Abstract
The anterior portion of the bed nucleus of the stria terminalis (BNST) modulates fear and stress responses. The anterodorsal BNST (adBNST) can be anatomically subdivided further into the lateral and medial divisions. Although output projections of BNST subregions have been studied, the local and global input connections to these subregions remain poorly understood. To further understand BNST-centered circuit operations, we have applied new viral-genetic tracing and functional circuit mapping to determine detailed synaptic circuit inputs to lateral and medial subregions of adBNST in the mouse. Monosynaptic canine adenovirus type 2 (CAV2) and rabies virus-based retrograde tracers were injected in the adBNST subregions. The amygdalar complex, hypothalamus and hippocampal formation account for the majority of overall inputs to adBNST. However, lateral versus medial adBNST subregions have distinct patterns of long-range cortical and limbic brain inputs. The lateral adBNST has more input connections from prefrontal (prelimbic, infralimbic, cingulate) and insular cortices, anterior thalamus and ectorhinal/perirhinal cortices. In contrast, the medial adBNST received biased inputs from the medial amygdala, lateral septum, hypothalamus nuclei and ventral subiculum. We confirmed long-range functional inputs from the amydalohippocampal area and basolateral amygdala to the adBNST using ChR2-assisted circuit mapping. Selected novel BNST inputs are also validated with the AAV axonal tracing data from the Allen Institute Mouse Brain Connectivity Atlas. Together, these results provide a comprehensive map of the differential afferent inputs to lateral and medial adBNST subregions, and offer new insight into the functional operations of BNST circuitry for stress and anxiety-related behaviors.
Keywords: BNST, CAV2, Rabies, Retrograde, Monosynaptic inputs
Abbreviations
- adBNST
bed nuclei of the stria terminalis, anterodorsal division
- alBNST
bed nuclei of the stria terminalis, anterodorsal division, lateral part
- BNSTal
bed nuclei of the stria terminalis, anterodorsal division, anterolateral area
- BNSTjc
bed nuclei of the stria terminalis, anterodorsal division, juxta-capsular nucleus
- BNSTov
bed nuclei of the stria terminalis, anterodorsal division, oval nucleus
- amBNST
bed nuclei of the stria terminalis, anterodorsal division, medial part
- AO
anterior olfactory nucleus
- FrA
frontal association cortex
- AC
anterior cingulate cortex
- PL
prelimbic cortex
- IL
Infralimbic cortex
- AI
agranular insular cortex
- Pir
piriform cortex
- NAc
nucleus accumbens
- fmi
forceps minor of the corpus callosum
- Cpu
caudate putamen
- VP
ventral pallidum
- M1
primary motor cortex
- M2
secondary motor cortex
- MS
medial septal nucleus
- DBB
diagonal band of broca
- LS
lateral septal nucleus
- PO
preoptic nucleus
- Sfi
septofimbrial nucleus
- S1
primary somatosensory cortex
- RSA
retrosplenial agranular cortex
- RSG
retrosplenial granular cortex
- amTH
anteromedial thalamic nucleus
- vTH
ventral thalamic nucleus
- aH
anterior hypothalamic area
- dmH
dorsomedial hypothalamic nucleus
- vmH
ventromedial hypothalamic nucleus
- pvH
paraventricular hypothalamic nucleus
- ARC
arcuate hypothalamic nucleus
- LH
lateral hypothalamic area
- Ce AMG
central amygdaloid nucleus
- BL AMG
basolateral amygdaloid nucleus
- Me AMG
medial amygdaloid nucleus
- Co AMG
cortical amygdaloid nucleus
- AHiA
amygdalohippocampal transition area
- APir
amygdalopiriform transition area
- Aud ctx
auditory cortex
- En
endopiriform nucleus
- Cl
caudal interstitial nucleus of the medial longitudinal fasciculus
- HPC
hippocampus
- Sub
subiculum
- ProS
prosubiculum
- Ent ctx
entorhinal cortex
- ECT
ectorhinal cortex
- PRh
perirhinal cortex
- PMM
premammillary nucleus
- SuMM
supramammillary nucleus, medial part
- MM
medial mammillary nucleus
- PAG
periaqueductal gray
- DR
dorsal raphe nucleus
- CLi
caudal linear nucleus of the raphe
- DpG
deep gray layer of the superior colliculus
1. Introduction
The bed nucleus of the stria terminalis (BNST) coordinates neuroendocrine, autonomic and somatomotor responses during affective and emotional behaviors (Crestani et al., 2013). Despite being a relatively small structure in the basal forebrain, the BNST consists of multiple subregions (Ju and Swanson, 1989; Moga et al., 1989). Early work on the BNST divided it into medial and lateral divisions based largely on cytoarchitecture (Bleier, 1961; Krettek and Price, 1978). These divisions are further supported based on their amygdalar inputs, with the proposed medial division characterized by inputs from the medial amygdalar nucleus and the lateral division characterized by inputs from the central and basolateral nuclei (de Olmos, 1972; Krettek and Price, 1978). This organizational model of medial versus lateral division is still widely used along with an later proposed anterior versus posterior (rostral/caudal) division of BNST, in which the gross anatomical and cytoarchitecture features were parsed into anterior and posterior portions marked by the anterior commissure (Dong et al., 2001; Wood and Swann, 2005).
The dorsal component of the anterior BNST (adBNST) can be further divided into two subregions: the lateral adBNST (alBNST), which include the anterolateral area (BNSTal), juxta-capsular (BNSTjc), and oval (BNSTov) subnuclei, as well as the medial adBNST (amBNST) (Dong and Swanson, 2004; Gungor and Pare, 2016; Ju and Swanson, 1989; Ju et al., 1989; Kim et al., 2013; Nguyen et al., 2016). Output projections of these anterior BNST subregions have been studied using traditional tracers, and some projections have been examined recently in functional and behavioral contexts. For example, the oval nucleus versus its surrounding subregions (termed as “adBNST” per Kim et al., 2013) modulate anxiogenesis and anxiolysis, respectively (Choi et al., 2007; Daniel and Rainnie, 2015; Kim et al., 2013; Walker et al., 2003), but see (Gungor and Pare, 2016). Three independent features of anxiolysis, 1) reduced risk-avoidance, 2) reduced respiratory rate, and 3) increased positive valence are functionally mediated by efferent projections from the lateral hypothalamus, parabrachial nucleus and ventral tegmental area to the adBNST, respectively (Kim et al., 2013). Furthermore, our recent cFos mapping study of brain regions activated by multi-modal and electric foot shock stress shows that there are pronounced anatomical differences in the density of cFos expression across BNST subregions (Lin et al., 2018). Although stress-induced cFos expression in the amBNST is significantly higher than that in the alBNST, there is a greater proportion of cFos-expressing CRH neurons in the alBNST. Recent studies showed that the alBNST mediates stress-induced anxiety-like behavior through the CRH neural network coordinating with CeA (Pomrenze et al., 2019; Yamauchi et al., 2018). The functional distinctions between the lateral and medial adBNST appear to go beyond stress and anxiety because opposing roles in motivated states via specific responses to aversive and rewarding stimuli are shown for BNST CRH and CCK neurons, respectively (Giardino et al., 2018; Giardino and Pomrenze, 2021). A specialized role in feeding regulation by PKC-δ neurons in the BNSTov is also reported (Schnapp et al., 2022; Wang et al., 2019). Thus, we hypothesized the observed functional distinctions between the lateral and medial adBNST are empowered by the differential circuit assemblies of these subregions. A comprehensive map of brain-wide circuit inputs to the lateral and medial adBNST subregions would further inform functional operations of BNST circuitry. Unfortunately, local and global input connections to different adBNST subregions remain poorly understood. To answer this question, we used novel viral-genetic tracing and functional circuit mapping methods to build a detailed map of synaptic circuit inputs onto the dorsolateral and dorsomedial subregions of the anterior BNST.
Genetically modified viruses provide powerful new tools to map and identify neural circuit connections. In this study, we applied two classes of neurotropic retrograde viruses injected into the lateral versus medial subregions of the adBNST (i.e., alBNST vs. amBNST), canine adenovirus type 2 (CAV2, which marks monosynaptic connections) (Carter et al., 2013; Gore et al., 2013; Schwarz et al., 2015; Sun et al., 2019) and glycoprotein-gene deleted mutant rabies virus (SADΔG-EGFP, which also marks monosynaptic connections) (Wickersham et al., 2007). This effectively identifies local and global afferent inputs to the lateral and medial subregions of adBNST. Our work features extensive, comparative characterizations of the differential input connections between the al and amBNST in a quantitative manner. Our data revealed distinct patterns of long-range cortical and limbic brain inputs to alBNST versus amBNST. We used ChR2-assisted circuit mapping to functionally validate significant inputs from the amydalohippocampal transition area (AHiA) and basolateral amygdala (BLA) to both al and amBNST subregions. Selected novel BNST inputs are also validated with the AAV axonal tracing data from the Allen Institute Mouse Brain Connectivity Atlas. These results provide important information on neural structures that provide the array of afferent inputs to the anterior BNST and our work will guide future functional studies of BNST subregions.
2. Results
2.1. Weak local connections between the lateral and medial adBNST
We used a genetically modified canine adenovirus type 2 (CAV2) to map the brain-wide monosynaptic input connections of lateral versus medial adBNST (alBNST vs. amBNST). By replacing the E1 gene from the viral genome with Cre recombinase (E1-deleted viral vector), the genetically modified CAV2 viral vector is replication incompetent. The Cre-expressing CAV2 virus (CAV2-Cre) can be taken efficiently by axon terminals in vivo, resulting in a monosynaptic, retrograde neuronal infection (Carter et al., 2013; Gore et al., 2013; Schwarz et al., 2015; Sun et al., 2019). We injected the CAV2-Cre virus into Ai9 Cre reporter mice (Madisen et al., 2009) to delineate the inputs onto alBNST versus amBNST. Four weeks after the injection, mice were perfused for histology processing. First, by examining the coronal mouse brain sections around the BNST injection site, tdTomato expressions demonstrated a precise targeting of alBNST (n = 7 mice, Fig. 1A), amBNST (n = 5 mice, Fig. 1B), or the entire adBNST (n = 3, Fig. 1C). From cases targeting the alBNST, we found the tdTomato expression was mainly restricted in the oval BNST with no or little labeling into its adjacent striatum and posterior BNST (Fig. 1A). Furthermore, there were essentially no labeled inputs in the adjacent amBNST (Fig. 1A). From cases targeting the amBNST, although sparse, there were labeled neurons in the adjacent alBNST. This result suggests that the alBNST may be connected to amBNST unidirectionally to some degree. To further confirm this connection, we searched the Allen Mouse Brain Connectivity Atlas (connectivity.brain-map.org), which performed AAV axonal tracing in targeted brain regions (Oh et al., 2014), and inspected an AAV tracing case targeting the BNSTov (a part of alBNST) with a Cre-dependent, EGFP-expressing AAV in a PKC-δ-Cre mouse (Haubensak et al., 2010). The spatially restricted Cre expression in the PKC-δ-Cre mouse allows the AAV infection localized in the BNSTov (Fig. 1D). We found EGFP-expressing axonal fibers in the adjacent amBNST; however, note much stronger axonal labeling in the ventral BNST (Fig. 1D).
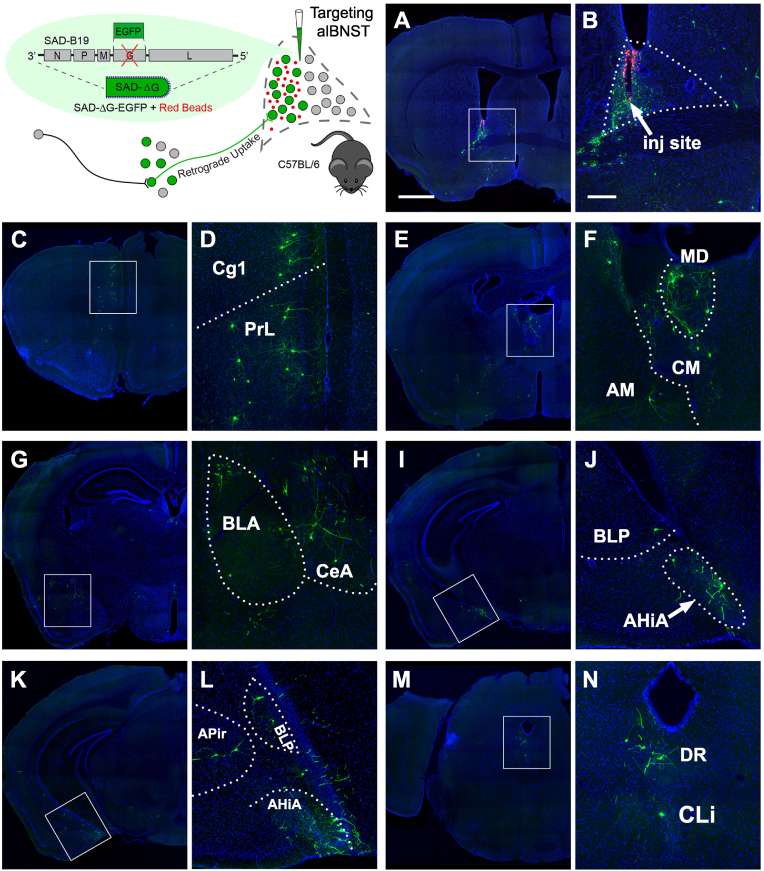
Fig. 1.
CAV2-Cre injections into subregions of the BNST in the Cre reporter Ai9 mice. (A) Top left: schematic illustration of the CAV2-Cre virus injection into the alBNST of an Ai9 mouse. CAV2-Cre-infected neurons near the injection site express tdTomato red fluorescent protein. Top right: injection site images from two representative mice targeting alBNST. The panels on the right are zoom-in views of the boxed regions in the panels on the left. The white dotted line shows the contours of the entire adBNST. Scale bar, 1 mm and 200 μm respectively and apply to similar panels in B and C. Bottom: example anterior to posterior images of the alBNST injection site from a third representative case. Scale bar: 200 μm. (B) Panels organized similar to A, but for targeting the amBNST. (C) Organized similar to A, but for targeting the entire adBNST. (D) Cre-dependent AAV injections targeting the oval region of the BNST (BNSTov) using the PKC-δ-Cre mouse revealed axonal projections to the nearby amBNST and ventral BNST. Scale bar: 200 μm. Data images were obtained and modified from Allen Mouse Brain Connectivity Atlas: connectivity. brain-map.org/projection/experiment/522078446. Image credit: Allen Institute for Brain Science (Oh et al., 2014). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.2. Overall input patterns of lateral and medial adBNST
Results from the alBNST injections showed strong inputs from the prefrontal cortex. Specifically, intense labeling was detected in the prelimbic (Fig. 2A–B) and anterior cingulate cortices (Fig. 2C–D). Other cortical inputs were also observed from the adjacent agranular insular cortex, piriform cortex, as well as ectorhinal and perirhinal cortices which reside in the more posterior part of the brain (Fig. 2E–F, Table 1). Inputs were also seen from the anteromedial nucleus of the thalamus (Fig. 2G–J), which is known to project to limbic cortex (van Groen et al., 1999). Consistent with previous reports (Dong et al., 2001; Krettek and Price, 1978; Weller and Smith, 1982), we observed strong innervations originated from the basolateral, central, medial and cortical amydaloid nuclei to alBNST (Fig. 2K-N), as well as from the amydalohippocampal (AHiA) and amygdalopiriform (APir) transition areas (Fig. 2M–P, Table 1). Finally, inputs from the subiculum (i.e., ventral prosubiculum) (Fig. 2P) and entorhinal cortex (Fig. 2Q) were also detected as well as from the raphe nuclei (Fig. 2R)(Matthews et al., 2016).
Fig. 2.
Limbic and cortical regions provide strong afferent inputto the alBNST. Schematic of E1-deleted CAV2 viral vector expressing the Cre recombinase and of CAV2-Cre-mediated retrograde tracing in the alBNST of Ai9 mice. The alBNST injection sites are shown in Fig. 1A. (A) An example of tdTomato-labeled neurons in the prelimbic cortex (PrL) providing monosynaptic afferent input into the alBNST. Scale bar, 1 mm. (B) High-magnification view of the PrL neurons showing the boxed region in panel A. Scale bar, 200 μm. (C–D) Labeled neurons in the cingulate cortex (Cg1) and PrL. (E) Labeled neurons in the angular insular (AI) and piriform (Pir) cortices. (F) Labeled neurons in the ectorhinal (ECT) and perirhinal (PRh) cortices. (G-H) Labeled neurons in the anteromedial thalamic nucleus (AM). (I-J) Labeled neurons in the thalamus, basolateral amygdaloid nucleus (BLA), lateral (CeL) and medial (CeM) subdivisions of the central amygdaloid nucleus (CeA). (K-L) Labeled neurons in the BLA and CeA. (M-N) Labeled neurons in the posterior part of the basolateral amygdaloid nucleus (BLP), amygdalohippocampal area (AHiA) and posteromedial cortical amygdaloid nucleus (PMCo). (O–P) Labeled neurons in the ventral subiculum/prosubiculum, APir, and PMCo. (Q–R) Labeled neurons in the lateral entorhinal cortex (LEnt), medial entorhinal cortex (MEnt), dorsal raphe (DR), and caudal linear nucleus of the raphe (CLi).
Table 1.
Percentage of total input connections to alBNST.
| Brain region | AO | FrA | Orbital ctx | AC/PL/IL | AI | Pir | NAc | fmi | Cpu | VP | M1 | M2 | MS/DBB | LS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MEAN% | 0.35 | 0.23 | 2.08 | 9.05 | 3.40 | 4.06 | 0.29 | 0.58 | 0.62 | 0.05 | 1.02 | 2.06 | 0.08 | 0.20 | |
| SE | 0.24 | 0.17 | 0.65 | 1.78 | 0.97 | 1.20 | 0.19 | 0.29 | 0.42 | 0.05 | 0.48 | 1.26 | 0.03 | 0.17 | |
| PO | Sfi | S1 | RSA/RSG | amTH | vTH | aH | dmH | vmH | pvH | ARC | LH | Ce AMG | BL AMG | Me AMG | |
| 0.41 | 0.00 | 0.17 | 0.41 | 10.52 | 0.52 | 0.56 | 0.52 | 0.23 | 0.00 | 0.13 | 2.13 | 4.07 | 20.53 | 2.51 | |
| 0.19 | 0.00 | 0.07 | 0.24 | 1.55 | 0.30 | 0.26 | 0.20 | 0.10 | 0.00 | 0.05 | 1.30 | 1.19 | 2.66 | 0.20 | |
| Co AMG | AHiA | APir | Aud ctx | Visual Ctx | En/Cl | HPC | Subiculum | Ent Ctx | PRh | PMM | SuMM/MM | PAG | DR/CLi | DpG | |
| 7.97 | 4.73 | 6.65 | 0.45 | 0.19 | 0.13 | 0.72 | 1.31 | 8.01 | 2.04 | 0.08 | 0.16 | 0.10 | 0.24 | 0.00 | N = 7 |
| 2.93 | 1.24 | 1.99 | 0.18 | 0.10 | 0.05 | 0.24 | 0.46 | 2.04 | 0.57 | 0.04 | 0.08 | 0.07 | 0.12 | 0.00 |
Injections into the amBNST revealed only weak inputs from the prelimbic and anterior cingulate cortices (Fig. 3A–D). Instead, we found strong labeling from the lateral septum and the adjacent preoptic nucleus of the hypothalamus, along with decent labeling from other hypothalamic nuclei (Fig. 3E–F, Table 2). Similarly, the predominant inputs of amBNST come from the amygdala and associated structures such as AHiA and APir (Fig. 3G-P), including basolateral, central, medial and cortical amygdaloid nuclei (Table 2). Decent labeling was also observed in the hippocampal formation, including the hippocampus proper, subiculum and entorhinal cortex (Fig. 3O–P). Weak labeling in the deep gray layer of the superior colliculus (DpG) and periaqueductal gray (PAG) was also seen (Fig. 3Q). Consistent with a recent report (Ding, 2022), we observed inputs from the posterior BNST to the amBNST (Fig. 3R), but not to the alBNST (Fig. 1A).
Fig. 3.
Amygdalar regions provide robust inputto the amBNST. Schematic of E1-deleted CAV2 viral vector expressing the Cre recombinase and of CAV2-Cre-mediated retrograde tracing in the amBNST of Ai9 mice. The amBNST injection site is shown in Fig. 1B. (A–B) Example of sparse labeling in the PrL and infralimbic cortex (IL). Scale bar, 1 mm in panel A and 200 μm in panel B. (C–D) Sparse labeling in the Cg1 and PrL. (E–F) Labeled neurons in the lateral hypothalamus (LH). (G–H) Labeled neurons in the anterior basomedial amygdaloid nucleus (BMA), the anterodorsal (CeMAD), anteroventral (CeMAV), and capsular (CeC) subdivisions of the central amygdaloid nucleus (CeA). Labeling of the BLA was sparse. (I–J) Labeled neurons in the CeA and the anterior part of the BMA. (K–L) Labeled neurons in the posterior part of the BLP and the AHiA. (M–N) Labeled neurons in the APir and PMCo. (O–P) Labeled neurons in the LEnt, APir, and ventral subiculum/prosubiculum. (Q) Weak labeling in the DpG and PAG. (R) Strong labeling in the posterior BNST.
Table 2.
Percentage of total input connections to amBNST.
| Brain region | AO | FrA | Orbital ctx | AC/PL/IL | AI | Pir | NAc | fmi | Cpu | VP | M1 | M2 | MS/DBB | LS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MEAN% | 0.37 | 0.01 | 0.51 | 2.27 | 0.25 | 3.39 | 0.42 | 0.37 | 0.20 | 0.19 | 0.10 | 0.30 | 0.64 | 3.31 | |
| SE | 0.15 | 0.01 | 0.08 | 0.41 | 0.04 | 1.09 | 0.10 | 0.14 | 0.08 | 0.15 | 0.04 | 0.10 | 0.35 | 0.75 | |
| PO | Sfi | S1 | RSA/RSG | amTH | vTH | aH | dmH | vmH | pvH | ARC | LH | Ce AMG | BL AMG | Me AMG | |
| 1.88 | 0.02 | 0.26 | 0.23 | 4.28 | 0.63 | 1.22 | 1.15 | 0.99 | 0.19 | 0.90 | 4.18 | 2.25 | 15.11 | 10.29 | |
| 0.14 | 0.02 | 0.02 | 0.16 | 1.00 | 0.10 | 0.19 | 0.10 | 0.10 | 0.03 | 0.06 | 0.24 | 0.20 | 2.89 | 1.01 | |
| Co AMG | AHiA | APir | Aud ctx | Visual Ctx | En/Cl | HPC | Subiculum | Ent Ctx | PRh | PMM | SuMM/MM | PAG | DR/CLi | DpG | |
| 14.10 | 4.92 | 5.48 | 0.05 | 0.05 | 0.25 | 1.70 | 3.69 | 9.97 | 0.20 | 0.20 | 0.19 | 0.25 | 0.09 | 0.34 | N = 5 |
| 3.44 | 0.54 | 2.31 | 0.01 | 0.01 | 0.05 | 0.37 | 0.51 | 4.84 | 0.04 | 0.05 | 0.07 | 0.08 | 0.03 | 0.09 |
As a means to corroborate the results observed in the above experiments, we injected CAV2-Cre virus into the entire extent of the dorsal BNST, targeting both alBSNT and amBNST, to confirm its inputs. As expected, major inputs identified by injections into subregions of BNST were labeled by injection into the entire adBNST, including the prelimbic and cingulate cortical regions (Fig. 4A–D), thalamus and hypothalamus (Fig. 4E–F), amydaloid nuclei (Fig. 4G-L), amygdalohippocampal and amygdalopiriform transition areas (Fig. 4K-P), and the hippocampal formation (Fig. 4Q–R).
Fig. 4.
Inputareasto the entireadBNST overlap with areas providing inputto the alBNST and amBNST. Schematic of E1-deleted CAV2 viral vector expressing the Cre recombinase and of CAV2-Cre-mediated retrograde tracing in the adBNST of Ai9 mice. The adBNST injection site is shown in Fig. 1C. (A–B) Labeled neurons in the PrL and Cg1. Scale bar, 1 mm in panel A and 200 μm in panel B. (C–D) Labeled neurons in the PrL. (E–F) Labeled neurons in the thalamus and LH. (G–H) Labeled neurons in the BLA, CeMAV, and BMA. (I–J) Labeled neurons in the BLA, BMP, BMA, and CeA. (K–N) Labeled neurons in the BLP, BMP, AHiA, and PMCo. (O–P) Labeled neurons in the Sub, APir, PMCo, and AHiA. (Q–R) Labeled neurons in the LEnt.
2.3. Quantitative comparisons of the input connections to alBNST and amBNST
Next, we quantified the anatomical distribution of the labeled inputs in each brain region for both alBNST and amBNST injected cases. Comparisons between alBNST and amBNST cases showed that alBNST received significantly more inputs from the angular insular cortex and prefrontal cortices, which include anterior cingulate, prelimbic and infralimbic regions (Fig. 5 and Table 1, Table 2, Table 3). alBNST also received more inputs from the anteromedial thalamus and ectorhinal/perirhinal cortices. On the other hand, amBNST received significantly more inputs from the lateral septum and multiple hypothalamic nuclei, including the preoptic, dorsomedial, ventromedial, paraventricular, and arcuate regions (Fig. 5 and Table 1, Table 2, Table 3). Most of the amygdala inputs were comparable between alBNST and amBNST except the inputs from medial amygdala were strongly biased toward amBNST (Fig. 5 and Table 1, Table 2, Table 3). In addition, amBNST also received stronger inputs from the hippocampus proper and subiculum (Fig. 5 and Table 1, Table 2, Table 3). Finally, inputs from the DpG selectively innervated amBNST, but not alBNST (Fig. 5 and Table 1, Table 2, Table 3). Together, our quantification suggests the inputs to alBNST are biased toward cortical regions while the inputs to amBNST are primarily from subcortical regions including the septum, hypothalamus, and hippocampus.
Fig. 5.
Quantification of afferent inputsto the alBNST versus amBNST. The graphs show the anatomical distribution (% of total inputs) of structures providing afferent input to the alBNST (black) and amBNST (red). The contribution of afferents from each anatomical brain region on the x-axis is shown as a percentage of the total number of labeled neurons providing afferent inputs into the BNST subregions. See Table 1, Table 2, Table 3 for detailed quantification and statistical comparisons. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 3.
| Brain region | Significance | P value | alBNST Mean | amBNST Mean | Difference | SE of difference | t ratio | df |
|---|---|---|---|---|---|---|---|---|
| AO | No | 0.9503 | 0.3477 | 0.3680 | −0.0203 | 0.3180 | 0.0639 | 10 |
| FrA | No | 0.3064 | 0.2346 | 0.0140 | 0.2206 | 0.2047 | 1.0780 | 10 |
| Orbital ctx | No | 0.0725 | 2.0810 | 0.5085 | 1.5730 | 0.7836 | 2.0070 | 10 |
| AC/PL/IL | *p < 0.05 | 0.0107 | 9.0530 | 2.2670 | 6.7860 | 2.1690 | 3.1290 | 10 |
| AI | * | 0.0222 | 3.4050 | 0.2492 | 3.1560 | 1.1670 | 2.7040 | 10 |
| Pir | No | 0.7039 | 4.0590 | 3.3930 | 0.6655 | 1.7010 | 0.3912 | 10 |
| NAc | No | 0.5811 | 0.2857 | 0.4240 | −0.1383 | 0.2426 | 0.5702 | 10 |
| fmi | No | 0.5777 | 0.5835 | 0.3689 | 0.2146 | 0.3729 | 0.5754 | 10 |
| Cpu | No | 0.4255 | 0.6236 | 0.2011 | 0.4224 | 0.5084 | 0.8308 | 10 |
| VP | No | 0.3049 | 0.0450 | 0.1890 | −0.1440 | 0.1332 | 1.0810 | 10 |
| M1 | No | 0.1437 | 1.0250 | 0.1044 | 0.9205 | 0.5802 | 1.5860 | 10 |
| M2 | No | 0.2709 | 2.0640 | 0.3037 | 1.7600 | 1.5100 | 1.1650 | 10 |
| MS/DBB | No | 0.0871 | 0.0790 | 0.6391 | −0.5601 | 0.2953 | 1.8970 | 10 |
| LS | ***p < 0.001 | 0.0007 | 0.1996 | 3.3090 | −3.1100 | 0.6503 | 4.7820 | 10 |
| PO | *** | 0.0002 | 0.4086 | 1.8790 | −1.4700 | 0.2562 | 5.7390 | 10 |
| Sfi | No | 0.2550 | 0.0000 | 0.0225 | −0.0225 | 0.0186 | 1.2080 | 10 |
| S1 | No | 0.3412 | 0.1676 | 0.2566 | −0.0890 | 0.0891 | 0.9994 | 10 |
| RSA/RSG | No | 0.5800 | 0.4070 | 0.2275 | 0.1795 | 0.3138 | 0.5720 | 10 |
| amTH | * | 0.0118 | 10.5200 | 4.2760 | 6.2480 | 2.0340 | 3.0710 | 10 |
| vTH | No | 0.7728 | 0.5207 | 0.6314 | −0.1107 | 0.3731 | 0.2966 | 10 |
| aH | No | 0.0900 | 0.5599 | 1.2240 | −0.6640 | 0.3537 | 1.8770 | 10 |
| dmH | * | 0.0298 | 0.5161 | 1.1470 | −0.6308 | 0.2492 | 2.5310 | 10 |
| vmH | *** | 0.0003 | 0.2318 | 0.9923 | −0.7605 | 0.1435 | 5.3010 | 10 |
| pvH | *** | 0.0000 | 0.0000 | 0.1887 | −0.1887 | 0.0232 | 8.1260 | 10 |
| ARC | *** | 0.0000 | 0.1338 | 0.8960 | −0.7622 | 0.0774 | 9.8460 | 10 |
| LH | No | 0.2220 | 2.1280 | 4.1760 | −2.0480 | 1.5720 | 1.3020 | 10 |
| Ce AMG | No | 0.2346 | 4.0660 | 2.2550 | 1.8110 | 1.4320 | 1.2650 | 10 |
| BL AMG | No | 0.2042 | 20.5300 | 15.1100 | 5.4180 | 3.9890 | 1.3580 | 10 |
| Me AMG | *** | 0.0000 | 2.5070 | 10.2900 | −7.7870 | 0.8724 | 8.9250 | 10 |
| Co AMG | No | 0.2050 | 7.9670 | 14.1000 | −6.1360 | 4.5260 | 1.3560 | 10 |
| AHiA | No | 0.9028 | 4.7280 | 4.9220 | −0.1944 | 1.5520 | 0.1252 | 10 |
| APir | No | 0.7102 | 6.6520 | 5.4810 | 1.1710 | 3.0620 | 0.3824 | 10 |
| Aud ctx | No | 0.0887 | 0.4523 | 0.0501 | 0.4023 | 0.2133 | 1.8860 | 10 |
| Visual Ctx | No | 0.2655 | 0.1883 | 0.0468 | 0.1415 | 0.1200 | 1.1800 | 10 |
| En/Cl | No | 0.1356 | 0.1317 | 0.2517 | −0.1200 | 0.0739 | 1.6230 | 10 |
| HPC | * | 0.0421 | 0.7190 | 1.6960 | −0.9773 | 0.4197 | 2.3290 | 10 |
| Subiculum | **p < 0.01 | 0.0066 | 1.3120 | 3.6940 | −2.3820 | 0.6966 | 3.4200 | 10 |
| Ent Ctx | No | 0.6849 | 8.0060 | 9.9700 | −1.9640 | 4.7010 | 0.4179 | 10 |
| PRh | * | 0.0227 | 2.0430 | 0.1952 | 1.8480 | 0.6868 | 2.6900 | 10 |
| PMM | No | 0.0889 | 0.0776 | 0.1975 | −0.1200 | 0.0637 | 1.8840 | 10 |
| SuMM/MM | No | 0.8126 | 0.1630 | 0.1894 | −0.0264 | 0.1084 | 0.2435 | 10 |
| PAG | No | 0.1990 | 0.1046 | 0.2531 | −0.1486 | 0.1080 | 1.3760 | 10 |
| DR/CLi | No | 0.3083 | 0.2441 | 0.0894 | 0.1546 | 0.1440 | 1.0740 | 10 |
| DpG | *** | 0.0007 | 0.0000 | 0.3426 | −0.3426 | 0.0711 | 4.8210 | 10 |
2.4. BNST connections confirmed with monosynaptic rabies tracing
Leveraging a different retrograde tracing approach, we injected an EGFP tagged, glycoprotein deficient rabies virus (SADΔG-EGFP) in either the alBNST or amBNST to verify our findings. We injected the rabies virus mixed with red fluorescent beads to unambiguously identify the injection site of alBNST and amBNST cases (Fig. 6A–B and Fig. 7A–B). As expected from the previous results using the CAV2-Cre virus, inputs of the alBNST (n = 2) were labeled in anterior cingulate and prelimbic cortices (Fig. 6C–D), thalamus (Fig. 6E–F), basal lateral and central amygdala (Fig. 6G–J), amygdalopiriform and amygdalohippocampal transition areas (Fig. 6I-L), and raphe nuclei (Fig. 6M–N). Our SADΔG-EGFP injections also targeted the amBNST successfully (n = 3) (Fig. 7A–B) and showed labeling consistent to the CAV2-Cre in the lateral septum, different regions of hypothalamus (Fig. 7C–G), and central amygdala (Fig. 7H–I). Strong labeling was also seen in the posterior part of the basolateral amygdala and amydalohippocampal transition area (Fig. 7J-M). Inputs from the hippocampal formation, including hippocampus, subiculum and entorhinal cortex (Fig. 7N–P) were also observed. While the number and intensity of labeled neurons varied between tracing methods (discussed in detail below), the results using the CAV2-Cre virus and G-deleted rabies virus are qualitatively the same.
Fig. 6.
Retrograde tracing of afferent inputsto the alBNST using a modified rabies virus. Schematic of SADΔG-EGFP virus injection into the alBNST of wild-type C57BL/6 mice. A modified rabies virus expressing EGFP was co-injected with red fluorescent microbeads for unambiguous identification of the injected area. The gray dotted line shows the contours of the entire adBNST. (A–B) A representative image of an injection site into the alBNST. Scale bar, 1 mm in panel A and 200 μm in panel B. (C–D) Retrogradely-labeled neurons expressing EGFP in the Cg1 and PrL. (E–F) Labeled neurons in the mediodorsal (MD), anteromedial (AM), and central medial (CM) thalamic nuclei. (G–H) Labeled neurons in the BLA and CeA. (I–J) Labeled neurons in the BLP and AHiA. (K–L) Labeled neurons in the APir, BLP, and AHiA. (M–N) Labeled neurons in the DR and CLi. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 7.
Retrograde tracing of afferent inputsto the amBNST using a modified rabies virus. Schematic of SADΔG-EGFP virus injection into the amBNST of wild-type C57BL/6 mice. A modified rabies virus expressing EGFP was co-injected with red fluorescent microbeads for unambiguous identification of the injected area. The gray dotted line shows the contours of the entire adBNST. (A–B) A representative image of an injection site into the amBNST. Scale bar, 1 mm in panel A and 200 μm in panel B. (C–E) Labeled neurons in the septohypothalamic nucleus (SHy), ventromedial preoptic nucleus (VMPO), and lateral septal nucleus (LS). (F–G) Labeled neurons in the hypothalamus. (H–I) Labeled neurons in the BLA, CeA, and BMA. (J–K) Labeled neurons in the BLP and AHiA. (L–M) Labeled neurons in the posterior part of the BLP. (N–P) Labeled neurons in hippocampus area CA1, Sub/ProS, LEnt, and PMCo. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.5. Verification of afferent inputs to the BNST using ChR2-assisted circuit mapping and anterograde tracing
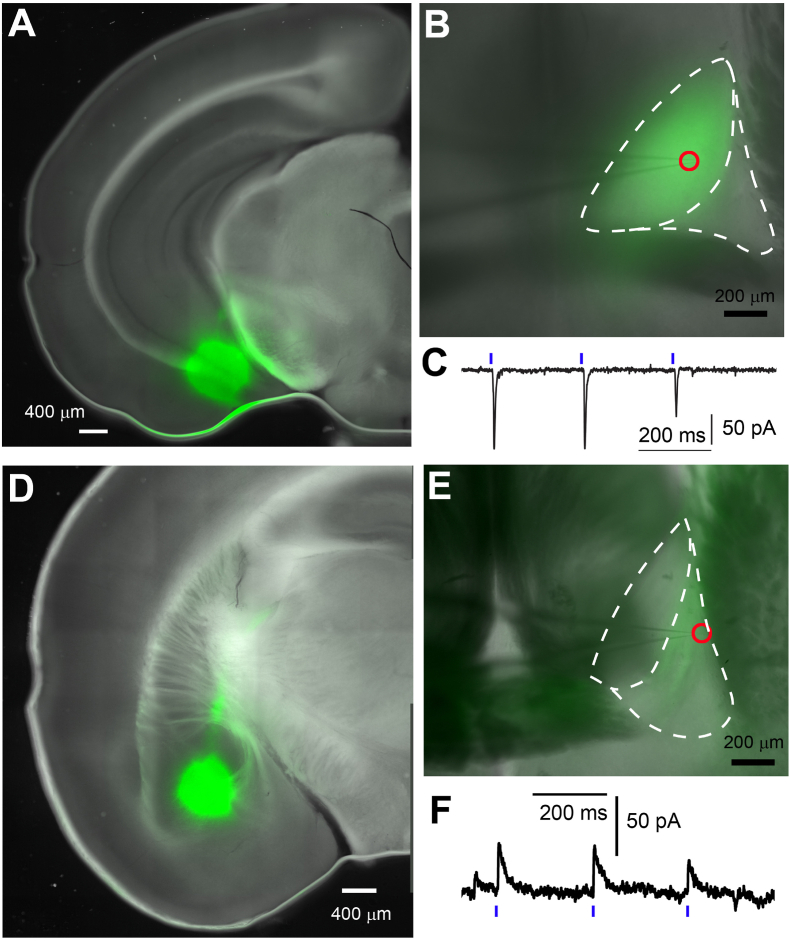
To functionally verify the long-range afferent inputs to BNST neurons, we performed ChR2-assisted circuit mapping using the whole-cell patch clamp recording and laser scanning photostimulations (Fig. 8) (Sun et al., 2016, 2019; Xu et al., 2016). We injected a ChR2-Venus expressing AAV virus into structures that provide anatomical inputs to the BNST subregions, guided by our tracing experiments. Focusing on the projection from the amydalohippocampal transition area (AHiA) to the adBNST (Fig. 8A) (Canteras et al., 1992; Weller and Smith, 1982), we first validated this connection by visualizing dense ChR2-Venus expressing axons in the amBNST (Fig. 8B). Following the photoactivation of intensely labeled ChR2-Venus axons from the AHiA, whole-cell recordings from the neurons in this subregion showed clear excitatory synaptic input responses (Fig. 8C). Finally, we validated another long-range input from the BLA to adBNST using the same method (Fig. 8D–F). In contrast to AHiA, photoactivation of the ChR2-Venus expressing axon terminals from the BLA evoked inhibitory postsynaptic responses to the recorded BNST neurons (Fig. 8D–F). Together, our results showed the adBNST receives functional long-range excitatory and inhibitory inputs from anatomically distinct brain regions.
Fig. 8.
Verification of functional synaptic input from amydalohippocampal transition area (AHiA)and basolateral amygdala (BLA)to theadBNST. (A) Bright field image showing AAV expression of ChR2-Venus at the injection site in the AHiA. (B) The BNST (dotted white line) superimposed on ChR2-Venus-expressing axons (green) originating from the viral injection shown in panel A. The red circle indicates the recorded cell location in the BNST. (C) Example of excitatory postsynaptic current (EPSC) responses recorded from an amBNST neuron after repeated optogenetic stimulation. (D) Bright field image showing AAV expression of ChR2-Venus at the injection site in the BLA. (E) The BNST (dotted white line) superimposed on ChR2-Venus-expressing axons (green) originating from the viral injection shown in panel D. The red circle indicates the recorded cell location in the BNST. (F) Example inhibitory postsynaptic current (IPSC) responses recorded from a BNST neuron in response to repeated optogenetic stimulation. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
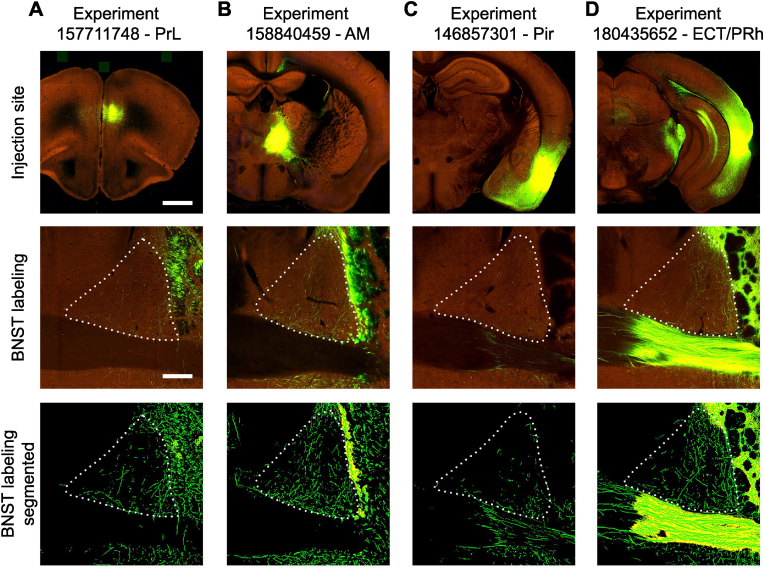
To further confirm the novel input connections from AM, Pir and ECT/PRh that revealed by the CAV2 tracing, we searched The Allen Mouse Brain Connectivity Atlas (connectivity.brain-map.org), which contains anterograde tracing data in the mouse brain using adeno-associated viruses (AAV) (Oh et al., 2014). We identified cases performed in wild type C57BL/6 mice with their injection sites (in PrL, AM, Pir, or Ect/PRh) that matched our retrograde labeling areas in the CAV2 cases and examined their axonal projections in the BNST. For validation purpose, first, we found significant axonal labeling in the adBNST in a case targeting the prelimbic cortex (Fig. 9A), a brain region that has known projections to the BNST. Furthermore, we found all the examined cases with injection sites in the AM, Pir, and ECT/PRh also showed axonal labeling in the adBNST, although some of them are relatively weak (Fig. 9B–D). Together, the anterograde tracing results further confirmed the novel input connections from the AM, Pir and ECT/PRh to the adBNST that revealed by the CAV2 tracing.
Fig. 9.
Verification of novel projections to the BNST using anterograde AAV tracing. (A) Top: the injection site of AAV-EGFP in the prelimbic cortex (PrL) in a wild type C57BL/6 mouse. The green color represents the signal from the AAV expression, while the maroon color represents the structure of the brain section. Scale bar: 1 mm and applies to all other panels on the top. Middle: the corresponding axonal projections in the BNST. Scale bar: 200 μm and applies to all other middle and bottom panels. Bottom: same as the middle panel, but segmented EGFP signals. Data images were obtained and modified from Allen Mouse Brain Connectivity Atlas: connectivity. brain-map.org/projection/experiment/157711748. (B) Same as A, but for an injection in the anteromedial thalamic nucleus (AM). Data images were obtained and modified from Allen Mouse Brain Connectivity Atlas: connectivity. brain-map.org/projection/experiment/158840459. (C) Same as A, but for an injection in the piriform cortex (Pir). Data images were obtained and modified from Allen Mouse Brain Connectivity Atlas: connectivity. brain-map.org/projection/experiment/146857301. (D) Same as A, but for an injection in the ectorhinal and perirhinal cortices (ECT/PRh). Data images were obtained and modified from Allen Mouse Brain Connectivity Atlas: connectivity. brain-map.org/projection/experiment/180435652. Image credit for this figure: Allen Institute for Brain Science (Oh et al., 2014). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3. Discussion
A mechanistic understanding of the neural circuit organization of different BNST subregions is critical for linking the circuit operations to its physiological functions. Taking advantage of two different retrograde viral-genetic tracing approaches (CAV2-Cre virus and rabies virus), we have mapped the brain-wide inputs to the alBNST and amBNST subregions of the dorsal component of the anterior BNST. Our results revealed a unidirectional local connection between alBNST and amBNST. Meanwhile, alBNST and amBNST received quantitatively different inputs, with cortical and thalamic inputs biased toward alBNST while inputs from the hypothalamus and medial amygdala biased toward amBNST. Our study provides a brain-wide input connectome to different subregions of the BNST that may underlie their complex functions.
The differential cortical inputs between the alBNST and amBNST from our quantitative assessment is highly consistent with previous qualitative studies. A previous study (McDonald et al., 1999) using PHA-L injections showed that the axons from the prefrontal cortex (PL and IL) primarily target the BNSTjc (a part of alBNST) and the axons from the agranular insular cortex (AI) primarily target the BNSTov, which is another part of alBNST. Similarly, they also reported the axons of the ventral subiculum primarily innervate the amBNST and axons of the amygdalopiriform transition area (APir) projects to the alBNST and amBNST in a more uniformed manner (Cullinan et al., 1993; Dong et al., 2001; McDonald et al., 1999). Interestingly, the ventral subiculum area that projects to the BNST in our study appears to match the location that was recently defined as the ventral prosubiculum which has distinct molecular and cellular features and provides specific innervations of the BNST (Ding et al., 2020). Importantly, our results extend these cortical inputs to the BNST by showing the ectorhinal and perirhinal cortices (ECT/PRh) preferentially innervate alBNST and the piriform cortex (Pir) also sends substantial inputs to both the alBNST and amBNST (Fig. 1, Fig. 5, Table 1, Table 2, Table 3).
Previous studies using anterograde and retrograde tracing showed that the basolateral and central amygdala projects primarily to the lateral division of the BNST while the medial amygdala projects primarily to the medial division of the BNST (Alheid and Heimer, 1988; Krettek and Price, 1978; Walker and Davis, 2008; Weller and Smith, 1982). This notion dominated the field until a more precise characterization was carried out by Dong et al. (2001), examining the topographic projections from the amygdala to the BNST (Dong et al., 2001). In this study, the authors called for a revision of the previous wiring diagram by showing three main findings: 1) The central amygdala projects to both the lateral and medial divisions of the anterior BNST. This finding is consistent with our quantification for Ce AMG (Fig. 5, Table 1, Table 2, Table 3). 2) The medial amygdala projects primarily to the medial division of the anterior BNST. This finding is consistent with the previous notion and was clearly corroborated from our results for Me AMG. (Fig. 5, Table 1, Table 2, Table 3). 3) The anterior basolateral amygdala (BLA) does not project significantly to the BNST. Although our quantification showed strong BNST innervations by the BL AMG (Fig. 5), this number includes both the BLA and the posterior basolateral amygdala (BLP). Our quantification was dominated by the BLP, as all the BLA labeling shown in our results were very weak (Fig. 2, Fig. 4, Fig. 6, Fig. 7). Thus, our results support the finding that the BLA weakly projects to the BNST. Note that our physiological mapping result showed a functional connection between the BLA and adBNST.
Another significant finding of our study is from the hypothalamus. Although the output projections of amBNST to the hypothalamus has been well characterized (Barbier et al., 2020; Dong and Swanson, 2006), few studies have characterized the hypothalamic inputs to the BNST in a systematic way (Conrad and Pfaff, 2004; Saper et al., 1976; Swanson, 1976). Here we found almost all the hypothalamic nuclei (except aH and LH) that shown in the quantification preferentially innervate amBNST, indicating a reciprocal connection between the amBNST and the hypothalamus. Given the function of the amBNST in response to stress, anxiety, and autonomic control (Crestani et al., 2013; Kim et al., 2013; Lin et al., 2018), the inputs from the hypothalamus to the BNST likely execute a feedback control of the Hypothalamic-Pituitary-Adrenal axis. Together, our results not only recapitulate the previous findings, but also provide novel discoveries regarding the topographic circuit connections of the medial and lateral adBNST.
3.1. Technical considerations
Although this study was not designed for comparing the tracing quality between CAV2-Cre and SADΔG-EGFP, we observed slight differences (non-statistical) in some aspects of the mapping. The first consideration is the variation between the total neurons counted across the cases using the two different viruses. On average, the CAV2-Cre virus labeled a total of 2199 neurons per case, while the SADΔG-EGFP virus labeled an average of 698 neurons per case (combined alBNST and amBNST cases). Furthermore, there was a fairly large variation from different cases within the SADΔG-EGFP virus experiments. For example, the total number of neurons labeled by the SADΔG-EGFP virus ranged from 281 to 1284 neurons per case. Finally, both viruses labeled similar areas, but with different percentages of the total number of labeled input neurons. For some brain regions, there was a greater percentage of inputs mapped by SADΔG-EGFP compared to the CAV2-Cre virus, despite the greater total cell count in the CAV2-Cre cases. One extreme example is, the SADΔG-EGFP labeled the periventricular, medial, and lateral hypothalamus at an average of 10.20 ± 5.56%, 23.96 ± 8.64%, and 15.79 ± 3.36% of the total cell count, respectively, comparing to the highest percentage recorded in the hypothalamus of CAV2-Cre cases (i.e., 4.18 ± 0.24% from LH, Table 1, Table 2). The CAV2-Cre virus also labeled ∼4% and 10% of the total cell count in the piriform cortex and anteromedial thalamus, but similar labeling was not seen in SADΔG-EGFP cases. Given that CAV2-Cre labeling in the Ai9 reporter mouse is highly sensitive, this is most likely due to the difference of labeling efficiency between the two approaches. However, despite the lack of direct evidence, we could not rule out the possibility that this may be due to the CAV2 uptake by the axons in passage situated right next to the alBNST. Overall, despite any influence of this technical limitation on the quantification or relative percentage of inputs, both viruses mapped similar areas in the prefrontal cortices, septum, thalamus, hypothalamus, amygdala, and hippocampal formation.
It is notable that the CAV2-Cre virus mapped more cell bodies than the SADΔG-EGFP. With respect to neuron viability, SADΔG-EGFP is more toxic to neurons than CAV2-Cre, with a survival time window of ∼14 days after infection (Osakada et al., 2011; Wickersham et al., 2007). To avoid this neural toxicity, mice injected with SADΔG-EGFP were perfused one week later to allow sufficient fluorescence expressions. In contrast, CAV2-Cre was allowed to express in the mice for one month, thus labeling a greater number of neurons in similar areas as the SADΔG-EGFP. The extended time allowed for the CAV2-Cre virus may enable infection of distant structures such as the piriform cortex and may also explain why the virus labeled almost twice as many neurons as the highest number of neurons mapped by the SADΔG-EGFP. However, because of this difference in the time allowed for each virus to replicate in the brain, one should not be deemed more efficient than the other.
In the ChR2-assisted circuit mapping experiments, the ChR2-expressing AAV was intended to target AHiA and BLA, respectively. However, our AAV injections in the AHiA may also cover a part of the ventral subiculum (Fig. 8A), which is known to send direct projections to the BNST. Thus, under our experimental conditions, we cannot rule out the possibility that the EPSCs that we recorded from the BNST slices may be contributed by both the AHiA and ventral subiculum connectivity.
3.2. Functional implications
The similarities between the inputs mapped by these viruses and those mapped in the literature outline subregion-specific feedback loops that can be further explored by behavioral studies. The projections from the BNST to the lateral hypothalamys (LH) have been shown to regulate appetitive and aversive behavioral states (Giardino et al., 2018; Kim et al., 2013; Wang et al., 2019). Reciprocally, our data showed projections from the LH to both the alBNST and amBNST. Future studies may find that the feedback control of different BNST subregions from the LH contribute to specific motivational states. Another structure with strong projections into the adBNST was the central amygdala, which has been shown to have reciprocal connections with the BNST and which plays a role in defense responses such as freezing to aversive stimuli (Walker and Davis, 2008; Walker et al., 2003, 2009). Therefore, a further exploration of adBNST circuits investigating basolateral and central amygdala feedback loops can place this subregion in a more specific behavioral context.
Further studies could also investigate the cell types that project to and from the adBNST and see whether they mediate specific functions or behaviors. For example, CRF expressing and CCK-expressing BNST neurons provide abundant and sparse inputs onto Hcrt-expressing neurons in the hypothalamus, respectively, and drive opposite emotionally valenced behaviors (Giardino et al., 2018). PKC-δ neurons in the BNSTov bidirectionally regulate feeding behaviors through the projection from the ventral BNST to the hypothalamus (Wang et al., 2019). Davis et al. (2009) also described how phasic and sustained types of fear were regulated by glutamatergic inputs from the basolateral amygdala and CRF inputs from the central amygdala to the lateral BNST(Davis et al., 2009). Furthermore, a recent anatomical study showed amBNST innervates proopiomelanocortin (POMC), agouti-related peptide (AgRP), melanin-concentrating-hormone (MCH) and orexin (ORX) neurons in the arcuate and the lateral hypothalamus (Barbier et al., 2020). These types of studies shed light on further characterizing the cell-type specific functions in different subregions of the BNST.
4. Methods
4.1. Subjects
All experiments were conducted according to National Institutes of Health guidelines for animal care and use and were approved by the Institutional Animal Care and Use Committee of the University of California, Irvine. For the CAV2-Cre tracing experiment, 15 Ai9 mice (Rosa-CAG-LSL-tdTomato-WPRE(Madisen et al., 2009), either sex) were used with 7 mice targeting alBNST, 5 targeting amBNST, and 3 targeting adBNST. For the G-deleted rabies (SADΔG-EGFP) tracing experiment, 6 C57BL/6 mice (either sex) were used with 3 mice targeting alBNST and 3 mice targeting amBNST. Mice were group housed with same-sex littermates and were kept on a 12-h light/dark cycle and had ad libitum access to food and water in their home cages at all times. Mice were 8–12 weeks old at the time of surgery.
4.2. Virus injections
The general procedure of virus injections has been previously described(Sun et al., 2014, 2017, 2018, 2019). In brief, mice were anesthetized for 10 min under 1.5% isoflurane with 0.8 L/min oxygen flow rate using an isoflurane table unit before securing their heads on a rodent stereotax (Leica Angle Two for mouse) for surgery. A three-axis micromanupulator and digital mouse brain atlas were used to find the injection site. A picospritzer and glass pipette were used to deliver approximately 0.2 μL of each virus at a rate of 20–30 nl/min with 10 ms pulse. The following injection coordinates targeting different BNST subfields were used (all values are given relative to the bregma): alBNST: lateromedial (ML) −1.00, anteroposterior (AP) 0.26, dorsoventral (DV) −4.10; amBNST: ML -0.75, AP 0.26, DV -4.10; adBNST: ML -0.90, AP 0.26, DV -4.10. To prevent backflow of virus, the pipette remained in the brain for 5 min after completion of the injection. Once the injection pipette was withdrawn, the mouse was removed from the stereotax, and the incision was closed with tissue adhesive (3M Vetbond). Mice were returned to their home cages to recover.
The replication-incompetent neurotropic canine adenovirus type 2 expressing Cre recombinase (CAV2-Cre) was obtained from Larry Zweifel's lab at the University of Washington, Seattle. CAV2-Cre was injected into Ai9 mice who were allowed to survive for four weeks post-injection to permit sufficient retrograde spread of the virus. Each injection of CAV2-Cre virus was comprised of 0.2 μl of ∼2.5 × 1012 physical particles/ml. The glycoprotein (G) deleted SAD B19 strain of rabies virus encoding enhanced green fluorescent protein (EGFP) (SADΔG-EGFP) was made at the University of California, Irvine, with required cell lines and seeding viruses from Edward Callaway's group at the Salk Institute for Biological Studies. The modification of G-deletion prevents trans-synaptic travel of the rabies virus (Wickersham et al., 2007). SADΔG-EGFP was injected into wild type mice and the mice were allowed to survive for one-week post-injection to allow for the virus to spread. Red microbeads were co-injected with the SADΔG-EGFP virus for unambiguous identification of the injected area. Both viruses have been cited as viable and specific retrograde tracers that can be used to clearly image cell morphology (Gore et al., 2013; Schwarz et al., 2015; Sun et al., 2014, 2019; Wickersham et al., 2007). An adeno-associated virus (AAV) expressing a Venus tagged channelrhodpsin-2 (ChR2) (AAV-ChR2-Venus) was also used to map long-range input connections and determine their functionality in optogenetic experiments.
4.3. Histology and quantification
Mice were deeply anesthetized with isoflurane and transcardially perfused with 5 ml of phosphate-buffered saline (PBS), followed by 25 ml of 4% paraformaldehyde-containing phosphate buffer. Brains were removed, post-fixed in paraformaldehyde overnight, and transferred into a 30% sucrose-PBS solution. Thirty-micron thick sections were cut on a freezing microtome and every third section was stained with 10 μL of DAPI before mounting and cover-slipping on slides in 1% PBS. Slides were visualized under a fluorescent microscope under 10× magnification (Olympus) using a high-sensitive Hamamatsu CCD camera and automated slide scanning and analysis software (Metamorph). GFP fluorescence was detected in tissue from SADΔG-EGFP-injected mice, and tdTomato red fluorescence was observed in CAV2-Cre infected tissue. Photoshop CS was used to visualize and count the number of labeled neurons in areas designated by The Mouse Brain in Stereotaxic Coordinates by Paxinos and Franklin. Quantification was performed manually by registering each brain section to the brain atlas. We only counted robust fluorescence-labeled cell bodies in each brain region. For studies using the CAV2-Cre, the number of neurons counted was summed and the percentage was calculated with respect to the total number of cells labeled by the red fluorescent protein. For the four cases using the SADΔG-EGFP virus, the number of neurons was summed and averaged per animal.
4.4. ChR2 photoactivation mediated circuit mapping
To complement virus-based circuit mapping, we used our established functional-mapping approach to perform CRACM using a previously described protocol (Sun et al., 2016, 2019; Xu et al., 2016). First, AAV1-CAG-ChR2-Venus was injected into either the AHiA or medial amygdala iontophoretically by using a Digital Midgard Precision Current Source (Stoelting). The virus was delivered by applying currents of positive 3 μA in an alternative mode with a cycle of 7 s on and 7 s off for 5 min. After recovery, mice were housed in the animal facility for 3 weeks before slice experiments.
Coronal sections of 400 μm were cut from BNST with a vibratome (VT1200S, Leica Biosystems, Buffalo Grove, IL) in sucrose containing ACSF (in mM: 85 NaCl, 75 sucrose, 2.5 KCl, 25 glucose, 1.25 NaH2PO4, 4 MgCl2, 0.5 CaCl2, and 24 NaHCO3). Slices were incubated for at least 30 min in normal ACSF (in mM: 126 NaCl, 2.5 KCl, 26 NaHCO3, 2 CaCl2, 2 MgCl2, 1.25 NaH2PO4, and 10 glucose) at 32 °C before transferred into slice recording chambers. Throughout the cutting, incubation and recording, the solutions were continuously supplied with 95% O2–5% CO2.
Whole-cell recordings were performed under a differential interference contrast/fluorescence microscope (BX51WI, Olympus). To target whole-cell recordings, cells were visualized at high magnification (60 × objective, 0.9 numerical aperture; LUMPlanFl/IR, Olympus). Cell bodies of recorded neurons were at least 50 μm below the surface of the slice. Patch pipettes (4–6 MΩ resistance) made of borosilicate glass were filled with an internal solution containing the following (in mM): 126 K-gluconate, 4 KCl, 10 HEPES buffer, 4 ATP-Mg, 0.3 GTP-sodium and 10 phosphocreatine (pH 7.2, 300 mOsm). Electrodes also contained 0.1% biocytin for post hoc cell labeling and further morphological identification. Once stable whole-cell recordings were achieved with good access resistance (usually <30 MΩ), basic electrophysiological properties were examined through hyperpolarizing and depolarizing current injections.
During CRACM experiments, TTX (1 μM) and 4-AP (100 μM) were added to the bath solution to block Na + channels and the K+ channels that are critical for repolarization of the axon so that spatially restricted activation of ChR2-expressing AHiA/amygdala axons with a 473-nm blue laser (∼3 mW/5.5 mm2, 0.25 ms; laser spot diameter, ∼50 μm) only occurred in the vicinity of the laser beam (with action potentials blocked), enabling functional assessments of direct and monosynaptic synaptic connectivity. We used sequential stimulation of the blue laser at 16 × 16 different sites arranged in a mapping grid covering most of the BNST slice in a nonraster, nonrandom sequence to avoid revisiting the vicinity of recently stimulated sites. Simultaneous whole-cell voltage-clamp recordings were made from BNST neurons to measure optogenetic stimulation-evoked excitatory postsynaptic current (EPSC) responses at the holding potential around −70 mV. The EPSC response from each stimulation site was the measurement of the sum of total EPSCs within the analysis window (10 ms–160 ms post photostimulation), with the baseline spontaneous response subtracted from the photostimulation response of the same site. The value was normalized against the duration of the analysis window (i.e., 150 ms) and expressed as the average integrated amplitudes in picoamperes (pA). Furthermore, fluorescent images of EGFP were taken in situ at the recording site to validate ChR2 expressions. After physiological assays had been completed, brain slices were fixed in 4% paraformaldehyde in PBS overnight and transferred to 30% sucrose solution in PBS. The slices were imaged under a fluorescent microscope under 10× magnification (Olympus) to further visualize the ChR2 expressions in both the injection and recording sites.
4.5. Statistical analyses
GraphPad Prism 9 was used for statistical analyses. All data were reported as mean ± standard error of the mean (SEM). We assumed all the data were normally distributed. We used a Student's t-test, when comparing the data between alBNST and amBNST groups. A p value (≤0.05) was considered statistically significant.
CRediT authorship contribution statement
Yanjun Sun: Investigation, Validation, Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. Larry S. Zweifel: Resources. Todd C. Holmes: Writing – review & editing. Xiangmin Xu: Conceptualization, Supervision, Funding acquisition, Investigation, Writing – review & editing.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by US National Institutes of Health grants MH113026 (XX), MH112085 (XX). TCH was supported by US National Institites of Health grant R35 GM127102.
Handling Editor: Rita Valentino
References
- Alheid G.F., Heimer L. New perspectives in basal forebrain organization of special relevance for neuropsychiatric disorders: the striatopallidal, amygdaloid, and corticopetal components of substantia innominata. Neuroscience. 1988;27:1–39. doi: 10.1016/0306-4522(88)90217-5. [DOI] [PubMed] [Google Scholar]
- Barbier M., González J.A., Houdayer C., Burdakov D., Risold P.Y., Croizier S. Projections from the dorsomedial division of the bed nucleus of the stria terminalis to hypothalamic nuclei in the mouse. J. Comp. Neurol. 2020;529:929–956. doi: 10.1002/cne.24988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleier R.H. 1961. The Hypothalamus of the Cat : a Cytoarchitectonic Atlas in the Horsley( -Clarke co-ordinate system) [Google Scholar]
- Canteras N.S., Simerly R.B., Swanson L.W. Connections of the posterior nucleus of the amygdala. J. Comp. Neurol. 1992;324:143–179. doi: 10.1002/cne.903240203. [DOI] [PubMed] [Google Scholar]
- Carter M.E., Soden M.E., Zweifel L.S., Palmiter R.D. Genetic identification of a neural circuit that suppresses appetite. Nature. 2013;503:111–114. doi: 10.1038/nature12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D.C., Furay A.R., Evanson N.K., Ostrander M.M., Ulrich-Lai Y.M., Herman J.P. Bed nucleus of the stria terminalis subregions differentially regulate hypothalamic-pituitary-adrenal Axis activity: implications for the integration of limbic inputs. J. Neurosci. 2007;27:2025–2034. doi: 10.1523/JNEUROSCI.4301-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad L.C.A., Pfaff D.W. Efferents from medial basal forebrain and hypothalamus in the rat. II. An autoradiographic study of the anterior hypothalamus. J. Comp. Neurol. 2004;169:221–261. doi: 10.1002/cne.901690206. [DOI] [PubMed] [Google Scholar]
- Crestani C., Alves F., Gomes F., Resstel L., Correa F., Herman J. Mechanisms in the bed nucleus of the stria terminalis involved in control of autonomic and neuroendocrine functions: a review. Curr. Neuropharmacol. 2013;11:141–159. doi: 10.2174/1570159X11311020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan W.E., Herman J.P., Watson S.J. Ventral subicular interaction with the hypothalamic paraventricular nucleus: evidence for a relay in the bed nucleus of the stria terminalis. J. Comp. Neurol. 1993;332:1–20. doi: 10.1002/cne.903320102. [DOI] [PubMed] [Google Scholar]
- Daniel S.E., Rainnie D.G. Stress modulation of opposing circuits in the bed nucleus of the stria terminalis. Neuropsychopharmacology. 2015;41:103–125. doi: 10.1038/npp.2015.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M., Walker D.L., Miles L., Grillon C. Phasic vs sustained fear in rats and humans: role of the extended amygdala in fear vs anxiety. Neuropsychopharmacology. 2009;35:105–135. doi: 10.1038/npp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Olmos J.S. vol. 2. 1972. pp. 145–204. (The Amygdaloid Projection Field in the Rat as Studied with the Cupric-Silver Method). [Google Scholar]
- Ding S.-L., Yao Z., Hirokawa K.E., Nguyen T.N., Graybuck L.T., Fong O., Bohn P., Ngo K., Smith K.A., Koch C., et al. Distinct transcriptomic cell types and neural circuits of the subiculum and prosubiculum along the dorsal-ventral Axis. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.L. A novel subdivision of the bed nucleus of stria terminalis in monkey, rat, and mouse brains. J. Comp. Neurol. 2022:1–25. doi: 10.1002/cne.25446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H.-W., Petrovich G.D., Swanson L.W. Topography of projections from amygdala to bed nuclei of the stria terminalis. Brain Res. Rev. 2001;38:192–246. doi: 10.1016/s0165-0173(01)00079-0. [DOI] [PubMed] [Google Scholar]
- Dong H.-W., Swanson L.W. Organization of axonal projections from the anterolateral area of the bed nuclei of the stria terminalis. J. Comp. Neurol. 2004;468:277–298. doi: 10.1002/cne.10949. [DOI] [PubMed] [Google Scholar]
- Dong H.-W., Swanson L.W. Projections from bed nuclei of the stria terminalis, anteromedial area: cerebral hemisphere integration of neuroendocrine, autonomic, and behavioral aspects of energy balance. J. Comp. Neurol. 2006;494:142–178. doi: 10.1002/cne.20788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardino W.J., Eban-Rothschild A., Christoffel D.J., Li S.-B., Malenka R.C., de Lecea L. Parallel circuits from the bed nuclei of stria terminalis to the lateral hypothalamus drive opposing emotional states. Nat. Neurosci. 2018;21:1084–1095. doi: 10.1038/s41593-018-0198-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardino W.J., Pomrenze M.B. Extended amygdala neuropeptide circuitry of emotional arousal: waking up on the wrong side of the bed nuclei of stria terminalis. Front. Behav. Neurosci. 2021;15 doi: 10.3389/fnbeh.2021.613025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore B.B., Soden M.E., Zweifel L.S. Manipulating gene expression in projection‐specific neuronal populations using combinatorial viral approaches. Current Protocols in Neuroscience. 2013;65 doi: 10.1002/0471142301.ns0435s65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gungor N.Z., Pare D. Functional heterogeneity in the bed nucleus of the stria terminalis. J. Neurosci. 2016;36:8038–8049. doi: 10.1523/JNEUROSCI.0856-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubensak W., Kunwar P.S., Cai H., Ciocchi S., Wall N.R., Ponnusamy R., Biag J., Dong H.-W., Deisseroth K., Callaway E.M., et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature. 2010;468:270–276. doi: 10.1038/nature09553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju G., Swanson L.W. Studies on the cellular architecture of the bed nuclei of the stria terminalis in the rat: I. cytoarchitecture. J. Comp. Neurol. 1989;280:587–602. doi: 10.1002/cne.902800409. [DOI] [PubMed] [Google Scholar]
- Ju G., Swanson L.W., Simerly R.B. Studies on the cellular architecture of the bed nuclei of the stria terminalis in the rat: II. chemoarchitecture. J. Comp. Neurol. 1989;280:603–621. doi: 10.1002/cne.902800410. [DOI] [PubMed] [Google Scholar]
- Kim S.-Y., Adhikari A., Lee S.Y., Marshel J.H., Kim C.K., Mallory C.S., Lo M., Pak S., Mattis J., Lim B.K., et al. Diverging neural pathways assemble a behavioural state from separable features in anxiety. Nature. 2013;496:219–223. doi: 10.1038/nature12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krettek J.E., Price J.L. Amygdaloid projections to subcortical structures within the basal forebrain and brainstem in the rat and cat. J. Comp. Neurol. 1978;178:225–253. doi: 10.1002/cne.901780204. [DOI] [PubMed] [Google Scholar]
- Lin X., Itoga C.A., Taha S., Li M.H., Chen R., Sami K., Berton F., Francesconi W., Xu X. c-Fos mapping of brain regions activated by multi-modal and electric foot shock stress. Neurobiol. Stress. 2018;8:92–102. doi: 10.1016/j.ynstr.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2009;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews Gillian A., Nieh Edward H., Weele Vander, Caitlin M., Halbert Sarah A., Pradhan Roma V., Yosafat Ariella S., Glober Gordon F., Izadmehr Ehsan M., Thomas Rain E., Lacy Gabrielle D., et al. Dorsal raphe dopamine neurons represent the experience of social isolation. Cell. 2016;164:617–631. doi: 10.1016/j.cell.2015.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald A.J., Shammah-Lagnado S.J., Shi C., Davis M. Cortical afferents to the extended amygdala. Ann. N. Y. Acad. Sci. 1999;877:309–338. doi: 10.1111/j.1749-6632.1999.tb09275.x. [DOI] [PubMed] [Google Scholar]
- Moga M.M., Saper C.B., Gray T.S. Bed nucleus of the stria terminalis: cytoarchitecture, immunohistochemistry, and projection to the parabrachial nucleus in the rat. J. Comp. Neurol. 1989;283:315–332. doi: 10.1002/cne.902830302. [DOI] [PubMed] [Google Scholar]
- Nguyen A.Q., Dela Cruz J.A.D., Sun Y., Holmes T.C., Xu X. Genetic cell targeting uncovers specific neuronal types and distinct subregions in the bed nucleus of the stria terminalis. J. Comp. Neurol. 2016;524:2379–2399. doi: 10.1002/cne.23954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.W., Harris J.A., Ng L., Winslow B., Cain N., Mihalas S., Wang Q., Lau C., Kuan L., Henry A.M., et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakada F., Mori T., Cetin Ali H., Marshel James H., Virgen B., Callaway Edward M. New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron. 2011;71:617–631. doi: 10.1016/j.neuron.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomrenze M.B., Tovar-Diaz J., Blasio A., Maiya R., Giovanetti S.M., Lei K., Morikawa H., Hopf F.W., Messing R.O. A corticotropin releasing factor network in the extended amygdala for anxiety. J. Neurosci. 2019;39:1030–1043. doi: 10.1523/JNEUROSCI.2143-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saper C.B., Swanson L.W., Cowan W.M. The efferent connections of the ventromedial nucleus of the hypothalamus of the rat. J. Comp. Neurol. 1976;169:409–442. doi: 10.1002/cne.901690403. [DOI] [PubMed] [Google Scholar]
- Schnapp, W.I., Kim, J., Wang, Y., Timilsena, S., Fang, C., and Cai, H. (2022). Central extended amygdala neurons in the development of activity-based anorexia. bioRxiv 2022.11.10.515977. [DOI] [PMC free article] [PubMed]
- Schwarz L.A., Miyamichi K., Gao X.J., Beier K.T., Weissbourd B., DeLoach K.E., Ren J., Ibanes S., Malenka R.C., Kremer E.J., et al. Viral-genetic tracing of the input–output organization of a central noradrenaline circuit. Nature. 2015;524:88–92. doi: 10.1038/nature14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Grieco S.F., Holmes T.C., Xu X. Local and long-range circuit connections to hilar mossy cells in the dentate gyrus. eneuro. 2017;4 doi: 10.1523/ENEURO.0097-17.2017. ENEURO.0097-0017.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Ikrar T., Davis Melissa F., Gong N., Zheng X., Luo Z.D., Lai C., Mei L., Holmes Todd C., Gandhi Sunil P., et al. Neuregulin-1/ErbB4 signaling regulates visual cortical plasticity. Neuron. 2016;92:160–173. doi: 10.1016/j.neuron.2016.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Jin S., Lin X., Chen L., Qiao X., Jiang L., Zhou P., Johnston K.G., Golshani P., Nie Q., et al. CA1-projecting subiculum neurons facilitate object–place learning. Nat. Neurosci. 2019;22:1857–1870. doi: 10.1038/s41593-019-0496-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Nguyen A.Q., Nguyen J.P., Le L., Saur D., Choi J., Callaway Edward M., Xu X. Cell-type-specific circuit connectivity of hippocampal CA1 revealed through cre-dependent rabies tracing. Cell Rep. 2014;7:269–280. doi: 10.1016/j.celrep.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Nitz D.A., Holmes T.C., Xu X. Opposing and complementary topographic connectivity gradients revealed by quantitative analysis of canonical and noncanonical hippocampal CA1 inputs. eneuro. 2018;5 doi: 10.1523/ENEURO.0322-17.2018. ENEURO.0322-0317.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson L.W. An autoradiographic study of the efferent connections of the preoptic region in the rat. J. Comp. Neurol. 1976;167:227–256. doi: 10.1002/cne.901670207. [DOI] [PubMed] [Google Scholar]
- van Groen T., Kadish I., Wyss J.M. Efferent connections of the anteromedial nucleus of the thalamus of the rat. Brain Research Reviews. 1999;30(1):1–26. doi: 10.1016/s0165-0173(99)00006-5. [DOI] [PubMed] [Google Scholar]
- Walker D.L., Davis M. Role of the extended amygdala in short-duration versus sustained fear: a tribute to Dr. Lennart Heimer. Brain Struct. Func. 2008;213:29–42. doi: 10.1007/s00429-008-0183-3. [DOI] [PubMed] [Google Scholar]
- Walker D.L., Miles L.A., Davis M. Selective participation of the bed nucleus of the stria terminalis and CRF in sustained anxiety-like versus phasic fear-like responses. Prog. Neuro Psychopharmacol. Biol. Psychiatr. 2009;33:1291–1308. doi: 10.1016/j.pnpbp.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D.L., Toufexis D.J., Davis M. Role of the bed nucleus of the stria terminalis versus the amygdala in fear, stress, and anxiety. Eur. J. Pharmacol. 2003;463:199–216. doi: 10.1016/s0014-2999(03)01282-2. [DOI] [PubMed] [Google Scholar]
- Wang Y., Kim J., Schmit M.B., Cho T.S., Fang C., Cai H. A bed nucleus of stria terminalis microcircuit regulating inflammation-associated modulation of feeding. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-10715-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller K.L., Smith D.A. Afferent connections to the bed nucleus of the stria terminalis. Brain Res. 1982;232:255–270. doi: 10.1016/0006-8993(82)90272-4. [DOI] [PubMed] [Google Scholar]
- Wickersham I.R., Lyon D.C., Barnard R.J.O., Mori T., Finke S., Conzelmann K.-K., Young J.A.T., Callaway E.M. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood R.I., Swann J.M. The bed nucleus of the stria terminalis in the Syrian hamster: subnuclei and connections of the posterior division. Neuroscience. 2005;135:155–179. doi: 10.1016/j.neuroscience.2005.05.029. [DOI] [PubMed] [Google Scholar]
- Xu X., Ikrar T., Sun Y., Santos R., Holmes T.C., Francesconi W., Berton F. High-resolution and cell-type-specific photostimulation mapping shows weak excitatory vs. strong inhibitory inputs in the bed nucleus of the stria terminalis. J. Neurophysiol. 2016;115:3204–3216. doi: 10.1152/jn.01148.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi N., Takahashi D., Sugimura Y.K., Kato F., Amano T., Minami M. Activation of the neural pathway from the dorsolateral bed nucleus of the stria terminalis to the central amygdala induces anxiety‐like behaviors. Eur. J. Neurosci. 2018;48:3052–3061. doi: 10.1111/ejn.14165. [DOI] [PubMed] [Google Scholar]