Abstract
Members of a group of marine bacteria that is numerically important in coastal seawater and sediments were characterized with respect to their ability to transform organic and inorganic sulfur compounds. Fifteen strains representing the Roseobacter group (a phylogenetic cluster of marine bacteria in the α-subclass of the class Proteobacteria) were isolated from seawater, primarily from the southeastern United States. Although more than one-half of the isolates were obtained without any selection for sulfur metabolism, all of the isolates were able to degrade the sulfur-containing osmolyte dimethyl sulfoniopropionate (DMSP) with production of dimethyl sulfide (DMS). Five isolates also degraded DMSP with production of methanethiol, indicating that both cleavage and demethylation pathways for DMSP occurred in the same organism, which is unusual. Five isolates were able to reduce dimethyl sulfoxide to DMS, and several isolates also degraded DMS and methanethiol. Sulfite oxygenase activity and methanesulfonic acid oxygenase activity were also present in some of the isolates. The ability to incorporate the reduced sulfur in DMSP and methanethiol into cellular material was studied with one of the isolates. A group-specific 16S rRNA probe indicated that the relative abundance of uncultured bacteria in the Roseobacter group increased in seawater enriched with DMSP or DMS. Because this group typically accounts for >10% of the 16S ribosomal DNA pool in coastal seawater and sediments of the southern United States, clues about its potential biogeochemical role are of particular interest. Studies of culturable representatives suggested that the group could mediate a number of steps in the cycling of both organic and inorganic forms of sulfur in marine environments.
Biogenic sulfur emissions from the oceans, mainly in the form of dimethyl sulfide (DMS), play an important role in the global sulfur cycle. DMS is oxidized in the atmosphere to sulfuric acid and methanesulfonic acid, two compounds that attract water molecules and promote cloud condensation (2). Important sources of DMS in marine environments include phytoplankton, macroalgae, and coastal vascular plants (e.g., Spartina alterniflora), which produce dimethyl sulfoniopropionate (DMSP), a precursor of DMS (15, 43). Organic sulfur compounds produced in paper industry plants and in wastewater treatment plants also find their way into marine ecosystems. The fate of DMS, DMSP, and other organic sulfur compounds in seawater is therefore important from both biogeochemical and biotechnological perspectives.
The role of marine bacteria in the generation of DMS and other volatile dissolved organic sulfur compounds from DMSP is well recognized (19, 20, 29), and several bacterial strains that use DMSP or DMS as a carbon source have been isolated from seawater (3, 5, 6, 19, 30, 40, 44). Recent studies have also suggested that marine bacteria may use DMSP as a sulfur source, incorporating DMSP sulfur during the synthesis of bacterial amino acids (24). However, despite these important connections to both carbon and sulfur cycling, little is known about the identity of the bacteria that carry out organic sulfur transformations in natural marine environments or about whether DMSP-degrading isolates are representative of the natural DMSP-degrading bacteria in the sea. In part, this reflects a broad lack of knowledge about the identity of ecologically important bacteria in the ocean, as well as problems in culturing bacteria representing many of the major oceanic lineages (4, 7, 32, 35).
One group of marine bacteria that may be a particularly appropriate focus for studies of organic sulfur cycling in the ocean is the Roseobacter group, which is also called the “marine alpha bacteria” (9). This phylogenetic cluster of organisms belonging to the α-subclass of the class Proteobacteria is abundant in coastal seawater in the southeastern United States, where it can account for up to 30% of the 16S ribosomal DNA (rDNA) (9). This group is also well represented in coastal and open-ocean 16S rDNA clone libraries obtained from a number of marine ecosystems (32, 39). Several cultured bacteria in this group have been found to mediate sulfur transformations; these bacteria include a DMSP-degrading bacterium isolated from the Sargasso Sea (29), a sulfite-oxidizing bacterium isolated from the Black Sea (36), and a methanesulfonic acid-utilizing bacterium isolated from Plymouth Sound (14) (Fig. 1). Preliminary characterizations of isolates from southeastern United States seawater likewise have indicated that growth at the expense of DMSP and other organic sulfur compounds might be a common characteristic of the culturable members of this group.
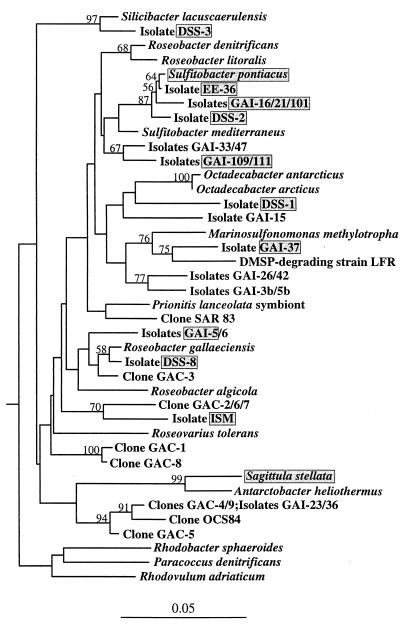
FIG. 1.
Phylogenetic tree for bacteria belonging to the Roseobacter group and representative bacteria not belonging to this group. Sequences whose designations begin with the prefixes GAI and GAC are isolates and 16S rDNA clones, respectively, obtained from southeastern United States seawater. The shaded boxes indicate strains characterized in this study. The tree is based on positions 50 to 410 (E. coli numbering) of the 16S rRNA gene and is unrooted; Hirschia baltica is the outgroup. Bootstrap values greater than 50% are indicated. The bar indicates a Jukes-Cantor distance of 0.05.
In this study we investigated the ability of culturable members of the Roseobacter group to transform DMSP and other sulfur compounds. Our intent was not to provide detailed physiological characterizations of individual isolates, but rather to assess in a broad way the abilities of isolates in this group to carry out transformations known to be important in the cycling of organic and inorganic sulfur in marine environments. We studied sulfur transformations by 15 isolates, including 13 isolates from southeastern United States seawater, 1 isolate from the Caribbean Sea, and 1 isolate from the Black Sea. All of the isolates were able to transform one or more sulfur compounds (including DMSP, DMS, methanethiol, dimethyl sulfoxide [DMSO], and inorganic sulfur); in some cases they did this by novel pathways and even when nonselective methods were used for isolation. A group-specific 16S rRNA-based probe indicated that the relative numbers of uncultured bacteria in the Roseobacter group increased in coastal seawater enriched with DMSP or DMS. This information, coupled with evidence of the abundance of members of this phylogenetic cluster in coastal marine habitats, provides a preliminary basis for linking this distinct taxonomic lineage of marine bacteria with an important biogeochemical function in the coastal ocean.
MATERIALS AND METHODS
Isolation of bacterial strains in the Roseobacter group.
The 15 isolates used in this study were obtained from several sources, although most of the organisms were isolated from coastal Georgia seawater by both selective and nonselective methods. Seven isolates were obtained nonselectively on low-nutrient seawater medium, which contained 10 mg of peptone (Difco Laboratories, Detroit, Mich.) per liter, 5 mg of yeast extract (Difco) per liter, and 1.5% purified agar (Difco) in filter-sterilized Sargasso Sea water that had been aged for more than 1 year in the dark (final salinity, 24‰) (9). The seawater samples used for nonselective isolation were collected in January 1997 (isolates GAI-5, GAI-16, GAI-21, and GAI-37) (9) and December 1997 (isolates GAI-101, GAI-109, and GAI-111). The isolates were determined to be members of the Roseobacter group by screening with probe MALF-1 (5′-GCCGGGGTTTCTTTACCA; positions 488 to 507 [Escherichia coli numbering system]) by using colony hybridization as previously described (9). Affiliations were confirmed by amplifying the 16S rRNA gene by PCR performed with α-subclass-specific primer 19F (31) and MALF-1 and sequencing the gene fragment by using MALF-1 (9). The sequences were aligned with sequences previously assigned to the Roseobacter cluster and representative sequences of members of the α-subclass of the class Proteobacteria. The sequences were also aligned with the most closely related sequence in the GenBank database by using the Genetics Computer Group package and running the program FASTA (8).
Four isolates (DSS-1, DSS-2, DSS-3, and DSS-8) were obtained by selective methods from a marine DMSP enrichment culture. Enrichment flasks were established with 6 liters of filter-sterilized coastal Georgia seawater (salinity, 14‰) amended with 10 μM DMSP hydrochloride (Research Plus Inc., Bayonne, N.J.) and were inoculated with 20 ml of filtered (pore size, 1 μm) seawater. After 2 weeks of incubation, 10-μl portions of the enrichment medium were plated onto low-nutrient seawater medium plates containing 10 μM DMSP, which were incubated in the dark for 15 days. Of the 14 isolates selected at random from the plates, 11 had 16S rRNA sequences consistent with placement in the Roseobacter group (based on amplification with primers 19F and 926R [28] and sequencing with primer 19F), although only four unique partial sequences were found.
Four additional strains used in this study were strain EE-36 (= DSM 11700), Sagittula stellata E-37 (10), Sulfitobacter pontiacus ChLG 10 (= DSM 10014 [37]), and strain ISM (7).
All 15 isolates were maintained on 10% YTSS agar (9). Complete 16S rRNA gene sequences were obtained for DSS-1, DSS-2, DSS-3, DSS-8, EE-36, GAI-5, GAI-21, GAI-109, and ISM (9). Because of phylogenetic affinities to phototrophic Roseobacter species, all of the isolates were tested for pigment production as described by Ledyard et al. (30).
Metabolism of organic sulfur compounds.
Cells were grown overnight in BM medium (1) modified by adding 5 mM glucose and a vitamin solution (10) and by substituting 0.1 mM FeEDTA (Sigma Chemical Co., St. Louis, Mo.) for FeSO4. Two milliliters of cell suspension (absorbance at 540 nm, 0.1 to 0.3) was harvested and placed in a 14-ml crimp top serum bottle sealed with a Teflon-faced septum and containing one of the following compounds: DMS, DMSP, DMSO, methanethiol, 3-methiolpropionate [obtained by alkaline hydrolysis of the methyl ester, methyl(3-methylthio)propionate], or α-ketomethiolbutyrate. The compounds were added at a concentration of 100 μM (DMSO) or 20 μmol per liter of medium (all other compounds).
Formation or degradation of the volatile gases DMS and methanethiol was monitored by using gas chromatography. Headspace gas (100 μl) was removed with a gas-tight syringe for analysis several times during the first day and once each day for the next 3 to 5 days. The amount of headspace gas removed was generally <5% of the total volume, and no corrections were made for this removal. The compounds were quantified by gas chromatography by using a Shimadzu model GC-9A gas chromatograph equipped with a flame photometric detector and a Teflon column (length, 2 m; diameter, 3 mm) filled with 60/80 Carbopak B 1% H3PO4–1.5% XE-60 (Supelco, Bellefonte, Pa.). The oven temperature was 110°C, and the carrier gas was helium at a flow rate of 50 ml min−1. The injector and detector temperatures were 175°C. DMS and methanethiol analyses were standardized by using permeation sources (VCI Metronics, Santa Clara, Calif.) for concentrations ranging from 10 to 1,000 nM and by using standards generated with DMSP after conversion to DMS by NaOH treatment for higher concentrations. Repeated injections of standards and samples yielded coefficients of variation of 2 to 5%.
In every case, the controls were inoculated vials (containing sulfur compounds) in which the organisms had been heat killed (80°C, 1 h). α-Ketomethiolbutyrate proved to be chemically unstable, and small amounts of methanethiol were detected in the killed controls in experiments performed with this compound. Strains that were recorded as positive for the formation of methanethiol from α-ketomethiolbutyrate produced from 20 to 400 times the amount of methanethiol detected in the killed controls.
Degradation of DMS and methanethiol was also tested in the absence of glucose. Cells were grown overnight in BM medium supplemented with 0.04% yeast extract and 0.025% tryptone. A 1-ml sample of the cell suspension was washed twice by centrifugation (8,000 × g, 2 min) in BM medium containing no carbon source. The pellet was resuspended in 2 ml of BM medium supplemented with FeEDTA and vitamins (but no glucose) and was incubated with DMS or methanethiol as described above. The negative control was a preparation to which neither DMS nor methanethiol was added.
Degradation of DMS was also tested by using a rich medium. Cells were grown overnight in BM medium supplemented with 0.04% yeast extract and 0.025% tryptone. A 2-ml sample was transferred to a 14-ml serum bottle and incubated with 20 μmol of DMS per liter of medium. An additional negative control to which no DMS was added was included since all of the cultures released methanethiol when they were grown in a yeast extract-tryptone medium. This did not interfere with the gas chromatographic detection of DMS, however.
Isolates were tested to determine whether they grew on DMSP, acrylate (the product of DMSP cleavage), and glycine betaine (an osmolyte and structural analog of DMSP) as sole carbon sources in liquid medium. The cells were first grown in BM medium supplemented with 0.04% yeast extract and 0.025% tryptone. After turbidity developed, 0.5 ml of each culture was transferred to 10 ml of BM medium containing FeEDTA, a vitamin solution, and 5 mM DMSP, 5 mM acrylate, or 5 mM glycine betaine as the sole carbon source. Samples (0.5 ml) were transferred to fresh medium after 3 to 5 days. A strain was recorded as positive for growth if turbidity persisted after three such transfers. Controls containing no carbon source did not become turbid.
Growth on methanesulfonic acid was tested by using solid BM medium containing FeEDTA, vitamins, 2% agar, and 5 mM methanesulfonic acid. Since isolate GAI-37 and S. stellata E-37 formed colonies on this medium after 2 weeks of incubation at room temperature, these organisms were also tested by using liquid medium containing methanesulfonic acid as described above. GAI-37 was tested to determine whether methanesulfonic acid oxygenase was present as described by Thompson et al. (42) by using extracts of cells that had been grown in a medium containing 10 mM glucose, 10 mM sodium acetate, and 5 mM methanesulfonic acid or in the same medium lacking MgSO4. In each case, 200 ml of medium was inoculated with 10 ml of culture that had been grown in BM medium supplemented with 0.04% yeast extract and 0.025% tryptone.
Transformation of DMS by S. stellata E-37.
Based on evidence that one strain (S. stellata E-37) was capable of oxidizing DMS to DMSO (i.e., DMS was rapidly consumed during the first hour of incubation), we tested this strain to determine whether it oxidized DMS to DMSO and metabolized DMSO further. Twenty milliliters of BM medium containing FeEDTA, vitamins, and 1 mM glucose was added to duplicate 160-ml flasks. The medium was inoculated with 1 ml of glucose medium containing cells that had been grown overnight. The flasks were sealed with Teflon-faced septa and aluminum seals, and DMS or DMSO was added to a concentration of 20 μmol per liter of medium. Noninoculated controls with and without added DMS or DMSO were run in parallel. All cultures were incubated in the dark.
To detect DMSO in the culture medium, we used a procedure adapted from a procedure of Kiene and Gerard (22). Two-milliliter samples of medium were removed from the flasks immediately after inoculation and on each subsequent day for 5 days. The samples were centrifuged to pellet the cells, and the supernatant was sparged with helium to remove the DMS. One milliliter of sparged culture medium was added to a 14-ml serum vial containing an equal volume of TiCl3. The vial was quickly sealed and heated at 50°C for 1 h, and the DMS that evolved was measured by gas chromatography as described above. Standards were prepared by adding known amounts of DMSO and treating vials in the same manner.
Sulfur incorporation by strain DSS-3.
We investigated the incorporation of sulfur from DMSP or methanethiol into the protein fraction of strain DSS-3. Strain DSS-3 was used as the model organism in these experiments because it formed methanethiol from a number of organic sulfur compounds and grew well in the laboratory. Two-milliliter subsamples of an overnight culture of DSS-3 in BM medium containing 5 mM glucose (absorbance at 540 nm, 0.2) were placed in a series of sterile 14-ml serum vials. These preparations were subsequently treated with four different amounts of unlabeled DMSP (0, 0.1, 1, and 10 μM) or three different amounts of methanethiol (0, 0.26, and 0.92 μM). For the DMSP-treated series, an additional 2.5 nM [35S]DMSP (6,000 dpm) was added to each vial; for the methanethiol-treated series an additional 29 nmol of [35S]methanethiol (30,000 dpm) per liter of medium was added (23, 25). Samples were incubated in the dark at 25°C for 6 h, and tests showed that during this time all of the added DMSP and methanethiol were degraded. The partitioning of 35S after incubation was then assessed as described below.
The methanethiol and DMS concentrations in the headspaces of the vials were monitored as described above approximately every hour for 6 h. At the end of the 6-h incubation period, the radioactivity incorporated into the macromolecular fraction of cells were determined by filtering 0.7 ml of culture from each vial through a 25-mm-diameter 0.2-μm-pore-size filter (Poretics Magna Nylon, Livermore, Calif.). The filters were rinsed three times with 0.5 ml of sterile medium, and then 5 ml of ice-cold 5% trichloroacetic acid (TCA) was added on top of each filter. After 5 min, the TCA was vacuumed through the filters, and the amounts of radioactivity remaining on the filters were determined by liquid scintillation methods. The DMSP or methanethiol sulfur assimilated into the TCA-insoluble fraction was expected to be primarily in the form of methionine residues in proteins (24).
For parallel non-TCA-treated samples, the filtrates were assayed to determine their total dissolved nonvolatile 35S and [35S]sulfate (a subset of the dissolved nonvolatile fraction) contents. Each filtrate was sparged with helium for 3 to 4 min to remove any 35S-labeled volatile compounds (DMS or methanethiol). Samples (0.8 ml) of the sparged filtrates were mixed with either 0.1 ml of water or 0.1 ml of 1 M BaCl2 in microcentrifuge tubes. The tubes were centrifuged for 3 min at 13,000 × g to sediment the BaSO4 precipitate. The amounts of radioactivity in the supernatants of samples that were mixed with water represented the total dissolved nonvolatile sulfur (corrected for dilution by water). The amounts of radioactivity in the supernatants of samples that were mixed with BaCl2 represented the total dissolved nonvolatile 35S minus 35SO42−. The amount of 35S in sulfate was calculated by difference.
The amounts of radioactivity in the headspaces (representing 35S-labeled volatile compounds) were determined by removing 5-ml portions of the headspace gases in incubation vials with a gas-tight syringe and injecting these samples into sealed vials which contained suspended cups (Kontes, Vineland, N.J.) filled with H2O2-soaked wicks (23). After 6 h the wicks were removed and counted by using Ecolume scintillation cocktail (ICN Biomedicals, Costa Mesa, Calif.).
Oxidation of inorganic sulfur compounds.
Isolates were screened for sulfite oxidation on solid BM medium containing FeEDTA, vitamins, 10 mM glucose, and 10 mM acetate to which a filter-sterilized solution of Na2SO3 (final concentration, 10 mM) had been added after autoclaving. Negative controls were prepared by using the same medium lacking Na2SO3. After 2 days of incubation, the plates were flooded with a solution containing 1 g of Ellman’s reagent [5,5′-dithiobis(2-nitrobenzoic acid)] per liter in 50 mM potassium phosphate buffer (pH 7.0). Ellman’s reagent reacts with sulfite and forms a bright yellow product. Colorless zones on the plates, therefore, indicated that sulfite was not present.
Ellman’s reagent was also used to monitor the oxidation of sulfite in liquid medium. The isolates were grown in 10 ml of BM medium containing FeEDTA, vitamins, 10 mM glucose, and 10 mM acetate to which Na2SO3 was added at a concentration of 5 mM. Samples (1 ml) were removed every day for 5 days and centrifuged to pellet the cells. A 20-μl portion of Ellman’s reagent (0.5 g/liter in 50 mM potassium phosphate buffer [pH 7.0]) was added to 350 μl of supernatant. After 30 min of incubation to allow for color development, the absorbance at 412 nm was measured.
Sulfite oxidase activity was assayed in cell extracts by using ferricyanide as an artificial electron acceptor (10, 17). Isolates were grown in BM medium containing FeEDTA, vitamins, 10 mM acetate, and 10 mM Na2SO3 or 10 mM Na2S2O3.
Enrichment cultures.
The relative abundance of uncultured bacteria belonging to the Roseobacter group in seawater enriched with 10 μM DMSP was assessed over time by using a 16S rRNA group-specific probe. The protocol used for the first enrichment experiment is described above. After the 2-week enrichment period, the enriched seawater was filtered through 47-mm-diameter, 0.2-μm-pore-size Nuclepore filters, and the DNA was extracted from the filters (see below). A 15-liter portion of the seawater that served as the inoculum was also filtered in order to extract DNA at the time that the enrichment was established; the water sample was prefiltered through 5- and 1-μm-pore-size filters, and then cells were collected on a 293-mm-diameter, 0.2-μm-pore-size polycarbonate filter. The contribution of bacteria in the Roseobacter group was determined by using a dot blot hybridization procedure with probe MALF-1, in which the hybridization signal from community DNA was compared with the hybridization signal from a positive control. The percentage was normalized based on binding of community DNA and positive control DNA to a universal probe (9). The bacterial cell numbers at the beginning and at the end of the enrichment procedure were determined by acridine orange direct counting (13).
The second seawater enrichment culture was set up by using flasks containing 2 liters of filtered (pore size, 3 μm) seawater collected from coastal Georgia (Skidaway River; salinity, 26‰). The medium was amended with 5 μM N as NH4NO3 and 1 μM P as KH2PO4. One of the following substrates was added to duplicate flasks at a concentration of 10 μmol per liter of medium: DMSP, DMS, Na2SO3, or glucose. The control contained seawater plus inorganic nutrient amendments without any of the substrates. The seawater was incubated for 1 week at 100 rpm in the dark at room temperature. Bacterial cells were filtered to determine the cell number and the contribution of the Roseobacter group to the total 16S rDNA (expressed as a percentage).
Community DNA samples.
The abundance of bacteria in the Roseobacter group in several coastal marine habitats was determined by using the MALF-1 group-specific probe. Samples were collected from the top 1 cm of intertidal sediments at low tide in the Altamaha, Satilla, and St. Marys estuaries in the southeastern United States. Sediment DNA was extracted from approximately 1 g of sediment by using the method of Zhou et al. (46). The DNA was further purified by using a Wizard DNA cleanup minicolumn (Promega Biological Research Products, Madison, Wis.).
Samples of S. alterniflora detritus were collected from the Altamaha River and Satilla River estuaries. The plant samples were ground with a blender, and DNA was then extracted as described below for water samples.
Coastal seawater samples were collected from the Atlantic coast of the southeastern United States and the Gulf of Mexico (Dauphin Island, Ala.). The samples were prefiltered immediately after collection through 5- and 1-μm-pore-size filters, and the cells were collected on 0.2-μm-pore-size polycarbonate filters. The filters were stored at −20°C, and the DNA was extracted as previously described (9).
Nucleotide sequence accession numbers.
The 16S rRNA sequences of the new isolates described in this study have been assigned the following GenBank numbers: DSS-1, AF098492; DSS-2, AF098490; DSS-3, AF098491; DSS-8, AF098493; GAI-109 and GAI-111, AF098494; and ISM, AF098495. The GenBank accession numbers for the sequences of the other isolates that were characterized in this study are as follows: GAI-5, AF007256; GAI-16, GAI-21, and GAI-101, AF007257; GAI-37, AF007260; EE-36, AF007254; S. stellata E-37, U58356; and S. pontiacus ChLG 10, Y13155. The GenBank accession numbers for the sequences of the organisms used to construct the phylogenetic trees are as follows: clone OCS84, U78943; clone SAR83, M63810; Antarctobacter heliothermus, Y11552; DMSP-degrading bacterium, L15345; Hirschia baltica, X52909; Marinosulfonomonas methylotropha, U62894; Octadecabacter antarcticus, U14583; Octadecabacter arcticus, U73725; Paracoccus denitrificans, X69159; Prionitis lanceolata gall symbiont, U37762; Rhodobacter capsulatus, D16427; Rhodovulum adriaticum, D16418; Roseobacter algicola, X78313; Roseobacter denitrificans, M96746; Roseobacter gallaeciensis, Y13244; Roseobacter litoralis, X78312; Roseovarius tolerans, Y11551; Silicibacter lacuscaerulensis, U77644; and Sulfitobacter mediterraneus, Y17387.
RESULTS
A high percentage of bacteria isolated from southeastern United States seawater on a nonselective, low-nutrient medium were members of the Roseobacter cluster. More than 40% of the isolates obtained from the January 1997 seawater sample belonged to this group based on dot blot hybridization results obtained with group-specific probe MALF-1 (9), while 57% of the isolates obtained from the December 1997 seawater sample belonged to this group based on colony hybridization results. The seven isolates in the Roseobacter group obtained from the nonselective plates that were characterized further in this study (GAI-5, GAI-16, GAI-21, GAI-37, GAI-101, GAI-109, and GAI-111) were chosen because they represented the phylogenetic diversity encompassed by the isolates (Fig. 1) and were easily maintained in the laboratory. In cases in which partial 16S rRNA gene sequences of isolates were identical (e.g., isolates GAI-16, GAI-21, and GAI-101 and isolates GAI-109 and GAI-111), substantial differences in colony morphology were evident.
Likewise, a high percentage of the bacteria isolated from the DMSP enrichment cultures (80%; 11 of 14 isolates) were found to belong to the Roseobacter cluster based on partial screening of the 16S rRNA gene. Only four unique partial sequences were identified among the 11 isolates, and one representative of each type (strains DSS-1, DSS-2, DSS-3, and DSS-8) was chosen for further study.
The remaining isolates chosen for characterization included two isolates obtained previously from seawater enrichment cultures prepared with pulp industry by-products (strain EE-36 and S. stellata E-37) (9), one isolate determined to be a dominant component of the bacterial community in seawater samples obtained from the Caribbean Sea (strain ISM) (7), and one isolate that had been well characterized previously with regard to inorganic sulfur metabolism (S. pontiacus ChLG 10) (36, 37).
Degradation of organic sulfur compounds.
All 15 strains were able to degrade DMSP when they were growing in glucose medium, as shown by the production of DMS in the headspaces of the vials (Fig. 2). DMS production began during the first hour of incubation in many cases, and all but three isolates converted 20 to 60% of the DMSP to DMS in the first 24 h of incubation. The three exceptions (strains GAI-109 and GAI-111 and S. stellata E-37) accumulated only 1 to 3% of the DMSP although independent tests performed with stationary-phase cultures revealed that the small amounts that accumulated were due to the rapid consumption of DMS that occurred simultaneously with DMS production (data not shown). There was evidence that all four strains isolated from the DMSP enrichment cultures (DSS-1, DSS-2, DSS-3, and DSS-8) further metabolized the accumulating DMS by 24 h. In the heat-killed controls, less than 0.2% of the DMSP was converted to DMS in 24 h.
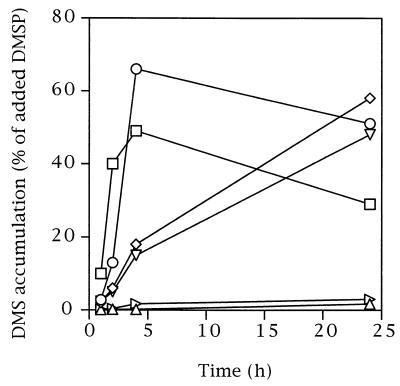
FIG. 2.
Time course for production of DMS from 20 μM DMSP for six representative isolates. The pattern exhibited by DSS-1 (○) and DSS-3 (□) was also exhibited by DSS-2 and DSS-8; the pattern exhibited by GAI-16 (◊) and GAI-37 (▿) was also exhibited by GAI-5, GAI-21, GAI-101, ISM, and S. pontiacus ChLG 10; and the pattern exhibited by GAI-109 (▵) and S. stellata E-37 (▹) was also exhibited by GAI-111. Independent tests showed that the lack of DMS accumulation in GAI-109, GAI-111, and S. stellata E-37 cultures was due to the rapid consumption of DMS that occurred simultaneously with DMS production.
Five strains were able to reduce a significant percentage of DMSO when they were growing in glucose medium. Isolates DSS-1, DSS-2, DSS-3, DSS-8, and GAI-5 converted 20 to 100% of the DMSO to DMS during the first 24 h. The remaining isolates converted only 0.03 to 3% of the DMSO to DMS, although these values were significantly greater than the values obtained with the heat-killed controls.
We also investigated the ability of the isolates to degrade and grow on DMSP when it was presented as a sole carbon source (i.e., in the absence of glucose or other substrate). The majority of the isolates grew on DMSP as a sole carbon source; the exceptions were GAI-21, GAI-101, GAI-109, GAI-111, and ISM (Table 1). A subset of the isolates that grew on DMSP also grew on acrylate, the product of DMSP cleavage. Glycine betaine, a nitrogen analog of DMSP which is also produced as an osmolyte, was used as a sole carbon source by eight of the strains (Table 1).
TABLE 1.
Growth of isolates on DMSP, glycine betaine, or acrylate (5 mM) as a sole carbon source in liquid medium
| Straina | Growth on:
|
||
|---|---|---|---|
| DMSP | Glycine betaine | Acrylate | |
| DSS-1 | + | + | + |
| DSS-2 | + | + | + |
| DSS-3 | + | + | − |
| DSS-8 | + | + | + |
| GAI-5 | + | − | + |
| GAI-16 | + | − | − |
| GAI-21 | − | − | − |
| GAI-37 | + | − | − |
| GAI-101 | − | − | − |
| GAI-109 | − | + | − |
| GAI-111 | − | − | − |
| EE-36 | + | + | − |
| S. stellata E-37 | + | + | + |
| ISM | − | + | − |
| S. pontiacus ChLG 10 | + | − | + |
Seven strains were capable of degrading DMS when they were grown in glucose medium (Table 2). S. stellata E-37 consumed 100% of the added DMS (20 μmol per liter of medium) during the first 24 h of incubation. Degradation of DMS by the other strains (DSS-1, DSS-2, DSS-3, DSS-8, GAI-5, and GAI-109) was considerably slower, but 50 to 90% of the DMS was consumed within 48 h. With the exception of S. stellata E-37, these strains were also capable of degrading methanethiol (20 μmol per liter of medium) when they were grown in glucose medium. One of these organisms (DSS-3) degraded more than 40% of the methanethiol (compared to abiotic controls) after just 4 h of incubation.
TABLE 2.
Degradation of DMS and methanethiol (20 μmol per liter of medium) by isolates under different growth conditions
| Strain | % Degradationa
|
||||
|---|---|---|---|---|---|
| DMS
|
Methanethiol
|
||||
| Glucose (48 h) | No carbon (48 h) | Yeast extract-tryptone (48 h) | Glucose (24 h) | No carbon (48 h) | |
| DSS-1 | 70 | 4 | 11 | 100 | 26 |
| DSS-2 | 59 | 16 | 100 | 63 | 39 |
| DSS-3 | 55 | 17 | 100 | 100 | 56 |
| DSS-8 | 48 | 6 | 40 | 100 | 0 |
| GAI-5 | 53 | 0 | 24 | 100 | 0 |
| GAI-16 | 0 | 0 | 0 | 0 | 0 |
| GAI-21 | 0 | 0 | 0 | 0 | 0 |
| GAI-37 | 0 | 0 | 0 | 0 | 0 |
| GAI-101 | 0 | 0 | 0 | 0 | 0 |
| GAI-109 | 90 | 17 | 29 | 100 | 71 |
| GAI-111 | 0 | 0 | 0 | 0 | 57 |
| EE-36 | 0 | 0 | 19 | 0 | 28 |
| S. stellata E-37 | 100 | 100 | 100 | 0 | 25 |
| ISM | 0 | 0 | 32 | 0 | 58 |
| S. pontiacus ChLG 10 | 0 | 4 | 10 | 0 | 0 |
Cells were grown in medium containing glucose (5 mM), suspended in a carbon-free medium, or grown in a yeast extract-tryptone medium. DMS and methanethiol contents in the headspace gas were measured by gas chromatography.
Resuspending cells in a basal medium without glucose (starving cells) or growing cells in a rich medium (containing yeast extract plus tryptone) changed the results slightly (Table 2), but a number of strains could rapidly degrade DMS or methanethiol under all conditions. With the seven isolates that degraded DMS in glucose medium, methanethiol (the expected product of DMS degradation) was not detected in the headspaces of DSS-3, GAI-5, GAI-109, and S. stellata E-37 cultures. This may have been due to rapid subsequent degradation of methanethiol, although in S. stellata E-37 oxidation of DMS to DMSO rather than to methanethiol is the likely reason (see below).
The isolates were also grown on glucose medium and tested to determine whether they released methanethiol from several potential methiol donor compounds (Table 3). Five isolates (DSS-3, DSS-8, GAI-5, GAI-109, and ISM) released methanethiol into the headspace when DMSP was added, suggesting that DMSP may be degraded by two pathways (one pathway through DMS and one pathway through methanethiol) in these organisms (Fig. 3). Isolates DSS-1, DSS-2, and DSS-8 released methanethiol when either DMS or DMSO was added. 3-Methiolpropionate and α-ketomethiolbutyrate were also donors of methiol groups for several isolates (Table 3).
TABLE 3.
Formation of methanethiol by bacteria in the Roseobacter group in the presence of several methiol donor compounds, each at a concentration of 20 μmol per liter of medium
| Strain | Formation of methanethiol in the presence ofa:
|
||||
|---|---|---|---|---|---|
| DMSP | DMS | DMSO | 3-Methiol-propionate | α-Ketomethiol-butyrate | |
| DSS-1 | − | + | + | + | − |
| DSS-2 | − | + | + | + | + |
| DSS-3 | + | − | − | + | + |
| DSS-8 | + | + | + | + | + |
| GAI-5 | + | − | − | + | + |
| GAI-16 | − | − | − | − | − |
| GAI-21 | − | − | − | − | − |
| GAI-37 | − | − | − | + | − |
| GAI-101 | − | − | − | − | − |
| GAI-109 | + | − | − | + | − |
| GAI-111 | − | − | − | − | − |
| EE-36 | − | − | − | + | + |
| S. stellata E-37 | − | − | − | + | + |
| ISM | + | − | − | + | − |
| S. pontiacus ChLG 10 | − | − | − | − | − |
Methanethiol in the headspace gas was detected by gas chromatography.
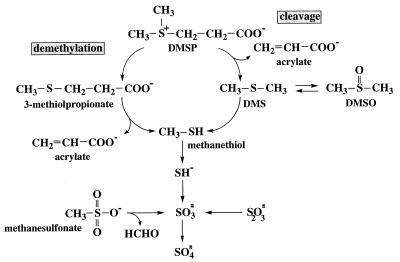
FIG. 3.
Pathways for degradation of organic sulfur compounds in members of the Roseobacter group. Most isolates were able to cleave DMSP to DMS, whereas five isolates also demethylated DMSP and produced methanethiol. Methanethiol was assimilated into cell material or was further degraded to inorganic sulfur compounds that could be enzymatically oxidized.
Whereas five isolates formed DMS from 0.1 mM DMSO during growth on glucose, S. stellata E-37 was the only strain that was able to reoxidize the DMS that was produced back to DMSO (trace levels of DMS [up to 0.1 μmol per liter of medium] were produced transiently during the first hour of incubation [data not shown]). In a medium containing 20 μmol of DMS per liter of medium, S. stellata E-37 completely oxidized DMS to DMSO within 1 h in the dark and then did not further degrade the DMSO.
Isolate GAI-37 and S. stellata E-37 formed colonies on plates containing methanesulfonic acid as the sole carbon source, although they did not produce turbidity in a liquid version of this medium. When methanesulfonic acid oxygenase was assayed with GAI-37 cells that had been grown on glucose, acetate, and methanesulfonic acid (without an alternative source of sulfate in the medium), methanesulfonic acid oxygenase activity was found (20 nmol of NADH mg of protein−1 min−1). S. stellata E-37 did not grow well enough in this medium to assay for enzyme activity.
Oxidation of inorganic sulfur compounds.
Since reduced inorganic sulfur compounds can be the end products of degradation of organic sulfur compounds, the isolates were tested to determine whether they oxidized sulfite and thiosulfate. When grown on a solid medium containing glucose, acetate, and sulfite, isolates DSS-1, DSS-2, DSS-3, DSS-8, and EE-36, S. stellata E-37, and S. pontiacus ChLG 10 converted sulfite to a compound that did not react with Ellman’s reagent (probably sulfate). When grown with glucose, acetate, and thiosulfate, most of these isolates (all except DSS-3 and EE-36) converted thiosulfate. Similar experiments performed in liquid medium containing glucose, acetate, and sulfite produced essentially the same results, except that DSS-1 did not show any evidence of sulfite oxidation even after 5 days of incubation.
The results of sulfite oxidase assays performed with isolates in the presence of sulfite or thiosulfate confirmed the results obtained by screening organisms on solid and liquid media. The highest specific activities were obtained with isolates DSS-3 and EE-36, and the rates were comparable to those found for S. pontiacus ChLG 10 and other strains studied previously (Table 4) (37). Isolate DSS-2 exhibited no activity in sulfite medium despite the positive screening results but did exhibit activity when it was grown in thiosulfate medium.
TABLE 4.
Sulfite oxidase activities of isolates
| Strain | Sulfite oxidase activity (nmol mg of protein−1 min−1) in the presence of a:
|
|
|---|---|---|
| Sulfite | Thiosulfate | |
| DSS-1 | 46 | 167 |
| DSS-2 | 0 | 88 |
| DSS-3 | 455 | 1,151 |
| DSS-8 | 62 | 155 |
| GAI-5 | 0 | 12 |
| GAI-37 | 0 | 0 |
| EE-36 | 882 | 960 |
| S. stellata E-37 | 0 | NDb |
| ISM | 15 | 78 |
| S. pontiacus ChLG 10 | 316 | 1,290 |
Strains were grown in medium containing acetate plus sulfite or medium containing acetate plus thiosulfate (each at a concentration of 10 mM).
ND, not determined.
Sulfur incorporation.
Direct incorporation of 35S from [35S]DMSP and [35S]methanethiol into cell material was studied by using strain DSS-3 as a representative of the five organisms that were found to convert DMSP to methanethiol (Table 3). Other work has shown that only organisms with the DMSP demethylation pathway (i.e., methanethiol-producing organisms) are able to incorporate sulfur from DMSP into cell biomass (24). 35S from either methanethiol or DMSP was assimilated by cells at all of the concentrations tested, and at lower concentrations assimilation was the primary fate of 35S. Essentially all of the 35S assimilated into cells from [35S]DMSP was found in the TCA-insoluble fraction, suggesting that the DMSP sulfur was incorporated into proteins (Table 5). Higher concentrations of DMSP resulted in a greater fraction of the added 35S accumulating in volatile degradation products than occurred with lower concentrations (Table 5), and most of the volatile sulfur in this case was in the form of DMS rather than methanethiol. Only a small amount of the 35S associated with the dissolved, nonvolatile fraction was present as sulfate.
TABLE 5.
Fate of 35S in DMSP and methanethiol in strain DSS-3, expressed as percentages of the [35S]DMSP addeda
| Compound | Concn (nmol per liter of medium) | % of 35S in:
|
% Recovery | ||||
|---|---|---|---|---|---|---|---|
| Volatile compounds | Dissolved nonvolatile compounds | 35SO42− | Filterable material (cells) | TCA-insoluble material | |||
| Methanethiol | 29 | 0 | 7 | 2.2 | 92 | 90 | 99 |
| 290 | 0 | 17 | 2.0 | 77 | 78 | 94 | |
| 950 | 0 | 29 | 1.9 | 67 | 67 | 96 | |
| DMSP | 2.5 | 2 | 12 | 1.3 | 84 | 75 | 98 |
| 100 | 2 | 13 | 1.1 | 78 | 81 | 93 | |
| 1,000 | 4 | 37 | 2.5 | 49 | 56 | 90 | |
| 10,000 | 38 | 38 | 4.3 | 15 | 12 | 91 | |
Levels of recovery were calculated based on the sum of 35S recovered as volatile compounds, dissolved nonvolatile compounds and filterable material (cells). A subset of the dissolved nonvolatile fraction was present as sulfate, and a subset of the filterable fraction was TCA-insoluble material; levels of recovery in these categories were also expressed as percentages of the [35S]DMSP added. Compounds were added to 2-ml portions of DSS-3 cultures that had been grown to an absorbance at 540 nm of 0.2 in seawater medium containing 5 mM glucose.
Pigment production.
None of the isolates produced bacteriochlorophyll a. Only GAI-5 and GAI-37 produced an absorption spectrum in methanol extracts typical of carotenoids (9).
Enrichment of the Roseobacter group.
Dot blot hybridizations performed with the MALF-1 probe and DNA from the first seawater enrichment culture indicated that the percentage of bacteria in the Roseobacter group increased following the addition of DMSP to seawater. The percentage of the 16S rRNA of bacteria belonging to this group increased from 11.5% (standard deviation, 0.4%) of the community 16S rDNA to 30.1% (standard deviation, 0.1%) after 2 weeks. The numbers of bacteria increased two- to threefold during the enrichment period based on acridine orange direct counts.
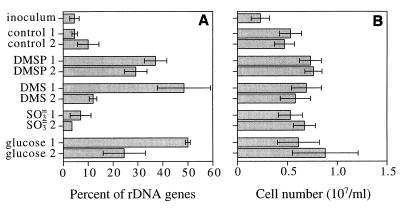
Dot blot hybridizations performed with the MALF-1 probe and DNA from the second seawater enrichment study likewise showed that the contributions of the 16S rDNA from members of the Roseobacter group to the total 16S rDNA pool increased when DMS, DMSP, and glucose were added to seawater. The percentage of 16S rDNA of members of the Roseobacter group increased from 4.5% in the inoculum to as much as 50% (in one DMS culture and one glucose enrichment culture) (Fig. 4). Seawater enriched with sulfite had approximately the same percentage of 16S rDNA of organisms belonging to the Roseobacter group as the controls (unamended seawater) had, and the final contributions were 10% or less (Fig. 4). The numbers of bacteria also increased two- to threefold during the enrichment procedure.
FIG. 4.
Contribution of bacteria in the Roseobacter group to enrichment community 16S rDNA, based on results of hybridizations with probe MALF-1 (A) and cell numbers in enrichment cultures as determined by acridine orange direct counting (B). Enrichment cultures were prepared in duplicate. The error bars indicate ±1 standard deviation.
Quantification of the Roseobacter group in marine habitats.
The Roseobacter group accounted for 3 to 11% of the 16S rDNA in coastal sediment communities (Table 6) and 2 to 24% of the 16S rDNA of communities associated with coastal plant detritus. The Roseobacter group was also an important component of the bacterial community in seawater from the southeastern United States (0 to 26% of the 16S rDNA pool) and coastal Gulf of Mexico (5 to 11%).
TABLE 6.
Contributions of bacteria in the Roseobacter group to 16S rDNA in coastal seawater and sediments and associated with decaying S. alterniflora
| Site | Sampling date (mo/yr) | % Contribution of Roseobacter group 16S rDNA | SD (%) | Salinity (‰) |
|---|---|---|---|---|
| Southeastern United States sediments | ||||
| Altamaha River | 12/97 | 8.1 | 1.8 | 16 |
| Altamaha River | 12/97 | 7.2 | 1.5 | 14 |
| Satilla River | 12/97 | 8.5 | 4.7 | 9 |
| Satilla River | 12/97 | 10.5 | 0.2 | 16 |
| St. Marys River pulp mill | 9/97 | 3.1 | 1.0 | 26 |
| Southeastern United States plant detritus | ||||
| Altamaha River | 12/97 | 7.2 | 7.4 | 16 |
| Altamaha River | 12/97 | 8.5 | 1.9 | 14 |
| Satilla River | 12/97 | 24.2 | 0.3 | 20 |
| Satilla River | 12/97 | 2.6 | 0.1 | 16 |
| Satilla River | 12/97 | 1.9 | 1.1 | 9 |
| Satilla River | 12/97 | 5.3 | 3.5 | 19 |
| Southeastern United States water column | ||||
| Skidaway River | 1/98 | 11.5 | 0.5 | 14 |
| Skidaway River | 6/97 | 4.5 | 1.9 | 26 |
| Jekyll Island | 4/97 | 4.2 | 0.4 | 28 |
| Satilla R. transect, 0 km | 12/97 | 16.5 | 7.8 | 22 |
| Satilla R. transect, 6 km | 12/97 | 25.5 | 0.7 | 27 |
| Satilla R. transect, 12 km | 12/97 | 11.3 | 1.0 | 30 |
| St. Marys River pulp mill | 9/97 | 0 | 0 | 26 |
| Gulf Coast water column | ||||
| Dauphin Island | 3/98 | 5.1 | 0.3 | 17 |
| Dauphin Island | 3/98 | 9.2 | 0.3 | 20 |
| Dauphin Island | 3/98 | 8.7 | 0.2 | 22 |
| Dauphin Island | 3/98 | 11.2 | 0.4 | 24 |
| Dauphin Island | 3/98 | 7.0 | 0.1 | 30 |
DISCUSSION
The abundance of the Roseobacter group in seawater is consistent with the results of a previous survey of the southeastern United States coast (9), as well as with the 16S rRNA clone libraries obtained from a number of coastal and open-ocean environments (32, 39). Members of the Roseobacter group have also been readily cultured from southeastern United States seawater (9 and this study), the Caribbean Sea (strain ISM) (7), the Black Sea (S. pontiacus) (36), the Mediterranean Sea (Sulfitobacter mediterraneus) (34), the Sargasso Sea (30), Plymouth Sound (14), and Antarctic Sea ice (Octadecabacter spp.) (11). 16S rDNA from members of the Roseobacter group was abundant in aerobic sediments and decaying plant material from coastal salt marshes, indicating that this group is not restricted to the water column. To our knowledge, members of the Roseobacter group are exclusively marine except for one moderate halophile that was isolated from a salt pond in Iceland (33) and members of two genera that were isolated from a hypersaline lake in Antarctica (26, 27).
The numerical importance of the Roseobacter group in many coastal marine environments strongly suggests that this phylogenetic group plays a major role in one or more central biogeochemical processes. Identifying this role, however, is an extremely difficult challenge, which is hampered by the low culturability of ecologically relevant bacteria and the difficulty of reproducing environmental conditions in the laboratory. While we acknowledge the potential pitfalls of moving from data from laboratory studies of culturable bacteria to conclusions about biogeochemical functions in nature, we believe that several lines of evidence suggest that the Roseobacter group plays an important role in the marine sulfur cycle. First, all of the bacteria in the Roseobacter group cultured from southeastern United States coastal environments exhibited significant abilities to transform sulfur compounds, despite the fact that many of the organisms were isolated nonselectively. Second, the metabolic capabilities of the strains isolated previously from other marine environments also suggest that the abilities of members of this phylogenetic cluster to transform organic and inorganic sulfur compounds are widespread (14, 30, 36). Third, uncultured bacteria in the Roseobacter group were readily enriched in coastal seawater by adding DMS and DMSP (but not sulfite). Thus, the group represents a numerically important lineage of marine bacteria which respond to increased concentrations of organic sulfur compounds in seawater and includes culturable members that are capable of mediating many key processes in the organic sulfur cycle.
All 15 isolates characterized in this study possessed DMSP lyase type activity, since DMS was produced from DMSP, and 5 isolates were able to reduce DMSO to DMS. Beyond this, subsets of isolates grew on DMSP, acrylate, or glycine betaine as a sole carbon source or degraded DMS or methanethiol. One isolate (S. stellata E-37) rapidly oxidized DMS to DMSO, apparently without further degradation of DMSO. The metabolic abilities of the isolates in many cases mimicked activities known to occur in natural seawater communities. For example, DMSP is degraded to DMS in seawater, and methanethiol is formed from both DMS and DMSP (15, 21, 43). DMSO is produced from DMS in seawater, primarily by photochemical reactions (12, 18), although organisms like S. stellata E-37 might also carry out this process.
Inorganic sulfur compounds are known to be produced during degradation of methanesulfonic acid (14), DMSP (24), and DMS by bacterial isolates. Eight of the strains characterized in this study were able to further oxidize inorganic intermediates in the form of sulfite or thiosulfate; these organisms included three isolates belonging to the Sulfitobacter cluster (strain EE-36, S. pontiacus ChLG 10, and strain DSS-2) and all of the isolates obtained from the DMSP enrichment cultures (Table 4). The oxidase activities of the isolates screened in this study were comparable to activities measured previously for S. pontiacus ChLG 10 and related sulfite oxidizers (37). Bacteria that are able to degrade DMS and DMSP in seawater could gain an energetic advantage by also deriving energy from inorganic intermediates produced during the degradation (38). For example, Johnston et al. (16) demonstrated that bacteria that degrade alkyl and aryl sulfonated compounds also exhibit sulfite oxidase activity and may derive energy from this reaction. Recent studies have shown that radiolabelled sulfur added to natural seawater in an organic form (DMSP, DMS, or methanethiol) can be recovered in a matter of hours as sulfate (24), suggesting that this pathway may indeed operate in seawater communities. Alternatively, bacteria may obtain energy from the oxidation of inorganic sulfur compounds in the absence of organic sulfur sources. S. pontiacus was isolated from the oxic-anoxic interface in the Black Sea, where reduced inorganic sulfur compounds derived from anaerobic metabolism may provide a ready source of energy via sulfite oxidation (36).
Previous studies of DMSP-degrading isolates and natural bacterial assemblages revealed two pathways for the degradation of DMSP by marine bacteria. The cleavage pathway involves splitting of the DMSP molecule into acrylate and DMS and may be particularly important in mediating organic sulfur emission into the atmosphere. The demethylation pathway involves the removal of a methyl group from DMSP to produce 3-methiolpropionate, which is then cleaved to methanethiol and probably acrylate or propionate (Fig. 3) (41). We obtained evidence that five isolates (DSS-3, DSS-8, GAI-5, GAI-109, and ISM) have both the demethylation and cleavage pathways. Visscher et al. (45) obtained evidence that there are two nonoverlapping groups of DMSP-degrading bacteria in the Caribbean Sea (the members of one group are able to cleave DMSP exclusively, and the members of the other group are able to demethylate exclusively), but to our knowledge this is the first report of marine bacteria that possess both pathways.
When the strains characterized in this study were grown on a number of compounds that could potentially serve as methiol donors (DMS, DMSP, DMSO, 3-methiolpropionate, and α-ketomethiolbutyrate) (Table 3), most isolates were able to form methanethiol from one or more of the compounds. The observation that several bacteria belonging to the Roseobacter group can demethylate DMSP and convert it to methanethiol is significant because this transformation seems to dominate DMSP metabolism in seawater (21) and may compete with the biogeochemically important transformation of DMS, even within the same organism (e.g., strain DSS-3). The potential role of methanethiol as a key intermediate in the assimilation of dissolved organic sulfur into methionine (through the activity of cystethionine γ-synthetase) has been described recently (24). We found that isolate DSS-3, one of the five strains in which the demethylation pathway was determined to be present (Table 3), incorporated the majority of radiolabelled sulfur (provided as either DMSP or methanethiol) into a TCA-insoluble fraction, most likely protein, after just a few hours of incubation.
It is noteworthy that the fraction of metabolized DMSP that was incorporated into cell mass decreased as the DMSP concentration increased (Table 5), while the fraction converted to volatile compounds (mostly DMS) increased. We speculate that this result may reflect saturation of the reduced sulfur demand by cells at the higher DMSP concentrations. The highest concentration of DMSP that we used in these experiments (10 μM) was low relative to the concentrations that other workers have used in metabolic or growth studies of DMSP-utilizing bacteria (3, 5, 30). It is possible that the demethylation pathway, which seems to be associated with sulfur assimilation, could be missed if very high DMSP substrate concentrations are used.
Additional detailed studies will be needed to assess the energetics of the sulfur transformations under environmentally relevant conditions. It is not known whether the isolates derive energy from all of the transformations described here, although S. pontiacus is able to gain energy from oxidation of inorganic sulfur compounds (36) and several isolates were capable of growing on DMSP as a sole energy source. Utilization of organic sulfur compounds as sources of reduced sulfur could also provide an energetic advantage (24). In the natural settings of the marine water column, organic sulfur compounds like those tested here are unlikely to be the major growth-supporting substrates utilized by bacteria. It is clear, however, that the bacteria characterized in this study carry the genetic material necessary to mediate a number of biogeochemically important transformations of sulfur compounds.
In aerobic marine environments, most of the reduced sulfur dissolved in seawater is expected to be in the organic form. DMSP concentrations range from 3 to 200 nM in oceanic and coastal seawater, and DMS concentrations are typically 1 to 20 nM; occasionally the concentrations are higher (15, 43). Reduced inorganic sulfur compounds produced by the activity of sulfate-reducing bacteria can also be found at the oxic-anoxic interface in marine sediments and seawater. Although metabolism of organic sulfur compounds is certainly not restricted to bacteria belonging to the Roseobacter group (5), the strains characterized in this study have the ability to transform organic and reduced inorganic sulfur compounds, as well as the ability to incorporate dissolved organic sulfur into cellular proteins, despite the fact that many of them were isolated by nonselective techniques. While these organisms represent only a culturable subset of the Roseobacter group, the fact that they nearly universally degrade a biogeochemically important substrate like DMSP and the fact that they are able to carry out many other key processes in the organic sulfur cycle strongly suggest that they play a major role in the transformation of sulfur compounds in nature. The abundance of the Roseobacter group in coastal marine systems and the responses of uncultured members to additions of DMS and DMSP to seawater provide ecological support for such a role. Definitively assigning a biogeochemical function to the Roseobacter group and other bacterial groups in marine environments remains a very difficult challenge but one that should ultimately lead to a better understanding of important microbially mediated transformations and how they are regulated.
ACKNOWLEDGMENTS
We thank Laura J. Linn for technical advice, Jed Fuhrman for furnishing isolate ISM, and Brent Hanson and Ronald DuBose (Gilman Paper Co.) for providing access to sampling sites on the St. Marys River in Georgia. The Dauphin Island Sea Lab provided housing to J.M.G.
This research was supported by NSF grants OCE-9730745 (Biological Oceanography) and OCE-95-30378 (Chemical Oceanography).
REFERENCES
- 1.Baumann P, Baumann L. The marine Gram-negative eubacteria: genera Photobacterium, Beneckea, Alteromonas, Pseudomonas, and Alcaligenes. In: Starr M P, Stolp H, Trüper H G, Balows A, Schlegel H G, editors. The prokaryotes. Berlin, Germany: Springer-Verlag; 1981. pp. 1302–1331. [Google Scholar]
- 2.Charlson R J, Lovelock J E, Andreae M O, Warren S G. Oceanic phytoplankton, atmospheric sulfur, cloud albedo and climate. Nature (London) 1987;326:655–661. [Google Scholar]
- 3.Dacey J W H, Blough N V. Hydroxide decomposition of dimethylsulfoniopropionate to form dimethylsulfide. Geophys Res Lett. 1987;14:1246–1249. [Google Scholar]
- 4.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 5.de Souza M P, Yoch D C. Purification and characterization of dimethylsulfoniopropionate lyase from an Alcaligenes-like dimethyl sulfide-producing marine isolate. Appl Environ Microbiol. 1995;61:21–26. doi: 10.1128/aem.61.1.21-26.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Zwart J M M, Kuenen J G. Aerobic DMS degradation in microbial mats: the use of a newly isolated species Methylophaga sulfidovorans in the mathematical description of sulfur fluxes in mats. In: Kiene R P, Visscher P T, Keller M D, Kirst G O, editors. Biological and environmental chemistry of DMSP and related sulfonium compounds. New York, N.Y: Plenum Press; 1996. pp. 413–426. [Google Scholar]
- 7.Fuhrman J A, Lee S H, Masuchi Y, Davis A A, Wilcox R M. Characterization of marine prokaryotic communities via DNA and RNA. Microb Ecol. 1994;28:133–145. doi: 10.1007/BF00166801. [DOI] [PubMed] [Google Scholar]
- 8.Genetics Computer Group. Program manual for the Wisconsin package, version 10.0. Madison, Wis: Genetics Computer Group; 1999. [Google Scholar]
- 9.González J M, Moran M A. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González J M, Mayer F, Moran M A, Hodson R E, Whitman W B. Sagittula stellata gen. nov., sp. nov., a lignin-transforming bacterium from a coastal environment. Int J Syst Bacteriol. 1997;47:773–780. doi: 10.1099/00207713-47-3-773. [DOI] [PubMed] [Google Scholar]
- 11.Gosink J J, Herwig R P, Staley J T. Octadecabacter arcticus gen. nov., sp. nov., and O. antarcticus sp. nov., nonpigmented, phychrophilic gas vacuolate bacteria from polar sea ice and water. Syst Appl Microbiol. 1997;20:356–365. [Google Scholar]
- 12.Hatton A D, Malin G, Turner S M, Liss P S. DMSO: a significant compound in the biogeochemical cycle of DMS. In: Kiene R P, Visscher P T, Keller M D, Kirst G O, editors. Biological and environmental chemistry of DMSP and related sulfonium compounds. New York, N.Y: Plenum Press; 1996. pp. 405–412. [Google Scholar]
- 13.Hobbie J E, Daley R J, Jasper S. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes A J, Kelly D P, Baker S C, Thompson A S, De Marco P, Kenna E M, Murrel J C. Methylosulfonomonas methylovora gen. nov., sp. nov., and Marinosulfonomonas methylotropha gen. nov., sp. nov.: novel methylotrophs able to grow on methanesulfonic acid. Arch Microbiol. 1977;167:46–53. doi: 10.1007/s002030050415. [DOI] [PubMed] [Google Scholar]
- 15.Iverson R L, Nearhoof F L, Andreae M O. Production of dimethylsulfonium propionate and dimethylsulfide by phytoplankton in estuarine and coastal water. Limnol Oceanogr. 1989;34:53–67. [Google Scholar]
- 16.Johnston J B, Murray K, Cain R B. Microbial metabolism of aryl sulphonates. A re-assessment of colorimetric methods for the determination of sulphite and their use in measuring desulphonation of aryl and alkylbenzene sulphonates. Antonie Leeunwenhoek. 1975;41:493–511. doi: 10.1007/BF02565092. [DOI] [PubMed] [Google Scholar]
- 17.Kelly D P, Wood A P. Enzymes involved in microbiological oxidation of thiosulfate and polythionates. Methods Enzymol. 1994;243:501–510. [Google Scholar]
- 18.Kieber D J, Jiao J, Kiene R P, Bates T S. Impact of dimethylsulfide photochemistry on methyl sulfur cycling in the equatorial Pacific Ocean. J Geophys Res. 1996;101:3715–3722. [Google Scholar]
- 19.Kiene R P. Dimethyl sulfide production from dimethylsulfoniopropionate in coastal seawater samples and bacterial cultures. Appl Environ Microbiol. 1990;56:3292–3297. doi: 10.1128/aem.56.11.3292-3297.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiene R P. Dynamics of dimethyl sulfide and dimethylsulfoniopropionate in oceanic water samples. Mar Chem. 1992;37:29–52. [Google Scholar]
- 21.Kiene R P. Production of methanethiol from dimethylsulfoniopropionate in marine surface waters. Mar Chem. 1996;54:69–83. [Google Scholar]
- 22.Kiene R P, Gerard G. Determination of trace levels of dimethylsulfoxide (DMSO) in seawater and rainwater. Mar Chem. 1994;47:1–12. [Google Scholar]
- 23.Kiene, R. P, and L. J. Linn. Unpublished data.
- 24.Kiene, R. P., L. J. Linn, J. M. González, M. A. Moran, and J. A. Bruton. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 25.Kiene R P, Hoffmann Williams L P, Walker J E. Seawater microorganisms have a high affinity glycine betaine uptake system which also recognizes dimethylsulfoniopropionate. Aquat Microb Ecol. 1998;15:39–51. [Google Scholar]
- 26.Labrenz M, Collins M D, Lawson P A, Tindall B J, Braker G, Hirsch P. Antarctobacter heliothermus gen. nov., sp. nov., a budding bacterium from hypersaline and heliothermal Ekho Lake. Int J Syst Bacteriol. 1998;48:1363–1372. doi: 10.1099/00207713-48-4-1363. [DOI] [PubMed] [Google Scholar]
- 27.Labrenz M, Collins M D, Lawson P A, Tindall B J, Schumann P, Hirsch P. Roseovarius tolerans gen. nov., sp. nov., a budding bacterium with variable bacteriochlorophyll a production from hypersaline Ekho Lake. Int J Syst Bacteriol. 1999;49:137–147. doi: 10.1099/00207713-49-1-137. [DOI] [PubMed] [Google Scholar]
- 28.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ledyard K M, Dacey J W H. Microbial cycling of DMSP and DMS in coastal and oligotrophic seawater. Limnol Oceanogr. 1996;41:33–40. [Google Scholar]
- 30.Ledyard K M, DeLong E F, Dacey J W H. Characterization of a DMSP-degrading bacterial isolate from the Sargasso Sea. Arch Microbiol. 1993;160:312–318. [Google Scholar]
- 31.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 32.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 33.Petursdottir S K, Kristjansson J K. Silicibacter lacuscaerulensis gen. nov., sp. nov., a mesophilic moderately halophilic bacterium characteristic of the Blue Lagoon geothermal lake in Iceland. Extremophiles. 1997;1:94–99. doi: 10.1007/s007920050020. [DOI] [PubMed] [Google Scholar]
- 34.Pukall R, Butefuß D, Frühling A, Rohde M, Kroppenstedt R M, Burghardt J, Lebaron P, Bernard L, Stackebrandt E. Sulfitobacter mediterraneus sp. nov., a new sulfite-oxidizing member of the alpha-Proteobacteria. Int J Syst Bacteriol. 1999;49:513–519. doi: 10.1099/00207713-49-2-513. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorokin D Y. Sulfitobacter pontiacus gen. nov., sp. nov.: a new heterotrophic bacterium from the Black Sea, specialized on sulfite oxidation. Microbiology (Engl Transl Mikrobiologiya) 1995;64:354–365. [Google Scholar]
- 37.Sorokin D Y, Lysenko A M. Heterotrophic bacteria from the Black Sea oxidizing reduced sulfur compounds to sulfate. Microbiology (Engl Transl Mikrobiologiya) 1993;62:1018–1031. [Google Scholar]
- 38.Suylen G M H, Stefess G C, Kuenen J G. Chemolithotrophic potential of a Hyphomicrobium species, capable of growth on methylated sulphur compounds. Arch Microbiol. 1986;146:192–198. [Google Scholar]
- 39.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Ströbel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor B F, Gilchrist D C. New routes for aerobic degradation of dimethylsulfoniopropionate. Appl Environ Microbiol. 1991;57:3581–3584. doi: 10.1128/aem.57.12.3581-3584.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor B F, Visscher P T. Metabolic pathways involved in DMSP degradation. In: Kiene R P, Visscher P T, Keller M D, Kirst G O, editors. Biological and environmental chemistry of DMSP and related sulfonium compounds. New York, N.Y: Plenum Press; 1996. pp. 265–276. [Google Scholar]
- 42.Thompsom A S, Owens N J P, Murrell J C. Isolation and characterization of methanesulfonic acid-degrading bacteria from the marine environment. Appl Environ Microbiol. 1995;61:2388–2393. doi: 10.1128/aem.61.6.2388-2393.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner S M, Malin G, Liss P S, Harbour D S, Holligan P M. The seasonal variation of dimethyl sulfide and dimethylsulfoniopropionate concentrations in nearshore waters. Limnol Oceanogr. 1988;33:364–375. [Google Scholar]
- 44.Visscher P T, Taylor B F. Demethylation of dimethylsulfoniopropionate to 3-mercaptopropionate by an aerobic marine bacterium. Appl Environ Microbiol. 1994;60:4617–4619. doi: 10.1128/aem.60.12.4617-4619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visscher P T, Diaz M R, Taylor B F. Enumeration of bacteria which cleave or demethylate dimethylsulfoniopropionate in the Caribbean Sea. Mar Ecol Prog Ser. 1992;89:293–296. [Google Scholar]
- 46.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]