Abstract
Mycobacterium tuberculosis is a specialized intracellular pathogen that must regulate gene expression to overcome stresses produced by host defenses during infection. SigH is an alternative sigma factor that we have previously shown plays a role in the response to stress of the saprophyte Mycobacterium smegmatis. In this work we investigated the role of sigH in the M. tuberculosis response to heat and oxidative stress. We determined that a M. tuberculosis sigH mutant is more susceptible to oxidative stresses and that the inducible expression of the thioredoxin reductase/thioredoxin genes trxB2/trxC and a gene of unknown function, Rv2466c, is regulated by sigH via expression from promoters directly recognized by SigH. We also determined that the sigH mutant is more susceptible to heat stress and that inducible expression of the heat shock genes dnaK and clpB is positively regulated by sigH. The induction of these heat shock gene promoters but not of other SigH-dependent promoters was markedly greater in response to heat versus oxidative stress, consistent with their additional regulation by a heat-labile repressor. To further understand the role of sigH in the M. tuberculosis stress response, we investigated the regulation of the stress-responsive sigma factor genes sigE and sigB. We determined that inducible expression of sigE is regulated by sigH and that basal and inducible expression of sigB is dependent on sigE and sigH. These data indicate that sigH plays a central role in a network that regulates heat and oxidative-stress responses that are likely to be important in M. tuberculosis pathogenesis.
Tuberculosis remains a major cause of human suffering, exacting an enormous toll of morbidity and mortality in much of the world (11). The cause of tuberculosis, the obligate pathogen Mycobacterium tuberculosis, is highly adapted for survival in the host organism. Following infection M. tuberculosis is ingested by macrophages and must persist in this environment in order to survive, either in a quiescent state or through active replication that results in tissue destruction and the disease of tuberculosis. In adapting to this intracellular environment, this bacterium must regulate its physiology to survive a variety of stresses produced by the macrophage, including reactive oxygen and reactive nitrogen species produced by these cells (1, 4, 28). In addition M. tuberculosis has been shown to alter the physiology of the macrophage to modulate host defenses (51). Although the sequencing of the M. tuberculosis genome and recent insights are beginning to shed light on pathogenic mechanisms of this organism (7, 8, 27, 30), the means by which M. tuberculosis adapts to survive and replicate in the host remain poorly understood.
Data from a number of laboratories have implicated several alternative sigma factors of mycobacteria, including SigB, SigE, SigF, and SigH, in the adaptation of the pathogen M. tuberculosis and the saprophyte Mycobacterium smegmatis to several stresses (5, 9, 12, 20, 29, 53). M. smegmatis SigH has been shown to be important in surviving organic peroxide stress and heat shock, and the transcription of sigH in M. tuberculosis is induced in response to a variety of stresses in vitro, including heat shock and oxidative stress, and following uptake by macrophages (12, 16, 29). In Streptomyces coelicolor, a member of the actinomycete family that includes mycobacteria, SigR, a close homologue of M. tuberculosis SigH, was recently shown to be important in responding to redox stress through regulation of the thioredoxin/thioredoxin reductase system (39).
The importance of heat stress and oxidative-stress responses in M. tuberculosis pathogenesis and immunity has been extensively investigated. Heat shock proteins, includng DnaK and GroEL, are induced during macrophage infection and have been shown to be major antigens recognized by the host immune system following infection by M. tuberculosis (2, 22, 25, 46). A recent report demonstrated enhanced protective immunity in mice overexpressing dnaK (49). The induction of heat shock proteins during infection and their function in maintenance of protein structure suggest that these proteins play a role protecting M. tuberculosis against oxidative and other stresses generated by host macrophages during infection. Among oxidative-stress response mechanisms, catalase-peroxidase activities were identified decades ago as important M. tuberculosis virulence factors that also play a role in susceptibility to the first-line antitubercular isoniazid (31, 32). More recent molecular investigation has linked these phenotypes to the genes encoding catalase/peroxidase (katG) and alkylhydroperoxidase (ahpC) in M. tuberculosis (48, 52, 55, 56). Strikingly, the gene encoding OxyR, a positive regulator of the peroxide stress response in other bacteria and in most other mycobacterial species, was found to be inactivated by multiple mutations in M. tuberculosis, suggesting altered regulation of the oxidative-stress response in this pathogen (10, 45).
Based on these observations, we sought to investigate the role of SigH in M. tuberculosis in survival and regulation of gene expression in response to oxidative and heat stresses. In this report we demonstrate that a sigH mutant of M. tuberculosis is impaired in its ability to survive different types of oxidative stress as well as heat stress. We further demonstrate that SigH, in response to these stresses, strongly induces the transcription of genes that have been shown to be of central importance in the response to these stresses. Among these are the genes encoding the only thioredoxin reductase of M. tuberculosis TrxB2 and the heat shock proteins DnaK and ClpB. In addition we demonstrate that SigH regulates the stress-inducible expression of the genes encoding the stress-responsive sigma factors SigE and SigB.
These data indicate that SigH plays an important role in regulating the response to heat and oxidative stress in M. tuberculosis through the regulation of major effectors and regulators of the response to these stresses. Together with published data, our results suggest that SigH plays a central role in the regulation of specific gene expression that allows the bacillus to adapt to and survive following exposure to host-generated stresses that this organism is likely to encounter during infection.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Escherichia coli DH5α (Life Technologies) and XL1Blue (Stratagene) were used as host strains for cloning experiments. M. tuberculosis H37Rv was used as the wild-type strain in the stress experiments and as the parental strain from which sigma factor mutant strains were constructed. For some primer extension experiments, RNA was isolated from Mycobacterium bovis BCG. E. coli was grown on L agar or in L broth. M. tuberculosis was grown in Middlebrook 7H9 broth supplemented with oleic acid, albumin, and dextrose complex (7H9-OADC) (Difco) plus 0.05% Tween 80 or on Middlebrook 7H9-OADC agar plates. Ampicillin (100 μg/ml), apramycin (30 μg/ml), and hygromycin (50 μg/ml for mycobacteria and 100 μg/ml for E. coli) were added to culture media as indicated.
Construction of mutant strains of M. tuberculosis.
The M. tuberculosis sigH and sigE mutants were constructed by allele exchange using the specialized transducing phage system as described elsewhere (14). Briefly, the sigH gene was disrupted by inserting a hygromycin resistance cassette into the coding region at codon 151, and the resulting disrupted sigH gene and flanking DNA were cloned into the cos vector pYUB572. The resulting plasmid was digested with PacI and ligated to PacI-digested pHAE87 DNA. The ligation mix was packaged in lambda in vitro packaging mix (Gigapack III; Stratagene), transduced into E. coli, and plated on L broth-hygromycin. Plasmid DNA was isolated from several colonies and restriction digested to verify the presence of the desired insert, and this DNA was electroporated into M. smegmatis strain mc2-155 (47) and plated for plaques at 30°C. Plate stocks were made from a temperature-sensitive phage, and M. tuberculosis H37Rv was infected, followed by plating at 37°C with hygromycin selection. Colonies were picked after 3 to 4 weeks of growth and were screened by PCR for disruption of sigH. Candidate clones were confirmed by Southern blotting to contain a single, disrupted copy of sigH, and one, designated RH349, was used as the sigH mutant strain in subsequent experiments. The M. tuberculosis sigE mutant, designated RH374, was constructed utilizing a similar strategy, except that the hygromycin resistance cassette was introduced into a 675-bp deletion (codons 1 to 225) in the sigE gene. The sigH/sigE double mutant, designated RH375, was generated by first disrupting sigH with a kanamycin resistance gene, utilizing the temperature-sensitive/counterselection strategy of Pelicic et al. (40), followed by disruption of sigE as described above.
Complemented sigH mutant strains were constructed by introducing sigH alone or sigH plus 3′ DNA including the coding sequence of rshA, in each case with 5′ flanking DNA that includes the sigH promoter regions, into the bacterial chromosome utilizing the integrating shuttle vector pMH94A (12, 26). Apramycin-resistant colonies obtained following electroporation of each construct into RH349 were screened by PCR and confirmed by Southern blotting to have both an intact copy and a disrupted copy of sigH. The strains complemented by sigH alone and by sigH plus rshA were designated RH377 and RH395, respectively.
Heat shock and oxidative-stress experiments.
Wild-type M. tuberculosis H37Rv, the sigH mutant strain RH349, and the sigH-complemented mutant strains RH377 and RH395 were compared for their ability to survive several stresses. Survival assays in liquid medium for chemical and heat stresses were performed as previously described (12, 53). Chemicals were added to log-phase cultures to a final concentration of 50 mM diamide–0.1 mM plumbagin. Heat shock was performed at 52°C. The number of viable cells was determined by plating serial dilutions on Middlebrook 7H9-OADC or by inoculating the contents of BACTEC 12B bottles and measuring the growth index (Becton Dickinson). Colonies on plates were counted after 3.5 to 4 weeks at 37°C for all strains except RH395, which requires 4.5 to 5 weeks to form colonies. For experiments using the BACTEC system, standard curves of CFU per milliliter (determined by plating serial dilutions) versus time to reach a growth index of 100 (T-100) were determined experimentally in the absence of stress for each strain. The growth index of 100 was then determined for each aliquot inoculated at the various time points in the stress experiments, and the number of bacilli inoculated was derived from the standard curve of that strain (13). For plating and BACTEC experiments, duplicate samples were assayed at each time point and each experiment was performed at least twice.
RNA isolation and primer extension.
RNA was isolated from mid-log-phase cultures. For experiments examining stress induction of transcription, the culture was divided into two aliquots, one of which was treated with 1 mM diamide for 20 min or 50°C heat shock for 15 min prior to RNA isolation. The cells were chilled, pelleted, resuspended in RNeasy lysis buffer (Qiagen), and transferred to a 2-ml tube containing ceramic and silica beads. The bacteria were lysed by shaking in an FP120 machine (BIO101), and the lysate was then removed to a fresh tube and centrifuged to remove the cell debris. The supernatant then was processed using the RNeasy kit to isolate total RNA according to the manufacturer's protocol (Qiagen).
For primer extension analysis, 0.5 pmol of γ-32P-labeled primer was mixed with 10 μg of RNA in a 6-μl volume. The mixture was heated at 65°C for 5 min and was cooled on ice. Reverse transcription mixture was prepared by mixing 2 μl of 5× first-strand synthesis buffer (Life Technologies), 1 μl of 0.1 M dithiothreitol, 1 μl of 10 mM deoxynucleoside triphosphate mix, and 10 U of Superscript II reverse transcriptase (Life Technologies). The 4-μl reverse transcription mixture was added to the primer-RNA mixture, and the reaction was incubated for 1 h at 42°C. The reaction was stopped by adding 5 μl of loading dye, and 5 μl of the reaction was loaded on a sequencing gel. Sequencing reactions, performed with the same primer used for primer extension, were run in adjacent lanes to determine the size of the transcripts. Quantification of 32P-labeled transcripts was performed using a PhosphorImager and ImageQuaNT software (Molecular Dynamics). In each case where the absence of a SigH-dependent transcript was observed in RNA isolated from the SigH mutant, the quality of the RNA was confirmed by using the same batch of RNA in a primer extension to determine the presence of a SigH-independent transcript.
Purification of M. tuberculosis SigH protein and in vitro transcription assays.
The M. tuberculosis sigH gene was cloned in pTYB1 (New England Biolabs) to overexpress SigH as a fusion protein with the intein and chitin binding domain tag from the vector. Following purification of the fusion protein on a chitin column (New England Biolabs), native SigH protein was released from the chitin-bound intein tag by self-cleavage of the tag in the presence of dithiothreitol. SigH protein was further purified by ion-exchange chromatography on a POROS 50 HQ column (PerSeptive Biosystems) and gel permeation chromatography on a Superdex-100 column (Amersham Pharmacia Biotech).
In vitro single-round runoff transcription analysis was performed using conditions modified from those described earlier by Kang et al. and Miyazaki et al. (23, 33). Purified SigH protein (0.5 μg) was incubated with 0.2 U of E. coli RNA polymerase core enzyme (Epicentre Technologies) at 37oC for 30 min in 30 μl of transcription buffer (50 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 50 mM KCl, 0.1 mM EDTA, 250 μg of bovine serum albumin per ml, and 0.5 U of SUPERase-In [Ambion] per μl). To the reconstituted EςH holoenzyme, 0.09 pmol of DNA template was added for 5 min at 37°C, followed by a mixture of [α-32P]UTP and three other nucleotides for 2 min at 37°C and then by a mixture of heparin and unlabeled UTP for 5 min at 37°C. Concentrations of nucleotides and heparin in 40 μl of final reaction volume were 0.15 mM ATP, 0.15 mM GTP, 0.15 mM UTP, 0.15 mM CTP, 4 μCi of [α-32P]UTP, and 200 μg heparin per ml. Samples were electrophoresed in a 6% denaturing polyacryamide gel containing 7 M urea and were analyzed by autoradiography. DNA templates for in vitro transcription reactions were prepared by PCR except for the sigH template, which was a ClaI restriction fragment containing DNA 5′ of the sigH coding sequence.
Computer database searching.
Searches of the M. tuberculosis H37Rv genome sequence for consensus promoter elements were performed utilizing the program Findpatterns in the GCG package of software and the “search pattern” program available on the TubercuList web site of the Pasteur Institute (http://genolist.pasteur.fr/TubercuList).
RESULTS
Growth and survival following oxidative and heat stresses.
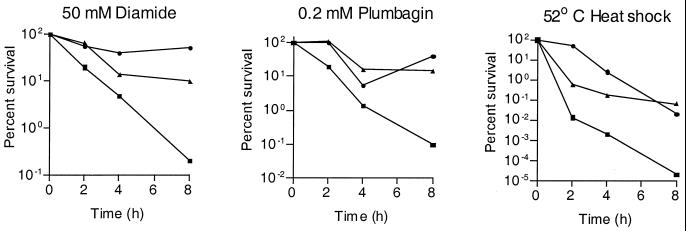
M. tuberculosis H37Rv, the sigH mutant strain RH349, and the mutant strain complemented with sigH alone (RH377) or complemented with sigH plus rshA (RH395) were assessed for in vitro growth and susceptibility to heat and oxidative stresses. H37Rv and RH349 were found to have very similar growth rates. In contrast, both RH377 and RH395 grew more slowly in liquid medium. These strains were tested for susceptibility to diamide, an agent that specifically oxidizes sulfhydryl groups (24), and the redox cycling compound plumbagin, which generates superoxide (19). The sigH mutant was found to be more susceptible to both diamide and plumbagin (Fig. 1). This strain was also more susceptible to 52°C heat stress (Fig. 1). For each stress tested, the sigH mutant was more susceptible than H37Rv in both the plating assay and in the BACTEC assay (not shown).
FIG. 1.
An M. tuberculosis sigH mutant (RH349) is more susceptible to heat and oxidative stress than the wild-type (H37Rv) parental strain. Mid-log-phase cultures of H37Rv, RH349, and RH395 (RH349 into which an intact copy of sigH plus rshA was reintroduced) were exposed to the indicated stress. Aliquots were removed at serial time points, and serial dilutions were plated in duplicate to determine the number of surviving bacteria. Results shown are those of one representative experiment. ●, H37Rv; ▪, RH349; ▴, RH395.
The sigH mutant strain in which a single copy of sigH had been reintroduced showed inconsistent complementation of the stress phenotypes of the mutant (not shown). In contrast, the sigH mutant strain complemented by sigH plus 3′ DNA containing rshA, a transcriptionally linked gene encoding a negative regulator of SigH (T. Song and R. N. Husson, unpublished), did show consistent complementation of the oxidative-stress and heat shock phenotypes of the sigH mutant (Fig. 1).
Regulation of stress response genes by SigH.
Two approaches were used to identify SigH-dependent promoters. First, based on the regulation of thioredoxin reductase by the SigH homologue SigR in S. coelicolor, the role of SigH in regulating transcription of this gene was examined. Second, based on the −10 and −35 region sequences of the M. tuberculosis and M. smegmatis sigH promoters previously demonstrated to be SigH dependent (12), we performed searches of the M. tuberculosis genome sequence for similar sequences positioned 5′ to the start codon of annotated open reading frames (7). Primer extension experiments, using RNA isolated from M. tuberculosis H37Rv and RH349, were performed to determine whether these sequences corresponded to in vivo promoters and whether they were dependent on SigH.
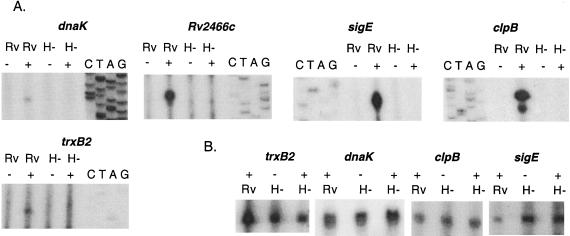
Figure 2 demonstrates the presence of a SigH-dependent promoter 5′ of trxB2 encoding thioredoxin reductase, Rv2466c encoding a protein of unknown function, dnaK encoding a heat shock protein/chaperone, clpB encoding a heat shock protein/chaperone that interacts with DnaK, and sigE encoding an alternative sigma factor involved in the response to stress. In each case, induction of transcription from these promoters was observed following exposure to diamide or heat stress and these promoters were not active in the sigH mutant strain. For each of these genes except Rv2466c, we observed, in the same reactions in which the SigH-dependent promoters were identified, at least one additional transcriptional start site that was active in the absence of stress and that was not SigH dependent (Fig. 2B). With the exception of dnaK and clpB, when induced by diamide, the SigH-dependent promoters became the most active promoters for each of these genes.
FIG. 2.
SigH regulates the transcription of genes involved in heat shock and oxidative-stress responses. Primer extension analysis was performed utilizing RNA isolated from the wild type (Rv) and a sigH mutant strain (H−) of M. tuberculosis, with (+) and without (−) stress induction. For stress induction in all cases except clpB, diamide was added to the culture at a final concentration of 1 mM for 20 min prior to RNA isolation. Because diamide induction of clpB transcription was difficult to detect, RNA was isolated following 50°C heat stress for 15 min for the clpB primer extension. Sequencing ladders generated with the same primer used in each primer extension are adjacent to the primer extension lanes and were used to identify the transcription start site of each gene. (A) SigH-dependent promoters observed observed in primer extension experiments for each of the indicated genes; (B) SigH-independent transcripts observed in primer extension experiments for each of the indicated genes. No SigH-independent transcript was identified for Rv2466c.
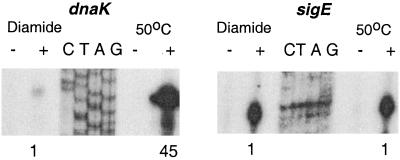
Further investigation of in vivo induction of SigH-dependent transcription showed that the level of transcription varied among the different promoters and in some cases was dependent on the inducing stimulus. The Rv2466, sigE, and trxB2 promoters showed similar levels of induction following diamide exposure and following heat shock. In contrast, the SigH-dependent promoters of dnaK and clpB were only weakly induced following diamide induction but were strongly induced by heat shock (Fig. 3).
FIG. 3.
Different effects of oxidative stress and heat stress on induction of dnaK and sigE gene transcription from their SigH-dependent promoters. RNA was isolated for primer extension analysis following exposure to 1 mM diamide for 20 min or following 50°C heat shock for 15 min. To compare the induction of transcription at the SigH-dependent promoters by these different stresses, the same amount of total RNA was used in each primer extension and the 32P-labeled transcript was quantified using a PhosphorImager (Molecular Dynamics). Quantification of the transcripts is shown below the autoradiogram, with intensity normalized to the signal observed following diamide exposure.
Regulation of sigB transcription by SigH and SigE.
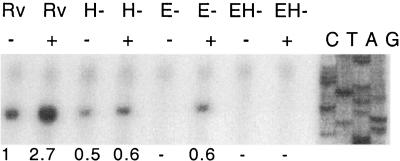
Having found that SigH regulates the inducible expression of sigE in response to heat shock and oxidative stress, we sought to determine whether there was additional cross-regulation of other sigma factors by SigH. Based on data indicating that sigB expression is increased in the response to several stresses, including heat stress in M. tuberculosis (29), we examined the transcription of this gene. A transcription start site, present in unstressed log-phase cells and induced in response to diamide stress, was observed in M. tuberculosis H37Rv (Fig 4). This transcription start site is 3 bases 5′ from the sigB transcription start previously identified. This difference is likely the result of the use of a heterologous sequence ladder generated from a non-GC-rich template, resulting in imprecise localization of the start site in the previous report, versus the use of a sigB sequence ladder to determine the exact start site in this work (Fig. 4) (20). In the sigH mutant strain, transcription from this promoter in log-phase growth was slightly decreased from that seen in the wild type. Induction of sigB transcription by diamide, however, was substantially blunted in the sigH mutant.
FIG. 4.
Transcription of sigB is regulated by SigE and SigH. Primer extension analysis was performed utilizing RNA from wild-type (Rv), sigH mutant (H−), sigE mutant (E−), and sigE/sigH double mutant (EH−) strains of M. tuberculosis. RNA was isolated from each strain in the absence of stress induction (−) and following exposure to 1 mM diamide (+). The intensity of each transcript band, quantified using a PhosphorImager (Molecular Dynamics), is given below the autoradiogram, expressed as relative signal normalized to the intensity of the signal originating from uninduced wild-type RNA.
Based on this observation and on our identification of the SigH-dependent promoter of SigE as responsible for the inducible expression of sigE, we undertook to determine whether this SigB promoter was SigE dependent. Figure 4 demonstrates the absence of basal sigB transcription from this promoter in the sigE mutant strain RH374 but demonstrates partial induction of transcription from this promoter following exposure to diamide. In a sigE/sigH double mutant, neither basal nor induced transcription from this promoter is observed. Thus, this promoter appears to be regulated by SigE and SigH, with both basal and inducible expression mediated by these two extracytoplasmic function sigma factors.
Direct recognition of SigH-dependent promoters by SigH.
To determine whether these promoters found to be SigH dependent in vivo were directly recognized by SigH, single-round in vitro transcription analysis was performed. Specific transcription requiring SigH, resulting in transcripts of the size expected based on the location of the promoter relative to the 3′ end of each template, was observed for all of the in vivo SigH-dependent promoters identified in this study (Fig. 5). In the context of the in vivo data, these results indicate that SigH regulates the transcription of these genes directly, via the recognition of promoter sequences by RNA polymerase holoenzyme containing SigH.
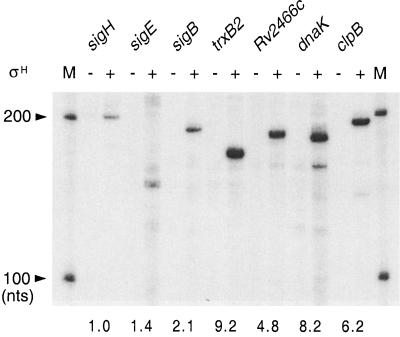
FIG. 5.
SigH-dependent promoters identified in vivo are directly recognized by purified SigH in vitro. Transcripts were synthesized from DNA templates containing SigH-dependent promoters of each gene by RNA polymerase core enzyme in the presence (+) or absence (−) of SigH as described in Materials and Methods. Size markers (lane M) were prepared by transcription of the RNA Century marker template set (Ambion) with the MAXIscript T7 in vitro transcription kit (Ambion). Predicted sizes of the transcripts are as follows: sigH, 209 nucleotides; sigE, 152 nucleotides; sigB, 194 nucleotides; trxB2, 171 nucleotides; Rv2466c, 186 nucleotides; dnaK, 186 nucleotides; and clpB, 198 nucleotides. The intensity of each transcript band, quantified using a PhosphorImager (Molecular Dynamics), is given below the autoradiogram, expressed as relative signal normalized to the intensity of sigH transcript band. nts, nucleotides.
Figure 6 shows an alignment of the SigH-dependent M. tuberculosis promoters identified in this study and of the previously identified SigH-dependent sigH promoter, arranged according to promoter strength in vitro. Strong consensus −35 and −10 region sequences are observed. The 8-base −35 region consensus includes 7 specific bases, plus one position that allows a pyrimidine (cytosine or thymine). The −10 region has a 5-base consensus in which the central GTT is conserved in all seven promoters. As seen in Fig. 5, differences in promoter activity were observed in the in vitro transcription assay using equal amounts of template under conditions where SigH was not limiting, as determined by titration of SigH. These differences show some correspondence to the extent of variation from consensus in the −35 region of these promoters, with trxB2, dnaK, clpB, and Rv2466c forming a group of stronger promoters and sigE, sigB, and sigH a group of weaker promoters. Examination of the promoter regions of these genes shows that the sigE, sigH, and sigB −35 sequences vary by at least 1 base from the consensus. Of interest, clpB differs from the consensus in the first two positions yet appears to be a strong promoter in vitro and in vivo.
FIG. 6.
Alignment of promoter regions of SigH-dependent genes. Sequences of the seven SigH-dependent promoters were compared to visualize the conserved features among the sequences. Consensus −10 and −35 sequences are capitalized and shown in the black boxes. The consensus sequence is shown above the alignment. The transcription start site of each gene is shown in boldface.
DISCUSSION
In this work we demonstrate that a sigH mutant of M. tuberculosis is more susceptible to heat and oxidative stresses and that this sigma factor directly regulates the expression of major effectors of the response to these stresses. We further show that sigH regulates the stress-inducible expression of sigE and sigB, two sigma factors previously shown to play a role in the mycobacterial response to these and other stresses (29, 53). Together with previous data indicating that sigH is autoregulated (12), these results suggest a major role for this alternative sigma factor in the regulation of M. tuberculosis gene expression in response to stresses generated by the host during infection by this pathogen. The observation that sigH transcription is induced following infection of macrophages (16) and preliminary data indicating attenuation of the sigH mutant in a mouse model of infection (W. R. Jacobs and R. N. Husson, unpublished) support a role for SigH in the regulation of gene expression required for M. tuberculosis virulence. The regulation by SigH of both stress response effector proteins and additional transcriptional regulators creates a regulatory network with the potential for modulation of gene expression, with responses varying quantitatively and in the specific components of the response that are induced by different classes of stress.
The inconsistent complementation of the sigH mutant strain by sigH alone and the consistent complementation by sigH plus rshA suggest that RshA is required for effective function of SigH in vivo. RshA is highly similar to RsrA, a redox-regulated negative regulator of SigR, a S. coelicolor homologue of SigH (23, 39), and functions as a negative regulator of SigH (Song and Husson, unpublished). In Streptomyces, rsrA mutant strains frequently acquire inactivating mutations in sigR (38), while in M. smegmatis it has not been possible to generate a rshA mutant (X. Puyang and R. N. Husson, unpublished). These data indicate that unregulated SigH or SigR activity, a situation that would result in an uncontrolled positive-feedback loop, is poorly tolerated by the bacterium.
The coordinate regulation of the heat shock genes dnaK and clpB makes physiologic sense in the context of recent insights into the interaction of the DnaK-DnaJ-GrpE complex with ClpB in the prevention of aggregate formation and insights into disaggregation and refolding of denatured proteins (15, 34, 35). The positive regulation of these heat shock genes by an alternative sigma factor, however, is a novel finding among the high-G+C-content, gram-positive bacteria. In these organisms and in most gram-positive bacteria, negative regulation by repressors, such as HscR, which regulates GroES/GroEL, and HrpA (HspR), which regulates the dnaK-dnaJ-grpE-hrpA and clpB operons in Streptomyces species and other species, has been described as the primary mechanism by which heat shock gene transcription is controlled (3, 17, 36, 44, 57). In contrast, in E. coli the alternative sigma factors ς32 (RpoH) and ς24 (RpoE) play a major role in regulating the heat shock response (18, 42–44).
The differences that we observed in induction of expression of the SigH-dependent promoters of dnaK and clpB by heat versus oxidative stress suggested the presence of a heat-labile repressor. The role of HspR (HrpA) as a repressor of dnaK expression in M. tuberculosis has recently been demonstrated (49). The differential effect of heat versus oxidative stress on induction of expression from the SigH-dependent dnaK and clpB promoters, presumably mediated by the different susceptibility of HspR to these stresses, allows for modulation of the SigH-mediated response to different classes of stress.
The response of M. tuberculosis to oxidative stress has been the subject of active investigation. OxyR is a major positive regulator of the oxidative-stress response in many bacteria, where it regulates the expression of genes, including katG encoding catalase peroxidase and ahpC encoding alkylhydroperoxidase, in response to peroxide stress (6, 50). OxyR is present in most mycobacteria and has been shown in Mycobacterium marinum to positively regulate expression of ahpC but not of katG (37). A striking finding was that in M. tuberculosis, oxyR is inactivated by multiple mutations, resulting in minimal basal or inducible expression of ahpC in this organism (10, 45). In M. smegmatis and M. tuberculosis, katG was recently shown to be cotranscribed with and regulated by furA, which encodes a repressor similar to Fur proteins in many bacteria (41, 54). The regulation of other known genes that play a role in the oxidative-stress response, e.g., sodA and sodC encoding the superoxide dismutases of M. tuberculosis, remains to be determined.
In this context, the susceptibility of the M. tuberculosis sigH mutant to diamide, which generates a redox stress, and plumbagin, which generates superoxide, suggests that SigH regulates another arm of the oxidative-stress response in mycobacteria. While we have demonstrated the SigH-mediated induction of thioredoxin reductase and thioredoxin expression, the exact mechanisms by which sigH mediates the response to these stresses is not known. No SigH consensus promoter sequences are present 5′ of sodA, sodC, or ahpC. The presence of a glutaredoxin motif in the protein encoded by Rv2466c, whose expression is SigH dependent, suggests that this gene may play a role in the SigH-regulated response to oxidative stress in M. tuberculosis.
A notable result of this work is the identification of a regulatory network that includes SigH, SigE, and SigB. Our findings indicate that the SigH-dependent sigE promoter is responsible for much of the increased transcription of sigE under high-heat and high-oxidative-stress conditions and that both SigE and SigH are required for maximal induction of sigB expression. The observation that transcription of sigH and sigE is induced following uptake of M. tuberculosis by macrophages (16) suggests that this SigH-dependent sigE promoter may be responsible for increased sigE transcription in the macrophage and that sigB transcription is also likely to be induced in this setting.
The consensus SigH-dependent promoter motifs identified in this report are similar to those defined for SigR-dependent promoters in S. coelicolor, with the potential expansion of the −35 consensus to 8 nucleotides in M. tuberculosis and a single-base difference in the 6-nucleotide, overlapping consensus between these species (39). Three of the strongest M. tuberculosis promoters share the exact consensus −35 sequence, while the weaker sigB, sigE, and sigH promoters vary by 1, 1, and 2 bases, respectively, from the consensus. The clpB promoter, despite varying from the consensus at two positions, is a strong promoter in vivo and in vitro. The relative strength of these promoters is consistent with the regulatory roles of these sigma factors, whose effect on cell physiology can be amplified through transcription of regulated genes, in contrast with the direct effector roles of thioredoxin reductase, DnaK, and ClpB.
The regulation of a single sigB promoter by SigE and SigH parallels the overlap in recognition of promoters by SigX and SigW in Bacillus subtilis (21). Definition of the importance of specific bases in the −10 and −35 regions through targeted mutagenesis should allow clearer understanding of the promoter elements involved in recognition by SigH and SigE. This insight, together with functional characterization of regulated genes, will allow further definition of the role of these sigma factors in the regulation of M. tuberculosis gene expression in response to stress and during infection.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (AI37901 and AI27150) and by a grant from the Potts Memorial Foundation.
S.R. and T.S. contributed equally to this work.
REFERENCES
- 1.Adams L B, Dinauer M C, Morgenstern D E, Krahenbuhl J L. Comparison of the roles of reactive oxygen and nitrogen intermediates in the host response to Mycobacterium tuberculosis using transgenic mice. Tuber Lung Dis. 1997;78:237–246. doi: 10.1016/s0962-8479(97)90004-6. [DOI] [PubMed] [Google Scholar]
- 2.Boom W H, Husson R N, Young R A, David J R, Piessens W F. In vivo and in vitro characterization of murine T-cell clones reactive to Mycobacterium tuberculosis. Infect Immun. 1987;55:2223–2229. doi: 10.1128/iai.55.9.2223-2229.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucca G, Hindle Z, Smith C P. Regulation of the dnaK operon of Streptomyces coelicolor A3(2) is governed by HspR, an autoregulatory repressor protein. J Bacteriol. 1997;179:5999–6004. doi: 10.1128/jb.179.19.5999-6004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan J, Xing Y, Magliozzo R, Bloom B. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med. 1992;175:1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen P, Ruiz R E, Li Q, Silver R F, Bishai W R. Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF. Infect Immun. 2000;68:5575–5580. doi: 10.1128/iai.68.10.5575-5580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christman M F, Morgan R W, Jacobson F S, Ames B N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985;41:753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 7.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 8.Cox J S, Chen B, McNeil M, Jacobs W R., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 9.DeMaio J, Zhang Y, Ko C, Young D, Bishai W. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1996;93:2790–2794. doi: 10.1073/pnas.93.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deretic V, Philipp W, Dhandayuthapani S, Mudd M H, Curcic R, Garbe T, Heym B, Via L E, Cole S T. Mycobacterium tuberculosis is a natural mutant with an inactivated oxidative-stress regulatory gene: implications for sensitivity to isoniazid. Mol Microbiol. 1995;17:889–900. doi: 10.1111/j.1365-2958.1995.mmi_17050889.x. [DOI] [PubMed] [Google Scholar]
- 11.Dye C, Scheele S, Dolin P, Pathania V, Raviglione M C. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 1999;282:677–686. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 12.Fernandes N, Wu Q-L, Kong D, Puyang X, Garg S, Husson R. A mycobacterial extracytoplasmic function sigma factor involved in survival following heat shock and oxidative stress. J Bacteriol. 1999;181:4266–4274. doi: 10.1128/jb.181.14.4266-4274.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fratazzi C, Arbeit R D, Carini C, Remold H G. Programmed cell death of Mycobacterium avium serovar 4-infected human macrophages prevents the mycobacteria from spreading and induces mycobacterial growth inhibition by freshly added, uninfected macrophages. J Immunol. 1997;158:4320–4327. [PubMed] [Google Scholar]
- 14.Glickman M S, Cox J S, Jacobs W R., Jr A novel mycolic acid cyclopropane synthetase is required for coding, persistence, and virulence of Mycobacterium tuberculosis. Mol Cell. 2000;5:717–727. doi: 10.1016/s1097-2765(00)80250-6. [DOI] [PubMed] [Google Scholar]
- 15.Goloubinoff P, Mogk A, Zvi A P, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci USA. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham J E, Clark-Curtiss J E. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS) Proc Natl Acad Sci USA. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grandvalet C, de Crecy-Lagard V, Mazodier P. The ClpB ATPase of Streptomyces albus G belongs to the HspR heat shock regulon. Mol Microbiol. 1999;31:521–532. doi: 10.1046/j.1365-2958.1999.01193.x. [DOI] [PubMed] [Google Scholar]
- 18.Grossman A, Erickson J, Gross C. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell. 1984;38:383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- 19.Hassan H, Fridovich I. Intracellular production of superoxide radical and of hydrogen peroxide by redox active compounds. Arch Biochem Biophys. 1979;196:385–395. doi: 10.1016/0003-9861(79)90289-3. [DOI] [PubMed] [Google Scholar]
- 20.Hu Y, Coates A R M. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J Bacteriol. 1999;181:469–476. doi: 10.1128/jb.181.2.469-476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X, Fredrick K, Helmann J. Promoter recognition by Bacillus subtilis ςW: autoregulation and partial overlap with the ςX regulon. J Bacteriol. 1998;180:3765–3770. doi: 10.1128/jb.180.15.3765-3770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Husson R, Young R. Genes for the major protein antigens of Mycobacterium tuberculosis: the etiologic agents of tuberculosis and leprosy share an immunodominant antigen. Proc Natl Acad Sci USA. 1987;84:1679–1683. doi: 10.1073/pnas.84.6.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang J G, Paget M S, Seok Y J, Hahn M Y, Bae J B, Hahn J S, Kleanthous C, Buttner M J, Roe J H. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 1999;18:4292–4298. doi: 10.1093/emboj/18.15.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosower N, Kosower E. Diamide: an oxidant probe for thiols. Methods Enzymol. 1995;251:123–133. doi: 10.1016/0076-6879(95)51116-4. [DOI] [PubMed] [Google Scholar]
- 25.Lee B Y, Horwitz M A. Identification of macrophage and stress-induced proteins of Mycobacterium tuberculosis. J Clin Investig. 1995;96:245–249. doi: 10.1172/JCI118028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee M H, Pascopella L, Jacobs W R, Jr, Hatfull G F. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci USA. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manabe Y C, Saviola B J, Sun L, Murphy J R, Bishai W R. Attenuation of virulence in Mycobacterium tuberculosis expressing a constitutively active iron repressor. Proc Natl Acad Sci USA. 1999;96:12844–12848. doi: 10.1073/pnas.96.22.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manca C, Paul S, Barry III C E, Freedman V H, Kaplan G. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect Immun. 1999;67:74–79. doi: 10.1128/iai.67.1.74-79.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manganelli R, Dubnau E, Tyagi S, Kramer F R, Smith I. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol Microbiol. 1999;31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 30.McKinney J D, Honer zu Bentrup K, Munoz-Elias E J, Miczak A, Chen B, Chan W T, Swenson D, Sacchettini J C, Jacobs W R, Jr, Russell D G. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 31.Middlebrook G, Cohn M. Some observations on the pathogenicity of isoniazid-resistant variants of tubercle bacilli. Science. 1953;118:297–299. doi: 10.1126/science.118.3063.297. [DOI] [PubMed] [Google Scholar]
- 32.Mitchison D, Selkon J, Lloyd J. Virulence in the guinea pig, susceptibility to hydrogen peroxide, and catalase activity of isoniazid-sensitive tubercle bacilli from South Indian and British patients. J Pathol Bacteriol. 1963;86:464–468. doi: 10.1002/path.1700860213. [DOI] [PubMed] [Google Scholar]
- 33.Miyazaki E, Chen J-M, Ko C, Bishai W R. The Staphylococcus aureus rsbW (orf159) gene encodes an anti-sigma factor of SigB. J Bacteriol. 1999;181:2846–2851. doi: 10.1128/jb.181.9.2846-2851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mogk A, Tomoyasu T, Goloubinoff P, Rudiger S, Roder D, Langen H, Bukau B. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motohashi K, Watanabe Y, Yohda M, Yoshida M. Heat-inactivated proteins are rescued by the DnaK·J-GrpE set and ClpB chaperones. Proc Natl Acad Sci USA. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narberhaus F. Negative regulation of bacterial heat shock genes. Mol Microbiol. 1999;31:1–8. doi: 10.1046/j.1365-2958.1999.01166.x. [DOI] [PubMed] [Google Scholar]
- 37.Pagan-Ramos E, Song J, McFalone M, Mudd M H, Deretic V. Oxidative stress response and characterization of the oxyR-ahpC and furA-katG loci in Mycobacterium marinum. J Bacteriol. 1998;180:4856–4864. doi: 10.1128/jb.180.18.4856-4864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paget M S, Bae J B, Hahn M Y, Li W, Kleanthous C, Roe J H, Buttner M J. Mutational analysis of RsrA, a zinc-binding anti-sigma factor with a thiol-disulphide redox switch. Mol Microbiol. 2001;39:1036–1047. doi: 10.1046/j.1365-2958.2001.02298.x. [DOI] [PubMed] [Google Scholar]
- 39.Paget M S, Kang J G, Roe J H, Buttner M J. sigmaR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2) EMBO J. 1998;17:5776–5782. doi: 10.1093/emboj/17.19.5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelicic V, Jackson M, Reyrat J-M, Jacobs W J, Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pym A S, Domenech P, Honoré N, Song J, Deretic V, Cole S T. Regulation of catalase-peroxidase (KatG) expression, isoniazid sensitivity and virulence by furA of Mycobacterium tuberculosis. Mol Microbiol. 2001;40:879–889. doi: 10.1046/j.1365-2958.2001.02427.x. [DOI] [PubMed] [Google Scholar]
- 42.Raina S, Missiakas D, Georgopoulos C. The rpoE gene encoding the ςE (ς24) heat shock sigma factor of Escherichia coli. EMBO J. 1995;14:1043–1055. doi: 10.1002/j.1460-2075.1995.tb07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rouviere P, De Las Penas A, Lu C, Rudd K, Gross C. rpoE, the gene encoding the second heat-shock sigma factor, ςE, in Escherichia coli. EMBO J. 1995;14:1032–1042. doi: 10.1002/j.1460-2075.1995.tb07084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segal R, Ron E Z. Regulation and organization of the groE and dnaK operons in Eubacteria. FEMS Microbiol Lett. 1996;138:1–10. doi: 10.1111/j.1574-6968.1996.tb08126.x. [DOI] [PubMed] [Google Scholar]
- 45.Sherman D R, Sabo P J, Hickey M J, Arain T M, Mahairas G G, Yuan Y, Barry III C E, Stover C K. Disparate responses to oxidative stress in saprophytic and pathogenic mycobacteria. Proc Natl Acad Sci USA. 1995;92:6625–6629. doi: 10.1073/pnas.92.14.6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silva C L, Lowrie D B. Identification and characterization of murine cytotoxic T cells that kill Mycobacterium tuberculosis. Infect Immun. 2000;68:3269–3274. doi: 10.1128/iai.68.6.3269-3274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 48.Sreevatsan S, Pan X, Zhang Y, Deretic V, Musser J M. Analysis of the oxyR-ahpC region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob Agents Chemother. 1997;41:600–606. doi: 10.1128/aac.41.3.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart G R, Snewin V A, Walzl G, Hussell T, Tormay P, O'Gaora P, Goyal M, Betts J, Brown I N, Young D B. Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection. Nat Med. 2001;7:732–737. doi: 10.1038/89113. [DOI] [PubMed] [Google Scholar]
- 50.Tartaglia L A, Storz G, Ames B N. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J Mol Biol. 1989;210:709–719. doi: 10.1016/0022-2836(89)90104-6. [DOI] [PubMed] [Google Scholar]
- 51.Ting L M, Kim A C, Cattamanchi A, Ernst J D. Mycobacterium tuberculosis inhibits IFN-gamma transcriptional responses without inhibiting activation of STAT1. J Immunol. 1999;163:3898–3906. [PubMed] [Google Scholar]
- 52.Wilson T M, de Lisle G W, Collins D M. Effect of inhA and katG on isoniazid resistance and virulence of Mycobacterium bovis. Mol Microbiol. 1995;15:1009–1015. doi: 10.1111/j.1365-2958.1995.tb02276.x. [DOI] [PubMed] [Google Scholar]
- 53.Wu Q-L, Kong D, Lam K, Husson R. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J Bacteriol. 1997;179:2922–2929. doi: 10.1128/jb.179.9.2922-2929.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zahrt T C, Song J, Siple J, Deretic V. Mycobacterial FurA is a negative regulator of catalase-peroxidase gene katG. Mol Microbiol. 2001;39:1174–1185. doi: 10.1111/j.1365-2958.2001.02321.x. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Dhandayuthapani S, Deretic V. Molecular basis for the exquisite sensitivity of Mycobacterium tuberculosis to isoniazid. Proc Natl Acad Sci USA. 1996;93:13212–13216. doi: 10.1073/pnas.93.23.13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 57.Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]