Abstract
The comet assay is gaining popularity as a means to assess DNA damage in cultured cells and tissues, particularly following exposure to chemicals or other environmental stressors. Use of the comet assay in regulatory testing for genotoxic potential in rodents has been driven by adoption of an Organisation for Economic Co-operation and Development (OECD) test guideline in 2014. Comet assay slides are typically prepared from fresh tissue at the time of necropsy; however, freezing tissue samples can avoid logistical challenges associated with simultaneous preparation of slides from multiple organs per animal and from many animals per study. Freezing also enables shipping samples from the exposure facility to a different laboratory for analysis, and storage of frozen tissue facilitates deferring a decision to generate DNA damage data for a given organ. The alkaline comet assay is useful for detecting exposure-related DNA double- and single-strand breaks, alkali-labile lesions, and strand breaks associated with incomplete DNA excision repair. However, DNA damage can also result from mechanical shearing or improper sample processing procedures, confounding the results of the assay. Reproducibility in collection and processing of tissue samples during necropsies may be difficult to control due to fluctuating laboratory personnel with varying levels of experience in harvesting tissues for the comet assay. Enhancing consistency through refresher training or deployment of mobile units staffed with experienced laboratory personnel is costly and may not always be feasible. To optimize consistent generation of high quality samples for comet assay analysis, a method for homogenizing flash frozen cubes of tissue using a customized tissue mincing device was evaluated. Samples prepared for the comet assay by this method compared favorably in quality to fresh and frozen tissue samples prepared by mincing during necropsy. Moreover, low baseline DNA damage was measured in cells from frozen cubes of tissue following prolonged storage.
Keywords: single cell gel electrophoresis, alkaline comet assay, chromosomal damage, DNA strand breaks, genotoxicity, in vivo, OECD
SUMMARY:
This protocol describes several procedures for preparing high quality frozen tissue samples at the time of necropsy for use in the comet assay to assess DNA damage: 1) minced tissue, 2) scraped epithelial cells from the gastrointestinal tract, and 3) cubed tissue samples, requiring homogenization using a tissue mincing device.
INTRODUCTION:
The comet assay is increasingly used as a means to evaluate DNA damage in cultured cells and tissues exposed to chemicals or other environmental stressors1. The assay can detect DNA double- and single-strand breaks, alkali-labile lesions, and single-strand breaks associated with incomplete DNA repair. The International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) guideline for pharmaceutical testing recommends a DNA strand breakage assay such as the comet assay as a second test to supplement the rodent erythrocyte micronucleus assay for assessing in vivo genotoxicity and as a follow-up test for assessing mode of action in target organs of tumor induction2. The European Food Safety Authority (EFSA) recommends the in vivo comet assay as a suitable follow-up test for investigating the relevance of a positive result in an in vitro genotoxicity test3. In 2014, an OECD test guideline was approved for the rodent comet assay, thus increasing the acceptability of the assay for use in regulatory testing of genotoxic potential. The assay is based on the electrophoretic separation of relaxed DNA loops and fragments that migrate from nucleoids of lysed cells. Basically, single cells are embedded in agarose that has been layered onto microscope slides. Slides are then immersed in lysis buffer followed by an alkaline (pH > 13) solution, which allows the tightly coiled nuclear DNA to relax and unwind. Slides are then placed in an electric field, which stimulates migration of negatively charged DNA toward the anode, creating images that resemble comets; the relative amount of DNA in the comet tail compared with the comet head is directly proportional to the amount of DNA damage; DNA content in the tail is typically quantified using digital imaging software.
Because the comet assay detects fragmented DNA, accurate quantification of exposure-induced DNA damage can be confounded by chromatin fragmentation associated with necrosis or apoptosis resulting from treatment-induced cytotoxicity or stress. Furthermore, DNA damage can occur as a result of mechanical shearing or improper sample processing4. The importance of maintaining harvested tissue chilled prior to slide preparation to minimize the baseline level of DNA damage has been previously demonstrated 5,6. Many laboratories prepare comet assay slides from fresh tissue; however, this can be logistically challenging when preparing slides from multiple tissue types per animal in a study with a large number of animals. Moreover, this presents a problem when slide preparation and analysis are to occur at a remote laboratory, necessitating shipment of samples. For example, the U.S. National Toxicology Program includes the comet assay as a component of its genetic toxicology testing program (https://ntp.niehs.nih.gov/testing/types/genetic/index.html) and sometimes incorporates the assay into 28- or 90-day repeat dose toxicity studies; this necessitates collection of tissue by the in-life laboratory and transfer of samples to another laboratory for analysis. To accomplish this, tissue pieces are minced, and/or epithelial cells of the gastrointestinal tract are scraped and cellular suspensions are flash frozen and stored in a freezer for subsequent shipment and storage by the receiving laboratory until analysis7. Proper handling of samples is crucial for obtaining high quality data using frozen tissue; however, reproducible manipulation of tissue samples during necropsies performed by ever-changing personnel is difficult to control, especially at in-life laboratories that do not routinely harvest tissues for the comet assay. Refresher training of necropsy staff or use of a mobile unit staffed by experienced laboratory personnel to collect fresh or frozen tissue samples is often too costly, not feasible, or simply undervalued.
To better ensure consistent generation of high quality tissue samples for transfer to a remote site for comet assay analysis, the utility of a published method6 of tissue preservation from flash frozen cubes of tissue was explored. In this method, frozen cubes of tissue are loaded into a stainless steel tissue mincing device (Figure 1) that is placed into a microcentrifuge tube containing cold buffer. The cube of tissue is then pushed through a small gauge mesh at the end of the device. Repeatedly forcing the tissue suspension through the mesh sieve in both directions several times results in a relatively uniform single cell suspension. Samples prepared by this method compared favorably in quality to both fresh and frozen tissue samples prepared by mincing. As an added benefit, unlike minced samples, tissue cubes can be stored frozen for prolonged periods of time and still yield high quality results in the comet assay.
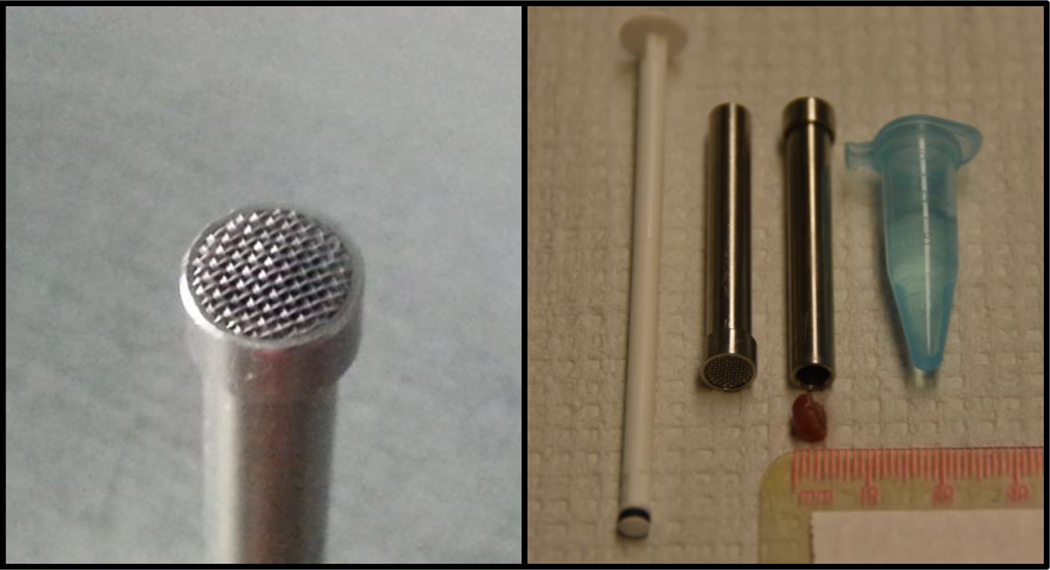
Figure 1: Tissue mincing device and plunger used to prepare a single cell suspension from frozen tissue.
The prototype device (4.5 mm inner diameter, 0.4 mm mesh size) was kindly provided by Dr. Gunnar Brunborg (NorGenotech, Oslo, Norway). The panel on the right provides an example of an appropriately sized cube of tissue for use with the device.
PROTOCOL:
Tissues were harvested during the conduct of studies performed at AAALAC-accredited facilities at NTP contract laboratories in accordance with Good Laboratory Practice regulations (21 CFR Part 58) and animal use protocols approved by the Institutional Animal Care and Use Committee (IACUC) at each laboratory.
1. Tissue harvest and processing
NOTE: It is useful to prepare duplicate sample tubes (e.g., liver) or transfer approximately half of a sample to another storage tube (e.g., duodenum, stomach) to enable re-analysis, if necessary. To minimize potential sample-to-sample variability for intestinal tract tissues, it is recommended that care be taken to sample the same region of tissue relative to the stomach for each animal, and a sample be divided to generate duplicate samples. Plastic forceps are recommended for transferring sticky tissues such as brain. As an option, minced, scraped or homogenized tissue preparations may be filtered to achieve homogeneous single cell suspensions using a 40 μm cell strainer attached to a conical tube.
1.1. Remove any attached connecting tissue, organs, and debris from organ(s) of interest. Immediately following removal, swish the organ vigorously in a medium-sized weigh boat containing ~7 mL of cold freshly prepared mincing solution (Mg++, Ca++ and phenol red-free Hank’s Balanced Salt Solution, 10% v/v DMSO, and 20 mM EDTA pH 7.5) to remove residual blood and debris.
1.2. Transfer each organ to another clean weigh boat containing sufficient (~7 mL) cold mincing solution to maintain the tissue submerged, on ice, until further processing.
1.3. Remove a section of each organ of interest and place it in an embedding cassette. Fix in 10% neutral buffered formalin, trim, and paraffin embed according to the laboratory’s standard procedures for possible future histopathology evaluation.
1.4. For the liver, cut a 5 mm longitudinal section of the left lobe and gently swish in ~7 mL of cold mincing solution. Place the strip of tissue into a clean medium-sized weigh boat containing ~7 mL of cold mincing solution and maintain on ice until ready to process further (step 1.8 or 1.11).
- 1.5. For the duodenum, cut a 10 mm portion of the duodenum proximal to the stomach. To remove debris and bacteria, insert a 21–25 G needle into one end of the duodenum and flush with 1 mL of cold mincing solution.
- 1.5.1 Flush in the other direction by flipping the duodenum over and repeating with another 1 mL of mincing solution. Discard the needle. Slice the duodenum open and rinse it in ~7 mL of mincing solution before putting it into a medium-sized weigh boat containing ~7 mL of clean mincing solution; maintain on ice until ready to process further (step 1.8 or 1.11).
- 1.6. For the brain, it may be useful to first divide the brain into two hemispheres; one hemisphere can be saved for possible histopathology (see step 1.2). Dissect the brain region(s) of interest and gently swish in ~7 mL of cold mincing solution.
- 1.6.1. Transfer tissue(s) to a clean medium-sized weigh boat containing ~7 mL of cold mincing solution and maintain on ice until ready to process further (step 1.8 or 1.11; note that small regions such as the cerebellum and hippocampus may not require any further trimming).
- 1.7. For the stomach
- 1.7.1. Remove and discard the forestomach. Cut open the glandular stomach and wash free from food using ~7 mL of cold mincing buffer in a medium-sized weigh boat.
- 1.7.2. Remove a 5 mm strip of glandular stomach proximal to the duodenum for fixation for possible histopathology evaluation (see step 1.2).
-
1.7.3. Place the remaining stomach into ~7 mL of cold mincing buffer in a medium-sized weigh boat and incubate on ice for 15–30 min.
- 1.7.4. Transfer the stomach to a clean piece of paraffin (or Petri dish or weigh boat) and gently scrape the surface epithelium two or more times using a scalpel blade or a polytetrafluoroethene (PTFE) scraper to remove debris. Pick up the gastric mucosa with forceps and rinse with 1 mL of cold mincing buffer using a pipet. Transfer the stomach tissue to a clean surface or dish.
- 1.7.5. Carefully scrape the stomach epithelium 4–5 times (more, if necessary) in mincing solution (typically 250 μL/mouse or 500–1000 μL/rat) with the back of a scalpel blade or PTFE scraper to release the cells. Optionally, rinse tissue with an approximately equal volume of mincing solution to recover more cells. Collect the mincing solution containing released cells using a pipet and transfer to a microcentrifuge tube on ice.
1.8. For mincing tissue samples, cut a 3–4 mm section of liver or brain tissue or ~5 mm of duodenum and transfer to a labeled 1.5 or 2.0 mL microcentrifuge tube containing 1 mL of cold mincing solution. Rapidly mince (≤30 s) with mincing scissors until finely dispersed, while keeping the sample cold. The sample should look slightly cloudy; however, it is common for some pieces to remain that will settle to the bottom of the tube.
1.9. To use the tissue fresh, maintain tubes containing minced tissue or scraped epithelial cells on ice; use the tissue to prepare comet slides within approximately 1 h of harvest (outlined in section 2).
- 1.10. For freezing the tissue samples, immediately following mincing or collection of scraped epithelial cells, secure the tube lid and drop tube into a Dewar flask containing liquid nitrogen; tubes may be maintained in liquid nitrogen for the duration of necropsy (≤3 h).
- 1.10.1. Retrieve tubes using a ladle and place on dry ice to sort. Transfer tubes to a freezer box on dry ice and store box in a −80 °C freezer.
1.11. For preparing frozen cubes of tissue, cut several small pieces of tissue (≤4 mm in diameter and ~6 mm in length; Figure 1) and individually drop them directly into a medium-sized weigh boat or other suitable vessel containing liquid nitrogen.
1.12. Wait a few seconds for the pieces to freeze completely and transfer individual frozen tissue cubes to a labeled microcentrifuge tube or cryotube maintained on dry ice; do not allow the pieces to clump together during the freezing process. Transfer tubes to a freezer box on dry ice and store box in a −80 °C freezer.
2. Slide preparation
- 2.1. For minced/scraped tissue
- 2.1.1. Maintain tubes containing fresh tissues on wet ice.
- 2.1.2. Place tubes of frozen tissue into a room temperature water bath until the samples are completely thawed, while ensuring they are kept cold. Immediately transfer the tubes onto ice.
- 2.1.3. Immediately prior to withdrawing cells for transfer to agarose (step 2.3), mix each cell suspension gently; for non-filtered samples, allow large chunks to settle to the bottom of the tube. Maintain all tubes on ice until slides have been prepared for all samples.
- 2.2 For cubed tissue
- 2.2.1. Retrieve tubes containing tissue cubes from the −80 °C freezer and maintain on dry ice until slide preparation.
- 2.2.2. Aliquot 1 mL of Merchant’s medium (0.14 M NaCl, 1.47 mM KH2PO4, 2.7 mM KCl, 8.1 mM Na2HPO4, 10 mM NaEDTA, pH 7.4) or mincing solution into a labeled 1.7 mL microcentrifuge tube for each sample and maintain on ice.
- 2.2.3. Working rapidly to avoid tissue thawing, place a cube of tissue into the open end of a room temperature tissue mincing device (Figure 1). Multiple tissue pieces may be used; if necessary, the frozen tissue may be cut or cracked into smaller pieces using a cold mortar and pestle to reduce the cube size to fit into the device.
- 2.2.4. Quickly insert a size-matched syringe plunger into the device to push the tissue to the mesh end which should then be placed into the microcentrifuge tube containing cold mincing solution; avoid forcing the liquid to overflow the tube. Immerse the mesh end of the device for approximately 5–10 s in the cold mincing solution to allow the tissue to thaw completely.
- 2.2.5. Completely depress the plunger until cells are seen extruding through the mesh pores. Then pull the plunger up approximately 2.5 cm and press down again completely; repeat the process several times until the mincing solution becomes turbid.
- 2.2.6. If necessary, the sample may be concentrated to increase cell density by refrigerated centrifugation for 5 min at 1100 rpm and removal of a portion of the supernatant.
- 2.2.7. Repeat the process for all samples using a clean tissue mincing device for each sample.
-
2.3. Transfer 50 μL of cell suspension to a tube containing 450 μL of 0.5% low melting point agarose at 37 °C.
NOTE: Volumes may be adjusted, but the amount of agarose should be ≤1%.
REPRESENTATIVE RESULTS:
Study 1
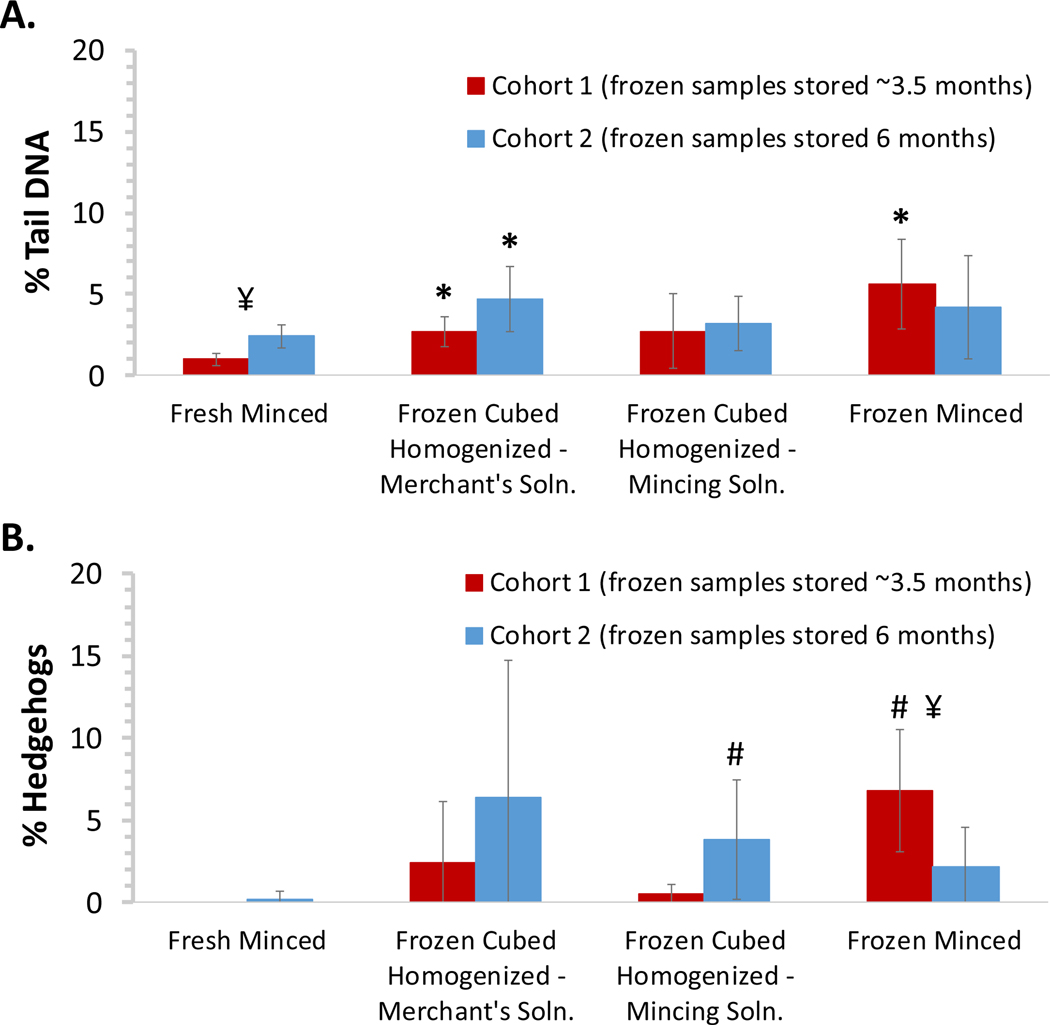
Liver was harvested from two cohorts of male Sprague Dawley rats administered corn oil for 4 days, staggered by one week. Slides were prepared from freshly minced tissue, frozen minced tissue, and frozen cubed tissue processed in Merchant’s medium or mincing solution using the tissue mincing device. Frozen tissues obtained from animals from the first cohort were evaluated after freezer storage for ~3.5 months. Frozen tissues obtained from animals from the second cohort were evaluated after storage for ~6 months. Comets exhibiting the characteristic “hedgehog” morphology, indicative of heavily damaged cells (of uncertain etiology that may include cytotoxicity or mechanical stress), were not included in the scoring of % tail DNA; the percentage of hedgehogs, identified solely upon visual inspection of morphology, was tabulated separately as per OECD TG 4899. The mean % tail DNA results for the animals in the two cohorts (based on scoring 100 cells/animal) are summarized in Figure 2A. All the methods using frozen tissue produced values higher, although not always statistically different, than fresh tissue. Considering that the recommended upper limit for % tail DNA in fresh rat liver is 6% 9, these results demonstrate that any of these methods can work well for evaluating frozen tissue. Mincing solution performed at least equally as well as Merchant’s medium; since the latter is more time-consuming to prepare, mincing solution was selected for subsequent studies. There were no statistically significant differences in damage in tissues from the second animal cohort that had been stored frozen for ~6 months prior to processing as compared to the tissues from the first cohort processed after ~3.5 months of storage. Overall, the % hedgehogs paralleled the % tail DNA although the % hedgehogs in the frozen tissues relative to fresh tissue was more variable than for % tail DNA (Figure 2B). The values for these endpoints in the fresh tissue provide a baseline against which the DNA damage artefactually introduced by the freezing methods can be gauged.
Figure 2: Comparison of DNA damage measured in fresh and frozen minced and homogenized mouse liver samples.
Liver was harvested from two staggered cohorts (n = 5/group/cohort) of male Sprague Dawley rats administered corn oil for 4 days. Slides were prepared from freshly minced tissue, frozen minced tissue, and frozen cubed tissue processed in Merchant’s medium or mincing solution using the tissue mincing device. Frozen tissues were analyzed ~3.5 and 6 months following necropsy for cohorts 1 and 2, respectively. Comets classified as hedgehogs based on morphology were not included in the scoring of % tail DNA. (A) Cohort mean % tail DNA results. (B) Cohort mean % hedgehog results. Error bars reflect standard deviation. Statistical analyses were conducted to compare frozen tissues against fresh tissue for each cohort and to compare the results between the two cohorts for each tissue preparation method. *Statistically different (p < 0.05) from fresh minced tissue within the same cohort using the Student’s t-test; #statistically different (p < 0.05) from fresh minced tissue within the same cohort using the Mann-Whitney test for non-normally distributed data; ¥ statistical difference (p < 0.05) between cohorts using the Student’s t-test.
Study 2
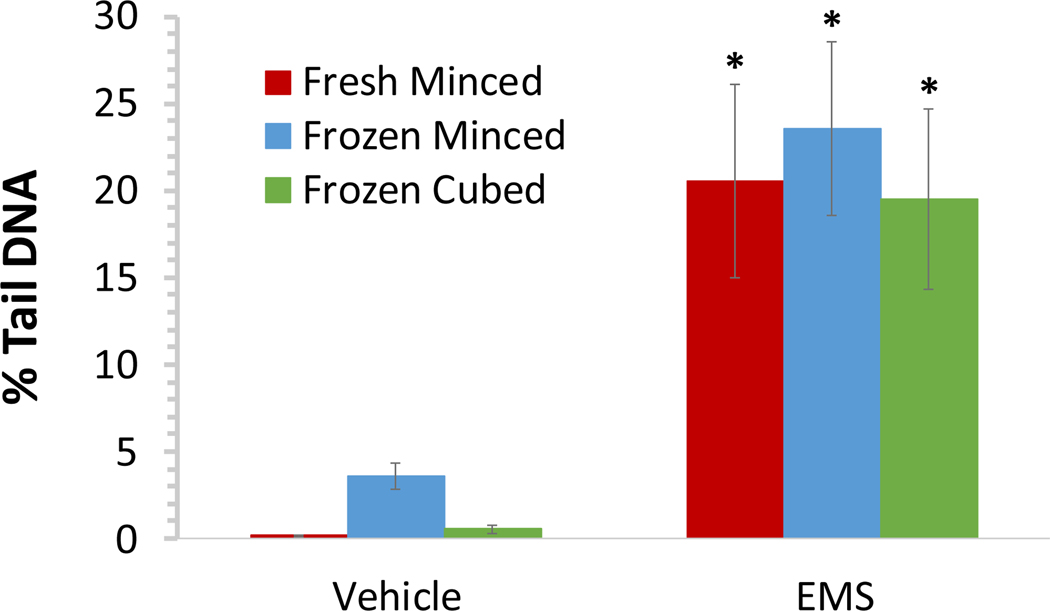
Female B6C3F1 mice were administered normal saline or the mutagen, ethyl methanesulfonate (EMS) at 200 mg/kg/day in normal saline for three days. Tissue was harvested 3 h after the final dose administration and processed fresh in the comet assay. Additional tissue was flash frozen after being minced in mincing solution or collected as cubes. Cubed samples were subsequently processed in mincing solution using the sieve device immediately prior to preparation of comet slides. On the premise that cells exhibiting the hedgehog morphology represent cells at the high end of the continuum of chemical-induced DNA damage, hedgehogs scorable by the imaging software were included in the automated scoring of % tail DNA. The % tail DNA results (based on scoring 150 cells/animal) are summarized in Figure 3. Liver tissue frozen as cubes and subsequently processed using the tissue mincing device yielded results very similar to those obtained for freshly minced tissue. The % tail DNA values were somewhat higher for the frozen minced tissue, but baseline values for the frozen minced tissue were below 6%, as recommended for fresh rat liver in the OECD test guideline for the comet assay9. A statistically significant increase in EMS-induced DNA damage was measured in tissue samples processed by all three methods.
Figure 3: Comparison of DNA damage induced by EMS in Liver tissue processed by different procedures.
Mean % tail DNA for female B6C3F1 mice administered vehicle or 200 mg/kg/day EMS for three days (n = 5/group). Livers were harvested 3 h following the final dose administration and processed fresh in the comet assay. Additional liver tissue was flash frozen after being minced in mincing solution or collected as cubes; cubed samples were homogenized using the tissue mincing device immediately prior to preparation of comet slides. To ensure cells at the high end of the continuum of EMS-induced DNA damage were not excluded from the analysis, comets that upon visual inspection appeared to meet the morphological description of hedgehogs but were found to be scorable by the imaging software were included. Error bars reflect standard deviation. A Student’s t-test was used to compare the amount of DNA damage in animals exposed to EMS against that in the vehicle control animals for each sample preparation method. *Statistically significant increase at p < 0.05.
Study 3
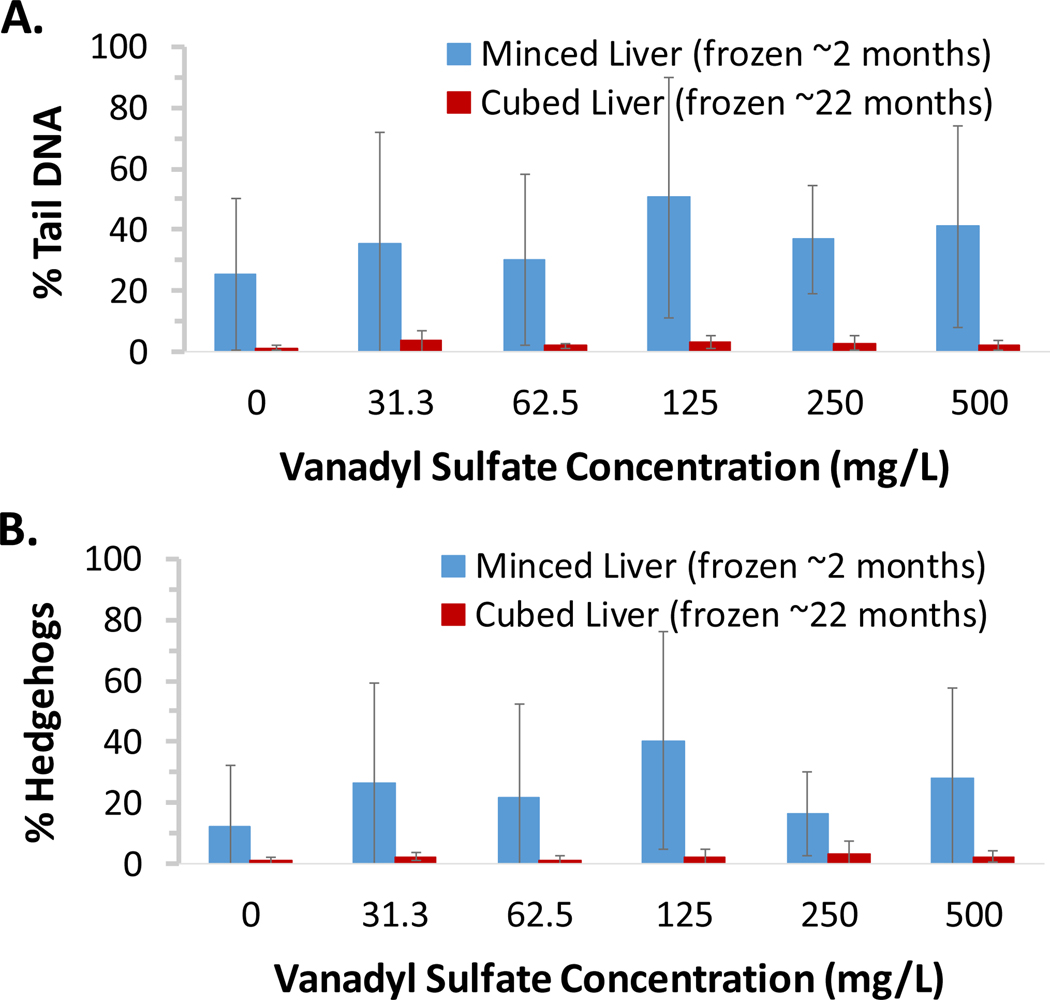
Portions of liver tissue collected from female B6C3F1 mice in a 90-day toxicity study of a test chemical conducted by a remote laboratory were minced or cut into cubes, flash frozen, and shipped overnight on dry ice to this laboratory for analysis. Minced tissue was analyzed following ~2 months of freezer storage (Figure 4). Cells that appeared upon visual inspection to be hedgehogs but were scorable by the imaging software were included in the % tail DNA measurements (150 cells scored/animal). The number of hedgehogs (whether scorable or nonscorable) identified solely upon visual inspection of morphology in a total of 150 cells/animal was tabulated separately to provide information on the frequency of these highly damaged cells. Overall, DNA damage (measured as % tail DNA) in frozen minced tissue was high across all the dose groups with substantial animal to animal variability, including in the vehicle control group. The % tail DNA results correlated with % hedgehogs (identified solely on the basis of morphology), indicating that hedgehogs contributed substantially to the high level of DNA damage that was observed in these samples. Based on the high background level of DNA damage measured in the minced tissue, frozen cubed tissue was subsequently analyzed after ~22 months of storage (Figure 4). Both DNA damage and % hedgehogs were very low in the flash frozen cubed tissue that had been prepared in parallel with the minced tissue samples. Thus, the highly damaged cells reflected by the hedgehog morphology in the minced tissue samples were clearly not the result of a biological process, but were likely introduced by mechanical disruption and/or tissue warming during the mincing procedure prior to being flash frozen; these data were considered to be unreliable. Fortunately, the analysis of frozen cubed tissue provided a clear result, namely the lack of a DNA damage response in the liver of female mice following three months of exposure to the test chemical. The results provided by this case study exemplify the risk associated with preparation of minced tissues by a relatively inexperienced laboratory and demonstrate the utility of the frozen cubed tissue method to circumvent this problem.
Figure 4. Case study demonstrating utility of frozen cube method and high quality data obtained after prolonged storage.
Portions of liver tissue collected from female B6C3F1 mice exposed to various concentrations of a test chemical for ~90 days (n = 5/group) were minced or cut into cubes and flash frozen at the in-life laboratory and subsequently shipped to this laboratory for analysis. Minced tissue was analyzed following ~2 months of storage; due to the poor quality of the data, frozen cubed tissue was subsequently analyzed following ~22 months of storage. To ensure cells at the high end of the continuum of chemical-induced DNA damage were not excluded from the analysis, data from scorable cells that, upon visual inspection appeared to be hedgehogs, were included. (A) Group mean % tail DNA results. Compared to minced tissue, high quality results were obtained from frozen cubed tissue, even after prolonged storage. (B) Group mean % hedgehog results. Poor mincing technique is evident from the extensive non-biological DNA damage observed as hedgehog comets in the minced tissue samples. Error bars reflect standard deviation. An ANOVA with Dunnett’s test was used to evaluate for a positive response to vanadyl sulfate for each sample preparation method. There were no statistically significant (p < 0.05) dose groups detected for either method.
DISCUSSION:
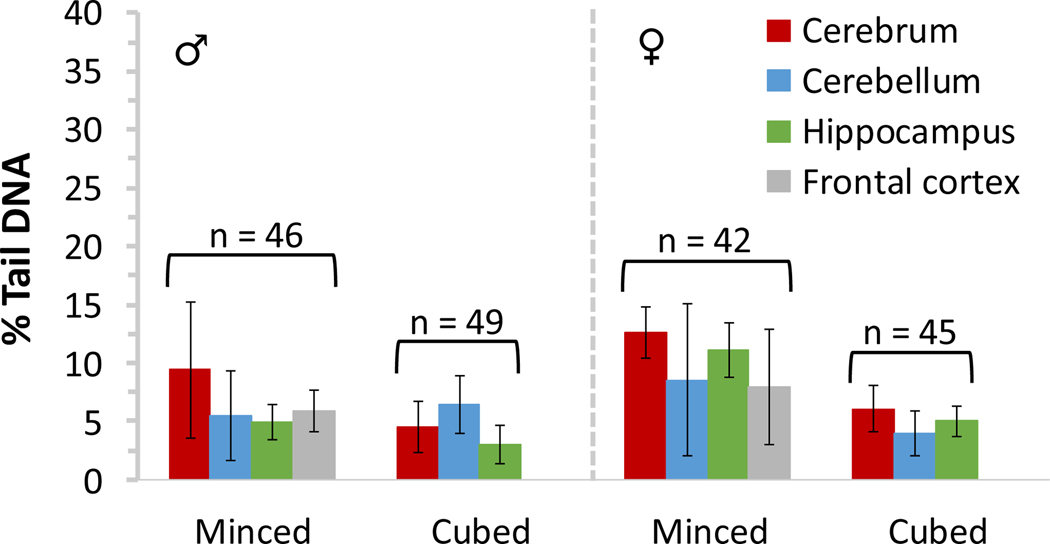
As demonstrated previously7,12,13, properly handled flash frozen minced tissue provides good results in the comet assay. In fact, baseline % tail DNA values for frozen minced rat and mouse liver prepared in our laboratory are typically ≤6%, as recommended by the OECD test guideline9 for freshly minced rat liver samples. Good results have been obtained by multiple laboratories using a variety of tissue types that have been minced and stored frozen; likewise, flash frozen epithelial cells scraped from organs in the gastrointestinal tract have been used successfully in the comet assay7,12–14. Frozen minced/scraped tissue appears to be relatively stable for at least several months. The method employing homogenization of flash frozen cubes of liver using a tissue mincing device originally described by Jackson et al.6, yields results in the comet assay at least comparable and typically better (i.e., lower baseline levels of DNA damage) than those obtained for frozen minced liver. This methodology is also applicable to other tissue types6; in our experience, excellent results have similarly been obtained using frozen cubed duodenum (data not shown) and brain tissue (Figure 5). However, the sieve device may not be amenable to homogenization of all tissue types. In a pilot study, we found that muscular heart tissue could not readily be forced through the sieve device, resulting in poor cell recovery; instead, mincing the tissue immediately upon thawing yielded cell suspensions with baseline % tail DNA values comparable to those minced prior to flash freezing (data not shown). Notably, when handled properly during necropsy (i.e., kept cold and moist; flash frozen as individual pieces and not allowed to partially thaw), flash frozen cubed tissue has yielded very low baseline DNA damage even after approximately two years of freezer storage (Figure 4).
Figure 5: Baseline DNA damage measured in frozen rat brain tissues.
Comet assay results are provided for frozen minced or cubed tissues representing several different regions of the brain of male and female Sprague Dawley rats. Data (generated by including scorable hedgehog comets) were collated over several studies; n = total number of brain tissues evaluated for each sample preparation method. Error bars reflect standard deviation.
Use of either frozen cell suspensions or cubed tissue in the comet assay offers several advantages over fresh tissue including: 1) facilitation of simultaneous collection of multiple tissue types for comet analysis; 2) facilitation of tissue collection at an in-life laboratory and transfer of samples to a different laboratory for analysis; 3) convenience to defer a decision to analyze a given tissue for months. Although the homogenization of frozen tissue at the time of slide preparation is more cumbersome than mincing fresh tissue at the time of harvest, use of frozen cubed tissue offers several additional benefits that include: 1) no requirement for personnel trained in the comet assay (e.g., mobile unit) or specialized supplies/reagents at necropsy; 2) less opportunity for technical errors (e.g., sample warming; insufficient release of cells; sub-optimal tissue sample size) during necropsy; 3) minimal opportunity to introduce mechanical DNA damage during sample processing (e.g., mincing shear); and 4) low baseline DNA damage, even after prolonged storage.
ACKNOWLEDGMENTS:
The authors are indebted to Lincoln Martin and Kelley Owens for expert technical assistance preparing and scoring comet slides and Dr. Carol Swartz for performing statistical analyses. The authors also acknowledge the supportive contributions of members of the Genetic Toxicology, Investigative Toxicology, and Necropsy programs at ILS.
DISCLOSURES:
This work was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (NIEHS) under contract HHSN273201300009C; however, the statements, opinions, or conclusions contained therein do not necessarily represent the statements, opinions, or conclusions of NIEHS, NIH, or the United States government.
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/59955.
REFERENCES:
- 1.van der Leede BJ et al. Performance and data interpretation of the in vivo comet assay in pharmaceutical industry: EFPIA survey results. Mutation Research/Genetic Toxicology and Environmental Mutagenesis. 775–776, 81–88 (2014). [DOI] [PubMed] [Google Scholar]
- 2.ICH. International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH) S2(R1): Guidance on Genotoxicity Testing and Data Interpretation for Pharmaceuticals Intended for Human Use. (2012). [Google Scholar]
- 3.EFSA. Scientific opinion on genotoxicity testing strategies applicable to food and feed safety assessment. EFSA Journal. 9 (9), 2379 (2011). [Google Scholar]
- 4.Lorenzo Y, Costa S, Collins AR, Azqueta A. The comet assay, DNA damage, DNA repair and cytotoxicity: hedgehogs are not always dead. Mutagenesis. 28 (4), 427–432 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Guerard M, Marchand C, Plappert-Helbig U. Influence of experimental conditions on data variability in the liver comet assay. Environmental and Molecular Mutagenesis. 55 (2), 114–121 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Jackson P. et al. Validation of freezing tissues and cells for analysis of DNA strand break levels by comet assay. Mutagenesis. 28 (6), 699–707 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Recio L, Kissling GE, Hobbs CA, Witt KL Comparison of Comet assay dose-response for ethyl methanesulfonate using freshly prepared versus cryopreserved tissues. Environmental and Molecular Mutagenesis. 53 (2), 101–113 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Uno Y. et al. JaCVAM-organized international validation study of the in vivo rodent alkaline comet assay for detection of genotoxic carcinogens: II. Summary of definitive validation study results. Mutation Research/Genetetic Toxicology and Environmental Mutagenesis. 786–788, 45–76 (2015). [DOI] [PubMed] [Google Scholar]
- 9.OECD. OECD Guideline for the Testing of Chemicals: In Vivo Mammalian Alkaline Comet Assay. 489 (2016). [Google Scholar]
- 10.Hobbs CA et al. Comet assay evaluation of six chemicals of known genotoxic potential in rats. Mutation Research/Genetetic Toxicology and Environmental Mutagenesis. 786–788, 172–181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding W, Bishop ME, Lyn-Cook LE, Davis KJ, Manjanatha MG In Vivo Alkaline Comet Assay and Enzyme-modified Alkaline Comet Assay for Measuring DNA Strand Breaks and Oxidative DNA Damage in Rat Liver. Journal of Visualized Experiments. (111), 10.3791/53833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hobbs CA, Chhabra RS, Recio L, Streicker M, Witt KL Genotoxicity of styrene-acrylonitrile trimer in brain, liver, and blood cells of weanling F344 rats. Environmental and Molecular Mutagenesis. 53 (3), 227–238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobbs CA et al. Comprehensive evaluation of the flavonol anti-oxidants, alpha-glycosyl isoquercitrin and isoquercitrin, for genotoxic potential. Food and Chemical Toxicology. 113, 218–227 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Pant K. et al. Vehicle and positive control values from the in vivo rodent comet assay and biomonitoring studies using human lymphocytes: historical database and influence of technical aspects. Environmental and Molecular Mutagenesis. 55 (8), 633–642 (2014). [DOI] [PubMed] [Google Scholar]