Abstract
Colorectal cancer (CRC) is one of the most common gastrointestinal malignancies around the world with high mortality. Accumulating evidences demonstrate that long non-coding RNAs (lncRNAs) play critical roles in CRC tumorigenesis by regulating different pathways of carcinogenesis. SNHG8 (small nucleolar RNA host gene 8), a lncRNA, is highly expressed in several cancers and acts as an oncogene that promotes cancer progression. However, the oncogenic role of SNHG8 in CRC carcinogenesis and the underlying molecular mechanisms remain unknown. In this study, we explored the role of SNHG8 in CRC cell lines by performing a series of functional experiments. Similar to the data reported in the Encyclopedia of RNA Interactome, our RT-qPCR results showed that SNHG8 expression was significantly upregulated in CRC cell lines (DLD-1, HT-29, HCT-116, and SW480) compared to the normal colon cell line (CCD-112CoN). We performed dicer-substrate siRNA transfection to knockdown the expression of SNHG8 in HCT-116 and SW480 cell lines which were expressing high levels of SNHG8. SNHG8 knockdown significantly reduced CRC cell growth and proliferation by inducing autophagy and apoptosis pathways through the AKT/AMPK/mTOR axis. We performed wound healing migration assay and demonstrated that SNHG8 knockdown significantly increased migration index in both cell lines, indicating reduced migration abilities of cells. Further investigation showed that SNHG8 knockdown suppresses epithelial to mesenchymal transition and reduces cellular migratory properties of CRC cells. Taken together, our study suggests that SNHG8 acts as an oncogene in CRC through the mTOR-dependent autophagy, apoptosis, and EMT pathways. Our study provides a better understanding the role of SNHG8 in CRC at molecular level and SNHG8 might be used as novel therapeutic target for CRC management.
Keywords: Long non-coding RNAs (lncRNAs), Small nucleolar RNA host gene 8 (SNHG8), Colorectal cancer (CRC), Autophagy, Apoptosis, Epithelial-mesenchymal transition (EMT)
Abbreviations
- CRC
Colorectal Cancer
- CCK-8
Cell counting kit-8
- COAD
Colon adeno ductal carcinoma
- DsiNC
Dicer-substrate negative control
- DsiSNHG8
Dicer-substrate SNHG8
- EMT
Epithelial to mesenchymal transition
- ENCORI
The Encyclopedia of RNA interactomes
- GEPIA
Gene expression profiling interactive analysis
- lncRNAs
Long non-coding RNAs
- ncRNAs
Non-coding RNAs
- SEM
Standard error of mean
- SNHG8
Small nucleolar RNA host gene 8
- TCGA
The cancer genome atlas
1. Introduction
Globally, colorectal cancer (CRC) remains one of the most frequently diagnosed gastrointestinal malignancies and the second leading cause of cancer-related death globally [1]. Despite the significant improvements in diagnosis and treatment protocols, the overall survival and clinical outcomes continue to fail to meet treatment expectations. Therefore, the study of molecular mechanisms associated with CRC pathogenesis and identification of effective diagnostic and therapeutic targets are crucial to minimize the global CRC-related mortality. So, the characterization of specific changes at the genomic or transcriptomic level may pave way the development of new therapeutic targets and prognostic biomarker for the management of CRC [2,3]. With the recent development in sequencing technologies, it has been established that nearly 2% of human genome is capable of code protein whereas the major part (>90%) is transcribed as non-coding RNAs (ncRNAs) [2]. The genome-wide transcriptional characterization results revealed that among total human genome, approximately 70% of ncRNAs belongs to its subclass long non-coding RNAs (lncRNAs) and 79% of it remains unannotated [3]. The samples from various cancer and their genome wide association study (GWAS) identified numerous oncogenic and tumour suppressive lncRNAs including CRC [4,5]. The abnormalities of lncRNAs expressions are mostly cells or tissue specific and therefore, it could be a prospective therapeutic targets for systemic CRC treatment [3].
The ncRNAs are primarily classified into two main sub-type including small non-coding RNAs and lncRNAs, based on their nucleotide contents in the transcript [6]. LncRNAs contain more than 200 nucleotides without protein-coding potential [6]. Previously, lncRNAs were considered to be junk RNA molecules, but ample research studies have proved their potential contribution to many cellular pathophysiological processes [[6], [7], [8]]. In addition to that, many studies have reported the central role of lncRNAs in CRC including, initiation, progression, metastasis, and drug resistance process of CRC. For example, our team has reported that upregulation of CASC9 and RAMS11 promotes CRC carcinogenesis through suppression of mTOR-dependent autophagy, apoptosis, and initiating epithelial to mesenchymal transition (EMT) [2,9]. Another study demonstrated that higher RAMS11 expression accelerates resistance to topoisomerase inhibitors chemotherapy via chromobox protein 4 mediated activation of topoisomerase II alpha [7]. Another example is that, Jiang et al. revealed that downregulation of RP11-468E2.5 activates JAK/STAT signaling to promote tumorigenesis whereas, upregulation suppresses malignancies by downregulating STAT5 and STAT6 expression [10]. Zhang et al. demonstrated that upregulated H19 correlated with poor prognosis of CRC patients where it directly binds with heterogenous nuclear ribonucleoprotein A2B1 to activate Raf-ERK pathway, resulting EMT induction to promote migration, and metastasis of CRC cells [11]. Another study illustrated the site-specific diverse regulatory mechanism of lncRNA in CRC. They have reported that Small nucleolar RNA host gene 1 (SNHG1) is highly upregulated in CRC patients compared to the normal tissues, upregulation is associated with tumour progression, development of metastasis and poor prognosis of the diseases [12]. In depth molecular study revealed that SNHG1 epigenetically silence kruppel like factor 2 and frequently mutated tumour suppressor gene cyclin dependent kinase inhibitor 2B expression in the nucleus to promote CRC cell growth and proliferation. Whereas in the cytoplasm, it reduces the suppressive activity of miR-154-5p in cyclin D2 to promote CRC cell proliferation [12]. The Cancer LncRNome Atlas (http://tcla.fcgportal.org/) reported that almost 900 lncRNAs are abnormally expressed in CRC [13]. However, only a limited number of them have been studied in detail. Thus, exploring more CRC associated lncRNAs and their molecular mechanism may provide a new insight of diagnosis, prognosis, and therapeutic targets.
Small nucleolar RNA host gene 8 (SNHG8) is a recently identified lncRNA. It is one of the SNHG family RNA containing 1062 nucleotides in the transcript [14]. SNHG8 was revealed to be abnormally expressed to promote pathogenesis of various cancer including CRC. In a study, Zhen et al. elucidated that expression of SNHG8 is highly upregulated in CRC tissue samples and cell lines to enhance CRC progression by regulating miR-663 [15]. Another study performed by He and colleagues reported that SNHG8 significantly upregulated to enhances CRC cells proliferation by regulating miR-588/ATG7 axis [16]. Both of these studies indicated a partial role of SNHG8 in CRC pathogenesis, but the underlying molecular mechanism in carcinogenesis remains unknown, especially, no studies associated with autophagy, apoptosis in AKT/AMPK/mTOR axis. Therefore, the aim of this study was to investigate the SNHG8 expression in CRC cells lines and explore its function in molecular events including, mTOR-regulated autophagy, apoptosis, and EMT pathways. Our study unveiled that expression of SNHG8 might be used to evaluate CRC prognosis and targeted silencing of SNHG8 could be a novel therapeutic approach for CRC management.
2. Materials and methods
The materials and methods are previously described in our published article [2,9].
2.1. Data mining and analysis
The differential expressions of SNHG8 in CRC tumour and the adjacent normal tissues were obtained from two publicly available dataset: The Encyclopedia of RNA Interactomes (ENCORI) and Gene Expression Profiling Interactive Analysis (GEPIA). The box-plot and overall survival curve were plotted using default settings of both data set.
2.2. Cell lines and culture conditions
The human CRC cell line HT-29-Red-Fluc was acquired from PerkinElmer, Inc. (Waltham, MA, USA). Three more CRC cell lines, DLD-1, HCT-116, and SW480 were kindly provided by Professor Jun YU, Department of Medicine and Therapeutics, Institute of Digestive Disease, The Chinese University of Hong Kong. The human normal colon cell line, CCD-112CoN was purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA). All CRC cell lines were cultured in Dulbecco modified eagle medium (DMEM; Gibco, USA), whereas, normal cells were cultured in Eagle's minimum essential medium (EMEM), (ATCC, VA, USA). Both medium were supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and culture condition was maintained at 37 °C in 5% CO2 in 100% humidity.
2.3. RNA isolation and RT-qPCR
The total RNA from different cell lines were extracted using RNeasy Mini Kit (Qiagen, Germany) according to standard protocol. The RNA concentration was measured by NanoDrop 200 (Thermo Scientific, USA). Following standard protocol, first-strand complementary DNA (cDNA) was synthesized using Superscript II and Random Hexamer primer (Invitrogen, USA). Master Mix LightCycler 480 SYBR Green I (Roche, Switzerland) was used to complete the quantitative reaction using LightCycler 480 Instrument II (Roche, Switzerland). A constant melting temperature and cycle number for amplification were maintained for each reaction. The specific target detection was evaluated by melting curve analysis. Here, GAPDH was used as the housekeeping gene, and relative expression was calculated by 2-ΔΔCt method. The following primer sequences were used in this study: GAPDH, forward: 5′-TGCCATCAATGACCCCTTC-3′ and reverse, 5′-CATCGCCCCACTTGATTTTG-3′; SNHG8, forward: 5′-CCCGAGAACCGTCAGTTTGA-3′ and reverse, 5′-ACACCCGTTTCCCCAACTAC-3′; HPRT1, forward: 5′-TGC TCGAGATGTGATGAAGG-3′, and reverse, 5′-TCCCCTGTT GACTGGTCATT-3′. All primers were purchased from IDT (United States).
2.4. Transfection with dicer-substrate RNA
To knockdown SNHG8, dicer-substrate mediated silencing was performed. Both HCT-116 and SW480 cell lines were seeded and cultured in a six-well plate. Transfection experiment was performed when the cell density reached to 60–70% confluence. A lipid-based in vitro transfection was carried out using Lipofectamine 2000 (Invitrogen, USA) following to the manufacturer's protocol. TriFECTa RNAi kits was purchased from Integrated DNA Technologies (IDT, USA), which contained a DsiNC, positive control (DsiHPRT-S1), transfection control (Ds-TYE 563), and predesigned DsiSNHG8 duplex. The duplex sequences for DsiSNHG8 were as follows: 5′-GAUGGAAACAUAAGACUAUCAAGAA-3′ and 3′-UUCUUGAUAGUCUUAUGUUUCCAUCAU-5′. The DsiNC and DsiHPRT-S1 sequences were not provided by the manufacturer.
2.5. Cell viability assay
After 24 h of transfection, cells were trypsinized and counted by hemocytometer for seeding and performing cell proliferation assay using Cell Counting Kit-8 (CCK-8) (Kumamoto, Japan). 5 × 103 cells in 100 μL of complete medium was seeded and cultured in a 96-well plate. According to the manufacturer's protocol, 10 μl of CCK-8 solution was added to the each well. After 4 h of incubation at 37 °C + 5% CO2, the amount of formazan that represents the number of live cells was measured at an absorbance of 450 nM at SPECTROstar Nano Microplate Reader (BMG Labtech, Germany).
2.6. Colony formation assay
The colony formation assay was performed to evaluate the cell proliferation potential in vitro. After being transfected for 24 h, 1 × 103 cells were seeded in a six-well plate in triplicates and cultured for around 2 weeks. Afterwards, the cell colonies were fixed with a 3:1 mixture of methanol and acetic acid. A solution of 0.5% crystal violet in methanol was used to stain and visualized the colonies. The images were taken, and the number of colonies were counted using the ImageJ software National Institutes of Health (NIH).
2.7. Migration assay
In the migration assay, 7 × 104 cells in 70 μL DMEM with 10% FBS were carefully placed in both compartments of the Culture-Insert 2 Well (Ibidi LLC, Germany). After 24 h of cell settling, the culture inserts were gently removed to create a gap of ∼500 μm for measuring the cell migration ability. Then, each well was filled with 1.5 mL of complete medium. The photographs of the wound areas were taken using an inverted microscope (Nikon, Japan) at various time points of 0, 24, and 48 h, respectively. The migration index indicating the size of the gap was measured using the MRI Wound Healing Tool in ImageJ (NIH).
2.8. Western blotting
Western blotting was performed using standard, established protocol as previously published [9]. Briefly, protein isolation was performed using RIPA lysis and extraction buffer (Thermo Fisher Scientific, United States) with a supplement of cOmplete ULTRA Tablets, Mini EDTA-free, Easy pack Protease Inhibitor Cocktail (Roche, Switzerland). Protein concentration was quantified using BCA Protein Assay Kit (Thermo Fisher Scientific, United States), and similar amounts of proteins were loaded and run on 8–12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis at ambient temperature. Proteins were then transferred onto Immun-Blot PVDF membrane (Bio-Rad Laboratories, Inc., United States), followed by 2-h blocking in 5% bovine serum albumin (BSA) (Hyclone BSA; GE Healthcare Life Science, United States) in Tris-buffered saline with a supplement of 0.1% Tween 20. Then, the blocked membrane was incubated overnight with primary antibodies: β-actin [#8457, Cell Signaling Technology, Inc., (CST, United States)], GAPDH (#2118, CST), AKT (#9272, AKT, Phosphor-AKT (#9271, CST), AMPKα (#5832, CST), phosphor-AMPKα (#2535, CST), E-cadherin (#3195, CST), N-cadherin (#13116S, CST), LC3B (#2775, CST), mTOR (#2972, CST), Phosphor-mTOR (#2535, CST), and vimentin (#5741S, CST) at 4 °C. The secondary anti-rabbit immunoglobulin G (IgG), horseradish peroxide (HRP)–linked, or anti-mouse IgG-HRP–linked (#7076, CST) antibody were added and incubated with the membrane for 2 h. Afterwards, Western Lightning Plus-Electrochemiluminescence (PerkinElmer, Inc., United States) was added to the membrane to visualize protein bands in a ChemiDoc MP Imaging System (Bio-Rad Laboratories, Inc., United States). The relative protein expressions were quantified using ImageJ software (NIH) with β-actin or GAPDH as internal control.
2.9. Statistical analysis
The mean ± standard error of mean (SEM) of at least three or more independent experiments were included for analysis. The statistical level of the experimental data was calculated by Student t-test or one-way analysis of variance using GraphPad Prism version 8.0 (GraphPad Software, Inc., San Diego, CA, United States). P < 0.05 is considered statistically significant.
3. Results
3.1. Abnormal expression of SNHG8 in CRC and dicer-substrate knockdown
Firstly, we analyzed the SNGH8 expression profile in an online based platform “The Encyclopedia of RNA Interactomes (ENCORI)”. The boxplot was plotted using the Cancer Genome Atlas Program (TCGA)-colon adeno ductal carcinoma (COAD) dataset containing 417 CRC tumour samples and 41 normal tissues. The ENCORI results shown that SNHG8 expression was markedly higher in CRC tissues samples compared to the adjacent normal tissues (Fig. 1A). Secondly, we evaluated the clinical outcome of CRC patients associated with SNHG8 expression. To do so, we used another publicly open web-based platform, called “Gene Expression Profiling Interactive Analysis (GEPIA). The TCGA-COAD dataset of GEPIA platform containing 275 CRC tumours and 349 normal tissues and overall survival curve was plotted considering log-rank P value less than 0.05, hazard ratio, and expression profiles of SNHG8 patient samples. The overall survival curve shown that higher SNHG8 expression is associated with worse prognosis of CRC (Fig. 1B).
Fig. 1.
SNHG8 overexpression in CRC cells and patient samples. (A) Boxplot analysis of SNHG8 was plotted using ENCORI-COAD dataset. The expression of SNHG8 was markedly upregulated in 417 CRC tumour samples compared with 41 normal tissues. (B) The overall survival was plotted using TCGA-COAD dataset in web-based platform the Gene Expression Profiling Interactive Analysis (GEPIA). The expression profile, TPM number, and hazard ratio were considered to plot the overall survival. Higher expression of SNHG8 associated with poor overall survival of patient. (C) The RT-qPCR was employed to evaluate the expression of SNHG8 in four CRC cell lines (DLD-1, HT-29, HCT-116 and SW480) and normal colon cells (CCD-112CoN). The data was shown as mean ± SEM compared to normal cells of six experiments. (D) The dicer-substrate DsiSNHG8 knockdown was performed to suppress the SNHG8 expression in most abundantly expressed HCT-116 and SW480 cells. The data was shown as mean ± SEM compared to negative control (DsiNC) group. (**P < 0.01, ***P < 0.001, and n = 4).
Then, the expression of SNHG8 was investigated in human normal colon cell line CCD-112CoN as well as four CRC cell lines namely DLD-1, HT-29, HCT-116, and SW480 using RT-qPCR (Fig. 1C). The in vitro results shown that SNHG8 level in CRC cells was significantly upregulated compared to normal cells, similar to our findings in the public database. To investigate the biological role of SNHG8 in CRC cells, we performed dicer mediated loss-off functions assay in HCT-116 and SW480 cell lines as SNHG8 was most endogenously expressed in these two cell lines. Moreover, our previous experiments and published data from other research group suggested that HCT-116 and SW480 cells are more resistance to phenotypic changes and are commonly used cell lines in CRC study. The knockdown efficiency of dicer-substrate SNHG8 (DsiSNHG8) compared to dicer-substrate negative control (DsiNC) in both cells were excellent (Fig. 1D) and validated by suppressing a gene of positive control (HPRT-1).
3.2. Knockdown of SNHG8 reduces CRC cell proliferation, colony formation and migration
To investigate the pathophysiological role of SNHG8 in CRC cells, HCT-116 and SW480 cell lines were transfected with DsiSNHG8 and DsiNC. Afterwards, CCK-8 assay, colony formation assay and wound healing migration assay were performed in HCT-116 and SW480 cell lines to evaluate their growth, proliferation and migration abilities (Fig. 2A–C). The results showed that DsiSNHG8 treated cells significantly reduced cell proliferation for 24, 48, and 72 h respectively (Fig. 2A). Furthermore, the results from the colony formation assay revealed that the number of colonies formed by DsiSNHG8 treated cells were significantly reduced compared to DsiNC treated cells (Fig. 2B). In concordance with the cell growth and proliferation, we investigated whether knockdown of SNHG8 promote or suppress CRC cells migration. To do so, both HCT-116 and SW480 cells were assessed for their migration ability using wound healing migration assays. After 24- and 48-h post-transfection with DsiSNHG8 in HCT-116 and SW480 cells, we found that knockdown of SNHG8 significantly increases the migration index compared to DsiNC group (Fig. 2C).
Fig. 2.
Knockdown of SNHG8 reduced CRC cell proliferation, colony formation and migration. (A) To evaluate the proliferation of HCT-116 and SW480 after SNHG8 knockdown, CCK-8 assay was performed. The silencing of SNHG8 significantly inhibited proliferation in both cell lines at 24, 48, 72, and 96 h after DsiSNHG8 treatment. (B) The number of colonies were also reduced significantly in both HCT-116 and SW480 cell lines after DsiSNHG8 transfection. (C) The wound healing migration assay was employed to evaluate migration index of HCT-116 and SW480 cells after DsiSNHG8 treatment. The results shown that DsiSNHG8 transfected cells significantly increased migration index of both cells for 24 and 48 h compared to negative control group. The data was shown as mean ± SEM compared to DsiNC. (*P < 0.05, **P < 0.01, ***P < 0.001, and n = 4).
3.3. Knockdown of SNHG8 induce autophagy
To investigate the molecular mechanism associated with cell proliferation, growth and migration, western blot was employed to assess the self-degradative process autophagy. Autophagy levels in DsiSNHG8 and DsiNC treated CRC cells were evaluated by measuring the expression levels of major autophagy markers, LC3B, p62, and Beclin-1. Our results showed that knockdown of SNHG8 significantly induce autophagy by promoting ratio of LC3-II/I in HCT-116 and SW480 cells (Fig. 3A and B). The knockdown of SNHG8 also significantly reduced the p62 expression in both cell lines (Fig. 3A and B). Moreover, western blotting results also shown that DsiSNHG8 transfected cells significantly enhanced Beclin-1 levels in HCT-116 and SW480 cells compared to DsiNC (Fig. 3A and B). Overall, knockdown of SNHG8 induce autophagy by increasing LC3B and Beclin-1 levels whereas trigger flux by reducing p62 expression.
Fig. 3.
The autophagy marker proteins LC3B, p62, and Beclin-1 expression were measured using western blotting in HCT-116 and SW480 cells. (A, B) The DsiSNHG8 treated cells increased the ratio of LC3-II/I and Beclin-1 with corresponding reduction of p62 expression. The data was shown as mean ± SEM compared to DsiNC using β-actin as housekeeping gene. (*P < 0.05, **P < 0.01, ***P < 0.001, and n = 4).
3.4. Knockdown of SNHG8 promote cellular apoptosis
Furthermore, we examined the role of SNHG8 in the process of cellular apoptosis. Similar to the autophagy, western blot of apoptosis markers Bcl-2 and Bcl-xL were used to assess the involvement of SNHG8 in the apoptosis pathway. Bcl-2 family proteins regulate intrinsic apoptosis pathway through mitochondrial outer membrane permeabilization. Therefore, downregulation of Bcl-2 family proteins promotes apoptosis to enhance cell death. As shown in Fig. 4A and B, knockdown of SNHG8 significantly reduced the expression of Bcl-2 and Bcl-xL in HCT-116 and SW48 cells. These overall results suggested that downregulation of SNHG8 induce intrinsic apoptosis pathway to accelerates programmed cell death process.
Fig. 4.
Protein expression of Bcl-2 and Bcl-xL in DsiSNHG8 transfected cells compared to NC group. (A, B) The western blotting in HCT-116 and SW480 cells revealed that knockdown of SNHG8 significantly promote cellular apoptosis by reducing Bcl-2 and Bcl-xL expression in both cell lines. The data was shown as mean ± SEM compared to DsiNC using β-actin as housekeeping gene. (*P < 0.05, **P < 0.01, ***P < 0.001, and n = 4).
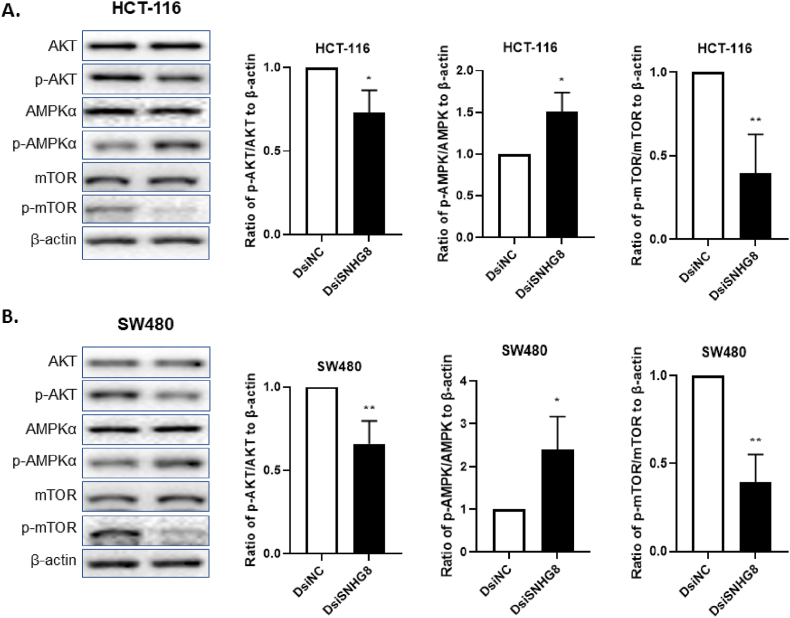
3.5. Knockdown of SNHG8 dephosphorylate AKT/mTOR axis through activation of AMPKα pathway
After revealing the oncogenic role of SNHG8 in CRC cells growth, proliferation and migration through autophagy and apoptosis pathways, we investigated the most frequently altered mTOR pathway associated with it. We analyzed the phosphorylation of mTOR signaling with its upstream and downstream effectors AKT and AMPKα. We found that the expression of AKT, AMPKα and mTOR in both HCT-116 and SW480 cells remained unchanged after transfecting with DsiNC and DsiSNHG8 (Fig. 5A and B). In the contrary, the expression of phosphorylated proteins p-AKT, and p-mTOR were significantly downregulated in both cell lines (Fig. 5A and B) after transfecting with DsiSNHG8. In addition, DsiSNHG8 treated cells significantly evaluated the expression of p-AMPKα compared to DsiNC (Fig. 5A and B). Taken all these together, our findings suggest that SNHG8 knockdown phosphorylate AMPK pathway through dephosphorylation of AKT and mTOR signaling.
Fig. 5.
The Knockdown of SNHG8 activate AMPK pathway by downregulating AKT/mTOR axis. (A, B) The ratio of p-AMPKα/AMPKα, p-AKT/AKT, and p-mTOR/mTOR to β-actin were measured using western blotting in HCT-116 and SW480 cells. The DsiSNHG8 transfected cell significantly inhibit phosphorylation of AKT and mTOR by activating AMPKα. The data was shown as mean ± SEM compared to DsiNC using β-actin as housekeeping gene. (*P < 0.05, **P < 0.01, and n = 4).
3.6. Knockdown of SNHG8 negatively regulates EMT pathway
To investigate the molecular mechanism associated with higher migration index for development of metastasis, western blotting of EMT markers, E-cadherin, N-cadherin, and Vimentin were performed. The results of our study showed that DsiSNHG8 transfected cells significantly promoted the epithelial marker E-cadherin in both HCT-116 and SW480 cell lines (Fig. 6A and B). Alternatively, SNHG8 knockdown displayed (Fig. 6A and B) significantly reduced expression of mesenchymal marker proteins N-cadherin and Vimentin after DsiSNHG8 treatment. Our observation indicated that SNHG8 is involved in the EMT pathway to promote migration and metastasis.
Fig. 6.
Investigation of EMT characteristics of HCT-116 and SW480 cells. (A, B) The EMT marker proteins E-cadherin, N-cadherin and Vimentin expression were evaluated by western blotting. The DsiSNGH8 medicated knockdown significantly increased E-cadherin and decreased N-cadherin and Vimentin expression in both cell lines compared to DsiNC group. The data was shown as mean ± SEM compared to DsiNC using β-actin as housekeeping gene. (*P < 0.05, **P < 0.01, and n = 4).
4. Discussion
CRC remains one of the most common malignancies and its morbidity and mortality dramatically increased in the recent decades. It is established that early diagnosis of CRC facilitate desired outcomes of patient. However, majority of the CRC cases are diagnosed in advanced distant metastatic stages with frustrating outcomes [17]. Despite the advances in CRC treatment protocols, the prognosis did not reach to the satisfactory level due to lack of reliable biomarkers and poor understanding of the underlying pathophysiological mechanism [18]. In recent years, rapid development of genomics technology focused on the potential role of lncRNAs to establish a new diagnostic and prognostic biomarkers for CRC patients [2,7,9,19,20]. The lncRNAs exert their oncogenic or tumour suppressive roles in CRC pathogenesis by making a network with host DNA, RNA and proteins [21,22]. Several studies confirmed that lncRNAs are able to control various proteins, genes, transcripts and miRNA expression during the tumour differentiation and developmental process [23]. Moreover, they participate in competitive endogenous network to regulate the gene expression profiles in transcriptional, post-transcriptional and in epigenetic levels [[20], [21], [22], [23]]. The research from past decades revealed that aberrant expression of lncRNAs promote or suppress CRC pathogenesis via modulating major oncogenic cascades including, p53, Wnt/β-catenin, EMT, PI3K/Akt, mTOR, AMPK, EGFR, NOTCH, DNA mismatch-repair, autophagy, apoptosis, and angiogenesis [9,21,[24], [25], [26]]. More importantly, lncRNAs expressions are also involved in CRC chemoresistance that regulate by genes, proteins, and miRNAs associated with proliferation, migration, invasion, and metastasis [21,27]. Although, the potential regulatory roles of lncRNAs have been discussed by many research groups, the molecular mechanism of each lncRNAs in individual cancer types are still largely underexplored. In the present study, we first evaluate the SNHG8 expression profile in online databases. Our result revealed SNHG8 upregulation indicating that it may be associated with CRC progression and metastasis. Furthermore, we explored how SNHG8 is involved in autophagy, apoptosis, phosphorylation of AKT/AMPK/mTOR and EMT axis.
There are many scientific investigations demonstrated that abnormal expression of lncRNAs promotes CRC carcinogenesis whereas silencing could potentially suppress the tumour pathogenesis [2,7,[10], [11], [12]]. Similarly, we performed in vitro SNHG8 loss-of function to assess its pathophysiological impact in CRC cell lines. The results from our study confirmed that SNHG8 knockdown significantly inhibits growth, proliferation, and migration of CRC cells. Indeed, these results are consistent with the previous demonstration where silencing of lncRNAs RAMS11, CASC9, H19 and SNHG1 remarkably increase chemo-sensitivity by suppressing CRC cell proliferation, invasion, migration, and metastasis [2,7,9,11,12]. Importantly, previous studies reported that SNHG8 associated with progression of several malignancies including, gastric cancer, ovarian cancer, nasopharyngeal carcinoma, and hepatocellular carcinoma [[28], [29], [30], [31], [32]]. All these findings and our demonstration suggested that SNHG8 might be a potential prognostic biomarker for different cancers.
Autophagy is known as type II cell death process of the cells where various damaged proteins and organelles are accumulated to form autophagosome and followed by degradation with lysosome to maintain cellular homeostasis [26,33]. During autophagy induction, the cytosolic form of LC3-I is conjugated to phosphatidylethanolamine to form LC3-II that covalently bind with autophagic membrane and serves as marker protein of the process. The microscopy-based LC3 puncta formation assay and western blots of LC3-II/LC3-I ratio are most commonly used methods to evaluate autophagy marker proteins [34]. Another, SQSTM1 encodes the protein p62, considered a marker for autophagy flux. The cargo adaptor protein p62 interact with autophagic substrate to deliver them for lysosomal degradation. In the process of autophagy induction, lysosomal degradation corresponds to the reduced p62 levels [35]. In addition, Beclin-1 is another autophagy regulators of mammalian cells. It positively mediate PI3KC3 to form cellular autophagosomes, resulting induction of autophagy [36]. We found that SNHG8 silencing promote ratio of LC3-II/LC3-I, Beclin-1, and reduced p62 levels in both HCT-116 and SW480 cells. Our findings suggested that SNHG8 knockdown induce autophagy and reduce CRC cell growth, proliferation and migration in vitro.
Apoptosis is one of the central pathways for most of the chemotherapeutics whereas chemoresistant cells remains nonresponding. The Bcl-2 family proteins regulate the process in various cellular stress condition such as nutritional stress, oncogene activation, growth factor deprivation, and DNA damage [37]. Functionally, Bcl-2 family proteins such as Bcl-2 and Bcl-xL prevents apoptotic cell death and induce chemoresistant through mitochondrial or intrinsic apoptosis pathway. As a result, excessive proliferation facilitate to the rapid development of tumour [2,37]. The current results indicated that knockdown of SNHG8 significantly induced the mitochondrial intrinsic cell death process by downregulating Bcl-2 and Bcl-xL proteins. Thus, these data are in agreement with previous demonstration where downregulation of Bcl-2 family proteins negatively contribute to the CRC development and progression in vitro.
The PI3K/AKT/mTOR signaling is a critical regulatory pathway associated with numerous physiological processes of CRC cells including, autophagy, apoptosis, growth, proliferation, migration, and metastasis [38]. Recent studies shown that AKT pathway respond differently in regard to different lncRNAs. For instance, LINC00470 phosphorylate AKT pathway to regulate autophagy and promote carcinogenesis in glioblastoma whereas, LINC00944 promote renal cell carcinoma by dephosphorylating AKT [39,40]. The studies have also shown that mTOR activity modulate autophagy by regulating PI3K/Akt signaling. Specifically, downregulation of mTOR activity or use of mTOR inhibitors induce autophagy in CRC [41]. The AMP-activate protein kinase (AMPK) is another important regulator of mTORC1 dependent autophagy. The AMPK and mTORC1 regulate expression of PI3K class protein complex VPS34 that participate in autophagy induction [42]. Alternatively, studies also shown that mTORC1 phosphorylate another autophagy-related proteins ATG14L in the VPS34 complex to inhibit autophagy by suppressing lipid kinase activity of VPS34 [43]. Our key findings revealed that knockdown of SNHG8 phosphorylate AMPK and suppress AKT and mTOR pathways to reduce CRC proliferation, growth and migration via induction of autophagy and apoptosis.
EMT is the collection of serial events where epithelial cells lose their adhesion and polarity to acquire mesenchymal migratory characteristics such as increased motility [2]. In the malignant tumours, cells acquire mesenchymal phenotypic characteristics that give them higher invasion, migration and metastatic potential [44]. The whole process is regulated by a group of EMT related proteins and its transcription factors. During EMT induction, the cellular adhesion is reduced by suppressing epithelial marker proteins cytokeratins and E-cadherin. Moreover, The expression of mesenchymal markers N-cadherin, vimentin, and fibronectin increases, stabilizing invasiveness and migration, thus contributing to the development of metastasis and chemoresistance [44,45]. Simultaneously, we also found that SNHG8 expression was positively correlated with CRC proliferation, migration, and metastasis. Our study found that knockdown of SNHG8 significantly increased E-cadherin expression whereas significantly suppressed N-cadherin and Vimentin in both HCT-116 and SW480 cells. These results further support that SNHG8 silencing could potentially suppress CRC tumorigenesis and might be a novel therapeutic target for CRC management.
5. Conclusions
Overall, we elucidated the potential oncogenic role of SNHG8 in CRC carcinogenesis. We revealed that knockdown of SNHG8 potentially inhibit CRC cells growth, proliferation, migration, and metastasis. It also regulates autophagy, apoptosis, AKT/AMPM/mTOR, and EMT pathways. These result guide us to have a better understanding of CRC pathogenesis and support our proposed that SNHG8 might be a potential biomarker for CRC management.
Author contributions
ZIK and HKL conceived and designed the project. ZIK conducted the experiments, analyzed data, and wrote the manuscript. HKL interpret the results and reviewed the manuscript. The authors read, approved and finalized the manuscript.
Funding
This project is partially supported by: (1) Research grant to HKL including Departmental Seeding Fund and Internal Institutional Research Fund (P0031318-UAHS). (2) Postgraduate studentship from The Hong Kong Polytechnic University for ZIK.
Declaration of competing interest
There were absences of commercial or financial relationship involved this research. The authors declared no potential conflict of interest.
Acknowledgements
We thank Prof. Jun Yu, Department of Medicine and Therapeutics, Institute of Digestive Diseases, The Chinese University of Hong Kong for providing DLD-1, HCT-116, and SW480 cells for this project.
Contributor Information
Md Zahirul Islam Khan, Email: zahir.islamkhan@connect.polyu.hk.
Helen Ka Wai Law, Email: hthelen@polyu.edu.hk.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021 doi: 10.3322/caac.21660. n/a(n/a) [DOI] [PubMed] [Google Scholar]
- 2.Islam Khan M.Z., Law H.K.W. RAMS11 promotes CRC through mTOR-dependent inhibition of autophagy, suppression of apoptosis, and promotion of epithelial-mesenchymal transition. Cancer Cell Int. 2021;21(1):321. doi: 10.1186/s12935-021-02023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo K., Geng J., Zhang Q., Xu Y., Zhou X., Huang Z., et al. LncRNA CASC9 interacts with CPSF3 to regulate TGF-β signaling in colorectal cancer. J. Exp. Clin. Cancer Res. 2019;38(1):249. doi: 10.1186/s13046-019-1263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhan A., Soleimani M., Mandal S.S. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.Can-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jing F., Jin H., Mao Y., Li Y., Ding Y., Fan C., et al. Genome-wide analysis of long non-coding RNA expression and function in colorectal cancer. Tumor Biol. 2017;39(5) doi: 10.1177/1010428317703650. [DOI] [PubMed] [Google Scholar]
- 6.Amin N., McGrath A., Chen Y.-P.P. Evaluation of deep learning in non-coding RNA classification. Nat. Mach. Intell. 2019;1(5):246–256. doi: 10.1038/s42256-019-0051-2. [DOI] [Google Scholar]
- 7.Silva-Fisher J.M., Dang H.X., White N.M., Strand M.S., Krasnick B.A., Rozycki E.B., et al. Long non-coding RNA RAMS11 promotes metastatic colorectal cancer progression. Nat. Commun. 2020;11(1):2156. doi: 10.1038/s41467-020-15547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Brien S.J., Bishop C., Hallion J., Fiechter C., Scheurlen K., Paas M., et al. Long non-coding RNA (lncRNA) and epithelial-mesenchymal transition (EMT) in colorectal cancer: a systematic review. Cancer Biol. Ther. 2020;21(9):769–781. doi: 10.1080/15384047.2020.1794239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Islam Khan M.Z., Law H.K.W. Cancer susceptibility candidate 9 (CASC9) promotes colorectal cancer carcinogenesis via mTOR-dependent autophagy and epithelial–mesenchymal transition pathways. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.627022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang L., Zhao X.-H., Mao Y.-L., Wang J.-F., Zheng H.-J., You Q.-S. Long non-coding RNA RP11-468E2.5 curtails colorectal cancer cell proliferation and stimulates apoptosis via the JAK/STAT signaling pathway by targeting STAT5 and STAT6. J. Exp. Clin. Cancer Res. 2019;38(1):465. doi: 10.1186/s13046-019-1428-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Huang W., Yuan Y., Li J., Wu J., Yu J., et al. Long non-coding RNA H19 promotes colorectal cancer metastasis via binding to hnRNPA2B1. J. Exp. Clin. Cancer Res. 2020;39(1):141. doi: 10.1186/s13046-020-01619-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu M., Chen X., Lin K., Zeng K., Liu X., Pan B., et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol. Cancer. 2018;17(1):141. doi: 10.1186/s12943-018-0894-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan X., Hu Z., Feng Y., Hu X., Yuan J., Zhao Sihai D., et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28(4):529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian X., Liu Y., Wang Z., Wu S. lncRNA SNHG8 promotes aggressive behaviors of nasopharyngeal carcinoma via regulating miR-656-3p/SATB1 axis. Biomed. Pharmacother. 2020;131 doi: 10.1016/j.biopha.2020.110564. [DOI] [PubMed] [Google Scholar]
- 15.Zhen Y., Ye Y., Wang H., Xia Z., Wang B., Yi W., et al. Knockdown of SNHG8 repressed the growth, migration, and invasion of colorectal cancer cells by directly sponging with miR-663. Biomed. Pharmacother. 2019;116 doi: 10.1016/j.biopha.2019.109000. [DOI] [PubMed] [Google Scholar]
- 16.He C., Fu Y., Chen Y., Li X. Long non-coding RNA SNHG8 promotes autophagy as a ceRNA to upregulate ATG7 by sponging microRNA-588 in colorectal cancer. Oncol. Lett. 2021;22(2):577. doi: 10.3892/ol.2021.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poursheikhani A., Abbaszadegan M.R., Kerachian M.A. Long non-coding RNA AC087388.1 as a novel biomarker in colorectal cancer. BMC Cancer. 2022;22(1):196. doi: 10.1186/s12885-022-09282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H., Li Q., Wu Y., Dong J., Lao Y., Ding Z., et al. Long non-coding RNA RP11-400N13.3 promotes the progression of colorectal cancer by regulating the miR-4722-3p/P2RY8 axis. Oncol. Rep. 2020;44(5):2045–2055. doi: 10.3892/or.2020.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saus E., Brunet-Vega A., Iraola-Guzmán S., Pegueroles C., Gabaldón T., Pericay C. Long non-coding RNAs as potential novel prognostic biomarkers in colorectal cancer. Front. Genet. 2016;7 doi: 10.3389/fgene.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galamb O., Barták B.K., Kalmár A., Nagy Z.B., Szigeti K.A., Tulassay Z., et al. Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J. Gastroenterol. 2019;25(34):5026–5048. doi: 10.3748/wjg.v25.i34.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siddiqui H., Al-Ghafari A., Choudhry H., Al Doghaither H. Roles of long non-coding RNAs in colorectal cancer tumorigenesis: a Review. Mol. Clin. Oncol. 2019;11(2):167–172. doi: 10.3892/mco.2019.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ragusa M., Barbagallo C., Statello L., Condorelli A.G., Battaglia R., Tamburello L., et al. Non-coding landscapes of colorectal cancer. World J. Gastroenterol. 2015;21(41):11709–11739. doi: 10.3748/wjg.v21.i41.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online. 2014;16:11. doi: 10.1186/1480-9222-16-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu M-d, Qi P., Du X. Long non-coding RNAs in colorectal cancer: implications for pathogenesis and clinical application. Mod. Pathol. 2014;27(10):1310–1320. doi: 10.1038/modpathol.2014.33. [DOI] [PubMed] [Google Scholar]
- 25.Wang H., Huang C., Yao X. The functions of long non-coding RNAs in colorectal cancer. Transl. Cancer Res. 2019;8(5):2192–2204. doi: 10.21037/tcr.2019.08.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Islam Khan M.Z., Tam S.Y., Law H.K.W. Autophagy-modulating long non-coding RNAs (LncRNAs) and their molecular events in cancer. Front. Genet. 2019;9 doi: 10.3389/fgene.2018.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lizarbe M.A., Calle-Espinosa J., Fernández-Lizarbe E., Fernández-Lizarbe S., Robles M., Olmo N., et al. Colorectal cancer: from the genetic model to posttranscriptional regulation by noncoding RNAs. BioMed Res. Int. 2017;2017 doi: 10.1155/2017/7354260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang P., Li S., Chen Z., Lu Y., Zhang H. LncRNA SNHG8 promotes proliferation and invasion of gastric cancer cells by targeting the miR-491/PDGFRA axis. Hum. Cell. 2020;33(1):123–130. doi: 10.1007/s13577-019-00290-0. [DOI] [PubMed] [Google Scholar]
- 29.Zou C., Liao J., Hu D., Su Y., Lin H., Lin K., et al. SNHG8 promotes the progression of Epstein-Barr virus-associated gastric cancer via sponging miR-512-5p and targeting TRIM28. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.734694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao W., Lu T., Liu X., Yin W., Zhang H. LncRNA SNHG8 induces ovarian carcinoma cells cellular process and stemness through Wnt/β-catenin pathway. Cancer Biomarkers. 2020;28(4):459–471. doi: 10.3233/cbm-190640. [DOI] [PubMed] [Google Scholar]
- 31.Tian X., Liu Y., Wang Z., Wu S. lncRNA SNHG8 promotes aggressive behaviors of nasopharyngeal carcinoma via regulating miR-656-3p/SATB1 axis. Biomed. Pharmacother. 2020;131 doi: 10.1016/j.biopha.2020.110564. [DOI] [PubMed] [Google Scholar]
- 32.Dong J., Teng F., Guo W., Yang J., Ding G., Fu Z. lncRNA SNHG8 promotes the tumorigenesis and metastasis by sponging miR-149-5p and predicts tumor recurrence in hepatocellular carcinoma. Cell. Physiol. Biochem. 2018;51(5):2262–2274. doi: 10.1159/000495871. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y., Hua Y., Li X., Arslan I.M., Zhang W., Meng G. Distinct types of cell death and the implication in diabetic cardiomyopathy. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sánchez-Martín P., Komatsu M. p62/SQSTM1 – steering the cell through health and disease. J. Cell Sci. 2018;131(21) doi: 10.1242/jcs.222836. [DOI] [PubMed] [Google Scholar]
- 36.Kang R., Zeh H.J., Lotze M.T., Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4):571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt L.H., Görlich D., Spieker T., Rohde C., Schuler M., Mohr M., et al. Prognostic impact of Bcl-2 depends on tumor histology and expression of MALAT-1 lncRNA in non–small-cell lung cancer. J. Thorac. Oncol. 2014;9(9):1294. doi: 10.1097/JTO.0000000000000243. 304. [DOI] [PubMed] [Google Scholar]
- 38.Porta C., Paglino C., Mosca A. Targeting PI3K/Akt/mTOR signaling in cancer. Front. Oncol. 2014;4 doi: 10.3389/fonc.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu C., Zhang Y., She X., Fan L., Li P., Feng J., et al. A cytoplasmic long noncoding RNA LINC00470 as a new AKT activator to mediate glioblastoma cell autophagy. J. Hematol. Oncol. 2018;11(1):77. doi: 10.1186/s13045-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C., Zheng H. LncRNA LINC00944 promotes tumorigenesis but suppresses Akt phosphorylation in renal cell carcinoma. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.697962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J., Liang D., Zhang X.P., He C.F., Cao L., Zhang S.Q., et al. Novel PI3K/Akt/mTOR signaling inhibitor, W922, prevents colorectal cancer growth via the regulation of autophagy. Int. J. Oncol. 2021;58(1):70–82. doi: 10.3892/ijo.2020.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim Y.C., Guan K.-L. mTOR: a pharmacologic target for autophagy regulation. J. Clin. Invest. 2015;125(1):25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan H.X., Russell R.C., Guan K.L. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy. 2013;9(12):1983–1995. doi: 10.4161/auto.26058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ieda T., Tazawa H., Okabayashi H., Yano S., Shigeyasu K., Kuroda S., et al. Visualization of epithelial-mesenchymal transition in an inflammatory microenvironment–colorectal cancer network. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-52816-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ribatti D., Tamma R., Annese T. Epithelial-mesenchymal transition in cancer: a historical overview. Translat. Oncol. 2020;13(6) doi: 10.1016/j.tranon.2020.100773. [DOI] [PMC free article] [PubMed] [Google Scholar]