Abstract
Upon recognition of foreign antigens, naïve B cells undergo rapid activation, growth, and proliferation. How B-cell growth and proliferation are coupled with activation remains poorly understood. Combining CRISPR/Cas9-mediated functional analysis and mouse genetics approaches, we found that Dhx33, an activation-induced RNA helicase, plays a critical role in coupling B-cell activation with growth and proliferation. Mutant mice with B-cell-specific deletion of Dhx33 exhibited impaired B-cell development, germinal center reactions, plasma cell differentiation, and antibody production. Dhx33-deficient B cells appeared normal in the steady state and early stage of activation but were retarded in growth and proliferation. Mechanistically, Dhx33 played an indispensable role in activation-induced upregulation of ribosomal DNA (rDNA) transcription. In the absence of Dhx33, activated B cells were compromised in their ability to ramp up 47S ribosomal RNA (rRNA) production and ribosome biogenesis, resulting in nucleolar stress, p53 accumulation, and cellular death. Our findings demonstrate an essential role for Dhx33 in coupling B-cell activation with growth and proliferation and suggest that Dhx33 inhibition is a potential therapy for lymphoma and antibody-mediated autoimmune diseases.
Keywords: RNA helicases, B cell activation, Germinal center reaction
Subject terms: Immunology, Cell biology
Introduction
B cells reside in lymphoid follicles of secondary lymphoid tissues such as the spleen and lymph nodes. They patrol the body by circulating through the blood and lymphatic vasculature. Naïve B cells are quiescent and do not secrete antibodies. Upon exposure to appropriate stimuli, B cells increase in size and start to proliferate, a process known as blasting. B-cell blasts can either form an extrafollicular focus or enter a specialized structure called the germinal center. In the extrafollicular response, B-cell blasts undergo further proliferation, class-switch recombination, and differentiation into short-lived plasma cells. B cells in the germinal center undergo extensive proliferation, class-switch recombination, and somatic hypermutation and eventually differentiate into plasma cells, some of which are long-lived, and memory B cells. While plasma cells are antibody-secreting cells, memory B cells maintain the B-cell phenotype but are programmed to rapidly differentiate into plasma cells upon encountering the same antigen. While cell surface receptors, their downstream signaling pathways and transcriptional networks that convert environmental stimuli into B-cell activation and differentiation programs have been extensively studied over the past decades, relatively little is known about how B cells, and lymphocytes in general, coordinate activation with growth and proliferation [1].
Proteins are the most abundant macromolecules in proliferating cells, and protein synthesis is of fundamental importance to cell growth and proliferation. Recent global quantification of transcriptomes and translatomes during lymphocyte activation has shed some light on the gene expression programs of naïve cells and activation-induced reprogramming that ramps up protein synthesis to meet the increasing needs of cell growth and proliferation [2, 3]. Naïve B cells are prepared to rapidly generate new mRNAs, as 90% of the promoters of genes to be expressed in activated B cells are preloaded with polymerase. In the first 24 h after activation, the total number of mRNA molecules increases by approximately 6-fold, largely due to rapid transcription of poised genes [2]. In addition to transcriptional readiness, naïve lymphocytes are also prepared for activation at the translational level. In naïve T cells, most highly expressed mRNAs are translationally repressed – those mRNAs mainly encode ribosomal proteins and translation initiation and elongation factors. In addition, there is a large pool of idling ribosomes, whose average translational output is maintained at a low level. Upon activation, their translational output is increased by 4-fold in the first 6 h, resulting in rapid synthesis of ribosomal proteins and translation factors, as well as proteins encoded by other preexisting and newly synthesized mRNAs [3]. The pool of preexisting ribosomes is not sufficient to sustain cell growth and proliferation. In the first 2 days of activation, the number of ribosomes per cell increases by 6-fold. This, together with elevated translational output of individual ribosomes, leads to a more than 20-fold increase in the total output of newly synthesized proteins. The drastic increase in protein synthesis is essential for the growth and proliferation of activated lymphocytes and the rapid expansion of antigen-specific lymphocyte clones. This forms the foundation of the clonal selection theory [4].
The rate of cell growth and proliferation is directly proportional to the rate of protein synthesis, which is intricately linked to ribosome biogenesis and is controlled at the level of ribosomal DNA (rDNA) transcription by RNA polymerase I. In mammalian cells, a single precursor of rRNA transcript, 47S rRNA (14.3 kb), is transcribed from rDNA by the RNA polymerase I complex. The 47S rRNA is subsequently processed into 18S, 5.8S, and 28S rRNAs, which form the major structural components of ribosomes. The generation of 47S rRNA is the initial and rate-limiting step in ribosome biogenesis and, as such, is controlled by multiple levels of regulation [5]. However, it is poorly understood how extracellular stimuli that activate lymphocytes are channeled to rDNA transcription and ribosome biogenesis.
RNA helicases participate in RNA metabolism and control gene expression by regulating different steps of RNA processing, including transcription, pre-mRNA splicing, nuclear export, cytoplasmic transport and storage, translation regulation, and degradation [6]. In a CRISPR/Cas9-mediated functional screening of RNA helicases for novel regulators of B-cell activation, proliferation, and plasma cell differentiation, we found that Dhx33, an RNA helicase induced upon B-cell activation, plays an indispensable role in B-cell growth and proliferation. Genetic studies showed that mutant mice with B-cell-specific deletion of Dhx33 exhibited impaired germinal center reactions, plasma cell differentiation, and antibody production. Mechanistically, Dhx33 associated with UBF, an rDNA architectural protein, to facilitate rDNA transcription and ribosome biogenesis. In the absence of Dhx33, B cells underwent normal activation but were compromised in their ability to ramp up 47S rRNA production and ribosome biogenesis, resulting in nucleolar stress, p53 accumulation, and cellular death. Our findings reveal an essential role of Dhx33 in coupling B-cell activation with growth and proliferation.
Results
Dhx33 identified as a critical regulator of B-cell proliferation via CRISPR/Cas9 screening
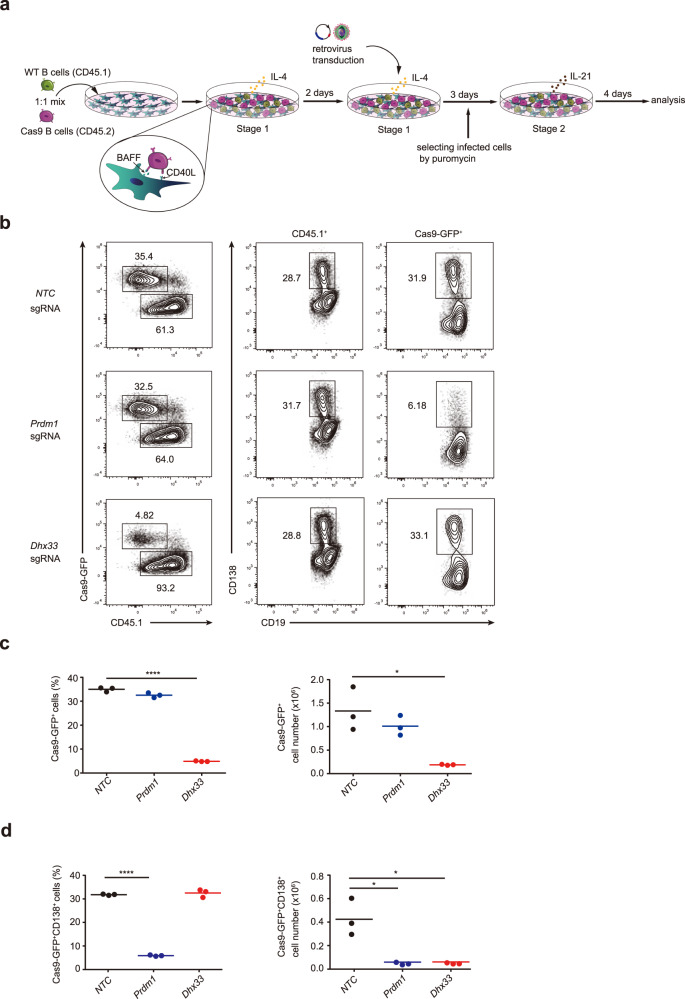
We recently performed CRISPR/Cas9-mediated functional screening of RNA helicases to identify novel regulators of B-cell activation, proliferation and differentiation. Toward this goal, B cells were cocultured with CD40L-and BAFF-expressing 40LB stromal cells in the presence of IL-4 and IL-21 to differentiate into germinal center B (iGCB, stage 1) cells and plasma cells (iPCs, stage 2), respectively [7]. Activated B cells were transduced with retroviruses encoding puromycin resistance genes and sgRNAs targeting 72 RNA helicases. Transduced B cells were positively selected with puromycin and replated on 40LB stromal cells in the presence of IL-21 to induce plasma cell differentiation. The number and percentage of plasma cells among sgRNA-transduced Cas9-GFP+ B cells were analyzed in the final population (manuscript in preparation). Dhx33 was identified as a positive regulator of B-cell proliferation in this screen (Supplementary Table 1). We further validated the roles of Dhx33 in B-cell proliferation and plasma cell differentiation in this in vitro culture system. As shown in Fig. 1a, Cas9-GFP (CD45.2+) and wild-type (CD45.1+) B cells were mixed in a 1:1 ratio, cocultured with 40LB cells plus IL-4 for iGCB differentiation, and transduced with retroviruses encoding sgRNA targeting Prdm1 (encoding Blimp-1, sgPrdm1), Dhx33, and nontargeting control (NTC). iGCB cells were replated on 40LB cells plus IL-21 for iPC differentiation. Consistent with the critical role of Prdm1 in controlling plasma cell differentiation, the percentage and number of Prdm1 sgRNA-transduced Cas9-GFP+ B cells were similar to those of their sgNTC-transduced counterparts, while those of CD138+ iPCs were drastically decreased among Prdm1 sgRNA-transduced Cas9-GFP+ B cells (Fig. 1b–d). In stark contrast, Dhx33 sgRNA-transduced Cas9-GFP+ B cells exhibited a markedly reduced frequency and number but a relatively normal percentage of CD138+ iPCs (Fig. 1b–d), suggesting that Dhx33 plays a critical role in B-cell proliferation and/or survival.
Fig. 1.
A critical role of Dhx33 in B-cell proliferation. a Experimental outline of CRISPR/Cas9 screening in the iGCB system. b Naive B cells from Cas9-GFP (CD45.2+) and CD45.1+ wild-type mice were mixed in a 1:1 ratio and transduced with retroviruses encoding sgRNA (BFP+) at Day 2 and treated with puromycin at Day 3. iGCB cells at Day 5 were cultured on 40LB cells with IL-21 to induce iPC differentiation for another 4 days. Flow cytometry analysis of Cas9-GFP+ and CD45.1+ B cells in NTC (nontargeted control), Prdm1 (Blimp1) and Dhx33 sgRNA-expressing (BFP+) iPC cells at Day 4 of iPC differentiation. c, d Percentage (left) and number (right) of Cas9-GFP+ (c) and Cas9-GFP+CD138+ (d) iPCs at Day 4 of iPC differentiation. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 (two-tailed unpaired Student’s t-test). The data are representative of two independent experiments
Impaired early B-cell development in the absence of Dhx33
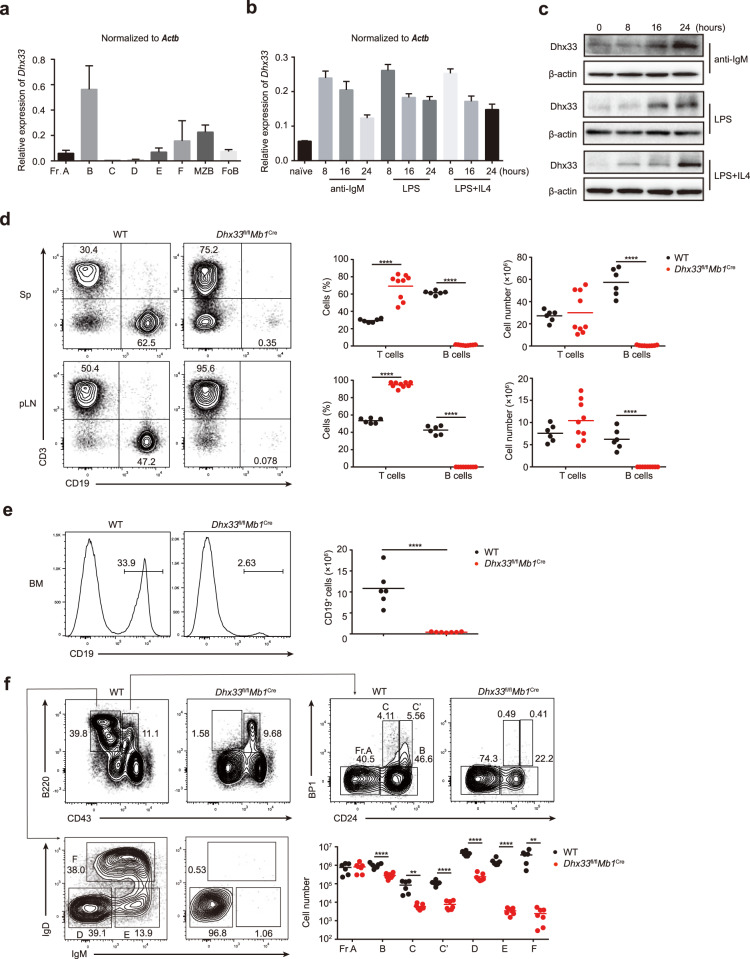
To study the function of Dhx33 in B cells, we first examined the expression of Dhx33 during B-cell development in the bone marrow and in various B-cell subsets in the periphery. As shown in Fig. 2a, Dhx33 mRNA expression was high in Fraction B (Fr. B), diminished afterward, and returned to marginal zone B (MZB) and follicular B (FoB) cells in the spleen as well as to mature B cells recirculated back to the bone marrow (Fr. F). Notably, robust expression of Dhx33 was induced in B cells upon stimulation with LPS, LPS plus IL-4, and anti-IgM (Fig. 2b, c), suggesting a potential role of Dhx33 during B-cell activation. To investigate the biological function of Dhx33, we generated a loxP-site flanked allele of Dhx33 (Supplementary Fig. 1a) and crossed the mutant mice with Mb1Cre mice in which the CD79a gene was replaced with a codon-optimized Cre recombinase gene [8]. The CD79a locus turns on Cre expression and drives efficient deletion of loxP-site flanked alleles at an early developmental stage of the B lineage (Supplementary Fig. 1b, c). Dhx33fl/flMb1Cre mice appeared healthy but had a complete absence of B cells in the spleen and peripheral lymph nodes (Fig. 2d). Loss of peripheral B cells may be caused by defective B-cell development. Thus, we assessed B-cell development in the bone marrow and found that the percentage and number of CD19+ B cells in the bone marrow were also dramatically reduced in Dhx33fl/flMb1Cre mice (Fig. 2e). Consistent with high Dhx33 expression in Fr. B (Fig. 2a), Dhx33fl/flMb1Cre mice exhibited a drastic developmental block at the Fr. A-to-B transition and a complete lack of B lineage cells after Fr. B (Fig. 2f). The lack of peripheral B cells in Dhx33fl/flMb1Cre mice prevented the study of Dhx33 function during immune responses. We therefore bred Dhx33fl/fl mice with CD19Cre and Cγ1Cre mice, in which the expression of Cre recombinase is under the control of the CD19 and Cγ1 loci, respectively [9, 10]. Cre expression is turned on at Frs. B and C in CD19Cre mice and after activation of mature B cells in Cγ1Cre mice, allowing the bypass of early B-cell developmental block in Dhx33fl/flMb1Cre mice. B-cell development was normal in Dhx33fl/flCD19Cre and Dhx33fl/flCγ1Cre mice. The proportions and numbers of various B-cell subsets in the spleen and peripheral lymph nodes of Dhx33fl/flCD19Cre and Dhx33fl/flCγ1Cre mice were also similar to those of wild-type mice (Supplementary Figs. 2 and 3). The number of MZB cells was decreased in the spleen of Dhx33fl/flCD19Cre mice, resulting in slight reductions in the percentage and number of total B cells in this organ (Supplementary Fig. 2d, e). Collectively, these results demonstrate that Dhx33 plays a critical role at the early stage of B-cell development and is dispensable afterward.
Fig. 2.
Dhx33 is indispensable for early B-cell development. a RT‒PCR analysis of Dhx33 mRNA expression at various stages of B-cell development in the spleen and bone marrow. MZB, marginal zone B cells FoB, follicular B cell. b, c Naïve B cells were treated with 25 μg/mL LPS, 25 μg/mL LPS + 5 ng/mL IL-4 or 10 μg/mL anti-IgM for the indicated amounts of time. Dhx33 mRNA and protein levels were determined by RT‒PCR (b) and immunoblot analysis (c). d Flow cytometry analysis of CD3+ T and CD19+ B cells in the spleens (Sp, upper) and peripheral lymph nodes (pLN, lower) of 7-week-old WT and Dhx33fl/flMb1Cre mice. Representative FACS plots (left). Summary of the percentage and number of CD3+ T and CD19+ B cells (right) (n ≥ 6 per group). e The percentage and number of CD19+ B lineage cells in the bone marrow were determined by flow cytometry. f Flow cytometry analysis of Frs. A-F of B lineage cells in the bone marrow of Dhx33fl/flMb1Cre and WT mice. Fr. A (B220hiCD43hiCD24loBP1lo), Fr. B (B220hiCD43hiCD24hiBP1lo), Fr. C (B220hiCD43hiCD24intBP1hi), Fr. C′ (B220hiCD43hiCD24hiBP1hi), Fr. D (B220hiCD43loIgDloIgMlo), Fr. E (B220hiCD43loIgDloIgMhi), Fr. F (B220hiCD43loIgDhiIgMhi). Each symbol represents an individual mouse. The small horizontal lines indicate the means ( ± s.e.m.). *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 (two-tailed unpaired Student’s t-test). The data are representative of three independent experiments
Dhx33 is required for germinal center formation and antibody responses
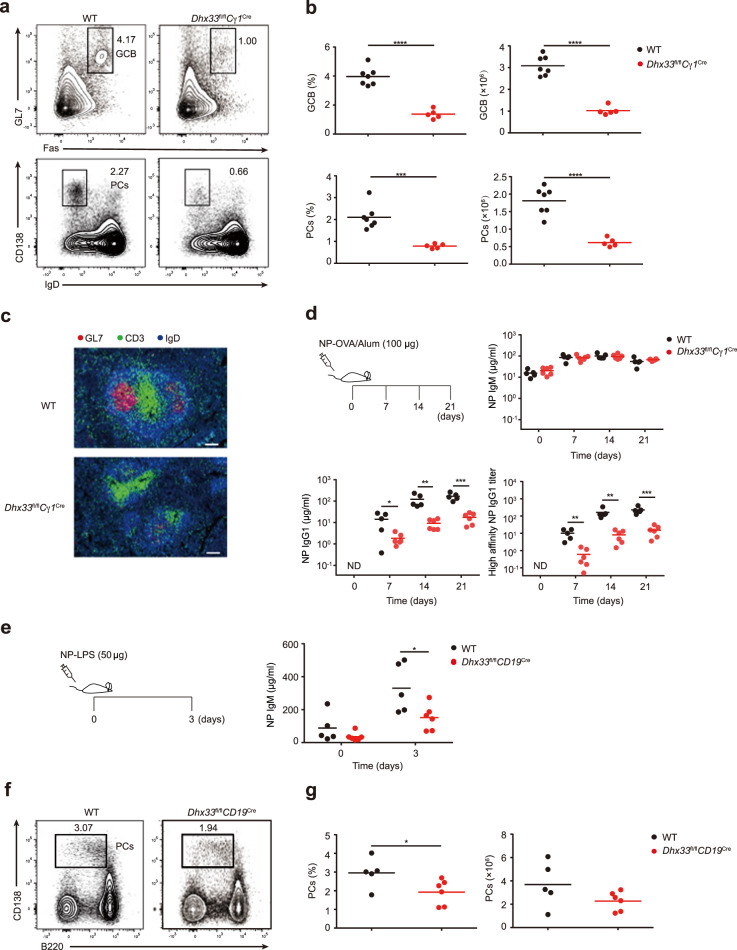
To determine the role of Dhx33 in the humoral immune response, we immunized Dhx33fl/flCγ1Cre and wild-type mice with ovalbumin (OVA) precipitated in alum together with lipopolysaccharide (LPS), a T-cell-dependent antigen. Flow cytometry analysis of immunized mice showed that the percentage and number of germinal center B and plasma cells were drastically decreased in OVA/alum/LPS-immunized Dhx33fl/flCγ1Cre mice (Fig. 3a, b). Immunohistochemical analysis confirmed that the size and number of germinal centers were largely reduced in the spleens of immunized Dhx33fl/flCγ1Cre mice (Fig. 3c and Supplementary Fig. 4). We then immunized Dhx33fl/flCγ1Cre and wild-type mice with 4-hydroxy-3-nitrophenyl hapten conjugated to ovalbumin (NP-OVA) precipitated in alum (NP-OVA/alum) and analyzed the NP-specific antibody response in the serum by ELISA. While NP-specific IgM concentrations were comparable in Dhx33fl/flCγ1Cre and wild-type mice, the serum levels of total and high-affinity NP-specific IgG1 were markedly decreased in Dhx33fl/flCγ1Cre mice (Fig. 3d). Similarly, Dhx33fl/flCD19Cre mice showed reductions in germinal center B and plasma cell formation and NP-specific IgG1 antibody production (Supplementary Fig. 5). We further immunized Dhx33fl/flCD19Cre and wild-type mice with the T-cell-independent antigen NP-LPS and measured plasma cell formation and the serum NP-IgM concentration. Similar to NP-OVA immunization, Dhx33fl/flCD19Cre mice exhibited a decreased percentage and number of plasma cells, as well as reduced serum NP-IgM levels (Fig. 3e–g). Taken together, our results demonstrate that Dhx33 is required for germinal center formation, plasma cell differentiation, and antibody responses.
Fig. 3.
Dhx33 plays a critical role in T-dependent and T-independent immune responses. a Flow cytometry analysis of germinal center B (GL7+Fas+, upper) and plasma (CD138+IgD-, lower) cells in the spleens of WT and Dhx33fl/flCγ1Cre mice at 6.5 days after immunization with OVA/alum/LPS. GCB germinal center B, PCs plasma cells. b Summary of the percentage and number of GC B (upper) and PCs (lower) from (a) (n = 5–7 per group). c Immunofluorescence images of the spleen from (a) stained for GL7 (red), CD3 (green) and IgD (blue). Scale bar, 100 μm. (d) The concentrations of NP-specific IgM, IgG1 and high affinity NP-IgG1 in the serum of WT and Dhx33fl/flCγ1Cre mice were determined by ELISA at Days 0, 7, 14 and 21 after immunization with 100 μg of NP-OVA in alum. e Titers of NP-specific IgM in the serum of WT and Dhx33fl/flCD19Cre mice were determined by ELISA at Days 0 and 3 after immunization with 50 μg NP-LPS (n = 5~6). f, g Representative FACS plots (f) and numbers (g) of CD138+ plasma cells in the spleen from mice in (e). Each symbol represents an individual mouse. The small horizontal lines indicate the means ( ± s.e.m.). *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 (two-tailed unpaired Student’s t-test). n = 5–6 mice. ND Not detected. The data are representative of two independent experiments
Dhx33 couples B-cell activation with growth and proliferation
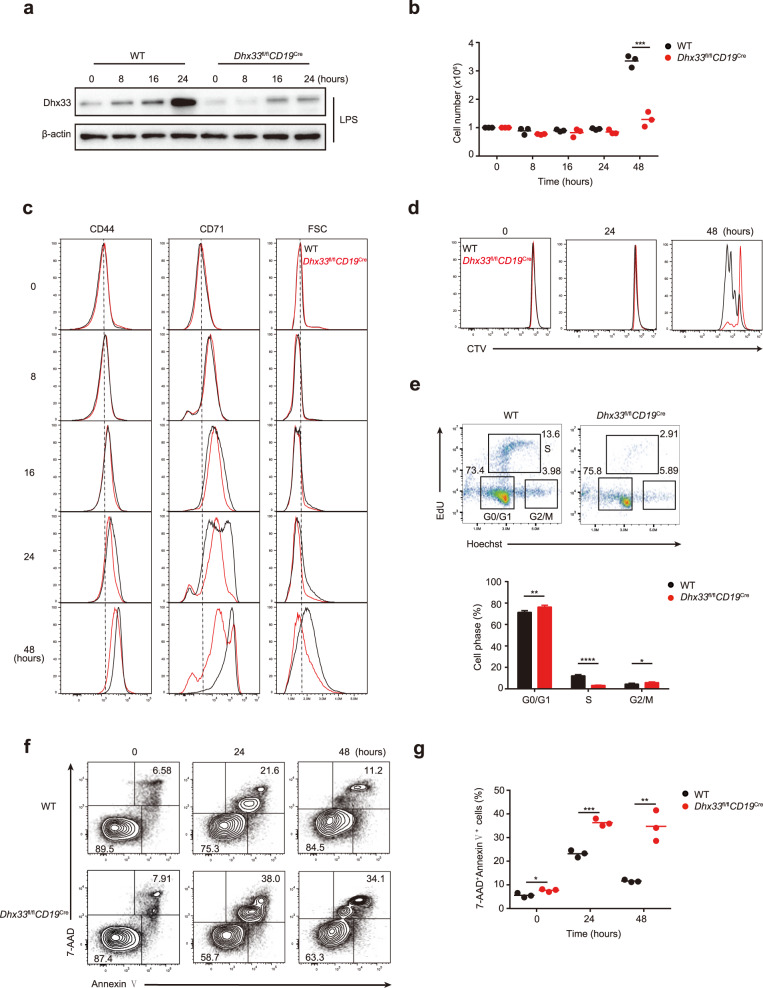
B cells can sense environmental cues, including antigen stimulation through B-cell receptors (BCRs), costimulatory signals and cytokines delivered by T cells, and pathogen-associated molecular patterns via innate immune system receptors such as Toll-like receptors (TLRs), to promote B-cell activation, growth, and proliferation. We wondered whether the reduced germinal center B-cell formation, plasma cell differentiation, and antibody production resulting from B-cell-specific deletion of Dhx33 were caused by impaired B-cell activation. To this end, B cells were purified from Dhx33fl/flCD19Cre and WT mice, treated with LPS in vitro, and examined for their activation, growth, and proliferation. Activation-induced Dhx33 expression was largely abrogated in Dhx33fl/flCD19Cre B cells (Fig. 4a). While wild-type B cells underwent a drastic expansion in numbers upon LPS stimulation, Dhx33fl/flCD19Cre B cells showed little increase in cell numbers (Fig. 4b). Flow cytometry analysis showed that upregulation of activation markers (CD44 and CD71) was normal in the first 16 h, but Dhx33fl/flCD19Cre B cells failed to grow in size, did not proliferate in 48 h, exhibited significant cell cycle arrest at the G1-to-S transition, and underwent apoptosis en masse (Fig. 4c–g). The same phenomenon was observed in Dhx33-deficient B cells stimulated with LPS plus IL-4 and anti-IgM (data not shown). We also examined Dhx33-deficient B cells in the in vitro differentiation system of iGCB culture and observed similar failures in growth and proliferation for Dhx33fl/flCD19Cre B cells (Supplementary Fig. 6a, b, c). We then investigated whether Dhx33 is required in later stages of B-cell activation and differentiation. For this purpose, we took advantage of Dhx33fl/flCγ1Cre mice in which Cre expression was turned on after B-cell activation and the Dhx33 gene was deleted at approximately Days 2-3 in iGCB culture (Supplementary Fig. 6d). Dhx33fl/flCγ1Cre B cells showed a 2-fold reduction in cell number at iGCB Day 4 and a complete absence of proliferation in the 2nd stage of culture (Supplementary Fig. 6e). Taken together, these results show that Dhx33 is not required for the initial activation of B cells but plays an indispensable role in B-cell growth and proliferation. In the absence of Dhx33, activated B cells are severely compromised in growth and proliferation and ultimately die. Therefore, Dhx33 couples B-cell activation with growth and proliferation.
Fig. 4.
Defective growth and proliferation of LPS-stimulated Dhx33-deficient B cells. Splenic B cells from control and Dhx33fl/flCD19Cre mice were stimulated with 25 μg/mL LPS for the indicated amounts of time. a Immunoblot analysis of Dhx33 and β-actin expression. b Summary of the total cell number. c Flow cytometry analysis of CD44 and CD71 expression and cell size (FSC). d Flow cytometry analysis of the proliferation of splenic B cells labeled with CTV. e Flow cytometry analysis of the cell cycle of B cells stimulated with 25 μg/mL LPS for 24 h (upper). Summary of the percentage of B cells at G0/G1, S and G2/M phases (lower). f Flow cytometry analysis of cell death by staining with 7-amino-actinomycin D (7-AAD) and Annexin V. g Summary of the percentage of apoptotic cells (Annexin V+7-AAD+) from (f). The small horizontal lines indicate the means ( ± s.e.m.). *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 (two-tailed unpaired Student’s t-test). The data are representative of three independent experiments
Dhx33 deficiency leads to nucleolar stress and global downregulation of translation
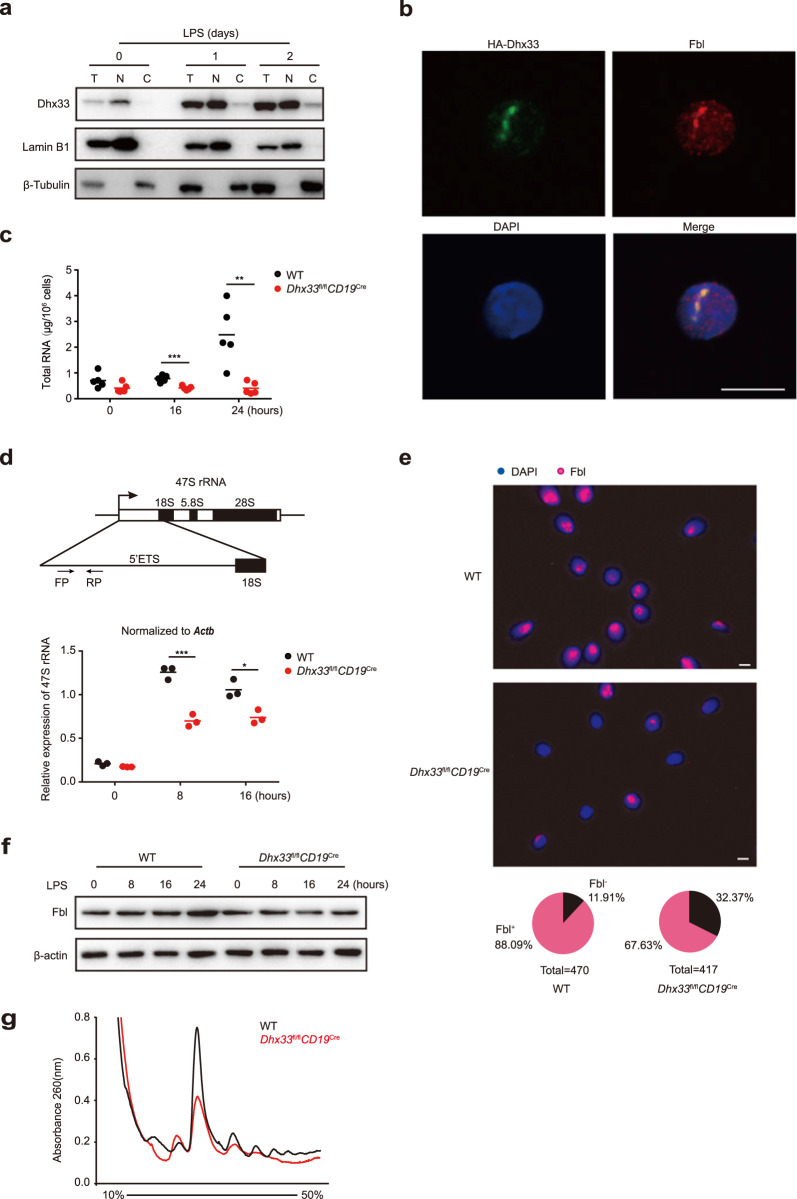
Dhx33 has been shown to be a nucleolar protein in cancer cells and fibroblasts [11]. Consistent with previous reports, we found that Dhx33 was mainly localized in the nuclei of naïve and activated B cells (Fig. 5a). To better understand the subnuclear localization of Dhx33, B cells from Dhx33fl/flCγ1Cre mice were transduced with retroviruses encoding HA-tagged Dhx33 during iGCB culture, subjected to iPC differentiation, and analyzed by immunofluorescence staining (Fig. 1a). Dhx33 showed punctate distribution and colocalization with the nucleolar protein fibrillarin (Fbl), suggesting nucleolar localization of Dhx33 in activated B cells (Fig. 5b). The nucleolus is the nuclear structure concentrated with DNA repeats encoding the 47S rRNA. The main functions of the nucleolus are transcription of rRNA by RNA polymerase I as well as subsequent processing and assembly into small and large ribosome subunits together with ribosome proteins translated in the cytoplasm and imported into the nucleus. Ribosome biogenesis is tightly coupled to protein synthesis, cell growth and proliferation, while transcription of rRNA by RNA polymerase I is the nexus of control for modulating ribosome biogenesis. As shown in Fig. 5c, d, LPS-stimulated Dhx33fl/flCD19Cre exhibited significant reductions in the levels of total RNA and 47 S rRNA. These reductions were accompanied by a drastic reduction in the number of intact nucleoli marked by Fbl staining (Fig. 5e), suggesting that Dhx33 is important for the maintenance of nucleolar structure and organization in activated B cells. Consistent with reduced rRNA production and compromised nucleolar structure, Dhx33 deficiency led to impaired activation-induced Fbl upregulation and global mRNA translation, as demonstrated by reduced amounts of mRNA in the monosome and polysome fractions in polysome profiling analysis of LPS-activated Dhx33fl/flCD19Cre B cells (Fig. 5f, g and Supplementary Fig. 7a). Therefore, Dhx33 deficiency leads to reduced rRNA production, nucleolar stress, and global downregulation of translation.
Fig. 5.
Dhx33 localizes in the nucleolus and is essential for the generation of 47S ribosomal RNA. a Immunoblot analysis of Dhx33, lamin B1 and β-tubulin in total cell lysates (T), nuclear (N) and cytosolic (C) extracts of B cells stimulated with 25 μg/mL LPS for 1 or 2 days. b Representative immunofluorescence images of iPCs transduced with retroviruses encoding HA-tagged Dhx33 and stained for HA (green), fibrillarin (Fbl, red) and DAPI (blue, nuclear staining). Scale bar, 8 μm. c, d Total RNA (c) and qRT‒PCR analysis of 47S rRNA (d) in Dhx33fl/flCD19Cre and WT B cells stimulated with LPS for the indicated amounts of time. Schematic representation of the 47S rRNA locus (upper panel in d). e Immunofluorescence analysis of nucleolar structure by staining with Fbl (red) and DAPI (blue) in Dhx33fl/flCD19Cre and WT B cells stimulated with LPS for 24 h. Pie chart representing the percentage of Fbl+ (red) and Fbl- (black) cells. f Immunoblot analysis of Fbl and β-actin in B cells stimulated by LPS for the indicated amounts of time. g Polysome profiling analysis of Dhx33fl/flCD19Cre and WT B cells stimulated with LPS for 24 h. The small horizontal lines indicate the mean ( ± s.e.m.). *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 (two-tailed unpaired Student’s t-test). The data are representative of three independent experiments
Dhx33 deficiency leads to p53 stabilization in activated B cells
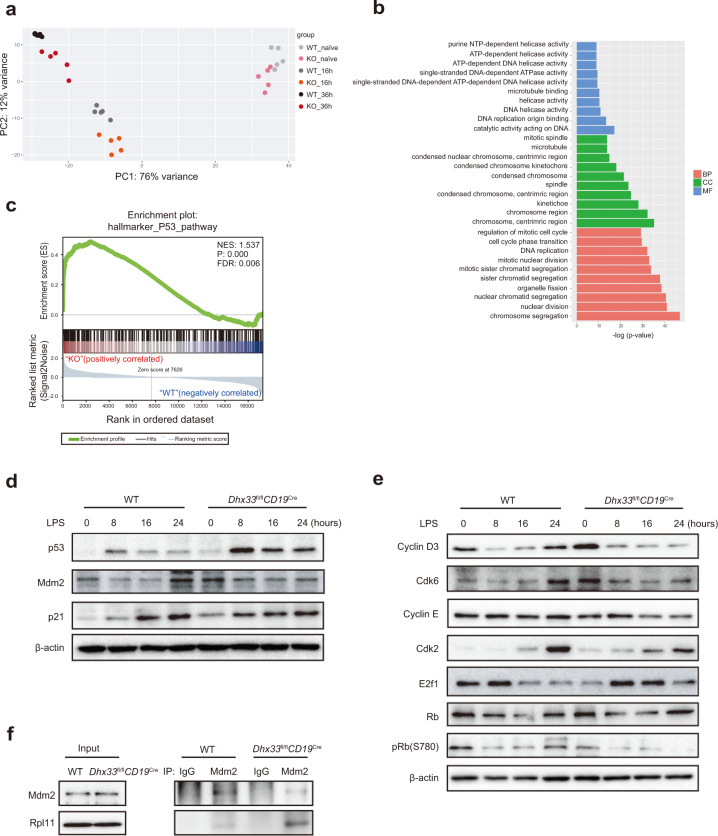
To gain molecular insights into the role of Dhx33 in activated B cells, LPS-stimulated WT and Dhx33fl/flCD19Cre B cells were analyzed by RNA-seq. Consistent with normal B-cell development in Dhx33fl/flCD19Cre mice (Supplementary Fig. 2), principal component analysis (PCA) of the transcriptomes of WT and Dhx33fl/flCD19Cre naïve B cells did not show any significant differences. Upon LPS stimulation, the transcriptomes of WT and Dhx33fl/flCD19Cre B cells seemed to move toward the same direction, but with a significant delay for Dhx33fl/flCD19Cre B cells (Fig. 6a). We then performed Gene Ontology (GO) analysis of differentially expressed genes and identified gene sets enriched in DNA replication and ATP-dependent DNA helicase activity (Fig. 6b). Gene set enrichment analysis (GSEA) revealed that the most upregulated genes were enriched with genes regulated by p53 (Fig. 6c). Indeed, Dhx33fl/flCD19Cre B cells exhibited significant upregulation of p53 protein levels upon LPS stimulation (Fig. 6d).
Fig. 6.
p53 stabilization in activated Dhx33-deficient B cells. a Principal component analysis (PCA) of transcriptomes of Dhx33fl/flCD19Cre and WT B cells treated with 25 μg/mL LPS for 16 and 36 h. b Gene Ontology (GO) analysis. BP, biological process. CC, cellular component. MF, molecular function. c Gene enrichment pathways identified by GSEA. NES, normalized enrichment score. P: nominal p value. FDR: false discovery rate q-value. d Immunoblot analysis of p53, Mdm2, p21 and β-actin in Dhx33fl/flCD19Cre and WT B cells stimulated with LPS for the indicated amounts of time. e Immunoblot analysis of the indicated cell cycle regulators in Dhx33fl/flCD19Cre and WT B cells stimulated with LPS for the indicated amounts of time. f B cells stimulated with LPS for 4 h were lysed, immunoprecipitated with Mdm2 antibodies and immunoblotted with Mdm2 and Rpl11 antibodies. The small horizontal lines indicate the means ( ± s.e.m.). *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 (two-tailed unpaired Student’s t test). The data are representative of three independent experiments
The cellular concentration of p53 is mainly regulated by its interaction partner MDM2, an E3 ubiquitin ligase. Cell cycle arrest and apoptosis are the two most prominent outcomes of p53 activation, and cell cycle arrest by p53 is mainly mediated by its transcriptional activation of p21.
Consistent with elevated p53 protein levels, Dhx33fl/flCD19Cre B cells exhibited increased mRNA and protein levels of p21 (Fig. 6d and Supplementary Fig. 7b). p21 binds to cyclin E/Cdk2 and cyclin D/Cdk6 complexes to cause G1 arrest in the cell cycle. The inhibition of Cdk2 and Cdk6 by p21 blocks pRb phosphorylation, promotes Rb binding to E2F1, and enhances transcription silencing of E2F1 targets critical for DNA replication and cell cycle progression. Accordingly, the protein levels of cyclin D3, cyclin E, Cdk2, and Cdk6 were reduced in Dhx33fl/flCD19Cre B cells after LPS stimulation, and these reductions were accompanied by a decrease in Rb phosphorylation (Fig. 6e). Previous studies have shown that perturbation of any steps of rDNA transcription, processing and assembly into ribosome subunits leads to nucleolar stress, triggering specific binding of several ribosomal proteins to Mdm2, which inhibits Mdm2’s E3 ubiquitin ligase function toward p53, leading to p53 stabilization and activation [12]. Indeed, LPS-stimulated Dhx33fl/flCD19Cre exhibited enhanced Mdm2 association with Rpl11, one of the ribosomal proteins able to bind to Mdm2 and inhibit its ubiquitination of p53 (Fig. 6f). This, together with our previous results showing impaired nucleolar integrity of LPS-stimulated Dhx33-deficient B cells (Fig. 5e), demonstrates that Dhx33 deficiency in B cells enhances ribosomal protein association with Mdm2 and, subsequently, p53 stabilization and accumulation.
UBF recruits Dhx33 to promote rDNA transcription
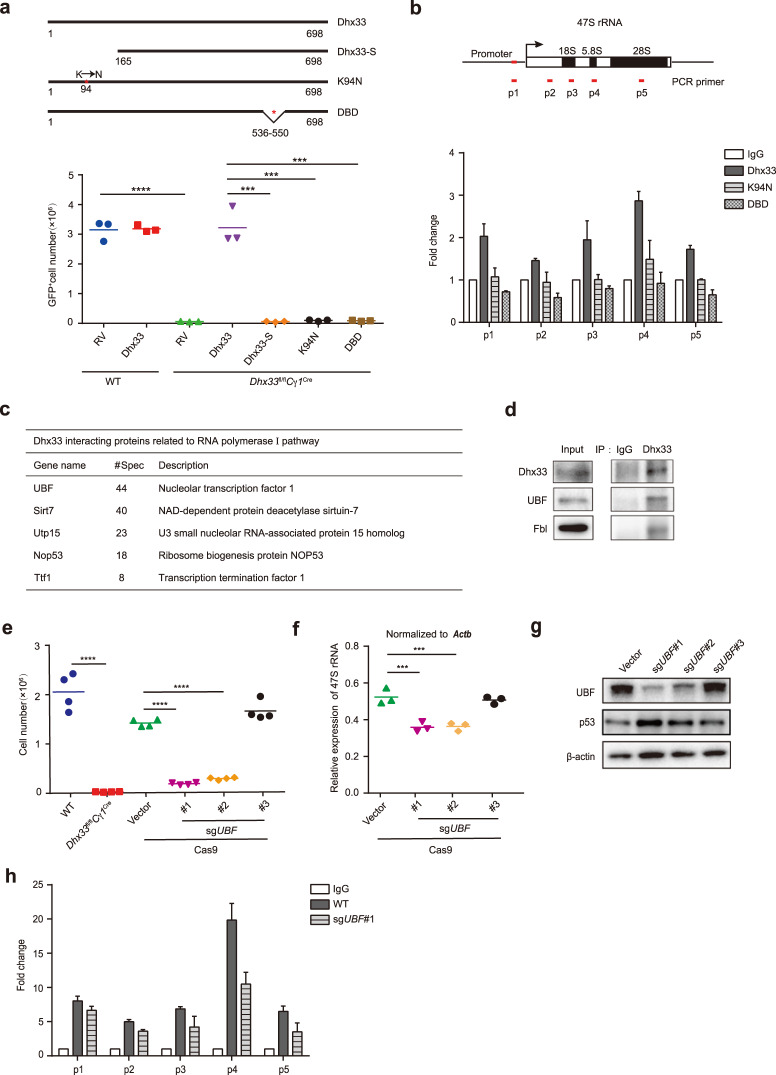
Dhx33 contains an ATP-binding site and a DNA-binding site [11]. A previous study also found the existence of a short isoform of Dhx33, which predominantly localizes in the cytoplasm and regulates mRNA translation [13]. To determine which domains of Dhx33 are important for its function in B cells, we generated retroviruses encoding wild-type and the short isoform of Dhx33 (Dhx33-S) as well as various mutants, including mutants with mutation of the ATP binding site (K94N) and deletion of the DNA-binding domain (DBD). Dhx33fl/flCγ1Cre B cells were transduced with those retroviruses during iGCB culture, and iPC differentiation was subsequently induced (Figs. 1a, 7a). Consistent with previous results (Supplementary Fig. 6e), Dhx33fl/flCγ1Cre B cells showed a drastic reduction in number during iPC differentiation. While retroviral expression of wild-type Dhx33 restored the number of Dhx33fl/flCγ1Cre B cells, none of the Dhx33 mutants (including Dhx33-S) showed any effect (Fig. 7a and Supplementary Fig. 8a), suggesting the functional importance of the ATPase activity and DNA-binding ability of Dhx33 in B cells. We speculated that DNA binding is crucial for Dhx33 regulation of rDNA transcription in B cells and therefore examined the association of Dhx33 with the promoter and other regions of rDNA by CUT&TAG assay [14]. As shown in Fig. 7b, Dhx33 bound multiple regulatory regions of the 47 S rDNA locus, and the K94N and DBD mutations abolished Dhx33 binding to those regions (Fig. 7b and Supplementary Fig. 8b, c), suggesting that ATPase activity and DNA binding are crucial for the function of Dhx33 in regulating rDNA transcription.
Fig. 7.
UBF recruits Dhx33 to promote rDNA transcription. a Restoration of proliferation of Dhx33-deficient B cells by full-length Dhx33 and its mutants and isoform. Upper panel, schematic representation of Dhx33 and its mutants. Lower panel, WT and Dhx33fl/flCγ1Cre B cells were transduced with retroviruses encoding HA-tagged Dhx33 or its mutants and isoform in iGCB culture and induced for iPC differentiation for 4 days. The cell numbers were calculated at terminal analysis. K94N, an ATP-binding site mutant. DBD, DNA-binding site deletion mutant. Dhx33-S, naturally occurring isoform of Dhx33 missing 15 amino acids at the N-terminus. b CUT&TAG assay of binding of Dhx33 and its mutants on rDNA. Upper panel, schematic outline of the 47S rRNA. Lower panel, CUT&TAG assay was performed with Dhx33fl/flCγ1Cre B cells transduced with retroviruses encoding HA-tagged Dhx33 or its mutants and isoform in iGCB culture and induced for iPC differentiation for 1 day. GAPDH was used as an internal control. c Wild-type B cells stimulated with LPS for 8 h were harvested for immunoprecipitation with anti-Dhx33 and mass spectrometry analysis. Dhx33-interacting proteins involved in the RNA polymerase I pathway are listed. d Total cell lysates of wild-type B cells stimulated with LPS for 8 h were immunoprecipitated with anti-Dhx33 or IgG isotype control antibodies and analyzed by immunoblotting with the indicated antibodies. e, f Cas9-GFP+ B cells were transduced with retroviruses encoding the indicated sgRNA in iGCB culture and induced for iPC differentiation for 4 days. e The cell numbers were calculated at terminal analysis. f 47S rRNA expression was measured by qRT‒PCR at Day 1 of iPC differentiation. g Immunoblot analysis of UBF, p53 and β-actin in Cas9-GFP+ B cells transduced with retroviruses encoding the indicated sgRNAs in iGCB culture at Day 5. h A CUT&TAG assay was performed in UBF-deleted (GFP+BFP+) and WT (GFP+BFP-) iPCs at Day 1 of iPC differentiation. Dhx33-bound regions were measured by PCR. GAPDH was used as an internal control. The small horizontal lines indicate the means ( ± s.e.m.). *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 (two-tailed unpaired Student’s t-test). The data are representative of two or three independent experiments
To gain insights into the molecular mechanisms underlying Dhx33 regulation of rDNA transcription, we performed immunoprecipitation–mass spectrometry analysis (IP–MS) to identify Dhx33-interacting proteins in LPS-stimulated B cells (Supplementary Table 2). UBF, Sirt7, Utp15, Nop53 and TTF1 are the most abundant Dhx33 binding proteins previously implicated in the RNA polymerase I pathway (Fig. 7c). Immunoprecipitation assays showed that Dhx33 was associated with UBF and Fbl in activated B cells (Fig. 7d). To test whether UBF, Sirt7 and TTF1 are involved in Dhx33 regulation of B-cell proliferation and rDNA transcription, we performed CRISPR-mediated functional screening of these genes, as they are directly involved in rDNA transcription in the iGCB culture system [15–17]. We found that only sgRNAs targeting UBF led to reductions in both cell number and 47S rRNA expression level, as well as p53 accumulation (Fig. 7e–g), suggesting that UBF and Dhx33 play similar roles in regulating B-cell proliferation and rDNA transcription. Similar to Dhx33 expression (Fig. 2c), UBF expression in B cells was induced upon B-cell activation by LPS, LPS plus IL-4, and anti-IgM (Supplementary Fig. 9a). UBF is a member of the HMG-box DNA-binding protein family and binds the upstream control element (UCE) of the rDNA promoter as well as many sites distributed across the entire transcribed rDNA repeats. UBF, RNA polymerase I, and selectivity factor SL1 form the transcription preinitiation complex (PIC), the core protein machinery that drives rDNA transcription [5]. UBF is essential for the recruitment of RNA polymerase I to the rDNA promoter region [5, 17]. It may also play a more general structural role over the rDNA repeat to facilitate transcription elongation [18]. Notably, Dhx33 association with rDNA was significantly decreased upon sgRNA-mediated deletion of UBF in iGCB culture (Fig. 7h and Supplementary Fig. 9b, c). Taken together, these results show that UBF recruits Dhx33 to rDNA to promote rDNA transcription.
Discussion
This study discovered a critical role of Dhx33 in B-cell development and immune responses. Genetic ablation of Dhx33 led to a complete developmental block at the early pro-B stage (Fr. B) in the bone marrow. In the periphery, Dhx33-deficient B cells appeared normal in the steady state and in the early stage of activation but were severely compromised in growth, proliferation, germinal center B-cell formation, plasma cell differentiation, and antibody production. Molecular and cellular analyses showed that Dhx33 was recruited to rDNA by association with UBF and promoted rDNA transcription. When Dhx33 was absent in activated B cells, the transcription of 47 S rRNA was drastically decreased, leading to nucleolar stress, p53 stabilization, cell cycle arrest, and cell death.
It was quite interesting that the absence of Dhx33 did not affect the steady state and early stages of activation of B cells but severely impaired B-cell growth and proliferation. A recent study on translational control in T-cell activation may shed some light on this intriguing observation [3]. Similar to naïve T cells, naïve B cells may harbor a large pool of idling ribosomes, whose translation rate can be rapidly ramped up to meet the early demand for protein synthesis upon B-cell activation. As shown in this study, B-cell activation upregulates the expression of UBF and Dhx33, which in turn promote rDNA transcription. This, together with increased production of ribosomal proteins and other factors, leads to the generation of millions of new ribosomes, which are essential for protein synthesis, cell growth and proliferation beyond the first 16–24 h after B-cell activation. Therefore, activation-induced upregulation of Dhx33 and UBF is dispensable for naïve B cells and their early stages of activation but plays a pivotal role in coupling B-cell activation with growth and proliferation.
A previous study has found that Dhx33 functions as both an RNA and DNA helicase [19]. Mass spectrometry analysis of Dhx33-interacting partners in B cells has shown that Dhx33 is associated with multiple proteins of the RNA polymerase I pathway, ranging from nucleosome positioning on rDNA, recruitment of RNA polymerase I to the rDNA promoter, deacetylation of RNA polymerase I subunits, rRNA processing, nucleolar retention of ribosomal proteins, and translational regulation of mRNA. It is conceivable that both the DNA and RNA helicase activity of Dhx33 can play key roles in those processes, whose molecular details warrant future investigations. Cancer cells often harbor mutations that inactivate tumor suppressors and hyperactivate oncogenes to promote rDNA transcription, ribosome biogenesis, and protein synthesis, thereby driving cell growth and proliferation [20]. Several inhibitors of rDNA transcription have been identified via high-throughput screening, including CX-5461 [21]. CX-5461 binds to SL-1 and prevents RNA polymerase I association with the promoter of rDNA genes, thereby inhibiting RNA polymerase I -mediated transcription. A phase I clinical trial in patients with relapsed or refractory hematological cancers was recently completed, and an objective clinical response was observed in approximately 30% of patients. Consistent with results from preclinical models, there was a positive correlation between wild-type p53 status and tumor response as well as patient outcome, suggesting that the therapeutic effect of CX-5461 is largely mediated by compromise of ribosome biogenesis and subsequent p53 stabilization. Upregulation of Dhx33 expression has been found in a broad spectrum of human cancers, including lymphomas [22–25]. It is worth noting that Dhx33 and UBF are both transcriptional targets of cMyc [22, 26], whose activation is one of the most commonly found oncogenic events in human cancers. Studies employing genetic deletion and shRNA knockdown have shown that Dhx33 plays critical roles in the proliferation and survival of cancer cells [22, 23, 25, 27–29]. This study demonstrated that Dhx33 is dispensable for naïve and resting B cells but essential for their growth and proliferation upon activation. It is conceivable that pharmacological inhibition of Dhx33 may impair B-cell activation while sparing naïve B cells. This may be explored to treat antibody-mediated autoimmune diseases. Similarly, Dhx33 inhibitors may be able to suppress the growth of tumor cells while sparing nonproliferating cells of normal tissues and, therefore, be useful for treating cancers.
Our RNA-seq analysis showed that p53-regulated genes are prominently enriched among genes differentially expressed between Dhx33-deficient and -sufficient activated B cells, suggesting that p53 stabilization is an important event downstream of Dhx33 ablation. Ribosome biogenesis requires coordinated actions of RNA polymerases I, II, and III. While RNA polymerase I transcribes 47S rDNA in the nucleolus, mRNAs encoding ribosomal proteins and 5S rRNA are transcribed in the nucleus by RNA polymerases II and III, respectively. Recent studies have shown that Rpl11 and Rpl5 form a preribosomal complex with 5S rRNA before integrating into the 60S ribosome. When the intricate balance between these three RNA polymerases is disrupted, as exemplified by Dhx33 ablation in this study, the nascent preribosomal Rpl5/Rpl11/5S rRNA complex is redirected from 60 S ribosome biogenesis in the nucleolus to the nucleoplasm, where it sequesters Mdm2 from p53, resulting in p53 stabilization, cell cycle arrest and cell death [30–32]. In a substantial portion of human cancers, Trp53, which encodes the p53 protein, is in the wild type form [33]. Studies in mouse models have demonstrated that p53 restoration causes regression of established tumors [34–36]. Therefore, Dhx33 may be the Achilles heel for both cancer and autoimmune diseases. Pharmacological inhibition of Dhx33 may lead to p53 stabilization, cell cycle arrest, and death of activated B and cancer cells. This can be explored for the design of novel therapeutics to treat debilitating diseases.
Methods and methods
Mice
Dhx33 mutant mice were purchased from KOMP/EMMA (EM:08228, KO first allele, reporter-tagged insertion with conditional potential). Cas9-GFP (Jax stock 024858) [37], Mb1Cre (Jax stock 020505) [8], CD19Cre (Jax stock 006785) [38] and Cγ1Cre (Jax stock 010611) [39] mice have been previously reported. B-cell development was analyzed at the age of 6–8 weeks. Mice were bled to measure immunoglobulin concentrations at the age of 8–12 weeks. All mice were bred and housed under specific pathogen-free conditions under a 12-hour light-dark cycle. All animal experiments were approved by the Animal Care and Use Committee of Xiamen University.
Antibodies and reagents
Anti-CD3 (145-2C11), anti-CD4 (GK1.5), anti-CD8 (53-6.7), anti-CD19 (eBio1D3), anti-CD38 (90), anti-IgM (II/41), anti-IgD (IA6-2) and anti-BP1 (6C3) were purchased from eBioscience. Anti-CD24 (M1/69), anti-CD44 (IM7), anti-CD95 (Jo2) and anti-CD138 (281-2) were purchased from BD. Anti-CD21 (7E9), anti-CD23 (B3B4), anti-CD43 (S11), anti-CD93 (AA4.1), anti-CD117 (2B8), anti-GL7 (GL7), anti-IgM (RMM-1) and anti-B220 (RA3-6B2) were purchased from BioLegend. Anti-Dhx33 (NB100-2581) was purchased from Novus Biologicals. Anti-Mdm2 (sc-965), anti-HA (sc-7392) and anti-UBF (sc-13125) were purchased from Santa Cruz. Anti-lamin B1 (66095-1-Ig), anti-Rpl11 (16277-1-AP) and anti-β-tubulin (66240-1-Ig) were purchased from Proteintech. Anti-FBL (A0850), anti-UBF (A9847), anti-E2f1 (A19579), anti-pRb (AP0444) and anti-p53 (A18803) were purchased from ABclonal. Anti-p21 (2947 S), anti-CyclinE (D7T3U), anti-CyclinD3 (DCS22), anti-Cdk2 (7882), anti-Cdk6 (DCS83), anti-Mdm2 (51541 S) and anti-β-actin (4970 L) were purchased from Cell Signaling Technology. A PE Annexin V apoptosis detection kit (559763) was purchased from BD Biosciences.
Flow cytometry
A single-cell suspension was freshly prepared from the spleen, draining lymph nodes and bone marrow, depleted of red blood cells by ACK buffer, stained for surface markers, and incubated at 4 °C for 15 min in the dark. For cell cycle analysis, cells were stained following the instructions of a BeyoClick™ EdU Cell Proliferation with Alexa Fluor 647 Kit (Beyotime; C0081S). Briefly, cells (1 × 106 cells/ml) were incubated with 10 μM EdU at 37 °C for 2 h in 6-well plates, fixed with BD Fix/Perm Solution (Cat No: 51-2090KZ) for 30 min at RT and then washed with PBS three times. EdU-stained cells were incubated in click reaction cocktail for 30 min in the dark at RT. Before flow cytometry analysis, cells were incubated with Hoechst 33342 for 10 min. All flow cytometry data were acquired on a BD Fortessa, Fortessa X20 (BD Biosciences), NovoCyte flow cytometer, or Quanteon (ACEA Biosciences, Agilent), and analyzed with FlowJo software (TreeStar).
Immunization and ELISA
Mice were immunized with 100 μg of NP-OVA/Alum (Biosearch Technologies; N-5051-100) intraperitoneally (i.p.). Serum samples were collected at 0, 7, 14, and 21 days post-immunization. The concentrations of NP-specific IgM (Southern Biotech; 1020-08) and IgG1 (Southern Biotech; 1070-08) were determined by ELISA. To measure TI immune responses, NP-LPS (Biosearch Technologies; N-5065-5) was injected intravenously (i.v.), and serum was collected at Days 0 and 3 post-immunization. To examine GCB and plasma cells, mice were immunized i.p. with 100 μg OVA precipitated with alum and 10 μg LPS. GCB and plasma cells were analyzed by flow cytometry. ELISA for NP-specific antibody production was performed as previously reported [40].
In vitro B-cell stimulation
Naïve B cells from the spleen were purified by negative selection with Beaver Beads (22307) and biotinylated antibodies (CD43, CD9, CD5, TER119 and CD93) (eBioscience and BioLegend) and stimulated at a density of 1 × 106 cells/ml with 25 μg/ml LPS (Sigma) in 6-well plates for Western blotting, CUT&TAG, and RNA-seq analysis. For assessment of cell activation, proliferation and death in vitro, B cells were activated with 10 μg/ml anti-IgM (SouthernBiotech), 25 μg/ml LPS, or 25 μg/ml LPS plus 5 ng/ml IL-4 (Novoprotein; CK74) in plates (Thermo Scientific Nunc) or cultured in the iGCB system [7]. For cell proliferation analysis, cells were labeled with Cell Trace Violet (Thermo Fisher; C34557) at room temperature for 10 min following the manufacturer’s instructions.
Immunohistochemistry
Spleens from immunized mice were fixed in BD Fix/Perm Solution (Cat No: 51-2090KZ) at 4 °C overnight. The fixative solution was discarded, and the fixed spleens were washed with PBS for 10 min on a shaker. Then, 30% sucrose (Sigma) was added to tissues at 4 °C. The tissues were incubated overnight and subsequently embedded in OCT (Tissue-Tek; Sakura). The dissected tissue sections were cut with a Leica CM1950 machine at a thickness of 10 μm. The sections were immunostained for IgD, CD3 and GL7 at RT in a dark box for 24 h. Images were acquired by using a Leica TCS SP8 confocal microscope (Leica, Germany) and analyzed by Imaris software.
RNA isolation, RT‒PCR and PCR analysis
Total RNA was isolated from B cells from Dhx33fl/flCD19Cre and WT mice by using a miRNeasy mini-Kit (Qiagen 217004) and quantified with a NanoDrop 2000 (Thermo Scientific). TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, AT311) was used for reverse transcription. The 47S rRNA primers have been described previously [41]. PCR primers: F 5’-TTTTTGGGGAGGTGGAGAGTC-3’ R 5’-CTGATACGGGCAGACACAGAAC-3’. Dhx33 primers: F 5’-TCCGAAACCTGGACAACG-3’ R 5’-AGAGGTACTGTGGGATCTGAG-3’. β-actin primers: F 5’-TGCTTCTAGGCGGACTGTTACTGAG-3’ R 5’-TTGCTCCAACCAACTGC TGTCG-3’. Quantitative real-time PCR was performed with ChamQ Universal SYBR qPCR Master Mix (Vazyme; Q711-02). All RT‒PCR data were acquired on a Bio-Rad CFX96 (Bio-Rad). Genomic DNA isolated from Fr. A and Fr. B cells from the bone marrow of control and Dhx33fl/flMb1Cre mice was used for PCR genotyping. PCR primers: F 5’-TTGCCAGTTAGTGAGGCAGG -3’ R 5’- TGAACTGATGGCGAGCTCAGACC-3’. The 1235-and 391-bp products corresponded to the control and deleted alleles, respectively.
RNA-seq and data analysis
After total RNA isolation, sequencing libraries were analyzed with an NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s recommendations. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) following the manufacturer’s recommendations. After cluster generation, the library preparations were used for sequencing on an Illumina NovaSeq platform (Novogene, Beijing). The reads were mapped to the mouse genome (mm10) using HISAT2. The resulting SAM files were sorted and converted to BAM files with Samtools. The read counts were extracted by StringTie. PCA and differential expression analyses were performed using DESeq2 (with a significance cutoff of p < 0.05 and log2(fold change)> 0.5 (or < (−0.5))). Differentially expressed genes were analyzed with GSEA v4.1.0 to find enriched biological processes and signaling pathways [42]. GO analysis was performed by clusterProfiler.
Cell fractionation to separate nucleoli and cytosol
Cells were washed with 1 ml PBS and centrifuged for 5 min at 500 × g and 4 °C. The cell pellets were resuspended in buffer A (10 mM HEPES, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA and 0.15% NP-40) supplemented with Halt Protease & Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific) and incubated on ice for 15 min. The homogenate was centrifuged for 30 s at 12000 rpm at 4 °C. The supernatant was collected as the cytosolic fraction. The pellet was the nuclear fraction. The nuclear pellets were resuspended in buffer A, incubated on ice for 2 min, and centrifuged for 30 s at 12000 rpm at 4 °C, after which the supernatant was removed. This washing step was repeated five times. The nuclear pellets were then lysed in lysis buffer (20 mM Tris-HCl pH 7.5, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 150 mM NaCl, 1 mM Na3VO4 and 1% SDS) supplemented with Halt Protease & Phosphatase Inhibitor Cocktail on ice for 10 min. The cytosolic and nuclear samples were boiled at 100 °C for 10 min, and the supernatants were stored at −20 °C or for analysis.
Retroviral transduction
HEK293T cells were transfected with 3 μg of MSCV-pU6-(BbsI)-CcdB-(BbsI)-Pgk-Puro-T2A-BFP vector and 1 μg of pcl-eco along with CaCl2 and 2× HBS. The targeting sequences of the single guide RNAs (sgRNAs) for NTC, Dhx33, Prdm1 and UBF were as follows (5′–3′): sgNTC: ACCGACGTTTAATCGAGGCTG; sgDhx33: ACTCGAAGCTGCTGCGAATAG; sgPrdm1: GTTATTGGCGTGGTAAGTAG; #1-sgUBF: ACTTGGAGCTGTCGTTGGAT; #2-sgUBF: ACAGGAGTTCGAGCGAAACC; and #3-sgUBF: TCTTACATGTCACAGACCGC. Dhx33 cDNA was inserted into the RV-3HA-IRES-EGFP vector. Viruses were produced by transfecting HEK293T cells. After 36–48 h, the virus-containing supernatant was collected and filtered. Naïve B cells cultured in a 6-well plate in the iGCB system at Day 2 were transduced with 1 ml/well retrovirus-containing supernatants with 6 μg/ml polybrene (EMD Millipore; TR-1003-G) and centrifuged at 1500 × g at 32 °C for 1 h.
Immunoblotting and immunoprecipitation
For immunoblotting, B cells and iPCs were lysed in lysis buffer. The cell lysates were resolved on SDS‒PAGE gels, and the proteins were transferred to PVDF membranes (Merck Millipore). The membranes were incubated overnight at 4 °C with primary antibodies diluted in NCM Universal Antibody Diluent (NCM Biotech; WB500D). After washing 3 times in TBS buffer with 0.5% Tween 20, each membrane was incubated with a horseradish peroxidase (HRP)-conjugated goat anti-rabbit or goat anti-mouse antibody (Abclonal). After washing 3 times in TBS buffer with 0.1% Tween 20, the protein bands were visualized with ECL Select Western Blotting Detection Reagent (GE Healthcare) or Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore) following the manufacturer’s instructions (GE Healthcare). Images were acquired with an Amersham Imager 600 (GE Healthcare). Immunoprecipitation was performed by using Protein A Dynabeads (Thermo; 10001D) washed twice with cell lysis buffer before use. The beads were incubated with an anti-Dhx33 antibody (NOVUS Biologicals, NB100–2581) and anti-rabbit IgG (Sigma; I5006) or an anti-Mdm2 antibody (Santa Cruz; sc-965) and anti-mouse IgG (Sigma; I5381) as a control and rotated overnight at 4 °C. Cell lysates were added and rotated with the bead-antibody mixture at 4 °C for another 8–10 h. After incubation, the beads were washed with cell lysis buffer five times before analysis. For mass spectrometry, samples were analyzed on an ion mobility mass spectrometer (Bruker timsTOF Pro).
Immunofluorescence microscopy
iPC cells were fixed in 4% paraformaldehyde at room temperature for 15 min and permeabilized with 0.1% Triton X-100 in PBS for 10 min in a 96-well plate. The cells were blocked with 5% FBS in PBS for 30 min and then stained with anti-HA and anti-FBL antibodies overnight at 4 °C. The cells were washed with PBS three times and then incubated with secondary antibodies for 2.5 h at RT. The secondary antibodies used for this assay were Alexa Fluor 647-conjugated donkey anti-mouse and Alexa Fluor 555-conjugated donkey anti-rabbit antibodies (Invitrogen). After rinsing three times with PBS, the cells were incubated with DAPI (Beyotime) diluted in PBS with 5% FBS for 15 min before microscopy analysis. Confocal images were obtained using a Leica TCS SP8 confocal microscope and analyzed with LAS AF software (Leica, Germany).
Polysome profiling
Polysome profiling was performed as described previously [43]. Briefly, naïve B cells were activated with LPS (25 μg/ml) for 24 h. Fifteen minutes prior to harvest, cells were treated with 100 μg/ml cyclohexamide (CHX). Cells were centrifuged for 5 min at 500 g and swelled in 10 ml of hypotonic buffer (1.5 ml KCl, 10 mM MgCl2, 5 mM Tris-HCl pH 7.4, and 100 μg/ml CHX). Cells were then resuspended in 400 μl hypotonic buffer for 10 min, and cytosolic fractions were extracted with 800 μl of a 1:1 mix of hypotonic buffer and hypotonic lysis buffer (2% sodium deoxycholate, 2% Triton X-100, 2.5 mM DTT, 10 units of RNase Inhibitors/ml, and 100 μg/ml CHX). Cells were incubated on ice for 20 min, and the nuclei were removed by centrifugation at 13,000 × g for 10 min. The supernatant was collected, loaded onto a linear (10–50%) RNase inhibitor-containing sucrose gradient and centrifuged at 38,000 rpm for 2 h using a Beckman SW41 rotor. The sucrose gradient was separated into 20 fractions from the top, and absorbance at 260 nm was monitored continuously by a Piston Gradient Fractionator (Biocomp, Canada).
CUT &Tag assay
The assay was performed following the manufacturer’s instructions for the Hyperactive In-Situ ChIP Library Prep Kit for Illumina (pA/G-Tn5) kit (Vazyme; TD903). Briefly, cultured cells were washed in Wash Buffer, resuspended in 100 μl of Wash Buffer, incubated with pretreated ConA beads, and harvested at RT for 10 min. The supernatant was discarded. The samples were incubated with an anti-HA antibody, anti-Dhx33 antibody, anti-mouse IgG or anti-rabbit IgG overnight at 4 °C. The supernatant was discarded. Goat anti-mouse IgG (Abcam; ab6702) or goat anti-rabbit IgG (Abcam; ab6708) was added, and the cells were incubated for 1 h at RT. The samples were washed with Dig-wash Buffer, resuspended in Dig-300 Buffer containing Hyperactive pA/G-Tn5 Transposon, and rotated for another hour. The samples were then washed with Dig-300 buffer, subjected to DNA extraction with MgCl2, and incubated at 37 °C for 1 h. Then, the reaction was blocked. DNA fragments were isolated for library building. Eluted DNA samples were analyzed by PCR, followed by PAGE for all the target genes. The primers for PCR were as follows:
P1: F 5’-GGTTCTTTTCGTTATGGGGTCAT-3’ R 5’-ACCTATCTCCAGGTCCAATAGG-3’; P2: F 5’-TCCGGTCGATGTCTACCTCC-3’ R 5’-CGCGAGAGATGGAAGGACG-3’; P3: F 5’-CGAGACTCTGGCATGCTAAC-3’ R 5’-CGGACATCTAAGGGCATCAC-3’; P4: F 5’-CGACTCTTAGCGGTGGATCACT-3’ R 5’-ACCGACGCTCAGACAGGC-3’; P5: F 5’-GAAGGCCGAAGTGGAGAAGG-3’ R 5’-GATCTGAACCCGACTCCCTT-3’ GAPDH: F 5’- CACTACAGACCCATGAGGAGT-3’ R 5’-ACCATATCAAGGGTGCCCG-3’.
Statistical analysis
The data were analyzed by using the unpaired two-tailed Student’s t test in IBM SPSS statistics 22. Statistical significance is displayed as *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. Graphs were analyzed with Prism software (GraphPad).
Supplementary information
Acknowledgements
We thank all members of the CX, W-HL, and Kairui Mao labs (Xiamen University) for technical assistance. This study was supported by the National Natural Science Foundation of China (31570882, 31770950 and 32070877 to W.-H. L, and 81961138008 to CX), the Fundamental Research Funds for the Central Universities of China-Xiamen University (20720170064 to CX), and the Sanofi Institute for Biomedical Research (SIBR).
Author contributions
CX, HYJ, and PC conceived this project. XH and JZ designed and performed most of the experiments, analyzed data, and generated figures; AA and PH performed some experiments and provided technical assistance; XL performed RNA sequencing analysis; PC, JX, YD, YL, and LL contributed to the experiments; CX and W-HL supervised the project. XH, JZ, W-HL, and CX wrote the manuscript with contributions from all authors.
Data availability
The RNA-seq data generated in this study have been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA850669.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Xiaoyu He, Jiayi Zhao.
Contributor Information
Wen-Hsien Liu, Email: whliu@xmu.edu.cn.
Changchun Xiao, Email: cxiao@scripps.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-022-00972-0.
References
- 1.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–8. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 2.Kouzine F, Wojtowicz D, Yamane A, Resch W, Kieffer-Kwon K-R, Bandle R, et al. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153:988–99. doi: 10.1016/j.cell.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf T, Jin W, Zoppi G, Vogel IA, Akhmedov M, Bleck CKE, et al. Dynamics in protein translation sustaining T cell preparedness. Nat Immunol. 2020;21:927–37. doi: 10.1038/s41590-020-0714-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnet FM. A modification of Jerne’s theory of antibody production using the concept of clonal selection. CA Cancer J Clin. 1976;26:119–21. doi: 10.3322/canjclin.26.2.119. [DOI] [PubMed] [Google Scholar]
- 5.Russell J, Zomerdijk JC. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourgeois CF, Mortreux F, Auboeuf D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat Rev Mol Cell Bio. 2016;17:426–38. doi: 10.1038/nrm.2016.50. [DOI] [PubMed] [Google Scholar]
- 7.Takuya N, Kei H, Tatsuya M, Moeko M, Sho M, Ikuo S, et al. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun. 2011;2:465. doi: 10.1038/ncomms1475. [DOI] [PubMed] [Google Scholar]
- 8.Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA. 2006;103:13789–94. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt-Supprian M, Rajewsky K. Vagaries of conditional gene targeting. Nat Immunol. 2007;8:665–8. doi: 10.1038/ni0707-665. [DOI] [PubMed] [Google Scholar]
- 10.Timm W, Jane S, Wiebke W, Tristan W, Van TC, Klaus R, et al. A novel allele for inducible Cre expression in germinal center B cells. Eur J Immunol. 2019;49:192–4. doi: 10.1002/eji.201847863. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y FJT, Miceli AP. Identification of DHX33 as a mediator of rRNA synthesis and cell growth. Mol Cell Biol. 2011;31:4676–91. doi: 10.1128/MCB.05832-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Lu H. Signaling to p53: Ribosomal proteins find their way. Cancer Cell. 2009;16:369–77. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su D, Yuan B, Su C, Zhang Y. A 54-kDa short variant of DHX33 functions in regulating mRNA translation. J Cell Physiol. 2019;234:15308–19. doi: 10.1002/jcp.28178. [DOI] [PubMed] [Google Scholar]
- 14.Kaya-Okur HS, Steven JW, Christine AC, Erica SP, Terri DB, Jorja GH, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10:1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumari P, Tarighi S, Braun T, Ianni A. SIRT7 acts as a guardian of cellular integrity by controlling nucleolar and extra-nucleolar functions. Genes (Basel) 2021;12:1361. doi: 10.3390/genes12091361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langst G, Becker PB, Grummt I. TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J. 1998;17:3135–45. doi: 10.1093/emboj/17.11.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grummt I, Langst G. Epigenetic control of RNA polymerase I transcription in mammalian cells. Bba-Gene Regul Mech. 1829;393-404:2013. doi: 10.1016/j.bbagrm.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Stefanovsky V, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol Cell. 2006;21:629–39. doi: 10.1016/j.molcel.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Ge W, Zhang Y. Recombinant DHX33 protein possesses dual DNA/RNA helicase activity. Biochemistry. 2019;58:250–8. doi: 10.1021/acs.biochem.8b00166. [DOI] [PubMed] [Google Scholar]
- 20.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: An emerging target for the treatment of cancer. Annu Rev Pharm Toxicol. 2010;50:131–56. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 21.Bursac S, Prodan Y, Pullen N, Bartek J, Volarevic S. Dysregulated ribosome biogenesis reveals therapeutic liabilities in Cancer. Trends Cancer. 2021;7:57–76. doi: 10.1016/j.trecan.2020.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Fu J, Liu Y, Wang X, Yuan B, Zhang Y. Role of DHX33 in c-Myc-induced cancers. Carcinogenesis. 2017;38:649–60. doi: 10.1093/carcin/bgx041. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Yu J, Wang X, Zhang Y. The RNA helicase DHX33 is required for cancer cell proliferation in human glioblastoma and confers resistance to PI3K/mTOR inhibition. Cell Signal. 2019;54:170–8. doi: 10.1016/j.cellsig.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Tian QH, Mei-F Z, Rong-G L, Jia F, Cong H, Gang H, et al. DHX33 expression is increased in hepatocellular carcinoma and indicates poor prognosis. Biochem Bioph Res Co. 2016;473:1163–9. doi: 10.1016/j.bbrc.2016.04.033. [DOI] [PubMed] [Google Scholar]
- 25.Zhu Y, Du Y, Zhang Y. DHX33 promotes colon cancer development downstream of Wnt signaling. Gene. 2020;735:144402. doi: 10.1016/j.gene.2020.144402. [DOI] [PubMed] [Google Scholar]
- 26.Poortinga G, Katherine MH, Hayley S, Carl RW, Anna J, Kerith S, et al. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 2004;23:3325–35. doi: 10.1038/sj.emboj.7600335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Weimin F, Cheng P, Shiyun C, Hongbin J, Hanbing Z, et al. Targeting RNA helicase DHX33 blocks Ras-driven lung tumorigenesis in vivo. Cancer Sci. 2020;111:3564–75. doi: 10.1111/cas.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Feng W, Yuan Z, Weber JD, Zhang Y. DHX33 Interacts with AP-2beta to regulate Bcl-2 gene expression and promote cancer cell survival. Mol Cell Biol. 2019;39:e00017-19. [DOI] [PMC free article] [PubMed]
- 29.Zhang Y, Saporita AJ, Weber JD. P19ARF and RasV(1)(2) offer opposing regulation of DHX33 translation to dictate tumor cell fate. Mol Cell Biol. 2013;33:1594–607. doi: 10.1128/MCB.01220-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sloan KE, Bohnsack MT, Watkins NJ. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 2013;5:237–47. doi: 10.1016/j.celrep.2013.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bursac S, Maja CB, Martin P, Ines O, Lior G, Yan Z, et al. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc Natl Acad Sci USA. 2012;109:20467–72. doi: 10.1073/pnas.1218535109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donati G, Peddigari S, Mercer CA, Thomas G. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 2013;4:87–98. doi: 10.1016/j.celrep.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–34. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Ventura A, David GK, Margaret EM, David AT, Jan G, Laura L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–5. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 36.Xue W, Lars Z, Cornelius M, Ross AD, Eva H, Valery K, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Platt RJ, Sidi C, Yang Z, Michael JY, Lukasz S, Hannah RK, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–55. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–8. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casola S, Giorgio C, Nathalie U, Sergei BK, Jane S, Zhenyue H, et al. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc Natl Acad Sci USA. 2006;103:7396–401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seung GK, Wen-H L, Peiwen L, Hyun YJ, Hyung WL, Jovan S. MicroRNAs of the miR-17~92 family are critical regulators of TFH differentiation. Nat Immunol. 2013;14:849–57. doi: 10.1038/ni.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuki H, Takao K, Hiroyuki K, Changshan W, Atsushi I, Keiji K. Downregulation of rRNA transcription triggers cell differentiation. PLoS One. 2014;9:e98586. doi: 10.1371/journal.pone.0098586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian A, Aravind S, Pablo T, Vamsi KM, Sayan M, Benjamin LE, Michael AG, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyun YJ, Hiroyo O, Pengda C, Chao Y, Xiaojuan Z, Seung GK, et al. Differential Sensitivity of Target Genes to Translational Repression by miR-17~92. Plos Genet. 2017;13:e1006623. doi: 10.1371/journal.pgen.1006623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated in this study have been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA850669.