Abstract
Genetic variants in filaggrin (FLG) are key in eczema and are less common in Africans than in Europeans and Asians. Here we examined the association between FLG Single Nucleotide Polymorphisms (SNPs) and eczema in a population of admixed Brazilian children and whether African ancestry modifies this association. We included 1010 controls and 137 cases and ran logistic regressions between SNPs in FLG and eczema in the studied population and also stratified the analyses according to the degree of African ancestry. In addition, we tested the replication of the findings on an independent set of individuals, as well as, we verified the impact on FLG expression according to each SNP genotype. The T allele of SNP rs6587666 was negatively associated with eczema in additive model (OR: 0.66, 95% CI: 0.47–0.93, P: 0.017). Moreover, African ancestry modifies the association between rs6587666 and eczema. The effect of the T allele was higher among individuals with higher African ancestry and the association with eczema was lost in individuals with lower African ancestry. In our analyses the expression of FLG in skin was slightly downregulated by the presence of the T allele of rs6587666. In our population, the T allele of rs6587666 in FLG was associated with protection to eczema and the degree of African ancestry was able to modify the observed association.
Keywords: Filaggrin, FLG, Atopic dermatitis, Skin, Allele
Highlights
-
•
The T allele of the rs6587666 SNP was negatively associated with eczema.
-
•
AsSociation between rs6587666 and eczema changed according to the African ancestry.
-
•
The higher is the negative association with eczema, higher is the African Ancestry.
-
•
In individuals with a lower African Ancestry, no association with eczema was found.
1. Introduction
Eczema is the most common chronic inflammatory skin disease worldwide, affecting up to 20% of children and 3% of adults [1] involving several targets, including interactions between skin barrier defects and immunological factors [2,3]. Moreover, approximately one‐third of patients with eczema develop asthma and two‐thirds develop allergic rhinitis [4].
The primary function of the skin is to act as a protective barrier between the host organism and its external environment, minimising water loss whilst at the same time preventing the entry of microorganisms and allergens [3,5]. A great number of genes have been linked to eczema susceptibility [[6], [7], [8]]. Interestingly, in several studies characterizing the filaggrin-deficient skin barrier and its consequences, mutations in the filaggrin gene (FLG) have been associated with the risk and severity of eczema [6,[9], [10], [11], [12], [13]].
Filaggrin (filament-aggregating-protein) is a key protein in the differentiation of the epidermis. Together with keratin filaments and other proteins, filaggrin forms a cornified cell envelope, which is critical to the skin's barrier function [14]. The role played by filaggrin explains why the lower expression of a single component of the epidermal differentiation complex might have a great influence on the function of the skin barrier.
Several studies have observed ethnic and regional differences in FLG variants [13,15]. Palmer and collaborators have shown that 17.5% of European individuals with eczema were carriers of FLG null alleles [10]. On the other hand, certain FLG variants common in Europeans and Asians are less common in African populations [6,9,12,13,[16], [17], [18]].
The Brazilian population is derived from more than 500 years of genetic admixture between Europeans, Native Americans, and Africans, with the proportions of these ancestral populations varying markedly among Brazilian regions [19]. The Northeast Brazil, mainly Bahia State, received thousands of slaves directly from Africa [20]. Therefore, the influence of African ancestry described in Bahia is different and much higher from other regions of Brazil. In this sense, the African ancestry component is the major contributor of the genetic structure of the population from Salvador, Bahia [21,22].
In this study, we examined the association between Single Nucleotide Polymorphisms (SNPs) in FLG and eczema considering whether the association would vary by the degree of African ancestry in a population of Brazilian children from the city of Salvador, Bahia.
2. Materials and methods
2.1. Subjects
Our population-based study design has been described in detail elsewhere [23,24]. The sample comprises 1307 children aged five to twelve years old in the city of Salvador in Northeast Brazil and enrolled in Social Change, Asthma and Allergy in Latin America (SCAALA) Program were included. Written informed consent was obtained from the parents of each child enrolled in the study. The parent/guardian answered the ISAAC phase II core questionnaires for eczema, translated into Portuguese and back-translated into English, and validated [25] as described by Strina and collaborators [23]. This study has been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) and was approved by the Institutional Review Board of the Collective Health Institute (register 003–05/CEP-ISC) of the Federal University of Bahia, Brazil.
2.2. Eczema definition
Eczema was defined according to ISAAC studies, as described by Strina and collaborators [23]. In this study, we considered the children as a case when the response was positive for the occurrence of: i) itchy rash that came and went for at least 6 months; ii) itchy rash at any time in the last 12 months and; iii) itchy rash at any time. A rash in the following places of the body was considered for the diagnosis of eczema: the folds of the elbows, behind the knees, in front of the ankles, under the buttocks, or around the neck, ears, or eyes (ISAAC, 1998). All other children whose answers were not positive to all three questions referred were classified as non-eczema controls.
2.3. Genotyping
DNA was extracted from peripheral blood samples using the Flexigene@ DNA Kit (Qiagen, Hilden, Germany), and genotyping was performed using the Illumina BeadChip HumanOmni 2.5 Kit (www.illumina.com) through EPIGEN-Brazil Consortium (https://epigen.grude.ufmg.br/). One individual was excluded from the analysis due to inconsistency between the sex registered and the genetic sex, based on X chromosome SNPs. Sixty-one individuals were removed from the sample due to the relationship determined by kinship coefficients for each possible pair of individuals. This method was implemented in the REAP software (Relatedness Estimation in Admixed Populations) [26]. We considered a pair of individuals as related if the estimated kinship coefficient between them was ≥0.1. This cut-off includes second-degree relatives such as a person's uncle/aunt, nephew/niece, grandparent/grandchild or half-sibling, and any closer pair of relatives. Ninety-eight people had missing values due to the absence of phenotype or covariates. A total of 1147 unrelated children were included in this study.
27 variants in FLG were extracted from chromosome 1 positions 152274641 to 152297715 (assembly: GRCh37.p13; location: NC_000001.10). For quality control, we applied the following filters: imbalance of Hardy–Weinberg equilibrium with P-value less than 0.05 and for the minor allele frequency – MAF of at least 1%. The genotyping call rate was greater than 99%. After quality control, a total of 18 SNPs were analyzed.
2.4. Stratification of the population according to the African ancestry
For our secondary analyses, subgroup analyses were conducted from the categorization based on different percentiles of the continuous distribution of individual African ancestry in our population. The calculation of ancestry was performed as described by Kehdy and collaborators [27], using the ADMIXTURE software [28] considering an tri-hybrid model (K = three). Quantitative genetic ancestry was estimated for individuals by including contemporary descendants of the approximate population of origin (pseudoancestors). As external panels samples of African and European individuals from the HapMap project and 93 Native American of the Human Genome Diversity (HGDP) project were used. A total of 370,539 SNPs shared by samples from the HapMap, HGDP and the study population were used for biogeographical ancestry estimate. African ancestry strata were then defined as “high” vs “low” (above and below the median ancestry, respectively), and by tertiles.
2.5. Replication from the Hartford - Puerto Rico (HPR)
Details for the HPR cohort recruitment and procedures have been previously described in detail [29]. Briefly, children 6–14 years old living with four Puerto Rican grandparents were recruited from Hartford, CT, and San Juan, PR. Written parental consent was obtained for participating children, from whom written assent was also obtained. The study was approved by the Institutional Review Boards of Connecticut Children's Medical Center (Hartford, CT), the University of Puerto Rico (San Juan, PR), Brigham and Women's Hospital (Boston, MA), and the University of Pittsburgh (Pittsburgh, PA). Genome-wide genotyping was performed using the Illumina HumanOmni2.5 BeadChip platform (Illumina, San Diego, CA). For this analysis, cases with eczema (n = 156) were defined as those reporting an itchy/scaly rash affecting the face, neck, arms, elbows, legs, or knees, and who either still had the rash or had it at least until age 4 years; controls (n = 514) were subjects who never had such rash.
2.6. Statistical analysis
Association analysis were performed using logistic regression model to calculate the odds ratio (OR) and 95% confidence intervals (CIs). The analyses were adjusted by sex, age, helminth infection, asthma symptoms and principal components (PC1, PC2 and PC3), as previously described [21]. The principal components were estimated from genome-wide genetic variants and used to adjust the effect of population stratification on the different multivariate models tested [30]. We calculated associations using additive, dominant and recessive genetic models. A computationally intensive procedure based on 1000 permutations was used to estimate the statistical significance of multiple correlation tests in the genetic association analysis [31]. Power of each genetic model was estimated. The genetic data and power of the genetic model were analyzed using PLINK 1.9 [32]. The analyses of SNP function were performed using the RegulomeDB Score portal (https://www.regulomedb.org/regulome-search/) [33]. The Expression Quantitative Trait Locus (eQTL) analysis on the skin were performed using Genotype-Tissue Expression (GTEx) Portal (www.gtexportal.org/home/) [34]. The forest plot was performed using GraphPad 6 (GraphPad Software, San Diego, CA, USA). Linkage disequilibrium (LD) was investigated with Haploview [35]. The validation analysis in the HPR cohort used the additive model adjusted for age, sex, study site, and the principal components: PC1, PC2, and PC3. Statistical significance was always defined as P < 0.05.
3. Results
Table 1 presents the sample summary information of the 1010 non-eczema and 137 eczema participants of the study. Participants with eczema were more likely to be male than those without eczema. There were no significant differences in age, helminth infection or African ancestry (high vs low or by tertiles) between eczema cases and controls. On the other hand, asthma symptoms were differently significant between eczema and non-eczema participants (P-value: <0.0001).
Table 1.
Sample summary information from the Social Change, Asthma and Allergy in Latin America (SCAALA) Brazilian cohort.
| Non-eczema |
Eczema |
P-value | |
|---|---|---|---|
| 1010 (88.06%) | 137 (11.94%) | ||
| Age | |||
| <6,0 | 397 (39.31) | 53 (38.69) | 0.947 |
| 6,0 to 7,0 | 339 (33.56) | 45 (32.85) | |
| >7,0 | 274 (27.13) | 39 (28.47) | |
| Sex | |||
| Female, No. (%) | 467 (46.24) | 58 (42.34) | 0.390 |
| Male, No. (%) | 543 (53.76) | 79 (57.66) | |
| Helminthic infection | |||
| Yes, No. (%) | 240 (23.76) | 29 (21.17) | 0.501 |
| No, No. (%) | 770 (76.24) | 108 (78.83) | |
| Asthma symptoms | |||
| Yes, No. (%) | 210 (20.79) | 47 (34.31) | <0.0001 |
| No, No. (%) | 800 (79.21) | 90 (65.69) | |
| Median for African ancestry | |||
| Lower than median, No. (%) | 494 (48.91) | 73 (52.28) | 0.337 |
| Higher than median, No. (%) | 516 (51.09) | 64 (46.72) | |
| Tertiles for African ancestry | |||
| 1st, No. (%) | 330 (32.67) | 51 (37.23) | 0.503 |
| 2nd, No. (%) | 344 (34.06) | 41 (29.93) | |
| 3rd, No. (%) | 336 (33.27) | 45 (32.85) | |
aP-value considering the Chi-Square test.
Table 2 showed that the FLG SNPs had MAF between 1% and 23% in non-eczema subjects and less than 1% up to 22% in eczema cases. The rs6587666 SNP is located in an intronic region and presented the largest difference in MAF: 16% in individuals with eczema and 23% in subjects from the control group. In the RegulomeDB Score [33], the values for the studied SNPs have ranged from “3” to “7”.
Table 2.
Characterization of the studied FLG SNPs in SCAALA Brazilian cohort.
| SNP | Position | A1 | A2 | Non-eczema | Eczema | Function | RegulomeDB Rank |
|---|---|---|---|---|---|---|---|
| rs115053459 | 152274909 | T | C | 0.02 | 0.01 | Utr variant 3 prime | 6 |
| rs76330665 | 152275273 | T | G | 0.02 | 0.01 | Missense | 3b |
| rs3814299 | 152275453 | A | G | 0.01 | 0.02 | Missense | 3a |
| rs3126065 | 152275559 | G | A | 0.20 | 0.22 | Missense | 5 |
| rs78733053 | 152275644 | A | G | 0.02 | 0.01 | Synonymous codon | 5 |
| rs62623409 | 152281745 | T | G | 0.01 | 0.01 | Missense | 4 |
| rs74129458 | 152281876 | C | T | 0.02 | <0.01 | Missense | 4 |
| rs12405278 | 152282267 | A | G | 0.14 | 0.11 | Missense | 5 |
| rs151103850 | 152282684 | A | G | 0.01 | 0.01 | Missense | 5 |
| rs76586335 | 152284673 | A | G | 0.02 | <0.01 | Missense, upstream variant 2 KB | 3a |
| rs77032592 | 152285002 | A | G | 0.02 | 0.01 | Missense, upstream variant 2 KB | 5 |
| rs114762657 | 152285290 | T | C | 0.04 | 0.02 | Missense, upstream variant 2 KB | 5 |
| rs74129464 | 152285759 | G | A | 0.02 | <0.01 | Missense, upstream variant 2 KB | 4 |
| rs11588170 | 152286032 | T | C | 0.14 | 0.11 | Missense | 5 |
| rs76438926 | 152287078 | A | G | 0.04 | 0.03 | Intron variant, missense | 4 |
| rs115489580 | 152287213 | C | T | 0.04 | 0.03 | Missense, nc transcript variant | 6 |
| rs76385639 | 152289573 | A | C | 0.02 | 0.01 | Intron variant | 3a |
| rs6587666 | 152296863 | T | C | 0.23 | 0.16 | Intron variant | 7 |
aSNP, single nucleotide polymorphism; bPosition, location (assembly GRCh37.p13); cA1, minor allele; dA2, ancestral allele; eMAF, minor allele frequency; fRegulomeDB rank refers to: 3a: TF binding + any motif + DNase peak; 3b: TF binding + matched TF motif; 4: TF binding + DNase peak; 5: TF binding or DNase peak; 6: Motif hit; 7: Other.
The data presented in Table 3 indicate that the T allele of the rs6587666 SNP was negatively associated with eczema in our Brazilian cohort in additive model (OR: 0.66, 95% CI: 0.47–0.93, P: 0.017). The power analysis performed for this study has demonstrated, approximately, 67%, 55% and 13% of power for additive, dominant and recessive models, respectively (data not shown).
Table 3.
Association between rs6587666 and eczema by additive model in SCAALA Brazilian cohort.
| Chr | Gene | SNP | Geno | Non-eczema | Eczema | OR (95% CI) | P-value* |
|---|---|---|---|---|---|---|---|
| 1q21 | FLG | rs6587666 | C/C | 609 (60.3%) | 96 (70.1%) | 0.66 (0.47–0.93) | 0.017 |
| T/C | 349 (34.5%) | 37 (27%) | |||||
| T/T | 52 (5.2%) | 4 (2.9%) |
aChr, Chromosome; bSNP, single nucleotide polymorphism; cADD, additive; dGeno, genotype; eOR, Odds ratio; fCI, confidence interval. *Adjusted by gender, age, helminthic infection, population structure, and asthma symptoms. *P-value considering adaptative permutations using the additive model.
The T allele of FLG rs6587666 was more frequent in non-eczema compared to eczema participants (Table 2). No other FLG SNP investigated were associated to eczema (data not shown) and also rs6587666 was not in LD with any other SNP evaluated here (see Supplemental Fig. S1).
We then moved to test whether the proportion of individual African ancestry could act as a potential effect modifier of the association between FLG variants and eczema in our population. To achieve that, we stratified the cohort according to tertiles of African ancestry. Interestingly, we observed that among participants with the greatest degree of African ancestry (highest tercile, 3rd), the frequency of the T allele of this SNP is higher in non-eczema compared to eczema participants (23% to 10%, respectively). Similarly, for the genotypic frequencies in non-eczema and eczema individuals, we find that the T/C and T/T genotypes are more frequent in controls compared to cases in both 1st and 3rd tertiles. This difference is optimal in the 3rd tercile (Table 4).
Table 4.
Genotype frequencies and Minor Allele Frequency (MAF) for T allele on rs6587666 in SCAALA Brazilian cohort, by African ancestry tertile.
| African ancestry | Genotype | Non-eczema | Eczema |
|---|---|---|---|
| Tercile, 1st | C/C | 190 (57.58%) | 34 (66.67%) |
| T/C | 121 (36.67%) | 15 (29.41%) | |
| T/T | 19 (5.76%) | 2 (3.92%) | |
| T allele MAF | 0.24 | 0.19 | |
| Tercile, 2nd | C/C | 222 (64.53%) | 25 (60.98%) |
| T/C | 105 (30.52%) | 15 (36.59%) | |
| T/T | 17 (4.94%) | 1 (2.44%) | |
| T allele MAF | 0.21 | 0.21 | |
| Tercile, 3rd | C/C | 197 (58.63%) | 37 (82.22%) |
| T/C | 123 (36.61%) | 7 (15.56%) | |
| T/T | 16 (4.76%) | 1 (2.22%) | |
| T allele MAF | 0.23 | 0.1 |
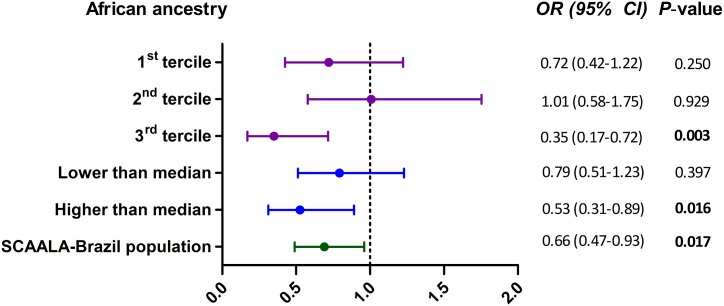
In Fig. 1, rs6587666 was negatively associated with eczema, the higher is the African ancestry contribution. Among the sujects in the highest tertile, we found an even stronger negative association between rs6587666 and eczema (OR: 0.35; 95% CI: 0.17–0.72; P: 0.003) than in the general cohort. Similarly, if we stratified at the median ancestry level, the association was significant in those with high African ancestry (OR: 0.53; 95% CI: 0.31–0.89; P: 0.016). On the other hand, the association was not significant in those with lower African ancestry, whether defined by tertiles or by the median. Thus, the lower the contribution of African ancestry in the total genome, the lower was the effect observed in both the general cohort analysis and 3rd tertile enriched for African ancestry. This analysis demonstrated a statistical power of 82% in the additive genetic model (data not shown).
Fig. 1.
Associations between rs6587666 and eczema in SCAALA Brazilian cohort. The association analyses were stratified by the degree of African ancestry in each individual using an additive model. Among the sujects in the 3rd tertile was found an stronger negative association between rs6587666 and eczema than in the general cohort. Stratified analyses were not adjusted. OR, Odds ratio; CI, confidence interval.
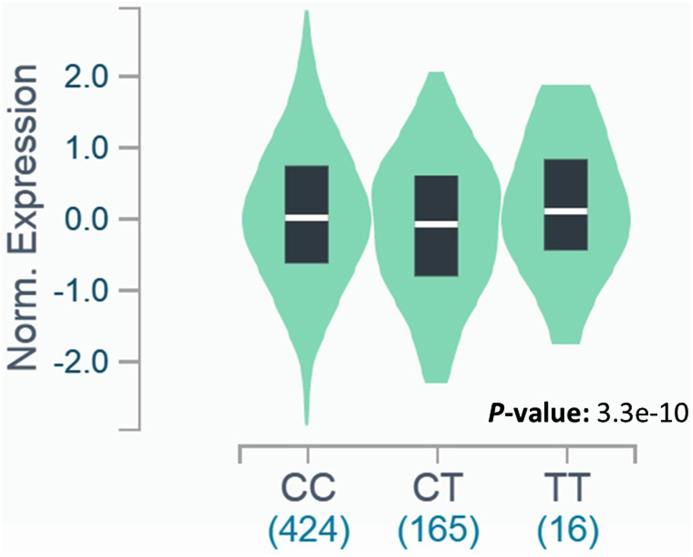
In our analyses from GTEX database (www.gtexportal.org/home/) [34], the expression of FLG in skin sun exposed was slightly downregulated by the presence of the T allele of rs6587666 (effect size: 0.19) (Table S1). For the CC genotype, the median of the gene expression was ∼0.01. For the TC and TT genotypes it was ∼0.08 and ∼0.11, respectively. Interestingly, for the TT genotype, the median expression was approximately 10 times greater when compared to the CC genotype (Fig. 2). In skin not sun exposed tissue we observed a similar effect.
Fig. 2.
Impact of the T allele in the rs6587666 on FLG expression in skin exposed sun tissue. For the TT genotype, the median expression of FLG was approximately 10 times greater when compared to the CC genotype (∼0.01 and ∼0.11, respectively). The gene expression and the respective figure were obtained from GTEx database (www.gtexportal.org/home/).
Other genes close to FLG may also have their expression affected by T allele of rs6587666. We observed that its associated with low expression of HRNR in skin not sun exposed (effect size: 0.33), as well as with high expression of the FLG-AS1 in skin sun exposed (effect size: 0.29) and skin not sun exposed (effect size: 0.36) (Table S1).
We were unable to replicate our finding with rs6587666 in Puerto Rican children/adolescents (Table S2).
4. Discussion
The loss of function of filaggrin may lead to the formation of fragile skin that contributes to an abnormal barrier function. This explains the relationship between mutations in FLG and an increased risk of eczema [36]. To the best of our knowledge, this is the first report of an association between the T allele of rs6587666 in the FLG gene and a lower risk of eczema in a Latin American population.
Several studies have related genetic variants in FLG with eczema in European and Asian populations [10,16]. In contrast, in African populations, FLG variants are less common [6,16]. Interestingly, we identified a new polymorphism rs6587666 in FLG, associated with protection to eczema in our population. This protection was as high as a ∼42%–∼62% in participants with higher African contribution in our stratified analyses (OR: 0.53 for those with African ancestry above the median; and OR: 0.35 for those in the highest tertile of African ancestry).
It is worth mentioning that even with a statistical power below 80% in the general population (data not shown), we found considerable significance in our association analysis. In addition, our study demonstrated a statistical power of 82% in the additive genetic models in the subpopulation of the 3rd tercile (data not shown). We believe that this probably is due to the increased effect of the SNP in this group of greater African ancestry.
The population of Salvador is admixed by Yoruba individuals from Ibadan, Nigeria and northern and western European origin populations [21,22]. In descriptive terms, the frequency of the T allele from this ancestral populations is lower than in our admixed population, according with 1000 Genomes Project Phase 3 (see Supplemental Fig. S2), especially when comparing with the controls in our study. Latin American people are the product of an admixture process that has generated chromosomes derived by a complex mixture of ancestry [19]. In relation to the differences in the frequency of alleles, this could explain the peculiar characteristics of our admixed population when compared to African and European ancestral populations.
The rs6587666 was not in LD with any other SNP analyzed herein (see Supplemental Fig. S1). We investigated the LD (r2) between rs6587666 and other SNPs using available databases. In the Ensembl platform (www.ensembl.org), we found that rs11584340, a missense FLG polymorphism, presented high LD (r2 ∼0.92) in the Yoruba in Ibadan (Nigeria) population. According to rSNPBase (rsnp.psych.ac.cn), rs11584340 in FLG had a high LD (r2 ∼0.97) in African populations. rs11584340 has been associated with greater eczema severity [37] and to functional parameters such as high levels of free fatty acids and high levels of mean corpuscular haemoglobin concentrations. It is relevant since the disruption of the permeability barrier seem to result in a marked increase in fatty acid synthesis in the skin [38]. However, other earlier studies found no differences or found reduced free fatty acids in atopic skin [39,40]. Kim and collaborators [38] reported that the increase in serum fatty acid levels perceived in their study could be due to other factors such as decreased metabolic utilization or decreased uptake by cells. Our findings indicate that perhaps the rs6587666 may have clinically singular relevant implications on the FLG in our population or that may be getting a signal from rs11584340.
The rs6587666 is an intronic polymorphism and as such may modify the gene expression level in the host gene in many different ways. The mechanism whereby it occurs remain unclear [41]. However, specific intron-hosted DNA elements have been identified to regulate transcription initiation affecting the gene expression. In fact, diseases related to genetic variants usually alter gene expression [42,43]. Some studies have observed the reduced expression of FLG in eczema as compared to normal subjects [[44], [45], [46]] and loss-of-function mutations in FLG associated with severity of atopic eczema [47]. The rs6587666 is intronic and have a functional mechanism other than null mutation. As such, it probably does not cause loss of protein function. In addition, the reduction in gene expression was not drastic from GTEx database in our findings. Therefore, we cannot know how this slight reduction in mRNA expression or whether FLG-AS1 could affect the expressed protein.
Due to ethical and legal implications we were unable to carry out the gene expression analysis in our own pediatric Brazilian population. The GTEx is a biorepository of samples collected from postmortem adult donors with no evidence of disease from several ancestry (https://biospecimens.cancer.gov/gtexbiobank/donors.asp). Therefore, the characteristics of the participants in GTEx database cannot represent our original population and this is a limitation of our work.
We were unable to replicate the association found to rs6587666 SNP using data from the Puerto Rican children/adolescents. The OR was the same direction (OR: 0.98). Perhaps, HPR cohort was underpowered for that effect size. Our results reinforce the need to investigate genetic factors to eczema in populations of African origin, which are still underrepresented in genetic association studies.
A limitation of this study is the sample size in the stratified analyses according to the contribution of the African ancestry. However, this was not a sufficient limitation to conceal a likely protective effect of the rs6587666 (T allele) against eczema when the analysis were performed in participants with a great African ancestry contribution. Indeed, to detect the effect size observed in the subgroup with greater individual African ancestry (3rd tercile), the study demonstrated a statistical power of 82% in the additive genetic models (data not shown). In addition, due to ethical and legal implications, we were unable to test gene expression in a pediatric population of Brazilian children similar to our original population.
To our knowledge, this is the first study showing an association between rs6587666 in FLG and protection to eczema in an admixed population in which the African ancestry may modify this association herein observed. Functional and selective pressure studies performed later may be able to show how the rs6587666 SNP may have clinically relevant implications on the FLG in the human skin, specialy those with high African ancestry contribution in their genome.
Author contribution statement
Raimon Rios: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Thiago Magalhães da Silva: Conceived and designed the experiments; Analyzed and interpreted the data
Agostino Strina: Contributed reagents, materials, analysis tools or data.
Erick Forno, Maurício L. Barreto: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Ryan Costa, Juan C. Celedón: Analyzed and interpreted the data.
Camila Alexandrina Figueiredo: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
Raimon Rios was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior [001].
Dra. Camila Alexandrina Viana Figueiredo was supported by Fundação de Amparo à Pesquisa do Estado da Bahia [009/2014; 8305/2014; PNE0003/2014; 008/2014; PNX0001/2014].
Data availability statement
Data will be made available on request.
Declaration of interest’s statement
The authors declare no competing interests.
Acknowledgements
We acknowledge the individuals who participated in this project and their families. This study is part of SCAALA (Social Change, Asthma and Allergy in Latin America) Program.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e13659.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Nutten S. Atopic dermatitis: global epidemiology and risk factors. Ann. Nutr. Metab. 2015;66(Suppl 1):8–16. doi: 10.1159/000370220. [DOI] [PubMed] [Google Scholar]
- 2.Peng W., Novak N. Pathogenesis of atopic dermatitis. Clin. Exp. Allergy. 2015;45(3):566–574. doi: 10.1111/cea.12495. [DOI] [PubMed] [Google Scholar]
- 3.Leung D.Y. New insights into atopic dermatitis: role of skin barrier and immune dysregulation. Allergol. Int. 2013;62(2):151–161. doi: 10.2332/allergolint.13-RAI-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spergel J.M. Epidemiology of atopic dermatitis and atopic march in children. Immunol. Allergy Clin. 2010;30(3):269–280. doi: 10.1016/j.iac.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Drislane C., Irvine A.D. The role of filaggrin in atopic dermatitis and allergic disease. Ann. Allergy Asthma Immunol. 2020;124(1):36–43. doi: 10.1016/j.anai.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Margolis D.J., Gupta J., Apter A.J., Hoffstad O., Papadopoulos M., Rebbeck T.R., et al. Exome sequencing of filaggrin and related genes in African-American children with atopic dermatitis. J. Invest. Dermatol. 2014;134(8):2272–2274. doi: 10.1038/jid.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai X.Y., Zheng X.D., Fang L., Zhou F.S., Sheng Y.J., Wu Y.Y., et al. A variant on chromosome 2p13.3 is associated with atopic dermatitis in Chinese Han population. Gene. 2017;628:281–285. doi: 10.1016/j.gene.2017.07.059. [DOI] [PubMed] [Google Scholar]
- 8.Loset M., Brown S.J., Saunes M., Hveem K. Genetics of atopic dermatitis: from DNA sequence to clinical relevance. Dermatology. 2019;235(5):355–364. doi: 10.1159/000500402. [DOI] [PubMed] [Google Scholar]
- 9.Margolis D.J., Apter A.J., Gupta J., Hoffstad O., Papadopoulos M., Campbell L.E., et al. The persistence of atopic dermatitis and filaggrin (FLG) mutations in a US longitudinal cohort. J. Allergy Clin. Immunol. 2012;130(4):912–917. doi: 10.1016/j.jaci.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer C.N., Irvine A.D., Terron-Kwiatkowski A., Zhao Y., Liao H., Lee S.P., et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 2006;38(4):441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 11.Kezic S., O'Regan G.M., Lutter R., Jakasa I., Koster E.S., Saunders S., et al. Filaggrin loss-of-function mutations are associated with enhanced expression of IL-1 cytokines in the stratum corneum of patients with atopic dermatitis and in a murine model of filaggrin deficiency. J. Allergy Clin. Immunol. 2012;129(4):1031–1039 e1. doi: 10.1016/j.jaci.2011.12.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margolis D.J., Mitra N., Gochnauer H., Wubbenhorst B., D'Andrea K., Kraya A., et al. Uncommon filaggrin variants are associated with persistent atopic dermatitis in african Americans. J. Invest. Dermatol. 2018;138(7):1501–1506. doi: 10.1016/j.jid.2018.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akiyama M. FLG mutations in ichthyosis vulgaris and atopic eczema: spectrum of mutations and population genetics. Br. J. Dermatol. 2010;162(3):472–477. doi: 10.1111/j.1365-2133.2009.09582.x. [DOI] [PubMed] [Google Scholar]
- 14.Osawa R., Akiyama M., Shimizu H. Filaggrin gene defects and the risk of developing allergic disorders. Allergol. Int. 2011;60(1):1–9. doi: 10.2332/allergolint.10-RAI-0270. [DOI] [PubMed] [Google Scholar]
- 15.Wang I.J., Lin T.J., Kuo C.F., Lin S.L., Lee Y.L., Chen P.C. Filaggrin polymorphism P478S, IgE level, and atopic phenotypes. Br. J. Dermatol. 2011;164(4):791–796. doi: 10.1111/j.1365-2133.2011.10212.x. [DOI] [PubMed] [Google Scholar]
- 16.Brown S.J., McLean W.H. One remarkable molecule: filaggrin. J. Invest. Dermatol. 2012;132(3 Pt 2):751–762. doi: 10.1038/jid.2011.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winge M.C., Hoppe T., Berne B., Vahlquist A., Nordenskjold M., Bradley M., et al. Filaggrin genotype determines functional and molecular alterations in skin of patients with atopic dermatitis and ichthyosis vulgaris. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0028254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winge M.C., Bilcha K.D., Lieden A., Shibeshi D., Sandilands A., Wahlgren C.F., et al. Novel filaggrin mutation but no other loss-of-function variants found in Ethiopian patients with atopic dermatitis. Br. J. Dermatol. 2011;165(5):1074–1080. doi: 10.1111/j.1365-2133.2011.10475.x. [DOI] [PubMed] [Google Scholar]
- 19.Salzano F.M., Sans M. Interethnic admixture and the evolution of Latin American populations. Genet. Mol. Biol. 2014;37(1 Suppl):151–170. doi: 10.1590/s1415-47572014000200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curtin P.D. University of Wisconsin Press; Madison: 1969. The Atlantic Slave Trade: a Census; p. 338. [Google Scholar]
- 21.Lima-Costa M.F., Rodrigues L.C., Barreto M.L., Gouveia M., Horta B.L., Mambrini J., et al. Genomic ancestry and ethnoracial self-classification based on 5,871 community-dwelling Brazilians (The Epigen Initiative) Sci. Rep. 2015;5:9812. doi: 10.1038/srep09812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magalhães da Silva T., Sandhya Rani M.R., de Oliveira Costa G.N., Figueiredo M.A., Melo P.S., Nascimento J.F., et al. The correlation between ancestry and color in two cities of Northeast Brazil with contrasting ethnic compositions. Eur. J. Hum. Genet. 2015;23(7):984–989. doi: 10.1038/ejhg.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strina A., Barreto M.L., Cunha S., de Fatima SPdOM., Moreira S.C., Williams H.C., et al. Validation of epidemiological tools for eczema diagnosis in Brazilian children: the ISAAC's and UK Working Party's criteria. BMC Dermatol. 2010;10:11. doi: 10.1186/1471-5945-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barreto M.L., Cunha S.S., Alcantara-Neves N., Carvalho L.P., Cruz A.A., Stein R.T., et al. Risk factors and immunological pathways for asthma and other allergic diseases in children: background and methodology of a longitudinal study in a large urban center in Northeastern Brazil (Salvador-SCAALA study) BMC Pulm. Med. 2006;6:15. doi: 10.1186/1471-2466-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada E., Vanna A.T., Naspitz C.K., Sole D. International Study of Asthma and Allergies in Childhood (ISAAC): validation of the written questionnaire (eczema component) and prevalence of atopic eczema among Brazilian children. J Investig. Allergol. Clin. Immunol. 2002;12(1):34–41. [PubMed] [Google Scholar]
- 26.Thornton T., Tang H., Hoffmann T.J., Ochs-Balcom H.M., Caan B.J., Risch N. Estimating kinship in admixed populations. Am. J. Hum. Genet. 2012;91(1):122–138. doi: 10.1016/j.ajhg.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kehdy F.S., Gouveia M.H., Machado M., Magalhaes W.C., Horimoto A.R., Horta B.L., et al. Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc. Natl. Acad. Sci. U. S. A. 2015;112(28):8696–8701. doi: 10.1073/pnas.1504447112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forno E., Sordillo J., Brehm J., Chen W., Benos T., Yan Q., et al. Genome-wide interaction study of dust mite allergen on lung function in children with asthma. J. Allergy Clin. Immunol. 2017;140(4):996–1003 e7. doi: 10.1016/j.jaci.2016.12.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 31.Doerge R.W., Churchill G.A. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142(1):285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 36.Thyssen J.P., Kezic S. Causes of epidermal filaggrin reduction and their role in the pathogenesis of atopic dermatitis. J. Allergy Clin. Immunol. 2014;134(4):792–799. doi: 10.1016/j.jaci.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 37.Lopes C., Rocha L., Sokhatska O., Soares J., Tavaria F., Correia O., et al. Filaggrin polymorphism Pro478Ser is associated with the severity of atopic dermatitis and colonization by staphylococcal aureus. J Investig. Allergol. Clin. Immunol. 2016;26(1):70–72. [PubMed] [Google Scholar]
- 38.Kim S.Y., Yang S.W., Kim H.L., Kim S.H., Kim S.J., Park S.M., et al. Association between P478S polymorphism of the filaggrin gene & atopic dermatitis. Indian J. Med. Res. 2013;138(6):922–927. [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto A., Serizawa S., Ito M., Sato Y. Stratum corneum lipid abnormalities in atopic dermatitis. Arch. Dermatol. Res. 1991;283(4):219–223. doi: 10.1007/BF01106105. [DOI] [PubMed] [Google Scholar]
- 40.Bleck O., Abeck D., Ring J., Hoppe U., Vietzke J.P., Wolber R., et al. Two ceramide subfractions detectable in Cer(AS) position by HPTLC in skin surface lipids of non-lesional skin of atopic eczema. J. Invest. Dermatol. 1999;113(6):894–900. doi: 10.1046/j.1523-1747.1999.00809.x. [DOI] [PubMed] [Google Scholar]
- 41.Chorev M., Carmel L. The function of introns. Front. Genet. 2012;3:55. doi: 10.3389/fgene.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bianchi M., Crinelli R., Giacomini E., Carloni E., Magnani M. A potent enhancer element in the 5'-UTR intron is crucial for transcriptional regulation of the human ubiquitin C gene. Gene. 2009;448(1):88–101. doi: 10.1016/j.gene.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 43.Beaulieu E., Green L., Elsby L., Alourfi Z., Morand E.F., Ray D.W., et al. Identification of a novel cell type-specific intronic enhancer of macrophage migration inhibitory factor (MIF) and its regulation by mithramycin. Clin. Exp. Immunol. 2011;163(2):178–188. doi: 10.1111/j.1365-2249.2010.04289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guttman-Yassky E., Suarez-Farinas M., Chiricozzi A., Nograles K.E., Shemer A., Fuentes-Duculan J., et al. Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. J. Allergy Clin. Immunol. 2009;124(6):1235–1244 e58. doi: 10.1016/j.jaci.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 45.Sugiura H., Ebise H., Tazawa T., Tanaka K., Sugiura Y., Uehara M., et al. Large-scale DNA microarray analysis of atopic skin lesions shows overexpression of an epidermal differentiation gene cluster in the alternative pathway and lack of protective gene expression in the cornified envelope. Br. J. Dermatol. 2005;152(1):146–149. doi: 10.1111/j.1365-2133.2005.06352.x. [DOI] [PubMed] [Google Scholar]
- 46.Esaki H., Ewald D.A., Ungar B., Rozenblit M., Zheng X., Xu H., et al. Identification of novel immune and barrier genes in atopic dermatitis by means of laser capture microdissection. J. Allergy Clin. Immunol. 2015;135(1):153–163. doi: 10.1016/j.jaci.2014.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bieber T., Cork M., Reitamo S. Atopic dermatitis: a candidate for disease-modifying strategy. Allergy. 2012;67(8):969–975. doi: 10.1111/j.1398-9995.2012.02845.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.