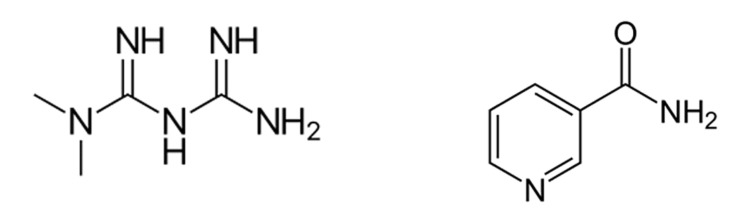Abstract
Whilst the pathophysiology at a cellular level has been defined, the cause of Parkinson’s disease (PD) remains poorly understood. This neurodegenerative disorder is associated with impaired dopamine transmission in the substantia nigra, and protein accumulations known as Lewy bodies are visible in affected neurons. Cell culture models of PD have indicated impaired mitochondrial function, so the focus of this paper is on the quality control processes involved in and around mitochondria.
Mitochondrial autophagy (mitophagy) is the process through which defective mitochondria are removed from the cell by internalisation into autophagosomes which fuse with a lysosome. This process involves many proteins, notably including PINK1 and parkin, both of which are known to be coded on genes associated with PD. Normally in healthy individuals, PINK1 associates with the outer mitochondrial membrane, which then recruits parkin, activating it to attach ubiquitin proteins to the mitochondrial membrane. PINK1, parkin, and ubiquitin cooperate to form a positive feedback system which accelerates the deposition of ubiquitin on dysfunctional mitochondria, resulting in mitophagy. However, in hereditary PD, the genes encoding PINK1 and parkin are mutated, resulting in proteins that are less efficient at removing poorly performing mitochondria, leaving cells more vulnerable to oxidative stress and ubiquitinated inclusion bodies, such as Lewy bodies. Current research that looks into the connection between mitophagy and PD is promising, already yielding potentially therapeutic compounds; until now, pharmacological support for the mitophagy process has not been part of the therapeutic arsenal. Continued research in this area is warranted.
Keywords: parkinson's disease, lewy body, parkin, pink1, mitophagy
Introduction and background
Parkinson’s disease and genetics
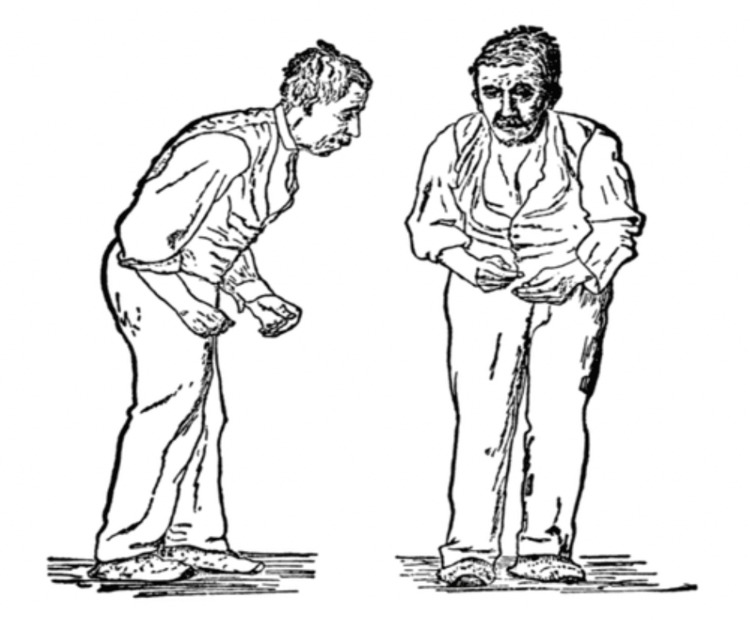
Parkinson’s disease (PD) is the second most common neurodegenerative disorder following Alzheimer’s, affecting 1% of adults over the age of 60. It is characterised by dopaminergic cell loss in substantia nigra pars compacta of the midbrain, and patients are presented with the classic symptoms of parkinsonism: bradykinesia, rigidity, resting tremor and postural instability (Figure 1) [1]. PD is a universal disorder present across the world, with a crude incidence rate of 5-19 people per 100,000 populations per year and a prevalence of 70-250 people per 100,000 populations, which rises with the increase in age [2,3]. In addition to placing enormous physical strain on the patients, PD also places a financial strain on the health service. Parkinson’s care costs the National Health Service (NHS) over £220 million per year and annual care for patients with advanced Parkinson’s (>75% of waking time in a state of decreased mobility) costs more than £60,000 per patient [4].
Figure 1. Figure showing Parkinson’s disease patient from the front and from the side. His face is expressionless, his posture is stooped forward, and he has difficulty walking.
Figure taken from [5], and permission was obtained from the original publisher to reproduce the content.
Although it is a high-profile disease and has attracted a great deal of research attention, the pathogenesis of PD remains poorly understood, with the disease being characterised mainly by its symptoms rather than its cause [6]. Most cases of PD (90%) are idiopathic, with late-onset. This form of PD is believed to be caused by a combination of genetic and environmental factors, with ageing being one of the most significant risk factors [7]. However, over the past 15 years a few Mendelian forms of PD have been discovered, comprising at least 10% of the disease burden. A number of distinct genome regions, known as PARK, have been identified as strongly associated with PD development. Mutations in genes encoding Parkinson juvenile disease protein 2-parkin (PARK2), phosphatase and tensin homolog (PTEN)-induced putative kinase 1-PINK1 (PARK6) and DJ-1 (PARK7) proteins are associated with autosomal recessive forms of PD, presumably by a loss-of-function mechanism, while gain-of-function mutations in genes that encode α-Syn (PARK1/4) and leucine-rich repeat kinase 2-LRRK2 (PARK8) are found in autosomal dominant forms [8]. These proteins, in particular PINK1 and parkin, are strongly linked to a pathway that negatively influences the degradation of mitochondria within the cell, known as mitophagy, which is thought to be related to the accumulation of defected mitochondria and dopaminergic neural cell death [9,10]. In this study, the proposed role of disrupted mitophagy in the pathogenesis of PD will be critically reviewed by drawing upon literatures, as well as holistically in the context of Parkinson’s therapy.
Diagnosis
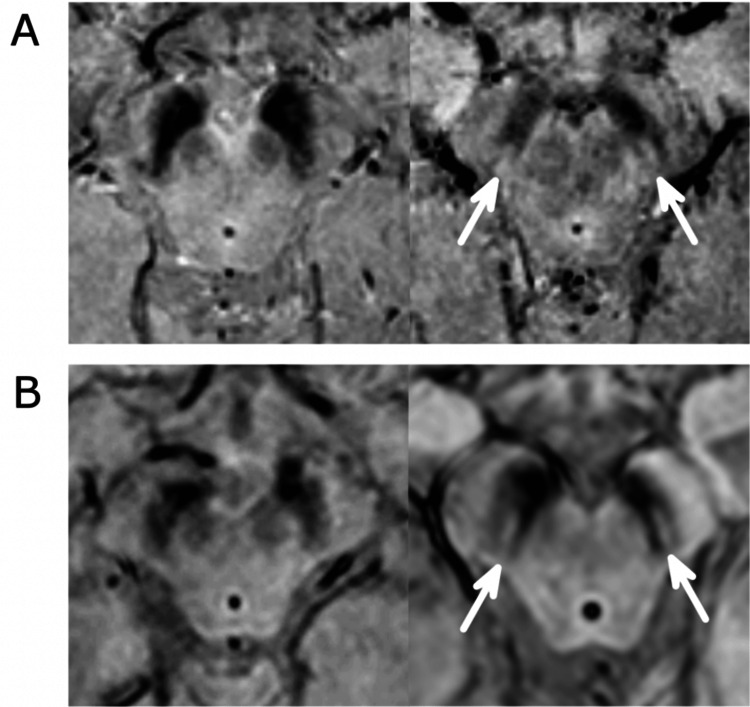
Diagnosis of PD can be challenging as early signs and symptoms may be dismissed as the effects of ageing, causing inaccurate diagnosis. This is complicated as other neurodegenerative disorders such as Creutzfeldt-Jakob disease can produce PD-like symptoms, referred to as Parkinson syndrome. There are, however, studies indicating potential advances in early diagnosis, such as the detection of hypokinetic dysarthria, speech disorders [11], and impaired olfaction, which can occur before motor symptoms develop [12]. Diagnosis may also be confirmed by using single-photon emission computerized tomography (SPECT) or MRI scan when PD cannot be differentiated from essential tremor. For healthy individuals, MRI examination of the substantia nigra will display a “swallow-tail” type appearance. However, such appearance is absent in many cases of PD [13]. These swallow-tails, and the lack thereof, are shown in Figure 2.
Figure 2. Swallow-tails indicated by arrows (right) present in a healthy female (A) and healthy male (B), or lack thereof in Parkinson’s disease patients (left), female (A) and male (B).
Figure taken from [14], and permission was obtained from the original publisher to reproduce the content.
In the UK, patients are referred to a specialist and diagnosis is based on the UK Parkinson’s Disease Society Brain Bank Clinical Diagnostic Criteria [15], which looks for the presence of motor symptomatology, particularly bradykinesia, and a number of non-motor symptoms such as dementia and depression.
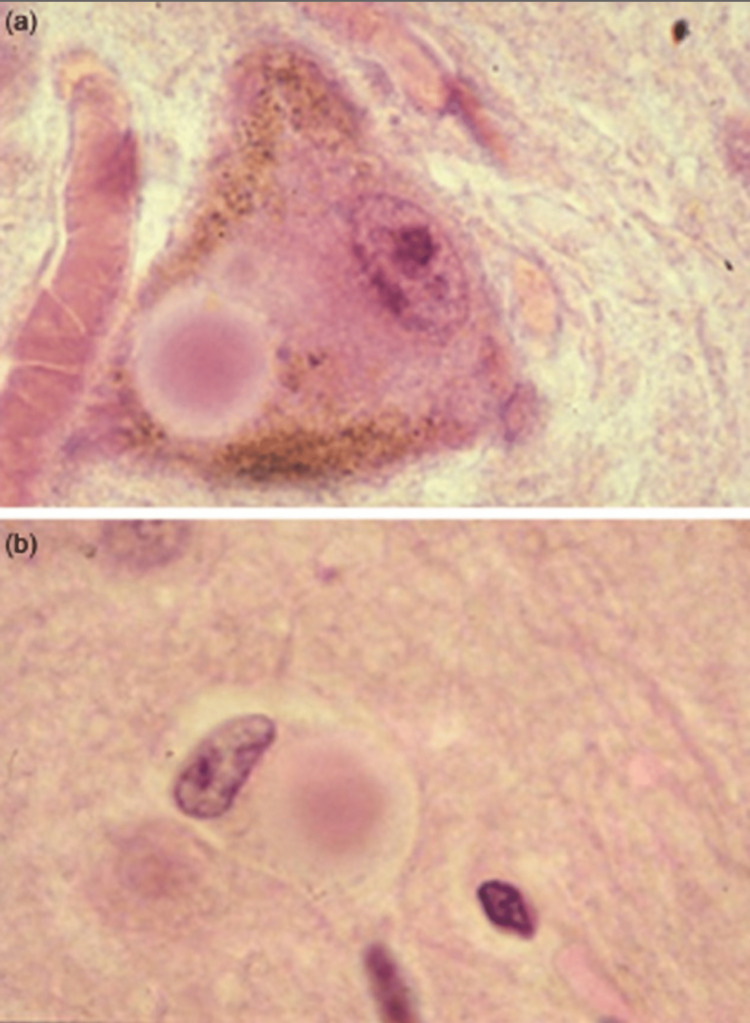
Post-mortem examination of the brain reveals abnormal accumulation of proteins into globular structures known as Lewy bodies (LBs) within the nerve cells, which can become large enough to displace other cellular components, as illustrated in Figure 3 [16,17]. LBs are composed of a combination of proteins including alpha-synuclein (α-Syn), ubiquitin, neurofilament protein, etc [18,19]. Tau proteins (which stabilise microtubules) and parkin can aggregate with α-Syn in the LBs [20,21]. It is highly likely that the aggregation of the misfolded proteins into LBs is an active process within the cell, with the aim of minimising the damage caused by aberrant proteins by condensing them into a single location rather than allowing their distribution throughout the entire cell. This active formation of LBs would classify as a type of aggresome [22,23]. However, it is this very attempt to prevent widespread cell damage that ultimately induces cell death via the hyperactivation of adenosine monophosphate (AMP)-activated protein kinase [24].
Figure 3. Lewy bodies (pink circular) present in brain tissue.
Figure taken from [19], and permission was obtained from the original publisher to reproduce the content.
Treatment
PD is currently incurable. Nonetheless, several symptomatic treatments are available with levodopa (L-DOPA) being the primary treatment in early PD [25,26]. L-DOPA is a precursor to dopamine, converted by the enzyme DOPA decarboxylase, and is administered to restore dopamine levels in the brain. L-DOPA is given instead of dopamine as the latter cannot cross the blood-brain barrier, and is often given in combination with a peripheral decarboxylase inhibitor to prevent premature conversion [27,28]. Unfortunately, long-term usage can lead to levodopa-induced dyskinesia as a side effect [29,30]. Various dopamine agonists such as aripiprazole, and MAO-B inhibitors can also be used to alleviate PD symptoms, but with lower efficacy [31]. In advanced PD, the development of motor complications necessitates adjuvant therapy with levodopa and either a Catechol-O-methyl transferase (COMT) inhibitor or dopamine agonist to ameliorate these complications [32]. If patients respond poorly to drug therapy, then surgical options such as deep brain stimulation (DBS) are also available. DBS with an electrode of the subthalamic nucleus and internal globus pallidus is conjectured to control the symptoms by either inhibiting or exciting local neurons [33].
Review
Disrupted mitophagy in Parkinson’s disease
Natural (Non-pathogenic) Mitophagy Processes
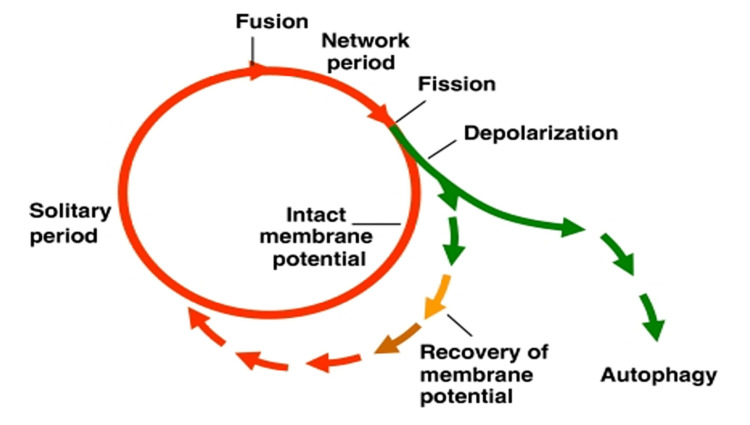
Mitophagy is defined as the specific and selective, targeted removal of excess or damaged mitochondria from the cell via macroautophagy, a process that utilises autophagosomes to deliver cellular structures to lysosomes for destruction [34]. Dysfunctional mitochondria accumulation as a result of impaired mitophagy has been implicated in several health conditions such as PD, metabolic syndrome, and cancer [35]. Mitophagy plays an important part in mitochondrial proliferation, i.e. enlarging and then a fission event produces two daughter mitochondria. The process is driven by AMP kinase (AMPK) which causes the mitochondria to elongate or fuse and proliferate, producing more ATP and reducing the production of reactive oxygen species (ROS) [36]. If daughter mitochondria have reduced membrane potential, they are selectively removed by mitophagy [37]. Mitochondria are also capable of temporary fusion with each other so that damaged or inefficient mitochondria may briefly receive electrochemical support from a larger mitochondrion, temporarily improving their performance [38,39]. Persistently depolarised mitochondria are targeted for mitophagy as illustrated in Figure 4.
Figure 4. Model of the life cycle of mitochondria.
Fusion events are rapidly followed by fission. Daughter mitochondria become solitary for a period before fusing again. Daughters with depolarised membranes may either recover their membrane potential, or are removed by mitophagy.
Figure taken from [37], and permission was obtained from the original publisher to reproduce the content.
Molecular mechanisms of PINK1/parkin-mediated mitophagy
The processes through which the molecular targeting of dysfunctional mitochondria takes place varies from one species to another, but in humans, the process involves the proteins PINK1, parkin and ubiquitin.
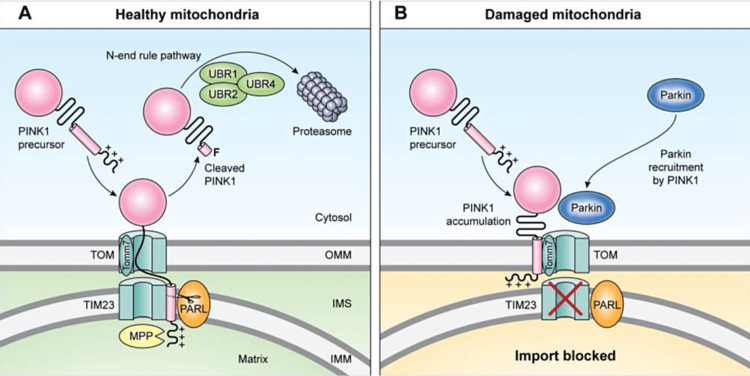
PINK1, a serine/threonine protein kinase, is a 63 kDa protein found throughout the cytosol and on the outer membranes of mitochondria (OMM), but generally only at low levels due to voltage-dependent proteolysis of PINK1 on healthy mitochondria, which can internalise and degrade PINK1 through a translocase of outer membrane (TOM) complex. The TOM complex then delivers PINK1 to the intermembrane space with the translocase of inner membrane (TIM) complex for cleavage by enzymes including mitochondrial processing peptidase (MPP) and presenilin-associated rhomboid-like protein (PARL) [40]. The resulting PINK1 fragments are then retro-translocated to the cytosol where they can be removed by lysosomal degradation. Due to the primary difference in the cytosolic presentation of PINK1 being an exposed and destabilised N-terminal amino acid, this process is known as the N-end pathway [10,41,42]. Dysfunctional mitochondria, which are not capable of maintaining the necessary inner membrane potential for this removal process, rapidly accumulate large amounts of PINK1 on the OMM which initiates the process of mitophagy [43,44]. This process of mitochondrial labelling is illustrated in Figure 5.
Figure 5. A comparison of healthy mitochondria (A) and damaged mitochondria (B) showing the processing of PINK1 by TOM, TIM, PARL, MPP, and the N-end rule pathway (A), or the accumulation of PINK1 on the OMM and recruitment of parkin (B).
Also indicated within the TOM complex is the minor subunit Tomm7, without which the functionality of the entire TOM ceases.
PINK1: PTEN-induced kinase 1; Parkin: Parkinson juvenile disease protein 2; UBR1: ubiquitin protein ligase E3 component N-recognin 1; UBR2: ubiquitin protein ligase E3 component N-recognin 2; UBR4: ubiquitin protein ligase E3 component N-recognin 4; TOM: translocase of outer-membrane complex; TIM23: mitochondrial import inner membrane translocase subunit; OMM: outer mitochondrial membrane; IMS: intermembrane space; IMM: inner mitochondrial membrane; Tomm7: translocase of outer mitochondrial membrane 7; MPP: mitochondrial processing peptidase; PARL: presenilin-associated rhomboid-like protein
Figure taken from [10], and permission was obtained from the original publisher to reproduce the content.
PINK1 is able to recruit parkin from the cytosol, associate and functionally cooperate on the surface of these compromised mitochondria to induce parkin-mediated mitophagy through phosphorylation of both parkin and ubiquitin (both at Ser65), with OMM-bound poly-ubiquitin chains acting as parkin receptors [45-47].
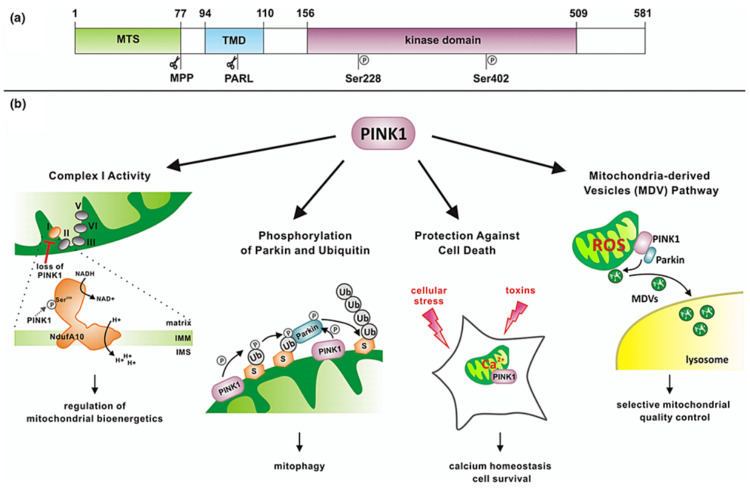
Research is ongoing into non-mitophagy-related functions of PINK1, as the protein has been associated with other cell signalling pathways promoting the transport of mitochondria and regulating dendritic morphogenesis [48], along with influencing cell survival under conditions of oxidative stress and regulating complex 1 activity [49,50]. Mutations in the gene coding for PINK1 (PARK6) can cause early onset PD while failing to protect dopaminergic neurons from stress-induced mitochondrial dysfunction [51]. Such mutations are associated with increased misfolding of mitochondrial proteins [52]. The involvement of PINK1 in cellular processes is shown in Figure 6.
Figure 6. : Domain architecture of PINK1 (a), showing mitochondrial targeting sequence (MTS), the transmembrane domain (TMD), and the kinase domain.
Some cellular processes influenced by PINK1 are shown (b) indicating a wide range of areas of activity.
MTS: mitochondrial targeting sequence; TMD: transmembrane domain; MPP: mitochondrial processing peptidase; PARL: presenilin-associated rhomboid-like protein; Ser228, Ser402: phosphorylation sites in PINK1; ROS: reactive oxygen species; Ub: ubiquitin; NAD: nicotinamide adenine dinucleotide; NADH: reduced form of NAD; NdufA10: NADH:ubiquinone oxidoreductase subunit A10; IMM: inner mitochondrial membrane; IMS: intermembrane space
Figure taken from [52], and permission was obtained from the original publisher to reproduce the content.
PINK1 has similarly been associated with mitochondrial trafficking. The OMM-bound GTPases Miro1 and Miro2 are integral to the migration of mitochondria in neurons, through a linking process attaching mitochondria to kinesin and dynein motor proteins [53,54]. Elevated levels of Miro are associated with dopaminergic neurone loss, but the levels can be returned to normal by increasing levels of PINK1. Overexpression of PINK1 was shown to inhibit the movement of mitochondria, resulting in perinuclear clustering [55]. This potentially implicates the Miro proteins and their interplay with PINK1 and parkin in PD pathogenesis.
Parkin is the next step in the PINK1-mediated identification of defective mitochondria. Encoded by the PARK2 gene [56,57], this E3 ligase mediates the deposition of ubiquitin on the surface of target mitochondria through the formation of a multicomponent ubiquitin ligase system which covalently binds ubiquitin to OMM surface proteins [58]. Interaction of parkin with PINK1 induces a change in the functionality of parkin (phosphorylation at Ser65), allowing the sequestration of ubiquitin from the cytoplasm, effectively labelling the dysfunctional mitochondria with a layer of ubiquitin proteins. Parkin is stabilised by neuregulin receptor degradation protein-1 (Nrdp1), which mediates the attachment of ubiquitin to mitochondria-bound parkin [59]. Little is known about Nrdp1 and its role in PD, it may be associated with neuronal apoptosis and protection from ROS [60], but it has been implicated in other areas such as gene expression in pancreatic cancer [61], and immunity [62], among others. PINK1 and parkin are also associated with the processes of mitochondrial fusion and fission, with both proteins having roles in the degradation of excess mitofusins via ubiquitination and subsequent proteasomal degradation, and PINK1 overexpression producing elongated and interconnected mitochondria [9,63]. Parkin has also been demonstrated to have a role in mitochondrial biogenesis through the ubiquitination of a transcriptional repressor called PARIS, which causes its lysosomal degradation. Removal of PARIS results in increased biogenesis of mitochondria [64].
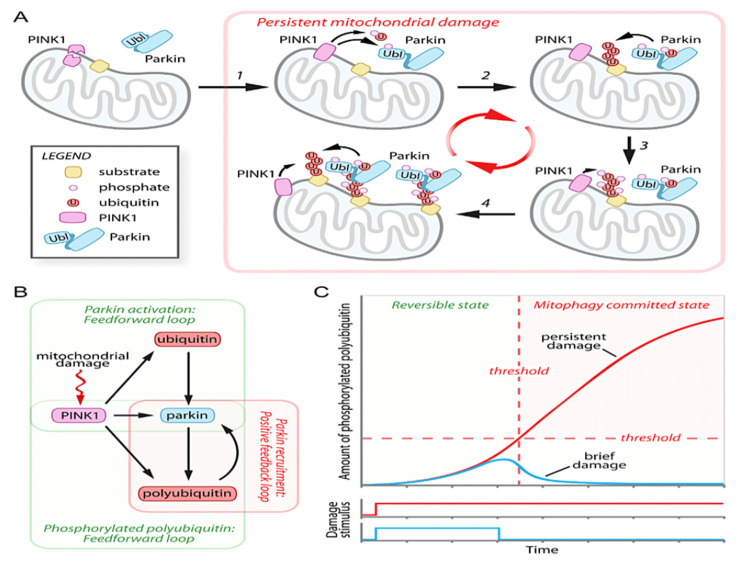
Whilst ubiquitin has several functions related to cell signalling and endocytosis, its relevant action is during the identification of cellular components for destruction [65,66]. First identified in 1975, this ubiquitously expressed 8.5 kDa protein has been identified in plants, yeasts and bacteria, indicating that it is evolutionarily conserved [67]. The ubiquitination of proteins occurs through several mechanisms. Examples of the chemical attachments include ester formation through serine or threonine residues, thioester formation through cysteine and peptide or isopeptide bonds at the N-terminus or lysine residues, respectively [68]. Once the process of PINK1-parkin-dependent ubiquitination has begun, a positive feedback process occurs to strengthen the accumulation of ubiquitin on the mitochondrial surface. Polyubiquitin chains are phosphorylated by PINK1. Phosphorylated polyubiquitin chains have a substantially higher affinity for phosphorylated parkin than for the native form (21 times the affinity) resulting in the recruitment of more active parkin to the mitochondria, its engulfing in an autophagosome, and subsequent delivery for lysosomal breakdown. In summary, ubiquitin is a substrate of parkin, and phosphorylated ubiquitin is an activator of parkin, through binding at an allosteric site. Therefore, ubiquitin is described as a homotropic allosteric regulator, thereby potentiating the effect of the parkin enzyme [10,41,69].
The feedback loop processes are shown in Figure 7, along with a hypothetical graph which models the levels of damage to targeted mitochondria, showing the point at which the process becomes irreversible.
Figure 7. Feedback loops as PINK1, ubiquitin and polyubiquitin enhance the migration of parkin.
Which then strengthens the action of PINK1 (A) and (B). Panel (C) shows a hypothetical graph indicating low phosphorylation of ubiquitin where reversal would be possible (blue line) and passing the threshold where damage becomes persistent and the mitochondrion is destroyed (red line).
Figure taken from [69], and permission was obtained from the original publisher to reproduce the content.
Mitochondrial recycling through mitophagy in differing cell types
In different cell types, the overall process remains similar, but the specific proteins involved vary from one species to another. In Saccharomyces cerevisiae (yeast), genes such as UTH1, an OMM protein, act as a marker to signal the mitochondrion to be removed by mitophagy, and knockout studies of this gene showed that in its absence there is a loss of selective mitophagy [70]. Other genes such as YME1, which encodes an ATPase, have the opposite effect that leads to an increase in mitophagy after they are knocked out. These findings in yeast demonstrated that mitophagy is closely controlled by a series of interconnected cellular proteins [71]. Recent research suggests that proteins anchored in the inner mitochondrial membrane (IMM) may trigger mitophagy as dysfunctional mitochondria have some compromise in the OMM leading to greater permeability and, therefore, unmasking IMM proteins, which in turn initiate mitophagy [72]. The IMM can become more permeable in the presence of increased levels of free calcium, increased ROS formation and decreased NADPH, resulting in a decrease of electrochemical stability and swelling of the mitochondrion, which leads to mitophagy. However, if mitophagy is impaired, this swelling can lead to OMM rupture and the release of pro-apoptotic proteins [73]. One of the proteins anchored to the IMM is phosphatidylserine decarboxylase, an enzyme that synthesises phosphatidylethanolamine (PE). PE modulates the solubility of α-Syn, and if low levels of PE are present and α-Syn is not controlled, causing mitochondrial defects, along with stress in the endoplasmic reticulum and a 3-fold increase in the levels of α-Syn [74]. This provides a better understanding of the mechanism of the prior observation that mitochondrial defects are associated with the formation of LBs [75].
Autophagy proteins (Atg) have been identified in yeasts as being critical to the mitophagy process, and a mammalian homologue has been found in the form of a Bcl-2-like protein 13 [76]. Atg32 is an OMM-bound 60 kDa protein that confers a level of selectivity to the process of mitophagy [77,78]. Atg32 can bind to Atg11, which then binds to Atg8, or Atg32 can bind directly to Atg8 as the tryptophan (W) and leucine (L) spacing in the binding region of Atg32 is sufficient for either interaction to take place (WXXL, where X represents other amino acids) [79,80]. The mammalian homologue of Atg8 is LC3 (microtubule-associated protein 1A/1B-light chain 3) [81]. Atg8 is bound to isolation membranes, so its interaction with mitochondrial membrane-bound Atg32 is thought to be the trigger for localising mitochondria into autophagosomes [82]. Atg32 is short-lived when expressed on the membrane of healthy mitochondria, and only becomes stable during the mitophagy process when it interacts with Atg11 or Atg8 [83]. While the two proteins are unrelated, this has certain similarity to the membrane instability in PINK1 as described previously, which is only stable on dysfunctional mitochondria.
An assortment of other proteins has involvements in various tissue types, including FUNDC1 (FUN14 domain-containing protein 1), a mammalian mitochondrial outer-membrane protein, which is a receptor for hypoxia-induced mitophagy. It interacts with LC3 via a YXXL binding motif, similar to the above-mentioned Atg32 sequence, but with tyrosine (Y) in place of tryptophan (W) [84]. The serine/threonine protein kinase ULK1 is thought to be involved in mitophagy through its interactions with FUNDC1, through a phosphorylation event [85,86]. Additionally, phosphoglycerate mutase family member 5 (PGAM5), a serine/threonine protein phosphatase, dephosphorylates FUNDC1 when under hypoxic conditions, which enhances its interaction with LC3 [87]. PGAM5 is thought to promote the PINK1/parkin pathway of mitophagy [88].
In the heart, cardiolipin is involved in the mitophagy process, as this IMM phospholipid has not only been detected migrating to the outer mitochondrial membrane and interacting with LC3, but knockout of this protein led to reduced capacity for mitophagy and upregulation of PINK1 in a bone marrow cell line [89].
Sequestosome-1 (ubiquitin-binding protein p62) is one of the most common markers to monitor mitophagy, along with LC3 [90]. Post-mortem analysis has found p62 in several neurodegenerative diseases, and experimentally induced upregulation of p62 has shown disruptions in mitochondrial structure and function [91], despite its known function as a PINK1/parkin-independent mitophagy agent, possibly with a role in immunomodulation [92]. Mutations leading to a failure to express p62, or heterozygous variants, have been associated with amyotrophic lateral sclerosis, Paget disease and frontotemporal dementia [93]. In some cases, healthy mitochondria are targeted for mitophagy. Mammalian red blood cells, for example, do not normally contain mitochondria. During the maturation process of reticulocytes, the OMM protein NIP3-like protein X (NIX) mediates the removal of mitochondria, as NIX-deficient mice have been shown to retain residual mitochondria in their red blood cells [94].
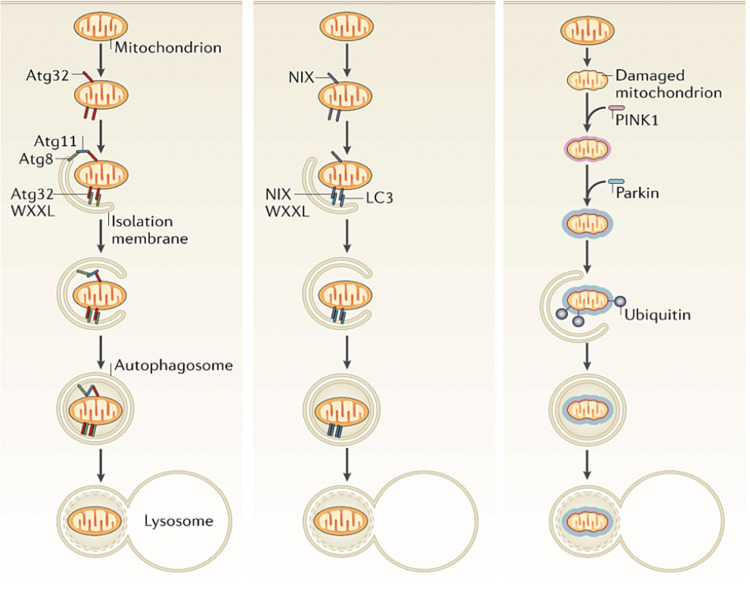
The mitophagy processes are complex and whilst the mechanics are gradually being elucidated, the specifics of the signalling stimuli are only partially understood, and significant further research in this area is required [95]. The basic principles and the main differences in the three mitophagy processes mentioned above are illustrated in Figure 8.
Figure 8. Mitophagy processes in yeast (left), showing Atg32 stabilisation on defective mitochondria and its subsequent interaction with Atg11 and/or Atg8 to associate with an isolation membrane for lysosomal degradation.
Red blood cells (centre) dispose of mitochondria through a mechanism thought to involve the membrane protein NIX. PINK1-parkin-induced mitophagy (right).
Atg32, Atg11, Atg8: autophagy proteins; WXXL: a conserved amino acid motif associated with LC3; NIX: NIP3-like protein X; LC3: microtubule-associated protein 1A/1B-light chain 3.
Figure taken from [82], and permission was obtained from the original publisher to reproduce the content.
In some instances, the targeting of healthy mitochondria may be of benefit. For example, in the ischaemic heart, once reperfusion has commenced, tissue can be damaged by the sudden generation of high levels of ROS, an effect which can be reduced given the removal of mitochondria prior to reperfusion [96]. Ischaemic preconditioning results in the localisation of parkin to mitochondria in isolated perfused rat hearts, with an associated greater cardiomyocyte survival than parkin-knockout mouse hearts [97]. Inhibition of mitophagy with bicarbonate has been shown to increase the risk of reperfusion injury [98]. The localisation of parkin to the mitochondria of cardiac myocytes is independent of PINK1, suggesting that there is another mechanism which could potentially compensate for PINK1 depletion in other cell types [99].
When the process of mitophagy is disrupted, dysfunctional mitochondria are allowed to accumulate in the affected cells. They are less likely to produce ATP and more likely to produce ROS, leading to cell damage or even cell death [100]. Conversely, excessive mitophagy can result in inadvertent apoptosis as the cell no longer has sufficient energy to survive [101,102]. In an effort to prevent excessive mitophagy, the mitochondrial membrane-bound apoptosis-regulating Bcl-2 family of proteins antagonises parkin-mediated mitophagy [103]. During the mitophagy process, Bcl-2 is able to escape from the degrading mitochondria [104]. Altogether, these points indicate the potential for a negative feedback system that would slow the mitophagy process following the accumulation of Bcl-2 in the cytosol after the successful degradation of mitochondria, thereby countering the above-mentioned PINK1-parkin-ubiquitin positive feedback system. Additionally, there are several ubiquitin-specific proteases which are capable of removing ubiquitin chains from parkin targets, thereby directly interfering with parkin-driven mitophagy [105].
As individuals age, the quality control systems that are usually at work within cells ensuring the fidelity of proteins and organelles become gradually compromised [106,107]. Age-related impairment in mitophagy increases the indications of ageing in the heart, such as becoming hypertrophic, fibrotic and increasing muscle stiffness. An accumulation of defective cellular organelles including mitochondria, which gives rise to increased production of ROS, are thought to be responsible for many of these symptoms. If cardiac mitophagy can be restored, it may offer treatments for heart disease.
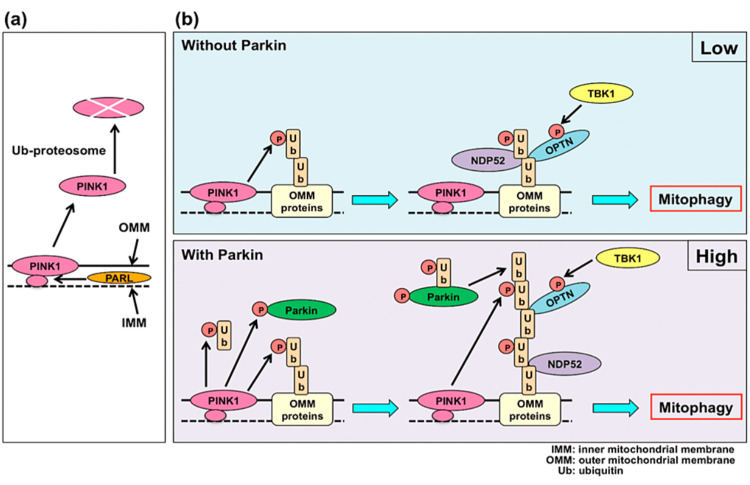
Mitophagy can also be disrupted in humans through genetic mutations which code for any of the proteins involved in this complex process, notably parkin and PINK1, which are known to be mutated in hereditary PD, specifically, mutations which abrogate the function of PINK1 result in early onset PD symptoms [43]. The mitophagy process through activation of PINK1 and parkin is shown in Figure 9, including the engagement of NDP52 and optineurin, parkin-independent mitophagy activators. These two proteins have been shown to be recruited by PINK1 even in the absence of parkin, which go on to recruit other mitophagy factors ULK1, DFCP1 and WIPI1. Together, this study indicates PINK1 accumulation on mitochondria as the trigger for mitophagy, with parkin acting to amplify the signal [108].
Figure 9. When mitochondria are healthy, PINK1 is degraded (a). When mitochondria are compromised, PINK1 and parkin recruit ubiquitin to label mitochondria for mitophagy (b).
PINK1: PTEN-induced kinase 1; PARL: presenilins-associated rhomboid-like protein; OMM: outer mitochondrial membrane; IMM: inner mitochondrial membrane; NDP52: calcium binding and coiled-coil domain 2; OPTN: optineurin; TBK1: TANK-binding kinase 1.
Figure taken from [109], and permission was obtained from the original publisher to reproduce the content.
Genetics and environmental disruptions to mitophagy in Parkinson’s disease
On the occasions of inherited PD, mutations are identifiable in the genes coding for a range of proteins, including α-Syn, parkin, PINK1, DJ-1, and LRRK2 [110]. An interesting starting point for this analysis is the differences seen in the genetics of PD patients of different ethnicities.
In a study of Czech patients, mutations were found in the gene coding for parkin, PARK2, in around 7% of subjects. Specifically, one patient presented with a point mutation while the others had exon deletions [111]. A similar Polish study identified around 4% of PD patients with mutations in PARK2, but was able to identify a range of heterozygous polymorphisms of PARK2 in 21% of patients [112].
A study that identified the genetic variants of the LRRK2 gene (PARK8) in a Taiwanese population of PD patients found higher expression of one allele (G2385RA) in PD patients in comparison to healthy controls. They also discovered two previously undocumented mutations that may affect the folding structure of LRRK2 [113]. Genetic studies of Nigerian patients with PD showed no pathogenic mutations when screened for changes in PARK2 or LRRK2 [114]. Similarly, a more recent Saudi study seeking mutations in SNCA (coding for α-Syn), PARK2, PINK1, PARK7 (DJ-1), LRRK2, and others, revealed only three mutations (two in PINK1 and one in PARK2) in a study involving 98 patients [115]. They went on to suggest that whilst very little was identified in the way of genetic mutations, gene expression and dosing may be a more pertinent issue, along with the possibility that there are undiscovered gene mutations involved in the development of PD. Mutations in LRRK2 are a major cause of familial PD and are known to occur in idiopathic PD [116], specifically the G2019S mutation which has been shown to occur in 6% of cases of idiopathic PD in Ashkenazi Jews, and over 29% in familial PD in the same population [117].
Mutations in PINK1 result in an inherited early onset PD [51]. The usual dimerised structure of PINK1 is not affected by mutations of PARK6, but its function is impaired sufficiently to result in deficits in mitochondrial respiration, impairment of proteasome function and increased aggregation of α-Syn [118]. However, mutation of PARK6 does not give rise to a single physiological change. A range of different mutations of PARK6 is possible, and each produces a slightly different pattern of abnormality in the resulting cells. Although, in all cases, reduced mitochondrial membrane potential and greater free radical production are evident [119]. Pesticides such as paraquat can induce parkin aggregation; an effect which is ameliorated by overexpression of LRRK2 [120]. Moreover, the G2019S mutation of LRRK2 has been shown to improve the survival of nematodes exposed to paraquat, which also results in the loss of dopaminergic neurons at an accelerated rate [121]. As dysfunctional LRRK2 is known to be one of the mutations giving rise to PD, this indicates that a combination of genetic and environmental factors may be involved in the onset of PD. Moreover, LRRK2 expression can be altered by mutated PINK1 in the absence of any genetic abnormality in the PARK8 gene [122].
Mutations in PARK2 give rise to altered parkin protein, which then produces autosomal recessive juvenile parkinsonism [57]. Mutations in parkin in ex vivo cell models resulted in higher respiration rates in mitochondria and greater growth of the cells as a whole, suggesting some kind of compensatory mechanism to preserve mitochondrial function in the absence of parkin [123]. In PD, parkin mutations specifically affecting Ser65 would explain the lack of phosphorylation by PINK1. In an experimental model, a mutated form of parkin (with Ser65 abnormality) is not activated by PINK1, whereas a similar mutation in ubiquitin allowed for ligase activation, demonstrating that phosphorylated ubiquitin was able to activate parkin allosterically [124]. Interestingly, the mitochondrial motility protein Miro1 has altered turnover on damaged mitochondria in PD, with ubiquitination of Miro1 being dependent on this same Ser65 residue within parkin [125].
Errors in these pathways do not only contribute to PD. Cutaneous melanoma occurs more frequently in PD patients than in the general population. A genetic analysis of these patients found a large degree of overlap between mutations in known PARK genes and the development of cutaneous melanoma, suggesting the possibility of a shared pathway in the two conditions [126].
Other possible underlying causes of mitophagy dysfunction
Environmental factors play a role in disrupted mitophagy. Exposure to certain pesticides such as rotenone can be a potential cause of PD, as it can impair mitophagy to damage neurons, but this is rescued by overexpression of DJ-1 [127]. Certain drugs such as amphetamines can induce mitochondrial dysfunction and DNA damage in cultured cells, resulting in increased oxidative stress [128]. Additionally, neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) can inhibit complex 1 of the mitochondrial respiratory chain and result in increased ROS production and DA degeneration, causing parkinsonism.
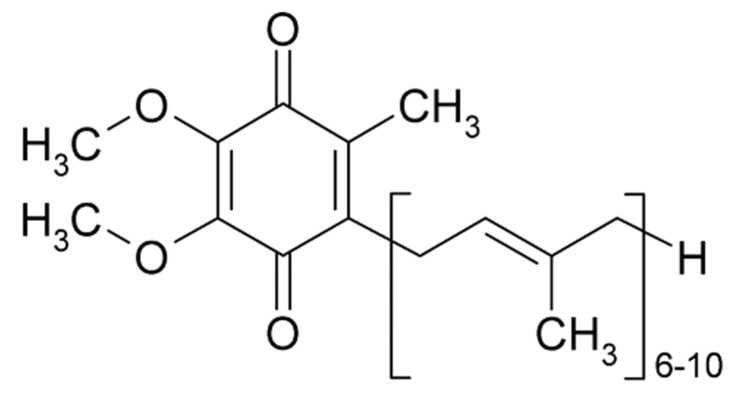
Besides chemical exposure, deficiencies of certain compounds have a role to play in the mitophagy process. Coenzyme Q10, or ubiquinone, due to its ubiquitous presence in most animals, is of fundamental value in the cellular respiration process, forming an integral part of the electron transport chain, and deficiency of it can trigger mitophagy [129,130]. Mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes (MELAS) is a genetic disorder affecting mitochondrial function and is characterised by muscle pain and weakness, seizures, and symptoms consistent with stroke [131]. In studies involving in vitro models, coenzyme Q10 has been shown to have significantly lower than normal levels, which is accompanied by increased oxidative stress and activation of mitochondrial permeability transition which should bring about mitophagy of the affected mitochondria. However, mitophagy is impaired in these cell models-an observation which is reversed following supplementation with coenzyme Q10 (Figure 10) [132]. Similar findings and benefits of coenzyme Q10 supplementation have been reported for other conditions, such as fibromyalgia [133], heart failure [134] and research is ongoing into the potential role of coenzyme Q10 in “mitochondrial resuscitation” for patients suffering septic shock [135], and as a neuroprotective agent to reduce secondary damage from the surge in ROS resulting from a traumatic brain injury [136].
Figure 10. Ubiquinone, also known as coenzyme Q10.
Coenzyme Q10 has even shown benefits in the management of PD with respect to patients’ ability to carry out “daily activities”, although a meta-analysis on the subject (concluding a benefit in the treatment of PD) was subsequently withdrawn from the Cochrane Library [137]. Patients treated with nicotinamide showed increased mitophagy and the associated decrease in the levels of mitochondria, yet the mitochondria remain to have significantly increased membrane potential and produce ATP at a greater rate than before treatment began [138].
Certain infections have also been shown to influence mitophagy. The hepatitis C virus, for example, once internalised causes the overexpression of PINK1 and parkin and induces perinuclear accumulation of mitochondria and subsequent increased levels of mitophagy [139].
Effect of mitophagy dysfunction within neurons on cell function
Dopaminergic neurons are more sensitive to stresses such as increased ROS than other types of neurons, probably due to their action to oxidise dopamine during its breakdown [105], however, they show a particular sensitivity to reduction in mortalin (mitochondrial 70 kDa heat shock protein-coded by the HSPA9 gene, and found on mitochondrial membranes or in the endoplasmic reticulum [140]. In mortalin-deficient Drosophila, similar effects are seen in established Drosophila models of PD, such as reduced ATP levels, and shortened life span, which are the result of abnormal mitochondrial function and morphology [141]. Point mutations in mortalin have been implicated in increasing endogenous oxidative stress and decreasing the tolerance of the cells. Mortalin is also known to inactivate p53, thus, preventing apoptosis [142].
Experimental knockout of PINK1 produces loss of the protein throughout the brain, however, the associated breakdown of mitophagy had heterogeneous effects, affecting the midbrain most severely while other areas of the brain seemed more adaptable or more resistant to oxidative stress [143].
When mitophagy is inhibited in mouse midbrains (through cell-specific deletion of Atg7), dendritic ubiquitinated inclusions appear in neurons, which themselves appear dystrophic and low in dopamine. When the experiment was expanded to include the whole brain, there was presynaptic accumulation of α-Syn and LRRK2 proteins, indicating a link between disrupted mitophagy and the physiological effects of PD [144].
Proposed model of mitophagy mechanisms leading to Parkinson’s disease
Impairment of various processes is well-established in ageing cells, e.g. including not only mitochondrial quality control as described previously, but mutations can often develop in other proteins produced by the endoplasmic reticulum. Indeed, impaired autophagy, mitochondrial dysfunction and metabolic distress are associated with cellular ageing [145]. This age-related inaccuracy in production of PINK1 could be potentially ameliorated by functional parkin, because, as described previously, parkin has been shown to induce mitophagy in the absence of PINK1 in some cell types. Errors in the production of parkin, however, could be more problematic. Disruption of both these proteins can interfere not only with mitophagy but also with mitochondrial fission and fusion events, and mitochondrial trafficking within the neuron.
Age-associated dysfunctional mitochondria can lead to increased levels of ROS, which in turn can make the IMM more permeable, causing swelling and further damage. Compromise of the inner membrane could result in increased levels of α-Syn through the action of the inner membrane-bound enzyme phosphatidylserine decarboxylase.
So, dysfunctional mitochondria can lead to overproduction of α-Syn, and its subsequent aggregation into LBs; yet increased levels of α-Syn can cause further disruption in mitochondrial function, creating an amplifying negative cycle [146]. LBs are composed primarily of α-Syn, a small portion of which is ubiquitinated. This ubiquitination may be important in the formation of LBs, possibly providing an aggregation seed [18].
Given larger concentrations of α-Syn in the cell, likely resulting in aggregation into LBs, coupled with the fact that cells deficient in PINK1 and parkin are more sensitive to oxidative stress, it is unsurprising that cellular damage possibly leading to apoptosis can occur in sensitive cells [9]. As described previously, the substantia nigra with its rich dopaminergic neuron population is more sensitive than other parts of the brain. Cell dysfunction in this area gives rise to the characteristic symptoms of parkinsonian syndromes, if not PD itself.
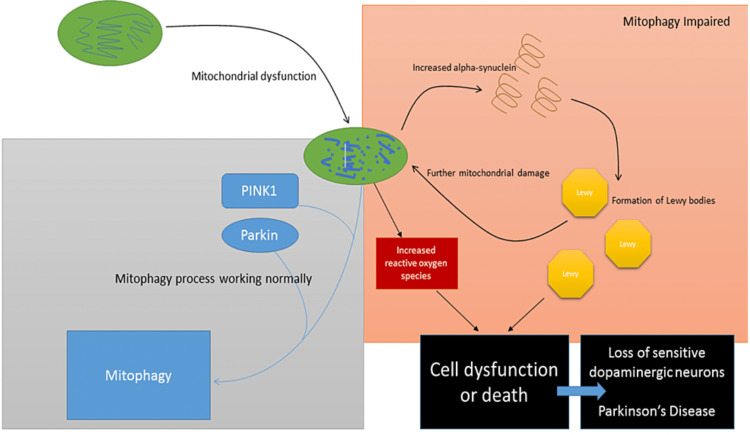
The model is summarised in Figure 11.
Figure 11. Model of dysfunctional mitophagy leading to increased reactive oxygen species, formation of Lewy bodies, cell dysfunction and death of sensitive dopaminergic neurons—leading to PD.
PINK1: PTEN-induced kinase 1.
Overall discussion and summary
The model described above lends itself to the pathogenesis of idiopathic PD, however, in familial PD the described process would simply be accelerated independently of the ageing process through mutations in the genes coding for parkin and PINK1. Indeed, these mutations are often associated with early onset variants of the condition (Table 1).
Table 1. Genetics of Parkinson's disease.
The table contains known PD-related loci and whether mutations result in classical PD or early onset PD.
Table taken from [147], and permission was obtained from the original publisher to reproduce the content.
| Symbol | Gene locus | Disorder | Inheritance | Gene | Status and remarks | Mode of identification |
| PARK1 | 4q21-22 | EOPD | AD | SNCA | Confirmed | Linkage analysis |
| PARK2 | 6q25.2–q27 | EOPD | AR | Parkin | Confirmed | Linkage analysis |
| PARK4 | 4q21–q23 | EOPD | AD | SNCA | Erroneous locus (identical to PARK1) | Linkage analysis |
| PARK5 | 4p13 | Classical PD | AD | UCHL1 | Unconfirmed (not replicated since described in 1998) | Functional candidate gene approach |
| PARK6 | 1p35–p36 | EOPD | AR | PINK1 | Confirmed | Linkage analysis |
| PARK7 | 1p36 | EOPD | AR | DJ-1 | Confirmed | Linkage analysis |
| PARK8 | 12q12 | Classical PD | AD | LRRK2 | Confirmed; variations in LRRK2 gene include risk-conferring variants and disease-causing mutations | Linkage analysis |
| PARK16 | 1q32 | Classical PD | Risk factor | Unknown | Confirmed susceptibility locus | Genome-wide association studies |
| PARK17 | 16q11.2 | Classical PD | AD | VPS35 | Confirmed | Exome sequencing |
| PARK18 | 3q27.1 | Classical PD | AD | EIF4G1 | Unconfirmed; recently published | Linkage analysis |
Yet, disrupted mitophagy and genetic predispositions towards it may not be the only issues to consider. Mutations in the protein deglycase DJ-1 (coded for on the PARK7 gene) are also known to be instrumental in the development of autosomal recessive early onset PD, specifically as its native form increases tolerance to ROS and inhibits the aggregation of α-Syn [148,149]. This appears to be entirely distinct from the mitophagy system, yet still protecting neurons from oxidative damage and subsequent cell death, while also reducing the formation of LBs [150]. Recent research in this area has yielded an antibody to the C-terminus of α-Syn which has been shown to inhibit fibril formation, which may potentially provide benefit in the immunotherapy of PD in years to come [151].
Other factors also come into play in the pathogenesis of PD. For example, the abnormal accumulation of α-Syn may not purely be a result of mitochondrial dysfunction. After the observation that the native enzyme responsible for the degradation of α-Syn (cathepsin D) is upregulated in some PD models, attention has been turned to glycosaminoglycans as regulators of cathepsin D [152]. Indeed, mutated forms of α-Syn can begin to cause PD-like non-motor symptoms in Drosophila prior to the formation of the characteristic fibrils that make up LBs [153].
The connection between disrupted mitophagy and PD has already been reviewed, but the authors quite rightly pointed out both a lack of available evidence in cell or animal models of PD and a lack of known pharmacological mitophagy enhancers, which would obviously be the goal of research aiming towards PD therapy [154]. Having said that, a mitophagy inducer was recently tested in cell culture that was found to successfully engage mitochondrial quality control and autophagy without requiring the recruitment of PINK1 or parkin. This showed none of the negative effects associated with usual experimental mitophagy inducers, however, this research is in its early stages [155]. Additionally, nicotinamide-related enhancement of mitophagy was briefly mentioned previously, with the authors speculating that increased mitophagy was resulting in decreased mitochondrial count, but increased efficiency in each mitochondrion. Further research in this area has allowed the elucidation of the mechanism involved: activation of an NAD-dependent deacetylase called SIRT1 (silent mating type information regulation 2 homolog). SIRT1 activators were subsequently shown to increase mitophagy but did not affect residual mitochondrial efficiency, which was dependent upon the metabolic state of the cell [156].
The established opinion which considers PD as a single condition pertaining to loss of dopaminergic transmission is currently being challenged as there is evidence that non-dopaminergic pathways can also be damaged in the condition. As there is a range of non-motor subtypes (e.g. cognitive disorders), and abnormal accumulations of α-Syn have been observed in the heart, gut, pancreas and skin, the theory has been put forward that PD would be better considered a syndrome rather than a single disease, with multiple dysfunctional neurotransmitter pathways in both central and peripheral regions of the nervous system [157].
Recommendations for future primary and secondary research
A pharmacological regulator of mitophagy is likely the next large step to be taken in the treatment of PD, so research to continue the work of Campanella and East and East et al. is important [155,158].
Interestingly, despite wide genetic heterogeneity, patients suffering from diabetes mellitus have a decreased incidence of PD, although the mechanism of this apparent resistance is unclear [159]. One possible research avenue that would be of particular interest is the restoration of dysfunctional parkin-mediated mitophagy that has been seen with the antidiabetic medication metformin [160]. They observed that treatment of mice with metformin resulted in a reduction in stress level in the endoplasmic reticulum combined with decreased expression of p53, a protein known to inhibit the translocation of parkin. Activation of AMPK in hepatocytes has been proposed as a potential mechanism for metformin (Figure 12).
Figure 12. Structures of metformin (left) and nicotinamide (right)—both of which have been associated with the enhancement of mitophagy.
PINK1 and parkin have also been shown to be upregulated in the vasculature of obese and diabetic mice, so research to elucidate the interplay between PINK1/parkin, mitophagy, diabetes and PD may help to clarify our understanding [161].
Potential therapeutic benefits of this research
Research in this area could revolutionise the treatment of PD. Patients who currently understand that the disease is progressive and incurable could be offered mitophagy-enhancing treatments which may confer a neuroprotective effect, thereby slowing disease progression. If the concurrent research into immunotherapy, with antibodies to α-Syn being developed, can be expanded upon, this offers the possibility of a double-edged attack on this devastating neurodegenerative condition.
Conclusions
Attempts to directly associate mitophagy with PD by discussing alterations in PINK1 or parkin would be a little short-sighted, as both proteins are not only involved in mitophagy but are also active in mitochondrial trafficking and fusion/fission events.
Having said that, there is a strong link between disruptions in mitophagy, the associated accumulation of dysfunctional mitochondria, the formation of LBs, and the destruction of dopaminergic neurons, so the mitophagy process is clearly a central player in PD pathophysiology. Whether particular proteins, or mutations thereof, can be identified as the initiator(s) of such dysfunction is unlikely, but remains to be seen. Rather, the mitophagy process as a whole should be thoroughly researched and understood with a view to the generation of therapeutic agents capable of restoring function, and in doing so, decelerating the destruction of dopaminergic neurons, slowing PD progression.
Acknowledgments
Tsz Ki Ko and Denise Jia Yun Tan contributed equally to the work and should be considered co-first authors.
Appendices
Atg Autophagy protein
α-Syn Alpha-synuclein
COMT Catechol-O-methyl transferase
FUNDC1 FUN14 domain-containing protein 1
LBs Lewy Bodies
LC3 Microtubule-associated protein 1A/1B-light chain 3
LRRK2 Leucine-rich repeat kinase 2
OMM Outer Mitochondrial Membrane
MELAS Mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes
NADP Nicotinamide adenine dinucleotide phosphate
Nrdp1 Neuregulin receptor degradation protein-1
NIX NIP3-like protein X
Parkin Parkinson juvenile disease protein 2
PD Parkinson’s disease
PGAM5 Phosphoglycerate mutase family member 5
PINK1 PTEN-induced putative kinase 1
PTEN Phosphatase and tensin homolog
ROS Reactive Oxygen Species
The content published in Cureus is the result of clinical experience and/or research by independent individuals or organizations. Cureus is not responsible for the scientific accuracy or reliability of data or conclusions published herein. All content published within Cureus is intended only for educational, research and reference purposes. Additionally, articles published within Cureus should not be deemed a suitable substitute for the advice of a qualified health care professional. Do not disregard or avoid professional medical advice due to content published within Cureus.
Footnotes
The authors have declared that no competing interests exist.
References
- 1.Parkinson's disease: genetics and pathogenesis. Shulman JM, De Jager PL, Feany MB. Annu Rev Pathol. 2011;6:193–222. doi: 10.1146/annurev-pathol-011110-130242. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Neurological Disorders: Public Health Challenges. Geneva, WHO. [ Dec; 2022 ]. 2006. https://www.who.int/publications/i/item/9789241563369 https://www.who.int/publications/i/item/9789241563369
- 3.The prevalence of Parkinson’s disease: a systematic review and meta-analysis. Pringsheim T, Jette N, Frolkis A, Steeves TD. Mov Disord. 2014;29:1583–1590. doi: 10.1002/mds.25945. [DOI] [PubMed] [Google Scholar]
- 4.Parkinson's UK, 2013 2013. Parkinson’s and the NHS in England: the cost of poor care. [ Dec; 2022 ]. 2013. http://www.innovationagencyexchange.org.uk/sites/default/files/Parkinson%E2%80%99s%20and%20the%20NHS%20in%20England%20-%20the%20cost%20of%20poor%20care.pdf http://www.innovationagencyexchange.org.uk/sites/default/files/Parkinson%E2%80%99s%20and%20the%20NHS%20in%20England%20-%20the%20cost%20of%20poor%20care.pdf
- 5.Gowers WR. London: J. & A. Churchill; 1886. A Manual of Diseases of the Nervous System. [Google Scholar]
- 6.Evidence for a role of adaptive immune response in the disease pathogenesis of the MPTP mouse model of Parkinson's disease. Martin HL, Santoro M, Mustafa S, Riedel G, Forrester JV, Teismann P. GLIA. 2016;64:386–395. doi: 10.1002/glia.22935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genetics of Parkinson's disease--state of the art, 2013. Bonifati V. Parkinsonism Relat Disord. 2014;20:0–8. doi: 10.1016/S1353-8020(13)70009-9. [DOI] [PubMed] [Google Scholar]
- 8.Mitophagy and Parkinson's disease: the PINK1-parkin link. Deas E, Wood NW, Plun-Favreau H. Biochim Biophys Acta. 2011;1813:623–633. doi: 10.1016/j.bbamcr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Narendra D, Walker JE, Youle R. Cold Spring Harb Perspect Biol. 2012;4:0. doi: 10.1101/cshperspect.a011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Pickrell AM, Youle RJ. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speech disorders in Parkinson's disease: early diagnostics and effects of medication and brain stimulation. Brabenec L, Mekyska J, Galaz Z, Rektorova I. J Neural Transm. 2017;124:303–334. doi: 10.1007/s00702-017-1676-0. [DOI] [PubMed] [Google Scholar]
- 12.Olfaction: new understandings, diagnostic applications. Ruggiero GF, Wick JY. Consult Pharm. 2016;31:624–632. doi: 10.4140/TCP.n.2016.624. [DOI] [PubMed] [Google Scholar]
- 13.Using 'swallow-tail' sign and putaminal hypointensity as biomarkers to distinguish multiple system atrophy from idiopathic Parkinson's disease: a susceptibility-weighted imaging study. Wang N, Yang H, Li C, Fan G, Luo X. Eur Radiol. 2017;27:3174–3180. doi: 10.1007/s00330-017-4743-x. [DOI] [PubMed] [Google Scholar]
- 14.The 'swallow tail' appearance of the healthy nigrosome - a new accurate test of Parkinson's disease: a case-control and retrospective cross-sectional MRI study at 3T. Schwarz ST, Afzal M, Morgan PS, Bajaj N, Gowland PA, Auer DP. PLoS One. 2014;9:0. doi: 10.1371/journal.pone.0093814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Accuracy of clinical diagnosis of idiopathic Parkinson's disease: a clinico-pathological study of 100 cases. Hughes AJ, Daniel SE, Kilford L, Lees AJ. J Neurol Neurosurg Psychiatry. 1992;55:181–184. doi: 10.1136/jnnp.55.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Specific patterns of neuronal loss in the pulvinar nucleus in dementia with lewy bodies. Erskine D, Thomas AJ, Attems J, Taylor JP, McKeith IG, Morris CM, Khundakar AA. Mov Disord. 2017;32:414–422. doi: 10.1002/mds.26887. [DOI] [PubMed] [Google Scholar]
- 17.Duration and pathologic correlates of Lewy body disease. Graff-Radford J, Aakre J, Savica R, et al. JAMA Neurol. 2017;74:310–315. doi: 10.1001/jamaneurol.2016.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ubiquitination of α-synuclein and autophagy in Parkinson's disease. Engelender S. Autophagy. 2008;4:372–374. doi: 10.4161/auto.5604. [DOI] [PubMed] [Google Scholar]
- 19.Latest concept of Lewy body disease. Kosaka K. Psychiatry Clin Neurosci. 2014;68:391–394. doi: 10.1111/pcn.12179. [DOI] [PubMed] [Google Scholar]
- 20.Cellular co-localization of phosphorylated tau- and NACP/alpha-synuclein-epitopes in lewy bodies in sporadic Parkinson's disease and in dementia with Lewy bodies. Arima K, Hirai S, Sunohara N, et al. Brain Res. 1999;843:53–61. doi: 10.1016/s0006-8993(99)01848-x. [DOI] [PubMed] [Google Scholar]
- 21.Colocalization of tau and alpha-synuclein epitopes in Lewy bodies. Ishizawa T, Mattila P, Davies P, Wang D, Dickson DW. J Neuropathol Exp Neurol. 2003;62:389–397. doi: 10.1093/jnen/62.4.389. [DOI] [PubMed] [Google Scholar]
- 22.Aggresomes formed by alpha-synuclein and synphilin-1 are cytoprotective. Tanaka M, Kim YM, Lee G, Junn E, Iwatsubo T, Mouradian MM. J Biol Chem. 2004;279:4625–4631. doi: 10.1074/jbc.M310994200. [DOI] [PubMed] [Google Scholar]
- 23.Corboy MJ, Thomas PJ, Wigley WC. Ubiquitin-Proteasome Protocols. Methods in Molecular Biology™. Vol. 301. Humana Press; 2005. Aggresome Formation; pp. 305–327. [DOI] [PubMed] [Google Scholar]
- 24.α-Synuclein binds and sequesters PIKE-L into Lewy bodies, triggering dopaminergic cell death via AMPK hyperactivation. Kang SS, Zhang Z, Liu X, et al. PNAS. 2017;114:1183–1188. doi: 10.1073/pnas.1618627114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkinson's disease in over 20s: diagnosis and management. [ Dec; 2022 ]. 2006. https://www.nice.org.uk/guidance/cg35 https://www.nice.org.uk/guidance/cg35
- 26.Vol. 113. Scottish Intercollegiate Guidelines Network (SIGN; [ Dec; 2022 ]. 2010. Diagnosis and pharmacological management of Parkinson’s disease: A national clinical guideline. [Google Scholar]
- 27.Use of PEGylated immunoliposomes to deliver dopamine across the blood-brain barrier in a rat model of Parkinson's disease. Kang YS, Jung HJ, Oh JS, Song DY. CNS Neurosci Ther. 2016;22:817–823. doi: 10.1111/cns.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Royal Pharmaceutical Society. Bnf. Vol. 70. London: BMJ Group; 2015. British National Formulary (BNF) 70; pp. 88–150. [Google Scholar]
- 29.Levodopa-induced dyskinesias and their management. Del Sorbo F, Albanese A. J Neurol. 2008;255:32–41. doi: 10.1007/s00415-008-4006-5. [DOI] [PubMed] [Google Scholar]
- 30.Diphasic dyskinesias during levodopa-carbidopa intestinal gel (LCIG) infusion in Parkinson's disease. Meloni M, Solla P, Mascia MM, Marrosu F, Cannas A. Parkinsonism Relat Disord. 2017;37:92–96. doi: 10.1016/j.parkreldis.2016.12.030. [DOI] [PubMed] [Google Scholar]
- 31.The MAO-B inhibitor deprenyl reduces the oral tremor and the dopamine depletion induced by the VMAT-2 inhibitor tetrabenazine. Podurgiel SJ, Yohn SE, Dortche K, Correa M, Salamone JD. Behav Brain Res. 2016;298:188–191. doi: 10.1016/j.bbr.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Systematic review of levodopa dose equivalency reporting in Parkinson's disease. Tomlinson CL, Stowe R, Patel S, Rick C, Gray R, Clarke CE. Mov Disord. 2010;25:2649–2653. doi: 10.1002/mds.23429. [DOI] [PubMed] [Google Scholar]
- 33.Mechanisms of deep brain stimulation. Herrington TM, Cheng JJ, Eskandar EN. J Neurophysiol. 2016;115:19–38. doi: 10.1152/jn.00281.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Lemasters JJ. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 35.Mitochondrial dysfunction in cancer. Boland ML, Chourasia AH, Macleod KF. Front Oncol. 2013;3:292. doi: 10.3389/fonc.2013.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.AMPK activation prevents and reverses drug-induced mitochondrial and hepatocyte injury by promoting mitochondrial fusion and function. Kang SWS, Haydar G, Taniane C, Farrell G, Arias IM, Lippincott-Schwartz J, Fu D. PLoS ONE. 2016;11:0. doi: 10.1371/journal.pone.0165638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Twig G, Elorza A, Molina AJ, et al. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Legros F, Lombès A, Frachon P, Rojo M. Mol Biol Cell. 2002;13:4343–4354. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitochondria: dynamic organelles in disease, aging, and development. Chan DC. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 40.Mitochondrial dysfunction in Parkinson's disease: molecular mechanisms and pathophysiological consequences. Exner N, Lutz AK, Haass C, Winklhofer KF. EMBO J. 2012;31:3038–3062. doi: 10.1038/emboj.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.PINK1- and Parkin-mediated mitophagy at a glance. Jin SM, Youle RJ. J Cell Sci. 2012;125:795–799. doi: 10.1242/jcs.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.PINK1 is degraded through the N-end rule pathway. Yamano K, Youle RJ. Autophagy. 2013;9:1758–1769. doi: 10.4161/auto.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. Narendra DP, Jin SM, Tanaka A, et al. PLoS Biol. 2010;8:0. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parkin structure and function. Seirafi M, Kozlov G, Gehring K. FEBS J. 2015;282:2076–2088. doi: 10.1111/febs.13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Regulation of PINK1-Parkin-mediated mitophagy. Springer W, Kahle PJ. Autophagy. 2011;7:266–278. doi: 10.4161/auto.7.3.14348. [DOI] [PubMed] [Google Scholar]
- 46.PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. Lazarou M, Narendra DP, Jin SM, Tekle E, Banerjee S, Youle RJ. J Cell Biol. 2013;200:163–172. doi: 10.1083/jcb.201210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. Wauer T, Swatek KN, Wagstaff JL, et al. EMBO J. 2015;34:307–325. doi: 10.15252/embj.201489847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beyond the mitochondrion: cytosolic PINK1 remodels dendrites through protein kinase A. Dagda RK, Pien I, Wang R, et al. J Neurochem. 2014;128:864–877. doi: 10.1111/jnc.12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The complex I subunit NDUFA10 selectively rescues Drosophila pink1 mutants through a mechanism independent of mitophagy. Pogson JH, Ivatt RM, Sanchez-Martinez A, Tufi R, Wilson E, Mortiboys H, Whitworth AJ. PLoS Genet. 2014;10:0. doi: 10.1371/journal.pgen.1004815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.The mitochondrial kinase PINK1: functions beyond mitophagy. Voigt A, Berlemann LA, Winklhofer KF. J Neurochem. 2016;139:232–239. doi: 10.1111/jnc.13655. [DOI] [PubMed] [Google Scholar]
- 51.Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Valente EM, Abou-Sleiman PM, Caputo V, et al. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 52.Genetic analysis of mitochondrial protein misfolding in Drosophila melanogaster. Pimenta de Castro I, Costa AC, Lam D, et al. Cell Death Differ. 2012;19:1308–1316. doi: 10.1038/cdd.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitochondrial trafficking in neurons and the role of the Miro family of GTPase proteins. Birsa N, Norkett R, Higgs N, Lopez-Domenech G, Kittler JT. Biochem Soc Trans. 2013;41:1525–1531. doi: 10.1042/BST20130234. [DOI] [PubMed] [Google Scholar]
- 54.Regulation of mitochondrial trafficking, function and quality control by the mitochondrial GTPases Miro1 and Miro2. Kittler J. http://4. SpringerPlus. 2015;4:0. doi: 10.1186/2193-1801-4-S1-L33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parkinson's disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. Liu S, Sawada T, Lee S, et al. PLoS Genet. 2012;8:0. doi: 10.1371/journal.pgen.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.A microdeletion of D6S305 in a family of autosomal recessive juvenile parkinsonism (PARK2) Matsumine H, Yamamura Y, Hattori N, Kobayashi T, Kitada T, Yoritaka A, Mizuno Y. Genomics. 1998;49:143–146. doi: 10.1006/geno.1997.5196. [DOI] [PubMed] [Google Scholar]
- 57.Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Kitada T, Asakawa S, Hattori N, et al. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 58.Ubiquitin ligases and cell cycle control. Teixeira LK, Reed SI. Annu Rev Biochem. 2013;82:387–414. doi: 10.1146/annurev-biochem-060410-105307. [DOI] [PubMed] [Google Scholar]
- 59.Parkin is ubiquitinated by Nrdp1 and abrogates Nrdp1-induced oxidative stress. Yu F, Zhou J. Neurosci Lett. 2008;440:4–8. doi: 10.1016/j.neulet.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 60.Nrdp1 is associated with neuronal apoptosis in lipopolysaccharide-induced neuroinflammation. Shen J, Song Y, Shen J, et al. Neurochem Res. 2015;40:971–979. doi: 10.1007/s11064-015-1552-y. [DOI] [PubMed] [Google Scholar]
- 61.The retinoic acid derivative, ABPN, inhibits pancreatic cancer through induction of Nrdp1. Byun S, Shin SH, Lee E, et al. Carcinogenesis. 2015;36:1580–1589. doi: 10.1093/carcin/bgv148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.K33-linked polyubiquitination of Zap70 by Nrdp1 controls CD8(+) T cell activation. Yang M, Chen T, Li X, et al. Nat Immunol. 2015;16:1253–1262. doi: 10.1038/ni.3258. [DOI] [PubMed] [Google Scholar]
- 63.Parkin and PINK1: much more than mitophagy. Scarffe LA, Stevens DA, Dawson VL, Dawson TM. Trends Neurosci. 2014;37:315–324. doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson's disease. Shin JH, Ko HS, Kang H, et al. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Proteasome-independent functions of ubiquitin in endocytosis and signaling. Mukhopadhyay D, Riezman H. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 66.The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Glickman MH, Ciechanover A. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 67.Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Goldstein G, Scheid M, Hammerling U, Schlesinger DH, Niall HD, Boyse EA. PNAS. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ubiquitin: structures, functions, mechanisms. Pickart CM, Eddins MJ. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 69.A polyubiquitin chain reaction: parkin recruitment to damaged mitochondria. Riley BE, Olzmann JA. PLoS Genet. 2015;11:0. doi: 10.1371/journal.pgen.1004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.The mitochondrial pathway in yeast apoptosis. Eisenberg T, Büttner S, Kroemer G, Madeo F. Apoptosis. 2007;12:1011–1023. doi: 10.1007/s10495-007-0758-0. [DOI] [PubMed] [Google Scholar]
- 71.Mitophagy. Tolkovsky AM. Biochim Biophys Acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 72.Mitophagy: permitted by prohibitin. Galluzzi L, Bravo-San Pedro JM, Kroemer G. Curr Biol. 2017;27:0–6. doi: 10.1016/j.cub.2016.11.055. [DOI] [PubMed] [Google Scholar]
- 73.Role of mitochondrial inner membrane permeabilization in necrotic cell death, apoptosis, and autophagy. Lemasters JJ, Qian T, He L, Kim JS, Elmore SP, Cascio WE, Brenner DA. Antioxid Redox Signal. 2002;4:769–781. doi: 10.1089/152308602760598918. [DOI] [PubMed] [Google Scholar]
- 74.Chemical compensation of mitochondrial phospholipid depletion in yeast and animal models of Parkinson's disease. Wang S, Zhang S, Xu C, et al. PLoS ONE. 2016;11:0. doi: 10.1371/journal.pone.0164465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mitochondrial dysfunction: the road to alpha-synuclein oligomerization in PD. Esteves AR, Arduíno DM, Silva DF, Oliveira CR, Cardoso SM. Parkinsons Dis. 2011;2011:1–20. doi: 10.4061/2011/693761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Murakawa T, Yamaguchi O, Hashimoto A, et al. Nat Commun. 2015;6:1–14. doi: 10.1038/ncomms8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Okamoto K, Kondo-Okamoto N, Ohsumi Y. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 79.Structural basis of target recognition by Atg8/LC3 during selective autophagy. Noda NN, Kumeta H, Nakatogawa H, et al. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 80.Atg8-family interacting motif crucial for selective autophagy. Noda NN, Ohsumi Y, Inagaki F. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 81.LC3 conjugation system in mammalian autophagy. Tanida I, Ueno T, Kominami E. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mechanisms of mitophagy. Youle RJ, Narendra DP. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.The degradation pathway of the mitophagy receptor Atg32 is re-routed by a posttranslational modification. Levchenko M, Lorenzi I, Dudek J. PLoS ONE. 2016;11:0. doi: 10.1371/journal.pone.0168518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Liu L, Feng D, Chen G, et al. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 85.Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignment. Kuroyanagi H, Yan J, Seki N, et al. Genomics. 1998;51:76–85. doi: 10.1006/geno.1998.5340. [DOI] [PubMed] [Google Scholar]
- 86.ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. Wu W, Tian W, Hu Z, et al. EMBO Rep. 2014;15:566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Chen G, Han Z, Feng D, et al. Mol Cell. 2014;54:362–377. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 88.Mitochondrial protein PGAM5 regulates mitophagic protection against cell necroptosis. Lu W, Sun J, Yoon JS, et al. PLoS ONE. 2016;11:0. doi: 10.1371/journal.pone.0147792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Chu CT, Ji J, Dagda RK, et al. Nat Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berezowska S, Galván JA. Histochemistry of Single Molecules. Methods in Molecular Biology. Vol. 1560. New York, NY: Humana Press; 2017. Immunohistochemical Detection of the Autophagy Markers LC3 and p62/SQSTM1 in Formalin-Fixed and Paraffin-Embedded Tissue; pp. 189–194. [DOI] [PubMed] [Google Scholar]
- 91.p62 pathology model in the rat substantia nigra with filamentous inclusions and progressive neurodegeneration. Jackson KL, Lin WL, Miriyala S, et al. PLoS ONE. 2017;12:0. doi: 10.1371/journal.pone.0169291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mycotoxin patulin suppresses innate immune responses by mitochondrial dysfunction and p62/sequestosome-1-dependent mitophagy. Tsai WT, Lo YC, Wu MS, et al. J Biol Chem. 2016;291:19299–19311. doi: 10.1074/jbc.M115.686683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Absence of the autophagy adaptor SQSTM1/p62 causes childhood-onset neurodegeneration with ataxia, dystonia, and gaze palsy. Haack TB, Ignatius E, Calvo-Garrido J, et al. Am J Hum Genet. 2016;99:735–743. doi: 10.1016/j.ajhg.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Schweers RL, Zhang J, Randall MS, et al. PNAS. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Molecular mechanisms of mitochondrial autophagy/mitophagy in the heart. Saito T, Sadoshima J. Circ Res. 2015;116:1477–1490. doi: 10.1161/CIRCRESAHA.116.303790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mitochondrial dysfunction in cardiac disease: ischemia--reperfusion, aging, and heart failure. Lesnefsky EJ, Moghaddas S, Tandler B, Kerner J, Hoppel CL. J Mol Cell Cardiol. 2001;33:1065–1089. doi: 10.1006/jmcc.2001.1378. [DOI] [PubMed] [Google Scholar]
- 97.Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. PLoS ONE. 2011;6:0. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bicarbonate increases ischemia-reperfusion damage by inhibiting mitophagy. Queliconi BB, Kowaltowski AJ, Gottlieb RA. PLoS ONE. 2016;11:0. doi: 10.1371/journal.pone.0167678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.PINK1 is dispensable for mitochondrial recruitment of parkin and activation of mitophagy in cardiac myocytes. Kubli DA, Cortez MQ, Moyzis AG, Najor RH, Lee Y, Gustafsson ÅB. PLoS ONE. 2015;10:0. doi: 10.1371/journal.pone.0130707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Targeting mitochondrial dysfunction in lung diseases: emphasis on mitophagy. Sureshbabu A, Bhandari V. Front Physiol. 2013;4:1–8. doi: 10.3389/fphys.2013.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Elgendy M, Sheridan C, Brumatti G, Martin SJ. Mol Cell. 2011;42:23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 102.Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Liu Y, Shoji-Kawata S, Sumpter RM Jr, et al. PNAS. 2013;110:20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bcl-2 family proteins participate in mitochondrial quality control by regulating Parkin/PINK1-dependent mitophagy. Hollville E, Carroll RG, Cullen SP, Martin SJ. Mol Cell. 2014;55:451–466. doi: 10.1016/j.molcel.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 104.Selective escape of proteins from the mitochondria during mitophagy. Saita S, Shirane M, Nakayama KI. Nat Commun. 2013;4:1–14. doi: 10.1038/ncomms2400. [DOI] [PubMed] [Google Scholar]
- 105.Mitochondrial dysfunction in Parkinson's disease. Hu Q, Wang G. Transl Neurodegener. 2016;5:1–8. doi: 10.1186/s40035-016-0060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Protein Quality Control in the Cytosol and the Endoplasmic Reticulum: Brothers in Arms. Buchberger A, Bukau B, Sommer T. Mol Cell. 2010;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 107.Hexosamine pathway metabolites enhance protein quality control and prolong life. Denzel M, Storm N, Gutschmidt A, et al. Cell. 2014;156:1167–1178. doi: 10.1016/j.cell.2014.01.061. [DOI] [PubMed] [Google Scholar]
- 108.The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Lazarou M, Sliter DA, Kane LA, et al. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.The function of autophagy in neurodegenerative diseases. Kiriyama Y, Nochi H. Int J Mol Sci. 2015;16:26797–26812. doi: 10.3390/ijms161125990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Parkinson's disease: from monogenic forms to genetic susceptibility factors. Lesage S, Brice A. Hum Mol Genet. 2009;18:0–59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- 111.Parkin (PARK 2) mutations are rare in Czech patients with early-onset Parkinson's disease. Fiala O, Zahorakova D, Pospisilova L, et al. PLoS ONE. 2014;9:0. doi: 10.1371/journal.pone.0107585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Analysis of PRKN variants and clinical features in Polish patients with Parkinson's disease. Oczkowska A, Florczak-Wyspianska J, Permoda-Osip A, Owecki M, Lianeri M, Kozubski W, Dorszewska J. Curr Genomics. 2015;16:215–223. doi: 10.2174/1389202916666150326002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Genetic variants ofLRRK2 in Taiwanese Parkinson's disease. Wu YR, Chang KH, Chang WT, et al. PLoS ONE. 2013;8:0. doi: 10.1371/journal.pone.0082001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Analysis of Nigerians with apparently sporadic Parkinson disease for mutations in LRRK2, PRKN and ATXN3. Okubadejo N, Britton A, Crews C, Akinyemi R, Hardy J, Singleton A, Bras J. PLoS ONE. 2008;3:0. doi: 10.1371/journal.pone.0003421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Parkinson's disease in Saudi patients: a genetic study. Al-Mubarak BR, Bohlega SA, Alkhairallah TS, et al. PLoS ONE. 2015;10:0. doi: 10.1371/journal.pone.0135950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.The role of the LRRK2 gene in Parkinsonism. Li JQ, Tan L, Yu JT. Mol Neurodegener. 2014;9:1–17. doi: 10.1186/1750-1326-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.The LRRK2 G2019S mutation as the cause of Parkinson's disease in Ashkenazi Jews. Thaler A, Ash E, Gan-Or Z, Orr-Urtreger A, Giladi N. J Neural Transm. 2009;116:1473–1482. doi: 10.1007/s00702-009-0303-0. [DOI] [PubMed] [Google Scholar]
- 118.PINK1 defect causes mitochondrial dysfunction, proteasomal deficit and alpha-synuclein aggregation in cell culture models of Parkinson's disease. Liu W, Vives-Bauza C, Acín-Peréz- R, et al. PLoS ONE. 2009;4:0. doi: 10.1371/journal.pone.0004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bioenergetic consequences of PINK1 mutations in Parkinson disease. Abramov AY, Gegg M, Grunewald A, Wood NW, Klein C, Schapira AHV. PLoS ONE. 2011;6:0. doi: 10.1371/journal.pone.0025622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Molecular mechanisms of pesticide-induced neurotoxicity: relevance to Parkinson's disease. Franco R, Li S, Rodriguez-Rocha H, Burns M, Panayiotidis MI. Chem Biol Interact. 2010;188:289–300. doi: 10.1016/j.cbi.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.LRRK2 modulates vulnerability to mitochondrial dysfunction in Caenorhabditis elegans. Saha S, Guillily MD, Ferree A, et al. J Neurosci. 2009;29:9210–9218. doi: 10.1523/JNEUROSCI.2281-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.LRRK2 expression is deregulated in fibroblasts and neurons from Parkinson patients with mutations in PINK1. Azkona G, López de Maturana R, Del Rio P, et al. Mol Neurobiol. 2018;55:506–516. doi: 10.1007/s12035-016-0303-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Altered mitochondrial respiration and other features of mitochondrial function in Parkin-mutant fibroblasts from Parkinson's disease patients. Haylett W, Swart C, van der Westhuizen F, et al. Parkinsons Dis. 2016;2016:1–11. doi: 10.1155/2016/1819209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ubiquitin is phosphorylated by PINK1 to activate parkin. Koyano F, Okatsu K, Kosako H, et al. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 125.Lysine 27 ubiquitination of the mitochondrial transport protein Miro is dependent on serine 65 of the Parkin ubiquitin ligase. Birsa N, Norkett R, Wauer T, et al. J Biol Chem. 2014;289:14569–14582. doi: 10.1074/jbc.M114.563031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Parkinson disease (PARK) genes are somatically mutated in cutaneous melanoma. Inzelberg R, Samuels Y, Azizi E, et al. Neurol Genet. 2016;2:0. doi: 10.1212/NXG.0000000000000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dysregulation of autophagy and mitochondrial function in Parkinson's disease. Wang B, Abraham N, Gao G, Yang Q. Transl Neurodegener. 2016;5:19. doi: 10.1186/s40035-016-0065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Amphetamines promote mitochondrial dysfunction and DNA damage in pulmonary hypertension. Chen PI, Cao A, Miyagawa K, et al. JCI Insight. 2017;2:0. doi: 10.1172/jci.insight.90427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Biochemical, physiological and medical aspects of ubiquinone function. Ernster L, Dallner G. http://www.ncbi.nlm.nih.gov/pubmed/7599208. Biochim Biophys Acta. 1995;1271:195–204. doi: 10.1016/0925-4439(95)00028-3. [DOI] [PubMed] [Google Scholar]
- 130.Coenzyme Q deficiency triggers mitochondria degradation by mitophagy. Rodríguez-Hernández A, Cordero M, Salviati L, et al. Autophagy. 2009;5:19–32. doi: 10.4161/auto.5.1.7174. [DOI] [PubMed] [Google Scholar]
- 131.Mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes: a distinctive clinical syndrome. Pavlakis SG, Phillips PC, DiMauro S, De Vivo DC, Rowland LP. Ann Neurol. 1984;16:481–488. doi: 10.1002/ana.410160409. [DOI] [PubMed] [Google Scholar]
- 132.Secondary coenzyme Q10 deficiency triggers mitochondria degradation by mitophagy in MELAS fibroblasts. Cotán D, Cordero MD, Garrido-Maraver J, et al. FASEB J. 2011;25:2669–2687. doi: 10.1096/fj.10-165340. [DOI] [PubMed] [Google Scholar]
- 133.Mitochondrial dysfunction and mitophagy activation in blood mononuclear cells of fibromyalgia patients: implications in the pathogenesis of the disease. Cordero MD, De Miguel M, Moreno Fernández AM, et al. Arthritis Res Ther. 2010;12:1–11. doi: 10.1186/ar2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Coenzyme Q10 and heart failure: a state-of-the-art review. Sharma A, Fonarow GC, Butler J, Ezekowitz JA, Felker GM. Circ Heart Fail. 2016;9:0. doi: 10.1161/CIRCHEARTFAILURE.115.002639. [DOI] [PubMed] [Google Scholar]
- 135.Metabolic resuscitation in sepsis: a necessary step beyond the hemodynamic? Leite HP, de Lima LF. J Thorac Dis. 2016;8:0–7. doi: 10.21037/jtd.2016.05.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.The use of antioxidants in the treatment of traumatic brain injury. Venegoni W, Shen Q, Thimmesch AR, Bell M, Hiebert JB, Pierce JD. J Adv Nurs. 2017;73:1331–1338. doi: 10.1111/jan.13259. [DOI] [PubMed] [Google Scholar]
- 137.WITHDRAWN: coenzyme Q10 for Parkinson's disease. Liu J, Wang LN, Zhan SY, Xia Y. Cochrane Database Syst Rev. 2012;5:0. doi: 10.1002/14651858.CD008150.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nicotinamide enhances mitochondria quality through autophagy activation in human cells. Kang HT, Hwang ES. Aging Cell. 2009;8:426–438. doi: 10.1111/j.1474-9726.2009.00487.x. [DOI] [PubMed] [Google Scholar]
- 139.Hepatitis C virus induces the mitochondrial translocation of Parkin and subsequent mitophagy. Kim SJ, Syed GH, Siddiqui A. PLoS Pathog. 2013;9:0. doi: 10.1371/journal.ppat.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cloning of the gene encoding peptide-binding protein 74 shows that it is a new member of the heat shock protein 70 family. Domanico SZ, DeNagel DC, Dahlseid JN, Green JM, Pierce SK. Mol Cell Biol. 1993;13:3598–3610. doi: 10.1128/mcb.13.6.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Knockdown of Hsc70-5/mortalin induces loss of synaptic mitochondria in a Drosophila Parkinson's disease model. Zhu JY, Vereshchagina N, Sreekumar V, et al. PLoS ONE. 2013;8:0. doi: 10.1371/journal.pone.0083714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Functional significance of point mutations in stress chaperone mortalin and their relevance to Parkinson disease. Wadhwa R, Ryu J, Ahn HM, Saxena N, Chaudhary A, Yun CO, Kaul SC. J Biol Chem. 2015;290:8447–8456. doi: 10.1074/jbc.M114.627463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Brain region specific mitophagy capacity could contribute to selective neuronal vulnerability in Parkinson's disease. Diedrich M, Kitada T, Nebrich G, et al. Proteome Sci. 2011;9:1–19. doi: 10.1186/1477-5956-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of α-synuclein and LRRK2 in the brain. Friedman LG, Lachenmayer ML, Wang J, et al. J Neurosci. 2012;32:7585–7593. doi: 10.1523/JNEUROSCI.5809-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mitochondria in the spotlight of aging and idiopathic pulmonary fibrosis. Mora AL, Bueno M, Rojas M. J Clin Invest. 2017;127:405–414. doi: 10.1172/JCI87440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.αSynuclein and mitochondrial dysfunction: A pathogenic partnership in Parkinson's disease? Protter D, Lang C, Cooper AA. Parkinsons Dis. 2012;2012:1–12. doi: 10.1155/2012/829207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Genetics of Parkinson's disease. Klein C, Westenberger A. Cold Spring Harb Perspect Med. 2012;2:0. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Bonifati V, Rizzu P, van Baren MJ, et al. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 149.DJ-1 is a redox-dependent molecular chaperone that inhibits α-synuclein aggregate formation. Shendelman S, Jonason A, Martinat C, Leete T, Abeliovich A. PLoS Biol. 2004;2:0. doi: 10.1371/journal.pbio.0020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.The oxidation state of DJ-1 regulates its chaperone activity toward alpha-synuclein. Zhou W, Zhu M, Wilson MA, Petsko GA, Fink AL. J Mol Biol. 2006;356:1036–1048. doi: 10.1016/j.jmb.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 151.Antibodies against the C-terminus of α-synuclein modulate its fibrillation. Sahin C, Lorenzen N, Lemminger L, et al. Biophys Chem. 2017;220:34–41. doi: 10.1016/j.bpc.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 152.New roles of glycosaminoglycans in α-synuclein aggregation in a cellular model of Parkinson disease. Lehri-Boufala S, Ouidja MO, Barbier-Chassefière V, et al. PLoS ONE. 2015;10:0. doi: 10.1371/journal.pone.0116641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pre-fibrillar α-synuclein mutants cause Parkinson's disease-like non-motor symptoms in Drosophila. Gajula Balija MB, Griesinger C, Herzig A, Zweckstetter M, Jäckle H. PLoS ONE. 2011;6:0. doi: 10.1371/journal.pone.0024701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Therapeutic potential of autophagy-enhancing agents in Parkinson's disease. Moors TE, Hoozemans JJ, Ingrassia A, Beccari T, Parnetti L, Chartier-Harlin MC, van de Berg WD. Mol Neurodegener. 2017;12:1–18. doi: 10.1186/s13024-017-0154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.PMI: a ΔΨm independent pharmacological regulator of mitophagy. East DA, Fagiani F, Crosby J, et al. Chem Biol. 2014;21:1585–1596. doi: 10.1016/j.chembiol.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nicotinamide-induced mitophagy: event mediated by high NAD+/NADH ratio and SIRT1 protein activation. Jang SY, Kang HT, Hwang ES. J Biol Chem. 2012;287:19304–19314. doi: 10.1074/jbc.M112.363747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Parkinson's: a syndrome rather than a disease? Titova N, Padmakumar C, Lewis SJ, Chaudhuri KR. J Neural Transm. 2017;124:907–914. doi: 10.1007/s00702-016-1667-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mitophagy and the therapeutic clearance of damaged mitochondria for neuroprotection. East DA, Campanella M. Int J Biochem Cell Biol. 2016;79:382–387. doi: 10.1016/j.biocel.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 159.Diabetes and risk of Parkinson's disease: an updated meta-analysis of case-control studies. Lu L, Fu DL, Li HQ, Liu AJ, Li JH, Zheng GQ. PLoS ONE. 2014;9:0. doi: 10.1371/journal.pone.0085781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Metformin restores Parkin-mediated mitophagy, suppressed by cytosolic p53. Song YM, Lee WK, Lee YH, Kang ES, Cha BS, Lee BW. Int J Mol Sci. 2016;17:122. doi: 10.3390/ijms17010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.PINK1-parkin-mediated mitophagy protects mitochondrial integrity and prevents metabolic stress-induced endothelial injury. Wu W, Xu H, Wang Z, et al. PLoS ONE. 2015;10:0. doi: 10.1371/journal.pone.0132499. [DOI] [PMC free article] [PubMed] [Google Scholar]