Abstract
Regulation of chromosome condensation 2 (RCC2) is associated with the cell cycle and is a crucial regulator of the chromatin condensation 1 (RCC1) family. The members of this family were normally regulators in the process of DNA replication and nucleocytoplasmic transport. RCC2 overexpression may lead to tumor formation and poor prognosis in some tumors including breast cancer and lung adenocarcinoma. However, the possible role of RCC2 in tumor formation and its prognostic function remains unclear. In this study, expression analysis from databases including The Cancer Genome Atlas (TCGA) and Clinical Proteomic Tumor Analysis Consortium (CPTAC) were combined to perform the first integrative and comprehensive analysis of RCC2 in human pan-cancer. RCC2 was highly expressed in most tumors which may lead to a poor prognosis. RCC2 expression was associated with immune/stromal infiltration, immune checkpoints, tumor mutational burden, and microsatellite instability. Thus, RCC2 could be a novel biomarker for prognosis and a promising cancer therapy target.
Keywords: RCC2, Prognosis, Immune infiltration, Enrichment analysis, Immune checkpoint inhibitor, Pan-cancer analysis
1. Introduction
Regulator of chromosome condensation 2 (RCC2) is a crucial member of the regulator of chromatin condensation 1 (RCC1) family and comprises of four RCC1-like domains. Genes in this family are normally involved in cell cycle, DNA replication, and nucleocytoplasmic transport [[1], [2], [3]]. RCC2 was first recognized as telophase disc-60 (TD60), a protein involved in mitosis [4]. RCC2 may be a crucial regulator of the cell cycle during interphase since it binds the nucleotide-free form of the small G protein Rac1 and is a Rac1guanine exchange factor, and regulates kinetochore-microtubule interactions during the G2 transition [[5], [6], [7], [8]].
RCC2 promotes breast cancer tumors by regulating Wnt signaling and inducing epithelial-mesenchymal transition (EMT) [9]. Similarly, it induces EMT in lung tissue and leads to lung adenocarcinoma (LUAD) formation and tumor metastasis [10]. RCC2 overexpression activates Rac1 and promotes tumor metastasis, and it was proposed that the p53/RCC2/Rac1 axis may be a possible cancer therapy target [11]. This was supported by a study showing that blocking Rac1 signaling overcomes drug resistance by RCC2 overexpression [12]. Meanwhile, RCC2 may be related to cisplatin resistance and contribute to tumor invasion in liver hepatocellular carcinoma (LIHC) [13,14]. These studies suggest that the RCC2 expression level may result in tumor formation, resistance to treatment, and tumor metastasis.
Although many studies have focused on special tumor types such as LIHC and LUAD, it is essential to perform a pan-cancer analysis of RCC2 to identify its general role in this disease. This study aimed to explore the potential role and mechanism of RCC2 in human pan-cancer through a systemic analysis of several databases to enable future studies. Data from The Cancer Genome Atlas (TCGA) and Clinical Proteomic Tumor Analysis Consortium (CPTAC) were combined to perform the first integrative and comprehensive analysis of RCC2 in human pan-cancer. Additionally, RCC2 alteration data, RCC2 expression at the transcriptional and translational levels, prognostic capability, and immune-related analysis were explored across various tumor categories. Furthermore, gene enrichment analysis of a combination of RCC2-related and RCC2-interacted genes was conducted to investigate the possible role of RCC2 in human tumors.
2. Materials and methods
2.1. Genetic alteration analysis and transcriptional and translational gene expression analyses
RCC2 genetic alteration analysis was performed using the cBioPortal tool (https://www.cbioportal.org/) [15]. The module “TCGA Pan cancer Atlas studies” was employed to achieve the RCC2 alteration data across various of tumors. In addition, data regarding the mutation sites of RCC2 were acquired from cBioPortal.
“RCC2” was inputted into the “Gene_DE” module of the Tumor Immune Estimation Resource version2 (TIMER2) website (http://timer.cistrome.org/) [16]. The difference between the RCC2 expression in the tumor tissues and the adjacent normal tissues in different tumors, or specific tumor subtypes, was evaluated in the TCGA database [17]. Moreover, the RCC2 expression data was also analyzed based on the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Breast cancer (GSE42568), ewing sarcoma (GSE17674, GSE6798 and GSE3526) and ovarian cancer (GSE12470) were chosen to validate the finding from TCGA cohort [[18], [19], [20], [21], [22]].
The difference in RCC2 expression between normal and tumor tissues at the translational level was analyzed using the user-friendly online tool UALCAN (http://ualcan.path.uab.edu/analysis-prot.html) which provides protein expression data based on the CPTAC dataset [23]. A total of ten available tumor types were analyzed: breast carcinoma (BRCA), colon adenocarcinoma (COAD), head and neck squamous cell carcinoma (HNSC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), kidney renal clear cell carcinoma (KIRC), glioblastoma multiforme (GBM), ovarian carcinoma (OV), pancreatic adenocarcinoma (PAAD), and uterine corpus endometrial carcinoma (UCEC). The Human Protein Atlas (HPA) cohort was used to explore RCC2 gene translational expression in different tissues, single cells, and tumors. RCC2 immunohistochemical staining across different tumor types was acquired from the HPA database.
2.2. Survival analysis
The overall survival (OS) and disease-free survival (DFS) of all tumors were analyzed using GEPIA2 (http://gepia2.cancer-pku.cn/), an online tool providing high-quality analysis of TCGA and the GTEx database [24]. The RCC2 expression cut-off was set at 50%–50% across all tumors to divide the patients into two groups: “high RCC2” and “low RCC2” expression. Prognosis analysis between these two groups was used to generate a survival heatmap and Kaplan-Meier survival curves. The log-rank p-value and hazard ratio (HR) were calculated using the GEPIA2 tool.
Survival outcome data for the TCGA database was obtained to further analyze the relationship between RCC2 expression and prognosis of various tumors [25]. The SangerBox (http://sangerbox.com/) online platform was used to perform the hazards model and analyze the relationship between RCC2 expression and prognosis across different tumors. Moreover, R package “survminer” and “survival” were used to achieve the Disease Specific Survival (DSS) analysis based on TCGA database.
2.3. Construction of related gene networks
The protein-protein interaction (PPI) network of RCC2 was constructed using the STRING tool (https://string-db.org/). Fifty RCC2 interacting proteins were harvested which were previously known or predicted. Additionally, the top 100 RCC2-correlated genes related to RCC2 genes were obtained based on the data from all the tumor tissues in the TCGA database using the GEPIA2 tool. The co-expression of RCC2 and the top five related genes in all patients were selected to further analyze the potential relationship between RCC2 expression and related genes using the GEPIA2 tool. The TIMER2 tool was used to investigate the co-expression status of RCC2 and the five genes in each tumor type.
2.4. Gene enrichment analysis
The R package “clusterProfiler” was used to analyze RCC2-related genes and RCC2-interaction genes using Gene Ontology (GO) enrichment and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. Gene set enrichment analysis (GSEA) was performed on the low- and high-expression groups based on the mean expression values of RCC2 in tumors of TCGA dataset. The R package “clusterProfiler” was used to perform C6 (oncogenic signature gene sets) enrichment analyses. The gene sets with |NES| > 1, FDR <0.25, and p. adjust <0.05, were considered to be significantly enriched.
2.5. Immune-related analysis
RCC2 expression was normalized and log2 (x+1) was transformed in all involved tumors. The SangerBox online platform was employed to gain the StromalScore, ImmuneScore, and ESTIMATEScore for all involved cancers [26]. The relationship between RCC2 expression and 60 immune checkpoint (ICP) genes (24 inhibitory genes and 36 stimulatory genes) was also analyzed [27]. The SangerBox platform was then used to calculate the tumor mutation burden (TMB) [28]. The microsatellite instability (MSI) score data for all tumors obtained from a previous study was re-analyzed to determine a correlation between RCC2 expression [29]. In addition, the TIMER2 tool was used to explore the relationship between RCC2 expression and cancer-associated fibroblasts (CAFs) across different cancers based on the TCGA database by using a purity-adjusted Spearman's rho.
3. Results
3.1. Genetic alteration analysis
The cBioPortal tool showed that RCC2 genetic variation type and frequency was significant with a deep delete rate of 5.56%, 2.25%, 1.15% and 0.67% in cholangiocarcinoma (CHOL), pheochromocytoma and paraganglioma (PCPG), mesothelioma (MESO) and testicular germ cell tumors (TGCT), respectively (Fig. 1A). There was no other kind of variation in these tumors. There is only one mutation type in diffuse large B-cell lymphoma (DLBC) and acute myeloid leukemia (LAML) with frequencies of 2.08% and 0.5%, respectively. The amplification rate was 1.96% in sarcoma (SARC), and the types of alterations were the combination of two or more types of alterations in other involved tumors. There were 101 variants of uncertain significance (VUS) in RCC2 in various tumors (Fig. 1B and Supplementary Table S1). The transcriptional expression of RCC2 categorized by different copy number and mutation in TCGA cohort was shown Supplementary Fig. S1.
Fig. 1.
Genetic alternation of regulation of chromosome condensation 2 (RCC2) in different tumors. (A) Alteration frequency with mutation type. (B) Mutation types, sites and case number of RCC2 genetic alternation.
3.2. Gene expression analysis
Comparison of RCC2 expression between different healthy tissues, single cell types, and tumors was performed based on the system analysis of HPA, GTEx, and functional annotation of mammalian genome 5 (FANTOM5) cohorts. RCC2 expression was highest in the esophagus in normal tissues; however, there was low RNA tissue specificity in all normal tissues (Supplementary Fig. S2). In addition, RCC2 expression showed low cell-type specificity in RNA single cell type analysis and low cancer specificity in RNA cancer category analysis (Supplementary Fig. S2).
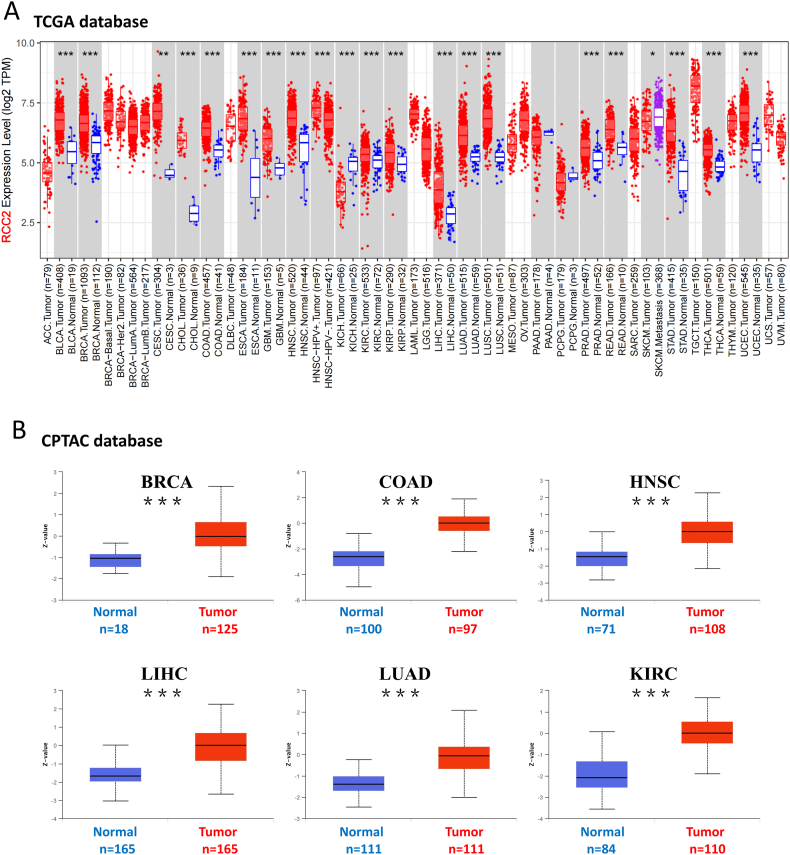
RCC2 transcriptional expression across different tumor types was analyzed according to the TCGA and GTEx databases. RCC2 was highly expressed in most tumors compared to that in the corresponding normal tissues (Fig. 2A). However, RCC2 expression was higher in normal tissues than in kidney chromophobe tumors (KICH) (p < 0.05). Additionally, RCC2 expression was similar in normal and tumor tissues in PAAD and PCPG. RCC2 protein expression within tumor tissues was significantly higher than the corresponding normal tissues in BRCA, COAD, HNSC, LIHC, LUAD, KIRC, GBM, OV, PAAD and UCEC based on the CPTAC database (Fig. 2B and Supplementary Fig. S3, all p < 0.001). Additionally, RCC2 was high expressed in tumor than normal tissue in breast cancer, ewing sarcoma and ovarian cancer based on GEO database (Supplementary Fig. S4). Immunohistochemical staining for the expression of RCC2 in COAD, LIHC, and stomach adenocarcinoma (STAD) based on HPA database is shown (Fig. 3).
Fig. 2.
RCC2 gene and protein expression in different tumors. (A) RCC2 expression in different tumors or specific tumor subtypes. (B) RCC2 total protein in normal tissue and tumorous tissue. ***p < 0.001.
Fig. 3.
RCC2 expression in different tumors at the translation level: three tumor types were shown (COAD, LIHC and STAD) compared to the correlating normal tissues (colon, live and stoma).
3.3. Survival analysis
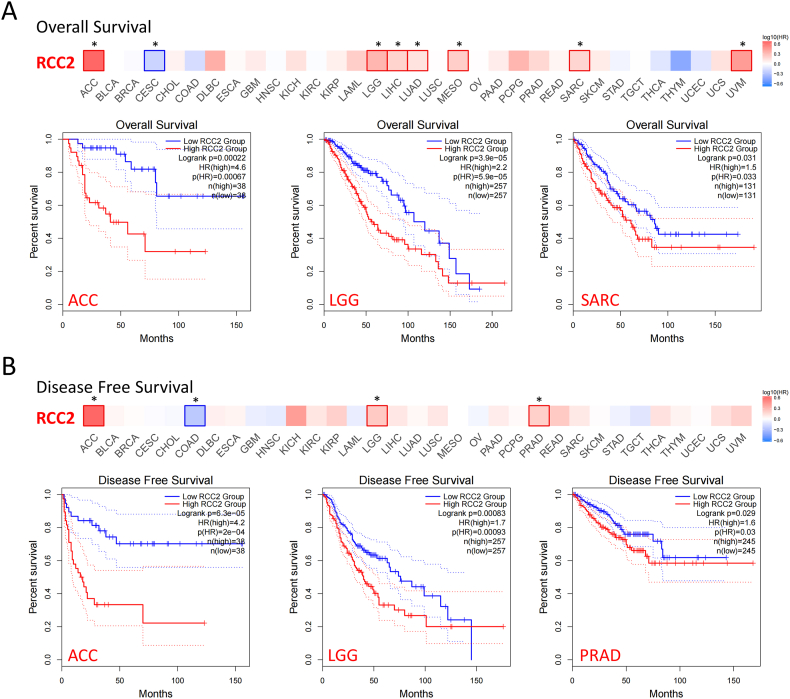
RCC2 expression from TCGA was positively correlated with OS in some tumors such as ACC, lower grade glioma (LGG), SARC, and UVM, but negatively correlated with OS in cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) (Fig. 4A, all p < 0.05). Individuals with higher RCC2 expression were linked to poor DFS in ACC, LGG, and prostate adenocarcinoma (PRAD), but linked to good DFS in COAD (Fig. 4B, all p < 0.05).
Fig. 4.
Correlation between RCC2 gene expression in tumors and survival prognosis in TCGA dataset. (A) Correlation between RCC2 expression and overall survival (OS) of different tumor patients. (B) Correlation between RCC2 expression and disease-free survival (DFS) of different tumor patients. *p < 0.05.
The relation between RCC2 expression and prognosis across various tumors based on TCGA cohort. RCC2 overexpression was associated with poor prognosis in LAML, PAAD, KIRC, and KICH for OS and linked to poor prognosis in LIHC and PAAD for DFS (Supplementary Figs. S5 and S6). Addtionnally, the high-expression of RCC2 was also linked to poor DSS in some tumors such as LIHC (Supplementary Fig. S7).
3.4. Gene enrichment analysis
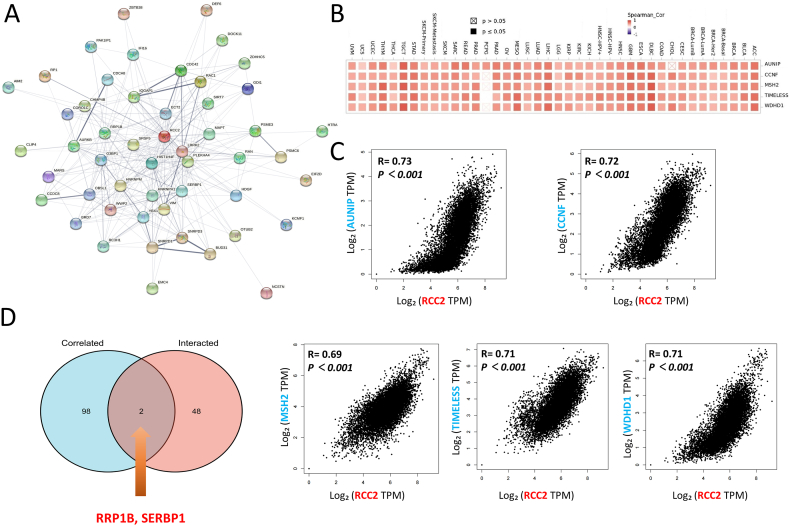
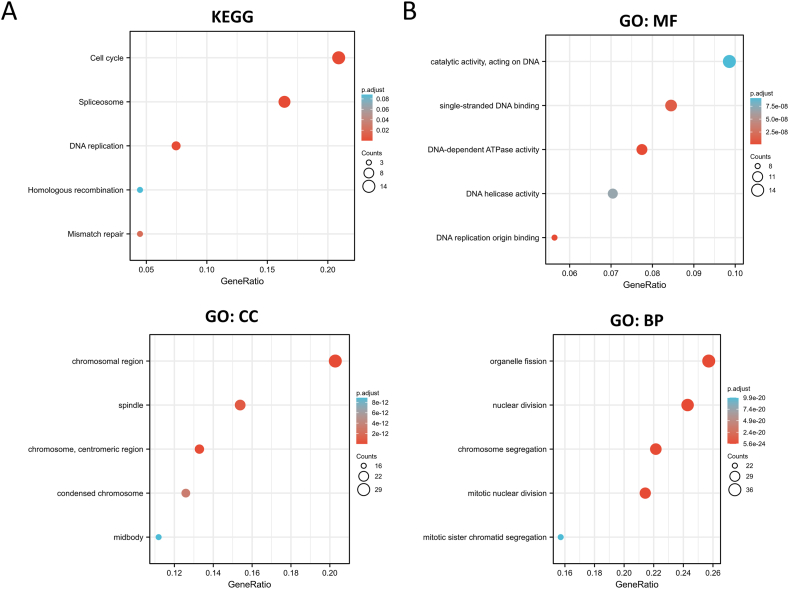
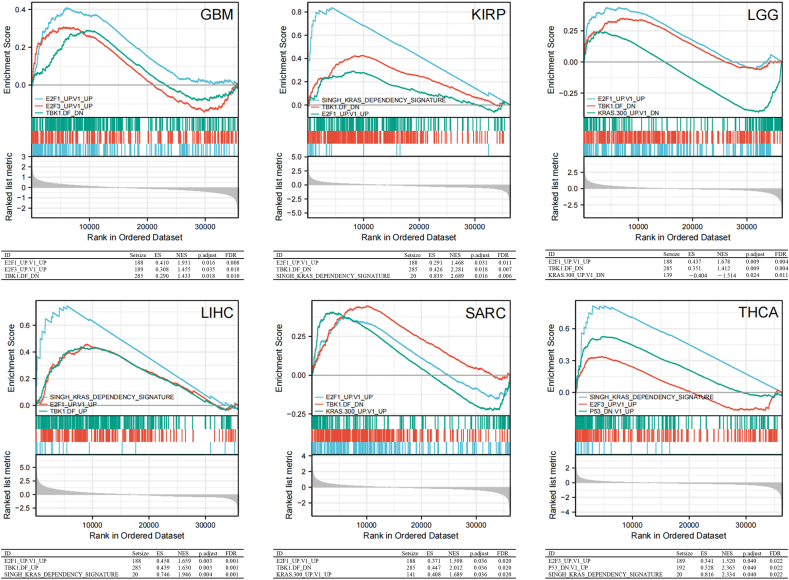
RCC2-interacting genes and RCC2-correlated genes were obtained using the STRING and GEPIA2 tools, respectively to investigate the possible mechanism of RCC2 in the progression of cancer formation and invasion (Supplementary Table S2). The STRING PPI network showed that the top five RCC2-correlated genes in cancer were aurora kinase A and ninein interacting protein (AUNIP), cyclin F (CCNF), MutS homolog 2 (MSH2), timeless circadian regulator (TIMELESS), and WD repeat and HMG-box DNA binding protein 1 (WDHD1) (Fig. 5A). RCC2 was positively related to these five genes across most of the involved tumors (Fig. 5B and C). In addition, the analysis across of RCC2-interacted and RCC2-correlated genes identified two genes: ribosomal RNA processing 1 B (RRP1B) and serpine1 mRNA binding protein 1 (SERBP1) (Fig. 5D). KEGG analysis of the RCC2-interacted and RCC2-correlated genes showed that RCC2 was associated with cell cycle, DNA replication, and mismatch repair (Fig. 6A). GO enrichment analysis indicated that RCC2 was associated with catalytic activity, acting on DNA and DNA helicase activity in the molecular function (MF) module and linked to chromosomal region, spindle in cellular component (CC) module, and related to organelle fission and nuclear division in the biological process (BP) module (Fig. 6B). In addition, the MSigDB C6 (oncogenic genes sets) analysis demonstrated that gene expression profile in patients of TCGA cohort with high expression of RCC2 was associated with signature oncogenes, such as E2F, KRAS, and TBK1 (Fig. 7).
Fig. 5.
RCC2-related gene enrichment analysis. (A) A protein-protein interaction network of 50 experimentally verified RCC2-interacting proteins. (B) Heatmap showing the correlation between RCC2 and the top 5 RCC2-correlated genes in the detailed tumor types. (C) Correlation between RCC2 and the five genes. (D) Cross analysis of the RCC2-interacted and RCC2-correlated genes identified RRP1B and SERBP1 genes.
Fig. 6.
(A) KEGG pathway analysis of the RCC2-interacting and RCC2-correlated genes. (B) GO analysis of the RCC2-interacting and RCC2-correlated genes.
Fig. 7.
Gene set enrichment analysis of RCC2. Oncogenic signature gene sets enriched in high RCC2 expression group. ES: enrichment score; NES: normalized enrichment score; FDR: false discovery rate.
3.5. Immune-related analysis
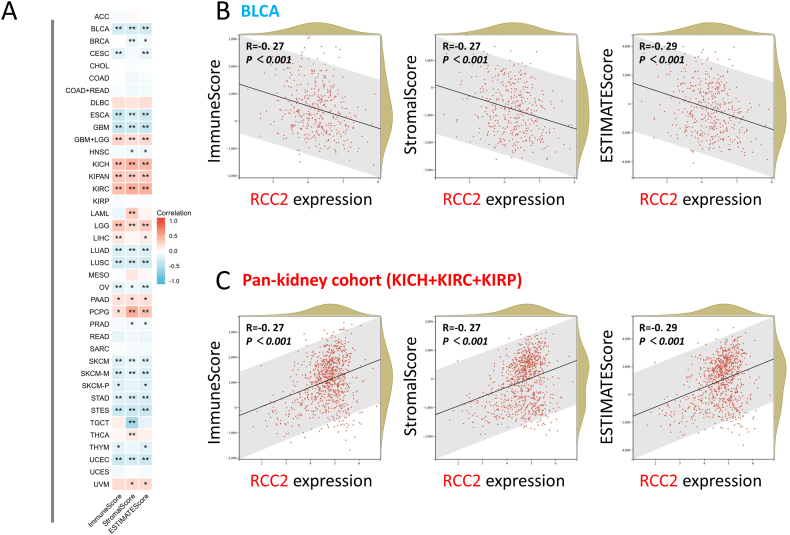
Significant differences were observed between RCC2 expression and the immune infiltration score (ImmuneScore, StromalScore, and ESTIMATEScore) in most of the tumors (Fig. 8). RCC2 expression was positively related to all three scores in KICH, KIRC, LGG, PAAD, and PCPG, but negatively related to bladder urothelial carcinoma (BLCA), esophageal carcinoma (ESCA), GBM, LUAD, lung squamous cell carcinoma (LUSC), OV, skin cutaneous melanoma (SKCM), STAD, and urothelial carcinoma (UCEC) (Fig. 8A, all p < 0.05). RCC2 expression was positively associated with CAF in some involved tumors and negatively associated with that in TGCT (Supplementary Fig. S8). Dot plots for the BLCA and pan-kidney cohorts are shown (Fig. 8B and C).
Fig. 8.
Correlation analysis between RCC2 expression and ImmuneScore, StromalScore and ESTIMATEScore. (A) Heatmap showing the correlation across the involved tumors. Two kinds of typical tumors were displayed: (B) BLCA and (C) Pan-kidney cohort (KICH + KIRC + KIRP). *p < 0.05; **p < 0.01.
RCC2 was positively related to MSI in GBM, LUAD, LUSC, and SARC, but negatively related to MSI in DLBC, HNSC, and PAAD. RCC2 expression was positively related to TMB in ACC, KICH, LUAD, rectal adenocarcinoma (READ), SARC, and STAD, but negatively related to TMB in CHOL and HNSC (Fig. 9A). ICP analysis showed that RCC2 was positively linked to most ICPs in thymoma (THYM), LIHC, PAAD, UVM, KICH, KIRC, LGG, and thyroid carcinoma (THCA). RCC2 is positively related to programmed cell death protein-1 (PDCD1, also known as PD-1), CD274 (also known as PD-L1), and cytotoxic T-lymphocyte-associated protein 4 (CTLA4, also known as CD152) in many tumors including BRCA, KIRC, and LIHC (Fig. 9B, Supplementary Fig. S9).
Fig. 9.
(A) Correlation between RCC2 expression and microsatellite instability (MSI) or tumor mutational burden (TMB). (B) Association between RCC2 expression and immune checkpoint (ICP) gene expression in different tumors. *p < 0.05.
4. Discussion
RCC2 regulates the cell cycle and may be associated with EMT [9,10,30]. Its overexpression can lead to tumor formation and poor prognosis in various cancers [3,13]. In this study, R software and several gene-related analysis online-tools such as cBioPortal were employed to analyze over 10,000 samples based on TCGA, GTEx, GEO and CPTAC cohorts across many cancer types. The data analyzed RCC2 genetic alteration, gene expression, prognosis, and immune infiltration. In addition, gene enrichment analysis was performed using the combined RCC2-interacted and RCC2-correlated genes, which could be helpful in exploring the possible mechanism between RCC2 expression and cancer formation.
RCC2 was overexpressed in most tumors including BRCA, ESCA, and LIHC, which correlated with previous studies [9,13,31]. However, RCC2 expression in normal tissues was higher than that in KICH and LAML tumorous tissues. Consequently, RCC2 might be a “double-edged sword” across all tumors because it may be an oncogene in most cancers, and a tumor suppressor gene in tumors such as KICH. Despite RCC2 was overexpressed in both lung cancer subtypes (LUAD and LUSC), it was linked to poor OS only in LUAD. This indicates that the prognostic ability of RCC2 may depend on the lung cancer subtype, or may also be present in other cancers. Further studies are required to confirm this hypothesis. OS was previously viewed as one of the best indices in cancer treatment as it might be used to evaluate treatment outcome, with DFS playing an important role in surgical treatment together with radiotherapy [25]. The present study indicated that RCC2 was positively associated with OS and DFS in some tumors such as ACC. Hence, RCC2 may be a promising biomarker for those tumors. Of note, the overexpression of RCC2 was associated with poor OS, DFS and DSS in some tumors such as LGG, which might suggest that RCC2 could be a potential target for the treatment for those tumors.
RCC2 was positively related to AUNIP, CCNF, MSH2, TIMELESS, and WDHD1 in most cancers. These five genes are linked to tumor formation, invasion, and DNA replication, with RCC2 and MSH2 previously regarded as molecular signatures for lung cancer malignancy [[32], [33], [34], [35], [36], [37]]. In this work, RRP1B and SERBP1 oncogenes were obtained via intersection analysis of RCC2-interacted and RCC2-correlated genes. These oncogenes may be linked to EMT in some tumors [[38], [39], [40], [41]]. This might reveal the potential mechanism by which RCC2 leads to cancer formation and metastasis. Our previous study showed that PTTG1 is associated with the cell cycle and could be linked to cancer formation via EMT [42]. RCC2 is a cell cycle regulator that promotes EMT in BRCA, LUAD and PAAD [9,10,43,44]. EMT appears in normal tissue during wound repair or fibrosis and might be associated with tumor formation or even cancer metastasis [45]. During EMT, normal epithelial cells invade and abandon polarity [44]. High RCC2 expression promoted cell proliferation, DNA replication, and mismatch repair. In addition, the GSEA demonstrated that high expression of RCC2 was associated with signature oncogenes, such as E2F, KRAS, and TBK1. The roles of E2F and KRAS in cancer have been extensively studied [45,46]. TBK1 was selectively essential in cells that harbor mutant KRAS [47]. Emerging evidence showed that TBK1 plays important role in the pathogenesis of cancer [48,49]. Thus, our findings suggested that RCC2 could be a potential oncogene in human cancer. And experimental based validation was needed to make this hypothesis more reliable.
Immune infiltration plays a crucial role in anti-cancer therapy because it rebuilds the tumor microenvironment and regulates anti-cancer responses [50,51]. This study indicated that RCC2 was positively related to ImmuneScore, StromalScore, and ESTIMATEScore in KICH, KIRC, LGG, PAAD, and PCPG. Thus, RCC2 overexpression was associated with the high immune and stromal cell infiltration and low tumor purity in the tumor mass. RCC2 expression was negatively correlated with all the scores in tumors such as BLCA, ESCA, and GBM indicating that there was poor immune infiltration. Consequently, RCC2-targeted treatment should depend on the tumor types rather than “one for all” therapy. Further research is required into immune infiltration and the tumor microenvironment.
Insights into the immune checkpoint (ICP) in tumor treatment may lead to revolutionary outcomes [[52], [53], [54]]. Cytotoxic T-lymphocyte-associated protein-4 (CTLA-4) and PD-1 are two of the best targets for tumor therapy [55,56]. Inhibitors of these two targets have achieved great success in tumor therapy [57,58]. This study showed that RCC2 expression was positively related to these two targets in many cancers, including STAD, PRAD, and BRCA, indicating that RCC2 could be a potential novel biomarker for PD-1 and CTLA-4 in those tumors. The latest findings show that TMB is a good biomarker for the assessment of the response to ICP inhibitor [59,60]. Thus, the relationship between RCC2 expression and MSI/TMB could be a promising biomarker for ICP inhibitors, such as PD-1 or CTLA-4. Consequently, the mixture of MSI and tumor microenvironment could be a new method for predicting the outcome of ICP inhibitor treatment for cancers [61,62]. In this study, RCC2 expression was positively associated with TMB/MSI in LUAD and SARC but negatively related to TMB/MSI in HNSC. Hence, RCC2 may be a promising biomarker for ICP inhibitor treatment of these tumors. However, more experimental validations were needed to further confirmed this finding.
This study presented the results of the systemic pan-cancer analysis of RCC2. It is the first analysis combining RCC2 expression, survival, gene enrichment, immune infiltration, ICP, and TMB/MSI analysis in human pan-cancer. This provides new insights into the possible mechanisms of RCC2 in cancer formation, invasion, and prognostic ability.
However, there are still some limitations in this study. Many databases were included in this study, so there may be some factors which may affect the result. Experiments base on in vivo or in vitro models could make our results more reliable and we would put our efforts on this field in the future.
Pan-cancer analysis of RCC2 showed that its expression is associated with tumor formation, immune infiltration, and prognosis. RCC2 may be used as a promising biomarker for ICP inhibitors across multiple tumors. This data may be helpful in understanding the potential role of RCC2 in carcinogenesis and antitumor therapy.
Author contributions
Siming Gong, Hao Wu, Changwu Wu, Yingjuan Duan, Juyu Tang and Jinfei Fu - Conceived and designed the studies; Siming Gong, Hao Wu and Panfeng Wu - Performed the studies; Siming Gong, Hao Wu and Changwu Wu - Analyzed and interpreted the data; Siming Gong, Changwu Wu and Jinfei Fu - Contributed analysis tools; Siming Gong, Hao Wu, Changwu Wu, Juyu Tang and Jinfei Fu - Wrote the paper. All authors approved the final manuscript and accounted for all aspects of work, read, and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (8220392, 81871577 and 82072194) and Hunan Provincial Natural Science Foundation of China (2022JJ40778). No funding bodies had any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability statement
The data provided in this study can be obtained in the method section of this manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
The results shown here are in part based upon data generated by TCGA Research Network (https://www.cancer.gov/tcga), GEO database (https://www.ncbi.nlm.nih.gov/geo/) and HPA database (https://www.proteinatlas.org/).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e13599.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Riddick G., Macara I.G. A systems analysis of importin-α-β mediated nuclear protein import. J. Cell Biol. 2005;168:1027–1038. doi: 10.1083/jcb.200409024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi H.J.C., Lin J.R., Vannier J.B., Slaats G.G., Kile A.C., Paulsen R.D., et al. NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol. Cell. 2013;51:423–439. doi: 10.1016/j.molcel.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu C., Duan Y., Gong S., Kallendrusch S., Schopow N., Osterhoff G. Integrative and comprehensive pancancer analysis of regulator of chromatin condensation 1 (RCC1) Int. J. Mol. Sci. 2021;22:7374. doi: 10.3390/ijms22147374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreassen P.R., Palmer D.K., Wener M.H., Margolis R.L. Telophase disc: a new mammalian mitotic organelle that bisects telophase cells with a possible function in cytokinesis. J. Cell Sci. 1991;99:523–534. doi: 10.1242/jcs.99.3.523. [DOI] [PubMed] [Google Scholar]
- 5.Yenjerla M., Panopoulos A., Reynaud C., Fotedar R., Margolis R.L. TD-60 is required for interphase cell cycle progression. Cell Cycle. 2013;12:837–841. doi: 10.4161/cc.23821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mollinari C., Reynaud C., Martineau-Thuillier S., Monier S., Kieffer S., Garin J., et al. The mammalian passenger protein TD-60 is an RCC1 family member with an essential role in prometaphase to metaphase progression. Dev. Cell. 2003;5:295–307. doi: 10.1016/S1534-5807(03)00205-3. [DOI] [PubMed] [Google Scholar]
- 7.Papini D., Langemeyer L., Abad M.A., Kerr A., Samejima I., Eyers P.A., et al. TD-60 links RalA GTPase function to the CPC in mitosis. Nat. Commun. 2015;6 doi: 10.1038/NCOMMS8678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu K., Li Y., Wu W., Chen H., Chen Z., Zhang Y., et al. High-performance gene expression and knockout tools using sleeping beauty transposon system. Mobile DNA. 2018;9 doi: 10.1186/s13100-018-0139-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z., Wu W., Huang Y., Xie L., Li Y., Chen H., et al. RCC2 promotes breast cancer progression through regulation of Wnt signaling and inducing EMT. J. Cancer. 2019;10:6837–6847. doi: 10.7150/jca.36430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pang B., Wu N., Guan R., Pang L., Li X., Li S., et al. Overexpression of RCC2 enhances cell motility and promotes tumor metastasis in lung adenocarcinoma by inducing epithelial–mesenchymal transition. Clin. Cancer Res. 2017;23:5598–5610. doi: 10.1158/1078-0432.CCR-16-2909. [DOI] [PubMed] [Google Scholar]
- 11.Song C., Liang L., Jin Y., Li Y., Liu Y., Guo L., et al. RCC2 is a novel p53 target in suppressing metastasis. Oncogene. 2018;37:8–17. doi: 10.1038/onc.2017.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu N., Ren D., Li S., Ma W., Hu S., Jin Y., et al. RCC2 over-expression in tumor cells alters apoptosis and drug sensitivity by regulating Rac1 activation. BMC Cancer. 2018;18 doi: 10.1186/s12885-017-3908-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Q., Jiang P., Jia B., Liu Y., Zhang Z. RCC2 contributes to tumor invasion and chemoresistance to cisplatin in hepatocellular carcinoma. Hum. Cell. 2020;33:709–720. doi: 10.1007/s13577-020-00353-7. [DOI] [PubMed] [Google Scholar]
- 14.Xu X., Zhou X., Zhang J., Li H., Cao Y., Tan X., et al. MicroRNA-191 modulates cisplatin-induced DNA damage response by targeting RCC2. Faseb. J. 2020;34:13573–13585. doi: 10.1096/fj.202000945R. [DOI] [PubMed] [Google Scholar]
- 15.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T., Fu J., Zeng Z., Cohen D., Li J., Chen Q., et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke C., Madden S.F., Doolan P., Aherne S.T., Joyce H., O'Driscoll L., et al. Correlating transcriptional networks to breast cancer survival: a large-scale coexpression analysis. Carcinogenesis. 2013;34:2300–2308. doi: 10.1093/carcin/bgt208. [DOI] [PubMed] [Google Scholar]
- 19.Savola S., Klami A., Myllykangas S., Manara C., Scotlandi K., Picci P., et al. High expression of complement component 5 (C5) at tumor site associates with superior survival in ewing's sarcoma family of tumour patients. ISRN Oncol. 2011;2011:1–10. doi: 10.5402/2011/168712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skov V., Glintborg D., Knudsen S., Jensen T., Kruse T.A., Tan Q., et al. Reduced expression of nuclear-encoded genes involved in mitochondrial oxidative metabolism in skeletal muscle of insulin-resistant women with polycystic ovary syndrome. Diabetes. 2007;56:2349–2355. doi: 10.2337/db07-0275. [DOI] [PubMed] [Google Scholar]
- 21.Roth F.P., Troia G.A., Worthington C.K., Handy D. Promoting Awareness of Sounds in Speech (PASS): the effects of intervention and stimulus characteristics on the blending performance of preschool children with communication impairments. Learn. Disabil. Q. 2006;29:67–88. doi: 10.1007/s10048-006-0032-6. [DOI] [Google Scholar]
- 22.Yoshihara K., Tajima A., Komata D., Yamamoto T., Kodama S., Fujiwara H., et al. Gene expression profiling of advanced-stage serous ovarian cancers distinguishes novel subclasses and implicates ZEB2 in tumor progression and prognosis. Cancer Sci. 2009;100:1421–1428. doi: 10.1111/j.1349-7006.2009.01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen F., Chandrashekar D.S., Varambally S., Creighton C.J. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 2019;10(1):5679. doi: 10.1038/s41467-019-13528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J., Lichtenberg T., Hoadley K.A., Poisson L.M., Lazar A.J., Cherniack A.D., et al. An integrated TCGA pan-cancer clinical data Resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4 doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beroukhim R., Mermel C.H., Porter D., Wei G., Raychaudhuri S., Donovan J., et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonneville R., Krook M.A., Kautto E.A., Miya J., Wing M.R., Chen H.-Z., et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis. Oncol. 2017;2017:1–15. doi: 10.1200/po.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calderon-Aparicio A., Bode A.M. Roles of regulator of chromosome condensation 2 in cancer: beyond its regulatory function in cell cycle. Onco Rev. 2021;15 doi: 10.4081/ONCOL.2021.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calderon-Aparicio A., Yamamoto H., de Vitto H., Zhang T., Wang Q., Bode A.M., et al. RCC2 promotes esophageal cancer growth by regulating activity and expression of the Sox2 transcription factor. Mol. Cancer Res. 2020;18:1660–1674. doi: 10.1158/1541-7786.MCR-19-1152. [DOI] [PubMed] [Google Scholar]
- 32.Ma C., Kang W., Yu L., Yang Z., Ding T. AUNIP expression is correlated with immune infiltration and is a candidate diagnostic and prognostic biomarker for hepatocellular carcinoma and lung adenocarcinoma. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.590006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang S.C., Hung C.S., Zhang B.X., Hsieh T.H., Hsu W., Ding J.L. A novel signature of ccnf-associated e3 ligases collaborate and counter each other in breast cancer. Cancers. 2021;13 doi: 10.3390/cancers13122873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu W., Ding K., Liao L., Ling Y., Luo X., Wang J. Analysis of the expression and prognostic value of MSH2 in pan-cancer based on bioinformatics. BioMed Res. Int. 2021;2021 doi: 10.1155/2021/9485273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chi L., Zou Y., Qin L., Ma W., Hao Y., Tang Y., et al. TIMELESS contributes to the progression of breast cancer through activation of MYC. Breast Cancer Res. 2017;19 doi: 10.1186/s13058-017-0838-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y., Chen J.J. STAT3 plays an important role in DNA replication by turning on WDHD1. Cell Biosci. 2021;11 doi: 10.1186/s13578-020-00524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujii K., Miyata Y., Takahashi I., Koizumi H., Saji H., Hoshikawa M., et al. Differential proteomic analysis between small cell lung carcinoma (SCLC) and pulmonary carcinoid tumors reveals molecular signatures for malignancy in lung cancer. Proteonomics Clin. Appl. 2018;12 doi: 10.1002/prca.201800015. [DOI] [PubMed] [Google Scholar]
- 38.Crawford N.P.S., Qian X., Ziogas A., Papageorge A.G., Boersma B.J., Walker R.C., et al. Rrp1b, a new candidate susceptibility gene for breast cancer progression and metastasis. PLoS Genet. 2007;3:2296–2311. doi: 10.1371/journal.pgen.0030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang T., Xu L., Jia R., Wei J. MIR-218 suppresses the metastasis and EMT of HCC cells via targeting SERBP1. Acta Biochim. Biophys. Sin. (Shanghai). 2017;49:383–391. doi: 10.1093/abbs/gmx017. [DOI] [PubMed] [Google Scholar]
- 40.Gong S., Wu C., Duan Y., Tang J., Wu P. A comprehensive pan-cancer analysis for pituitary tumor-transforming gene 1. Front. Genet. 2022:353. doi: 10.3389/FGENE.2022.843579. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin H., Zhang X., Feng N., Wang R., Zhang W., Deng X., et al. LncRNA LCPAT1 mediates smoking/Particulate matter 2.5-induced cell autophagy and epithelial-mesenchymal transition in lung cancer cells via RCC2. Cell. Physiol. Biochem. 2018;47:1244–1258. doi: 10.1159/000490220. [DOI] [PubMed] [Google Scholar]
- 42.Mukherjee M., Goswami S. Identification of key deregulated RNA-binding proteins in pancreatic cancer by meta-analysis and prediction of their role as modulators of oncogenesis. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.713852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stone R.C., Pastar I., Ojeh N., Chen V., Liu S., Garzon K.I., et al. Epithelial-mesenchymal transition in tissue repair and fibrosis. Cell Tissue Res. 2016;365:495–506. doi: 10.1007/s00441-016-2464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kent L.N., Leone G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer. 2019;19(6):326–338. doi: 10.1038/s41568-019-0143-7. [DOI] [PubMed] [Google Scholar]
- 46.Tang D., Kroemer G., Kang R. Oncogenic KRAS blockade therapy: renewed enthusiasm and persistent challenges. Mol. Cancer. 2021;20(1):128. doi: 10.1186/s12943-021-01422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu L., Xie H., Liu X., Potjewyd F., James L.I., Wilkerson E.M., et al. TBK1 is a synthetic lethal target in cancer with VHL loss. Cancer Discov. 2020;10(3):460–475. doi: 10.1158/2159-8290.CD-19-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Runde A.P., Mack R., SJ PB. Zhang J. The role of TBK1 in cancer pathogenesis and anticancer immunity. J. Exp. Clin. Cancer Res. 2022;41(1):135. doi: 10.1186/s13046-022-02352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gajewski T.F., Schreiber H., Fu Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J., Bae J.S. Tumor-associated macrophages and neutrophils in tumor microenvironment. Mediat. Inflamm. 2016;2016 doi: 10.1155/2016/6058147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Topalian S.L., Taube J.M., Anders R.A., Pardoll D.M. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abril-Rodriguez G., Ribas A. SnapShot: immune checkpoint inhibitors. Cancer Cell. 2017;31:848–848.e1. doi: 10.1016/j.ccell.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 54.Mezquita L., Auclin E., Ferrara R., Charrier M., Remon J., Planchard D., et al. Association of the lung immune prognostic index with immune checkpoint inhibitor outcomes in patients with advanced non-small cell lung cancer. JAMA Oncol. 2018;4:351–357. doi: 10.1001/jamaoncol.2017.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rotte A. Combination of CTLA-4 and PD-1 blockers for treatment of cancer. J. Exp. Clin. Cancer Res. 2019;38 doi: 10.1186/s13046-019-1259-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shukla S.A., Bachireddy P., Schilling B., Galonska C., Zhan Q., Bango C., et al. Cancer-germline antigen expression discriminates clinical outcome to CTLA-4 blockade. Cell. 2018;173:624–633.e8. doi: 10.1016/j.cell.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowshanravan B., Halliday N., Sansom D.M. CTLA-4: a moving target in immunotherapy. Blood. 2018;131:58–67. doi: 10.1182/blood-2017-06-741033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dermani F.K., Samadi P., Rahmani G., Kohlan A.K., Najafi R. PD-1/PD-L1 immune checkpoint: potential target for cancer therapy. J. Cell. Physiol. 2019;234:1313–1325. doi: 10.1002/jcp.27172. [DOI] [PubMed] [Google Scholar]
- 59.Chalmers Z.R., Connelly C.F., Fabrizio D., Gay L., Ali S.M., Ennis R., et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9 doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan T.A., Yarchoan M., Jaffee E., Swanton C., Quezada S.A., Stenzinger A., et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann. Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goodman A.M., Sokol E.S., Frampton G.M., Lippman S.M., Kurzrock R. Microsatellite-stable tumors with high mutational burden benefit from immunotherapy. Cancer Immunol. Res. 2019;7:1570–1573. doi: 10.1158/2326-6066.CIR-19-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luchini C., Bibeau F., Ligtenberg M.J.L., Singh N., Nottegar A., Bosse T., et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann. Oncol. 2019;30:1232–1243. doi: 10.1093/annonc/mdz116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data provided in this study can be obtained in the method section of this manuscript.