Abstract
A shallow-water hydrothermal vent system in the Aegean Sea close to the island of Milos (Greece) was chosen to study the diversity and distribution of the chemolithoautotrophic sulfur-oxidizing bacterium Thiomicrospira. Cell numbers in samples from different regions around a solitary vent decreased toward the center of the vent (horizontal distribution), as well as with depth (vertical distribution), corresponding to an increase in temperature (from ca. 25 to 60°C) and a decrease in pH (from ca. pH 7 to 5). Thiomicrospira was one of the most abundant culturable sulfur oxidizers and was even dominant in one region. Phylogenetic analysis of Thiomicrospira spp. present in the highest most-probable-number (MPN) dilutions revealed that most of the obtained sequences grouped in two new closely related clusters within the Thiomicrospira branch. Two different new isolates, i.e., Milos-T1 and Milos-T2, were obtained from high-dilution (10−5) enrichments. Phylogenetic analysis indicated that isolate Milos-T1 is related to the recently described Thiomicrospira kuenenii and Hydrogenovibrio marinus, whereas isolate Milos-T2 grouped with the MPN sequences of cluster 2. The predominance of strain Milos-T2 was indicated by its identification in several environmental samples by hybridization analysis of denaturing gradient gel electrophoresis (DGGE) patterns and by sequencing of one of the corresponding bands, i.e., ML-1, from the DGGE gel. The results shown in this paper support earlier indications that Thiomicrospira species are important members of hydrothermal vent communities.
Hydrothermal vents are located at tectonically active regions all over the world; the most spectacular are those at the deep-sea floor, where very unusual light-independent ecosystems based on chemolithoautotrophic sulfur-oxidizing bacteria exist (see, e.g., references 16 and 17). However, hydrothermal vents have also been found in several shallow-water areas (see, e.g., references 15, 26, 28, 30, and 31). These shallow-water vents differ from their deep-sea counterparts mainly by the presence of light. Accordingly, the biology of these ecosystems is different too; e.g., at shallow-water vents, photosynthetic organisms, such as benthic microalgae and cyanobacteria, can be present (see, e.g., references 9 and 26).
Off the Greek island of Milos in the Aegean Sea, shallow-water vents were found at depths ranging from the littoral zone down to 115 m (9). A rich macrofauna depending on endosymbiotic bacteria is absent at the Milos vents (9, 29). Until now, the only sulfur-oxidizing bacteria (SOB) described for the hydrothermal vents around Milos were thiobacilli and Achromatium volutans at vents less than 30 m deep (9) and Thioploca at a vent 46 m deep (11). These identifications were based on microscopic observations. For Achromatium volutans and Thioploca, this is appropriate because of the particular morphological features of these organisms. For thiobacilli, however, microscopic identification is more problematic, because the rod-shaped morphology might easily be confused with, e.g., the recently described rod-shaped isolates of Thiomicrospira (5, 6).
Members of the genus Thiomicrospira are chemolithoautotrophic bacteria, which use reduced sulfur compounds as electron donors and obtain carbon from CO2 (7, 19). Although Thiomicrospira spp. were detected in and isolated from several different environments (see, e.g., references 3 and 34), their ecological importance remained obscure for most habitats. Several isolates were obtained from intertidal mud flats (3, 18, 35, 36). However, in a recent study it was found that Thiomicrospira constituted only a minor fraction of the total sulfur-oxidizing bacterial community in this habitat, pointing to a minor role of these microorganisms (4). On the other hand, an analysis by molecular methods, i.e., denaturing gradient gel electrophoresis (DGGE) and sequencing of PCR amplified 16S rDNA fragments, demonstrated that Thiomicrospira spp. were dominant community members of hydrothermal vent sites at the Mid-Atlantic Ridge (21).
Here, we present the results of an investigation of the diversity and distribution of Thiomicrospira populations at a shallow-water hydrothermal vent off Milos. For this study, a combination of molecular and microbiological methods was used in an attempt to understand the ecological importance of these microorganisms.
MATERIALS AND METHODS
Sampling.
Samples were taken in June and September 1996 and in June 1997 at a solitary gaseous hydrothermal vent, located at a water depth of 8 m in Palaeochori Bay (24°31.220′E, 36°40.391′N), a sandy bay in the southeastern part of the Greek island of Milos in the Aegean Sea. The mean composition of the discharged gases from the different seeps was 80.5% CO2, 1.2% H2S, 0.8% CH4, and 0.4% H2 (10). The reduced hydrothermal fluid has an increased salinity of up to 58‰, compared to 39‰ for the ambient seawater (29).
Sampling was carried out along a transect from the center of the almost circular vent to the surrounding area. Sediment cores were taken with polycarbonate cylinders (36 mm in diameter), by SCUBA divers, along the transect at several distances from the vent center. In June 1996, samples were taken at 10, 123, 165, and 235 cm from the vent center, and in September 1996, samples were taken at 30, 117, and 200 cm. The distances for sampling were chosen according to different physicochemical parameters around the vent, e.g., increasing temperature and a decreasing pH towards the center and with sediment depth (25). For the 200- and 235-cm distances, moderate environmental conditions were described (25°C and pH 7.0 in all layers). For the 117-, 123-, and 165-cm distances, the temperature and pH were moderate at the surface (25°C and pH 6.5), but increased and decreased with depth, respectively (40°C and pH 5.5 at a sediment depth of 3 cm). For the cores taken at 117 and 123 cm, the surface consisted of a white precipitate that was 2 to 5 mm thick. This conspicuous white precipitate was formed on the sediment surface and increased in thickness under calm weather conditions. In September 1996, a second core was taken at 117 cm from the vent center, 1 week later than the other samples, at which time no white precipitate was present. At 10 and 30 cm from the center, respectively, temperatures of about 30°C at the surface and 60°C at a sediment depth of 3 cm were measured, while the pH values were between 5.7 at the surface and about 5.0 at a sediment depth of 3 cm. After sampling, the cores were brought to the shore, sliced immediately, and further processed for most-probable-number (MPN) counts and DNA extraction.
Subsampling of sediment cores.
For all samples taken in June 1996, the following layers were investigated: water directly from above the sediment surface and sediment samples from depths of 0 to 10, 10 to 20, and 20 to 30 mm. For samples taken in September 1996, cores were sliced at a finer resolution as follows: water directly from above the sediment surface and sediment from depths of 0 to 5, 8 to 13, and 16 to 26 mm. Water from just above the surface was sampled with a syringe. Sediment cores were subsampled by extruding the sediment from the polycarbonate tubes and slicing each core. Dilutions for the MPN series were performed directly after slicing. Subsamples of the same layers for DNA extraction were frozen immediately in liquid N2 until used in further processing.
MPN counts of sulfur-oxidizing bacteria.
The MPN technique was used to estimate the abundance of SOB in the different subsamples taken in September 1996. Sediment cores were subsampled as described above and serially diluted (1:10 steps) with mineral medium without a substrate. Between every dilution step, the samples were vigorously shaken on a vortex apparatus to dislodge the bacteria from the sediment particles. From each dilution, three replicate tubes containing growth medium were inoculated and incubated at their approximate in situ temperature. Mineral medium with 20 mM thiosulfate (added by sterile filtration to the autoclaved medium) as the sole electron donor was used. The composition of the mineral medium was (per 1,000 ml of H2O) 29 g of NaCl, 1 g of (NH4)2SO4, 1.5 g of MgSO4 · 7H2O, 0.42 g of CaCl2 · 2H2O, 0.5 g of K2HPO4, 0.7 g of KCl, 0.05 g of vitamin B12, and 1 ml trace element solution with EDTA (32). Bromthymol blue was added as a pH indicator at a concentration of 4 mg liter−1. K2HPO4 was autoclaved separately and added to the medium after autoclaving.
With the second core taken at a distance of 117 cm, an MPN series, in which the salinity of the medium was increased to 55‰ by adding NaCl, was performed. This salinity was similar to the salinity of the outflowing brine (29). This parallel MPN series was carried out to investigate whether there were SOB specifically adapted to the higher salt concentration.
The cultures were incubated in the dark, to avoid growth of phototrophic bacteria, and at their approximate in situ temperatures (see Fig. 1). Growth was monitored by observing a color change of the pH indicator and by microscopic observation. The presence of Thiomicrospira cells in the MPN cultures was determined by using a Thiomicrospira-specific primer set in a PCR with cells taken directly from the cultures (3). The numbers of SOB and Thiomicrospira cells were determined by using the MPN index of the American Public Health Association (1) in three parallel determinations.
FIG. 1.
MPN counts of chemolithoautotrophic SOB (dark bars) in sediment samples from the shallow-water hydrothermal vent system off Milos. The Thiomicrospira cell number (light bars) were determined by enzymatic amplification of the 16S rDNA with genus-specific primers. The error bars indicate the 95% confidence intervals. Percentages shown next to the Thiomicrospira bar are the Thiomicrospira numbers given as percentages of total SOB. The incubation temperatures are also given.
Isolation and cultivation of bacteria.
High-dilution enrichments were performed with samples taken in June 1996 and June 1997, with the intention of obtaining the most abundant SOB. The medium and the culture conditions were the same as for the MPN cultures. Aliquots from the tubes were transferred on solid agar plates, containing the same culture medium and 1% (wt/vol) agar (Difco). All known Thiomicrospira species and strains isolated so far form intensely yellow colonies due to sulfur precipitation. Additionally, they all produce acid. Colonies with these characteristics were chosen for identification by a Thiomicrospira-specific PCR (3). The positive colonies obtained were transferred at least three times before being considered pure.
Nucleic acid extraction.
DNA was extracted from sediment and water samples by the method described by Zhou et al. (37) and modified as described by Sievert et al. (25).
Oligonucleotides used for PCR.
Oligonucleotides TMS128F and TMS849R are specific for the 16S rDNA of bacteria belonging to the genus Thiomicrospira (3). With these primers, about 700-bp 16S rDNA fragments are obtained. The primers have been used for the identification of Thiomicrospira isolates and for the detection of bacteria belonging to this genus in the MPN cultures. The PCR products obtained from the highest MPN dilutions in which Thiomicrospira could be detected were sequenced to determine the phylogenetic affiliation of the respective organisms.
Oligonucleotides GM3F and GM4R are specific for the 16S rDNA of members of the domain Bacteria and were used as primers in a PCR to amplify the nearly complete (1,500-bp) genes of the new isolates. The PCR products obtained with these primers were used for sequencing.
The primer pair GM5F and 907R amplifies the 16S rDNA of members belonging to the domain Bacteria and was used to obtain 550-bp fragments for DGGE analysis. The sequences of both primer pairs (GM3F plus GM4R and GM5F plus 907R) have been published by Muyzer et al. (21).
PCR amplification of 16S rDNA fragments.
PCR amplifications were performed as described by Muyzer et al. (21). A touchdown PCR (12) was performed for primer pair GM5F plus 907R (annealing temperature from 65 to 55°C in 20 cycles). For primer pairs TMS128F plus TMS849R (annealing temperature of 44°C), and GM3F plus GM4R (annealing temperature of 40°C), no touchdown PCR was used. The amplification products were analyzed as described by Muyzer et al. (21) before being subjected to further characterization by DGGE analysis or DNA sequencing.
DGGE analysis of PCR products.
DGGE was performed with the D-Gene system (Bio-Rad Laboratories, Inc.). The protocol as described by Brinkhoff and Muyzer (3) was used: 1-mm-thick, 6% (wt/vol) polyacrylamide gels, 1× TAE electrophoresis buffer (40 mM Tris-HCl [pH 8.3], 20 mM acetic acid, 1 mM EDTA), 20 to 70% denaturant, and an electrophoresis time of 20 h at a constant voltage of 100 V. After electrophoresis, the gels were stained with ethidium bromide and photographed as described previously (22).
Hybridization analysis of blotted DGGE patterns.
DGGE patterns were transferred to nylon membranes (Hybond-N+; Amersham, Little Chalfont, United Kingdom) by electroblotting and hybridized with the digoxigenin-labeled Thiomicrospira-specific probe TMS849R, as previously described (3).
Sequencing of PCR products.
PCR products were purified by using the Qiaquick Spin PCR purification kit (Qiagen Inc., Chatsworth, Calif.). The Taq Dyedeoxy Terminator cycle-sequencing kit (Applied Biosystems, Foster City, Calif.) was used to sequence the 16S rDNA fragments. The sequencing primers for the nearly complete 16S rDNA of bacterial isolates were GM3F, GM1F, and GM4R (8). The sequencing primers for Thiomicrospira 16S rDNA fragments obtained from MPN cultures with the specific primer pair were the same Thiomicrospira-specific oligonucleotides, TMS128F and TMS849R (3). One band in the DGGE gel, which hybridized with the Thiomicrospira-specific probe TMS849R, was excised from the gel, reamplified with the primer pair GM5F and 907R, and sequenced with the Thiomicrospira-specific primer TMS849R. The sequence reaction mixtures were electrophoresed on an Applied Biosystems 373S DNA sequencer.
Comparative analysis of 16S rRNA sequences.
The 16S rRNA sequences were aligned with those obtained from the Ribosomal Database Project (20) and GenBank (2). Sequence alignments were prepared with the sequence editor SEQAPP (14). Phylogenetic trees were created by using the neighbor joining algorithm with maximum-likelihood correction as implemented in the test version of PAUP 4 developed by Swofford.
Nucleotide sequence accession numbers.
The sequences obtained in this study are available from the EMBL nucleotide sequence database under accession nos. AJ237757 to AJ237769.
RESULTS
MPN counts of SOB and Thiomicrospira cells.
Numbers of cultivable aerobic chemolithoautotrophic SOB and Thiomicrospira cells counted by MPN serial dilution were variable for the different zones and layers (Fig. 1). Where Thiomicrospira spp. were present, they accounted for between 2 and 100% of the total numbers of cultivable SOB, which indicates that Thiomicrospira spp. belonged to the dominant SOB in this habitat. Generally, the numbers of SOB varied between not detectable and 1.4 × 106 cells g (wet weight) of sediment−1. Thiomicrospira cell numbers varied between not detectable and 2.7 × 105 cells g (wet weight) of sediment−1.
In the first zone, at 30 cm from the vent center, no or only small numbers of SOB, up to 2.4 × 102 cells g (wet weight) of sediment−1, could be obtained. Thiomicrospira cells were not detectable in the MPN cultures of this zone (Fig. 1A).
In the second zone, at 117 cm from the vent center, the numbers of SOB and Thiomicrospira cells were different for the two cores examined. The surface of the first core (core I, Fig. 1B) contained the white precipitate mentioned above, which was absent at the surface of the second core (core II, Fig. 1C). Core II was used for the MPN counts with the increased salinity. Whereas the cell numbers of SOB obtained for the three sediment layers for core II were only slightly smaller than those obtained for core I, the numbers of Thiomicrospira spp. were appreciable smaller in the sediment samples of core II than of core I. This is also indicated by the finding that Thiomicrospira spp. constituted a much smaller fraction of the total numbers of SOB in core II than in core I. The largest numbers of SOB and Thiomicrospira at 117 cm were obtained with the sample from the water above the sediment surface of core I, which contained the white precipitate (4.1 × 104 and 1.9 × 104 cells ml of water−1, respectively [Fig. 1B]).
For the third zone, at 200 cm from the vent center, the smallest numbers of SOB and Thiomicrospira cells were obtained for the water above the sediment surface (Fig. 1D). The largest numbers of SOB (1.4 × 106 cells g−1) were found in the second sediment layer (8 to 13 mm deep), while the largest numbers of Thiomicrospira cells (2.7 × 105 cells g−1) were found in the first sediment layer (0 to 5 mm deep). Here Thiomicrospira species were also one of the dominant SOB, since they accounted for between 2 and 100% of the total numbers of SOB. Comparison of the different zones revealed generally the largest numbers of SOB and Thiomicrospira cells in the sediment layers of the third zone.
Detection of Thiomicrospira by DGGE and hybridization analysis.
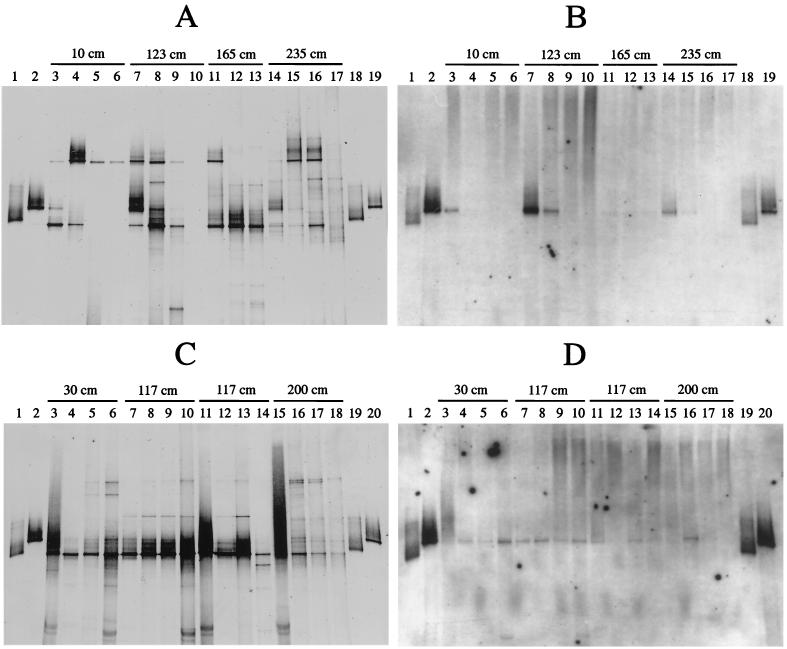
DGGE profiles of 16S rDNA fragments obtained from sediment and water samples from June and September 1996, respectively (Fig. 2A and C), were hybridized with a probe specific for the genus Thiomicrospira (Fig. 2B and D). Positive signals were obtained with the bands from two isolates and with bands from samples of different zones and layers. The results for the samples from June 1996 are described first. For the first zone, at 10 cm from the vent center, a signal was obtained for the water above the sediment (Fig. 2B, lane 3). For the second zone (123 cm from the vent center), signals were obtained for the water above the sediment and the first sediment layer (Fig. 2B, lanes 7 and 8). No signals were obtained for the third zone (Fig. 2B, 165 cm from the vent center). For the fourth zone (235 cm) a signal was obtained for the water above the sediment (Fig. 2B, lane 14) and a weak signal was obtained for the first sediment layer (Fig. 2B, lane 15).
FIG. 2.
Hybridization analysis of DGGE profiles of 16S rDNA fragments obtained with primers specific for Bacteria and template DNA from environmental samples taken along a transect from the shallow-water hydrothermal vent system off Milos and Thiomicrospira isolates obtained from the same location. (A) DGGE pattern of samples taken in June 1996. Lanes: 1 and 18, isolate Milos T-1; 2 and 19, isolate Milos T-2; 3 to 6, samples taken at 10 cm from the vent center (lane 3, surface; lane 4, 0 to 10 mm deep; lane 5, 10 to 20 mm deep; lane 6, 20 to 30 mm deep); 7 to 10, equivalent samples taken at 123 cm from the vent center; 11 to 13, samples taken at 165 cm from the vent center (lane 11, 0 to 10 mm deep; lane 12, 10 to 20 mm deep; lane 13, 20 to 30 mm deep); 14 to 17, equivalent samples taken at 235 cm to those taken at 10 cm. (B) Hybridization analysis of the pattern in panel A with a Thiomicrospira-specific digoxigenin-labeled probe. (C) DGGE pattern of samples taken in September 1996. Lanes: 1 and 19, isolate Milos T-1; 2 and 20, isolate Milos T-2; 3 to 6, samples taken at 30 cm from the vent center (lane 3, surface; lane 4, 0 to 5 mm depth; lane 5, 8 to 13 mm depth; lane 6, 16 to 26 mm depth); 7 to 10, equivalent samples taken at 117 cm from the vent center (sediment core number 1); 11 to 14, equivalent samples taken at 117 cm from the vent center (sediment core number 2); 15 to 18, equivalent samples taken at 200 cm from the vent center. (D) Hybridization analysis of the pattern in panel C with the Thiomicrospira-specific probe.
Positive signals were also obtained for the bands from two Thiomicrospira isolates and from samples taken in September 1996. For the first zone, at 30 cm from the vent center, signals were obtained for the first, second, and third sediment layers (Fig. 2D, lanes 4, 5, and 6, respectively) but not for the water above the sediment (lane 3). For the two cores taken in the second zone (117 cm from the vent center), slightly different results were obtained. For the first core (lanes 7 to 10), signals for the surface (lane 7), and all sediment layers (lanes 8 to 10) were obtained (although for the second layer [lane 9] only a weak signal was found). For the second core (lanes 11 to 14), only weak signals were obtained for the surface and the second sediment layer (lanes 11 and 13, respectively). For the third zone (200 cm from the vent center), a signal was obtained only for the first sediment layer (lane 16). A signal from the water above the sediment might have been obscured by a poor PCR, as indicated by the smear in Fig. 2C, lane 15). All hybridization signals obtained with the Thiomicrospira-specific probe run in both gels at the same position as the bands belonging to strain Milos T-2 (Fig. 2B, lanes 2 and 19, and Fig. 2D, lanes 2 and 20), which indicates identical or nearly identical sequences.
Phylogenetic analysis of Thiomicrospira from MPN cultures.
Although the Thiomicrospira species from the highest dilutions of the MPN counts performed in September 1996 were not isolated, partial sequences of their 16S rDNA could be determined by using the Thiomicrospira-specific primer pair for amplification and sequencing. With the exception of the sequence of Tms-MPN/Milos-CIV1, all the sequences from the MPN cultures were very similar (ca. 99%) and grouped in two clusters, i.e., clusters 2 and 3 (Fig. 3). The sequence of Tms-MPN/Milos-CIV1, obtained from the MPN culture from the deepest sediment layer (16 to 26 mm deep) of the second zone (117 cm from the vent center) and cultured at 55‰ salinity, was very different from all others and was shown to be the deepest-branching Thiomicrospira sequence known so far. For one MPN culture (Tms-MPN/Milos-DIV5), a mixed sequence was obtained, indicating that more than one Thiomicrospira sequence type was present in this culture.
FIG. 3.
Neighbor-joining tree showing the phylogenetic affiliation of the Thiomicrospira isolates (strain Milos-T1 and Milos-T2), and of Thiomicrospira spp. present in MPN cultures (indicated by the annotation Tms-MPN/Milos), both from the shallow-water hydrothermal vent site near Milos. ML-1 is the sequence from the excised DGGE band, which had the same position in the gel as the band of Thiomicrospira sp. strain Milos-T2. Tms-MPN/JB sequences were obtained from Thiomicrospira spp. present in MPN cultures of an intertidal mud flat of Jadebusen Bay, Germany, and are published elsewhere (3). The tree is created after comparison of partial (700-nucleotide) 16S rRNA sequences. Sequences marked with an asterisk are molecular isolates. The sequences determined in this study are shown in bold; A to D refer to the different zones around the vent (30 cm, 117 cm [core I], 117 cm [core II] and 200 cm [Fig. 1]); Roman numerals I to IV refer to the different layers (from surface to 16 to 26 mm deep); and Arabic numbers refer to the different MPN dilutions.
Characterization of Thiomicrospira isolates.
Two isolates, which were both obtained from 10−5 dilutions of the enrichments, gave a positive signal with the Thiomicrospira-specific PCR. Strain Milos T-1 was received from a sample taken in June 1996 from the upper sediment layer, underneath the white precipitate, at 123 cm from the vent center. Strain Milos T-2 was isolated directly from the white precipitate in June 1997. While isolate Milos T-1 is vibrio shaped, like most other Thiomicrospira spp., strain Milos T-2 is rod-shaped, like two recently described new Thiomicrospira species, T. frisia (5) and T. chilensis (6).
Phylogenetic analysis confirmed the affiliation of the isolates to the genus Thiomicrospira (Fig. 3). The two new isolates are not closely related to each other and are located in different subclusters of the Thiomicrospira lineage. Strain Milos T-1 is related to Thiomicrospira kuenenii and Hydrogenovibrio marinus (Fig. 3, cluster 1). The latter differs from Thiomicrospira spp. only in the ability to use hydrogen as an electron donor (23). Strain Milos T-2 grouped with sequences from the MPN cultures and the excised DGGE band ML-1, all obtained during this study (Fig. 3, cluster 2). Band ML-1, which runs in the DGGE gel at the same position as the band from strain Milos T-2 and hybridized with the Thiomicrospira-specific probe, was excised from a DGGE gel and subsequently sequenced. Comparison of the sequence of the excised DGGE band ML-1 with that of strain Milos T-2 resulted in a 1-bp difference in a total of 470 bp (99.8% sequence similarity).
DISCUSSION
The high abundance of Thiomicrospira cells in some zones of the shallow-water hydrothermal vent off the isle of Milos can probably be attributed to the presence of high concentrations of reduced sulfur compounds and CO2 (10), which both favor the growth of chemolithoautotrophic sulfur-oxidizing bacteria.
The distribution pattern of Thiomicrospira as determined by the MPN counts probably reflects the influence of environmental parameters, such as temperature, pH, and oxygen concentration. These environmental parameters were determined concurrently along the same transect (25). Temperatures of about 30 to 60°C and pH values between 5.7 and 5.0 were measured for the layers of the first zone (30 cm from the center of the vent). This might explain the small numbers of SOB in general and the absence of Thiomicrospira.
At 117 cm from the center of the vent, the numbers of SOB and Thiomicrospira cells were decreasing with depth (Fig. 1B). While the environmental conditions of this zone are moderate at the sediment surface (25°C and pH 6.5), temperatures of about 40°C and pH values of 5.5 and lower at the deeper layers, as well as a decreasing oxygen concentration with depth, seemed to inhibit the growth of SOB and Thiomicrospira. The numbers of SOB as determined in the MPN count with an increased salinity of 55‰ (Fig. 1C) were slightly smaller for the sediment layers than were those obtained with 30‰ salinity, even though the higher value reflected the in situ concentration of the outflowing brine. This indicates that SOB adapted to a higher salinity were not dominant. The finding that Thiomicrospira spp. constituted a smaller fraction of the total numbers of SOB in the sediment samples from core II than in those from core I might be an indication that Thiomicrospira spp. were more strongly affected by the increased salinity. However, it could also be related to natural variability. The second possibility is supported by the observation that bands hybridizing with the Thiomicrospira-specific probe were less abundant and weaker for the samples from core II than for those from core I (Fig. 2B and D). The significantly smaller numbers of SOB and Thiomicrospira cells determined for the surface layer of the MPN counts with the increased salinity might be due to the absence of the white precipitate of the respective sample (25).
At 200 cm from the center of the vent, moderate conditions (25°C and pH 7.0) and a deeper oxygen penetration seemed to allow the presence of larger numbers of SOB and Thiomicrospira cells in deeper layers (Fig. 1D). The predominance of Thiomicrospira cells in the first sediment layer of this zone indicates that the environmental parameters favored the growth of members of this genus. It is worth noting, however, that Thiomicrospira spp. were not the only SOB present in the highest positive dilutions (24). At 200 cm, Thiomicrospira cells were also found in deeper layers at nearly the same order of magnitude as in the upper layers, although oxygen was not present (25). This phenomenon was described previously for this genus in sediment of an intertidal mud flat (4). However, in the latter habitat, Thiomicrospira populations were found to be metabolically active only in the oxic part of the sediment.
Comparison of the results of the hybridization patterns with the MPN results showed differences for the distribution of Thiomicrospira, especially for the first zone, i.e., at 10 and 30 cm from the vent center. No Thiomicrospira cells were detected here by the MPN counts. On the other hand, hybridization analysis of both DGGE patterns gave positive signals for this zone (Fig. 2B and D). The physicochemical parameters should actually allow the growth of Thiomicrospira spp. at the surface of this region, as indicated by the positive band from the samples taken in June 1996 (Fig. 2B, lane 3). However, the presence of Thiomicrospira in the sediment samples taken in September 1996 (Fig. 2D, lanes 4 to 6) is in contrast to the environmental parameters. It is unlikely that Thiomicrospira spp. grew in these layers, since the in situ temperature (40 to 60°C) was at or above the upper growth limit of known species (7) and since oxygen, which is needed as electron donor (7), was not present (25). That these Thiomicrospira populations were not detected by the MPN determination is probably a result of their special adaptation to the extreme environmental conditions that were not imitated in the MPN culture. This possibility would be supported by the finding of the sequence Tms-MPN/Milos-CIV1, which was obtained from the MPN cultures with a salinity of 55‰. Its phylogenetic position and the extreme environment where the sequence was found (temperature around 40°C, pH 5.0 to 5.5, and no oxygen) might mean that the corresponding organism has a different physiology from known Thiomicrospira species. However, the position of the Thiomicrospira-positive bands for the samples obtained at 30 cm from the vent center were at the same position in the gel as was the band for the isolate Milos-T2, which argues against a unique population. Most probably, resuspension of the upper sediment layers due to a storm, before to the sampling in September 1996, was responsible for the occurrence of Thiomicrospira populations in deeper sediment layers (25). This would also account for the observation that bands belonging to Thiomicrospira spp. were confined to the upper sediment layers and the water above the sediment in June 1996 (Fig. 2B, lanes 3, 7, 8, 14, and 15), which is in agreement with the known physiological capabilities of this genus (7, 19), whereas they were more homogeneously distributed with no clear trend in September 1996 (Fig. 2D).
By using a PCR-DGGE-hybridization assay, it was found that a particular bacterial population representing 0.1% of the total community could still be detected (27). In the present study, we could detect Thiomicrospira populations, even though their contribution to the total cell numbers (determined by acridine orange direct counts [25]) was well below 0.1%. Different explanations, which are not mutually exclusive, could account for this. First, we do not know the extent to which Archaea contributed to the total cell counts. Thus, the contribution of Thiomicrospira cells to the total bacterial cells might be higher. Second, the association of cells in clumps or with particles, as well as the insensitivity of the detection method (33), might have led to an underestimation of Thiomicrospira cell numbers. In addition, it is conceivable that different and highly related Thiomicrospira populations were present in the highest dilutions. Third, a bias toward Thiomicrospira might have been caused by the DNA extraction and/or the PCR; i.e., Thiomicrospira cells were more efficiently lysed and/or their 16S rRNA might have been specifically overamplified compared to other sequences. Furthermore, not all cells stained with acridine orange might have contained enough DNA for amplification. The hybridization analysis of DGGE patterns with the environmental samples obtained in June and September 1996 showed that all bands, which gave positive signals, ran at the same position in the denaturing gel as those from the new isolated Thiomicrospira strain Milos T-2. Based on these data, it can be seen that this is apparently the dominant Thiomicrospira sequence type of this ecosystem. This was confirmed by the isolation of this organism from a high-dilution enrichment of a sample taken in June 1997 and additionally by the sequence similarity of a band excised from a DGGE gel, having 99.8% sequence homology to strain Milos T-2. No bands corresponding to the band of strain Milos T-1 were found in the DGGE patterns, and no similar sequence was obtained from the MPN cultures, even though strain Milos T-1 was also isolated from a high-dilution enrichment, like strain Milos T-2. This finding cannot be explained by the present data.
Almost all the sequences presented in this study show differences among each other of less than 2%. Therefore, physiological adaptations to the different zones of the studied vent system might be possible without greater differences in the 16S rDNA. An indication for this is that all sequences obtained from the MPN cultures of the second zone (117 cm from the vent center), grown at a salinity of 30‰, are identical. On the other hand, most sequences obtained from the MPN counts of the same zone, grown at a salinity of 55‰, formed another cluster. This might reflect an adaptation of specific Thiomicrospira populations to a higher salinity. This would be similar to the finding that highly related cyanobacterial populations are adapted to different temperatures according to their occurrence in a thermal gradient along an outflow of a hot spring (13). It was generally observed that the more moderate conditions in the outer zone of the vent correlated with a higher phylogenetic diversity of the Thiomicrospira sequences obtained from this zone. This reflects a trend that is also apparent for the total bacterial community (25).
Due to the presence of light, chemolithoautotrophic SOB were found not to be the only primary producers of the shallow-water vent system off Milos (9, 29). However, results from this study clearly indicate that even though differences between the deep-sea and shallow-water hydrothermal vent systems exist, Thiomicrospira is in both cases an important member of the sulfur-oxidizing community and must be taken into account in further ecological and microbiological investigations of these ecosystems.
ACKNOWLEDGMENTS
We thank Wiebke Ziebis, Susanne Menger, and Guido Lützenkirchen for SCUBA diving, sampling, and help with field work, and we thank the mechanical workshop of the MPI for building the sampling devices. We are grateful for support by other participants of the EU-funded project Hydrothermal Fluxes and Biological Production in the Aegean Sea. We acknowledge the Greek authorities for permission to undertake SCUBA diving and field work.
This research was financially supported by the Max-Planck-Society, Munich, Germany, and by the European Union under MAST CT-95-0021.
REFERENCES
- 1.American Public Health Association. Standard methods for the examination of water and wastewater, including bottom sediments and sludge. Washington, D.C: American Public Health Association; 1969. pp. 604–609. [Google Scholar]
- 2.Benson D A, Boguski M S, Lipman D J, Ostell J. GenBank. Nucleic Acids Res. 1997;25:1–6. doi: 10.1093/nar/25.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinkhoff T, Muyzer G. Increased species diversity and extended habitat range of sulfur-oxidizing Thiomicrospira spp. Appl Environ Microbiol. 1997;63:3789–3796. doi: 10.1128/aem.63.10.3789-3796.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinkhoff T, Santegoeds C M, Sahm K, Kuever J, Muyzer G. A polyphasic approach to study the diversity and vertical distribution of sulfur-oxidizing Thiomicrospira species in coastal sediments of the German Wadden Sea. Appl Environ Microbiol. 1998;64:4650–4657. doi: 10.1128/aem.64.12.4650-4657.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinkhoff T, Muyzer G, Wirsen C O, Kuever J. Characterization of Thiomicrospira kuenenii sp. nov. and Thiomicrospira frisia sp. nov., two mesophilic obligately chemolithoautotrophic sulfur-oxidizing bacteria isolated from an intertidal mud flat. Int J Syst Bacteriol. 1999;49:385–392. doi: 10.1099/00207713-49-2-385. [DOI] [PubMed] [Google Scholar]
- 6.Brinkhoff T, Muyzer G, Wirsen C O, Kuever J. Characterization of Thiomicrospira chilensis sp. nov., a mesophilic obligately chemolithoautotrophic sulfur-oxidizing bacterium isolated from a Thioploca mat. Int J Syst Bacteriol. 1999;49:875–879. doi: 10.1099/00207713-49-2-875. [DOI] [PubMed] [Google Scholar]
- 7.Brinkhoff, T., J. Kuever, G. Muyzer, and H. W. Jannasch. The genus Thiomicrospira. In D. J. Brenner (ed.), Bergey’s manual of systematic bacteriology, 2nd ed., vol. 2, in press. The Williams & Wilkins Co., Baltimore, Md.
- 8.Buchholz-Cleven B E E, Rattunde B, Straub K L. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole cell hybridization. Syst Appl Microbiol. 1997;20:301–309. [Google Scholar]
- 9.Dando P R, Hughes J A, Thiermann F. Preliminary observations on biological communities at shallow hydrothermal vents in the Aegean Sea. In: Parson L M, Walker C L, Dixon D R, editors. Hydrothermal vents and processes. Special Publication 87. London, United Kingdom: Geological Society; 1995. pp. 303–317. [Google Scholar]
- 10.Dando P R, Hughes J A, Leahy Y, Niven S J, Taylor L J, Smith C. Rates of gas venting from submarine hydrothermal areas around the island of Milos in the Hellenic Volcanic Arc. Continental Shelf Res. 1995;15:913–929. [Google Scholar]
- 11.Dando P R, Hooper L E. Hydrothermal Thioploca—more unusual mats from Milos. BRIDGE Newsl. 1997;1997:45–47. [Google Scholar]
- 12.Don R H, Cox P T, Wainwright B, Baker K, Mattick J S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert D G. SeqApp—a biosequence analysis application. Bloomington: Indiana University; 1992. [Google Scholar]
- 15.Hashimoto J, Miura T, Fujikura K, Ossaka J. Discovery of vestimentiferan tube-worms in the euphotic zone. Zool Sci. 1993;10:1063–1067. [Google Scholar]
- 16.Jannasch H W, Wirsen C O. Chemosynthetic primary production at East Pacific sea floor spreading centers. BioScience. 1979;29:592–598. [Google Scholar]
- 17.Jannasch H W, Mottl M J. Geomicrobiology of deep-sea hydrothermal vents. Science. 1985;229:717–725. doi: 10.1126/science.229.4715.717. [DOI] [PubMed] [Google Scholar]
- 18.Kuenen J G, Veldkamp H. Thiomicrospira pelophila, gen. nov., sp. nov., a new obligately chemolithotrophic colorless sulfur bacterium. Antonie Leeuwenhoek. 1972;38:241–256. doi: 10.1007/BF02328096. [DOI] [PubMed] [Google Scholar]
- 19.Kuenen J G, Robertson L A, Tuovinen O H. The genera Thiobacillus, Thiomicrospira, and Thiosphaera. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. 2nd ed. Berlin, Germany: Springer-Verlag KG; 1992. pp. 2638–2657. [Google Scholar]
- 20.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muyzer G, Teske A, Wirsen C O, Jannasch H W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 22.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. 3rd ed. 3.4.4. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–27. [Google Scholar]
- 23.Nishihara H, Igarashi Y, Kodama T. Hydrogenovibrio marinus gen. nov. sp. nov., a marine obligately chemolithoautotrophic hydrogen-oxidizing bacterium. Int J Syst Bacteriol. 1991;41:130–133. [Google Scholar]
- 24.Sievert, S. M., and J. Kuever. 1999. Unpublished data.
- 25.Sievert S M, Brinkhoff T, Muyzer G, Ziebis W, Kuever J. Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos island (Greece) Appl Environ Microbiol. 1999;65:3834–3842. doi: 10.1128/aem.65.9.3834-3842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorokin D Y. Oxidation of reduced sulphur compounds in volcanic regions in the Bay of Plenty (New Zealand) and Matupy Harbour (New Britian, Papua-New Guinea) Proc USSR Acad Sci Ser B. 1991;3:376–387. [Google Scholar]
- 27.Straub K L, Buchholz-Cleven B E E. Enumeration and detection of anaerobic ferrous iron-oxidizing, nitrate-reducing bacteria from diverse European sediments. Appl Environ Microbiol. 1998;64:4846–4856. doi: 10.1128/aem.64.12.4846-4856.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarasov V G, Propp M V, Propp L N, Zhirmunsky A V, Namsaraev B B, Gorlenko V M, Starynin D A. Shallow-water gasohydrothermal vents of Ushishir Volcano and the ecosystem of Kraternaya Bight (the Kurile Islands) PSZNI Mar Ecol. 1990;11:1–23. [Google Scholar]
- 29.Thiermann F, Akoumianaki I, Hughes J A, Giere O. Benthic fauna of a shallow-water gasohydrothermal vent area in the Aegean Sea (Milos, Greece) Mar Biol. 1997;128:149–159. [Google Scholar]
- 30.Trager G C, DeNiro M J. Chemoautotrophic sulfur bacteria as a food source for mollusks at intertidal hydrothermal vents: evidence from stable isotopes. Veliger. 1990;33:359–362. [Google Scholar]
- 31.Vidal V M, Vidal F M, Isaacs J D. Coastal submarine hydrothermal activity off northern Baja California. J Geophys Res. 1978;83:1757–1774. [Google Scholar]
- 32.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. Berlin, Germany: Springer-Verlag KG; 1991. pp. 3352–3378. [Google Scholar]
- 33.Wirsen C O, Tuttle J H, Jannasch H W. Activities of sulfur-oxidizing bacteria at the 21°N East Pacific Rise vent site. Mar Biol. 1986;92:449–456. [Google Scholar]
- 34.Wirsen C O, Brinkhoff T, Kuever J, Muyzer G, Molyneaux S, Jannasch H W. A new Thiomicrospira strain from the Mid-Atlantic ridge compared to known hydrothermal vent isolates. Appl Environ Microbiol. 1998;64:4057–4059. doi: 10.1128/aem.64.10.4057-4059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood A P, Kelly D P. Isolation and physiological characterization of Thiobacillus thyasiris sp. nov., a novel marine facultative autotroph and the putative symbiont of Thyasira flexuosa. Arch Microbiol. 1989;152:160–166. [Google Scholar]
- 36.Wood A P, Kelly D P. Reclassification of Thiobacillus thyasiris as Thiomicrospira thyasirae comb. nov., an organism exhibiting pleomorphism in response to environmental conditions. Arch Microbiol. 1993;159:45–47. [Google Scholar]
- 37.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]