Abstract
Organ transplantation has been linked to certain gene polymorphisms. The effect of gene polymorphisms–associated organ transplantation gene on infection, on the other hand, is yet unknown. The research studying the link between the CTLA-4 rs5742909, rs733618, rs4553808, rs231775, and polymorphisms of the organ transplantation gene and infection were found in PubMed, Web of Science, Scopus, and Embase, and the published articles from 2012 to 2020 were gathered. For the best estimation of the intended results, a random-effects model was used in this meta-analysis. In this study, 1,567 studies were initially included and 9 eligible studies were eligible for further analyses. A significant correlation between CTLA4+49 [A/G-231775 odds ratio (OR) = 077, 0.59–0.95] and CTLA4 [rs5742909TT OR: 0.09, 0.27–0.45] gene polymorphism with infection in organ transplantation was observed. Also, no significant association was found between other CTLA4 gene polymorphisms with infection in organ transplantation. Further studies involving gene–gene and gene–diet interactions should be conducted to investigate this association with infection.
Keywords: CTLA-4, transplantation, polymorphism
Introduction
Polymorphisms in the cytotoxic T-lymphocyte antigen-4 (CTLA4) gene, which may influence CTLA4’s role in regulating the immune response, have been postulated to affect disease susceptibility and chronicity in individuals with HBV infection; however, the results are still contentious. CD28, like CTLA4, binds to B7 family receptors (CD80 and CD86), but after being produced on T cells, it gives a positive costimulatory signal for T-cell proliferation1,2. CTLA4 (cytotoxic T lymphocyte-associated antigen 4) is an important negative regulator of the T cell-mediated immune response and a vital component in the immune system’s induction of immunological tolerance3. It is also produced constitutively on the surface of regulatory T cells (Tregs); it may be detected on roughly 50% of Tregs but just 1% of naive helper T cells4. In mice, ligation of CTLA4 on Tregs leads to a considerable reduction in antigen-presenting cell presentation capability and effector T cell downregulation5. CTLA4 plays a critical function in the immune response’s downregulation.
In 172 chronic HBV-infected individuals, Duan and colleagues looked at the CTLA-4 49A/G and 318 T/C polymorphisms. In chronic HBV-infected individuals, the AA genotype and A allele of the CTLA-4 49A/G polymorphisms, as well as the genotype CC of the CTLA-4 -318 C/T polymorphisms, were observed more often6. Thio and colleagues discovered a link between the CTLA-4 49A/G gene and HBV infection clearance7. The rs231775 (+49A/G) single nucleotide polymorphism (SNP) is found inside the molecule’s signal peptide and affects full-length isoform expression on the T cell surface. The rs3087243 (+6230G/A) SNP is discovered within the 39 untranslated regions of the CTLA-4 gene and has been linked to autoimmune disease susceptibility8. Furthermore, CTLA4 SNPs like 21772T/C (rs733618), +49A/G (rs231775), and +6230 G/A (rs3087243) have a role in graft rejection and the long-term clinical outcome of organ transplantation9,10–14. It has been proposed that the CTLA4 gene variant influences infection following juvenile heart transplantation. The SNP CTLA4 +49(rs231775) has been linked to late post-transplantation viral infection in pediatric heart transplant patients in the United States, according to Ohmann et al. The relevance of CTLA4 SNPs in T cell-mediated immunity and their connection with infection following renal transplantation is uncertain. As a result, the goal of this study was to look at the links between infection and five CTLA4 SNPs (rs733618 C/T, rs4553808 A/G, rs5742909 C/T, rs231775 A/G, and rs3087243 G/A) in Chinese kidney transplant recipients.
The SNP CTLA4 +49(rs231775) has been linked to late post-transplantation viral infection in pediatric heart transplant patients in the United States, according to Ohmann et al.15 Several studies have found that the CC genotype is related to viral illness and persistent HBV infection for the SNP rs5742909 C/T16. The SNPs rs5742909 and rs231775, on the other hand, did not show any statistically significant relationships in the research. This lack of correlation might be attributed to the small sample size and insufficient power to detect a link; also, the frequency of the G allele at the CTLA4 +49 (rs231775) locus is substantially greater in the Chinese population than in other populations17. This might suggest that genetic bias has little impact on infection susceptibility. The CTLA4+49(rs231775)GG genotype was also linked to enhanced interferon-c production following immunological activation, according to recent research.
Several SNPs in the CTLA4 gene may affect gene expression, leading to amino acid substitution and mRNA splicing, which might influence T-cell activation and, eventually, host immunological state. These genetic polymorphisms have been linked to post-transplant infections15,18, and they might lead to interindividual variances in immunotherapy targeting this protein. There are numerous polymorphic markers in the CTLA4 gene. The most commonly examined SNPs in the CTLA4 3′ untranslated region (UTR) are at locations 1722, 1661, 1147, 658, 318, + 49, and + 6230, as well as dinucleotide (AT)n repeats13,14,19,20. Cytomegalovirus (CMV) infection is controlled in part by innate and adaptive immune responses. CTLA4 is a critical component of both the innate and adaptive immune systems. CTLA4 is a cell surface molecule that is only found on CD4+ and CD8+ T lymphocytes. CD4+ T cells have been found to be able to reduce initial systemic CMV infection21, limit persistent replication in specific organs22, and boost antibody responses23 in experimental CMV infection models. CD8+ cells, on the other hand, can protect immunocompromised people and animals from CMV infection by T cells by limiting the viral reactivation from a state of delay24,25.
Materials and Methods
Search Strategy
A literature search was conducted using combinations of the keywords “CTLA-4 or cytotoxic T-lymphocyte antigen-4 associated AND polymorphism or SNP or single nucleotide polymorphisms or rs5742909 or rs733618 or rs4553808 or rs231775 or genotype AND organ Transplantation or Transplantation AND viral infection or bacterial infection” in PubMed, Google Scholar, Web of Science, EMBASE, Google Scholar, Wanfang, China National Knowledge Infrastructure (CNKI), Islamic World Science Citation Center (ISC), and Scientific Information Database (SID) databases. All research that looked at the link between organ transplantation gene polymorphisms and infection risk was gathered. The literature search has no language restrictions. To find possibly relevant papers, the reference lists of the eligible studies, reviews, and prior meta-analysis publications were manually examined. All procedures were carried out in accordance with the 1975 Helsinki Protocol and its subsequent amendments. It was also approved by the Ethics Committee of Shiraz University of Medical Sciences.
Inclusion and Exclusion Criteria
The following studies were included in the study: The study described the link between rs5742909, rs733618, rs4553808, and rs231775 polymorphisms and organ transplantation risk, using a case–control design in which the cases were organ transplantation patients and the controls were healthy participants and offered odds ratios (ORs) and 95% confidential intervals (CIs). Studies with no control group, letters to the editor, editorials, opinion pieces, animal studies, case reports, and case series were excluded.
Data Extraction
I searched the databases separately and retrieved data from the papers that were included, using a uniform procedure. Each study’s first author’s last name, year of publication, race of origin, sample size (cases/controls), frequency of genotypes in both case and control samples, ethnicity, mean age, mean OT, source of controls (population-based or hospital-based), odds ratios (OR), and 95% confidence intervals for homozygote, heterozygote, recessive, and dominant models were collected.
Quality Assessment
The source of the control group, ethnicity, odds ratios (OR), 95% confidence intervals among the controls, and sample size were all evaluated as methodological components that may bias the link between organ transplantation gene polymorphism and obesity risk. Due to the short number of studies included, the effect sizes were computed using both random-effect models and the DerSimonian–Laird approach. Q and I2 techniques were used to analyze study heterogeneity. Significant heterogeneity was defined as an I2 value of more than 50% at a significance level of 0.1. The Egger’s test and contour-enhanced funnel plot were used to examine publication bias and small study impact.
Result
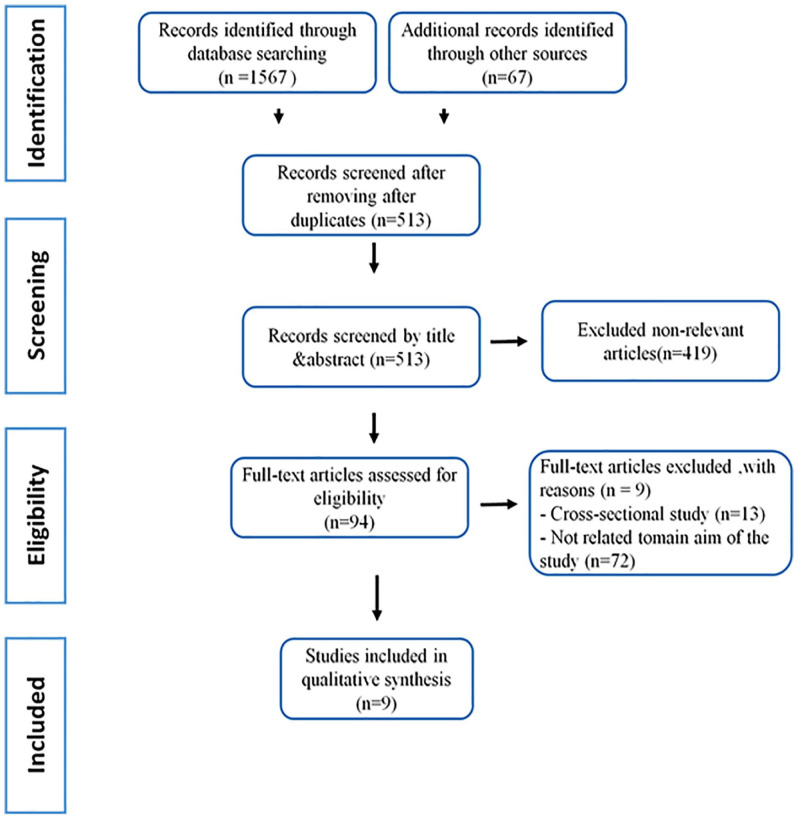
A total of 1,567 studies were found to be suitable after extensive database searches. In total, 513 publications were omitted due to duplicated records, 912 papers were excluded following the primary screening, and 154 papers were excluded after full-text evaluation after screening the collected studies based on inclusion criteria for the current meta-analysis. The methodological technique for including research and obtaining important data from eligible papers are depicted in Fig. 1. Finally, nine studies found an association between viral infection and rs5742909, rs733618, rs4553808, and rs231775 polymorphisms in organ transplantation.
Figure 1.
Flow chart of the literature search strategy and study selection.
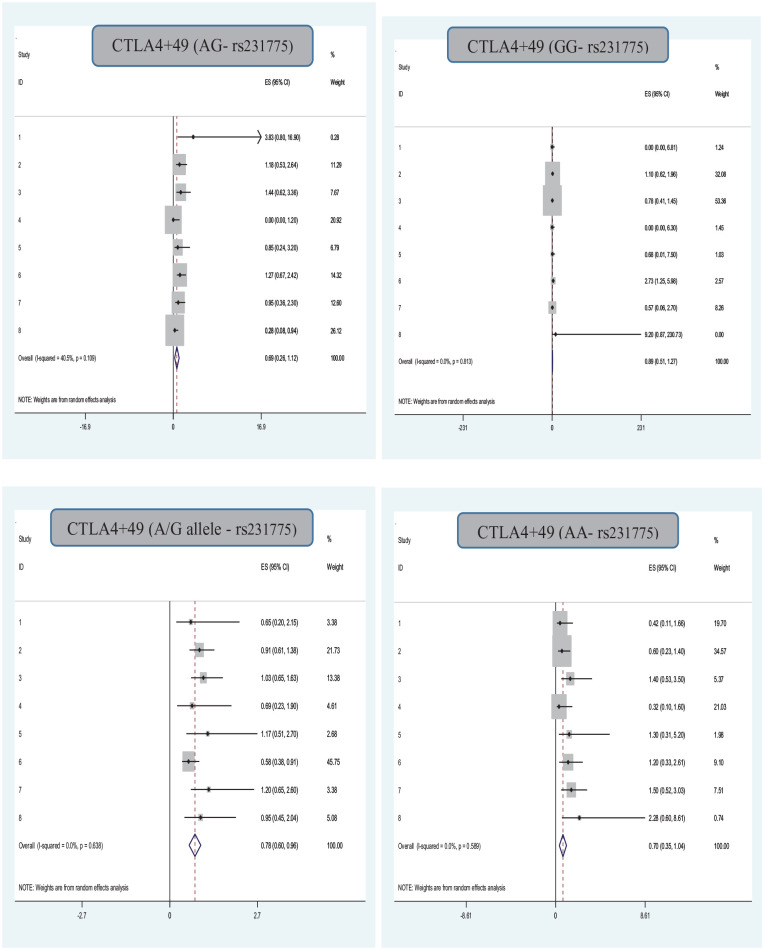
Association of the CTLA4+49 (A/G- 231775) Gene Polymorphism With Infection in Organ Transplantation
The association between rs231775 gene polymorphism and infection in organ transplantation risk was investigated in nine qualified studies, using a random-effects DerSimonian–Laird approach, and no significant correlation between the dominant, recessive, heterozygous, homozygous, infection in organ transplantation risk was observed. Also, a significant correlation between the allelic with infection in organ transplantation risk was observed (Table 1 and Fig. 2). The results also indicated that there was no significant evidence of publication bias determined by assessing the included studies based on the Egger’s tests for the allele (P Egger’s test = 0.33), CTLA-4(AA-P Egger’s test = 0.75), CTLA-4(Ag-P P Egger’s test = 0.88), and CTLA-4(gg P Egger’s test = 0.54) models. The publication bias determined by assessing the included studies based on Egger’s tests for the CTLA-4(AG-P Egger’s test = 0.04) (Fig. 3).
Table 1.
The Pooled Results of CTLA4+49 (A/G-231775) Gene Polymorphism, Using Random-Effects and DerSimonian–Laird Approach Model.
| DL-POOLED ES | OR | 95% CI | %Weight | Overall (I2) | P |
|---|---|---|---|---|---|
| CTLA4(231775)AG | 0.68 | 0.25–1.11 | 100.0 | 40.5 | 0.10 |
| CTLA4(231775)GG | 0.89 | 0.51–1.27 | 100.0 | 0.0 | 0.81 |
| CTLA4(231775)AA | 0.69 | 0.35–1.04 | 100.0 | 0.0 | 0.58 |
| CTLA4(231775)A allele | 0.77 | 0.59–0.95 | 100.0 | 0.0 | 0.63 |
OR: odds ratio; CI: confidence interval.
Figure 2.
Association of the CTLA4 (rs231775) gene polymorphism with infection in organ transplantation.
Figure 3.
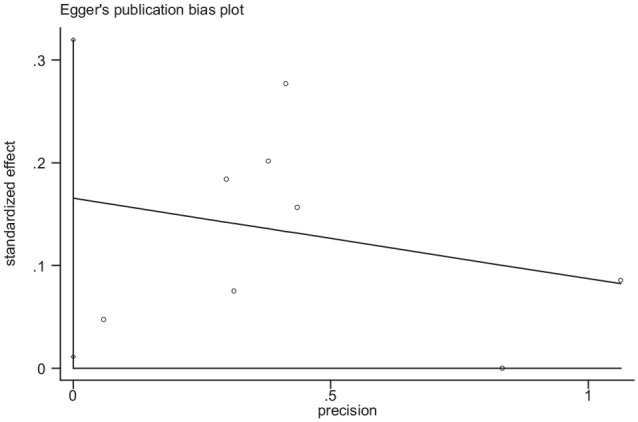
Funnel plot for publication bias in the meta-analysis of CTLA-4(rs231775-AG) gene polymorphism.
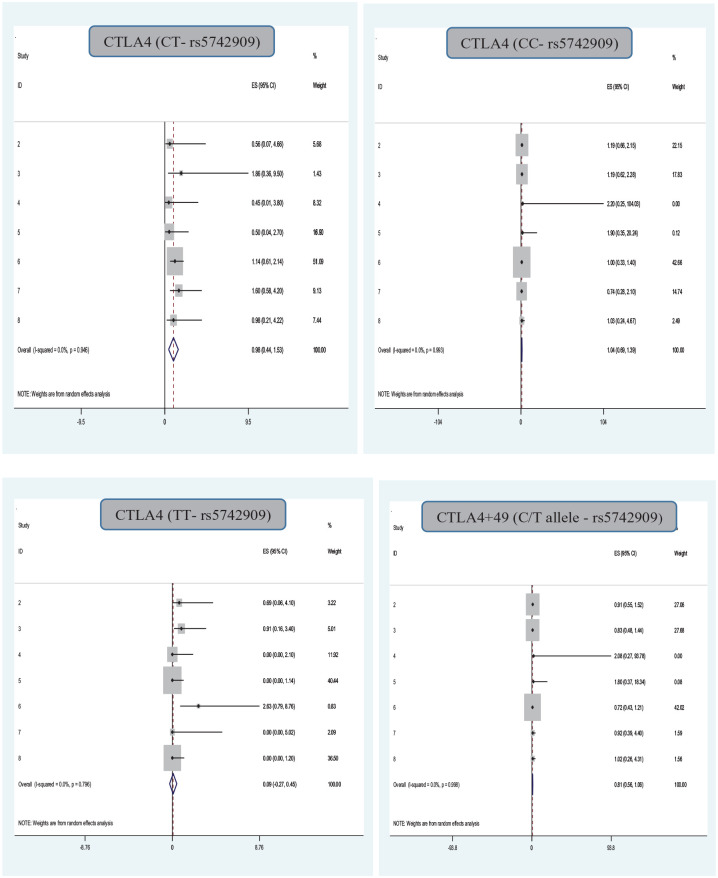
Association of the CTLA4 (318- rs5742909) Gene Polymorphism With Infection in Organ Transplantation
The association between rs5742909 gene polymorphism and infection in organ transplantation risk was investigated in nine qualified studies, using a random-effects DerSimonian–Laird approach, and no significant correlation between the dominant, recessive, heterozygous, infection in organ transplantation risk was observed. Also, a significant correlation between the CTLA4(rs5742909)TT gene polymorphisms with infection in organ transplantation risk were observed (Table 2 and Fig. 4). The results also indicated that there was no significant evidence of publication bias determined by assessing across the included studies based on the Egger’s tests for the allele (P Egger’s test = 0.37), CTLA-4(CC-P Egger’s test = 0.81), CTLA-4(CT-P P Egger’s test = 0.62), and CTLA-4(TT P Egger’s test = 0.54) models.
Table 2.
The Pooled Results of CTLA4 (318-5742909) Gene Polymorphism, Using Random-Effects and DerSimonian–Laird Approach Model.
| DL-POOLED ES | OR | 95% CI | %Weight | Overall (I2) | P |
|---|---|---|---|---|---|
| CTLA4(rs5742909)CT | 0.98 | 0.43–1.52 | 100.0 | 0.0 | 0.94 |
| CTLA4(rs5742909)TT | 0.09 | 0.27–0.45 | 100.0 | 0.0 | 0.79 |
| CTLA4(rs5742909)CC | 1.04 | 0.69–1.38 | 100.0 | 0.0 | 0.99 |
| CTLA4(rs5742909)C allele | 0.81 | 0.55–1.06 | 100.0 | 0.0 | 0.99 |
OR: odds ratio; CI: confidence interval.
Figure 4.
Association of the CTLA4(rs5742909) gene polymorphism with infection in organ transplantation.
Association of the CTLA4(-1661-rs4553808) Gene Polymorphism With Infection in Organ Transplantation
The association between rs4553808 gene polymorphism and infection in organ transplantation risk was investigated in nine qualified studies, using a random-effects DerSimonian–Laird approach, and no significant correlation between the allelic, dominant, recessive, heterozygous, homozygous, infection in organ transplantation risk was observed (Table 3 and Fig. 5). The results also indicated that there was no significant evidence of publication bias determined by assessing across the included studies based on the Egger’s tests for the allele (P Egger’s test = 0.26), CTLA-4(AA-P Egger’s test = 0.46), CTLA-4(Ag-P P Egger’s test = 0.88), and CTLA-4(gg P Egger’s test = 0.87) models.
Table 3.
The Pooled Results of CTLA4(-1661-rs4553808) Gene Polymorphism, Using Random-Effects and DerSimonian–Laird Approach Model.
| DL-POOLED ES | OR | 95% CI | %Weight | Overall (I2) | P |
|---|---|---|---|---|---|
| CTLA4(4553808)AA | 0.95 | 0.60–1.29 | 100.0 | 0.0 | 0.98 |
| CTLA4(4553808)AG | 0.82 | 0.41–1.24 | 100.0 | 0.0 | 0.87 |
| CTLA4(4553808)GG | 1.16 | 0.33–1.98 | 100.0 | 0.0 | 0.90 |
| CTLA4(4553808)A allele | 0.79 | 0.58–1.007 | 100.0 | 0.0 | 0.86 |
OR: odds ratio; CI: confidence interval.
Figure 5.
Association of the CTLA4(rs4553808) gene polymorphism with infection in organ transplantation.
Association of the CTLA4 (1722-733618) Gene Polymorphism With Infection in Organ Transplantation
The association between rs733618 gene polymorphism and infection in organ transplantation risk was investigated in nine qualified studies, using a random-effects DerSimonian–Laird approach, and no significant correlation between the dominant, recessive, heterozygous, homozygous with infection in organ transplantation risk was observed. Also, a significant correlation between the CTLA4(rs5742909)TT gene polymorphisms with infection in organ transplantation risk were observed (Table 4 and Fig. 6). The results also indicated that there was no significant evidence of publication bias determined by assessing across the included studies based on the Egger’s tests for the allele (P Egger’s test = 0.95, CTLA-4(CC-P Egger’s test = 0.20), CTLA-4(CT-P P Egger’s test = 0.58), and CTLA-4(TT P Egger’s test = 0.97) models.
Table 4.
The Pooled Results of CTLA4 (1722- 733618) Gene Polymorphism, Using Random-Effects and DerSimonian–Laird Approach Model.
| DL-POOLED ES | OR | 95% CI | %Weight | Overall (I2) | P |
|---|---|---|---|---|---|
| CTLA4(rs733618)TT | 0.85 | 0.47–1.23 | 100.0 | 0.0 | 0.98 |
| CTLA4(rs733618)TC | 1.04 | 0.43–1.69 | 100.0 | 0.0 | 0.99 |
| CTLA4(rs733618)CC | 1.15 | 0.31–1.99 | 100.0 | 0.0 | 0.99 |
| CTLA4(rs733618)C allele | 0.87 | 0.59–1.15 | 100.0 | 0.0 | 0.72 |
OR: odds ratio; CI: confidence interval.
Figure 6.
Association of the CTLA4(rs733618) gene polymorphism with infection in organ transplantation.
Discussion
In this meta-analysis, a significant correlation between the CTLA4(rs5742909)TT, CTLA4(rs5742909)TT, and CTLA4+49 (A/G- 231775) allelic gene polymorphisms with infection in organ transplantation risk was observed. Also, no significant association was found between the allelic, dominant, recessive, heterozygous, and homozygous CTLA4(-1661-rs4553808) gene polymorphism with infection in organ transplantation. CTLA4 polymorphism locations have been shown to alter the outcome of human cytomegalovirus (HCMV) infection in transplant patients. Karimi et al.26 found a significant difference in genotype frequencies of CTLA4 (rs231775A>G) variants between HCMV-positive and HCMV-negative patients, indicating an association with risk for HCMV cases among allograft recipients in an Iranian population, similar to our findings among symptomatic HCMV cases in transplant recipients. In a Brazilian population, the mutant genotypes GG of CTLA4 rs231775 and rs3087243 SNPs were related to vulnerability to chronic hepatitis C infection27. In a Chinese population, however, no significant connection between CTLA4 rs231775 and rs3087243 SNPs and viral susceptibility was found at the genotypic/allelic level18. Alizadeh et al.28 and Schott et al.16 have both demonstrated that CTLA-4-318 C/T has a strong relationship, having a proclivity for HBV infection (chronic). Jiang and colleagues discovered that the GG genotype of mice CTLA-4 49A/G expression in Chinese liver transplant recipients has been linked to a lower incidence of HBV infection recurrence29. Han and his allies have demonstrated that CTLA-4 GG genotype vs CTLA-4 GG genotype tumor necrosis factor and interferon levels are lower in 49A/G30 people who have been infected with HBV for a long time. In allogeneic hematopoietic stem cell transplantation (HSCT) patients compared with autologous transplant patients, a substantial correlation between the T allele of the CTLA-4 318 T/C polymorphisms and HBV infection was detected, similar to previous results. The SNPs rs4553808 (21661A/G) and rs5742909 (2318C/T) in the CTLA4 gene promoter were identified to change the expression pattern of the protein produced by CTLA431. Similarly, the rs733618 (21772T) allele was discovered to reduce CTLA4 transcription through altering transcription factor binding32.
Environmental and/or ethnic variables probably played a role in the disparities in the results. Furthermore, because CTLA4 has such a wide geographical distribution, different SNPs may be important in different populations, and while this may not be important in identifying individual transplant patients at risk universally, it may still be relevant in identifying at-risk patients in certain populations and have an impact on pretransplant HCMV monitoring and management of these patients. In various disorders, the promoter region SNPs of CTLA4rs5742909, rs11571317, rs16840252, and rs4553808 have demonstrated no protective or susceptible effect12,14.
There is also a great role of oral health and hygiene who receive an organ transplant. The patients should be referred to a dental clinic beforehand, to remove any potential sources of oral infection33–35. Educating physicians and the patients about the importance of early dental screening and pre-transplant dental treatment is important. Careful oral examination with evaluation of the patient, including laboratory tests, ensure correct oral preparation and control of oral disease prior to liver transplantation. Patients with CTLA4+ 49(rs231775) AA and A had a greater prevalence of chronic HBV infection in Chinese Han populations, and haplotype+49A-318C was considerably overrepresented36. The +49A allele appears to increase the likelihood of developing viral and parasite illnesses (e.g., dengue fever, Chagas disease, and American cutaneous leishmaniasis) while conferring resistance against autoimmune disorders (MG, PE)37. The SNP CTLA4 +49(rs231775) has been linked to late post-transplantation viral infection in pediatric heart transplant patients in the United States, according to Ohmann et al.15 Several studies have found that the CC genotype is related to viral illness and persistent HBV infection for the SNP rs5742909 C/T16. The SNPs rs4553808 (21661A/G) and rs5742909 (2318C/T) in the CTLA4 gene promoter were identified to change the expression pattern of the protein encoded by CTLA4. Similarly, the rs733618 (21772T) allele was discovered to reduce CTLA4 transcription through altering transcription factor binding10,32. The rs231775 (+49A/G) SNP is found inside the molecule’s signal peptide and affects full-length isoform expression on the T cell surface. The rs3087243 (+6230G/A) SNP is discovered within the 39 untranslated regions of the CTLA-4 gene and has been linked to autoimmune disease susceptibility8,11. Furthermore, CTLA4 SNPs like 21772T/C (rs733618), +49A/G (rs231775), and +6230 G/A (rs3087243) have a role in graft rejection and the long-term clinical outcome of organ transplantation9–14. It has been proposed that the CTLA4 gene variant influences infection following juvenile heart transplantation. The SNP CTLA4 +49(rs231775) has been linked to late post-transplantation viral infection in pediatric heart transplant patients in the United States, according to Ohmann et al. Viral infection worsens outcomes after HSCT by reactivating latent virus or introducing primary viral infection for the first time38,39. Previously published research has linked costimulatory molecule gene polymorphisms to a range of post-HSCT clinical outcomes, including viral infections such as CMV and hepatitis viruses16,40,41. The existence of susceptible alleles at various promoter locations of the CTLA4 gene, for example, has been shown to have a substantial association with the occurrence or recurrence of GVHD42. CTLA4 gene polymorphisms, on the other hand, have a significant impact on a range of clinical outcomes associated with CMV infection. PD-1 is involved in the modulation of T cells during CMV infection43. CTLA4, also known as CD152, is a protein receptor present on the surface of T cells that downregulates the immune system, and an attack may be activated by activating the CD28 receptor on T cells44. CTLA4 is an immunoglobulin superfamily member that is related to CD2845. CTLA4 is present intracellularly in regulatory T cells and may have a role in their activity. Increased expression of CTLA4, an inhibitory receptor for B7 molecules, occurs when T cells are activated via the T cell receptor and CD28. Earlier studies have found discrepancies in the correlations between CTLA4 gene polymorphisms and viral infection susceptibility, chronicity, and clearance, particularly HBV7,28,46. The incidence of CTLA4?49 A/G, -318 C/T, -1661 A/G genotypes were observed to differ significantly between active CMV infection in rejected and non-rejected liver transplant patients in a previous study26. In another research, the AG genotype of CTLA4?49 A/G (dbSNP: rs231775) reduced T cells’ capacity to clear HBV and increased the risk of HBV-related hepatocellular cancer46. In additional studies, significant relationships between CTLA4?49 A/G, -318 C/T polymorphisms, and HBV infection chronicity and clearance were discovered28,46. Other studies have found that having the G allele and GG genotype of the CT60 polymorphism in a bone marrow transplant donor reduces the risk of high-grade aGVHD, and having this polymorphic allele and genotype at the CTLA4 leader sequence has a higher risk of chronic GVHD47,48. The GG genotype at the leader sequences and the A/G heterozygote at -1661 of the CTLA4 promoter region have also been linked to gastric cancer and oral squamous cell carcinoma49. The G allele and GG genotype of the CTLA4 -1661 A/G were considerably more common in active CMV infected allogeneic HSCT patients with low-grade aGVHD, similar to previous results50,51, also there is a significant association between GA genotype (CTLA4-1661) in a male group with GVHD than without GVHD52. In our earlier report, significant differentiation was found in the frequency of CTLA4 ?49 A/G, -318 C/T, -1661 A/G genotypes between active CMV infection in rejected and non-rejected liver transplant patients53. In our other information, the GG genotype of the cytotoxic T-lymphocyte antigen 4 +49 A/G was substantially more prevalent in transplanted individuals infected with torque teno virus, but the AG genotype was more common in those who did not have this infection, according to our findings. Furthermore, among transplanted individuals infected with the virus, the -1661 AA and GA genotypes, as well as the -318 TC genotypes, were much more common54.
Conclusion
This meta-analysis indicated that a significant correlation between CTLA4+49 (A/G- 231775) and CTLA4(rs5742909TT) gene polymorphism with infection in organ transplantation risk was observed. Further studies involving gene–gene and gene–diet interactions should be conducted to investigate this association.
Acknowledgments
We thank Dr. Ramzi (Hematology, Oncology and Bone Marrow Transplantation Department, Shiraz University of Medical Sciences, Shiraz, Iran) for providing us with technical support.
Footnotes
Author Contributions: All authors have substantially contributed to the conception and design, acquisition of data, or analysis and interpretation of data; drafted the article or revised it critically for important intellectual content; and have approved the final version to be submitted for publication.
Availability of Data and Material: Available upon request from the corresponding author.
Ethical Approval: This study was approved by our institutional review board.
Statement of Human and Animal Rights: This article does not contain any studies with human or animal subjects.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Research Ethics and Patient Consent: All authors have complied with research ethics and patient satisfaction in this project.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study was supported by a grant from Shiraz University of Medical Sciences, Shiraz, Iran.
ORCID iD: Mahdiyar Iravani Saadi  https://orcid.org/0000-0002-7292-4435
https://orcid.org/0000-0002-7292-4435
References
- 1. Sperling AI, Bluestone JA. The complexities of T-cell co-stimulation: CD28 and beyond. Immunol Rev. 1996;153:155–82. [DOI] [PubMed] [Google Scholar]
- 2. Chen L. Co-inhibitory molecules of the B7–CD28 family in the control of T-cell immunity. Nat Rev Immunol. 2004;4(5):336–47. [DOI] [PubMed] [Google Scholar]
- 3. Alegre M-L, Frauwirth KA, Thompson CB. T-cell regulation by CD28 and CTLA-4. Nat Rev Immunol. 2001;1(3):220–28. [DOI] [PubMed] [Google Scholar]
- 4. Grohmann U, Orabona C, Fallarino F, Vacca C, Calcinaro F, Falorni A, Candeloro P, Belladonna ML, Bianchi R, Fioretti MC, Puccetti P. CTLA-4–Ig regulates tryptophan catabolism in vivo. Nat Immunol. 2002;3(11):1097–101. [DOI] [PubMed] [Google Scholar]
- 5. Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322(5899):271–75. [DOI] [PubMed] [Google Scholar]
- 6. Margam VM, Coates BS, Ba MN, Sun W, Binso-Dabire CL, Baoua I, Ishiyaku MF, Shukle JT, Hellmich RL, Covas FG, Ramasamy S, et al. Geographic distribution of phylogenetically-distinct legume pod borer, Maruca vitrata (Lepidoptera: Pyraloidea: Crambidae). Mol Biol Rep. 2011;38(2):893–903. [DOI] [PubMed] [Google Scholar]
- 7. Thio CL, Mosbruger TL, Kaslow RA, Karp CL, Strathdee SA, Vlahov D, O'Brien SJ, Astemborski J, Thomas DL. Cytotoxic T-lymphocyte antigen 4 gene and recovery from hepatitis B virus infection. J Virol. 2004;78(20):11258–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ueda H, Howson JM, Esposito L, Heward J, Chamberlain G, Rainbow DB, Hunter K, Smith AN, Di Genova G, Herr MH. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423(6939):506–11. [DOI] [PubMed] [Google Scholar]
- 9. Gorgi Y, Sfar I, Abdallah TB, Abderrahim E, Ayed SJ, Aouadi H, Bardi R, Ayed K. Ctla-4 exon 1 (+ 49) and promoter (− 318) gene polymorphisms in kidney transplantation. Transplant P. 2006;38:2303–305. [DOI] [PubMed] [Google Scholar]
- 10. Wu J, Tang JL, Wu SJ, Lio HY, Yang YC. Functional polymorphism of CTLA-4 and ICOS genes in allogeneic hematopoietic stem cell transplantation. Clin Chim Acta. 2009;403(1–2):229–33. [DOI] [PubMed] [Google Scholar]
- 11. Tapirdamaz O, Pravica V, Metselaar HJ, Hansen B, Moons L, van Meurs JB, Hutchinson IV, Shaw J, Agarwal K, Adams DH, Day CP, et al. Polymorphisms in the T cell regulatory gene cytotoxic T lymphocyte antigen 4 influence the rate of acute rejection after liver transplantation. Gut. 2006;55(6):863–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Reuver P, Pravica V, Hop W, Boor P, Metselaar HJ, Hutchinson IV, Tilanus HW, Kwekkeboom J. Recipient CTLA-4+ 49 G/G genotype is associated with reduced incidence of acute rejection after liver transplantation. Am J Transplant. 2003;3(12):1587–94. [DOI] [PubMed] [Google Scholar]
- 13. Kusztal M, Kościelska-Kasprzak K, Drulis-Fajdasz D, Magott-Procelewska M, Patrzałek D, Janczak D, Chudoba P, Klinger M. The influence of CTLA-4 gene polymorphism on long-term kidney allograft function in Caucasian recipients. Transpl Immunol. 2010;23(3):121–24. [DOI] [PubMed] [Google Scholar]
- 14. Gao JW, Guo YF, Fan Y, Qiu JX, Bao ED, Liu Y, Qin Y, Zhang F. Polymorphisms in cytotoxic T lymphocyte associated antigen-4 influence the rate of acute rejection after renal transplantation in 167 Chinese recipients. Transpl Immunol. 2012;26(4):207–11. [DOI] [PubMed] [Google Scholar]
- 15. Ohmann EL, Brooks MM, Webber SA, Girnita DM, Ferrell RE, Burckart GJ, Chinnock R, Canter C, Addonizio L, Bernstein D, Kirklin JK, et al. Association of genetic polymorphisms and risk of late post-transplantation infection in pediatric heart recipients. J Heart Lung Transplant. 2010;29(12):1342–51. [DOI] [PubMed] [Google Scholar]
- 16. Schott E, Witt H, Pascu M, van Boemmel F, Weich V, Bergk A, Halangk J, Müller T, Puhl G, Wiedenmann B. Association of CTLA4 single nucleotide polymorphisms with viral but not autoimmune liver disease. Eur J Gastroenterol Hepatol. 2007;19(11):947–51. [DOI] [PubMed] [Google Scholar]
- 17. Lee CS, Lee YJ, Liu HF, Su CH, Chang SC, Wang BR, Chen TL, Liu TL. Association of CTLA4 gene AG polymorphism with rheumatoid arthritis in Chinese. Clin Rheumatol. 2003;22(3):221–24. [DOI] [PubMed] [Google Scholar]
- 18. Guo Y, Guo F, Wei C, Qiu J, Liu Y, Fang Y, Gao J. CTLA4 gene polymorphisms influence the incidence of infection after renal transplantation in Chinese recipients. Plos One. 2013;8(8):e70824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim HJ, Jeong KH, Lee SH, Moon JY, Lee TW, Kang SW, Park SJ, Kim YH, Chung JH. Polymorphisms of the CTLA4 gene and kidney transplant rejection in Korean patients. Transpl Immunol. 2010;24(1):40–44. [DOI] [PubMed] [Google Scholar]
- 20. Hunt KA, McGovern DP, Kumar PJ, Ghosh S, Travis SP, Walters JR, Jewell DP, Playford RJ, van Heel DA. A common CTLA4 haplotype associated with coeliac disease. Eur J Hum Genet. 2005;13(4):440–44. [DOI] [PubMed] [Google Scholar]
- 21. Jonjić S, Mutter W, Weiland F, Reddehase MJ, Koszinowski UH. Site-restricted persistent cytomegalovirus infection after selective long-term depletion of CD4+ T lymphocytes. J Exp Med. 1989;169(4):1199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jonjić S, Pavić I, Lucin P, Rukavina D, Koszinowski UH. Efficacious control of cytomegalovirus infection after long-term depletion of CD8+ T lymphocytes. J Virol. 1990;64(11):5457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jonjić S, Pavić I, Polić B, Crnković I, Lucin P, Koszinowski UH. Antibodies are not essential for the resolution of primary cytomegalovirus infection but limit dissemination of recurrent virus. J Exp Med. 1994;179(5):1713–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Riddell SR, Greenberg PD. T cell therapy of human CMV and EBV infection in immunocompromised hosts. Rev Med Virol. 1997;7(3):181–92. [DOI] [PubMed] [Google Scholar]
- 25. Reddehase MJ, Simon CO, Seckert CK, Lemmermann N, Grzimek NK. Murine model of cytomegalovirus latency and reactivation. Curr Top Microbiol Immunol. 2008;325:315–31. [DOI] [PubMed] [Google Scholar]
- 26. Karimi MH, Motazedian M, Geramizadeh B, Nikeghbalian S, Yaghobi R, Abedi F, Hossin Aghdaee M, Azarpira N, Arabpour M, Malekpour Z, Namayandeh M. Association of the co-stimulatory molecules polymorphisms with CMV infection in liver transplant recipients. Int J Organ Transplant Med. 2011;2(4):171–77. [PMC free article] [PubMed] [Google Scholar]
- 27. Danilovic DL, Mendes-Correa MC, Lima EU, Zambrini H, K Barros R, Marui S. Correlations of CTLA-4 gene polymorphisms and hepatitis C chronic infection. Liver Int. 2012;32(5):803–808. [DOI] [PubMed] [Google Scholar]
- 28. Alizadeh AHM, Hajilooi M, Ranjbar M, Fallahian F, Mousavi SM. Cytotoxic T-lymphocyte antigen 4 gene polymorphisms and susceptibility to chronic hepatitis B. World J Gastroenterol. 2006;12(4):630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang Z, Feng X, Zhang W, Gao F, Ling Q, Zhou L, Xie H, Chen Q, Zheng S. Recipient cytotoxic T lymphocyte antigen-4+ 49 G/G genotype is associated with reduced incidence of hepatitis B virus recurrence after liver transplantation among Chinese patients. Liver Int. 2007;27(9):1202–208. [DOI] [PubMed] [Google Scholar]
- 30. Han Q, Duan S, Zhang G, Li Z, Li N, Zhu Q, Lv Y, Chen J, Liu Z. Associations between cytotoxic T lymphocyte-associated antigen-4 polymorphisms and serum tumor necrosis factor-α and interferon-γ levels in patients with chronic hepatitis B virus infection. Inflamm Res. 2011;60(11):1071–78. [DOI] [PubMed] [Google Scholar]
- 31. Bouqbis L, Izaabel H, Akhayat O, Pérez-Lezaun A, Calafell F, Bertranpetit J, Comas D. Association of the CTLA4 promoter region (− 1661G allele) with type 1 diabetes in the South Moroccan population. Genes Immun. 2003;4(2):132–37. [DOI] [PubMed] [Google Scholar]
- 32. Hudson LL, Rocca K, Song YW, Pandey JP. CTLA-4 gene polymorphisms in systemic lupus erythematosus: a highly significant association with a determinant in the promoter region. Hum Genet. 2002;111(4–5):452–55. [DOI] [PubMed] [Google Scholar]
- 33. Santos PSDS, Fernandes KS, Gallottini MH. Assessment and management of oral health in liver transplant candidates. J Appl Oral Sci. 2012;20(2):241–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kwak EJ, Kim DJ, Choi Y, Joo DJ, Park W. Importance of oral health and dental treatment in organ transplant recipients. Int Dent J. 2020;70(6):477–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bogusławska-Kapała A, Hałaburda K, Rusyan E, Gołąbek H, Strużycka I. Oral health of adult patients undergoing hematopoietic cell transplantation. Pre-transplant assessment and care. Ann Hematol. 2017;96(7):1135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duan S, Zhang G, Han Q, Li Z, Liu Z, Chen J, Lv Y, Li N, Wang Y, Li M, Lou S, et al. CTLA-4 exon 1+ 49 polymorphism alone and in a haplotype with −318 promoter polymorphism may confer susceptibility to chronic HBV infection in Chinese Han patients. Mol Biol Rep. 2011;38(8):5125–32. [DOI] [PubMed] [Google Scholar]
- 37. Fernández-Mestre M, Sánchez K, Balbás O, Gendzekhzadze K, Ogando V, Cabrera M, Layrisse Z. Influence of CTLA-4 gene polymorphism in autoimmune and infectious diseases. Hum Immunol. 2009;70(7):532–35. [DOI] [PubMed] [Google Scholar]
- 38. Husain S, Pietrangeli CE, Zeevi A. Delayed onset CMV disease in solid organ transplant recipients. Transpl Immunol. 2009;21(1):1–9. [DOI] [PubMed] [Google Scholar]
- 39. Boeckh M, Bowden RA, Goodrich JM, Pettinger M, Meyers JD. Cytomegalovirus antigen detection in peripheral blood leukocytes after allogeneic marrow transplantation. Blood. 1992;80:1358–64. [PubMed] [Google Scholar]
- 40. Zhang G, Liu Z, Duan S, Han Q, Li Z, Lv Y, Chen J, Lou S, Li N. Association of polymorphisms of programmed cell death–1 gene with chronic hepatitis B virus infection. Hum Immunol. 2010;71(12):1209–13. [DOI] [PubMed] [Google Scholar]
- 41. Pérez-García A, De la Cámara R, Román-Gómez J, Jiménez-Velasco A, Encuentra M, Nieto JB, de la Rubia J, Urbano-Ispizúa A, Brunet S, Iriondo A. CTLA-4 polymorphisms and clinical outcome after allogeneic stem cell transplantation from HLA-identical sibling donors. Blood. 2007;110(1):461–67. [DOI] [PubMed] [Google Scholar]
- 42. Blazar BR, Carreno BM, Panoskaltsis-Mortari A, Carter L, Iwai Y, Yagita H, Nishimura H, Taylor PA. Blockade of programmed death-1 engagement accelerates graft-versus-host disease lethality by an IFN-γ-dependent mechanism. J Immunol. 2003;171(3):1272–77. [DOI] [PubMed] [Google Scholar]
- 43. Pons Miñano JA, Ramírez Romero P, Robles Campos R, Sánchez Bueno F, Parrilla Paricio P. Tolerance and chimerism in liver transplantation. Rev Esp Enferm Dig. 2007;99(6):343–50. [DOI] [PubMed] [Google Scholar]
- 44. Dariavach P, Mattéi MG, Golstein P, Lefranc MP. Human Ig superfamily CTLA-4 gene: chromosomal localization and identity of protein sequence between murine and human CTLA-4 cytoplasmic domains. Eur J Immunol. 1988;18(12):1901–905. [DOI] [PubMed] [Google Scholar]
- 45. Magistrelli G, Jeannin P, Herbault N, Benoit De, Coignac A, Gauchat JF, Bonnefoy JY, Delneste Y. A soluble form of CTLA-4 generated by alternative splicing is expressed by nonstimulated human T cells. Eur J Immunol. 1999;29(11):3596–602. [DOI] [PubMed] [Google Scholar]
- 46. Gu X, Qi P, Zhou F, Ji Q, Wang H, Dou T, Zhao Y, Gao C. + 49G> A polymorphism in the cytotoxic T-lymphocyte antigen-4 gene increases susceptibility to hepatitis B-related hepatocellular carcinoma in a male Chinese population. Hum Immunol. 2010;71(1):83–87. [DOI] [PubMed] [Google Scholar]
- 47. Azarian M, Busson M, Lepage V, Charron D, Toubert A, Loiseau P, de Latour RP, Rocha V, Socié G. Donor CTLA-4+ 49 A/G* GG genotype is associated with chronic GVHD after HLA-identical haematopoietic stem-cell transplantations. Blood. 2007;110(13):4623–24. [DOI] [PubMed] [Google Scholar]
- 48. Pérez-García A, Brunet S, Berlanga JJ, Tormo M, Nomdedeu J, Guardia R, Ribera JM, Heras I, Llorente A, Hoyos M, Esteve J, et al. CTLA-4 genotype and relapse incidence in patients with acute myeloid leukemia in first complete remission after induction chemotherapy. Leukemia. 2009;23(3):486–91. [DOI] [PubMed] [Google Scholar]
- 49. Hou R, Cao B, Chen Z, Li Y, Ning T, Li C, Xu C, Chen Z. Association of cytotoxic T lymphocyte-associated antigen-4 gene haplotype with the susceptibility to gastric cancer. Mol Biol Rep. 2010;37(1):515–20. [DOI] [PubMed] [Google Scholar]
- 50. Yaghobi R, Saadi MI, Karimi MH, Geramizadeh B, Ramzi M, Zakerinia M. Relation between costimulatory molecule polymorphism and hepatitis B infections in hematopoietic stem cell transplant recipients. Exp Clin Transplant. 2014;12(4):357–66. [DOI] [PubMed] [Google Scholar]
- 51. Saadi MI, Yaghobi R, Karimi MH, Geramizadeh B, Ramzi M, Zakerinia M. Association of the costimulatory molecule gene polymorphisms and active cytomegalovirus infection in hematopoietic stem cell transplant patients. Mol Biol Rep. 2013;40(10):5833–42. [DOI] [PubMed] [Google Scholar]
- 52. Iravani-Saadi M, Karimi MH, Yaghobi R, Geramizadeh B, Ramzi M, Niknam A, Pourfathollah A. Polymorphism of costimulatory molecules (CTLA4, ICOS, PD.1 and CD28) and allogeneic hematopoietic stem cell transplantation in Iranian patients. Immunol Invest. 2014;43(4):391–404. [DOI] [PubMed] [Google Scholar]
- 53. Niknam A, Karimi MH, Yaghobi R, Geramizadeh B, Roozbeh J, Salehipour M, Iravani M. The association between viral infections and co-stimulatory gene polymorphisms in kidney transplant outcomes. Jundishapur J Microbiol. 2016;9(8):e31338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ramzi M, Iravani M, Zarei T, Yaghobi R, Arandi N. Association between cytotoxic T-lymphocyte antigen 4 gene polymorphisms and torque teno virus infection after hematopoietic stem cell transplantation. Exp Clin Transplant. 2017;19:259–63. [DOI] [PubMed] [Google Scholar]