Abstract
Escherichia coli O157 antigen-specific bacteriophages were isolated and tested to determine their ability to lyse laboratory cultures of Escherichia coli O157:H7. A total of 53 bovine or ovine fecal samples were enriched for phage, and 5 of these samples were found to contain lytic phages that grow on E. coli O157:H7. Three bacteriophages, designated KH1, KH4, and KH5, were evaluated. At 37 or 4°C, a mixture of these three O157-specific phages lysed all of the E. coli O157 cultures tested and none of the non-O157 E. coli or non-E. coli cultures tested. These results required culture aeration and a high multiplicity of infection. Without aeration, complete lysis of the bacterial cells occurred only after 5 days of incubation and only at 4°C. Phage infection and plaque formation were influenced by the nature of the host cell O157 lipopolysaccharide (LPS). Strains that did not express the O157 antigen or expressed a truncated LPS were not susceptible to plaque formation or lysis by phage. In addition, strains that expressed abundant mid-range-molecular-weight LPS did not support plaque formation but were lysed in liquid culture. Virulent O157 antigen-specific phages could play a role in biocontrol of E. coli O157:H7 in animals and fresh foods without compromising the viability of other normal flora or food quality.
Serotype O157:H7 Shiga toxin-producing Escherichia coli is commonly associated with hemorrhagic colitis and its secondary systemic sequelae (15, 32). Few therapeutic alternatives and poor prognoses for severe sequelae have led to intensive research targeting elimination of this human pathogen from its sources (15, 32). Cattle and sheep transiently harbor E. coli O157:H7, and many disease outbreaks have been linked to contaminated bovine food products (16, 20, 30). In addition, deer, horses, dogs, and birds can also transiently harbor these bacteria (16, 19, 26, 37). Feces from these animals or from humans may contaminate food or water and pose a risk for human infection (15, 32, 33).
Elimination of E. coli O157:H7 at the preharvest stage could play a significant role in preventing the introduction of this human pathogen into the food chain (16, 36). Diet and probiotic therapy have been evaluated as preharvest management strategies that may reduce the risk of culture-positive animals (16, 22, 40). Other more direct alternatives include vaccination or the use of colicins (16, 24). Recolonization of previously infected animals with the same or different strains of E. coli O157:H7 has been demonstrated and may limit these approaches (13, 22). In addition, serotype specificity is not a characteristic of colicin activity (24). Elimination of E. coli O157:H7 during food processing is effective, and pasteurization is a common technique that can assure the safety of dairy products and apple cider (7, 8, 18, 33). In addition, agents that can effectively eliminate E. coli O157:H7 from water and vegetables without compromising the quality of these materials are being evaluated (33).
Bactericidal bacteriophages (phages) may provide a natural, nontoxic, feasible approach for controlling several human pathogens (2). Phages are parts of both gastrointestinal and environmental ecosystems (34). In fact, early in this century, phages were used to treat bacterial infections before the advent of chemical antibiotics (2). Early studies suggested that phage therapy may be effective against a broad range of human infections caused by members of the genera Staphylococcus, Salmonella, Klebsiella, Escherichia, Proteus, and Pseudomonas (2, 4). In addition, in experimental animal studies workers analyzed conditions that influence the in vivo activities of various phages (2, 4). The lack of techniques to counter phage sequestration, resistance, and conversion eventually led to replacement of phage therapy with antibiotic treatment (2, 4). However, with our present knowledge of phage and bacterial genetics, it may be possible to circumvent the problems encountered in previous attempts to use phages as natural antimicrobial agents (2, 4).
In this study, we isolated phages and evaluated the ability of these phages to lyse laboratory cultures of E. coli O157. O-antigen-specific virulent phages may provide an economical tool for controlling E. coli O157:H7 in environmental settings without compromising food quality, health, or the viability of other normal flora. Only one other report has described the isolation and characterization of a phage capable of lysing E. coli O157:H7, and this coliphage (AR1) is toxin specific and lyses other Shiga toxin-producing E. coli and Shigella dysenteriae (27). Therefore, we attempted to isolate O157-specific phages and assess the ability of these phages to cause bacterial death.
MATERIALS AND METHODS
Bacterial strains.
E. coli O157:H7 strain ATCC 43894 (stx1+/stx2+) (American Type Culture Collection, Manassas, Va.) was used as a representative E. coli O157:H7 strain for phage isolation, propagation, and evaluation. E. coli O111:NM and O5:NM were used along with ATCC 43894 to isolate phages that require the O157 antigen for adsorption. Fifty-seven bacterial strains (see Table 1) were tested for phage sensitivity. These strains included a variety of E. coli strains that have O157 and non-O157 serotypes and express a variety of flagellar H antigens and bacteria that belong to different genera and have O antigens similar to O157. In addition, nalidixic acid-resistant (Nalr) E. coli O157:H7 strain 86-24 Nalr and O-antigen-deficient derivatives of this strain were tested (see Table 2).
TABLE 1.
Evaluation of phage specificity with various bacteria
| Bacterial isolate(s) | Serotype | Reference(s) | Plaque
formation
|
Lytic efficiency of phage mixture
ata:
|
||||
|---|---|---|---|---|---|---|---|---|
| KH1 | KH4 | KH5 | 37°C
|
4°Cb | ||||
| Aerated | Nonaerated | |||||||
| E. coli isolates | ||||||||
| ATCC 43894 | O157:H7 | 12, 39 | + | + | + | + | − | + |
| ATCC 43890 | O157:H7 | 12 | + | + | + | + | − | + |
| ATCC 43889 | O157:H7 | 12, 23 | + | + | + | + | − | + |
| ATCC 43888 | O157:H7 | 12 | + | + | + | + | − | + |
| A-9 | O157:H7 | 20 | + | + | + | + | − | + |
| G-13 | O157:H7 | 20 | + | + | + | + | − | + |
| I-1 | O157:H7 | 20 | + | + | + | + | − | + |
| FDA 13A81 | O157:H16 | 9 | + | + | + | + | − | + |
| FDA 13A82 | O157:H16 | 9 | + | + | + | + | − | + |
| CDC 3005-89 | O157:H38 | 9 | + | + | + | + | − | + |
| FDA 13A83 | O157:H45 | 9 | + | + | + | + | − | + |
| CDC 3004-89 | O157:H3 | 9 | − | − | − | + | − | + |
| CDC G5933 | O157:H12 | 9 | − | − | − | + | − | + |
| FDA 13A80 | O157:H16 | 9 | − | − | − | + | − | + |
| 7A, 7C, 7D, 7E | O157:H43 | 9 | − | − | − | + | − | + |
| 5A, 5B, 5C, 5D, 5E | O55:H7 | 9 | − | − | − | − | − | − |
| Enteropathogenic isolatesc | 10 | − | − | − | − | − | − | |
| 3812-3 | O5:NMd | 20 | − | − | − | − | − | − |
| Other species | ||||||||
| Yersinia enterocolitica | O9 | 12, 38 | − | − | − | − | − | − |
| Citrobacter freundii | NAe | 12 | − | − | − | − | − | − |
| Vibrio cholerae 569B | O1 | 12, 38 | − | − | − | − | − | − |
| Vibrio cholerae O395 | O1 | 12, 38 | − | − | − | − | − | − |
The MOI was 103 PFU/CFU.
Data for aerated and nonaerated cultures.
A total of 33 different enteropathogenic E. coli isolates of assorted serotypes were tested. The serotypes included ON:HN, O8:H41, O12:NM, O15:HN, O18:HN, O23:H15, O25:H2, O26:H2, O26:HN, O26:NM, O46:HN, O55:H7, O66:HN, O75:HN, O85:NM, O86:H34, O96:HN, O103:H6, O111:HN, O115:NM, O118:H8, O125:H6, O127:H40, O128:H2, O131:HN, O145:HN, O153:HN, and O156:H8.
NM, nonmotile.
NA, not applicable.
TABLE 2.
Phage infection of E. coli O157:H7 strain 86-24 Nalr and its O-antigen-deficient derivatives
| E. coli O157:H7 straina | Description | Latex agglutination test (O157 antigen) | Plaque
formation
|
Lytic efficiency of phage mixture
atb:
|
||||
|---|---|---|---|---|---|---|---|---|
| KH1 | KH4 | KH5 | 37°C
|
4°Cc | ||||
| Aerated | Nonaerated | |||||||
| 86-24 Nalr | Nalidixic acid-resistant mutant of E. coli O157:H7 strain 86-24 | + | + | + | + | + | − | + |
| F12 | O157 antigen-deficient mutant derived from strain 86-24 Nalr | − | − | − | − | − | − | − |
| F12(pSK+) | F12 transformed with plasmid Bluescript SK+ | − | − | − | − | − | − | − |
| F12(pF12) | F12 transformed with rfbEECO157:H7 cloned into pSK+ | + | − | − | − | − | − | − |
Phage isolation.
To isolate E. coli O157:H7-specific phages, 53 bovine or ovine fecal samples were collected from four separate cattle and sheep farms in Cottonwood and Moscow, Idaho, and Pullman, Wash. One-gram portions of each sample were inoculated into 5-ml exponential cultures of E. coli ATCC 43894 (2 × 108 CFU/ml), and the cultures were grown in Luria-Bertani (LB) medium supplemented with 5 mM MgSO4 (LBM) overnight at 37°C. After overnight growth, 0.5 ml of chloroform was added to each culture, and the cultures were vortexed, centrifuged at 15,000 × g for 10 min, and filtered (pore size, 0.45 μm) to remove cellular debris and fecal material. Single plaques were isolated from aliquots of the filtrates that were plated on lawns of E. coli ATCC 43894. Based on a preliminary screening for O157 specificity, three phages were selected for further analysis and designated KH1, KH4, and KH5. A derivative of KH1 has been deposited in the American Type Culture Collection under accession no. ATCC 55952.
Preparation of phage stocks.
E. coli ATCC 43894 in lambda, R, or LB medium, each with and without 5 mM MgSO4, was evaluated for propagation of O157-specific phages (28). Distinct plaques and high-titer stocks were obtained only with LBM. Sterile salt-magnesium (SM) buffer was used to prepare all phage dilutions and suspensions.
Low-titer phage stocks were prepared by using the soft agar overlay technique (28). Each phage isolate was diluted and plated onto LBM along with 107 CFU of E. coli O157:H7 suspended in a soft agar (0.75% LBM agar) overlay. After overnight incubation at 37°C, single plaques were suspended in SM buffer, incubated at 37°C for 1 h, and serially diluted. Phage suspensions were mixed with 107 CFU of E. coli O157:H7, adsorbed at 37°C for 15 min, and plated. After overnight incubation at 37°C, SM buffer was added to plates exhibiting semiconfluent lysis, and the top agar was harvested. The soft agar slurry was centrifuged twice at 12,000 × g for 20 min to collect the phage-rich supernatant (lysate), which was then treated with 0.05 volume of chloroform. High-titer phage stocks were prepared from the lysates by liquid infection (28). For each phage, 1 ml of low-titer lysate was mixed with 107 CFU of E. coli O157:H7, adsorbed, added to 200 ml of LBM broth, and incubated overnight at 37°C with aeration. After additional incubation at 25°C without aeration for 24 h, the cultures were each treated with 0.05 volume of chloroform. Lysates were harvested by centrifugation (12,000 × g, 20 min) twice and by filtration with 0.2-μm-pore-size filters. Phage stocks were stored in 0.05 volume of chloroform at 4°C. Cultures prepared to determine whether there were bacteria in phage stocks were consistently negative.
Phage infection.
To determine the optimal multiplicity of infection (MOI) for phage infection, E. coli O157:H7 strain ATCC 43894 was mixed with KH1 at ratios ranging from 10−3 to 103 PFU/CFU. All assays were done in duplicate. KH1 was selected as the test phage in these experiments because it formed clear, medium-sized plaques on the ATCC 43894 host cells. Bacteria were grown in LBM broth at 37°C without agitation to an absorbance at 600 nm of 1.0 to 1.3 (∼108 CFU/ml). Suspensions were adsorbed and incubated at 37 or 4°C with or without aeration, and the viable cell counts were determined in triplicate by serial dilution and spread plate culturing on LBM agar. Phage-free cultures (containing only bacteria) and cell-free cultures (containing only phage) were used as controls in all experiments to demonstrate the absence of contamination. The deduced optimal MOI (103 PFU/CFU) was used in subsequent tests performed with phages KH4 and KH5 and phage mixtures and with ATCC 43894 and all of the other bacteria tested.
E. coli O157:H7 isolates recovered after phage treatment were analyzed for phage resistance by spotting the surface of a bacterial lawn with 10 μl of phage stock. Plaque formation and the absence of plaque formation were taken to indicate susceptibility and resistance to phage, respectively.
Determination of bacterial and phage concentrations.
Survival of E. coli O157 in phage-infected cultures was determined by plating serial dilutions of cultures onto LB agar. At various times after phage infection, the surviving E. coli O157:H7 cells were enumerated and compared to the number of bacteria at the start of the experiment and the number of bacteria in a phage-free E. coli O157:H7 control culture. After overnight incubation at 37°C, colonies were confirmed to have the O157 serotype serologically (ProLab Diagnostics, Richmond Hill, Ontario, Canada). Cultures that produced no colonies were analyzed further by transferring 1 ml of the bacterium-phage mixture into 5 ml of an enrichment medium and incubating the preparation with aeration at 37°C; colonies were confirmed to have the O157 serotype serologically, as previously described (21).
Phages were enumerated by using the soft agar overlay technique described above. One milliliter of each culture was treated with 0.05 volume of chloroform and centrifuged. The supernatants were serially diluted with sterile SM buffer, mixed with 107 CFU of E. coli O157:H7, and overlaid on LBM plates. After overnight incubation at 37°C, the plates were examined for plaques.
Preparation of LPS.
Overnight cultures of bacteria were grown in LBM at 37°C without aeration. The viable cell count was determined for each culture as described above. Lipopolysaccharide (LPS) was extracted by using the protocol described by Inzana (17). Bacterial cells were pelleted and washed in phosphate-buffered saline (PBS) containing 0.15 mM CaCl2 and 0.5 M MgCl2 prior to hot phenol extraction of LPS. The final LPS pellet obtained after ethanol precipitation was resuspended in 50 μl of sterile distilled water (17).
Immunoblot analysis of LPS.
The LPS of E. coli O157 strains that were resistant and susceptible to plaque formation by the three phages were compared by performing an immunoblot analysis. The following strains were analyzed: CDC 3004-89, CDC G5933, FDA 13A80, 7A, 7C, 7D, and 7E, which were resistant to plaque formation, and ATCC 43894, ATCC 43888, ATCC 43889, FDA 13A81, and 86-24 Nalr, which were susceptible to plaque formation (Tables 1 and 2).
LPS were separated on sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis gels and stained with silver (35) (results not shown) or transferred electrophoretically to Immobilon polyvinylidene difluoride membranes (Millipore Corp., Bedford, Mass.). Following this transfer, the membranes were incubated overnight at 4°C in blocking solution containing 5% (wt/vol) nonfat dry milk, 0.05% (vol/vol) Tween 20 (Sigma), and 0.02% (wt/vol) sodium azide (Sigma). The membranes were then placed in PBS containing 0.05% (vol/vol) Tween 20 and 1:100 (vol/vol) rabbit anti-O157 antibody (Difco Laboratories, Detroit, Mich.) for 1 h at room temperature on a rocking platform. After the membranes were washed three times in PBS containing 0.05% (vol/vol) Tween 20 at room temperature with rocking, they were incubated in PBS containing 0.05% (vol/vol) Tween 20 and goat anti-rabbit immunoglobulin G peroxidase (1:12,000 dilution; Boehringer-Mannheim, Indianapolis, Ind.). Immunoreactive LPS were visualized with the SuperSignal chemiluminescent Western blot substrate (Pierce, Rockford, Ill.) used according to the manufacturer’s instructions, and the membranes were exposed to X-ray film, which was then developed.
RESULTS
Isolation of phages that form plaques on an E. coli O157 host.
Fifty-three independent bovine and ovine fecal samples were enriched for phage, and five samples contained lytic phages that produced plaques on lawns of E. coli O157:H7 strain ATCC 43894. Three phages (KH1, KH4, and KH5) produced plaques on this strain but not on Shiga toxin-producing E. coli strains O111:NM and O5:NM. KH1 formed medium-size (0.8-mm-diameter) clear plaques on E. coli O157:H7 strain ATCC 43894, whereas KH4 and KH5 formed smaller (0.4- to 0.5-mm-diameter) clear plaques. High-titer stocks prepared for each phage contained phage particles at concentrations between 109 and 1010 PFU/ml.
KH1, KH4, and KH5 coliphages are O157 antigen specific.
Fifty-seven E. coli isolates of human or animal origin were tested to determine the O-antigen specificity of the phages (Table 1). KH1, KH4, and KH5 lysed all E. coli isolates with the O157 serotype and did not lyse any of the non-O157 strains tested (Table 1). Likewise, they did not form plaques on any non-O157 strain tested; however, 7 of the 57 E. coli O157 strains tested did not support plaque formation (Table 1).
To examine the possibility that the length of the O157 LPS influences plaque formation, we tested the phage susceptibility of an O157 antigen-deficient mutant of E. coli O157:H7 strain 86-24 Nalr, designated F12, and derivatives of this mutant (Table 2). The F12 mutant maintained a transposon insertion in rfbEECO157:H7 that codes for an enzyme required for synthesis of perosamine, a subunit of the O157 LPS (9). Although strain F12(pF12) makes the O157 antigen, as demonstrated serologically, it expresses a shorter O157 LPS chain than the parent strain expresses (9). KH1, KH4, and KH5 did not lyse F12 or its derivatives, F12(pSK+) (F12 transformed with the Bluescript SK+ plasmid) and F12(pF12) (F12 transformed with pSK+ containing the cloned rfbEECO157:H7 gene) (Table 2). The three phages lysed only E. coli O157:H7 parent strain 86-24 Nalr, which produced the full-length O157 antigen.
LPS from E. coli O157 strains that were resistant and susceptible to plaque formation were different.
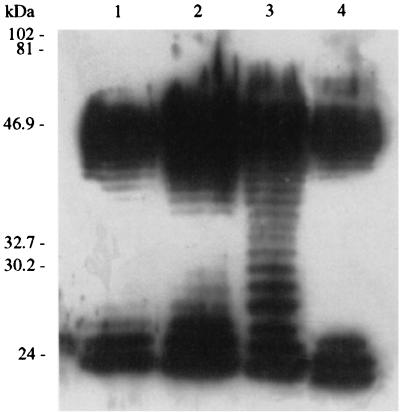
The immunoreactive LPS found in plaque-sensitive E. coli strains 86-24 Nalr, ATCC 43888, ATCC 43889, ATCC 43894, and FDA 13A81 were high-molecular-weight (> ∼40-kDa) and low-molecular-weight (∼24- to 26-kDa) O157-antigenic LPS, whereas plaque-resistant strains CDC G5933, CDC 3004-89, FDA 13A80, 7A, 7C, 7D, and 7E produced high-molecular-weight, low-molecular-weight, and abundant mid-range-molecular-weight (∼26- to 40-kDa) O157 antigens. The molecular weight distributions of the O157 antigens of representative strains are shown in Fig. 1.
FIG. 1.
Immunoblot analysis of LPS produced by strains resistant and susceptible to plaque formation by phage. High- and low-molecular-weight immunoreactive LPS, but not intermediate-molecular-weight immunoreactive LPS, were observed in the strains susceptible to plaque formation. Lanes 1 and 4, E. coli O157:H7 strain ATCC 43894; lane 2, E. coli O157:H7 strain 86-24 Nalr. Abundant immunoreactive LPS at all molecular weights were observed in strains resistant to plaque formation. Lane 3 contained representative strain E. coli O157:H16 FDA 13A80. The positions of molecular size standards are indicated on the left.
High MOI is required for efficient killing by O157-specific phages.
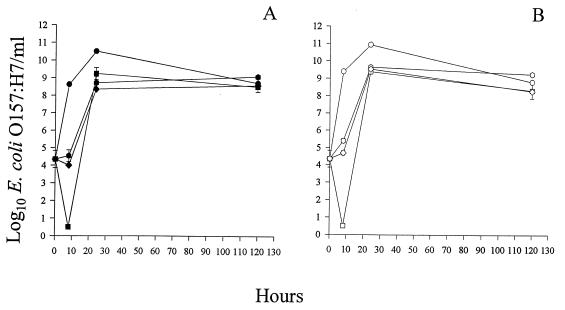
Infection of E. coli O157:H7 with KH1, KH4, or KH5 at various MOIs with and without aeration was monitored for 5 days. At MOIs ranging from 10−3 to 102 PFU/CFU, a minimal decline in bacterial titer was observed (data not shown). A significant reduction in the number of CFU per milliliter was observed at an MOI of 103 PFU/CFU (Fig. 2). Infections with KH1 resulted in a 105-fold decrease in the number of viable E. coli O157:H7 CFU/ml after 8 h (Fig. 2). The E. coli O157:H7 concentration (3 CFU/ml) was >108-fold less than the concentration in the phage-free bacterial control (3.58 × 108 CFU/ml) after 8 h (Fig. 2A). In all subsequent experiments, an MOI of 103 PFU/CFU was used.
FIG. 2.
Comparison of the lytic efficiencies of phages. Cultures were incubated at 37°C without aeration (A) or with aeration (B). E. coli O157:H7 (3 × 104 CFU) was treated with KH1 (■ and □), KH4 ( and ), or KH5 (⧫ and ◊). The MOI was 103 PFU/CFU. The control contained only E. coli O157:H7 (● and ○). The standard errors of the E. coli O157:H7 concentrations ranged from 0.03 to 0.5 log10 CFU/ml (n = 4).
O157-specific phages differ in the ability to kill their hosts.
The efficiencies of the phages in reducing the titer of viable E. coli O157:H7 varied (Fig. 2). There was a steady increase in the E. coli O157:H7 concentrations over time in the phage-free bacterial control (Fig. 2). The bacterial titers in cultures infected with KH1 decreased significantly after 8 h of incubation (see above), while KH4 and KH5 infections resulted in minimal declines 8 h after phage infection (Fig. 2). Thus, at 37°C, although KH1 was more efficient in reducing the number of viable bacteria than KH4 or KH5 was, no single phage was able to eliminate all bacteria.
Bacterial growth was observed in cultures 8, 24, and 48 h after phage infection, but the surviving E. coli O157:H7 cells were sensitive to all three phages, which indicated that they had not acquired resistance to phage (data not shown). However, at 5 days after phage infection, the surviving E. coli O157:H7 cells were resistant to all three phages (data not shown). The bacterial isolates that survived infection continued to express the O157 antigen, because they were agglutinated with anti-O157 sera.
The phage concentrations in cultures infected at an MOI of 103 PFU/CFU consistently increased 10-fold after 8 h of incubation (KH4) or 24 h of incubation (KH1 or KH5), suggesting that the phages replicated (data not shown). The phage concentrations in the cell-free controls decreased about 10-fold after 8 h of incubation at 37°C.
Similar experiments performed at 4°C with KH1, KH4, or KH5 resulted in 0.5- to 0.2-log10 decreases in the titers of viable bacteria compared to the phage-free bacterial control. The phage titers in all of the experiments remained constant throughout the time course of infection (data not shown). At 4°C, no bacterial growth was observed in any culture, and the E. coli O157:H7 concentration remained 3 × 104 CFU/ml in the phage-free bacterial control (data not shown).
Phage mixtures can eliminate E. coli O157.
The ability of serial additions of the same phage or different phages to eliminate E. coli O157:H7 was assessed. E. coli O157:H7 infected with KH1 at an MOI of 103 PFU/CFU was superinfected with 108 PFU of KH1 per ml (KH1 + KH1), 108 PFU of KH4 per ml (KH1 + KH4), or 108 PFU of KH5 per ml (KH1 + KH5) after 24 h. The cultures were incubated at 37°C without aeration and were assayed 48 h after the second phage addition. The E. coli O157:H7 titers in these superinfected cultures declined by 102-fold (KH1 + KH1) to 104-fold (KH1 + KH4 or KH1 + KH5) (data not shown). Thus, superinfection with either the same phage or a different phage resulted in a decrease in bacterial survival.
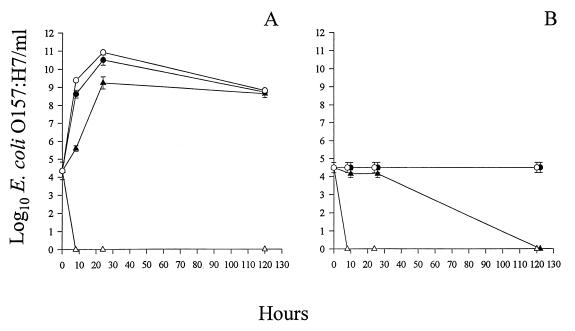
Finally, we studied simultaneous infection of E. coli O157:H7 with all three phages at an MOI of 103 PFU/CFU. The infected cultures were incubated at 37 or 4°C with or without aeration. In aerated cultures at 37 or 4°C, complete bacterial lysis was observed within 8 h after infection (Fig. 3). No growth occurred on LB agar, and even enrichment of the suspension did not yield viable E. coli O157:H7 cells (data not shown). The bacterial concentrations 8 h after phage infection were >109- and >104-fold less than the concentrations in the aerated phage-free bacterial controls at 37 and 4°C, respectively (Fig. 3). In nonaerated cultures at 37°C, the bacterial concentrations decreased 10- to 1,000-fold compared to the concentrations in the nonaerated phage-free bacterial controls by 8 to 24 h after phage infection. However, at 5 days after phage infection, the E. coli O157:H7 survivors had grown to a high density (Fig. 3A). In the nonaerated cultures at 4°C, only slight decreases in the bacterial concentrations were observed until 5 days after phage infection, when complete bacterial death occurred (Fig. 3B). In experiments in which all of the E. coli O157:H7 cells were killed, the phage titers increased 10-fold or remained the same (data not shown). The phage concentrations in the nonaerated cultures at 37°C decreased 10-fold after 24 h of incubation and then remained at that level (data not shown).
FIG. 3.
E. coli O157:H7 lysis by a mixture of phages. Cultures were incubated at 37°C (A) or 4°C (B). E. coli O157:H7 (3 × 104 CFU) was infected with a mixed phage suspension (107 PFU) containing equal concentrations of KH1, KH4, and KH5. Cultures were incubated with (▵) or without (▴) aeration. The controls contained only E. coli O157:H7 and were either aerated (○) or not aerated (●). The standard errors of the E. coli O157:H7 concentrations ranged from 0 to 0.5 log10 CFU/ml (n = 4).
Under similar incubation conditions, the KH1-KH4-KH5 phage mixture killed all of the E. coli O157 strains tested (Tables 1 and 2). In contrast, none of the non-O157 E. coli, O-antigen-deficient E. coli mutant, or non-E. coli cultures were killed by the phage mixture (Tables 1 and 2).
DISCUSSION
We found that a mixture of three O157-specific phages is able to eliminate E. coli O157 from cultures. The factors that are critical for rapid cell lysis include aeration, incubation at 37°C, a high MOI, and simultaneous infection with the three phages. Phage treatment at 4°C without aeration resulted in bacterial death only after 5 days. The three O157-specific phages are not identical isolates with the same genotype because they behave differently when they kill E. coli O157.
Our decision to evaluate phages as anti-E. coli O157 agents was based on attempts by other workers to use phages as antibacterial agents. After the discovery of phages in 1915 to 1917, the use of bacterial viruses in human clinical settings became common in Europe (2, 4). Phage therapy was used to combat infections of the skin, bone, gastrointestinal tract, chest, abdomen, head, neck, and other organ systems (2). In fact, phages were also inadvertently responsible for controlling the spread of cholera in several parts of the Indian subcontinent a century ago (4). However, phage therapy was plagued by several controversies, primarily due to the inability to manipulate phage genetics (2, 4). More recently, studies conducted by Smith et al. have shown that phages may have immense potential for controlling E. coli infections in cattle (29). These researchers cured or prevented enteropathogenic E. coli (109 CFU) diarrhea in calves with strain-specific phages administered in a single oral dose (105 PFU) or sprayed on the litter (102 PFU) (29). However, efficacy was observed only when a phage was administered before or together with the infective bacteria. If the phage was administered after the onset of diarrhea, the disease intensity was decreased but the disease was not cured (29). Similarly, Berchieri et al. controlled an experimental Salmonella typhimurium (109 CFU) infection in poultry with Salmonella phage (109 PFU) (6).
Encouraged by these studies and the need for alternative methods of E. coli O157:H7 control, we isolated E. coli O157-specific phages. Adsorption of phage particles to bacterial cells, the initial step of phage infection, is dependent on the presence of specific receptors on the cell wall (34). Many cell wall receptors can be shared by different bacterial strains and serotypes (34). To obtain O157 antigen-specific phages, we screened for phages that bind to the O157 antigen and against phages that bind to common E. coli receptors, such as pili, fimbriae, flagella, LPS cores, and other outer membrane proteins (10, 34). In this way, we hoped to find phages that are lethal to E. coli O157 and not to other normal flora. In the one previous report of an E. coli O157:H7-specific coliphage, Ronner and Cliver described a phage that was toxin specific and thus lysed other Shiga toxin-producing bacteria, including S. dysenteriae (27).
The evidence that supports the conclusion that KH1, KH4, and KH5 are specific for serotype O157 includes the fact that these phages lysed all of the E. coli O157 strains tested and did not lyse non-O157 E. coli, non-E. coli, or O157-deficient mutant E. coli strains. In addition, phage infection was dependent on the nature of the O157 LPS. The complement of the E. coli O157-deficient mutant, E. coli F12(pF12), which produces a truncated O157 LPS (9), was resistant to infection.
Interestingly, although the mixture of phages KH1, KH4, and KH5 lysed (in liquid culture) all of the natural E. coli O157 strains tested, we found a few O157 strains that were resistant to plaque formation by individual phages. Like phage infection, plaque formation appears to be influenced by the nature of the O157 LPS. The E. coli O157 plaque-resistant strains produced significantly more mid-range-molecular-weight LPS than the strains susceptible to plaque formation produced (Fig. 1). The excess mid-range-molecular-weight LPS made by the plaque-resistant E. coli O157 strains may accumulate around cells in soft agar and influence phage attachment but diffuse from cells in liquid culture. The phenomenon of an LPS requirement for plaque formation is not unique. For example, the Salmonella strains susceptible to phage P22 contain around 20 repeats of the common O-antigen trisaccharide unit; strains with shorter O antigens do not support P22 plaque formation (5). Therefore, an appropriate length of the O side chains and an optimal LPS concentration may be necessary to make the receptor available for phage interactions and/or to allow irreversible phage binding (11). It is tempting to speculate that abundant or size-specific immunoreactive O157 LPS competitively inhibits the adherence of the phages under plaquing conditions, but elucidation of the mechanism was beyond the scope of this study. Alternatively, the O157 plaque-resistant strains may not possess auxiliary mechanisms for phage adherence and uptake that are critical for plaque formation. The plaque-resistant O157 strains were non-H7 isolates and belong to lineages quite distant from the lineage to which E. coli O157:H7 belongs. It is, however, unlikely that expression of H7 antigen plays a role in plaque resistance since several other O157 non-H7 strains were susceptible to plaque formation by the phages studied.
The presence of magnesium is important for propagation of all three O157-specific phages. Mg2+ or Ca2+ ions are required for adsorption of a subset of phages (11). Like other coliphages, the O157-specific phages are lytic, and a high MOI favors killing of host cells. Presumably, a high MOI is necessary to ensure that every bacterium is infected by at least one phage. However, even at the optimal MOI (103 PFU/CFU), no single phage could clear an E. coli O157:H7 culture. Complete bacterial elimination was observed only when cultures were infected with all three phages, indicating that the combined effects of the phages were more than additive. In addition, culture aeration played a critical role in phage induction of rapid (within 8 h postinfection) and complete bacterial death. Aeration may increase the opportunity for phage-bacterium interactions. In nonaerated cultures, complete elimination of bacteria occurred only 5 days after phage infection and only at 4°C. We hypothesize that a low temperature and an absence of bacterial growth may favor better phage adsorption and infection. In contrast, a higher temperature (37°C), cell growth, and the potential for phenotypic variability in expression of the O antigen may favor survival of phage-resistant cells (14).
All of the bacteria that survived phage infection were tested for phage resistance (2, 4), and among the cells that had been exposed to phages for 5 days, we found E. coli O157:H7 cells that were resistant to plaque formation with these phages. The factors that contribute to phage resistance include alteration or loss of receptors (2, 4, 11). The finding that all phage-resistant bacteria agglutinate with the O157 antiserum indicates that these organisms have not lost the O side chains completely. These strains, like E. coli F12(pF12), may have a shorter O157 antigen. Consistent with this idea, preliminary results indicated that in the phage-resistant E. coli O157:H7 strains there may be significant alterations in the length or regulation of LPS production (data not shown). Thus, efficient use of phage to control E. coli O157:H7 infections may require isolation of mutant O157-specific phage that can adsorb to hosts that make shorter O side chains.
The phages that we isolated in this study may be used for biocontrol of E. coli O157. Based on our finding that killing occurs at 4°C, trials are under way to use these phages to eliminate E. coli O157:H7 from fresh vegetables. Several disease outbreaks in the United States have been linked to lettuce contaminated with E. coli O157:H7 (1). In addition, contaminated radish sprouts were the likely source of an E. coli O157:H7 outbreak in Japan in which more than 10,000 people were infected and 13 people died (25, 31). Because our phages kill efficiently at 4°C, they may be used to eliminate E. coli O157:H7 from these types of foods under refrigerated conditions. We have shown that O157-specific phages are prevalent in nature, and therefore it should be possible to isolate additional O157-specific phages or mutants of these phages that can kill under anaerobic conditions and may be used to eliminate E. coli O157 from the gastrointestinal tracts of carrier ruminants.
ACKNOWLEDGMENTS
This work was supported in part by U.S. Department of Agriculture NRICGP grants 92-04350 and 95-37201-1979 (to C.J.H.), by Public Health Service grant AI33981 from the National Institutes of Health (to C.J.H.), and by a grant from the M. J. Murdock Charitable Trust (to C.J.H. and P.Y.). This work was also supported by USDA grant 96-01601 (to P.I.T.)
We acknowledge R. England for skillful technical assistance.
REFERENCES
- 1.Ackers M, Mahon B E, Leahy E, Goode B, Damrow T, Hayes P S, Bibb W F, Rice D H, Barrett T J, Hutwagner L, Griffin P M, Slutsker L. An outbreak of Escherichia coliO157:H7 infections associated with leaf lettuce consumption. J Infect Dis. 1998;177:1588–1593. doi: 10.1086/515323. [DOI] [PubMed] [Google Scholar]
- 2.Alisky J, Iczkowski K, Rapoport A, Troitsky N. Bacteriophages show promise as antimicrobial agents. J Infect. 1998;36:5–15. doi: 10.1016/s0163-4453(98)92874-2. [DOI] [PubMed] [Google Scholar]
- 3.American Type Culture Collection. Catalogue of bacteria and bacteriophages. 18th ed. Rockville, Md: American Type Culture Collection; 1992. [Google Scholar]
- 4.Barrow P A, Soothill J S. Bacteriophage therapy and prophylaxis: rediscovery and renewed assessment of potential. Trends Genet. 1997;5:268–271. doi: 10.1016/S0966-842X(97)01054-8. [DOI] [PubMed] [Google Scholar]
- 5.Baxa U, Steinbacher S, Miller S, Weintraub A, Huber R, Seckler R. Interactions of phage P22 tails with their cellular receptor, SalmonellaO-antigen polysaccharide. Biophys J. 1996;71:2040–2048. doi: 10.1016/S0006-3495(96)79402-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berchieri A J, Lovell M A, Barrow P A. The activity in the chicken alimentary tract of bacteriophages lytic for Salmonella typhimurium. Res Microbiol. 1991;142:541–549. doi: 10.1016/0923-2508(91)90187-f. [DOI] [PubMed] [Google Scholar]
- 7.Besser R E, Lett S M, Weber J T, Doyle M P, Barrett T J, Wells J G, Griffin P M. An outbreak of diarrhea and hemolytic uremic syndrome from Escherichia coliO157:H7 in fresh-pressed apple cider. JAMA. 1993;269:2217–2220. [PubMed] [Google Scholar]
- 8.Bielaszewska M, Janda J, Blahova K, Minarikova H, Jikova E, Karmali M A, Laubova J, Sikulova J, Preston M A, Khakhria R, Karch H, Klazarova H, Nyc O. Human Escherichia coliO157:H7 infection associated with the consumption of unpasteurized goat’s milk. Epidemiol Infect. 1997;119:299–305. doi: 10.1017/s0950268897008297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bilge S S, Vary J C J, Dowell S F, Tarr P I. Role of Escherichia coli O157:H7 O side chain in adherence and analysis of an rfblocus. Infect Immun. 1996;64:4795–4801. doi: 10.1128/iai.64.11.4795-4801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bokete T N, Whittam T S, Wilson A W, Clausen C R, O’Callahan C M, Moseley S L, Fritsche T R, Tarr P I. Genetic and phenotypic analysis of Escherichia coliwith enteropathogenic characteristics isolated from Seattle children. J Infect Dis. 1997;175:1382–1389. doi: 10.1086/516470. [DOI] [PubMed] [Google Scholar]
- 11.Calendar R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Press; 1988. [Google Scholar]
- 12.Chart H, Said B, Stokes N, Rowe B. Heterogeneity in expression of lipopolysaccharides by strains of Escherichia coliO157. J Infect. 1993;27:237–241. doi: 10.1016/0163-4453(93)91952-l. [DOI] [PubMed] [Google Scholar]
- 13.Cray W C, Jr, Moon H W. Experimental infection of calves and adult cattle with Escherichia coliO157:H7. Appl Environ Microbiol. 1995;61:1586–1590. doi: 10.1128/aem.61.4.1586-1590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dodds K L, Perry M B, McDonald I J. Alterations in lipopolysaccharide produced by chemostat-grown Escherichia coliO157:H7 as a function of growth rate and growth-limiting nutrient. Can J Microbiol. 1987;33:452–458. doi: 10.1139/m87-075. [DOI] [PubMed] [Google Scholar]
- 15.Griffin P M. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli. In: Blaser M F, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press, Ltd.; 1995. pp. 739–761. [Google Scholar]
- 16.Hancock D D, Besser T E, Rice D H. Ecology of Escherichia coli O157:H7 in cattle and impact of management practices. In: Kaper J B, O’Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C: ASM Press; 1998. pp. 85–91. [Google Scholar]
- 17.Inzana T. Electrophoretic heterogeneity and interstrain variation of the lipopolysaccharide of Haemophilus influenzae. J Infect Dis. 1983;148:492–499. doi: 10.1093/infdis/148.3.492. [DOI] [PubMed] [Google Scholar]
- 18.Keene W E, Hedberg K, Herriott D E, Hancock D D, McKay R W, Barrett T J, Fleming D W. A prolonged outbreak of Escherichia coliO157:H7 infections caused by commercially distributed raw milk. J Infect Dis. 1997;176:815–818. doi: 10.1086/517310. [DOI] [PubMed] [Google Scholar]
- 19.Keene W E, Sazie E, Kok J, Rice D H, Hancock D D, Balan V K, Zhao T, Doyle M P. An outbreak of Escherichia coliO157:H7 infections traced to jerky made from deer meat. JAMA. 1997;277:1229–1231. doi: 10.1001/jama.1997.03540390059036. [DOI] [PubMed] [Google Scholar]
- 20.Kudva I T, Hatfield P G, Hovde C J. Characterization of Escherichia coli O157:H7 and other Shiga toxin-producing E. coliserotypes isolated from sheep. J Clin Microbiol. 1997;35:892–899. doi: 10.1128/jcm.35.4.892-899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kudva I T, Hatfield P G, Hovde C J. Effect of diet on the shedding of Escherichia coliO157:H7 in a sheep model. Appl Environ Microbiol. 1995;61:1363–1370. doi: 10.1128/aem.61.4.1363-1370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kudva I T, Hunt C W, Williams C J, Nance U M, Hovde C J. Evaluation of dietary influences on Escherichia coliO157:H7 shedding by sheep. Appl Environ Microbiol. 1997;63:3878–3886. doi: 10.1128/aem.63.10.3878-3886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marques L R M, Moore M A, Wells J G, Wachsmuth I K, O’Brien A D. Production of Shiga-like toxin by Escherichia coli. J Infect Dis. 1986;154:338–341. doi: 10.1093/infdis/154.2.338. [DOI] [PubMed] [Google Scholar]
- 24.Murinda S E, Roberts R F, Wilson R A. Evaluation of colicins for inhibitory activity against diarrheagenic Escherichia colistrains, including serotype O157:H7. Appl Environ Microbiol. 1996;62:3196–3202. doi: 10.1128/aem.62.9.3196-3202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nathan R. American seeds suspected in Japanese food poisoning epidemic. Nat Med. 1997;3:705–706. doi: 10.1038/nm0797-705b. [DOI] [PubMed] [Google Scholar]
- 26.Rice D H, Hancock D D, Besser T E. Verotoxigenic E. coliO157 colonization of wild deer and range cattle. Vet Rec. 1995;137:524. doi: 10.1136/vr.137.20.524. [DOI] [PubMed] [Google Scholar]
- 27.Ronner A B, Cliver D O. Isolation and characterization of a coliphage specific for Escherichia coliO157:H7. J Food Prot. 1990;53:944–947. doi: 10.4315/0362-028X-53.11.944. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Smith H W, Huggins M B, Shaw K M. The control of experimental Escherichia colidiarrhea in calves by means of bacteriophages. J Gen Microbiol. 1987;133:1111–1126. doi: 10.1099/00221287-133-5-1111. [DOI] [PubMed] [Google Scholar]
- 30.Su C Y, Brandt L J. Escherichia coliO157:H7 infection in humans. Ann Intern Med. 1995;123:698–714. doi: 10.7326/0003-4819-123-9-199511010-00009. [DOI] [PubMed] [Google Scholar]
- 31.Swinbanks D. Outbreak of E. coliinfection in Japan renews concerns. Nature. 1996;382:290. doi: 10.1038/382290c0. [DOI] [PubMed] [Google Scholar]
- 32.Tarr P I. Escherichia coliO157:H7: clinical, diagnostic, and epidemiological aspects of human infection. Clin Infect Dis. 1995;20:1–10. doi: 10.1093/clinids/20.1.1. [DOI] [PubMed] [Google Scholar]
- 33.Tauxe R V. Emerging foodborne diseases: an evolving public health challenge. Emerg Infect Dis. 1997;3:425–434. doi: 10.3201/eid0304.970403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Topley W W C, Wilson G S. Principles of bacteriology, virology and immunity. B.C. London, United Kingdom: Decker Publisher; 1990. [Google Scholar]
- 35.Tsai C M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 36.U.S. Department of Agriculture, Animal and Plant Health Inspection Service, Veterinary Services. An update: Escherichia coliO157:H7 in humans and cattle. Rep Cen Epidemiol Anim Health. 1997;1982:1–28. [Google Scholar]
- 37.Wallace J S, Cheasty T, Jones K. Isolation of Vero cytotoxin-producing Escherichia coliO157 from wild birds. J Appl Microbiol. 1997;82:399–404. doi: 10.1046/j.1365-2672.1997.00378.x. [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Reeves P R. Organization of Escherichia coliO157 O antigen gene cluster and identification of its specific genes. Infect Immun. 1998;66:3545–3551. doi: 10.1128/iai.66.8.3545-3551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wells J G, Davis B R, Wachsmuth I K, Riley L W, Remis R, Sokolow R, Morris G K. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coliserotype. J Clin Microbiol. 1983;18:512–520. doi: 10.1128/jcm.18.3.512-520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao T, Doyle M P, Harmon B G, Brown C A. VTEC’97, 3rd International Symposium and Workshop on Shiga Toxin (Verotoxin)-Producing Escherichia coli Infections. 1997. Reduction of Escherichia coli O157:H7 in dairy cattle by selected probiotic bacteria, abstr. V63/II; p. 32. [Google Scholar]