Abstract
Previously, we constructed a set of mutants from which eight penicillin binding protein (PBP) genes were deleted in 192 combinations from Escherichia coli (S. A. Denome, P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young, J. Bacteriol. 181:3981-3993, 1999). Although these mutants were constructed correctly as determined by restriction mapping and the absence of relevant protein products, we recently discovered by PCR mapping that strains from which mrcA (PBP 1a) was deleted were also missing two neighboring genes of unknown function (yrfE and yrfF). We created a new deletion mutation in mrcA and reconstructed 63 strains lacking PBP 1a and other PBP mutant combinations. The new mrcA mutants do not exhibit mucoidy, phage resistance, temperature sensitivity, growth rate defects, or antibiotic resistance, suggesting that these phenotypes require the loss of either yrfE or yrfF alone or in combination with the absence of multiple PBPs.
Four high-molecular-weight penicillin binding proteins (PBPs) of Escherichia coli (PBPs 1a, 1b, 2, and 3) are responsible for synthesizing and assembling the peptidoglycan sacculus that forms the rigid bacterial cell wall (5, 6). However, E. coli also possesses at least seven low-molecular-weight PBPs (PBPs 4, 5, 6, and 7 and DacD, AmpC, and AmpH), the biological functions of which are either poorly characterized or completely unknown (2, 5, 6).
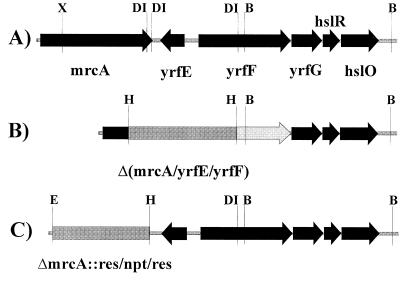
To address this question of physiological function, we constructed a set of multiply mutated strains in which one to seven PBPs were deleted in every viable combination (2). At the time of construction, each strain was tested by restriction mapping and by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to confirm that the correct genes and protein products had been deleted. Recently, we found we were unable to PCR amplify the mutated mrcA gene (encoding PBP 1a) from chromosomal preparations when using oligonucleotide primers hybridizing to sequences just upstream and downstream of the putative deletion endpoints. Primers further away from the mutated site did give an amplification product (data not shown), suggesting that a larger fragment had been deleted than was reported previously. DNA sequencing confirmed that one open reading frame (ORF) (yrfE) and the promoter and 5′ end of a second open reading frame (yrfF) were deleted in addition to most of the mrcA gene (data not shown). The extent of the deletion is pictured schematically in Fig. 1B. Thus, every strain designated as ΔmrcA in our previous publication (2) is actually a Δ(mrcA-yrfE-yrfF) deletion. All other PBP gene deletions were correct as reported (data not shown).
FIG. 1.
Deletions of mrcA and neighboring genes in E. coli. (A)Gene order downstream of mrcA in the parental strain, E. coli CS109. (B) Extent of deletion created in E. coli CS13-2K, previously reported to encompass only mrcA (2). The segment from the XhoI site in mrcA to the BspDI site in yrfF was replaced by the res-npt-res cassette (4). (C) Extent of the new mrcA deletion in E. coli BMCS04-1K reported in this work. The entire mrcA gene from the initiation codon to the termination codon was replaced by the res-npt-res cassette (4). Abbreviations: B, BamHI; E, EcoRI; DI, BspDI; H, HindIII; X, XhoI.
The data in Fig. 1 illustrates why the deletion was not correctly characterized earlier. When creating the original mrcA mutation we observed a single XhoI-BspDI DNA fragment, leading us to believe that one BspDI site existed in the cloned segment. However, after the mutants were constructed, the complete genomic sequence of E. coli (1) revealed there were three BspDI sites—two so close together that they could not be distinguished as separate sites and the third in the yrfF gene. Because the lengths of the two XhoI-BspDI and BspDI-BspDI fragments are almost identical, they appeared as a single band on our gels. Thus, our original digestions actually created a deletion from the XhoI site in mrcA to the BspDI site in yrfF (Fig. 1B).
To correct this situation, we deleted mrcA by using the λ recombination system described by Yu et al. (8). The res-npt-res cassette of plasmid pCK155 (4) was amplified by PCR using two primers homologous to each end of the cassette and containing at their 5′ ends chromosomal sequences homologous to those preceding the AUG start codon of mrcA or sequences following the UGA stop codon of mrcA. The primer sequences were, respectively, ACCGCGCGTTTGTTTATAAACTGCCCAAATGAAACTAAATGGAATTCGAGCTCTGCAGTCCC and CACTTTGTCAGCAAACTGAAAAGGCGCCGAAGCGCCTTTTTAAGATAAGCTTGCATGCCTGCAG. The resulting PCR product was electroporated into E. coli DY329 (8), and the cells were plated onto Luria-Bertani agar plates plus kanamycin (50 μg/ml) and incubated for 2 days at 32°C. Kanamycin-resistant colonies were screened for the correct mutation by PCR amplification, and the new mrcA deletion was confirmed by PCR amplification with combinations of internal and external primers and by SDS-PAGE of 125I-labeled PBPs (3) (data not shown). The mutation was moved into E. coli CS109 by P1 transduction to form strain BMCS04-1K. The extent of this new mrcA deletion is pictured schematically in Fig. 1C.
The new ΔmrcA::res-npt-res mutation was moved into selected E. coli strains to recreate the set of multiple mutants lacking PBP 1a in combination with every possible combination of six other PBPs (Table 1). Therefore, these strains are replacements for and should be used instead of the mrcA mutants described previously (2).
TABLE 1.
New mrcA (PBP 1a) mutantsa
| Strain | PBPs Deleted | Parent | Strain | PBPs Deleted | Parent | Strain | PBPs Deleted | Parent | ||
|---|---|---|---|---|---|---|---|---|---|---|
| CS229-1K | 1a 7 | CS9-19 | CS456-1K | 1a 4 5 7 | CS315-1 | CS541-1K | 1a 4 7 H C | CS441-1 | ||
| CS230-1K | 1a H | CS15-3 | CS457-1K | 1a 4 6 7 | CS316-1 | CS542-1K | 1a 4 5 6 H | CS442-3 | ||
| CS231-1K | 1a 6 | CS17-1 | CS458-1K | 1a 4 7 H | CS317-3 | CS543-1K | 1a 5 6 7 H | CS443-3 | ||
| CS232-1K | 1a 5 | CS12-7 | CS459-1K | 1a 4 7 C | CS318-1 | CS544-1K | 1a 4 5 6 C | CS445-1 | ||
| CS233-1K | 1a 4 | CS11-2 | CS460-1K | 1a 4 C H | CS320-1 | CS545-1K | 1a 4 5 6 7 | CS446-1 | ||
| CS234-1K | 1a C | CS14-2 | CS461-1K | 1a 4 5 6 | CS322-1 | CS546-1K | 1a 4 6 7 C | CS447-1 | ||
| CS367-1K | 1a 4 7 | CS203-1B | CS462-1K | 1a 4 6 H | CS323-3 | CS547-1K | 1a 4 5 7 H | CS448-3 | ||
| CS368-1K | 1a 4 H | CS221-3 | CS463-1K | 1a 4 6 C | CS324-1 | CS548-1K | 1a 4 5 7 C | CS449-2 | ||
| CS369-1K | 1a 4 5 | CS219-1 | CS464-1K | 1a 4 5 H | CS326-1 | CS549-1K | 1a 5 6 7 C | CS450-1 | ||
| CS370-1K | 1a 4 6 | CS220-1 | CS465-1K | 1a 4 5 C | CS327-1 | CS550-1K | 1a 5 7 C H | CS452-2 | ||
| CS375-1K | 1a 6 7 | CS205-1 | CS466-1K | 1a 5 6 7 | CS331-1 | CS551-1K | 1a 4 6 C H | CS453-1 | ||
| CS376-1K | 1a 6 H | CS213-1 | CS467-1K | 1a 6 7 H | CS332-3 | CS552-1K | 1a 4 5 C H | CS454-1 | ||
| CS377-1K | 1a 5 7 | CS204-1 | CS468-1K | 1a 6 C H | CS334-1 | CS553-1K | 1a 6 7 C H | CS455-1 | ||
| CS378-1K | 1a 5 H | CS215-3 | CS469-1K | 1a 5 6 H | CS336-3 | CS554-1K | 1a 5 6 C H | CS470-1 | ||
| CS379-1K | 1a 7 H | CS206-3 | CS470-1K | 1a 5 6 C | CS337-1 | CS614-1K | 1a 4 5 6 7 H | CS531-3 | ||
| CS380-1K | 1a 4 C | CS222-1 | CS471-1K | 1a 7 C H | CS342-1 | CS615-1K | 1a 4 5 6 7 C | CS533-1 | ||
| CS381-1K | 1a 5 C | CS216-2 | CS472-1K | 1a 6 7 C | CS343-1 | CS616-1K | 1a 4 5 7 C H | CS534-1 | ||
| CS382-1K | 1a 6 C | CS213-1 | CS473-1K | 1a 5 7 H | CS345-3 | CS617-1K | 1a 4 6 7 C H | CS535-1 | ||
| CS383-1K | 1a 7 C | CS207-2 | CS474-1K | 1a 5 7 C | CS346-1 | CS618-1K | 1a 5 6 7 C H | CS536-1 | ||
| CS384-1K | 1a C H | CS209-1 | CS475-1K | 1a 5 C H | CS349-1 | CS619-1K | 1a 4 5 6 C H | CS539-1 | ||
| CS385-1K | 1a 5 6 | CS211-2 | CS540-1K | 1a 4 6 7 H | CS440-3 | CS703-1K | 1a 4 5 6 7 C H | CS612-1 |
The parental strain from which individual and multiple PBP genes were deleted was E. coli CS109 (W1485 rpoS rph). Each mutant (“Strain” columns) was created by P1 transduction of mrcA::res-npt-res from BMCS04-1K (CS109 mrcA::res-npt-res) into the respective parental strain (“Parent” columns) and selection for kanamycin resistance. The parental strains are described by Denome et al (2). PBPs deleted from individual E. coli strains are abbreviated as follows (PBP, followed by gene name): 1a, = PBP 1a, mrcA; 4 = PBP 4, dacB; 5 = PBP 5, dacA; 6 = PBP 6, dacC; 7 = PBP 7, pbpG; C = AmpC, ampC; H = AmpH, ampH.
After screening the original mrcA mutants (now known to be ΔmrcA-yrfEF) (2), we reported preliminary observations that deletion of PBP 1a alone or in combination with other PBP mutations resulted in expression of a colanic acid capsule, phage resistance, temperature sensitivity, and resistance to lysis by certain β-lactams (7). The new mutant combinations described in this work exhibited none of these phenotypes (data not shown), suggesting that loss of either yrfE or yrfF alone or in combination with the absence of multiple PBPs is responsible for these characteristics. We are currently examining the relationship among these genes and phenotypes.
REFERENCES
- 1.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 2.Denome S A, Elf P K, Henderson T A, Nelson D E, Young K D. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol. 1999;181:3981–3993. doi: 10.1128/jb.181.13.3981-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henderson T A, Dombrosky P M, Young K D. Artifactual processing of penicillin-binding proteins 7 and 1b by the OmpT protease of Escherichia coli. J Bacteriol. 1994;176:256–259. doi: 10.1128/jb.176.1.256-259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kristensen C S, Eberl L, Sanchez-Romero J M, Givskov M, Molin S, de Lorenzo V. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J Bacteriol. 1995;177:52–58. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsuhashi M. Utilization of lipid-precursors and the formation of peptidoglycan in the process of cell growth and division: membrane enzymes involved in the final steps of peptidoglycan synthesis and the mechanism of their regulation. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science BV; 1994. pp. 55–71. [Google Scholar]
- 6.Spratt B G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young K D. Approaching the physiological functions of penicillin-binding proteins in Escherichia coli. Biochimie. 2001;83:99–102. doi: 10.1016/s0300-9084(00)01205-0. [DOI] [PubMed] [Google Scholar]
- 8.Yu D, Ellis H M, Lee E-C, Jenkins N A, Copeland N G, Court D L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]