With Immunotherapy Using Fecal microbiome Transplantation | Recruiting | Renal Cell Carcinoma | Drug: Fecal microbiome Transplantation | Phase 1 |
| 12 | NCT03819296 | Role of Gut Microbiome and Fecal Transplant on Medication‐Induced GI Complications in Patients With Cancer | Recruiting |
Clinical Stage 0 Cutaneous Melanoma AJCC v8 Clinical Stage I Cutaneous Melanoma AJCC v8 Clinical Stage IA Cutaneous Melanoma AJCC v8 |
Other: Best Practice Other: Biospecimen Collection Procedure: Endoscopic Procedure |
Phase 1 Phase 2 |
| 13 | NCT03353402 | Fecal microbiome Transplantation (FMT) in Metastatic Melanoma Patients Who Failed Immunotherapy | Unknown |
Melanoma Stage Iv Unresectable Stage III Melanoma |
Procedure: Fecal microbiome Transplant (FMT) | Phase 1 |
Probiotics are healthy, living bacteria that can be used as adjuvant therapy in tumor treatment to enhance immunity. Probiotics have been shown to regulate intestinal microflora and have demonstrated antitumor activity in several studies. 153 , 260 Animal study results showed that Lactobacillus acidophilus and Bifidobacterium upregulated miR‐26b and miR18a and downregulated miR135b, miR155, and KRAS expression, inhibiting colon cancer progression. 155 Clinical results show that probiotics can maintain intestinal flora balance after CRC surgery and effectively protect the intestinal mucosal barrier, prevent postoperative inflammation, reduce postoperative complications, and reduce the incidence of diarrhea caused by 5‐fluorouracil and postoperative radiotherapy. 156 Probiotics derived from Akkermansia muciniphila (A. muciniphila) are considered to be promising candidates. 261 Four metastatic melanoma patients whose immunotherapy for anti‐PD‐1 led to clinical response were found to have abundant A. muciniphila . 262 Also, Routy et al. investigated the relationship between immunocheckpoint inhibitors and gut microbiota in cancer patients. According to their findings, patients receiving PD‐1 antibodies reported a significantly higher intestinal level of A. muciniphila. 263 A. muciniphila multiplied in response to the applied density. 264 Based on these findings, it can be concluded that cancer immunotherapy in combination with A. muciniphila as one of the important probiotics in selective microbiota transplantation will produce better outcomes for patients in the near future. 265 , 266 Accordingly, it is reported one patient with high‐grade metastatic urothelial carcinoma experienced ICIs‐associated colitis following combined CTLA‐4 and CTLA‐3 therapies. The patient's intestinal tract showed evidence of good colonization of donor‐derived bacteria after treatment with FMT for ICI‐associated colitis, as evidenced by the presence of a higher level of A. muciniphila. 267
Prebiotics support selective fermentation including oligomers of fructose (FOS), xylan, galactose (GOS), inulin, and fructan. Prebiotic compounds can increase the proportion of beneficial intestinal bacteria, stimulate SCFA synthesis, regulate immune responses, modify microbial gene expression, increase cecum and colon absorption of micronutrients, and regulate metabolic enzymes. 25 It was found that FOS and inulin‐rich prebiotics combined with Lactobacillus rhamnosus and Bifidobacterium lactis inhibited azoxymethanase‐induced proliferation in rat colon cancer cells. 25 GOS produced by galactosidase or glucosidase conversion can increase the concentration of intestinal lactate and SCFAs, reduce the concentration of secondary bile acid and lactic acid in the feces, inhibit the activity of intestinal nitroreductase and glucuronidase, and may prevent the occurrence of CRC. Probiotic microorganisms consume prebiotic fibers for energy metabolism and growth. Moreover, prebiotic supplementation resulted in significant increases in IgG and IgM levels in perioperative CRC patients. 268
Combining probiotics and prebiotics, synbiotics have been used to treat a variety of tumor types, including CRC. Clinical studies show that synbiotics significantly reduce postoperative infection in CRC patients. In addition, the combined supportive treatment regimen of L. acidophilus, L. rhamnosus, Lactobacillus casei, Bifidobacterium, and FOS significantly reduced surgical site infections in CRC patients. 26 , 269 Animal experiments have confirmed that a cocktail of acidophiles (64 × 1011 CFU) composed of FOS, maltose dextrin, and lactic acid bacteria inhibited tumor growth, promoted apoptosis, and inhibited inflammation 270 (Figure 4).
FIGURE 4.
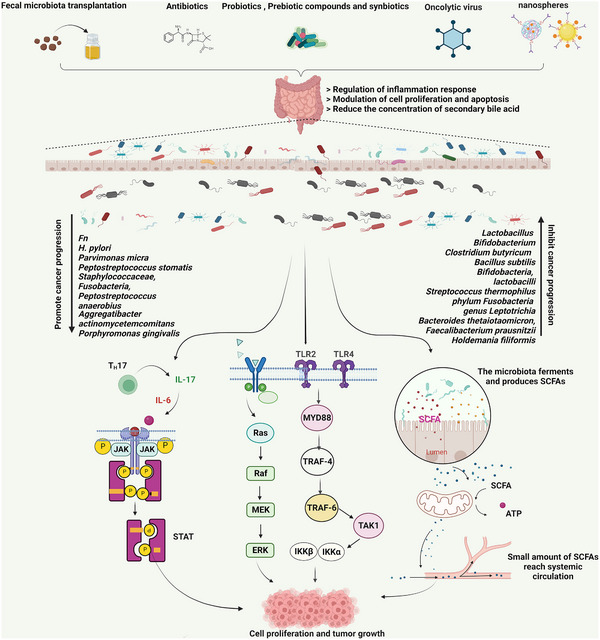
Microbiome treatment for cancer. Possible therapeutic approaches for cancer include fecal microbiome transplantation (FMT), probiotics, prebiotic compounds, synbiotics, antibiotics, oncolyitc virus and nanospheres, which are processes through which the intestinal microbiome can regulate inflammation, modulate cell proliferation and apoptosis, and reduce the concentration of secondary bile acids. By activating the JAK/STAT, PI3K/AKT, Ras/MEK, and TLR2/4‐MYD88 pathways, which are involved in tumorigenesis, microbial dysbiosis and special bacteria can influence cancer development and progression. The microbes that ferment and produce SCFAs also affect cell proliferation and tumor growth. Several microbes, such as Fn, Helicobacter pylori, Fusobacterium, Staphylococcus aureus, Peptostreptococcus anaerobius, and Porphyromonas gingivalis, promote cancer progression. Other microbes, such as Lactobacillus, Bifidobacterium, Clostridium butyricum, Bacillus subtilis, Bifidobacteria, Lactobacoilli, Streptococcus thermophilus, Leptotrichia, Bacteroides thetaiotaomicron, and Faecalibacterium prausnitzii, can inhibit cancer progression.
Wide‐spectrum antibiotics have adverse effects on patients receiving cancer immunotherapy. Therefore, the use of specific antibiotics may be beneficial to patients. Antibiotics targeting specific microbial flora can regulate immune capacity. For example, depletion of bacteria sensitive to vancomycin enhanced tumor sensitivity to radiation therapy and significantly inhibited tumor growth. 271 , 272 Bacteriophages are viruses that can infect and kill bacteria, and they naturally exist in the microbiome where they play a key role in maintaining community balance. Phages can reduce environmental pathogenic bacteria; however, no phage is currently available for the treatment of clinical disease. 273
In recent years, oncolytic viruses have become an increasingly popular method of treating cancer. Adenovirus is the most potent and first virus used in oncolytic virotherapy. Several studies have recently indicated that other viruses can also be candidates for cancer therapies, such as herpes simplex virus (HSV) and measles virus. There has been a successful Phase III clinical trial for T‐VEC, an HSV‐based oncolytic virus approved by the United States Food and Drug Administration for use in biological cancer therapies. Preclinical and clinical trials have demonstrated impressive results for the vaccine strain of the measles virus. With their therapeutic efficacy, safety, and reduced side effects, such engineered viruses may represent a major breakthrough in cancer treatment. 274 Radiation‐induced antitumor immune responses are regulated by intestinal fungi in breast cancer and melanoma mouse models. 275
Microbiota intervention techniques, such as antibiotics, probiotics, and microbiota transplantation, have been used traditionally to boost cancer treatments’ efficacy. Moreover, new microbial treatments, such as nanotechnology, have been developed for antitumor therapy. A new approach for tumor therapy is provided by nanotechnology, which alters the microbiome's size and intervenes in microbiological molecules from a microscale perspective.
It is widely known that nanotechnology can be used to treat cancer. The first generation of nanotechnology has numerous advantages for clinical applications. These benefits include to deliver the drug targeted to the correct tissues, control its release, improve the permeability and solubility, improve the effectiveness, and decrease the toxicity of the drug. Second nanotechnology is in the process of being clinically tested. As a result of its current advantages, other functions of nanoscale particles have been enhanced, including targeting tissues, improved drug delivery, and stimulation of the immune system. There are more advantages to nanotechnology in the third generation, such as the regulation of the immune system and the ability to penetrate biological barriers. An anticancer particle based on nanotechnology could enhance the tumor microenvironment, promote local immunity cell proliferation, increase the ability of cancer microorganisms to penetrate tissues, and inhibit their growth. In addition to intervene microbiomes‐tumor microenvironment, Nanotechnology‐based anticancer particle can target cancer‐promoting metabolites secreting by microbiomes and improve locally hypoxic in the tumor microenvironment.
5. CONCLUSIONS AND PERSPECTIVE
The human microbiome is a complex network that interacts with the host in multiple body parts. In recent years, the role of the microbiome in disease development has become increasingly important, affecting the development, progression, and metastasis of many types of cancers (Tables 2 and 3).
TABLE 3.
Human bacteria with putative anticancer properties.
| Bacteria | cancer | Molecular mechanism |
|---|---|---|
| Lactobacillus | Colon cancer | Upregulated caspase‐3, caspase‐9, and Bax and down‐regulated Bcl2, cyclin D1, cyclin E, and ERBB2 genes. 26 , 269 |
| Bifidobacterium | Colon cancer | Induced the tumor suppressor miRNAs (miR‐145 and miR‐15a) expression. 26 , 269 |
| Clostridium butyricum | Colon cancer | Inhibited NF‐κB pathway and promoted apoptosis. 115 |
| Bacillus subtilis | Oral cancer | Inhibited PI3K/Akt/ NF‐κB and AP‐1/IL‐6 signaling pathways. 276 |
| Bifidobacteria | Colon cancer | Down‐regulated antiapoptotic genes and upregulated proapoptotic genes. 277 |
| Lactobacoilli | Cervical cancer | Upregulated E‐cadherin. 278 |
| Streptococcus thermophilus | Colorectal cancer | Secreted β‐galactosidase. 279 |
| Faecalibacterium prausnitzii | Colon cancer | Suppressed lipid peroxidation levels. 280 |
Recombinant Listeria monocytogenes vaccine showed a good inhibitory effect against tumor cell proliferation in mouse. As a kinds of phytotoxins vector, bacteria can harbor ricin, saporin, or pseudomonal exotoxins to suppress protein synthesis, induce cancer cells apopotosis. Investigators struggled with regulating microbiome to enhance cancer treatment response and eliminate toxic side effects of cancer therapy. Numerous studies indicated that interaction between external (diet, antigen exposure, drug, and mental stress) and host inherent factors can regulate cancer‐promoting microbioma and exert an antitumor effect. Fecal microbiota transplantation, probiotics, next‐generation biotherapeutics, designer microbial consortia, and targeting the tumor microbiota are commonly performed therapeutic for the management of cancer. However, the complex of the microbiomes promoting cancer mechanisms is still far from being completely understood. For example, there is a widely bidirectional feedback between microbiomes and neuroendocrine system. The microbiomes anticancer exact mechanism is unclear. Moreover, commercially available probiotic formulas generally is not clear. There are also differences in reactivity to tumor suppression. Commercially available probiotic can also have adverse effects such as diarrhea.
However, this is still a novel and young field. Many questions remain to open especially the exact anticancer mechanism of a certain microbioma. It is important to clarify the complex ecosystem in cancer development and is involved in many aspects of tumorigenesis from basic, epidemiological and clinical research. Consequently, it is imperative to understand how microorganisms and their secretions contribute to cancer occurrence and progression. Their advantages and limitations in the treatment of tumors could help to elucidate the mechanisms underlying tumor occurrence and progression. It will provide new strategies for the development of microbial‐related personalized drugs to improve treatment efficacy.
AUTHOR CONTRIBUTION
C. L. X., J. Y. S., C. L., Z. K. M., S. H. Y., and C. S. Y. collected the related reports. C. L. X. drafted the manuscript. J. Y. S., C. L., L. W. F., and C. L. X. revised the manuscript. L. W. F. and C. L. X. participated in designing the review. All authors read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ETHICS STATEMENT
Not applicable.
ACKNOWLEDGMENTS
We use biorender (https://biorender.com/) to create our figures. This work was financially supported by the National Natural Science Foundation of China (81902709).
Xia C, Su J, Liu C, et al. Human microbiomes in cancer development and therapy. MedComm. 2023;4:e221. 10.1002/mco2.221
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Grice EA, Segre JA. The human microbiome: our second genome. Annu Rev Genomics Hum Genet. 2012;13:151‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375(24):2369‐2379. [DOI] [PubMed] [Google Scholar]
- 3. Dominguez‐Bello MG, Godoy‐Vitorino F, Knight R, Blaser MJ. Role of the microbiome in human development. Gut. 2019;68(6):1108‐1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19(1):55‐71. [DOI] [PubMed] [Google Scholar]
- 5. Jeon YJ, Gil CH, Won J, Jo A, Kim HJ. Symbiotic microbiome Staphylococcus aureus from human nasal mucus modulates IL‐33‐mediated type 2 immune responses in allergic nasal mucosa. BMC Microbiol. 2020;20(1):301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hamilton SE, Griffith TS. A wild microbiome improves mouse modeling of the human immune response. Lab Anim (NY). 2019;48(11):337‐338. [DOI] [PubMed] [Google Scholar]
- 7. Lau K, Srivatsav V, Rizwan A, et al. Bridging the gap between gut microbial dysbiosis and cardiovascular diseases. Nutrients. 2017;9(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahn J, Hayes RB. Environmental influences on the human microbiome and implications for noncommunicable disease. Annu Rev Public Health. 2021;42:277‐292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rodricks J, Huang Y, Mantus E, Shubat P. Do interactions between environmental chemicals and the human microbiome need to be considered in risk assessments. Risk Anal. 2019;39(11):2353‐2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7‐33. [DOI] [PubMed] [Google Scholar]
- 11. Rajagopala SV, Vashee S, Oldfield LM, et al. The human microbiome and cancer. Cancer Prev Res (Phila). 2017;10(4):226‐234. [DOI] [PubMed] [Google Scholar]
- 12. Doocey CM, Finn K, Murphy C, Guinane CM. The impact of the human microbiome in tumorigenesis, cancer progression, and biotherapeutic development. BMC Microbiol. 2022;22(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Katongole P, Sande OJ, Joloba M, Reynolds SJ, Niyonzima N. The human microbiome and its link in prostate cancer risk and pathogenesis. Infect Agent Cancer. 2020;15:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Akbar S, Gu L, Sun Y, et al. Understanding host‐microbiome‐environment interactions: insights from Daphnia as a model organism. Sci Total Environ. 2022;808:152093. [DOI] [PubMed] [Google Scholar]
- 15. Bäckhed F. Host responses to the human microbiome. Nutr Rev. 2012;70(Suppl 1):S14‐17. [DOI] [PubMed] [Google Scholar]
- 16. Liu Q, Zhang Y, Xu C, et al. Dentists are at a higher risk for oral helicobacter pylori infection. Biomed Res Int. 2020;2020:3945189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knoll LJ, Hogan DA, Leong JM, Heitman J, Condit RC. Pearls collections: what we can learn about infectious disease and cancer. PLoS Pathog. 2018;14(3):e1006915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tokunaga K, Suzuki C, Hasegawa M, Fujimori I. Cost analysis in helicobacter pylori eradication therapy based on a database of health insurance claims in Japan. Clinicoecon Outcomes Res. 2021;13:241‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Piscione M, Mazzone M, Di Marcantonio MC, Muraro R, Mincione G. Eradication of helicobacter pylori and gastric cancer: a controversial relationship. Front Microbiol. 2021;12:630852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ito M, Tanaka S, Chayama K. Characteristics and early diagnosis of gastric cancer discovered after helicobacter pylori eradication. Gut Liver. 2021;15(3):338‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farrell PJ. Epstein‐barr virus and cancer. Annu Rev Pathol. 2019;14:29‐53. [DOI] [PubMed] [Google Scholar]
- 22. Jin C, Lagoudas GK, Zhao C, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell. 2019;176(5):998‐1013.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oliva M, Mulet‐Margalef N, Ochoa‐De‐Olza M, et al. Tumor‐associated microbiome: where do we stand. Int J Mol Sci. 2021;22(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lutfia A, Munir E, Yurnaliza Y, Basyuni M. Chemical analysis and anticancer activity of sesterterpenoid from an endophytic fungus Hypomontagnella monticulosa Zg15SU and its host Zingiber griffithii Baker. Heliyon. 2021;7(2):e06292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wen Y, Jin R, Chen H. Interactions between gut microbiota and acute childhood leukemia. Front Microbiol. 2019;10:1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Panebianco C, Andriulli A, Pazienza V. Pharmacomicrobiomics: exploiting the drug‐microbiota interactions in anticancer therapies. Microbiome. 2018;6(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Setlai BP, Mkhize‐Kwitshana ZL, Mehrotra R, Mulaudzi TV, Dlamini Z. Microbiomes, epigenomics, immune response, and splicing signatures interplay: potential use of combination of regulatory pathways as targets for malignant mesothelioma. Int J Mol Sci. 2022;23(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Woodhams DC, Bletz MC, Becker CG, et al. Host‐associated microbiomes are predicted by immune system complexity and climate. Genome Biol. 2020;21(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sambrani R, Abdolalizadeh J, Kohan L, Jafari B. Recent advances in the application of probiotic yeasts, particularly Saccharomyces, as an adjuvant therapy in the management of cancer with focus on colorectal cancer. Mol Biol Rep. 2021;48(1):951‐960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344‐355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guevarra LA Jr, Afable A, Belza P, et al. Immunogenicity of a Fap2 peptide mimotope of Fusobacterium nucleatum and its potential use in the diagnosis of colorectal cancer. Infect Agent Cancer. 2018;13:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gur C, Maalouf N, Shhadeh A, et al. Fusobacterium nucleatum supresses anti‐tumor immunity by activating CEACAM1. Oncoimmunology. 2019;8(6):e1581531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaakoush NO, Deshpande NP, Man SM, et al. Transcriptomic and proteomic analyses reveal key innate immune signatures in the host response to the gastrointestinal pathogen Campylobacter concisus. Infect Immun. 2015;83(2):832‐845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen J, Liu F, Lee SA, et al. Detection of IL‐18 and IL‐1β protein and mRNA in human oral epithelial cells induced by Campylobacter concisus strains. Biochem Biophys Res Commun. 2019;518(1):44‐49. [DOI] [PubMed] [Google Scholar]
- 35. Nagata N, Takeuchi T, Masuoka H, et al. Human gut microbiota and its metabolites impact immune responses in COVID‐19 and its complications. Gastroenterology. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abdulla MH, Agarwal D, Singh JK, et al. Association of the microbiome with colorectal cancer development (Review). Int J Oncol. 2021;58(5). [DOI] [PubMed] [Google Scholar]
- 37. Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. Dynamics of human gut microbiota and short‐chain fatty acids in response to dietary interventions with three fermentable fibers. mBio. 2019;10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goossens JF, Bailly C. Ursodeoxycholic acid and cancer: from chemoprevention to chemotherapy. Pharmacol Ther. 2019;203:107396. [DOI] [PubMed] [Google Scholar]
- 39. Zeng H, Umar S, Rust B, Lazarova D, Bordonaro M. Secondary bile acids and short chain fatty acids in the colon: a focus on colonic microbiome, cell proliferation, inflammation, and cancer. Int J Mol Sci. 2019;20(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fang Y, Yan C, Zhao Q, et al. The roles of microbial products in the development of colorectal cancer: a review. Bioengineered. 2021;12(1):720‐735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Muhealdeen DN, Shwan A, Yaqo RT, et al. Epstein‐Barr virus and Burkitt's lymphoma. Associations in Iraqi Kurdistan and twenty‐two countries assessed in the International Incidence of Childhood Cancer. Infect Agent Cancer. 2022;17(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bayda N, Tilloy V, Chaunavel A, et al. Comprehensive Epstein‐Barr virus transcriptome by RNA‐sequencing in angioimmunoblastic T cell lymphoma (AITL) and other lymphomas. Cancers (Basel). 2021;13(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vasudevan HN, Yom SS. Nasopharyngeal carcinoma and its association with Epstein‐Barr virus. Hematol Oncol Clin North Am. 2021;35(5):963‐971. [DOI] [PubMed] [Google Scholar]
- 44. Kanno‐Okada H, Takahashi K, Katano H, et al. A case of Epstein‐Barr virus‐associated lymphoepithelioma‐like carcinoma of the colon. Pathol Int. 2021;71(6):420‐426. [DOI] [PubMed] [Google Scholar]
- 45. Yang J, Liu Z, Zeng B, Hu G, Gan R. Epstein‐Barr virus‐associated gastric cancer: a distinct subtype. Cancer Lett. 2020;495:191‐199. [DOI] [PubMed] [Google Scholar]
- 46. Rizzo G, Cabibbo G, Craxì A. Hepatitis B virus‐associated hepatocellular carcinoma. Viruses. 2022;14(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li M, Du M, Cong H, et al. Characterization of hepatitis B virus DNA integration patterns in intrahepatic cholangiocarcinoma. Hepatol Res. 2021;51(1):102‐115. [DOI] [PubMed] [Google Scholar]
- 48. Li M, Shen Y, Chen Y, et al. Characterization of hepatitis B virus infection and viral DNA integration in non‐Hodgkin lymphoma. Int J Cancer. 2020;147(8):2199‐2209. [DOI] [PubMed] [Google Scholar]
- 49. Liu X, Zhang ZH, Jiang F. Hepatitis B virus infection increases the risk of pancreatic cancer: a meta‐analysis. Scand J Gastroenterol. 2021;56(3):252‐258. [DOI] [PubMed] [Google Scholar]
- 50. Mert D, Merdin A, Ceken S, Dal MS, Ertek M, Altuntas F. Evaluation of hepatitis B virus, hepatitis C virus, and human immunodeficiency virus seroprevalence in patients with diffuse large B cell lymphoma and Hodgkin's lymphoma. J Cancer Res Ther. 2021;17(4):951‐955. [DOI] [PubMed] [Google Scholar]
- 51. Khatun M, Ray R, Ray RB. Hepatitis C virus associated hepatocellular carcinoma. Adv Cancer Res. 2021;149:103‐142. [DOI] [PubMed] [Google Scholar]
- 52. Lam JO, Hurley LB, Lai JB, et al. Cancer in people with and without hepatitis C virus infection: comparison of risk before and after introduction of direct‐acting antivirals. Cancer Epidemiol Biomarkers Prev. 2021;30(12):2188‐2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rattotti S, Ferretti VV, Rusconi C, et al. Lymphomas associated with chronic hepatitis C virus infection: a prospective multicenter cohort study from the Rete Ematologica Lombarda (REL) clinical network. Hematol Oncol. 2019;37(2):160‐167. [DOI] [PubMed] [Google Scholar]
- 54. Wang H, Liu Y, Zhao Y. The association of hepatitis C virus infection and thyroid disease: a systematic review and meta‐analysis. Int J Biol Markers. 2021;36(4):3‐9. [DOI] [PubMed] [Google Scholar]
- 55. Lange P, Damania B. Kaposi sarcoma‐associated herpesvirus (KSHV). Trends Microbiol. 2020;28(3):236‐237. [DOI] [PubMed] [Google Scholar]
- 56. Iftode N, Rădulescu MA, Aramă ȘS, Aramă V. Update on Kaposi sarcoma‐associated herpesvirus (KSHV or HHV8) ‐ review. Rom J Intern Med. 2020;58(4):199‐208. [DOI] [PubMed] [Google Scholar]
- 57. Lidenge SJ, Tso FY, Ngalamika O, et al. Similar immunological profiles between African endemic and human immunodeficiency virus type 1‐associated epidemic kaposi sarcoma (KS) patients reveal the primary role of KS‐associated herpesvirus in KS pathogenesis. J Infect Dis. 2019;219(8):1318‐1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shmakova A, Germini D, Vassetzky Y. HIV‐1, HAART and cancer: a complex relationship. Int J Cancer. 2020;146(10):2666‐2679. [DOI] [PubMed] [Google Scholar]
- 59. Gavegnano C, Savarino A, Owanikoko T, Marconi VC. Crossroads of cancer and HIV‐1: pathways to a cure for HIV. Front Immunol. 2019;10:2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Latini A, Alei L, Magri F, et al. Nonmelanoma skin cancer and melanoma in HIV‐1‐infected patients. AIDS. 2020;34(10):1570‐1572. [DOI] [PubMed] [Google Scholar]
- 61. Vigano S, Bobisse S, Coukos G, Perreau M, Harari A. Cancer and HIV‐1 infection: patterns of chronic antigen exposure. Front Immunol. 2020;11:1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mulherkar TH, Gómez DJ, Sandel G, Jain P. Co‐infection and cancer: host‐pathogen interaction between dendritic cells and HIV‐1, HTLV‐1, and other oncogenic viruses. Viruses. 2022;14(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Morsica G, Galli L, Messina E, et al. Levels of alpha‐fetoprotein and association with mortality in hepatocellular carcinoma of HIV‐1‐infected patients. J Oncol. 2022;2022:3586064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Svensson JP. Targeting epigenetics to cure HIV‐1: lessons from (and for) cancer treatment. Front Cell Infect Microbiol. 2021;11:668637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Farisyi MA, Sufiawati I. Detection of Epstein‐Barr virus DNA in saliva of HIV‐1‐infected individuals with oral hairy leukoplakia. Oral Dis. 2020;26(Suppl 1):158‐160. [DOI] [PubMed] [Google Scholar]
- 66. Flores‐Miramontes MG, Olszewski D, Artaza‐Irigaray C, et al. Detection of alpha, beta, gamma, and unclassified human papillomaviruses in cervical cancer samples from Mexican women. Front Cell Infect Microbiol. 2020;10:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xu L, Selk A, Garland SM, et al. Prophylactic vaccination against human papillomaviruses to prevent vulval and vaginal cancer and their precursors. Expert Rev Vaccines. 2019;18(11):1157‐1166. [DOI] [PubMed] [Google Scholar]
- 68. Xia C, Li S, Long T, Chen Z, Chan P, Boon SS. Current updates on cancer‐causing types of human papillomaviruses (HPVs) in East, Southeast, and South Asia. Cancers (Basel). 2021;13(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gheit T, Rollo F, Brancaccio RN, et al. Oral infection by mucosal and cutaneous human papillomaviruses in the men who have sex with men from the OHMAR study. Viruses. 2020;12(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sias C, Salichos L, Lapa D, et al. Alpha, beta, gamma human papillomaviruses (HPV) detection with a different sets of primers in oropharyngeal swabs, anal and cervical samples. Virol J. 2019;16(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Galati L, Brancaccio RN, Robitaille A, et al. Detection of human papillomaviruses in paired healthy skin and actinic keratosis by next generation sequencing. Papillomavirus Res. 2020;9:100196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ramqvist T, Ortiz‐Villalon C, Brandén E, et al. Analysis of human papillomaviruses and human polyomaviruses in lung cancer from Swedish never‐smokers. Acta Oncol. 2020;59(1):28‐32. [DOI] [PubMed] [Google Scholar]
- 73. Gupta I, Jabeen A, Al‐Sarraf R, et al. The co‐presence of high‐risk human papillomaviruses and Epstein‐Barr virus is linked with tumor grade and stage in Qatari women with breast cancer. Hum Vaccin Immunother. 2021;17(4):982‐989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang Y, Zhang X, Wang H, Shen D. Stage IA1 HPV‐associated cervical squamous cell carcinoma metastasizing to ovary by special pathway: a case report and literature review. J Ovarian Res. 2022;15(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sadri Nahand J, Esghaei M, Hamidreza Monavari S, et al. The assessment of a possible link between HPV‐mediated inflammation, apoptosis, and angiogenesis in Prostate cancer. Int Immunopharmacol. 2020;88:106913. [DOI] [PubMed] [Google Scholar]
- 76. Ohadian Moghadam S, Mansori K, Nowroozi MR, Afshar D, Abbasi B, Nowroozi A. Association of human papilloma virus (HPV) infection with oncological outcomes in urothelial bladder cancer. Infect Agent Cancer. 2020;15:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Giam CZ. HTLV‐1 replication and adult T cell leukemia development. Recent Results Cancer Res. 2021;217:209‐243. [DOI] [PubMed] [Google Scholar]
- 78. Su C, Bai HX, Xiao R. HTLV‐1 status should be recorded in cases of T cell lymphomas/lymphoproliferative disorders ‐ cases of adult T cell leukaemia lymphoma masquerading as other T cell lymphomas/lymphoproliferative disorders could explain some of the apparent ethnic disparities ‐ response to Lo Bello and Naresh. Br J Haematol. 2019;185(2):327‐328. [DOI] [PubMed] [Google Scholar]
- 79. Shimizu Y, Arima K, Noguchi Y, et al. Possible mechanisms underlying the association between human T‐cell leukemia virus type 1 (HTLV‐1) and hypertension in elderly Japanese population. Environ Health Prev Med. 2021;26(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Loilome W, Dokduang H, Suksawat M, Padthaisong S. Therapeutic challenges at the preclinical level for targeted drug development for Opisthorchis viverrini‐associated cholangiocarcinoma. Expert Opin Investig Drugs. 2021;30(9):985‐1006. [DOI] [PubMed] [Google Scholar]
- 81. Mbanefo EC, Agbo CT, Zhao Y, et al. IPSE, an abundant egg‐secreted protein of the carcinogenic helminth Schistosoma haematobium, promotes proliferation of bladder cancer cells and angiogenesis. Infect Agent Cancer. 2020;15:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tetteh‐Quarcoo PB, Akuetteh BK, Owusu IA, et al. Cytological and wet mount microscopic observations made in urine of schistosoma haematobium‐infected children: hint of the implication in bladder cancer. Can J Infect Dis Med Microbiol. 2019;2019:7912186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gaber DA, Wassef RM, El‐Ayat WM, et al. Role of a schistosoma haematobium specific microRNA as a predictive and prognostic tool for bilharzial bladder cancer in Egypt. Sci Rep. 2020;10(1):18844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Alipour M. Molecular mechanism of helicobacter pylori‐induced gastric cancer. J Gastrointest Cancer. 2021;52(1):23‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Liou JM, Malfertheiner P, Lee YC, et al. Screening and eradication of Helicobacter pylori for gastric cancer prevention: the Taipei global consensus. Gut. 2020;69(12):2093‐2112. [DOI] [PubMed] [Google Scholar]
- 86. Holleczek B, Schöttker B, Brenner H. Helicobacter pylori infection, chronic atrophic gastritis and risk of stomach and esophagus cancer: results from the prospective population‐based ESTHER cohort study. Int J Cancer. 2020;146(10):2773‐2783. [DOI] [PubMed] [Google Scholar]
- 87. Kornerup LS, Jepsen P, Bartels LE, Dahlerup JF, Vilstrup H. Lower incidence of hepatobiliary cancer in helicobacter pylori‐infected persons: a Cohort study of 53.633 persons. J Clin Exp Hepatol. 2022;12(3):793‐799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cho J, Prashar A, Jones NL, Moss SF. Helicobacter pylori infection. Gastroenterol Clin North Am. 2021;50(2):261‐282. [DOI] [PubMed] [Google Scholar]
- 89. Kunovsky L, Dite P, Jabandziev P, et al. Helicobacter pylori infection and other bacteria in pancreatic cancer and autoimmune pancreatitis. World J Gastrointest Oncol. 2021;13(8):835‐844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Durazzo M, Adriani A, Fagoonee S, Saracco GM, Pellicano R. Helicobacter pylori and respiratory diseases: 2021 update. Microorganisms. 2021;9(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lu YT, Hsin CH, Lu YC, et al. Risk of head and neck cancer in patients with peptic ulcers and the effect of Helicobacter pylori treatment. Sci Rep. 2021;11(1):6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Larfors G, Richter J, Själander A, Stenke L, Höglund M. Increased risk of chronic myeloid leukemia following gastric conditions indicating helicobacter pylori infection: a case‐control study. Cancer Epidemiol Biomarkers Prev. 2020;29(1):151‐156. [DOI] [PubMed] [Google Scholar]
- 93. Xi Y, Yuefen P, Wei W, et al. Analysis of prognosis, genome, microbiome, and microbial metabolome in different sites of colorectal cancer. J Transl Med. 2019;17(1):353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Inamura K, Hamada T, Bullman S, Ugai T, Yachida S, Ogino S. Cancer as microenvironmental, systemic and environmental diseases: opportunity for transdisciplinary microbiomics science. Gut. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Olsen I, Yilmaz Ö. Possible role of Porphyromonas gingivalis in orodigestive cancers. J Oral Microbiol. 2019;11(1):1563410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sun L, Cai J, Gonzalez FJ. The role of farnesoid X receptor in metabolic diseases, and gastrointestinal and liver cancer. Nat Rev Gastroenterol Hepatol. 2021;18(5):335‐347. [DOI] [PubMed] [Google Scholar]
- 97. Yin J, Dong L, Zhao J, et al. Composition and consistence of the bacterial microbiome in upper, middle and lower esophagus before and after Lugol's iodine staining in the esophagus cancer screening. Scand J Gastroenterol. 2020;55(12):1467‐1474. [DOI] [PubMed] [Google Scholar]
- 98. Yu Y, Dunaway S, Champer J, Kim J, Alikhan A. Changing our microbiome: probiotics in dermatology. Br J Dermatol. 2020;182(1):39‐46. [DOI] [PubMed] [Google Scholar]
- 99. Minakata T, Nakano Y, Tamura S, et al. Tuberculous spondylitis caused by intravesical bacillus calmette‐guerin therapy. Intern Med. 2020;59(5):733‐737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bi H, Tian Y, Song C, et al. Urinary microbiota ‐ a potential biomarker and therapeutic target for bladder cancer. J Med Microbiol. 2019;68(10):1471‐1478. [DOI] [PubMed] [Google Scholar]
- 101. Jani K, McMillen T, Morjaria S, Babady NE. Performance of the sōna aspergillus galactomannan lateral flow assay in a cancer patient population. J Clin Microbiol. 2021;59(9):e0059821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Deng J, Yuan W, Tan Q, Wei X, Ma J. Non‐absorbable antibiotic treatment inhibits colorectal cancer liver metastasis by modulating deoxycholic acid metabolism by intestinal microbes. J Cancer. 2022;13(3):764‐774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tan X, Mao L, Huang C, et al. Comprehensive analysis of lncRNA‐miRNA‐mRNA regulatory networks for microbiota‐mediated colorectal cancer associated with immune cell infiltration. Bioengineered. 2021;12(1):3410‐3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Feng X, Han L, Ma S, et al. Microbes in tumoral in situ tissues and in tumorigenesis. Front Cell Infect Microbiol. 2020;10:572570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Erdman SE, Poutahidis T, Tomczak M, et al. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2‐deficient mice. Am J Pathol. 2003;162(2):691‐702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Swartz MA, Iida N, Roberts EW, et al. Tumor microenvironment complexity: emerging roles in cancer therapy. Cancer Res. 2012;72(10):2473‐2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zackular JP, Baxter NT, Iverson KD, et al. The gut microbiome modulates colon tumorigenesis. mBio. 2013;4(6):e00692‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zackular JP, Rogers MA, Ruffin MT 4th, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila). 2014;7(11):1112‐1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early‐stage detection of colorectal cancer. Mol Syst Biol. 2014;10(11):766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yu L, Zhao G, Wang L, et al. A systematic review of microbial markers for risk prediction of colorectal neoplasia. Br J Cancer. 2022;126(9):1318‐1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16(11):690‐704. [DOI] [PubMed] [Google Scholar]
- 112. Ternes D, Karta J, Tsenkova M, Wilmes P, Haan S, Letellier E. Microbiome in colorectal cancer: how to get from meta‐omics to mechanism. Trends Microbiol. 2020;28(5):401‐423. [DOI] [PubMed] [Google Scholar]
- 113. Ranjbar M, Salehi R, Haghjooy Javanmard S, et al. The dysbiosis signature of Fusobacterium nucleatum in colorectal cancer‐cause or consequences? A systematic review. Cancer Cell Int. 2021;21(1):194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Okumura S, Konishi Y, Narukawa M, et al. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat Commun. 2021;12(1):5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Chen D, Jin D, Huang S, et al. Clostridium butyricum, a butyrate‐producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 2020;469:456‐467. [DOI] [PubMed] [Google Scholar]
- 116. Ballard J, Towarnicki SG. Mitochondria, the gut microbiome and ROS. Cell Signal. 2020;75:109737. [DOI] [PubMed] [Google Scholar]
- 117. Li C, Gu Y, He Q, et al. Integrated analysis of microbiome and transcriptome data reveals the interplay between commensal bacteria and fibrin degradation in endometrial cancer. Front Cell Infect Microbiol. 2021;11:748558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ruuskanen MO, Vats D, Potbhare R, et al. Towards standardized and reproducible research in skin microbiomes. Environ Microbiol. 2022;24(9):3840‐3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Schreurs M, de Vos van Steenwijk PJ, Romano A, Dieleman S, Werner H. How the gut microbiome links to menopause and obesity, with possible implications for endometrial cancer development. J Clin Med. 2021;10(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kotakeyama Y, Nakamura R, Kurosawa M, et al. Development of a skin microbiome diagnostic method to assess skin condition in healthy individuals: application of research on skin microbiomes and skin condition. Int J Cosmet Sci. 2021;43(6):677‐690. [DOI] [PubMed] [Google Scholar]
- 121. Ren Z, Li A, Jiang J, et al. Gut microbiome analysis as a tool towards targeted non‐invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68(6):1014‐1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Mrázek J, Mekadim C, Kučerová P, et al. Melanoma‐related changes in skin microbiome. Folia Microbiol (Praha). 2019;64(3):435‐442. [DOI] [PubMed] [Google Scholar]
- 123. Fu C, Yang Z, Yu J, Wei M. The interaction between gut microbiome and anti‐tumor drug therapy. Am J Cancer Res. 2021;11(12):5812‐5832. [PMC free article] [PubMed] [Google Scholar]
- 124. Chung M, Zhao N, Meier R, et al. Comparisons of oral, intestinal, and pancreatic bacterial microbiomes in patients with pancreatic cancer and other gastrointestinal diseases. J Oral Microbiol. 2021;13(1):1887680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Gugnacki P, Sierko E. Is there an interplay between oral microbiome, head and neck carcinoma and radiation‐induced oral mucositis. Cancers (Basel). 2021;13(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Lee WH, Chen HM, Yang SF, et al. Bacterial alterations in salivary microbiota and their association in oral cancer. Sci Rep. 2017;7(1):16540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zhao H, Chu M, Huang Z, et al. Variations in oral microbiota associated with oral cancer. Sci Rep. 2017;7(1):11773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Di Pilato V, Freschi G, Ringressi MN, Pallecchi L, Rossolini GM, Bechi P. The esophageal microbiota in health and disease. Ann N Y Acad Sci. 2016;1381(1):21‐33. [DOI] [PubMed] [Google Scholar]
- 129. Gillespie MR, Rai V, Agrawal S, Nandipati KC. The role of microbiota in the pathogenesis of esophageal adenocarcinoma. Biology (Basel). 2021;10(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Dan W, Peng L, Yan B, Li Z, Pan F. Human microbiota in esophageal adenocarcinoma: pathogenesis, diagnosis, prognosis and therapeutic implications. Front Microbiol. 2021;12:791274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zhou J, Sun S, Luan S, et al. Gut microbiota for esophageal cancer: role in carcinogenesis and clinical implications. Front Oncol. 2021;11:717242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Chen C, Chen L, Lin L, Jin D, Du Y, Lyu J. Research progress on gut microbiota in patients with gastric cancer, esophageal cancer, and small intestine cancer. Appl Microbiol Biotechnol. 2021;105(11):4415‐4425. [DOI] [PubMed] [Google Scholar]
- 133. Kaakoush NO, Lecomte V, Maloney CA, Morris MJ. Cross‐talk among metabolic parameters, esophageal microbiota, and host gene expression following chronic exposure to an obesogenic diet. Sci Rep. 2017;7:45753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Cheng ES, Weber M, Steinberg J, Yu XQ. Lung cancer risk in never‐smokers: an overview of environmental and genetic factors. Chin J Cancer Res. 2021;33(5):548‐562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Shah DR, Masters GA. Precision medicine in lung cancer treatment. Surg Oncol Clin N Am. 2020;29(1):15‐21. [DOI] [PubMed] [Google Scholar]
- 136. Georgiou K, Marinov B, Farooqi AA, Gazouli M. Gut microbiota in lung cancer: where do we stand. Int J Mol Sci. 2021;22(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Najafi S, Abedini F, Azimzadeh Jamalkandi S, Shariati P, Ahmadi A, Gholami Fesharaki M. The composition of lung microbiome in lung cancer: a systematic review and meta‐analysis. BMC Microbiol. 2021;21(1):315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Lee YP, Jeong BH, Eun Y, et al. Extrapulmonary tuberculosis in patients with RET fusion‐positive non‐small cell lung cancer treated with pralsetinib: a Korean single‐centre compassionate use experience. Eur J Cancer. 2021;159:167‐173. [DOI] [PubMed] [Google Scholar]
- 139. Boesch M, Baty F, Albrich WC, et al. Local tumor microbial signatures and response to checkpoint blockade in non‐small cell lung cancer. Oncoimmunology. 2021;10(1):1988403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Goto T. Airway microbiota as a modulator of lung cancer. Int J Mol Sci. 2020;21(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Zhuang H, Cheng L, Wang Y, et al. Dysbiosis of the gut microbiome in lung cancer. Front Cell Infect Microbiol. 2019;9:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Markowski MC, Boorjian SA, Burton JP, et al. The microbiome and genitourinary cancer: a collaborative review. Eur Urol. 2019;75(4):637‐646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Nicolaro M, Portal DE, Shinder B, Patel HV, Singer EA. The human microbiome and genitourinary malignancies. Ann Transl Med. 2020;8(19):1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Taghinezhad‐S S, Keyvani H, Bermúdez‐Humarán LG, Donders G, Fu X, Mohseni AH. Twenty years of research on HPV vaccines based on genetically modified lactic acid bacteria: an overview on the gut‐vagina axis. Cell Mol Life Sci. 2021;78(4):1191‐1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Tajdozian H, Seo H, Kim S, Rahim MA, Lee S, Song HY. Efficacy of lactobacillus fermentum isolated from the vagina of a healthy woman against carbapenem‐resistant klebsiella infections in vivo. J Microbiol Biotechnol. 2021;31(10):1383‐1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Nami Y, Abdullah N, Haghshenas B, Radiah D, Rosli R, Yari Khosroushahi A. A newly isolated probiotic Enterococcus faecalis strain from vagina microbiota enhances apoptosis of human cancer cells. J Appl Microbiol. 2014;117(2):498‐508. [DOI] [PubMed] [Google Scholar]
- 147. Castanheira CP, Sallas ML, Nunes R, Lorenzi N, Termini L. Microbiome and cervical cancer. Pathobiology. 2021;88(2):187‐197. [DOI] [PubMed] [Google Scholar]
- 148. Brooks RA, Fleming GF, Lastra RR, et al. Current recommendations and recent progress in endometrial cancer. CA Cancer J Clin. 2019;69(4):258‐279. [DOI] [PubMed] [Google Scholar]
- 149. Boutriq S, González‐González A, Plaza‐Andrades I, et al. Gut and endometrial microbiome dysbiosis: a new emergent risk factor for endometrial cancer. J Pers Med. 2021;11(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Łaniewski P, Herbst‐Kralovetz MM. Bacterial vaginosis and health‐associated bacteria modulate the immunometabolic landscape in 3D model of human cervix. NPJ Biofilms Microbiomes. 2021;7(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Morikawa A, Kawabata A, Shirahige K, Akiyama T, Okamoto A, Sutani T. Altered cervicovaginal microbiota in premenopausal ovarian cancer patients. Gene. 2022;811:146083. [DOI] [PubMed] [Google Scholar]
- 152. Xu J, Peng JJ, Yang W, Fu K, Zhang Y. Vaginal microbiomes and ovarian cancer: a review. Am J Cancer Res. 2020;10(3):743‐756. [PMC free article] [PubMed] [Google Scholar]
- 153. Bilski K, Dobruch J, Kozikowski M, Skrzypczyk MA, Oszczudłowski M, Ostrowski J. Urobiome in gender‐related diversities of bladder cancer. Int J Mol Sci. 2020;21(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Bajic P, Wolfe AJ, Gupta GN. The urinary microbiome: implications in bladder cancer pathogenesis and therapeutics. Urology. 2019;126:10‐15. [DOI] [PubMed] [Google Scholar]
- 155. Ohadian Moghadam S, Momeni SA. Human microbiome and prostate cancer development: current insights into the prevention and treatment. Front Med. 2021;15(1):11‐32. [DOI] [PubMed] [Google Scholar]
- 156. Wheeler KM, Liss MA. The microbiome and prostate cancer risk. Curr Urol Rep. 2019;20(10):66. [DOI] [PubMed] [Google Scholar]
- 157. Polakowski CB, Kato M, Preti VB, Schieferdecker M, Ligocki Campos AC. Impact of the preoperative use of synbiotics in colorectal cancer patients: a prospective, randomized, double‐blind, placebo‐controlled study. Nutrition. 2019;58:40‐46. [DOI] [PubMed] [Google Scholar]
- 158. Girardi B, Principi M, Pricci M, et al. Chemoprevention of inflammation‐related colorectal cancer by silymarin‐, acetyl‐11‐keto‐beta‐boswellic acid‐, curcumin‐ and maltodextrin‐enriched dietetic formulation in animal model. Carcinogenesis. 2018;39(10):1274‐1282. [DOI] [PubMed] [Google Scholar]
- 159. Ortiz A, Sansinenea E. Macrolactin antibiotics: amazing natural products. Mini Rev Med Chem. 2020;20(7):584‐600. [DOI] [PubMed] [Google Scholar]
- 160. Curth HM, Pelc A, Kütting F, Steffen HM. Augmented renal vancomycin clearance in cancer patients: a case report and review of the literature. Oncol Res Treat. 2015;38(4):182‐184. [DOI] [PubMed] [Google Scholar]
- 161. Veeranarayanan S, Azam AH, Kiga K, Watanabe S, Cui L. Bacteriophages as solid tumor theragnostic agents. Int J Mol Sci. 2021;23(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Fan J, Bellon M, Ju M, et al. Clinical significance of FBXW7 loss of function in human cancers. Mol Cancer. 2022;21(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. 2018;17(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Yeh CH, Bellon M, Wang F, Zhang H, Fu L, Nicot C. Loss of FBXW7‐mediated degradation of BRAF elicits resistance to BET inhibitors in adult T cell leukemia cells. Mol Cancer. 2020;19(1):139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Wang J, Wang T, Bishop MA, et al. Collaborating genomic, transcriptomic and microbiomic alterations lead to canine extreme intestinal polyposis. Oncotarget. 2018;9(49):29162‐29179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Cheng W, Zheng T, Wang Y, et al. Activation of Notch1 signaling by HTLV‐1 Tax promotes proliferation of adult T‐cell leukemia cells. Biochem Biophys Res Commun. 2019;512(3):598‐603. [DOI] [PubMed] [Google Scholar]
- 167. Bai XT, Yeh CH, Nicot C. NOTCH1 activation depletes the pool of side population stem cells in ATL. J Cancer Sci. 2017;4(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Song NJ, Yun UJ, Yang S, et al. Notch1 deficiency decreases hepatic lipid accumulation by induction of fatty acid oxidation. Sci Rep. 2016;6:19377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Bellon M, Moles R, Chaib‐Mezrag H, Pancewicz J, Nicot C. JAG1 overexpression contributes to Notch1 signaling and the migration of HTLV‐1‐transformed ATL cells. J Hematol Oncol. 2018;11(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Becker F, Gavins F, Fontenot J, et al. Dynamic gut microbiome changes following regional intestinal lymphatic obstruction in primates. Pathophysiology. 2019;26(3‐4):253‐261. [DOI] [PubMed] [Google Scholar]
- 171. Tanaka T, Matsuno Y, Torisu T, et al. Gastric microbiota in patients with Helicobacter pylori‐negative gastric MALT lymphoma. Medicine (Baltimore). 2021;100(38):e27287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Zeze K, Hirano A, Torisu T, et al. Mucosal dysbiosis in patients with gastrointestinal follicular lymphoma. Hematol Oncol. 2020;38(2):181‐188. [DOI] [PubMed] [Google Scholar]
- 173. Wong L, Bozhilov K, Hernandez B, et al. Underlying liver disease and advanced stage liver cancer are associated with elevated neutrophil‐lymphocyte ratio. Clin Mol Hepatol. 2019;25(3):305‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Dong R, Zhang B, Zhang X. Liver organoids: an in vitro 3D model for liver cancer study. Cell Biosci. 2022;12(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Liao Y, Hu X, Chen J, et al. Precore mutation of hepatitis B virus may contribute to hepatocellular carcinoma risk: evidence from an updated meta‐analysis. PLoS One. 2012;7(6):e38394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Zhuo Z, Rong W, Li H, et al. Long‐read sequencing reveals the structural complexity of genomic integration of HBV DNA in hepatocellular carcinoma. NPJ Genom Med. 2021;6(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Xu J, Xiao X, Yan B, et al. Green tea‐derived theabrownin induces cellular senescence and apoptosis of hepatocellular carcinoma through p53 signaling activation and bypassed JNK signaling suppression. Cancer Cell Int. 2022;22(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Lun YZ, Cheng J, Chi Q, Wang XL, Gao M, Sun LD. Transactivation of proto‐oncogene c‐Myc by hepatitis B virus transactivator MHBs(t167). Oncol Lett. 2014;8(2):803‐808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Tsai MC, Huang CC, Wei YC, et al. Combined chibby and β‐catenin predicts clinical outcomes in patients with hepatocellular carcinoma. Int J Mol Sci. 2020;21(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Nagornykh AM, Tyumentseva MA, Tyumentsev AI, Akimkin VG. Protocol for chronic hepatitis B virus infection mouse model development by patient‐derived orthotopic xenografts. PLoS One. 2022;17(2):e0264266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Li W, Cui X, Huo Q, et al. Profile of HBV integration in the plasma DNA of hepatocellular carcinoma patients. Curr Genomics. 2019;20(1):61‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Roden R, Stern PL. Opportunities and challenges for human papillomavirus vaccination in cancer. Nat Rev Cancer. 2018;18(4):240‐254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Canfell K, Kim JJ, Brisson M, et al. Mortality impact of achieving WHO cervical cancer elimination targets: a comparative modelling analysis in 78 low‐income and lower‐middle‐income countries. Lancet. 2020;395(10224):591‐603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Łaniewski P, Ilhan ZE, Herbst‐Kralovetz MM. The microbiome and gynaecological cancer development, prevention and therapy. Nat Rev Urol. 2020;17(4):232‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Lo Cigno I, Calati F, Albertini S, Gariglio M. Subversion of host innate immunity by human papillomavirus oncoproteins. Pathogens. 2020;9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Cao M, Wang Y, Wang D, et al. Increased high‐risk human papillomavirus viral load is associated with immunosuppressed microenvironment and predicts a worse long‐term survival in cervical cancer patients. Am J Clin Pathol. 2020;153(4):502‐512. [DOI] [PubMed] [Google Scholar]
- 187. Yuan Y, Cai X, Shen F, Ma F. HPV post‐infection microenvironment and cervical cancer. Cancer Lett. 2021;497:243‐254. [DOI] [PubMed] [Google Scholar]
- 188. Yuan L, Li S, Chen Q, et al. EBV infection‐induced GPX4 promotes chemoresistance and tumor progression in nasopharyngeal carcinoma. Cell Death Differ. 2022;29(8):1513‐1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Wang HY, Chang YL, To KF, et al. A new prognostic histopathologic classification of nasopharyngeal carcinoma. Chin J Cancer. 2016;35:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Chen YP, Chan A, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394(10192):64‐80. [DOI] [PubMed] [Google Scholar]
- 191. Kang D, Skalsky RL, Cullen BR. EBV BART MicroRNAs target multiple pro‐apoptotic cellular genes to promote epithelial cell survival. PLoS Pathog. 2015;11(6):e1004979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Tsang CM, Lui V, Bruce JP, Pugh TJ, Lo KW. Translational genomics of nasopharyngeal cancer. Semin Cancer Biol. 2020;61:84‐100. [DOI] [PubMed] [Google Scholar]
- 193. Wang H, Zhang K, Wu L, Qin Q, He Y. Prediction of pathogenic factors in dysbiotic gut microbiomes of colorectal cancer patients using reverse microbiomics. Front Oncol. 2022;12:882874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Imai J, Kitamoto S, Kamada N. The pathogenic oral‐gut‐liver axis: new understandings and clinical implications. Expert Rev Clin Immunol. 2021;17(7):727‐736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Krueger A, Zaugg J, Chisholm S, et al. Secreted toxins from staphylococcus aureus strains isolated from keratinocyte skin cancers mediate pro‐tumorigenic inflammatory responses in the skin. Front Microbiol. 2021;12:789042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Nemattalab M, Shenagari M, Taheri M, et al. Co‐expression of interleukin‐17A molecular adjuvant and prophylactic Helicobacter pylori genetic vaccine could cause sterile immunity in Treg suppressed mice. Cytokine. 2020;126:154866. [DOI] [PubMed] [Google Scholar]
- 197. Sanaii A, Shirzad H, Haghighian M, et al. Role of Th22 cells in Helicobacter pylori‐related gastritis and peptic ulcer diseases. Mol Biol Rep. 2019;46(6):5703‐5712. [DOI] [PubMed] [Google Scholar]
- 198. Jost M, Wehkamp U. The skin microbiome and influencing elements in cutaneous T‐cell lymphomas. Cancers (Basel). 2022;14(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Jeon JI, Lee KH, Kim JM. Bacteroides fragilis enterotoxin upregulates matrix metalloproteinase‐7 expression through MAPK and AP‐1 activation in intestinal epithelial cells, leading to syndecan‐2 release. Int J Mol Sci. 2021;22(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Arthur JC, Perez‐Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer‐inducing activity of the microbiota. Science. 2012;338(6103):120‐123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Arima K, Zhong R, Ugai T, et al. Western‐style diet, pks Island‐carrying Escherichia coli, and colorectal cancer: analyses from two large prospective cohort studies. Gastroenterology. 2022;163(4):862‐874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Wilson MR, Jiang Y, Villalta PW, et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science. 2019;363(6428). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Akkız H. The gut microbiome and hepatocellular carcinoma. J Gastrointest Cancer. 2021;52(4):1314‐1319. [DOI] [PubMed] [Google Scholar]
- 204. Wu Y, Wu J, Chen T, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis in mice via a toll‐like receptor 4/p21‐activated kinase 1 cascade. Dig Dis Sci. 2018;63(5):1210‐1218. [DOI] [PubMed] [Google Scholar]
- 205. Queen J, Domingue JC, White JR, et al. Comparative analysis of colon cancer‐derived fusobacterium nucleatum subspecies: inflammation and colon tumorigenesis in murine models. mBio. 2022;13(1):e0299121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Perera M, Al‐Hebshi NN, Speicher DJ, Perera I, Johnson NW. Emerging role of bacteria in oral carcinogenesis: a review with special reference to perio‐pathogenic bacteria. J Oral Microbiol. 2016;8:32762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Kim J, Lee HK. Potential role of the gut microbiome in colorectal cancer progression. Front Immunol. 2021;12:807648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Chattopadhyay I, Verma M, Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol Cancer Res Treat. 2019;18:1533033819867354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Elmaghrawy K, Hussey S, Moran GP. The oral microbiome in pediatric IBD: a source of pathobionts or biomarkers. Front Pediatr. 2020;8:620254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Yin C, Chen J, Wu X, et al. Preterm birth is correlated with increased oral originated microbiome in the gut. Front Cell Infect Microbiol. 2021;11:579766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Sami A, Elimairi I, Stanton C, Ross RP, Ryan CA. The role of the microbiome in oral squamous cell carcinoma with insight into the microbiome‐treatment axis. Int J Mol Sci. 2020;21(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Degenhardt R, Sobral Marques Souza D, Acordi Menezes LA, et al. Detection of enteric viruses and core microbiome analysis in artisanal colonial salami‐type dry‐fermented sausages from Santa Catarina, Brazil. Foods. 2021;10(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Yamamura K, Baba Y, Nakagawa S, et al. Human microbiome fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res. 2016;22(22):5574‐5581. [DOI] [PubMed] [Google Scholar]
- 214. Somineni HK, Weitzner JH, Venkateswaran S, et al. Site‐ and taxa‐specific disease‐associated oral microbial structures distinguish inflammatory bowel diseases. Inflamm Bowel Dis. 2021;27(12):1889‐1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Vuppala S, Kim J, Joo BS, Choi JM, Jang J. A combination of pharmacophore‐based virtual screening, structure‐based lead optimization, and DFT study for the identification of S. epidermidis TcaR inhibitors. Pharmaceuticals (Basel). 2022;15(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Gopinath D, Kunnath Menon R, Veettil SK, George Botelho M, Johnson NW. Periodontal diseases as putative risk factors for head and neck cancer: systematic review and meta‐analysis. Cancers (Basel). 2020;12(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Migliorati CA. Periodontal diseases and cancer. Lancet Oncol. 2008;9(6):510‐512. [DOI] [PubMed] [Google Scholar]
- 218. Ray K. Gut microbiota: oral microbiome could provide clues to CRC. Nat Rev Gastroenterol Hepatol. 2017;14(12):690. [DOI] [PubMed] [Google Scholar]
- 219. Atanasova KR, Yilmaz O. Looking in the Porphyromonas gingivalis cabinet of curiosities: the microbium, the host and cancer association. Mol Oral Microbiol. 2014;29(2):55‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Epeldegui M, Hussain SK. The role of microbial translocation and immune activation in AIDS‐associated non‐hodgkin lymphoma pathogenesis: what have we learned. Crit Rev Immunol. 2020;40(1):41‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Talotta R, Sarzi‐Puttini P, Atzeni F. Microbial agents as putative inducers of B cell lymphoma in Sjögren's syndrome through an impaired epigenetic control: the state‐of‐the‐art. J Immunol Res. 2019;2019:8567364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Loke P, Lee SC, Oyesola OO. Effects of helminths on the human immune response and the microbiome. Mucosal Immunol. 2022;15(6):1224‐1233. [DOI] [PubMed] [Google Scholar]
- 223. Sharon I, Quijada NM, Pasolli E, et al. The core human microbiome: does it exist and how can we find it? A critical review of the concept. Nutrients. 2022;14(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Licht P, Mailänder V. Transcriptional heterogeneity and the microbiome of cutaneous T‐cell lymphoma. Cells. 2022;11(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Blümel E, Munir Ahmad S, Nastasi C, et al. Staphylococcus aureus alpha‐toxin inhibits CD8(+) T cell‐mediated killing of cancer cells in cutaneous T‐cell lymphoma. Oncoimmunology. 2020;9(1):1751561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Willerslev‐Olsen A, Buus TB, Nastasi C, et al. Staphylococcus aureus enterotoxins induce FOXP3 in neoplastic T cells in Sézary syndrome. Blood Cancer J. 2020;10(5):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Coker OO, Nakatsu G, Dai RZ, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 2019;68(4):654‐662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Khodaverdi N, Zeighami H, Jalilvand A, Haghi F, Hesami N. High frequency of enterotoxigenic Bacteroides fragilis and Enterococcus faecalis in the paraffin‐embedded tissues of Iranian colorectal cancer patients. BMC Cancer. 2021;21(1):1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Long X, Wong CC, Tong L, et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol. 2019;4(12):2319‐2330. [DOI] [PubMed] [Google Scholar]
- 230. Tsoi H, Chu E, Zhang X, et al. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology. 2017;152(6):1419‐1433.e5. [DOI] [PubMed] [Google Scholar]
- 231. Sheng X, Zuo X, Liu X, Zhou Y, Sun X. Crosstalk between TLR4 and Notch1 signaling in the IgA nephropathy during inflammatory response. Int Urol Nephrol. 2018;50(4):779‐785. [DOI] [PubMed] [Google Scholar]
- 232. Zhang J, Xia Y, Sun J. Breast and gut microbiome in health and cancer. Genes Dis. 2021;8(5):581‐589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Ingman WV. The gut microbiome: a new player in breast cancer metastasis. Cancer Res. 2019;79(14):3539‐3541. [DOI] [PubMed] [Google Scholar]
- 234. Vostrikova SM, Grinev AB, Gogvadze VG. Reactive oxygen species and antioxidants in carcinogenesis and tumor therapy. Biochemistry (Mosc). 2020;85(10):1254‐1266. [DOI] [PubMed] [Google Scholar]
- 235. Audirac‐Chalifour A, Torres‐Poveda K, Bahena‐Román M, et al. Cervical microbiome and cytokine profile at various stages of cervical cancer: a pilot study. PLoS One. 2016;11(4):e0153274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Zhang Y, Zhang L, Zheng S, et al. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF‐κB/ICAM1 axis. Gut Microbes. 2022;14(1):2038852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Gu Y, Raja V, Lee HM, Hong H, Prestwich G, Ryan ME. Therapeutic potential of a novel semi‐synthetic‐sulfated‐polysaccharide to suppress inflammatory mediators in P. gingivalis LPS stimulated human monocytes/macrophages. J Inflamm (Lond). 2021;18(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Parvez MM, Basit A, Jariwala PB, et al. Quantitative investigation of irinotecan metabolism, transport, and gut microbiome activation. Drug Metab Dispos. 2021;49(8):683‐693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Propper DJ, Balkwill FR. Harnessing cytokines and chemokines for cancer therapy. Nat Rev Clin Oncol. 2022;19(4):237‐253. [DOI] [PubMed] [Google Scholar]
- 240. Ozga AJ, Chow MT, Luster AD. Chemokines and the immune response to cancer. Immunity. 2021;54(5):859‐874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Falvo P, Orecchioni S, Hillje R, et al. Cyclophosphamide and vinorelbine activate stem‐like CD8(+) T cells and improve anti‐PD‐1 efficacy in triple‐negative breast cancer. Cancer Res. 2021;81(3):685‐697. [DOI] [PubMed] [Google Scholar]
- 242. Cheng WT, Kantilal HK, Davamani F. The mechanism of bacteroides fragilis toxin contributes to colon cancer formation. Malays J Med Sci. 2020;27(4):9‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Pietroiusti A, Magrini A, Campagnolo L. New frontiers in nanotoxicology: gut microbiota/microbiome‐mediated effects of engineered nanomaterials. Toxicol Appl Pharmacol. 2016;299:90‐95. [DOI] [PubMed] [Google Scholar]
- 244. Nagao T, Nakayama‐Imaohji H, Elahi M, et al. L‑histidine augments the oxidative damage against Gram‑negative bacteria by hydrogen peroxide. Int J Mol Med. 2018;41(5):2847‐2854. [DOI] [PubMed] [Google Scholar]
- 245. Wang Y, Malkmes MJ, Jiang C, et al. Antibacterial mechanism and transcriptome analysis of ultra‐small gold nanoclusters as an alternative of harmful antibiotics against Gram‐negative bacteria. J Hazard Mater. 2021;416:126236. [DOI] [PubMed] [Google Scholar]
- 246. Lewis ZT, Totten SM, Smilowitz JT, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Fransen F, van Beek AA, Borghuis T, et al. Aged gut microbiota contributes to systemical inflammaging after transfer to germ‐free mice. Front Immunol. 2017;8:1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Cheung LS, Fu J, Kumar P, et al. Second‐generation IL‐2 receptor‐targeted diphtheria fusion toxin exhibits antitumor activity and synergy with anti‐PD‐1 in melanoma. Proc Natl Acad Sci U S A. 2019;116(8):3100‐3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Donovan C, Liu G, Shen S, et al. The role of the microbiome and the NLRP3 inflammasome in the gut and lung. J Leukoc Biol. 2020;108(3):925‐935. [DOI] [PubMed] [Google Scholar]
- 250. Hakozaki T, Richard C, Elkrief A, et al. The gut microbiome associates with immune checkpoint inhibition outcomes in patients with advanced non‐small cell lung cancer. Cancer Immunol Res. 2020;8(10):1243‐1250. [DOI] [PubMed] [Google Scholar]
- 251. Chioma OS, Hesse LE, Chapman A, Drake WP. Role of the microbiome in interstitial lung diseases. Front Med (Lausanne). 2021;8:595522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Li H, Chen X, Zhou SJ. Dauricine combined with clindamycin inhibits severe pneumonia co‐infected by influenza virus H5N1 and Streptococcus pneumoniae in vitro and in vivo through NF‐κB signaling pathway. J Pharmacol Sci. 2018;137(1):12‐19. [DOI] [PubMed] [Google Scholar]
- 253. Wang L, Zhang X, Wu G, et al. Streptococcus pneumoniae aminopeptidase N contributes to bacterial virulence and elicits a strong innate immune response through MAPK and PI3K/AKT signaling. J Microbiol. 2020;58(4):330‐339. [DOI] [PubMed] [Google Scholar]
- 254. Zitvogel L, Daillère R, Roberti MP, Routy B, Kroemer G. Anticancer effects of the microbiome and its products. Nat Rev Microbiol. 2017;15(8):465‐478. [DOI] [PubMed] [Google Scholar]
- 255. Lee SH, Cho SY, Yoon Y, et al. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat Microbiol. 2021;6(3):277‐288. [DOI] [PubMed] [Google Scholar]
- 256. Li X, Zhang S, Guo G, Han J, Yu J. Gut microbiome in modulating immune checkpoint inhibitors. EBioMedicine. 2022;82:104163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Crump C, Sundquist J, Sieh W, Winkleby MA, Sundquist K. Fetal growth and subsequent maternal risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2015;24(8):1184‐1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Delaune V, Orci LA, Lacotte S, et al. Fecal microbiota transplantation: a promising strategy in preventing the progression of non‐alcoholic steatohepatitis and improving the anti‐cancer immune response. Expert Opin Biol Ther. 2018;18(10):1061‐1071. [DOI] [PubMed] [Google Scholar]
- 259. Chen Z, Qian X, Chen S, Fu X, Ma G, Zhang A. Akkermansia muciniphila enhances the antitumor effect of cisplatin in lewis lung cancer mice. J Immunol Res. 2020;2020:2969287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Prasetya RA, Metselaar‐Albers M, Engels F. Concomitant use of analgesics and immune checkpoint inhibitors in non‐small cell lung cancer: a pharmacodynamics perspective. Eur J Pharmacol. 2021;906:174284. [DOI] [PubMed] [Google Scholar]
- 261. Zhang T, Li Q, Cheng L, Buch H, Zhang F. Akkermansia muciniphila is a promising probiotic. Microb Biotechnol. 2019;12(6):1109‐1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti‐PD‐1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD‐1‐based immunotherapy against epithelial tumors. Science. 2018;359(6371):91‐97. [DOI] [PubMed] [Google Scholar]
- 264. Zhai Q, Feng S, Arjan N, Chen W. A next generation probiotic, Akkermansia muciniphila. Crit Rev Food Sci Nutr. 2019;59(19):3227‐3236. [DOI] [PubMed] [Google Scholar]
- 265. Gharaibeh RZ, Jobin C. Microbiota and cancer immunotherapy: in search of microbial signals. Gut. 2019;68(3):385‐388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Wu X, Zhang T, Chen X, Ji G, Zhang F. Microbiota transplantation: targeting cancer treatment. Cancer Lett. 2019;452:144‐151. [DOI] [PubMed] [Google Scholar]
- 267. Wang J, Chen L, Zhao N, Xu X, Xu Y, Zhu B. Of genes and microbes: solving the intricacies in host genomes. Protein Cell. 2018;9(5):446‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Xie X, He Y, Li H, et al. Effects of prebiotics on immunologic indicators and intestinal microbiota structure in perioperative colorectal cancer patients. Nutrition. 2019;61:132‐142. [DOI] [PubMed] [Google Scholar]
- 269. De Pietri S, Ingham AC, Frandsen TL, et al. Gastrointestinal toxicity during induction treatment for childhood acute lymphoblastic leukemia: the impact of the gut microbiota. Int J Cancer. 2020;147(7):1953‐1962. [DOI] [PubMed] [Google Scholar]
- 270. Galloway‐Peña JR, Shi Y, Peterson CB, et al. Gut microbiome signatures are predictive of infectious risk following induction therapy for acute myeloid leukemia. Clin Infect Dis. 2020;71(1):63‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Masetti R, Zama D, Leardini D, et al. Microbiome‐derived metabolites in allogeneic hematopoietic stem cell transplantation. Int J Mol Sci. 2021;22(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Vicente‐Dueñas C, Janssen S, Oldenburg M, et al. An intact gut microbiome protects genetically predisposed mice against leukemia. Blood. 2020;136(18):2003‐2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Hakim H, Dallas R, Wolf J, et al. Gut microbiome composition predicts infection risk during chemotherapy in children with acute lymphoblastic leukemia. Clin Infect Dis. 2018;67(4):541‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Mondal M, Guo J, He P, Zhou D. Recent advances of oncolytic virus in cancer therapy. Hum Vaccin Immunother. 2020;16(10):2389‐2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Shiao SL, Kershaw KM, Limon JJ, et al. Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy. Cancer Cell. 2021;39(9):1202‐1213.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Vo T, Lee CW, Wu CZ, et al. Surfactin from Bacillus subtilis attenuates ambient air particulate matter‐promoted human oral cancer cells metastatic potential. J Cancer. 2020;11(20):6038‐6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Faghfoori Z, Faghfoori MH, Saber A, Izadi A, Yari Khosroushahi A. Anticancer effects of bifidobacteria on colon cancer cell lines. Cancer Cell Int. 2021;21(1):258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Pawar K, Aranha C. Lactobacilli metabolites restore E‐cadherin and suppress MMP9 in cervical cancer cells. Curr Res Toxicol. 2022;3:100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Li Q, Hu W, Liu WX, et al. Streptococcus thermophilus inhibits colorectal tumorigenesis through secreting β‐galactosidase. Gastroenterology. 2021;160(4):1179‐1193.e14. [DOI] [PubMed] [Google Scholar]
- 280. Dikeocha IJ, Al‐Kabsi AM, Chiu HT, Alshawsh MA. Faecalibacterium prausnitzii ameliorates colorectal tumorigenesis and suppresses proliferation of HCT116 colorectal cancer cells. Biomedicines. 2022;10(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


