Abstract
Human strongyloidiasis is an important neglected tropical disease primarily caused by the nematode Strongyloides stercoralis, and to a lesser extent Strongyloides fuelleborni which mainly infects non-human primates. Zoonotic sources of infection have important implications for control and prevention of morbidity and mortality caused by strongyloidiasis. Recent molecular evidence suggests that for S. fuelleborni, primate host specificity is variable among genotypes across the Old World, and consequently that these types likely vary in their capacity for human spillover infections. Populations of free-roaming vervet monkeys (Chlorocebus aethiops sabaeus), introduced to the Caribbean Island of Staint Kitts from Africa, live in close contact with humans, and concern has arisen regarding their potential to serve as reservoirs of zoonotic infections. In this study, we sought to determine the genotypes of S. fuelleborni infecting St. Kitts vervets to explore whether they are potential reservoirs for human-infecting S. fuelleborni types. Fecal specimens were collected from St. Kitts vervets and S. fuelleborni infections were confirmed microscopically and by PCR. Strongyloides fuelleborni genotypes were determined from positive fecal specimens using an Illumina amplicon sequencing-based genotyping approach targeting the mitochondrial cox1 locus and 18S rDNA hypervariable regions I and IV of Strongyloides species. Phylogenetic analysis of resultant genotypes supported that S. fuelleborni from St. Kitts vervets is of an exclusively African variety, falling within the same monophyletic group as an isolate which has been detected previously in a naturally infected human from Guinea-Bissau. This observation highlights that St. Kitts vervets may serve as potential reservoirs for zoonotic S. fuelleborni infection, which warrants further exploration.
Keywords: Strongyloides fuelleborni, Strongyloidiasis, Vervet, Saint Kitts, Genotyping, Population structure
Graphical abstract
Highlights
-
•
Free-roaming vervets on the Caribbean Island of Saint Kitts are infected with S. fuelleborni.
-
•
We genotyped S. fuelleborni from St. Kitts vervets to assess the zoonotic potential of these worms.
-
•
St. Kitts vervets are infected with an African S. fuelleborni type that has been found previously in a human.
-
•
St. Kitts vervets may therefore represent a reservoir of human S. fuelleborni infection.
1. Introduction
Human strongyloidiasis is an important disease caused by the intestinal nematode Strongyloides stercoralis, and to a lesser extent Strongyloides fuelleborni. The clinical characteristics of strongyloidiasis are of variable severity, but generally include abdominal pain, diarrhea, skin rash, eosinophilia, and respiratory symptoms; disseminated strongyloidiasis or hyperinfections caused by S. stercoralis infection can be life-threatening because of autoinfection (Nutman, 2017). Strongyloides cycles between parasitic and free-living stages (Hunt et al., 2016), and the most common route of transmission is via direct penetration of exposed skin by larvae in contaminated soil. Infected human hosts will then pass larvae or eggs in the feces. Human strongyloidiasis is cosmopolitan in its distribution, and has an estimated global burden of approximately 600 million human infections (Nutman, 2017; Buonfrate et al., 2020). The largest burden of disease occurs in low socioeconomic contexts, due to the lack of access to clean water, sanitation and hygiene (i.e., WASH) interventions (Strunz et al., 2014; Garn et al., 2022).
Historically, S. stercoralis has been considered capable of infecting both humans and domestic dogs (Canis lupus familiaris), while S. fuelleborni has been recognized as a cause of strongyloidiasis in humans and non-human primates throughout the Old World (Barratt and Sapp, 2020). More recently, molecular evidence has revealed that multiple genetic subpopulations exist within S. fuelleborni and S. stercoralis, supporting that these two species may comprise species complexes (Barratt and Sapp, 2020). Two subspecies of S. fuelleborni are currently recognized; S. fuelleborni fuelleborni (S. f. fuelleborni) which has been reported in sub-Saharan Africa, South Asia, and southeast Asia (Barratt and Sapp, 2020), and S. fuelleborni kellyi (S. f. kellyi), which is an enigmatic species that possesses an ambiguous relationship with S. f. fuelleborni and is only found in Papua New Guinea (Dorris et al., 2002). Strongyloides f. fuelleborni infects Old-World primates, including pig-tailed macaques, Bornean orangutans, gorillas, chimpanzees, among others (Hasegawa et al., 2009, 2010, 2016; Kooriyama et al., 2012; Thanchomnang et al., 2017, 2018).
Strongyloides fuelleborni is recognized as a possible source of zoonotic strongyloidiasis via transmission from primates into nearby human populations or humans who encroach on the primates’ habitat. For Asian S. fuelleborni types, this spillover seems the most important route of infection, as documented cases involve people residing in close proximity to monkey troops harboring S. fuelleborni, or keeping monkeys as pets, in the absence of other likely exposures (Thanchomnang et al., 2018; Janwan et al., 2020). Anthroponotic transmission of Asian/southeast Asian types of S. fuelleborni is rare or absent. Regarding S. fuelleborni types from Africa, it is not known whether anthroponotic transmission is an important route of infection. However, prior analysis has shown close genetic clustering of some human-derived African S. fuelleborni types which might suggest that anthroponotic transmission occurs for certain African types (Barratt and Sapp, 2020). Among African varieties of S. fuelleborni, distinct genetic sub-structuring has been noted, with discrete clustering of Tanzanian types (East African) separately from Central and West African isolates, with human infections caused by both East and West African varieties (Hasegawa et al., 2010, 2016; Barratt and Sapp, 2020; Ko et al., 2023). Asian varieties also possess a genetic population substructure based on geography, with isolates from Borneo, mainland Southeast Asia, and Japan clustering discretely from one another (Barratt and Sapp, 2020; Ko et al., 2023).
African vervet monkeys (Chlorocebus aethiops sabaeus) exist as free-roaming populations on the Caribbean island of Saint Kitts, descended from a small group originally introduced by Europeans during the 17th century (McGuire, 1974). Since their introduction their numbers have exploded, and due to the damage they cause to a variety of crops/fruits, native flora, and wildlife, St. Kitts vervets are considered pests by local farming communities contrary to other government agencies who consider their presence on the Island an important tourist attraction. Recent surveys have shown that St. Kitts vervets are a host for multiple human-pathogenic helminths including Trichuris trichiura, Schistosoma mansoni, and S. fuelleborni, in addition to other protozoal, bacterial, and viral pathogens (Gallagher et al., 2019; Ketzis et al., 2020; Cruz et al., 2021). It is possible that vervets originally introduced to St. Kitts brought an African variety of S. fuelleborni to the island, although a detailed molecular analysis of S. fuelleborni types infecting St. Kitts vervets has not been performed. The large number of vervets on the island of St. Kitts and their curiosity causes them to regularly spill into human occupied areas of the island which could place the islands’ human inhabitants at risk of S. fuelleborni infection. Determining whether St. Kitts vervets are infected with S. fuelleborni types identified previously in humans might provide greater insight into the possible risk of zoonotic S. fuelleborni transmission on the island.
We sought to explore whether free-roaming vervets on the island of St. Kitts are reservoirs of S. fuelleborni types that have previously been reported to cause human strongyloidiasis. Using a next-generation sequencing (NGS)-based multi-locus-sequence-typing (MLST) method targeting the mitochondrial cox1 locus, and hypervariable regions (HVRs) –I and -IV of the 18S rDNA (Barratt et al., 2019a; Beknazarova et al., 2019), we genetically characterized S. fuelleborni from vervets inhabiting several regions of St. Kitts. Using a machine-learning-based genetic distance computation method to facilitate phylogenetic reconstructions (Barratt and Sapp, 2020; Jacobson et al., 2022), the genotypes were clustered alongside previously described S. fuelleborni genotypes from Africa and Asia, including types previously isolated from humans.
2. Materials and methods
2.1. Study population and specimen collection
The study population included multiple troops of free-roaming African vervet monkeys (Chlorocebus aethiops sabaeus) from the island of Saint Kitts. Fecal specimens were collected from the transport cages of vervets trapped for other purposes (relocating, contract research) and stored at 4 °C until an initial screening (direct smear with Lugol's iodine; within 3–5 days of collection). Samples identified with Strongyloides in this initial screen (n = 34) were placed directly into 90% ethanol (1 g–3 mL) and stored at −20 °C until shipment to the Parasitic Diseases Branch (PDB) at the US Centers for Disease Control and Prevention (CDC) for all subsequent analyses, including S. fuelleborni genotyping, and a secondary microscopic screen (400× magnification) of wet mounts for the presence of Strongyloides eggs and/or larvae. The specimens were stored at 4 °C prior to subsequent molecular analyses. A single bronchoalveolar lavage (BAL) fluid specimen from a human patient who tested positive for S. stercoralis in the parasitology reference laboratory at CDC (by PCR and microscopy) was included in this study as a positive control.
2.2. Ethics
The collection of fecal specimens from St. Kitts vervets was approved by the Ross University School of Veterinary Medicine Institutional Animal Care and Use Committee, Tissue/Sample Use application #TSU10.23.19. Ethics approval for the use of anonymized, de-identified, human samples as non-engaged research was granted by the CDC Center for Global Health Human Subjects Review, approval number 2016-314.
2.3. Extraction of DNA from vervet fecal specimens
DNA was extracted from vervet fecal specimens using a DNeasy PowerSoil Kit (Qiagen, Germantown, Maryland, USA) with some minor modifications. As the fecal specimens had been preserved in 70% ethanol, they were washed in PBS prior to DNA extraction. To do this, approximately 0.25 g of feces suspended in ethanol was placed in a 2 mL Eppendorf tube and centrifuged in a benchtop centrifuge (Eppendorf Centrifuge 5424) at maximum speed (15,000 RPM). The supernatant was discarded. Next, the tube was filled to 2 mL with PBS and vortexed to disrupt the pellet. The suspension was centrifuged at maximum speed once more and the supernatant was removed again. The resulting pellet was transferred to a PowerBead Tube and subjected to Bead beating for 3 min at 1400 RPM on an Eppendorf Thermomixer. After adding solution C1 to the tube containing the PowerBeads and feces, we also added 20 mL of proteinase K taken from a DNEasy Blood & Tissue Kit (Qiagen). The solution was gently vortexed and left to digest at 65 °C for 30 min. The digested specimen was vortexed again following digestion. The DNA was eluted in a volume of 50 μL of Solution C6.
2.4. Polymerase chain reaction
Three genotyping loci were amplified from each specimen by PCR; Cytochrome c oxidase subunit 1 (cox1), 18S rDNA hypervariable region I (HVR-I), and 18S rDNA hypervariable region IV (HVR-IV). The primer sequences used to amplify each locus are provided in Table 1. Amplification reactions were prepared to contain 2.5 μL of each forward and reverse primer (10 μM solutions), 18 μL of PCR grade water, 25 μL of NEB Next Q5 Hot Start HiFi PCR Master Mix (New England Biolabs, Ipswich, MA, USA), and 2 μL of template DNA. For cox1 and HVR-I, temperature cycling conditions were as follows; initial melting at 98 °C for 2 min followed by 45 cycles of 98 °C for 10 s, annealing at 65 °C for 10 s, and extension 72 °C for 10 s. The protocol concluded with a final extension of 72 °C for 2 min, and holding at 4 °C. For HVR-IV, temperature cycling conditions were as follows: initial melting at 98 °C for 2 min followed by 45 cycles of 98 °C for 10 s, annealing at 63 °C for 10 s, and extension 72 °C for 10 s. The protocol concluded with a final extension step of 72 °C for 2 min, and holding at 4 °C. All PCR runs were accompanied by a negative template control (PCR grade water added to a reaction in place of DNA), and a positive template control comprising DNA extracted from a bronchoalveolar lavage (BAL) fluid specimen that had previously tested positive by PCR and microscopy for S. stercoralis in the diagnostic parasitology reference laboratory at CDC.
Table 1.
PCR primers and reaction conditions.
| Target locus | Primer | Amplicon Length α | Sequence | Annealing temperature |
|---|---|---|---|---|
| COXI | SSP_COX1_F* | 217 bp | 5′-TTTGATCCTAGTTCTGGTGGTAATCC-3′ | 65 °C |
| SSP_COX1_R* | 5′-GTAGCAGCAGTAAAATAAGCACGAGA-3′ | |||
| 18S rDNA HVR-I | NEW_HVR_I_F | ∼434 bp β | 5′-GCTCATTATAACAGCTATAGACTACACGGTA-3′ | 65 °C |
| NEW_HVR_I_R | 5′-CCACAACAATCATTTTATGCACTTGG-3′ | |||
| 18S rDNA HVR-IV | NEW_HVR_IV_F | ∼255 bp β | 5′-CGGGCCGGACACTATAAGG-3′ | 63 °C |
| NEW_HVR_IV_R | 5′-ATCTCTAAACAGGAACATAATGATCACTAC-3′ |
α Excluding PCR primer sequences from the amplicon.
β Length varies depending on genotype.
∗ Broadly specific primers for generation of Strongyloides sp. cox1 amplicons as well as those of several strongyles.
2.5. Illumina amplicon sequencing and bioinformatic analysis
The sequencing methods employed here were modified from those described by Beknazarova et al. (2019) and Barratt et al. (2019a). Briefly, each amplicon was separately purified and normalized using a SequalPrep Normalization Kit (Thermo Fisher Scientific Waltham, Massachusetts, USA) with an elution volume of 20 μL. After purification and normalization, equal volumes of each amplicon from each fecal specimen were pooled and the resulting normalized pool of amplicons was subjected to library preparation using Nextera XT DNA Library Prep Kit in accordance with the manufacturer's instructions (Illumina, San Diego, California, USA). Next, DNA concentration and DNA molecular size for the resultant Illumina library were assessed using a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific) and a High Sensitivity D1000 ScreenTape on the 2200 TapeStation (Agilent, Santa Clara, California, USA), respectively. The quantified library was diluted to 10–15 pM and sequenced on the MiSeq Platform using the MiSeq Reagent Kits V2 (500 cycle) (Illumina). Analysis of Illumina data was performed using a custom Geneious workflow (Geneious Prime, version 2022: www.geneious.com). Briefly, this workflow performed read quality control and identification of the haplotype composition of each specimen at the three loci sequenced. This workflow is described in detail in the studies by Beknazarova et al. (2019) and Barratt et al. (2019a). A detailed description of the haplotype naming conventions used in this study is provided in later sections.
2.6. Reference dataset of published Strongyloides genotypes
We used 128 S. fuelleborni genotypes compiled as part of previous work (Barratt et al., 2019a; Barratt and Sapp, 2020), plus 18 from an undefined Strongyloides species identified in Bornean slow lorises (Frias et al., 2018). Our dataset included 24 cox1 sequences and HVR-IV sequences from Thai isolates of S. fuelleborni: one from a human and 23 from pig-tailed macaques (Macaca nemestrina) (Janwan et al., 2020). We also included 39 S. fuelleborni cox1 sequences generated by Ko et al. (2023) from a range of Asian non-human primates including rhesus macaques from Myanmar (Macaca mulatta), Japanese macaques (Macaca fuscata), and sequences from four primate species housed in zoological parks in Japan; siamang (Symphalangus syndactylus), red-shanked douc (Pygathrix nemaeus), Francois’ langur (Trachypithecus francoisi), and Bornean Orangutan (Pongo pygmaeus) (Ko et al., 2023). To serve as an outgroup for our phylogenetic analysis, we sequenced our S. stercoralis PCR positive control amplicons in the same Illumina library as the vervet samples, and included 28 previously published S. stercoralis genotypes (Barratt and Sapp, 2020), plus a cox1 sequence extracted from the mitochondrial genome of S. stercoralis reference strain PV001 (GenBank [GB] - accession: NC_028624.1). A detailed overview of all specimens within this dataset, including information on hosts, genotype, and GenBank accession numbers, is provided in File S1, Tab A.
2.7. Assigning haplotype names for construction of genotypes
To characterize the genotypes from our reference isolates (and from the St. Kitts vervets), all cox1, 18S HVR-I, and HVR-IV sequences were assigned a haplotype name following earlier haplotype naming conventions developed for Strongyloides sp. (Jaleta et al., 2017; Barratt et al., 2019a; Barratt and Sapp, 2020), with some modifications for cox1 (discussed in a later section). Haplotype names were assigned by BLASTN comparison against the fasta sequences provided in File S2. These BLASTN searches were executed using the Geneious Prime interface, requiring hits of 100% sequence identity. BLASTN results were exported from Geneious in text format, where each text file contained the list of haplotype names detected in a given isolate. These text files were used to construct a haplotype data sheet (HDS); a condensed format for representing haplotype data (File S1, Tab B and Tab D) which is the required input format for computation of genetic distances using Barratt's heuristic (Barratt et al., 2021; Jacobson et al., 2022) – see https://github.com/Joel-Barratt/Eukaryotyping. In all, the HDS contained 30 genotypes from S. stercoralis and 18 genotypes from the loris-derived Strongyloides sp., plus 191 published S. fuelleborni genotypes, and 48 S. fuelleborni genotypes generated here from St Kitts vervets (see results).
2.8. Establishing minimum data requirements and cox1 haplotype definitions
Barratt's heuristic was used to compute pairwise genetic distances for phylogenetic tree construction (Barratt et al., 2019b; Nascimento et al., 2020; Jacobson et al., 2022). Barratt's heuristic was used because it can compute distances for datasets comprising isolates sequenced at different but overlapping combinations of markers. Many of the genotypes examined here were sequenced as part of separate, unrelated studies, so the markers sequenced were not always the same. Barratt's heuristic accommodates such datasets by imputing missing distance values when comparing isolates that have not been sequenced at the same loci (Jacobson et al., 2022). This method was designed for large datasets, and as the number of isolates with mismatched markers increases within a dataset, and/or as the number of shared loci sequenced between a given isolate pair decreases, the more tenuous these imputations become (Barratt and Sapp, 2020; Jacobson et al., 2022). Consequently, realistic minimum data requirements and maximum limits on the number of markers for which imputation is attempted must be established prior to analysis (Jacobson et al., 2022). A detailed description of how these minimum data requirements and maximum imputation limits were established, is provided in Supplementary File S1, Appendix A.
2.9. Distance computation and construction of phylogenetic trees
Genetic distance computation was performed using Barratt's heuristic via the scripts and instructions available here: https://github.com/Joel-Barratt/Eukaryotyping. Our final calibrated HDS was used as input for distance computation and subsequent tree construction. The resultant pairwise matrix was clustered via the ‘agnes’ R package using Ward's method (Ward, 1963) to generate a hierarchical tree. We also generated a neighbor-joining tree (Saitou and Nei, 1987) from the same distance matrix using the ‘nj’ function available in the ‘ape’ R package. The ‘root’ function in the ‘ape’ R package was used to root the tree at the S. stercoralis/Strongyloides sp. ‘Loris’ clade – establishing this clade as the outgroup. The ‘ggtree’ R package was used to visualize and annotate the resultant trees. Images of relevant hosts were obtained from PhyloPic (http://phylopic.org) or prepared in-house for annotation of dendrograms. Maps were generated in R using ggplot. Images were rendered using the GNU Image manipulation program (https://www.gimp.org).
3. Results
3.1. Morphological examination and amplicon sequencing
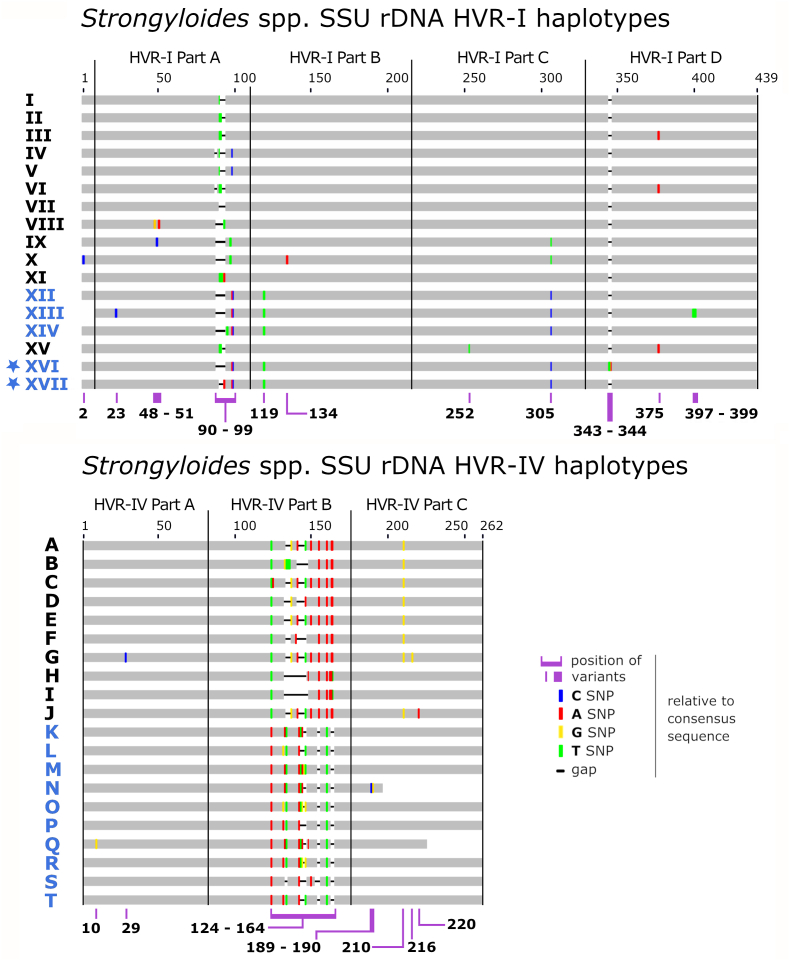
Larvated eggs consistent with Strongyloides sp. were observed microscopically in 29 of the 34 fecal specimens during the secondary microscopic screening performed at CDC (Table 2). HVR-I and HVR-IV sequences were obtained from all 34 specimens, while a cox1 sequence was not obtained for 6 of these. Between one and three Strongyloides sp. cox1 sequences of 217 bp were detected in these 28 specimens. Haplotype XII of HVR-I, which is consistent with S. fuelleborni from African primates (Table 3), was detected in all 34 specimens. Additionally, two novel HVR-I haplotypes (XVI and XVII) were detected. These two sequences were confirmed as belonging to S. fuelleborni based on BLASTN searches against the NCBI nucleotide database where the nearest matches (∼99.5% identity) included hits to S. fuelleborni 18S sequences. Haplotypes XVI and XVII possessed a ‘T’ SNP at position 119, and a ‘C’ SNP at position 305 relative to the alignment in Fig. 1; these SNP's are characteristic of all known HVR-I sequences from S. fuelleborni. The HVR-IV haplotypes L, T, M, O, R, and P detected in these samples are also characteristic of S. fuelleborni from African primates (Table 3).
Table 2.
Results of morphological screen for Strongyloides sp. forms from trapped vervets†
| Animal ID |
Specimen number |
Trapping location |
Strongyloides sp. (eggs) |
Strongyloides-like larvae* |
Cox1 (# of sequences)∗∗ |
HVR-I (haplotypes detected)∗∗∗ |
HVR-IV (haplotypes detected)∗∗∗ |
|---|---|---|---|---|---|---|---|
| CDC morphological screen | PCR and sequencing results (positive: + or negative:) | ||||||
| 9808 | Specimen 1 | Cayon | + | - | + (2)c,f | XII, XVI, XVII | T, M, O R, P |
| 9772 | Specimen 2 | Not recorded‡ | + | - | + (1)f | XII, XVI, XVII | L, T, M, O, R, P |
| 9809 | Specimen 3 | Cayon | + | - | – | XII, XVI, XVII | L, T, M, O, R, P |
| 9749 | Specimen 4 | Not recorded‡ | + | - | – | XII, XVI, XVII | L, T, M, R, P |
| 9767 | Specimen 5 | Not recorded‡ | + | - | + (3)a,e,f | XII, XVI, XVII | L, T, M, R, P |
| 9862 | Specimen 6 | Phillips | + | - | – | XII, XVI, XVII | L, T, M, R, P |
| 9852 | Specimen 7 | Dieppe Bay | - | - | – | XII, XVI, XVII | L, T, M, R, P |
| 9855 | Specimen 8 | Cedar Grove | + | - | + (1)f | XII, XVI, XVII | L, T, M, R, P |
| 9859 | Specimen 9 | Cedar Grove | + | - | – | XII, XVI, XVII | L, T, M, O, R, P |
| 9861 | Specimen 10 | Cedar Grove | + | - | – | XII, XVI, XVII | L, T, M, O, R, P |
| 9858 | Specimen 11 | Cedar Grove | + | - | + (1)f | XII, XVI, XVII | L, T, M, R, P |
| 9863 | Specimen 12 | Phillips | - | - | + (1)f | XII, XVI, XVII | L, T, M, R, P |
| 9788 | Specimen 13 | Cedar Grove | + | - | + (2)d,f | XII, XVI, XVII | L, T, M, R, P |
| 9786 | Specimen 14 | Cedar Grove | - | - | + (1)f | XII, XVI, XVII | L, T, M, R, P |
| 9868 | Specimen 15 | Cedar Grove | + | - | + (2)d,f | XII, XVI, XVII | L, T, M, R, P |
| 9873 | Specimen 16 | Cedar Grove | + | - | + (1)f | XII, XVI, XVII | L, T, M, R, P |
| 9875 | Specimen 17 | Monkey Hill | + | - | + (3)d,e,f | XII, XVI, XVII | L, T, M, R, P |
| 9876 | Specimen 18 | Cedar Grove | + | - | + (1)f | XII, XVI, XVII | L, T, M, R, P |
| 9867 | Specimen 19 | Cedar Grove | + | - | + (2)a,f | XII, XVI, XVII | L, T, M, R, P |
| 9871 | Specimen 20 | Cedar Grove | + | - | + (2)a,f | XII, XVI, XVII | L, T, M, R, P |
| 9866 | Specimen 21 | Cedar Grove | + | - | + (1)f | XII, XVI, XVII | L, T, M, O, R, P |
| 9893 | Specimen 22 | Cedar Grove | + | - | + (1)f | XII, XVI, XVII | L, T, M, R, P |
| 9894 | Specimen 23 | Cedar Grove | + | + | + (2)e,f | XII, XVI, XVII | L, T, M, R, P |
| 9896 | Specimen 24 | Cedar Grove | - | - | + (1)f | XII, XVI, XVII | L, T, M, R, P |
| 9898 | Specimen 25 | Cedar Grove | + | - | + (3)a,b,f | XII, XVI, XVII | L, T, M, R, P |
| 9897 | Specimen 26 | Cedar Grove | + | - | + (3)a,e,f | XII, XVI, XVII | L, T, M, O, R, P |
| 9750 | Specimen 27 | Not recorded‡ | + | + | + (1)f | XII, XVI, XVII | L, T, M, R, P |
| 9758 | Specimen 28 | Not recorded‡ | + | - | + (1)f | XII, XVI, XVII | L, T, M, O, R, P |
| 9845 | Specimen 29 | Dale Mountain | - | - | + (2)e,f | XII, XVI, XVII | L, T, M, R, P |
| 9853 | Specimen 30 | Sir Gillian, Old Road | + | - | + (3)a,d,f | XII, XVI, XVII | L, T, M, R, P |
| 9844 | Specimen 31 | Dale Mountain | + | - | + (2)a,f | XII, XVI, XVII | L, T, M, O, R, P |
| 9842 | Specimen 32 | Cedar Grove | + | - | + (1)f | XII, XVI, XVII | L, T, M, R, P |
| 9850 | Specimen 33 | Lodge | + | - | + (1)f | XII, XVI, XVII | L, T, M, O, R, P |
| 9849 | Specimen 34 | Lodge | + | + | + (3)a,e,f | XII, XVI, XVII | L, T, M, O, R, P |
Notes: HVR-I haplotypes XVI and XVII are novel sequences detected for the first time in this study. Additionally, please note that all sequencing results were obtained from individual fecal samples from individual monkeys (not pooled samples). Therefore, individual vervets harbored multiple genotypes of S. fuelleborni.
These are the results obtained for the secondary microscopic screen performed at CDC after specimens were received there and following the initial screen performed on St. Kitts shortly after specimen collection.
The vervets that produced these specimens are thought to have been trapped at Greenhill, though this is uncertain.
The precise Cox1 sequence/s detected in each specimen can be determined using the footnotes (a-f) below, where GenBank accession numbers (GB) are listed for each Cox1 sequence.
Type 1 - GB: OQ190184.
Type 2 - GB: OQ190185.
Type 3 - GB: OQ190186.
Type 4 - GB: OQ190187.
Type 5 - GB: OQ190188.
Type 6 - GB: OQ190189.
See Table 3 to obtain GenBank accession numbers for HVR sequences.
Table 3.
HVR-I and HVR-IV haplotypes of Strongyloides fuelleborni.
| Haplotypes | GenBank Accession/s | Hosts |
|---|---|---|
| HVR-I haplotypes of S. fuelleborni | ||
| XII | AB453320.1, AB453321.1, AB821045.1, AB821046.1, MN076377 – MN076380, OQ190413 | gorillas, chimpanzees, humans, baboons, vervets∗∗ |
| XIII | AB453322.1 | Gorilla |
| XIV | AB677955.1, AB272235.1, AB453317.1, AB453318.1, AB453319.1 | Japanese macaque |
| XVI∗ | OQ190414 | vervets |
| XVII∗ | OQ190415 | vervets |
| HVR-IV haplotypes of S. fuelleborni | ||
| K | LC085496.1, LC085493.1, LC085492.1, LC085491.1, LC085484.1, LC085494.1, AB526820.1, AB453320.1, AB526823.1, AB526821.1 | chimpanzees and human |
| L∗∗ | LC085490.1, LC085489.1, OQ190398 | gorilla, vervets |
| M∗∗ | LC085486.1, MN076407, OQ190400 | human, vervets |
| N | LC085497.1 | Chimpanzee |
| O∗∗ | AB453322.1, OQ190401 | gorilla, vervets |
| P∗∗ | LC085488.1, AB526824.1, AB526825.1, OQ190402 | chimpanzee, gorilla, vervets |
| Q | AB526822.1 | baboon |
| R∗∗ | AB453321.1, OQ190403 | chimpanzee, vervets |
| S | KY081222.1, AB453317.1, AB453318.1, AB453319.1, AB272235.1, MH045486.1, MH045487.1, MH045487.1, MN076415, MT484268, MT484269 | human, long-tailed macaque, pig-tailed macaque |
| T∗∗ | MN076408, OQ190399 | human, vervets |
Note: haplotype XV of HVR-I was identified in an earlier study (Barratt and Sapp, 2020) and is intentionally excluded from this table as that haplotype belongs to S. stercoralis (table only shows S. fuelleborni haplotypes).
Novel haplotypes detected for the first time in the present study in African vervets on St Kitts.
Haplotypes that have been identified in previous studies from primates in Africa that were also identified here in African vervets from St. Kitts.
Fig. 1.
Schematic of the Strongyloides genotyping scheme referenced here
Graphical representation a Strongyloides sp. genotyping scheme after the description of Barratt et al. (Barratt et al., 2019a; Barratt and Sapp, 2020). This scheme was expanded here to include haplotypes XVI and XVII of 18S HVR-I (indicated by a star) identified here from feral vervet monkeys living on the island of St Kitts (GenBank accessions in Table 3). Haplotype names shown in blue belong to S. fuelleborni and those shown in black belong to other Strongyloides species in accordance with this typing scheme. Haplotype sequences are provided in File S1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Constructing S. fuelleborni genotypes using NGS data from St Kitts vervets
The S. fuelleborni genotypes obtained from the vervets were mixed, where three HVR-I haplotypes (XII, XVI, XVII), and 5 or 6 HVR-IV haplotypes (L, T, M, R, P or L, T, M, O, R, P) were detected in all specimens (Table 2). Therefore, it was not possible to determining the HVR-I and HVR-IV haplotype combinations that belonged to a given cox1 sequence to construct individual underlying genotypes. However, segments B and C of HVR-I are identical for haplotypes XII, XVI and XVII (Fig. 1). Additionally, segments A and C are identical across haplotypes L, T, M, O, R and P (Fig. 1). Subsequently, to maximize genotype completeness for distance computation, all cox1 sequences from vervets were linked to HVR-I segments B and C, plus HVR-IV segments A and C, when constructing genotypes (i.e., haplotype lists) for entry into the HDS. In all, 48 S. fuelleborni cox1 sequences of 217 bp (i.e., segments J3 through O1) were generated from St. Kitts vervets and each was used to construct a single genotype, including the cox1 sequence and the conserved segments of HVR-I and HVR-IV.
3.3. Phylogenetic analysis
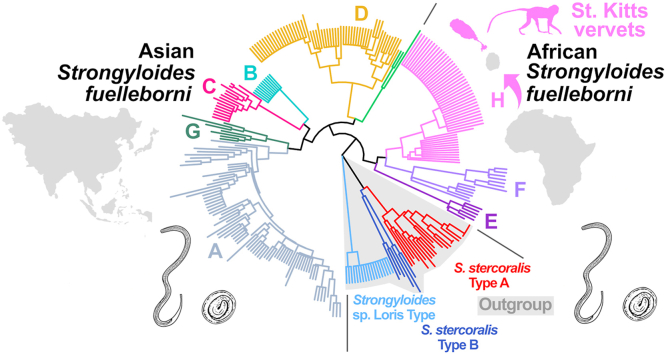
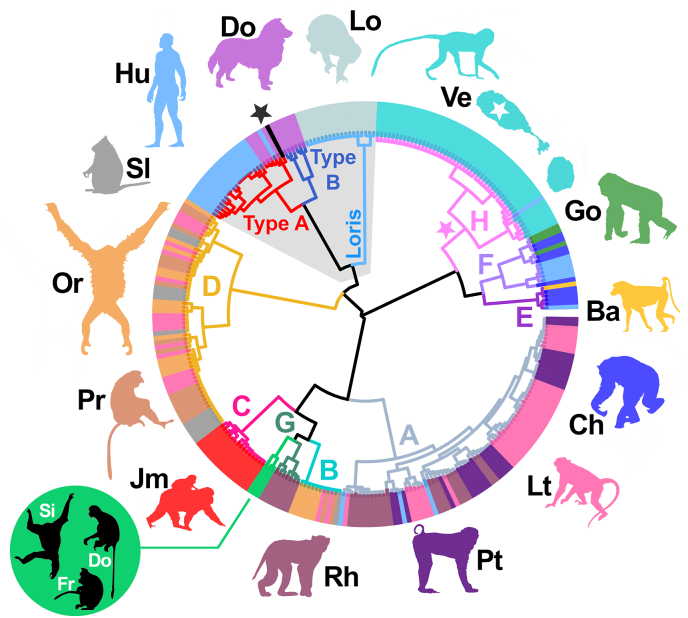
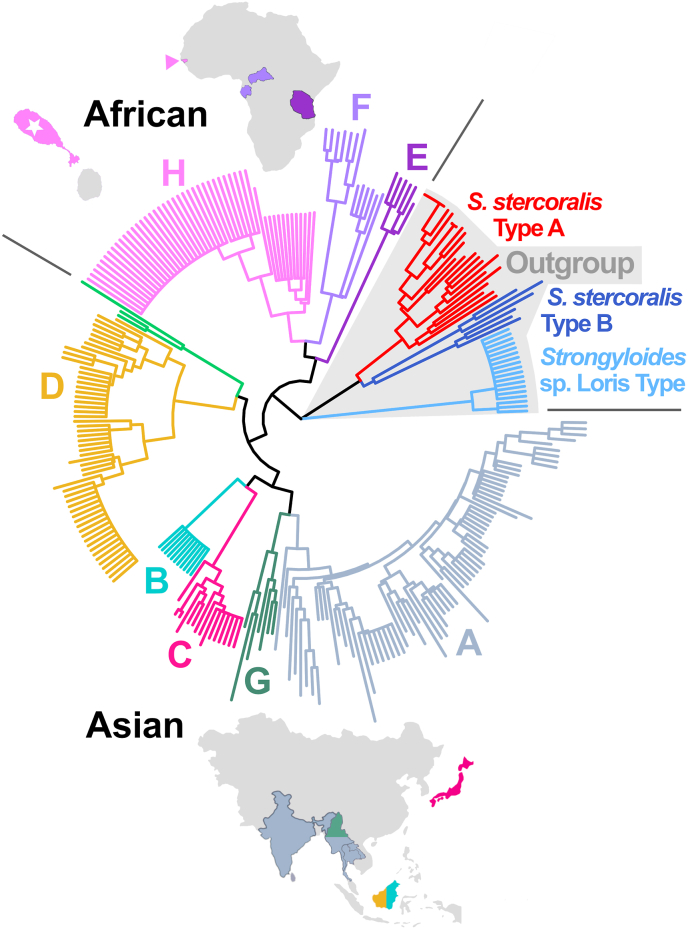
Two of the 287 genotypes failed to meet the minimum data requirements for distance computation (fewer than 14 cox1 segments) and were excluded, leaving 285 genotypes for phylogenetic analysis. A robust phylogenetic reconstruction was achieved by analyzing any HVR data available for these 285 types plus cox1 segments I1 through O1 (i.e., 19 segments, or 285 bp), which constitutes an additional five cox1 segments beyond the 14 segments defined within the 217 bp amplicon (see HDS, File S2, Tab B). The pairwise distance matrix (File S2, Tab C) generated from this HDS was clustered using Wards method to construct a hierarchical tree (Fig. 2), and using the Neighbor-Joining method to produce a phylogeny (Fig. 3). These trees supported that the 48 S. fuelleborni genotypes from St. Kitts vervets were of an African variety, forming a unique cluster within the larger monophyletic African clade. This African clade includes S. fuelleborni types causing infections in chimpanzees, gorillas, baboons, and humans (Figs. 1 and 2). Additionally, an S. fuelleborni genotype sequenced previously from a human in Guinea-Bissau (Barratt et al., 2019a) clustered within the St. Kitts group (cluster H, Fig. 2). The Neighbor-Joining tree revealed a population structure that was consistent with that presented by Ko et al. (2023), which included many of the same reference isolates. As such, these trees were annotated (Figs. 2 and 3) following the convention of Ko et al. (2023) who introduced S. fuelleborni types A through G. Cluster H (introduced here) includes S. fuelleborni from St. Kitts vervets and the type detected in a human in Guinea-Bissau.
Fig. 2.
Hierarchical tree of clustered distances generated from genotyped S. fuelleborni from St Kitts vervets and other Strongyloides sp. genotypes
This unrooted tree was generated using Wards method (Ward, 1963) to cluster a pairwise distance matrix computed from 285 Strongyloides genotypes, including 48 from St. Kitts vervets. Branches are colored according to their cluster membership (A through H). We introduce S. fuelleborni type H (pink star) identified from St. Kitts vervets, noting that an H-type isolate was found previously in a human from Guinea-Bissau. Colored peripheral bars reflect the host species from which isolates were derived; dog (Do), human (Hu), chimpanzee (Ch), lorises (Lo), long-tailed macaques (Lt), pig-tailed macaques (Pt), Japanese macaques (Jm), proboscis monkeys (Pr), silvered leaf monkeys (Sl), orangutans (Or), Rhesus macaques (Rh), St Kitts (white star) vervets (Ve), gorilla (Go), and baboon (Ba). Divergent S. fuelleborni types described by Ko et al. (2023) were also clustered (green circle and branches) from Siamang (Si), Douc (Do), and Francois' langur (Fr) housed in zoological parks in Japan. The black bar and black star indicate S. stercoralis reference strain PV001. Strongyloides stercoralis types A and B, are shown with red and blue branches respectively. The loris clade is shown in light blue. The S. stercoralis and loris clades clustered here for comparison are shaded in a gray background. A version of this same tree with isolate names shown on the branch tips is provided in File S3; Tree A. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
Neighbor-Joining tree generated from St Kitts S. fuelleborni genotypes and other Strongyloides types
This tree was generated by applying the Neighbor-Joining clustering method (Saitou and Nei, 1987) to a pairwise distance matrix computed using Barratt's heuristic from 285 Strongyloides genotypes, including 48 from St. Kitts vervets. Branches are colored according to their cluster membership (A through H) as defined by Ko et al. (2022). We also introduce S. fuelleborni type H (light pink) from St. Kitts vervets, noting that an H-type isolate was found previously in a human from Guinea-Bissau (pink triangle). Divergent S. fuelleborni types described by Ko et al. (2022) were also analyzed (bright green branches without a cluster letter) from Siamang, Douc, and Francois' langur housed in zoological parks in Japan. A version of this same tree with isolate names shown on the branch tips is provided in File S3; Tree B. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
We genetically characterized S. fuelleborni from St. Kitts vervets and confirmed that they are genetically similar to a S. fuelleborni type found previously in a human from Guinea-Bissau (Barratt et al., 2019a). This supports that S. fuelleborni types infecting St. Kitts vervets were brought to the island with the small group of vervets originally introduced by Europeans from Africa during the 17th century (McGuire, 1974). Clustering of S. fuelleborni genotypes from St. Kitts alongside genotypes of other S. fuelleborni varieties demonstrated that St. Kitts S. fuelleborni, in addition to the isolate from Guinea-Bissau, represent a novel variety which we designate as S. fuelleborni Type H, following the convention of Ko et al. (2023).
As S. fuelleborni Type H was identified here in St. Kitts vervets and previously in a naturally infected human, it is possible that these vervets may serve as a reservoir for human infection via zoonotic spillover as vervet populations encroach on human-occupied areas of the island. While human infections with S. stercoralis on St. Kitts have been documented (Ketzis and Conan, 2017), data on S. fuelleborni infections in the human inhabitants of St. Kitts are lacking. However, S. fuelleborni infections in humans are likely underreported globally, due to misidentification of the eggs as those of other nematodes and because the eggs are typically shed in low numbers (Potters et al., 2020). Additionally, rhabditiform larvae may hatch from eggs in the stool if processing is delayed, and the likely outcome of this would be a misidentification as S. stercoralis larvae (which are morphologically identical to those of S. fuelleborni) (Ashford and Barnish, 1989). Therefore, the possibility remains that human infections are occurring or could occur on the island, and isolates from humans in St. Kitts are required to investigate this hypothesis. On the other hand, our data support that St. Kitts vervets are unlikely to be important reservoirs of S. stercoralis infection; given the sensitive deep amplicon sequencing methodology employed, it seems possible that low-level coinfections with S. stercoralis may have been encountered if this were the case, though we observed no evidence for this in the present study.
The findings of Ko et al. (2023) are mirrored here, indicating that globally distributed S. fuelleborni populations display genetic population sub-structuring based largely on geography, with Asian and African varieties being monophyletic. Sub-structuring based on the distribution of specific hosts seems of lesser importance versus geographic origin, suggestive of allopatric divergence. Strongyloides fuelleborni includes a diverse array of geographically and genetically distinct subpopulations (A through H), lending further support to the hypothesis that S. fuelleborni is not a single species but rather exists as a species complex as previously proposed (Barratt and Sapp, 2020). Within these subpopulations, the proportion of human infections appears to vary from rare, such as in S. fuelleborni complex type A to an unusual sub-cluster of S. fuelleborni complex type F which was isolated exclusively from humans. Studies involving broad sampling of both human and non-human primate hosts within regions known to be S. fuelleborni-endemic will be necessary to better understand the predilection for zoonotic versus anthroponotic transmission within the S. fuelleborni complex.
Our choice of Barratt's heuristic for genetic distance computation is noteworthy and has proven useful for investigating relationships among complex groups of sexually reproducing parasites (Jacobson et al., 2022). This approach was first applied to a Strongyloides MLST dataset by Barratt and Sapp (2020) to maximize the variety of Strongyloides types (and the number of loci) that could be included in a clustering meta-analysis, despite that the combinations of loci sequenced varied between studies included in this meta-analysis (Sato et al., 2007; Hasegawa et al., 2009, 2010, 2016; Makouloutou et al., 2014; Huong Giang et al., 2017; Thanchomnang et al., 2017, 2018; Barratt et al., 2019a; Janwan et al., 2020). As discussed above, Barratt's heuristic implements routines that impute distance values for each missing marker when comparing isolates sequenced at different but overlapping marker combinations (Jacobson et al., 2022). It can also compute distances between isolates possessing heterozygosity at one or more loci, where all alleles contribute to resultant tree structures (Jacobson et al., 2022). Despite its unique benefits, Barratt's heuristic possesses some important limitations that are best understood in the context of the rationale underpinning the method, which includes steps that scale dissimilarity scores generated for each locus by frequentist probabilities and entropy (Nascimento et al., 2020). Consequently, this method is best suited to massive datasets where isolates number in the hundreds to thousands (Barratt and Sapp, 2020; Nascimento et al., 2020). This is because the method assumes a wide, representative sampling to help ensure that isolates being analyzed are more likely to represent the diversity of isolates in nature. In line with this, the present study includes numerous isolates of S. fuelleborni from a broad range of geographic locations.
Though Barratt's heuristic allows comparison of sequences with missing data, for accurate imputation, at least some isolates should possess data for all segments of the loci being considered. As detailed above, the imputation steps have limits and attempting to impute values beyond these limits can lead to tenuous tree structures. Therefore, it was necessary to systematically reduce the ‘frame’ of cox1 considered for distance computation to avoid exceeding these imputation limits, which eventually yielded a tree that conformed well to established relationships between key S. fuelleborni varieties included in our reference population (Jaleta et al., 2017; Frias et al., 2018; Janwan et al., 2020; Ko et al., 2023). Calibration of our analysis based on established relationships previously observed among our reference genotypes ensures that our novel S. fuelleborni type H would be placed accurately within our final phylogenetic tree structure.
The prior studies by Ko et al. (2023) and Janwan et al. (2020) present Neighbor-Joining haplotype networks generated by aligning a 710 bp fragment of cox1 from all isolates. As haplotype network analysis (and other alignment-based phylogenetic techniques) requires isolates to be sequenced at precisely the same loci (Jacobson et al., 2022), performing a similar analysis that includes the same isolates analyzed here would require trimming of all cox1 sequences to 217 bp, and exclusion of all HVR data. As our MLST method was designed specifically for adaptation to the Illumina MiSeq platform, the cox1 amplicons generated are intentionally short to ensure they would fit within the span of a single paired-end Illumina read (Barratt et al., 2019a; Beknazarova et al., 2019). Using Barratt's heuristic in this study facilitated a phylogenetic meta-analysis that included almost all S. fuelleborni MLST genotypes currently available in GenBank despite inconsistencies in the loci (including the region of cox1) sequenced between studies, highlighting the utility and potential benefits of this method.
Our genetic characterization of S. fuelleborni from St. Kitts vervets sheds further light on the global population structure of the S. fuelleborni complex while also revealing preliminary evidence of zoonotic risk. Our phylogenetic reconstructions conformed with prescient knowledge of the global population structure of the S. fuelleborni complex. Though reconstruction of complete genotypes based on Illumina sequenced PCR products from fecal DNA was not possible, our data successfully confirmed the African origin of S. fuelleborni from St Kitts and was sufficient to identify S. fuelleborni type H as a previously unrecognized sub-population of the S. fuelleborni complex. Notably, a member of S. fuelleborni type H was observed previously in a human from Guinea-Bissau, highlighting the potential for zoonotic spillover of S. fuelleborni from St. Kitts vervets. Surveillance for human infections on St. Kitts and genotyping of isolates will help to further elucidate the role of vervets as potential reservoirs of human S. fuelleborni infection on the island. Overall, these data add to the growing understanding of S. fuelleborni as a species complex with more nuanced geographic and host relationships than what was assumed prior to the molecular era.
Disclaimer
The findings and conclusions in this manuscript are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention/the Agency for Toxic Substances and Disease Registry.
Author contributions
JB: Conception, study design, PCR, sequencing, data analysis, bioinformatics, data interpretation, manuscript editing. TR: Study design, PCR, sequencing, drafting of manuscript. JKK: Collection and screening of samples, review of manuscript SM: Conception and review of manuscript. SS: microscopy, manuscript editing. YQ: study coordination and funding, review of manuscript.
Declaration of competing interest
The authors of this manuscript have no conflicts of interest to disclose.
Acknowledgements
This research was supported by the Division of Parasitic Diseases and Malaria of the US Centers for Disease Control and Prevention.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2023.02.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- Ashford R., Barnish G. In: Strongyloidiasis: A Major Roundworm Infection of Man. Grove D.e., editor. Taylor & Francis Group; 1989. Strongyloides fuelleborni. [Google Scholar]
- Barratt J., Houghton K., Richins T., Straily A., Threlkel R., Bera B., Kenneally J., Clemons B., Madison-Antenucci S., Cebelinski E., Whitney B.M., Kreil K.R., Cama V., Arrowood M.J., Qvarnstrom Y. Investigation of US Cyclospora cayetanensis outbreaks in 2019 and evaluation of an improved Cyclospora genotyping system against 2019 cyclosporiasis outbreak clusters. Epidemiol. Infect. 2021;149:e214. doi: 10.1017/S0950268821002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt J.L.N., Lane M., Talundzic E., Richins T., Robertson G., Formenti F., Pritt B., Verocai G., Nascimento de Souza J., Mato Soares N., Traub R., Buonfrate D., Bradbury R.S. A global genotyping survey of Strongyloides stercoralis and Strongyloides fuelleborni using deep amplicon sequencing. PLoS Neglected Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt J.L.N., Park S., Nascimento F.S., Hofstetter J., Plucinski M., Casillas S., Bradbury R.S., Arrowood M.J., Qvarnstrom Y., Talundzic E. Genotyping genetically heterogeneous Cyclospora cayetanensis infections to complement epidemiological case linkage. Parasitology. 2019;146:1275–1283. doi: 10.1017/S0031182019000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt J.L.N., Sapp S.G.H. Machine learning-based analyses support the existence of species complexes for Strongyloides fuelleborni and Strongyloides stercoralis. Parasitology. 2020;147:1184–1195. doi: 10.1017/S0031182020000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beknazarova M., Barratt J., Bradbury R., Lane M., Whiley H., Ross K. Detection of classic and cryptic Strongyloides genotypes by deep amplicon sequencing: a preliminary survey of dog and human specimens collected from remote Australian communities. PLoS Neglected Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonfrate D., Bisanzio D., Giorli G., Odermatt P., Furst T., Greenaway C., French M., Reithinger R., Gobbi F., Montresor A., Bisoffi Z. The global prevalence of Strongyloides stercoralis infection. Pathogens. 2020;9 doi: 10.3390/pathogens9060468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz K., Corey T.M., Vandenplas M., Trelis M., Osuna A., Kelly P.J. Case report: control of intestinal nematodes in captive Chlorocebus sabaeus. Onderstepoort J. Vet. Res. 2021;88:e1–e5. doi: 10.4102/ojvr.v88i1.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorris M., Viney M.E., Blaxter M.L. Molecular phylogenetic analysis of the genus Strongyloides and related nematodes. Int. J. Parasitol. 2002;32:1507–1517. doi: 10.1016/s0020-7519(02)00156-x. [DOI] [PubMed] [Google Scholar]
- Frias L., Stark D.J., Lynn M.S., Nathan S.K., Goossens B., Okamoto M., MacIntosh A.J.J. Lurking in the dark: cryptic Strongyloides in a bornean slow loris. Int J Parasitol Parasites Wildl. 2018;7:141–146. doi: 10.1016/j.ijppaw.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher C., Beierschmitt A., Cruz K., Choo J., Ketzis J. Should monkeys wash their hands and feet: a pilot-study on sources of zoonotic parasite exposure. One Health. 2019;7 doi: 10.1016/j.onehlt.2019.100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garn J.V., Wilkers J.L., Meehan A.A., Pfadenhauer L.M., Burns J., Imtiaz R., Freeman M.C. Interventions to improve water, sanitation, and hygiene for preventing soil-transmitted helminth infection. Cochrane Database Syst. Rev. 2022;6 doi: 10.1002/14651858.CD012199.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H., Hayashida S., Ikeda Y., Sato H. Hyper-variable regions in 18S rDNA of Strongyloides spp. as markers for species-specific diagnosis. Parasitol. Res. 2009;104:869–874. doi: 10.1007/s00436-008-1269-9. [DOI] [PubMed] [Google Scholar]
- Hasegawa H., Kalousova B., McLennan M.R., Modry D., Profousova-Psenkova I., Shutt-Phillips K.A., Todd A., Huffman M.A., Petrzelkova K.J. Strongyloides infections of humans and great apes in Dzanga-Sangha Protected Areas, Central African Republic and in degraded forest fragments in Bulindi, Uganda. Parasitol. Int. 2016;65:367–370. doi: 10.1016/j.parint.2016.05.004. [DOI] [PubMed] [Google Scholar]
- Hasegawa H., Sato H., Fujita S., Nguema P.P., Nobusue K., Miyagi K., Kooriyama T., Takenoshita Y., Noda S., Sato A., Morimoto A., Ikeda Y., Nishida T. Molecular identification of the causative agent of human strongyloidiasis acquired in Tanzania: dispersal and diversity of Strongyloides spp. and their hosts. Parasitol. Int. 2010;59:407–413. doi: 10.1016/j.parint.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Hunt V.L., Tsai I.J., Coghlan A., Reid A.J., Holroyd N., Foth B.J., Tracey A., Cotton J.A., Stanley E.J., Beasley H., Bennett H.M., Brooks K., Harsha B., Kajitani R., Kulkarni A., Harbecke D., Nagayasu E., Nichol S., Ogura Y., Quail M.A., Randle N., Xia D., Brattig N.W., Soblik H., Ribeiro D.M., Sanchez-Flores A., Hayashi T., Itoh T., Denver D.R., Grant W., Stoltzfus J.D., Lok J.B., Murayama H., Wastling J., Streit A., Kikuchi T., Viney M., Berriman M. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat. Genet. 2016;48:299–307. doi: 10.1038/ng.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huong Giang N.T., Hoan T.D., Thu Huyen N.T., Kim Lan N.T., Doanh P.N. Morphological and molecular characterisation of Strongyloides ransomi (Nematoda: strongyloididae) collected from domestic pigs in Bac Giang province, Vietnam. Tap. Chi Sinh Hoc. 2017;39:270–275. doi: 10.15625/0866-7160/v39n3.10796. [DOI] [Google Scholar]
- Jacobson D., Zheng Y., Plucinski M.M., Qvarnstrom Y., Barratt J.L.N. Evaluation of various distance computation methods for construction of haplotype-based phylogenies from large MLST datasets. Mol. Phylogenet. Evol. 2022;177 doi: 10.1016/j.ympev.2022.107608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleta T.G., Zhou S., Bemm F.M., Schar F., Khieu V., Muth S., Odermatt P., Lok J.B., Streit A. Different but overlapping populations of Strongyloides stercoralis in dogs and humans-Dogs as a possible source for zoonotic strongyloidiasis. PLoS Neglected Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janwan P., Rodpai R., Intapan P.M., Sanpool O., Tourtip S., Maleewong W., Thanchomnang T. Possible transmission of Strongyloides fuelleborni between working Southern pig-tailed macaques (Macaca nemestrina) and their owners in Southern Thailand: molecular identification and diversity. Infect. Genet. Evol. 2020;85 doi: 10.1016/j.meegid.2020.104516. [DOI] [PubMed] [Google Scholar]
- Ketzis J.K., Conan A. Estimating occurrence of Strongyloides stercoralis in the Caribbean island countries: implications for monitoring and control. Acta Trop. 2017;171:90–95. doi: 10.1016/j.actatropica.2017.03.037. [DOI] [PubMed] [Google Scholar]
- Ketzis J.K., Lejeune M., Branford I., Beierschmitt A., Willingham A.L. Identification of Schistosoma mansoni infection in a nonhuman primate from St. Kitts more than 50 Years after interruption of human transmission. Am. J. Trop. Med. Hyg. 2020;103:2278–2281. doi: 10.4269/ajtmh.20-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko P.P., Haraguchi M., Hara T., Hieu D.D., Ito A., Tanaka R., Tanaka M., Suzumura T., Ueda M., Yoshida A., Maruyama H., Nagayasu E. Population genetics study of Strongyloides fuelleborni and phylogenetic considerations on primate-infecting species of Strongyloides based on their mitochondrial genome sequences. Parasitol. Int. 2023;92 doi: 10.1016/j.parint.2022.102663. [DOI] [PubMed] [Google Scholar]
- Kooriyama T., Hasegawa H., Shimozuru M., Tsubota T., Nishida T., Iwaki T. Parasitology of five primates in mahale mountains national park, Tanzania. Primates. 2012;53:365–375. doi: 10.1007/s10329-012-0311-9. [DOI] [PubMed] [Google Scholar]
- Makouloutou P., Mbehang Nguema P., Fujita S., Takenoshita Y., Hasegawa H., Yanagida T., Sato H. Prevalence and genetic diversity of Oesophagostomum stephanostomum in wild lowland gorillas at Moukalaba-Doudou National Park, Gabon. Helminthologia. 2014;51:83–93. [Google Scholar]
- McGuire M.T. The history of the St Kitts vervet. Caribb. Q. 1974;20:37–52. [Google Scholar]
- Nascimento F.S., Barratt J., Houghton K., Plucinski M., Kelley J., Casillas S., Bennett C.C., Snider C., Tuladhar R., Zhang J., Clemons B., Madison-Antenucci S., Russell A., Cebelinski E., Haan J., Robinson T., Arrowood M.J., Talundzic E., Bradbury R.S., Qvarnstrom Y. Evaluation of an ensemble-based distance statistic for clustering MLST datasets using epidemiologically defined clusters of cyclosporiasis. Epidemiol. Infect. 2020;148:e172. doi: 10.1017/S0950268820001697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutman T.B. Human infection with Strongyloides stercoralis and other related Strongyloides species. Parasitology. 2017;144:263–273. doi: 10.1017/S0031182016000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potters I., Micalessi I., Van Esbroeck M., Gils S., Theunissen C. A rare case of imported Strongyloides fuelleborni infection in a Belgian student. Clinical Infection in Practice. 2020:7–8. doi: 10.1016/j.clinpr.2020.100031. [DOI] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sato H., Torii H., Une Y., Ooi H.K. A new rhabditoid nematode species in Asian sciurids, distinct from Strongyloides robustus in North American sciurids. J. Parasitol. 2007;93:1476–1486. doi: 10.1645/GE-1106.1. [DOI] [PubMed] [Google Scholar]
- Strunz E.C., Addiss D.G., Stocks M.E., Ogden S., Utzinger J., Freeman M.C. Water, sanitation, hygiene, and soil-transmitted helminth infection: a systematic review and meta-analysis. PLoS Med. 2014;11 doi: 10.1371/journal.pmed.1001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanchomnang T., Intapan P.M., Sanpool O., Rodpai R., Sadaow L., Phosuk I., Somboonpatarakun C., Laymanivong S., Tourtip S., Maleewong W. First molecular identification of Strongyloides fuelleborni in long-tailed macaques in Thailand and Lao People's Democratic Republic reveals considerable genetic diversity. J. Helminthol. 2018:1–8. doi: 10.1017/S0022149X18000512. [DOI] [PubMed] [Google Scholar]
- Thanchomnang T., Intapan P.M., Sanpool O., Rodpai R., Tourtip S., Yahom S., Kullawat J., Radomyos P., Thammasiri C., Maleewong W. First molecular identification and genetic diversity of Strongyloides stercoralis and Strongyloides fuelleborni in human communities having contact with long-tailed macaques in Thailand. Parasitol. Res. 2017;116:1917–1923. doi: 10.1007/s00436-017-5469-z. [DOI] [PubMed] [Google Scholar]
- Ward J.H. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963;58:236–244. doi: 10.1080/01621459.1963.10500845. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.