Summary
SARS-CoV-2 Omicron BA.2.75 subvariant has evolved to a series of progeny variants carrying several additional mutations in the receptor-binding domain (RBD). Here, we investigated whether and how these single mutations based on BA.2.75 affect the neutralization of currently available anti-RBD monoclonal antibodies (mAbs) with well-defined structural information. Approximately 34% of mAbs maintained effective neutralizing activities against BA.2.75, consistent with those against BA.2, BA.4/5, and BA.2.12.1. Single additional R346T, K356T, L452R, or F486S mutations further facilitated BA.2.75-related progeny variants to escape from broadly neutralizing antibodies (bnAbs) at different degree. Only LY-CoV1404 (bebtelovimab) displayed a first-class neutralization potency and breadth against all tested Omicron subvariants. Overall, these data make a clear connection between virus escape and antibody recognizing antigenic epitopes, which facilitate to develop next-generation universal bnAbs against emerging SARS-CoV-2 variants.
Subject areas: Immunology, Virology
Graphical abstract
Highlights
-
•
RBD mAbs are re-classified into 8 groups based on their diverse binding epitopes
-
•
Additional mutations broaden the antibody evasion of Omicron BA.2.75 variants
-
•
LY-CoV1404 displays a first-class neutralization against tested Omicron subvariants
-
•
S2H97-site mAbs show relatively moderate but broad-spectrum neutralization
Immunology; Virology
Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has never stopped worldwide since the end of 2019. To date, SARS-CoV-2 has infected over 665 million people and caused more than 6.7 million deaths. As SARS-CoV-2 spread rapidly across the world, a large number of variants emerged including Alpha, Beta, Gamma, Delta, Kappa, Lambda, Mu, and Omicron, etc., posing great challenges to the current COVID-19 vaccines and therapeutic monoclonal antibodies (mAbs).1,2,3,4,5,6 In addition, since 2022, a series of Omicron subvariants including BA.1, BA.2, BA.3, BA.4, and BA.5 have emerged, leading the waves of COVID-19 around the world.7,8,9,10 What is worse, the appearance of Omicron subvariants with additional mutations in the spike protein further strengthened the COVID-19 pandemic. For example, BA.2.12.1, carrying additional L452Q and S704L mutations as compared to BA.2, was first discovered in the United States and spread quickly due to the increased immune evasion.10,11,12
Recently, a new subvariant of BA.2, BA.2.75, was first detected in India, appeared to be increasing in prevalence, and subsequently reported in many other countries.13,14,15 On July 7, 2022, BA.2.75 has been classified as a variant of concern lineage under monitoring by the World Health Organization (WHO). Continuous monitoring of submitted BA.2.75 sequences showed that BA.2.75 had been divided into several progeny variants, carrying one or two additional mutations in the region of receptor-binding domain (RBD). However, it is largely unknown whether and how these single substitutions based on BA.2.75 affect the neutralization of currently available monoclonal neutralizing antibodies (nAbs).
In this study, we summarized a total of 59 mAbs with well-defined structural information and re-classified them into 8 groups based on their diverse binding epitopes on the RBD. We comprehensively evaluated the antibody evasion of BA.2.75 and its subvariants from the neutralization of 44 available mAbs. These results showed that BA.2.75 variant escaped most of the nAbs, and additional mutation could further enhance its antibody escape.
Results
Mutations in the spike protein and prevalence of BA.2.75 subvariants
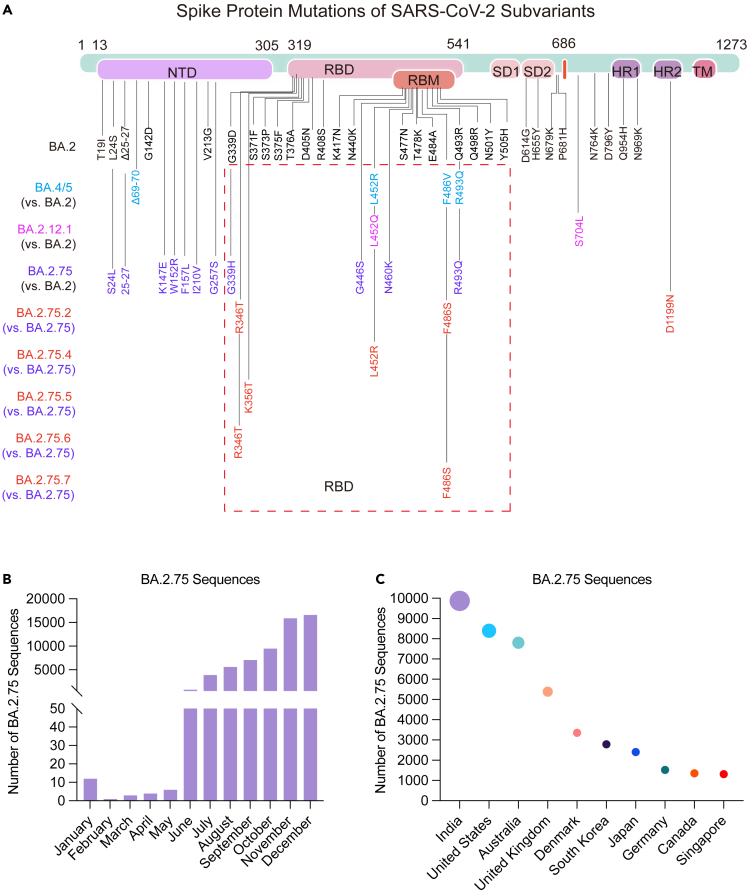
Compared with BA.2, BA.2.75 harbored many special mutations, especially in the RBD, which were mainly recognized by most of potent nAbs (Figure 1A). Further sequence analysis showed that some of BA.2.75 progeny variants (BA.2.75.2, BA.2.75.4, BA.2.75.5, BA.2.75.6, and BA.2.75.7) carry one or two additional mutations in the RBD including R346T, K356T, L452R, and F486S associated with potential antibody escape, which may be the main reason for the rapid spread of BA.2.75 around the world over time (Figures 1B and 1C).
Figure 1.
Overall display of mutations in the spike protein and prevalence of BA.2.75 subvariant
(A) The mutations in the spike protein of BA.2, BA.4/5, BA.2.12.1, and BA.2.75 subvariants as compared to the reference wild-type isolate (NC_045512). The mutations of BA.4/5, BA.2.12.1, and BA.2.75 were also compared with BA.2. The mutations of BA.2.75.2, BA.2.75.4, BA.2.75.5, BA.2.75.6, and BA.2.75.7 were also compared with BA.2.75 and highlighted in red. BA.4 and BA.5 sharing the same spike protein sequence were represented as BA.4/5.
(B and C) The number of BA.2.75 sequences collected in the GISAID and cov-spectrum in 2022 were shown for countries (n = 55,933) (B) and for temporal distributions (n = 59,413) (C).
Overall structural complexes of SARS-CoV-2 RBD binding with angiotensin-converting enzyme 2 (ACE2) and 59 mAbs
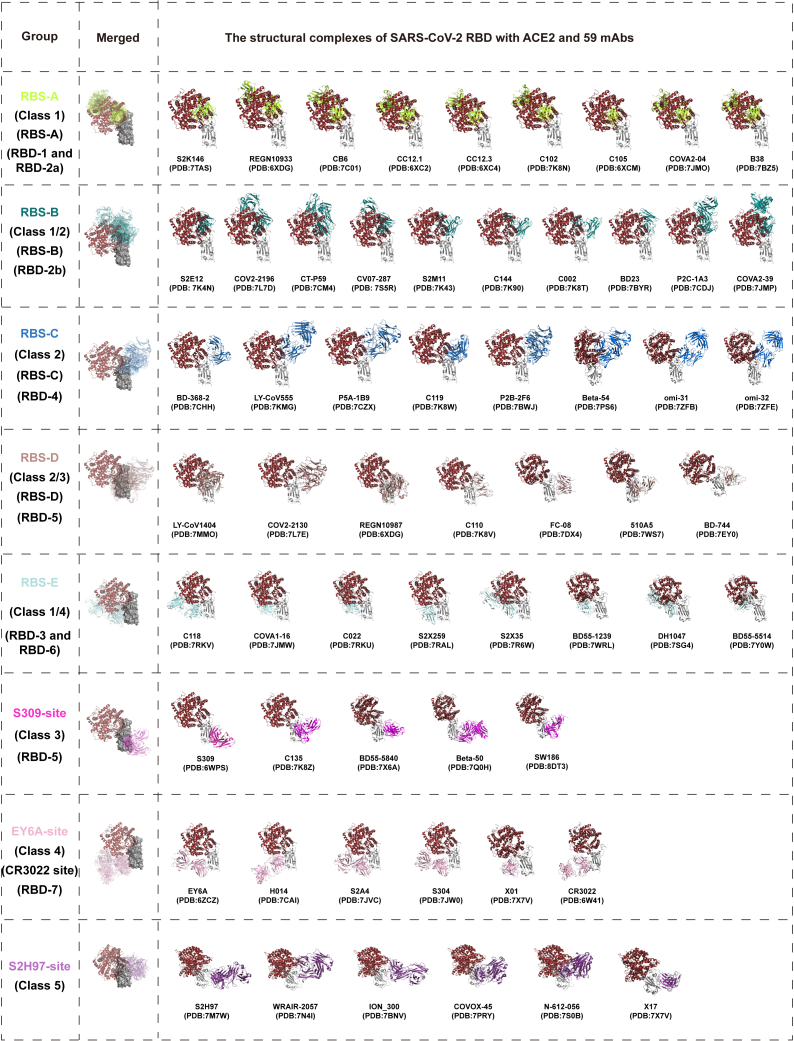
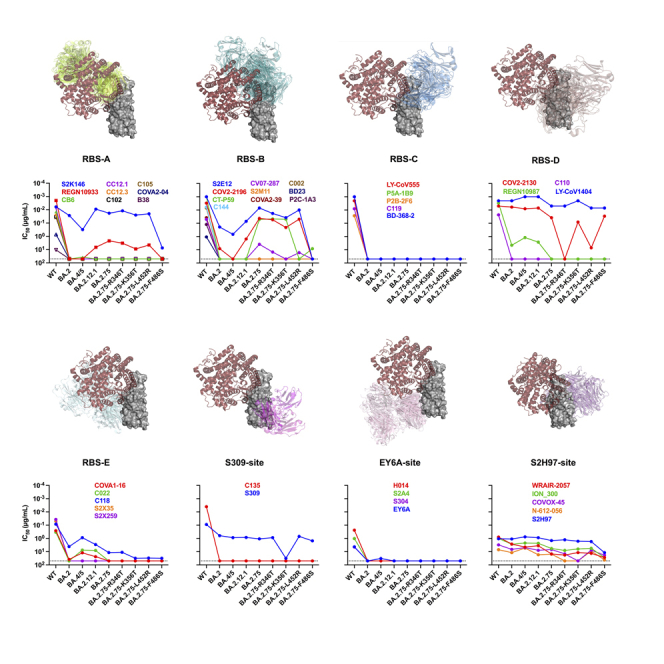
Several previous studies usually categorized RBD-specific mAbs based on the germline gene, structural information, and competition between each other.16,17,18,19 The classification of class 1 to class 4 was established earlier according to the angles of recognition of mAbs on the RBD, which is not comprehensive enough now.16 Similarly, while the classification of receptor-binding site (RBS)-A to RBS-D, CR3022 site, and S309 site was relatively straightforward, it was also summarized at the early stage.17,18 What is more, CR3022 with a cross-recognition ability to SARS-CoV and SARS-CoV-2 could not neutralize SARS-CoV-2, which might not be an appropriate representative of this group of mAbs. The classification of RBD-1 to RBD-7 based on the competitive relationship between mAbs failed to give us a clear understanding of their differences.17,18 Therefore, we summarized and re-classified 59 RBD-specific mAbs according to the structure information and previous classification methods (Figure 2). These mAbs were categorized as eight groups according to their binding epitopes on the RBD and competitions with the cellular receptor (human angiotensin-converting enzyme 2, hACE2). Most of mAbs (42/59) directly competed with hACE2 and targeted to the RBD with different angles of approach. According to the relationship between binding epitopes and the RBS, these mAbs could be divided into 5 groups including RBS-A, RBS-B, RBS-C, RBS-D, and RBS-E. With hACE2 located on the upper left side of the RBD as a reference, mAbs of RBS-A, RBS-B, and RBS-C groups bound to the left, middle, and right regions on the top of RBD, respectively. RBS-D mAbs recognized the outer face on the right side of RBD, while RBS-E mAbs bound to the inner face on the left side of RBD. By contrast, the rest of 17 mAbs were not competitors with hACE2 and distributed over the bottom half of RBD belonging to S309-site (right), EY6A-site (left), and S2H97-site (back) groups.
Figure 2.
Overall structure of complexes of ACE2 and 59 mAbs with RBD
A total of 59 RBD-specific mAbs were categorized into 8 groups (RBS-A, RBS-B, RBS-C, RBS-D, RBS-E, S309-site, EY6A-site, and S2H97-site) based on their binding epitopes and whether competing with ACE2 or not. RBS-related mAbs bind to the regions overlapping with RBS and compete with ACE2. The binding epitopes of S309-site, EY6A-site, and S2H97-site mAbs are far away from RBS, which do not compete with ACE2 directly. The merged figures show an overview of the complexes of RBD with ACE2 and all mAbs in each class. The antibody fragments in each class are drawn as cartoons in different colors. The ACE2 is shown as cartoon in brown. The RBD is shown in gray. All protein data bank (PDB) codes involved in this study are provided in the figure.
Neutralization of 44 mAbs against SARS-CoV-2 BA.2.75 subvariants
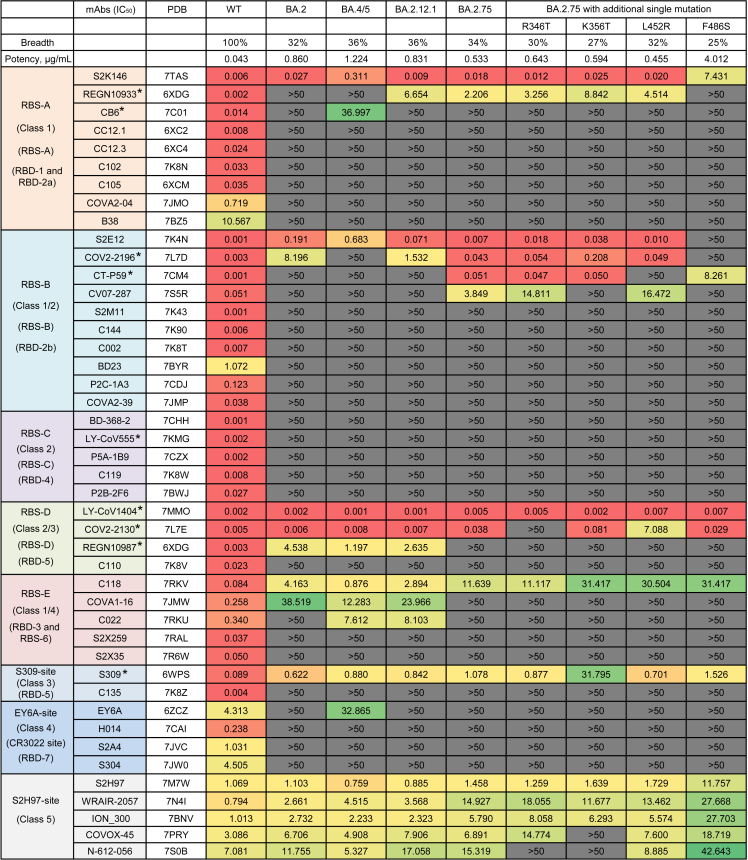
To make a head-to-head comparison, we also constructed wild-type (WT), BA.2, BA.4/5 (BA.4 and BA.5 sharing the same protein sequence in the spike), and BA.2.12.1 pseudoviruses in parallel (Figures 1A and S1). All of these 44 mAbs showed potent or effective neutralizing activities against the WT SARS-CoV-2 (Figure 3). Only 34% of mAbs (15/44) retained effective neutralizations against BA.2.75, whose 50% inhibitory concentrations (IC50s) were below 50 μg/mL. Similar with BA.2, BA.4/5, and BA.2.12.1, the neutralizing breadth and potency against BA.2.75 were largely lower than those against WT (geometric mean IC50 = 0.043 μg/mL). Of note, 6 of 9 tested approved antibody drugs could effectively neutralize BA.2.75 with IC50s ranging from 0.005 μg/mL to 2.206 μg/mL, including REGN10933 (casirivimab), COV2-2196 (tixagevimab), CT-P59 (regdanvimab), LY-CoV1404 (bebtelovimab), COV2-2130 (cilgavimab), and S309 (sotrovimab). From the point of view of antibody classification, S2K146 of RBS-A, S2E12 of RBS-B, and LY-CoV1404 and COV2-2130 of RBS-D maintained similar neutralizing activities against BA.2, BA.4/5, BA.2.12.1, and BA.2.75 with those against WT. Of note, the neutralizations of S2K146 and S2E12 against BA.4/5 were slightly decreased than those against WT, BA.2, BA.2.12.1, and BA.2.75. The sequence alignment showed that F486V was the unique mutation in BA.4/5, which might affect its susceptibility to S2K146 and S2E12 (Figure 1A). Although C118 of RBS-E and S309 still neutralized these Omicron subvariants, their neutralizing potencies were largely decreased comparing to those against WT. By contrast, nearly all mAbs of RBS-C and EY6A-site groups totally lost their neutralizing activities. Differently, all 5 tested mAbs of S2H97 site showed relatively moderate but broad-spectrum neutralization against most of Omicron and its sub-lineages. These results indicated that a few RBS-related and S309-site mAbs and most of S2H97-site mAbs could contribute to the maintained neutralization of existing mAbs elicited by the prototype SARS-CoV-2 infection or vaccination.
Figure 3.
The neutralization of 44 RBD-specific mAbs against WT SARS-CoV-2 and Omicron subvariants
The neutralization breadth is defined as the percentage of mAbs with effective neutralizing activities against each pseudovirus among all 44 tested mAbs. The neutralization potency is calculated by the geometric mean of neutralizing value of less than 50 μg/mL. The data shown are means of two or three independent experiments. The IC50 values are highlighted in different colors to show distinct neutralizing activities. Red: high, Yellow: moderate, Green: weak, Dark gray: non-neutralization. Symbol “∗” indicates some approved antibody drugs: REGN10933 (casirivimab), CB6 (etesevimab), COV2-2196 (tixagevimab), CT-P59 (regdanvimab), LY-CoV555 (bamlanivimab), LY-CoV1404 (bebtelovimab), COV2-2130 (cilgavimab), REGN10987 (imdevimab), and S309 (sotrovimab). See also Figures S1 and S2.
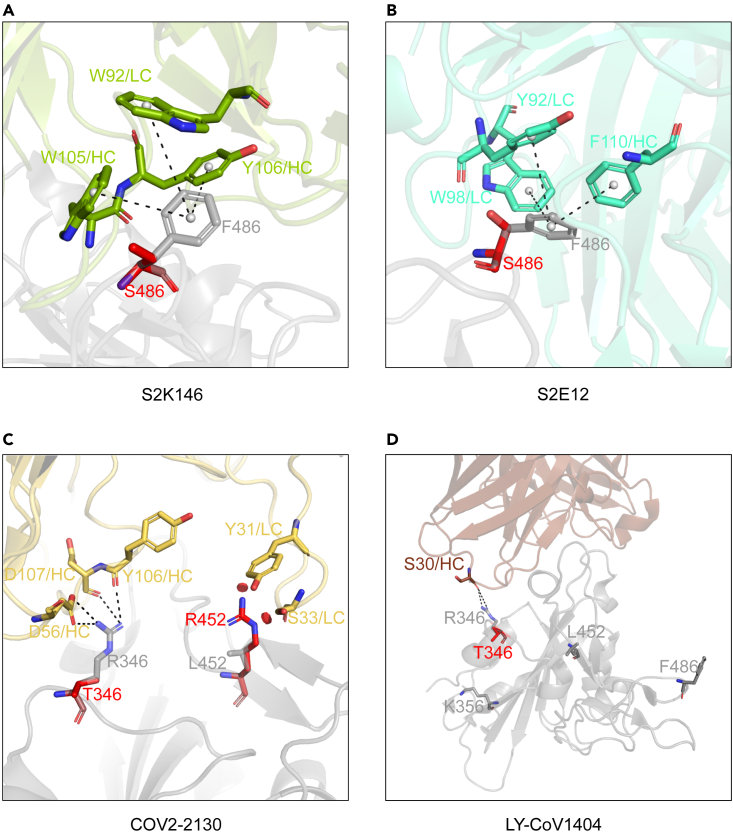
To explore the broad neutralization mechanism of these broad nAbs (bnAbs) in different groups, we further performed a detailed analysis of the interactions between bnAbs and RBDs based on their available structure data (Figure S2). According to the position relationships between the binding epitopes of bnAbs and mutated residues of Omicron subvariants, there are two main situations for these bnAbs. One is bnAbs like S2K146 and S2E12, which can tolerate F486V and/or Q493R mutations appearing in their binding epitopes to some extent. A kind of cation-π interactions with several residuals of S2K146 or S2E12 around F486 of RBD is disappeared when it mutates to V486, which partially accounts for the slight reductions of S2K146 and S2E12 neutralizing against BA.4/5. The other one is S2H97-site bnAbs, whose binding epitopes do not contain any key mutations. For this reason, we proposed a hypothesis that additional mutations even single substitutions appearing in recently emerged BA.2.75 might mediate its further evasion from currently available bnAbs.
Additional single mutations cause further antibody escape of BA.2.75
Based on the submitted sequences of BA.2.75 subvariants in the real world, 4 representative single mutations (R346T, K356T, L452R, and F486S) in the RBD were selected, and we evaluated their sensibilities to 44 existing mAbs. As shown in Figure 3, the additional F486S mutation largely reduced or even abolished the neutralization of S2K146 (7.431 μg/mL) and S2E12 (>50 μg/mL), comparing to that against BA.2.75 (0.018 μg/mL and 0.007 μg/mL). The F486S mutation leads to the loss of cation-π interactions between bnAbs and RBDs. In addition, the mutated serine (S) is a polar amino acid and really different from the original phenylalanine (F), which is an aromatic nonpolar amino acid (Figures 4A and 4B). The neutralization of COV2-2130 was obviously affected by single R346T (inactivated) and L452R (186-fold reduction) mutations. Interaction details of COV2-2130 binding to the RBD show that the R346T mutation makes a loss of extensive hydrogen-bonding interactions with D56, Y106, and D107 residues of heavy chain. Meanwhile, L452R mutation creates a steric clash to affect the recognition of COV2-2130 to the RBD (Figure 4C). However, C118, S309, and some S2H97-site bnAbs retained relatively weak or moderate neutralizing activities against these BA.2.75 subvariants carrying R346T, K356T, L452R, or F486S. Most notably, only 1 of 44 mAbs, LY-CoV1404, could neutralize all tested Omicron subvariants with really potent neutralization, whose IC50 values ranged from 0.001 μg/mL to 0.007 μg/mL. The structure modeling showed that K356T, L452R, and F486S mutations are not located in the binding epitopes of LY-CoV1404 (Figure 4D). While R346T mutation slightly affects the interaction at the edge of interface between LY-CoV1404 and RBD, BA.2.75 + R346T is still neutralized by LY-CoV1404 with a high potency.
Figure 4.
Structural analysis of bnAbs binding to WT and mutated RBDs
SARS-CoV-2 RBD is shown in gray. Fabs of S2K146 (A), S2E12 (B), COV2-2130 (C), and LY-CoV1404 (D) are shown in green, cyan, yellow, and pink, respectively. Mutated S486, T346, and R452 are represented by red sticks. Hydrogen bonds and salt bridges are represented by black dashed lines.
Discussion
In this study, we summarized and re-analyzed the structure data of 59 available RBD-specific mAbs and classified them into 8 groups on the basis of previous studies.7,16,17,18,20 Using 44 representative mAbs, we mainly evaluated the antibody evasion of Omicron BA.2.75 and its subvariants. The majority of mAbs lost their neutralizing activities against BA.2.75, with similar trends as those against BA.2, BA.4/5, and BA.2.12.1. However, the structural analyses of BA.2.75 showed its increased frequency of RBD in up conformation under acidic conditions, which suggested its enhanced low-pH-endosomal pathway usage.21 Meanwhile, BA.2.75 also exhibited higher binding affinity to the hACE2 than other Omicron subvariants.21,22,23 Consistent with the neutralization results in several studies,21,22,23,24 BA.2.75 totally escaped some clinically approved antibody drugs, such as CB6 (etesevimab), LY-CoV555 (bamlanivimab), and REGN10987 (imdevimab). Collectively, the increased infectivity and antibody evasion of BA.2.75 caused its enhanced transmissibility. Besides, our results demonstrated that additional single mutations based on BA.2.75 could further abolish some existing bnAbs, such as R346T for COV2-2130, L452R for CT-P59, and F486S for S2E12.
Several studies reported that the mutation occurring at the R346, L452, or F486 position largely affected the susceptibilities of SARS-CoV-2 variants to mAbs. Except R346T, other mutations at R346 had been found in several variants, such as R346K in Mu and BA.1.1, R346I in BA.5.9, and R346S in BA.4.7, which had been proved to be important reasons for their enhanced antibody-escape capacities.6,25,26 The L452R or L452Q was one of featured mutations in Delta or Lambda variant, respectively, accounting for its resistance to class 2 and class 3 mAbs.4,27,28,29 The F486V mutation was a characteristic of BA.4/BA.5 variant, mainly destroying the recognition of some class 1 and class 2 mAbs to RBD.30 Our data demonstrated that F486S mediated a more serious antibody evasion than F486V. Recent studies also evaluated the effect of different combinations of these additional mutations, such as BA.2.75 + R346T + F486S (BA.2.75.2) and BA.2.75 + R346T + L452R + F486S (CA.1), showing extensive escape from mAbs and polyclonal plasma samples.31,32 Fortunately, LY-CoV1404 (bebtelovimab) still maintained the potent neutralization against a series of concerned Omicron subvariants including BA.2.75.2 and CA.1. Besides, researchers also continued to identify novel bnAbs to fight against the current and future variants. BD55-5514 (SA55) and BD55-5840 (SA58) were two non-competing bnAbs isolated from vaccinated SARS convalescents, demonstrating high neutralizing potency against numerous Omicron subvariants.32,33 Meanwhile, hundreds of Omicron-specific mAbs were identified from BA.1, BA.2, and BA.5 breakthrough-infected donors, also showing good cross-neutralization against BA.2.75 and progeny variants.22,32,34
Of course, structure-based epitope analysis is sometimes not comprehensive for studying the mechanism of virus escaping from bnAbs because the binding and neutralizing of mAbs usually need multiple interactions with viral epitopes. The deep mutational scanning (DMS) assay has been extensively investigated to reveal the key immune escape profiles of most available monoclonal nAbs. Jesse D. Bloom’s group developed a yeast-display system to map all antibody-escape mutations in the SARS-CoV-2 RBD and to predict their antibody-escape abilities and the evolution trends of future variants.35,36,37 Xiaoliang Sunney Xie’s group also performed the DMS assay to screen the potential escape mutations and demonstrated that humoral imprinting could promote convergent evolution of SARS-CoV-2 in the RBD region.32 Above-mentioned four convergent sites in BA.2.75 subvariants, R346, K356, L452, and F486, were successfully inferred, which predicted their antibody-evasion risks in advance. In addition, several site mutations were screened and designed to evaluate their resistances to existing nAbs. For example, K444 N/T, V445A, and N450D mutations largely affected the neutralization of class 2 and class 3 mAbs.32
Overall, our data reveal the effectiveness of a wide range of RBD-specific mAbs against BA.2.75 subvariants and provide some favorable help for developing universal therapeutic bnAbs to combat the ongoing COVID-19 pandemic. Only by combining multiple analysis methods, we can define the antibody-escape mechanism and predict the virus evolution direction more comprehensively and clearly under the current infection and vaccination background in people.
Limitations of the study
While a total of 44 RBD-specific mAbs were selected to evaluate the antibody evasion of BA.2.75 subvariants, the sample size was still small as compared to thousands of submitted mAbs so far, might causing an incomplete description and evaluation. We observed that S2H97-site nAbs showed relatively weak but broad-spectrum neutralizing activities, however, potential mechanisms behind which were not confirmed in this study.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat# 15140163 |
| Puromycin dihydrochloride hydrate | Sangon Biotech | Cat# A610593-0025 |
| Polyethylenimines (PEIs) 25K | PolySciences | Cat# 23966 |
| Trypsin | Gibco | Cat# 25200-072 |
| EZ transfection reagent | Life-iLab | Cat# AC04L092 |
| Fetal bovine serum | Gibco | Cat# 10099-141C |
| Dulbecco’s Modified Eagle Medium | Gibco | Cat# 11965-092 |
| HEPES (1M) Buffer Solution | Gibco | Cat# 15630-080 |
| Opti-MEM Reduced Serum Medium | Gibco | Cat# 51985034 |
| DEAE-Dextran hydrochloride | Sigma Aldrich | Cat# D9885-10G |
| Critical commercial assays | ||
| Bright-Lite Luciferase Assay System | Vazyme Biotech | Cat# DD1204-03 |
| Mut Express II Fast Mutagenesis Kit V2 | Vazyme Biotech | Cat# C214-01 |
| Experimental models: Cell lines | ||
| 293 F | Gibco | Cat# R79007 |
| Human: HEK-293T | ATCC | Cat# CRL-3216 |
| HEK-293T hACE2 | YEASEN Biotech | Cat# 41107ES03 |
| Recombinant DNA | ||
| pNL4-3.Luc.R-E− | NIH AIDS Reagent Program | Cat# 3418 |
| Software and algorithms | ||
| Graphpad Prism 9 | GraphPad | https://www.graphpad.com/ |
| PyMOL Molecular Graphics System 2.5.4 | PyMOL | http://www.pymol.org |
| PISA v1.52 | PDBePISA | https://www.ebi.ac.uk/pdbe/pisa/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zheng Zhang (zhangzheng1975@aliyun.com).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Material Transfer Agreement.
Experimental model and subject details
Cell lines
The 293 F (Gibco) cells were incubated in FreeStyle 293 expression medium (Gibco) at 37 °C with 8% CO2 in flasks at 130 rpm. HEK-293T (ATCC) and HEK-293T-hACE2 (YEASEN Biotech) cells were cultured at 37 °C with 5% CO2 in Dulbecco’s modified eagle medium (DMEM, Gibco) containing 10% (v/v) fetal bovine serum (Gibco), 1% HEPES (1M) buffer (Gibco) and 1% penicillin-streptomycin (Gibco). The culture of HEK-293T-ACE2 cells also required a final concentration of 0.8 μg/mL of puromycin (Sangon Biotech).
Method details
Data acquisition and analysis of Omicron BA.2.75
The available sequence information of BA.2.75 in the world was obtained from the GISAID (http://www.gisaid.org) and cov-spectrum (https://cov-spectrum.org) until December 31, 2022. The sequences were further analyzed and classified according to the collected countries and the time of collection. Graphs were plotted using GraphPad Prism 9 software.
Structural analysis of complexes of RBD with ACE2 and 59 RBD-specific monoclonal antibodies (mAbs)
All structure data were downloaded from the protein databank (PDB) and PDB codes were provided in Figure 2. The potential interactions between mAb and RBD were calculated by the PISA v1.52 (https://www.ebi.ac.uk/pdbe/pisa/). The structural models were made using PyMOL Molecular Graphics System 2.5.4.
Expression and purification of 44 RBD-specific mAbs
All antibody gene sequences were downloaded from the National Center of Biotechnology Information (NCBI) and the PDB, which were synthesized and then cloned into the full-length IgG1 expression vectors by Sangon Biotech and GenScript. Paired heavy- and light-chain plasmids were co-transfected into 293 F cells. Using Protein-A columns, mAbs were purified from cell supernatants after a five-day culture according to the manufacturer’s instructions (Senhui Microsphere Technology). Purified mAbs were quantified by NanoDrop.
Generation of SARS-CoV-2 pseudoviruses
The spike genes of wild-type (WT) SARS-CoV-2 and variants (BA.2, BA.4/5, BA.2.12.1, and BA.2.75) were synthesized by GenScript and then inserted into the pVAX1 vector. Additional R346T, K356T, L452R, and F486S mutations were constructed by point mutation based on the BA.2.75 spike expression vector using the Mut Express II Fast Mutagenesis Kit V2 (Vazyme Biotech). SARS-CoV-2 pseudovirus was generated by co-transfection of HEK-293T cells with each spike plasmid and env-deficient HIV-1 backbone vector (pNL4-3.Luc.R-E−) using EZ transfection reagent according to the manufacturer’s instruction (Life-iLab). After 2-day co-transfection, all culture supernatants were harvested, clarified by centrifugation, and stored at −80 °C. The optimal infectious titer in further neutralization assay was determined by measuring the luciferase activity in the HEK-293T-hACE2 cells using Bright-Lite Luciferase reagent (Vazyme Biotech).
Detailed sequence information of spike protein in this study was listed below, respectively.
Wild-type (WT) SARS-CoV-2: accession number: NC_045512;
SARS-CoV-2 BA.2: accession number: EPI_ISL_9652748:
T19I, L24S, del25/27, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K;
SARS-CoV-2 BA.4/5: accession number: EPI_ISL_11542550:
T19I, L24S, del25/27, del69/70, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K; BA.4 and BA.5 shared the same sequence of spike protein, so we used BA.4/5 to represent the sequences of BA.4 and BA.5 in this study.
SARS-CoV-2 BA.2.12.1: accession number: EPI_ISL_9767878:
T19I, L24S, del25/27, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452Q, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, S704L, N764K, D796Y, Q954H, N969K;
SARS-CoV-2 BA.2.75: accession number: EPI_ISL_13502576:
T19I, G142D, K147E, W152R, F157L, I210V, V213G, G257S, G339H, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, G446S, N460K, S477N, T478K, E484A, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K.
SARS-CoV-2 pseudovirus-based neutralization assay
To measure the neutralizing activities, RBD-specific mAbs were serially diluted by 5-fold from the highest concentration of 5 μg/mL or 50 μg/mL. WT or mutated pseudoviruses were then mixed with these diluted mAbs and incubated at 37 °C for 1 h. Pseudovirus without mAbs was used as the virus control. The mixture was added into the HEK-293T-hACE2 cells in 96-well white cell plates and then incubated for 48 h at 37 °C. The culture medium was removed and 100 μL of the Bright-Lite Luciferase reagent (Vazyme Biotech) was added. After shaking at room temperature for 3 min, the luciferase activity was measured using the Varioskan LUX multimode microplate reader (Thermo Fisher Scientific). The inhibition was determined by comparing with the virus control. The value of 50% inhibitory concentration (IC50) was calculated using Graphpad Prism 9 software by log (inhibitor) vs. normalized response – Variable slope (four parameters) model. The cut-off value of neutralization was set as 50 μg/mL.
Quantification and statistical analysis
The independent experiment replicates were indicated in the figure legends. The inhibition was determined by comparing with the virus control. The value of 50% inhibitory concentration (IC50) was calculated using Graphpad Prism 9 software by log (inhibitor) vs. normalized response – Variable slope (four parameters) model. The cut-off value of neutralization was set as 50 μg/mL. We did not perform any statistical analysis in this study.
Acknowledgments
We thank the scientists around the world for sharing their antibody sequences and structure information on the public database in time. This work was funded by the National Key Plan for Scientific Research and Development of China (2021YFC2301900, 2021YFC0864500), the National Science Fund for Distinguished Young Scholars (82025022), the National Natural Science Foundation of China (82002140, 92169204, 82171752), the Guangdong Basic and Applied Basic Research Foundation (2021B1515020034), the Shenzhen Science and Technology Program (RCYX20200714114700046), the Science and Technology Innovation Committee of Shenzhen Municipality (JSGG20220226085550001, JSGG20200207155251653, JSGG20210901145200002), and the Shenzhen Natural Science Foundation (JCYJ20190809115617365, JCYJ20220530163413031, JCYJ20200109144201725).
Author contributions
Z.Z. and B.J. conceived and designed the study. H.G., J.J., S.S., and X.G. performed all experiments and analyzed the data together with assistance from Q.F., B.Z., and L.C. Z.Z., B.J., and H.G. wrote the manuscript, and all authors read and approved this version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: February 27, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106283.
Contributor Information
Bin Ju, Email: jubin2013@163.com.
Zheng Zhang, Email: zhangzheng1975@aliyun.com.
Supplemental information
Data and code availability
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021 doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 2.Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., Ho D.D. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29:747–751.e4. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236.e13. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo H., Fan Q., Song S., Shen S., Zhou B., Wang H., Cheng L., Ge X., Ju B., Zhang Z. Increased resistance of SARS-CoV-2 Lambda variant to antibody neutralization. J. Clin. Virol. 2022;150-151:105162. doi: 10.1016/j.jcv.2022.105162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uriu K., Kimura I., Shirakawa K., Takaori-Kondo A., Nakada T.A., Kaneda A., Nakagawa S., Sato K., Genotype to Phenotype Japan G2P-Japan Consortium Neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine serum. N. Engl. J. Med. 2021;385:2397–2399. doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu L., Iketani S., Guo Y., Chan J.F.W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/d41586-021-03826-3. [DOI] [PubMed] [Google Scholar]
- 7.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/d41586-021-03825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iketani S., Liu L., Guo Y., Liu L., Chan J.F.W., Huang Y., Wang M., Luo Y., Yu J., Chu H., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ai J., Wang X., He X., Zhao X., Zhang Y., Jiang Y., Li M., Cui Y., Chen Y., Qiao R., et al. Antibody evasion of SARS-CoV-2 Omicron BA.1, BA.1.1, BA.2, and BA.3 sub-lineages. Cell Host Microbe. 2022;30:1077–1083.e4. doi: 10.1016/j.chom.2022.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, & BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu P., Faraone J., Evans J.P., Zou X., Zheng Y.M., Carlin C., Bednash J.S., Lozanski G., Mallampalli R.K., Saif L.J., et al. Neutralization of the SARS-CoV-2 omicron BA.4/5 and BA.2.12.1 subvariants. N. Engl. J. Med. 2022;386:2526–2528. doi: 10.1056/NEJMc2206725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hachmann N.P., Miller J., Collier A.R.Y., Ventura J.D., Yu J., Rowe M., Bondzie E.A., Powers O., Surve N., Hall K., Barouch D.H. Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022;387:86–88. doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q., Iketani S., Li Z., Guo Y., Yeh A.Y., Liu M., Yu J., Sheng Z., Huang Y., Liu L., Ho D.D. Antigenic characterization of the SARS-CoV-2 Omicron subvariant BA.2.75. bioRxiv. 2022 doi: 10.1101/2022.07.31.502235. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamasoba D., Kimura I., Kosugi Y., Uriu K., Fujita S., Ito J., Sato K. Neutralization sensitivity of Omicron BA.2.75 to therapeutic monoclonal antibodies. bioRxiv. 2022 doi: 10.1101/2022.07.14.500041. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheward D.J., Kim C., Fischbach J., Muschiol S., Ehling R.A., Björkström N.K., Karlsson Hedestam G.B., Reddy S.T., Albert J., Peacock T.P., Murrell B. Evasion of neutralizing antibodies by Omicron sublineage BA.2.75. bioRxiv. 2022 doi: 10.1101/2022.07.19.500716. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hastie K.M., Li H., Bedinger D., Schendel S.L., Dennison S.M., Li K., Rayaprolu V., Yu X., Mann C., Zandonatti M., et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: a global consortium study. Science. 2021;374:472–478. doi: 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan M., Huang D., Lee C.C.D., Wu N.C., Jackson A.M., Zhu X., Liu H., Peng L., van Gils M.J., Sanders R.W., et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373:818–823. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui Z., Liu P., Wang N., Wang L., Fan K., Zhu Q., Wang K., Chen R., Feng R., Jia Z., et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell. 2022;185:860–871.e13. doi: 10.1016/j.cell.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan R., Wang R., Ju B., Yu J., Zhang Y., Liu N., Wang J., Zhang Q., Chen P., Zhou B., et al. Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies. Cell Res. 2021;31:517–525. doi: 10.1038/s41422-021-00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Y., Song W., Wang L., Liu P., Yue C., Jian F., Yu Y., Yisimayi A., Wang P., Wang Y., et al. Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75. Cell Host Microbe. 2022;30:1527–1539.e5. doi: 10.1016/j.chom.2022.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q., Iketani S., Li Z., Guo Y., Yeh A.Y., Liu M., Yu J., Sheng Z., Huang Y., Liu L., Ho D.D. Antigenic characterization of the SARS-CoV-2 Omicron subvariant BA.2.75. Cell Host Microbe. 2022;30:1512–1517.e4. doi: 10.1016/j.chom.2022.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito A., Tamura T., Zahradnik J., Deguchi S., Tabata K., Anraku Y., Kimura I., Ito J., Yamasoba D., Nasser H., et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2.75 variant. Cell Host Microbe. 2022;30:1540–1555.e15. doi: 10.1016/j.chom.2022.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheward D.J., Kim C., Fischbach J., Muschiol S., Ehling R.A., Björkström N.K., Karlsson Hedestam G.B., Reddy S.T., Albert J., Peacock T.P., Murrell B. Evasion of neutralising antibodies by omicron sublineage BA.2.75. Lancet Infect. Dis. 2022;22:1421–1422. doi: 10.1016/S1473-3099(22)00524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halfmann P.J., Kuroda M., Armbrust T., Theiler J., Balaram A., Moreno G.K., Accola M.A., Iwatsuki-Horimoto K., Valdez R., Stoneman E., et al. Characterization of the SARS-CoV-2 B.1.621 (Mu) variant. Sci. Transl. Med. 2022;14:eabm4908. doi: 10.1126/scitranslmed.abm4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jian F., Yu Y., Song W., Yisimayi A., Yu L., Gao Y., Zhang N., Wang Y., Shao F., Hao X., et al. Further humoral immunity evasion of emerging SARS-CoV-2 BA.4 and BA.5 subvariants. Lancet Infect. Dis. 2022;22:1535–1537. doi: 10.1016/s1473-3099(22)00642-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 28.Kimura I., Kosugi Y., Wu J., Zahradnik J., Yamasoba D., Butlertanaka E.P., Tanaka Y.L., Uriu K., Liu Y., Morizako N., et al. The SARS-CoV-2 Lambda variant exhibits enhanced infectivity and immune resistance. Cell Rep. 2022;38:110218. doi: 10.1016/j.celrep.2021.110218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng L., Song S., Fan Q., Shen S., Wang H., Zhou B., Ge X., Ju B., Zhang Z. Cross-neutralization of SARS-CoV-2 Kappa and Delta variants by inactivated vaccine-elicited serum and monoclonal antibodies. Cell Discov. 2021;7:112. doi: 10.1038/s41421-021-00347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheward D.J., Kim C., Fischbach J., Sato K., Muschiol S., Ehling R.A., Björkström N.K., Karlsson Hedestam G.B., Reddy S.T., Albert J., et al. Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies. Lancet Infect. Dis. 2022;22:1538–1540. doi: 10.1016/s1473-3099(22)00663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao Y., Jian F., Wang J., Yu Y., Song W., Yisimayi A., Wang J., An R., Chen X., Zhang N., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature. 2023;614:521–529. doi: 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao Y., Jian F., Zhang Z., Yisimayi A., Hao X., Bao L., Yuan F., Yu Y., Du S., Wang J., et al. Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents. Cell Rep. 2022;41:111845. doi: 10.1016/j.celrep.2022.111845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huo J., Dijokaite-Guraliuc A., Liu C., Zhou D., Ginn H.M., Das R., Supasa P., Selvaraj M., Nutalai R., Tuekprakhon A., et al. A delicate balance between antibody evasion and ACE2 affinity for Omicron BA.2.75. Cell Rep. 2023;42:111903. doi: 10.1016/j.celrep.2022.111903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.