Abstract
The molecular and biological consequences of UV-B radiation were investigated by studying five species of marine bacteria and one enteric bacterium. Laboratory cultures were exposed to an artificial UV-B source and subjected to various post-UV irradiation treatments. Significant differences in survival subsequent to UV-B radiation were observed among the isolates, as measured by culturable counts. UV-B-induced DNA photodamage was investigated by using a highly specific radioimmunoassay to measure cyclobutane pyrimidine dimers (CPDs). The CPDs determined following UV-B exposure were comparable for all of the organisms except Sphingomonas sp. strain RB2256, a facultatively oligotrophic ultramicrobacterium. This organism exhibited little DNA damage and a high level of UV-B resistance. Physiological conditioning by growth phase and starvation did not change the UV-B sensitivity of marine bacteria. The rates of photoreactivation following exposure to UV-B were investigated by using different light sources (UV-A and cool white light). The rates of photoreactivation were greatest during UV-A exposure, although diverse responses were observed. The differences in sensitivity to UV-B radiation between strains were reduced after photoreactivation. The survival and CPD data obtained for Vibrio natriegens when we used two UV-B exposure periods interrupted by a repair period (photoreactivation plus dark repair) suggested that photoadaptation could occur. Our results revealed that there are wide variations in marine bacteria in their responses to UV radiation and subsequent repair strategies, suggesting that UV-B radiation may affect the microbial community structure in surface water.
Marine bacteria may account for up to 90% of the cellular DNA in oceanic environments (11, 42). These organisms play a central role in the cycling of nutrients in aquatic ecosystems and are a fundamental link in the carbon transfer process (i.e., the microbial loop [3]). Therefore, the study of factors that control bacterial growth in the environment is of primary importance. Among the different factors that affect bacterial growth, solar UV radiation (UVR) (wavelengths, 290 to 400 nm) has only recently received attention. UVR could be particularly deleterious for bacteria because these organisms have simple haploid genomes with little or no functional redundancy and are small, which precludes effective cellular shading or protective pigmentation (13). The last conclusion is supported by the observation that more photodamage is induced in bacterioplankton than in larger eukaryotic plankton by the same amount of solar UVR (20, 21). The detrimental effects of sunlight on bacterioplankton are manifested by reduced DNA and protein synthesis (1, 18), reduced exoenzymatic activity (36), reduced amino acid uptake (4), reduced oxygen consumption (39), and a decrease in bacterial abundance (36, 39).
DNA photodamage by UVR is wavelength dependent. UV-A (wavelengths, 320 to 400 nm) causes only indirect damage to DNA, proteins, and lipids through reactive oxygen intermediates. UV-B (wavelengths, 290 to 320 nm) causes both indirect and direct damage because of the strong absorption of wavelengths below 320 nm by DNA. The most abundant products formed during irradiation with UV-B are the cyclobutane pyrimidine dimers (CPDs) (35). A CPD can be lethal if the lesion blocks DNA synthesis and RNA transcription or can be mutagenic if the lesion is bypassed by DNA polymerase. Induction of CPD formation in marine bacterioplankton under laboratory and field UVR conditions has been studied by Jeffrey and coworkers (20, 21).
In response to UV damage, bacteria have developed different repair pathways, including photoenzymatic repair (PER), nucleotide excision repair (NER) (also called dark repair), and recombinational repair (postreplication repair). PER involves direct monomerization of CPDs by a single enzyme (photolyase) with near-UV or visible light as a source of energy (45). PER has been observed in marine bacterioplankton exposed to artificial UV-B after secondary irradiation with UV-A or photosynthetically active radiation (PAR) (22). The existence of PER in natural populations has been inferred from the results of CPD kinetic analyses performed with bacterioplankton samples obtained from the marine water column throughout the solar day (21). Similar field studies have also shown that marine bacterioplankton repair much of the DNA damage at night after diurnal exposure to sunlight, indicating that NER is a fundamental repair mechanism as well (21, 33, 39). Species-specific differences in UV-B sensitivity have been found in algae (23), cyanobacteria (51), ciliates (53), and metazooplankton (54). Studies of photobiological responses of marine bacterial species are very rare (17), and little is known about the diverse responses of marine bacteria during UV exposure and subsequent recovery. Understanding these responses is crucial for predicting the extent to which UVR may affect bacterial diversity by selecting phototolerant species or by inducing photoadaptation. Therefore, we studied the molecular and biological responses of five marine bacteria to UV-B radiation under standardized experimental conditions. By measuring survival and induction and repair of CPDs under different light conditions, we estimated the relative contributions of dark- and light-dependent repair systems to recovery.
MATERIALS AND METHODS
Bacterial strains and growth procedures.
Sphingomonas sp. strain RB2256, a facultatively oligotrophic ultramicrobacterium, was isolated from natural seawater from Resurrection Bay, Seward, Alaska, by an extinction seawater method; this organism was a numerically important member of the total community at the time of sampling (46). Strain RB2256 shares several features with indigenous marine bacteria, including a small cell size and a low apparent DNA content (46). Other marine bacteria, including Vibrio natriegens ATCC 14048, Pseudoalteromonas haloplanktis CIP 103197T (Collection Institut Pasteur, Paris, France), Deleya aquamarina CIP 74.8T, and Pseudomonas stutzeri (49), were chosen because they are frequently isolated from marine environments on high-nutrient media. The responses of marine bacteria were compared to the responses of Salmonella typhimurium CIP 60.62T, an enteric bacterium that can experience UV stress outside its normal habitat (intestinal tracts), including wastewater discharge events. S. typhimurium was maintained on tryptone soy broth (TSB) (Becton Dickinson) solidified with 15 g of Bacto Agar (Difco) liter−1. The marine bacteria were maintained on a complex marine medium containing artificial seawater (ASWJP) (41) supplemented with 1 g of yeast extract liter−1 and 5 g of Bacto Peptone (Difco) liter−1 (ASWJP+PY); this medium was solidified with 15 g of Bacto Agar liter−1. Cultures were grown at 37°C (S. typhimurium) or 25°C (marine bacteria) in 200-ml flasks containing 100 ml of TSB (S. typhimurium) or 100 ml of ASWJP+PY (marine bacteria) with shaking (200 rpm). One-milliliter portions of overnight cultures were used as inocula. The growth phases of cultures were determined from growth curves obtained by measuring the optical density at 600 nm (OD600). Stationary-phase cells were obtained after growth for 6 h (S. typhimurium), 18 h (V. natriegens, P. haloplanktis, D. aquamarina, and P. stutzeri), or 48 h (strain RB2256). Depending on the experiment, cells were harvested in the mid-log phase (maximum OD600/2) or at the beginning of the stationary phase (maximum OD600) by centrifugation at 6,000 × g for 10 min at 10°C. The pellets were washed twice in 0.9% (wt/vol) NaCl (S. typhimurium) or ASWJP (marine bacteria). Bacterial suspensions were diluted in order to obtain a concentration of 108 cells per ml, and the resulting preparations were used for irradiation experiments.
UV-B irradiation.
A portion of each cell suspension was transferred to a sterile plastic tissue culture dish (100 by 20 or 150 by 25 mm; Corning) so that the depth of the liquid was not greater than 2 mm. The petri dish (without a lid) was then placed in an incubator and incubated at 20°C with slow shaking (50 rpm). The petri dish was covered with a sheet of Kodacel (Kodak, Rochester, N.Y.) to block out UV-C (50% transmittance at 295 nm) and then exposed to four 20-W UV-B lamps (Philips). UV-B irradiance was quantified with a radiometer (model IL-1400A; International Light, Newburyport, Mass.) coupled to a UV-B detector (26) (with maximum sensitivity at 295 nm). The average intensity was 2.3 W m−2. The UV dose (in joules per square meter) was calculated by multiplying the intensity by the exposure time (in seconds). The amount of PAR was determined with a model GUV511 radiometer (Biospherical Instruments, San Diego, Calif.).
Photoreactivation and liquid holding.
UV-B-irradiated cell suspensions were exposed to different types of photoreactivating light, including UV-A light (20-W bulbs [Philips]) and visible light (20-W cool white bulbs [General Electric]). The petri dishes containing UV-B-irradiated cells were covered with a sheet of Mylar-D to block out UV-B (cutoff, 320 nm) (1) and placed under two photoreactivating lamps. The UV-A fluence rate was measured with a model IL-1400A radiometer coupled to a UV-A detector (with maximal sensitivity at 355 nm). The UV-A intensities used for the photoreactivating treatments were 2.6 W m−2 (UV-A treatment) and 0.026 W m−2 (0.0027 microeinsteins of PAR/cm2/s) (visible light treatment). The cell suspensions were continuously shaken at 50 rpm during photoreactivation. The numbers of CFU per milliliter were corrected for possible evaporation during exposure by using the following formula: CFUcorr ml−1 = (CFU × final volume)/initial volume. Tests to determine possible liquid holding recovery in the dark were performed by placing UV-B-irradiated cell suspensions in plastic tubes in the dark and incubating them without agitation at room temperature.
Starvation conditions.
The effects of starvation on both UV-B resistance and PER capacity were assessed by recording the numbers of CFU of S. typhimurium and V. natriegens exposed to radiation for different times in nutrient-depleted solutions. Erlenmeyer flasks (capacity, 500 ml) containing 200 ml of 0.9% NaCl (S. typhimurium) or 200 ml of ASWJP (V. natriegens) were autoclaved (120°C, 15 min), cooled, and inoculated with cells from stationary-phase cultures washed as described above. The flasks were incubated in the dark at 20°C. Subsamples were removed each day for 4 days. The cells were exposed to UV-B radiation (950 and 1,650 J m−2 for V. natriegens and S. typhimurium, respectively, resulting in about 0.1% survival for each strain) and then to UV-A for 2 h (photoreactivation). The numbers of CFU were determined before and after each radiation treatment.
Culturable counts.
A 100-μl portion of each cell suspension was removed in order to prepare serial dilutions in 0.9% NaCl (S. typhimurium) or ASWJP (marine bacteria). Portions (100 μl) of the appropriate serial dilutions were spread in triplicate onto TSB plates (S. typhimurium) or ASWJP+PY plates (marine bacteria). The numbers of CFU were determined after 24 h of incubation in the dark at 37°C for S. typhimurium and after 2 to 7 days of incubation in the dark at 25°C for the marine bacteria. The dilution and plating procedures were carried out under low-luminosity conditions to avoid any stray photoreactivating light. The fraction of surviving cells was calculated by dividing the number of CFU in the treated sample by the number of CFU in the unirradiated sample at time zero.
DNA photoproducts.
Culture subsamples (5 to 15 ml) were filtered through 0.22-μm-pore-size polysulfone membranes (Gelman), and the membranes were then stored at −20°C in microcentrifuge tubes. Five hundred microliters of STE buffer (10 mM Tris [pH 8], 1 mM EDTA, 100 mM NaCl) containing 0.5% sodium dodecyl sulfate was added to each tube, and then the tubes were capped, vortexed for 30 s, and placed in a boiling water bath for 2 min. After they were chilled on ice for 5 min, the filters were removed, and the DNA was extracted with 0.8 ml of chloroform-isoamyl alcohol (49:1). The aqueous phase was collected after centrifugation at 3,000 × g at 4°C for 30 min. To precipitate the nucleic acids, 1 μl of a 25-mg ml−1 glycogen solution, 0.1 volume of 3 M sodium acetate (pH 5), and an equal volume of isopropanol were added to each preparation, and the preparations were incubated overnight at −20°C. Each precipitate was collected by centrifugation at 13,000 × g at 4°C for 30 min. The pellet was washed with 0.5 ml of cold 70% ethanol and resuspended in 0.5 ml of STE buffer. DNA was quantified by fluorometry with Hoechst 33258 dye (43).
CPDs were quantified by using a highly sensitive radioimmunoassay described by Mitchell (34). This assay measures the binding of radiolabeled UV-irradiated DNA to antibody raised in rabbits against triplet (acetone)-sensitized UV-B-irradiated DNA, conditions which induce cyclobutane dimers exclusively. The quantity of CPDs was determined by comparison to standard DNA (pUC19) for which the rates of photoproduct formation were known. Samples were analyzed in duplicate, and the amount of CPD that accumulated was compared to the amount in a solution of calf thymus DNA previously exposed to the same UV-B dose.
Flow cytometric detection of DNA content.
Bacterial samples were fixed with 2% (vol/vol) (final concentration) formaldehyde and stored in the dark at 4°C until they were analyzed. The DNA content was determined as described by Lebaron et al. (27) by using SYBR-I (Molecular Probes Inc., Eugene, Oreg.). SYBR-I was added at a final concentration of 10−4 M to the stock solution, and the preparation was incubated for 15 min in the dark with 30 mM (final concentration) potassium citrate (pH 7.4). All experiments were performed with a FACS-Calibur flow cytometer (Becton Dickinson) equipped with an air-cooled laser providing 15 mW at 488 nm and the standard filter setup. All parameters were measured as logarithmic signals. Green fluorescence was determined in the FL1 channel (530 ± 15 nm). Relative cellular sizes were estimated by using the side scatter signal. Yellow-green fluorescent microspheres (2-μm-diameter fluorescent size standard beads; Polysciences Inc., Warrington, Pa.) were systematically added to each sample as an internal reference to normalize cell fluorescence and side scatter values.
RESULTS
Diversity of UV-B resistance.
Although the UV-B intensity used in this study (2.3 W m−2) was higher than the natural intensity, the UV-B doses applied (up to 3,000 J m−2) were similar to daily UV-B doses recorded in the field at the surface of water (8). To ensure that the measured biological responses were reliable, we previously tested the principle of reciprocity, which requires that a response be a function of the dose and not a function of the dose rate. For reciprocity to be satisfied, different combinations of dose rate and exposure time, which result in the same dose, should yield the same response. Stationary-phase cultures of S. typhimurium were exposed to three UV-B intensities (0.5, 1.1, and 2.3 W m−2) for different times, which resulted in eight doses (0 to 2,000 J m−2) for each intensity. The inactivation rates (loss of CFU) obtained for the three UV-B intensities did not differ (data not shown). The high cell densities used in this study did not introduce bias through a protective effect; the inactivation rates obtained with different S. typhimurium cell densities (106 to 108 cells per ml) were comparable, indicating that shielding did not occur (data not shown).
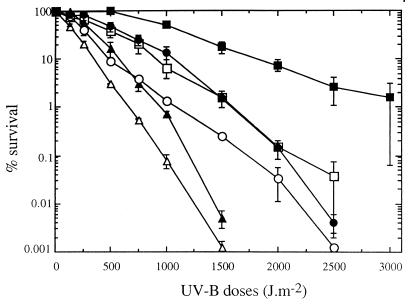
With stationary-phase cells of most bacteria exposed to artificial UV-B radiation there was an exponential decrease in the number of CFU. The only exception was strain RB2256; a shoulder was observed at doses up to 500 J m−2 with this strain (Fig. 1). Large variations in the UV-B survival rates were observed with the different organisms. The marine bacteria could be divided into the following three resistance categories: sensitive (V. natriegens and P. haloplanktis), intermediate (P. stutzeri and D. aquamarina), and resistant (strain RB2256). Ninety-nine percent inactivation was observed after 800-, 1,600-, and 3,000-J m−2 UV-B doses with members of the sensitive, intermediate, and resistant groups, respectively. The response of S. typhimurium was similar to the response of the sensitive strains at doses up to 1,000 J m−2 but resembled the response of the intermediate strains at higher UV-B doses.
FIG. 1.
Effects of UV-B radiation on the survival of different bacteria collected during the stationary phase. The data are expressed as percentages of the initial counts (∼108 CFU ml−1) and are means (± standard deviations) based on the data from two different experiments. Symbols: ▵, V. natriegens; ▴, P. haloplanktis; ○, S. typhimurium; ●, P. stutzeri; □, D. aquamarina; ■, strain RB2256.
Accumulation of DNA photoproducts.
Because DNA is the primary target for UV-B radiation, it was important to estimate variations in DNA content and distribution in the different bacteria. Flow cytometric DNA histograms showed that bacterial populations harvested in the stationary phase were mainly composed of cells containing a single genome (Table 1). On the basis of mean fluorescence values for cells containing a single genome, we determined that the chromosome of strain RB2256 was 1.5 to 2 times smaller than the chromosomes of the other strains (Table 1). The cells of strain RB2256 were also smaller, as estimated by the side scatter signal (Table 1).
TABLE 1.
Flow cytometric characteristics of cells containing a single genome in stationary-phase cultures
| Bacterium | % of cells containing a single genome | DNA fluores-cencea | Side scatter signala |
|---|---|---|---|
| Salmonella typhimurium | 90 | 0.088 | 0.036 |
| Pseudoalteromonas haloplanktis | 87 | 0.077 | 0.031 |
| Vibrio natriegens | 92 | 0.089 | 0.026 |
| Pseudomonas stutzeri | 83 | 0.064 | 0.025 |
| Deleya aquamarina | 86 | 0.083 | 0.021 |
| Sphingomonas sp. strain RB2256 | 56 | 0.043 | 0.012 |
Relative units were normalized by using 2-μm-diameter fluorescent beads (see text).
To determine if the diversity of UV-B resistance resulted mainly from effective protection or DNA repair, CPDs were quantified after two doses of irradiation (1,000 and 2,000 J m−2) (Fig. 2). DNA damage frequencies (expressed as the number of CPDs per megabase of DNA) were determined for stationary-phase cells immediately following UV-B irradiation and prior to initiation of any DNA repair processes. The number of CPDs per megabase of DNA in a solution of calf thymus DNA was comparable to the values recorded for purified DNA exposed to 1 solar day at the surface of marine water (between 500 and 1,750 CPDs/Mb of DNA) (1, 20, 29, 44), indicating that the UV-B doses used in this study were comparable to natural daily doses. V. natriegens, P. haloplanktis, and D. aquamarina accumulated the same amounts of CPDs after different UV-B doses despite their different levels of resistance. Surprisingly, the levels of CPDs obtained for these strains were equivalent to the level obtained for the DNA solution exposed to the same UV-B dose, suggesting that no cellular photoprotective mechanisms were present. In contrast, strain RB2256 accumulated very small amounts of CPDs after different UV-B doses. The amount of CPDs in strain RB2256 was fourfold less than the amounts in the other marine bacteria after a 2,000-J m−2 dose. After a 1,000-J m−2 UV-B dose, the level of DNA damage in S. typhimurium was intermediate between the levels of DNA damage in strain RB2256 and the other marine bacteria. With higher UV-B doses, the S. typhimurium damage frequencies were comparable to the damage frequencies observed in the DNA solution.
FIG. 2.
Accumulation of CPDs in marine bacteria and S. typhimurium during UV-B irradiation. The data are means (± standard deviations) based on the data from duplicate analyses. Symbols: ▵, V. natriegens; ▴, P. haloplanktis; ○, S. typhimurium; ●, D. aquamarina; □, strain RB2256; ■, DNA solution.
Photoreactivation and liquid holding recovery.
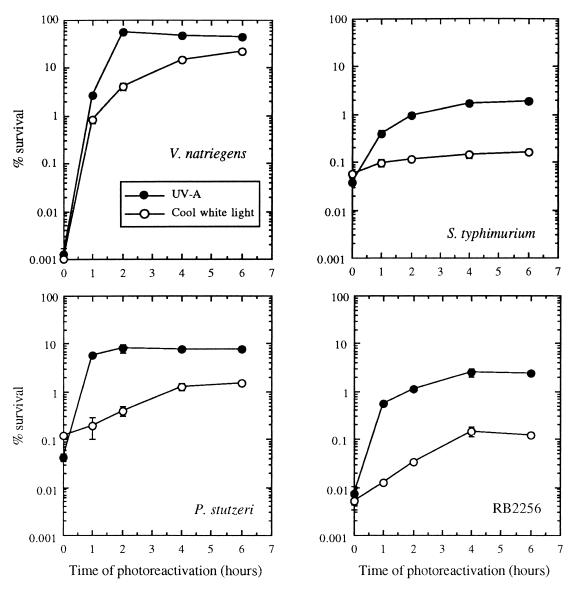
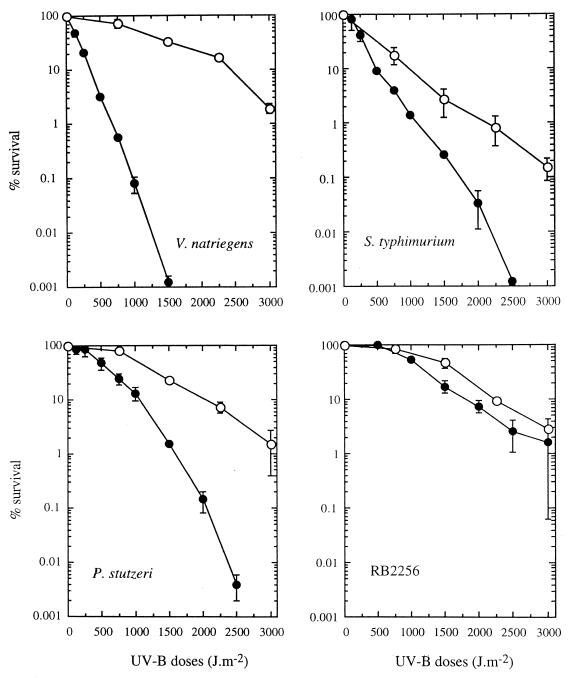
PER of bacteria after UV-B irradiation was investigated by using different light sources (UV-A and cool white light). With all strains, photoreactivation was greatest after UV-A exposure; maximum survival was observed after 2 h of exposure, which corresponded to a dose of 19 kJ m−2 (Fig. 3). Under photoreactivating light conditions, V. natriegens, P. stutzeri, and strain RB2256 exhibited similar levels of resistance to UV-B, and a shoulder was observed at doses up to 500 J m−2 (Fig. 4). Under these conditions 99% inactivation was obtained after a 3,000-J m−2 UV-B dose. The very sensitive organism V. natriegens had a very effective photorepair mechanism. For this strain, the number of cells that survived after a 1,500-J m−2 UV-B dose increased by more than four orders of magnitude when the cells were allowed to photorepair via exposure to UV-A. In contrast to the marine bacteria, S. typhimurium exhibited effective photoreactivation only under UV-A irradiation conditions, and the level of photorepair was less than the levels observed in marine bacteria and exhibited no shoulder (i.e., repair capacity). With S. typhimurium, 99% inactivation was observed after a 2,250-J m−2 UV-B dose.
FIG. 3.
Effects of different light sources on photoreactivation kinetics after UV-B exposure. V. natriegens, S. typhimurium, P. stutzeri, and strain RB2256 were exposed to 1,500-, 1,650-, 2,000-, and 3,500-J m−2 UV-B doses, respectively, before they were exposed to photoreactivating light. The data are expressed as percentages of the initial counts (∼108 CFU ml−1) and are means (± standard deviations) based on data obtained from triplicate plates.
FIG. 4.
Effects of UV-B radiation without (●) and with (○) exposure to UV-A radiation for 2 h after UV-B treatment for V. natriegens, S. typhimurium, P. stutzeri, and RB2256. The data are expressed as percentages of the initial counts (∼108 CFU ml−1) and are means (± standard deviations) based on the data from two different experiments.
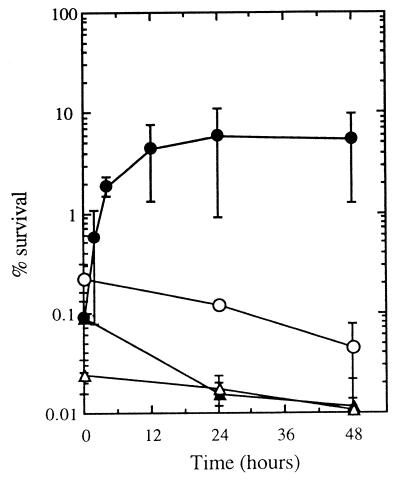
The principle of reciprocity was also verified under photoreactivation conditions. No differences in photorecovery were observed when stationary-phase cultures (S. typhimurium was used as the test organism) were exposed to UV-A light after equivalent UV-B doses were delivered by two UV-B intensities (0.5 and 2.3 W m−2) (data not shown). To determine whether de novo protein synthesis was necessary for photoreactivation, we conducted experiments with V. natriegens (the most photoreactive organism in this study) in the presence of 50 μg of chloramphenicol ml−1 (the MIC of chloramphenicol for this strain is approximately 2.5 μg ml−1). After UV-B exposure, chloramphenicol was added, and the cells were exposed to UV-A for 2 h. Under these conditions, photoreactivation was not altered (data not shown). Of the different strains tested and within the time frame used (2 days), only V. natriegens benefited from a liquid holding period after UV-B inactivation (Fig. 5). Maximum recovery of V. natriegens occurred after 12 h of incubation in the dark. When liquid holding recovery was taken into account, the resistance of V. natriegens was improved and more closely resembled the resistance of the intermediate organisms (i.e., D. aquamarina and P. stutzeri) (data not shown). Under these conditions 99% inactivation was observed after a 1,500-J m−2 UV-B dose (data not shown).
FIG. 5.
Kinetics of liquid holding recovery after UV-B irradiation. V. natriegens, S. typhimurium, P. stutzeri, and strain RB2256 were exposed to 950-, 1,650-, 2,000-, and 4,000-J m−2 UV-B doses, respectively, before liquid holding. The data are expressed as percentages of the initial counts (∼108 CFU ml−1) and are means (± standard deviations) based on the data from two different experiments. Symbols: ●, V. natriegens; ○, S. typhimurium; ▴, P. stutzeri; ▵, strain RB2256.
Effects of growth rate on UV-B resistance and photoreactivation capacity.
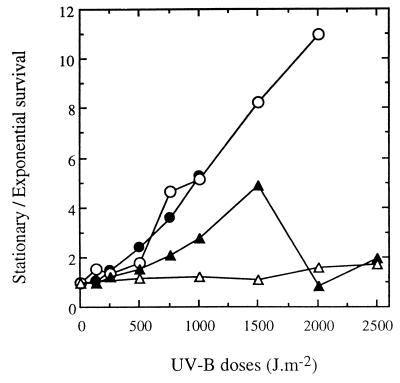
We compared the UV-B resistance of V. natriegens, P. stutzeri, S. typhimurium, and strain RB2256 under exponential- and stationary-phase growth conditions. Figure 6 shows the ratios of stationary-phase cells to exponential-phase cells that survived after different doses of UV-B light. In both cases, the percentages of survival were expressed as a function of the initial CFU counts. The resistance of stationary-phase cells was slightly greater than (ratio, less than 5) or comparable to the resistance of exponential-phase cells for the marine bacteria. A greater difference between stationary- and exponential-phase cells was observed with S. typhimurium exposed to UV-B doses higher than 1,000 J m−2.
FIG. 6.
Influence of growth phase on the survival of V. natriegens, S. typhimurium, P. stutzeri, and strain RB2256 after exposure to UV-B radiation. The data are ratios of stationary-phase cells to exponential-phase cells that survived after different UV-B doses. The percentages of survival at both growth phases used to calculate ratios were determined relative to the initial counts (∼108 CFU ml−1) and are means based on the data from two different experiments. Symbols: ●, V. natriegens; ○, S. typhimurium; ▴, P. stutzeri; ▵, strain RB2256.
In addition, the effects of starvation on UV-B resistance and photoreactivation of V. natriegens and S. typhimurium were examined. During 4 days of starvation, the percentage of viable S. typhimurium cells did not change, and the percentage of viable V. natriegens cells decreased slightly (data not shown). For both strains, the number of CFU after UV-B exposure and UV-A exposure did not change significantly during starvation, indicating that these conditions did not alter UV-B resistance or photoreactivation capacity (data not shown).
Potential for photoadaptation.
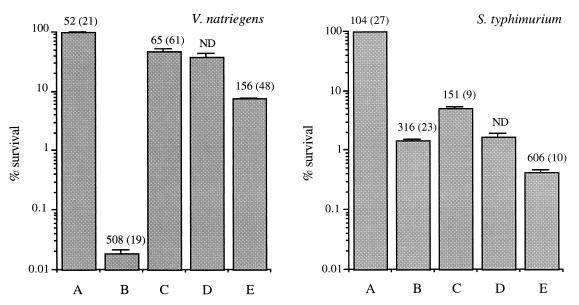
In order to identify UV-B photoadaptive responses, we treated V. natriegens and S. typhimurium with 1,000 J of UV-B light m−2; this was followed by 2 h of exposure to photoreactivating UV-A light, 10 h of incubation in the dark, and a second exposure to 1,000 J of UV-B light m−2. At each point during this protocol, the numbers of CFU and CPDs were determined (Fig. 7). Under these conditions, all of the CPDs that were accumulated by V. natriegens cells were removed after photoreactivation, and CFU were recovered up to 46% of the initial count. During the dark period, no change in the number of culturable cells was observed. The DNA damage and sensitivity after the second UV-B exposure were considerably less than the DNA damage and sensitivity after the first exposure. When this protocol was used, the S. typhimurium cells removed about one-half of the accumulated CPDs during the photoreactivation phase, but the level of recovery of CFU was only 5%. Liquid holding slightly decreased the number of CFU. The second exposure to UV-B resulted in a less significant loss of CFU than the first exposure but greater cumulative DNA damage (the number of CPDs was twice the number after the initial exposure).
FIG. 7.
Changes in CFU and CPDs under the following conditions: bars A, control (stationary-phase cells); bars B, treatment A plus UV-B radiation (1,000 J m−2); bars C, treatment B plus photoreactivation (UV-A radiation for 2 h); bars D, treatment C plus liquid holding (10 h); bars E, treatment D plus UV-B radiation (1,000 J m−2). The CFU data are expressed as percentages of the initial counts (∼108 CFU ml−1) and are means (± standard deviations) based on data obtained from triplicate plates. The numbers above the bars are the numbers of CPDs per megabase of DNA and are means (± standard deviations) based on data obtained from duplicate analyses. ND, not determined.
DISCUSSION
UV-B resistance and DNA damage.
Significant species-specific differences in susceptibility to UV-B irradiation were observed among bacterial strains, particularly when photodependent repair mechanisms were excluded. When culture media were used to measure bacterial resistance, sufficient nutrients were supplied for the energy requirements of NER. In the case of V. natriegens, we found that maintenance of cells in a nutrient-depleted solution after irradiation and prior to plating resulted in increased survival. Such liquid holding recovery has been demonstrated previously for Escherichia coli strains after UV-C irradiation and, more recently, for the fish pathogen Vibrio anguillarum (31). The mechanisms of this response are not yet understood, but Ganesan and Smith (12) observed that liquid holding recovery occurs in E. coli K-12 only when excision repair, but not recombination repair, is operative. From these results it is evident that in order to understand how low concentrations of dissolved organic carbon in seawater constitute a limiting factor for optimal repair, the precise energy requirements of NER must be determined.
In contrast to previous observations of Antarctic diatoms exposed to artificial UV-B radiation (23), equivalent DNA damage frequencies did not correlate with cell killing in the sensitive and intermediate organisms. The bacteria investigated have different G+C contents, as follows: 41 to 45% for P. haloplanktis (6), 46 to 47% for V. natriegens (5), 50 to 53% for S. typhimurium (28), 56% for D. aquamarina (2), 61 to 66% for P. stutzeri (40), and 61.6 to 67.8% for Sphingomonas sp. strain RB2256 (50). Although a high G+C content is known to protect DNA against damage by thymine dimerization (48), the differences in G+C content among the bacteria studied here did not appear to affect the quantity of CPDs induced. Likewise, differences in the amount of DNA per cell could not explain the differences in resistance observed between intermediate and sensitive organisms since most cells of all of the organisms tested in the stationary phase had one genome and the genomes were similar sizes.
The fact that comparable levels of CPDs per megabase of DNA were accumulated by sensitive and intermediate organisms and calf thymus DNA in solution indicated that there was a complete lack of photoprotection in these bacteria. In contrast, the high level of resistance of strain RB2256 cells to UV-B light was associated with low CPD frequencies and indicated that a constitutive photoprotective mechanism was present in these cells. The yellow pigmentation in RB2256 cells is thought to be insufficient to explain the resistance of the cells to UV-B irradiation since the small size of the cells probably precludes effective protection by pigmentation (13). Consistent with this hypothesis, Gascón et al. (14) observed that strains of Rhodobacter sphaeroides with high levels of pigment (associated with phototrophic growth) were more sensitive to UV-C irradiation than strains with less pigment (associated with heterotrophic growth) were. In addition to pigments, DNA binding proteins have been implicated in photoprotection (47). Small, acid-soluble proteins bind to the DNA of Bacillus and Clostridium spores and provide strong protection against DNA damage due to UV irradiation and other stresses (e.g., heat and hydrogen peroxide). It has also been shown that certain E. coli strains which lack specific DNA binding proteins (protein HU) are sensitive to UV-C and γ irradiation (7, 29). Since these proteins mitigate DNA cleavage by γ rays (7) but do not reduce CPD frequencies after UV-C irradiation (29), they are probably not involved in the photoprotection observed in RB2256 cells. The mechanism behind the multiple resistance of strain RB2256 to a variety of exogenous stress-inducing agents, such as heat shock, H2O2, and ethanol (9), is intriguing and worthy of future research efforts.
Photoreactivation in marine bacteria.
Photoreactivation occurs in each of the three marine organisms studied and appears to be an important mechanism of DNA repair, especially for the sensitive bacterium V. natriegens. When PER is taken into account, the diversity of resistance is largely reduced. The different levels of photoreactivation observed with marine bacteria may result from differences in the specific activities of the individual DNA photolyases or from differences in the intracellular concentrations of the enzymes (37). Different photoreactivation capacities may also occur in members of the same genus, as described previously for six Bacillus species (37). Observations made with V. natriegens treated with chloramphenicol indicated that the initial photolyase content is sufficient to ensure total photoreactivation. Moreover, the photoreactivation capacity was not attenuated by prior starvation with both V. natriegens and S. typhimurium cells. Unlike NER and postreplication repair, PER does not require energy mobilization and may therefore be very important in aquatic microbial populations that are nutrient limited. Indeed, photoreactivation may be the main repair mechanism that operates immediately after exposure to solar UVR.
The rates of photoreactivation were greatest during UV-A exposure, although diverse responses were observed. The marine organisms responded to a broader spectrum for photoreactivation than S. typhimurium, and efficient PER was observed under UV-A and cool white light conditions. Photolyases are categorized on the basis of the chromophore involved in energy transfer, and S. typhimurium photolyase belongs to the folate class (30) and exhibits maximum activity at 384 nm (i.e., UV-A), which is consistent with our results. Other known photolyases fall into either a deazaflavin class (maximum activity at 440 nm) (19) or a “no-second-chromophore” class (maximum activity at both 370 and 450 nm) (24). The patterns of photoreactivation observed in marine strains suggest that the efficiency of photoreactivation in marine water should correlate with UV-A penetration. Our results agree with those of Kaiser and Herndl (22), who found that UV-A radiation produced more efficient photoreactivation than PAR produced in both aged and freshly collected seawater.
Physiological conditioning and photoadaptation.
Our results show that growth phase has little impact on the resistance of marine bacteria to UV-B irradiation. Significant interstrain variability in growth phase responses has been observed in different bacteria exposed to UV-C radiation (10, 15, 16, 25), although the types of damage induced by UV-C radiation were essentially the same as the types of damage induced by UV-B light. Some workers observed increased resistance (10, 16) but other studies revealed increased sensitivity when cells entered the stationary phase (25). Our results agree with the results of other studies and show that the sensitivities of cells harvested from exponential- and stationary-phase cultures did not differ (15). Our results show that starvation of stationary-phase cells in ASWJP or an NaCl solution did not change the UV-B resistance of the cells. In contrast, Nyström et al. (38) showed that marine Vibrio sp. strain S14 kept in seawater depleted of C, N, and P exhibited increased resistance to UV-C and UV-B (302-nm) radiation. The lengths of starvation time required by this strain to reach the maximal level of stress resistance were 30 h for UV-C radiation and about 70 h for UV-B radiation. In the same way, Enterococcus faecalis cells in the exponential growth phase and the early stationary phase became progressively more resistant to UV-C radiation when they were incubated in tap water (an oligotrophic environment); maximum survival occurred after 35 days (16). Many studies have shown that stationary-phase or starved bacteria are far more resistant to stresses, such as starvation, abnormal temperatures, hydrogen peroxide, and UV-A radiation (photooxidation), than their exponentially growing counterparts are. Expression of the genes responsible for this increased resistance is controlled by the stationary-phase-specific sigma factor ςS, which is encoded by rpoS (32). To our knowledge, genes controlled by rpoS responsible for UV-C or UV-B resistance have never been described. It has been shown that mutations in the E. coli rpoS gene (initially named nur) may reduce the efficiency of repair after exposure to 313-nm UV-B radiation but not after exposure to 290-nm UV-B radiation (52). This result may be explained by the increase in the proportion of photooxidations to dimerizations in the upper part of the UV-B spectrum (i.e., near the UV-A spectrum).
As demonstrated with V. natriegens, photoadaptation could occur after UV-B irradiation and subsequent recovery (photoreactivation and dark holding). This photoadaptation resulted in increased resistance and decreased CPD accumulation during a second UV-B irradiation period. The reduced CPD frequency observed after the second irradiation suggests that V. natriegens could mitigate the DNA damage by some inducible photoprotective mechanism. Whether bacterial photoadaptation occurs in aquatic environments has not been clearly established. Pakulski et al. (39) observed recovery of bacterial production and respiration in subtropical coral reef bacteria incubated at a fixed depth during a second day of exposure to natural sunlight. These authors explained these findings by either photoinduced selection for light-tolerant cells or physiological adaptation to ambient light regimens that occurred during exposure. Herndl et al. (18) showed that no photoadaptation occurred in bacterioplankton obtained from surface water (depth, 0.5 m) that were as sensitive (as determined by thymidine incorporation) to surface UV-B radiation as subpycnocline bacteria were (depth, 20 m). Based on the diversity of the photobiological responses of different marine bacterioplankton to solar UVR observed in this study, it is apparent that an integrated approach involving molecular, physiological, and taxonomic end points will be required to better understand the impact of UV-B radiation on bacterioplankton populations. Our results demonstrate the variability of the responses of marine bacteria that are exposed to UV-B radiation and emphasize the difficulties in deriving models that predict UV effects in the environment from laboratory studies. The clear differences between the photobiology of culturable marine bacteria isolated on high-nutrient media and the photobiology of the facultatively oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256 should encourage isolation of the latter type of organisms, which may constitute a significant part of the open ocean bacterial community.
ACKNOWLEDGMENTS
We thank Staffan Kjelleberg (School of Microbiology and Immunology, The University of New South Wales, Sydney, Australia) for providing Sphingomonas sp. strain RB2256 and Jason Kase for critically reviewing the manuscript.
This work was supported by a postdoctoral fellowship to F.J. from the Conseil Régional Languedoc-Roussillon (France) and by National Science Foundation Office of Polar Programs grant OPP-9419037 to W.H.J.
REFERENCES
- 1.Aas P, Lyons M M, Pledger R, Mitchell D L, Jeffrey W H. Inhibition of bacterial activities by solar radiation in nearshore waters and the Gulf of Mexico. Aquat Microb Ecol. 1996;11:229–238. [Google Scholar]
- 2.Akagawa M, Yamasato K. Synonymy of Alcaligenes aquamarina, Alcaligenes faecalis subsp. homari, and Deleya aesta: Deleya aquamarina comb. nov. as the type species of the genus Deleya. Int J Syst Bacteriol. 1989;39:462–466. [Google Scholar]
- 3.Azam F, Fenchel T, Field J G, Gray J S, Meyer-Reil L A, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- 4.Bailey C A, Neihof R A, Tabor P S. Inhibitory effect of solar radiation on amino acid uptake in Chesapeake Bay bacteria. Appl Environ Microbiol. 1983;46:44–49. doi: 10.1128/aem.46.1.44-49.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumann P, Furniss A L, Lee J V. Genus Vibrio. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 518–538. [Google Scholar]
- 6.Baumann P, Gauthier M J, Baumann L. Genus Alteromonas. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 342–352. [Google Scholar]
- 7.Boubrik F, Rouvière-Yaniv J. Increased sensitivity to γ irradiation in bacteria lacking protein HU. Proc Natl Acad Sci USA. 1995;92:3958–3962. doi: 10.1073/pnas.92.9.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demers S, Belzile C, Lean D R S, Mostajir B, Roy S, De Mora S, Bird D, Gosselin M, Chanut J-P, Levasseur M. An experimental tool to study the effects of ultraviolet radiations on planktonic communities: a mesocosm approach. Environ Technol. 1998;19:667–682. [Google Scholar]
- 9.Eguchi M, Nishikawa T, MacDonald K, Cavicchioli R, Gottschal J C, Kjelleberg S. Responses to stress and nutrient availability by the marine ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol. 1996;62:1287–1294. doi: 10.1128/aem.62.4.1287-1294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freedman M L, Bruce A K. The relationship of radioresistance to balanced growth-rate in Micrococcus radiodurans. Int J Radiat Biol. 1971;19:111–121. doi: 10.1080/09553007114550161. [DOI] [PubMed] [Google Scholar]
- 11.Fuhrman J A, Sleeter T D, Carlson C A, Proctor L M. Dominance of bacterial biomass in the Sargasso Sea and its ecological implications. Mar Ecol Prog Ser. 1989;57:207–217. [Google Scholar]
- 12.Ganesan A K, Smith K C. Dark recovery processes in Escherichia coli irradiated with ultraviolet light. J Bacteriol. 1968;96:365–371. doi: 10.1128/jb.96.2.365-373.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Pichel F. A model for internal self-shading in planktonic organisms and its implications for the usefulness of ultraviolet sunscreen. Limnol Oceanogr. 1994;39:1704–1717. [Google Scholar]
- 14.Gascón J, Oubiña A, Pérez-Lezaun A, Urmeneta J. Sensitivity of selected bacterial species to UV radiation. Curr Microbiol. 1995;30:177–182. doi: 10.1007/BF00296205. [DOI] [PubMed] [Google Scholar]
- 15.Hartke A, Bouche S, Gansel X, Boutibonnes P, Auffray Y. Starvation-induced stress resistance in Lactococcus lactis subsp. lactis IL1403. Appl Environ Microbiol. 1994;60:3474–3478. doi: 10.1128/aem.60.9.3474-3478.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartke A, Giard J-C, Laplace J M, Auffray Y. Survival of Enterococcus faecalis in an oligotrophic microcosm: changes in morphology, development of general stress resistance, and analysis of protein synthesis. Appl Environ Microbiol. 1998;64:4238–4245. doi: 10.1128/aem.64.11.4238-4245.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helbling E W, Marguet E R, Villafañe V E, Holm-Hansen O. Bacterioplankton viability in Antarctic waters as affected by solar ultraviolet radiation. Mar Ecol Prog Ser. 1995;126:293–298. [Google Scholar]
- 18.Herndl G J, Müller-Niklas G, Frick J. Major role of ultraviolet-B in controlling bacterioplankton growth in the surface layer of the ocean. Nature. 1993;361:717–719. [Google Scholar]
- 19.Jagger J, Tabeke H, Snow J. Photoreactivation and killing in Streptomyces: action spectra and kinetic studies. Photochem Photobiol. 1970;12:185–196. doi: 10.1111/j.1751-1097.1970.tb06050.x. [DOI] [PubMed] [Google Scholar]
- 20.Jeffrey W H, Aas P, Lyons M M, Coffin R B, Pledger R J, Mitchell D L. Ambiant solar radiation-induced photodamage in marine bacterioplankton. Photochem Photobiol. 1996;64:419–427. [Google Scholar]
- 21.Jeffrey W H, Pledger R J, Aas P, Hager S, Coffin R B, Von Haven R, Mitchell D L. Diel and depth profiles of DNA photodamage in bacterioplankton exposed to ambiant solar ultraviolet radiation. Mar Ecol Prog Ser. 1996;137:283–291. [Google Scholar]
- 22.Kaiser E, Herndl G J. Rapid recovery of marine bacterioplankton activity after inhibition by UV radiation in coastal waters. Appl Environ Microbiol. 1997;63:4026–4031. doi: 10.1128/aem.63.10.4026-4031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karentz D, Cleaver J E, Mitchell D L. Cell survival characteristics and molecular responses of Antarctic phytoplankton to ultraviolet-B radiation. J Phycol. 1991;27:326–341. [Google Scholar]
- 24.Kato R, Hasegawa K, Hidaka Y, Kuramitsu S, Hoshino T. Characterization of a thermostable DNA photolyase from an extremely thermophilic bacterium, Thermus thermophilus HB27. J Bacteriol. 1997;179:6499–6503. doi: 10.1128/jb.179.20.6499-6503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller L C, Maxcy R B. Effect of physiological age on radiation resistance of some bacteria that are highly radiation resistant. Appl Environ Microbiol. 1984;47:915–918. doi: 10.1128/aem.47.5.915-918.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirk J T O, Hargreaves B R, Morris D P, Coffin R B, David B, Frederickson D, Karentz D, Lean D R S, Lesser M P, Madronich S, Morrow J H, Nelson N B, Scully N M. Measurement of UV-B in two freshwater lakes: an instrument intercomparison. Arch Hydrobiol Beih. 1994;43:71–99. [Google Scholar]
- 27.Lebaron P, Parthuisot N, Catala P. Comparison of blue nucleic acid dyes for flow cytometric enumeration of bacteria in aquatic systems. Appl Environ Microbiol. 1998;64:1725–1730. doi: 10.1128/aem.64.5.1725-1730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Minor L. Genus Salmonella. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 427–458. [Google Scholar]
- 29.Li S, Waters R. Escherichia coli strains lacking protein HU are UV sensitive due to a role for HU in homologous recombination. J Bacteriol. 1998;180:3750–3756. doi: 10.1128/jb.180.15.3750-3756.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y F, Sancar A. Cloning, sequencing, expression and characterization of DNA photolyase from Salmonella typhimurium. Nucleic Acids Res. 1991;19:4885–4890. doi: 10.1093/nar/19.18.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liltved H, Landfald B. Influence of liquid holding recovery and photoreactivation on survival of ultraviolet-irradiated fish pathogenic bacteria. Water Res. 1996;30:1109–1114. [Google Scholar]
- 32.Loewen P C, Hu B, Strutinsky J, Sparling R. Regulation in the rpoS regulon of Escherichia coli. Can J Microbiol. 1998;44:707–717. doi: 10.1139/cjm-44-8-707. [DOI] [PubMed] [Google Scholar]
- 33.Lyons M M, Aas P, Pakulski J D, Van Waasbergen L, Mitchell D L, Miller R V, Jeffrey W H. Ultraviolet radiation induced DNA damage in coral reef microbial communities. Mar Biol. 1998;130:537–543. [Google Scholar]
- 34.Mitchell D L. Radioimmunoassay of DNA damaged by ultraviolet light. In: Pfeiffer G, editor. Technologies for DNA damage and mutations. New York, N.Y: Plenum Publishing Corp.; 1996. pp. 73–83. [Google Scholar]
- 35.Mitchell D L, Karentz D. The induction and repair of DNA photodamage in the environment. In: Young A R, Bjorn L O, Moan J, Nultsch W, editors. Environmental UV photobiology. New York, N.Y: Plenum; 1993. pp. 345–377. [Google Scholar]
- 36.Müller-Niklas G, Heissenberger A, Puskeric S, Herndl G J. Ultraviolet-B radiation and bacterial metabolism in coastal waters. Aquat Microb Ecol. 1995;9:111–116. [Google Scholar]
- 37.Nicholson W L. Photoreactivation in the genus Bacillus. Curr Microbiol. 1995;31:361–364. doi: 10.1007/BF00294700. [DOI] [PubMed] [Google Scholar]
- 38.Nyström T, Olsson R M, Kjelleberg S. Survival, stress resistance, and alterations in protein expression in the marine Vibrio sp. strain S14 during starvation for different individual nutrients. Appl Environ Microbiol. 1992;58:55–65. doi: 10.1128/aem.58.1.55-65.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pakulski J D, Aas P, Jeffrey W, Lyons M, Von Waasenbergen L, Mitchell D, Coffin R. Influence of light on bacterioplankton production and respiration in a subtropical coral reef. Aquat Microb Ecol. 1998;14:137–148. [Google Scholar]
- 40.Palleroni N J. Genus Pseudomonas. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 141–199. [Google Scholar]
- 41.Paul J H. The use of Hoechst dyes 33258 and 33342 for enumeration of attached and planktonic bacteria. Appl Environ Microbiol. 1982;43:939–944. doi: 10.1128/aem.43.4.939-944.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paul J H, Carlson D. Genetic material in the marine environment: implication for bacterial DNA. Limnol Oceanogr. 1984;29:1091–1097. [Google Scholar]
- 43.Paul J H, Myers B. Fluorimetric determination of DNA in aquatic microorganisms by use of Hoechst 33258. Appl Environ Microbiol. 1982;43:1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Regan J D, Carrier W L, Gucinski H, Olla B L, Yoshida H, Fujimura R K, Wicklund R I. DNA as a solar dosimeter in the ocean. Photochem Photobiol. 1992;56:35–42. doi: 10.1111/j.1751-1097.1992.tb09599.x. [DOI] [PubMed] [Google Scholar]
- 45.Sancar A. Structure and function of DNA photolyase. Biochemistry. 1994;33:2–9. doi: 10.1021/bi00167a001. [DOI] [PubMed] [Google Scholar]
- 46.Schut F, De Vries E J, Gottschal J C, Robertson B R, Harder W, Prins R A, Button D K. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl Environ Microbiol. 1993;59:2150–2160. doi: 10.1128/aem.59.7.2150-2160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Setlow P. I will survive: protecting and repairing spore DNA. J Bacteriol. 1992;174:2737–2741. doi: 10.1128/jb.174.9.2737-2741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Setlow R B. Cyclobutane-type pyrimidin dimers in polynucleotides. Science. 1966;153:379–386. doi: 10.1126/science.153.3734.379. [DOI] [PubMed] [Google Scholar]
- 49.Stewart G J, Carlson C A, Ingraham J L. Evidence for an active role of donor cells in natural transformation of Pseudomonas stutzeri. J Bacteriol. 1983;156:30–35. doi: 10.1128/jb.156.1.30-35.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeuchi M, Sawada H, Oyaizu H, Yokota A. Phylogenetic evidence for Sphingomonas and Rhizomonas as nonphotosynthetic members of the alpha-4 subclass of the proteobacteria. Int J Syst Bacteriol. 1994;44:308–314. doi: 10.1099/00207713-44-2-308. [DOI] [PubMed] [Google Scholar]
- 51.Vincent W F, Quesada A. Ultraviolet radiation effects on cyanobacteria: implications for Antarctic microbial ecosystems. Antarct Res Ser. 1994;62:111–124. [Google Scholar]
- 52.Webb R B, Tuveson R W. Differential sensitivity to inactivation of nur and nur+ strains of Escherichia coli at six selected wavelengths in the UVA, UVB, and UVC ranges. Photochem Photobiol. 1982;36:525–530. doi: 10.1111/j.1751-1097.1982.tb04411.x. [DOI] [PubMed] [Google Scholar]
- 53.Wickman S, Carsten M. Effects of ultraviolet-B radiation on two Arctic microbial food webs. Aquat Microb Ecol. 1998;16:163–171. [Google Scholar]
- 54.Williamson C E, Zagarese H E, Schule P C, Hargreaves B R, Seva J. The impact of short-term exposure to UV-B radiation on zooplankton communities. Limnol Oceanogr. 1994;41:1024–1034. [Google Scholar]