Abstract
In this study, we designed aptamer-based self-assemblies for the delivery of quinine. Two different architectures were designed by hybridizing quinine binding aptamers and aptamers targeting Plasmodium falciparum lactate dehydrogenase (PfLDH): nanotrains and nanoflowers. Nanotrains consisted in controlled assembly of quinine binding aptamers through base-pairing linkers. Nanoflowers were larger assemblies obtained by Rolling Cycle Amplification of a quinine binding aptamer template. Self-assembly was confirmed by PAGE, AFM and cryoSEM. The nanotrains preserved their affinity for quinine and exhibited a higher drug selectivity than nanoflowers. Both demonstrated serum stability, hemocompatibility, low cytotoxicity or caspase activity but nanotrains were better tolerated than nanoflowers in the presence of quinine. Flanked with locomotive aptamers, the nanotrains maintained their targeting ability to the protein PfLDH as analyzed by EMSA and SPR experiments. To summarize, nanoflowers were large assemblies with high drug loading ability, but their gelating and aggregating properties prevent from precise characterization and impaired the cell viability in the presence of quinine. On the other hand, nanotrains were assembled in a selective way. They retain their affinity and specificity for the drug quinine, and their safety profile as well as their targeting ability hold promise for their use as drug delivery systems.
Keywords: Nanotrain, Nanoflower, Targeted delivery, DNA aptamer, Quinine, Malaria
Graphical abstract
Highlights
-
•
Drug-binding aptamers were introduced to enlarge the potential transported drugs.
-
•
Nanotrains and nanoflowers were designed to carry quinine and target malaria.
-
•
Nanotrains preserve the structure, affinity and selectivity of quinine aptamers.
-
•
The affinity of nanotrains for its target protein was determined and confirmed.
-
•
Nanoflowers exhibit high loading capacity but stability and quantification issues.
1. Introduction
Delivering a specific drug to specific tissues or cells remains a great challenge in drug delivery. Conjugating a drug to a specific ligand, e.g. an antibody fragment, a small molecule or an aptamer, improves the accumulation of the drug into specific tissue or cells and limits undesired effects due to the uptake in non-targeted cells, overall increasing the maximum tolerated dose and the therapeutic effect (Xuan et al., 2018). Nucleic acid aptamers have been largely explored as specific ligands due to their unique conformation and ability to bind their target with high affinity and selectivity (He et al., 2020). As compared to antibodies, aptamers offer the advantage of small size, easy chemical synthesis and modification, low immunogenicity and higher tissue penetration (Xuan et al., 2018). Since the first Aptamer−Drug Conjugate reported by Huang et al., composed of the anticancer drug doxorubicin and the sgc8 aptamer targeting PTK7 overexpressing cancer cells (Huang et al., 2009), numerous systems have been developed (Xuan et al., 2018). Most systems use covalent conjugation between the drug and the aptamer and control the release using various stimuli-responsive linkers. However, this strategy is limited by the drug/carrier ratio (usually 1:1). Alternatively, non-covalent conjugation has been explored for intercalating agents such as doxorubicin (Bagalkot et al., 2006). Thanks to the programmable nature of DNA aptamers, they can be easily included into rationally designed bottom-up DNA nanostructures for drug delivery (Madhanagopal et al., 2018). For instance, DNA nanoflowers (NF) result from the rolling cycle amplification (RCA) of a template, usually incorporating targeting and cell imaging elements, allowing the display of multiple targeting elements. Such systems, initially developed by Guizhi Zhu and coll., were explored for their imaging (Zhu et al., 2013a) or drug loading properties (Hu et al., 2014) for cancer cell targeting (Pan et al., 2020) in vitro and in vivo (Yue et al., 2021; Zhu et al., 2013b). Although these nanoflowers exhibited a high versatility, they have only been reported for anthracycline drugs, their dense packing prevents from quick release and their accurate quantification is difficult (Lv et al., 2015).
In parallel, Zhu et al. reported the self-assembled double-strand DNA Nanotrain (NT), formed by the hybridization chain reaction (HCR) of single strand “boxcars” triggered by a targeting “locomotive” aptamer (Zhu et al., 2013a). The resulting nanotrains allowed high loading capacity of doxorubicin and induced selective cytotoxicity in a leukemia model in vitro and in vivo (Zhu et al., 2013a). Nanotrains were further developed using modified nucleotides (Zhang et al., 2020) or a therapeutic peptide (Xu et al., 2019). However, their length was not controlled and only intercalating agents could be loaded on the double strand DNA. Here, we intended to enlarge the design of nanotrains and nanoflowers in order to carry other drugs than DNA intercalators. We incorporated drug-binding aptamers in the boxcar or template design of nanotrains or nanoflowers, respectively, since these aptamers are able to bind a small molecule drug with high affinity and selectivity via non-covalent interactions (Ruscito and DeRosa, 2016). As the aptamer/drug system, we selected the quinine aptamer since this three-way junction aptamer has been extensively described, it displays high affinity for quinine (QN) (KD = 0.2 μM) and quinine binding can be monitored by fluorescence (Neves et al., 2017a). In addition, quinine is an antimalaria drug that needs to be directed to the red blood cells infected by Plasmodium falciparum, to reach schizont-stage of the parasite and prevent from its replication by interfering their hemoglobin digestion ability (Achan et al., 2011). In this work, we hypothesize that aptamer nanotrains and/or nanoflowers could be engineered to carry quinine in a specific and targeted manner. Both DNA aptamer-based drug delivery systems (DDS) have been compared in aspects of characterization, stabilities, drug loading abilities, safety and targeting ability for quinine delivery.
2. Materials and methods
2.1. Chemical and reagents
Oligonucleotides (Table S1) were purchased from Eurogentec (Seraing, Belgium) and were purified by denaturing PAGE (Table 1). The secondary structures of aptamers were predicted by Predict1 (https://rna.urmc.rochester.edu/RNAstructureWeb/Servers/Predict1/Predict1.html). Quinine sulphate monohydrate (QN) and xylene cyanol (XC) were purchased from Sigma Aldrich Chemical Co (Milwaukee, USA). Fetal bovine serum (FBS) was purchased from HyClone (Kremplstrasse, Austria). T4 DNA ligase, Phi29 DNA polymerase, dNTP, recombinant albumin was purchased from New England Biolabs (Evry, France). DNase I (RQ1) was purchased from Promega (Madison, USA), SYBR™Gold was purchased from Invitrogen (ThermoFisher, Bordeaux, France). Ethidium bromide (EB) was purchased from EurobioScientific (Les Ulis, France). ImageLab software (Bio-Rad, freeware) was used for band quantification. Equations used in this study are detailed in Supporting Information.
Table 1.
Dissociation constants and thermodynamic values determined by ITC analysis for the binding between the boxcar A or the NT0 and quinine in PBS (pH 7.4), 5 mM MgCl2 at 15 °C, n = 2.
| Aptamer | KD (μM) | ΔH(kcal/mol) | −TΔS (kcal/mol) | Binding sites N |
|---|---|---|---|---|
| A | 0.61 ± 0.07 | −12.6 ± 0.6 | 4.4 ± 0.7 | 1 |
| NT0 | 0.53 ± 0.09 | −11.8 ± 0.9 | 3.5 ± 1.0 | 4 |
2.2. Preparation of nanotrains and nanoflowers
2.2.1. Construction of nanotrains
All oligonucleotides were heated at 95 °C for 3 min and cooled on ice for 3 min prior to use. To assemble the nanotrains, aptamers were assembled in the desired ratio in PBS supplemented with 5 mM MgCl2 to get a final concentration of 1 μM nanotrain. The mixture was left at room temperature overnight to form the assembly, which was then confirmed by native PAGE (8%, 19:1 Acrylamide: Bis Acrylamide in TB buffer 4 W, 3 h, 4 °C, SYBR™Gold) and visualized by fluorescence (PharosFX™ system, external laser 488, Bio-Rad, Hercules, USA).
2.2.2. Production of nanoflowers by rolling circle amplification (RCA)
Nanoflowers were prepared according to a previous protocol (Lv et al., 2015). Briefly, 0.6 μM of phosphorylated linear template and 1.2 μM of primer were mixed and annealed in DNA ligation buffer (5 mM Tris-HCl pH 7.5, 1 mM MgCl2, 0.1 mM ATP, 1 mM dithiothreitol) by heating at 95 °C for 2 min, followed by gradual cooling to room temperature over 3 h. The annealed product was incubated with T4 DNA ligase (10 U/μL) at room temperature for 3 h to form circularized templates. Then, 0.3 μM of circularized template was incubated with Phi29 DNA polymerase (1 U/μL), dNTP (2 mM/μL), and 0.1 mg/mL recombinant albumin in buffer solution (50 mM Tris-HCl (pH 7.4), 10 mM (NH4)2SO4, 10 mM MgCl2, 4 mM dithiothreitol) at 30 °C for 3, 6 or 24 h. The enzymes were inactivated by heating at 65 °C for 10 min. The formation of nanoflowers was confirmed by agarose gel electrophoresis (2%, 100 V, 30 min). The DNA bands were stained with EB and visualized under UV irradiation (G:Box, Syngene, Biocon, India).
The raw solution of RCA product was sonicated, washed with water (EurobioScientific, DNase/RNase Free), centrifugated at 14,000 g for 10 min, and stored in DNase/RNase free water as stock solution at 4 °C for future use. The mass concentration of the formed nanoflower was determined using a Nanodrop (DS-11+ spectrophotometer, Denovix, mode ssDNA, Wilmington, USA).
2.3. Characterization of nanotrains and nanoflowers
2.3.1. Atomic Force Microscopy (AFM)
AFM imaging was performed on a Bruker Dimension FastScan AFM in liquid using PeakForce Tapping imaging mode with a Bruker MSNL-E probe (nominal spring constant of 0.1 N/m, resonant frequency around 38 kHz and nominal curvature radius of a few nm) (Bruker, Billerica, USA). The PeakForce setpoint was set to 200–300 pN of maximum force to minimize the interactions with the analyzed objects. For sample deposition, a substrate of freshly cleaved muscovite mica was first incubated with 20 μL of 5 mM NiCl2 for 15 min, then rinsed with MilliQ water and air dried to optimize the structure's attachment on the negatively charged mica substrate. For nanotrains the sample could also be prepared without NiCl2 pre-coating since MgCl2 ions are present in the stock solution. For the nanotrain NT0, the stock solution (25 μM) was diluted 5 times before deposition of 10 μL of sample on the mica substrate. For the nanoflowers NF0_6 h stock solution was diluted 3 times before deposition of 20 μL of sample on the mica substrate. All samples were incubated 15 min on mica before rinsing with imaging buffer (HEPES 10 mM pH 7.4, NiCl2 5 mM). To calculate the frequency distributions, the lengths of nanotrains and nanoflowers were measured using the Gwyddion software.
2.3.2. Scanning electron cryomicroscopy (CryoSEM)
The samples were prepared by filling a rivet with the NF0 samples (3 h, 6 h and 24 h), itself mounted on a stub, at room temperature. Freezing was carried out by plunging the stub in liquid nitrogen then vacuum pumping until nitrogen slush formed. Afterwards, the frozen sample was transferred to the preparation chamber (−140 °C), fractured and sublimed (−95 °C). The sublimation was conducted for 7 min (NF0_3 h or 24 h) or 8 min (NF0_6 h), following by sputtering Platinum for the sample metallization for 35 s (NF0_3 h) or 40 s (NF0_6 h or 24 h) to increase conductivity. Specimens were then put into the SEM cryo stage (−140 °C) to be observed, using CRYO-SEM PP3010T (Quorum Technologies, Lewes, England). Observations were done at 3 kV, in high vacuum mode using a GeminiSEM 300 FESEM (ZEISS, Dresden, Germany).
2.4. UV thermal stability
Thermal stability of aptamer assemblies was investigated by UV-spectrophotometry (Cary 100, Agilent Technologies) using 10 mm quartz cuvettes (Hellma Analytics, France, Paris). Aptamer was diluted in 20 mM sodium cacodylate buffer (pH 7.4), 5 mM MgCl2, to reach a final concentration of 1 μM/aptamer. Heating was programmed at 0.2 °C/min from 10 °C to 90 °C then from 90 °C to 10 °C for 3 ramps. The absorbance was recorded at 260 nm and 320 nm. For data analysis, the absorbance was normalized using eq. S13. To determine the Tm values, the first derivative of the melting curve was plotted against temperature using GraphPad Prism 7.0 (n = 3 ramps).
2.5. Isothermal titration calorimetry (ITC)
The ITC method was adapted from Slavkovic et al. (Slavkovic et al., 2018). ITC experiments were performed using a MicroCal VP-ITC instrument (Malvern Panalytical, Malvern, UK). Samples were degassed for at least 10 min at 15 °C prior to use. The binding experiments were corrected for the heat of dilution of the titrant. Titrations were performed in aptamer buffer (PBS complemented with 5 mM MgCl2) at 15 °C with 1.8 mL of 20 μM nanotrain (or aptamer A) in the cell and 300 μL of 312 μM quinine as the titrant. The binding experiments were programmed with an initial delay of 60 s, followed with a first injection of 2 μL. The subsequent 34 injections were 8 μL, spaced every 300 s. The first point was removed from all datasets due to the different injection volume and delay parameters. Data were analyzed by fitting to a one-site binding model using Origin 7 software.
2.6. Binding affinity by fluorescence
Dissociation equilibrium constants (KD) for the aptamer-quinine complexes were obtained by monitoring the quenching of quinine fluorescence (0.1 μM quinine concentration in all samples) upon increasing aptamer concentrations. Different concentrations of nanotrains ([Aptamer] = 8.3, 5, 2, 1, 0.5, 0.25, 0.1, 0.02, 0.004, 0.0008, 0.00016 μM) in PBS, 5 mM MgCl2, containing 0.1 μM quinine buffer were prepared. Fluorescence was measured at 20 °C (λexc = 315 nm, λem = 330–600 nm) using an Edinburgh FS5 Spectrofluorometer (Livingston, England) and 10 mm quartz cuvettes (Quartz SUPRASIL, Hellma Analytics). Control experiments were run using the same aptamer concentrations without quinine in the buffer. For data analysis, fluorescence of each sample was corrected by the background of aptamer without quinine at the same concentration and plotted. To determine the KD value of aptamer assembly, fluorescence at maximum (λexc = 315 nm, λem = 384 nm) was extracted from the spectra for each aptamer concentration and plotted against aptamer concentrations. The binding curves were fitted with model of [inhibitor] vs. response (three parameters) or [Inhibitor] vs. response – Variable slope (four parameters) using GraphPad Prism 7.0.
2.7. Encapsulation Efficiency (EE %) and Drug Loading Capacity (DLC %)
50 μL of nanoflowers (NF0_6 h) raw solution was incubated with 150 μL of 134 μM quinine (final quinine concentration: 100 μM) overnight (Total mass of oligonucleotides: 50 μg, total mass of quinine: 9 μg). Then the mixture was centrifuged at 14,000g for 10 min. The concentration of free quinine was determined by UV–vis spectrophotometry (331 nm, Cary 100, Agilent, Santa Clara, USA) and corrected by subtracting the A331 of NF0_6 h alone. The mass of free oligos in the supernatant was determined by using Nanodrop (Denovix). The mass of formed nanoflower was calculated by subtracting the mass of free oligos from the total oligos, resulting in 20 μg of NF0.
To determine the EE % and DLC % of nanotrains, NT0 (20 μg, 1.3 μM, 200 μL) and quinine (9 μg, 100 μM) were incubated overnight and subsequently centrifuged 14,000 g for 10 min on NanoSep (3 K cut-off, PALL). The absorbance of quinine in the filtrate (331 nm) was measured and corrected by the filter adsorption of quinine according to Eq. S1.
Xylene cyanol was used as a negative control for drug loading, since it exhibits a similar structure and molecular weight than quinine, its aromatic structure allows the UV–vis detection (403 nm) distinctively from DNA (260 nm), and it does not exhibit specific interaction with DNA. To determine the EE % and DLC % of nanoflower and nanotrain for XC, XC (9 μg) was used instead of quinine in the same procedures as above for nanotrains and nanoflowers. The free XC was determined by UV–vis absorbance at 403 nm using the same procedures.
The Encapsulation Efficiency (EE %) and Drug Loading Capacity (DLC %) were calculated by Eq. S2. The stoichiometry was calculated by Eq. S3.
2.8. In vitro stability studies
To assess the stability of NT0 in FBS, NT0 containing 14.2 μM of aptamers was prepared in aptamer buffer (PBS supplemented with 5 mM MgCl2). This NT0 stock was diluted in aptamer buffer as the 0 h unreacted control containing 7.1 μM aptamers or was diluted in 100% FBS to reach a final aptamer concentration of 7.1 μM containing 10%, 50% or 80% FBS. The mixtures were incubated at 37 °C for 24 h. 10 μL of each sample was collected from the tube at 0.5, 2, 6, 24 h and stored at −20 °C until agarose analysis (2% agarose in TB buffer, 100 V, 30 min) or native PAGE (8% Acry/Bis 19:1, 4 W, 2 h, stained by StainsAll™).
The stability of NF0 in FBS was assessed by nearly the same procedures as the NT0, except that the molar concentration of aptamer was replaced by the mass concentration of NF0 quantified by Nanodrop (see EE% section), since the molar concentration of NF was difficult to determine due to their nature of gel and polydispersity.
To obtain the quinine release profile in 10% of FBS, 100 μL of NT0 containing 7.1 μM of aptamers and 7.1 μM of quinine was placed in 96-well plate (Costar 96 Flat Bottom Black) to monitor the quinine release by fluorescence reader (TECAN infinite M1000 pro, Salzburg, Austria) (λexc = 321 nm, λem = 358 nm). After a reading at t = 0 h, 11 μL of 100% FBS (10% final FBS) was added in 100 μL sample. Then the plate was read at 0.5, 1, 2, 6, 24 h.
To obtain the quinine release profile in 50% of FBS, 10 μL of NT0 containing 14.2 μM of aptamers and 14.2 μM of quinine was incubating with 10 μL of 100% FBS (final concentration in the system: 7.1 μM aptamer, 7.1 μM quinine in 50% FBS) in a 384 well plate (Gre384 fb black plate, Greiner, Kremsmünster, Austria). The fluorescence was monitored at 0.5, 1, 2, 6 h (λexc = 321 nm, λem = 358 nm). The data at 0 h was obtained by reading the fluorescence of 20 μL of NT0 containing 7.1 μM of aptamer and 7.1 μM of quinine. The release of quinine % was calculated by Eq. S4.
For DNase degradation studies, nanotrains containing 7.1 μM aptamers were incubated with DNase I (0.01, 0.03, 0.06, 0.09 U/μL, DNase I stock: 1 U/μL RQ1, Promega) at room temperature. 30 μL of samples were collected at 0, 0.5, 1, 2, 4 h and transferred into a 384 well plate (Gre384 fb black plate, Greiner) for quinine fluorescence measurement as described above.
2.9. Cell viability, cytotoxicity and apoptosis assays
HepG2 cells (ATCC) were maintained routinely in RPMI1640 medium supplemented with 10% FBS, 1% PS (100 U/mL penicillin and 100 μg/mL streptomycin sulphate (Gibco, New York, USA)) at 37 °C under a 5% CO2 atmosphere. The cell density was determined by an automated cell counter (BioRad, TC20™) before the experiment.
Cytotoxicity was tested via LDH Assay Kit (Biovision, Waltham, USA). Cell viability was evaluated via PrestoBlue™ reagent (Invitrogen, Eugene, USA). Apoptosis level was determined using Caspase 3/7 activity kit (Promega, Madison, USA). The protocols were adapted from NCL Assay Cascade Protocols (https://www.cancer.gov/nano/research/ncl/protocols-capabilities). Briefly, HepG2 cells were seeded at 2 × 104 cells per well into 96-well plates for 24 h. The nanotrain NT0 (0–962 ng/μL) or nanoflower NF0_6 h (0–96.2 ng/μL) in presence or absence of quinine (0–1.5 μM) were prepared in FBS-free RPMI 1640 medium. The cells were treated with 100 μL of prepared carriers (NT0 ± QN, NF0 ± QN) or quinine alone (50, 10, 2, 0.4, 0.8, 0.016, 0.0032 μM) in FBS-free medium at 37 °C for 2 h. Then, the supernatant medium was removed and replaced by fresh medium (10% FBS + 1% PS, 100 μL) for 24 h. The positive groups (10 mM acetaminophen (APAP) and 5% DMSO) were prepared in complete medium (10% FBS) and were incubated directly with cells for 26 h. The third positive control, 10 μL of 1% Triton X-100, was added to lyse the cells and as LDH high control at 37 °C for 10 min before the LDH assay. Blank controls were cell-free medium treated with the same samples.
To evaluate the cytotoxicity, 10 μL of medium from plate was transferred in a new 96-well plate containing 100 μL of LDH assay buffer, following by incubation for 30 min at room temperature and reading absorbance of the plate at 450 nm (Tecan, Infinite M1000, reference wavelength 650 nm, Mannedorf, Switzerland). Each concentration was tested in triplicate for nanotrains and in duplicate for nanoflowers. Cell cytotoxicity was calculated by Eq. S5.
To evaluate the cell viability, 10 μL of PrestoBlue™ was added in cells, followed by incubation at room temperature for 30 min. The fluorescence was recorded (λexc = 560 nm, λem = 590 nm, 10 nm bandwidth). Each concentration was tested in triplicate for nanotrains and in duplicate for nanoflowers. Cell viability was calculated by Eq. S6.
To evaluate the apoptosis level, 100 μL of Apo-ONE® Caspase-3/7 Reagent (1× Caspase subtract Z-DEVD-R110) was added in the plate, following by incubation at room temperature for 30 min. The fluorescence was measured (λexc = 499 nm, λem = 521 nm, 10 nm bandwidth). Each concentration was tested in duplicate. Cell apoptosis level was calculated by Eq. S7.
2.10. Hemolysis assay
Hemolysis was performed to assess the hemocompatibility of nanotrains and nanoflowers using the procedures described before (Viricel et al., 2015). Briefly, the fresh human red blood cells (HRBC, provided by University Hospital Center of Bordeaux) were rinsed twice by PBS and diluted 1:20 in PBS. 200 μL of HRBC suspension was incubated with NT0 (0–962 ng/μL) or NF0 (0–96.2 ng/μL). Experiments were run in triplicate, including positive (H2O) and negative (PBS) controls. After 45 min incubation at 37 °C, samples were centrifuged at 6000 g during 5 min and the absorbance of the supernatant was measured at 540 nm to determine hemoglobin release. Results were expressed as a percentage of the positive control hemolysis.
2.11. Targeted nanotrains analysis
2.11.1. PfLDH protein production and purification
The plasmid of PfLDH was a kind gift from Pr Tanner (Li Ka Shing Faculty of Medicine, Hong Kong University) and the production process was adapted from his work (Cheung et al., 2013). The plasmid (PfLDH-pET-28a) was amplified in E. coli (XL1 blue) and transformed in Rosetta2 E. coli cells by electroporation for protein expression. Transformed bacterial culture was grown in TB at 37 °C up to an OD600 > 2 and cooled down to 15 °C for an extra 2 h. Induction was performed with 1 mM Isopropyl β- d-1-thiogalactopyranoside (IPTG) followed by overnight expression at 15 °C. The cell culture was centrifuged at 5000 rpm for 15 min. Pellets containing proteins were lysed using 3 passes through the emulsifier in the lysis buffer (1.5 X PBS, 1 mM MgAc2, 0.1% NP-40, 20 mM imidazole, 10% glycerol). After centrifugation at 50,000 g for 1 h, the supernatant was incubated with Cobalt-affinity resin (Sigma-Aldrich) for 20 min. The resin was loaded on a XK16 column (Sigma-Aldrich) and washed with 20 volumes column with a buffer containing 25 mM Tris-HCl, pH 7.5, 100 mM NaCl. The protein was eluted with a linear gradient of buffer containing 250 mM imidazole in 25 mM Tris-HCl, pH 7.5, 100 mM NaCl. Buffer was exchanged by gel filtration (buffer: 25 mM Tris-HCl, pH 7.5, 100 mM NaCl). The purity of each protein was analyzed using SDS-PAGE. The purified proteins were stored in 40% glycerol at −20 °C.
2.11.2. Electrophoresis Mobility Shift Assay (EMSA)
The method of EMSA was adapted from literature (Cheung et al., 2013). EMSA experiments were carried out by incubating 25 nM locomotive (L2 or L3) or nanotrain (NT2 or NT3) with PfLDH (tetramer) at concentrations ranging from 0 to 6.2 μM in 25 mM Tris-HCl at pH 7.5 containing 0.1 M NaCl and 20 mM imidazole at room temperature for 1 h. Reactions were loaded on native PAGE (12%, 37.5:1 acrylamide: bis acrylamide in TB buffer 4 W, 3 h, 4 °C, SYBR™Gold and visualized by Chemidoc Gel Imaging system (Bio-Rad). Band intensities were analyzed using ImageLab software and data were fitted with a model of [inhibitor] vs. response (three parameters) using GraphPad Prism 7.0 for the determination of KD.
2.11.3. Surface plasmon resonance (SPR) binding assays
The SPR experiments were performed at 25 °C with a Biacore T200 instrument (Cytiva), using streptavidin sensor chips (SA) conditioned according to the manufacturer's instructions (Cytiva). Biotinylated L2 and D were prepared in HBS-EP+ running buffer (Cytiva). Thirteen and 230 resonance units (RU) of L2 and D, respectively, were immobilized on one flow cell, at 5 μL/min. A flow cell left blank was used for double referencing of the sensorgrams. The DNA oligonucleotides C, B, A1 and L2, were prepared at 500 nM in the running buffer. Before injection, they were denatured at 95 °C, cooled on ice 3 min and left at least 10 min at room temperature prior to use. The nanotrain was assembled on the sensor chip by injecting on immobilized D successively C, B, A1 and L2 (or buffer) at 5 μL/min for 1 min (see Fig. S14). The pre-assembled mix containing C, B, A1 and L2 (or buffer) was captured on D in similar conditions (5 μL/min, 1 min, see Fig. S15). The protein PfLDH samples were prepared from the stock solution in the running buffer. They were injected at 25 μL/min simultaneously on biotinylated L2 and on the captured nanotrains, using the single cycle kinetics (SCK) or multiple cycle kinetics (MCK) methods. All samples were injected in duplicate (same solution) and at least three independent experiments were performed. To check for specificity of the interaction, streptavidin (SA, IBA-Lifesciences) and PfLDH were injected on the nanotrains, with and without L2 aptamer, respectively. Due to rebinding during the dissociation phase (see Fig. S16), it was impossible to determine the dissociation equilibrium constant, KD, for PfLDH-L2 by direct fitting of the sensorgrams to a Langmuir 1:1 model of interaction, either using the SCK or the MCK methods (see Fig. S17). Therefore KD (value is the average and SD of six independent experiments) was determined by steady-state analysis of the MCK sensorgrams using the Biacore T200 Bia-Evaluation software. Despite double referencing of the SPR signals spikes were still present at the beginning and at the end of the injections. We did not remove them because they did not affect the results. The regeneration of the sensor chip surface was achieved with a 1 min pulse of 20 mM sodium hydroxide.
3. Results
3.1. Construction and characterization of nanotrains and nanoflowers
Two types of DDS were designed for quinine delivery: nanotrains and nanoflowers. First, nanotrains NT0 were designed as the controlled assemblies of boxcars (A, B, C, D). Each boxcar is composed of the quinine loading aptamer MN4 (Slavkovic et al., 2018) flanked with complementary 15-nt sequences, which hybridize to assemble the nanotrain (Fig. 1A, full sequences of aptamers are detailed in Table S1). Importantly, the linker sequences were designed to be specific to each boxcar and did not impact the folding of quinine loading aptamer (as predicted by Predict1). The formation of nanotrain products after mixing equimolar boxcars was monitored by native PAGE (Fig. 1B). Compared to the boxcars alone (A, B, C and D), the bands of dimer (AB), trimer (ABC) and tetramer (ABCD = NT0) shifted to higher positions, indicating the successful stepwise formation of nanotrains. The band quantification shows that the preparation yielded the longest nanotrain as the main product, with >50% yield, while few by-products were observed (Fig. S1). As by-products, we could identify single boxcar A left, suggesting that an underestimation of the aptamer concentration, residual dimers in ABC and residual trimers in ABCD. Specific assembly of boxcars was demonstrated since the incubation of boxcars AC, BD or AD did not yield any dimer formation (Fig. S2). NT0 were observed using AFM imaging in liquid environment revealing a filamentous structure (Fig. 1C). The nanotrains exhibit 2 nm in height, which matches the diameter of double strand DNA, and a mean length of 23.9 ± 5.5 nm (Fig. S3), which agrees with a theoretical length of 20 nm, corresponding to 4 × 15 nucleotides linkers.
Fig. 1.
Formation and characterization of Nanotrains (NT) and Nanoflowers (NF). (A) Scheme of aptamer self-assembled nanotrain NT0. (B) Characterization of the assembly of nanotrains by native PAGE (8%, Acry/Bis 19:1, 4 W, 2 h, total [Aptamer] = 1 μM). (C) AFM image using PeakForce Tapping mode of NT0 in liquid environment. Scale bar is 50 nm. (D) Scheme of aptamer nanoflower NF0. (E) Agarose gel (2%, 100 V, 30 min, 10 μL of NF0 raw solution, [Template], [Ligated] = 0.3 μM, [Primer] = 0.6 μM) after NF0_6 h formation. (F) AFM image in liquid environment using PeakForce Tapping mode of NF0_6h. Scale bar is 2 μm. Diameter range: 0.4–0.7 μM, height range: 0.3–0.7 nm. (G) CryoSEM images of NF0 formed in 6 h (NF0_6 h). Mean diameter 0.8 ± 0.1 μm (n = 10, estimated by Image J software). The samples were analyzed by Zeiss Gemini 300 SEM operating system at a maximum voltage of 3.0 kV. Scale bar is 300 nm.
In parallel, nanoflowers 0 (NF0) were generated by RCA which allows the formation of very long single-strand DNA, as compared to synthetic approaches (Lv et al., 2015). As for quinine delivery, we hypothesized that the longer DNA strands and higher molecular weight of nanoflowers could prolong the blood circulation time as compared to nanotrains, thereby improving the red blood cells targeting. First, the linear template containing the reverse complementary sequence of quinine loading aptamer was hybridized with the primer and ligated by T4 ligase (Fig. 1D, Table S1). Then, the resulting circularized template was amplified by Phi 29 DNA polymerase to form a nanoflower NF0 (Fig. 1D).
The formation of NF0 was confirmed by agarose gel electrophoresis (Fig. 1E), since PAGE could not immobilize NFs. The RCA products (NF0) were trapped in the well whereas the primer, the linear template and the circularized template could migrate, indicating the successful formation of large nanoflowers. NF0 was observed by AFM in liquid environment (Fig. 1F) and CryoSEM (Fig. 1G). Nanoflowers observed by AFM images range from 0.4 μm to 0.7 μm in diameter, and from 0.3 μm to 0.7 μm in height, which is much larger than nanotrains (Fig. S4A). However, they display a large polydispersity and appear to be more aggregated than nanotrains (Fig. S4B). CryoSEM revealed their spherical structures with a petal-like surface and densely-packed core, with a mean diameter of 0.8 ± 0.1 μm (Fig. 1G), in agreement with AFM measurements. Unexpectedly, the size of nanoflowers could not be controlled by the reaction time as previously reported (Lv et al., 2015), since 3 h, 6 h or 24 h reaction time yielded NFs with similar diameters (Fig. S5). However, filamentous structures observed of at 3 h of reaction time suggested that the reaction was not completed. Pursuing the reaction for 24 h yielded aggregated, gelifying NFs which were very difficult to resuspend. Therefore, we selected 6 h as the optimal formation time for the following experiments.
3.2. Thermal stability and quinine binding affinity of nanotrains
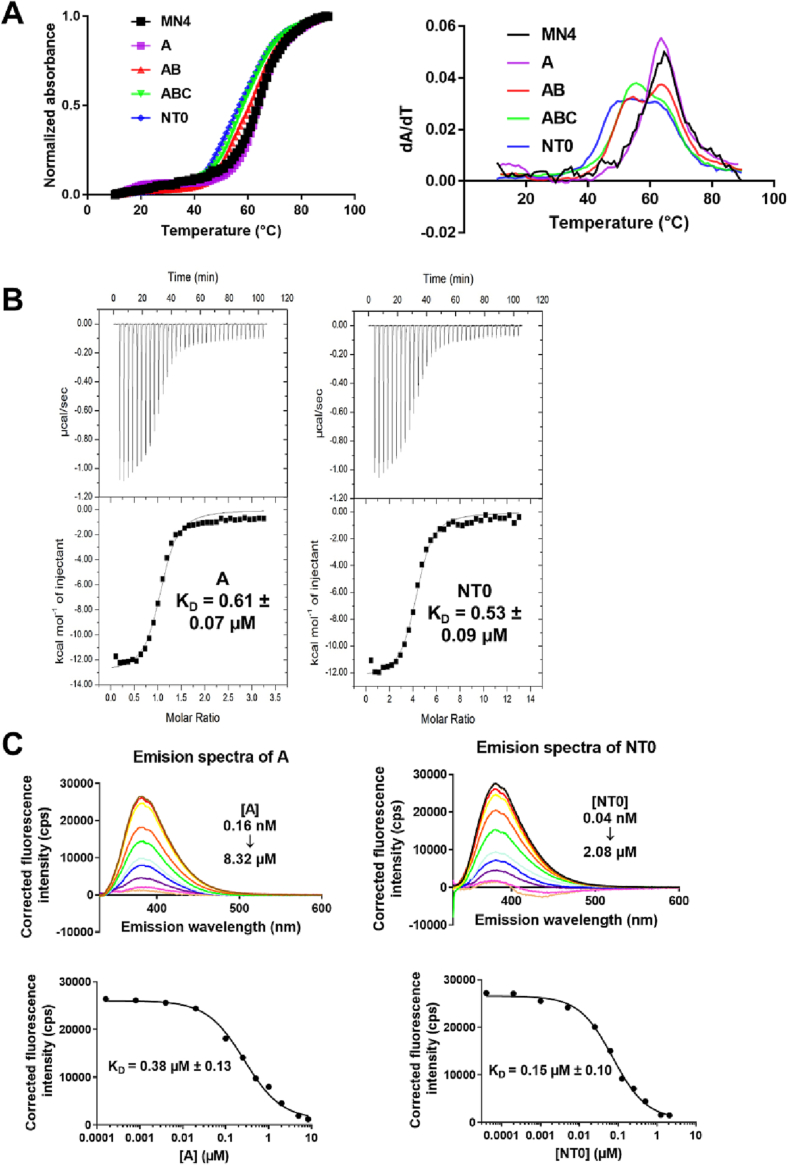
To compare the structure of nanotrains to the single quinine loading aptamer, the thermal stability of assemblies was monitored by UV (Fig. 2A). The single boxcar A exhibited a melting temperature (Tm) of 63.5 °C, in agreement with the quinine loading aptamer MN4 (Fig. 2A). Nanotrains exhibited similar intramolecular Tm (around 63 °C), as well as a second transition temperature ranging from 46 to 57 °C. This latter corresponds to the intermolecular linkers (Table S2), and confirms the hybridization of the nanotrains (Fig. 2A). As expected, magnesium ions (5 mM MgCl2 in PBS) did stabilize the structure of boxcar A as demonstrated by a 10 °C increase of its Tm (Fig. S6A). Interestingly, the presence of quinine did not significantly impact the Tm values (Fig. S6B), probably because MN4 is not a ligand-induced switchable aptamer (Neves et al., 2017b).
Fig. 2.
(A) Thermal stability of aptamer nanotrains. Left: normalized UV absorbance of nanotrains. Right: First derivatives of normalized absorbances for Tm quantification. [Aptamer] = 1 μM/boxcar, 20 mM sodium cacodylate buffer (pH 7.4) with 5 mM MgCl2, 10 to 90 °C, 0.2 °C/min. (B) ITC analysis of quinine (312 μM) binding by the boxcar A or nanotrain NT0 ([Aptamer] = 20 μM) (15 °C in PBS, pH 7.4, 5 mM MgCl2). (C) Top: Fluorescence analysis of quinine binding by boxcar A and nanotrain NT0 ([Quinine] = 0.1 μM, λexc = 315 nm, in PBS pH 7.4, 5 mM MgCl2 at 20 °C). Bottom: binding curves of corrected fluorescence intensity (λexc = 315 nm, λem = 384 nm) fitted with the [Inhibitor] vs. response model (three parameters).
Then, the quinine binding affinity of nanotrains was determined using ITC and fluorescence quenching titration. ITC experiments were performed by titrating quinine into aptamers (Fig. 2B, Table 1). The single boxcar A and nanotrain NT0 exhibited similar values of dissociation constant (KD = 0.61 ± 0.07 μM and 0.53 ± 0.09 μM, respectively), which is consistent with the reported values of MN4 (KD = 0.15 μM) (Neves et al., 2017b). Of note, both enthalpy, entropy and overall free energy were negative and similar to single aptamer, demonstrating the binding of quinine binding was still thermodynamically favourable for the nanotrains. Importantly, the boxcar A displays only one binding site, while the nanotrain NT0 exhibited 4 binding sites without cooperativity, in agreement with its designed structure (Fig. 1A). To confirm the binding affinity values, the intrinsic fluorescence of quinine was monitored upon binding to the nanotrains. As shown in Fig. 2C, fluorescence of quinine was quenched upon addition of boxcar A, leading to a binding curve (using maximum fluorescence at 384 nm). Boxcar A and Nanotrains NT0 exhibited KD = 0.38 ± 0.13 μM and 0.14 ± 0.16 μM, respectively, which are consistent with MN4 aptamer (KD = 0.28 ± 0.06 μM), similarly to dimer AB and trimer ABC (Table S3). No cooperativity could be detected in the binding mechanism, as demonstrated by Hill coefficients (Table S3), which is consistent with the independent nature of the binding sites. The negative control SS1 (MN4 mutation) (Shoara et al., 2017) did not show any binding affinity for quinine (Table S3), demonstrating the specificity of quinine binding on nanotrains. Altogether, thermal stability and affinity studies demonstrate that single boxcar A and nanotrains NT0 display similar thermal stability and binding affinity as quinine loading aptamer MN4 alone, suggesting that the secondary structure of the quinine loading aptamer was retained after being flanked with linkers and assembled into nanotrains. Unfortunately, these studies could not be conducted on nanoflowers since their polydispersity and aggregation impairs UV thermal stability and ITC measurements as well as the precise determination of aptamer concentration within NFs. Therefore, we focused on the drug loading ability of both DDS.
3.3. Drug loading ability of nanotrains and nanoflowers
The drug encapsulation efficiency (EE %) and drug loading capacity (DLC %) are crucial parameters for drug delivery systems. Therefore, the EE % and DLC % of nanotrain NT0 and nanoflower NF0 were determined by UV after incubation with a large excess of quinine or xylene cyanol (XC). This latter is a dye with similar properties to quinine, which exhibits low DNA interaction, and is therefore used as a negative control of quinine to probe non-specific drug loading. As shown in Table 2, both EE % and DLC % were higher for nanoflowers NF0 (24%, 10%) as compared to nanotrains NT0 (7%, 3%) for a total quinine concentration of 100 μM. This EE% value of nanotrains is consistent with the theoretical value of quinine binding efficiency according to its KD (EEtheo% = 5.2%, calculated from eqs. S9 and S10 using KD = 0.5 μM, total quinine 100 μM and total aptamer 5.2 μM). However, the stoichiometry calculation demonstrated that NF exhibited a higher aptamer/drug ratio than expected (5:1 for NF vs 1:1 for NT, details in eq. S3). This suggested that the larger and porous structure of nanoflowers might accommodate a higher amount of quinine in a non-specific fashion. This was confirmed by XC binding, which was not bound by NT0, but was retained by NF0 (EE % = 4% and DLC % = 3%). Altogether, these results demonstrate that both nanoflowers NF0 and nanotrains NT0 can load quinine, however nanotrains NT0 exhibited a better selectivity for quinine compared to nanoflowers NF0.
Table 2.
Encapsulation efficiency and drug loading ability of NF0_6 h and NT0 determined in 200 μL mix solution containing 9 μg quinine (QN) or xylene cyanol (XC) and 20 μg of formed nanocarriers (NF0 or NT0) (n = 3).
| NF0 |
NT0 |
|||
|---|---|---|---|---|
| QN | XC | QN | XC | |
| EE%a | 24 ± 3 | 4 ± 3 | 7 ± 4 | n.d. |
| DLC%b | 10 ± 1 | 3 ± 3 | 3 ± 2 | n.d. |
| Stoichiometryc (Aptamer: drug) | 1:5 | / | 1:1 | / |
Calculated by Eq. S2. The Abs331 of supernatant and nanocarriers alone were determined by UV–vis spectrophotometry.
Calculated by Eq. S2. The mass of formed NF0_6 h was determined using Nanodrop (20 μg).
Molar ratio between aptamer and drug. Calculated by the Eq. S3.
3.4. Serum stability of nanotrains and nanoflowers
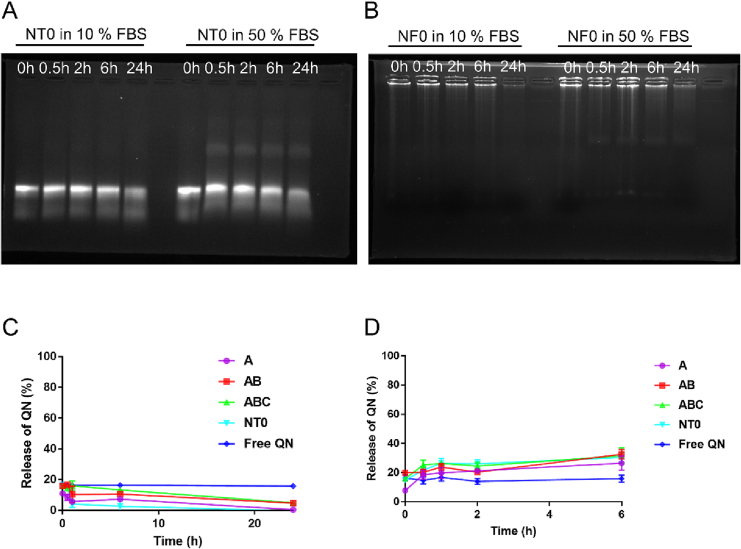
Since DDS are bound to be evaluated in vitro (see next section) or administered in vivo by intravenous administration, their stability towards biological media needs to be investigated. Nanotrains NT0 and nanoflowers NF0 loaded with quinine were incubated in 10% or 50% FBS and their presence was checked by agarose gel. Both NT0 and NF0 were still detected after 6 h in 10% and 50% FBS, whereas their band intensity decreased at 24 h for 10% FBS and after 6 h for 50% FBS, suggesting a partial degradation (Fig. 3A and B). Increasing the FBS concentration decreased the stability time (Fig. S8). In parallel, the quinine release from NT0 was monitored by fluorescence in the same samples (Fig. 3C and D). Considering KD = 0.5 μM, the free quinine concentration was estimated to 1.7 μM (i.e. 23%, eq. S10 and S11), which was used as the baseline concentration (free quinine in Fig. 3C and D). Nevertheless, no fluorescence increase could be detected in 10% FBS as compared to free quinine, confirming the stability of NT0 and its binding to quinine. In 50% FBS, a slightly higher fluorescence suggested that quinine was partially released from NT0 after 2 h, which is likely due to the partial degradation observed on native PAGE (Fig. S7). The eventual biodegradability and quinine release from NT0 were further investigated by a DNase I susceptibility assay (Fig. S9A). Interestingly, NT0 did not release quinine after 4 h in low concentration of DNase I (< 0.01 U/μL), but required 0.03 U/μL of DNase I to initiate quinine release (50% in 4 h). Using this latter DNase I concentration, agarose gel confirmed that NT0 was degraded after 6 h and NF0 after 24 h (Fig. S9B). This suggests a higher stability of nanoflowers in high DNase concentrations (≥ 0.03 U/μL), consistently with previous reports (Lv et al., 2015; Zhu et al., 2013a). Altogether, these results demonstrated that both nanotrains and nanoflowers remain stable in the time course of an in vitro (6 h, 10% FBS) or in vivo (2 h, 50% FBS) experiment, without significant quinine release. Nevertheless, both would eventually be degraded by DNase I (0.03 U/μL). This confirms the biodegradability of both systems and the associated quinine release.
Fig. 3.
Serum stability of nanotrains and nanoflowers. The integrity of (A) Nanotrain NT0 ([Total aptamer] = 7.1 μM, equivalent to 137 ng/μL) or (B) Nanoflower NF0 (137 ng/μL) in 10% or 50% FBS within 24 h characterized by agarose gel (2%, 100 V, 30 min). Quinine (7.1 μM) release profiles of NT0 ([Total aptamer] = 7.1 μM) were monitored in (C) 10% FBS and (D) 50% FBS by fluorescence.
3.5. Cell viability, cytotoxicity, apoptosis and hemocompatibility of nanotrains and nanoflowers
We next investigated nanotrains and nanoflowers on three aspects: cell viability (presto Blue assay), cytotoxicity (LDH assay) and apoptosis (Caspase 3/7 assay) in HepG2 cells, since most toxicity of nanomedicines is detected in the liver after intravenous injection. As shown in Fig. 4, nanotrains NT0 in presence or absence of quinine showed good cell viability up to 1 μg/L (> 80%, Fig. 4A) without significant cytotoxicity (< 20%, Fig. 4B) nor apoptosis (< 3, Fig. S10A), as compared to positive controls 5% of DMSO or 0.1% of Triton X-100. The DNA nature of the blank nanocarriers might even promote cell proliferation. Surprisingly, nanoflowers NF0 loaded with quinine impaired cell viability compared to nanoflowers alone (< 80%, Fig. 4C), while exhibiting low cytotoxicity (< 20%, Fig. 4D) and nearly no apoptosis (Fig. S10B). However, the nanoflowers alone (Fig. 4C and D, Fig. S10B) or the quinine alone (Fig. S11) at the same concentration range were well tolerated on the three criteria. This suggested that the combination of quinine and nanoflowers impaired HepG2 viability cells without triggering membrane permeation nor apoptosis in 48 h. In addition, hemocompatibility was investigated through hemolysis assay, since these DDS are bound to be injected in the systemic circulation to reach infected red blood cells. Both nanotrains NT0 and nanoflowers NF0 exhibited good hemocompatibility since there was no observable hemolysis at effective concentrations of NT0 or NF0 in normal red blood cells (Fig. S12). Altogether, these results suggested that nanotrains are more suitable for quinine delivery than nanoflowers since they are well tolerated by hepatic cells and red blood cells, in presence or absence of drug.
Fig. 4.
Cell viability and cytotoxicity of nanotrain NT0 and nanoflower NF0. Cell viability of HepG2 cells treated with increasing concentrations of (A) NT0 and (C) NF0 using PrestoBlue™ reagent. Cytotoxicity of (B) NT0 or (D) NF0 assessed through LDH leakage. DMSO is a positive control for cell viability and Triton X-100 for LDH release. QN: quinine. APAP: acetaminophen. Results are reported in mass concentration to compare nanotrains and nanoflowers, since NF polydispersity prevents from precise molar concentration determination. Data are presented as mean ± SD (n = 3 for NT0 and n = 2 for NF0).
3.6. Targeting ability of nanotrain and nanoflower
To deliver the drug at the target site is one of the main requirements of DDS. In the case of malaria, several aptamers have been reported for diagnostic purposes. 2008s is a DNA aptamer with hairpin-bulge structures selected by SELEX to recognize Plasmodium falciparum lactate dehydrogenase (PfLDH) (Cheung et al., 2013). Since one advantage of our system is the modular and controlled assembly of aptamers, a “locomotive” aptamer was designed by flanking a complementary 15-nt linker on aptamer 2008s to the extremity of nanotrains NT0 (Fig. 5A). The resulted nanotrains were named NT2. For the nanoflowers, the sequence of 2008s was included in the template and amplified using RCA, yielding nanoflowers (denoted as NF2) with one targeting agent for each quinine loading aptamers (Fig. 5C).
Fig. 5.
Design and preparation of targeted nanotrains and nanoflowers. (A) Scheme of targeted nanotrains flanked with locomotives aptamers L2 or L3. (B) Characterization of the assembly of NT2 by native PAGE (8%, Acry/Bis 19:1, 4 W, 3.5 h, total [aptamer]: 1 μM). (C) Scheme of targeted nanoflowers NF2 (2008s + MN4) formation, by incorporation of targeted sequence into the primer (see full sequence in Table S1). (D) Characterization of formation of NF2 by agarose gel (2%, 100 V, 30 min, 10 μL of NF2 raw solution, [Template], [Ligated] = 0.3 μM, [Primer] = 0.6 μM).
The formation of nanotrains NT2 and nanoflowers NF2 was assessed as described for NT0 and NF0, by PAGE and agarose gel, respectively. As shown in Fig. 5B, the successful formation of nanotrains was demonstrated by a higher shift as compared to the NT0 band. Similarly, NF2 were retarded in the wells as NF0 after RCA, demonstrating the formation of nanoflowers (Fig. 5D).
Next, the binding affinity of targeted nanotrains was checked by EMSA. Since NF were too large to migrate in agarose gels, EMSA was not suited for assessing their binding ability. Therefore, only the binding affinities of locomotive L2 or nanotrains NT2 for PfLDH were determined (Fig. 6 and Table S4). Both displayed a retardation shift on native PAGE, showing their affinity for PfLDH. Binding affinities were in the same range than reported values ((KD 2008s ∼ 56 nM,(Cheung et al., 2013)). In order to verify the versality of our design, we applied this design to another targeting aptamers recognizing PfLDH, pL1 (Cheung et al., 2018) resulting in locomotive L3 and nanotrains NT3 (Fig. S13). The successful formation of the latter was checked by native PAGE (Fig. S13A) and the binding affinity of L3 and NT3 determined by EMSA (Fig. S13B, C and Table S4) suggests that locomotives and nanotrains preserved the target binding ability to PfLDH.
Fig. 6.
Electrophoretic mobility shift assay (EMSA, 8% native PAGE, Acry/Bis 37.5:1, 4 W, 3 h) for locomotive aptamer L2 and nanotrain NT2 ([Locomotive] = 25 nM) binding to PfLDH (tetramer) in range of 0–6250 nM protein. Image obtained by ChemiDoc. EMSA data was fitted with the model of [Inhibitor] vs. response (three parameters) in GraphPad.
This recognition ability was further investigated by SPR. SPR was performed by immobilizing biotinylated locomotive L2 or the biotinylated nanotrain NT2 on the chip following by PfLDH injection. Whether the nanotrain was assembled on the chip (Fig. S14) or pre-assembled before immobilization (Fig. S15), SPR results indicates that NT2 recognized tightly and specifically PfLDH. Indeed, the interaction between NT2 and PfLDH generated nearly 60 RU response compared to the interaction between NT2 and streptavidin (SA, negative control for the protein) or NT0 (without the targeting aptamer) with PfLDH that generated nearly negligible RU (Fig. 7A). The locomotive L2 showed a similar sensorgram as the NT2 (Fig. 7B), demonstrating the targeting ability of nanotrain NT2. The KD of L2 was determined by the method of steady-state affinity analysis, since the 1:1 interaction model could not fit adequately the results (Fig. S17) due to a rebinding phenomenon (Fig. S16). Affinity was determined to be 66 ± 7 nM (Fig. 7C and D), which was consistent with the KD value (59 nM) of aptamer 2008s for PfLDH (Cheung et al., 2013). Importantly, these experiments demonstrate that nanotrains NT2 maintain their binding ability to the target protein PfLDH despite the hybridization of boxcars to the aptamers 2008s.
Fig. 7.
SPR analysis of PfLDH binding to the nanotrains or L2. PfLDH binding to the nanotrain assembled on the sensor chip (A) and to biotinylated L2 (B). The analysis was carried out by the SCK method which consisted in injecting successively increasing concentrations of the analyte (12.5, 50 and 200 nM) without regeneration step between each injection (binding cycles with injections in duplicate). The red, blue and grey curves represent the injections of PfLDH on the nanotrain, on the nanotrain without L2 (buffer injected), and of streptavidin (SA), respectively. The arrows represent the beginning of the injections. (C) Kinetic analysis of PfLDH binding to biotinylated L2 immobilized on the SA sensor chip by the MCK method. After each injection of the analytes (in duplicate) a regeneration step was performed with 1 min pulse of 20 mM sodium hydroxide. KD was determined by steady-state analysis (D) using the Biacore T200 Bia-Evaluation software by plotting as the function of the protein concentration the RU values averaged over 5 s, 1 min and 30 s before the end of the injections. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
DNA aptamers are attracting building blocks to construct nanoscale drug delivery systems. The high binding affinity and specificity of DNA aptamers to their target, their programmability has been exploited for targeting purposes, responsive linker design, as well as drug loading units, in several types of micro and nanocarrier systems (He et al., 2020; Vázquez-González and Willner, 2021).
Nanotrains and nanoflowers have been developed to carry intercalating drugs, e.g. doxorubicin, thanks to their GC-rich sequences (Zhu et al., 2013a, Zhu et al., 2013b). In this work, we introduced drug binding aptamers in the boxcar and template sequences of nanotrains and nanoflowers, respectively, in order to carry selectively other drugs and target other diseases than cancer. As a model system, we selected the antimalaria drug quinine and used an aptamer targeting Plasmodium falciparum (Cheung et al., 2013). We intended to tackle the blood stage of the parasite, by designing a supramolecular assembly exhibiting serum stability and prolonged circulation time after intravenous injection. Such properties are related to the DNA sequence, length, chemistry, and its supramolecular conformation, among other parameters (He et al., 2020). Therefore, we compared two assemblies, namely nanotrains and nanoflowers, since both have demonstrated nanoscale assemblies with i.v. injection ability (Yue et al., 2021; Zhu et al., 2013b).
Including drug-binding sequences in nanotrain design prevented the use of hybridization chain reaction (HCR), reported for nanotrain preparation (Zhu et al., 2013b). Nevertheless, we developed an orthogonal linker design for each boxcar that allowed to control the sequential assembly and length of nanotrains up to 4 boxcars and 1 locomotive. Our design resulted in smaller assemblies than reported nanotrains (20 nm vs. 200 nm by AFM), but allowed a better control over the specific assembly of boxcars and a high production yield (Fig. S1). In addition, the nanotrains NT0 maintained the drug binding affinity and thermal stability as compared to the single aptamer MN4 (Neves et al., 2017b), which suggested that the secondary structure of the three-way junction aptamer MN4 was preserved in the supramolecular assembly. Similarly, the PfLDH targeting aptamer 2008s maintained its targeting properties once flanked with boxcars, as demonstrated by EMSA and SPR for NT2, suggesting that its conformation was preserved. As far as we know, this is the first study determining the affinity constant of nanotrains with their target protein, as previous reports confirmed the targeting ability of nanotrains directly on cells (Pei et al., 2020; Xu et al., 2019; Zhang et al., 2020; Zhu et al., 2013b). We did not observe cooperativity in drug binding for nanotrains, probably because the binding sites of aptamers were independent.
To further increase the lengths of assemblies, which might in turn increase the serum stability and blood circulation time, we designed DNA nanoflowers as reported by Tan et al. (Ali et al., 2014; Thevendran et al., 2020; Zhu et al., 2013a). Unlike reported studies, we were not able to control the size by tuning the reaction time (Lv et al., 2015). Issues in caking and aggregation were consistent with reported studies (Song et al., 2019), and are likely related to the Pi-Pi stacking interactions between DNA bases, but strongly impaired the characterization. Obviously, the large and porous structure allowed high drug loading capacity, although non-specific. Therefore, the preparation process requires optimization before progressing to in vivo administration.
In this study, we demonstrate that a drug binding aptamer can be included in the nanotrain design and its drug binding affinity can be preserved. Since aptamers can be raised against a variety of targets, this strategy could be applied to other small molecules, but also peptides, nucleic acids, sugars or proteins as long as a specific aptamer has been reported to the desired therapeutics. As demonstrated recently, the free or bound drug concentrations can be tuned thanks to the KD, the total aptamer and the total drug concentration (Desrosiers et al., 2022). Thus, these parameters could be tuned to load the nanotrain with a therapeutic concentration of drugs (close to IC50) while keeping the free drug concentration at a non-toxic level for the untargeted cells (Eq. S12). In our model case, the IC50 of quinine for resistant Plasmodium falciparum strains is around 0.5 μM (Ehrhardt et al., 2016). According to the eq. S9 and S10, if 0.5 μM quinine is loaded on nanotrains with 99% of quinine bound fraction, the total aptamer required is 50 μM, which is quite large in a production perspective. Drugs exhibiting lower KD and/or lower IC50 would result in lower aptamer amount, resulting in higher feasibility. Since aptamer MN4 has demonstrated binding affinity for other antimalaria drugs such as chloroquine or artemisinin, with lower KD 37 nM and 30 nM (Slavkovic et al., 2022; Slavkovic et al., 2018), respectively, further studies could involve the loading of these drugs on the nanotrains to reduce the aptamer amount required for therapeutic concentrations and/or evaluate the combination of drugs on the same nanotrain. If required, in vivo stability of DNA nanoassemblies could be improved by chemical modifications of bases (Thevendran et al., 2020) or conjugation to PEG (Mishra et al., 2016) or lipids (Xu et al., 2013), which would increase the circulatory half-life.
5. Conclusion
In this study, we investigated nanotrains as drug delivery systems for other drugs than intercalating agents and how an optimal system can be designed. Our results confirm that DNA aptamers can be assembled in a programmed manner. Importantly, their binding affinity could be maintained in supramolecular assemblies, at least up to 4 boxcars and 1 locomotive for nanotrains. To increase the length of these assemblies, we prepared nanoflowers by Rolling Cycle Amplification. The assemblies were much larger and exhibited a high quinine loading capacity. However, their aggregating properties prevented from thorough characterization. Nanotrains exhibited suitable serum stability and hemocompatibility for intravenous injection, as well as a safety profile on hepatocytes. Notably, the targeting ability of the locomotive aptamer 2008s for PfLDH was maintained after being flanked with drug binding aptamers. In further studies, we will focus on the therapeutic efficiency of such systems, multiple drug loading, as well as their pharmacokinetic profile after iv administration.
Funding
This work was supported by Région Nouvelle-Aquitaine [grant AAPR2020A-2019-8508010], Université de Bordeaux [Idex] and ARNA [Collaborative grants].
CRediT authorship contribution statement
Mengyuan Cao: Investigation, Formal analysis, Visualization, Writing – original draft. Anthony Vial: Investigation, Visualization. Laetitia Minder: Investigation, Visualization. Aurore Guédin: Validation, Writing – review & editing. Sébastien Fribourg: Investigation, Validation, Writing – review & editing. Laurent Azéma: Writing – review & editing. Cécile Feuillie: Methodology, Supervision. Michael Molinari: Methodology, Writing – review & editing, Resources. Carmelo Di Primo: Methodology, Formal analysis, Writing – review & editing, Resources, Funding acquisition. Philippe Barthélémy: Project administration. Leblond Chain Jeanne: Conceptualization, Methodology, Validation, Writing – original draft, Writing – review & editing, Resources, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank Isabelle Svahn (Bordeaux Imaging Center) for cryoSEM images, Pr Julian Tanner (The University of Hong Kong) for PfLDH plasmid, Anne Bourdoncle (ARNA) for ITC training, Sladjana Slavkovic (York University) for ITC data analysis, Sebastien Imbert (CRCTB) for providing blood for hemocompatibility assay.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpx.2023.100172.
Contributor Information
Mengyuan Cao, Email: mengyuan.cao@inserm.fr.
Leblond Chain Jeanne, Email: jeanne.leblond-chain@inserm.fr.
Appendix A. Supplementary data
Supplementary materials containing additional figures, tables and equations
Data availability
Data will be made available on request.
References
- Achan J., Talisuna A.O., Erhart A., Yeka A., Tibenderana J.K., Baliraine F.N., Rosenthal P.J., D’Alessandro U. Quinine, an old anti-malarial drug in a modern world: role in the treatment of malaria. Malar. J. 2011;10:144. doi: 10.1186/1475-2875-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M.M., Li F., Zhang Z., Zhang K., Kang D.-K., Ankrum J.A., Le X.C., Zhao W. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014;43:3324–3341. doi: 10.1039/C3CS60439J. [DOI] [PubMed] [Google Scholar]
- Bagalkot V., Farokhzad O.C., Langer R., Jon S. An aptamer–doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew. Chem. Int. Ed. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- Cheung Y.-W., Kwok J., Law A.W.L., Watt R.M., Kotaka M., Tanner J.A. Structural basis for discriminatory recognition of Plasmodium lactate dehydrogenase by a DNA aptamer. Proc. Natl. Acad. Sci. 2013;110:15967–15972. doi: 10.1073/pnas.1309538110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung Y.-W., Dirkzwager R.M., Wong W.-C., Cardoso J., D’Arc Neves Costa J., Tanner J.A. Aptamer-mediated Plasmodium-specific diagnosis of malaria. Bioch. Aptamer Technol. Appl. 2018;145:131–136. doi: 10.1016/j.biochi.2017.10.017. [DOI] [PubMed] [Google Scholar]
- Desrosiers A., Derbali R.M., Hassine S., Berdugo J., Long V., Lauzon D., De Guire V., Fiset C., DesGroseillers L., Leblond Chain J., Vallée-Bélisle A. Programmable self-regulated molecular buffers for precise sustained drug delivery. Nat. Commun. 2022;13:6504. doi: 10.1038/s41467-022-33491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt K., Deregnaucourt C., Goetz A.-A., Tzanova T., Gallo V., Arese P., Pradines B., Adjalley S.H., Bagrel D., Blandin S., Lanzer M., Davioud-Charvet E. The redox cycler plasmodione is a fast-acting antimalarial lead compound with pronounced activity against sexual and early asexual blood-stage parasites. Antimicrob. Agents Chemother. 2016;60:5146–5158. doi: 10.1128/AAC.02975-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Wen N., Xiao D., Yan J., Xiong H., Cai S., Liu Z., Liu Y. Aptamer-based targeted drug delivery systems: current potential and challenges. Curr. Med. Chem. 2020;27:2189–2219. doi: 10.2174/0929867325666181008142831. [DOI] [PubMed] [Google Scholar]
- Hu R., Zhang X., Zhao Z., Zhu G., Chen T., Fu T., Tan W. DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery. Angew. Chem. Int. Ed. 2014;53:5821–5826. doi: 10.1002/anie.201400323. [DOI] [PubMed] [Google Scholar]
- Huang Y.-F., Shangguan D., Liu H., Phillips J.A., Zhang X., Chen Y., Tan W. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. Chembiochem. 2009;10:862–868. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y., Hu R., Zhu G., Zhang X., Mei L., Liu Q., Qiu L., Wu C., Tan W. Preparation and biomedical applications of programmable and multifunctional DNA nanoflowers. Nat. Protoc. 2015;10:1508–1524. doi: 10.1038/nprot.2015.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhanagopal B.R., Zhang S., Demirel E., Wady H., Chandrasekaran A.R. DNA nanocarriers: programmed to deliver. Trends Biochem. Sci. 2018;43:997–1013. doi: 10.1016/j.tibs.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Mishra P., Nayak B., Dey R.K. PEGylation in anti-cancer therapy: an overview. Asian J. Pharm. Sci. 2016;11:337–348. doi: 10.1016/j.ajps.2015.08.011. [DOI] [Google Scholar]
- Neves Miguel A.D., Shoara A.A., Reinstein O., Borhani O.A., Martin T.R., Johnson P.E. Vol. 7. ACS Sensors; 2017. Optimizing Stem Length to Improve Ligand Selectivity in a Structure-Switching Cocaine-Binding Aptamer. [DOI] [PubMed] [Google Scholar]
- Neves Miguel A.D., Slavkovic S., Churcher Z.R., Johnson P.E. Salt-mediated two-site ligand binding by the cocaine-binding aptamer. Nucleic Acids Res. 2017;45:1041–1048. doi: 10.1093/nar/gkw1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M., Jiang Q., Sun J., Xu Z., Zhou Y., Zhang L., Liu X. Programming DNA Nanoassembly for Enhanced Photodynamic Therapy. Angew. Chem. Int. Ed. 2020;59:1897–1905. doi: 10.1002/anie.201912574. [DOI] [PubMed] [Google Scholar]
- Pei W., Liu M., Wu Y., Zhao Y., Liu T., Sun B., Liu Y., Wang Q., Han J. High payload and targeted release of anthracyclines by aptamer-tethered DNA nanotrains — Thermodynamic and release kinetic study. Eur. J. Pharm. Sci. 2020;148 doi: 10.1016/j.ejps.2020.105319. [DOI] [PubMed] [Google Scholar]
- Ruscito A., DeRosa M.C. Small-molecule binding aptamers: selection strategies, characterization, and applications. Front. Chem. 2016;4:14. doi: 10.3389/fchem.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoara A., Slavkovic S., Donaldson L., Johnson P. Analysis of the interaction between the cocaine-binding aptamer and its ligands using fluorescence spectroscopy. Can. J. Chem. 2017;95 doi: 10.1139/cjc-2017-0380. [DOI] [Google Scholar]
- Slavkovic S., Churcher Z.R., Johnson P.E. Nanomolar binding affinity of quinine-based antimalarial compounds by the cocaine-binding aptamer. Bioorg. Med. Chem. 2018;26:5427–5434. doi: 10.1016/j.bmc.2018.09.017. [DOI] [PubMed] [Google Scholar]
- Slavkovic S., Shoara A.A., Churcher Z.R., Daems E., de Wael K., Sobott F., Johnson P.E. DNA binding by the antimalarial compound artemisinin. Sci. Rep. 2022;12:133. doi: 10.1038/s41598-021-03958-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Zhang Y., Cheng P., Chen X., Luo Y., Xu W. A rapidly self-assembling soft-brush DNA hydrogel based on RCA products. Chem. Commun. 2019;55:5375–5378. doi: 10.1039/C9CC01022J. [DOI] [PubMed] [Google Scholar]
- Thevendran R., Sarah S., Tang T.-H., Citartan M. Strategies to bioengineer aptamer-driven nanovehicles as exceptional molecular tools for targeted therapeutics: a review. J. Control. Release. 2020;323:530–548. doi: 10.1016/j.jconrel.2020.04.051. [DOI] [PubMed] [Google Scholar]
- Vázquez-González M., Willner I. Aptamer-functionalized micro- and nanocarriers for controlled release. ACS Appl. Mater. Interfaces. 2021;13:9520–9541. doi: 10.1021/acsami.0c17121. [DOI] [PubMed] [Google Scholar]
- Viricel W., Mbarek A., Leblond J. Switchable lipids: conformational change for fast pH-triggered cytoplasmic delivery. Angew. Chem. Int. Ed. Eng. 2015;54:12743–12747. doi: 10.1002/anie.201504661. [DOI] [PubMed] [Google Scholar]
- Xu W., Siddiqui I.A., Nihal M., Pilla S., Rosenthal K., Mukhtar H., Gong S. Aptamer-conjugated and doxorubicin-loaded unimolecular micelles for targeted therapy of prostate cancer. Biomaterials. 2013;34:5244–5253. doi: 10.1016/j.biomaterials.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Ni R., Chen Y. Targeting breast cancer stem cells by a self-assembled, aptamer-conjugated DNA nanotrain with preloading doxorubicin. Int. J. Nanomedicine. 2019;14:6831–6842. doi: 10.2147/IJN.S200482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W., Peng Y., Deng Z., Peng T., Kuai H., Li Y., He J., Jin C., Liu Y., Wang R., Tan W. A basic insight into aptamer-drug conjugates (ApDCs) Biomaterials. 2018;182:216–226. doi: 10.1016/j.biomaterials.2018.08.021. [DOI] [PubMed] [Google Scholar]
- Yue S., Li Y., Qiao Z., Song W., Bi S. Rolling circle replication for biosensing, bioimaging, and biomedicine. Trends Biotechnol. 2021 doi: 10.1016/j.tibtech.2021.02.007. [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang S., Yang Z., Hoshika S., Xie S., Li J., Chen X., Wan S., Li L., Benner S.A., Tan W. An aptamer-nanotrain assembled from six-letter DNA delivers doxorubicin selectively to liver cancer cells. Angew. Chem. Int. Ed. 2020;59:663–668. doi: 10.1002/anie.201909691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Guizhi, Hu R., Zhao Z., Chen Z., Zhang X., Tan W. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J. Am. Chem. Soc. 2013;135:16438–16445. doi: 10.1021/ja406115e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Zheng J., Song E., Donovan M., Zhang K., Liu C., Tan W. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc. Natl. Acad. Sci. 2013;110:7998–8003. doi: 10.1073/pnas.1220817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials containing additional figures, tables and equations
Data Availability Statement
Data will be made available on request.