Abstract
This review presents a comprehensive summary of the latest research in the field of bioremediation with filamentous fungi. The main focus is on the issue of recent progress in remediation of pharmaceutical compounds, heavy metal treatment and oil hydrocarbons mycoremediation that are usually insufficiently represented in other reviews. It encompasses a variety of cellular mechanisms involved in bioremediation used by filamentous fungi, including bio-adsorption, bio-surfactant production, bio-mineralization, bio-precipitation, as well as extracellular and intracellular enzymatic processes. Processes for wastewater treatment accomplished through physical, biological, and chemical processes are briefly described. The species diversity of filamentous fungi used in pollutant removal, including widely studied species of Aspergillus, Penicillium, Fusarium, Verticillium, Phanerochaete and other species of Basidiomycota and Zygomycota are summarized. The removal efficiency of filamentous fungi and time of elimination of a wide variety of pollutant compounds and their easy handling make them excellent tools for the bioremediation of emerging contaminants. Various types of beneficial byproducts made by filamentous fungi, such as raw material for feed and food production, chitosan, ethanol, lignocellulolytic enzymes, organic acids, as well as nanoparticles, are discussed. Finally, challenges faced, future prospects, and how innovative technologies can be used to further exploit and enhance the abilities of fungi in wastewater remediation, are mentioned.
Keywords: bioremediation, removal efficiency, pharmaceutical compounds, heavy metals -, oil hydrocarbons, pollutants
Graphical Abstract
1 Introduction: History and benefits of using filamentous fungi for wastewater treatment
The microbial world are well-known for the vast diversity of functionalities they present (Dunlap, 2001; Shu and Huang, 2022). These functionalities have been exploited by humans to produce novel products (Stahl et al., 2013; Vuong et al., 2022), but also to degrade human produced waste (Raziyafathima et al., 2016; Chiang et al., 2020; Gupta et al., 2022). Miraculously microbes have always been discovered that could degrade most of the novel products that humans synthesized and that become waste. This is besides those that can already degrade naturally occurring organic compounds that occur in higher concentrations than normal due to human activities. Surprisingly these attributes of microbes are still, relatively speaking, vastly underutilized in industry and rehabilitation activities, despite a number of success stories that have already been applied (Gupta et al., 2022; Rafeeq et al., 2022). The same holds true for the industrial application of microbes that forms the topic of this review, namely the sustainable rehabilitation of specifically wastewater, which is a very complex field.
Fungi are, similar to other microbes such as bacteria and algae, incredibly useful in bio-industries (Dhiman et al., 2022; Hadibarata et al., 2022). Numerous applications using fungi exist, including several in the field of wastewater rehabilitation. In fact, in some cases fungi can be synergistically used together with other microbes, and not just on their own (Gultom and Hu, 2013; Chu et al., 2021). Their abilities stem from the fact that they form the foundation of every ecosystem on Earth, and that they occupy every possible niche and geographical area where they are saprobes, parasites or symbionts (Chaudhary V. B. et al., 2022b; Tedersoo et al., 2022). In order to survive, they thus need a wide range of enzymatic abilities, such as pectinases, cellulases and phosphatases, and can produce diverse types of compounds, such as siderophores (Haripriyan et al., 2022). Their ability to withstand a range of environmental conditions, of which some are extreme similar to those in waste waters, makes them highly beneficial. However, ironically the majority of applications in waste water rehabilitation uses algae and bacteria.
The biodiversity of fungi that has thus far been used in waste water rehabilitation is staggering. The fungi used encompasses various phyla of the fungi, and within phyla, they are also diverse (Bhunjun et al., 2022). The ease by which fungi can be grown in fermentation, selected from nature or where known cultures from culture collections can be tested for their ability to degrade or convert a specific compound, gives them incredible potential to be used in green technology in an eco-friendly manner (Yang and Peng, 2022). This is especially so when native fungi are exploited with possible better or novel properties. Often, fungi can be isolated from environments mimicking waste areas, or the target environment itself, because they are already present, for example as sediment fungi, metallophyles or fungi from extreme environments (Chaudhary P. et al., 2022a; Talukdar et al., 2022). They can then be made into a product that can be easily added to the problem area.
Compounds that must be degraded, or converted, in waste waters for rehabilitation are challenging because they are so incredibly diverse. These compounds range from metals and other elements that are not supposed to be present in specific environments, various types of oils such as crude oils, derived products, and food based oils, and biocides. Human made products such as plastics, varnish, foams, dyes, disinfectants, hygiene products and pharmaceuticals, add further to this list of xenobiotics (Gultom and Hu, 2013; Chu et al., 2021). Due to the saprobic nature of fungi, they are also useful where too high concentrations of organic matter, such as cattle farms with manure, crop based agricultural wastes, fisheries or abattoirs, are found (Camenzind et al., 2022; Ediriweera et al., 2022). Moreover, often beneficial by-products can be developed by the fungi from these problem areas (Guo et al., 2022).
This review presents a comprehensive summary of the latest research that has been mentioned above in waste water bioremediation utilizing specifically fungi. It covers cellular mechanisms, processes for wastewater treatment, a summary of the fungi involved and how they are utilized for the various types of pollutants, and beneficial byproducts made by fungi. Lastly, challenges faced, future prospects, and how innovative technologies can be used to further exploit and enhance the abilities of fungi in wastewater remediation, are mentioned.
2 Mycoremediation mechanisms
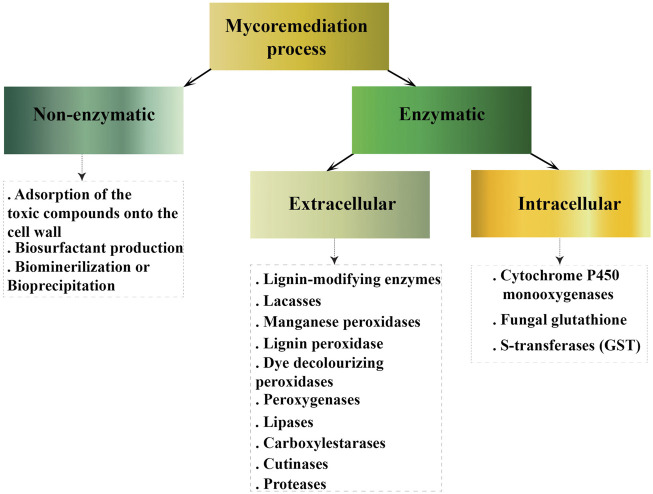
2.1 Non-enzymatic processes
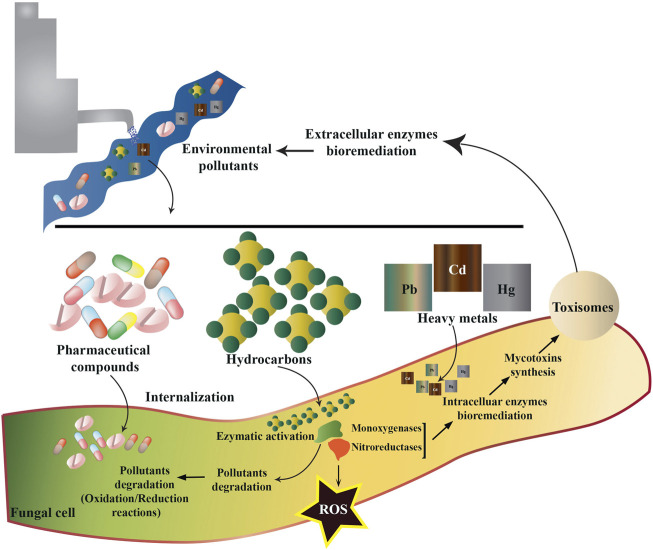
There are several mechanisms of bioremediation described for filamentous fungi (Figure 1). The first one is adsorption of the toxic compounds into the cell wall. Bioadsorption is a physico-chemical process in which the concentration of toxic compounds is adsorbed on the bio-surface of fungi. Absorption takes place via specific aminopolysaccharides that are part of the composition of the cell wall, such as chitosan or chitin (Kumari et al., 2016). The functional groups of glucosamine in chitosan act as metal absorbent sites for heavy metal chelation or complexation facilitating the reduction and recovery of heavy metal from the polluted environment (Alsharari et al., 2018; Geetha et al., 2021). Absorption is the predominant mechanism of bioremediation of the heavy metals Copper (Cu) and Cobalt (Co.) described for Trichoderma, Penicillium, and Aspergillus (Dusengemungu et al., 2020). In filamentous fungi Phoma sp. biosorption into fungal mycelia has an important role for some pharmaceutical compounds removal, until this bioadsorption reaches equilibrium (Hofmann and Schlosser, 2016). Filamentous fungi Mucor shows considerable promise to be applied as a mycoremediation agent for the removal of cyanobacterial toxins from aquatic environments due to excellent toxin uptake into the fungal cell (Balsano et al., 2015). Removal of Fe3+ and Co2+ by fungi Aspergillus sp. AHM69 and Penicillium sp. AHM96 was shown via biosorption and bioaccumulation on the biomass surface (El-Bondkly and El-Gendy, 2022).
FIGURE 1.
Cellular mechanisms involve in mycoremediation.
The other mechanism of bioremediation is associated with biosurfactants production by filamentous fungi. These biomolecules reduce the surface/interfacial tension between fluid phases (da Silva et al., 2021). Lipopeptides, molecules consisting of a short amino acid chain, covalently modified with lipids are well known class of biosurfactants, proved suitable for bioremediation of crude oil (Carolina et al., 2021). The biotechnological potential of Penicillium, Aspergillus and Fusarium for Lipopeptides production is described (Gautam et al., 2014; Qazi et al., 2014; Bills et al., 2016). The produced crude biosurfactant or fungal strain Mucor circinelloides could be directly used for the mycoremediation of crude oil contamination in oil fields (Hasanizadeh et al., 2017). However, microbial biosurfactant production is still quite restricted, since the production costs are currently higher than chemical surfactants. Besides most bioprocesses show low yield and productivity.
The other mechanism of filamentous fungi to counteract with a toxic compounds is biomineralization or bio-precipitation. Oxalate is a key metabolite that plays a significant role in many metal and mineral transformations mediated by fungi. Oxalic acid, (COOH)2 is a relatively strong organic di-acid with the ability of the oxalate anion to complex metals including those of Co, Cd, Cu, Mn, Mg, Ni, Sr, Pb and Zn (Gadd, 1999). It is generally stated that metal oxalate formation may confer protection from the potentially toxic effects of such metals (Johansson et al., 2008; Gadd et al., 2014). Oxalic acid produced by a variety of filamentous fungi (Gadd et al., 2014).
It is well described that in Aspergillus niger, oxalic acid synthesized by conversion of oxaloacetate to oxalate and acetate by a cytoplasmic oxaloacetate acetylhydrolyase. Oxaloacetate in turn synthesized from pyruvate, coming from glycolysis, by the action of pyruvate carboxylase (Pedersen et al., 2000a; Pedersen et al., 2000b). This pathway may be widely distributed in fungi (Gadd et al., 2014). Oxalate is potentially toxic in high concentration and needs to efflux from cells. It was demonstrated copper oxalates formation by A. niger (Frank-Kamenetskaya et al., 2021) that can be used to develop of copper bioremediation from soil. A strain of Aspergillus nomius, isolated from a heavily-polluted soil from a zinc smelting region, was able to precipitate zinc oxalate (Sutjaritvorakul et al., 2016).
Lead is one of the most widely found toxic metal contaminants in the environment arising from industrial activities (Flora et al., 2012). Complete removal of Pb from solution and extensive precipitation of lead oxalate (PbC2O4) around the mycelium of A. niger and Paecilomyces javanicus was demonstrated (Liang et al., 2016). Other study also demonstrated reduction of Pb toxicity by Aspergillus niger and Penicillium oxalicum via formation of insoluble Pb minerals, primarily lead oxalate (Tian et al., 2019). Penicillium chrysogenum was effectively utilized for the biomineralization of chromate and lead from an aqueous solution and contaminated soil. It was shown efficient fungal-mediated calcite precipitation of heavy metals in the remediation (Qian et al., 2017).
2.2 Enzymatic processes
The above mentioned mechanisms include non-enzymatic process. The other strategies include biotransformation and biodegradation mediated by enzymatic systems. Enzymatic fungal-mediated biotransformation in turn, can be carried out by means of extracellular and intracellular enzymes. Extracellular enzymes catalyze variety of reactions (e.g., oxidative coupling products, hydroxylation, CO2 emission, ether cleavage, aromatic ring fission, quinoid products formation and other) resulting in biotransformation of toxic compounds facilitating bioremediation processes (Singh et al., 2021). The biodegradation is particularly committed for filamentous fungi since they can produce the extracellular non-specific lignin-modifying enzymes, laccases, manganese peroxidases, lignin peroxidase, dye decolorizing peroxidases, peroxygenases (El-Gendi et al., 2022).
Decolourization is a process in which filamentous fungi are particularly effective (Al-Ansari et al., 2022; Dhir, 2022). This process involves the removal of color from a solution, often by breaking down the colored compounds present in the solution. Filamentous fungi have been shown to accomplish this through the production of laccase and manganese peroxidase enzymes (Bilal et al., 2016a; Bilal et al., 2016b). Laccase is a type of enzyme that can break down a wide range of colored compounds through oxidation, acting on a variety of colored compounds such as azo dyes and natural pigments (Srinivasan and Sadasivam, 2018). Therefore, fungal laccases were widely applied for the bioremediation of complex synthetic dyes by altering the dye molecules into non-colored, safer, and ecofriendly structures (Sanghi et al., 2009; El-Fakharany et al., 2016). Manganese peroxidase (MnP) is another enzyme that can be involved in decolorization by filamentous fungi. MnP uses hydrogen peroxide as a cofactor to oxidize a wide range of compounds, particularly lignin-derived compounds which are found in industrial effluents and are responsible for the color of the solution (Bilal et al., 2016b). The use of filamentous fungi in bioremediation processes can take place in different types of reactors, including solid-state fermentation, submerged fermentation, or adsorption-biodegradation (Svobodová and Novotný, 2018; Zapana-Huarache et al., 2020; Usuldin et al., 2021). The process can be improved by optimizing the environmental conditions, such as pH, temperature, and oxygen availability, and by selecting the appropriate fungal strains. It is also important to note that decolorization alone may not remove other pollutants in the solution, thus the process should be combined with other remediation techniques, such as biodegradation, in order to achieve complete remediation of the contaminated site (Bilal et al., 2016a; Bilal et al., 2016b).
On the other hand, polycyclic aromatic hydrocarbons are abundant and widespread contaminants for the environment. Aspergillus oryzae and Mucor irregularis isolated from crude oil efficiently degraded hydrocarbons from oil by the action of secreted laccase, manganese peroxidase, and lignin peroxidase (Asemoloye et al., 2020). Plastic pollution poses serious environmental problems, in part due to the extremely stable and durable nature of this polymer. Fungal secreted hydrolases (lipases, carboxylesterases, cutinases and proteases) able to modify the plastic surface, increasing its hydrophilicity (Vertommen et al., 2005). These enzymes were successfully used to degrade polyethylene terephthalate and polyurethane hydrolyzing chemical bonds in the polymer structures (Cregut et al., 2013; Webb et al., 2013). Highly stable carbon–carbon bonds in plastic polymers require oxidation before the depolymerisation process (Restrepo-Florez et al., 2014). These reactions efficiently facilitate secreted oxidoreductases (laccases and peroxidases) which are involved in plastic degradation into smaller molecules such as oligomers, dimers and monomers (Alvarez-Barragan et al., 2016). Fungal laccases and peroxidases are very promising enzymes in degrading polyethylene and polyvinyl chloride (Temporiti et al., 2022). Secreted fungal laccases and peroxidases enzymes can be effectively used for detoxification of heavy metals and pharmaceuticals (Olicon-Hernandez et al., 2017; Mousavi et al., 2021).
Intracellular enzymes participating in bioremediation of toxic compounds can be divided in two groups. First–cytochromes and second–glutathione transferases. The cytochrome P450 system is a large family of enzymes, mainly monooxygenases catalyzing reactions of hydroxylation, oxygenation, dealkylation, epoxidation of C=C bonds, reduction, and dehalogenation (Díaz-Cruz et al., 2014). It was shown that P450 play an indirect but crucial role in the process of clearance of cadmium and lead in filamentous fungi Phanerochaete chrysosporium belonging to the Basidiomycota phylum (Zhang et al., 2015). CYP63A2 P450 monooxygenase from this fungus oxidized polycyclic aromatic hydrocarbons, alkylphenols, and alkanes from crude oil (Syed et al., 2013). Cytosolic and mitochondrial iso-forms of P450 from Fusarium oxysporum and other fungi are used in degradation of dioxins (Guengerich and Munro, 2013; Sakaki et al., 2013). F. oxysporum P450 monooxygenases are promising catalysts in production of omega hydroxy fatty acids when heterologously expressed in Saccharomyces cerevisiae (Durairaj et al., 2015). It was demonstrated that several Aspergillus species are capable of degrading a carcinogenic contaminant Benzo [a]pyrene by the action of the cytochrome P450 monooxygenase (Loss et al., 2019).
Fungal glutathione S-transferases (GST) are detoxification enzymes which can catalyze the conjugation of glutathione to non-polar compounds containing an electrophilic carbon, nitrogen, or sulfur atom. By this mechanism, glutathione S-transferases are able to metabolize drugs, pesticides, xenobiotics, heavy metals and other toxic compaunds (Pocsi et al., 2004). GST have been reported to contribute to the defense against heavy metal stress in different filamentous fungi including A. nidulans (Fraser et al., 2002). Glutathione transferases play an important role in the intracellular reduction of Cr (VI) in fungi (Garcia-Hernandez et al., 2017). It was shown that the detoxification of penicillin side-chain precursors might depend on microsomal GST in P. chrysogenum (Emri et al., 2003). Fungal cytochromes can catalase a lot of different reactions, which enables them to degrade xenobiotics and pollutants (Syed et al., 2013; Chen et al., 2019; Wang et al., 2019; Carstens et al., 2020; Hu et al., 2020). Fungal enzymes, isolated from Basidiomycetes and Ascomycetes, show high ability of plastic waste biodegradation. Fungal laccases and peroxidases, generally used by fungi to degrade lignin, polyethylene and polyvinyl chloride, while esterases such as cutinases and lipases were successfully used to degrade polyethylene terephthalate and polyurethane (Temporiti et al., 2022).
3 Processes involved in wastewater treatment
Wastewater treatment necessitates the expenditure of energy, time, and money. Any solution that can enhance the wastewater treatment process is critical since one of the sustainable development goals is to minimise energy consumption and strategic processes globally (Shojaei and Shojaei, 2021). Treatment of wastewater could be accomplished through physical, biological, and chemical processes.
3.1 Physical wastewater treatment process
Physical wastewater treatment process refers to separating particle matter and solids in sanitary and industrial effluents (Pirzadeh, 2022). Depending on the sewage type, fabric fragments, sand, plastic bits, and tree foliage may be in the wastewater entering the treatment plants. These particles can cause damage to the equipment (including pipes, pumps, and fittings) used in wastewater treatment. Furthermore, failing to remove these things puts a lot of strain on the machinery in biological and chemical wastewater treatment, lowering the output quality.
In the physical wastewater treatment process, mechanical procedures based on physical rules are employed to remove pollutants (Woodard, 2001). Physical operations are typically simpler and more quantitatively effective than other wastewater treatment methods. Physical wastewater treatment techniques remove pollutants using naturally occurring forces, including electrostatic attraction, gravity, van der Waal forces, and physical barriers (Tripathi and Misra, 2019). In general, the techniques involved in physical treatment do not lead in changes in chemical composition of the target compounds. In some cases, the physical state is altered, as in vaporization, and scattered substances are often made to agglomerate, as in filtration (Crittenden et al., 2012). The first phase in every wastewater treatment system is filtration (Gerba and Pepper, 2019). This procedure requires the separation of non-biodegradable contaminants from a wastewater treatment plant. Sedimentation, skimming, adsorption, aeration, and flotation are physical methods of wastewater treatment (Saravanathamizhan and Perarasu, 2021), as are barriers such as deep bed filters, screens, bar racks, and membranes.
3.2 Chemical wastewater treatment process
Chemical wastewater treatment is another process of wastewater treatment. It is considered a tertiary treatment process that incorporates chemical treatment (Ahmed et al., 2021). Chemicals are used in this process, which involves separating or converting pollutants because of their chemical reaction. Chemical precipitation, neutralization, disinfection (ozone, chlorine, UV light), ion exchange, and adsorption are the most commonly used chemical treatment processes (Samer, 2015).
3.3 Biological wastewater treatment
Biological wastewater treatment involves the use of microorganisms in wastewater treatment, a poorly understood process that combines biology and biochemistry (Sahu et al., 2019). This process is used in removing organic, soluble, biodegradable, organic and nutrient-containing compounds, and colloids, present in wastewater. The wastewater is introduced into a specifically designed bioreactor, where organic matter is utilised by bacteria (anaerobically or aerobically), fungi and algae (aerobically) (Samer, 2015). The bioreactor provides suitable environmental conditions for microorganisms to grow and utilize dissolved organic materials as a source of energy. The biological oxidation of organic substances will continue as long as the microorganisms receive oxygen and food from collected wastewater. Bacteria, which form the fundamental trophic level of the food chain in the bioreactor, carry out the majority of the biological process.
The conversion of dissolved organic molecules into thick bacterial biomass via bioconversion has the potential to purify wastewater (Meena et al., 2021). Following that, sedimentation is used to segregate the microbial biomass from the treated wastewater. Microorganisms breakdown organic materials via two distinct biological processes: biosynthesis and biological oxidation (Samer, 2015). Biosynthesis converts colloidal and dissolved organic materials into dense biomass, which then can be recovered via sedimentation. In this case colloidal and dissolved organic molecules are converted into dense biomass, which can then be removed via sedimentation. Some biological oxidation by-products, such as minerals, remain in the solution and are eliminated with the effluent.
4 Species diversity of filamentous fungi used in pollutant removal
Filamentous fungi are organism belonging to a large group of eukaryotes that includes some yeasts, moulds and some mushrooms. Filamentous fungi, like fungi in general, belong to a separate kingdom. They are unable to photosynthesis, possess urea metabolism and glycogen storage. Fungi reveal the predominance of the osmotrophic nutrition over the phagotrophic and reproduce using spores. The main difference between fungi and other organisms is the peculiarity of the structure of the cell wall containing chitin. However, key characteristic contributing to classifying fungi as filamentous fungi is the ability to possess hyphae. These hyphae, in turn, form branches making up fungal mycelia, growing like threadlike structures.
Filamentous fungi are phylogenetically diverse; however, members of three groups, namely Ascomycetes, Basidiomycetes, and Zygomycetes are mostly found in association with bioremediation research studies, or commercial exploitation (Troiano et al., 2020).
A huge number of known filamentous fungi belong to the phylum Ascomycota (Moretti, 2009). Filamentous fungi encompass many genera with Aspergillus, Penicillium, Fusarium, Verticillium and some other more investigated than the other genera for their suitability of use in bioremediation (Olicon-Hernandez et al., 2017) (Table 1).
TABLE 1.
Species diversity of filamentous fungi used in pollutant removal.
| Pollutants | Species | References |
|---|---|---|
| Ascomycota | ||
| Cd | Aspergillus niger | Barros et al. (2003) |
| A. clavatus DESM | Cernansky et al. (2007) | |
| Cu,Pb,As | A. niger | Clausen (2004), Mukherjee et al. (2010) |
| Cr | A. niger, A. foetidus | Prasenjit and Sumathi (2005), Srivastava and Thakur (2006) |
| Ni, Pb bioleaching | A. niger | Le et al. (2006), Chakraborty et al. (2014) |
| Crude oil | A. ramosus, A. flavus | ((Davies and Westlake, 1979) |
| Gasoline | A. oryzae | (Binsadiq, 1996; Oboh et al., 2006) |
| PAH | A. niger, A. terreus | Capotorti et al. (2004), Okoro, 2008; Ali et al. (2012) |
| Dye | Aspergillus spp. | Devi and Kaushik (2005), Ponraj et al. (2011) |
| Acidic and basic dyes | A. foetidus, A. niger | Ryu and Weon (1992), Sumathi and Manju (2000), Fu and Viraraghavan (2002) |
| Cd | Penicillium simplicissimum | Fan et al. (2008) |
| P. chrysogenum | Niu et al. (1993), Holan and Volesky (1995), Skowronski et al. (2001) | |
| P. canescens | Say et al. (2003) | |
| Zn | P. digitatum | Galun et al. (1987) |
| P. simplicissimum | Fan et al. (2008) | |
| P. chrysogenum | Niu et al. (1993) | |
| P. spinulosum | Townsley and Ross (1985) | |
| Pb | P. simplicissimum | ((Fan et al., 2008) |
| P. chrysogenum | Niu et al. (1993) | |
| P. canescens | Say et al. (2003) | |
| Cu | P. chrysogenum | Skowronski et al. (2001) |
| P. cyclopium | Ianis et al. (2006) | |
| P. spinulosum | Townsley and Ross (1985) | |
| P. italicum | Mendil et al. (2008) | |
| As, Hg, Cr | P. canescens | Say et al. (2003) |
| Mn, Fe, Ni, Co. | P. italicum | Mendil et al. (2008) |
| Fluorene | P. canescens, P. janczewskii, P. montanense, P. simplicissimum, P. restrictum | Garon et al. (2004) |
| P. chrysogenum, P. italicum | Garon et al. (2002) | |
| Pyrene | P. harzianum, P. terrestre | Saraswathy and Hallberg (2002) |
| P. ochrochloron | Saraswathy and Hallberg (2005) | |
| P. chrysogenum, P. aurantiogriseum, P. crustosum, P. rugulosum | Ravelet et al. (2000) | |
| Phenol | P. simplicissimum | Marr et al. (1996) |
| P. chrysogenum | Leitao et al. (2007) | |
| 2-, 3- and 4-Chlorophenol | P. frequentans Bi 7/2, P. simplicissimum and P. simplicissimum | Hofrichter et al. (1994), Marr et al. (1996) |
| Olive mill wastewater | P. decumbens | Borja et al. (1992) |
| Vinasses | Penicillium spp. | Jimenez et al. (2005) |
| Coffee residues | P. curstosum, P. verrucosum, P. crustosum, P. restrictum, P. implicatum, P. citrinum | Fujii and Takeshi (2007) |
| Tl | Fusarium sp. FP, Arthrinium sp. FB, and Phoma sp. FR | Mo et al. (2022) |
| PAH | Fusarium sp. ZH-H2 | Zhao et al. (2019) |
| Chlorpyrifos | Verticillium sp | Farhan et al. (2021) |
| Pb, Zn | V. insectorum J3 | Feng et al. (2018) |
| Basidiomycota | ||
| Anthracene | Phanerochaete chrysosporium | Vyas et al. (1994), Novotny et al. (2004) |
| Naphthalene | Ph. chrysosporium | Mollea et al. (2005) |
| Phenanthrene | Ph. chrysosporium | Bumpus (1989) |
| Pyrene | Ph. chrysosporium | Novotny et al. (2004) |
| Pentachlorophenol | Ph. chrysosporium | Bumpus (1989), Lamar and Dietrich, 1990 (1992) |
| DTT, lindane | Ph. chrysosporium | Bumpus (1989) |
| 2,4,6-Trinitrotoluene | Ph. chrysosporium | Fernando et al. (1990) |
| PAH | Xerotus discolor | Acevedo et al. (2011) |
| Bjerkandera specie | Matsubara et al. (2006) | |
| Irpex lacteus | Bhatt et al. (2002), Cajthaml et al. (2006) | |
| Phellinus sp. | Arun and Eyini (2011) | |
| Schizophyllum commune | Matsubara et al. (2006) | |
| Stropharia coronilla | Steffen (2000) | |
| Pentachlorophenol | Agrocybe perfecta, T. villosa, T. hirsuta | Machado et al. (2005) |
| Ceriporiopsis subvermispora | (Lamar and Dietrich, 1990; 1992) | |
| AH | Allescheriella sp. | D'Annibale et al. (2006), D'Annibale et al. (2007) |
| Nitrobenzene, anthracene | Coriolopsis polyzona | Vyas et al. (1994) |
| C. trogii | Levin et al. (2003) | |
| Zygomycota | ||
| Diazinon | Cunninghamella elegans ATCC36112 | Zhao et al. (2020) |
| DiclofenacR | Mucor hiemalis | Esterhuizen-Londt et al. (2017) |
| Malachite greenR | Cunninghamella elegansR | Cha et al. (2001) |
| CopperR | Mortierella spp. | Fu et al. (2022) |
| PyreneR | Mucor mucedo | Hou et al. (2015) |
| PAHsR | Мucor spp.R | Gupta et al. (2015) |
| Crude oilR | Мucor spp.R | Okoro et al. (2013) |
The genus of Aspergillus consists of different species applied in bioremediation of virtually all types of environmental hazards. This genus is of particular interest due to its rich species variety, with various ecological functions. Aspergillus contain some unique enzymes, rarely produced by other microorganism, some of them help the species to counteract toxic substances and some are responsible for bioremediation of a wide range of toxic substances.
A lot of strains of Aspergillus work as excellent biosorbents for heavy metals removal, including cadmium from oil field water (Barros et al., 2003) and from aqueous solution (Cernansky et al., 2007) copper, lead, arsenic (Clausen, 2004; Mukherjee et al., 2010), chromium (Prasenjit and Sumathi, 2005; Srivastava and Thakur, 2006). A. niger is using to produce a variety of organic acids for the leaching of heavy metals from contaminated soils (Wasay et al., 1998; Ren et al., 2009) and is effective in the bioleaching of nickel laterite ores and Pb (Le et al., 2006; Chakraborty et al., 2014). Dead biomass of Aspergillus can be used as an effective biosorbent of heavy metals (Rostami and Joodaki, 2002). Pretreated and modification of the fungal cell surface increased the sorption of heavy metals (Kapoor et al., 1999; Rao et al., 2005). Strains of Aspergillus are capable of degrading hydrocarbons, such as crude oil (Aspergillus ramosus) (Davies and Westlake, 1979), gasoline (Binsadiq, 1996; Oboh et al., 2006), polycyclic aromatic hydrocarbons (Capotorti et al., 2004; Okoro, 2008; Ali et al., 2012), crude oil (Adegunlola et al., 2012). Some of the Aspergillus sp. are well adapted to wastewater cleaning (Devi and Kaushik, 2005; Ponraj et al., 2011) and are able to decolorizing acidic and basic dyes (Ryu and Weon, 1992; Sumathi and Manju, 2000; Fu and Viraraghavan, 2002).
Penicillium spp. are very important for the processes of environmental remediation. They can work as a naturally biosorbents for heavy metal environmental reduction. Some strains of Penicillium [P. simplicissimum (Fan et al., 2008), P. chrysogenum (Niu et al., 1993; Holan and Volesky, 1995; Skowronski et al., 2001)], P. canescens (Say et al., 2003) are able to remediate cadmium. A lot of strains can absorb zinc [P. digitatum (Galun et al., 1987), P. simplicissimum (Fan et al., 2008), P. chrysogenum (Niu et al., 1993), P. spinulosum (Townsley and Ross, 1985)], Pb [P. simplicissimum (Fan et al., 2008), P. chrysogenum (Niu et al., 1993), P. canescens (Say et al., 2003)], Cu [P. chrysogenum (Skowronski et al., 2001), P. cyclopium (Ianis et al., 2006), P. spinulosum (Townsley and Ross, 1985), P. italicum (Mendil et al., 2008)], As, Hg, Cr [P. canescens (Say et al., 2003)], Mn, Fe, Ni, Co. (P. italicum) (Mendil et al., 2008).
Polycyclic aromatic hydrocarbons comprise a large group of organic compounds with two or more fused aromatic rings, that occur naturally in coal, crude oil, and gasoline. A lot of Penicillium spp can remediate these chemicals. Fluorene is an ortho-fused tricyclic hydrocarbon that is a major component of fossil fuels and their derivatives. This chemical can be remediated by P. canescens, P. janczewskii, P. montanense, P. simplicissimum, P. restrictum (Garon et al., 2004), P. chrysogenum, P. italicum (Garon et al., 2002). Pyrene is a polycyclic aromatic hydrocarbon consisting of four fused benzene rings, resulting in a flat aromatic system. It can be degraded by P. harzianum, P. terrestre (Saraswathy and Hallberg, 2002), P. ochrochloron (Saraswathy and Hallberg, 2005), P. chrysogenum, P. aurantiogriseum, P. crustosum, P. rugulosum (Ravelet et al., 2000). Such polycyclic aromatic hydrocarbons like benz [a]pyrene, phenanthrene, fluoranthene also can be utilized by fungi (Sack and Gunther, 1993).
Several Penicillium strains have the ability to transform phenol and its toxic derivatives into products that are less mutagenic. For example, P. simplicissimum (Marr et al., 1996) and P. chrysogenum (Leitao et al., 2007) can degrade phenol compounds. 2-, 3- and 4-Chlorophenol can be metabolized by P. frequentans Bi 7/2, P. simplicissimum and P. simplicissimum (Hofrichter et al., 1994; Marr et al., 1996).
P. decumbens have high catabolic enzyme activity and can utilize a lot of simple aromatic compounds, including detoxification of olive mill wastewater (Borja et al., 1992). Penicillius also take part in the remediation of vinasses (Jimenez et al., 2005) and coffee residues (P. curstosum, P. verrucosum, P. crustosum, P. restrictum, P. implicatum, P. citrinum) (Fujii and Takeshi, 2007) in liquid wastes.
Different Fusarium strains are adapted to higher concentrations of heavy metals (Co, Cu, and Mn) and can be used as natural biosorbents for bioremediation (Poonam et al., 2016). Fusarium sp. FP, Arthrinium sp. FB, and Phoma sp. FR can remove Thallium Tl (I) from aqueous solutions (Mo et al., 2022). Combination of Fusarium sp. ZH-H2 and starch offers a suitable alternative for bioremediation of aged polycyclic aromatic hydrocarbon (PAH)-contaminated soil in coal mining areas (Zhao et al., 2019).
Fungi Verticillium sp. exhibited the successful degradation of pesticides, such as chlorpyrifos in different mediums (Farhan et al., 2021). Verticillium insectorum J3 have efficient biosorption mechanism for Pb (II) and Zn (II) and can be used for the development of biotreatment technologies for heavy metal-polluted waste (Feng et al., 2018).
The phylum Basidiomycota include filamentous fungi with enormous bioremediation potential. Phanerochaete chrysosporium is perhaps the most widely used basidiomycete species for bioremediation. A wide spectrum of hydrocarbons can be bioremediated by Phanerochaete chrysosporium thanks to their various enzyme systems (Paszczynski and Crawford, 1995; Cameron et al., 2000). These fungi are able to degrade anthracene (Vyas et al., 1994; Novotny et al., 2004), naphthalene (Mollea et al., 2005), phenanthrene (Bumpus, 1989), pyrene (Novotny et al., 2004) and pentachlorophenol (Bumpus, 1989; Lamar and Dietrich, 1990; 1992). Such pollutants as DTT, lindane (Bumpus, 1989) and 2,4,6-Trinitrotoluene (Fernando et al., 1990) can also be completely degraded by P. chrysosporium. A high level of polycyclic aromatic hydrocarbon biodegradation was reported for such fungi as Xerotus discolor (Acevedo et al., 2011), Bjerkandera species (Matsubara et al., 2006), Irpex lacteus (Bhatt et al., 2002; Cajthaml et al., 2006), Phellinus sp. (Arun and Eyini, 2011), Schizophyllum commune Fr (Matsubara et al., 2006), Stropharia coronilla (Steffen, 2000). Pentachlorophenol can be degraded by Agrocybe perfecta, T. villosa (Sw.) (Machado et al., 2005), T. hirsuta and Ceriporiopsis subvermispora (Lamar and Dietrich, 1990; 1992), aromatic hydrocarbons (AH) can be remediated by Allescheriella sp. (D'Annibale et al., 2006; D'Annibale et al., 2007), nitrobenzene and anthracene are reported to be degraded by Coriolopsis polyzona (Vyas et al., 1994) and C. trogii (Levin et al., 2003).
Some well-studied species of filamentous fungi representatives of Zygomycota are used for bioremediation (Olicon-Hernandez et al., 2017). Cunninghamella elegans have been extensively used as model fungi in different studies on the metabolism of xenobiotics (Asha and Vidyavathi, 2009). Cunnninghamella spp., have the ability to degrade insecticides and dyes (Cha et al., 2001; Zhao et al., 2020). It was shown that Cunninghamella elegans ATCC36112 could effectively degrade the pesticide (diazinon) mediated by cytochrome P450 (Zhao et al., 2020). Another genus of the fungus Zygomycota, Mortierella, obviosly сould considered as additional bioremediation agent of cooper contaminated soil (Fu et al., 2022). Content of hydrocarbons of a crude oil and PAHs was decreased with Mucor spp., (Okoro et al., 2013; Gupta et al., 2015). The degradation of pyrene and polycyclic aromatic hydrocarbons (PAHs) residues was shown by Mucor mucedo immobilizing on carrier-corncob of contaminated agricultural soil (Hou et al., 2015). M. hiemalis decrease concentrations of diclofenac in the media (Esterhuizen-Londt et al., 2017). Thus, filamentous fungi of all groups Ascomycota, Basidiomycota, and Zygomycota have a good remedial potential and can be suitable for removing from polluted environments both pharmaceutical compounds and heavy metals, as well as oil hydrocarbons and other pollutants.
5 Recent progress in bioremediation by filamentous fungi
Anthropogenic activity leads to increasing pollution of the environment with a rising set of organic and inorganic compounds, deteriorating the state of the ecosystem. Filamentous fungi with a wide range of extracellular and intracellular enzymes, surfactant production and, biosorption properties and the ability to symbiosis can change the approaches to bioremediation and wastewater treatment. Long remediation time and slow growth serves as limit factors the application mycoremediation, however, recent research offers an opportunity to overcome these challenges.
5.1 Wastewater treatment of pharmaceutical compounds
The excessive amount of pharmaceutical compounds released into environments poses a long-term risk to humans and wildlife, even under low concentration (Angeles et al., 2020; Akerman-Sanchez and Rojas-Jimenez, 2021). Large range of common pharmaceutical compounds were found in wastewater treatment plants and effluent waters, including analgesics, antidiabetics, anti-inflammatories, antibiotics, anti-hypertensives, beta-blockers, diuretics, and lipid regulators negativelly. These compounds negatively impact on environmental and aquatic ecosystems health (Olicon-Hernandez et al., 2019; Akerman-Sanchez and Rojas-Jimenez, 2021). Filamentous fungi belong to the microorganisms responsible for the degradation of most organic compounds polluting environment (Bulkan et al., 2020; Ferreira et al., 2020). Increasing studies indicate that filamentous fungi can transform many of pharmaceutical compounds, including antibiotics, anti-inflammatories, β-blockers, etc., (Bulkan et al., 2020; Alamo et al., 2021; Kasonga et al., 2022). Recent studies have shown prospects of biotechnology based on filamentous fungi for almost complete elimination of a wide range of pharmaceutical compounds that may have ecotoxic effects in the environment (Table 2). Reports (Dalecka et al., 2020a; Dhiman et al., 2022) have shown that filamentous fungi can efficiently remove pharmaceutical compounds at level of 78%–100% from a synthetic wastewater media. Research of past years mostly have shown a slow rate of degradation of pharmaceutical compounds with a treatment duration of more than 26 days (Li et al., 2015; Badia-Fabregat et al., 2016; Mir-Tutusaus et al., 2017). However, studies with the white-rot fungus Trametes versicolor (Shreve et al., 2016) and recent results with species of Aspergillus, Mucor, Penicillium, Rhizopus, Trametes and Trichoderma demonstrate (Mohapatra et al., 2022; Shourie and Vijayalakshmi, 2022) demonstrated significant degradation with high removal rates in 4 h–10 days (Olicon-Hernandez et al., 2019; Dalecka et al., 2020b; Alamo et al., 2021; Kasonga et al., 2022). Using adsorption to solid supports, specifically, polyurethane foam carriers for immobilized white rot fungus Trametes versicolor resulted in 99.9% diclofenac removal after even 4 h during batch experiments (Stenholm et al., 2019). The choice of carrier and the level of cell immobilization are importance in this process. Whereas, experiments that utilized polyethylene-carriers with negligible immobilization of T. versicolor, a 98% total diclofenac removal was achieved only after one week investigation. To optimize the elimination of pharmaceuticals, the type of bioreactor acts as another important factors. The fungal fixed-bed reactor could be more suitable than the stirred tank reactor to remove pharmaceutical compounds from wastewater, since it was able to remove PhACs to a great extent and simultaneously detoxify real wastewater. A trickle-bed bioreactor (TBB) using fungal biomass immobilized on rice husks, achieved an elimination of 88.6% and 89.8% in synthetic and real wastewater, respectively (Tormo-Budowski et al., 2021). Additionally, toxicological tests showed a decrease in the hospital wastewater’s toxicity after the treatment in the TBB. The exploring with focusing on optimizing the conditions inside the bioreactors reveals as meangfull for the effective mycoremediation process (Akhtar and Mannan, 2020). A novel idea with effective and fast treatment of actual pharmaceutical wastewater containing β-lactam antibiotics was presented in recent research (Ji et al., 2021). Aspergillus niger mycelial pellets were used as biological carriers for displaying β-lactamase on the cell surface of fungi. This A. niger-Bla system significantly improved the removal of antibiotics for > 60% and demonstrated complete degradation of amoxicillin and ampicillin within only 1 h, and cefamezin at 80.45%.
TABLE 2.
Pharmaceutical compounds (PhCs) removal efficiency by filamentous fungi.
| Strain | Pollutant | Removal efficiency % | Time | References |
|---|---|---|---|---|
| Aspergillus luchuensis | Diclofenac | > 99.9% | 10 days | Dalecka et al. (2020b) |
| Aspergillus niger | diclofenac | 78–100 | 24 h | Kasonga et al. (2022) |
| Mucor circinelloides | diclofenac | 78–100 | 24 h | Kasonga et al. (2022) |
| ibuprofen | 98 | 2 days | ||
| Penicillium oxalicum | diclofenac | 99 | 24 h | Olicon-Hernandez et al. (2019) |
| Rhizopus microspores | carbamazepine | 87 | 10 days | Kasonga et al. (2022) |
| diclofenac | 78–100 | 24 h | ||
| Rhizopus sp. | carbamazepine | 100 | 9 days | Kasonga et al. (2022) |
| diclofenac | 100 | |||
| ibuprofen | 100 | |||
| Trametes versicolor | 45–79 of different PhCs | 69 | – | Alamo et al. (2021) |
| 16 of different PhCscarbamazepine | 88.6–89.8 | 14 days | Tormo-Budowski et al. (2021) | |
| trimethoprim | 34–82 | 48 h | Alharbi et al. (2019) | |
| sulfamethoxazole | 39–95 | 48 h | Stenholm et al. (2019) | |
| diclofenac | 49–56 | 48 h | ||
| 98–99 | 4 h - 1 week | |||
| Trametes polyzona | diclofenac | 78–100 | 24 h | Kasonga et al. (2022) |
| Trichoderma longibrachiatum | diclofenac | 78–100 | 24 h | Kasonga et al. (2022) |
Degradation of individual pharmaceuticals was more efficient than their elimination from а mixture. For example, diclofenac (5 mg/L) was completely removed by Trametes versicolor to below its detection limit (1 mg/L) within 8 h in the individual experiment vs. after 24 h in a mixture with the other pharmaceuticals (Alharbi et al., 2019). A similar trend was visible with trimethoprim (TMP), carbamazepine (CBZ), and sulfamethoxazole (SMX), with 95% vs. 39%, 82% vs. 34%, and 56% vs. 49% removal after 48 h with 5 mg/L of TMP, CBZ, and SMX individually or as mixtures, respectively.
Thе ability of filamentous fungi to degrade a wide variety of pharmaceutical compounds and their easy handling make them excellent tool for the bioremediation of emerging contaminants of pharmaceutical origin. The study of different carriers, such as solid ones for fungal cells or biological ones for their enzymes, type of fungal reactor serves as important role that can accelerate mycoremediation time and removal effeciency of pharmaceutical compounds and bring closer the time of their large-scale application for wastewater treatment.
5.2 Heavy metal treatment
Filamentous fungi can be considered as an important factor in bioremediation and heavy metal treatment due to their ability of biosorption, bioacumulation and metal recovery. Filamentous fungi capable to tolerating high metal concentrations, can decontaminate the environment encumbered with heavy metal pollutants (Manguilimotan and Bitacura, 2018; Kapahi and Sachdeva, 2019; Dusengemungu et al., 2021). Filamentous fungy can be used in recovery of many essential metals from various metal sources, including solid mine wastes and polluted soils, mine tailings and electronic waste, spent catalysts and low-grade ores solubilizing metals through the secretion of organic acids (Ghosh et al., 2020; Dusengemungu et al., 2021) (Table 3).
TABLE 3.
Heavy metal removal efficiency by filamentous fungi reported recently.
| Strain | Metal | Removal efficiency % | Waste | References |
|---|---|---|---|---|
| Aspergillus carbonarius | Al | 70–80 | Spent Catalyst | Das et al. (2019) |
| Aspergillus foetidus | Al | 88.43 | Spent Catalyst | Das et al. (2019) |
| Aspergillus fumigatus | Cd | 79.0 | Polluted soil | Khan et al. (2014) |
| Aspergillus nidulans mutant | Cu | 70.0 | Industrial effluents | Rissoni Toledo et al. (2021) |
| Aspergillus niger | Al | 80–81 | Spent Catalyst | Das et al. (2019) |
| Aspergillus niger and Penicillium simplicissium | Al | ≥ 98 | Bauxite | Shah et al. (2020) |
| Aspergillus niger | Cd | 98 | Polluted soil | Khan et al. (2014) |
| Aspergillus niger | U | 80 | Uranium mine | Sun et al. (2020a) |
| Aspergillus niger | Pb | 98.4 | Aqueous Solution | Nuban et al. (2021) |
| Pb | 100 | Sewage | Ghyadh et al. (2019) | |
| Aspergillus niger | Cr | 100 | Xu et al. (2021) | |
| Aspergillus tubingensis | Pb | 90.8 | Polluted soil | Tang et al. (2021) |
| Zn | 68.4 | |||
| Cr | 64.5 | |||
| Cladosporium sp. A | Mg | 74 | Mine tailings | Huerta-Rosas et al. (2020) |
| Ag | 67 | |||
| Cladosporium sp. B | Mg | 58 | Mine tailings | Huerta-Rosas et al. (2020) |
| Ag | 40 | |||
| Mucor hiemalis | Al, Cd, Co., Cr, Cu, Hg, Ni, Pb | > 81%–99% | Polluted water | Hoque and Fritscher (2019) |
| U, Zn | ||||
| Ag, Au, Ti | ||||
| Penicilium chrysogenum | Mg | 69 | Mine tailings | Huerta-Rosas et al. (2020) |
| Ag | 53 | |||
| Penicillium simplicissimum | Cu | 90 | Waste electrical equipment | Arshadi et al. (2019) |
| Ni | 89 | |||
| Penicillium rubens | Cd | 98 | Polluted soil | Khan et al. (2014) |
| Rhizopus oligosporium | Cd | 65 | Sewage | Ghyadh et al. (2019) |
One of the most studied in this regard is the genus Aspergillus which demonstrates high leaching and removal capacity, as well as multi-metal tolerant properties. The removal capacity of A. niger was reached at 100% from Cd and Pb polluted sewage in 6 days and only 65% was reached when using Rhizopus oligosporium (Ghyadh et al., 2019). Aspergillus species are known for their ability to tolerate different metals such as Al, Cd, Co, Cr, Cu, Fe, Mn, Mg, Ni, Pb, Zn, and U (Dusengemungu et al., 2021; Naveen Kumar and Prakash, 2021). A unique microbial biotechnology for simultaneous bioremediation and biomining of twelve ionic metals overcoming the obstacles of multimetal toxicity to microbes were described. Three new strains of Mucor hiemalis was demonsrated multimetal-resistance, hyper-accumulation and elicitation power, intracellular fixing and deposition of mercury as nanospheres in sporangiospores (Hoque and Fritscher, 2019). Microbiomes, germinated spores and dead insoluble cellwalls of these strains removed > 81%–99% of applied Al, Cd, Co, Cr, Cu, Hg, Ni, Pb, U, and Zn simultaneously and furthermore enriched precious Ag, Au and Ti from water all within 48 h (Hoque and Fritscher, 2019).
The eco-friendliness and cost-effectiveness of recent advances in metal biorecovery and bioremediation by fungi show that filamentous fungi have an exceptional impact for industrial-scale bioremediations in future operations. Problems of optimization of metal recovery technology, how to extract metals from the cells after bioaccumulation/biosorption, as well as exploration of optimal alternative sources of energy for fungal growth are challenges for the current research community, that will be necessary to overcome for commercialization fungal bioremediation.
5.3 Oil hydrocarbons bioremediation
The use of microorganisms for bioremediation and restoration of oil hydrocarbons contaminated soils and aquatic ecosystems have the challenge that a huge amount of petroleum hydrocarbons accumulated in environments that are of great toxicity (Varjani, 2017; Chicca et al., 2022). Bioremediation of crude oil polluted environments is often limited by the scarcity microorganisms with complementary substrate specificity required for degrading different hydrocarbons (Varjani, 2017; Anno et al., 2021; Chicca et al., 2022). Filamentous fungi are reported for their ability to degrade hydrocarbon pollutants, similar to many bacteria (Widdel and Rabus, 2001; Xu et al., 2018; Zahri et al., 2021), yeasts (Rusyn et al., 2003; Farag and Soliman, 2011; Al-Dhabaan, 2021) and algae (Al-Hussieny et al., 2020; Anno et al., 2021; Ghodrati et al., 2021). Depending on the strain of filamentous fungi characterized the rate of crude oil removal efficiencies ranged from 30.4% to 98.1% during 7–10 days (Table 4). Consortia of filamentous fungi and bacteria e.g. Aspergillus and Bacillus, was more effective in bioremediation and reached 99.0% in comparison with monocultures which utilized 98.0% and 20.0% in 7 days (Perera et al., 2021). The ability of Penicillium sp. to tolerate oil pollutants from 2.5% to 10% and degradate more than 50% suggest that it could be employed as an bioremediation agent for restoring the ecosystem when contaminated by oil (Vanishree et al., 2014; Dirisu et al., 2018). Among 84 filamentous fungal isolates belonging to eight different genera, Penicillium polonicum AMF16, P. chrysogenum AMF47 and two isolates affiliated to P. cyclopium, were determined as the most promising isolates for bioremediation of crude oil pollution (1%–5%) in the marine environment within the frame of bioaugmentation or biostimulation processes (Maamar et al., 2020). Degradation of petroleum hydrocarbons both by single filament strains and in synergism with bacteria allows progressive increases in bioremediation of contaminated sites.
TABLE 4.
Bioremediation of crude oil and hydrocarbons by filamentous fungi reported recently.
| Strain | Pollutant | Removal efficiency % | Time | References |
|---|---|---|---|---|
| Aspergillus sp. RFC- 1 | crude oil (1%) | 60.3 | 7 days | Al-Hawash et al. (2019) |
| naphthalene | 97.4 | |||
| phenanthrene | 84.9 | |||
| pyrene | 90.7 | |||
| Aspergillus ustus HM3 | crude oil (2%) | 30.4 | 10 days | Benguenab and Chibani (2021) |
| Aspergillus sp. MM1 and Bacillus sp. MM1 | crude oil (8 g/L) | 99.0 | 7 days | Perera et al. (2021) |
| Mucor sp | crude oil | 65.0 | 3 weeks | Dirisu et al. (2018) |
| Penicilium sp | crude oil | 55.0 | 3 weeks | Dirisu et al. (2018) |
| Purpureocillium lilacinum HM4 | crude oil (2%) | 44.5 | 10 days | Benguenab and Chibani (2021) |
6 Fungal byproduct recovery
6.1 Traditional fungal byproducts
Wastewaters containing pollutants such as pharmaceutically active compounds, heavy metals, aliphatic and polycyclic aromatic hydrocarbons, herbicides, pesticides, surfactants, dyes, personal care products and others can be purified by filamentous fungi (Ferreira et al., 2020; Morsi et al., 2020). Waste or side streams generated from the agricultural, forest, industrial, food and municipal sectors can be converted to value-added components by these organisms. Side streams are generally rich in carbohydrates (lignocellulose, starch, sugar), proteins, fat, minerals, etc., that can be converted to a range of value-added components such as fungal biomass, ethanol, hydrolytic enzymes, and organic acids.
Wastewaters, in contrast to side streams, contain minor amounts of carbohydrates and nitrogen sources commonly used by filamentous fungi. Thus, for removal of pollutants, additional nutrients should be spent to support growth of filamentous fungi. As a result, fungal biomass and enzymes are only obtained value-added products. The use of fungal biomass extracts as nutrients can be applied to enrich wastewaters or side streams where nutrient supplementation is required (Nair et al., 2017). It is necessary to classify side streams aimed for bioremediation, focused on removal of pollutants, and those aimed for valorization into products, for integration into anthropogenic activities. Some strategies were proposed for integration of valorization of side streams with removal of pollutants from wastewaters by filamentous fungi. For example, paper and pulp industries produce side streams that have been considered in both perspectives (Ferreira et al., 2020). However, there are no commercial processes presently known.
Filamentous fungal biomass is the main value-added component. It is generated independently of the intended valuable product to be produced. Fungal biomass grown in different side streams contains 40%–60% protein, as well as profiles of amino acids and polyunsaturated fatty acids similar to those in fishmeal and soybean meal, which are the main protein sources for animal feed. Several representatives of filamentous fungi, e.g., A. oryzae, Fusarium venenatum, Monascus purpureus, Neurospora intermedia, Rhizopus microsporas, Rhizopus oligosporus, and Rhizopus oryzae have been used for production of fermented foods, and are recognized as GRAS (Generally recognized as safe) microorganisms (Ferreira et al., 2020; Sar et al., 2022b). Their GRAS status is a necessary point for consideration of fungal biomass for feed applications. Filamentous fungi have been used for biomass production on a variety of food industry side-streams such as fish industry wastewaters, vinasse, olive oil mill wastewater, thin stillage, etc., (Ferreira et al., 2015; Karimi et al., 2019; Sar et al., 2020a; Sar et al., 2020b; Sar et al., 2021). It was described that fungal biomass production by Rhizopus delemar at an industrial scale contained 53% crude protein from edible potato protein liquor (Sar et al., 2022b). The R. delemar fungal biomass can be a promising raw material for feed and food production, since its protein and fatty acid profiles include 41% essential amino acids and 33% polyunsaturated fatty acids (Sar et al., 2022a).
Despite the fact that fungal biomass first of all is considered as a promising material for feed and food production, other potential applications of residual fungal biomass are proposed. The cell wall of fungal biomass contain chitosan, chitin, and β-glucans, with confirmed immune-stimulation activities when used in feed recipes (Karimi et al., 2018). Additionally, potential applications of chitin and chitosan in pharma, cosmetics, bioplastics, biopolymers, and agricultural sectors are well described (Isaza-Perez et al., 2020).
The filamentous fungus Mucor indicus provided promising results in heavy metal removal from wastewaters (Javanbakht et al., 2011). The biomass of this fungus has shown a great potential to be used as a rich nutritional source. Cells of Mucor indicus are great sources of chitosan and polyunsaturated fatty acids particularly γ-linolenic acid (Karimi and Zamani, 2013). Moreover, this fungus produced high ethanol production levels during fermentation of glucose and lignocellulosic hydrolysates, which is an important value-added product. Increase of ethanol yield from glucose was reached by supplementation with fungal biomass autolysates (Asachi et al., 2011).
Aspergillus oryzae and Neurospora intermedia were employed for the production of ethanol and high-protein biomass by cultivation on enzymatically liquefied bread-waste medium (Kawa-Rygielska et al., 2022). The cultivation of Neurospora intermedia in wheat-derived thin stillage resulted in extra ethanol production (Ferreira et al., 2015). Fusarium oxysporum is among the few filamentous fungi able to ethanol production directly from lignocellulose biomass. This fungus revealed the ability to produce ethanol from Ficus fruits employing a mild hydrothermal pretreatment without supplementing any extraneous enzymes using (Nongthombam et al., 2022). A cellulolytic thermophilic filamentous fungus Myceliophthora thermophila revealed huge potential for ethanol production from glucose and cellobiose. Recombinant strains expressing the cellodextrin transport system from N. crassa and alcohol dehydrogenase from Saccharomyces cerevisiae produced increased amounts of ethanol from cellobiose during fermentation at 48°C (Li et al., 2020).
Filamentous fungi play a significant role in the production of a wide range of lignocellulolytic enzymes, which are important value-added products (Ghosh, 2015; Ghosh et al., 2021). Such hydrolytic and oxidative enzymes are used for the production of important metabolites. Wild type strains have limited efficiency in enzyme production. Despite great progress in optimizing the cultivation conditions for enzyme production (Ferreira et al., 2016), molecular engineering methods were used for more pronounced improvement of the fungal production of lignocellulolytic enzymes. Constitutive production of pectinases (Alazi et al., 2019) and arabinolytic enzymes (Reijngoud et al., 2019) in Aspergillus niger were reached by point mutation in GaaR and AraR regulators, respectively. Deletion of the negative regulator CreA resulted in higher production of hemicellulase (Robl et al., 2018) in A. niger. Overexpression of mutated AraR in Penicillium oxalicum lead to constitutive production of α-L-arabinofuranosidase (Gao et al., 2019). Deletion of the regulatory gene Atf1 increased cellulase and xylanase production in P. oxalicum (Zhao et al., 2019). Deletion of repressors SxlR and Rce1 in Trichoderma reesei significantly stimulated xylanase (Liu et al., 2017) and cellulase (Cao et al., 2017) activities, respectively. Constitutive and increased production of xylanases and cellulases was also reached using the recombinant Trichoderma reesei overexpressing an artificial transcription activator Xyr1-Cre1b (Zhang et al., 2017).
Organic acids, predominantly citric, gluconic and itaconic acids, are value-added compounds produced by filamentous fungi (Ferreira et al., 2016). Citric acid production is the second largest fermentation product after ethanol production (Dhillon et al., 2013). Citric acid is mostly used in the food industry. Currently, A. niger is used for large scale production of this acid from sucrose or glucose containing substrates such as molasses or glucose syrup (Tsay and To, 1987; Show et al., 2015). To decrease citric acid production costs, other cost-effective substrates were investigated, e.g. apple pomace, orange processing waste, starch, rape seed oil (Behera, 2020). Huge efforts were made to optimize the fermentation process (Barrington et al., 2009). A significant increase of citric acid production was reached with metabolic engineering approaches of A. niger directed to carbon utilization and respiratory chain improvement, precursor biosynthesis enhancement, by-product removal and feedback inhibition reduction improvement (Tong et al., 2019).
Gluconic acid is an important biotechnological product used in food, feed, beverage, textile, pharmaceutical and construction industries. Gluconic acid produced with A. niger from glucose can be done at an industrial scale (Ramachandran et al., 2006; Yan et al., 2022). Other representatives of Aspergillus and Penicillium spp. have also been recognized as robust producers of this acid (Ma et al., 2022). Environmental and economical friendly processes of gluconic acid production trended to replace those using glucose and sucrose form waste products (e.g., hydrolysates of sugarcane, corn stover, corn cob, tea waste, starch, inulin, whey, figs, bananas, grapes, surpluses, wastepaper and lignocellulose) (Ferreira et al., 2016).
Itaconic acid is widely used in chemical synthesis industries (Wierckx et al., 2020). It was reported that itaconic acid suitable for the synthesis of antimicrobial biopolymers, drug carriers, intelligent food packaging, superabsorbent polymers, hydrogels in water treatment and analysis (Teleky and Vodnar, 2021). Itaconic acid production is carried out by fermentation more commonly using Aspergillus terreus from molasses or glucose (Huang et al., 2021). But currently several filamentous fungi are genetically engineered to produce this acid in high quantities and on different bio-wastes substrates (Sun L. et al., 2020b). Overall, filamentous fungi have great potential for further applications as robust producers of value-added products.
6.2 Mycosynthesis of nanoparticles
Furthermore, fungal biomass grown in heavy metal-rich media can be used as a source of nanocatalysts and nanoparticles (NPs) with potential application in chemical industries and micropollutant removal approaches (Huang et al., 2018; Kratosova et al., 2019). Mycosynthesis of metal-containing nanoparticles is alternative to conventional physico-chemical processes (Sebesta et al., 2022). This is a innovative biological approach to nanoparticle synthesis which leads its beginning from 2001 (Mukherjee et al., 2001). Then, formation of silver and gold nanoparticles using the fungus Verticillium was shown below the cell wall surface due to reduction of the metal ions by enzymes present in the cell wall membrane. NPs can be produced extracellular, outside the cell walls, which makes their separation from the biomass a simple process (Yadav et al., 2015; Priyadarshini et al., 2021). Some NPs synthesize intracellular, on the cell walls and even on the inner side of the cell walls (Mukherjee et al., 2001; Priyadarshini et al., 2021). Currently, nanoparticles of a wide range of different chemical compounds are known which are synthesised with the help of а filamentous fungi, such as metal Ag, Au, Cu, Pb, Pt, and bimetallic Ag-Au NPs, and also oxides, such as BaTiO3, Bi2O3, CoFe2O4, Co3O4, Fe2O3, Fe3O4, NiO, TiO2, ZnO, and ZrO2 (Mousa et al., 2021; Priyadarshini et al., 2021; Sebesta et al., 2022). Myconanoparticles can be used in various fields, i.e., in biomedicine, antimicrobial applications, catalysis, biosensing, mosquito control, and precision agriculture (Sebesta et al., 2022).
7 Problems associated with the fungal treatment systems and future prospects
Microscopic examination of hundreds of biomass samples from industrial effluent treatment plants and municipal wastewater treatment plants have revealed different operational issues. One of the main disadvantages of fungal reactors is that their wastewater treatment is not cost-effective (Espinosa-Ortiz et al., 2016). Others include filamentous bulking, toxicity, and dispersed growth, while fungal scattered mycelium typically causes bioreactor operation problems, including foaming, higher mixing and oxygen supply requirements, and growth on reactor walls and agitators.
Currently, various technologies are used to determine the toxicological mechanism of action of mycotoxins of these filamentous fungi. Among them, molecular biology techniques allow understanding the basic structure of nucleic acids and other molecules and help mimic their natural functions in both in vivo and in vitro studies. They are used to assess various aspects of cytotoxicity, cellular responses, gene expression, and the activation of specific signaling pathways and transcription factors (Manzoni et al., 2018). Notably, the use of zebrafish for in vivo toxicological research has been rapidly expanding (de Castro et al., 2016; Della Torre et al., 2018; Parenti et al., 2019; Taufek et al., 2020; Wan-Mohtar et al., 2020; Wan-Mohtar et al., 2021). Due to their easy bred, reared, easy laboratory maintenance, stability and ease of stable genetic manipulations, zebrafish are positioned as an ideal vertebrate model for in vivo studies compared to other vertebrates (Tonelli et al., 2020). For example, a toxicity study of exopolysaccharides, obtained from medicinal mushroom mycelial extracts, conducted on zebra fish embryo has been used as safety screening approach prior to pre-clinical testing and considered as national and international standards (Usuldin et al., 2021).
Fungi are not typically found in substantial amounts in aerobic wastewater treatment systems, but given the correct set of growth or environmental condition, fungi can grow out of control, negatively affecting treatments and effluent quality (Mir-Tutusaus et al., 2018). Fungi, on average, require a longer hydraulic retention time to remove pollutants from wastewater than bacteria. This complicates incorporating a fungal treatment step into a normal wastewater treatment system (Mir-Tutusaus et al., 2018). Some wastewaters cannot be treated with fungi. For example, fungal wastewater treatment systems are unsuitable for anoxic treatment of groundwater where oxygen is lacking. Fungi require aerobic conditions to grow and carry out other cellular activities (Trueba-Santiso et al., 2017). In addition, fungal treatment systems need additional nutrients, even while organic micro pollutants include carbon, some fungi require an extra assimilable carbon source for survival and growth (Mir-Tutusaus et al., 2018).
Bacteria outperform fungi and not just in natural settings but also in bioreactors. When compared to fungi, bacteria can survive a wider range of environmental conditions, proliferate faster, and destroy a wider range of contaminants (Espinosa-Ortiz et al., 2016). Another problem associated with the fungal treatment systems is in controlling bacterial contamination, which is one of the most common issues in the non-aseptic fungal treatment of wastewater (Espinosa-Ortiz et al., 2016). The proliferation of bacteria during fungal treatment of wastewater causes strong competition for the limited organic substrate, which impacts fungal metabolism because bacterial growth occurs on fungal filaments as support media (Badia-Fabregat et al., 2017; Hu et al., 2022). In addition, in industrial applications, wastewater sterilization is quite expensive. As a result, sustaining fungal biomass dominance in wastewater necessitates developing technologies to boost fungal competitive advantages over the bacteria microflora, while inhibiting growth of bacteria.
Fungal-bacterial mix for pollutant removal in fungal-pelleted bioreactors has been found in some situations to even improve the breakdown of some pollutants. As a result, further research is needed in the creation of fungal-bacterial symbiosis consortia for wastewater treatment. Additionally, more research is needed to expand the practicability of fungi in wastewater treatment, evaluate the economic, environmental, and technical aspects of fungal biofilm technologies in removing recalcitrant chemicals, and build a fungal biofilm immobilization technology for wastewater treatment.
8 Conclusion
The research that has been done up to now, and the applications that could be developed, indicate that only the surface of using fungi in sustainable wastewater remediation has been scratched. Research has only been done in a relatively limited number of geographical locations compared to other regions of the globe, and only a small percentage of fungi of the number of species out there, have been tested or exploited. Actually, most likely candidates for rehabilitation of specific problems were selected from a limited pool. Numerous opportunities exist, including the production of revenue in the form of byproducts contributes to the sustainability of these fungal applications, especially for a circular bio-economy model. A better understanding of the genes involved in these properties of used fungi, further opens up opportunities, enhances understanding, could solve some of the current problems experienced, and opens up recombinant DNA solutions. The ease by which fungi can be integrated in nanoparticle applications also enhances their abilities. It is thus likely that with the correct mindset solutions to the current problems faced in this field can be found in future.
Acknowledgments
The authors are grateful to the University of the Free State, South Africa, the Polish National Science Center (NCN) project nos 2021/41/B/NZ1/01224 and 2020/37/B/NZ1/02232, grant N3_053, National Research Foundation of Ukraine grant 2020.01/0090, State Scientific and Technical Program of Uzbekistan (2021–2024) and Ministry of Innovative Development of the Republic of Uzbekistan (Projects no. AL 2021090820) for supporting this study. The authors are thankful to Dr. Swagata Ghosh, Assistant Professor of English, Symbiosis Institute of Technology, Symbiosis International University, Pune, India for editing this manuscript.
Author contributions
SG and IR conceptualized the idea. SG, IR, OD, KD, HO, MG, and YG collected, analyzed the data and drafted the manuscript. All the authors read, edit and approved the manuscript for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Acevedo F., Pizzul L., Castillo M. D., Cuevas R., Diez M. C. (2011). Degradation of polycyclic aromatic hydrocarbons by the Chilean white-rot fungus Anthracophyllum discolor . J. Hazard. Mater. 185, 212–219. 10.1016/j.jhazmat.2010.09.020 [DOI] [PubMed] [Google Scholar]
- Adegunlola G. A., Oloke J. K., Majolagbe O. N., Adebayo E. A. (2012). Microbial desulphurization of crude oil using Aspergillus flavus . Eur. J. Exp. Biol. 2, 400–403. [Google Scholar]
- Ahmed S. F., Mofijur M., Nuzhat S., Chowdhury A. T., Rafa N., Uddin M. A., et al. (2021). Recent developments in physical, biological, chemical, and hybrid treatment techniques for removing emerging contaminants from wastewater. J. Hazard. Mater. 416, 125912. 10.1016/j.jhazmat.2021.125912 [DOI] [PubMed] [Google Scholar]
- Akerman-Sanchez G., Rojas-Jimenez K. (2021). Fungi for the bioremediation of pharmaceutical-derived pollutants: A bioengineering approach to water treatment. Environ. Adv. 4, 100071. 10.1016/j.envadv.2021.100071 [DOI] [Google Scholar]
- Akhtar N., Mannan M. A. (2020). Mycoremediation: Expunging environmental pollutants. Biotechnol. Rep. (Amst) 26, e00452. 10.1016/j.btre.2020.e00452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ansari M. M., Li Z., Masood A., Rajaselvam J. (2022). Decolourization of azo dye using a batch bioreactor by an indigenous bacterium Enterobacter aerogenes ES014 from the waste water dye effluent and toxicity analysis. Environ. Res. 205, 112189. 10.1016/j.envres.2021.112189 [DOI] [PubMed] [Google Scholar]
- Al-Dhabaan F. A. (2021). Isolation and identification of crude oil-degrading yeast strains from Khafji oil field, saud Arabia. Saudi J. Biol. Sci. 28, 5786–5792. 10.1016/j.sjbs.2021.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hawash A. B., Zhang X. Y., Ma F. Y. (2019). Removal and biodegradation of different petroleum hydrocarbons using the filamentous fungus Aspergillus sp. RFC-1. Microbiologyopen 8 (1), e00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hussieny A. A., Imran S. G., Jabur Z. A. (2020). The use of local blue-green algae in the bioremediation of hydrocarbon pollutants in wastewater from oil refineries. Plant Arch. 20, 797–802. [Google Scholar]
- Alamo A. C., Pariente M. I., Sanchez-Bayo A., Puyol D., Rodriguez R., Orales V. M., et al. (2021). Assessment of trametes versicolor, isochrysis galbana, and purple phototrophic bacteria for the removal of pharmaceutical compounds in hospital wastewater. Adv. Environ. Eng. Res. 02, 1. 10.21926/aeer.2104027 [DOI] [Google Scholar]
- Alazi E., Niu J., Otto S. B., Arentshorst M., Pham T. T. M., Tsang A., et al. (2019). W361R mutation in GaaR, the regulator of D-galacturonic acid-responsive genes, leads to constitutive production of pectinases in Aspergillus niger . Microbiologyopen 8, e00732. 10.1002/mbo3.732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alharbi S. K., Nghiem L. D., Van De Merwe J. P., Leusch F. D. L., Asif M. B., Hai F. I., et al. (2019). Degradation of diclofenac, trimethoprim, carbamazepine, and sulfamethoxazole by laccase from Trametes versicolor: Transformation products and toxicity of treated effluent. Biocatal. Biotransformation 37, 399–408. 10.1080/10242422.2019.1580268 [DOI] [Google Scholar]
- Ali M. I. A., Khalil N. M., Abd El-Ghany M. N. (2012). Biodegradation of some polycyclic aromatic hydrocarbons by Aspergillus terreus . Afr. J. Microbiol. Res. 6, 3783–3790. [Google Scholar]
- Alsharari S. F., Tayel A. A., Moussa S. H. (2018). Soil emendation with nano-fungal chitosan for heavy metals biosorption. Int. J. Biol. Macromol. 118, 2265–2268. 10.1016/j.ijbiomac.2018.07.103 [DOI] [PubMed] [Google Scholar]
- Alvarez-Barragan J., Dominguez-Malfavon L., Vargas-Suarez M., Gonzalez-Hernandez R., Aguilar-Osorio G., Loza-Tavera H. (2016). Biodegradative activities of selected environmental fungi on a polyester polyurethane varnish and polyether polyurethane foams. Appl. Environ. Microbiol. 82, 5225–5235. 10.1128/aem.01344-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles L. F., Mullen R. A., Huang I. J., Wilson C., Khuniar W., Sirotkin H. I., et al. (2020). Assessing pharmaceutical removal and reduction in toxicity provided by advanced wastewater treatment systems. Environ. Science-Water Res. Technol. 6, 62–77. 10.1039/c9ew00559e [DOI] [Google Scholar]
- Anno F. D., Rastelli E., Sansone C., Brunet C., Ianora A., Anno A. D. (2021). Bacteria, fungi and microalgae for the bioremediation of marine sediments contaminated by petroleum hydrocarbons in the omics era. Microorganisms 9, 1695. 10.3390/microorganisms9081695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshadi M., Nili S., Yaghmaei S. (2019). Ni and Cu recovery by bioleaching from the printed circuit boards of mobile phones in non-conventional medium. J. Environ. Manag. 250, 109502. 10.1016/j.jenvman.2019.109502 [DOI] [PubMed] [Google Scholar]
- Arun A., Eyini M. (2011). Comparative studies on lignin and polycyclic aromatic hydrocarbons degradation by basidiomycetes fungi. Bioresour. Technol. 102, 8063–8070. 10.1016/j.biortech.2011.05.077 [DOI] [PubMed] [Google Scholar]
- Asachi R., Karimi K., Taherzadeh M. J. (2011). Fungal autolysate as a nutrient supplement for ethanol and chitosan production by Mucor indicus. Biotechnol. Lett. 33, 2405–2409. 10.1007/s10529-011-0725-2 [DOI] [PubMed] [Google Scholar]
- Asemoloye M. D., Tosi S., Dacco C., Wang X., Xu S. H., Marchisio M. A., et al. (2020). Hydrocarbon degradation and enzyme activities of Aspergillus oryzae and mucor irregularis isolated from Nigerian crude oil-polluted sites. Microorganisms 8, 1912. 10.3390/microorganisms8121912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asha S., Vidyavathi M. (2009). Cunninghamella - a microbial model for drug metabolism studies - a review. Biotechnol. Adv. 27, 16–29. 10.1016/j.biotechadv.2008.07.005 [DOI] [PubMed] [Google Scholar]
- Badia-Fabregat M., Lucas D., Pereira M. A., Alves M., Pennanen T., Fritze H., et al. (2016). Continuous fungal treatment of non-sterile veterinary hospital effluent: Pharmaceuticals removal and microbial community assessment. Appl. Microbiol. Biotechnol. 100, 2401–2415. 10.1007/s00253-015-7105-0 [DOI] [PubMed] [Google Scholar]
- Badia-Fabregat M., Lucas D., Tuomivirta T., Fritze H., Pennanen T., Rodríguez-Mozaz S., et al. (2017). Study of the effect of the bacterial and fungal communities present in real wastewater effluents on the performance of fungal treatments. Sci. total Environ. 579, 366–377. 10.1016/j.scitotenv.2016.11.088 [DOI] [PubMed] [Google Scholar]
- Balsano E., Schwartz K., Esterhuizen-Londt M., Hoque E., Lima S. P. (2015). Toxin resistance in aquatic fungi poses environmentally friendly remediation possibilities: A study on the growth responses and biosorption potential of mucor hiemalis EH5 against cyanobacterial toxins. Mycoses 58, 52–53. [Google Scholar]
- Barrington S., Kim J. S., Wang L., Kim J. W. (2009). Optimization of citric acid production by Aspergillus niger NRRL 567 grown in a column bioreactor. Korean J. Chem. Eng. 26, 422–427. 10.1007/s11814-009-0071-4 [DOI] [Google Scholar]
- Barros L. M., Macedo G. R., Duarte M. M. L., Silva E. P., Lobato A. K. C. L. (2003). Biosorption of cadmium using the fungus Aspergillus niger . Braz. J. Chem. Eng. 20, 229–239. 10.1590/s0104-66322003000300003 [DOI] [Google Scholar]
- Behera B. C. (2020). Citric acid from Aspergillus niger: A comprehensive overview. Crit. Rev. Microbiol. 46, 727–749. 10.1080/1040841x.2020.1828815 [DOI] [PubMed] [Google Scholar]
- Benguenab A., Chibani A. (2021). Biodegradation of petroleum hydrocarbons by filamentous fungi (Aspergillus ustus and Purpureocillium lilacinum) isolated from used engine oil contaminated soil. Acta Ecol. Sin. 41, 416–423. 10.1016/j.chnaes.2020.10.008 [DOI] [Google Scholar]
- Bhatt M., Cajthaml T., Sasek V. (2002). Mycoremediation of PAH-contaminated soil. Folia Microbiol. (Praha) 47, 255–258. 10.1007/bf02817647 [DOI] [PubMed] [Google Scholar]
- Bhunjun C. S., Niskanen T., Suwannarach N., Wannathes N., Chen Y. J., Mckenzie E. H. C., et al. (2022). The numbers of fungi: Are the most speciose genera truly diverse? Fungal Divers. 114, 387–462. 10.1007/s13225-022-00501-4 [DOI] [Google Scholar]
- Bilal M., Asgher M., Iqbal M., Hu H., Zhang X. (2016a). Chitosan beads immobilized manganese peroxidase catalytic potential for detoxification and decolorization of textile effluent. Int. J. Biol. Macromol. 89, 181–189. 10.1016/j.ijbiomac.2016.04.075 [DOI] [PubMed] [Google Scholar]
- Bilal M., Iqbal M., Hu H., Zhang X. (2016b). Mutagenicity and cytotoxicity assessment of biodegraded textile effluent by Ca-alginate encapsulated manganese peroxidase. Biochem. Eng. J. 109, 153–161. 10.1016/j.bej.2016.01.020 [DOI] [Google Scholar]
- Bills G. F., Yue Q., Chen L., Li Y., An Z., Frisvad J. C. (2016). Aspergillus mulundensis sp. nov., a new species for the fungus producing the antifungal echinocandin lipopeptides, mulundocandins. J. Antibiot. (Tokyo) 69, 141–148. 10.1038/ja.2015.105 [DOI] [PubMed] [Google Scholar]
- Binsadiq A. (1996). Biodegradation of gasoline by fungal flora isolated from Saudi Arabia soil. Geobios 23, 185–188. [Google Scholar]
- Borja R., Martin A., Maestro R., Alba J., Fiestas J. A. (1992). Enhancement of the anaerobic digestion of olive mill wastewater by the removal of phenolic inhibitors. Process Biochem. 27, 231–237. 10.1016/0032-9592(92)80023-v [DOI] [Google Scholar]
- Bulkan G., Ferreira J. A., Taherzadeh M. J. (2020). “Removal of organic micro-pollutants using filamentous fungi,” in Current developments in biotechnology and bioengineering: Emerging organic micro-pollutants. Editors Varjani S., Pandey A., Tyagi R. D., Ngo H. H., Larroche C. (Elsevier; ), 363–395. [Google Scholar]
- Bumpus J. A. (1989). Biodegradation of polycyclic hydrocarbons by Phanerochaete chrysosporium . Appl. Environ. Microbiol. 55, 154–158. 10.1128/aem.55.1.154-158.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajthaml T., Erbanova P., Sasek V., Moeder M. (2006). Breakdown products on metabolic pathway of degradation of benz[a]anthracene by a ligninolytic fungus. Chemosphere 64, 560–564. 10.1016/j.chemosphere.2005.11.034 [DOI] [PubMed] [Google Scholar]
- Camenzind T., Weimershaus P., Lehmann A., Aguilar‐Trigueros C., Rillig M. C. (2022). Soil fungi invest into asexual sporulation under resource scarcity, but trait spaces of individual isolates are unique. Environ. Microbiol. 24, 2962–2978. 10.1111/1462-2920.16012 [DOI] [PubMed] [Google Scholar]
- Cameron M. D., Timofeevski S., Aust S. D. (2000). Enzymology of Phanerochaete chrysosporium with respect to the degradation of recalcitrant compounds and xenobiotics. Appl. Microbiol. Biotechnol. 54, 751–758. 10.1007/s002530000459 [DOI] [PubMed] [Google Scholar]
- Cao Y., Zheng F., Wang L., Zhao G., Chen G., Zhang W., et al. (2017). Rce1, a novel transcriptional repressor, regulates cellulase gene expression by antagonizing the transactivator Xyr1 in Trichoderma reesei . Mol. Microbiol. 105, 65–83. 10.1111/mmi.13685 [DOI] [PubMed] [Google Scholar]
- Capotorti G., Digianvincenzo P., Cesti P., Bernardi A., Guglielmetti G. (2004). Pyrene and benzo(a)pyrene metabolism by an Aspergillus terreus strain isolated from a polycylic aromatic hydrocarbons polluted soil. Biodegradation 15, 79–85. 10.1023/b:biod.0000015612.10481.e6 [DOI] [PubMed] [Google Scholar]
- Carolina C. F., Kumar P. S., Ngueagni P. T. (2021). A review on new aspects of lipopeptide biosurfactant: Types, production, properties and its application in the bioremediation process. J. Hazard. Mater. 407, 124827. 10.1016/j.jhazmat.2020.124827 [DOI] [PubMed] [Google Scholar]
- Carstens L., Cowan A. R., Seiwert B., Schlosser D. (2020). Biotransformation of phthalate plasticizers and bisphenol A by marine-derived, freshwater, and terrestrial fungi. Front. Microbiol. 11, 317. 10.3389/fmicb.2020.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernansky S., Urik M., Sevc J., Littera P., Hiller E. (2007). Biosorption of arsenic and cadmium from aqueous solutions. Afr. J. Biotechnol. 6, 1932–1934. 10.5897/ajb2007.000-2293 [DOI] [Google Scholar]
- Cha C. J., Doerge D. R., Cerniglia C. E. (2001). Biotransformation of malachite green by the fungus Cunninghamella elegans . Appl. Environ. Microbiol. 67, 4358–4360. 10.1128/aem.67.9.4358-4360.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Mukherjee A., Khuda-Bukhsh A. R., Das T. K. (2014). Cadmium-induced oxidative stress tolerance in cadmium resistant Aspergillus foetidus: Its possible role in cadmium bioremediation. Ecotoxicol. Environ. Saf. 106, 46–53. 10.1016/j.ecoenv.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Chaudhary P., Beniwal V., Sharma P., Goyal S., Kumar R., Alkhanjaf A. a. M., et al. (2022a). Unloading of hazardous Cr and Tannic Acid from real and synthetic waste water by novel fungal consortia. Environ. Technol. Innovation 26, 102230. 10.1016/j.eti.2021.102230 [DOI] [Google Scholar]
- Chaudhary V. B., Aguilar-Trigueros C. A., Mansour I., Rillig M. C. (2022b). Fungal dispersal across spatial scales. Annu. Rev. Ecol. Evol. Syst. 53, 69–85. 10.1146/annurev-ecolsys-012622-021604 [DOI] [Google Scholar]
- Chen Z. H., Yin H., Peng H., Lu G. N., Liu Z. H., Dang Z. (2019). Identification of novel pathways for biotransformation of tetrabromobisphenol A by Phanerochaete chrysosporium, combined with mechanism analysis at proteome level. Sci. Total Environ. 659, 1352–1361. 10.1016/j.scitotenv.2018.12.446 [DOI] [PubMed] [Google Scholar]
- Chiang Y. R., Wei S. T. S., Wang P. H., Wu P. H., Yu C. P. (2020). Microbial degradation of steroid sex hormones: Implications for environmental and ecological studies. Microb. Biotechnol. 13, 926–949. 10.1111/1751-7915.13504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicca I., Becarelli S., Di Gregorio S. (2022). Microbial involvement in the bioremediation of total petroleum hydrocarbon polluted soils: Challenges and perspectives. Environments 9, 52. 10.3390/environments9040052 [DOI] [Google Scholar]
- Chu R., Li S., Zhu L., Yin Z., Hu D., Liu C., et al. (2021). A review on co-cultivation of microalgae with filamentous fungi: Efficient harvesting, wastewater treatment and biofuel production. Renew. Sustain. Energy Rev. 139, 110689. 10.1016/j.rser.2020.110689 [DOI] [Google Scholar]
- Clausen C. (2004). Improving the two-step remediation process for CCA-treated wood: Part I. Evaluating oxalic acid extraction. Waste Manag. 24, 401–405. 10.1016/j.wasman.2003.11.008 [DOI] [PubMed] [Google Scholar]
- Cregut M., Bedas M., Durand M.-J., Thouand G. (2013). New insights into polyurethane biodegradation and realistic prospects for the development of a sustainable waste recycling process. Biotechnol. Adv. 31, 1634–1647. 10.1016/j.biotechadv.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Crittenden J. C., Trussell R. R., Hand D. W., Howe K. J., Tchobanoglous G. (2012). MWH's water treatment: Principles and design. John Wiley & Sons. [Google Scholar]
- D'annibale A., Leonardi V., Federici E., Baldi F., Zecchini F., Petruccioli M. (2007). Leaching and microbial treatment of a soil contaminated by sulphide ore ashes and aromatic hydrocarbons. Appl. Microbiol. Biotechnol. 74, 1135–1144. 10.1007/s00253-006-0749-z [DOI] [PubMed] [Google Scholar]
- D'annibale A., Rosetto F., Leonardi V., Federici F., Petruccioli M. (2006). Role of autochthonous filamentous fungi in bioremediation of a soil historically contaminated with aromatic hydrocarbons. Appl. Environ. Microbiol. 72, 28–36. 10.1128/aem.72.1.28-36.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva A. F., Banat I. M., Giachini A. J., Robl D. (2021). Fungal biosurfactants, from nature to biotechnological product: Bioprospection, production and potential applications. Bioprocess Biosyst. Eng. 44, 2003–2034. 10.1007/s00449-021-02597-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalecka B., Juhna T., Rajarao G. (2020a). Constructive use of filamentous fungi to remove pharmaceutical substances from wastewater. J. Water Process Eng. 33, 100992. 10.1016/j.jwpe.2019.100992 [DOI] [Google Scholar]
- Dalecka B., Oskarsson C., Juhna T., Rajarao G. K. (2020b). Isolation of fungal strains from municipal wastewater for the removal of pharmaceutical substances. Water 12. [Google Scholar]
- Das S., Naik Deshavath N., Goud V. V., Dasu V. V. (2019). Bioleaching of Al from spent fluid catalytic cracking catalyst using Aspergillus species. Biotechnol. Rep. 23, e00349. 10.1016/j.btre.2019.e00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. S., Westlake D. W. S. (1979). Crude-oil utilization by fungi. Can. J. Microbiol. 25, 146–156. 10.1139/m79-023 [DOI] [PubMed] [Google Scholar]
- De Castro M. E. G., Dulay R. M. R., Enriquez M. (2016). Toxic and teratogenic effects of medicinal and culinary mushroom, Termitomyces clypeatus, collected from the termite mound in mt. makiling forest reserve, los baños, laguna, Philippines on developing embryos of zebrafish (Danio rerio). Der Pharm. Lett. 8, 237–242. [Google Scholar]
- Della Torre C., Maggioni D., Ghilardi A., Parolini M., Santo N., Landi C., et al. (2018). The interactions of fullerene C(60) and Benzo(α)pyrene influence their bioavailability and toxicity to zebrafish embryos. Environ. Pollut. 241, 999–1008. 10.1016/j.envpol.2018.06.042 [DOI] [PubMed] [Google Scholar]
- Devi M., Kaushik B. (2005). Decolourization of textile dyes and dye effluent by Aspergillus spp. Indian J. Microbiol. 45, 41. [Google Scholar]
- Dhillon G. S., Brar S. K., Kaur S., Verma M. (2013). Bioproduction and extraction optimization of citric acid from Aspergillus niger by rotating drum type solid-state bioreactor. Industrial Crops Prod. 41, 78–84. 10.1016/j.indcrop.2012.04.001 [DOI] [Google Scholar]
- Dhiman N., Chaudhary S., Singh A., Chauhan A., Kumar R. (2022). Sustainable degradation of pharmaceutical waste using different fungal strains: Enzyme induction, kinetics and isotherm studies. Environ. Technol. Innovation 25, 102156. 10.1016/j.eti.2021.102156 [DOI] [Google Scholar]
- Dhir B. (2022). “Degradation of dyes using filamentous fungi,” in Dye biodegradation, mechanisms and techniques: Recent advances. Editors Muthu S. S., Khadir A. (Singapore: Springer Singapore; ), 51–66. [Google Scholar]
- Díaz-Cruz M. S., Gago-Ferrero P., Badia-Fabregat M., Caminal G., Vicent T., Barceló D. (2014). Fungal-mediated biodegradation of ingredients in personal care products. Personal Care Prod. Aquatic Environ., 295–317. [Google Scholar]
- Dirisu C. G., Ogbulie J. N., Orji J. C., Eresia-Eke R., Olowu M. D., Neighbour R. (2018). Crude oil degradation by penicilium and mucor species isolated from fresh water swamp. Ann. Ecol. Environ. Sci. 2, 76–81. [Google Scholar]
- Dunlap P. V. (2001). “Microbial diversity,” in Encyclopedia of biodiversity. Editor Scheiner S. M. Second ed, 280–291. [Google Scholar]
- Durairaj P., Malla S., Nadarajan S. P., Lee P. G., Jung E., Park H. H., et al. (2015). Fungal cytochrome P450 monooxygenases of Fusarium oxysporum for the synthesis of omega-hydroxy fatty acids in engineered Saccharomyces cerevisiae . Microb. Cell. Factories 14, 45. 10.1186/s12934-015-0228-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusengemungu L., Kasali G., Gwanama C., Mubemba B. (2021). Overview of fungal bioleaching of metals. Environ. Adv. 5, 100083. 10.1016/j.envadv.2021.100083 [DOI] [Google Scholar]
- Dusengemungu L., Kasali G., Gwanama C., Ouma K. O. (2020). Recent advances in biosorption of copper and Cobalt by filamentous fungi. Front. Microbiol. 11, 582016. 10.3389/fmicb.2020.582016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ediriweera A. N., Karunarathna S. C., Yapa P. N., Schaefer D. A., Ranasinghe A. K., Suwannarach N., et al. (2022). Ectomycorrhizal mushrooms as a natural bio-indicator for assessment of heavy metal pollution. Agronomy 12, 1041. 10.3390/agronomy12051041 [DOI] [Google Scholar]
- El-Bondkly A. M. A., El-Gendy M. M. a. A. (2022). Bioremoval of some heavy metals from aqueous solutions by two different indigenous fungi Aspergillus sp. AHM69 and Penicillium sp. AHM96 isolated from petroleum refining wastewater. Heliyon 8, e09854. 10.1016/j.heliyon.2022.e09854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Fakharany E. M., Hassan M. A., Taha T. H. (2016). Production and application of extracellular laccase produced by Fusarium oxysporum EMT. Int. J. Agric. Biol. 18, 939–947. 10.17957/ijab/15.0190 [DOI] [Google Scholar]
- El-Gendi H., Saleh A. K., Badierah R., Redwan E. M., El-Maradny Y. A., El-Fakharany E. M. (2022). A comprehensive insight into fungal enzymes: Structure, classification, and their role in mankind's challenges. J. Fungi 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emri T., Oláh B., Sámi L., Pócsi I. (2003). Does the detoxification of penicillin side‐chain precursors depend on microsomal monooxygenase and glutathione S‐transferase in Penicillium chrysogenum? J. Basic Microbiol. Int. J. Biochem. Physiology, Genet. Morphol. Ecol. Microorg. 43, 287–300. 10.1002/jobm.200390032 [DOI] [PubMed] [Google Scholar]
- Espinosa-Ortiz E. J., Rene E. R., Pakshirajan K., Van Hullebusch E. D., Lens P. N. L. (2016). Fungal pelleted reactors in wastewater treatment: Applications and perspectives. Chem. Eng. J. 283, 553–571. 10.1016/j.cej.2015.07.068 [DOI] [Google Scholar]
- Esterhuizen-Londt M., Hendel A. L., Pflugmacher S. (2017). Mycoremediation of diclofenac using Mucor hiemalis . Toxicol. Environ. Chem. 99, 795–808. 10.1080/02772248.2017.1296444 [DOI] [Google Scholar]
- Fan T., Liu Y. G., Feng B. Y., Zeng G. M., Yang C. P., Zhou M., et al. (2008). Biosorption of cadmium(II), zinc(II) and lead(II) by Penicillium simplicissimum: Isotherms, kinetics and thermodynamics. J. Hazard. Mater. 160, 655–661. 10.1016/j.jhazmat.2008.03.038 [DOI] [PubMed] [Google Scholar]
- Farag S., Soliman N. A. (2011). Biodegradation of crude petroleum oil and environmental pollutants by Candida tropicalis strain. Braz. Archives Biol. Technol. 54, 821–830. 10.1590/s1516-89132011000400023 [DOI] [Google Scholar]
- Farhan M., Ahmad M., Kanwal A., Butt Z. A., Khan Q. F., Raza S. A., et al. (2021). Biodegradation of chlorpyrifos using isolates from contaminated agricultural soil, its kinetic studies. Sci. Rep. 11, 10320. 10.1038/s41598-021-88264-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C. L., Li J., Li X., Li K. L., Luo K., Liao X. S., et al. (2018). Characterization and mechanism of lead and zinc biosorption by growing Verticillium insectorum J3. Plos One 13, e0203859. 10.1371/journal.pone.0203859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando T., Bumpus J. A., Aust S. D. (1990). Biodegradation of tnt (2,4,6-trinitrotoluene) by Phanerochaete-chrysosporium . Appl. Environ. Microbiol. 56, 1666–1671. 10.1128/aem.56.6.1666-1671.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira J. A., Lennartsson P. R., Taherzadeh M. J. (2015). Production of ethanol and biomass from thin stillage by Neurospora intermedia: A pilot study for process diversification. Eng. Life Sci. 15, 751–759. 10.1002/elsc.201400213 [DOI] [Google Scholar]
- Ferreira J. A., Mahboubi A., Lennartsson P. R., Taherzadeh M. J. (2016). Waste biorefineries using filamentous ascomycetes fungi: Present status and future prospects. Bioresour. Technol. 215, 334–345. 10.1016/j.biortech.2016.03.018 [DOI] [PubMed] [Google Scholar]
- Ferreira J. A., Varjani S., Taherzadeh M. J. (2020). A critical review on the ubiquitous role of filamentous fungi in pollution mitigation. Curr. Pollut. Rep. 6, 295–309. 10.1007/s40726-020-00156-2 [DOI] [Google Scholar]
- Flora G., Gupta D., Tiwari A. (2012). Toxicity of lead: A review with recent updates. Interdiscip. Toxicol. 5, 47–58. 10.2478/v10102-012-0009-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Kamenetskaya O. V., Zelenskaya M. S., Izatulina A. R., Vereshchagin O. S., Vlasov D. Y., Himelbrant D. E., et al. (2021). Copper oxalate formation by lichens and fungi. Sci. Rep. 11, 24239. 10.1038/s41598-021-03600-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J. A., Davis M. A., Hynes M. J. (2002). A gene from Aspergillus nidulans with similarity to URE2 of Saccharomyces cerevisiae encodes a glutathione S-transferase which contributes to heavy metal and xenobiotic resistance. Appl. Environ. Microbiol. 68, 2802–2808. 10.1128/aem.68.6.2802-2808.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Zhang L., Dong P. C., Wang J., Shi L., Lian C. L., et al. (2022). Remediation of copper-contaminated soils using Tagetes patula L., earthworms and arbuscular mycorrhizal fungi. Int. J. Phytoremediation 24, 1107–1119. 10.1080/15226514.2021.2002809 [DOI] [PubMed] [Google Scholar]
- Fu Y. Z., Viraraghavan T. (2002). Dye biosorption sites in Aspergillus niger . Bioresour. Technol. 82, 139–145. 10.1016/s0960-8524(01)00172-9 [DOI] [PubMed] [Google Scholar]
- Fujii K., Takeshi K. (2007). Penicillium strains as dominant degraders in soil for coffee residue, a biological waste unsuitable for fertilization. J. Appl. Microbiol. 103, 2713–2720. 10.1111/j.1365-2672.2007.03529.x [DOI] [PubMed] [Google Scholar]
- Gadd G. M., Bahri-Esfahani J., Li Q., Rhee Y. J., Wei Z., Fomina M., et al. (2014). Oxalate production by fungi: Significance in geomycology, biodeterioration and bioremediation. Fungal Biol. Rev. 28, 36–55. 10.1016/j.fbr.2014.05.001 [DOI] [Google Scholar]
- Gadd G. M. (1999). Fungal production of citric and oxalic acid: Importance in metal speciation, physiology and biogeochemical processes. Adv. Microb. physiology 41, 47–92. 10.1016/s0065-2911(08)60165-4 [DOI] [PubMed] [Google Scholar]
- Galun M., Galun E., Siegel B., Keller P., Lehr H., Siegel S. (1987). Removal of metal ions from aqueous solutions by Penicillium biomass: Kinetic and uptake parameters. Water, Air, Soil Pollut. 33, 359–371. 10.1007/bf00294204 [DOI] [Google Scholar]
- Gao L., Li S., Xu Y., Xia C., Xu J., Liu J., et al. (2019). Mutation of a conserved alanine residue in transcription factor AraR Leads to HYPERPRODUCTION of α‐l‐arabinofuranosidases in Penicillium oxalicum . Biotechnol. J. 14, 1800643. 10.1002/biot.201800643 [DOI] [PubMed] [Google Scholar]
- Garcia-Hernandez M. A., Villarreal-Chiu J. F., Garza-Gonzalez M. T. (2017). Metallophilic fungi research: An alternative for its use in the bioremediation of hexavalent chromium. Int. J. Environ. Sci. Technol. 14, 2023–2038. 10.1007/s13762-017-1348-5 [DOI] [Google Scholar]
- Garon D., Krivobok S., Wouessidjewe D., Seigle-Murandi F. (2002). Influence of surfactants on solubilization and fungal degradation of fluorene. Chemosphere 47, 303–309. 10.1016/s0045-6535(01)00299-5 [DOI] [PubMed] [Google Scholar]
- Garon D., Sage L., Seigle-Murandi F. (2004). Effects of fungal bioaugmentation and cyclodextrin amendment on fluorene degradation in soil slurry. Biodegradation 15, 1–8. 10.1023/b:biod.0000009934.87627.91 [DOI] [PubMed] [Google Scholar]
- Gautam G., Mishra V., Verma P., Pandey A. K., Negi S. (2014). A cost effective strategy for production of bio-surfactant from locally isolated Penicillium chrysogenum SNP5 and its applications. J. Bioprocess. Biotech. 4, 1. 10.4172/2155-9821.1000177 [DOI] [Google Scholar]
- Geetha N., Bhavya G., Abhijith P., Shekhar R., Dayananda K., Jogaiah S. (2021). Insights into nanomycoremediation: Secretomics and mycogenic biopolymer nanocomposites for heavy metal detoxification. J. Hazard. Mater. 409, 124541. 10.1016/j.jhazmat.2020.124541 [DOI] [PubMed] [Google Scholar]
- Gerba C. P., Pepper I. L. (2019). “Municipal wastewater treatment,” in Environmental and pollution science (Elsevier; ), 393–418. [Google Scholar]
- Ghodrati M., Kosari-Nasab M., Zarrini G., Movafeghi A. (2021). Crude oil contamination enhances the lipoxygenase gene expression in the green microalga scenedesmus dimorphus. Biointerface Res. Appl. Chem. 11, 11431–11439. [Google Scholar]
- Ghosh S., Godoy L., Anchang K. Y., Achilonu C. C., Gryzenhout M. (2021). “Fungal cellulases: Current research and future challenges,” in Industrially important fungi for sustainable development. Fungal biology. Editors Abdel-Azeem A. M., Yadav A. N., Sharma M. (Cham: Springer; ). [Google Scholar]
- Ghosh S. K., Sen R., Chanakya H. N., Pariatamby A. (2020). Bioresource utilization and bioprocess. Springer. [Google Scholar]
- Ghosh S. (2015). Metagenomic Screening of cell wall hydrolases, their anti-fungal activities and potential role in wine fermentation. South Africa: Ph.D. thesis, Stellenbosch University. [Google Scholar]
- Ghyadh B. A., Al-Ashoor A., Ibrahim Z. M. (2019). Biotechnology of waste water treatment with fungi (Aspergillus niger and Rhizopus oligosporium). Plant Arch. 19, 1546–1549. [Google Scholar]
- Guengerich F. P., Munro A. W. (2013). Unusual cytochrome P450 enzymes and reactions. J. Biol. Chem. 288, 17065–17073. 10.1074/jbc.r113.462275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gultom S. O., Hu B. (2013). Review of microalgae harvesting via co-pelletization with filamentous fungus. Energies 6, 5921–5939. 10.3390/en6115921 [DOI] [Google Scholar]
- Guo J., Zhang M., Fang Z. (2022). Valorization of mushroom by‐products: A review. J. Sci. Food Agric. 102, 5593–5605. 10.1002/jsfa.11946 [DOI] [PubMed] [Google Scholar]
- Gupta P., Pandey K., Verma N. (2022). Augmented complete mineralization of glyphosate in wastewater via microbial degradation post CWAO over supported Fe-CNF. Chem. Eng. J. 428, 132008. 10.1016/j.cej.2021.132008 [DOI] [Google Scholar]
- Gupta S., Pathak B., Fulekar M. H. (2015). Molecular approaches for biodegradation of polycyclic aromatic hydrocarbon compounds: A review. Rev. Environ. Sci. Bio/Technology 14, 241–269. 10.1007/s11157-014-9353-3 [DOI] [Google Scholar]
- Hadibarata T., Kristanti R. A., Bilal M., Al-Mohaimeed A. M., Chen T. W., Lam M. K. (2022). Microbial degradation and transformation of benzo [a] pyrene by using a white-rot fungus Pleurotus eryngii F032. Chemosphere 307, 136014. 10.1016/j.chemosphere.2022.136014 [DOI] [PubMed] [Google Scholar]
- Haripriyan U., Gopinath K. P., Arun J., Govarthanan M. (2022). Bioremediation of organic pollutants: A mini review on current and critical strategies for wastewater treatment. Archives Microbiol. 204, 286. 10.1007/s00203-022-02907-9 [DOI] [PubMed] [Google Scholar]
- Hasanizadeh P., Moghimi H., Hamedi J. (2017). Biosurfactant production by Mucor circinelloides on waste frying oil and possible uses in crude oil remediation. Water Sci. Technol. 76, 1706–1714. 10.2166/wst.2017.338 [DOI] [PubMed] [Google Scholar]
- Hofmann U., Schlosser D. (2016). Biochemical and physicochemical processes contributing to the removal of endocrine-disrupting chemicals and pharmaceuticals by the aquatic ascomycete Phoma sp UHH 5-1-03. Appl. Microbiol. Biotechnol. 100, 2381–2399. 10.1007/s00253-015-7113-0 [DOI] [PubMed] [Google Scholar]
- Hofrichter M., Bublitz F., Fritsche W. (1994). Unspecific degradation of halogenated phenols by the soil fungus Penicillium-frequentans-Bi-7/2. J. Basic Microbiol. 34, 163–172. 10.1002/jobm.3620340306 [DOI] [PubMed] [Google Scholar]
- Holan Z. R., Volesky B. (1995). Accumulation of cadmium, lead, and nickel by fungal and wood biosorbents. Appl. Biochem. Biotechnol. 53, 133–146. 10.1007/bf02788603 [DOI] [Google Scholar]
- Hoque E., Fritscher J. (2019). Multimetal bioremediation and biomining by a combination of new aquatic strains of Mucor hiemalis . Sci. Rep. 9, 10318. 10.1038/s41598-019-46560-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W., Zhang L., Li X. J., Gong Z. Q., Yang Y. W., Li Z. (2015). Influence of Mucor mucedo immobilized to corncob in remediation of pyrene contaminated agricultural soil. Environ. Eng. Res. 20, 149–154. 10.4491/eer.2015.013 [DOI] [Google Scholar]
- Hu K. D., Toran J., Lopez-Garcia E., Barbieri M. V., Postigo C., De Alda M. L., et al. (2020). Fungal bioremediation of diuron-contaminated waters: Evaluation of its degradation and the effect of amendable factors on its removal in a trickle-bed reactor under non-sterile conditions. Sci. Total Environ. 743, 140628. 10.1016/j.scitotenv.2020.140628 [DOI] [PubMed] [Google Scholar]
- Hu L., Wang Z., He J., Lv S., Zhou B., Hrynsphan D., et al. (2022). Co-culturing fungus Penicillium citrinum and strain Citrobacter freundii improved nitrate removal and carbon utilization by promoting glyceride metabolism. Bioresour. Technol. 360, 127563. 10.1016/j.biortech.2022.127563 [DOI] [PubMed] [Google Scholar]
- Huang D. L., Guo X. Y., Peng Z. W., Zeng G. M., Xu P., Gong X. M., et al. (2018). White rot fungi and advanced combined biotechnology with nanomaterials: Promising tools for endocrine-disrupting compounds biotransformation. Crit. Rev. Biotechnol. 38, 671–689. 10.1080/07388551.2017.1386613 [DOI] [PubMed] [Google Scholar]
- Huang X. N. A., Men P., Tang S., Lu X. F. (2021). Aspergillus terreus as an industrial filamentous fungus for pharmaceutical biotechnology. Curr. Opin. Biotechnol. 69, 273–280. 10.1016/j.copbio.2021.02.004 [DOI] [PubMed] [Google Scholar]
- Huerta-Rosas B., Cano-Rodriguez I., Gamino-Arroyo Z., Gomez-Castro F. I., Carrillo-Pedroza F. R., Romo-Rodriguez P., et al. (2020). Aerobic processes for bioleaching manganese and silver using microorganisms indigenous to mine tailings. World J. Microbiol. Biotechnol. 36, 124. 10.1007/s11274-020-02902-6 [DOI] [PubMed] [Google Scholar]
- Ianis M., Tsekova K., Vasileva S. (2006). Copper biosorption by Penicillium cyclopium: Equilibrium and modelling study. Biotechnol. Biotechnol. Equip. 20, 195–201. 10.1080/13102818.2006.10817332 [DOI] [Google Scholar]
- Isaza-Perez F., Ramirez-Carmona M., Rendon-Castrillon L., Ocampo-Lopez C. (2020). Potential of residual fungal biomass: A review. Environ. Sci. Pollut. Res. 27, 13019–13031. 10.1007/s11356-020-08193-6 [DOI] [PubMed] [Google Scholar]
- Javanbakht V., Zilouei H., Karimi K. (2011). Lead biosorption by different morphologies of fungus Mucor indicus . Int. Biodeterior. Biodegrad. 65, 294–300. 10.1016/j.ibiod.2010.11.015 [DOI] [Google Scholar]
- Jimenez A. M., Borja R., Martin A., Raposo F. (2005). Mathematical modelling of aerobic degradation of vinasses with Penicillium decumbens . Process Biochem. 40, 2805–2811. 10.1016/j.procbio.2004.12.011 [DOI] [Google Scholar]
- Johansson E. M., Fransson P. M. A., Finlay R. D., Van Hees P. a. W. (2008). Quantitative analysis of exudates from soil-living basidiomycetes in pure culture as a response to lead, cadmium and arsenic stress. Soil Biol. Biochem. 40, 2225–2236. 10.1016/j.soilbio.2008.04.016 [DOI] [Google Scholar]
- Kapahi M., Sachdeva S. (2019). Bioremediation options for heavy metal pollution. J. Health Pollut. 9, 191203. 10.5696/2156-9614-9.24.191203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Viraraghavan T., Cullimore D. R. (1999). Removal of heavy metals using the fungus Aspergillus niger . Bioresour. Technol. 70, 95–104. 10.1016/s0960-8524(98)00192-8 [DOI] [Google Scholar]
- Karimi K., Zamani A. (2013). Mucor indicus: Biology and industrial application perspectives: A review. Biotechnol. Adv. 31, 466–481. 10.1016/j.biotechadv.2013.01.009 [DOI] [PubMed] [Google Scholar]
- Karimi S., Soofiani N. M., Lundh T., Mahboubi A., Kiessling A., Taherzadeh M. J. (2019). Evaluation of filamentous fungal biomass cultivated on vinasse as an alternative nutrient source of fish feed: Protein, lipid, and mineral composition. Fermentation-Basel 5, 99. 10.3390/fermentation5040099 [DOI] [Google Scholar]
- Karimi S., Soofiani N. M., Mahboubi A., Taherzadeh M. J. (2018). Use of organic wastes and industrial by-products to produce filamentous fungi with potential as aqua-feed ingredients. Sustainability 10, 3296. 10.3390/su10093296 [DOI] [Google Scholar]
- Kasonga T. K., Kamika I., Ngole-Jeme V. M. (2022). Ligninolytic enzyme activity and removal efficiency of pharmaceuticals in a water matrix by fungus Rhizopus sp. Isolated from cassava. Environ Technol 10, 1-14. 10.1080/09593330.2021.2024885 [DOI] [PubMed] [Google Scholar]
- Kawa-Rygielska J., Pietrzak W., Lennartsson P. R. (2022). High-efficiency conversion of bread residues to ethanol and edible biomass using filamentous fungi at high solids loading: A biorefinery approach. Appl. Sciences-Basel 12, 6405. 10.3390/app12136405 [DOI] [Google Scholar]
- Khan S. A., Uddin I., Moeez S., Ahmad A. (2014). Fungus-mediated preferential bioleaching of waste material such as fly - ash as a means of producing extracellular, protein capped, fluorescent and water soluble silica nanoparticles. Plos One 9, e107597. 10.1371/journal.pone.0107597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratosova G., Holisova V., Konvickova Z., Ingle A. P., Gaikwad S., Skrlova K., et al. (2019). From biotechnology principles to functional and low-cost metallic bionanocatalysts. Biotechnol. Adv. 37, 154–176. 10.1016/j.biotechadv.2018.11.012 [DOI] [PubMed] [Google Scholar]
- Kumari D., Qian X. Y., Pan X. L., Achal V., Li Q. W., Gadd G. M. (2016). Microbially-induced carbonate precipitation for immobilization of toxic metals. Adv. Appl. Microbiol. 94, 79–108. 10.1016/bs.aambs.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Lamar R. T., Dietrich D. M. (1990). In situ depletion of pentachlorophenol from contaminated soil by Phanerochaete spp. Appl. Environ. Microbiol. 56, 3093–3100. 10.1128/aem.56.10.3093-3100.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar R. T., Dietrich D. M. (1992). Use of lignin-degrading fungi in the disposal of pentachlorophenol-treated wood. J. Industrial Microbiol. 9, 181–191. 10.1007/bf01569622 [DOI] [Google Scholar]
- Le L., Tang J., Ryan D., Valix M. (2006). Bioleaching nickel laterite ores using multi-metal tolerant Aspergillus foetidus organism. Miner. Eng. 19, 1259–1265. 10.1016/j.mineng.2006.02.006 [DOI] [Google Scholar]
- Leitao A. L., Duarte M. P., Oliveira J. S. (2007). Degradation of phenol by a halotolerant strain of Penicillium chrysogenum . Int. Biodeterior. Biodegrad. 59, 220–225. 10.1016/j.ibiod.2006.09.009 [DOI] [Google Scholar]
- Levin L., Viale A., Forchiassin A. (2003). Degradation of organic pollutants by the white rot basidiomycete Trametes trogii . Int. Biodeterior. Biodegrad. 52, 1–5. 10.1016/s0964-8305(02)00091-4 [DOI] [Google Scholar]
- Li J., Zhang Y., Li J., Sun T., Tian C. (2020). Metabolic engineering of the cellulolytic thermophilic fungus Myceliophthora thermophila to produce ethanol from cellobiose. Biotechnol. biofuels 13, 23–15. 10.1186/s13068-020-1661-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Q., Xu J. M., De Toledo R. A., Shim H. (2015). Enhanced removal of naproxen and carbamazepine from wastewater using a novel countercurrent seepage bioreactor immobilized with Phanerochaete chrysosporium under non-sterile conditions. Bioresour. Technol. 197, 465–474. 10.1016/j.biortech.2015.08.118 [DOI] [PubMed] [Google Scholar]
- Liang X. J., Kierans M., Ceci A., Hillier S., Gadd G. M. (2016). Phosphatase-mediated bioprecipitation of lead by soil fungi. Environ. Microbiol. 18, 219–231. 10.1111/1462-2920.13003 [DOI] [PubMed] [Google Scholar]
- Liu R., Chen L., Jiang Y. P., Zou G., Zhou Z. H. (2017). A novel transcription factor specifically regulates GH11 xylanase genes in Trichoderma reesei . Biotechnol. Biofuels 10, 194. 10.1186/s13068-017-0878-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loss E. M. O., Lee M. K., Wu M. Y., Martien J., Chen W. P., Amador-Noguez D., et al. (2019). Cytochrome P450 monooxygenase-mediated metabolic utilization of benzo[a]Pyrene by Aspergillus species. mBio 10 (3), e00558-19. 10.1128/mbio.00558-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Li B., Zhang X. Y., Wang C., Chen W. (2022). Production of gluconic acid and its derivatives by microbial fermentation: Process improvement based on integrated routes. Front. Bioeng. Biotechnol. 10, 864787. 10.3389/fbioe.2022.864787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar A., Lucchesi M. E., Debaets S., Van Long N. N., Quemener M., Coton E., et al. (2020). Highlighting the crude oil bioremediation potential of marine fungi isolated from the port of oran (Algeria). Diversity-Basel 12, 196. 10.3390/d12050196 [DOI] [Google Scholar]
- Machado K. M. G., Matheus D. R., Monteiro R. T. R., Bononi V. L. R. (2005). Biodegradation of pentachorophenol by tropical basidiomycetes in soils contaminated with industrial residues. World J. Microbiol. Biotechnol. 21, 297–301. 10.1007/s11274-004-3693-z [DOI] [Google Scholar]
- Manguilimotan L. C., Bitacura J. G. (2018). Biosorption of cadmium by filamentous fungi isolated from coastal water and sediments. J. Toxicol. 2018, 1–6. 10.1155/2018/7170510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoni C., Kia D. A., Vandrovcova J., Hardy J., Wood N. W., Lewis P. A., et al. (2018). Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Briefings Bioinforma. 19, 286–302. 10.1093/bib/bbw114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr J., Kremer S., Sterner O., Anke H. (1996). Transformation and mineralization of halophenols by Penicillium simplicissimum SK9117. Biodegradation 7, 165–171. 10.1007/bf00114628 [DOI] [PubMed] [Google Scholar]
- Matsubara M., Lynch J., De Leij F. (2006). A simple screening procedure for selecting fungi with potential for use in the bioremediation of contaminated land. Enzyme Microb. Technol. 39, 1365–1372. 10.1016/j.enzmictec.2005.04.025 [DOI] [Google Scholar]
- Meena M., Sonigra P., Yadav G., Barupal T. (2021). “Wastewater treatment techniques: An introduction,” in Removal of emerging contaminants through microbial processes (Springer; ), 161–182. [Google Scholar]
- Mendil D., Tuzen M., Soylak M. (2008). A biosorption system for metal ions on Penicillium italicum loaded on Sepabeads SP 70 prior to flame atomic absorption spectrometric determinations. J. Hazard. Mater. 152, 1171–1178. 10.1016/j.jhazmat.2007.07.097 [DOI] [PubMed] [Google Scholar]
- Mir-Tutusaus J. A., Baccar R., Caminal G., Sarra M. (2018). Can white-rot fungi be a real wastewater treatment alternative for organic micropollutants removal? A review. Water Res. 138, 137–151. 10.1016/j.watres.2018.02.056 [DOI] [PubMed] [Google Scholar]
- Mir-Tutusaus J. A., Parlade E., Llorca M., Villagrasa M., Barcelo D., Rodriguez-Mozaz S., et al. (2017). Pharmaceuticals removal and microbial community assessment in a continuous fungal treatment of non-sterile real hospital wastewater after a coagulation-flocculation pretreatment. Water Res. 116, 65–75. 10.1016/j.watres.2017.03.005 [DOI] [PubMed] [Google Scholar]
- Mo J. Y., Liu Y. H., Gao X. N., Zhou S. Y., Deng Y. R., Ke Y. Y., et al. (2022). Potential application of Fusarium fungal strains (Fusarium sp. FP, Arthrinium sp. FB, and Phoma sp. FR) for removal of Tl (I) ions from water. Environ. Sci. Pollut. Res. 29, 46049–46063. 10.1007/s11356-022-18791-1 [DOI] [PubMed] [Google Scholar]
- Mohapatra D., Rath S. K., Mohapatra P. K. (2022). Soil fungi for bioremediation of pesticide toxicants: A perspective. Geomicrobiol. J. 39, 352–372. 10.1080/01490451.2021.2019855 [DOI] [Google Scholar]
- Mollea C., Bosco F., Ruggeri B. (2005). Fungal biodegradation of naphthalene: Microcosms studies. Chemosphere 60, 636–643. 10.1016/j.chemosphere.2005.01.034 [DOI] [PubMed] [Google Scholar]
- Moretti A. N. (2009). Taxonomy of Fusarium genus: A continuous fight between lumpers and splitters. Zb. Matice Srp. za Prir. nauke, 7–13. 10.2298/zmspn0917007m [DOI] [Google Scholar]
- Morsi R., Bilal M., Iqbal H. M. N., Ashraf S. S. (2020). Laccases and peroxidases: The smart, greener and futuristic biocatalytic tools to mitigate recalcitrant emerging pollutants. Sci. Total Environ. 714, 136572. 10.1016/j.scitotenv.2020.136572 [DOI] [PubMed] [Google Scholar]
- Mousa S. A., El-Sayed E. S. R., Mohamed S. S., El-Seoud M. a. A., Elmehlawy A. A., Abdou D. a. M. (2021). Novel mycosynthesis of Co3O4, CuO, Fe3O4, NiO, and ZnO nanoparticles by the endophytic Aspergillus terreus and evaluation of their antioxidant and antimicrobial activities. Appl. Microbiol. Biotechnol. 105, 741–753. 10.1007/s00253-020-11046-4 [DOI] [PubMed] [Google Scholar]
- Mousavi S. M., Hashemi S. A., Moezzi S. M. I., Ravan N., Gholami A., Lai C. W., et al. (2021). Recent advances in enzymes for the bioremediation of pollutants. Biochem. Res. Int. 2021, 1–12. 10.1155/2021/5599204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Das D., Mondal S. K., Biswas R., Das T. K., Boujedaini N., et al. (2010). Tolerance of arsenate-induced stress in Aspergillus niger, a possible candidate for bioremediation. Ecotoxicol. Environ. Saf. 73, 172–182. 10.1016/j.ecoenv.2009.09.015 [DOI] [PubMed] [Google Scholar]
- Mukherjee P., Ahmad A., Mandal D., Senapati S., Sainkar S. R., Khan M. I., et al. (2001). Fungus-mediated synthesis of silver nanoparticles and their immobilization in the mycelial matrix: A novel biological approach to nanoparticle synthesis. Nano Lett. 1, 515–519. 10.1021/nl0155274 [DOI] [Google Scholar]
- Nair R. B., Lundin M., Lennartsson P. R., Taherzadeh M. J. (2017). Optimizing dilute phosphoric acid pretreatment of wheat straw in the laboratory and in a demonstration plant for ethanol and edible fungal biomass production using Neurospora intermedia . J. Chem. Technol. Biotechnol. 92, 1256–1265. 10.1002/jctb.5119 [DOI] [Google Scholar]
- Naveen Kumar K., Prakash J. (2021). Bioremoval of different heavy metals in industrial effluent by the resistant fungal strain Aspergillus niger . Nat. Environ. Pollut. Technol. 20 (4). [Google Scholar]
- Niu H., Xu X. S., Wang J. H., Volesky B. (1993). Removal of lead from aqueous-solutions by Penicillium biomass. Biotechnol. Bioeng. 42, 785–787. 10.1002/bit.260420615 [DOI] [PubMed] [Google Scholar]
- Nongthombam G. D., Sarangi P. K., Singh T. A., Sharma C. K., Talukdar N. C. (2022). Bioethanol production from Ficus fruits (Ficus cunia) by Fusarium oxysporum through consolidated bioprocessing system. 3 Biotech. 312, 178. 10.1007/s13205-022-03234-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny C., Svobodova K., Erbanova P., Cajthaml T., Kasinath A., Lang E., et al. (2004). Ligninolytic fungi in bioremediation: Extracellular enzyme production and degradation rate. Soil Biol. Biochem. 36, 1545–1551. 10.1016/j.soilbio.2004.07.019 [DOI] [Google Scholar]
- Nuban A. A., Safitri A., Andayani U. (2021). Biosorption of lead (Pb) from aqueous solution by immobilized Aspergillus niger: A green technology for heavy metals removal. Indonesian Green Technol. J. 10. [Google Scholar]
- Oboh B. O., Ilori M. O., Akinyemi J. O., Adebusoye S. A., Yaba A. (2006). Hydrocarbon degrading potentials of bacteria isolated from a Nigerian bitumen (tarsand) deposit. Nat. Sci. 4 (3), 51-57. [Google Scholar]
- Okoro B. C., Nwadike O. A., Agunwamba J. C. (2013). Prediction of remediation rates of microbes in polluted crude oil soil samples. Environ. Nat. Resour. Res. 3. 10.5539/enrr.v3n3p89 [DOI] [Google Scholar]
- Okoro C. C. (2008). Biodegradation of hydrocarbons in untreated produce water using pure fungal cultures. Afr. J. Microbiol. Res. 2, 217–223. [Google Scholar]
- Olicon-Hernandez D. R., Camacho-Morales R. L., Pozo C., Gonzalez-Lopez J., Aranda E. (2019). Evaluation of diclofenac biodegradation by the ascomycete fungus Penicillium oxalicum at flask and bench bioreactor scales. Sci. Total Environ. 662, 607–614. 10.1016/j.scitotenv.2019.01.248 [DOI] [PubMed] [Google Scholar]
- Olicon-Hernandez D. R., Gonzalez-Lopez J., Aranda E. (2017). Overview on the biochemical potential of filamentous fungi to degrade pharmaceutical compounds. Front. Microbiol. 8, 1792. 10.3389/fmicb.2017.01792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenti C. C., Ghilardi A., Della Torre C., Magni S., Del Giacco L., Binelli A. (2019). Evaluation of the infiltration of polystyrene nanobeads in zebrafish embryo tissues after short-term exposure and the related biochemical and behavioural effects. Environ. Pollut. 254, 112947. 10.1016/j.envpol.2019.07.115 [DOI] [PubMed] [Google Scholar]
- Paszczynski A., Crawford R. L. (1995). Potential for bioremediation of xenobiotic compounds by the white-rot fungus Phanerochaete-Chrysosporium . Biotechnol. Prog. 11, 368–379. 10.1021/bp00034a002 [DOI] [Google Scholar]
- Pedersen H., Christensen B., Hjort C., Nielsen J. (2000a). Construction and characterization of an oxalic acid nonproducing Strain of Aspergillus niger . Metab. Eng. 2, 34–41. 10.1006/mben.1999.0136 [DOI] [PubMed] [Google Scholar]
- Pedersen H., Hjort C., Nielsen J. (2000b). Cloning and characterization of oah, the gene encoding oxaloacetate hydrolase in Aspergillus niger . Mol. General Genet. 263, 281–286. 10.1007/s004380051169 [DOI] [PubMed] [Google Scholar]
- Perera M., Chinthaka S. D. M., Wijayarathna C. D., Wijesundera S., Seneviratne G., Jayasena S. (2021). Reduction of lag in crude oil degradation by Aspergillus when it is in synergy with Bacillus in biofilm mode. Bioprocess Biosyst. Eng. 44, 1501–1510. 10.1007/s00449-021-02534-6 [DOI] [PubMed] [Google Scholar]
- Pirzadeh B. (2022). “Physical wastewater treatment,” in Wastewater treatment (IntechOpen. [Google Scholar]
- Pocsi I., Prade R. A., Penninckx M. J. (2004). Glutathione, altruistic metabolite in fungi. Adv. Microb. Physiology 49, 1–76. 10.1016/S0065-2911(04)49001-8 [DOI] [PubMed] [Google Scholar]
- Ponraj M., Gokila K., Zambare V. (2011). Bacterial decolorization of textile dye-Orange 3R. Int. J. Adv. Biotechnol. Res. 2, 168–177. [Google Scholar]
- Poonam V., Sanjay S., Verma R. (2016). Heavy metal biosorption by Fusarium strains isolated from iron ore mines overburden soil. Int. J. Environ. Sci. Toxicol. Res. 4, 61–69. [Google Scholar]
- Prasenjit B., Sumathi S. (2005). Uptake of chromium by Aspergillus foetidus . J. Material Cycles Waste Manag. 7, 88–92. 10.1007/s10163-005-0131-8 [DOI] [Google Scholar]
- Priyadarshini E., Priyadarshini S. S., Cousins B. G., Pradhan N. (2021). Metal-Fungus interaction: Review on cellular processes underlying heavy metal detoxification and synthesis of metal nanoparticles. Chemosphere 274, 129976. 10.1016/j.chemosphere.2021.129976 [DOI] [PubMed] [Google Scholar]
- Qazi M. A., Kanwal T., Jadoon M., Ahmed S., Fatima N. (2014). Isolation and characterization of a biosurfactant-producing Fusarium sp BS-8 from oil contaminated soil. Biotechnol. Prog. 30, 1065–1075. 10.1002/btpr.1933 [DOI] [PubMed] [Google Scholar]
- Qian X. Y., Fang C. L., Huang M. S., Achal V. (2017). Characterization of fungal-mediated carbonate precipitation in the biomineralization of chromate and lead from an aqueous solution and soil. J. Clean. Prod. 164, 198–208. 10.1016/j.jclepro.2017.06.195 [DOI] [Google Scholar]
- Rafeeq H., Qamar S. A., Nguyen T. A., Bilal M., Iqbal H. M. N. (2022). “Microbial degradation of environmental pollutants,” in Biodegradation and biodeterioration at the nanoscale (Elsevier; ), 509–528. [Google Scholar]
- Ramachandran S., Fontanille P., Pandey A., Larroche C. (2006). Gluconic acid: Properties, applications and microbial production. Food Technol. Biotechnol. 44. [Google Scholar]
- Rao K. R., Rashmi K., Latha J. N. L., Mohan P. M. (2005). Bioremediation of toxic metal ions using biomass of Aspergillus fumigatus from fermentative waste. [Google Scholar]
- Ravelet C., Krivobok S., Sage L., Steiman R. (2000). Biodegradation of pyrene by sediment fungi. Chemosphere 40, 557–563. 10.1016/s0045-6535(99)00320-3 [DOI] [PubMed] [Google Scholar]
- Raziyafathima M. P. P. K., Praseetha P. K., Rimal I. R. S. (2016). Microbial degradation of plastic waste: A review. Chem. Biol. Sci. 4, 231–242. [Google Scholar]
- Reijngoud J., Deseke M., Halbesma E. T. M., Alazi E., Arentshorst M., Punt P. J., et al. (2019). Correction to: Mutations in AraR leading to constitutive expression of arabinolytic genes in Aspergillus niger under derepressing conditions. Appl. Microbiol. Biotechnol. 103, 5063. 10.1007/s00253-019-09853-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W. X., Li P. J., Geng Y., Li X. J. (2009). Biological leaching of heavy metals from a contaminated soil by Aspergillus niger . J. Hazard. Mater. 167, 164–169. 10.1016/j.jhazmat.2008.12.104 [DOI] [PubMed] [Google Scholar]
- Restrepo-Florez J. M., Bassi A., Thompson M. R. (2014). Microbial degradation and deterioration of polyethylene - a review. Int. Biodeterior. Biodegrad. 88, 83–90. 10.1016/j.ibiod.2013.12.014 [DOI] [Google Scholar]
- Rissoni Toledo A. G., Reyes Andrade J. C., Palmieri M. C., Bevilaqua D., Pombeiro Sponchiado S. R. (2021). Innovative method for encapsulating highly pigmented biomass from Aspergillus nidulans mutant for copper ions removal and recovery. PloS one 16, e0259315. 10.1371/journal.pone.0259315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robl D., Benoit I., Padilla G., Da Cruz Pradella J. G., De Vries R. P. (2018). Improved hemicellulase production by genetic modification of carbon catabolite repression and xylanolitic activation in Aspergillus niger . Curr. Biotechnol. 7, 10–18. 10.2174/2211550105666160722120556 [DOI] [Google Scholar]
- Rostami K., Joodaki M. R. (2002). Some studies of cadmium adsorption using Aspergillus niger, Penicillium austurianum, employing an airlift fermenter. Chem. Eng. J. 89, 239–252. 10.1016/s1385-8947(02)00131-6 [DOI] [Google Scholar]
- Rusyn I. B., Moroz O. M., Karabyn V. V., Kulachkovs'kiĭ O. R., Hudz S. P. (2003). Biodegradation of oil hydrocarbons by Candida yeast. Mikrobiol. Z 65, 36–42. [PubMed] [Google Scholar]
- Ryu B. H., Weon Y. D. (1992). Decolourisation of azo dyes by Aspergillus sojae B10. J. Microbiol. Biotechnol. 2, 215–219. [Google Scholar]
- Sack U., Gunther T. (1993). Metabolism of PAH by fungi and correlation with extracellular enzymatic-activities. J. Basic Microbiol. 33, 269–277. 10.1002/jobm.3620330411 [DOI] [PubMed] [Google Scholar]
- Sahu J., Zabed H., Karri R. R., Shams S., Qi X. (2019). “Applications of nano-biotechnology for sustainable water purification,” in Industrial applications of nanomaterials (Elsevier; ), 313–340. [Google Scholar]
- Sakaki T., Yamamoto K., Ikushiro S. (2013). Possibility of application of cytochrome P450 to bioremediation of dioxins. Biotechnol. Appl. Biochem. 60, 65–70. 10.1002/bab.1067 [DOI] [PubMed] [Google Scholar]
- Samer M. (Editor) (2015). Wastewater treatment engineering. InTech. 10.5772/59384 [DOI] [Google Scholar]
- Sanghi R., Dixit A., Verma P., Puri S. (2009). Design of reaction conditions for the enhancement of microbial degradation of dyes in sequential cycles. J. Environ. Sci. 21, 1646–1651. 10.1016/s1001-0742(08)62468-7 [DOI] [PubMed] [Google Scholar]
- Sar T., Ferreira J. A., Taherzadeh M. J. (2020a). Bioprocessing strategies to increase the protein fraction of Rhizopus oryzae biomass using fish industry sidestreams. Waste Manag. 113, 261–269. 10.1016/j.wasman.2020.06.005 [DOI] [PubMed] [Google Scholar]
- Sar T., Ferreira J. A., Taherzadeh M. J. (2021). Conversion of fish processing wastewater into fish feed ingredients through submerged cultivation of Aspergillus oryzae . Syst. Microbiol. Biomanufacturing 1, 100–110. 10.1007/s43393-020-00009-5 [DOI] [Google Scholar]
- Sar T., Larsson K., Fristedt R., Undeland I., Taherzadeh M. J. (2022a). Demo-scale production of protein-rich fungal biomass from potato protein liquor for use as innovative food and feed products. Food Biosci. 47, 101637. 10.1016/j.fbio.2022.101637 [DOI] [Google Scholar]
- Sar T., Larsson K., Fristedt R., Undeland I., Taherzadeh M. J. (2022b). Demo-scale production of protein-rich fungal biomass from potato protein liquor for use as innovative food and feed products. Food Biosci. 47, 101637. 10.1016/j.fbio.2022.101637 [DOI] [Google Scholar]
- Sar T., Ozturk M., Taherzadeh M. J., Ferrei J. A. (2020b). New insights on protein recovery from olive oil mill wastewater through bioconversion with edible filamentous fungi. Processes 8, 1210. 10.3390/pr8101210 [DOI] [Google Scholar]
- Saraswathy A., Hallberg R. (2002). Degradation of pyrene by indigenous fungi from a former gasworks site. FEMS Microbiol. Lett. 210, 227–232. 10.1111/j.1574-6968.2002.tb11185.x [DOI] [PubMed] [Google Scholar]
- Saraswathy A., Hallberg R. (2005). Mycelial pellet formation by Penicillium ochrochloron species due to exposure to pyrene. Microbiol. Res. 160, 375–383. 10.1016/j.micres.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Saravanathamizhan R., Perarasu V. T. (2021). “Improvement of biodegradability index of industrial wastewater using different pretreatment techniques,” in Wastewater treatment (Elsevier; ), 103–136. [Google Scholar]
- Say R., Yilmaz N., Denizli A. (2003). Removal of heavy metal ions using the fungus Penicillium canescens . Adsorpt. Sci. Technol. 21, 643–650. 10.1260/026361703772776420 [DOI] [Google Scholar]
- Sebesta M., Vojtkova H., Cyprichova V., Ingle A. P., Urik M., Kolencik M. (2022). Mycosynthesis of metal-containing nanoparticles-fungal metal resistance and mechanisms of synthesis. Int. J. Mol. Sci. 23, 14084. 10.3390/ijms232214084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S. S., Palmieri M. C., Sponchiado S. R. P., Bevilaqua D. (2020). Enhanced bio-recovery of aluminum from low-grade bauxite using adapted fungal strains. Braz. J. Microbiol. 51, 1909–1918. 10.1007/s42770-020-00342-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojaei S., Shojaei S. (2021). “Optimization of process conditions in wastewater degradation process,” in Soft computing techniques in solid waste and wastewater management (Elsevier; ), 381–392. [Google Scholar]
- Shourie A., Vijayalakshmi U. (2022). Fungal diversity and its role in mycoremediation. Geomicrobiol. J. 39, 426–444. 10.1080/01490451.2022.2032883 [DOI] [Google Scholar]
- Show P. L., Oladele K. O., Siew Q. Y., Aziz Zakry F. A., Lan J. C. W., Ling T. C. (2015). Overview of citric acid production from Aspergillus niger . Front. life Sci. 8, 271–283. 10.1080/21553769.2015.1033653 [DOI] [Google Scholar]
- Shreve M. J., Brockman A., Hartleb M., Prebihalo S., Dorman F. L., Brennan R. A. (2016). The white-rot fungus Trametes versicolor reduces the estrogenic activity of a mixture of emerging contaminants in wastewater treatment plant effluent. Int. Biodeterior. Biodegrad. 109, 132–140. 10.1016/j.ibiod.2016.01.018 [DOI] [Google Scholar]
- Shu W. S., Huang L. N. (2022). Microbial diversity in extreme environments. Nat. Rev. Microbiol. 20, 219–235. 10.1038/s41579-021-00648-y [DOI] [PubMed] [Google Scholar]
- Singh A., Kumari R., Yadav A. N. (2021). “Fungal secondary metabolites for bioremediation of hazardous heavy metals,” in Recent trends in mycological research: Volume 2: Environmental and industrial perspective. Editor Yadav A. N. (Cham: Springer International Publishing; ), 65–98. [Google Scholar]
- Skowronski T., Pirszel J., Pawlik-Skowronska B. (2001). Heavy metal removal by the waste biomass of Penicillium chrysogenum . Water Qual. Res. J. Can. 36, 793–803. 10.2166/wqrj.2001.042 [DOI] [Google Scholar]
- Srinivasan S., Sadasivam S. K. (2018). Exploring docking and aerobic-microaerophilic biodegradation of textile azo dye by bacterial systems. J. water process Eng. 22, 180–191. 10.1016/j.jwpe.2018.02.004 [DOI] [Google Scholar]
- Srivastava S., Thakur I. S. (2006). Evaluation of bioremediation and detoxification potentiality of Aspergillus niger for removal of hexavalent chromium in soil microcosm. Soil Biol. Biochem. 38, 1904–1911. 10.1016/j.soilbio.2005.12.016 [DOI] [Google Scholar]
- Stahl D. A., Flowers J. J., Hullar M., Davidson S. (2013). “Structure and function of microbial communities,” in The prokaryotes: Prokaryotic communities and ecophysiology. Editors Rosenberg E., Delong E. F., Lory S., Stackebrandt E., Thompson F. (Berlin, Heidelberg: Springer Berlin Heidelberg; ), 3–30. [Google Scholar]
- Steffen K. T. (2000). Degradation of recalcitrant biopolymers and polycyclic aromatic hydrocarbons by litter-decomposing basidiomycetous fungi. Academic Dissertation in Microbiology, Viikki Biocenter, University of Helsinki. [Google Scholar]
- Stenholm A., Hedeland M., Arvidsson T., Pettersson C. E. (2019). Removal of diclofenac from a non-sterile aqueous system using Trametes versicolor with an emphasis on adsorption and biodegradation mechanisms. Environ. Technol. 40, 2460–2472. 10.1080/09593330.2018.1444098 [DOI] [PubMed] [Google Scholar]
- Sumathi S., Manju B. S. (2000). Uptake of reactive textile dyes by Aspergillus foetidus . Enzyme Microb. Technol. 27, 347–355. 10.1016/s0141-0229(00)00234-9 [DOI] [PubMed] [Google Scholar]
- Sun J., Li G. Y., Li Q., Wang Y. D., Ma J. H., Pang C. Y., et al. (2020a). Impacts of operational parameters on the morphological structure and uranium bioleaching performance of bio-ore pellets in one-step bioleaching by Aspergillus niger . Hydrometallurgy 195, 105378. 10.1016/j.hydromet.2020.105378 [DOI] [Google Scholar]
- Sun L., Gong M. Y., Lv X. Q., Huang Z. Y., Gu Y., Li J. H., et al. (2020b). Current advance in biological production of short-chain organic acid. Appl. Microbiol. Biotechnol. 104, 9109–9124. 10.1007/s00253-020-10917-0 [DOI] [PubMed] [Google Scholar]
- Sutjaritvorakul T., Gadd G. M., Whalley A. J. S., Suntornvongsagul K., Sihanonth P. (2016). Zinc oxalate crystal formation by Aspergillus nomius . Geomicrobiol. J. 33, 289–293. 10.1080/01490451.2015.1048395 [DOI] [Google Scholar]
- Svobodová K., Novotný Č. (2018). Bioreactors based on immobilized fungi: Bioremediation under non-sterile conditions. Appl. Microbiol. Biotechnol. 102, 39–46. 10.1007/s00253-017-8575-z [DOI] [PubMed] [Google Scholar]
- Syed K., Porollo A., Lam Y. W., Grimmett P. E., Yadav J. S. (2013). CYP63A2, a catalytically versatile fungal P450 monooxygenase capable of oxidizing higher-molecular-weight polycyclic aromatic hydrocarbons, alkylphenols, and alkanes. Appl. Environ. Microbiol. 79, 2692–2702. 10.1128/aem.03767-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar D., Jasrotia T., Umar A., Kumar R., Kumar R., Alkhanjaf A. a. M., et al. (2022). Mechanistic and analytical understanding of biological immobilization of chromium metal ions from waste-sites. J. Environ. Chem. Eng. 10, 107498. 10.1016/j.jece.2022.107498 [DOI] [Google Scholar]
- Tang A. X., Lu Y. H., Li Q. Y., Zhang X. L., Cheng N., Liu H. B., et al. (2021). Simultaneous leaching of multiple heavy metals from a soil column by extracellular polymeric substances of Aspergillus tubingensis F12. Chemosphere 263, 127883. 10.1016/j.chemosphere.2020.127883 [DOI] [PubMed] [Google Scholar]
- Taufek N. M., Harith H. H., Abd Rahim M. H., Ilham Z., Rowan N., Wan-Mohtar W. a. Q. I. (2020). Performance of mycelial biomass and exopolysaccharide from Malaysian Ganoderma lucidum for the fungivore red hybrid Tilapia (Oreochromis sp.) in Zebrafish embryo. Aquac. Rep. 17, 100322. Aquaculture Reports . 10.1016/j.aqrep.2020.100322 [DOI] [Google Scholar]
- Tedersoo L., Mikryukov V., Zizka A., Bahram M., Hagh-Doust N., Anslan S., et al. (2022). Global patterns in endemicity and vulnerability of soil fungi. Glob. Change Biol. 28, 6696–6710. 10.1111/gcb.16398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleky B. E., Vodnar D. C. (2021). Recent Advances in Biotechnological Itaconic Acid Production, and Application for a Sustainable Approach. Polymers 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temporiti M. E. E., Nicola L., Nielsen E., Tosi S. (2022). Fungal enzymes involved in plastics biodegradation. Microorganisms 10, 1180. 10.3390/microorganisms10061180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Jiang Z. Q., Jiang L., Su M., Feng Z. Y., Zhang L., et al. (2019). A new insight into lead (II) tolerance of environmental fungi based on a study of Aspergillus niger and Penicillium oxalicum . Environ. Microbiol. 21, 471–479. 10.1111/1462-2920.14478 [DOI] [PubMed] [Google Scholar]
- Tonelli F., Bek J. W., Besio R., De Clercq A., Leoni L., Salmon P., et al. (2020). Zebrafish: A resourceful vertebrate model to investigate skeletal disorders. Front. Endocrinol. 11, 489. 10.3389/fendo.2020.00489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z. Y., Zheng X. M., Tong Y., Shi Y. C., Sun J. B. (2019). Systems metabolic engineering for citric acid production by Aspergillus niger in the post-genomic era. Microb. Cell. Factories 18, 28. 10.1186/s12934-019-1064-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo-Budowski R., Cambronero-Heinrichs J. C., Duran J. E., Masis-Mora M., Ramirez-Morales D., Quiros-Fournier J. P., et al. (2021). Removal of pharmaceuticals and ecotoxicological changes in wastewater using Trametes versicolor: A comparison of fungal stirred tank and trickle-bed bioreactors. Chem. Eng. J. 410, 128210. 10.1016/j.cej.2020.128210 [DOI] [Google Scholar]
- Townsley C. C., Ross I. S. (1985). Copper uptake by Penicillium-spinulosum . Microbios 44, 125–134. [Google Scholar]
- Tripathi P., Misra S. R. (2019). Recently developed technologies for waste water treatment in industries. Agrobios Newsl. 36. [Google Scholar]
- Troiano D., Orsat V., Dumont M. J. (2020). Status of filamentous fungi in integrated biorefineries. Renew. Sustain. Energy Rev. 117, 109472. 10.1016/j.rser.2019.109472 [DOI] [Google Scholar]
- Trueba-Santiso A., Parlade E., Rosell M., Lliros M., Mortan S. H., Martinez-Alonso M., et al. (2017). Molecular and carbon isotopic characterization of an anaerobic stable enrichment culture containing Dehalobacterium sp during dichloromethane fermentation. Sci. Total Environ. 581, 640–648. 10.1016/j.scitotenv.2016.12.174 [DOI] [PubMed] [Google Scholar]
- Tsay S. S., To K. Y. (1987). Citric acid production using immobilized conidia of Aspergillus niger TMB 2022. Biotechnol. Bioeng. 29, 297–304. 10.1002/bit.260290302 [DOI] [PubMed] [Google Scholar]
- Usuldin S. R. A., Wan-Mohtar W. a. Q. I., Ilham Z., Jamaludin A. A., Abdullah N. R., Rowan N. (2021). In vivo toxicity of bioreactor-grown biomass and exopolysaccharides from Malaysian tiger milk mushroom mycelium for potential future health applications. Sci. Rep. 11, 23079. 10.1038/s41598-021-02486-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanishree M., Thatheyus A., Ramya D. (2014). Biodegradation of petrol using the fungus Penicillium sp. Sci. Int. 2, 26–31. 10.17311/sciintl.2014.26.31 [DOI] [Google Scholar]
- Varjani S. J. (2017). Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 223, 277–286. 10.1016/j.biortech.2016.10.037 [DOI] [PubMed] [Google Scholar]
- Vertommen M. a. M. E., Nierstrasz V. A., Van Der Veer M., Warmoeskerken M. M. C. G. (2005). Enzymatic surface modification of poly(ethylene terephthalate). J. Biotechnol. 120, 376–386. 10.1016/j.jbiotec.2005.06.015 [DOI] [PubMed] [Google Scholar]
- Vuong P., Wise M. J., Whiteley A. S., Kaur P. (2022). Small investments with big returns: Environmental genomic bioprospecting of microbial life. Crit. Rev. Microbiol. 48, 641–655. 10.1080/1040841x.2021.2011833 [DOI] [PubMed] [Google Scholar]
- Vyas B. R. M., Bakowski S., Sasek V., Matucha M. (1994). Degradation of anthracene by selected white-rot fungi. Fems Microbiol. Ecol. 14, 65–70. 10.1111/j.1574-6941.1994.tb00091.x [DOI] [Google Scholar]
- Wan-Mohtar W. a. Q. I., Halim-Lim S. A., Kamarudin N. Z., Rukayadi Y., Abd Rahim M. H., Jamaludin A. A., et al. (2020). Fruiting-body-base flour from an Oyster mushroom waste in the development of antioxidative chicken patty. J. Food Sci. 85, 3124–3133. 10.1111/1750-3841.15402 [DOI] [PubMed] [Google Scholar]
- Wan-Mohtar W. a. Q. I., Ilham Z., Jamaludin A. A., Rowan N. (2021). Use of zebrafish embryo assay to evaluate toxicity and safety of bioreactor-grown exopolysaccharides and endopolysaccharides from European Ganoderma applanatum mycelium for future aquaculture applications. Int. J. Mol. Sci. 22, 1675. 10.3390/ijms22041675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Q., Tanaka Y., Ohno H., Jia J. B., Mori T., Xiao T. F., et al. (2019). Biotransformation and detoxification of the neonicotinoid insecticides nitenpyram and dinotefuran by Phanerochaete sordida YK-624. Environ. Pollut. 252, 856–862. 10.1016/j.envpol.2019.06.022 [DOI] [PubMed] [Google Scholar]
- Wasay S. A., Barrington S. F., Tokunaga S. (1998). Using Aspergillus niger to bioremediate soils contaminated by heavy metals. Bioremediation J. 2, 183–190. 10.1080/10889869891214303 [DOI] [Google Scholar]
- Webb H. K., Arnott J., Crawford R. J., Ivanova E. P. (2013). Plastic degradation and its environmental implications with special reference to poly(ethylene terephthalate). Polymers 5, 1–18. 10.3390/polym5010001 [DOI] [Google Scholar]
- Widdel F., Rabus R. (2001). Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12, 259–276. 10.1016/s0958-1669(00)00209-3 [DOI] [PubMed] [Google Scholar]
- Wierckx N., Agrimi G., Lubeck P. S., Steiger M. G., Mira N. P., Punt P. J. (2020). Metabolic specialization in itaconic acid production: A tale of two fungi. Curr. Opin. Biotechnol. 62, 153–159. 10.1016/j.copbio.2019.09.014 [DOI] [PubMed] [Google Scholar]
- Woodard F. (2001). Industrial waste treatment handbook. Elsevier. [Google Scholar]
- Xu H. M., Hao R. X., Xu X. Y., Ding Y., Lu A. H., Li Y. H. (2021). Removal of hexavalent chromium by Aspergillus niger through reduction and accumulation. Geomicrobiol. J. 38, 20–28. 10.1080/01490451.2020.1807659 [DOI] [Google Scholar]
- Xu X., Liu W., Tian S., Wang W., Qi Q., Jiang P., et al. (2018). Petroleum hydrocarbon-degrading bacteria for the remediation of oil pollution under aerobic conditions: A perspective analysis. Front. Microbiol. 9, 2885. 10.3389/fmicb.2018.02885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A., Kon K., Kratosova G., Duran N., Ingle A. P., Rai M. (2015). Fungi as an efficient mycosystem for the synthesis of metal nanoparticles: Progress and key aspects of research. Biotechnol. Lett. 37, 2099–2120. 10.1007/s10529-015-1901-6 [DOI] [PubMed] [Google Scholar]
- Yan Y., Liu X., Jiang X., Zhang W., Wang Y., Wang Y., et al. (2022). Surface charge modifications modulate glucose oxidase pH-activity profiles for efficient gluconic acid production. J. Clean. Prod. 372, 133817. 10.1016/j.jclepro.2022.133817 [DOI] [Google Scholar]
- Yang C., Peng B. (2022). Biodegradation characteristics of patulin by Saccharomyces cerevisiae during fermentation. Food Control 145, 109463. [Google Scholar]
- Zahri K. N. M., Zulkharnain A., Gomez-Fuentes C., Sabri S., Ahmad S. A. (2021). Study of growth kinetics of Antarctic bacterial community for biodegradation of waste canola oil. Desalin. Water Treat. 213, 128–138. 10.5004/dwt.2021.26692 [DOI] [Google Scholar]
- Zapana-Huarache S. V., Romero-Sánchez C. K., Dueñas Gonza A. P., Torres-Huaco F. D., Lazarte Rivera A. M. (2020). Design and testing of a cost-efficient bioremediation system for tannery effluents using native chromium-resistant filamentous fungi. Int. J. Environ. Sci. Technol. 17, 3825–3834. 10.1007/s13762-020-02726-9 [DOI] [Google Scholar]
- Zhang Q. H., Zeng G. M., Chen G. Q., Yan M., Chen A. W., Du J. J., et al. (2015). The effect of heavy metal-induced oxidative stress on the enzymes in white rot fungus Phanerochaete chrysosporium . Appl. Biochem. Biotechnol. 175, 1281–1293. 10.1007/s12010-014-1298-z [DOI] [PubMed] [Google Scholar]
- Zhang X. Y., Li Y. H., Zhao X. Q., Bai F. W. (2017). Constitutive cellulase production from glucose using the recombinant Trichoderma reesei strain overexpressing an artificial transcription activator. Bioresour. Technol. 223, 317–322. 10.1016/j.biortech.2016.10.083 [DOI] [PubMed] [Google Scholar]
- Zhao M. A., Gu H., Zhang C. J., Jeong H., Kim E. O. H., Zhu Y. Z. (2020). Metabolism of insecticide diazinon by Cunninghamella elegans ATCC36112. RSC Adv. 10, 19659–19668. 10.1039/d0ra02253e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Liao X. Z., Wang J. X., Ning Y. N., Li C. X., Liao L. S., et al. (2019). Transcription factor Atf1 regulates expression of cellulase and xylanase genes during solid-state fermentation of ascomycetes. Appl. Environ. Microbiol. 85 (24), e01226-19. 10.1128/aem.01226-19 [DOI] [PMC free article] [PubMed] [Google Scholar]