Abstract
The 24th North American International Society for the Study of Xenobiotics (ISSX) meeting, held virtually from September 13 to 17, 2021, embraced the theme of “Broadening Our Horizons.” This reinforces a key mission of ISSX: striving to share innovative science related to drug discovery and development. Session speakers and the ISSX New Investigator Group, which supports scientific and professional development of student and early career ISSX members, elected to highlight the scientific content presented during the captivating session titled, “Epigenetics in Drug Disposition & Drug Therapy”. The impact genetic variation has on drug response is well established; however, this session underscored the importance of investigating the role of epigenetics in drug disposition and drug discovery. Session speakers, Drs. Ning, McClay, and Lazarus, detailed mechanisms by which epigenetic players including long non-coding RNA (lncRNAs), microRNA (miRNAs), DNA methylation, and histone acetylation can alter the expression of genes involved in pharmacokinetics, pharmacodynamics, and toxicity. Dr. Ning detailed current knowledge about miRNAs and lncRNAs and the mechanisms by which they can affect expression of drug metabolizing enzymes (DMEs) and nuclear receptors. Dr. Lazarus discussed the potential role of miRNAs on UDP-glucuronosyltransferase (UGT) expression and activity. Dr. McClay provided evidence that aging alters methylation and acetylation of DMEs in the liver, affecting gene expression and activity. These topics, compiled by the symposium organizers, presenters, and the ISSX New Investigators Group, are herein discussed, along with exciting future perspectives for epigenetics in drug disposition and drug discovery research.
Introduction
The ISSX New Investigator Group provides opportunities for student and early career members of ISSX to be actively engaged in the Society by organizing webinars, fireside chats, networking and mentorship, and career development events. The ISSX New Investigator Group selected a symposium from the 24th North American ISSX Meeting held virtually from September 13–17, 2021, to share the most up-to-date knowledge on a cutting-edge topic with unique insights from the speakers. Our objective is to provide a high-level summary from the different presentations, allowing for attendees to recall the session highlights while also promoting dissemination of learnings to those who were unable to attend. The ISSX New Investigator Group selected the “Epigenetics in Drug Disposition and Drug Therapy” symposium since it is a rapidly evolving expanding field that can be used as a tool in precision medicine (Rasool et al. 2015).
In studying genomics and pharmacogenomics, we look at the effects of specific mutations or polymorphisms in a gene; i.e., changes in the DNA sequence, which alter the expression level, localization, and/or activity of proteins such as a drug metabolizing enzymes (DMEs), and nuclear receptors (NR) (Sim et al. 2013; Ahmed et al. 2016). These genetic variations are broadly consistent across the lifespan and are generally considered unresponsive to environmental factors. On the other hand, the study of epigenetics does not refer to alterations to the DNA sequence itself; epigenetics refers to certain signature marks placed upon areas of the genome in response to a variety of environmental factors such as stress, diet, drug exposure, and changes across the lifespan (Heerboth et al. 2014). The epigenetic marks include DNA methylation of the carbon 5 position of cytosine residues (5-methylcytosine, 5mC) and several different posttranslational modifications to histone proteins, which make up the nucleosomes around which DNA is coiled. Histone modifications include methylation and acetylation of histone lysine residues. Typically, closed or “silent” chromatin results from the addition of 5mC and removal of histone acetylation marks. Conversely, open, active chromatin results from the addition of histone acetylation marks and the removal of 5mC (Handel et al. 2010; Stricker et al. 2017). MicroRNA (miRNA) and long noncoding RNA (lncRNA) regulation also fall under the umbrella of epigenetics, and there has been increasing interest in using circulating miRNAs and lncRNAs as disease biomarkers (Bolha et al., 2017; Condrat et al., 2020). Epigenetic modifications including DNA methylation, chromatin remodeling, histone modification, miRNAs, and lncRNAs contribute to global gene expression including genes relevant to xenobiotic metabolism and transport, as well as genes involved in disease processes such as cancer.
The session chair, Ann Daly (Newcastle University, Newcastle upon Tyne, United Kingdom), recognized that although considerable progress has been made in the field, this is still a relatively poorly understood area, and that studying changes in epigenetic regulation due to disease and environmental factors are of particular importance for maximizing effective drug therapy. Drugs that epigenetically modulate gene expression, especially those relevant to oncology, are increasingly being investigated and approved, and the scientific community is just beginning to scratch the surface in terms of understanding implications on biological processes. Pharmaceutical companies have invested heavily into epigenetics research with a particular interest in oncology. However, epigenetics is also of increasing interest in disease areas outside of oncology including metabolic diseases such as metabolic associated fatty liver disease (Bayoumi et al. 2020), central nervous system pathologies including Alzheimer’s Disease, and inflammatory diseases such as asthma (Prachayasittikul et al. 2017). The global epigenetics market was valued at $1.0 billion USD in 2020 and is projected to reach $4.1 billion USD by 2030, growing at a compound annual growth rate of 14.1% from 2021 to 2030 (Balkrishna and Sumant 2022). The major classes of epigenetic drugs currently in use are DNA methylation inhibiting drugs, bromodomain inhibitors, histone acetyltransferase inhibitors, histone deacetylase inhibitors, protein methyltransferase inhibitors, and histone methyltransferase inhibitors (Heerboth et al. 2014).
The speakers for the session represented diverse research expertise from both academia and government. Dr. Baitang Ning (National Center for Toxicological Research/FDA, Jefferson, Arkansas, USA) provided detailed current knowledge about how miRNAs and lncRNAs can regulate the expression of drug metabolizing enzymes and nuclear receptors. Dr. Philip Lazarus (Washington State University College of Pharmacy, Spokane, Washington, USA) discussed the potential role of miRNAs on UDP-glucuronosyltransferase (UGT) expression and activity. Dr. Joseph McClay (Virginia Commonwealth University, Richmond, Virginia, USA) described epigenetic profiles in drug response and provided evidence that DMEs in the liver are subject to epigenetic aging via altered DNA methylation and histone acetylation. Data presented by Dr. McClay suggests epigenetic state is a better predictor of drug metabolism than chronological age.
The symposium offered detailed presentations about the current landscape for epigenetics in drug disposition and drug discovery, and there was an engaging question and answer session after the presentations. The consensus was that there is still much more to be discovered about epigenetics to continue to advance human health, but current research is paving the way for these insights. The fast-developing advancements in genomics and epigenomics research provide the groundwork necessary to improve current drug treatment, while also breaking ground for novel therapeutics for difficult-to-treat diseases such as rare diseases, cancer, cardiovascular, and neurological diseases (Nguyen 2019). Similar to how far genomics research has helped advance drug discovery in the past 20 years (Russell et al. 2021), we anticipate that in the coming 20 years, epigenetics will play a critical role in discovering next-generation therapeutics with improved efficacy and safety, and will provide the ability to harness novel targets.
Noncoding RNAs Affect Expression of Cytochrome P450 and Nuclear Receptor Genes
Dongying Li, PhD and Baitang Ning, PhD
Disclaimer: The views presented in this article do not necessarily reflect those of the U.S. Food and Drug Administration. Any mention of commercial products is for clarification and is not intended as an endorsement.
Cytochrome P450 (CYP) expression can be influenced by many factors, including genetic and epigenetic elements. Previously, extensive research has demonstrated that various types of genetic variations affect CYP gene expression, such as pseudogenes, copy number variations, and single nucleotide polymorphisms (SNPs) (Savolainen et al. 1997; Agundez 2004; Zhou et al. 2009). Transcriptional regulators, such as transcription factors and co-regulators, play important roles in transcriptional control of the expression of CYPs and nuclear receptors (NRs). Many NRs serve as key transactivators for CYPs and NRs, including hepatic nuclear factor (HNF) 1A, HNF4α, pregnane X receptor (PXR), constitutive androstane receptor (CAR), aryl hydrocarbon receptor (AHR), and vitamin D receptor (VDR) (Honkakoski and Negishi 2000). It has been shown that epigenetic factors, including DNA methylation and histone modification, also have critical regulatory functions in CYP and NR gene expression (Zanger and Schwab 2013; Tang and Chen 2015). For example, DNA methylating agents may cause hypermethylation and widespread transcriptional inhibition of CYP genes in major organs such as the liver (Dannenberg and Edenberg 2006; Zanger and Schwab 2013; Cheng 2015; Habano et al. 2015; Li Y et al. 2018). In the past couple of decades, non-coding RNAs (ncRNAs) including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) have emerged as epigenetic regulators of CYP and NR gene expression. miRNAs and lncRNAs respond to environmental and pharmaceutical chemicals at their expression levels; meanwhile, they may influence xenobiotic metabolism and toxicity by regulating the expression of DMEs, transporters, and NRs (Ning et al. 2019) (Figure 1).
Figure 1.
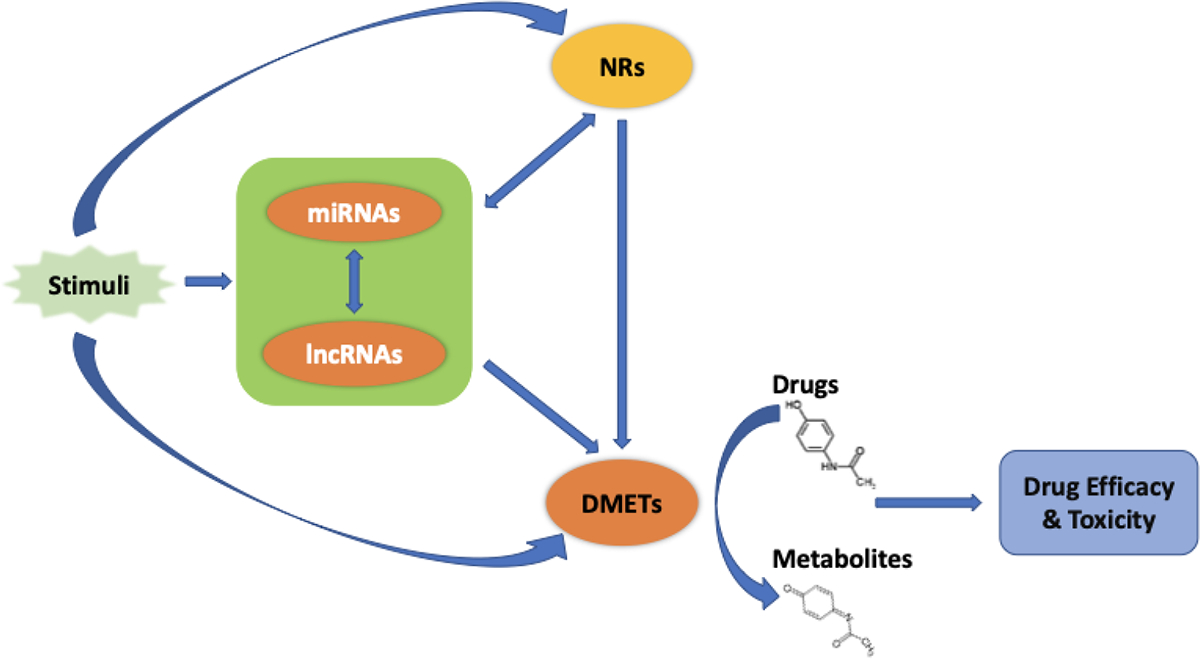
Crosstalk: miRNAs, lncRNAs, NRs and DMETs work as a network. The interaction among miRNAs, lncRNAs, NRs and DMETs constitutes a network that works together to respond to endogenous and exogenous stimuli. The network is a key modulator contributing to drug efficacy and safety. Figure reproduced with permission from Elsevier.
miRNAs are a type of small ncRNAs that primarily inhibit gene expression post-transcriptionally by binding to target mRNAs and recruiting RNA-induced silencing complexes (RISC) for mRNA degradation or translational repression (Huntzinger and Izaurralde 2011). In 2006, Tsuchiya et al. demonstrated that CYP1B1 mRNA was targeted by miR-27b for gene suppression. This was a pioneering breakthrough in epigenetics of DMEs at the time, identifying that CYP genes can regulated by miRNAs (Tsuchiya et al. 2006). Following this study, more findings were reported on miRNAs regulating CYP expression by directly binding to the mRNAs of target genes (Yu et al. 2015; Jin et al. 2016; Chen Y et al. 2017; Wang Y et al. 2017; Zeng et al. 2017; Li D, Tolleson, et al. 2019; Ning et al. 2019). miRNAs can also mediate CYP expression by targeting transcriptional activators including NRs or repressors of CYP expression. For example, multiple miRNAs may downregulate CYP3A4 indirectly by targeting mRNAs of NRs that activate CYP3A4, such as HNF4α, PXR, and VDR (Takagi et al. 2008; Pan YZ et al. 2009; Yu et al. 2018). In contrast, miRNA-mediated silencing of a transcriptional repressor of CYPs will increase CYP expression. For instance, miR-142-3p targets and inhibits small heterodimer partner (SHP), a repressor of CYP2D6, leading to an increase in expression levels of CYP2D6 in mice (Pan X et al. 2017). miRNA-mediated regulation of CYP and NR expression shows further complexity when considering the functions of lncRNAs, particularly as miRNA sponges.
LncRNAs modulate gene expression by various mechanisms and often by interacting with regulatory proteins; the regulatory effects of lncRNAs on gene expression depend on their subcellular localization and the functions of the proteins they associate with (Li D, Tolleson, et al. 2019). Emerging studies have reported regulatory roles of lncRNAs in CYP and NR expression. LncRNAs may regulate CYP expression in association with miRNAs and NRs. For instance, LINC00844 modulates the expression of CYP3A4 and CYP2E1 in a molecular network that consists of miR-486-5p, PXR, and HNF4α via multiple cross-talking pathways (Li D et al. 2020). Further, lncRNAs in neighboring genomic positions with NR genes may mediate CYP expression that is targeted by those NRs. For example, HNF1α-AS1 and HNF4α-AS1 regulate the expression of several transactivation targets of HNF1α and HNF4α, including several CYPs and NRs, and influence the susceptibility to hepatotoxicity induced by acetaminophen, as shown in HepaRG cells (Chen L et al. 2018; Chen L et al. 2020). Despite recent efforts, many questions remain regarding the roles of lncRNAs in influencing xenobiotic metabolism and toxicity via the regulation of CYPs and NRs (Li D, Knox, et al. 2019).
Increasing evidence shows that CYP expression is mediated by different regulators (e.g., miRNAs, lncRNAs, and NRs), at multiple levels (e.g., transcriptional and post-transcriptional), via diverse mechanisms (e.g., genetic and epigenetic), in different subcellular compartments, and with different regulatory outcomes (e.g., gene activation vs. suppression) (Li D, Tolleson, et al. 2019). Human CYP2E1 serves as a good example for the diversity and complexity of ncRNA regulation of CYP expression (Figure 2). miRNAs may regulate CYP2E1 expression by a) targeting different regions of CYP2E1 DNA and mRNA (Mohri et al. 2010; Miao et al. 2016; Wang Y et al. 2017), b) silencing transactivators of CYP2E1 such as HNF1α and NR1I2 (Yu et al. 2018) and c) functionally interacting with lncRNAs that target CYP2E1 (Li D et al. 2020). The regulatory effect of miRNAs on CYP2E1 may be affected by genetic variations, such as SNPs residing in the miRNA response element (Nakano et al. 2015). NcRNA regulation of CYP2E1 expression is critical to xenobiotic-induced CYP2E1-mediated liver toxicity and carcinogenesis, although the underlying mechanisms are complex and remain elusive.
Figure 2.
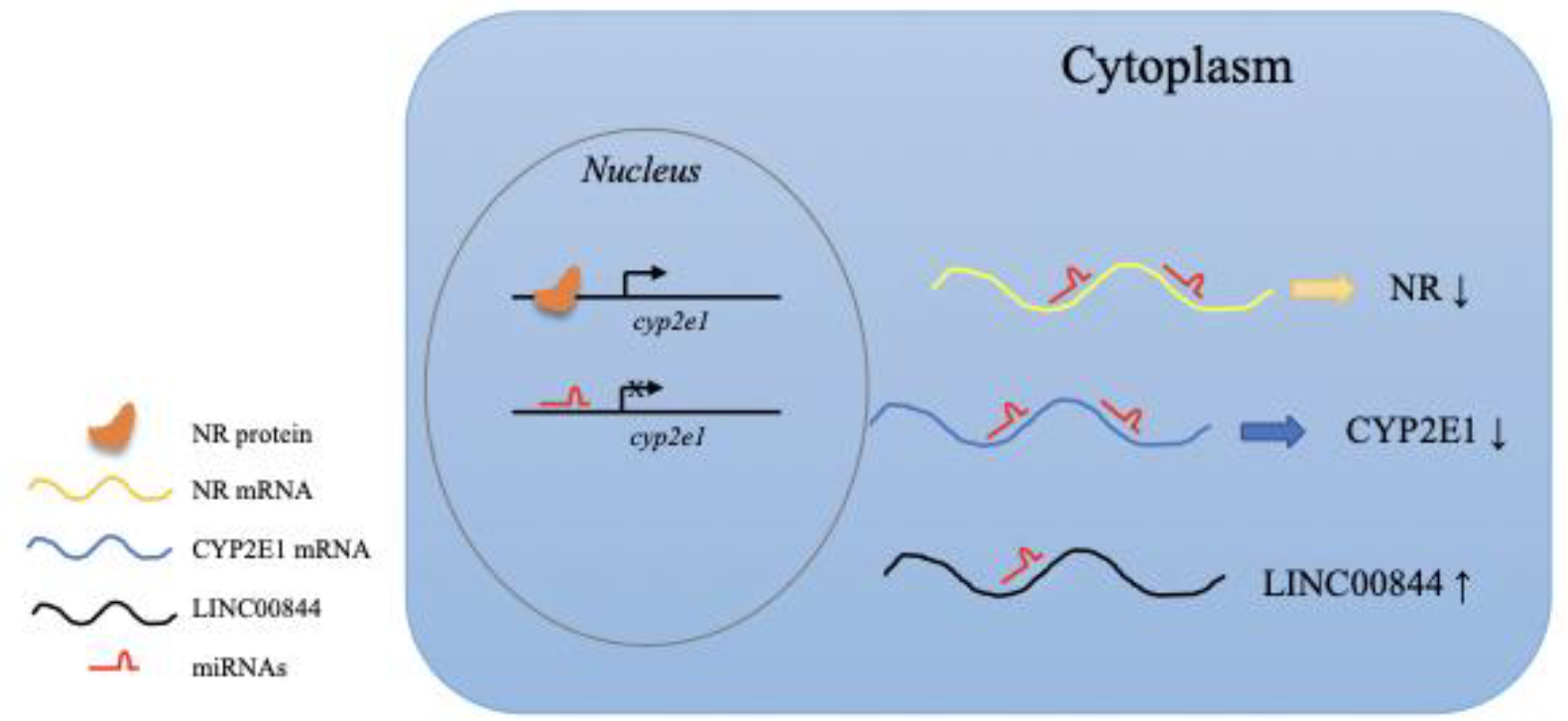
Diverse mechanisms for ncRNA regulation of CYP2E1 expression. Multiple miRNAs may recognize and bind to different regions of CYP2E1 DNA and mRNA and thus directly inhibit or promote CYP2E1 expression. miRNAs may also downregulate CYP2E1 by directly binding to mRNAs of CYP2E1 transactivators such as HNF1? and PXR and repressing the expression of the transactivators. miR-486-5p indirectly increases the expression of CYP2E1 by upregulating lncRNA LINC00844.
To better understand the complicated and diverse mechanisms of ncRNAs in regulating CYP and NR expression, an integrated strategy that combines in silico, in vitro, and in vivo approaches is needed to generate multi-dimensional, high-confidence data that support novel findings. Various computational analyses may greatly enhance the efficiency of target screening and tremendously reduce time, cost, and labor needed for validation and mechanistic investigation. Several databases and computational tools are freely available to the public for ncRNA target prediction, gene expression correlation, and functional characterization. To verify ncRNA targeting site and regulatory effects on CYPs and NRs, a series of in vitro molecular and cellular techniques can be used, including luciferase reporter assays, gain- and loss-of-function assays, fluorescent-based RNA electrophoretic mobility shift assays, and cellular toxicity assays (Li D, Knox, et al. 2019; Yu et al. 2020). Animal models are particularly useful for examining ncRNA and target gene expression as well as systemic responses (e.g., blood biochemistry and organ histology) to assess chemical-induced toxicity at multiple levels in vivo (Li D et al. 2021).
Current evidence indicates that ncRNA regulation of CYP and NR expression is common across different species, even though regulatory potency or efficiency may vary. Great advances have been made in understanding miRNA regulation of some major CYPs and NRs; however, studies detailing lncRNA modulation of CYPs and NRs remain elusive due to the vastly diverse mechanisms that lncRNAs utilize for gene regulation. NcRNA regulation of CYP expression remains a critical research subject as it has significant clinical implications, particularly concerning drug-drug interactions (DDI) and drug-food interactions. In response to certain drug or food intake, the levels of ncRNAs may change, which in turn increases or decreases the expression of their target CYP and NR genes, thus influencing the efficacy and toxicity of drugs that are metabolized by the affected CYPs. Additionally, ncRNAs are being explored as drug targets and agents for human diseases as a new type of RNA therapeutics (Winkle et al. 2021; Zeng et al. 2021). In conclusion, ncRNAs show great promise in enhancing drug development and drug safety and risk assessment, and they hold a bright future to be further utilized in personalized medicine.
Potential Role of miRNA in the Regulation of UGT Expression and Activity
Philip Lazarus, PhD
microRNAs (miRNAs) are dynamic epigenetic regulators of gene expression and play a role in both normal tissue development as well as tumor biology. miRNA post-transcriptionally repress protein expression primarily by binding to target messenger RNA (mRNA), usually within the 3’-untranslated region (UTR), with target recognition predominantly driven by the hybridization of the miRNA ‘seed sequence’ to the mRNA target (Carthew and Sontheimer 2009; Guo et al. 2010). Acting as a genetic switch and/or fine-tuner of protein expression, miRNA regulation can account for not only mild (e.g., two-fold) protein repression, but can also cause mRNA destabilization to dramatically reduce protein expression (Baek et al. 2008; Selbach et al. 2008; Mukherji et al. 2011).
The impact of miRNAs on drug response has not been studied extensively. miRNAs regulate expression of several human cytochrome P450 (CYP) phase I DMEs, including the major drug and hormone metabolizing CYPs such as 3A4, 2E1, 1B1, and 24 (Tsuchiya et al. 2006; Komagata et al. 2009; Pan YZ et al. 2009; Mohri et al. 2010), however, fewer studies have aimed to characterize effects of miRNAs on additional phase I enzymes as well as phase II DMEs.
The UDP-glucuronosyltransferase (UGT) phase II metabolic enzyme family primarily consists of two large subfamilies, the UGT1As and 2Bs. The UGTs are responsible for the metabolism and excretion of numerous endogenous compounds including bilirubin (Bosma et al. 1994) and steroid hormones (Belanger et al. 1998), as well as exogenous compounds including drugs, chemotherapeutic agents, and carcinogens (Nagar and Remmel 2006; Balliet et al. 2009; Sun et al. 2013). UGT family members exhibit extensive interindividual variability of expression that may contribute to variability in patient response and toxicity (Court et al. 2001). The focus of the present seminar was to describe the miRNAs that are potentially important in regulating the UGT 1A and 2B enzymes.
After in silico approaches suggested that several miRNAs potentially interacted with the UGT1A family of enzymes, the UGT1A 3’-UTR was cloned into the luciferase pGL3-promoter vector immediately 3’ of the luciferase open reading frame. After co-transient transfection of this plasmid together with miR-491-3p miRNA mimic into HEK293 (human embryonic kidney) cells, luciferase activity was significantly repressed at both 1 nM (P<0.05) and 2 nM (P<0.05) miR-491-3p concentrations, as compared to the scrambled miRNA-transfected control. No significant difference in luciferase activity was observed between a miR-491-3p seed deletion mutant and the negative scrambled miRNA control, and no significant alteration in luciferase activity was observed in co-transfections with mimics of two other miRNA, miR-148a and miR-136, predicted in silico to bind the UGT1A 3’-UTR.
miR-491-3p demonstrated significant decreases in the endogenous mRNA levels of UGTs 1A1, 1A3, and 1A6 as compared to scrambled miRNA-transfected controls in HuH-7 (human hepatoma-derived) cells. This corresponded with significantly decreased formation of raloxifene glucuronides in HuH-7 cell homogenates after transfection with miR-491-3p mimic; raloxifene is a known substrate of several UGT1A enzymes including UGT1A1 (Sun et al. 2013). In contrast, endogenous UGT1A1 mRNA levels were significantly elevated in the presence of a specific miR-491-3p inhibitor as compared to the scrambled miRNA inhibitor control. This increase corresponded with significant increases in formation of raloxifene-6-glucuronide and raloxifene-4’-glucuronide in HuH-7 homogenates with repressed miR-491-3p levels as compared to scrambled controls. No alteration in mRNA levels or enzyme activity was observed for endogenous UGT2B7, which has its own unique 3’-UTR different from the common UGT1A 3’-UTR and is not predicted to bind miR-491-3p, after transfection with either miR-491-3p mimic or the mMiR-491-3p-specific inhibitor.
Interestingly, an opposite trend was observed in a panel of normal human liver specimens, where a significant inverse correlation (r = −0.487; P < 0.01) was observed between UGT1A6 mRNA and miR-491-3p expression levels in 37 normal human liver specimens that expressed UGT1A6 (Figure 3). A significantly (P < 0.05) higher level of UGT1A6 expression was observed in miR-491-3p non-expressing liver specimens as compared to miR-491-3p expressers. While similar results were also observed for other UGT1A enzymes including UGT1A3, no correlation existed between UGT1A1 or UGT1A9 mRNA levels and miR-491-3p expression in the same panel of liver specimens.
Figure 3.
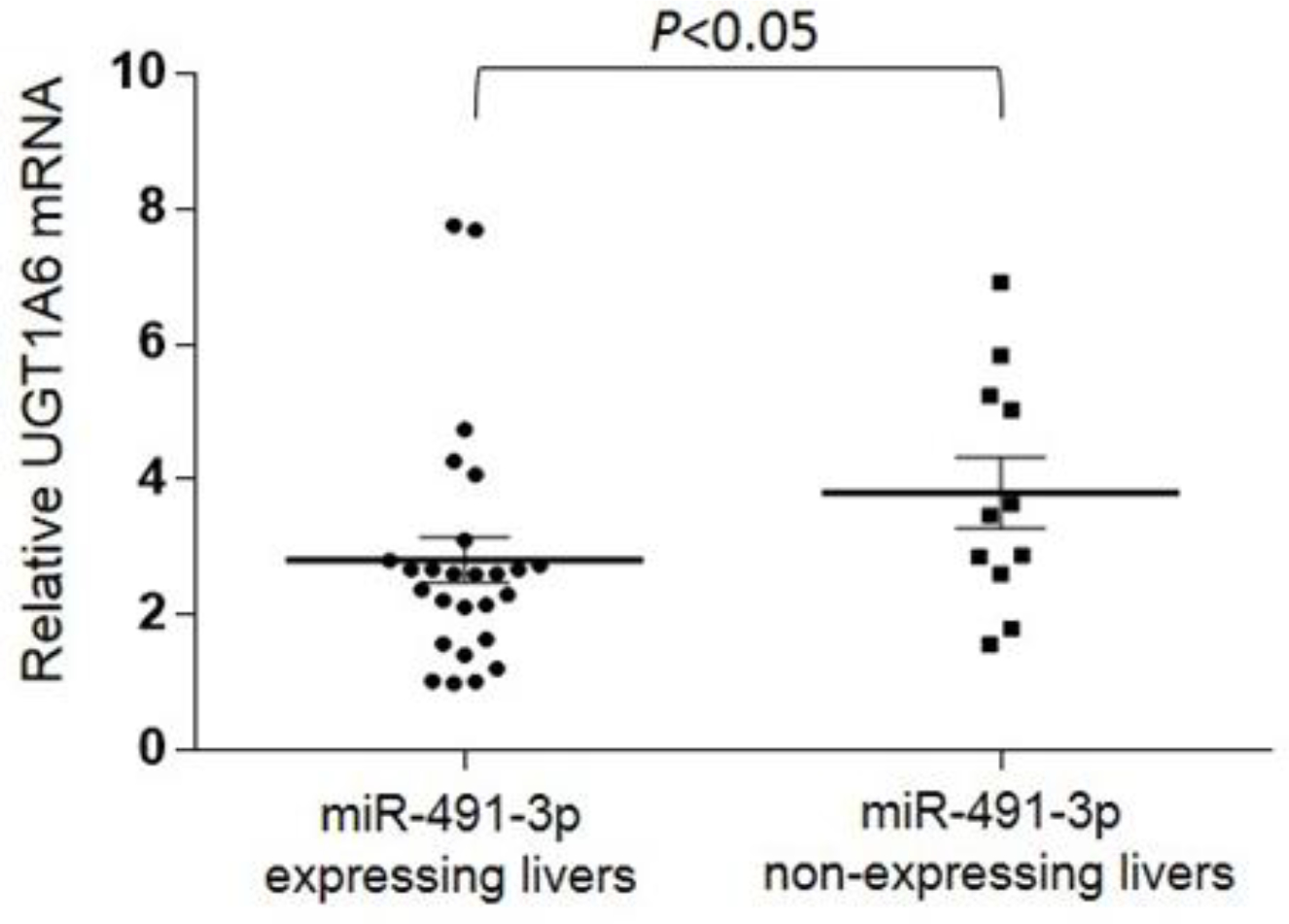
Expression of UGT1A6 mRNA versus miR-491-3p in human liver specimens. mRNA levels of UGT1A6 (n=37 mRNA vs. miR-491-3p) were quantified in normal liver samples via RT-qPCR and normalized to RPLPO and RNU6B, respectively. UGT1A6 mRNA expression levels were examined in liver specimens stratified by expression vs. no expression of miR-491-3p. Dots represent the mean ± S.E. of three independent replicates. Reproduced with permission from Dluzen et al., 2014; J Pharmacol Exper Ther.
A similar approach was used to identify miRNA that potentially regulated UGT2B enzymes (Dluzen et al. 2016). In silico and luciferase data suggested the presence of a functional binding motif for miR-216b-5p within the 3′ untranslated regions of UGTs 2B4, 2B7, and 2B10. Overexpression of a miR-216b-5p mimic significantly repressed UGT2B7 and UGT2B10 mRNA levels in HuH-7 cells, and UGT2B4 and UGT2B10 mRNA in Hep3B cells. UGT2B7 protein levels were repressed in both HuH-7 and Hep3B cells in the presence of increasing miR-216b-5p mimic concentrations, corresponding with a significant decrease in glucuronidation of the UGT2B7-specific substrate, epirubicin. Inhibition of endogenous miR-216b-5p levels significantly increased UGT2B7 mRNA levels in HuH-7 and Hep3B cells, and increased epirubicin glucuronidation by 85% and 50% for HuH-7 and Hep3B cells, respectively, compared to scramble controls. UGT2B4-mediated glucuronidation of codeine and UGT2B10-mediated glucuronidation of nicotine were significantly decreased in both HuH-7 and Hep3B cells after overexpression of a miR-216b-5p mimic.
To further characterize other miRNAs that may play a role in UGT2B regulation, a novel high resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS) method was employed to measure UGT2B protein levels in a panel of human liver microsomal samples (n=62) (Sutliff et al. 2019). Concurrent in silico analysis identified eight candidate miRNAs as potential regulators of UGT2B enzymes. Comparison of UGT2B protein expression and candidate miRNA levels from human liver samples demonstrated a significant inverse correlation between UGTs 2B10 (P = 0.047, r = −0.214) and 2B15 (P = 0.038, r = −0.228) and one of these candidate miRNAs, miR-485-5p. A near-significant correlation was also observed between UGT2B7 and miR-485-5p expression. In vitro analysis using luciferase-containing vectors suggested an interaction of miR-485-5p within the 3′-UTRs of UGT2B10 and UGT2B7 but not with the 3’-UTR of UGT2B15. A significant reduction in luciferase activity was also observed for a luciferase vector containing the UGT2B7 3’-UTR; this was not observed for the UBT2B15 3’-UTR. Subsequently, miR-485-5p mimic was overexpressed in HuH-7 and HepB3 cells, resulting in decreased UGT2B10 and UGT2B7 activities measured using were probed using nicotine and aminobenzotriazole, respectively, and a significant decrease in glucuronidation activity were observed for both substrates in HuH-7 and Hep3B cells upon overexpression of miR-485-5p mimic.
In summary, using several different approaches, we demonstrated that several miRNAs may be important in the regulation of different UGT enzymes. It appears that several of these miRNAs may be exerting a coordinated regulation of multiple UGT genes, which is consistent with the high homology of 3’-UTR sequences observed for UGT2B genes and the fact that the 3’-UTR is the same for all UGT1A genes. Further studies are necessary to determine whether this regulation is important in individual response to drugs and carcinogen exposures in vivo.
Epigenetic profiles and drug response
Joseph L. McClay, PhD
In recent years, evidence has accumulated that epigenetic factors play a major role in the regulation of genes involved in xenobiotic metabolism (Fisel et al. 2016; Kronfol et al. 2017). There are many potential sources of variation in the levels of epigenetic marks such as DNA methylation (5-methylcytosine or 5mC) and histone acetylation. Environmental factors such as stress, diet and exposure to drugs and toxins may all affect epigenetic states (Feinberg 2007; Feil and Fraga 2012). In this regard, epigenetics differs substantially from DNA sequence, which is invariant in response to environmental factors except for mutagens. Epigenetic states can also vary by sex and by age. Considering aging, it has been established for several decades that epigenetically-driven changes to gene expression occur in early development (in utero into childhood) and are intrinsic to processes such as cell differentiation (Li E 2002). However, more recently and particularly in the last decade, it has become apparent that sweeping changes occur to the epigenome through adulthood and into old age (Fraga and Esteller 2007; Horvath 2013; McClay et al. 2014; Benayoun et al. 2015). These changes 1) are non-random, 2) co-localize with binding sites of chromatin binding factors of regulatory importance, 3) occur in genes belonging to specific biological pathways and 4) often have significant effects on gene expression (McClay et al. 2014; Steegenga et al. 2014; Peters et al. 2015). These observations led us to hypothesize that expression of genes involved in xenobiotic metabolism may be affected by epigenetic aging. It is known that drug clearance, mediated by factors including altered DME activity, expression, or hepatic blood flow, changes with age and the 65+ age group is at higher risk of severe adverse drug reactions (Routledge et al. 2004; Budnitz et al. 2011). We decided to test this hypothesis by selecting genes with evidence of epigenetic aging and testing levels of 5mC and histone 3 lysine 9 acetylation (H3K9ac) at these genes for association with gene function and rates of drug metabolism in the liver.
Our first step in the process was to identify genes that show some evidence of epigenetic aging. At the time that we started this project, in 2016, there were no epigenome-wide studies of aging in the liver of humans or model organisms, to the best of our knowledge. However, several studies of epigenetic aging had been conducted in human blood and some other tissues. We therefore collected information on epigenome-wide studies of aging in humans and focused on those conducted using Illumina 5mC microarrays because of the ease of comparing results across studies. We found three studies where the CYP2E1 gene changed with age (Horvath 2013; Steegenga et al. 2014; Peters et al. 2015). It is also notable that CYP2E1 was one of the loci comprising the epigenetic clock of Horvath (Horvath 2013), a highly accurate multi-locus predictor of biological aging. To study epigenetic aging of CYP2E1 in the liver, we examined the mouse ortholog Cyp2e1 in liver tissue from genetically homogenous mice aged under controlled conditions in the National Institute on Aging rodent colonies. Using these mice minimizes extraneous genetic and environmental variation that could hamper our study if using human post-mortem tissue. Our mouse samples ranged from 4 to 32 months of age, spanning young adulthood to very old age (approximately 20 to 80+ years old in human terms).
We first confirmed that 5mC levels at the Cyp2e1 promoter, specifically the mouse region homologous to the human region identified by Horvath (Horvath 2013), did in fact change with age in mouse liver. In the cohort of mice aged 4 to 32 months, we observed significant hypermethylation in the Cyp2e1 promoter of aged mice (P=0.002). We then proceeded to assay Cyp2e1 mRNA expression and protein expression levels in the same liver samples using quantitative polymerase chain reaction (PCR) and western blotting, respectively. Both mRNA and protein expression were significantly (P<0.05) suppressed in the older samples relative to the younger samples. This is the expected direction of effect given that 5mC, typically associated with gene repression, increased with age at the Cyp2e1 promoter. We also assayed histone 3 lysine 9 acetylation (H3K9ac) and histone 3 lysine 27 acetylation (H3K27ac) using chromatin immunoprecipitation and RT-qPCR. While no effect was detected with H3K27ac, we observed significant changes with age in H3K9ac levels. Finally, we tested if these epigenetic changes were associated with Cyp2e1-mediated metabolism. Therefore, we extracted microsomes from the same livers and assayed the rate of chlorzoxazone metabolism. Chlorzoxazone is a muscle relaxant that is almost exclusively metabolized by CYP2E1 and is therefore used as a CYP2E1 probe substrate (Court et al. 1997). We measured the Cyp2e1-mediated conversion of chlorzoxazone to 6-hydroxychlorzoxazone and tested for association between age-related epigenetic changes (5mC and H3K9ac) at Cyp2e1 and chlorzoxazone intrinsic clearance. We found significant (P<0.05) associations for both 5mC and H3K9ac. Notably the effect sizes were quite large, with Cyp2e1 5mC and H3K9ac levels showing correlations with chlorzoxazone intrinsic clearance of −0.3 and 0.5 respectively. Overall, this analysis demonstrated a substantial effect of epigenetic aging on the expression and function of CYP2E1 (Kronfol et al. 2020).
Following our analysis of the Phase I drug metabolism enzyme CYP2E1, we decided to focus our attention on enzymes involved in Phase II drug metabolism. Once again, we looked to human studies to identify candidate Phase II (conjugative) drug metabolism genes potentially subject to epigenetic aging. We identified sulfotransferase 1A1 (SULT1A1) as the human Phase II gene showing most evidence for epigenetic aging (Reynolds et al. 2014; Steegenga et al. 2014) and we tested its epigenetic states and expression in aged mouse livers, as carried out for Cyp2e1 above. For Sult1a1, we found significant hypomethylation with aging coupled to increased H3K9ac. We also found that H3K9ac explained almost one quarter of the variation in Sult1a1 expression levels across all ages. Therefore, we were once again able to detect a substantial influence of epigenetic aging on expression of this drug metabolism gene (Kronfol et al. 2021).
Our studies were limited to the mouse and a few specific genes. Future studies should repeat these analyses for the entire epigenome using human liver tissue, if available. Moreover, our studies were correlational and we were unable to manipulate the epigenetic levels to unambiguously show the causal influence of epigenetic marks. Nevertheless, the effect sizes of epigenetic aging on expression and function of drug metabolizing enzymes appear to be substantial and further work is warranted. Our aspirational goal for this research program is to identify epigenetic biomarkers of drug metabolism in aging that could be used to adjust dosing and reduce the incidence of adverse drug reactions in older patients (Kronfol et al. 2017).
Conclusion
Recent advances in the field of epigenetics have uncovered several regulatory mechanisms that control aspects of DMEs, including miRNA, epigenetic aging, and lncRNA.
Dr. Ning discussed the types of ncRNAs, such as miRNAs and lncRNAs, and their various roles in regulating ADME (absorption, distribution, metabolism, and excretion) processes. miRNAs repress gene expression via direct interaction with target mRNA thus leading to RISC-mediated degradation. For example, miR-27b targets CYP1B1 mRNA, leading to lowered gene expression. Alternatively, lncRNAs modulate gene expression via several mechanisms, including direct interaction with regulatory proteins. For instance, LINC00844 is known to alter gene expression of multiple CYP enzymes by interacting with regulatory proteins, miRNAs, and nuclear receptors. Additionally, lncRNAs and miRNAs may interact directly thus adding further layers of complexity to post-transcriptional epigenetic regulation.
Dr. Lazarus demonstrated that miRNA, specifically miR-491-3p, exerts a family-wide repressive effect on the expression of UGT1A enzymes due to a conserved 3’-UTR binding site across each protein isoform. The repressive effect results in measurable decreases in raloxifene glucuronidation in vitro and these results may translate to in vivo, thus partially explaining the variable UGT activity observed in the clinic. Additional miRNAs were also discussed that may similarly regulate other UGTs and DMEs.
Finally, Dr. McClay discussed epigenetic aging, which is the phenomenon of non-random epigenetic modification of specific genes associated with increased age that impacts the activity of certain biological pathways that occurs with increased age. CYP2E1 was cited as an example in mice because the Cyp2e1 gene promoter region can be repressed or activated by methylation or acetylation, respectively. Gene promoter region hypermethylation was observed in elderly mice, resulting in decreased mRNA and protein expression. Across the entire mouse population, microsomal Cyp2e1 intrinsic clearance was positively correlated with promoter region acetylation and negatively correlated with promoter region methylation. Screening efforts also identified Sult1a1 as a potential target of epigenetic aging with increased age being correlated with gene promoter hypomethylation. Further experimentation is required to identify additional DMEs impacted by epigenetic aging as well as assessing clinical relevance in humans. Overall, both Phase I and Phase II metabolism have been shown to be controlled, in part, by epigenetic modulation of gene expression. Future work incorporating in silico, in vitro, and in vivo models will help expand our understanding of the clinical relevance of epigenetics in drug ADME. These advances will enable more accurate predictions of drug ADME and safety in years to come.
Future perspectives
Different regulators (e.g., miRNAs, lncRNAs, and NRs), act via several mechanisms which can have different outcomes, such as gene activation or gene suppression (Dluzen and Lazarus 2015; Li D, Knox, et al. 2019; Wang J et al. 2020). As increasing evidence emerges about the mechanisms involved in the process of epigenetic regulation of DMEs, these processes will begin to be used in drug development to improve the drug screening and selection process. However, screening for and evaluating epigenetic modifications and their impact on ADME and pharmacokinetics is not currently routine practice. Furthermore, although the current conference report highlights epigenetics in DMEs, effects of epigenetics on drug transporters should be further studied to understand their influence in ADME processes.
As seen in the examples discussed herein and from other studies, in vitro evidence clearly demonstrates that epigenetic modifications contribute to the differential expression of DMEs. These epigenetic changes entail different mechanisms with various external and environmental influences (Kringel et al. 2021). Given the direct interplay between DME expression and activity and pharmacokinetic parameters such as bioavailability, volume of distribution, and half-life, epigenetic modifications can alter clearance of drugs leading to variable exposure (Ingelman-Sundberg et al. 2013). Additional studies are required to evaluate the clinical relevance of such impacts on the pharmacokinetics of drugs. One barrier to directly study epigenetic signatures and their implications in pharmacokinetics and pharmacodynamics is that they are often tissue specific. Blood is commonly sampled as a surrogate tissue, and extracellular vesicles including exosomes and microvesicles released by cancer and immune cells containing epigenetic-related players may help serve as a biomarker of epigenetic regulation in disease or drug therapy (Lorico et al. 2015). However, it remains critical to obtain a biopsy from the tissue of interest to adequately assess epigenetic regulation of other specific genes (Bonder et al. 2014; Hannon et al. 2015; Lowe et al. 2015; Lauschke et al. 2019). Furthermore, an unmet need in drug development is to develop quantitative models to predict the magnitude of the effect epigenetic modifications will have on pharmacokinetics in specific patient populations. Although in silico and in vitro studies have been conducted to evaluate epigenetic mechanisms and their impacts on DME expression and activity, quantitative predictive models need to be developed to account for these changes during drug development and can allow for a precision medicine approach (Stern et al. 2016). Development of quantitative systems pharmacology (QSP) models or PKPD relationships which account for epigenetic mechanisms would be useful in predicting time-dependent changes in the human dose for compounds which modulate epigenetics. The predictive models would also be useful to evaluate the impact of factors which influence epigenetics like diet, age, and comedications, which will be useful in calculating personalized human dose. However, a current barrier is the lack of robust datasets detailing the different mechanisms and pathways that regulate DMEs through epigenetic mechanisms, which would be a prerequisite for development of QSP models. Epigenetic regulations which affect the expression levels of different DMEs as seen above might also explain the observed clinical variability in pharmacokinetics.
Certain clinical compounds which modulate epigenetics can lead to changes in DME expression which can cause and exacerbate DDIs. Evaluation of induction- and/or inhibition-mediated changes in DMEs have become a routine during drug development, which are useful to predict clinical drug-drug interactions. Reversible inhibition mediated DDIs are predicted with reasonable accuracy; however, prediction of irreversible and induction mediated DDIs are less accurate (Einolf et al. 2014; Fowler et al. 2017; Yadav J et al. 2018; Lu and Di 2020; Yadav J. et al. 2020). There are several factors that cause misprediction (Treyer et al. 2019; Tseng et al. 2021). Although in vitro studies have suggested that epigenetic mechanisms could potentially lead to DDIs, these have not been evaluated or remain poorly understood. Moreover, drugs which modulate enzyme activity via inhibition or induction could also modulate apparent activity through epigenetic mechanisms. Consideration of epigenetic mechanisms along with other mechanisms like induction and inhibition might be useful to improve the prediction accuracy. Since there is limited clinical evidence of DDIs due to epigenetic mechanisms, such evaluations are rarely performed. More systematic studies need to be performed to assess the role of epigenetic mediated changes in DDIs.
Despite current challenges, the future of epigenetics in drug discovery and development is bright. We anticipate key discoveries contributing to advancing the current understanding of epigenetic modulations and their downstream effects on pharmacokinetics and pharmacodynamics. As we continue to gather epigenomic data and understand its contribution to disease and pharmacotherapy, these learnings may also contribute to the understanding of drug resistance (Cascorbi and Schwab 2016). We foresee that the optimization and implementation of current epigenetic assays in drug discovery will help close knowledge gaps and contribute to more robust datasets, which will be used to train predictive models to understand implications of epigenetics on ADME genes, pharmacokinetics, drug response, and toxicity – working toward precision medicine to benefit all patients.
Figure 4.
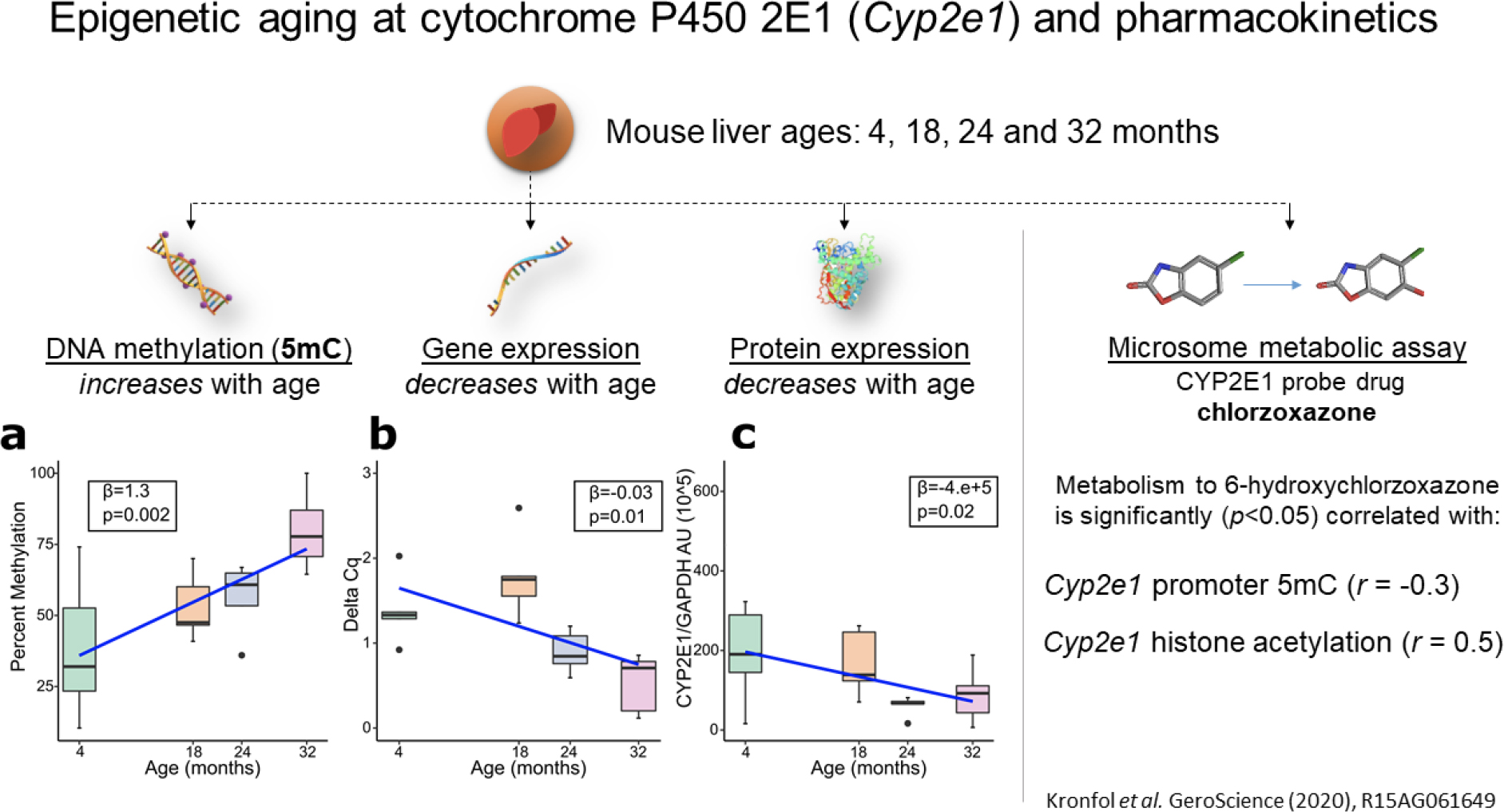
Summary of a study of epigenetic aging effects on regulation of CYP2E1. Aged mouse liver samples were studied using several different assays. In panel a, we show that DNA methylation (5mC) levels significantly increased with age at the Cyp2e1 gene promoter in mouse liver, resulting in a concomitant decrease in gene (panel b) and protein (panel c) expression levels. We also assayed the rate of chlorzoxazone metabolism in microsomes from the same livers. Chlorzoxazone is a CYP2E1 probe drug, meaning that it is almost exclusively metabolized by that CYP450 enzyme. We found a substantial negative correlation between intrinsic clearance of chlorzoxazone and Cyp2e1 5mC levels and a substantial positive correlation with Cyp2e1 histone acetylation levels, indicating a substantial effect of epigenetic aging on the function of CYP2E1. Reproduced with permission from Kronfol et al., GeroScience (2020).
Abbreviations
- 5mC
5-methylcytosine, methylated cytosine typically associated with gene repression
- ADME
absorption, distribution, metabolism, excretion
- AHR
Aryl hydrocarbon receptor
- CAR
Constitutive androstane receptor
- CYP
Cytochrome P450, phase I drug metabolizing enzymes
- DDI
Drug-drug interaction
- DME
Drug metabolizing enzymes
- H3K9ac
Histone 3 Lysine 9 Acetylation
- H3K27ac
Histone 3 Lysine 27 Acetylation
- HNF
Hepatic nuclear factor
- HuH
human hepatoma-derived cells
- LC-MS/MS
Liquid chromatography-tandem mass spectrometry
- lncRNA
long non-coding RNA
- mRNA
messenger RNA
- miRNA
microRNA
- ncRNA
Non-coding RNA
- NR
Nuclear receptors
- PCR
Polymerase chain reaction
- PK
Pharmacokinetics
- PD
Pharmacodynamics
- PXR
Pregnane X factor
- QSP
Quantitative systems pharmacology
- RISC
RNA-induced silencing complexes
- SHP
Small heterodimer partner
- SNP
Single nucleotide polymorphism
- SULT1A1
Sulfotransferase 1A1
- UGT
UDP-glucuronosyltransferases, phase II drug metabolizing enzymes
- UTR
Untranslated region
- VDR
Vitamin D receptor
References
- Agundez JA. 2004. Cytochrome P450 gene polymorphism and cancer. Current drug metabolism. 5(3):211–224. eng. [DOI] [PubMed] [Google Scholar]
- Ahmed S, Zhou Z, Zhou J, Chen SQ. 2016. Pharmacogenomics of Drug Metabolizing Enzymes and Transporters: Relevance to Precision Medicine. Genomics, proteomics & bioinformatics. 14(5):298–313. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. 2008. The impact of microRNAs on protein output. Nature. 455(7209):64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkrishna D, Sumant O. 2022. Epigenetics market by products (kits, reagents, enzymes, instruments), by application (oncology, non oncology), by end user (academic and research institutes, pharmaceutical and biotechnology companies, contract research organizations): global opportunity analysis and industry forecast, 2020–2030. Portland, OR: Allied Market Research; [accessed 2022 April 28]. https://www.alliedmarketresearch.com/epigenetics-market. [Google Scholar]
- Balliet RM, Chen G, Gallagher CJ, Dellinger RW, Sun D, Lazarus P. 2009. Characterization of UGTs active against SAHA and association between SAHA glucuronidation activity phenotype with UGT genotype. Cancer research. 69(7):2981–2989. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayoumi A, Grønbæk H, George J, Eslam M. 2020. The Epigenetic Drug Discovery Landscape for Metabolic-associated Fatty Liver Disease. Trends Genet. 36(6):429–441. eng. [DOI] [PubMed] [Google Scholar]
- Belanger A, Hum DW, Beaulieu M, Levesque E, Guillemette C, Tchernof A, Belanger G, Turgeon D, Dubois S. 1998. Characterization and regulation of UDP-glucuronosyltransferases in steroid target tissues. J Steroid Biochem Mol Biol. 65(1–6):301–310. [DOI] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Brunet A. 2015. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol. 16(10):593–610. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder MJ, Kasela S, Kals M, Tamm R, Lokk K, Barragan I, Buurman WA, Deelen P, Greve JW, Ivanov M et al. 2014. Genetic and epigenetic regulation of gene expression in fetal and adult human livers. BMC Genomics. 15(1):860. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma PJ, Seppen J, Goldhoorn B, Bakker C, Oude Elferink RP, Chowdhury JR, Chowdhury NR, Jansen PL. 1994. Bilirubin UDP-glucuronosyltransferase 1 is the only relevant bilirubin glucuronidating isoform in man. J Biol Chem. 269(27):17960–17964. [PubMed] [Google Scholar]
- Budnitz DS, Lovegrove MC, Shehab N, Richards CL. 2011. Emergency hospitalizations for adverse drug events in older Americans. N Engl J Med. 365(21):2002–2012. eng. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. 2009. Origins and Mechanisms of miRNAs and siRNAs. Cell. 136(4):642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascorbi I, Schwab M. 2016. Epigenetics in Drug Response. Clin Pharmacol Ther. 99(5):468–470. eng. [DOI] [PubMed] [Google Scholar]
- Chen L, Bao Y, Piekos SC, Zhu K, Zhang L, Zhong XB. 2018. A Transcriptional Regulatory Network Containing Nuclear Receptors and Long Noncoding RNAs Controls Basal and Drug-Induced Expression of Cytochrome P450s in HepaRG Cells. Mol Pharmacol. 94(1):749–759. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang P, Manautou JE, Zhong XB. 2020. Knockdown of Long Noncoding RNAs Hepatocyte Nuclear Factor 1α Antisense RNA 1 and Hepatocyte Nuclear Factor 4α Antisense RNA 1 Alters Susceptibility of Acetaminophen-Induced Cytotoxicity in HepaRG Cells. Mol Pharmacol. 97(4):278–286. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zeng L, Wang Y, Tolleson WH, Knox B, Chen S, Ren Z, Guo L, Mei N, Qian F et al. 2017. The expression, induction and pharmacological activity of CYP1A2 are post-transcriptionally regulated by microRNA hsa-miR-132–5p. Biochemical pharmacology. 145:178–191. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G 2015. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev. 81:75–93. eng. [DOI] [PubMed] [Google Scholar]
- Court MH, Duan SX, von Moltke LL, Greenblatt DJ, Patten CJ, Miners JO, Mackenzie PI. 2001. Interindividual variability in acetaminophen glucuronidation by human liver microsomes: identification of relevant acetaminophen UDP-glucuronosyltransferase isoforms. J Pharmacol Exp Ther. 299(3):998–1006. [PubMed] [Google Scholar]
- Court MH, Von Moltke LL, Shader RI, Greenblatt DJ. 1997. Biotransformation of chlorzoxazone by hepatic microsomes from humans and ten other mammalian species. Biopharm Drug Dispos. 18(3):213–226. eng. [DOI] [PubMed] [Google Scholar]
- Dannenberg LO, Edenberg HJ. 2006. Epigenetics of gene expression in human hepatoma cells: expression profiling the response to inhibition of DNA methylation and histone deacetylation. BMC Genomics. 7:181. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dluzen DF, Lazarus P. 2015. MicroRNA regulation of the major drug-metabolizing enzymes and related transcription factors. Drug Metab Rev. 47(3):320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dluzen DF, Sutliff AK, Chen G, Watson CJ, Ishmael FT, Lazarus P. 2016. Regulation of UGT2B Expression and Activity by miR-216b-5p in Liver Cancer Cell Lines. J Pharmacol Exp Ther. 359(1):182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einolf H, Chen L, Fahmi O, Gibson C, Obach R, Shebley M, Silva J, Sinz M, Unadkat J, Zhang L. 2014. Evaluation of various static and dynamic modeling methods to predict clinical CYP3A induction using in vitro CYP3A4 mRNA induction data. Clinical Pharmacology & Therapeutics. 95(2):179–188. [DOI] [PubMed] [Google Scholar]
- Feil R, Fraga MF. 2012. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 13(2):97–109. eng. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. 2007. Phenotypic plasticity and the epigenetics of human disease. Nature. 447(7143):433–440. eng. [DOI] [PubMed] [Google Scholar]
- Fisel P, Schaeffeler E, Schwab M. 2016. DNA Methylation of ADME Genes. Clin Pharmacol Ther. 99(5):512–527. eng. [DOI] [PubMed] [Google Scholar]
- Fowler S, Morcos PN, Cleary Y, Martin-Facklam M, Parrott N, Gertz M, Yu L. 2017. Progress in prediction and interpretation of clinically relevant metabolic drug-drug interactions: a minireview illustrating recent developments and current opportunities. Current pharmacology reports. 3(1):36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Esteller M. 2007. Epigenetics and aging: the targets and the marks. Trends Genet. 23(8):413–418. eng. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 466(7308):835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habano W, Kawamura K, Iizuka N, Terashima J, Sugai T, Ozawa S. 2015. Analysis of DNA methylation landscape reveals the roles of DNA methylation in the regulation of drug metabolizing enzymes. Clinical epigenetics. 7:105. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel AE, Ebers GC, Ramagopalan SV. 2010. Epigenetics: molecular mechanisms and implications for disease. Trends in molecular medicine. 16(1):7–16. eng. [DOI] [PubMed] [Google Scholar]
- Hannon E, Lunnon K, Schalkwyk L, Mill J. 2015. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 10(11):1024–1032. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerboth S, Lapinska K, Snyder N, Leary M, Rollinson S, Sarkar S. 2014. Use of epigenetic drugs in disease: an overview. Genetics & epigenetics. 6:9–19. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkakoski P, Negishi M. 2000. Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochem J. 347(Pt 2):321–337. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S 2013. DNA methylation age of human tissues and cell types. Genome Biol. 14(10):R115. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. 2011. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 12(2):99–110. eng. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Zhong XB, Hankinson O, Beedanagari S, Yu AM, Peng L, Osawa Y. 2013. Potential role of epigenetic mechanisms in the regulation of drug metabolism and transport. Drug Metab Dispos. 41(10):1725–1731. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Yu D, Tolleson WH, Knox B, Wang Y, Chen S, Ren Z, Deng H, Guo Y, Ning B. 2016. MicroRNA hsa-miR-25–3p suppresses the expression and drug induction of CYP2B6 in human hepatocytes. Biochemical pharmacology. 113:88–96. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komagata S, Nakajima M, Takagi S, Mohri T, Taniya T, Yokoi T. 2009. Human CYP24 catalyzing the inactivation of calcitriol is post-transcriptionally regulated by miR-125b. Mol Pharmacol. 76(4):702–709. [DOI] [PubMed] [Google Scholar]
- Kringel D, Malkusch S, Lötsch J. 2021. Drugs and Epigenetic Molecular Functions. A Pharmacological Data Scientometric Analysis. International Journal of Molecular Sciences. 22(14):7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronfol MM, Abudahab S, Dozmorov MG, Jahr FM, Halquist MS, McRae M, Wijesinghe DS, Price ET, Slattum PW, McClay JL. 2021. Histone acetylation at the sulfotransferase 1a1 gene is associated with its hepatic expression in normal aging. Pharmacogenet Genomics. 31(9):207–214. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronfol MM, Dozmorov MG, Huang R, Slattum PW, McClay JL. 2017. The role of epigenomics in personalized medicine. Expert review of precision medicine and drug development. 2(1):33–45. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronfol MM, Jahr FM, Dozmorov MG, Phansalkar PS, Xie LY, Aberg KA, McRae M, Price ET, Slattum PW, Gerk PM et al. 2020. DNA methylation and histone acetylation changes to cytochrome P450 2E1 regulation in normal aging and impact on rates of drug metabolism in the liver. GeroScience. 42(3):819–832. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauschke VM, Zhou Y, Ingelman-Sundberg M. 2019. Novel genetic and epigenetic factors of importance for inter-individual differences in drug disposition, response and toxicity. Pharmacol Ther. 197:122–152. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Knox B, Chen S, Wu L, Tolleson WH, Liu Z, Yu D, Guo L, Tong W, Ning B. 2019. MicroRNAs hsa-miR-495–3p and hsa-miR-486–5p suppress basal and rifampicin-induced expression of human sulfotransferase 2A1 (SULT2A1) by facilitating mRNA degradation. Biochemical pharmacology. 169:113617. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Knox B, Gong B, Chen S, Guo L, Liu Z, Tong W, Ning B. 2021. Identification of Translational microRNA Biomarker Candidates for Ketoconazole-Induced Liver Injury Using Next-Generation Sequencing. Toxicol Sci. 179(1):31–43. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Tolleson WH, Yu D, Chen S, Guo L, Xiao W, Tong W, Ning B. 2019. Regulation of cytochrome P450 expression by microRNAs and long noncoding RNAs: Epigenetic mechanisms in environmental toxicology and carcinogenesis. Journal of environmental science and health Part C, Environmental carcinogenesis & ecotoxicology reviews. 37(3):180–214. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wu L, Knox B, Chen S, Tolleson WH, Liu F, Yu D, Guo L, Tong W, Ning B. 2020. Long noncoding RNA LINC00844-mediated molecular network regulates expression of drug metabolizing enzymes and nuclear receptors in human liver cells. Archives of toxicology. 94(5):1637–1653. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 3(9):662–673. eng. [DOI] [PubMed] [Google Scholar]
- Li Y, Li Y, Zheng G, Zhu L, Wang J, Mu S, Ren Q, Feng F. 2018. Cytochrome P450 1A1 and 1B1 promoter CpG island methylation regulates rat liver injury induced by isoniazid. Molecular medicine reports. 17(1):753–762. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- mLorico A, Corbeil D, Pawelek JM, Alessandro R. 2015. Transmission of information in neoplasia by extracellular vesicles. Hindawi. [DOI] [PMC free article] [PubMed]
- Lowe R, Slodkowicz G, Goldman N, Rakyan VK. 2015. The human blood DNA methylome displays a highly distinctive profile compared with other somatic tissues. Epigenetics. 10(4):274–281. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Di L. 2020. In vitro and in vivo methods to assess pharmacokinetic drug- drug interactions in drug discovery and development. Biopharm Drug Dispos. 41(1–2):3–31. eng. [DOI] [PubMed] [Google Scholar]
- McClay JL, Aberg KA, Clark SL, Nerella S, Kumar G, Xie LY, Hudson AD, Harada A, Hultman CM, Magnusson PK et al. 2014. A methylome-wide study of aging using massively parallel sequencing of the methyl-CpG-enriched genomic fraction from blood in over 700 subjects. Hum Mol Genet. 23(5):1175–1185. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, Yao H, Li C, Pu M, Yao X, Yang H, Qi X, Ren J, Wang Y. 2016. A dual inhibition: microRNA-552 suppresses both transcription and translation of cytochrome P450 2E1. Biochim Biophys Acta. 1859(4):650–662. eng. [DOI] [PubMed] [Google Scholar]
- Mohri T, Nakajima M, Fukami T, Takamiya M, Aoki Y, Yokoi T. 2010. Human CYP2E1 is regulated by miR-378. Biochem Pharmacol. 79(7):1045–1052. [DOI] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. 2011. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 43(9):854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar S, Remmel RP. 2006. Uridine diphosphoglucuronosyltransferase pharmacogenetics and cancer. Oncogene. 25(11):1659–1672. [DOI] [PubMed] [Google Scholar]
- Nakano M, Mohri T, Fukami T, Takamiya M, Aoki Y, McLeod HL, Nakajima M. 2015. Single-Nucleotide Polymorphisms in Cytochrome P450 2E1 (CYP2E1) 3’-Untranslated Region Affect the Regulation of CYP2E1 by miR-570. Drug Metab Dispos. 43(10):1450–1457. eng. [DOI] [PubMed] [Google Scholar]
- Nguyen KV. 2019. Potential epigenomic co-management in rare diseases and epigenetic therapy. Nucleosides, Nucleotides and Nucleic Acids. 38(10):752–780. [DOI] [PubMed] [Google Scholar]
- Ning B, Yu D, Yu AM. 2019. Advances and challenges in studying noncoding RNA regulation of drug metabolism and development of RNA therapeutics. Biochemical pharmacology. 169:113638. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Kent R, Won KJ, Jeong H. 2017. Cholic Acid Feeding Leads to Increased CYP2D6 Expression in CYP2D6-Humanized Mice. Drug Metab Dispos. 45(4):346–352. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YZ, Gao W, Yu AM. 2009. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos. 37(10):2112–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, Reinmaa E, Sutphin GL, Zhernakova A, Schramm K et al. 2015. The transcriptional landscape of age in human peripheral blood. Nature communications. 6:8570. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachayasittikul V, Prathipati P, Pratiwi R, Phanus-Umporn C, Malik AA, Schaduangrat N, Seenprachawong K, Wongchitrat P, Supokawej A, Prachayasittikul V et al. 2017. Exploring the epigenetic drug discovery landscape. Expert opinion on drug discovery. 12(4):345–362. eng. [DOI] [PubMed] [Google Scholar]
- Rasool M, Malik A, Naseer MI, Manan A, Ansari S, Begum I, Qazi MH, Pushparaj P, Abuzenadah AM, Al-Qahtani MH et al. 2015. The role of epigenetics in personalized medicine: challenges and opportunities. BMC Med Genomics. 8 Suppl 1(Suppl 1):S5. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds LM, Taylor JR, Ding J, Lohman K, Johnson C, Siscovick D, Burke G, Post W, Shea S, Jacobs DR Jr. et al. 2014. Age-related variations in the methylome associated with gene expression in human monocytes and T cells. Nature communications. 5:5366. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routledge PA, O’Mahony MS, Woodhouse KW. 2004. Adverse drug reactions in elderly patients. British journal of clinical pharmacology. 57(2):121–126. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell LE, Zhou Y, Almousa AA, Sodhi JK, Nwabufo CK, Lauschke VM. 2021. Pharmacogenomics in the era of next generation sequencing - from byte to bedside. Drug Metab Rev. 53(2):253–278. eng. [DOI] [PubMed] [Google Scholar]
- Savolainen VT, Pajarinen J, Perola M, Penttilä A, Karhunen PJ. 1997. Polymorphism in the cytochrome P450 2E1 gene and the risk of alcoholic liver disease. J Hepatol. 26(1):55–61. eng. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature. 455(7209):58–63. [DOI] [PubMed] [Google Scholar]
- Sim SC, Kacevska M, Ingelman-Sundberg M. 2013. Pharmacogenomics of drug-metabolizing enzymes: a recent update on clinical implications and endogenous effects. Pharmacogenomics J. 13(1):1–11. eng. [DOI] [PubMed] [Google Scholar]
- Steegenga WT, Boekschoten MV, Lute C, Hooiveld GJ, de Groot PJ, Morris TJ, Teschendorff AE, Butcher LM, Beck S, Müller M. 2014. Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. Age (Dordrecht, Netherlands). 36(3):9648. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern AM, Schurdak ME, Bahar I, Berg JM, Taylor DL. 2016. A Perspective on Implementing a Quantitative Systems Pharmacology Platform for Drug Discovery and the Advancement of Personalized Medicine. Journal of biomolecular screening. 21(6):521–534. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker SH, Köferle A, Beck S. 2017. From profiles to function in epigenomics. Nat Rev Genet. 18(1):51–66. eng. [DOI] [PubMed] [Google Scholar]
- Sun D, Jones NR, Manni A, Lazarus P. 2013. Characterization of raloxifene glucuronidation: potential role of UGT1A8 genotype on raloxifene metabolism in vivo. Cancer Prev Res (Phila). 6(7):719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutliff AK, Shi J, Watson CJW, Hunt MS, Chen G, Zhu HJ, Lazarus P. 2019. Potential Regulation of UGT2B10 and UGT2B7 by miR-485-5p in Human Liver. Mol Pharmacol. 96(6):674–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S, Nakajima M, Mohri T, Yokoi T. 2008. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem. 283(15):9674–9680. [DOI] [PubMed] [Google Scholar]
- Tang X, Chen S. 2015. Epigenetic Regulation of Cytochrome P450 Enzymes and Clinical Implication. Current drug metabolism. 16(2):86–96. eng. [DOI] [PubMed] [Google Scholar]
- Treyer A, Ullah M, Parrott N, Molitor B, Fowler S, Artursson P. 2019. Impact of intracellular concentrations on metabolic drug-drug interaction studies. The AAPS journal. 21(5):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng E, Eng H, Lin J, Cerny MA, Tess DA, Goosen TC, Obach RS. 2021. Static and Dynamic Projections of Drug-Drug Interactions Caused by Cytochrome P450 3A Time-Dependent Inhibitors Measured in Human Liver Microsomes and Hepatocytes. Drug Metabolism and Disposition. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. 2006. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 66(18):9090–9098. [DOI] [PubMed] [Google Scholar]
- Wang J, Yu L, Jiang H, Zheng X, Zeng S. 2020. Epigenetic Regulation of Differentially Expressed Drug-Metabolizing Enzymes in Cancer. Drug Metab Dispos. 48(9):759–768. eng. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yu D, Tolleson WH, Yu LR, Green B, Zeng L, Chen Y, Chen S, Ren Z, Guo L et al. 2017. A systematic evaluation of microRNAs in regulating human hepatic CYP2E1. Biochemical pharmacology. 138:174–184. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkle M, El-Daly SM, Fabbri M, Calin GA. 2021. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 20(8):629–651. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav J, Korzekwa K, Nagar S. 2018. Improved predictions of drug–drug interactions mediated by time-dependent inhibition of CYP3A. Molecular pharmaceutics. 15(5):1979–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav J, Paragas E, Korzekwa K, Nagar S. 2020. Time-dependent enzyme inactivation: Numerical analyses of in vitro data and prediction of drug-drug interactions. Pharmacol Ther. 206:107449. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Chen S, Li D, Knox B, Guo L, Ning B. 2020. FREMSA: A Method That Provides Direct Evidence of the Interaction between microRNA and mRNA. Methods Mol Biol. 2102:557–566. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Green B, Marrone A, Guo Y, Kadlubar S, Lin D, Fuscoe J, Pogribny I, Ning B. 2015. Suppression of CYP2C9 by microRNA hsa-miR-128–3p in human liver cells and association with hepatocellular carcinoma. Sci Rep. 5:8534. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Wu L, Gill P, Tolleson WH, Chen S, Sun J, Knox B, Jin Y, Xiao W, Hong H et al. 2018. Multiple microRNAs function as self-protective modules in acetaminophen-induced hepatotoxicity in humans. Archives of toxicology. 92(2):845–858. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, Schwab M. 2013. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 138(1):103–141. eng. [DOI] [PubMed] [Google Scholar]
- Zeng L, Chen Y, Wang Y, Yu LR, Knox B, Chen J, Shi T, Chen S, Ren Z, Guo L et al. 2017. MicroRNA hsa-miR-370–3p suppresses the expression and induction of CYP2D6 by facilitating mRNA degradation. Biochemical pharmacology. 140:139–149. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Li D, Tong W, Shi T, Ning B. 2021. Biochemical features and mutations of key proteins in SARS-CoV-2 and their impacts on RNA therapeutics. Biochemical pharmacology. 189:114424. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SF, Liu JP, Chowbay B. 2009. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 41(2):89–295. [DOI] [PubMed] [Google Scholar]


