Abstract
We screened for transposon-generated mutants of Synechocystis sp. strain PCC 6803 that exhibited aberrant phototactic movement. Of the 300 mutants generated, about 50 have been partially characterized; several contained transposons in genes encoding chemotaxis-related proteins, while others mapped to novel genes. These novel genes and their possible roles in motility are discussed.
Cyanobacteria such as Synechocystis sp. strain PCC 6803 exhibit surface-dependent phototactic motility which requires type IV pili (2, 4). To identify structural and regulatory components important for motility, we screened for mutants with aberrant phototactic movement (i.e., nonmotile or negatively phototactic under conditions in which wild-type cells show positive phototaxis).
A Synechocystis genomic library was constructed that contained approximately 105 clones with an average insert size of 6 to 8 kbp (3). The library of clones was used for in vitro transposon mutagenesis, and mutagenized clones were used for transformation of wild-type Synechocystis cells. Following transformation, cells were spread on 0.6% agar plates (10 μg of kanamycin/ml) and placed under a directional light source. Colonies that were nonmotile or exhibited an inverted response were picked, rescreened, and maintained for further analysis. Approximately 300 independent mutants (90% were nonmotile and 10% exhibited negative phototaxis) from a population of 8,000 transformants were identified. This screen required robust cell growth, thus eliminating mutants that might have appeared nonmotile because of growth defects. To identify the site of transposon insertion, we used plasmid rescue or direct sequencing of genomic DNA using primers that anneal to the ends of the transposon-kanamycin resistance gene cassette. Of the 50 or so mutants characterized so far, several contained transposons in genes encoding chemotaxis-related proteins (3). We also identified transposons in several novel genes, described in this report, that provide us with clues concerning functional and possibly architectural features that are important for motility.
Although we have not yet confirmed the phenotypic results by insertionally inactivating all of the genes found to be disrupted by transposons in the mutant strains, a number of findings strongly argue that the insertion of the transposon cassette is responsible for the mutant phenotype. First, based on Southern blot hybridizations, all of the mutants examined appeared to have a single transposon insertion (data not shown). Second, in several cases, independent transposition events led to the insertion of the transposon into different positions within the same gene or genetic locus; in all cases, the insertions always yielded the same phenotype. Since the entire Synechocystis genome has been sequenced, the positions of the inserted transposon can be easily mapped, and the phenotypes of all of the mutants generated can be readily confirmed by targeted inactivations. The third and most convincing argument is that in all cases in which we have specifically inactivated a target gene identified by the transposon mutagenesis (10 cases to date), the targeted and transposon-generated mutants yielded identical phenotypes (2, 3, 4).
Table 1 shows the phenotypes of various mutants isolated from our screen (it excludes the chemotaxis-related genes which are described in Bhaya et al. [3]) and gives the positions of the independent transposon insertions (relative to the open reading frame), the size of the disrupted open reading frames (in amino acids and nucleotides), the gene designation (as given in Cyanobase), and the putative function of the gene product. Three nonmotile mutants were shown to harbor transposons in the pilT1 and pilC genes, which are located adjacent to each other on the genome. We have recently demonstrated that disruption of either of these genes results in a nonmotile phenotype (2). This provides evidence that the method of mutant generation is robust and can be used to isolate novel motility mutants. Four independent transposon insertions led to the disruption of a gene encoding a putative DnaK-like chaperone (sll0058). There are three DnaK-like genes on the Synechocystis genome, of which sll0170 and sll1932 are likely to be the major chaperones (based on theoretical predictions), while sll0058 may be a minor chaperone dedicated to particular aspects of pilus biogenesis (10). In Myxococcus xanthus it has been shown that a nonessential DnaK-like chaperone is required for social motility and the production of fibrils (15), although its exact role in these processes has yet to be established.
TABLE 1.
Mutants generated by transposon mutagenesis
| Phenotype | Position(s) of transposon insertion(s) | Size of gene, amino acids (nucleotides) | Gene designation | Putative gene function and/or protein domain |
|---|---|---|---|---|
| Nonmover | −13, 776 | 369 (1,107) | slr0161 | PilT1 |
| Nonmover | 521 | 405 (1,215) | slr0162 | PilC |
| Nonmover | 99, 448, 534, 1107 | 692 (2,076) | sll0058 | Chaperone |
| Nonmover | 617 | 354 (1,062) | sll0415 | ABC transporter |
| Nonmover | 276, 606, 664 | 324 (972) | sll0564 | Ubiquinone methyltransferase-like |
| Nonmover | 270 | 133 (399) | sll0565 | E/Q-rich protein downstream of ubiquinone methyltransferase |
| Nonmover | 911 | 416 (1,248) | sll1575 | Eukaryotic-type serine/threonine protein kinase |
| Nonmover | 953 | 337 (1,011) | slr1991 | Adenylyl cyclase (cya1) |
| Nonmover | 89 | 158 (474) | slr2016 | Part of a gene locus (slr2015-2019) which includes an ABC transporter (slr2019) and pilA-like gene (slr2016) |
| Nonmover | 83, 244, 583, 859 | 360 (1,080) | slr2017 | Part of a gene locus (slr2015-2019) |
| Nonmover | 78, 2280 | 799 (2,397) | slr2018 | Part of a gene locus (slr2015-2019) |
| Slowmover | 437 | 374 (1,122) | slr0358 | E/P-rich |
| Nonmover | −3, 443, 1057 | 535 (1,605) | slr1301 | E/Q-rich coiled coil |
| Nonmover | 147 | 134 (402) | slr1964 | Q-rich |
| Nonmover | 64, 348, 502 | 259 (777) | sll0183 | Pentapeptide repeat domain and tetratricopeptide domain |
| Slowmover | 342 | 286 (858) | sll0414 | Pentapeptide repeat domain |
| Nonmover | 97 | 169 (507) | sll0301 | Pentapeptide repeat domain |
We identified a nonmotile mutant disrupted for an ABC transporter gene (sll0415), and the phenotype of the mutant strain was confirmed by specific targeted inactivation. Interestingly, the mutant strain appears to have a normal complement of thin and thick pili (P. Shahi, D. Bhaya, and A. R. Grossman, unpublished results). Of the approximately 42 putative ABC transporters of Synechocystis (http://www.biology.ucsd.edu/≈ipaulsen/transport/), the one encoded by sll0415 has the highest similarity to an ABC transporter (PilH) required for motility in M. xanthus (38% identity and 60% similarity), although the role of the M. xanthus protein in motility is not known (16).
Three transposition events have been mapped to sll0564, whose gene product shows low but significant homology (25% identity and 36% similarity) to a methyltransferase of the ubiquinone methyltransferase type which uses S-adenosylmethionine as a methyl donor. The putative polypeptide encoded by sll0564 does not resemble the CheR polypeptide, which is a methyltransferase involved in methylation of the methyl-accepting chemoreceptor proteins (11). Therefore, we cannot assign a function to this putative polypeptide without detailed biochemical analyses. It is also notable that a transposon which mapped to the gene (sll0565) downstream of the putative methyltransferase also produced a nonmotile phenotype, raising the possibility that both genes are important for motility. The putative sll0565 protein does not show similarity to any protein in the database with an assigned function but has a relatively high proportion of glutamine (Q) and glutamic acid (E) residues (see below).
Transposons have also been mapped to genes encoding polypeptides with homology to a eukaryotic-type serine-threonine kinase (sll1575) and an adenylyl cyclase (slr1991); both of these polypeptides may participate in signal transduction events required for phototactic movement. It has been shown previously that a lesion in the adenylyl cyclase gene resulted in a nonmotile strain (13). There are three genes on the Synechocystis genome that encode polypeptides with homology to adenylyl cyclase; it is not known if the other two genes (sll1161 and sll0646) function in the regulation of motility. Kamei et al. have also demonstrated that sll1575 is required for motility; however, the precise function of this kinase in motility is unclear (8). It is interesting that a eukaryotic-type kinase is involved in this process; this suggests that signal transduction events that govern phototaxis may involve complex interactions.
A novel locus containing five genes (slr2015 to slr2019) is interesting because transposons mapped to three of the genes (slr2016, slr2017, and slr2018). The polypeptide encoded by slr2016 shows weak but significant homology to the family of PilA-like polypeptides (4), while the slr2017 and slr2018 gene products do not show obvious homologies to other polypeptides in the databases, nor do they contain obvious functional motifs. We are currently examining the function of this locus by generating specific, nonpolar gene disruptions in each open reading frame and analyzing the effect of the disruptions on pilus morphology and biogenesis.
Table 2 lists seven genes encoding putative novel polypeptides that we have identified as being required for motility, based on phenotypes of strains mutated in these genes. These polypeptides fall into two distinct categories. The first category, represented by slr0358, sll0565, slr1964, and slr1301, are polypeptides that are relatively rich (defined here as being higher than 10%, usual levels being between 3 and 5%) in glutamic acid (E) or glutamine (Q) residues and that do not appear to have any recognizable motifs (based on analysis by Pfam [http://pfam.wustl.edu/index.html] or COG [http://www.ncbi.nlm.nih.gov/COG/] [1, 12]). While these four putative polypeptides appear to lack a signal sequence, sll0565 and slr1964 have potential transmembrane helices at the amino (N) or carboxy (C) terminus, respectively, based on DAS analysis (http://www.sbc.su.se/≈miklos/DAS/) (7). Furthermore, all of these polypeptides except slr0358, which is proline (P) rich, have the possibility of forming coiled-coil structures, based on the predictions of Coils (http://www.ch.embnet.org/software/COILSform.html) (9). The coiled-coil configuration of the putative slr1301 polypeptide may be especially extensive (data not shown). It has recently been demonstrated that FrzS of M. xanthus has an extensive coiled-coil structure and an FrzS mutant is incapable of social motility (14). Proteins rich in glutamate and glutamine residues and with the ability to form coiled coils may be involved in protein-protein interactions, often being part of a large macromolecular complex (6).
TABLE 2.
Novel mutants with high E, Q, and P percentages or repeated motifs
| Gene | Size
|
Insertion position(s) | Amino acid composition (% E/% Q/% P)a | Signal peptide | Coils | Transmembrane helices | PFAM designation(s) | |
|---|---|---|---|---|---|---|---|---|
| Residues | Nucleotides | |||||||
| slr0358 | 374 | 1,122 | 437 | 13/4.3/10.4 | No | No | No | No hits |
| sll0565 | 133 | 399 | 270 | 10.5/11.3/0.8 | No | Yes | Yes | No hits |
| slr1964 | 134 | 402 | 147 | 5.2/10.4/0.75 | No | Yes | Yes | No hits |
| slr1301 | 535 | 1,605 | 3, 443, 1057 | 13.6/13.6/1.9 | No | Yes | No | No hits |
| sll0183 | 259 | 777 | 64, 348, 502 | 5/4.6/3.1 | Yes (18/19)b | No | Yes | 00515, 00805 |
| sll0414 | 286 | 858 | 342 | 4.5/9.1/3.5 | No | No | No | 00805 |
| sll0301 | 169 | 507 | 97 | 3/2.4/1.2 | Yes (32/33) | No | Yes | 00805 |
Numbers in bold represent amino acid percentages higher than 10% (usual levels are 3 to 5%).
Numbers in parentheses represent putative signal peptide cleavage sites.
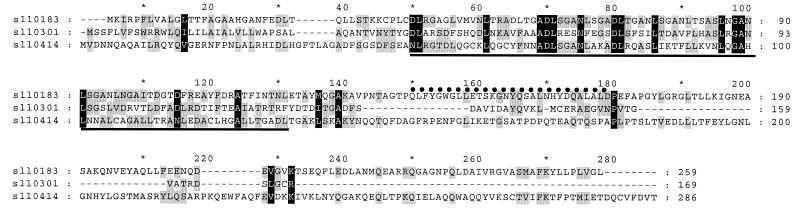
The putative polypeptides encoded by sll0183, sll0414, and sll0301 are in a second category that is distinguished by the presence of a repeated pentapeptide (XGAD/NL) motif categorized in the Pfam database as pfam 00805 (ANLSGADLTGADLRGADLSGADLTGANLSGANLSGADLSG). The repeat of a leucine residue every fifth residue is reminiscent of a leucine zipper, but this region is not predicted to form an alpha helix (Fig. 1). There are 15 polypeptides encoded by the Synechocystis genome that contain this domain (slr1819, sll1446, sll0414, slr1851, slr1152, slr1519, slr0516, slr0967, sll0274, slr0719, sll0183, sll0577, sll0301, sll1011, and slr1697; genes in italic are transposon mutants). The proteins encoded by sll0183 and sll0301 may also contain signal sequences (the putative cleavage sites are shown in Table 2). The protein encoded by sll0183 contains a second motif which is a tetratricopeptide repeat (amino acid 150 to 180) (Fig. 1). This motif (pfam 00515) represents an ancient module that functions in protein-protein interactions and participates in many cellular processes, including cell cycle progression, transcription, chaperone action, and protein transport (5). These findings raise the intriguing possibility that pilus biogenesis or function requires several polypeptides critical for protein-protein interactions, possibly having a scaffold or architectural function. We are currently addressing these questions.
FIG. 1.
Comparison of sll0183, sll0301, and sll0414, with black boxes indicating identical or conserved residues in all polypeptides and gray boxes indicating residues conserved in two polypeptides. The region with pentapeptide repeats is underlined; the tetratricopeptide motif of sll0183 is indicated by black circles.
The results of the mutant screen highlight two intriguing findings. First, there appear to be many polypeptides involved in cyanobacterial motility that have specific biochemical functions (based on homologies with other proteins), but their role in motility is relatively unexplored. We have evidence that two loci (tax1 and tax3) which contain several che-like genes are involved in pilus-mediated phototaxis (3). These loci include histidine kinases as well as a chemoreceptor with a putative phytochrome-like chromophore-binding domain. We postulate that these proteins are involved in a complex signal transduction pathway that regulates phototaxis (3). In some cases analogous proteins have been found to be required for motility in other organisms, such as M. xanthus. Second, groups of polypeptides with distinct structural characteristics are required for motility, and although their role in motility is not known, they may serve to facilitate interactions among proteins in large complexes or be part of an intracellular matrix that is required to assemble and/or anchor the pilus-associated motor complex.
Acknowledgments
The work presented in this manuscript was supported in part by National Science Foundation grant MCB 9727836.
Footnotes
Carnegie Institution of Washington publication no. 1488.
REFERENCES
- 1.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaya D, Bianco N R, Bryant D, Grossman A. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp PCC 6803. Mol Microbiol. 2000;37:941–951. doi: 10.1046/j.1365-2958.2000.02068.x. [DOI] [PubMed] [Google Scholar]
- 3.Bhaya D, Takahashi A, Grossman A. Light regulation of Type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC 6803. Proc Natl Acad Sci USA. 2001;98:7540–7545. doi: 10.1073/pnas.131201098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaya D, Watanabe N, Ogawa T, Grossman A R. The role of an alternate sigma factor in motility and pili formation in the cyanobacterium Synechocystis sp. strain PCC 6803. Proc Natl Acad Sci USA. 1999;96:3188–3193. doi: 10.1073/pnas.96.6.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blatch G L, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 6.Burkhard P, Strelkov S V, Stetefeld J. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 7.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 8.Kamei A, Yuasa T, Orikawa K, Geng X, Ikeuchi M. A eukaryotic-type protein kinase, SpkA, is required for normal motility of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2001;183:1505–1510. doi: 10.1128/JB.183.5.1505-1510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 10.Mrazek J, Bhaya D, Grossman A R, Karlin S. Highly expressed and alien genes of the Synechocystis genome. Nucleic Acids Res. 2001;29:1590–1601. doi: 10.1093/nar/29.7.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simms S A, Stock A M, Stock J B. Purification and characterization of the S-adenosylmethionine:glutamyl methyltransferase that modifies membrane chemoreceptor proteins in bacteria. J Biol Chem. 1987;262:8537–8543. [PubMed] [Google Scholar]
- 12.Tatusov R L, Natale D A, Garkavtsev I V, Tatusova T A, Shankavaram U T, Rao B S, Kiryutin B, Galperin M Y, Fedorova N D, Koonin E V. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terauchi K, Ohmori K. An adenylate cyclase, cya1: regulates cell motility in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 1999;40:248–251. doi: 10.1093/oxfordjournals.pcp.a029534. [DOI] [PubMed] [Google Scholar]
- 14.Ward M J, Lew H, Zusman D R. Social motility in Myxococcus xanthus requires FrzS, a protein with an extensive coiled-coil domain. Mol Microbiol. 2000;37:1357–1371. doi: 10.1046/j.1365-2958.2000.02079.x. [DOI] [PubMed] [Google Scholar]
- 15.Weimer R M, Creighton C, Stassinopoulos A, Youderian P, Hartzell P L. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. J Bacteriol. 1998;180:5357–5368. doi: 10.1128/jb.180.20.5357-5368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S S, Wu J, Cheng Y L, Kaiser D. The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol Microbiol. 1998;29:1249–1261. doi: 10.1046/j.1365-2958.1998.01013.x. [DOI] [PubMed] [Google Scholar]