Abstract
Many questions remain unanswered concerning the long-term effects of COVID-19 on children. In this report, we describe a computable phenotyping algorithm for identifying children and adolescents with postacute sequelae of COVID-19 (PASC) and pilot this tool to characterize the clinical epidemiology of pediatric PASC in a large healthcare delivery network.
Keywords: COVID-19, pediatrics, postacute sequelae of COVID-19 (PASC)
INTRODUCTION
Since the SARS-CoV-2 pandemic began in late 2019, over 13 million children in the United States have been infected with the virus [1]. Although many of these acute infections have not resulted in severe morbidity or mortality, a subset of children and adolescents have experienced recurrent or persistent symptoms beyond the typical recovery period [2]. The constellation of findings that occur postinfection is known as postacute sequelae of SARS-CoV-2 (PASC), or colloquially as “long-Covid.” The U.S. Centers for Disease Control and Prevention (CDC) defines PASC as a wide range of health problems that linger for more than 4 weeks following an acute COVID-19 infection [3]. Although this is an area of active research, relatively little is currently known about its clinical epidemiology in the pediatric population.
Considering the large number of children who have been affected by COVID-19, it is critical that we monitor the rates, trends, and outcomes of PASC in this population. An important first step toward these efforts is the development of a tool that can quickly and easily identify cases in large clinical populations. With the widespread adoption of electronic health records (EHR), it is now possible to develop computable phenotypes using data that are collected for clinical care, which can be used for population-level analysis to inform the public health response [4, 5]. In this report, we describe a novel phenotyping algorithm to define the burden, clinical spectrum, and outcomes of pediatric PASC using real-world data.
METHODS
Source Population
For this cross-sectional study, encounter- and patient-level data were extracted from the EHR of patients who were ≤21 years old and had at least one healthcare visit in the Yale New Haven Health System (YNHHS) between 5/1/21 and 9/30/21, corresponding to the 5-month period after the peak of the alpha variant of SARS-CoV-2 in Connecticut (Figure S1). The YNHHS is the largest health system in Connecticut, with close to 4 million outpatient encounters per year, and consists of 5 integrated delivery networks, 6 multispecialty centers, and over 100 ambulatory clinics. The YNHHS covers a geographic area of roughly 650 square miles and serves a diverse patient population that closely mirrors the national demographics in terms of race/ethnicity and socioeconomic status. All clinics, emergency departments, and hospitals in the YNHHS use a single EHR system [6]. Clinical data from the EHR are mapped to standard vocabularies and stored in a data warehouse using the Observational Medical Outcomes Partnership common data model.
PHENOTYPING ALGORITHM
We utilized a rule-based EHR phenotyping algorithm to identify patients who had a medical encounter (ambulatory, telemedicine, or emergency room) for PASC. This algorithm searched administrative billing records, patient problem lists, and encounter diagnoses for PASC-specific and PASC-related clinical terms mapped to Systematized Nomenclature of Medicine Clinical Terms, or International Classification of Diseases, 10th Revision, Clinical Modification codes (see Figures S2 and S3). The pattern of these PASC-related terms (eg, “personal history of COVID-19” plus “chronic fatigue”) was used to infer PASC in the absence of a more specific diagnosis code (eg, ICD-10: U09.9). Boolean logic was used to process the various clinical terms for inclusion and exclusion criteria. A patient was defined as having “computable PASC” if, at any encounter during the study period, a clinician noted one of the PASC-related clinical terms as either a visit or billing diagnosis or as an active problem on their problem list, excluding encounters related to clearance to return to sports or patients with multisystem inflammatory syndrome in children. To test the performance of the computable phenotype, the medical records of all phenotype-identified PASC cases plus a set of 100 predicted-negative controls were manually reviewed and abstracted using standardized forms by two clinicians (TAF and CMM). Predicted-negative controls were randomly selected from the subgroup of patients who had a medical visit during the study period, had a recent diagnosis of COVID-19 (1–6 months prior to their visit), and were not classified by the computable phenotype as PASC. After a manual review of medical records, patients were classified as having EHR-verified PASC (true positives) based on the CDC case definition, which entailed documentation of at least one clinical manifestation associated with PASC that (1) lasted for ≥4 weeks, (2) followed a confirmed or suspected COVID-19 infection, and (3) could not be explained by any other diagnosis (eg, neurologic, oncologic, or autoimmune diseases). Study definitions are further detailed in Table S1. The performance of the computable phenotype was evaluated using standard metrics of accuracy, such as precision, recall, specificity, and F-measure, as previously described [7]. Descriptive statistics were used to summarize the manually abstracted demographic and clinical characteristics of PASC cases and predicted-negative controls. All patients who met inclusion criteria and had not opted out of research were included in the analysis. Stata V.17 was used for statistical analyses. The institutional review board at the Yale University School of Medicine approved the study and waived the requirement for informed consent.
RESULTS
A total of 41,312 children had at least one medical encounter between 5/1/21 and 9/30/21 in the YNHHS. Among those with an encounter, 1,641 had a recent history of COVID-19, and 43 met the computable phenotype definition for PASC. Two patients with computable PASC (5%) did not have available encounter notes for review and were excluded from subsequent analyses. Out of the 141 patients whose medical records were reviewed, the computable PASC diagnosis was verified by manual review in 37/41. One of the 100 predicted-negative controls met the criteria for PASC on manual review, yielding an overall accuracy of 0.96 (95% confidence interval [95% CI], 0.92–0.99), a precision of 0.90 (95% CI, 0.77–0.97), a recall of 0.97 (95% CI, 0.86–0.99), a specificity of 0.96 (95% CI, 0.90–0.99), and an F1 score of 0.94 (95% CI, 0.88–0.99). The four false positives consisted of a patient with COVID-19 vaccine-related complications, two patients who did not meet the criteria for the duration of symptoms (<4 weeks), and one patient being evaluated for recurrent fevers post-COVID-19, which was determined to be due to an infection with another respiratory virus. The one false negative consisted of an adolescent who was undergoing cardiac evaluation post-COVID-19 for persistent chest pain.
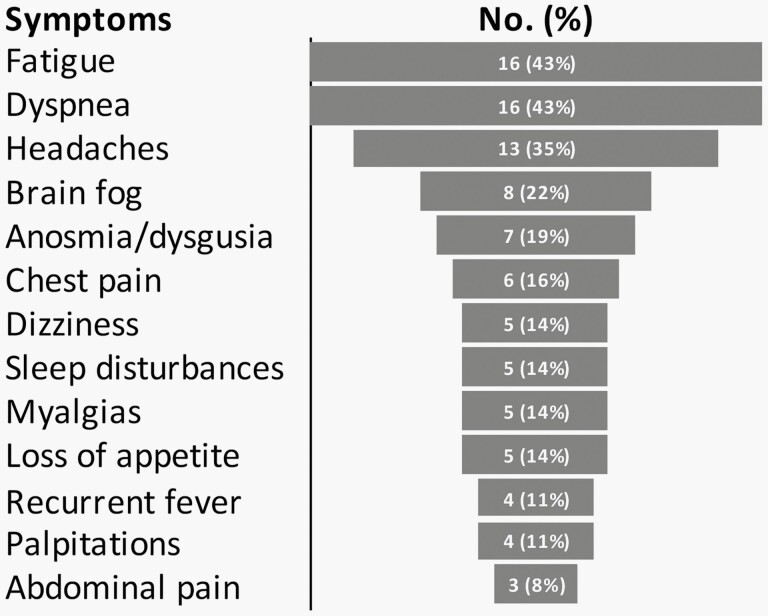
Demographic characteristics of the EHR-verified PASC cases and selected controls are shown in Table 1. Among controls, most encounters were either for a routine physical exam (35%), a new infection (26%), or management of an injury (9%). The median age for the 37 EHR-verified PASC cases was 17 years (range 4–21 years), and 68% (25/37) were female. Cases identified their race/ethnicity as either non-Hispanic Black (2/37; 5%), Hispanic (12/37; 32%), non-Hispanic White (22/37; 56%), or non-Hispanic other (1/37; 3%). Nearly one-third (11/37; 30%) of patients with EHR-verified PASC had no prior comorbidities. The majority (35/37; 95%) had mild or asymptomatic COVID-19 during the acute phase of the disease. The median time between acute COVID-19 and their PASC encounter was 16 weeks (range 4–60 weeks). The most prevalent documented symptoms are shown in Figure 1.
Table 1.
Characteristics of Children <21 Years Of age with an Encounter at YNHHS Between 5/1/21 and 9/30/21
| Predicted-Negative Controls, N = 100 | EHR-Verified PASC, N = 37 | |||
|---|---|---|---|---|
| Age, years | N | % | N | % |
| <5 years | 32 | 32% | 2 | 5% |
| 5–10 years | 26 | 26% | 1 | 3% |
| 11–15 years | 34 | 34% | 8 | 22% |
| 16–21 years | 8 | 8% | 26 | 70% |
| Sex | ||||
| Male | 41 | 41% | 12 | 32% |
| Female | 59 | 59% | 25 | 68% |
| Race/Ethnicity | ||||
| Hispanic | 44 | 44% | 12 | 32% |
| Non-Hispanic White | 32 | 32% | 22 | 59% |
| Non-Hispanic Black | 21 | 21% | 2 | 6% |
| Non-Hispanic Other | 3 | 3% | 1 | 3% |
| Epidemiological Weeks* | ||||
| Weeks 18–24 | 34 | 34% | 10 | 27% |
| Weeks 25–31 | 51 | 51% | 9 | 24% |
| Weeks 32–38 | 15 | 15% | 18 | 49% |
| Comorbidities | ||||
| Asthma | 15 | 15% | 8 | 22% |
| Eczema/Allergy | 23 | 23% | 13 | 35% |
| Obesity | 11 | 11% | 4 | 11% |
| Anxiety/Depression | 3 | 3% | 4 | 11% |
*CDC epidemiological week of the index medical visit.
Other race/ethnicity: Asian, American Indian or Alaska Native, Native Hawaiian or Pacific Islander.
Abbreviations: EHR: electronic health record; PASC: postacute sequelae of COVID-19; YNHHS: Yale New Haven Health System.
Figure 1.
Symptoms of Patients with EHR-Verified PASC, N = 37. Self-reported Symptoms Documented on Routine Medical Visits. Captured by Manual Review of Clinician Notes.
DISCUSSION
This study leveraged real-world data from a large healthcare system to define a computable phenotype for PASC and explore the clinical spectrum and outcomes of children and adolescents experiencing symptoms post-COVID-19. This work has several important findings. First, it presents a PASC monitoring approach that is feasible to implement and resource efficient. Second, it highlights the burden of PASC and the wide range of clinical symptoms that children and adolescents can experience post-COVID-19, a reminder of the often-overlooked consequences of the pandemic in children. Third, it builds on the body of evidence showing how significant complications can occur after mild COVID-19 infections and even in previously healthy children, underscoring the importance of vaccination in this age group [8].
Most of the work on pediatric PASC thus far has relied on self-reported data from patient questionnaires or laborious chart reviews, which are inefficient when considering large-scale or long-term surveillance [9]. Recent studies have used a case-finding approach that relies on ICD-10 billing codes [10, 11]. Although easy to implement, billing codes may underestimate cases when used alone, as PASC could be recorded in the EHR but not coded as a billable diagnosis. Our approach entailed merging billing codes with other EHR elements, such as problem lists and SNOMED-CT codes. On evaluation, we found a high level of concordance between our computable phenotype and manual chart reviews (accuracy = 96%). A key strength of this work is its use of a nomenclature that is widely used across EHR and more extensive than ICD-10 codes, which may facilitate interoperability and enhance the accuracy of phenotyping [12]. However, because the billing and documentation standards are still evolving, it was not possible to compare the performance of our computable phenotype to an established gold standard. We also restricted our analysis to the period immediately preceding the introduction of the Omicron variant. More research will be needed to ensure this computable phenotype remains valid for the surveillance of PASC following infections with more recent variants of concern.
These data have other potential limitations. The clinical definition of pediatric PASC has not yet been established; as a result, clinicians may hold varying diagnostic criteria for PASC. The two patients with false-positive PASC who had symptoms for <4 weeks at diagnosis highlight how this surveillance approach is dependent on patterns of diagnosis by clinicians. As is the case with most studies that repurpose data generated primarily for health care, missing data from inconsistent documentation and overrepresentation of patients who are more severely affected, or have more access to healthcare could have affected the results. Further, the sample size of PASC cases was relatively small, so the findings should be interpreted with caution. However, the demographics and clinical spectrum of PASC in this study are comparable to those of earlier studies in children [13]. Last, as some of the codes used in the study were new and inconsistently used (eg, ICD-10 code U09.9 applied retrospectively to encounters), our algorithm may be underestimating the true burden of the disease.
CONCLUSIONS
In this study, we describe the design and application of an EHR-based monitoring system for PASC in children. Our evaluation provides further evidence of the utility of EHRs and computable phenotypes for public health disease surveillance.
Supplementary Material
Funding support: This work was supported in part by the National Institutes of Health (NIH) grant number K23AI159518 (C.R.O) R01HD103512 (E.P.) and D43TW011526 (E.P) from the National Institute of Allergy and Infectious Diseases (NIAID). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIH. The funders of the study had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. The corresponding author had final responsibility for the decision to submit for publication.
Potential conflict of interests: Dr. Niccolai has served as a Scientific Advisor for Janssen and Moderna (both unrelated to this work). All other authors declare no conflicts of interest.
Contributor Information
Tomini A Fashina, Yale University School of Public, New Haven, Connecticut, USA.
Christine M Miller, Department of Pediatrics, Section of Infectious Diseases and Global Health, Yale University School of Medicine, New Haven, Connecticut, USA.
Elijah Paintsil, Department of Pediatrics, Section of Infectious Diseases and Global Health, Yale University School of Medicine, New Haven, Connecticut, USA; Department of Pharmacology, Yale University School of Medicine, New Haven, Connecticut, USA; Department of Epidemiology of Microbial Diseases, Yale University School of Public, New Haven, Connecticut, USA.
Linda M Niccolai, Department of Epidemiology of Microbial Diseases, Yale University School of Public, New Haven, Connecticut, USA.
Cynthia Brandt, Department of Biostatistics, Section of Health Informatics, Yale University School of Public Health, New Haven, Connecticut, USA, USA.
Carlos R Oliveira, Department of Pediatrics, Section of Infectious Diseases and Global Health, Yale University School of Medicine, New Haven, Connecticut, USA; Department of Biostatistics, Section of Health Informatics, Yale University School of Public Health, New Haven, Connecticut, USA, USA.
Author Contributions
Dr Carlos Oliveira had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors contributed to data collection and acquisition, database development, discussion and interpretation of the results, and to the writing of the manuscript. All authors have read and approved the final manuscript.
References
- 1. American Academy of Pediatrics. Children and COVID-19: State-Level Data Report 2022 [6/17/22]. https://www.aap.org/en/pages/2019-novel-coronavirus-covid-19-infections/children-and-covid-19-state-level-data-report/.
- 2. Ashkenazi-Hoffnung L, Shmueli E, Ehrlich S, et al. Long COVID in children: Observations from a designated pediatric clinic. Pediatr Infect Dis J 2021; 40:e509–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. CDC. Long COVID or Post-COVID Conditions. [11/3/22]. https://www.cdc.gov/coronavirus/2019-ncov/long-term-effects/index.html.
- 4. Khera R, Mortazavi BJ, Sangha V, et al. A multicenter evaluation of computable phenotyping approaches for SARS-CoV-2 infection and COVID-19 hospitalizations. NPJ Digit Med 2022; 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yakely AE, Niccolai LM, Oliveira CR.. Trends in anogenital wart diagnoses in Connecticut, 2013-2017. JAMA Netw Open 2020; 3:e1920168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peaper DR, Murdzek C, Oliveira CR, Murray TS.. Severe acute respiratory syndrome coronavirus 2 testing in children in a large regional us health system during the coronavirus disease 2019 pandemic. Pediatr Infect Dis J 2021; 40:175–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oliveira CR, Niccolai P, Ortiz AM, et al. Natural language processing for surveillance of cervical and anal cancer and precancer: Algorithm development and split-validation study. JMIR Med Inform 2020; 8:e20826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oliveira CR, Niccolai LM, Sheikha H, et al. ; Yale SARS-CoV-2 Genomic Surveillance Initiative. Assessment of clinical effectiveness of BNT162b2 COVID-19 vaccine in US adolescents. JAMA Netw Open 2022; 5:e220935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Behnood SA, Shafran R, Bennett SD, et al. Persistent symptoms following SARS-CoV-2 infection amongst children and young people: A meta-analysis of controlled and uncontrolled studies. J Infect 2022; 84:158–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duerlund LS, Shakar S, Nielsen H, Bodilsen J.. Positive predictive value of the ICD-10 diagnosis code for long-COVID. Clin Epidemiol 2022; 14:141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pfaff ER, Madlock-Brown C, Baratta JM, et al. Coding Long COVID: Characterizing a new disease through an ICD-10 lens. medRxiv. 2022:2022.04.18.22273968. doi: 10.1101/2022.04.18.22273968. PMID: 36093345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wei WQ, Teixeira PL, Mo H, Cronin RM, Warner JL, Denny JC.. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J Am Med Inform Assoc 2016; 23:e20–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zimmermann P, Pittet LF, Curtis N.. How common is long COVID in children and adolescents? Pediatr Infect Dis J 2021; 40:e482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.