Abstract
Background
Pediocin PA-1 is a bacteriocin of recognized value with applications in food bio-preservation and the medical sector for the prevention of infection. To date, industrial manufacturing of pediocin PA-1 is limited by high cost and low-performance. The recent establishment of the biotechnological workhorse Corynebacterium glutamicum as recombinant host for pediocin PA-1 synthesis displays a promising starting point towards more efficient production.
Results
Here, we optimized the fermentative production process. Following successful simplification of the production medium, we carefully investigated the impact of dissolved oxygen, pH value, and the presence of bivalent calcium ions on pediocin production. It turned out that the formation of the peptide was strongly supported by an acidic pH of 5.7 and microaerobic conditions at a dissolved oxygen level of 2.5%. Furthermore, elevated levels of CaCl2 boosted production. The IPTG-inducible producer C. glutamicum CR099 pXMJ19 Ptac pedACDCg provided 66 mg L−1 of pediocin PA-1 in a two-phase batch process using the optimized set-up. In addition, the novel constitutive strain Ptuf pedACDCg allowed successful production without the need for IPTG.
Conclusions
The achieved pediocin titer surpasses previous efforts in various microbes up to almost seven-fold, providing a valuable step to further explore and develop this important bacteriocin. In addition to its high biosynthetic performance C. glutamicum proved to be highly robust under the demanding producing conditions, suggesting its further use as host for bacteriocin production.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12934-023-02044-y.
Keywords: Corynebacterium glutamicum, Pediocin PA-1, Bioprocess, Food additive, Bacteriocin, Antimicrobial peptide, Acidic pH, Oxygen limitation, Calcium supplementation, Listeria sp
Background
Antimicrobial peptides (AMPs) are a group of bioactive substances that induce transmembrane pores and attack other intracellular targets, capable of rapidly killing microorganisms [1]. The typically small molecules are ribosomally synthesized by various cell types and tissues, including invertebrates, plants, animals, and last not least, a wide range of bacteria, AMPs of the latter being classified as bacteriocins [2, 3]. In contrast to most currently available antibiotics, many bacteriocins have only narrow spectra of target organisms and are therefore regarded attractive agents for precision therapy and prevention of infection [4]. Furthermore, bacteriocins have become important and safe agents for food preservation, because they efficiently act against food borne pathogens, while their peptide structures are easily digested in the human body [5]. Bacteriocins have recognized commercial relevance, as foodborne illness causes to 420,000 annual deaths worldwide [6] and two-third of food related diseases are caused by bacterial contamination [7].
Pediocins are commercially relevant bacteriocins of the class IIa type [8, 9]. They have been shown to efficiently bind to their receptors in bacterial membranes resulting in pore formation and killing of the target cell by destruction of the proton motive force [10, 11]. They efficiently act against Listeria monocytogenes, a food-borne pathogen of increasing concern to the food industry which may be found in raw milk, dairy products, vegetables, and meat products and can grow under refrigeration temperatures (growth has been reported at temperatures as low as − 1 °C), high salt concentrations (up to 10%), low pH (pH 5.0), and high temperatures (44 °C) [12]. Native pediocin producers are lactic acid bacteria including species of Pediococcus [13] and Lactobacillus [14]. The most prominent pediocin is pediocin PA-1, produced by Pediococcus acidilactici and extensively studied over the past decades due to its unique properties and application potential [15], including identification and sequencing of the encoding pediocin PA-1 cluster pedABCD [13, 14, 16, 17].
A commercial pediocin formulation (non-purified fermentates of native food grade producers with low levels of the antimicrobial peptide) has been introduced under the trade name ALTA (Quest International, Irvine, CA, USA) [18]. However, pure pediocin PA-1 is not commercially available [19]. For research purposes, concentrated bacteriocin solutions are generally produced by costly cultivation of natural producers and subsequent peptide purification [20, 21]. The efficiency of production is limited by the need for expensive media (that meet the multiple auxotrophies of the native producer strains) and the ultimately low titer [22]. To enable a broader access to pediocins for food applications but also clinical trials, production processes with higher pediocin titers and reduced costs appear crucial [23–25]. For further development, the successful demonstration of heterologous pediocin PA-1 production in the Gram-positive soil bacterium Corynebacterium glutamicum (upon episomal expression of a codon optimized and shortened version of the pediocin PA-1 cluster (pedACD) under induction by IPTG) recently provided a valuable and promising starting point [23].
Here, we advanced the production of pediocin PA-1 using the recently developed producer C. glutamicum CR099 pXMJ19 Ptac pedACDCg. The microbe turned out to be highly robust to the production and the presence of the peptide. Several rounds of medium and bioprocess optimization finally provided an optimum operation mode at slightly acidic pH, limiting oxygen supply, and elevated levels of calcium ions in a lean production medium. Benchmarked in a batch process, the microbe produced pediocin PA-1 up to 135,700 BU mL−1 (66 mg L−1), almost seven-fold more than any other producers studied so far. A novel C. glutamicum strain that expressed the cluster under control of the constitutive Ptuf promotor enabled pediocin PA-1 production up to 82,800 BU mL−1 without the need for costly IPTG. Our work displays a valuable demonstration of using C. glutamicum in an advanced process setting for high-level pediocin PA-1 production, providing this important bioactive for further exploration and promising to use the host also for other bacteriocins in the future.
Results
C. glutamicum appears metabolically unaffected by production of the antimicrobial peptide pediocin PA-1
Recently, we demonstrated production of the antimicrobial peptide pediocin PA-1 using C. glutamicum CR099 pXMJ19 Ptac pedACDCg that expressed a codon-optimized version of the pediocin PA-1 cluster from P. acidilactici under control of the inducible Ptac promotor [23]. The cluster encoded the pre-pediocin PA‐1 (PedA), the accessory export protein PedC, and the ABC transporter PedD (involved in the excision of the signal peptide and the secretion of pediocin) respectively [26]. Incubated on TY medium, the mutant started to grow from early on (Fig. 1A). After induction with IPTG, it took about 5 h until pediocin PA-1 became detectable in the culture broth. The level of the peptide reached a maximum of 3450 BU mL−1 after 16 h (i. e. 11 h after induction). During later stages, the concentration of the peptide dropped to finally below 828 BU mL−1. Throughout the process, the cells grew linearly up to a final OD660 of 28.4. Meanwhile, glucose was continuously consumed. After 32 h, the sugar was fully depleted. The control strain expressing the empty plasmid, did not produce any pediocin PA-1, as expected (Fig. 1B). In terms of growth and substrate consumption, it behaved exactly as strain C. glutamicum CR099 pXMJ19 Ptac pedACDCg. On a first glance, this suggested that the microbe seemed not affected by the recombinant pathway and, also, not by the antimicrobial product itself.
Fig. 1.
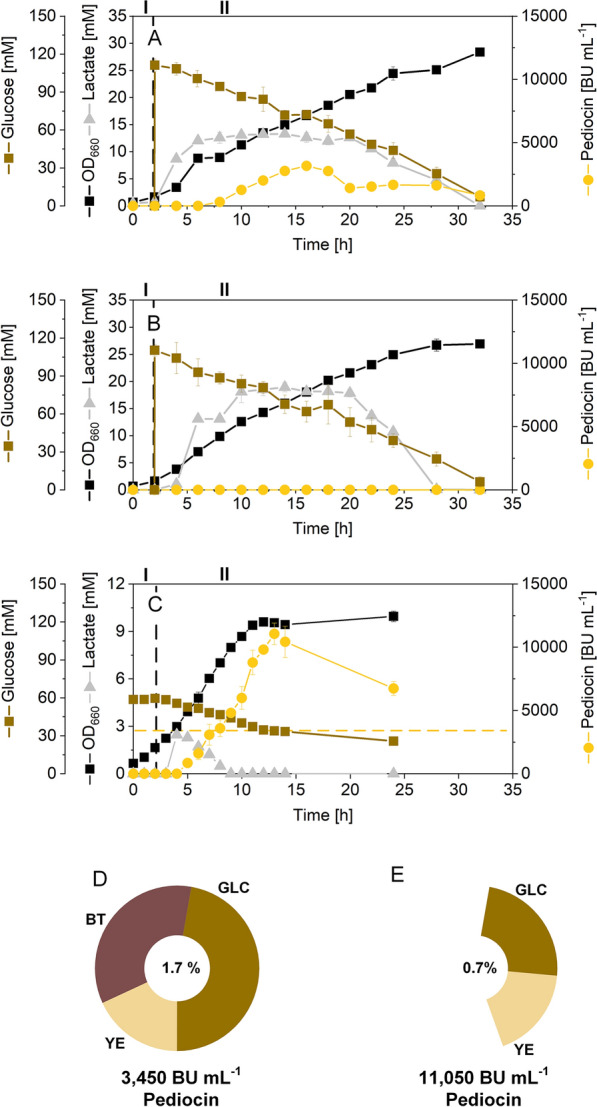
Pediocin production in shake flasks using recombinant C. glutamicum CR099 pXMJ19 Ptac pedACDCg. The data show growth and production on TY medium, containing 20 g L−1 of yeast extract, 16 g L−1 of bacto-tryptone, additionally supplemented with 20 g L−1 of glucose and 0.2 mM of IPTG after 2 h, indicated by a vertical dashed line A. The non-producing reference strain C. glutamicum CR099 pXMJ19, expressing the empty vector, is shown for comparison (B). In addition, GY medium was used to produce pediocin (C). The medium contained 10 g L−1 of glucose, 10 g L−1 of yeast extract, mineral salts, and vitamins. It was supplemented with IPTG (0.2 mM), two hours after the start. In addition, the corresponding total carbon content of the TY (D) and the GY medium (E) is given. For the complex ingredients, the total carbon content was estimated from the composition given by the supplier, whereby carbohydrates were considered as glucose units [34]. n = 3
We now were interested to assess the influence of pediocin PA-1 on C. glutamicum in more detail and compared the transcriptome of the producer to that of the control strain. The two strains were sampled at 13 h of the process, reflecting a time point, where the producing cells actively synthetized pediocin PA-1 and already faced a higher level of the product. The transcriptome data revealed excellent quality and reproducibility among the biological replicates (Additional file 1: Fig. S1). The pediocin cluster genes were strongly up regulated in the producer, indicating efficient transcription of the heterologous pathway (Table 1). Regarding the native metabolism of C. glutamicum, the impact of pediocin PA-1 production on gene expression was rather small. Only very few out of more than 3000 genes were significantly changed in expression in the producer, as compared to the reference strain (Table 1). The gene CGL_RS05840, encoding a pspC binding domain containing protein from the group of phage shock proteins as response to cell envelope stress [27, 28] was up regulated. In contrast, bioY, encoding a biotin transporter, was found down regulated. A few other genes were weakly affected in expression. Taken together, C. glutamicum CR099 pXMJ19 Ptac pedACDCg tolerated the implemented pediocin PA-1 production well. The microbe appeared vital in terms of growth and, also, the global transcriptional machinery remained almost unchanged. Severe bottlenecks in metabolism did not show up, suggesting focusing on the bioprocess level for further improvement rather than on engineering the cell factory.
Table 1.
Strains and plasmids used in this study
| Strain | Description | Refs. |
|---|---|---|
| E. coli | ||
| DH10B | Cloning host | [98] |
| L. innocua | ||
| pIMK2 | Sensor strain expressing the plasmid pIMK2 kanR | [23] |
| pNZ44 | Sensor strain expressing the plasmid pNZ44 cmR | [23] |
| C. glutamicum | ||
| CR099 | Genome reduced derivative of strain ATCC 13,032 with deletion of ΔCGP1-3 and ΔISCg1-2 | [96] |
| CR099 pEKEx2 Ptac pedACDCg | CR099 with episomal expression of codon-optimized pediocin operon pedACDCg from Pediococcus acidilactici under control of Ptac | [23] |
| CR099 pXMJ19 Ptac pedACDCg | CR099 with episomal expression of the codon-optimized pediocin operon from P. acidilactici under control of Ptac | [23] |
| CR099 pXMJ19 | CR099 with episomal expression of empty pXMJ19 | [23] |
| CR099 pClik 5α Ptuf pedACDCg | CR099 with episomal expression of the codon-optimized pediocin operon from P. acidilactici under control of Ptuf | This work |
| CR099 pEKEx2 Ptac pedAM31LCDCg | CR099 with episomal expression of the codon-optimized pediocin operon from P. acidilactici under control of Ptac including mutated pedAM31L | This work |
| CR099 pClik 5α | CR099 with episomal expression of empty pClik 5α | This work |
| CR099 pClik 5α vgb | CR099 with episomal expression of the heterologous gene vgb from Vitreoscilla spp. [52] under control of Ptuf | This work |
| CR099 pClik 5α dps | CR099 with episomal expression of the native C. glutamicum gene dps [49] under control of Ptuf | This work |
| CR099 pClik 5α mcbR | CR099 with episomal expression of the native C. glutamicum gene mcbR [49] under control Ptuf | This work |
| CR099 pClik 5α mpx | CR099 with episomal expression of the native C. glutamicum gene mpx [47] under control of Ptuf | This work |
| CR099 pClik 5α mshA | CR099 with episomal expression of the native C. glutamicum gene mshA [48] under control of Ptuf | This work |
| CR099 pClik 5α katA | CR099 with episomal expression of the native C. glutamicum gene katA [49] under control of Ptuf | This work |
| CR099 pClik 5α zwf | CR099 with episomal expression of the native C. glutamicum gene zwf [99] under control of Ptuf | This work |
| CR099 pClik 5α zwf ATG | CR099 with episomal expression of the native C. glutamicum gene zwf under control of the translational start codon ATG [99] and Ptuf | This work |
| CR099 pClik 5α ATG zwf ATG, A243T | CR099 with episomal expression of the mutated variant zwf A243T [56] under control of the translational start codon ATG [99] and Ptuf | This work |
| CR099 pClik 5α PH30 DR1558 | CR099 with episomal expression of the heterologous gene DR1558 from Deinococcus radiodurans [50] under control of the synthetic promotor PH30 | This work |
| CR099 pClik 5α Ptuf DR1558 | CR099 with episomal expression of the heterologous gene DR1558 from Deinococcus radiodurans [50] under control of Ptuf | This work |
| Plasmids | ||
| pClik5α MCS | Episomal vector, ORICg, ORIEc, kanR | [97] |
| pXMJ19 | Episomal vector, PtacI lacI, ORICg, ORIEc, cmR | [23] |
| pXMJ19 Ptac pedACDCg | Episomal expression of pedACDCg under control of Ptac | [23] |
| pClik 5α Ptuf pedACDCg | Episomal expression of pedACDCg under control of Ptuf | This work |
| pClik 5α Ptuf vgb | Episomal expression of vgb [52] under control of Ptuf | This work |
| pClik 5α Ptuf dps | Episomal expression of dps [49] under control of Ptuf | This work |
| pClik 5α Ptuf mcbR | Episomal expression of mcbR [49] under control of Ptuf | This work |
| pClik 5α Ptuf mpx | Episomal expression of mpx [47] under control of Ptuf | This work |
| pClik 5α Ptuf mshA | Episomal expression of mshA [48] under control of Ptuf | This work |
| pClik 5α Ptuf katA | Episomal expression of katA [49] under control of Ptuf | This work |
| pClik 5α Ptuf zwf | Episomal expression of zwf under control of Ptuf | This work |
| pClik 5α Ptuf zwf ATG | Episomal expression of zwf under control of the start codon ATG [12] and Ptuf | This work |
| pClik 5α Ptuf zwf ATG, A243T | Episomal expression of the mutated variant zwf A243T [56] under control of the start codon ATG [12] and Ptuf | This work |
| pClik 5α PH30 DR1558 | Episomal expression of DR1558 [50] under control of PH30 | This work |
| pClik 5α Ptuf DR1558 | Episomal expression of DR1558 [50] under control of Ptuf | This work |
Fine-tuning of oxygen supply is crucial to preserve active pediocin PA-1 while enabling growth of the aerobic microbe
Pediocin PA-1 contained an oxygen-sensitive l-methionine residue at position 31, and oxidation of this amino acid residue was known to cause a loss of antimicrobial activity [19]. To overcome this problem, we first followed a previous strategy that had demonstrated increased stability of mutated peptide variants in which the l-methionine residue had been replaced by small nonpolar amino acids, including l-leucine [29]. In short, we created the pediocin PA-1 variant pedAM31L using a mutated primer that carried the desired point mutation. After validation by sequencing, the novel strain pEKEx2 Ptac pedAM31LCDCg, based on a well-established shuttle vector for expression in C. glutamicum [30], was cultivated as described above. Unfortunately, the alteration of the peptide did not provide any detectable active pediocin PA-1 (Additional file 1: Fig. S3).
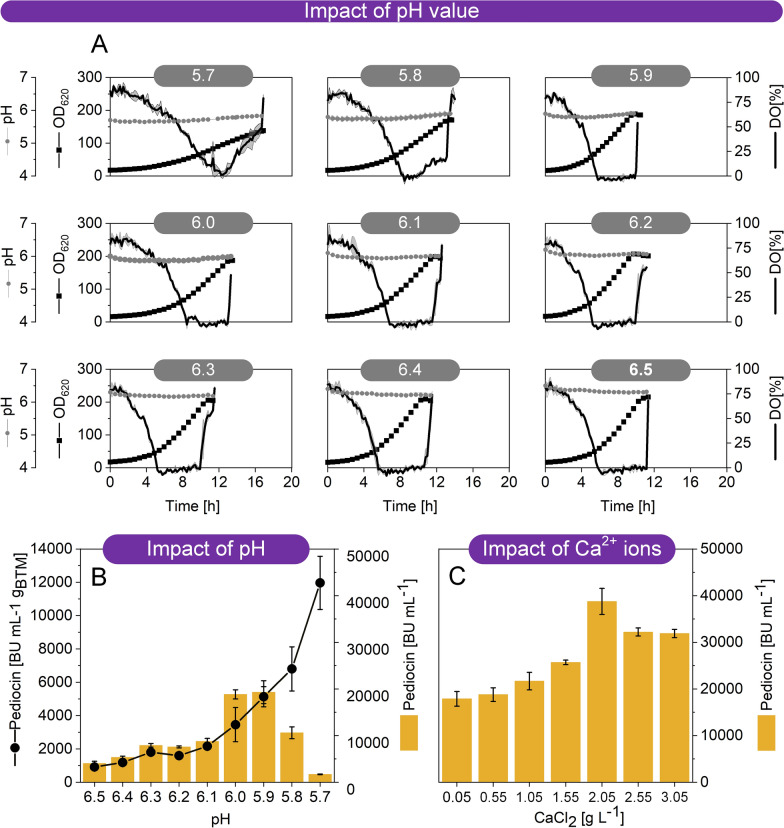
At this point, we decided to keep producing the native pediocin PA-1 molecule and optimize the oxygen supply instead. For this purpose, the production process was transferred to lab scale bioreactors, which allowed a precise adjustment of the dissolved oxygen (DO) level by the stirring and the aeration rate. In addition to a well-supplied reactor (DO level of 30%), two set-ups were conducted at lower DO levels (5% and 2.5%) (Fig. 2A–C). As expected, the oxygen availability strongly affected the growth of C. glutamicum CR099 pXMJ19 Ptac pedACDCg. The cells grew best at the highest oxygen supply (µ = 0.45 h−1), still managed well at 5% (µ = 0.41 h−1) but were found reduced in growth at 2.5% (µ = 0.15 h−1). Under the latter condition, the cells, furthermore, reached a lower biomass concentration.
Fig. 2.
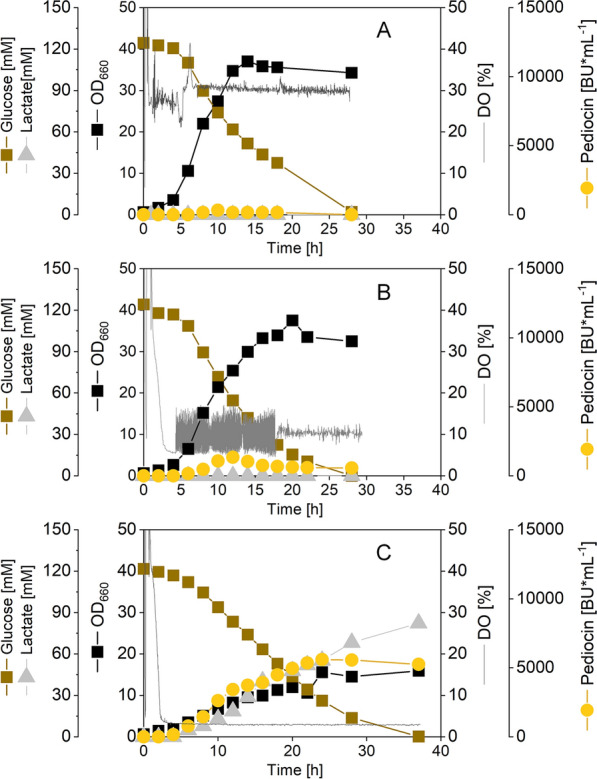
Impact of the dissolved oxygen (DO) level on pediocin production in recombinant C. glutamicum CR099 pXMJ19 Ptac pedACDCg. The process was conducted in lab scale bioreactors using rich TY medium, supplemented after two h with 20 g L−1 of glucose (pH 6.5 and 30 °C). The DO level was controlled at 30% (A), 10% (B), and 2.5% (C). The experiments were performed as single replicate each. The conducted processes, according to experience, have a standard deviation of less than 10%
The opposite effect of the dissolved oxygen level was found regarding formation of the peptide. The set-up, operated at 30%, produced the least amount (320 BU mL−1), even ten-fold less than observed in shake flasks (Fig. 1A). In contrast, the highest amount of pediocin PA-1 (5,590 BU mL−1) was formed at the lowest dissolved oxygen level (2.5%). The results identified the control of the DO at 2.5% as optimal for pediocin PA-1 production. Because, growth was already strongly inhibited under these conditions, an even further reduction appeared not promising.
Medium optimization provides elevated levels of pediocin PA-1 at reduced amount of complex ingredients
So far, pediocin PA-1 production in C. glutamicum was based on rich TY medium that contained high levels of expensive ingredients such as yeast extract and tryptone plus glucose. The composition was based on complex media, used before to produce bacteriocins in lactic acid bacteria, well-known hosts to provide this type of product [31]. Towards a simplified and cheaper recipe, we upgraded a commonly used minimal medium for C. glutamicum that contained glucose (10 g L−1) as carbon source plus mineral salts and vitamins [32] for pediocin PA-1 production. Because it had been reported that the synthesis of bacteriocins benefits from supplementation with yeast extract, whereas tryptone supplements have weaker effects [33], we decided to add 10 g L−1 of yeast extract to the minimal formulation. The obtained GY medium exhibited a total carbon content of 0.7%, as inferred from inspection of the individual components [34], 60% less than the original TY medium (total carbon content 1.7%) [23, 34]. When growing strain CR099 pXMJ19 Ptac pedACDCg in the new GY medium, pediocin PA-1 production was enhanced to 11,056 BU mL−1 (Fig. 1C), almost 2.5-fold more than in the original medium. In addition, the novel medium enabled significantly faster growth. The GY medium was therefore kept for further optimization.
Pediocin PA-1 production in C. glutamicum is boosted by acidic pH value
Previously, the pH value was shown to influence the adsorption of bacteriocins to producing cells and, also, the post-translational maturation processes for bacteriocin activation [35]. As example, bacteriocin production was found to be triggered by a pH decrease, and this phenomenon has been also observed for the native pediocin PA-1 producer P. acidilactici [36] as well as for other bacteriocins and their native producers [20]. Of note, we recently were able to establish recombinant production of garvicin Q, another class II bacteriocin, with C. glutamicum. Considerable levels of garvicin Q were only achieved in minimal medium without urea allowing acidification during cultivation [37]. The pediocin PA-1 production in C. glutamicum, conducted in shake flasks on TY medium, revealed the same trend (Additional file 1: Fig. S2A). The initially neutral pH decreased to values below pH 6 during the first 7 h, where it stabilized for the rest of the process. Interestingly, pediocin PA-1 only accumulated in the medium when the pH had become slightly acidic. The same picture resulted for the production in GY medium (Additional file 1: Fig. S2B). The pH decrease was associated to the formation of lactic acid that accumulated in the medium (Fig. 1). Furthermore, online monitoring of the DO indicated that the cells were oxygen-limited during long phases of the process (Additional file 1: Fig. S2A), which apparently triggered formation of the acid [38, 39].
To explore the potential of acidic pH for improved pediocin PA-1 formation, the cell factory CR099 pXMJ19 Ptac pedACDCg was grown on GY medium, buffered with MES at different pH values between 5.7 and 6.5 (Fig. 3A, B). Pre-tests had shown that growth was not possible at more acidic conditions. The cultures were conducted in a microbioreactor using 48-well plates with integrated OD620, pH, and DO sensors that allowed precise online monitoring of these parameters. As shown, the pH could be well controlled at each desired set point. Remarkably, the pH value had a strong effect on the final pediocin PA-1 titer, and acidic conditions were found optimal. The specific pediocin PA-1 production, normalized to the phase of oxygen limitation and the corresponding average amount of biomass, was dramatically increased with decreasing pH value. It was highest at pH 5.7, the lowest value that the cells still tolerated. Production was also decreased at more neutral pH values, although the cells grew better under these conditions. On the other hand, cell growth was impaired at two most acidic conditions, pH 5.8 and 5.7. This caused a lower concentration of producing cells and, inter alia, an extended period of oxygen excess, negatively affecting the total amount of product formed. That was why, all in all, the maximum production was obtained at pH 5.9 and pH 6.0. Here, a pediocin PA-1 titer of approximately 20,000 BU mL−1 was achieved, almost twice as high as before.
Fig. 3.
Improvement of pediocin production in recombinant C. glutamicum CR099 pXMJ19 Ptac pedACDCg by medium optimization. The cultures were conducted in a miniaturized microtiter plate system with online sensing of cell concentration, pH value, and DO level (A). In a first series of experiments, the pH was adjusted to different values between 5.7 and 6.5 using 200 mM MES (A). Impact of the pH value on pediocin production (B). The data display the final pediocin activity in culture supernatant. In addition, the specific production efficiency is given, normalized to the period of oxygen limitation (required for pediocin accumulation) and the average cell concentration during this phase. In a second series of experiments, the impact of CaCl2 on pediocin production was assessed (C). Different levels of were supplemented to the medium. The pH was kept at 5.9. The data display the final pediocin activity in culture supernatant. n = 3
Calcium chloride supplementation at low pH enhances the accumulation of pediocin PA-1 in the culture supernatant
Ca2+ ions were shown to improve the secretion of heterologous proteins by C. glutamicum [40, 41]. Moreover, we recently observed a pronounced effect of Ca2+ ions on the tolerance of C. glutamicum to nisin [42]. The addition of CaCl2 (2 g L−1) to the culture medium significantly increased the resistance of the microbe against the bacteriocin, and it was concluded that this effect was attributed to the Ca2+ ions, that masked negative charges of cell wall constituents and thus prevented binding of nisin. Although, C. glutamicum was apparently not sensitive to pediocin PA-1 [23], we hypothesized that reduced (or blocked) binding of pediocin PA-1 to the cells could increase the titer in the supernatant. The original GY medium contained 55 mg L−1 CaCl2. This low level, typically used in defined media to support growth of C. glutamicum [43–45], was almost 40-fold lower than the concentration, found effective to affect resistance of the microbe to nisin [42]. Therefore, a next series of cultures was conducted at microbioreactor scale in GY medium, this time buffered at pH 5.9 and containing different levels of CaCl2 (0.05–3.05 g L−1) (Fig. 3C). Beneficially, pediocin PA-1 production was enhanced up to two-fold by increased CaCl2 levels. Maximum pediocin PA-1 activity in the culture supernatant (40,000 BU mL−1) was observed at 2 g L−1 of CaCl2 and above. At this point, we regarded the slightly acidic (pH 5.9) medium, containing glucose, yeast extract, 2 g L−1 CaCl2 plus other mineral salts and selected vitamins suitable for pediocin PA-1 production in C. glutamicum.
The overexpression of different stress-protecting genes does not further promote growth of C. glutamicum at low pH and low oxygen
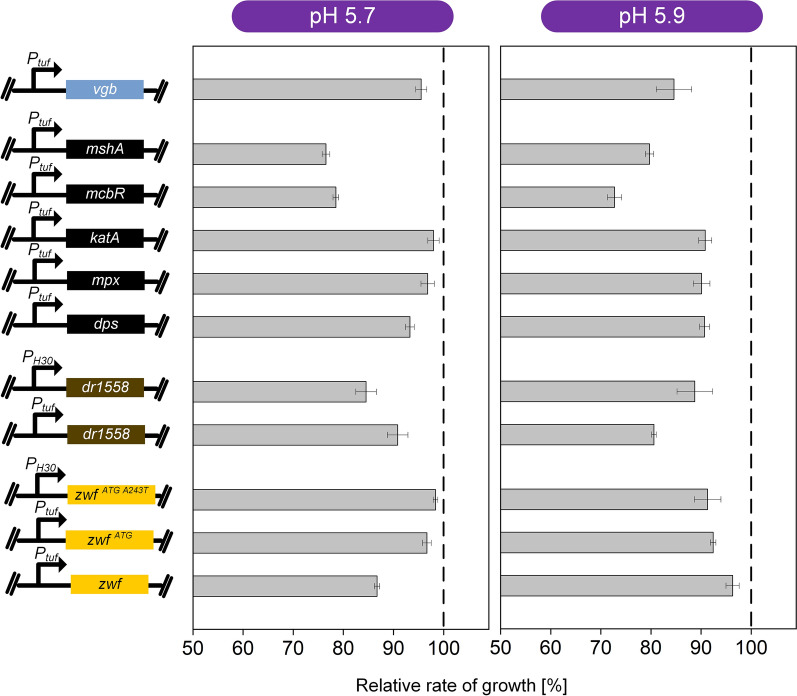
Although the pediocin PA-1-producing cell factory could grow well at acidic pH and low oxygen level, the optimum production conditions clearly differed from the growth optimum of the microbe which naturally prefers neutral pH and sufficient oxygen supply [46]. We were interested, if growth under pediocin PA-1-producing conditions could eventually be stimulated by the expression of genes, previously found helpful to grow other C. glutamicum strains under stress conditions. With regard to low pH, the selected candidates included the C. glutamicum genes dps, mpx, katA, mcbR, and mshA [47–49], as well as the heterologous gene DR1558 from D. radiodurans [50], encoding for homologous and heterologous proteins that had been found beneficial for growth under acidic conditions (Fig. 4). With regard to low oxygen, we chose the gene vgb from Vitreoscilla spp., encoding a bacterial hemoglobin, shown to increase oxygen uptake in C. glutamicum [51, 52]. Furthermore, we aimed to enhance the supply of the reducing equivalent NADPH, well-known for its importance in the defense of C. glutamicum to stress [53]. The latter was realized by over expression of zwf, encoding glucose 6-phosphate dehydrogenase at the entry into the PP pathway, inter alia, displaying the major route of NADPH supply in C. glutamicum [54]. In addition to the native zwf gene, we tested two variants for enhanced translation efficiency and enhanced NADPH supply [55, 56]. Each of the ten candidates was individually overexpressed in the chassis strain C. glutamicum CR099, using a strain that expressed the empty vector as a control.
Fig. 4.
Evaluation of ten gene candidates to support growth of the chassis strain C. glutamicum CR099 at pH 5.9 and low oxygen supply. The cultures were conducted in a miniaturized microtiter plate system with online sensing of cell concentration. The different gene candidates were previously studied to increase the tolerance of C. glutamicum and encoded for homologous and heterologous proteins: dps, iron-storage protein Dps; mpx, mycothiol peroxidase; katA, catalase; mcbR, transcriptional repressor McbR; mshA, D-inositol 3-phosphate glycosyltransferase [47–49]; DR1558, transcriptional response regulator [50] (Table 2). Each gene was episomally expressed under control of the constitutive promoters Ptuf and PH30. A strain expressing the empty vector served as control. n = 3
All eleven strains were grown in the newly developed production medium at pH 5.7 and pH 5.9 with online monitoring of cell growth. None of the expressed genes, however, could improve the specific growth rate or the biomass formed (Fig. 4). The expression of several genes in fact even reduced the growth ability. For example, overexpression of the native genes zwf, mshA, and mcbR reduced growth. Likewise, DR1558 expression negatively affected cell vitality. Taken together, the unmodified CR099 chassis seemed to work best and was therefore kept.
Benchmarking the recombinant cell factory under optimum process conditions enables high-efficiency pediocin PA-1 production
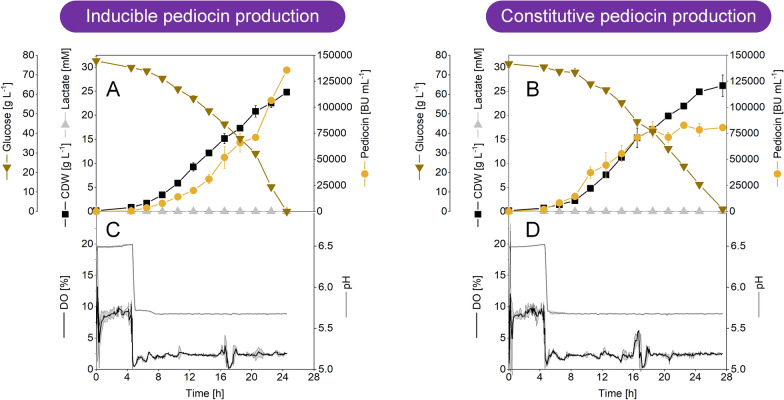
Finally, all individual improvements were combined in an optimized process setting. The developed process strategy was benchmarked in lab-scale bioreactors, operated in batch mode with the inducible pediocin PA-1 producer C. glutamicum CR099 pXMJ19 Ptac pedACDCg (Fig. 5A, B). To maximize production, a few changes were made in comparison to the shake flask set-up. First, the initial glucose concentration was set to 80 g L−1 to enable growth to higher cell concentrations, given the fact the C. glutamicum copes well with high sugar levels [57]. In addition, the concentration of yeast extract was slightly increased (15 g L−1). Second, the medium was buffered, after initial test fermentations had shown that the use of an unbuffered medium resulted in larger fluctuations of the pH (data not shown). Third, the pH value was set to 6.5, while the DO level was controlled at 10% to support cell growth during the initial process phase. Both parameters were supposed to be shifted at a later stage. Fourth, the inducer IPTG was present in the initial medium, after it had turned out that the induced cluster did not affect strain vitality (Table 1).
Fig. 5.
Optimized pediocin PA-1 production in lab scale bioreactors in batch mode using recombinant C. glutamicum. As carbon source, the medium contained 80 g L−1 of glucose and 15 g L−1 of yeast extract. After an initial growth phase (10% DO, pH 6.5), the process was shifted to pediocin PA-1 production (2.5% DO, pH 5.7). The data comprise C. glutamicum CR099 pXMJ19 Ptac pedACDCg which expressed the cluster under control of the Ptac promotor and was induced by 0.2 mM IPTG at process start (A, B). In addition, C. glutamicum CR099 pClik 5α Ptuf pedACDCg was used, expressing the cluster under control of the constitutive Ptuf promotor, so that no inducer was required (C, D). n = 2
As intended, the producer CR099 pXMJ19 Ptac pedACDCg started to grow from early on (Fig. 5A). Within 5 h, the biomass concentration increased to 0.7 g L−1, about four-fold. As expected for pH 6.5, pediocin PA-1 formation was negligible during this initial phase. After 5 h, the process conditions were shifted to pH 5.7 and 2.5% DO, because the cells had shown the highest specific production performance under these conditions. It turned out that this set-up allowed highly efficient pediocin PA-1 production. Within only 24 h, the activity in the culture supernatant increased to 135700 BU mL−1 (66 mg L−1). Towards the end of the process, the cells continued to grow and reached a final biomass concentration of 25 g L−1, while glucose was completely consumed. Even though oxygen was limiting conditions, the cells did not form any lactate. This was a remarkable finding given the fact that lactate is the predominant by-product of C. glutamicum during growth with glucose under oxygen limiting conditions [58]. Notably, the mutant accumulated high amounts of lactate under the same limiting oxygen level, however at an elevated pH of 6.4 (Fig. 2C). A pH dependence of lactate formation was also obvious from the shake flask cultures, where cells stopped to excrete the acid at a pH below 6.0 (Fig. 1). We conclude that the low pH value of the production process (pH 5.7) abolished lactate formation. From a process point of view, the absence of lactate, a short chain organic acid that is well known for its toxicity to microbes, appeared highly beneficial.
Given the fact, that the immediate induction of the pedACDCg cluster at process start apparently had no negative effects, we created the new strain CR099 pClik 5α Ptuf pedACDCg, an alternative producer that expressed the genes constitutively. For expression, we selected the promotor Ptuf, the promotor of the gene of elongation factor TU, known to efficiently support gene expression in the microbe [59–61]. When grown under the same conditions, the constitutive producer performed well (Fig. 5C, D). Cells started to grow immediately and handled the shift to acidic pH and low oxygen availability very well. Again, no lactate was formed throughout the whole process. During the first 18 h, pediocin PA-1 production was even stronger than that for the inducible strain. However, during the final stage of the process, the formation of pediocin PA-1 slowed down, despite the cells were still growing and consumed glucose. Finally, an activity of 82,800 BU mL−1 (42 mg L−1) was achieved.
Discussion
Under optimized conditions, C. glutamicum provides 66 mg L−1 of active pediocin PA-1 surpassing previous efforts to derive the bacteriocin almost seven-fold
Bacteriocins are a group of antimicrobial peptides that exhibit remarkable activities against dangerous pathogens, including Staphylococcus, Bacillus, Listeria, and Enterococcus [62–64]. Pediocin PA-1 is a prominent, commercially relevant bacteriocin [65]. It efficiently combats L. monocytogenes, a food-borne pathogen that causes invasive Listeriosis, a severe infection of the bloodstream or the brain in older adults and people with weakened immune systems, leading to death in one out of five cases [7, 66–68]. When listeriosis occurs during pregnancy, it can cause abortion, fetal death, and neonatal morbidity [69]. Although research results regarding the efficiency of pediocins, among other bacteriocins, as bio-preservatives are remarkable and promising, there is substantial reluctance by the industry to commit financially in developing commercial preparations, due to the costly production (low production rates, unstable products and expensive downstream processing) [65]. Here, we demonstrate high-efficiency production of pediocin PA-1 (Fig. 5). As shown, using special producing conditions of low pH, low oxygen level, and elevated amounts of calcium ions, recombinant strains of C. glutamicum accumulated pediocin PA-1 up to 66 mg L−1, surpassing previous efforts in various producers almost seven-fold (Fig. 6). Hereby, the simplification of the production medium from a costly complex nutrient mixture to a glucose-based formulation with only minor shares of yeast extract enabled production at significantly lower costs (Fig. 1). Beneficially, the creation of a cell factory that expressed the biosynthetic pedACD cluster under control of the constitutive Ptuf promotor made it possible to dispense with the addition of IPTG to the production process (Fig. 5), previously needed [23]. IPTG is an efficient molecular inducer for regulating transcriptional activity but suffers from limitations due to toxicity, cost, and culture monitoring [70] so that the demonstrated IPTG-free production of pediocin PA-1 appears attractive. Future fine-tuning of the promotor strength might help to further optimize performance when using constitutive promotors [71]. Taken together, the developed process (Fig. 6) provides a valuable improvement to overcome the present challenges linked to commercial pediocin production [65].
Fig. 6.
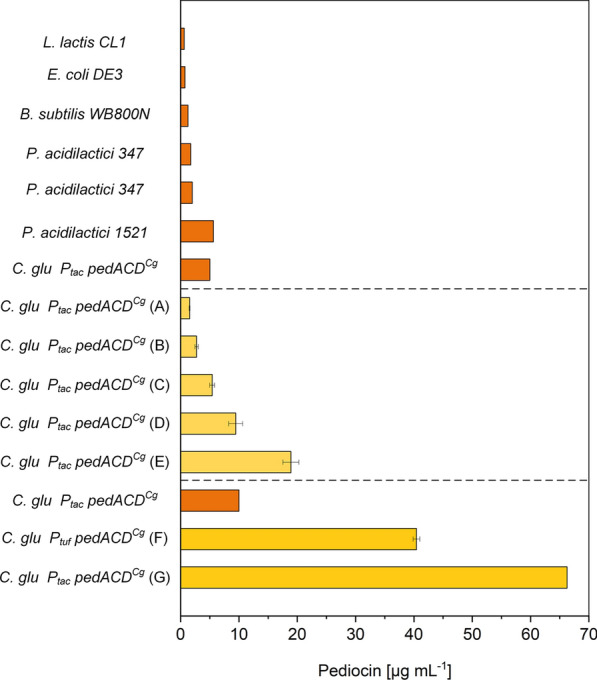
Benchmarking of microbial pediocin PA-1 production in natural and recombinant cell factories. The data show previous efforts (from top to bottom in the upper part) using L. lactis CL1 [107], B. subtilis [108], P. acidilactici 347 [109], E. coli producing the M31L version of the peptide [110], P. acidilactici 347 [111], C. glutamicum CR099 pXMJ19 Ptac pedACDCg [23], and P. acidilactici 1521 [112] in shake flasks and test tubes. In addition, the results of the optimization from this work are shown, using C. glutamicum CR099 pXMJ19 Ptac pedACDCg. The data comprise (from top to bottom in the middle part) production in in shake flasks using TY medium (A), TY medium, at a reduced DO level of 2.5% (B), GY medium at reduced nutrient content (C), GY medium at pH 5.9 (D), and GY medium at pH 5.9 and 2 g L−1 CaCl2 (E). Furthermore, the performance of pediocin PA-1 production in bioreactors is shown. This includes a fed-batch process in TY medium using C. glutamicum CR099 pXMJ19 Ptac pedACDCg [23] and two batch processes in GY medium with 80 g L−1 of glucose and 2 g L−1 CaCl2 from this work, that comprised an initial phase of growth (pH 6.5, 10% DO) and a switch to production (pH 5.7, 2.5% DO) after 4 h using C. glutamicum CR099 pClik Ptuf pedACDCg CR099 with constitutive cluster expression (F) and the inducible producer C. glutamicum CR099 pXMJ19 Ptac pedACDCg (G). The medium of the letter additionally contained 2 mM IPTG. The pediocin PA-1 concentration obtained with B. subtilis was estimated from the reported biological activity [108] using the recently obtained conversion factor [23]
A special combination of environmental conditions drives the production of active pediocin PA-1 in C. glutamicum
Different to most of the products that are made using C. glutamicum under aerobic conditions and neutral pH [56, 72, 73], the production of pediocin PA-1 under these conditions was very weak (Fig. 1). In fact, it required the use of a special set-up to drive production of the peptide to high efficiency (Figs. 2, 3, 5). Due to its oxygen sensitivity, active pediocin could not be produced at high aeration, commonly used for the microbe, but required limiting conditions [23] with optimum performance at 2.5% DO (Figs. 2, 5). These microaerobic conditions promoted cell growth better than the simplified control via the shaking rate during flask-based and microbioreactor-based production that always ended up in DO levels of 0% (Figs. 1, 3). A bit surprising, the expression of a mutated M31L-pediocin PA-1 under high oxygen supply provided the peptide only at low level (Additional file 1: Fig. S3), although the variant had been shown to be stable, when stored under these conditions [29]. Eventually, the expression machinery of the host did not well provide the mutant peptide in its active form, or the process environment negatively affected the peptide structure, but more work is needed to clarify this outcome.
An important outcome was the observation that acidic pH and elevated levels of bivalent calcium ions boosted pediocin PA-1 production (Fig. 3). At neutral pH, the cationic peptide [74] interacted with negatively charged residues on the cell wall of C. glutamicum [75]. It is well understandable that this interaction caused massive absorption of the peptide to the cells, resulting in apparently low levels of free pediocin PA-1 in the culture supernatant. Indeed, we recently observed that, when incubated C. glutamicum ATCC13032 at neutral pH, more than 90% of garvicin Q, another class II bacteriocin, was adsorbed to biomass [37]. A low pH reduced the negative charge of the cell wall by the elevated proton concentrations. In the same direction, the bivalent calcium ions paired with the negative charges on the cell surface. Thus, both low pH and bivalent cations may result in reduced interaction between the cell envelope and cationic peptides. Although definite proof for the exact mechanistic effect of low pH and Ca2+ ions on peptide adsorption is still missing, these conditions were highly beneficial for pediocin production. As shown, the specific pediocin production rate, normalized to the biomass, increased almost exponentially with reduced pH and was highest at pH 5.7, the lowest value tested (Fig. 3B). Notably, most small bacteriocins are cationic [76] and therefore similarly absorb, so that acidic conditions appear generally crucial when producing such molecules in C. glutamicum. It should, however, be noticed that the acidic environment posed a challenge to the microbe. Growth of C. glutamicum was negatively affected already at pH 6, as shown here (Fig. 3) and before [77], while maintenance of the cell structure obviously requires an extracellular pH value above 5.5 [78]. Fortunately, the precise pH control in the bioreactor process and the implementation of an initial phase at pH 6.5 to support growth, allowed to successfully operate at pH 5.7 during later production, close to the growth minimum of the bacterium. Beneficially these conditions fully abolished the formation of lactate, an otherwise undesired by-product.
C. glutamicum shows favorable robustness under the special conditions required for recombinant PA-1 production
The recombinant host was found almost unaffected by the overexpression of the pediocin PA-1 cluster (Fig. 1A,B, Table 1) and, appeared highly vital even in late stages of the production process, where it faced high levels of the antimicrobial peptide, low oxygen availability, and an inhibitory acidic environment (Fig. 5). Notably, C. glutamicum has been shown to be robust for the production of a range of demanding chemicals at high concentration, sometimes using quite toxic feedstocks [32, 59, 61, 79, 80]. The microbe performed well under suboptimal growth conditions such as oxygen limitation, required to synthetize lactate [81] and succinate [82], and acidic pH, promoting the production of diaminopentane [50]. In addition, our work demonstrates substantial robustness of C. glutamicum to challenging conditions, and that this high naturally existing tolerance makes the bacterium, also, a superior producer of bacteriocins.
It appeared a bit surprising, however, that none of the tested stress-protecting gene candidates promoted growth of the chassis strain CR099 in the acidic oxygen limiting environment but partially even negatively affected growth. An interesting observation was the reduced growth upon stronger expression of the glucose 6-phosphate dehydrogenase gene (Fig. 4, Table 2). As shown before for C. glutamicum, this design pushes extra carbon into the pentose phosphate (PP) pathway while reducing the flux through the Emden-Meyerhof-Parnas (EMP) pathway [83]. Efficient growth of C. glutamicum under oxygen limitation, as applied here, however, relies on a high EMP pathway flux [81, 84]. It therefore seems that the negative growth effect might be due to an unfavorable pathway flux for the specific conditions used here. In contrast, a higher PP pathway flux was found beneficial under oxidative stress conditions (in the presence of sufficient oxygen), obviously posing a different demand on the cell [53].
Table. 2.
Impact of recombinant pediocin production in C. glutamicum on global gene expression
| Locus tag | Gene | Gene description | log2-fold change |
|---|---|---|---|
| Up regulated | |||
| pedA | Pediocin biosynthesis | 12.8 | |
| pedD | Pediocin biosynthesis | 12.8 | |
| CGL_RS05840 | PspC-domain containing protein | 3.0 | |
| CGL_RS03930 | pdxS | Pyridoxal 5'-phosphate synthase lyase | 0.8 |
| CGL_RS04960 | aroF | 3-Deoxy-7-phosphoheptulonate synthase | 0.7 |
| Down regulated | |||
| CGL_RS09745 | bioY | Biotin transporter | 1.6 |
| CGL_RS09750 | ABC transporter ATP-binding protein | 0.6 | |
| CGL_RS10490 | ABC transporter substrate-binding protein | 0.6 | |
C. glutamicum CR099 pXMJ19 Ptac pedACDCg was compared with CR099 pXMJ19 expressing the empty plasmid as control. The cultures were sampled after 13 h (Fig. 1). The data comprise genes that found significantly changed in expression (log2-fold change > 1.0). In addition, weakly affected genes are included (1 > log2-fold > 0.6). Data significance was verified by a t test (p < 0.1). n = 3
In addition, the overexpression of the gene DR1558 from D. radiodurans slowed down growth, different to the growth-promoting effect during diaminopentane production in engineered C. glutamicum KTCT 1857 [50]. Admittedly, the two set-ups used here, differed in terms of strain background and culture conditions, indicating a complex, yet to be fully explored, impact of the heterologous gene. oge.
Conclusions
Here, we demonstrated high level production of the antimicrobial peptide pediocin PA-1, a bioactive compound of significant relevance as agent for precision therapy and prevention of infection [4] and, also, for food preservation [5]. The latter covers applications against Listeria monocytogenes, a food-borne pathogen of increasing concern [87], explaining the huge interest in pediocin PA-1 production. At present, the compound is not available in pure form, and existing production processes are complex, expensive, and inefficient. After several rounds of optimization, we could show that metabolically engineered C. glutamicum accumulated up to 66 mg L−1 of active pediocin PA-1, surpassing previous efforts in various microbes to derive the molecule almost seven-fold (Fig. 6). In this regard, our development sets a benchmark towards industrial pediocin PA-1 production.
Traditionally, the microbe is an established workhorse for amino acid manufacturing [88]. Over the years, metabolic engineering efforts have expanded its product portfolio to a range of bulk chemicals [89], materials [90], and fuels [91]. More recent developments, additionally, demonstrated an impressive capability of C. glutamicum to synthetize high-value active ingredients for food, feed, human health, and well-being [73], including cell-protective extremolytes [92], flavor compounds [93], pharmaceuticals [94] and nutraceuticals [95]. As shown in this work, C. glutamicum appears also capable to take a leading role for the production antimicrobial peptides. The GRAS status, granted to products that are made with the microbe, together with a inherent high metabolic flexibility, genetic accessibility, and process robustness are excellent traits to further drive its development into an antimicrobial peptide production platform and provide many more of these important bioactive compounds.
Materials and methods
Microorganisms, plasmids, and genes
C. glutamicum ATCC 13032 and the genome-reduced strain C. glutamicum CR099 [96], the recombinant pediocin PA-1 producer CR099 pXMJ19-Ptac pedACDCg, and the pediocin PA-1-sensitive assay strains Listeria innocua LMG2785 pIMK2 (kanR) and LMG2785 pNZ44 (cmR) were obtained from previous work [23]. Plasmids, based on the episomal vector pClik5α MCS [97], were amplified in E. coli DH10B (Invitrogen, Carlsbad, CA, USA) [98]. A set of genes was episomally expressed for increased acid tolerance [47–52, 55, 56, 99]. The corresponding plasmids, containing the constitutive promotors PH30 (86 bp) [100] and Ptuf (200 bp) [101], the heterologous genes DR1558 from D. radiodurans [50] and vgb from Vitreoscilla spp. [52], the C. glutamicum genes mcbR, katA, mshA, zwf, the translational start codon variant zwf ATG [55, 56], and the additionally feedback resistant variant zwf ATG, A243T [83], respectively, were synthesized from digital sequence information (GenScript, Piscataway, NJ, USA). All strains and plasmids are listed in Table 2.
Genetic and molecular engineering
The software SnapGene 5.3.2 was used to design strategies for genetic engineering (GSL Biotech, Chicago, IL, USA). For episomal gene expression in C. glutamicum, the amplification and assembly of DNA fragments and the amplification, purification, and transformation of the obtained plasmids into E. coli and C. glutamicum was performed as described previously [57, 71, 102]. Two novel pediocin PA-1 producers were constructed. C. glutamicum pClik Ptuf pedACDCg was derived by replacing the inducible promotor Ptac by the constitutive Ptuf promotor [61]. C. glutamicum pXMJ19 Ptac pedAM31LCDCg expressed a mutated pedAM31L gene that exhibited a substitution of l-methionine by l-leucine at position 31 [29]. The desired derivative was created via PCR using primers that contained the point mutation [60]. The primers used are given in the supplementary information (Additional file 1: Table S1).
Growth media
Different media were used in this work. Pre-cultures were conducted in BHI medium (37 g L−1, Brain Heart Infusion, Becton Dickinson, Heidelberg, Germany). For plate cultures, the medium was supplemented with 12 g L−1 of agar (Becton Dickinson). For initial pediocin PA-1 production, complex TY medium was used, as described before [23]. It contained 20 g L−1 of yeast extract (Becton Dickinson), 15 g L−1 of Bacto- tryptone (Becton Dickinson), and 5 g L−1 of NaCl. The TY medium was supplemented with 20 g L−1 of glucose after 2 h of cultivation [23]. Towards optimized production, a lean medium (designated as GY medium) was developed from a minimal medium recipe [32]. At the start it contained per liter: 10 g of glucose, 15 g of (NH4)2SO4, 10 g of yeast extract (Becton Dickinson), 1 g of NaCl, 0.2 g of MgSO4 7H2O, 55 mg of CaCl2, 20 mg of FeSO4 7H2O, 0.5 mg of biotin,1 mg of thiamin HCl, 1 mg of calcium panthothenate, 10 mL of a 100 × trace element solution (200 mg L−1 FeCl3 6H2O, 200 mg L−1 MnS04 H2O, 50 mg L−1 ZnSO4 7H2O, 20 mg L−1 CuCl2 2H2O, 20 mg L−1 Na2B4O7 10H2O, 10 mg L−1 (NH4)6Mo7O24 4H2O, pH 1.0), and 30 µg L−1 of 3,4-dihydroxybenzoic acid. During the study, the GY medium was optimized, including the use of 200 mM MES buffer at pH values between 5.7 and 6.5 and supplementation with CaCl2 up to 3 g L−1. Further details are given below. For plasmid maintenance, the medium was supplemented with kanamycin (50 µg mL−1) or chloramphenicol (12.5 µg mL−1). When growing inducible production strains, IPTG was added to the culture from a filter-sterilized stock after 2 h to a final level of 0.2 mM.
Medium screening in a microbioreactor
Medium tests were conducted in a microbioreactor with on-line sensing of growth, dissolved oxygen (DO), and pH-value (BioLector I, 700 rpm, 30 °C, 85% humidity, Beckman Coulter, Baesweiler, Germany) using 48-well flower plates, filled with 1 mL medium per well (MTP-48-BOH1, Beckman Coulter) [80]. The experiments (n = 3) were inoculated from a pre-culture in BHI medium, grown overnight in baffled shake flasks (10% filling volume) at 30 °C and 230 rpm (Infors HT Multitron, Bottmingen, Switzerland) and harvested by centrifugation (3 min, 8800 ×g, room temperature). When growing inducible production strains, IPTG was added to the culture from a filter-sterilized stock after 2 h to a final level of 0.2 mM.
Process development in lab scale bioreactors
The impact of the dissolved oxygen (DO) level on production was studied in 1 L stirred tank bioreactors, controlled by the DASGIP control software (SR0700ODLS, Eppendorf SE, Hamburg, Germany). Each reactor was filled with 300 mL TY medium. The inoculum was prepared in two steps. First, cells were grown overnight in BHI medium, harvested as described above, and used to inoculate a second pre-culture in TY medium, which was incubated under the same conditions, harvested during the mid-exponential phase (3 min, 8800 ×g, room temperature) and used to inoculate the process to an initial OD660 of 0.5. The temperature was set to 30 °C (CWD4 Bioblock, Eppendorf SE). Electrodes were used to monitor the DO (VisiFerm DO 225, Hamilton, Höchst, Germany) and the pH value (405-DPAS-SC-K8S/225, Mettler Toledo, Giessen, Germany). During the process, the pH value was maintained at pH 6.5 by automatic addition of 6 M NaOH and 6 M HCl, respectively. In three different setups, the DO level was controlled at 2.5%, 5%, and 30%, respectively, by adjustment of the stirrer speed and the rate of aeration. Pediocin PA-1 production was induced 2 h after inoculation (0.2 mM IPTG). At the same time point, glucose (20 g L−1) was added. Each condition was evaluated as single replicate.
Pediocin PA-1 production in shake flasks
The inoculum was prepared using two subsequent steps. The first pre-culture, inoculated from a BHI agar plate pre-incubated for two days at 30 °C, was grown overnight in BHI medium in baffled shake flasks (10% filling volume) at 30 °C and 230 rpm (Infors HT Multitron), harvested (3 min, 8800 ×g, room temperature) and used to inoculate the second pre-culture which was based on the medium that was later used for the main culture (TY or GY medium) and was grown under the same conditions. Cells were harvested during the mid-exponential phase (3 min, 8800 ×g, room temperature) and used to inoculate the main cultures to an initial OD660 of 0.5. The main cultures were incubated in unbaffled shake flasks on an orbital shaker at a low shaking rate (30 °C, 130 rpm, 10% filling volume, Infors HT Multitron) [23]. In selected cases, the cultures were grown in unbaffled flasks with optical sensors for on-line monitoring of pH and DO [103] using the PreSens SFR system (PreSens, Regensburg, Germany). Pediocin PA-1 production was induced 2 h after inoculation (0.2 mM IPTG). When growing cells in TY medium, glucose (20 g L−1) was additionally supplemented at this time point. All experiments were conducted in triplicate.
Pediocin PA-1 production in lab-scale bioreactors
The production of pediocin PA-1 was demonstrated in 1 L bioreactors (SR0700ODLS, Eppendorf), controlled by the DASGIP control software (Eppendorf). Each reactor was filled with 300 mL medium, buffered at pH 6.5 with 200 mM MES. The medium contained per liter: 80 g of glucose, 15 g of (NH4)2SO4, 15 g of yeast extract (Becton Dickinson), 1 g of NaCl, 0.2 g of MgSO4 7H2O, 2 g CaCl2, 20 mg of FeSO4 7H2O, 0.5 mg of biotin,1 mg of thiamin HCl, 1 mg of calcium panthothenate, 10 mL of a 100 × trace element solution (200 mg L−1 FeCl3 6H2O, 200 mg L−1 MnS04 H2O, 50 mg L−1 ZnSO4 7H2O, 20 mg L−1 CuCl2 2H2O, 20 mg L−1 Na2B4O7 10H2O, 10 mg L−1 (NH4)6Mo7O24 4H2O, pH 1.0), 30 µg L−1 of 3,4-dihydroxybenzoic acid, and 0.2 mM IPTG. The temperature was controlled at 30 °C (CWD4 Bioblock, Eppendorf). Electrodes were used to monitor the DO level (VisiFerm DO 225, Hamilton) and the pH value (405-DPAS-SC-K8S/225, Mettler Toledo). The two parameters were controlled by the automatic addition of 7.3 M NH4OH and 6 M NaOH and the adjustment of stirrer speed and aeration rate, respectively. During the first 4 h, the process was operated at pH 6.5 and a DO level of 10%. For the rest of the process, the pH was controlled at 5.7, while the DO level was reduced to 2.5%. The inoculum was prepared via two pre-culture steps, as described above, and used to inoculate the process to an initial OD660 of 0.5. The processes were conducted as duplicate.
Quantification of the cell concentration
The optical density (OD660 ) in shake flask and bioreactor cultures was monitored off-line at 660 nm, using a spectrophotometer. A correlation between cell dry weight (CDW) and optical density (CDW [g L−1] = 0.32 × OD660) was taken from previous work [44] to infer CDW concentrations from OD660 readings. In microbioreactor cultures, the photometric analysis was conducted on-line at 620 nm.
Quantification of glucose
Glucose was quantified by HPLC (1260 Infinity Series, Agilent Technologies, Santa Clara, CA, USA), equipped with a MetaCarb 87 C column (7,8 × 300 mm, Agilent Technologies) as stationary phase that was heated to 80 °C. Deionized water served as mobile phase (1 mL min−1) [104]. The refractive index was used for detection (1260 RID G1362A, Agilent Technologies), and external standards were used for quantification.
Quantification of lactate
Lactate was quantified by HPLC (1260 Infinity Series, Agilent Technologies, Santa Clara, CA, USA), using an ion-moderated partition column (Aminex HPX-87H, 300 × 7.8 mm, 45 °C, Bio-Rad, München, Germany) and isocratic elution with 12 mM H2SO4 (0.5 mL min−1). The refractive index was used for detection (1260 RID G1362A, Agilent Technologies), and external standards were used for quantification.
Quantification of pediocin PA-1 activity
The biological activity of pediocin PA-1 was determined using a growth inhibition assay [23, 105]. The sensor strains L. innocua pIMK2 and L. innocua pNZ44 were grown overnight in glass tubes (filled 20% with BHI medium) on a rotary shaker (37 °C, 230 rpm, Infors HT Multitron). The samples (culture supernatant) were stepwise diluted with BHI medium in a 96-well microtiter plate. This yielded a twofold dilution series (100 µL for each dilution). In parallel, the L. innocua cultures were diluted 1:25-fold in fresh BHI medium, and the obtained suspension was mixed 1:1 with each diluted sample. The filled microtiter plate was incubated for 6 h (37 °C, 230 rpm, Infors HT Multitron). Afterwards, the cell concentration in each well was quantified at 595 nm (Labsystems iEMS Reader MF, Thermo Fisher, Waltham, MA, USA). The pediocin PA-1 activity was then estimated from the growth inhibition data [105], using software-based parameter fitting with the sigmoidal dose response tool (Origin 2021, Northampton, UK). Throughout this study, the activity is given as bacteriocin units per mL (BU mL−1). Using the recently determined specific biological activity of purified, commercial pediocin PA-1 [23], allowed to infer peptide concentrations from biological activity measurement.
Global gene expression profiling
One-color DNA microarrays were used for global gene expression profiling of C. glutamicum [106]. In short, custom-made microarrays (8 ×15 K) with 8990 probes were designed with the eArray online tool (Agilent Technologies). The final array covered the entire genome of C. glutamicum ATCC 13,032 (entry number: # T00102, KEGG database), the genes pedA and pedD from the heterologous pediocin PA-1 operon, and 60 Agilent-based quality control probes (SurePrint G3 Custom GE 8 × 60 K, Agilent Technologies). Each gene was represented by three different 45–60 bp probes. Each probe was applied as 6 replicates and randomized to feature locations on the slide. Prior to analysis, cells were quickly harvested (1 min, 13,000 ×g, RT). The obtained pellet was immediately transferred into liquid nitrogen, followed by RNA isolation (RNeasy Mini Kit, Qiagen, Hilden, Germany). Concentration and purity of the isolated total RNA were analyzed by absorption measurement (NanoDrop 1000, PEQLAB Biotechnology GmbH, Erlangen, Germany). Furthermore, the RNA integrity was determined using semi-automatized chip-based electrophoresis (RNA 6000 Nano Kit, 2100 Bioanalyzer System, Agilent Technologies). All samples exhibited an RNA integrity number of > 9.8. Subsequently, 50 ng of total RNA was converted into Cy3-labeled antisense cRNA (Low Input Quick Amp WT Labeling kit, Agilent Technologies). In all cases, the reaction yielded > 825 ng cRNA with a Cy3-activity of > 15 pmol per µg RNA (NanoDrop 1000, PEQLAB Biotechnology GmbH). Then, 600 ng of labeled cRNA was fragmented and hybridized (Gene Expression Hybridization Kit (Agilent Technologies). For hybridization onto the microarray, 40 µL of the prepared cRNA solution was applied in between a gasket slide and the array slide using the SureHyb chamber (G2534A, Agilent Technologies). The paired slides were incubated in a hybridization oven at 65 °C for 17 h (G2545A, Agilent Technologies). Slide holders (G2505-60525, Agilent Technologies) were used to transfer the paired slides into the SureScan microarray scanner cassette (G2600D, Agilent Technologies). Afterwards, the array was scanned (SureScan Microarray Scanner G4900DA, SureScan Microarray Scanner Control Software, Agilent Technologies). Signals were recorded according to the AgilentG3_GX_1 color scanner protocol. Data were extracted by the microarray and feature extraction software (version 12.1.1.1, Agilent Technologies). The software GeneSpring (version 14.9, Agilent Technologies) was used for data visualization. For statistical analysis, a moderated t test was applied (Smyth, 2004), considering asymptotic computation of p-values adjusted for multiple testing according to the Benjamini–Hochberg method and a q-value cut-off of 0.05 (Benjamini and Hochberg, 1995). The data were then filtered for genes with a log2-fold change ≥ 1 (p-value ≤ 0.1). RNA extraction and analysis were conducted in biological triplicate for each strain. The entire transcriptome data set is available at GEO (GSE220713).
Supplementary Information
Additional file 1 Figure S1. Principal component analysis of the transcriptome data set comprising the pediocin producer C. glutamicum CR099 pXMJ19 Ptac pedACD and its reference C. glutamicum CR09 pXMJ19, expressing the empty plasmid. n=3. Figure S2. Pediocin production in recombinant C. glutamicum CR099 pXMJ19 Ptac pedACDCg using TY medium (A). Glucose (20 g L-1) was added after 2 h. Pediocin production was induced, also after 2 h, by the addition of 0.2 mM IPTG. During the process, pH value and DO were monitored online. GY medium based pediocin production with additional pH monitoring (B). n=3. Figure S3. Pediocin production in recombinant C. glutamicum CR099 pEKEx2 Ptac pedAM31LCDCg using TY medium in baffled shake flasks (aerobic conditions). Glucose (20 g L-1) was added after 2 h. Pediocin production was induced, also after 2 h, by the addition of 0.2 mM IPTG. The used plasmid is a well-established vector for C. glutamicum [1]. n=3. Table S1. Primers for strain construction.
Author contributions
JC: Conceptualization, Methodology, Validation, Investigation, Formal analysis, Writing—original draft, Writing—review and editing. PC: Methodology, Investigation, Writing—review and editing. JB: Conceptualization, Methodology, Writing—review and editing. CKD: Conceptualization, Methodology, Writing—review and editing. OG: Methodology, Investigation, Writing—review and editing. CUR: Conceptualization, Funding acquisition, Methodology, Validation, Formal analysis, Writing—review and editing. MK: Conceptualization, Methodology, Validation, Formal analysis, Writing—review and editing. CW: Conceptualization, Funding acquisition, Methodology, Validation, Formal analysis, Writing—original draft, Writing—review & editing. All authors read and approved the final munscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This project has received funding from the Bio Based Industries Joint Undertaking under the European Union’s Horizon 2020 research and innovation program under grant agreement No 790507. CW furthermore acknowledges funding by the German Ministry of Education and Research through the grants Explomare (031BB0866D) and EXTRA (031B0822A). JB and MK acknowledge funding by the Hans-and-Ruth-Giessen-Stiftung.
Availability of data and materials
The dataset(s) supporting the conclusions of this article are all included within the article.
Declarations
Ethics approval and consent to participate
Not applicable. The manuscript does not contain data collected from humans or animals.
Consent for publication
Not applicable.
Competing interests
OG, CR, and CW are co-inventors on a patent application related to this research. The other authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jens Christmann, Email: jens.christmann@uni-saarland.de.
Peng Cao, Email: peng.cao@uni-saarland.de.
Judith Becker, Email: judith.becker@uni-saarland.de.
Christian K. Desiderato, Email: christian.desiderato@uni-ulm.de
Oliver Goldbeck, Email: oliver.goldbeck@uni-ulm.de.
Christian U. Riedel, Email: christian.riedel@uni-ulm.de
Michael Kohlstedt, Email: michael.kohlstedt@uni-saarland.de.
Christoph Wittmann, Email: christoph.wittmann@uni-saarland.de.
References
- 1.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 2.Chikindas ML, Weeks R, Drider D, Chistyakov VA, Dicks LM. Functions and emerging applications of bacteriocins. Curr Opin Biotechnol. 2018;49:23–28. doi: 10.1016/j.copbio.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cotter PD, Ross RP, Hill C. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol. 2013;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 4.Heilbronner S, Krismer B, Brotz-Oesterhelt H, Peschel A. The microbiome-shaping roles of bacteriocins. Nat Rev Microbiol. 2021;19:726–739. doi: 10.1038/s41579-021-00569-w. [DOI] [PubMed] [Google Scholar]
- 5.Mills S, Stanton C, Hill C, Ross R. New developments and applications of bacteriocins and peptides in foods. Annu Rev Food Sci Technol. 2011;2:299–329. doi: 10.1146/annurev-food-022510-133721. [DOI] [PubMed] [Google Scholar]
- 6.Organization WH: WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007–2015. World Health Organization. 2015.
- 7.Abebe E, Gugsa G, Ahmed M. Review on major food-borne zoonotic bacterial pathogens. J Trop Med. 2020;2020:4674235. doi: 10.1155/2020/4674235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cintas L, Casaus P, Fernández M, Hernández P. Comparative antimicrobial activity of enterocin L50, pediocin PA-1, nisin A and lactocin S against spoilage and foodborne pathogenic bacteria. Food Microbiol. 1998;15:289–298. [Google Scholar]
- 9.Nes IF, Diep DB, Håvarstein LS, Brurberg MB, Eijsink V, Holo H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Van Leeuwenhoek. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 10.Ríos Colombo NS, Chalón MC, Navarro SA, Bellomio A. Pediocin-like bacteriocins: new perspectives on mechanism of action and immunity. Curr Genet. 2018;64:345–351. doi: 10.1007/s00294-017-0757-9. [DOI] [PubMed] [Google Scholar]
- 11.Zhu L, Zeng J, Wang C, Wang J. Structural basis of pore formation in the mannose Phosphotransferase system by pediocin PA-1. Appl Environ Microbiol. 2022;88:e01992–e1921. doi: 10.1128/AEM.01992-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Keeffe T, Hill C. Bacteriocins|potential in food preservation. In: Robinson RK, editor. Encyclopedia of Food Microbiology. Oxford: Elsevier; 1999. pp. 183–191. [Google Scholar]
- 13.Marugg JD, Gonzalez CF, Kunka BS, Ledeboer AM, Pucci MJ, Toonen MY, Walker SA, Zoetmulder LC, Vandenbergh PA. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, and bacteriocin from Pediococcus acidilactici PAC1. 0. Appl Environ Microbiol. 1992;58:2360–2367. doi: 10.1128/aem.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ennahar S, Aoude-Werner D, Sorokine O, Van Dorsselaer A, Bringel F, Hubert J-C, Hasselmann C. Production of pediocin AcH by Lactobacillus plantarum WHE 92 isolated from cheese. Appl Environ Microbiol. 1996;62:4381–4387. doi: 10.1128/aem.62.12.4381-4387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chikindas ML, Garcia-Garcera MJ, Driessen AJ, Ledeboer AM, Nissen-Meyer J, Nes IF, Abee T, Konings WN, Venema G. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl Environ Microbiol. 1993;59:3577–3584. doi: 10.1128/aem.59.11.3577-3584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motlagh A, Bukhtiyarova M, Ray B. Complete nucleotide sequence of pSMB 74, a plasmid encoding the production of pediocin AcH in Pediococcus acidilactici. Lett Appl Microbiol. 1994;18:305–312. doi: 10.1111/j.1472-765x.1994.tb00876.x. [DOI] [PubMed] [Google Scholar]
- 17.Venema K, Kok J, Marugg JD, Toonen MY, Ledeboer AM, Venema G, Chikindas ML. Functional analysis of the pediocin operon of Pediococcus acidilactici PAC1. 0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol Microbiol. 1995;17:515–522. doi: 10.1111/j.1365-2958.1995.mmi_17030515.x. [DOI] [PubMed] [Google Scholar]
- 18.López-Cuellar MdR, Rodríguez-Hernández AI, Chavarría-Hernández N. LAB bacteriocin applications in the last decade. Biotechnol Biotechnol Equip. 2016;30:1039–1050. [Google Scholar]
- 19.Bédard F, Hammami R, Zirah S, Rebuffat S, Fliss I, Biron E. Synthesis, antimicrobial activity and conformational analysis of the class IIa bacteriocin pediocin PA-1 and analogs thereof. Sci Rep. 2018;8:9029. doi: 10.1038/s41598-018-27225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abbasiliasi S, Tan JS, Ibrahim TAT, Bashokouh F, Ramakrishnan NR, Mustafa S, Ariff AB. Fermentation factors influencing the production of bacteriocins by lactic acid bacteria: a review. RSC Adv. 2017;7:29395–29420. [Google Scholar]
- 21.Johnson EM, Jung YG, Jin YY, Jayabalan R, Yang SH, Suh JW. Bacteriocins as food preservatives: challenges and emerging horizons. Crit Rev Food Sci Nutr. 2018;58:2743–2767. doi: 10.1080/10408398.2017.1340870. [DOI] [PubMed] [Google Scholar]
- 22.Teusink B, Molenaar D. Systems biology of lactic acid bacteria: for food and thought. Current Opin Syst Biol. 2017;6:7–13. doi: 10.1016/j.coisb.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldbeck O, Desef DN, Ovchinnikov KV, Perez-Garcia F, Christmann J, Sinner P, Crauwels P, Weixler D, Cao P, Becker J, et al. Establishing recombinant production of pediocin PA-1 in Corynebacterium glutamicum. Metab Eng. 2021;68:34–45. doi: 10.1016/j.ymben.2021.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mesa-Pereira B, Rea MC, Cotter PD, Hill C, Ross RP. Heterologous expression of biopreservative bacteriocins with a view to low cost production. Front Microbiol. 2018;9:1654. doi: 10.3389/fmicb.2018.01654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ongey EL, Neubauer P. Lanthipeptides: chemical synthesis versus in vivo biosynthesis as tools for pharmaceutical production. Microb Cell Fact. 2016;15:1–16. doi: 10.1186/s12934-016-0502-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venema K, Kok J, Marugg JD, Toonen MY, Ledeboer AM, Venema G, Chikindas ML. Functional analysis of the pediocin operon of Pediococcus acidilactici PAC1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol Microbiol. 1995;17:515–522. doi: 10.1111/j.1365-2958.1995.mmi_17030515.x. [DOI] [PubMed] [Google Scholar]
- 27.Brissette JL, Russel M, Weiner L, Model P. Phage shock protein, a stress protein of Escherichia coli. Proc Natl Acad Sci. 1990;87:862–866. doi: 10.1073/pnas.87.3.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darwin AJ. The phage-shock-protein response. Mol Microbiol. 2005;57:621–628. doi: 10.1111/j.1365-2958.2005.04694.x. [DOI] [PubMed] [Google Scholar]
- 29.Johnsen L, Fimland G, Eijsink V, Nissen-Meyer J. Engineering increased stability in the antimicrobial peptide pediocin PA-1. Appl Environ Microbiol. 2000;66:4798–4802. doi: 10.1128/aem.66.11.4798-4802.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bakkes PJ, Ramp P, Bida A, Dohmen-Olma D, Bott M, Freudl R. Improved pEKEx2-derived expression vectors for tightly controlled production of recombinant proteins in Corynebacterium glutamicum. Plasmid. 2020;112:102540. doi: 10.1016/j.plasmid.2020.102540. [DOI] [PubMed] [Google Scholar]
- 31.Kim WS, Hall RJ, Dunn NW. The effect of nisin concentration and nutrient depletion on nisin production of Lactococcuslactis. Appl Microbiol Biotechnol. 1997;48:449–453. doi: 10.1007/s002530051078. [DOI] [PubMed] [Google Scholar]
- 32.Kind S, Neubauer S, Becker J, Yamamoto M, Völkert M, Abendroth GV, Zelder O, Wittmann C. From zero to hero—production of bio-based nylon from renewable resources using engineered Corynebacterium glutamicum. Metab Eng. 2014;25:113–123. doi: 10.1016/j.ymben.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 33.Aasen IM, Møretrø T, Katla T, Axelsson L, Storrø I. Influence of complex nutrients, temperature and pH on bacteriocin production by Lactobacillus sakei CCUG 42687. Appl Microbiol Biotechnol. 2000;53:159–166. doi: 10.1007/s002530050003. [DOI] [PubMed] [Google Scholar]
- 34.Biosciences B: BD Bionutrients technical manual. 4 edn. Sparks. 2006.
- 35.Biswas SR, Ray P, Johnson MC, Ray B. Influence of growth-conditions on the production of a bacteriocin, Pediocin Ach, by Pediococcus-Acidilactici H. Appl Environ Microbiol. 1991;57:1265–1267. doi: 10.1128/aem.57.4.1265-1267.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerra N, Pastrana L. Influence of pH drop on both nisin and pediocin production by Lactococcus lactis and Pediococcus acidilactici. Lett Appl Microbiol. 2003;37:51–55. doi: 10.1046/j.1472-765x.2003.01346.x. [DOI] [PubMed] [Google Scholar]
- 37.Desiderato CK, Hasenauer KM, Reich SJ, Goldbeck O, Holivololona L, Ovchinnikov KV, Reiter A, Oldiges M, Diep DB, Eikmanns BJ, Riedel CU. Garvicin Q: characterization of biosynthesis and mode of action. Microb Cell Fact. 2022;21:236. doi: 10.1186/s12934-022-01952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inui M, Murakami S, Okino S, Kawaguchi H, Vertes AA, Yukawa H. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol. 2004;7:182–196. doi: 10.1159/000079827. [DOI] [PubMed] [Google Scholar]
- 39.Tsuge Y, Uematsu K, Yamamoto S, Suda M, Yukawa H, Inui M. Glucose consumption rate critically depends on redox state in Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol. 2015 doi: 10.1007/s00253-015-6540-2. [DOI] [PubMed] [Google Scholar]
- 40.Teramoto H, Watanabe K, Suzuki N, Inui M, Yukawa H. High yield secretion of heterologous proteins in Corynebacterium glutamicum using its own Tat-type signal sequence. Appl Microbiol Biotechnol. 2011;91:677–687. doi: 10.1007/s00253-011-3281-8. [DOI] [PubMed] [Google Scholar]
- 41.Freier L, Hemmerich J, Scholer K, Wiechert W, Oldiges M, von Lieres E. Framework for Kriging-based iterative experimental analysis and design: optimization of secretory protein production in Corynebacterium glutamicum. Eng Life Sci. 2016;16:538–549. [Google Scholar]
- 42.Weixler D, Goldbeck O, Seibold GM, Eikmanns BJ, Riedel CU: Towards improved resistance of Corynebacterium glutamicum against nisin. bioRxiv. 2021.
- 43.Wittmann C, Kim HM, Heinzle E. Metabolic network analysis of lysine producing Corynebacterium glutamicum at a miniaturized scale. Biotechnol Bioeng. 2004;87:1–6. doi: 10.1002/bit.20103. [DOI] [PubMed] [Google Scholar]
- 44.Rohles CM, Giesselmann G, Kohlstedt M, Wittmann C, Becker J. Systems metabolic engineering of Corynebacterium glutamicum for the production of the carbon-5 platform chemicals 5-aminovalerate and glutarate. Microb Cell Fact. 2016;15:154. doi: 10.1186/s12934-016-0553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vassilev I, Giesselmann G, Schwechheimer SK, Wittmann C, Virdis B, Krömer JO. Anodic electro-fermentation: Anaerobic production of L-Lysine by recombinant Corynebacterium glutamicum. Biotechnol Bioeng. 2018;6:1490. doi: 10.1002/bit.26562. [DOI] [PubMed] [Google Scholar]
- 46.Limberg MH, Joachim M, Klein B, Wiechert W, Oldiges M. pH fluctuations imperil the robustness of C. glutamicum to short term oxygen limitation. J Biotechnol. 2017;259:248–260. doi: 10.1016/j.jbiotec.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 47.Wang T, Gao F, Kang Y, Zhao C, Su T, Li M, Si M, Shen X. Mycothiol peroxidase MPx protects Corynebacterium glutamicum against acid stress by scavenging ROS. Biotech Lett. 2016;38:1221–1228. doi: 10.1007/s10529-016-2099-y. [DOI] [PubMed] [Google Scholar]
- 48.Liu YB, Yang XB, Yin YJ, Lin JS, Chen C, Pan JF, Si MR, Shen XH. Mycothiol protects Corynebacterium glutamicum against acid stress via maintaining intracellular pH homeostasis, scavenging ROS, and S-mycothiolating MetE. J Gen Appl Microbiol. 2016;62:144–153. doi: 10.2323/jgam.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Xu N, Lv H, Wei L, Liang Y, Ju J, Liu J, Ma Y. Impaired oxidative stress and sulfur assimilation contribute to acid tolerance of Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2019;103:1877–1891. doi: 10.1007/s00253-018-09585-y. [DOI] [PubMed] [Google Scholar]
- 50.Kang SB, Choi JI. Enhanced cadaverine production by recombinant Corynebacterium glutamicum with a heterologous DR1558 regulator at low pH condition. Pro Biochem. 2021;111:63–70. [Google Scholar]
- 51.Liu Q, Zhang J, Wei X-X, Ouyang S-P, Wu Q, Chen G-Q. Microbial production of l-glutamate and l-glutamine by recombinant Corynebacterium glutamicum harboring Vitreoscilla hemoglobin gene vgb. Appl Microbiol Biotechnol. 2008;77:1297–1304. doi: 10.1007/s00253-007-1254-8. [DOI] [PubMed] [Google Scholar]
- 52.Stark BC, Dikshit KL, Pagilla KR. The Biochemistry of Vitreoscilla hemoglobin. Comput Struct Biotechnol J. 2012;3:e201210002. doi: 10.5936/csbj.201210002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krömer JO, Bolten CJ, Heinzle E, Schröder H, Wittmann C. Physiological response of Corynebacterium glutamicum to oxidative stress induced by deletion of the transcriptional repressor McbR. Microbiology. 2008;154:3917–3930. doi: 10.1099/mic.0.2008/021204-0. [DOI] [PubMed] [Google Scholar]
- 54.Wittmann C, Heinzle E. Genealogy profiling through strain improvement by using metabolic network analysis: metabolic flux genealogy of several generations of lysine-producing corynebacteria. Appl Environ Microbiol. 2002;68:5843–5859. doi: 10.1128/AEM.68.12.5843-5859.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Becker J, Buschke N, Bucker R, Wittmann C. Systems level engineering of Corynebacterium glutamicum—reprogramming translational efficiency for superior production. Eng Life Sci. 2010;10:430–438. [Google Scholar]
- 56.Becker J, Rohles CM, Wittmann C. Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products. Metab Eng. 2018;50:122–141. doi: 10.1016/j.ymben.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 57.Rohles CM, Glaser L, Kohlstedt M, Giesselmann G, Pearson S, del Campo A, Becker J, Wittmann C. A bio-based route to the carbon-5 chemical glutaric acid and to bionylon-6,5 using metabolically engineered Corynebacterium glutamicum. Green Chem. 2018;20:4662–4674. [Google Scholar]
- 58.Zahoor A, Lindner SN, Wendisch VF. Metabolic engineering of Corynebacterium glutamicum aimed at alternative carbon sources and new products. Comput Struct Biotechnol J. 2012;3:e201210004. doi: 10.5936/csbj.201210004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohles CM, Gläser L, Kohlstedt M, Gießelmann G, Pearson S, del Campo A, Becker J, Wittmann C. A bio-based route to the carbon-5 chemical glutaric acid and to bionylon-6,5 using metabolically engineered Corynebacterium glutamicum. Green Chem. 2018;20:4662–4674. [Google Scholar]
- 60.Hoffmann SL, Kohlstedt M, Jungmann L, Hutter M, Poblete-Castro I, Becker J, Wittmann C. Cascaded valorization of brown seaweed to produce l-lysine and value-added products using Corynebacterium glutamicum streamlined by systems metabolic engineering. Metab Eng. 2021;67:293–307. doi: 10.1016/j.ymben.2021.07.010. [DOI] [PubMed] [Google Scholar]
- 61.Rohles C, Pauli S, Giesselmann G, Kohlstedt M, Becker J, Wittmann C. Systems metabolic engineering of Corynebacterium glutamicum eliminates all by-products for selective and high-yield production of the platform chemical 5-aminovalerate. Metab Eng. 2022;73:168–181. doi: 10.1016/j.ymben.2022.07.005. [DOI] [PubMed] [Google Scholar]
- 62.Ovchinnikov KV, Chi H, Mehmeti I, Holo H, Nes IF, Diep DB. Novel group of leaderless multipeptide bacteriocins from gram-positive bacteria. Appl Environ Microbiol. 2016;82:5216–5224. doi: 10.1128/AEM.01094-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tosukhowong A, Zendo T, Visessanguan W, Roytrakul S, Pumpuang L, Jaresitthikunchai J, Sonomoto K. Garvieacin Q, a novel class II bacteriocin from Lactococcus garvieae BCC 43578. Appl Environ Microbiol. 2012;78:1619–1623. doi: 10.1128/AEM.06891-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tymoszewska A, Diep DB, Wirtek P, Aleksandrzak-Piekarczyk T. The non-lantibiotic bacteriocin garvicin Q targets man-PTS in a broad spectrum of sensitive bacterial genera. Sci Rep. 2017;7:1–14. doi: 10.1038/s41598-017-09102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Papagianni M, Anastasiadou S. The bacteriocins of Pediococci Sources, production, properties and applications. Microb Cell Fact. 2009;8:3. doi: 10.1186/1475-2859-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Authority EFS Prevention ECfD Control The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018;16:e05500. doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rothrock MJ, Jr, Davis ML, Locatelli A, Bodie A, McIntosh TG, Donaldson JR, Ricke SC. Listeria occurrence in poultry flocks: detection and potential implications. Front Vet Sci. 2017;4:125. doi: 10.3389/fvets.2017.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tahoun AB, Abou Elez RM, Abdelfatah EN, Elsohaby I, El-Gedawy AA, Elmoslemany AM. Listeria monocytogenes in raw milk, milking equipment and dairy workers: molecular characterization and antimicrobial resistance patterns. J Global Antimicrob Resist. 2017;10:264–270. doi: 10.1016/j.jgar.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 69.Mateus T, Silva J, Maia RL, Teixeira P. Listeriosis during pregnancy: a public health concern. ISRN Obstet Gynecol. 2013;2013:851712. doi: 10.1155/2013/851712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Briand L, Marcion G, Kriznik A, Heydel JM, Artur Y, Garrido C, Seigneuric R, Neiers F. A self-inducible heterologous protein expression system in Escherichia coli. Sci Rep. 2016;6:33037. doi: 10.1038/srep33037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Giesselmann G, Dietrich D, Jungmann L, Kohlstedt M, Jeon EJ, Yim SS, Sommer F, Zimmer D, Muhlhaus T, Schroda M, et al. Metabolic engineering of Corynebacterium glutamicum for high-level ectoine production: design, combinatorial assembly, and implementation of a transcriptionally balanced heterologous ectoine pathway. Biotechnol J. 2019;14:e1800417. doi: 10.1002/biot.201800417. [DOI] [PubMed] [Google Scholar]
- 72.Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels—Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol. 2012;23:631–640. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 73.Wolf S, Becker J, Tsuge Y, Kawaguchi H, Kondo A, Marienhagen J, Bott M, Wendisch VF, Wittmann C. Advances in metabolic engineering of Corynebacterium glutamicum to produce high-value active ingredients for food, feed, human health, and well-being. Essays Biochem. 2021;65:197–212. doi: 10.1042/EBC20200134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karpiński TM, Szkaradkiewicz AK. Characteristic of bacteriocines and their application. Pol J Microbiol. 2013;62:223–235. [PubMed] [Google Scholar]
- 75.Burkovski A. Cell envelope of corynebacteria: structure and influence on pathogenicity. Int Sch Res Not. 2013 doi: 10.1155/2013/935736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Uteng M, Hauge HH, Brondz I, Nissen-Meyer J, Fimland G. Rapid two-step procedure for large-scale purification of pediocin-like bacteriocins and other cationic antimicrobial peptides from complex culture medium. Appl Environ Microbiol. 2002;68:952–956. doi: 10.1128/AEM.68.2.952-956.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Täuber S, Blöbaum L, Wendisch VF, Grünberger A. Growth response and recovery of Corynebacterium glutamicum colonies on single-cell level upon defined pH stress pulses. Frontiers Microbiol. 2021;12:711893. doi: 10.3389/fmicb.2021.711893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jakob K, Satorhelyi P, Lange C, Wendisch VF, Silakowski B, Scherer S, Neuhaus K. Gene expression analysis of Corynebacterium glutamicum subjected to long-term lactic acid adaptation. J Bacteriol. 2007;189:5582–5590. doi: 10.1128/JB.00082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bolten CJ, Schröder H, Dickschat J, Wittmann C. Towards methionine overproduction in Corynebacterium glutamicum–methanethiol and dimethyldisulfide as reduced sulfur sources. J Microbiol Biotechnol. 2010;20:1196–1203. doi: 10.4014/jmb.1002.02018. [DOI] [PubMed] [Google Scholar]
- 80.Becker J, Kuhl M, Kohlstedt M, Starck S, Wittmann C. Metabolic engineering of Corynebacterium glutamicum for the production of cis, cis-muconic acid from lignin. Microb Cell Fact. 2018;17:115. doi: 10.1186/s12934-018-0963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsuge Y, Yamamoto S, Kato N, Suda M, Vertes AA, Yukawa H, Inui M. Overexpression of the phosphofructokinase encoding gene is crucial for achieving high production of D-lactate in Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol. 2015;99:4679–4689. doi: 10.1007/s00253-015-6546-9. [DOI] [PubMed] [Google Scholar]
- 82.Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol. 2008;81:459–464. doi: 10.1007/s00253-008-1668-y. [DOI] [PubMed] [Google Scholar]
- 83.Becker J, Klopprogge C, Herold A, Zelder O, Bolten CJ, Wittmann C. Metabolic flux engineering of L-lysine production in Corynebacterium glutamicum–over expression and modification of G6P dehydrogenase. J Biotechnol. 2007;132:99–109. doi: 10.1016/j.jbiotec.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 84.Tsuge Y, Yamamoto S, Suda M, Inui M, Yukawa H. Reactions upstream of glycerate-1,3-bisphosphate drive Corynebacterium glutamicum (D)-lactate productivity under oxygen deprivation. Appl Microbiol Biotechnol. 2013;97:6693–6703. doi: 10.1007/s00253-013-4986-7. [DOI] [PubMed] [Google Scholar]
- 85.Wittmann C, Weber J, Betiku E, Krömer J, Bohm D, Rinas U. Response of fluxome and metabolome to temperature-induced recombinant protein synthesis in Escherichia coli. J Biotechnol. 2007;132:375–384. doi: 10.1016/j.jbiotec.2007.07.495. [DOI] [PubMed] [Google Scholar]
- 86.Michel A, Koch-Koerfges A, Krumbach K, Brocker M, Bott M. Anaerobic growth of Corynebacterium glutamicum via mixed-acid fermentation. Appl Environ Microbiol. 2015;81:7496–7508. doi: 10.1128/AEM.02413-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reda WW, Abdel-Moein K, Hegazi A, Mohamed Y, Abdel-Razik K. Listeria monocytogenes: an emerging food-borne pathogen and its public health implications. J Infect Dev Ctries. 2016;10:149–154. doi: 10.3855/jidc.6616. [DOI] [PubMed] [Google Scholar]
- 88.Becker J, Wittmann C. Systems and synthetic metabolic engineering for amino acid production - the heartbeat of industrial strain development. Curr Opin Biotechnol. 2012;23:718–726. doi: 10.1016/j.copbio.2011.12.025. [DOI] [PubMed] [Google Scholar]
- 89.Tsuge Y, Hasunuma T, Kondo A. Recent advances in the metabolic engineering of Corynebacterium glutamicum for the production of lactate and succinate from renewable resources. J Ind Microbiol Biotechnol. 2015;42:375–389. doi: 10.1007/s10295-014-1538-9. [DOI] [PubMed] [Google Scholar]
- 90.Kind S, Kreye S, Wittmann C. Metabolic engineering of cellular transport for overproduction of the platform chemical 1,5-diaminopentane in Corynebacterium glutamicum. Metab Eng. 2011;13:617–627. doi: 10.1016/j.ymben.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 91.Yamamoto S, Suda M, Niimi S, Inui M, Yukawa H. Strain optimization for efficient isobutanol production using Corynebacterium glutamicum under oxygen deprivation. Biotechnol Bioeng. 2013;110:2938–2948. doi: 10.1002/bit.24961. [DOI] [PubMed] [Google Scholar]
- 92.Becker J, Schäfer R, Kohlstedt M, Harder BJ, Borchert NS, Stöveken N, Bremer E, Wittmann C. Systems metabolic engineering of Corynebacterium glutamicum for production of the chemical chaperone ectoine. Microb Cell Fact. 2013;12:110. doi: 10.1186/1475-2859-12-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dickschat J, Wickel S, Bolten CJ, Nawrath T, Schulz S, Wittmann C. Pyrazine biosynthesis in Corynebacterium glutamicum. Eur J Org Chem. 2010;2010:2687–2695. [Google Scholar]
- 94.Milke L, Kallscheuer N, Kappelmann J, Marienhagen J. Tailoring Corynebacterium glutamicum towards increased malonyl-CoA availability for efficient synthesis of the plant pentaketide noreugenin. Microb Cell Fact. 2019;18:71. doi: 10.1186/s12934-019-1117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Henke NA, Heider SA, Peters-Wendisch P, Wendisch VF. Production of the marine carotenoid astaxanthin by metabolically engineered Corynebacterium glutamicum. Mar Drugs. 2016;14(7):124. doi: 10.3390/md14070124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baumgart M, Unthan S, Rückert C, Sivalingam J, Grünberger A, Kalinowski J, Bott M, Noack S, Frunzke J. Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl Environ Microbiol. 2013;79:6006–6015. doi: 10.1128/AEM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Buschke N, Schröder H, Wittmann C. Metabolic engineering of Corynebacterium glutamicum for production of 1,5-diaminopentane from hemicellulose. Biotechnol J. 2011;6:306–317. doi: 10.1002/biot.201000304. [DOI] [PubMed] [Google Scholar]
- 98.Grant SG, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Becker J, Buschke N, Bücker R, Wittmann C. Systems level engineering of Corynebacterium glutamicum–reprogramming translational efficiency for superior production. Eng Life Sci. 2010;10:430–438. [Google Scholar]
- 100.Yim SS, An SJ, Kang M, Lee J, Jeong KJ. Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnol Bioeng. 2013;110:2959–2969. doi: 10.1002/bit.24954. [DOI] [PubMed] [Google Scholar]
- 101.Becker J, Zelder O, Häfner S, Schröder H, Wittmann C. From zero to hero—design-based systems metabolic engineering of Corynebacterium glutamicum for l-lysine production. Metab Eng. 2011;13:159–168. doi: 10.1016/j.ymben.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 102.Hoffmann SL, Jungmann L, Schiefelbein S, Peyriga L, Cahoreau E, Portais JC, Becker J, Wittmann C. Lysine production from the sugar alcohol mannitol: design of the cell factory Corynebacterium glutamicum SEA-3 through integrated analysis and engineering of metabolic pathway fluxes. Metab Eng. 2018 doi: 10.1016/j.ymben.2018.04.019. [DOI] [PubMed] [Google Scholar]
- 103.Wittmann C, Kim HM, John G, Heinzle E. Characterization and application of an optical sensor for quantification of dissolved O2 in shake-flasks. Biotechnol Lett. 2003;25:377–380. doi: 10.1023/a:1022402212537. [DOI] [PubMed] [Google Scholar]
- 104.Gläser L, Kuhl M, Stegmuller J, Ruckert C, Myronovskyi M, Kalinowski J, Luzhetskyy A, Wittmann C. Superior production of heavy pamamycin derivatives using a bkdR deletion mutant of Streptomyces albus J1074/R2. Microb Cell Fact. 2021;20:111. doi: 10.1186/s12934-021-01602-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Holo H, Nilssen O, Nes IF. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J Bacteriol. 1991;173:3879–3887. doi: 10.1128/jb.173.12.3879-3887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kohlstedt M, Weimer A, Weiland F, Stolzenberger J, Selzer M, Sanz M, Kramps L, Wittmann C. Biobased PET from lignin using an engineered cis, cis-muconate-producing Pseudomonas putida strain with superior robustness, energy and redox properties. Metab Eng. 2022;72:337–352. doi: 10.1016/j.ymben.2022.05.001. [DOI] [PubMed] [Google Scholar]
- 107.Reviriego C, Fernández A, Horn N, Rodrıguez E, Marın M, Fernández L. Rodríguez J: Production of pediocin PA-1, and coproduction of nisin A and pediocin PA-1, by wild Lactococcus lactis strains of dairy origin. Int Dairy J. 2005;15:45–49. [Google Scholar]
- 108.Wang G, Guo Z, Zhang X, Wu H, Bai X, Zhang H, Hu R, Han S, Pang Y. Gao Za: Heterologous expression of pediocin/papA in Bacillus subtilis. Microb Cell Fact. 2022;21:1–11. doi: 10.1186/s12934-022-01829-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martínez JM, Kok J, Sanders JW, Hernández PE. Heterologous coproduction of enterocin A and pediocin PA-1 by Lactococcus lactis: detection by specific peptide-directed antibodies. Appl Environ Microbiol. 2000;66:3543–3549. doi: 10.1128/aem.66.8.3543-3549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kuniyoshi TM, O’Connor PM, Lawton E, Thapa D, Mesa-Pereira B, Abulu S, Hill C, Ross RP, Oliveira RP, Cotter PD. An oxidation resistant pediocin PA-1 derivative and penocin a display effective anti-Listeria activity in a model human gut environment. Gut Microbes. 2022;14:2004071. doi: 10.1080/19490976.2021.2004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Horn N, Martinez MI, Martinez JM, Hernandez PE, Gasson MJ, Rodriguez JM, Dodd HM. Enhanced production of pediocin PA-1 and coproduction of nisin and pediocin PA-1 by Lactococcus lactis. Appl Environ Microbiol. 1999;65:4443–4450. doi: 10.1128/aem.65.10.4443-4450.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guyonnet D, Fremaux C, Cenatiempo Y, Berjeaud J. Method for rapid purification of class IIa bacteriocins and comparison of their activities. Appl Environ Microbiol. 2000;66:1744–1748. doi: 10.1128/aem.66.4.1744-1748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Figure S1. Principal component analysis of the transcriptome data set comprising the pediocin producer C. glutamicum CR099 pXMJ19 Ptac pedACD and its reference C. glutamicum CR09 pXMJ19, expressing the empty plasmid. n=3. Figure S2. Pediocin production in recombinant C. glutamicum CR099 pXMJ19 Ptac pedACDCg using TY medium (A). Glucose (20 g L-1) was added after 2 h. Pediocin production was induced, also after 2 h, by the addition of 0.2 mM IPTG. During the process, pH value and DO were monitored online. GY medium based pediocin production with additional pH monitoring (B). n=3. Figure S3. Pediocin production in recombinant C. glutamicum CR099 pEKEx2 Ptac pedAM31LCDCg using TY medium in baffled shake flasks (aerobic conditions). Glucose (20 g L-1) was added after 2 h. Pediocin production was induced, also after 2 h, by the addition of 0.2 mM IPTG. The used plasmid is a well-established vector for C. glutamicum [1]. n=3. Table S1. Primers for strain construction.
Data Availability Statement
The dataset(s) supporting the conclusions of this article are all included within the article.