Abstract
Polyphosphate (polyP) is a ubiquitous biopolymer whose function and metabolism are incompletely understood. The polyphosphate kinase (PPK) of Acinetobacter sp. strain ADP1, an organism that accumulates large amounts of polyP, was purified to homogeneity and characterized. This enzyme, which adds the terminal phosphate from ATP to a growing chain of polyP, is a 79-kDa monomer. PPK is sensitive to magnesium concentrations, and optimum activity occurs in the presence of 3 mM MgCl2. The optimum pH was between pH 7 and 8, and significant reductions in activity occurred at lower pH values. The greatest activity occurred at 40°C. The half-saturation ATP concentration for PPK was 1 mM, and the maximum PPK activity was 28 nmol of polyP monomers per μg of protein per min. PPK was the primary, although not the sole, enzyme responsible for the production of polyP in Acinetobacter sp. strain ADP1. Under low-phosphate (Pi) conditions, despite strong induction of the ppk gene, there was a decline in net polyP synthesis activity and there were near-zero levels of polyP in Acinetobacter sp. strain ADP1. Once excess phosphate was added to the Pi-starved culture, both the polyP synthesis activity and the levels of polyP rose sharply. Increases in polyP-degrading activity, which appeared to be mainly due to a polyphosphatase and not to PPK working in reverse, were detected in cultures grown under low-Pi conditions. This activity declined when phosphate was added.
Polyphosphates (polyP) are energy- and phosphorus-rich biopolymers that accumulate in a variety of organisms (8). Although their function is not entirely clear, these biopolymers have been implicated as potential energy and phosphorus reservoirs for cells, and they may also contribute to the chelation of ions, the formation of transmembrane channels for DNA or Ca2+ entry, and stress regulation (9). polyP synthesis and degradation occur within the complex framework of phosphorus metabolism, in which complicated regulatory systems that allow organisms to cope with phosphate limitations have evolved (21). How and under what circumstances polyP metabolism is regulated are not fully understood.
The metabolism of polyP plays a key role in the responses of many organisms to phosphate starvation. In Escherichia coli, the phosphate starvation response can be dampened by degradation of intracellular supplies of polyP (17). In a number of bacteria and some eukaryotes, polyP accumulates only after the organism is shifted from conditions in which phosphate starvation occurs to conditions in which there is a phosphate surplus (9). One of the primary enzymes involved in the synthesis of polyP is polyP kinase (PPK), which transfers a phosphate group from ATP to the end of the polyP chain. The PPK reaction is reversible and thus can facilitate degradation of polyP in order to supply the cell with ATP. In several organisms, the ppk gene appears to be regulated by extracellular phosphate levels. The ppk genes of E. coli and Klebsiella aerogenes are preceded by putative pho boxes, which are promoter regions that are regulated as part of the cellular response to phosphate starvation (6, 18). Recently, the ppk gene of Acinetobacter sp. strain ADP1, whose promoter does not contain an E. coli consensus pho box, was shown to be induced by phosphate starvation (4).
In addition to its role in the phosphate starvation response, polyP plays a key role in the biological removal of phosphorus from wastewater, a process known as enhanced biological phosphorus removal (EBPR). In EBPR wastewater treatment systems, bacteria are cycled through two different zones. In the first zone, which is anaerobic and carbon rich, polyP is broken down and phosphate is released. In the second zone, which is aerobic but carbon poor, polyP is accumulated, and the external phosphate concentration is decreased to levels below those set by state and federal standards. While it is well-documented that intracellular polyP levels fall and rise over the course of the EBPR cycle (5, 10, 13), how polyP metabolism is regulated within the cells is not understood. Understanding polyP regulation could lead to identification of critical parameters that could be monitored to predict, and hopefully prevent, process upsets in EBPR treatment systems.
The PPK of several organisms, including E. coli, Propionibacterium shermanii, and Neisseria meningitidis, have been purified and characterized (1, 15, 19). However, none of these organisms is known to reside in wastewater treatment systems. In this paper, we describe purification of the PPK of Acinetobacter sp. strain ADP1. A number of Acinetobacter species have been isolated from different treatment plants; the most notable of these species are Acinetobacter johnsonii, Acinetobacter calcoaceticus, and Acinetobacter lwoffi. Like many Acinetobacter species, Acinetobacter sp. strain ADP1 accumulates large amounts of polyP. Following isolation of the ppk gene, the PPK protein was overexpressed in E. coli and purified to homogeneity. A preliminary characterization of the enzyme was conducted, and the assay conditions were optimized. In addition, we studied the response of Acinetobacter sp. strain ADP1 polyP metabolism to phosphate starvation and surplus conditions.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used were E. coli DH10B [F′-mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara,leu)7697 galU galK l− rspL nupG), which was purchased as electrocompetent cells from Gibco, Acinetobacter sp. strain ADP1 (= BD413), Acinetobacter sp. strain ADP4002 (Strr/Spcr), and Acinetobacter sp. strain WH435 (ppk::lacZ Kmr). Strains ADP4002 and WH435 were both derived from Acinetobacter sp. strain ADP1. WH435 has a lacZ reporter gene and a kanamycin resistance cassette inserted into the chromosomal ppk gene of ADP1, which inactivates that gene (4). The plasmids used were pBluescript SK− (Stratagene), pMMB206 (11), pPLT7b (pBSK− with Acinetobacter sp. strain ADP1 ppk), and pPLT8 (pMMB206 with Acinetobacter sp. strain ADP1 ppk behind the Ptaclac promoter).
Media.
Luria broth (LB) was used for cloning and protein purification. All metabolic studies were performed in MOPS (morpholinepropanesulfonic acid)-buffered minimal medium (12) containing various concentrations of phosphate. The minimal medium was supplemented with 19 amino acids (no cysteine was used) at the concentrations recommended by Neidhardt et al. (12). The following antibiotics were used: streptomycin (10 μg/ml), chloramphenicol [68 μg/ml in LB; 17 μg/ml in MOPS medium for WH435(pPLT8)], kanamycin (10 μg/ml), and ampicillin (100 μg/ml).
Enzyme and polyP assays. (i) Forward PPK activity.
Levels of polyP-producing PPK activity were determined by measuring the amount of polyP formed over a 10-min period by using the method of Tinsley et al. (19). Because of the inhibitory nature of ADP, an ATP regeneration system consisting of 6 mM phosphoenolpyruvate and 20 U of pyruvate kinase per ml was included in the assay mixture. Sodium polyP (chain length, 75; Sigma) was used as a standard. Levels of activity were calculated by determining the amount of polyP formed during the 10-min reaction; 1 U of activity was defined as the activity which produced 1 pmol of polyP (in monomers) per min. Duplicate or triplicate samples were examined. Typical standard deviations were 20% of the values obtained.
(ii) polyP-degrading activity.
The forward PPK assay was modified to measure polyP-degrading activity. The ATP was replaced with ADP, and the phosphoenolpyruvate was replaced with pyruvate to form an ADP regeneration system. In addition, 0.5 mM sodium polyP was added. The reaction volume was 500 μl, and the reaction was started by adding 50 μl of cell lysate (50 to 100 μg of protein). The reaction mixture was mixed for a few seconds before 220 μl of the mixture was transferred into 60 μl of cetyltrimethylammonium bromide reagent. After 10 min of incubation, another 220 μl of the mixture was removed. Levels of polyP were measured as they were in the forward PPK activity assay. polyP-degrading activity was calculated by determining the amount of polyP consumed; 1 U of activity was defined as the activity which consumed 1 pmol of polyP (in monomers) per min.
(iii) Processivity assays.
The processivity of PPK was determined by using the methods of Keasling et al. (7). Purified E. coli PPK was used as a positive control.
(iv) β-gal activity.
β-Galactosidase (β-gal) activity was measured by using standard methods (16).
(v) polyP.
Intracellular polyP levels were measured by the method described by Ault-Riche et al. (2). polyP was quantified by digesting it with purified E. coli PPK (purified by S. Van Dien [Department of Chemical Engineering, University of California, Berkeley] essentially as described by Ahn and Kornberg [1]) in a reaction mixture (50 mM Tris-HCl [pH 7.5], 40 mM ammonium sulfate, 4 mM MgCl2, 5 μM ADP) for 45 min at 37°C. The amount of ATP produced by degradation of polyP was measured by using a luciferin-luciferase assay system (Sigma) and a luminometer (model 2020-000; Turner Designs, Sunnyvale, Calif.).
Cloning of ppk from Acinetobacter sp. strain ADP1.
Acinetobacter sp. strain ADP1 ppk was cloned by using a partial ppk sequence that has been submitted to GenBank by Geißdörfer et al. (GenBank accession no. Z46863) (4). The partial sequence was used to design an oligonucleotide primer, primer PPK3 (5′-AAC AGA ATT CTA AGC GAG GGA ACG GAT G-3′), which was labeled at the 3′ end with digoxigenin (DIG) by using a DIG oligonucleotide 3′ end labeling kit (Boehringer Mannheim, Indianapolis, Ind.); the probe was used in a Southern hybridization analysis to show that the ppk gene was located on a 10-kb PstI fragment of ADP4002 genomic DNA (which was prepared by the method of Pospiech and Neumann [14] and was purified by adsorption to silica resin minicolumns [Blood and Cell Culture DNA Mini-kit; QIAGEN, Chatsworth, Calif.]). Plasmid pPLT5, which harbored this 10-kb PstI fragment cloned into the PstI site of pBSK−, was isolated by colony hybridization of clones that were transformed with a partial gene library by using DIG-labeled PPK3 and standard procedures (16).
Construction of inducible PPK system.
Using the complete ppk coding sequence reported by Geißdörfer et al. (4), we constructed two primers, primer PPK6 (5′-GGC TGC AGG ATA GGA TTA GCG CAT GAA-3′) and primer PPK7 (5′-CTC CTG CAG AAA GAG TAC CCC GCT-3′). PPK6 contained the 5′ end of the ppk gene, while PPK7 was located after the end of the putative termination sequence. PCR DNA amplification was performed with the High Fidelity PCR System (Boehringer Mannheim) and was used to amplify a ppk gene that contained an optimized Shine-Dalgarno sequence but lacked its own promoter. The gene was initially cloned into pBSK− to produce plasmid pPLT7. All but 200 bp of the ppk gene in pPLT7 was replaced with the same fragment excised from chromosome-derived plasmid pPLT5 in order to remove any errors that the PCR may have introduced. Sequencing indicated that the remaining 200 bp was also error free. The resulting plasmid was designated pPLT7b. The gene was then inserted into the broad-host-range vector pMMB206 under control of the Ptaclac promoter, and the resulting plasmid was designated pPLT8.
Purification of PPK.
E. coli DH10B containing pPLT7b was used to overproduce PPK for purification; the methods of Ahn and Kornberg (1) were used to do this. E. coli DH10B contains the E. coli ppk gene; however, the gene is natively expressed at very low levels. E. coli DH10B(pPLT7b) growing exponentially was used to inoculate 8 liters of LB containing 100 μg of ampicillin per ml to an optical density at 600 nm (OD600) of 0.01. The culture was harvested at an OD600 of 0.8, and the resulting pellet was resuspended in 20 ml of Tris-sucrose solution (50 mM Tris-HCl [pH 7.5], 10% sucrose), frozen in liquid nitrogen, and stored at −87°C overnight. The cells were thawed, and an equal volume (40 ml) of lysis buffer (50 mM Tris-HCl, 10% sucrose, 300 mM NaCl, 90 mM EDTA, 3 mg of lysozyme per ml) was added. The mixture was incubated on ice for 1 h before it was frozen in liquid N2 and thawed at 30°C three times. DNase I was added to a concentration of 20 μg/ml, and MgCl2 was added to a concentration of 5 mM. The mixture was sonicated by using five 10-s pulses while the temperature was maintained at or below 4°C. KCl was added to a concentration of 1 M, and Na2CO3 was added to a concentration of 0.1 M. After the protein mixture was gently mixed for 80 min, it was sonicated again and then centrifuged (31,400 × g, 4°C, 1 h). Ammonium sulfate (15 g) was added to the supernatant (75 ml) over a 1-h period at 4°C. Following centrifugation (31,400 × g, 4°C, 1 h), the resulting pellet was resuspended in Tris-sucrose solution, and this preparation was designated fraction I (25 ml). The supernatant (70 ml) was precipitated again with ammonium sulfate (7.5 g) over a 30-min period at 4°C and then centrifuged (31,400 × g, 4°C, 1 h).
Fraction I (25 ml) was dialyzed overnight against 4 liters of buffer B (1 mM EDTA, 1 mM dithiothreitol, 10% glycerol) containing 10 mM potassium phosphate (pH 7.0) by using 10,000-molecular-weight cutoff Slide-A-Lyzers (Pierce, Rockford, Ill.). Dialyzed fraction I (postdialysis volume, 35 ml) was applied to a 90-ml type P11 phosphocellulose (Whatman, Maidstone, England) column that had been equilibrated with buffer B containing 10 mM Pi. The column was washed with 180 ml of buffer B containing 10 mM Pi. The enzyme was eluted with a 450-ml gradient (buffer B containing 10 to 500 mM Pi) at a flow rate of 1.4 ml/min. Fractions (4.5 ml) were collected, and the activities of the fractions were tested. The most active fractions were pooled, and the resulting preparation was designated fraction II (volume, 33 ml). Fraction II was dialyzed against 3.6 liters of buffer C (25 mM Tris-HCl [pH 7.5], 15% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol) containing 10 mM KCl. The dialyzed PPK was applied to a 6.5-ml DEAE-Sepharose (Pharmacia) column that had been equilibrated with buffer C containing 10 mM KCl. The column was washed with 45 ml of buffer C containing 10 mM KCl. The column was then eluted with 32 ml of buffer C containing 120 mM KCl to remove contaminating proteins. PPK was eluted with 32 ml of buffer C containing 180 mM KCl. Finally, the remaining protein was eluted with 20 ml of buffer C containing 2 M KCl. All washes and elutions were done at a flow rate of 1 ml/min. The 9.5-ml sample containing the most PPK activity was designated fraction III.
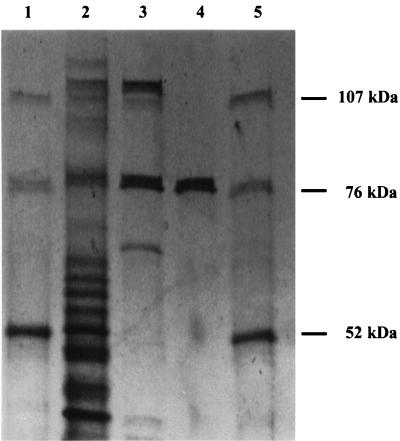
The purity of PPK in each of the fractions was determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Samples (0.7 μg of protein for fraction I, 0.6 μg of protein for fraction II, and 0.2 μg of protein for fraction III) were loaded onto a 7.5% polyacrylamide gel (Bio-Rad). Bio-Rad low-range silver-stained SDS-PAGE molecular weight standards were used. The purity of PPK was confirmed by native PAGE. Samples (0.8 μg of protein for fraction III) were loaded onto a 7.5% polyacrylamide gel (Bio-Rad). The proteins in a Nondenatured Protein Molecular Weight Marker kit (Sigma) were used as standards. The gels were stained with a silver stain (Bio-Rad) as recommended by the manufacturer.
The size of the native enzyme was determined by using gel filtration chromatography performed with 5 μ silica powder (TOSOHAAS A3435) and phosphate buffer (0.3 M NaCl, 0.05 M Na3PO4; pH 6.9). The size was confirmed by native PAGE. Bio-Rad gel filtration standards were used to determine the size of the enzyme.
Experiments in which cultures were shifted from phosphate starvation conditions to phosphate surplus conditions.
Shifts from phosphate starvation conditions to phosphate surplus conditions were performed essentially as described by Van Bogelen and coworkers (20); the methods used were similar to those used by Geißdörfer et al. (4). Cultures of ADP1 and WH435 were grown in high-phosphate minimal medium to the mid-exponential phase before they were centrifuged and washed twice in medium containing no phosphate. Each culture was then resuspended in medium containing 100 μM Pi to an OD600 of 0.04. (This Pi concentration was chosen so that the cells would become limited for Pi at an OD600 significantly below the maximum OD600 supported by this medium containing surplus Pi.) The samples used for the PPK activity and polyP analyses were taken from the ADP1 culture, while the samples used for the β-gal activity analysis were taken from the WH435 culture over the course of the experiment. Monitoring for PPK activity, polyP, and β-gal activity began at an OD600 of approximately 0.2. As the cultures became starved for phosphate (at an OD600 of approximately 1.0), excess phosphate was added (to a Pi concentration of 13.2 mM). (Care was taken to ensure that cells were not starved for Pi any longer that was necessary in order to observe changes in polyP and PPK.) We continued to take samples as the cultures reentered the exponential growth phase.
RESULTS
Analysis of PPK.
The PPK of Acinetobacter sp. strain ADP1 was purified from a culture in which it was overexpressed via plasmid pPLT7b in E. coli. The specific activity in the crude lysate was 334 U/μg of protein, and the enzyme was purified 36-fold to a final specific activity of 11,900 U/μg of protein (Table 1). An SDS-PAGE gel revealed the increase in purity from the ammonium sulfate precipitation step to the pure fractions obtained from the final DEAE column (Fig. 1); 4% of the total activity was recovered as pure PPK. The results of both gel filtration chromatography and native PAGE suggested that the native PPK is the 79-kDa monomer (data not shown).
TABLE 1.
PPK purification
| Fraction | Description | Total activity | Total protein (mg) | Sp act (U/μg of protein) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Lysate | 83 | 249 | 334 | 100 | ||
| FI | Ammonium sulfate | 44 | 76.7 | 573 | 1.72 | 53 |
| FII | Phosphocellulose | 19 | 4.15 | 4,580 | 13.7 | 23 |
| FIII | DEAE-Sepharose | 3.4 | 0.285 | 11,900 | 36.6 | 4 |
One unit of PPK activity was defined as the amount of PPK that polymerized 1 pmol of polyP (in monomers) per min.
FIG. 1.
SDS-PAGE analysis of PPK purification results. Lanes 1 and 5 contained molecular weight standards. Lane 2, fraction I (ammonium sulfate); lane 3, fraction II (phosphocellulose); lane 4, fraction III (DEAE-Sepharose).
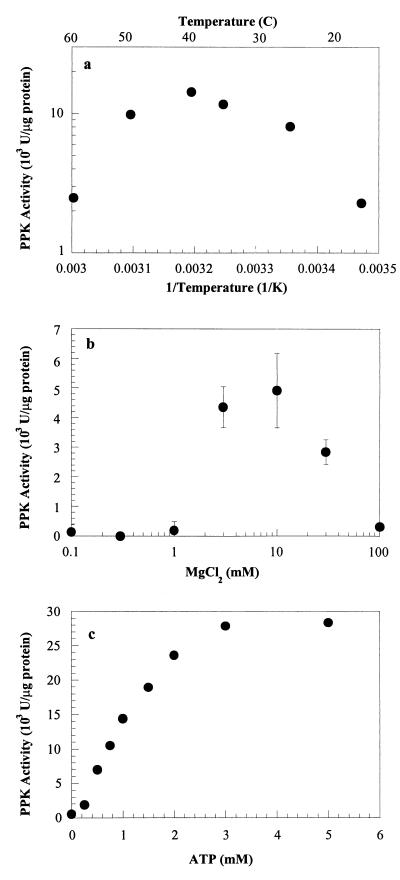
The PPK activity of Acinetobacter sp. strain ADP1 depended on temperature. The enzyme was most active at 40°C (Fig. 2a). Using an Arrhenius plot, we estimated that the activation energy was 54 kJ/mol. The enzyme was also strongly dependent on the MgCl2 concentration (Fig. 2b) and the pH (data not shown). The enzyme was most active at pH 7 to 8 (the lowest pH examined was pH 5); at higher pHs, the PPK assay mixture precipitated, and so higher pHs could not be investigated. Dependence on ATP was also studied. While the maximum PPK activity was 28,000 U/μg of protein at an ATP concentration greater than or equal to 3 mM and the half-saturation ATP concentration was 1 mM, a Michaelis-Menten model did not fit the data (Fig. 2c).
FIG. 2.
PPK characteristics. (a) Arrhenius plot of PPK activity dependence on temperature. (b) PPK activity dependence on magnesium concentration. (c) PPK activity dependence on ATP concentration. Dashed line, best fit to Michaelis-Menten equation (Vmax = 43.4 × 103 U per μg of protein; Km = 2.05 mM); solid line, Michaelis-Menten equation with a Vmax of 28 × 103 U per μg of protein and a Km of 1 mM.
Purified PPK appeared to work only in the forward direction (i.e., it produced polyP but did not degrade it) under the conditions used in the PPK activity and polyP-degrading activity assays. In the polyP-degrading activity assays, E. coli PPK was used as a control, and this enzyme readily degraded sodium polyP. Little or no degradation was observed when the Acinetobacter sp. strain ADP1 PPK was used.
PPK is a processive enzyme. Samples taken during synthesis of polyP and separated by size on polyacrylamide gels produced two major bands corresponding to ATP and long-chain polyP at all times during synthesis (data not shown). Similar bands were observed when E. coli PPK was used. On the basis of these results it appears that the polyP produced by this enzyme activity is approximately the same length as the polyP produced by the E. coli PPK.
Comparison of a ppk mutant, WH435, and the wild type, ADP1.
Because of the presence of a strong polyP-degrading activity in cell lysates, measurements of PPK activity actually yielded a net positive value (PPK activity minus polyP-degrading activity). We distinguished this combined activity from pure PPK activity by referring to it as “net PPK activity.”
When starved for phosphate, Acinetobacter sp. strain ADP1 produced eightfold more polyP and net PPK activity than the ppk mutant WH435 (Table 2). However, the levels of both polyP and net PPK activity were extremely low. Once the cultures were shifted to high-Pi-concentration conditions, the net PPK activities and polyP levels of both cultures rose. Eight hours after the shift, the net PPK activity of ADP1 was 10-fold higher than the activity before the shift, and the polyP level had increased 150-fold. The net PPK activity of WH435 also increased. However, ADP1 had 14-fold more polyP and 3.5-fold more net PPK activity than WH435. Under excess-phosphate conditions with no Pi shift, the polyP levels of both ADP1 and WH435 were very low (data not shown). The net PPK activities were similarly low (data not shown).
TABLE 2.
Comparison of wild-type strain ADP1 and ppk mutant WH435
| Culture
|
polyP concn (μmol of P/g of dry cell weight) | Net PPK activity (U/μg of protein) | |
|---|---|---|---|
| Strain | Conditions | ||
| ADP1 | Low Pi | 0.04 | 6.0 |
| WH435 | Low Pi | 0.005 | 0.7 |
| ADP1 | Low Pi → high Pi | 60 | 60 |
| WH435 | Low Pi → high Pi | 4.4 | 17 |
Induction of ppk transcription and PPK activity during shifts from low to high Pi levels.
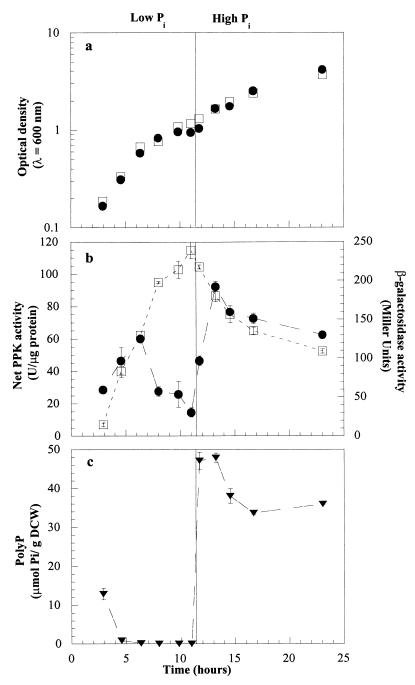
Cell growth, ppk gene expression, and net PPK activity were monitored in WH435 and ADP1 during shifts in the extracellular phosphate concentrations. The shift conditions were essentially the same as those described by Van Bogelen et al. (20) and Geißdörfer et al. (4) and were chosen so that we could monitor changes in polyP levels, PPK activity, gene expression, and cell growth as the extracellular phosphate level changed. The two strains grew at similar rates in the presence of both high and low phosphate concentrations (Fig. 3a). Following a shift from a high Pi level to a low Pi level, the β-gal activity increased steadily from 15 to 240 Miller units, indicating that there was a steady increase in induction of the ppk gene (Fig. 3b). After phosphate was added, the β-gal activity slowly declined over the remainder of the experiment. Prior to a decrease in the growth rate due to exhaustion of the Pi in the medium (at an OD600 of ∼0.6), the net PPK activity of ADP1 rose slightly; it increased from an initial level of 28 U/μg of protein to 60 U/μg of protein (Fig. 3b). However, as the cells became increasingly Pi starved, the net activity began to drop. By the time that growth of the culture had nearly ceased and the culture was shifted to high-phosphate conditions after another 4.5 h, the net activity had fallen to 15 U/μg of protein. When excess phosphate was added, the level of activity immediately rose, increasing to 92 U/μg of protein in 2.25 h, before it slowly declined over the remainder of the experiment. The polyP levels began to drop during the early stages of phosphate starvation (Fig. 3c). Five hours after the initial shift to low-Pi conditions, the level of polyP was less than 1 μmol of P/g of dry cell weight. Within 45 min after phosphate was added, the polyP level had risen to 47 μmol of P/g of dry cell weight. After that it dropped slowly to about 35 μmol of P/g of dry cell weight, where it remained.
FIG. 3.
Shift from phosphate starvation conditions to surplus conditions. The shift from low-Pi to high-Pi conditions occurred at 11.5 h. (a) Growth. Symbols: ●, wild-type strain ADP1; □, ppk mutant WH435. (b) Induction of ppk and net PPK activity. Symbols: ●, net PPK activity in ADP1; □, β-gal activity in WH435. (c) polyP levels in ADP1. DCW, dry cell weight.
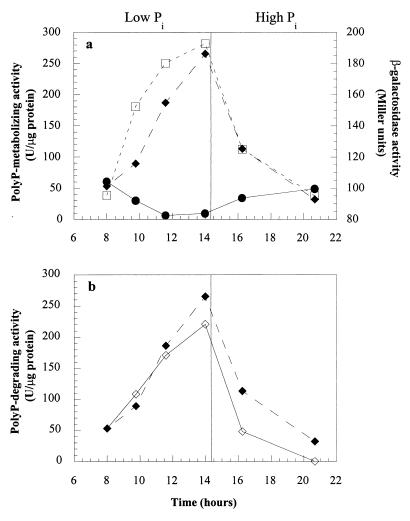
In a similar experiment, samples were analyzed to determine their polyP-degrading activities, as well as their net PPK and β-gal activities. The polyP-degrading activity in ADP1 increased during the phosphate starvation period and decreased following the shift to Pi surplus conditions, while the net PPK activity in ADP1 declined and β-gal activity in WH435 rose, as described above (Fig. 4a). When WH435 samples were analyzed, a comparable amount of polyP-degrading activity was observed (Fig. 4b).
FIG. 4.
polyP-degrading and net PPK activities during the shift from phosphate starvation conditions to surplus conditions. (a) Symbols: □, β-gal activity in WH435; ●, net PPK activity in ADP1; ⧫, polyP-degrading activity in ADP1. (b) Symbols: ⧫, polyP-degrading activity in ADP1; ◊, polyP-degrading activity in WH435.
The polyP-degrading activities of Pi-starved WH435 and ADP1 cultures were assayed without Pi in the assay mixtures, which allowed Pi production to be monitored over time. Over a 20-min period, the Pi levels in the reaction mixtures rose as the polyP levels declined, indicating that the polyP-degrading activity was probably due to a polyphosphatase (PPX).
Induction studies.
The plasmid containing the inducible ppk system, pPLT8, was transformed into WH435. During growth in MOPS minimal medium containing excess Pi, WH435 without pPLT8 accumulated no polyP, and the net PPK activity was very low (data not shown). The net PPK activity and polyP level in the inducible organism WH435(pPLT8) were directly correlated with the amount of inducer used. The net PPK activity rose from 50 to 900 U/μg of protein and the polyP level rose from 0 to 17 μmol/g of dry cell weight with increasing levels of induction. WH435 containing parental plasmid pMMB206 or no plasmid was induced with 264 μg of isopropyl-β-d-thiogalactopyranoside (IPTG) per ml, and very low levels of PPK activity and polyP were observed.
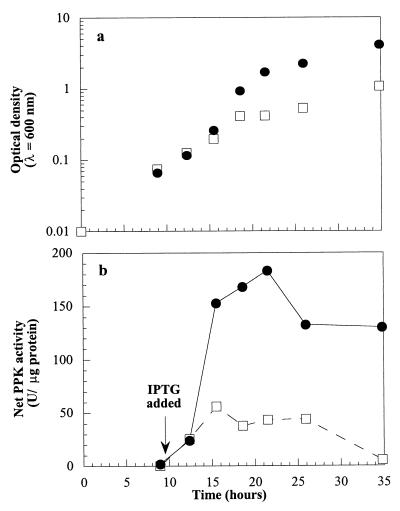
The effects of the extracellular Pi concentration on net PPK activity in cells containing pPLT8 were examined by inducing a culture of WH435(pPLT8) with 8 μg of IPTG per ml while it was growing in medium containing 100 μM Pi. This culture was compared to a culture growing in medium containing 1.32 mM Pi. The two cultures grew similarly until the low-Pi culture reached an OD600 of 0.2, when its growth slowed compared to the growth of the high-Pi culture. At that point, the net PPK activity of the low-Pi culture began to level off at approximately 40 U/μg of protein, while the net PPK activity of the high-Pi culture continued to rise to 180 U/μg of protein (Fig. 5). When both cultures reached the stationary phase, the activity of the low-Pi culture was 5 U/μg of protein, while the activity of the high-Pi culture was 130 U/μg of protein.
FIG. 5.
Effect of phosphate starvation on induced net PPK activity in WH435(pPLT8). Cultures were induced with 8 μg of IPTG per ml. Symbols: ●, high-Pi (1.32 mM Pi) culture; □, low-Pi (100 μM Pi) culture. (a) Growth. (b) Net PPK activity.
DISCUSSION
Acinetobacter sp. strain ADP1 PPK was purified to apparent homogeneity and characterized. ADP1 PPK appeared to be active in its monomer form, as shown by gel filtration chromatography and native PAGE. The polyP-AMP phosphotransferase of A. johnsonii 210a and the PPK of P. shermanii also appear to be active as monomers (3, 15); however, the PPK of E. coli functions as a tetramer (1). Like the A. johnsonii 210a polyP-AMP phosphotransferase and the N. meningitidis PPK, ADP1 PPK had a broad pH optimum. However, it was active at somewhat higher pHs than the pHs at which the N. meningitidis PPK was active. Like E. coli PPK and N. meningitidis PPK, ADP1 PPK was most active at Mg concentrations ranging from 5 to 10 mM. At Mg concentrations below 1 mM, there was little or no activity. Unlike the PPK of E. coli and the PPK of P. shermanii, ADP1 PPK did not follow Michaelis-Menten kinetics, although saturation kinetics was observed. Instead, the enzyme appeared to become saturated earlier than the Michaelis-Menten kinetics would predict. The ATP half-saturation concentration, 1 mM, is somewhat lower than the Km values observed for E. coli PPK (3 mM ATP) and N. meningitidis PPK (1.5 mM ATP). In addition, the purified enzyme did not work in reverse to degrade polyP under several different assay conditions. E. coli PPK, on the other hand, readily functions in reverse in vitro.
It has been observed that many organisms accumulate large amounts of polyP when they are shifted to a phosphate-rich medium following phosphate starvation (9). However, in a number of organisms, it is phosphate starvation and not phosphate surplus that appears to induce the ppk gene. The ppk genes of both E. coli and K. aerogenes have two promoter regions, one of which contains a putative pho box which is associated with the cellular response to phosphate starvation (6, 18). It has been shown that in both of these organisms transcription of ppk increases when the organisms are starved for phosphate (6). Geißdörfer et al. (4) have shown that even though its promoter does not contain the E. coli consensus pho box, the Acinetobacter sp. strain ADP1 ppk gene is induced under phosphate starvation conditions. Our results suggest that the polyP metabolism of ADP1 is even more complicated. While phosphate starvation induces ppk expression and addition of phosphate represses gene expression, the net PPK activity that produces polyP from ATP appears to follow an opposite trend. Phosphate starvation causes this activity to decrease. It is not until excess phosphate is added that the net PPK activity rises sharply. In addition, the ppk mutant WH435 had a minor secondary PPK activity that was also expressed following a shift from phosphate starvation conditions to surplus conditions. It has recently been reported that Pseudomonas aeruginosa also contains an alternate pathway for polyP synthesis (22).
These results were confirmed by the results of studies performed with the inducible ppk system (pPLT8) in the ppk mutant WH435. When 8 μg of IPTG per ml was added to a culture growing in phosphate-rich medium, the net PPK activity rose over time. However, when the same amount of IPTG was added to a culture growing in low-Pi medium, the activity began to level off and then declined as the culture became starved for phosphate (as indicated by a slower rate of growth).
There are several possible explanations for the difference between the patterns of ppk gene induction and net PPK activity. Geißdörfer et al. showed that ppk was induced by measuring both β-gal activity in WH435 and ppk mRNA levels in ADP1. It is probably safe to assume that PPK is produced at the time of ppk transcription and that the PPK protein levels in ADP1 mirror the β-gal activity in WH435. Because we determined net PPK activity by measuring the production of polyP, we would have missed reverse PPK activity that resulted in the consumption of polyP. To investigate if reverse PPK activity was present, we measured polyP-degrading activity during the shift from phosphate starvation conditions to surplus conditions. A pattern that was very similar to the pattern of induction of the ppk gene was observed. However, the polyP-degrading activity was severalfold greater than the polyP-producing net PPK activity. Also, when the polyP-degrading activities of the ppk mutant WH435 were measured, similar levels of activity were observed. Since the major PPK activity had been eliminated in WH435, the polyP-degrading activity appeared to be due to an enzyme other than PPK. polyP-degrading activity assays performed without Pi in the assay mixture yielded levels of Pi that increased as the amount of polyP decreased, suggesting that a PPX was responsible for much of the polyP-degrading activity. This PPX appeared to be induced by phosphate starvation conditions, since its activity increased during phosphate starvation conditions and declined very quickly once phosphate was added to the medium. Because of the high activity of PPX under phosphate starvation conditions, it was impossible to determine if there was also weak polyP-degrading PPK activity. The lack of any reverse PPK activity in vitro under conditions in which the E. coli PPK was reversible indicated that the enzyme was not reversible under physiological conditions or that there was some regulation of its activity that could not be determined under the assay conditions used.
The decline in net PPK activity under low-Pi conditions may have been an artifact of a cell lysate that contained both PPX and PPK. Indeed, the PPX activity was significantly higher than the PPK activity during phosphate starvation conditions. Any polyP produced by PPK could be immediately broken down by PPX.
Despite the confusion introduced by the unexpected presence of a strong polyP-degrading activity, our results have some implications for research on the role of polyP in phosphate starvation and for EBPR research. Our findings suggest that formation of polyP-producing enzymes is linked to formation of polyP-degrading enzymes. Thus, the same conditions (Pi starvation in this study) that trigger induction of PPX lead to induction of PPK. When the conditions change (e.g., when Pi is added), PPK is ready and available to form polyP. Obviously, this system should produce polyP only if the change in conditions results in differential expression of the two enzymes. Examples of such differential expression could include activation of PPK, deactivation of PPX, and production of a PPX that is less stable than PPK and thus is removed from the cell at a faster rate than PPK is removed. Such a system could provide an explanation for a puzzling aspect of EBPR metabolism. The success of the EBPR process depends on organisms that store polyP at one time for use at some future time. While it is readily apparent why an organism would produce an enzyme to degrade an existing pool of polyP in order to ameliorate the effects of immediate phosphate- or energy-depleting conditions, it is not as apparent why the organism would produce an enzyme to make polyP when the polyP would not be used until some future time when the extracellular phosphate supply might be low. The linkage of formation of polyP-synthesizing enzymes to formation of polyP-degrading enzymes provides an explanation for the presence of these enzymes in the EBPR system.
In addition, our findings suggest that EBPR researchers may want to consider the role of Pi levels in EBPR metabolism, as well as carbon availability and anaerobic-aerobic shifts, in order to understand the metabolism involved. Typical standards that regulate phosphorus levels in wastewater treatment effluent are 1 to 2 mg of P per liter (32 to 64 μM Pi). At these concentrations, many organisms, including Acinetobacter sp. strain ADP1, experience phosphate starvation effects, which could lead to expression of key genes in EBPR metabolism.
ACKNOWLEDGMENTS
This work was supported by grants from the National Science Foundation (grant BES-9612840) and the National Institutes of Health (grant 2T32GM08352) to J.D.K. and by a National Science Foundation graduate fellowship to P.L.T.
We thank Rob Frankenberg for much helpful advice and Deborah Hong for her work on the processivity of PPK.
REFERENCES
- 1.Ahn K, Kornberg A. Polyphosphate kinase from Escherichia coli. J Biol Chem. 1990;265:11734–11739. [PubMed] [Google Scholar]
- 2.Ault-Riche D, Fraley C D, Tzeng C-M, Kornberg A. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J Bacteriol. 1998;180:1841–1847. doi: 10.1128/jb.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonting C F C, Kortstee G J J, Zehnder A J B. Properties of polyphosphate:AMP phosphotransferase of Acinetobacter strain 210a. J Bacteriol. 1991;173:6484–6488. doi: 10.1128/jb.173.20.6484-6488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geißdörfer W, Ratajczak A, Hillen W. Transcription of ppk from Acinetobacter sp. strain ADP1, encoding a putative polyphosphate kinase, is induced by phosphate starvation. Appl Environ Microbiol. 1998;64:896–901. doi: 10.1128/aem.64.3.896-901.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenkins D, Tandoi V. The applied microbiology of enhanced biological phosphorus removal—accomplishments and needs. Water Res. 1991;25:1471–1478. [Google Scholar]
- 6.Kato J, Yamamoto T, Yamada K, Ohtake H. Cloning, sequence and characterization of the polyphosphate kinase-encoding gene (ppk) of Klebsiella aerogenes. Gene. 1993;137:237–242. doi: 10.1016/0378-1119(93)90013-s. [DOI] [PubMed] [Google Scholar]
- 7.Keasling J D, Bertsch L, Kornberg A. Guanosine pentaphosphate phosphohydrolase from Escherichia coli is a long-chain polyphosphatase. Proc Natl Acad Sci USA. 1993;90:7029–7033. doi: 10.1073/pnas.90.15.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kornberg A. Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J Bacteriol. 1995;177:491–496. doi: 10.1128/jb.177.3.491-496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulaev I S, Vagabov V M. Polyphosphate metabolism in microorganisms. Adv Microb Physiol. 1983;24:83–171. doi: 10.1016/s0065-2911(08)60385-9. [DOI] [PubMed] [Google Scholar]
- 10.Mino T, Kawakami T, Matsuo T. Behavior of intracellular polyphosphate in the biological phosphate removal process. Water Sci Technol. 1985;17:11–21. [Google Scholar]
- 11.Morales V M, Backman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- 12.Neidhardt F C, Bloch P L, Smit D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira H, Lemos P C, Reis M A M, Crespo J P S G, Carrondo M J T, Santos H. Model for carbon metabolism in biological phosphorus removal processes based on in vivo13C-NMR labelling experiments. Water Res. 1996;30:2128–2138. [Google Scholar]
- 14.Pospiech A, Neumann B. A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 15.Robinson N A, Clark J E, Wood H G. Polyphosphate kinase from Propionibacterium shermanii. J Biol Chem. 1987;262:5216–5222. [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Sharfstein S T, Dien S J V, Keasling J D. Modulation of the phosphate-starvation response in Escherichia coli by genetic manipulation of the polyphosphate pathways. Biotechnol Bioeng. 1996;51:434–438. doi: 10.1002/(SICI)1097-0290(19960820)51:4<434::AID-BIT6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 18.Sharfstein S T, Keasling J D. Polyphosphate metabolism in Escherichia coli. Ann N Y Acad Sci. 1994;745:77–91. doi: 10.1111/j.1749-6632.1994.tb44365.x. [DOI] [PubMed] [Google Scholar]
- 19.Tinsley C R, Manjula B N, Gotschlich E C. Purification and characterization of polyphosphate kinase from Neisseria meningitidis. Infect Immun. 1993;61:3703–3710. doi: 10.1128/iai.61.9.3703-3710.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Bogelen R A, Olson E R, Wanner B L, Neidhardt F C. Global analysis of proteins synthesized during phosphate restriction in Escherichia coli. J Bacteriol. 1996;178:4344–4366. doi: 10.1128/jb.178.15.4344-4366.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wanner B L. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1357–1381. [Google Scholar]
- 22.Zago A, Chugani S, Chakrabarty A M. Cloning and characterization of polyphosphate kinase and exopolyphosphatase genes from Pseudomonas aeruginosa 8830. Appl Environ Microbiol. 1999;65:2065–2071. doi: 10.1128/aem.65.5.2065-2071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]