Abstract
Magnetic resonance imaging is the primary method used to diagnose canine glial cell neoplasia and non-infectious inflammatory meningoencephalitis. Subjective differentiation of these diseases can be difficult due to overlapping imaging characteristics. This study utilizes texture analysis of intra-axial lesions both as a means to quantitatively differentiate these broad categories of disease and to help identify glial tumor grade/cell type and specific meningoencephalitis subtype in a group of 119 dogs with histologically confirmed diagnoses. Fifty-nine dogs with gliomas and 60 dogs with non-infectious inflammatory meningoencephalitis were retrospectively recruited and randomly split into training (n=80) and test (n=39) cohorts. Forty-five of 120 texture metrics differed significantly between cohorts after correcting for multiple testing (false discovery rate <0.05). After training the random forest algorithm, the classification accuracy for the test set was 85% (sensitivity 89%, specificity 81%). Texture analysis was only partially able to differentiate the inflammatory subtypes (Granulomatous Meningoencephalitis (GME), Necrotizing Meningoencephalitis (NME), and Necrotizing Leukoencephalitis (NLE)) (out-of-bag error rate of 35.0%) and was unable to identify metrics that could correctly classify glioma grade or cell type (out-of-bag error rate of 59.6% and 47.5%, respectively). Multiple demographic differences, such as patient age, sex, weight, and breed were identified between disease cohorts and subtypes which may be useful in prioritizing differential diagnoses. Texture analysis of MR images with a random forest algorithm provided classification accuracy of inflammatory and neoplastic brain disease approaching the accuracy of previously reported subjective radiologist evaluation.
Keywords: Radiomics, MRI, MUO, TA
Introduction
Magnetic resonance imaging (MRI) is one of the most important diagnostic tests available for assessing meningoencephalitis of unknown origin/etiology (MUO/MUE) and primary brain tumors in dogs. MUO is a term used to collectively refer to a number of recognized idiopathic non-infectious meningoencephalomyelitides. The term MUO is used to refer to three specific disease subtypes including: granulomatous meningoencephalitis (GME), necrotizing meningoencephalomyelitis (NME), and necrotizing leukoencephalitis (NLE); other non-infectious meningoencephalitides, such as steroid responsive meningitis-arteritis (SRMA) and eosinophilic meningoencephalomyelitis (EME) have characteristic diagnostic features and are not included with this collective term.1 On MR imaging, these encephalitides have differing grey and white matter involvement and anatomic predilection sites but tend to have a similar pattern of focal to multifocal, variably contrast enhancing, T2 weighted (T2W) hyperintense and T1 weighted (T1W) isointense to hypointense intra-axial lesions (compared to surrounding brain parenchyma). Vasogenic edema and meningeal enhancement may also accompany the primary lesions.2,3 Glial cell tumors form a focal intra-axial mass that is T1W isointense to hypointense, T2W hyperintense, and can have variable contrast enhancement with or without peritumoral edema.4 Common similarities between inflammatory and neoplastic lesions include: a mass effect, T1W/T2W intensity pattern, vasogenic edema, and contrast enhancement. These shared imaging features can make differentiation between granulomatous disease, specifically MUO, and gliomas challenging5. Tissue biopsy is required to confirm a specific histopathologic diagnosis; however, this is not often performed due to the high cost and risk of the procedure, resulting in most patients having a presumed clinical diagnosis based off of a combination of clinical signs, imaging features, and CSF evaluation.
Tumor grade can be difficult to predict from imaging characteristics and is clinically important for prognosis. Several studies have described certain MR features which help partially differentiate oligodendrogliomas from astrocytomas or low-grade from high-grade gliomas; however, there are no characteristics which entirely or repeatably distinguish these groups.6,7 A prior study identified that MR is both highly sensitive and specific for identifying brain lesions in dogs but accuracy decreased when reviewers were required to categorize the abnormality into specific subcategories of neoplastic, inflammatory, or cerebrovascular diseases.8 Quantitative analysis of images may provide additional information regarding the imaging diagnosis, type of encephalitis, and tumor grade.
Texture analysis (TA) is a component of radiomics, the quantitative study of image data. Texture analysis works by inputting an image, or portion of an image (e.g. an ROI), into one of several available software programs in order to evaluate the distribution of pixel intensities. There are several methods to quantify a pixel intensity and, therefore, several methods to perform TA. Methods of performing TA include: statistical, transformative, wavelet, and auto-regressive/model based; however, a complete description of each method is beyond the scope of this paper. The statistical class is most commonly used in image analysis.9 In order to turn a subjective interpretation of a pixel intensity into something that can be mathematically calculated, the intensity or grey-level of each pixel is quantified into a numerical value. An example of a basic CT TA metric that is commonly used by clinicians in everyday practice is mean pixel intensity (Hounsfield Units) within a specified region of interest (ROI). Metrics like mean pixel intensity that evaluate the pixel grey level, but not the spatial relationship of the pixels within the image, are termed first order variables (or histogram features). Second order features are those that consider the spatial relationships between neighboring pixels and the distribution of intensities within the image. Together, these first and second order variables are a way of evaluating the image texture. In other words, TA is the quantification of image grey-level, homogeneity, and coarseness within an image.9 MRI is primarily used to image gross pathologic features in vivo; however, some disease processes may only induce histologic changes which may still be detectable and quantified by texture analysis.10 TA provides the opportunity to extract more information from the data contained in the MR images than is seen visually.
In people, the majority of publications on MR texture analysis involve the brain.11,12 Some examples of how TA has been used with magnetic resonance imaging include early identification of multiple sclerosis,13 discrimination between benign and malignant tumors,14 classifying brain tumor type and grade,15,16,17 and classification of Alzheimer’s disease.18 In veterinary medicine, texture analysis has been used to differentiate dogs with muscular dystrophy, a model of Duchenne Muscular Dystrophy in people,19,20 and to predict the histologic grade of meningiomas.21 A prior study in people found that TA could distinguish normal brain tissues as well as having limited ability to differentiate certain brain tumors on T1W and T2W sequences.22 Based on the finding from these prior studies, TA has the potential to add additional quantitative diagnostic information to the qualitative imaging characteristics of inflammatory and neoplastic brain disorders on MR images in dogs.
The objective of this study was to apply texture analysis to MR examinations of the brain in dogs with known MUO or intra-axial glial cell neoplasm to determine if quantitative methods could accurately predict the underlying etiology. Our hypothesis was that there would be statistically significant differences between these etiologies using first and second order statistical variables. Further, we hypothesized that texture analysis would be able to correctly classify different glioma types within the neoplasia subpopulation and different encephalitides within the MUO subpopulation. If TA proves beneficial, it may result in earlier accurate diagnosis or reduction in the need for potentially invasive and expensive diagnostic testing, such as tissue biopsy. Future artificial intelligence programs, in the form of predictive computer aided diagnostics, could then be developed to assist in disease evaluation on clinical cases.
Methods:
Cohort selection
In this retrospective randomized diagnostic accuracy study, the medical records of a single veterinary academic institution (the University of California, Davis) were reviewed from 2003 to 2018 for MR studies of the canine brain in which a diagnosis of an idiopathic non-infectious meningoencephalomyelitis or an intra-axial glial cell neoplasm was made. Patients were included in the study only if a necropsy or biopsy derived histopathologic diagnosis was made within one year of the MR exam. Histopathology was performed by a pathologist certified by the American College of Veterinary Pathology (DACVP) or by a resident under their supervision. Extra-axial neoplasms or neoplasms primarily arising from within the ventricular system were excluded. In cases where multiple MR studies were performed, images were collected from the first MR exam performed at our institution in which abnormalities were identified. Patients that had previously undergone tissue biopsy or a surgical procedure within the calvarium were excluded so as to avoid including any abnormal signal that was the result of iatrogenic intervention. All studies were performed on a 1.5 T Signa MR unit (General Electric Medical Systems, Milwaukee, WI). Postcontrast images were acquired immediately after intravenous administration of 0.2 mL/kg gadopentetate dimeglumine delivered via manual hand injection (Magnevist, 469mg/mL, Bayer HealthCare Pharmaceuticals Inc., Whippany, NJ).
Image segmentation
All image segmentation and decisions on patient inclusion were performed by a single observer, a third-year radiology resident [MWW]. A two-dimensional ROI was manually drawn at the level of the largest transverse-sectional diameter of the intracalvarial lesion (Figure 1). The number of pixels in each ROI varied based on multiple factors, including the size of the patient, image matrix, and lesion size. The ROI was saved for each dog after accounting for lesion extent in each image sequence. In cases where there was more than one visible lesion, the largest lesion was selected. Contour delineation of the lesion was manually drawn to include both contrast-enhancing and non-enhancing regions. Large regions of perilesional vasogenic edema and meningeal enhancement were excluded where possible. The ROI was applied to matched single transverse (axial) images on the T1-weighted fast spin echo precontrast, T1-weighted fast spin echo postcontrast, T2-weighted fast spin echo, and T2-weighted fluid-attenuated inversion recovery (FLAIR) sequences. Image acquisition parameters were not standardized for all patients as these parameters varied based on the coil used and patient size. In twelve patients, movement had occurred during the scan necessitating shifting of the saved ROI (10) or redrawing the ROI (2) to accommodate for patient rotation. This ROI was then applied to each sequence individually to perform the texture analysis calculations ensuring, as best as possible, that the same tissue was included within the ROI regardless of the sequence evaluated. The radiomics freeware program LIFEx (Institut Curie Centre De Recherche, Orsay, France; v4) was used to perform the texture analysis for each sequence on each individual patient.23
Figure 1.
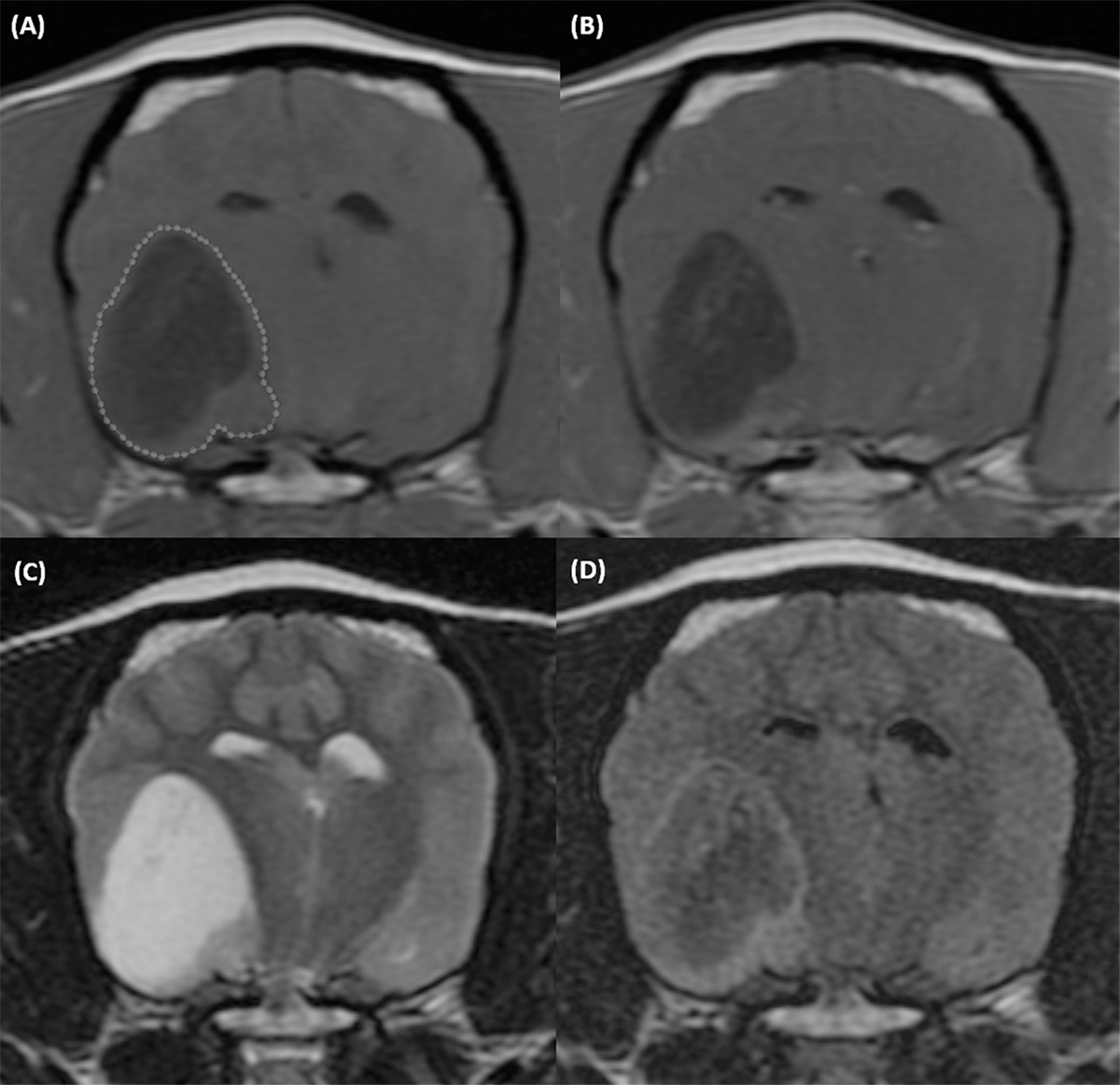
T1-weighted precontrast (A), T1-weighted postcontrast (B), T2-weighted (C), and T2 FLAIR (D) transverse MR images of the brain of a 9-year-old female intact Boston Terrier with a grade 2 (low grade) oligodendroglioma. An ROI outlines the lesion on the T1-weighted precontrast image (dotted line). Acquisition parameters: transverse, slice thickness 3mm, slice spacing 3.3mm. T1W precontrast: FSE T1, TR 567ms, TE 11ms, NEX 4. T1W postcontrast: FSE T1, TR 567ms, TE 11ms, NEX 4. T2W: FSE T2, TR 3500ms, TE104ms, NEX 3. T2 FLAIR: IR, TR 8002ms, TE126ms, TI 2000ms, NEX 1.
Texture analysis
Texture analysis data acquisition parameters were selected by a nuclear medicine physician scientist (MD, PhD) with 12 years of clinical and research experience involving medical image analysis (YGA). MR grey levels within ROIs were normalized between mean ± 3 standard deviations, then discretized into 64-bins (grey levels) to construct histograms and run-length and co-occurrence texture matrices. The histogram values are independent of the spatial localization of the pixel in the image. Histogram values evaluated included mean pixel intensity, variance, skewness, kurtosis, histogram entropy, and histogram energy. Second order features, those that evaluate pairs or groups of spatially related pixels, such as run-length and co-occurrence matrices were also evaluated.10,24 A total of 30 histogram and second order texture analysis metrics were included (Supplement 1).
Statistical analysis:
Statistical analysis was performed by the principal biostatistician (SLT) for the UC Davis Clinical and Translational Science Center (PhD, MSc). The data set was randomly split into training and test sets with approximately two-thirds of the cases (80) in the training set and one-third of the cases (39) in the test set. This random split maintained the relative distribution of the disease type and tumor grade between groups. Five patients had incomplete studies (lacking one or several of the four MR sequences evaluated) in the training set, and one patient had an incomplete study in the test set. For these patients, missing values were imputed using the k-nearest neighbor algorithm with k set to 10. 25 Imputation was done separately for the training and test sets using all texture analysis metrics using the impute.knn function in the R package impute (R Core Team, 2014).26
Discriminating neoplastic from inflammatory disease
Using only the training set, texture analysis metrics were compared between the neoplastic and inflammatory disease groups. Because the distribution of many of the metrics deviated from normality, Wilcoxon’s rank sum test was used to test for median differences of texture metrics between cohorts. False discovery rates (FDR) were calculated to account for multiple testing with significance set at <0.05. The random forest algorithm (in the R package Random Forest version 4.6–14) was then used to construct a classifier to discriminate neoplastic and inflammatory disease in the training set.27 A forest of 1000 trees was constructed. Ten variables were evaluated at each node split. The random forest classifier was then evaluated by applying it to the test set.
Evaluating differences across tumor grade and tumor/inflammatory cell type
Differences in texture analysis metrics across neoplastic disease grades (2, 3, and 4), neoplastic disease cell types (oligodendroglioma, astrocytoma/glioblastoma multiforme, mixed glial/oligoastrocytoma), and inflammatory disease cell types (GME, NME, and NLE) were evaluated using Kruskal-Wallis tests. FDR were calculated to control for multiple testing. Glioblastoma multiforme cases were included as astrocytomas for this portion of the evaluation. Random forest classifiers were then constructed to discriminate disease grades, neoplastic disease cell types and inflammatory disease cell types. Due to the smaller sample sizes associated with the subgroup analyses, the training and test sets were combined to create a complete data set. The entire data set was used in constructing the random forest classifiers; thus, evaluation of the classifier on a separate test set was not conducted.
Evaluation of Demographic Characteristics
Analysis of Variance followed by pairwise comparisons using Tukey’s Honestly Significance Difference procedure was used to evaluate age difference among cell types (GME, NLE and NME) and a two-sample t-test with unequal variances was used to compare age between dogs with inflammatory and neoplastic disease. For weight which was right-skewed, a Kruskal-Wallis test was used to evaluate differences among cell types followed by Dunn’s Test with a Holm’s adjustment to control the type I error rate. Wilcoxon’s rank sum test was used to compare weights between inflammatory and neoplastic disease. Sex distribution by disease type (inflammatory vs. neoplastic) was tested with a Chi-square test. Age was summarized as mean ± standard deviation and weight as median [25th, 75th].
Results:
Study population
One hundred and nineteen dogs met the inclusion criteria; 59 dogs with an intra-axial glial cell neoplasm and 60 with an MUO. Of the dogs with neoplastic disease, 33 dogs were diagnosed with oligodendrogliomas, 21 dogs were diagnosed with astrocytomas, and 5 dogs with mixed oligoastrocytomas. Histologic evaluation was able to provide a specific tumor grade in 52 of the 59 cases with the distribution as follows: 17 low grade (grade 2), 24 high grade (grade 3), and 11 glioblastoma multiforme (GBM, grade 4) (Table 1). Mean age of the neoplastic group was 7.7 years (median: 8.2 years) with a range of 1.4–12.9 years. Sex distribution was as follows: 23 spayed females (39%), 1 intact female (2%), 34 neutered males (58%), and 1 intact male (2%). Seventeen different breeds were represented (12 Boxers, 8 Boston Terriers, 6 French Bulldogs, 5 Pit Bull Terriers, 4 Bulldogs, 4 Mastiffs, 3 Australian Shepherds, 3 Golden Retrievers, 3 Labrador Retrievers, 2 Staffordshire Terriers, and one each of: Doberman Pinscher, Great Dane, Jack Russel Terrier, Rhodesian Ridgeback, Soft Coated Wheaten Terrier, Toy Poodle, and West Highland White Terrier) in addition to 2 mixed breed dogs. Mean and median weight of the dogs in the neoplastic group were 24.8 kg and 25.0 kg, respectively. No age, weight, sex, or apparent breed differences were identified between tumor subtype or grade.
Table 1.
Histopathology derived tumor grade of the neoplastic subpopulation. GBM, Glioblastoma Multiforme; NR, No grade reported
| Glioma Type | Grade 2 | Grade 3 | Grade 4 | NR |
|---|---|---|---|---|
|
| ||||
| Oligodendroglioma | 8 | 22 | - | 3 |
| Astrocytoma/GBM | 8 | 0 | 11 | 2 |
| Oligoastrocytoma (mixed) | 1 | 2 | - | 2 |
|
| ||||
| Total | 17 | 24 | 11 | 7 |
In the MUO cohort, 26 dogs had a final diagnosis of NME, 24 dogs were diagnosed with GME, and 10 with NLE. Mean age of the inflammatory group was 4.7 ± 2.4 years with a range of 8 months to 10.4 years. Further subdivision showed a mean age of 6.0 ± 1.8 years in dogs with GME, 3.9 ± 2.3 years in dogs with NME, and 3.4 ± 2.0 years in dogs with NLE. GME dogs were significantly older than the NME (adj. p=0.002) and NLE (adj. p=0.004) groups; there was no difference in age between NME and NLE dogs (adj. p=0.761). Sex distribution was as follows: 29 spayed females (48%), 10 intact females (17%), 17 neutered males (28%), and 4 intact males (7%). Nineteen different breeds were represented (10 Pugs, 9 Chihuahuas, 6 Maltese, 6 Yorkshire Terriers, 4 Golden Retrievers, 2 Airedale Terriers, and one each of: Belgian Malinois, Boston Terrier, Brussels Griffon, Dachshund, Jack Russel Terrier, Miniature Pinscher, Miniature Poodle, Miniature Schnauzer, Pekingese, Pembroke Welsh Corgi, Pomeranian, Schipperke, Shih Tzu) in addition to 10 mixed breed dogs. Median weight of the dogs in the inflammatory group was 4.9 [2.9, 6.1] kg, with the GME dogs weighing significantly more (median: 6.4 [ 3.8, 28.4] kg) than the NME (6.0 [2.4, 8.7] kg) (adj. p = 0.04) and NLE (2.0 [1.7, 2.7] kg) dogs (adj. p < 0.001). Ten dogs in the inflammatory population weighed greater than 13 kg, with 9/10 of these dogs having a final diagnosis of GME, the only other dog in this group was an Airedale Terrier diagnosed with NLE and weighing 25 kg.
Mean age (p<0.001) and median weight (P<0.001) of dogs in the inflammatory group were lower than the neoplastic group (age: 4.7 ± 2.4 vs. 7.7 ± 3.0; weight: 4.8 [2.6, 9.4] vs. 25.0 [11.5, 34.0]). There were more females in the inflammatory group compared to the neoplastic group (p=0.013). Due to the relatively low numbers of each individual breed, statistical comparison of breed differences could not be performed; however, apparent breed differences exist. The inflammatory group primarily consisted of small or toy breed dogs as evidenced by significantly lower mean weight in this group. Within the encephalitis types, additional predispositions were seen with 60% (6/10) of the NLE cases being Yorkshire Terriers or Yorkshire Terrier mixes and nearly half (12/26, 46%) of NME cases being Pug or Pug crosses. No pug or pug cross in this study was diagnosed with any other inflammatory encephalitis other than NME.
Time from the MR study to histopathologic diagnosis varied from 0 to 296 days (median: 2 days). Diagnosis was made via necropsy in 101 cases, biopsy with subsequent necropsy in 5 cases, and biopsy alone in 13 cases (needle biopsy or surgical biopsy). All dogs had T2-weighted images, 118/119 dogs had T1-weighted precontrast images, 116/119 dogs had FLAIR images, and 114/119 dogs had T1-weighted postcontrast images. Slice thickness varied from 3.3–5.5mm while in-plane voxel size varied from 0.20–0.74mm3. At the time of writing, there is no standard recommendation for data discretization in texture analysis either by number of bins or by fixed bin size.28 In texture analysis it is recommended to optimize the number of grey levels for each specific application with inherent advantages and disadvantages of using lower (e.g. 8–16) or higher (e.g. 64–128) bin numbers.29 For the purpose of data quantification, we chose to use 64 bins as a compromise between signal-to-noise ratio and discretization power.
Differentiation between MUO and glioma
Of the 120 variables evaluated (30 texture analysis metrics on 4 different sequences), 67 metrics had a raw p-value <0.05 indicating differential expression between MUO and glioma diagnoses with 45 having an FDR <0.05, when controlling for multiple testing. All statistically significant metrics are displayed in Supplement 1. The T2W sequences (T2W and FLAIR) yielded larger numbers of significant metrics (17 and 14, respectively) compared with the T1W sequences (8 for T1W postcontrast and 6 for T1W precontrast). A variable importance plot can be used to identify the most important discriminatory variables for a random forest classifier and is provided as Figure 2 for this analysis. For the training set, the out-of-bag error rate, an estimate of the mean prediction error, was 24%. Random forest classifier accuracy was 76% with a sensitivity of 76% and specificity of 77%. The trained random forest classifier was then applied to the test set where the out-of-the-bag classification accuracy was 85% with a sensitivity of 89% and a specificity of 81%.
Figure 2.
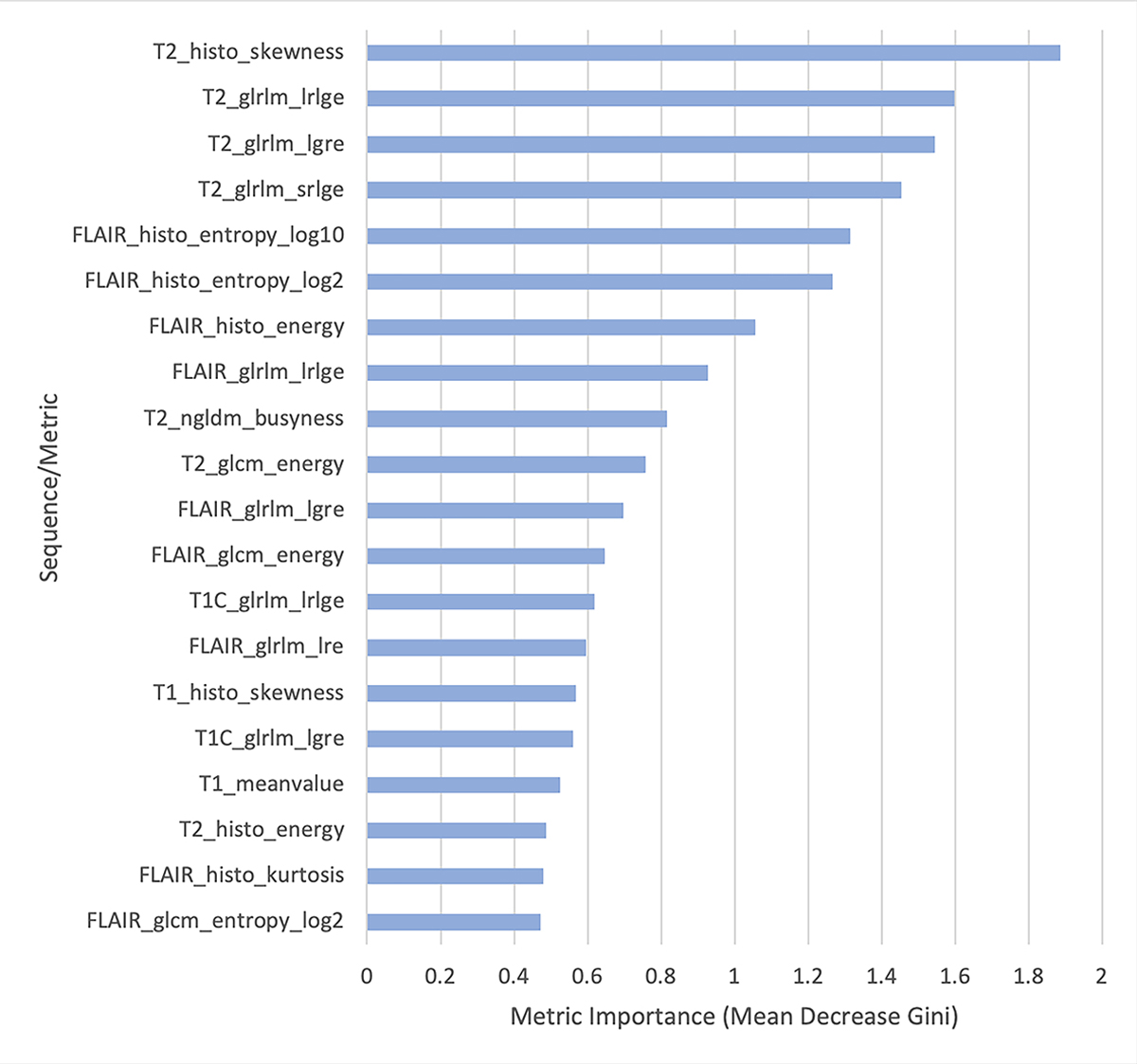
Variable importance plots from the random forest classifier differentiating inflammatory versus neoplastic brain disease in order of decreasing importance (top to bottom). This graph displays the 20 most discriminatory metrics in constructing the random forest classifier. histo, histogram; glrlm, grey-level run length matrix; ngldm, neighborhood grey-level different matrix; glcm, grey-level co-occurrence matrix; lrlge, long run low grey-level emphasis; lgre, low grey-level run emphasis; srlge, short run low grey-level emphasis; lre, long run emphasis. Magnetic resonance imaging (MR) sequences include T2 weighted (T2), fluid attenuated inversion recovery (FLAIR), T1 weighted (T1), and T1 plus intravenous contrast administration (T1C).
Differentiation between tumor grades
In discriminating between different tumor grades, analyses were first conducted to differentiate grade 2, 3, and 4 glial cell tumors. Dogs without a tumor grade reported were not included in this analysis. No TA metrics were found to have a significant difference across grades. The random forest classifier performed poorly with an out-of-bag error rate of 59.6%. Subsequently, grades 3 and 4 were combined and compared to grade 2 tumors. Again, no significant metrics were identified and the out-of-bag error rate only modestly improved (40.4%).
In discriminating between different tumor cell types, the glioblastoma multiforme cases were included with astrocytomas, consistent with the historical classification system, for a combined 21 astrocytomas. No texture analysis metrics were found to have a significant difference (FDR <0.05) across tumor cell types and the out-of-bag error rate for the random forest classifier was 47.5%.
Differentiation between types of encephalitis
In discriminating between different inflammatory diseases, 46 texture metrics differed significantly (FDR <0.05) between encephalitis type. Despite the number of significant metrics, the out-of-bag error rate remained relatively high (35.0%). Texture analysis was particularly poor at classifying NLE, correctly classifying only 2/10 cases (Table 2).
Table 2.
Predictive results of the random forest algorithm in classifying subtypes of noninfectious meningoencephalitis compared with the true histopathologic diagnosis. GME, Granulomatous Meningoencephalitis; NME, Necrotizing Meningoencephalitis; NLE, Necrotizing Leukoencephalitis
| True | Predicted | ||
|---|---|---|---|
|
| |||
| NME | GME | NLE | |
|
| |||
| NME | 19 | 5 | 2 |
| GME | 5 | 18 | 1 |
| NLE | 6 | 2 | 2 |
Discussion:
The texture analysis procedure performed in the current study was able to identify 45 metrics with statistical significance that helped in differentiation of non-infectious inflammatory encephalitides from intra-axial glial cell neoplasms. A prior study described a sensitivity of 87.4% and a specificity of 91.7% for observers correctly classifying neoplastic disease and a sensitivity of 86.0% and specificity of 93.1% for correctly classifying inflammatory disease into broad categories on MR studies.8 The current analysis approaches this accuracy without including any observational components. It would be interesting to investigate whether a combination of observational and quantitative methods would further improve classification above current subjective classification accuracy alone.
Despite identifying a large number of discriminatory variables, overall random forest classifier accuracy was 76% for the training set and 85% for the test set. The classification errors were due to variability within the neoplastic and inflammatory groups having some degree of data overlap for every evaluated metric. This result is perhaps expected given the subjective overlap in appearance of these disease processes (Figure 3). The reason why the algorithm performed better on the test set compared to the training set is uncertain, but it may be the result of decreased variability of the discriminatory metrics within the randomly assigned test set cohort.
Figure 3.

Transverse T2-weighted (A, B, C, G, H, I) and T1-weighted post contrast (D, E, F, J, K, L) MR images of the brain of a: 9 year old Staffordshire Terrier with a grade 2 (low grade) astrocytoma (A+D), 13 year old Boston Terrier with a grade 3 (high grade) oligodendroglioma (B+E), 6 year old Pit Bull Terrier with a GBM (grade 4, high grade astrocytoma; C+F), 8 year old Airedale Terrier with GME (G+J), 1 year old Chihuahua with NME (H+K), and 6 year old Yorkshire Terrier with NLE (I+L). Similarities exist between the distribution and imaging characteristics of neoplastic and inflammatory diseases. T2 FSE, TR 2967ms, TE 81ms, thickness 4mm (A). T2 FSE, TR 3500ms, TE 104ms, thickness 3mm (B). T2 FSE, TR 3500ms, TE 100ms, thickness 3mm (C). T1 SE, TR 400ms, TE 10ms, thickness 4mm (D). T1 FSE, TR 517ms, TE 11ms, thickness 3mm (E). T1 FSE, TR 667ms, TE 11ms, thickness 3mm (F). T2 FSE, TR 3500ms, TE 100ms, thickness 3mm (G). T2 FSE, TR 3500ms, TE 104ms, thickness 3mm (H). T2 FSE, TR 3500ms, TE 102ms, thickness 3mm (I). T1 FSE, TR 717ms, TE 11ms, thickness 3mm (J). T1 FSE, TR 683ms, TE 11ms, thickness 3mm (K). T1 FSE, TR 650ms, TE 17ms, thickness 3mm (L).
Previously, it has been suggested that access to demographic data did not improve the accuracy of radiologist interpretation in classifying intracranial lesions in dogs and may have actually reduced inter-rater agreement in some instances.8 Nevertheless, as is further supported in our data, certain signalment data such as patient age, size, sex, and breed are features that have been associated with specific intra-axial neoplasms or types of inflammatory brain disease and are factors which may play a role in radiologist evaluation.4,30,31 As random forest algorithms can include additional nonparametric variables, repeating our analysis with inclusion of patient age, size, and sex may further improve predictive accuracy. Due to the small numbers of dogs in each specific breed, it would be unlikely that this variable could be meaningfully included in a repeat analysis unless a much larger data set was utilized.
Our texture analysis metrics failed to identify a difference between grade 2, 3, and 4 glial tumors (out-of-bag error of 59.6%). Recently, it has been proposed that glial cell tumors should be graded as low grade (grade 2) or high grade (grade 3 and grade 4) instead of three distinct grades as has been done previously.30 Even when this new classification method was adopted, the classification accuracy remained too low to be clinically useful (out-of-bag error 40.4%). When comparing different glial tumor cell types, texture analysis was again unable to accurately differentiate astrocytomas, oligodendrogliomas, and mixed oligoastrocytomas. Therefore, at this time, we must reject our second hypothesis that texture analysis would be beneficial in identifying glial tumor cell type or glial tumor grade. Although our results displayed some ability to differentiate the specific type of meningoencephalitis, overall predicted accuracy for this test remained relatively low (65%).
Multiple demographic differences were appreciated between groups and within group subtypes. Perhaps as expected, Boxers, Boston Terriers, French Bulldogs, and Pit Bull Terriers comprised the four largest breed groups in the glioma subpopulation. Similarly, toy breed dogs (Pugs, Chihuahuas, Maltese, and Yorkshire Terriers) comprised the four largest breed groups in the MUO subpopulation. Additionally, although no specific breed predispositions were identified within the subtypes of the glioma population, Pug and Pug crosses were over-represented in our NME population and Yorkshire Terriers were over-represented in our NLE population. Dogs with GME were significantly larger than dogs in the other two inflammatory subtypes and subjectively had a wider variety of affected breeds. GME dogs were also significantly older than the NME and NLE dogs. Overall, dogs in the MUO subpopulation were significantly younger, smaller, and more likely to be female than the neoplastic population.
There are few limitations of this study. Multiple studies in the human medical literature have identified that texture analysis performs better when performed on a 3-dimensional volume of interest (VOI) instead of a 2-dimensional region of interest.32,33 Indeed, performing an analysis on a 3D volume may have further improved our classification accuracy in this study. The decision to perform 2D ROIs was due to feasibility and the time required to draw an accurate VOI on the large number of cases in this study. This remains a potential future application of texture analysis in veterinary medicine. Secondly, all ROIs were drawn by the same observer; however, due to variable contrast enhancement and lesion conspicuity accidental omission of affected tissue or inclusion of non-affected tissue is possible. Our MR image acquisition parameters varied by patient size and the receiver coil used; since texture analysis relies heavily on tissue contrast, pixel size, signal to noise ratios, and signal intensity, such as from proximity to a surface coil, these parameters can alter texture features. For the same reason, texture metrics cannot be completely standardized between facilities with different MR scanners and magnetic field strengths. That being said, a multicenter study found that the differences in texture features between different centers had only a small impact on results.34 Not having standardized image acquisition parameters is a limitation of this study, but is expected in a retrospective study and is representative of true clinical cases. All appropriate texture metrics available through the LIFEx freeware program were included, though it should be noted that additional texture features and texture filters have been described with other software programs that were not evaluated in this study. The inclusion of these additional features may have further improved classification accuracy. Lastly, although all cases were confirmed with histopathology and the median delay to histopathologic results from the time of imaging was only 2 days, in a minority of cases this delay was up to 296 days. This delay introduces a potential limitation in that a tumor grade may change in the interim between the imaging study and the time of presentation to histopathology.
Texture analysis is capable of differentiating intra-axial glial cell neoplasia from MUO with a sensitivity of 76–89% and a specificity of 77–81%, approaching reported observational results. Our texture analysis did not prove beneficial in discriminating glial tumor grade or cell type and was only partially discriminatory in determining meningoencephalitis subtype. Texture analysis could be included as part of a future computer aided diagnostic to integrate quantitative analysis into case interpretation alongside both visual interpretation and consideration of demographic data.
Supplementary Material
Results of Wilcoxon’s Rank Sum Test comparing texture features in dogs with Neoplastic and Inflammatory brain disease. Medians [25th, 75th] are shown for each group. Statistically significant variables (FDR<0.05) are displayed in green.
Abbreviations: FDR, false discovery rate; Histo, histogram; GLCM, grey-level co-occurrence matrix; GLRLM, grey level run length matrix; NGLDM, neighborhood grey level different matrix; SRE, short run emphasis; LRE, long run emphasis; LGRE, low grey level run emphasis; HGRE, high grey level run emphasis; SRLGE, short run low grey level emphasis; SRHGE, short run high grey level emphasis; LRLGE, long run low grey level emphasis; LRHGE, long run high grey level emphasis; GLNU, grey level non-uniformity; RLNU, run length non-uniformity; RP, run percentage.
Funding sources:
The imaging studies in this publication were performed by the In Vivo Translational Imaging Shared Resource which is supported by the UC Davis Comprehensive Cancer Center’s NCI Cancer Center Support Grant (CCSG; NCI P30CA093373). The project described was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through grant number UL1 TR001860. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations
- MUO
Meningoencephalitis of Unknown Origin
- GME
Granulomatous Meningoencephalitis
- NME
Necrotizing Meningoencephalitis
- NLE
Necrotizing Leukoencephalitis
- TA
Texture Analysis
- ROI
Region of Interest
- FDR
False Discovery Rate
Footnotes
Conflicts of interest: None.
Study Design: Retrospective Randomized Diagnostic Accuracy Study
EQUATOR network disclosure: None.
References:
- 1.Cornelis I, Van Ham L, Gielen I, De Decker S, Bhatti SFM. Clinical presentation, diagnostic findings, prognostic factors, treatment and outcome in dogs with meningoencephalomyelitis of unknown origin: A review. Vet J 2019;244:37–44. [DOI] [PubMed] [Google Scholar]
- 2.Hecht S, Adams WH. MRI of brain disease in veterinary patients part 2: acquired brain disorders. Vet Clin N Am - Small 2010;40(1):39–63. [DOI] [PubMed] [Google Scholar]
- 3.Young BD, Fosgate GT, Holmes SP, et al. Evaluation of standard magnetic resonance characteristics used to differentiate neoplastic, inflammatory, and vascular brain lesions in dogs. Vet Radiol Ultrasound 2014;55(4):399–406. [DOI] [PubMed] [Google Scholar]
- 4.Wisner ER, Dickinson PJ, Higgins RJ. Magnetic resonance imaging features of canine intracranial neoplasia. Vet Radiol Ultrasound 2011;52(suppl. 1):52–61. [DOI] [PubMed] [Google Scholar]
- 5.Diangelo L, Cohen-Gadol A, Heng HG, et al. Glioma Mimics: Magnetic Resonance Imaging Characteristics of Granulomas in Dogs. Front Vet Sci. 2019;6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young BD, Levine JM, Poter BF, et al. Magnetic resonance imaging features of intracranial astrocytomas and oligodendrogliomas in dogs. Vet Radiol Ultrasound 2011;52(2):132–141. [DOI] [PubMed] [Google Scholar]
- 7.Bentley RT, Ober CP, Anderson KL, et al. Canine intracranial gliomas: relationship between magnetic resonance imaging criteria and tumor type and grade. Vet J 2013;198(2):463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolff CA, Holmes SP, Young BD, et al. Magnetic resonance imaging for the differentiation of neoplastic, inflammatory, and cerebrovascular brain disease in dogs. J Vet Intern Med 2012;26(3):589–597. [DOI] [PubMed] [Google Scholar]
- 9.Lubner MG, Smith AD, Sandrasegaran K, Sahani DV., Pickhardt PJ. CT Texture Analysis: Definitions, Applications, Biologic Correlates, and Challenges. RadioGraphics. 2017;37(5):1483–1503. [DOI] [PubMed] [Google Scholar]
- 10.Castellano G, Bonilha L, Li LM, Cendes F. Texture analysis of medical images. Clin Radiol 2004;59(12):1061–1069. [DOI] [PubMed] [Google Scholar]
- 11.Larroza A, Bodí V, Moratal D. Texture analysis in magnetic resonance imaging: review and considerations for future applications. Assessment of Cellular and Organ Function and Dysfunction Using Direct and Derived MRI Methodologies, IntechOpen. 2016:75–106. [Google Scholar]
- 12.Kjaer L, Ring P, Thomsen C, Henriksen O. Texture analysis in quantitative MR imaging. Tissue characterization of normal brain and intracranial tumors at 1.5T. Acta radiol 1995;36(2):127–135. [PubMed] [Google Scholar]
- 13.Mathias JM, Tofts PS, Losseff NA. Texture analysis of spinal cord pathology in multiple sclerosis. Magn Reson Med 1999;42(5):929–935. [DOI] [PubMed] [Google Scholar]
- 14.Juntu J, Sijbers J, De Backer S, Rajan J, Van Dyck D. Machine learning study of several classifiers trained with texture analysis features to differentiate benign from malignant soft-tissue tumors in T1-MRI images. J Magn Reson Im 2010;31(3):680–689. [DOI] [PubMed] [Google Scholar]
- 15.Zacharaki EI, Wang S, Chawla S, et al. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 2009;62(6):1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skogen K, Schulz A, Dormagen JB, Ganeshan B, Helseth E, Server A. Diagnostic performance of texture analysis on MRI in grading cerebral gliomas EurJ Radiol 2016;85(4):824–829. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh KL, Chen C, Lo C. Quantitative glioma grading using transformed gray-scale invariant textures of MRI. Comput Biol Med 2017;83:102–108. [DOI] [PubMed] [Google Scholar]
- 18.de Oliveira MS, Balthazar ML, D’Abreu A, et al. MR imaging texture analysis of the corpus callosum and thalamus in amnestic mild cognitive impairment and mild Alzheimer disease. Am J Neuroradiol 2011;32(1):60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang G, Lalande V, Chen L, et al. MRI texture analysis of GRMD dogs using orthogonal moments: a preliminary study. Irbm 2015;36(4):213–219. [Google Scholar]
- 20.Duda D, Kretowski M, Azzabou N, de Certaines JD. MRI texture analysis for differentiation between healthy and golden retriever muscular dystrophy dogs at different phases of disease evolution. Lect Notes Comput Sci 2015;9339:255–266. [Google Scholar]
- 21.Banzato T, Bernardini M, Cherubini GB, Zotti A. Texture analysis of magnetic resonance images to predict histologic grade of meningiomas in dogs. Am J Vet Res 2017;78(10):1156–1162. [DOI] [PubMed] [Google Scholar]
- 22.Herlidou-Meme S, Constans JM, Carsin B, et al. MRI texture analysis on texture test objects, normal brain and intracranial tumors. Magn Reson Imaging 2003;21(9):989–993. [DOI] [PubMed] [Google Scholar]
- 23.Nioche C, Orlhac F, Boughdad S, et al. LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res 2018;78(16):4786–4789. [DOI] [PubMed] [Google Scholar]
- 24.Alobaidli S, McQuaid S, South C, Prakash V, Evans P, Nisbet A. The role of texture analysis in imaging as an outcome predictor and potential tool in radiotherapy treatment planning. Br J Radiol 2014;87:20140369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Troyanskaya O, Cantor M, Sherlock G, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17(6):520–525. doi: 10.1093/bioinformatics/17.6.520 [DOI] [PubMed] [Google Scholar]
- 26.Hastie T, Tibshirani R, Narasimhan B, Chu G. impute: impute: Imputation for microarray data. R package version 1.56.0. 2018. [Google Scholar]
- 27.Liaw A, Wiener M. Classification and Regression by randomForest. R News 2002;2:18–22. [Google Scholar]
- 28.Zwanenburg A, Vallières M, Abdalah MA, et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020;295(2):328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. arXiv preprint arXiv:1612.07003. [Google Scholar]
- 30.Koehler JW, Miller AD, Miller CR, et al. A revised diagnostic classification of canine glioma: towards validation of the canine glioma patient as a naturally occurring preclinical model for human glioma. J Neuropath Exp Neur 2018;77(11):1039–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flegel T Breed-specific magnetic resonance imaging characteristics of necrotizing encephalitis in dogs. Front Vet Sci 2017;4:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahmoud-Ghoneim D, Toussaint G, Constans JM, de Certaines JD. Three dimensional texture analysis in MRI: a preliminary evaluation in gliomas. Magn Reson Imaging 2003;21(9):983–987. [DOI] [PubMed] [Google Scholar]
- 33.Wagner F, Gryanik A, Schulz-Wendtland R, Fasching PA, Wittenberg T. 3D characterization of texture: evaluation for the potential application in mammographic mass diagnosis. Biomed Eng-Biomed Te 2012;57(suppl. 1):490–493. [Google Scholar]
- 34.Mayerhoefer ME, Breitenseher MJ, Kramer J, Aigner N, Hofmann S, Materka A. Texture analysis for tissue discrimination on T1-weighted MR images of the knee joint in a multicenter study: transferability of texture features and comparison of feature selection methods and classifiers. J Magn Reson Im 2005;22(5):674–680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results of Wilcoxon’s Rank Sum Test comparing texture features in dogs with Neoplastic and Inflammatory brain disease. Medians [25th, 75th] are shown for each group. Statistically significant variables (FDR<0.05) are displayed in green.
Abbreviations: FDR, false discovery rate; Histo, histogram; GLCM, grey-level co-occurrence matrix; GLRLM, grey level run length matrix; NGLDM, neighborhood grey level different matrix; SRE, short run emphasis; LRE, long run emphasis; LGRE, low grey level run emphasis; HGRE, high grey level run emphasis; SRLGE, short run low grey level emphasis; SRHGE, short run high grey level emphasis; LRLGE, long run low grey level emphasis; LRHGE, long run high grey level emphasis; GLNU, grey level non-uniformity; RLNU, run length non-uniformity; RP, run percentage.


