Abstract
2,4-Di-tert-butylphenol (2,4-DTBP) is an important commercial antioxidant and a toxic natural secondary metabolite that has been detected in humans. However, there is scant information regarding its toxicological effects. We asked whether 2,4-DTBP is a potential obesogen. Using a human mesenchymal stem cell adipogenesis assay, we found that exposure to 2,4-DTBP led to increased lipid accumulation and expression of adipogenic marker genes. Antagonist assays revealed that 2,4-DTBP increased lipid accumulation by activating the peroxisome proliferator-activated receptor (PPAR) γ-retinoid X receptor (RXR) heterodimer. 2,4-DTBP likely activated the PPARγ/RXRα heterodimer by activating RXRα but not directly binding to PPARγ. We confirmed that 2,4-DTBP directly bound to RXRα by solving the crystal structure of this complex, then predicted and demonstrated that related compounds could also activate RXRα. Our study demonstrated that 2,4-DTBP and related chemicals could act as obesogens and endocrine disruptors via RXRs. These data showed that 2,4-DTBP belongs to a family of compounds whose endocrine-disrupting and obesogenic effects can be strongly modulated by their chemical composition. Structure–activity studies such as the present one could help guide the rational development of safer antioxidants that do not interact with important nuclear receptors having broad effects on human development and physiology.
Keywords: endocrine disruptor, RXR, PPAR, MSC, obesogen, adipogenesis
Obesity has become a serious health problem worldwide in infants, children, adolescents, and adults (1). Approximately 13% of the world population is clinically obese (body mass index ≥ 30 kg/m2) and 39% of adults are overweight (body mass index ≥25 kg/m2) (2). Obesity increases the risk of many chronic diseases such as type 2 diabetes, heart disease, hypertension, stroke, and certain cancers (3) and elevates risk of serious illness from COVID-19. According to the World Health Organization, at least 2.8 million deaths annually are related to overweight or obesity worldwide (4). Obesity is an even greater problem in the United States where 42% of adults are currently obese (5).
Obesity is considered to be a result of the combined effects of both genetic and environmental factors (6). Traditional risk factors include lack of physical activity, excessive consumption of food and genetic susceptibility (7). However, these factors cannot fully explain the dramatic increase in the obesity rate worldwide. An increasing number of epidemiological studies together with experimental evidence in animal models indicated that some environmental chemicals could be important contributors to obesity (6). Such “obesogens” include natural chemicals, pharmaceutical chemicals, and xenobiotic chemicals that can promote obesity (8). Known obesogens include organotins, bisphenols, many agrochemicals, perfluoroalkyl substances, phthalates, and polybrominated diphenyl ethers (reviewed in 9). Obesogens can contribute to obesity by increasing white adipocyte cell number or increasing lipid storage in existing adipocytes (10). These chemicals can also lead to obesity by affecting appetite and satiety, changing basal metabolic rate, altering energy balance, or influencing gut microbiota (6). Identifying which environmental pollutants are obesogens and revealing the underlying mechanisms will contribute to ameliorating the obesity pandemic and related diseases.
2,4-Di-tert-butyl phenol (2,4-DTBP) is an important commercial antioxidant that is mainly used as an intermediate to produce other high molecular weight antioxidants and UV stabilizers for hydrocarbon-based products and plastics (11). 2,4-DTBP has been detected in indoor dust (12), river sediment (13), wastewater, and sludge (14). 2,4-DTBP is also a natural toxic secondary metabolite produced by bacteria, fungi, plants, and animals (15). Human exposure can result from ingestion of dust (12), packaged food intake because tris(2,4-di-tert-butylphenol) phosphite in food packaging materials can break down to 2,4-DTBP (16), and intake from foods containing 2,4-DTBP (17). A recent study analyzing synthetic phenolic antioxidants (SPAs) in humans detected 2,4-DTBP in urine samples from the United States (18). 2,4-DTBP was detected in 100% of investigated human urine samples and was the predominant congener, contributing 88.2% to total target concentrations of SPAs in the urine samples. The concentration of total 2,4-DTBP (7.3-130 ng/mL [0.03 μM-0.63 μM]; geometric mean, 25.8 ng/mL; 0.12 μM) was much higher than 2,6-di-tert-butyl-4-methylphenol (0.85 ng/mL, 3.8 nM) and 3-tert-butyl-4-hydroxyanisole (0.39 ng/mL, 2 nM), which were 2 other SPAs of concern. The relatively high levels of 2,4-DTBP found in human urine led us to investigate its possible adverse impacts.
In the present study, we assessed the capability of 2,4-DTBP to induce adipogenesis in human multipotent mesenchymal stromal stem cells (a.k.a. mesenchymal stem cells or MSCs) and explored its mechanism of action. We found that 2,4-DTBP, but not its structural analog 2,6-di-tert-butylphenol (2,6-DTBP), which is also used industrially as a UV stabilizer and antioxidant for hydrocarbon-based products (11), induced adipogenesis and expression of white adipocyte marker genes in MSCs. We used receptor selective agonist and antagonist studies to link 2,4-DTBP exposure with activation of the peroxisome proliferator activated receptor gamma (PPARγ) and its heterodimeric partner, the retinoid X receptor (RXR) to promote adipogenesis in MSCs. Reporter gene assays revealed that 2,4-DTBP, but not 2,6-DTBP, activated RXRα, the RXRα subunit of the PPARγ/RXRα heterodimer and RXRα in the context of 2 other nuclear receptor complexes: the liver X receptor alpha (LXRα)/RXRα heterodimer and the thyroid hormone receptor β (TRβ)/RXRα heterodimer. We solved the crystal structure of 2,4-DTBP complexed with RXRα and identified the mechanistic basis underlying binding of 2,4-DTBP to RXRα and activation of RXRα by 2,4-DTBP. This analysis suggested that other related chemicals such as 2,4,6-tri-tert-butyl phenol and 1,3,5-tri-tert-butylbenzene would activate RXRα whereas 2,6-DTBP and 1,3-di-tert-butylbenzene would not. Reporter gene assays confirmed these predictions. Overall, our studies identified 2,4-DTBP as an RXRα activator and therefore a potential endocrine disrupting chemical (EDC) and obesogen.
Materials and Methods
Chemicals and Reagents
Dexamethasone, isobutylmethylxanthine, Nile red, Hoechst 33342, HX531, LG100268 (LG), 2,4-di-tert-butylphenol (2,4-DTBP), 2,6-di-tert-butylphenol (2,6-DTBP), 2,4,6-tri-tert-butylphenol (2,4,6-TTBP), 1,3-di-tert-butylbenzene (1,3-DTBB), 1,3,5-tri-tert-butylbenzene (1,3,5-TTBB), LG100268, GW3065, T3, and TTNPB were purchased from Sigma-Aldrich. CD3254 was purchased from Tocris Bioscience. Rosiglitazone (ROSI) was purchased from Cayman Chemical Company. T0070907 was from Enzo Life Sciences. Dimethylsulfoxide (DMSO) was purchased from Thermo Fisher Scientific.
Human MSC Adipogenesis Assay
Human bone marrow MSCs were purchased from PromoCell at passage 2 (lot 429Z001-Female). Human MSCs were cultured in alpha Modification of Eagle’s minimal essential medium (αMEM) (Thermo Fisher Scientific) supplemented with 15% fetal bovine serum (FBS) (Gemini Bio-Products), 2% penicillin, 5000 IU/mL-streptomycin 5000 μg/mL (Corning), and 10 μM HEPES (Fisher Chemical). The cells were split when the confluency reached 90% and used at passage 6 for adipogenesis assays. A total of 40 000 cells per well were seeded in a 24-well plate or 80 000 cells per well were seeded in a 12-well plate. When cells reached 100% confluency, media were replaced with differentiation media (αMEM, 15% FBS supplemented with adipogenic induction cocktail [500 μM isobutylmethylxanthine, 1 μM dexamethasone, 5 μg/mL human recombinant insulin]) and ligands. Specific ligands (dissolved in DMSO) were administered twice weekly for 2 weeks. Antagonist assays were performed similarly, except that T0070907 was added twice daily (to 10 μM). DMSO concentrations in the medium were identical between vehicle controls and test chemicals and never exceeded 0.1%. At the end of each assay, cells in the 24-well plates were fixed in 3.7% formaldehyde in phosphate-buffered saline for 30 minutes at 25 °C for lipid staining; cells grown in 12-well plates were homogenized and total RNA extracted for gene expression analysis.
Gene Expression Analysis
Total RNA was isolated from cells using Trizol reagent as recommended by the manufacturer (Invitrogen Life Technologies). Reverse transcription and quantitative real-time reverse transcriptase polymerase chain reaction were performed using Transcriptor reverse transcriptase and Sybr Green Master Mix (Roche Diagnostics Corp., Indianapolis, IN). The following genes were examined: FABP4 (fatty acid binding protein 4), FSP27 (fat-specific protein of 27 kDa), PPARγ2, and LPL (lipoprotein lipase). The details of the experiment and sequences of primers used for quantitative real-time reverse transcriptase polymerase chain reaction were described in our previous studies (19, 20).
Transient Transfection and Luciferase Reporter Gene Assays
The CMX-GAL4 effector plasmids using pCMX-GAL4 DNA-binding domain (DBD) fused to nuclear receptor ligand-binding domains (LBDs) for human PPARγ (pCMX-GAL4-PPARγ), human RXRα (pCMX-GAL4-RXRα), human LXRα (pCMX-GAL4-LXRα), and human TRβ (pCMX-GAL4-TRβ) were previously described (21-23). Transient transfections were performed in COS-7 cells (ATCC CRL-1,651TM) as described (20). Briefly, COS-7 cells were seeded at 15 000 cells/well in 96-well tissue culture plates in Dulbecco’s modified Eagle’s medium (HyClone) supplemented with 10% calf bovine serum (Atlanta Biologicals). Twenty-four hours later, cells were transfected in Opti-MEM reduced-serum medium (Invitrogen Life Technologies, Grand Island, NY) when they reached ∼90% confluency (∼24 hours after seeding). Cells were transfected with 1 μg of CMX-GAL4 effector plasmids (0.3 μg for pCMX-GAL4-PPARγ plasmid) with or without 1 μg of pCMX-LBD of RXRα (L-RXRα) (0.3 μg of pCMX-L-RXRα for pCMX-GAL4-PPARγ plasmid) together with 5 μg of tk-(MH100)4-luciferase reporter and 5 μg of CMX-β-galactosidase transfection control plasmids in Opti-MEM using Lipofectamine 2000 reagent (Invitrogen Life Technologies, Grand Island, NY), following the manufacturer’s recommended protocol. After 24 hours of incubation, the medium was replaced with phenol red–free Dulbecco’s modified Eagle’s medium (Invitrogen Life Technologies)/10% resin charcoal-stripped FBS plus ligands at concentrations indicated in the figure legends for an additional 24 hours before luciferase and β-galactosidase assays. 2,4-DTBP and 2,6-DTBP were tested from 1.2 μM to 40 μM with 2-fold dilutions. No noticeable cytotoxicity was produced at 40 μM, as judged by reduction of β-galactosidase activity (data not shown). The ligands were dissolved in DMSO. The DMSO concentrations in the medium were identical between vehicle controls and test chemicals and never exceeded 0.1%. Twenty-four hours after adding the ligands to the media, cells were lysed in 165 μL of lysis buffer and allowed to shake for 30 minutes at 25 °C. For luciferase assay, 35 μL of the lysate from each well was transferred to a well in a clean, nontreated, white, flat-bottom, polystyrene, 96-well plate (Costar). Additionally, 100 μL of freshly made luciferase solution was added to each well with the cell lysate, and that plate was shaken for 30 seconds at room temperature. Plates were placed in a Dynatech ML3000 luminometer, and data were acquired with luminometer ML3000 (version 3.07) software. For β-galactosidase assays, 35 μL of the cell lysate were transferred to a clear, flat-bottom, 96-well plate (Thermo Fisher Scientific) and 100 μL of β-galactosidase solution added to each well. Plates were shaken for 30 seconds, then incubated at 25 °C for 15 minutes. Absorbance was measured at 405 nm on a Versamax microplate reader (Molecular Devices) using SOFTmaxPRO 4.0 software. Each luciferase read was normalized with the corresponding β-galactosidase read coming from the same transfection well and multiplied by the number of minutes the β-galactosidase plate was incubated (usually 15 minutes). Data are reported as relative light units (Luc/Gal). The median effective concentration and maximum activations were calculated using GraphPad Prism 9.0 (GraphPad Software, Inc.). Transfections were performed in triplicate and reproduced in multiple experiments.
Statistical Analysis
The data are expressed as the mean value ± standard error of the mean (SEM) (n = 3). Unpaired t-test was used to determine the significance of effects elicited by the chemicals relative to the control group in human MSC adipogenesis assays and to determine the significance of differences comparing +T0070907 vs −T0070907 or +HX531 vs −HX531 with each other. In the transfection assays, the significance of the experimental data was analyzed using 1-way analysis of variance, followed by a least significant difference multiple comparisons test. A P value of <.05 was considered to be statistically significant. Statistical analysis used GraphPad Prism 9.0 (GraphPad Software Inc., San Diego, CA).
Retinoic Acid Receptor β Reporter Cell Line
The HELN retinoic acid receptor (RAR) β cell line was already described (24). Briefly, we firstly generated the HELN cell line by transfecting HeLa cells with the p-ERE-βGlob-Luc-SVNeo plasmid. Secondly, we transfected HELN cells with pRARβ(ERαDBD)-puro plasmid. pERE-βGlob-Luc-SVNeo contains a luciferase gene driven by an estrogen receptor binding site in front of the β-globin promoter and a neomycin phosphotransferase gene under the control of the SV40 promoter. In pRARβ(ERαDBD), the encoded chimeric RARβ protein contains the DBD of the estrogen receptor α. Puromycin N-acetyl transferase selection marker expression confers resistance to puromycin.
To measure the activity of the chemicals, HELN RARβ cells were seeded at a density of 40 000 cells per well in 96-well white opaque tissue culture plates (Dutscher, Brumath, France). Compounds were added 8 hours later alone in presence of the pan RAR-agonist TTNPB 100 nM or the pan RXR-agonist CD3254 100 nM, and cells were incubated for 16 hours. At the end of incubation, the culture medium was replaced by a medium containing 0.3 mM luciferin. Luciferase activity was measured for 2 seconds in intact living cells using a microBeta Wallac luminometer (Perkin Elmer, Villebon sur Yvette, France). Tests were performed in quadruplicate in at least 3 independent experiments and data were expressed as mean ± SEM. Results are expressed in percentage of the maximal activity obtained in presence of TTNP 100 nM. Dose–response curves were fitted using the sigmoid dose–response function of a graphics and statistics software package (Graph-Pad Prism, version 4, 2003, Graph-Pad Software Inc., San Diego, CA, USA).
Crystallographic Analysis
The histidine-tagged LBD of human RXRα (residues 223-462 in a pET15b vector) was expressed in Escherichia coli BL21(DE3). Cells were grown in ZYM-5052 autoinduction medium supplemented with 100 µg/mL ampicillin. After 24 hours of incubation at 25 °C, cell cultures were harvested by centrifugation at 8000g for 20 minutes. The cell pellet from 2 L of culture was resuspended in 250 mL of buffer A (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 5 mM imidazole) supplemented with 100 µg/mL of lysozyme and a protease inhibitor cocktail (Complete, mini, EDTA-free tablet, Roche Applied Science) then incubated with shaking for 30 minutes at 4 °C. The suspension was lysed by sonication and centrifuged at 35 000g for 30 minutes at 4 °C. The supernatant was loaded onto a 5-mL Ni2+ affinity column (HiTrap chelating column, GE healthcare), pre-equilibrated with buffer A, using the Äkta pure system (GE Healthcare). The column was washed with 10 volumes of buffer A. Protein was eluted in 0 to 500 mM imidazole gradient using 25 column volumes of the buffer. The fractions containing RXRα LBD were pooled and incubated overnight at 4 °C with thrombin to remove the histidine tag. The protein was further purified using a Superdex 75 26/60 gel filtration column (GE healthcare) pre-equilibrated with buffer C (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 5% glycerol, 5 mM dithiothreitol).
5wFractions containing the purified receptor were pooled, mixed with 1:3 molar ratio of 2,4-DTBP and 1:2 molar ratio of the TIF2 coactivator peptide and concentrated to 8 mg/mL. Crystals were obtained by vapor diffusion at 293 °K by mixing 1 µL of protein with 2 µL of precipitant solution. The well buffer contained 20% polyethylene glycol 3350 and 0.2 M sodium formate. A single crystal was mounted from the mother liquor onto a cryoloop (Hampton Research, Aliso Viejo, CA, USA), soaked in the reservoir solution supplemented with 20% glycerol and 40 nM 2,4-DTBP and then frozen in liquid nitrogen. Diffraction data were collected on the ID30A-1 at the European Synchrotron Radiation Facility (Grenoble, France). Data were processed and scaled with XDS (25) and Scala (26). Crystals belong to space group P212121. The structure was refined using REFMAC5 (27) and COOT (28). Figures were generated with PyMol (https://pymol.org/2/). Structural data were deposited in the Worldwide Protein Data Bank with accession number 7NKE (29). Data collection and refinement statistics can be found elsewhere (Table S1 (30)).
Results
2,4-DTBP Increased Adipogenesis in Human MSCs
MSCs have been widely used to study the obesogenic properties of environmental chemicals (19, 20, 31, 32). We tested the potential obesogenic properties of 2,4-DTBP and 2,6-DTBP using a human MSC adipogenesis assay. The selective PPARγ agonist, ROSI was used as a positive control, DMSO vehicle (0.1% v/v) was the negative control. Treatment of MSCs with adipogenesis cocktail supplemented with 500 nM ROSI for 14 days elicited a significant and substantial increase in lipid accumulation, as measured by Nile Red fluorescence normalized to cellular DNA content measured by Hoechst 33342 fluorescence. After treatment with 10 μM 2,4-DTBP for 14 days, adipogenesis was increased significantly whereas lower doses of 2,4-DTBP or all doses of 2,6-DTBP did not (Fig. 1A). Induction of adipogenesis by LG100268 was similar to that of 2,4-DTBP (Fig. 1A).
Figure 1.
Effect of ROSI, LG100268, 2,4-DTBP, and 2,6-DTBP on lipid accumulation and adipogenic marker gene expression in human MSCs. Cells were treated with 500 nM ROSI, 100 nM LG100268, or different concentrations of 2,4-DTBP or 2,6-DTBP as indicated. Three replicate wells were included for each group in a 24-well plate for lipid accumulation assays or in 12-well plates for adipogenic marker gene expression assay. (A) effects of chemical exposure on adipogenesis in hMSCs. Data are expressed as lipid accumulation normalized to cellular DNA content. (B-E) effects of chemical exposure on expression of adipogenic markers in hMSCs. Data are expressed as fold induction over vehicle control. Error bars represent the standard error of the mean for 3 replicates. *P ≤ .05, **P ≤ .01, ***P ≤ .001 compared with cells treated with 0.1% DMSO vehicle control.
We confirmed the adipogenic effects by evaluating the expression of white adipocyte marker genes in response to chemical exposure. Levels of mRNAs encoding FABP4, FSP27, LPL, and PPARγ-2 were significantly increased in the 500 nM ROSI–treated cells compared with vehicle (DMSO) (Fig. 1B and 1E). Levels of FSP27, LPL, FABP4, and PPARγ2 mRNAs were increased in cells treated with 10 μM 2,4-DTBP compared with 2,6-DTBP and vehicle control, both of which were inactive.
2,4-DTBP-Induced Adipogenesis Was Blocked by the PPARγ Antagonist T0070907 or the RXRα Antagonist HX531
PPARγ plays a key role in regulating the transcription of genes involved in adipogenesis in MSCs (33). Therefore, we explored whether antagonizing PPARγ activation with the specific antagonist, T0070907 affected 2,4-DTBP-mediated increases in lipid accumulation in human MSCs. We found that cotreatment with 10 μM T0070907 (34) significantly (P < .05) inhibited lipid accumulation induced by treatment with 100 nM ROSI or by 10 μM 2,4-DTBP (Fig. 2A).
Figure 2.
Study of the inhibitory effects of PPARγ antagonist T0070907 and RXRα antagonist HX531 on lipid accumulation induced by 2,4-DTBP in human MSCs. (A) Cells were treated with 100 nM ROSI or 10 μM 2,4-DTBP in the absence or presence of 10 μM T0070907. (B) Cells were treated with 100 nM LG100268 or 10 μM 2,4-DTBP in the absence or presence of 1 μM HX531. Data are expressed as lipid accumulation normalized to cellular DNA content. Error bars represent the standard error of the mean for 3 replicates. *P ≤ .05, **P ≤ .01, ***P ≤ .001 compared with cell samples treated with 0.1% DMSO vehicle.10 μM T0070907 or 1 μM HX531.
PPARγ regulates the transcription of adipogenic genes as a heterodimer with RXRs (33). To assess the extent of RXR involvement in 2,4-DTBP-induced lipid accumulation, we conducted an RXR antagonist assay by comparing the effects of 2,4-DTBP in the presence or absence of the specific antagonist HX531 (35). Cotreatment with 1 μM HX531 significantly (P < .05) inhibited lipid accumulation induced by 100 nM of the specific RXR agonist, LG100268 (22), and by 10 μM 2,4-DTBP (Fig. 2B). Taken together, these results implicate the PPARγ-RXR heterodimer in 2,4-DTBP-induced adipogenesis, but do not identify which partner is responsible.
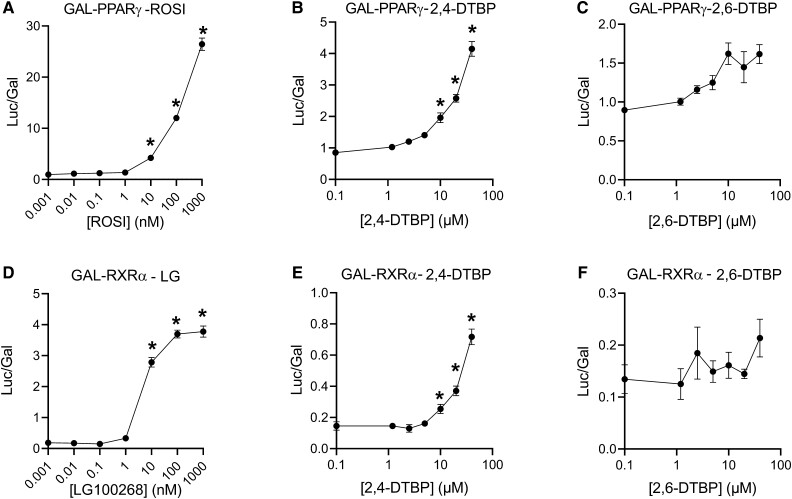
2,4-DTBP Showed Agonistic Activity Toward GAL4-PPARγ and GAL4-RXRα
We next assessed whether 2,4-DTBP could activate human PPARγ or RXRα using transient transfection in COS-7 cells with GAL4-receptor chimeras. ROSI enhanced the activity of a GAL4-PPARγ in a dose-dependent manner with a lowest effective concentration (LOEC) value of ∼10 nM (Fig. 3A). Similarly, 2,4-DTBP enhanced GAL4-PPARγ activation in a dose-dependent manner with a LOEC value of ∼10 μM (Fig. 3B). 2,6-DTBP did not activate GAL4-PPARγ between 1.2 and 40 μM (Fig. 3C).
Figure 3.
The effect of ROSI, LG100268, 2,4-DTBP, and 2,6-DTBP on GAL4-PPARγ- and GAL4-RXRα-regulated luciferase activity in COS-7 cells. (A-C) GAL4-PPARγ or (D-F) GAL4-RXRα were transfected into COS-7 cells. Plasmid transfected cells were treated with different concentrations of chemicals for 24 hours. Three replicated wells were included for each group in a 96-well plate. Data are reported as relative light units (Luc/Gal). Each luciferase read was normalized with the corresponding β-galactosidase read coming from the same transfection well and multiplied by the number of minutes the β-galactosidase plate was incubated (15 minutes). Error bars represent the standard error of the mean for 3 replicates. *P ≤ .05, compared with cell samples treated with 0.1% DMSO vehicle.
Interestingly, 2,4-DTBP also showed agonistic activity toward human RXRα. Treatment with LG100268 strongly induced activity by GAL4-RXRα in transient transfection assays also with a LOEC of ∼10 nM (Fig. 3D). 2,4-DTBP also enhanced GAL4-RXRα activity with a LOEC value of ∼10 μM (Fig. 3E). 2,6-DTBP was inactive (Fig. 3F). Taken together, these results confirm that 2,4-DTBP appeared to have agonistic activity toward both PPARγ and RXRα.
RXRα Mediated the Agonistic Activity of 2,4-DTBP Toward PPARγ
We next asked whether 2,4-DTBP could interfere with the ability of ROSI to activate GAL4-PPARγ as would be expected by a bona fide PPARγ ligand. We tested the ability of different concentrations of 2,4-DTBP to affect activation of GAL4-PPARγ by 500 nM ROSI and found no significant increase or decrease in activity (Fig. 4A), suggesting that 2,4-DTBP and ROSI did not compete for the same binding site in PPARγ. Since 2,4-DTBP did not further activate GAL4-PPARγ, we inferred that it does not directly interact with PPARγ.
Figure 4.
The effect of LG100268 and 2,4-DTBP on GAL4-PPARγ- or GAL4-PPARγ/L-RXRα-regulated luciferase activity in COS-7 cells. (A-C) ROSI interference assays evaluating the ability chemicals to interfere with ROSI-induced reporter activity on GAL4-PPARγ (A), or GAL4-PPARγ/L-RXRα-induced reporter activity (B, C). (D-F) Agonist assays evaluating the effects of chemicals on ability of GAL4-PPARγ (D), or GAL4-PPARγ/L-RXRα (E, F) to elicit luciferase reporter activity. The concentration of ROSI were 500 and 25 nM in the GAL4-PPARγ- and GAL4-PPARγ/L-RXRα-regulated luciferase reporter gene assay, respectively. Transfected cells were treated with indicated concentrations of chemicals for 24 hours in triplicate in 96-well plates. Data are reported as relative light units (Luc/Gal). Error bars represent the standard error of the mean for 3 replicates. *P ≤ .05, compared with cell samples treated with 0.1% DMSO vehicle. (D-F) or with 500 nM ROSI (A) or 25 nM ROSI (B, C).
Since PPARγ/RXR is a permissive heterodimer, activation of RXR in the heterodimer could lead to activation of the complex or further enhance the activity of the agonist-bound PPARγ (23). Because COS-7 cells express endogenous RXRα (36), it is possible that there is some interaction between endogenous full-length RXRα and the GAL4-PPARγ heterodimer in the transient transfection assays. Since 2,4-DTBP had agonistic activity toward RXRα (Fig. 3E), we hypothesized that 2,4-DTBP might activate GAL4-PPARγ via a heterodimer with endogenous RXRs. In support of this hypothesis, we found that LG100268 could also elicit modest (∼2-fold) activation of GAL4-PPARγ (Fig. 4D).
To further explore the involvement of RXRα in activation of the PPARγ/RXRα heterodimer by 2,4-DTBP, we cotransfected COS-7 cells with 2 plasmids: 1 expressing GAL4-PPARγ and the other expressing only the LBD of RXRα (L-RXRα). L-RXRα lacks the ability to bind DNA and must exert any observed actions via its heterodimeric partner (23). Cotransfection with L-RXRα (Fig 4E) strongly potentiated the ability of LG100268 to activate GAL4-PPARγ (compare Fig. 4E with Fig. 4D). Similarly, 2,4-DTBP was now able to induce robust activation of the GAL4-PPARγ-L-RXRα heterodimer at 1 μM (Fig. 4F). Both LG100268 (Fig 4B) and 2,4-DTBP (Fig 4C) potentiated reporter gene activity induced by 25 nM ROSI in cells cotransfected with GAL4-PPARγ and L-RXRα, although LG100268 potentiation was only significant at 10 nM and higher. We inferred that 2,4-DTBP action on the PPARγ-RXR heterodimer is mediated through RXR since expression of exogenous RXRα LBD potentiated action of 2,4-DTBP and the RXR-selective agonist LG100268 on GAL4-PPARγ.
2,4-DTBP Activated GAL4-LXRα and GAL4-TRβ Through RXRα in the Heterodimers
In addition to PPARγ, RXRα also heterodimerizes with many other nuclear receptors, including LXR, TR, RAR, constitutive androstane receptor, farnesoid X receptors, pregnane X receptor, and vitamin D receptor (37, 38). Since we found that 2,4-DTBP activation of RXRα in the heterodimers could affect the function of PPARγ, we next asked whether 2,4-DTBP could also modulate the function of other nuclear receptors by activating RXRα in the heterodimers.
To test this hypothesis, we evaluated the effects of 2,4-DTBP on human GAL4-LXRα and GAL4-TRβ. The results obtained for GAL4-LXRα were similar to those shown for GAL4-PPARγ in Fig. 4. In COS-7 cells transfected with GAL4-LXRα alone, LG100268 (Fig. S1A) and 2,4-DTBP (Fig. S1B) increased reporter gene activity significantly (30). This increase was strongly potentiated by cotransfection with L-RXRα (Fig. S1E and S1F (30)). Neither LG100268 (Fig. S1C) nor 2,4-DTBP (Fig. S1D) (30) showed any interference with activation of GAL4-LXRα by its specific agonist GW3965 (39). When cells were additionally cotransfected with L-RXRα, activation by GW3965 was potentiated by LG100268 (Fig. S1G) and 2,4-DTBP (Fig. S1H) (30). We inferred that LG100268 and 2,4-DTBP activated GAL4-LXRα heterodimer via its RXRα component, rather than LXRα directly.
We next tested the effects of these chemicals on TRβ and observed that LG100268 (Fig. S2A, C) and 2,4-DTBP (Fig. S2B, D) neither activated nor interfered with activation of GAL4-TRβ by T3 (30). However, cotransfection with L-RXRα allowed LG100268 (Fig. S2E, G) and 2,4-DTBP (Fig. S2F, H) to activate GAL4-TRβ in the absence (Fig. S2E, F) or presence (Fig. S2G, H) of its specific ligand, T3 (30). These results further supported a model in which 2,4-DTBP alters the function of nuclear receptor heterodimers via activation of the RXRα partner.
Crystallographic Analysis of the Binding of 2,4-DTBP to RXRα
Prototypical synthetic or natural rexinoids (the subclass of retinoids targeting RXRα such as 9-cis-retinoic acid) contain a carboxylic head group involved in a salt bridge with an arginine residue and a long aliphatic/aromatic chain making numerous van der Waals contacts with essentially non-polar residues distributed all over the ligand-binding pocket (LBP) of RXRα. 2,4-DTBP neither structurally nor chemically resembles classical rexinoids. To gain structure-based insight into its mode of action, we solved the crystal structure of the RXRα LBD (hereafter RXR) bound to 2,4-DTBP and a short LxxLL-containing peptide derived from the transcriptional intermediary factor 2 (TIF2), at 2.35 Å resolution (Table S1(30)). The structure reveals a RXR homodimer with 2,4-DTBP and the coactivator peptide bound to both protomers (Fig. 5A). Each subunit displayed the canonical active conformation with the C-terminal helix H12 (also termed AF-2 or activation helix) capping the LBP, and the 2,4-DTBP could be precisely placed in its electron density. Interestingly, the density map allowed the unambiguous identification of 2 alternative orientations of 2,4-DTBP that can be reached through rotation by 180° around the hydroxyl axis (Fig. 5A, inset).
Figure 5.
Structural basis for RXR interaction with and activation by 2,4-DTBP. (A) Overall structure of RXR LBD homodimer in complex with 2,4-DTBP (yellow sticks) and TIF2-NR2 peptide (orange). The activation helix H12 is highlighted in red. The inset shows the 2Fo-Fc electron density map of 2,4-DTBP. (B) Close-up view of the ligand-binding pocket of RXR bound to 2,4-DTBP. Key RXR residues in contact with the tert-butyl moieties at positions 2, 2′ and 4 are depicted as green and blue sticks, respectively. (C) Superposition of the 2,4-DTBP- and CD3254-bound RXR structures (PDB code 3E94). As a prototypical rexinoid, CD3254 (magenta sticks) contains an acidic head group and a long aliphatic/aromatic chain bridging arginine 316 (H5) on 1 side and H11 on the other. (D) Superposition of the 2,4-DTBP- and TBT-bound RXR structures (PDB code 3FUG) showing that the binding sites of the 2 compounds overlap. TBT is displayed as orange sticks.
2,4-DTBP bound between helices H3 and H11, and in close proximity to C432. While not directly in contact with the AF-2 helix, the tert-butyl group at position 2 (and the alternative 2′ site) plays an important role in stabilizing key residues of helices H3, H5 and H11 to which helix H12 docks in the transcriptionally active conformation (green residues in Fig. 5B). On the other hand, the tert-butyl at position 4 makes extensive van der Waals contacts with hydrophobic residues deeply buried in the LBP (Fig. 5B, residues are blue). These interactions are critical for the binding affinity of 2,4-DTBP as well as for its activity, by maintaining the tert-butyl group at position 2 in close proximity to residues involved in receptor activation. The hydroxyl moiety of 2,4-DTBP appears to be involved in a hydrogen bond with the main chain carbonyl group of C432.
Comparison of the present structure with that of RXRα bound to the reference synthetic agonist CD3254 (40) shows that 2,4-DTBP occupies only a small fraction of the LBP, but that interactions with residues crucial for stabilization of the receptor-active form are preserved in both complexes (Fig. 5C). Interestingly, similar observations were previously made with the potent RXR environmental agonist tributyltin (41). Indeed, superposition of the 2,4-DTBP- and TBT-bound RXRα structures revealed that the binding sites of the 2 compounds largely overlap (Fig. 5D). Overall, it appears that, as for TBT, although 2,4-DTBP interacts with only a subset of binding pocket residues in the H11 region, it is engaged in enough essential contacts to stabilize RXRα in its active conformation. However, the high-affinity binding of TBT is ensured by a covalent bond linking the metal atom of the organotin and cysteine residue 432 in helix H11 (masked by the ligands in Fig. 5D). This covalent bond does not exist for 2,4-DTBP, which, as a consequence, binds much less avidly than TBT to RXRα.
Structure–Activity Analysis of 2,4-DTBP and Congeners on RXRα Based on Stably Transfected HELN RARβ Reporter Cells
The crystallographic data revealed the structural basis for the agonistic activity of 2,4-DTBP toward RXRα and could allow a better understanding of the binding and activation properties of its congeners. We next investigated the activity of 2,4-DTBP and closely related compounds toward RXRα/RARβ heterodimer by using a stably transfected HELN RARβ reporter cell line in which both RXR and RAR agonist are able to induce luciferase expression. We monitored the agonist potential of 2,4-DTBP and 4 related compounds (Fig. S3A (30)), namely the 2,4,6-tri-tert-butylphenol bearing an additional tert-butyl group at position 6 (2,4,6-TTBP), the 2,6-DTBP containing 2 tert-butyl groups in ortho positions, and the 2 corresponding benzene derivatives lacking the hydroxyl moiety, the 1,3-DTBB and 1,3,5-tri-tert-butylbenzene (1,3,5-TTBB). We found that 2,4,6-TTBP is, by far, the most potent RXRα agonist, followed by 1,3,5-TTBB and 2,4-DTBP, while 1,3-DTBB and 2,6-DTBP are inactive (Fig. S3A (30)). To assess the specific effects of 2,4-DTBP, 2,4,6-TTBP, and 1,3,5-TTBB, HELN RARβ cells were coincubated with saturating concentrations of the RAR and RXR agonists CD3254 or TTNPB. 2,4-DTBP, 2,4,6-TTBP, and 1,3,5-TTBB further activated the TTNPB-saturated RXR-RARβ heterodimer (Fig. S3B (30)). However, 2,4-DTBP, 2,4,6-TTBP, and 1,3,5-TTBB appear unable to act in conjunction with CD3254 to enhance the activity of RXR-RARβ (Fig. S3C (30)). These transactivation experiments confirmed the ability of 2,4-DTBP, 2,4,6-TTBP, and 1,3,5-TTBB to activate the RXRα-RARβ heterodimer through RXRα. These activity profiles are in full agreement with our structural analysis and confirm that the tert-butyl groups at positions 2, 4 and 6 (2′ site in the 2,4-DTBP complex structure) are important for compound affinity and activity. With 3 tert-butyl groups, 2,4,6-TTBP displays the best potency and efficacy values followed by 1,3,5-TTBB differing from the latter by the lack of the hydroxyl moiety. The specific role of the tert-butyl at position 4 suggested by the crystal structure was also confirmed by cellular assays since the 2,6-DTBP containing only 2 tert-butyl groups on both sides of the hydroxyl moiety shows no detectable activity whereas with 2 tert-butyl groups in ortho and para positions, 2,4-DTBP acts an effective RXRα activator.
Discussion
2,4-DTBP has been detected in urine samples from the general US population at high concentrations (18); however, information about its toxicity is very limited. Since 2,4-DTBP is widely used as an antioxidant in plastics, is known to interact with nuclear hormone receptors (42), and has been detected in humans, we asked whether 2,4-DTBP is a potential obesogen and, if so, what is its underlying mechanism of action.
We used an established MSC assay (19, 20, 22, 34, 43) to evaluate the potential of 2,4- and 2,6-DTBP for obesogenic activity. We found that 2,4-DTBP exposure induced lipid accumulation in human MSCs with a corresponding increase in the induction of marker genes for white adipocyte differentiation (Fig. 1). In contrast, 2.6-DTBP was inactive in these assays, indicating that 2,4-DTBP is a potential obesogen. We further found that 2,4-DTBP activated the PPARγ-RXRα heterodimer—the so-called “master regulator” of adipogenesis (33). Activation of PPARγ is a well-established predictor of adipogenic activity in vivo, and many chemicals have been shown to promote adipogenesis by activating PPARγ (reviewed in (31, 32)). We used specific PPARγ and RXRα antagonist assays to confirm the requirement for this heterodimer in promoting adipogenesis (Fig. 2). Transient transfection assays suggested that 2,4-DTBP specifically modulated activation of this heterodimer (Fig. 3).
It is well known that the PPARγ/RXRα heterodimer is permissive because it can be activated by ligands for either partner (38). Using a series of GAL4-receptor LBD chimeric receptors and cotransfection with the RXRα LBD, we showed that 2,4-DTBP specifically activated the RXRα subunit of the PPARγ-RXR, LXRα-RXRα, and TRβ-RXR heterodimers (Figs. 3 and 4; Figs S1, S2 (30)). Previously, studies showed that activating the RXRα in the PPARγ/RXRα heterodimer played an important role in mediating the obesogenic effect of some environmental chemicals. For example, we found that the agrochemical fludioxonil induced adipogenesis in 3T3-L1 preadipocytes and in MSCs via the PPARγ/RXRα heterodimer by activating RXRα but not PPARγ (20). Moreover, we demonstrated that signaling through the RXRα subunit of the PPARγ/RXRα heterodimer was critical to commit MSCs to the adipocyte lineage (22). Combining the results of human MSC adipogenesis assay and receptor activation assays, we inferred that 2,4-DTBP is a potential obesogen that acts via RXRα in the context of the PPARγ/RXRα heterodimer.
Unlike the PPARγ/RXRα heterodimer, TRβ/RXRα was thought to be a nonpermissive heterodimer (23). However, RXRα can act as a “nonsilent” partner of TR in particular cellular environments (high ratio of coactivator to corepressor), allowing activation of TR signaling by RXR agonists (44, 45). The well-studied environmental obesogen TBT can act as an RXRα agonist to potentiate TR-regulated gene expression and resultant morphological programs directed by thyroid hormone signaling in vitro and in vivo (46). Our studies showed that 2,4-DTBP activated the TRβ/RXRα heterodimer by activating RXRα. Overall, our results demonstrated that the 2,4-DTBP activated the RXRα component of PPARγ/RXRα, LXRα/RXRα, and TRβ/RXR heterodimers.
We next solved the crystal structure of 2,4-DTBP bound to RXRα and identified its specific mode of binding (Fig. 5). The structure explained the RXR binding and activation properties of 2,4-DTBP and allowed us to predict the activity of various congeners. Notably, we predicted that 2,4,6-TTBP would be a more potent activator of RXR, and receptor activation assays confirmed this prediction (Fig. S3 (30)). In a recent study, Kodama et al explored the structure–activity relationship of 1,3-bis-tert-butyl monocyclic benzene derivatives for RXR activation by in vitro and in silico analyses. Their results also indicated that addition of a bulky substituent at the 6-position of 2,4-DTBP is favorable for agonistic activity toward human RXR (47).
Because 2,4-DTBP received little attention previously, limited information was available about its toxicity. 2,4-DTBP elicited hepatic toxicity (related to centrilobular hypertrophy of hepatocytes which resulted in heavier livers) and renal toxicity (tubular basophilia and an increased incidence of proteinaceous and granular casts) in rats after oral administration at 300 mg/kg/day for 28 days (48). 2,4-DTBP also increased cholesterol and phospholipid in female rats at the dose of 300 mg/kg/day. Susceptibility of newborn rats to 2,4-DTBP was 4 to 5 times higher than that of young rats. In a 1-generation fertility and repeated dose toxicity study, parental generation rats were administrated 2,4-DTBP orally for 4 weeks before mating and throughout mating, gestation, and lactation. No treatment-related gross lesions or impairment of reproductive capability were found in the parental generation animals treated with 2,4-DTBP at up to 300 mg/kg/day. However, dietary administration of 2,4-DTBP at 300 mg/kg daily for 13 weeks to F1 generation rats elicited a toxic effect on the growth rate, which was secondary to reduced diet palatability (49). Based on several in vitro assays, 2,4-DTBP was shown to be a possible EDC. 2,4-DTBP bound to the rainbow trout estrogen receptor (50). Using an effect-directed analysis-based approach to identify EDCs in multicontaminated river sediment, 2,4-DTBP was identified as a new environmental human estrogen receptor ligand by using gas chromatography coupled with mass spectrometry (13).
The molecular mechanisms underlying these observations were unclear. Our data showing that 2,4-DTBP disrupted nuclear receptor function via RXRα and previous studies linking 2,4-DTBP to estrogen receptor signaling (13) provided potential clues to identifying mechanisms underlying these toxic effects. There is considerable evidence linking EDCs and obesity and other health problems (10, 51, 52). Many EDCs exert their effects by disrupting the functions of nuclear receptors including PPARs, RXRα, TRs, LXRs, and RARs (10, 53). PPARγ, RXRs, LXRα, TRβ, and RARs play important roles in many physiological and pathological processes such as development, differentiation, and growth (54). The ability of 2,4-DTBP and related chemicals to activate RXRα and RXRα heterodimers implicates it as an EDC. Comprehensive in vivo and in vitro toxicology studies are warranted to evaluate the potential effects of 2,4-DTBP and related chemicals.
2,4-DTBP concentrations in human urine samples were in the range of 7.3 to 130 ng/mL (0.03 μM-0.63 μM). 2,4-DTBP promoted adipogenesis in human MSCs at concentrations of ∼10 μM, which is somewhat higher, but not so far from the concentrations observed in limited studies of human urine. Receptor activation studies showed that 2,4-DTBP and 2,4,6-TTBP activated RXRs in the low micromolar to submicromolar range, which is an environmentally relevant level according to the limited human biomonitoring data (18). Considering the ability of tert butyl phenols to target RXRα in the context of multiple receptor heterodimers, as well as their ability to activate ERS, additional large-scale surveys on the burden to the human body of 2,4-DTBP are warranted.
Taken together, the data presented here demonstrated that 2,4-DTBP belongs to a family of compounds whose potential endocrine-disrupting and obesogenic effects can be strongly modulated by their chemical composition. That the United States, and presumably other populations, can be exposed to endocrine active chemicals such as DTBPs in the absence of specific knowledge of these effects by regulatory agencies means that the population is vulnerable to such chemicals and that current regulatory approaches are not providing protection from their endocrine-disrupting effects. Structure–activity studies such as the present one could help the rational development of safer antioxidants.
Acknowledgments
We thank Dr. Jane Muncke (Food Packaging Forum Foundation) for suggesting 2,4-DTBP as a compound of interest. We acknowledge experimental assistance from the staff of the European Synchrotron Radiation Facility (Grenoble, France) during crystallographic data collection.
Glossary
Abbreviations
- DBD
DNA-binding domain
- DMSO
dimethylsulfoxide
- DTBB
di-tert-butylbenzene
- DTBP
di-tert-butyl phenol
- EDC
endocrine disrupting chemical
- FABP4
fatty acid binding protein 4
- FBS
fetal bovine serum
- FSP27
fat-specific protein of 27 kDa
- LBD
ligand-binding domain
- LBP
ligand-binding pocket
- LOEC
lowest effective concentration
- LPL
lipoprotein lipase
- LXR
liver “X” receptor
- MSC
multipotent mesenchymal stromal stem cell
- PPAR
peroxisome proliferator activated receptor
- PPARγ2
peroxisome proliferator activated receptor gamma 2
- RAR
retinoic acid receptor
- ROSI
rosiglitazone
- RXR
retinoid “X” receptor
- SPA
synthetic phenolic antioxidant
- TIF2
transcriptional intermediary factor 2
- TR
thyroid hormone receptor
- TTBB
tri-tert-butylbenzene
- TTBP
tri-tert-butyl-phenol
Contributor Information
Xiao-Min Ren, Department of Developmental and Cell Biology, University of California, Irvine, CA 92697-2300, USA; Faculty of Environmental Science and Engineering, Kunming University of Science and Technology, Kunming 650500, China.
Richard C Chang, Department of Developmental and Cell Biology, University of California, Irvine, CA 92697-2300, USA.
Yikai Huang, Department of Developmental and Cell Biology, University of California, Irvine, CA 92697-2300, USA.
Angélica Amorim Amato, Department of Developmental and Cell Biology, University of California, Irvine, CA 92697-2300, USA.
Coralie Carivenc, Centre de Biologie Structurale, Université de Montpellier, CNRS, Inserm, Montpellier, France.
Marina Grimaldi, Institut de Recherche en Cancérologie de Montpellier (IRCM), Inserm U1194, Université Montpellier, Institut régional du Cancer de Montpellier (ICM), Montpellier, France.
Yun Kuo, Department of Developmental and Cell Biology, University of California, Irvine, CA 92697-2300, USA.
Patrick Balaguer, Institut de Recherche en Cancérologie de Montpellier (IRCM), Inserm U1194, Université Montpellier, Institut régional du Cancer de Montpellier (ICM), Montpellier, France.
William Bourguet, Centre de Biologie Structurale, Université de Montpellier, CNRS, Inserm, Montpellier, France.
Bruce Blumberg, Department of Developmental and Cell Biology, University of California, Irvine, CA 92697-2300, USA; Department of Pharmaceutical Sciences, University of California, Irvine, CA 92697-2300, USA; Department of Biomedical Engineering, University of California, Irvine, CA 92697-2300, USA.
Funding
Supported by grants from the National Institutes of Health (ES023316, ES031139) to B.B., by the European Union's Horizon 2020 research and innovation program under grant agreement GOLIATH (825489) to P.B., W.B., and B.B., by the ANSES TOXCHEM (2018/1/020) to P.B. and W.B., by the European Union's Horizon 2021 research and innovation PARC (101057014) programs to P.B. and W.B., and by a grant from China Scholarship Council to X.R. This work was supported by the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INBS-05.
Author Contributions
X.-M.R., R.C.C., Y.H., Y.K., M.G., C.C., and W.B. performed experiments. Xi.-M.R., R.C.C., W.B., PB., and B.B. designed experiments and interpreted data. X.-M.R., W.B., P.B., and B.B. wrote the manuscript. W.B., P.B., and B.B. acquired funding and supervised the experiments. All authors have given approval to the final version of the manuscript.
Disclosures
B.B. is a named inventor on US patents 5,861,274, 6,200,802, 6,815,168, and 7,250,273 related to PPARγ. All other authors declare they have nothing to disclose.
Data Availability
Original data generated and analyzed during this study are included in this published article or in the data repositories listed in References. The atomic coordinates and structure factors have been deposited in the PDB under accession code 7NKE (29) and data collection and refinement statistics can be found in Table S1 (30). Supplemental Figures S1-S3 and Table S1 can be found online at Figshare (30).
References
- 1. Hales CM, Fryar CD, Carroll MD, Freedman DS, Ogden CL. Trends in obesity and severe obesity prevalence in US youth and adults by sex and age, 2007-2008 to 2015-2016. JAMA. 2018;319(16):1723–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. WHO . Obesity and overweight; 2018. Accessed August 16, 2022. https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight.
- 3. Picon-Ruiz M, Morata-Tarifa C, Valle-Goffin JJ, Friedman ER, Slingerland JM. Obesity and adverse breast cancer risk and outcome: mechanistic insights and strategies for intervention. CA Cancer J Clin. 2017;67(5):378–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. WHO . Obesity and Overweight Fact Sheet No. 311. World Health Organization, 2015. [Google Scholar]
- 5. Stierman B, Afful J, Carroll MD, et al. National health and nutrition examination survey 2017–March 2020 prepandemic data files development of files and prevalence estimates for selected health outcomes. National Health Statistics Reports (NHSR No. 158). 10.15620/cdc:106273 [DOI]
- 6. Heindel JJ, Blumberg B. Environmental obesogens: mechanisms and controversies. Annu Rev Pharmacol Toxicol. 2019;59(1):89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turcot V, Lu Y, Highland HM, et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat Genet. 2018;50(1):26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Janesick AS, Blumberg B. Obesogens: an emerging threat to public health. Am J Obstet Gynecol. 2016;214(5):559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heindel JJ, Howard S, Agay-Shay K, et al. Obesity II: establishing causal links between chemical exposures and obesity. Biochem Pharmacol. 2022;199:115015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heindel JJ, Blumberg B, Cave M, et al. Metabolism disrupting chemicals and metabolic disorders. Reprod Toxicol. 2017;68:3–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. ChemView . United States Environmental Protection Agency; 2016. Accessed August 22, 2022. https://chemview.epa.gov/chemview
- 12. Liu R, Lin Y, Ruan T, Jiang G. Occurrence of synthetic phenolic antioxidants and transformation products in urban and rural indoor dust. Environ Pollut. 2017;221:227–233. [DOI] [PubMed] [Google Scholar]
- 13. Creusot N, Budzinski H, Balaguer P, Kinani S, Porcher JM, Aït-Aïssa S. Effect-directed analysis of endocrine-disrupting compounds in multi-contaminated sediment: identification of novel ligands of estrogen and pregnane X receptors. Anal Bioanal Chem. 2013;405(8):2553–2566. [DOI] [PubMed] [Google Scholar]
- 14. Sebok A, Vasanits-Zsigrai A, Helenkár A, Záray G, Molnár-Perl I. Multiresidue analysis of pollutants as their trimethylsilyl derivatives, by gas chromatography-mass spectrometry. J Chromatogr A. 2009;20(12):2288–2301. [DOI] [PubMed] [Google Scholar]
- 15. Zhao F, Wang P, Lucardi RD, Su Z, Li S. Natural sources and bioactivities of 2,4-Di-tert-butylphenol and its analogs. Toxins (Basel). 2020;12(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang Y, Hu C, Zhong H, Chen X, Chen R, Yam KL. Effects of ultraviolet (UV) on degradation of irgafos 168 and migration of its degradation products from polypropylene films. J Agric Food Chem. 2016;64(41):7866–7873. [DOI] [PubMed] [Google Scholar]
- 17. Choi SJ, Kim JK, Kim HK, et al. 2,4-Di-tert-butylphenol From sweet potato protects against oxidative stress in PC12 cells and in mice. J Med Food. 2013;16(11):977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu R, Mabury SA. Unexpectedly high concentrations of 2,4-di-tert-butylphenol in human urine. Environ Pollut. 2019;252(Pt B):1423–1428. [DOI] [PubMed] [Google Scholar]
- 19. Chamorro-García R, Kirchner S, Li X, et al. Bisphenol A diglycidyl ether induces adipogenic differentiation of multipotent stromal stem cells through a peroxisome proliferator-activated receptor gamma-independent mechanism. Environ Health Perspect. 2012;120(7):984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Janesick AS, Dimastrogiovanni G, Vanek L, et al. On the utility of ToxCast™ and ToxPi as methods for identifying new obesogens. Environ Health Perspect. 2016;124(8):1214–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grün F, Watanabe H, Zamanian Z, et al. Endocrine-disrupting organotin compounds are potent inducers of adipogenesis in vertebrates. Mol Endocrinol. 2006;20(9):2141–2155. [DOI] [PubMed] [Google Scholar]
- 22. Shoucri BM, Martinez ES, Abreo TJ, et al. Retinoid X receptor activation alters the chromatin landscape to commit mesenchymal stem cells to the adipose lineage. Endocrinology. 2017;158(10):3109–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Forman BM, Umesono K, Chen J, Evans RM. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81(4):541–550. [DOI] [PubMed] [Google Scholar]
- 24. Balaguer P, Boussioux AM, Demirpence E, Nicolas JC. Reporter cell lines are useful tools for monitoring biological activity of nuclear receptor ligands. Luminescence. 2001;16(2):153–158. [DOI] [PubMed] [Google Scholar]
- 25. Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(2):125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(1):72–82. [DOI] [PubMed] [Google Scholar]
- 27. Murshudov GN, Skubak P, Lebedev AA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(4):355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(12):2126–2132. [DOI] [PubMed] [Google Scholar]
- 29. Carivenc C, Bourguet W. Crystal structure of human RXRalpha ligand binding domain in complex with 2,4-Di-tert-butylphenol and a coactivator fragment. March 2, 2022 ed. Worldwide PDB Protein Data Bank. March 2, 2022. 10.2210/pdb7nke/pdb [DOI]
- 30. Ren X-M, Chang RC, Huang Y, et al. 2,4-Di-tert-butylphenol induces adipogenesis in human mesenchymal stem cells by activating retinoid X Receptors. Figshare. Deposited January 18, 2023. 10.6084/m9.figshare.21919368 [DOI] [PMC free article] [PubMed]
- 31. Kassotis CD, Vom Saal FS, Babin PJ, et al. Obesity III: obesogen assays: limitations, strengths, and new directions. Biochem Pharmacol. 2022;199:115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mohajer N, Du CY, Checkcinco C, Blumberg B. Obesogens: how they are identified and molecular mechanisms underlying their action. Front Endocrinol (Lausanne). 2021;12:780888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77(1):289–312. [DOI] [PubMed] [Google Scholar]
- 34. Kirchner S, Kieu T, Chow C, Casey S, Blumberg B. Prenatal exposure to the environmental obesogen tributyltin predisposes multipotent stem cells to become adipocytes. Mol Endocrinol. 2010;24(3):526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ebisawa M, Umemiya H, Ohta K, et al. Retinoid X receptor-antagonistic diazepinylbenzoic acids. Chem Pharm Bull (Tokyo). 1999;47(12):1778–1786. [DOI] [PubMed] [Google Scholar]
- 36. Kahlen JP, Carlberg C. Allosteric interaction of the 1alpha,25-dihydroxyvitamin D3 receptor and the retinoid X receptor on DNA. Nucleic Acids Res. 1997;25(21):4307–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81(3):1269–1304. [DOI] [PubMed] [Google Scholar]
- 38. Evans RM, Mangelsdorf DJ. Nuclear receptors, RXR, and the big bang. Cell. 2014;157(1):255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Collins JL, Fivush AM, Watson MA, et al. Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J Med Chem. 2002;45(10):1963–1966. [DOI] [PubMed] [Google Scholar]
- 40. Santin EP, Germain P, Quillard F, et al. Modulating retinoid X receptor with a series of (E)-3-[4-hydroxy-3-(3-alkoxy-5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-y l)phenyl]acrylic acids and their 4-alkoxy isomers. J Med Chem. 2009;52(10):3150–3158. [DOI] [PubMed] [Google Scholar]
- 41. le Maire A, Grimaldi M, Roecklin D, et al. Activation of RXR-PPAR heterodimers by organotin environmental endocrine disruptors. EMBO Rep. 2009;10(4):367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Creusot N, Tapie N, Piccini B, et al. Distribution of steroid- and dioxin-like activities between sediments, POCIS and SPMD in a French river subject to mixed pressures. Environ Sci Pollut Res Int. 2013;20(5):2784–2794. [DOI] [PubMed] [Google Scholar]
- 43. Li X, Pham HT, Janesick AS, Blumberg B. Triflumizole is an obesogen in mice that acts through peroxisome proliferator activated receptor gamma (PPARγ). Environ Health Perspect. 2012;120(12):1720–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Castillo AI, Sánchez-Martínez R, Moreno JL, Martínez-Iglesias OA, Palacios D, Aranda A. A permissive retinoid X receptor/thyroid hormone receptor heterodimer allows stimulation of prolactin gene transcription by thyroid hormone and 9-cis-retinoic acid. Mol Cell Biol. 2004;24(2):502–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fattori J, Campos JL, Doratioto TR, et al. RXR Agonist modulates TR: corepressor dissociation upon 9-cis retinoic acid treatment. Mol Endocrinol. 2015;29(2):258–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mengeling BJ, Murk AJ, Furlow JD. Trialkyltin rexinoid-X receptor agonists selectively potentiate thyroid hormone induced programs of Xenopus laevis metamorphosis. Endocrinology. 2016;157(7):2712–2723. [DOI] [PubMed] [Google Scholar]
- 47. Kodama S, Matsumoto S, Takamura Y, et al. Structural characterization of 1,3-bis-tert-butyl monocyclic benzene derivatives with agonistic activity towards retinoid X receptor alpha. Toxicol Lett. 2022;373:76–83. [DOI] [PubMed] [Google Scholar]
- 48. Hirata-Koizumi M, Hamamura M, Furukawa H, et al. Elevated susceptibility of newborn as compared with young rats to 2-tert-butylphenol and 2,4-di-tert-butylphenol toxicity. Congenit Anom. 2005;45(4):146–153. [DOI] [PubMed] [Google Scholar]
- 49. ECHA . Registration profile for 2,4-di-tert butylphenol; 2018. Accessed August 22, 2022. Available from, as of July 27, 2018: https://echa.europa.eu/registration-dossier/-/registered-dossier/14828/14827/14821.
- 50. Tollefsen KE, Julie Nilsen A. Binding of alkylphenols and alkylated non-phenolics to rainbow trout (Oncorhynchus mykiss) hepatic estrogen receptors. Ecotoxicol Environ Saf. 2008;69(2):163–172. [DOI] [PubMed] [Google Scholar]
- 51. Gore AC, Chappell VA, Fenton SE, et al. EDC-2: the endocrine Society's Second scientific statement on endocrine-disrupting chemicals. Endocr Rev. 2015;36(6):E1–E150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Papalou O, Kandaraki EA, Papadakis G, Diamanti-Kandarakis E. Endocrine disrupting chemicals: an occult mediator of metabolic disease. Front Endocrinol. 2019;10:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hall JM, Greco CW. Perturbation of nuclear hormone receptors by endocrine disrupting chemicals: mechanisms and pathological consequences of exposure. Cells. 2019;9(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yen PM. Classical nuclear hormone receptor activity as a mediator of complex biological responses: a look at health and disease. Best Pract Res Clin Endocrinol Metab. 2015;29(4):517–528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original data generated and analyzed during this study are included in this published article or in the data repositories listed in References. The atomic coordinates and structure factors have been deposited in the PDB under accession code 7NKE (29) and data collection and refinement statistics can be found in Table S1 (30). Supplemental Figures S1-S3 and Table S1 can be found online at Figshare (30).