Abstract
The cell cycle is modulated by ubiquitin ligases, including CRL4, which facilitate degradation of the chromatin-bound substrates involved in DNA replication and chromosome segregation. One of the members of the CRL4 complex, RepID (DCAF14/PHIP), recognizes kinetochore-localizing BUB3, known as the CRL4 substrate, and recruits CRL4 to the chromatin/chromosome using the WD40 domain. Here, we show that the RepID WD40 domain provides different platforms to CRL4 and BUB3. Deletion of the H-box or exon 8 located in the RepID WD40 domain compromises the interaction between RepID and CRL4, whereas BUB3 interacts with the exon 1–2 region. Moreover, deletion mutants of other exons in the WD40 domain lost chromatin binding affinity. Structure prediction revealed that the RepID WD40 domain has two beta-propeller folds, linked by loops, which are possibly crucial for chromatin binding. These findings provide mechanistic insights into the space occupancy of the RepID WD40 domain to form a complex with CRL4, BUB3, or chromatin.
Keywords: RepID, WD40 domain, CRL4, BUB3, Cell cycle, DCAF
1. Introduction
The WD40 repeat-containing domain, one of the most abundant in the human genome, consists of approximately 40–60 amino acid motifs with a tryptophan and aspartic acid (WD) end [1]. Each WD40 repeat has a four-stranded antiparallel β-sheet with the protein folded into a β-propeller architecture [2–4]. The WD40 domain serves as a rigid scaffold for protein-protein interactions to coordinate downstream events, such as histone methylation and ubiquitination. Therefore, WD40 domain-containing proteins are involved in various cellular functions, including cell cycle control, apoptosis, transcriptional regulation, and chromatin dynamics.
Cullin-RING E3 ubiquitin ligase 4 (CRL4), a crucial mediator for ubiquitination of proteins required for DNA replication as well as cell cycle control, forms its structure through WD40 domain-dependent interactions [5–8]. CRL4 contains Cullin 4A/B (CUL4A/B) as molecular scaffolds, and DDB1 adaptor protein, E2-conjugating enzyme RBX, and DDB1-CUL4-associated factors (DCAFs) as substrate receptors [7,8]. The DDB1 protein has 21 WD40-like repeats that fold into a three-β propeller configuration (BPA, BPB, BPC) and forms a complex with the arc-shaped helical amino-terminal domain of CUL4A/B using BPB [9,10]. The BPA-BPC double propeller of DDB1 binds to the WD40 domain of DCAFs, leading to bridge formation between CUL4A/B and DCAFs [11–13].
The replication origin binding protein RepID (DCAF14; PHIP) is a member of the DCAF family. RepID facilitates interaction with DDB1 in part via the H-box located in the WD40 domain [14]. RepID is larger than other DCAFs and includes chromatin-recognition domains containing bromodomains and cryptic tudor domains [13,15,16]. Previous reports have shown that WD40-containing proteins are regarded as a new class of histone code readers that recognize methylated lysine and arginine residues [17,18]. Recently, we reported that RepID acts as a structural DCAF by recruiting the CRL4 complex to chromatin/chromosome during cell cycle progression [19,20]. Once CRL4 is recruited to the chromatin/chromosome by the structural DCAF, another DCAF is incorporated into the DCAF pocket within CRL4 instead of structural DCAF to catalyze its substrates. For example, RepID recruits CRL4 to kinetochore-localizing BUB3, which is the subunit of spindle assembly checkpoint (SAC) proteins and prevents premature chromosome segregation by inhibiting anaphase-promoting complex/cyclosome ubiquitin E3 ligase (APC/C), and then another DCAF RBBP7 localized on mitotic spindle rapidly incorporates into CRL4 complex to ubiquitinate BUB3 [19]. This “DCAF switch mechanism” explains loading of the CRL4 complex onto chromatin and dynamics of the CRL4 assembly based on WD40 domain-dependent interactions in space and time [21]. However, functional research on the RepID WD40 domain, including its structure, chromatin binding activity, and how CRL4 and its substrate BUB3 occupy the WD40 domain of the RepID at specific time points is unclear.
Here, we report that WD40 domain has nine blades showing three- or four-stranded antiparallel β-sheets in each blade folded into two β-propeller configuration, connected via a loop generated by the exon 3-containing region. Except for the H-box, loop, and blade 9 portions, we observed that most parts of the WD40 domain are crucial for the chromatin binding of RepID, which recruits CRL4 on chromatin. Moreover, we identified that RepID utilizes the H-box as well as the 9th blade to bind to CRL4. Reduced chromatin binding of RepID or failure of interaction between RepID and CRL4 by exon deletion leads to low amounts of chromatin-bound CRL4, resulting in resistance to the neddylation inhibitor pevonedistat. Compared to CRL4, BUB3 could bind with the 2nd and 3rd blades located in the first β-propeller of the RepID WD40 domain, showing that CRL4 and BUB3 may occupy different portions of the RepID WD40 domain. These observations provide insights into the functional dynamics of the CRL4-substrate based on RepID and the role of the RepID WD40 domain, which could act as a possible target for cancer therapy.
2. Materials and methods
2.1. Cell culture and chemicals
Human U2OS cells with or without RepID/RepID mutants were incubated in Dulbecco’s modified Eagle’s medium (Invitrogen, 10569-010) supplemented with 10% heat-inactivated fetal bovine serum in a 37 °C/5% CO2 humidified incubator. Original U2OS osteosarcoma cell lines were obtained from the American Type Culture Collection (ATCC; www.atcc.org). All cell lines tested negative for mycoplasmas (Lonza, LT07-418). MLN4924 (Pevonedistat) was purchased from Cayman Chemicals (Cat. 15217-1). Drugs were added to the medium at the indicated concentrations.
2.2. RepID-depleted cell lines
2.3. Constructs
The exon deletion mutants in the RepID WD40 domain were constructed using the Q5 site-directed mutagenesis kit (New England Biolabs, Cat. E0554S) using pCMV-FLAG-tagged RepID full-length (FL) plasmid DNA as a template. The following primers were used: ΔH-box, forward, 5′-GAGCGACTTGTTCCAACTGC-3′, reverse, 5′-GCT GGG TGG GCT ACC ATA-3’; ΔExon1, forward, 5′-GGA CAT GCT GAA ATA TC-3′, reverse, 5′-AAG AAT TCG TTT ATG CAT TTT C-3’; ΔExon2, forward, 5′-CTT CAG GGC CAT AGT GCA-3′, reverse, 5′-TCC TCT TAA GGT AGC TAA CAA C-3’; ΔExon3, forward, 5′-CCT GCA AAA TTT ACA GAG-3′, reverse, 5′-CTG AAG AAC AGC CAA AGG3’; ΔExon4, forward, 5′-TTG GAG TTT CAT ACT GAC AAA G-3′, reverse, 5′-AGG GCG CTC TGT AAA TTT TG-3’; ΔExon5, forward, 5′-TTG TTG GAT ATG GCT ACT C-3′, reverse, 5′-CTC CAA TTC TGA TAT TTT CTC-3’; ΔExon6, forward, 5′-CTG ATG GGT CAT GAA GAT G-3′, reverse, 5′-TTT ATC TTC TAT TCC TTG AAG G-3’; ΔExon7, forward, 5′-TAT TTC AAT ATG ATT GAA GGC-3′, reverse, 5′-CAT CAG GAC ATG AAT TAG TTG-3’; ΔExon8, forward, 5′-ATA GCA GAT CAG ATG TTC-3′, reverse, 5′-GCC TTC AAT CAT ATT GAA ATA AG-3’.
2.4. Chromatin fractionation, co-IP, and immunoblotting
Extraction of the chromatin-bound proteins, immunoprecipitation and immunoblotting was performed as described previously [19,20].
2.5. Immunofluorescence analysis
Immunofluorescence was performed as described previously [19,20].
2.6. Clonogenic survival assay
Clonogenic survival assay was done as described previously [19,20].
2.7. Model building
FASTA sequences of the WD40 domain (1–551 amino acids) of RepID were entered into the website http://swissmodel.expasy.org. Built models were downloaded, and the highest identity score was chosen (ID: 6zqc.17.A). For the model, a cartoon representation of the structure was preferred with the DSSP secondary structure and NGL 3D viewer. The RaptorX server (http://raptorx.uchicago.edu/StructurePropertyPred/predict/) was used to build the secondary structure of the RepID WD40 domain. The model built by using RaptorX was then optimized by calculating the involvement of amino acid numbers in each exon based on 8-state secondary structures.
3. Results
3.1. WD40 domain of the RepID contributes to its chromatin binding
To examine the function of the RepID WD40 domain, we dissected the structure of RepID. It contains nine exons in the WD40 domain that involves the H-Box, which is crucial for binding to DDB1 in the CRL4 complex [14]. Other functional domains, including cryptic Tudor or bromodomains, are located next to the WD40 domain (Fig. 1A). We constructed mutants of the WD40 domain by deleting each exon. Next, we measured the levels of chromatin-bound RepID mutants in RepID-depleted U2OS cells in which we overexpressed these mutants. In wild-type cells, most of the endogenous RepID was detected in chromatin-bound fractions, whereas overexpressed FL in RepID-deficient cells showed a somewhat high fraction of soluble nuclear fractions (approximately 17.2%) (Fig. 1B). Notably, some of the RepID mutants, including exon 1, 2, 4, 5, 6, or 7-deleted forms were detected even in cytosolic fractions (Fig. 1B and C), consistent with a lower chromatin-bound ratio (Fig. 1E). Exon 5 and 6-depleted mutants showed the lowest chromatin binding by increasing soluble nuclear fractions (Fig. 1D and E). Deletion of the H-box, exon 3, or exon 8 had no effect on chromatin binding affinity or cellular localization (Fig. 1B–E). These results were consistent with immunofluorescence analysis, in which FL and mutants deleting H-box, exon 3, or exon 8 were present in pre-extracted nuclei, whereas other mutants were lower due to the dissociation of RepID from chromatin (Fig. 1F and G). These observations suggest that the RepID WD40 domain contributes to chromatin binding.
Fig. 1.
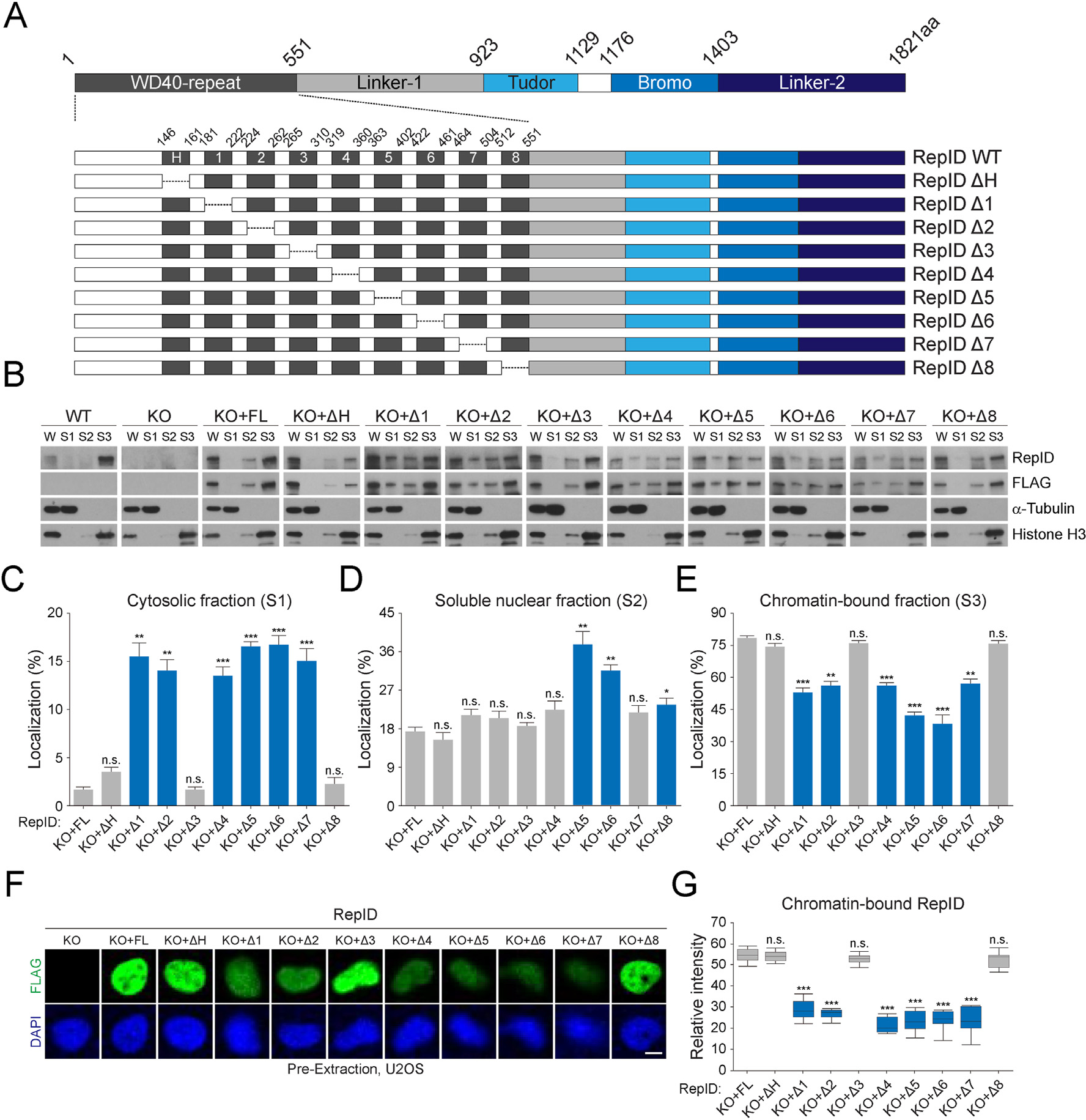
RepID WD40 domain contributes to its chromatin binding. (A) RepID domains and WD40 deletion mutants. WD40 domain consisting of 9 exons including H-box and deletion constructs of each exon is represented with amino acid composition. (B) Levels of RepID full-length (FL) or its deletion mutants from U2OS cells transfected with RepID fragments are indicated. W, whole cell lysates; S1, cytosolic fractions; S2, soluble nuclear fractions; S3, chromatin-bound fractions. Histone H3 and α-tubulin were used as loading controls and fraction markers. (C–E) Quantification of relative RepID signals in cytosolic (C), soluble nuclear (D), and chromatin-bound fractions (E) analyzed as shown in B after normalization with respect to histone H3 or α-tubulin signal intensities, followed by normalization with intensities in whole cell lysates in each cell lines. Error bars represent standard deviations from three independent experiments (*p value < 0.05, **p < 0.01, ***p < 0.001, n.s., not significant, Student’s t-test). (F) U2OS cells were pre-extracted and chromatin-bound RepID using FLAG antibodies (green) and DNA content (DAPI; blue) were detected using immunofluorescence analysis. Scale bar indicates 10 μm. (G) Quantification of relative intensity of each RepID mutant and p-values were calculated using a two-tailed t-test (***p value < 0.001, n.s., not significant, n = 10). Significant changes are indicated as blue bars.
3.2. RepID WD40 domain provides different binding platforms to CRL4 and its substrate
We investigated whether RepID mutations in the WD40 domain affect the CRL4 association on chromatin. As previously reported, the RepID WD40 domain interacts with the CRL4 and recruits CRL4 to the chromatin [20]. Consistent with this notion, components of CRL4, including CUL4A, CUL4B, and DDB1, were detected in the chromatin fraction of cells with rescued FL of RepID but not in cells overexpressing exon 1, 2, 4, 5, 6, or 7-deletion mutants, which show lower chromatin binding of RepID (Fig. 2A and B). Moreover, other mutants including H-box or exon 8-depletion showing similar chromatin-bound RepID ratios as FL also showed reduced chromatin-bound CRL4, except for the exon 3-deletion mutant (CUL4A: 11.046-fold, CUL4B: 6.087-fold, DDB1: 12.148-fold increase compared to average chromatin-bound CRL4 in other mutants). In contrast to the CRL4, the chromatin association of BUB3 was not affected by the RepID mutants (Fig. 2A and B).
Fig. 2.
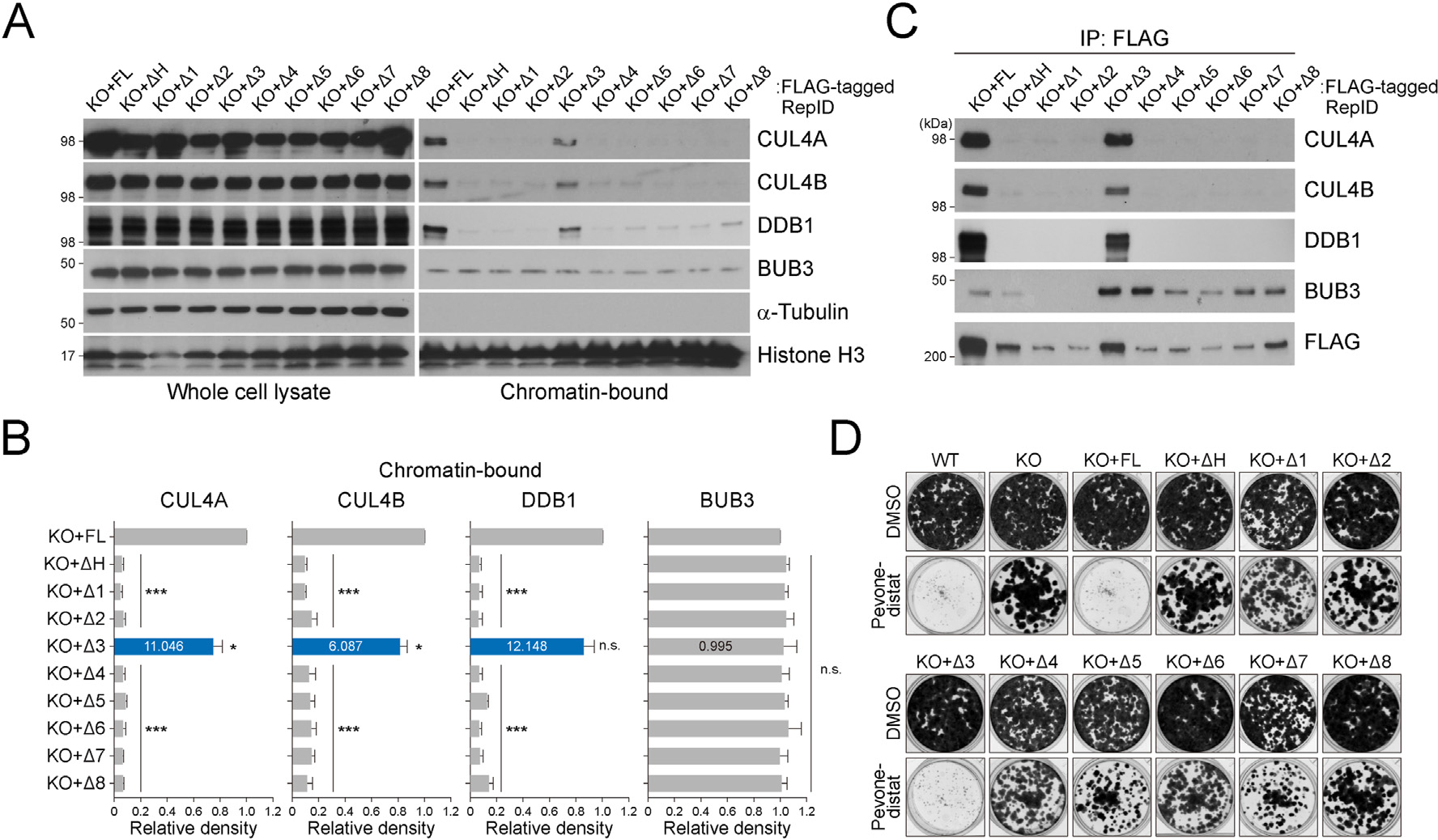
CRL4 and its substrate BUB3 occupy different sites of the RepID WD40 domain. (A) CRL4 and BUB3 levels in whole cell lysates and chromatin-bound fractions from U2OS cells transfected as indicated. (B) Quantification of chromatin-bound CUL4A, CUL4B, DDB1, and BUB3 levels. Error bars represent standard deviations from three independent experiments (*p value < 0.05, ***p < 0.001, n.s., not significant, Student’s t-test). Intensity of the exon 3-deletion mutant is indicated as blue bars and inside numbers represent fold change compared to average intensities of CRL4 and BUB3 in other mutants. (C) Immunoprecipitation assay using a FLAG antibody with chromatin-bound fractions was performed and co-precipitated CRL4 and BUB3 were analyzed via immunoblotting. (D) Colony-formation assay with U2OS cells transfected with various RepID mutants in untreated or MLN4924 (pevonedistat)-treated condition.
We have previously shown that RepID associates with CRL4 and BUB3 using its WD40 domain [19]. Here, we determined how these two proteins interact with the RepID WD40 domain. We generated stable cell lines expressing FLAG-tagged RepID FL or the above mutants in a RepID KO background and immunoprecipitated chromatin proteins using FLAG-agarose beads. As expected, the interaction between CRL4 and RepID mutants was not evident in cells with mutations that cause dissociation of the RepID from chromatin due to the lower chromatin fraction of CRL4 (Fig. 2C). Exon 3-depleted RepID, which showed no significant differences for chromatin-bound RepID, interacted with CRL4, whereas the exon-8-deleted mutant of RepID could not interact with CRL4, consistent with the deletion of the H-box. In contrast to CRL4, the interaction between RepID and BUB3 was evident in most of the mutants, except for exon 1- and 2-deleted forms (Fig. 2C). These results show that CRL4 (H-Box, exon 8) and BUB3 (exons 1 and 2) occupy different regions of the WD40 domain when they bind to RepID, suggesting that the RepID WD40 domain provides different binding platforms to CRL4 and its substrate.
The addition of NEDD8 to CRL4, called neddylation, is essential for CRL4 activation, and we previously reported that reduced CRL4-chromatin binding in the RepID depletion decreases the sensitivity of cancer cells to MLN4924 (pevonedistat) [22,23]. Consistent with a previous report, cells that can recruit CRL4 on chromatin (WT, KO + FL, or KO + deleted exon 3) were sensitive to pevonedistat, whereas cells with faulty CRL4-chromatin loading showed resistance to CRL4 inhibition (Fig. 2D). This result suggests that the expression level of RepID might regulate the sensitivity of cancer cells to drugs targeting CRL4.
3.3. WD40 domain of the RepID consists of two β-propeller folds linked by loop
To determine the molecular characteristics of the RepID WD40 domain, we modeled the structure of its WD40 domain based on its sequence homology using the Swiss-Model algorithm. This domain was predicted to be two β-propeller folds (Fig. 3A). The first bpropeller labeled with RepID-BPA consists of four propeller blades composed of a four-stranded antiparallel β-sheet, except for 4th blade (Exon 3) that contains a three-stranded antiparallel β-sheet with a loop (Fig. 3A and B). This loop positioning Asp301 to Gln323 connects RepID-BPA to the second β-propeller labeled with RepID-BPB, which involves five propeller blades, and the 6th blade (Exon 5) and 9th blade (Exon 8) contain a three-stranded β-sheet (Fig. 3A and B). Moreover, the proximity between blade 1 (H-box) at the N-terminal region and blade 9 (Exon 8) at the C-terminal region was consistent with our above observation, in which CRL4 interacts with the H-box and possibly exon 8 of the RepID WD40 domain.
Fig. 3.
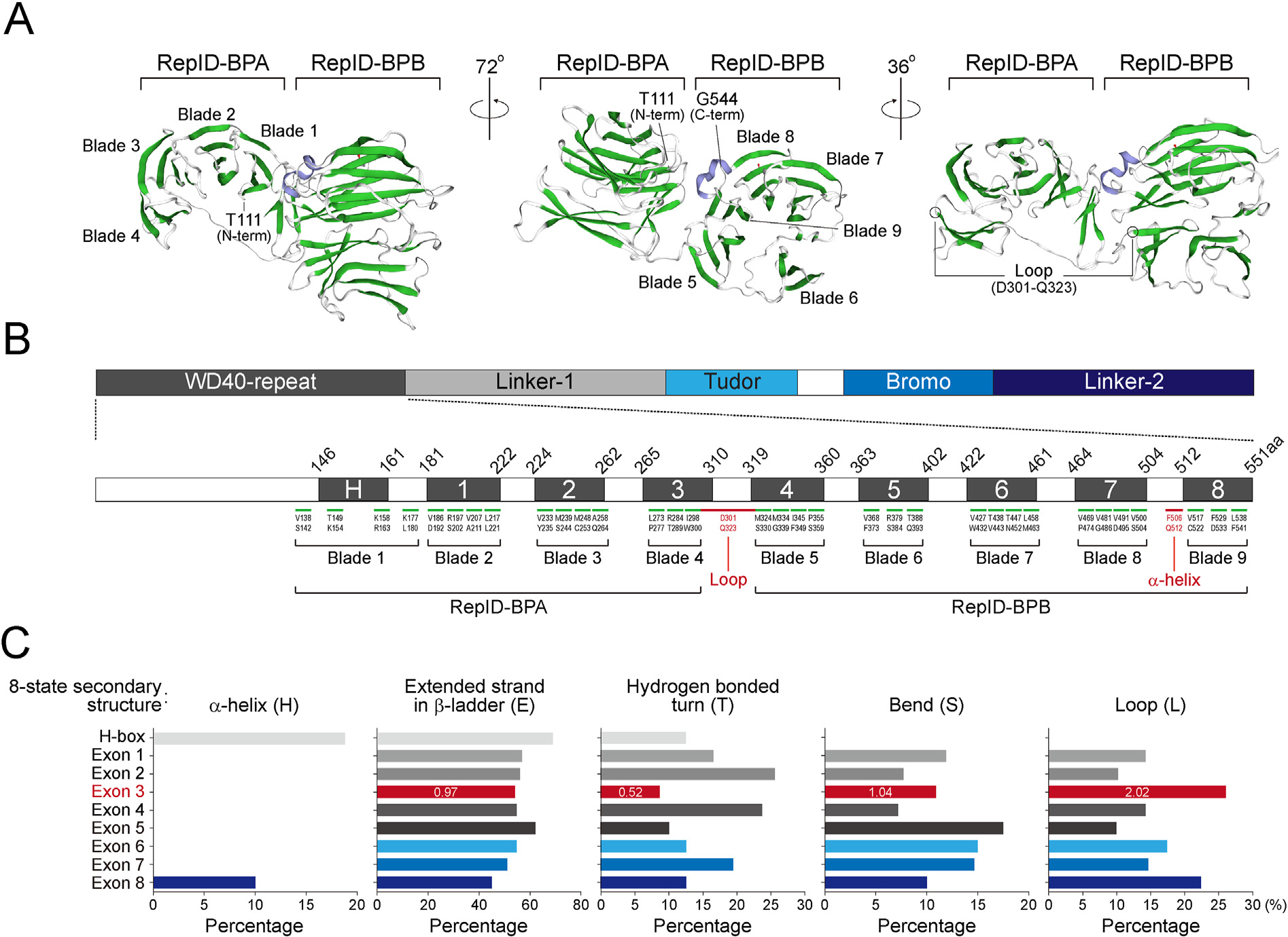
RepID WD40 domain consists of two β-propeller folds linked by a loop. (A) Molecular modeling of the RepID WD40 domain was performed using Swiss-Model (http://swissmodel.expasy.org). The positions of N-terminal, C-terminal regions with two β-propeller folds (BPA and BPB) involving 9 blades composed of a three- or four-stranded antiparallel β-sheet are indicated (left and middle panel). The loop region is indicated with amino acid positions (right panel). β-helix and β-sheets are represented as purple and green, respectively. (B) Amino acid positions of each β-sheet in the 9 blades located in 9 exon-containing WD40 domains. Predicted loop and α-helix is indicated in red. (C) RaptorX server-based prediction (http://raptorx.uchicago.edu/StructurePropertyPred/predict/). Bar graph represents percentage of 8-state secondary structures (H, α-helix; G, 3helix; I, 5-helix; E, extended strand in β-ladder; B, isolated β-bridge; T, hydrogen bonded turn; S, bend; L, loop) in each exon. Numbers in exon 3 (red) represents fold change compared to average percentage of other mutants.
WD40 repeats play a role in regulating protein-protein interactions [17]. Based on the 8-state secondary structure, all nine exons showed no 3-helix (G), 5-helix (I), or isolated β-bridge (B); all had a similar ratio of the extended strand in β-ladder (E), and showed no significant differences in bend structure (S) (Fig. 3C, section E and S). RaptorX predicted the presence of α-helix in 3 amino acids of the H-box (19.0%) and 4 amino acids of exon 8 (10.0%) (Fig. 3C, section H). Exon 3, containing 46 amino acids had a low hydrogen-bonded turn (T) structure around 0.52-fold lower compared to the average of the hydrogen-bonded turn in other exons (Fig. 3C, section T). Consistent with the Swiss-Model prediction, exon 3 region was predicted to have a high presence of the loop structure (26.0%; 12 amino acids), approximately 2.02-fold higher compared to the average of loops in the other exons (Fig. 3C, section L). These results suggest that exon 3 links RepID-BPA and BPB and does not affect CRL4 and BUB3 binding or the chromatin binding ability of RepID.
4. Discussion
WD40 domain-containing proteins provide binding platforms for various proteins that control a several cellular processes. Here, we demonstrate that the WD40 domain of RepID forms two β-propeller folds connected by a loop formation, which are crucial for molecule assembly involving CRL4 and its substrate by providing different binding platforms as well as by maintaining the chromatin binding ability.
Our findings suggest that RepID binds chromatin via the WD40 domain. RepID can recognize methylated or acetylated histones via cryptic tudors or bromodomains, respectively. Even if our various mutants contain these functional domains, observation of highly reduced chromatin-bound RepID by deletion of segments within the WD40 domain suggests that chromatin binding of the RepID may be determined by three functional domains: bromo-, cryptic tudor-, and WD40-domains. Consistent with these results, many WD40 proteins have been previously identified as histone code readers. WDR5, a component of the MLL subfamily known as histone H3K4 methyltransferases, is required for the conversion of histone H3K4 dimethylation to trimethylation [24–26]. LRWD1 uses its WD40 domain to recognize repressive marks, including H3K9me3, H3K27me3, and H4K20me3, with the ORC complex [27,28]. The histone code recognized might be different or not depending on RepID choice, and which code reader domain is used. Further studies are required to address the question of how RepID utilizes the three domains to interpret various histone codes that can be modified by cell cycle progression or cellular environmental changes.
Several DCAF proteins, including DCAF4–6, DCAF9, and DDB2, use the H-box located in the WD40 domain as a major fraction to bind with DDB1 [14]. However, the truncation mutant of DDB2, which lacks an H-box, still maintains its interaction with DDB1 [29], suggesting that DCAFs might interact with DDB1 through multiple interfaces besides the H-box. In concordance, we observed that RepID uses the H-box as well as the C-terminal region encoded in exon 8 within the WD40 domain to interact with DDB1. Our predicted structure of the RepID WD40 domain showing close proximity between the H-box and C-terminal region is in line with previous reports demonstrating a multi-surface interaction between the RepID WD40 domain and DDB1.
Our results propose the following model for RepID’s various platforms in regulating its chromatin binding and assembly of different molecules including CRL4 and BUB3 in the WD40 domain (Fig. 4). First, two β-propeller folds acts as “molecular double clips” for chromatin binding. Depletion of most parts of the clips causes low chromatin binding of RepID, leading to decreased chromatin-bound CRL4. Loop formation, which connects each clip, has no effect on chromatin binding ability and the interaction with CRL4 or BUB3. The closely located N-terminal (H-box) and C-terminal (exon 8) regions provide binding sites for DDB1, whereas BUB3 binding occurs next to the H-box (exons 1 and 2). This model suggests how the WD40 interacting molecules, including chromatin or proteins, can overcome space occupancy without physical collisions.
Fig. 4.
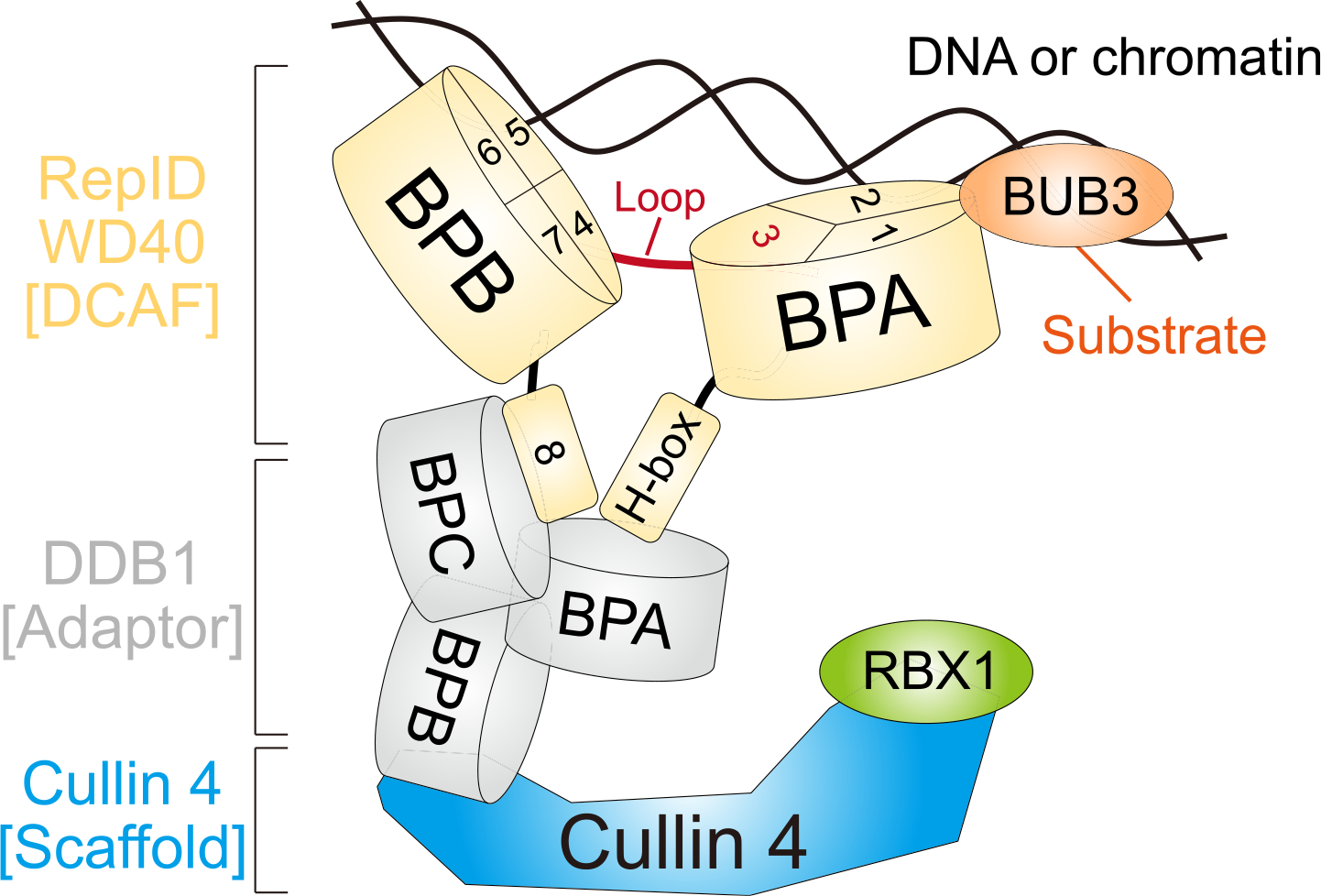
Schematic model. RepID WD40 domain consists of two β-propeller folds (RepID-BPA and BPB) and the loop structure positioned at exon 3 connects them. H-box locating N-terminal and exon 8 (blade 9) at the C-terminal region does not affect DNA/chromatin binding of the RepID but provides the platform for binding to BPA and BPC of DDB1 connected with CUL4 using BPB region. Two molecular clips of RepID (BPA and BPB) possibly recognize and interact with DNA/chromatin via exon 1–2 in BPA (blade 2–3) and exon 4–7 in BPB (blade 5–8). Therefore, the deleted form of BPA or BPB in RepID loses its DNA/chromatin binding ability, leading to compromised CRL4 recruitment. Exon 3 consists of blade 4, but also contains high amounts of loop structure which does not affect DNA/chromatin and CRL4 binding of RepID. BUB3 is located on chromatin in a RepID-independent manner and binds to exon 1 and 2 region in RepID-BPA.
Our previous studies proposed that targeting the WD40 domain might have significant clinical implications. Cancer cells that have low amounts of chromatin-bound CRL4 mediated by depletion of structural DCAF show high sensitivity to inhibition of CRL1, which is a compensative ubiquitin E3 ligase instead of CRL4 [20]. Paclitaxel shows a synergistic effect in cells that have issues in mitosis, such as RepID-depleted cells with a metaphase-anaphase transition delay via failure of BUB3 degradation [19]. Further, reduced CRL4 loading onto chromatin leads to resistance to pevonedistat [20]. Therefore, we propose that identification of RepID WD40 domain-targeting drugs may be necessary for synergistic effects with other anti-cancer drugs in combination therapy. Our findings uncover important interactions of the versatile WD40 domain of RepID, laying the foundation for new strategies to target this domain in tumor treatment.
Supplementary Material
Acknowledgements
This research was supported by Chungbuk National University Korea National University Development Project (2020). We would like to thank Editage (www.editage.com) for English language editing.
Footnotes
Declaration of competing interest
The authors have no potential conflict of interest to declare in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found s at https://doi.org/10.1016/j.bbrc.2021.06.047.
References
- [1].Xu C, Min J, Structure and function of WD40 domain proteins, Protein Cell 2 (2011) 202–214, 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fulop V, Jones DT, Beta propellers: structural rigidity and functional diversity, Curr. Opin. Struct. Biol. 9 (1999) 715–721, 10.1016/s0959-440x(99)00035-4. [DOI] [PubMed] [Google Scholar]
- [3].Paoli M, Protein folds propelled by diversity, Prog. Biophys. Mol. Biol. 76 (2001) 103–130, 10.1016/s0079-6107(01)00007-4. [DOI] [PubMed] [Google Scholar]
- [4].Pons T, Gomez R, Chinea G, Valencia A, Beta-propellers: associated functions and their role in human diseases, Curr. Med. Chem. 10 (2003) 505–524, 10.2174/0929867033368204. [DOI] [PubMed] [Google Scholar]
- [5].Zhong W, Feng H, Santiago FE, Kipreos ET, CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing, Nature 423 (2003) 885–889, 10.1038/nature01747. [DOI] [PubMed] [Google Scholar]
- [6].Petroski MD, Deshaies RJ, Function and regulation of cullin-RING ubiquitin ligases, Nat. Rev. Mol. Cell Biol. 6 (2005) 9–20, 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- [7].Jang SM, Redon CE, Aladjem MI, Chromatin-bound cullin-ring ligases: regulatory roles in DNA replication and potential targeting for cancer therapy, Front Mol Biosci 5 (2018) 19, 10.3389/fmolb.2018.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jang SM, Redon CE, Thakur BL, Bahta MK, Aladjem MI, Regulation of cell cycle drivers by Cullin-RING ubiquitin ligases, Exp. Mol. Med. 52 (2020) 1637–1651, 10.1038/s12276-020-00508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li T, Chen X, Garbutt KC, Zhou P, Zheng N, Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase, Cell 124 (2006) 105–117, 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- [10].Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N, Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery, Nature 443 (2006) 590–593, 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- [11].Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H, CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation, Nat. Cell Biol. 8 (2006) 1277–1283, 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- [12].He YJ, McCall CM, Hu J, Zeng Y, Xiong Y, DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases, Genes Dev. 20 (2006) 2949–2954, 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lee J, Zhou P, DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase, Mol Cell 26 (2007) 775–780, 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- [14].Li T, Robert EI, van Breugel PC, Strubin M, Zheng N, A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery, Nat. Struct. Mol. Biol. 17 (2010) 105–111, 10.1038/nsmb.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Morgan MAJ, Rickels RA, Collings CK, He X, Cao K, Herz HM, Cozzolino KA, Abshiru NA, Marshall SA, Rendleman EJ, Sze CC, Piunti A, Kelleher NL, Savas JN, Shilatifard A, A cryptic Tudor domain links BRWD2/PHIP to COMPASS-mediated histone H3K4 methylation, Genes Dev. 31 (2017) 2003–2014, 10.1101/gad.305201.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang Y, Huang L, Fu H, Smith OK, Lin CM, Utani K, Rao M, Reinhold WC, Redon CE, Ryan M, Kim R, You Y, Hanna H, Boisclair Y, Long Q, Aladjem MI, A replicator-specific binding protein essential for site-specific initiation of DNA replication in mammalian cells, Nat. Commun. 7 (2016) 11748, 10.1038/ncomms11748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Migliori V, Mapelli M, Guccione E, On WD40 proteins: propelling our knowledge of transcriptional control? Epigenetics 7 (2012) 815–822, 10.4161/epi.21140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B, Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive, Nature 449 (2007) 933–937, 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- [19].Jang SM, Nathans JF, Fu H, Redon CE, Jenkins LM, Thakur BL, Pongor LS, Baris AM, Gross JM, O’Neill MJ, Indig FE, Cappell SD, Aladjem MI, The RepID-CRL4 ubiquitin ligase complex regulates metaphase to anaphase transition via BUB3 degradation, Nat. Commun. 11 (2020) 24, 10.1038/s41467-019-13808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jang SM, Zhang Y, Utani K, Fu H, Redon CE, Marks AB, Smith OK, Redmond CJ, Baris AM, Tulchinsky DA, Aladjem MI, The replication initiation determinant protein (RepID) modulates replication by recruiting CUL4 to chromatin, Nat. Commun. 9 (2018) 2782, 10.1038/s41467-018-05177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jang SM, Redon CE, Aladjem MI, Switching DCAFs: beyond substrate receptors, Bioessays (2021), e2100057, 10.1002/bies.202100057. [DOI] [PubMed] [Google Scholar]
- [22].Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP, An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer, Nature 458 (2009) 732–736, 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- [23].Zhao Y, Sun Y, Cullin-RING Ligases as attractive anti-cancer targets, Curr. Pharmaceut. Des. 19 (2013) 3215–3225, 10.2174/13816128113199990300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, Plotnikov AN, Arrowsmith CH, Min J, Structural basis for molecular recognition and presentation of histone H3 by WDR5, EMBO J. 25 (2006) 4245–4252, 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, Verdine GL, Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex, Nat. Struct. Mol. Biol. 13 (2006) 704–712, 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD, WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development, Cell 121 (2005) 859–872, 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- [27].Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M, Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers, Cell 142 (2010) 967–980, 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- [28].Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T, Nucleosome-interacting proteins regulated by DNA and histone methylation, Cell 143 (2010) 470–484, 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jin J, Arias EE, Chen J, Harper JW, Walter JC, A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1, Mol Cell 23 (2006) 709–721, 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


