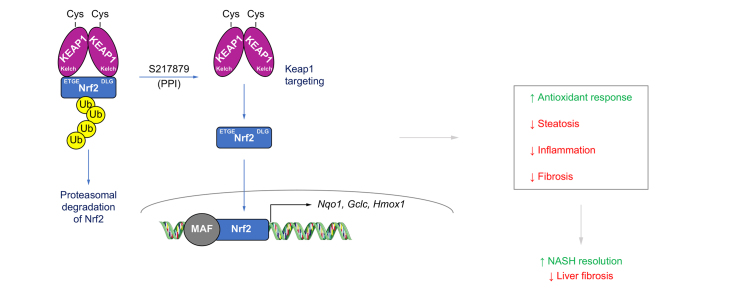
Abstract
Background & Aims
Oxidative stress is recognized as a major driver of non-alcoholic steatohepatitis (NASH) progression. The transcription factor NRF2 and its negative regulator KEAP1 are master regulators of redox, metabolic and protein homeostasis, as well as detoxification, and thus appear to be attractive targets for the treatment of NASH.
Methods
Molecular modeling and X-ray crystallography were used to design S217879 – a small molecule that could disrupt the KEAP1-NRF2 interaction. S217879 was highly characterized using various molecular and cellular assays. It was then evaluated in two different NASH-relevant preclinical models, namely the methionine and choline-deficient diet (MCDD) and diet-induced obesity NASH (DIO NASH) models.
Results
Molecular and cell-based assays confirmed that S217879 is a highly potent and selective NRF2 activator with marked anti-inflammatory properties, as shown in primary human peripheral blood mononuclear cells. In MCDD mice, S217879 treatment for 2 weeks led to a dose-dependent reduction in NAFLD activity score while significantly increasing liver Nqo1 mRNA levels, a specific NRF2 target engagement biomarker. In DIO NASH mice, S217879 treatment resulted in a significant improvement of established liver injury, with a clear reduction in both NAS and liver fibrosis. αSMA and Col1A1 staining, as well as quantification of liver hydroxyproline levels, confirmed the reduction in liver fibrosis in response to S217879. RNA-sequencing analyses revealed major alterations in the liver transcriptome in response to S217879, with activation of NRF2-dependent gene transcription and marked inhibition of key signaling pathways that drive disease progression.
Conclusions
These results highlight the potential of selective disruption of the NRF2-KEAP1 interaction for the treatment of NASH and liver fibrosis.
Impact and implications
We report the discovery of S217879 – a potent and selective NRF2 activator with good pharmacokinetic properties. By disrupting the KEAP1-NRF2 interaction, S217879 triggers the upregulation of the antioxidant response and the coordinated regulation of a wide spectrum of genes involved in NASH disease progression, leading ultimately to the reduction of both NASH and liver fibrosis progression in mice.
Keywords: NASH, fibrosis, NRF2, oxidative stress
Abbreviations: ARE, antioxidant response element; DIO, diet-induced obesity; HEC, hydroxyethyl cellulose; hPBMCs, human peripheral blood mononuclear cells; KEAP1, Kelch-like ECH associated protein 1; LPS, lipopolysaccharide; MCDD, methionine- and choline-deficient diet; NAFLD, non-alcoholic fatty liver disease; NAS, NAFLD activity score; NASH, non-alcoholic steatohepatitis; NRF2, nuclear factor erythroid 2–related factor 2; PSR, Picrosirius red; ROS, reactive oxygen species; GSEA, Gene Set Enrichment Analysis; HSCs, Hepatic Stellate Cells; PPI, Protein-protein interaction; 4-HNE, 4-hydroxynonenal
Graphical abstract
Highlights
-
•
S217879 is a potent and selective compound that disrupts the KEAP1-NRF2 interaction leading to robust NRF2 pathway activation.
-
•
S217879 treatment prevents NASH progression in mice fed a methionine- and choline-deficient diet.
-
•
S217879 treatment significantly reduces both NAS score and fibrosis in DIO-NASH mice.
-
•
NRF2 activation triggers upregulation of the antioxidant response and coordinated regulation of a wide spectrum of genes.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is a common and progressive disease mainly characterized by hepatic fat accumulation in the absence of alcohol consumption. NAFLD is strongly associated with obesity, metabolic syndrome, type 2 diabetes and dyslipidemia. NAFLD is subdivided into non-alcoholic fatty liver (NAFL) and non-alcoholic steatohepatitis (NASH) based on histological examination of liver biopsy and defined by the presence of inflammation and hepatocyte ballooning with various degrees of fibrosis.1,2 NAFLD is the most common cause of chronic liver disease worldwide, with an estimated prevalence of 25%. It is therefore considered as a global health problem associated with a significant socioeconomic burden.3,4 In contrast to NAFL, which is considered as a benign and reversible disease state, NASH accounts for an increased number of patients with cirrhosis, liver failure and hepatocellular carcinoma.5 NAFLD further increases the risk of mortality in a population with a high cardiovascular risk. Long-term follow-up studies revealed that fibrosis is the main driver of mortality in NASH.6,7 NAFLD remains an unmet medical need since there are no approved therapies despite significant R&D efforts (See8 for review). NAFLD disease phenotype results from the chronic exposure to environmental factors on a susceptible polygenic background comprising multiple independent modifiers.9 Several genes have been associated with the development of NAFLD and more generally liver diseases using genome-wide association studies (see10 for review). From a mechanistic standpoint, it is believed that the accumulation of both triglycerides and proinflammatory and cytotoxic lipid oxidation side-products results in the formation of a necro-inflammatory milieu which triggers the activation of the main fibrogenic hepatic cell population, namely hepatic stellate cells. Oxidative stress is largely recognized as a major driver of NASH progression.11 Patients with NAFLD usually display increased levels of lipid peroxidation products and systemic oxidative stress markers.12,13 Furthermore, histological analyses confirmed that patients with NAFLD exhibit a significant increase in 4-hydroxynonenal-positive cells in the liver.14,15 4-hydroxynonenals are formed during lipid peroxidation of polyunsaturated fatty acids. Finally, there is a marked decrease of glutathione and hepatic antioxidant enzymes in patients with NAFLD.16 Indeed, the insulin resistance-mediated increase in oxidative phosphorylation is a major source of oxidative stress which triggers hepatocellular damage and further exacerbates insulin resistance. Satapati and colleagues have shown that hepatic oxidative stress and inflammation are associated with an elevated oxidative metabolism of saturated fatty acids in NAFLD.17 Adaptation of mitochondrial function during NAFL is lost in NASH, with increased reactive oxygen species (ROS), lipid peroxidation products and decreased ATP content leading to necrosis and fueling the development of steatohepatitis.18 Since oxidative stress plays a major role in NASH disease progression, as well as the development of liver fibrosis,11,19 it is an attractive target for the treatment of NASH.
The nuclear factor erythroid 2–related factor 2 (NRF2), a member of the cap’ n’ collar basic leucine zipper transcription factor family, and its negative regulator, the E3 ligase adaptor Kelch-like ECH associated protein 1 (KEAP1), are master regulators of cellular resistance to oxidants (See Dodson et al. for review20). Under resting conditions, NRF2 is sequestered within the cytoplasm. The N-terminal domain of the KEAP1 homodimer binds one molecule of NRF2 leading to ubiquitination by the E3 ligase complex, namely CUL3/RBX1 and ultimately to proteasomal degradation.21,22 Under oxidative conditions, increased levels of electrophilic molecules result in the covalent modifications of highly reactive cysteine residues within KEAP1 triggering conformational changes and the release of CUL3.[23], [24], [25], [26] NRF2 is then released and translocates into the nucleus where it accumulates and dimerizes with small MAF proteins to activate transcription of genes containing the so-called antioxidant response element (ARE).27,28 NRF2 regulates the transcription of more than 250 genes bearing an ARE involved in antioxidant cellular defense, xenobiotic metabolism and detoxification, carbohydrate and lipid metabolism, protein degradation as well as inflammation.29 It is therefore not surprising that the NRF2 signaling pathway is considered as a valid target for a number of acute and chronic diseases for which oxidative stress and inflammation are key biological drivers.30 Increased oxidative stress is a hallmark of chronic liver diseases which explains why NRF2 biology has been extensively probed in the liver (See31 for review 2014). Interestingly, Nrf2-deficient mice display an increased susceptibility to the development of NASH and fibrosis when placed on a high-fat diet for 24 weeks.32 Biochemical and molecular analyses revealed that NRF2 protects against NASH, at least in part, by regulating oxidative stress and suppressing de novo lipogenesis, endoplasmic reticulum stress and inflammation.32 Similar findings, namely marked increases in steatosis, inflammation and oxidative stress have been reported in Nrf2 knockout mice fed a methionine- and choline-deficient diet (MCDD).33,34 Conversely, sustained NRF2 activation in Keap1 gene knockdown mice prevented MCDD-induced liver injury.35 Furthermore, NRF2 pharmacological activation of using potent electrophilic compounds such as 1-[2-cyano3-,12-dioxooleana-1,9(11)-dien-28-oyl] imidazole (CDDO-Im), TBE-31, sulforaphane and omaveloxolone were shown to limit NASH progression and fibrosis in multiple preclinical models.[35], [36], [37], [38] Finally, NRF2 protein levels have been shown to be significantly reduced in livers from obese patients with NASH but not NAFL.39,40 Taken together, all these studies strongly support NRF2 targeting for the treatment of NASH and liver fibrosis. While a number of electrophilic NRF2 agonists delivered initially promising results for various therapeutic indications in preclinical models, they all suffer from a reduced safety margin, mainly due to their limited selectivity and pleiotropic pharmacology, preventing their clinical evaluation in randomized trials for NASH. Bardoxolone has been shown to interact with more than 500 molecular species41 and to inhibit the endothelin-1 signaling pathway and increase cardiovascular risk in patients with chronic kidney disease.42,43 The availability of KEAP1 X-ray structure44 opened the door to the discovery of second-generation NRF2 activators with a completely different way of engaging the target via the direct disruption of its interaction with KEAP1.45 This led to the identification of potent and selective compounds but with limited oral bioavailability.46 Herein, we report the discovery and characterization of S217879, a potent and selective small molecule disrupting the KEAP1-NRF2 interaction with good pharmacokinetic properties upon oral administration in rodents. Then, we evaluated its potential as a novel treatment for NASH and liver fibrosis in two different preclinical models.
Materials and methods
S217879
S217879 was synthesized by Servier medicinal chemistry department and was determined to be >98% pure by high-performance liquid chromatography and/or nuclear magnetic resonance analysis (chemical synthesis described within the supplementary information).
LPS-induced cytokine secretion assay
Human peripheral blood mononuclear cells (hPBMCs) were provided by Lonza (#CC-2702). hPBMCs (150,000 cells/well) were plated in 96 well plates in RPMI1640 medium supplemented with 10% FCS, 2% Glutamax and 2% penicillin-streptomycin cocktail (Life Technologies). Cells were then pre-incubated at 37 °C for 4 h with various concentrations of S217879 or vehicle (DMSO, 0.1%). hPBMCs were then stimulated with lipopolysaccharide (LPS: 10 ng/ml) for 4 h. At the end of the stimulation period, cytokine secretion was evaluated using MagPix multiplex kits (Biorad) according to manufacturer’s instructions.
Animals
Mice were maintained on a 12:12 h light/dark cycle at 21 ± 2 °C and had ad libitum access to tap water and standard (A04) or MCD or AMLN diets. All procedures were performed according to the ethical protocol that has been approved by the Servier Institutional Animal Care and Use Committee in accordance with the French regulations (Decree n° 2013-118 from 01 February 2013 relative to the protection of animals used for scientific purposes and 4 for orders).
MCDD models
8-week-old male C57BL6/J mice (Janvier labs, France) were randomly assigned to either control diet (A04 diet, SAFE) or the MCDD (BROGAARDEN, open source diets) for 2 weeks. Mice on MCDD immediately received either S217879 (3 or 30 mg/kg/day) or vehicle (hydroxyethyl cellulose [HEC] 1%) by gavage for 2 weeks. At the end of the study, mice were fasted for 4 h before termination. Intracardiac blood samples were collected on anesthetized animals (using isoflurane). Liver samples were collected for biochemical, histological and gene expression analyses.
DIO NASH model
5–6-week-old C57BL/6JRj mice were fed with an AMLN diet (D09100301, Research Diet, US) (40% fat [18% trans-fat], 40% carbohydrate [20% fructose], and 2% cholesterol) for 33 weeks prior to initiation of the treatment protocol. Prior to treatment, all mice underwent liver biopsy for confirmation and stratification of liver steatosis and fibrosis, using the NAFLD activity score (NAS) and fibrosis staging system as previously described.47 Only mice with fibrosis stage ≥1 and steatosis score ≥2, were included in the study. DIO NASH mice were kept on an AMLN diet and received either S217879 (30 mg/kg, PO, QD) or vehicle (HEC 1%) for 8 weeks. A terminal blood sample was collected from the tail vein in non-fasted mice and used for plasma biochemistry. Animals were sacrificed by cardiac puncture under isoflurane anesthesia. Liver samples were processed as described below.
Histological analyses
Formalin-fixed, paraffin-embedded livers were sliced into 3 μm sections. H&E staining was performed to investigate liver histology and Picrosirius red (PSR) staining was used for liver fibrosis. Type 1 collagen (Southern Biotech, #1310-01), galectin-3 and alpha smooth muscle actin (αSMA, Abcam, Ab124964) immunohistochemistry were performed using standard procedures. NAS and fibrosis stage were determined by two double-blinded persons using the NASH Clinical Research Network scoring system.48 For hepatocellular steatosis, livers were classified into scores 0 to 3 (0: <5% of hepatocytes presenting steatosis, 1: 5 to 33% of hepatocytes presenting steatosis, 2: 33 to 66% of hepatocytes presenting steatosis and 3: >66% of hepatocytes presenting steatosis). For inflammation, livers were scored into grades 0 to 3 (0: non-inflammatory foci, 1: 1 inflammatory focus, 2: 2 to 4 inflammatory foci, 3: >4 inflammatory foci). Fibrosis was scored into stages from 0 to 4 (0: no fibrosis, 1: perisinusoidal or periportal fibrosis, 2: perisinusoidal and periportal fibrosis, 3: bridging fibrosis or septa, 4: cirrhosis).
Biochemical analyses
Plasma parameters were determined with an automatic biochemical analyzer (Indiko Clinical Chemistry Analyzer, Thermofisher). Liver triglyceride levels were measured using a commercially available kit (Roche Diagnostics) after homogenization and extraction as described.49 For hydroxyproline levels, liver samples are homogenized in 6 M HCl and hydrolyzed to degrade collagen. Samples are centrifuged and the hydroxyproline content is measured in duplicates in the supernatant, using a colorimetric assay (Quickzyme Biosciences) according to the manufacturer’s instructions.
Gene expression studies by reverse-transcription quantitative PCR
Total RNA was extracted using Qiagen RNeasy Lipid extraction kit following manufacturer’s instructions. Total RNA was treated with DNase I (Qiagen) at 37 °C for 30 min, followed by inactivation at 75 °C for 5 min. Reverse-transcription quantitative PCR assays were performed using an Applied Biosystems QuantStudio 7 Flex System. Total RNA (1 μg) was reverse transcribed with random hexamers using Hight-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Applied Biosystems, ThermoFisher Scientific) following the manufacturer’s protocol. Gene expression levels were determined with the Taqman Universal Master mix (2x) using Taqman assays (Applied Biosystems). The 18S transcript was used as an internal control to normalize the variations for RNA amounts. Gene expression levels are expressed relative to 18S mRNA levels. All primers used in this study were provided by Thermofisher.
Liver RNA-sequencing
Total RNA was extracted using Qiagen RNA extraction kits following manufacturer’s instructions. RNA concentrations were obtained using nanodrop or a fluorometric Qubit RNA assay (Life Technologies, Grand Island, New York, USA). The quality of the RNA (RNA integrity number) was determined on the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) as per the manufacturer’s instructions. To construct the libraries, 400 ng of high-quality total RNA sample (RNA integrity number >8) was processed using TruSeq Stranded mRNA kit (Illumina) according to the manufacturer’s instructions. Briefly, after purification of poly-A-containing mRNA molecules, mRNA molecules are fragmented and reverse transcribed using random primers. Replacement of dTTP with dUTP during the second strand synthesis will enable the achievement of strand specificity. Addition of a single A base to the cDNA is followed by ligation of Illumina adapters. Libraries were quantified by qPCR using the KAPA Library Quantification Kit for Illumina Libraries (KapaBiosystems, Wilmington, MA) and library profiles were assessed using the DNA High Sensitivity LabChip kit on an Agilent Bioanalyzer. Libraries were sequenced on an Illumina NovaSeq 6000 instrument using 150 base-lengths read V2 chemistry in a paired-end mode. Sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36. The trimmed reads were mapped to the Mus musculus GRCm38 reference genome available on ENSEMBL using the STAR aligner v.2.5.2b. Unique gene hit counts were calculated by using the featureCounts from the Subread package v.1.5.2. Only unique reads that fell within exon regions were counted. Counts have been normalized in transcripts per million using the standard formula.
RNA-seq data analyses
Data analyses were carried out using R system software (http://www.R-project.org, V4.0.2) packages including those of Bioconductor or original R code. Gene hit counts were used for downstream differential expression analysis. Using DESeq2, a comparison of gene expression between the S217879-treated samples and vehicle samples was performed. The Wald test was used to generate p values and log2 fold changes. Genes with an adjusted p value <0.05 and absolute log2 fold-change >1 (fold-change >2) were considered as differentially expressed genes between conditions. Enrichment analysis was performed by applying the fast gene set-enrichment analysis (fgsea function from fgsea R package v.1.16.0, pre-ranked mode) against two separated genesets collections, HALLMARK and a combination of KEGG- and Reactome-curated pathways. Representations of data and results have been generated with ComplexHeatmap v.2.6.2 and ggplot2 v.3.3.5 packages. Data are available under the GSE212644 accession number.
Statistical analysis
For comparison of two groups, an unpaired Student’s t test was used (GraphPad Prism software) after verification of the normal distribution of data. For more than two groups, a one-way ANOVA was performed followed by a Dunnett’s multiple comparison test. For body weight, a two-way ANOVA was performed followed by a Tukey’s test. For histological parameters, a statistical model ANOVA was performed with strain and diet as fixed effects on a score parameter (NAS, steatosis, fibrosis or inflammation), followed by least-square mean estimations (SAS software). Significance threshold was 5%. Results are expressed as mean ± SEM.
Results
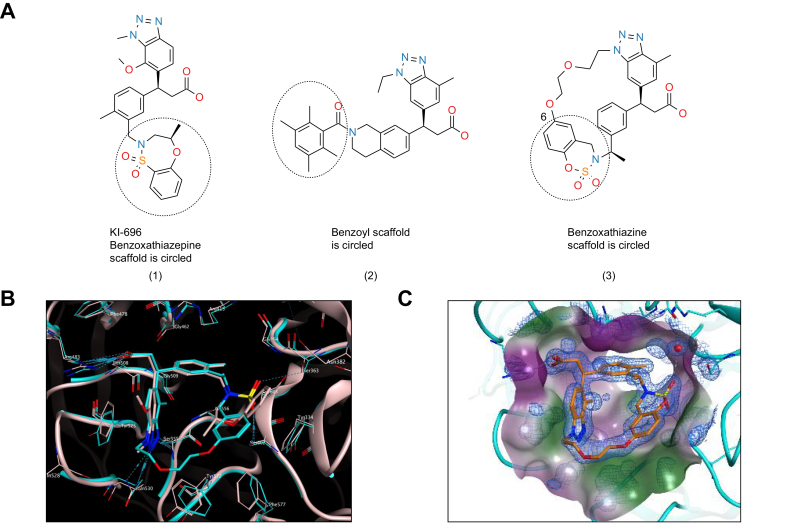
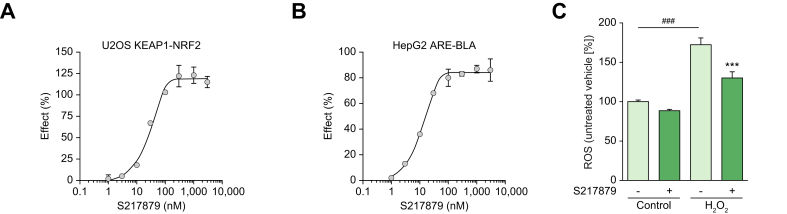
Non-covalent inhibitors of the KEAP1 Kelch/NRF2 protein-protein interaction may increase selectivity with reduced cytotoxicity, translating into a larger therapeutic index. Several groups have reported the discovery of such molecules. Most of them exhibit promising binding affinities. However, they usually suffer from poor ADME-pharmacokinetic properties limiting their use for probing NRF2 biology in vivo. Davies and colleagues reported the discovery of KI-696 (1) (Fig. 1), a potent compound which activates the NRF2 antioxidant response in cellular models. In vivo target engagement required i.v. infusion due to its poor oral bioavailability.46 KI-696 binding to the Kelch domain has been confirmed by X-ray crystallography.50 More recently, the discovery of a novel non-covalent chemotype with an acceptable pharmacokinetic profile for oral dosing was reported.51 Preliminary in vivo results of the best compound (2) (Fig. 1) revealed target engagement as measured by NRF2-driven gene induction from 10 mg/kg onwards.51 The X-ray structure of compound 1 and compound 2 Kelch complexes revealed the possibility of macrocyclization which is generally known to modify the entropic contribution to binding.52 In addition, variation of the linker’s position and substitution allows for the fine-tuning of the physicochemical properties without disrupting the interactions with the protein. We decided to replace the benzoxathiazepine and benzoyl scaffolds of 1 and 2 with benzoxathiazine in the hope that the binding mode would exhibit the same key interactions as the parent molecules, with improved hydrophobic interactions (Tyr334, Phe557). We hypothesized that position 6 of the benzoxathiazine’s benzene ring would provide a suitable vector for linking the aromatic ring with benztriazole’s N-alkyl group and would tolerate diverse substitution patterns on the linker. Substutition of the benzyl position next to the benzoxathiazine nitrogene atom also proved to be an effective tool to fine-tune the interaction between benzoxathiazine oxygenes and the protein. Optimization of the macrocyclics’ structure was focused on the linker part and led to the synthesis and subsequent selection of compound 3 (See 0; Compound 3 is referred to as S217879 throughout the manuscript) as a candidate. Binding to KEAP1 Kelch domain was confirmed with a direct surface plasmon resonance assay (Kd = 4.15 nM; Table S1). The X-ray cocrystal structure confirmed that the benzoxathiazine ring’s position allowed for improved interactions with hydrophobic residues (Tyr334, Phe557, Fig. 1C). Then, we assessed the ability of S217879 to trigger NRF2 nuclear translocation in U2OS cells using a beta galactosidase complementation assay. Andrographolide, a plant-derived diterpenoid was used at 10 μM as a positive control. Andrographolide triggers NRF2 nuclear translocation by forming adducts with cysteine 151 within KEAP1.53 S217879 induced a concentration-dependent increase in NRF2 translocation in U2OS cells with an EC50 of 23 nM (Fig. 2A). In line with these results, S217879 significantly activated a reporter gene driven by an ARE in HepG2 cells in a concentration-dependent manner with an EC50 of 18 nM (Fig. 2B). Since NRF2 is a master regulator of cellular resistance to oxidants (See Dodson et al. for review0), we next tested S217879’s ability to reduce H2O2-stimulated ROS production in HepG2 cells. As expected, H2O2 triggered a significant increase in ROS production which was significantly reduced by S217879 treatment (Fig. 2C). Taken together, these results indicate that S217879 is a potent NRF2 activator in vitro.
Fig. 1.
Identification of S217879 as a novel NRF2 activator.
(A) Chemical structure of Kelch-NRF2 interaction disruptors: (1) PPI compound with in vivo target engagement established upon i.v. infusion46 (2) PPI compound with in vivo target engagement established upon oral administration51 (3) Chemical structure of novel PPI compound with good developability profile aka S217879. (B) Overlay of X-ray structures 5GNU (compound 1, grey) and compound 3 (cyan). (C) Crystal structure of the Kelch domain of KEAP1 bound to S217879 (resolution: 1.3 Angström). PPI, protein-protein interaction.
Fig. 2.
S217879 is a potent NRF2 activator in cell-based assays.
(A) S217879 triggers NRF2 translocation in U2OS cells in a concentration-dependent manner (2 experiments performed in triplicates; data expressed as mean ± SEM). (B) Activation of ARE-driven transcription in HepG2 cells in response to S217879 (2 experiments performed in triplicates; data expressed as mean ± SEM). (C) S217879 pretreatment (1 mM overnight, DMSO 0.1% used as vehicle) limits H2O2-triggered ROS generation in HepG2 cells (n = 5 per condition, data expressed as mean ± SEM). Statistical significance was assessed by one-way ANOVA followed by Dunnett’s multiple comparison test. ###p <0.001 H2O2-triggered vs. untriggered cells, ∗∗∗p <0.001 S217879-treated vs. vehicle.
Next, S217879 was evaluated in a broad selectivity panel comprising 110 targets. In this panel, S217879 was tested at 10 μM, a roughly 2,000-fold higher concentration with respect to KEAP1 binding. Interestingly, none of these targets were significantly activated or inhibited in response to S217879, strongly supporting its selectivity (Table S2).
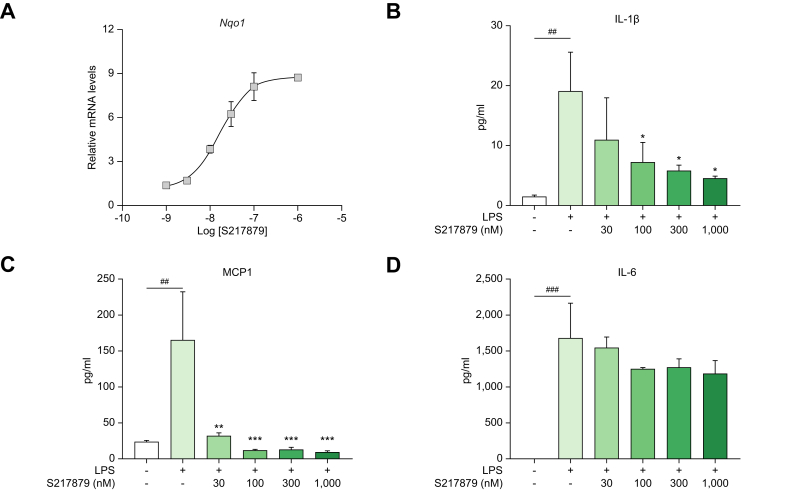
Since NRF2 activation has been shown to inhibit LPS-driven inflammation independently of redox control in both monocytes and macrophages,54 we next evaluated the anti-inflammatory properties of S217879 in primary hPBMCs. In hPBMCs, S217879 treatment for 6 h resulted in a concentration-dependent increase in Nqo1 gene expression with an EC50 of 16 nM (Fig. 3A) in line with its cell-based potency determined in both HepG2 and U2OS cells (Fig. 2). As expected, LPS triggered a robust and significant increase in IL-1β, IL-6 and MCP-1 secretion (Fig. 3B-D). Pretreatment for 4 h with S217879 led to a significant and concentration-dependent inhibition of LPS-induced cytokine release as measured by IL1β, MCP-1 and IL6 MagPix multiplex assays (Fig. 3B-D). Of note, both IL-1β and MCP-1 secretion were significantly reduced by S217879 treatment (IC50 <30 nM in line with NRF2 cell-based potency) in contrast to IL-6 which was slightly and not significantly reduced at higher concentrations (Fig. 3B-D). NRF2 has been shown to negatively interfere with the NF-κB pathway which regulates the expression of a wide spectrum of inflammatory genes.54 Why IL6 regulation seems to be less sensitive to NRF2 remains unknown. Nevertheless, these data indicate that S217879 displays significant anti-inflammatory properties in hPBMCs (Fig. 3B-D) while strongly engaging the NRF2 signaling pathway (Fig. 3A).
Fig. 3.
Inhibition of LPS-driven inflammatory response by S217879 in PBMCs.
(A) Nqo1 gene upregulation in human PBMCs in response to increasing concentrations of S217879 (6 h incubation; DMSO 0.1% used as vehicle). (B-D) Inhibition of LPS-stimulated cytokine secretion in human PBMCs exposed to increasing concentrations of S217879 (4 h pre-incubation; DMSO 0.1% used as vehicle). Data expressed as mean ± SEM. Statistical significance was assessed by one-way ANOVA followed by Dunnett’s multiple comparison test. ##p <0.01 and ###p <0.001: LPS-triggered vs. untriggered cells. ∗p <0.05, ∗∗p <0.01 and ∗∗∗p <0.001: S2178789-treated vs. vehicle.
In vitro ADME and safety parameters were profiled comprehensively to identify an orally available and safe compound (Table S3). The ability of S217879 to activate the NRF2 signaling pathway in vivo was next evaluated in C57BL6 mice. Male C57BL6 mice received a single administration of S217879 (30 mg/kg) or vehicle (HEC 1%) by gavage. Drug plasma exposure was quantified over time and NRF2 target engagement was assessed in the liver by measuring Nqo1 mRNA levels by reverse-transcription quantitative PCR. S217879 drug levels rise rapidly following oral administration with Cmax reaching 3.2 μM (Fig. S1). Plasma concentrations declined rapidly (AUC = 3.8 μM h) while Nqo1 mRNA levels in the liver increased in a time-dependent manner, reaching an up to 10-fold increase over vehicle 24 h post oral administration (Fig. S1A). Similar results were obtained in the kidney (data not shown). Of note, S217879 treatment (single or chronic administration) was found to increase the hepatic expression of numerous NRF2 target genes such as Gclc, Gstm1, Gpx2 (Fig. S1B, Fig. 7, Figs. S7–8). These results indicate that S217879 oral administration leads to NRF2 pathway activation in vivo.
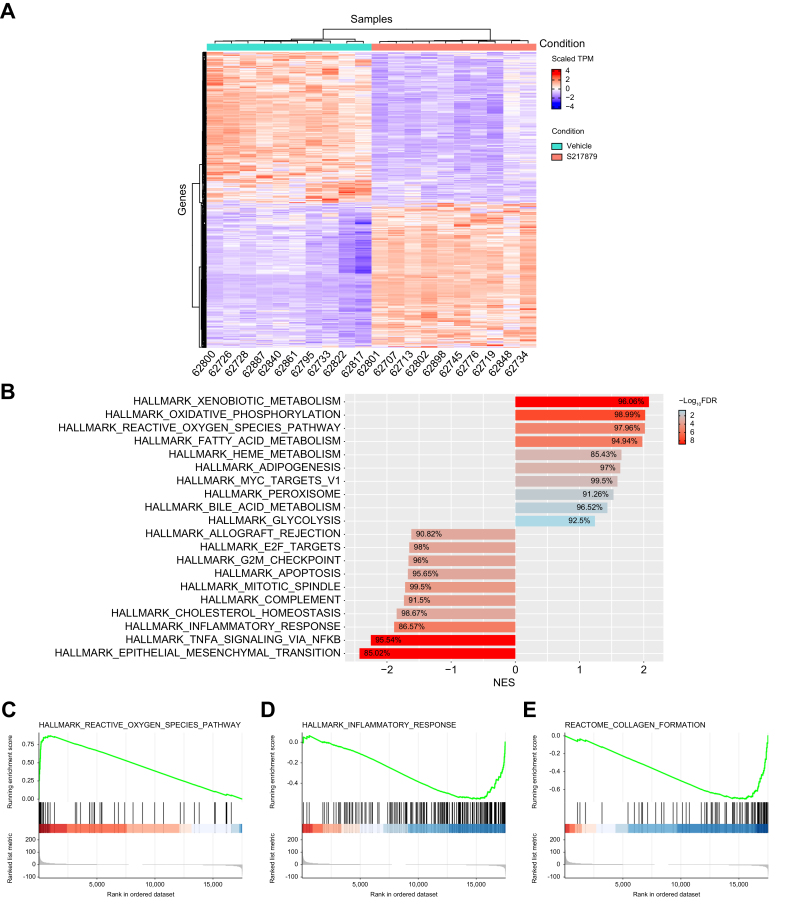
Fig. 7.
S217879-mediated NRF2 activation leads to upregulation of the antioxidant response and suppression of a wide spectrum of genes involved in disease progression.
(A) Heatmap of differentially expressed genes from S217879-treated DIO NASH mice vs. vehicle (Log2 fold-change cut-off >1; FDR-adjusted p value cut-off: 0.05). (B) Gene set-enrichment analysis of differentially expressed genes from DIO NASH mice treated with S217879 or vehicle (HEC 1%). Enriched pathways from gene ontology analysis. (C-E) Gene sets from Hallmark and Reactome revealed upregulated and downregulated pathways (enrichment plots). (C) Reactive oxygen species, (D) inflammatory response and (E) collagen formation. NES, normalized enrichment score.
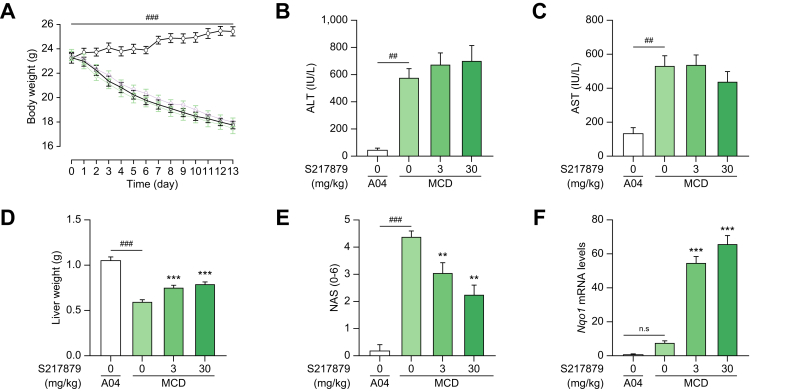
Having characterized S217879 as a potent and selective NRF2 activator in vitro and in vivo, we next evaluated its potential for the treatment of NASH using two well-established preclinical models. First, the MCDD mouse model was selected as a screening model to test the ability of S217879 to prevent or limit disease progression. This model is highly reproducible and mice rapidly develop a NASH-like liver phenotype after just a few weeks (see55 for review). Furthermore, this model was initially used to genetically validate NRF2 as a potential target for NASH.[33], [34], [35] As expected, mice fed the MCDD rapidly lost weight compared to mice receiving the control diet (Fig. 4A). Two weeks of exposure to MCDD led to significant liver injury with a sharp increase in plasma liver enzymes (Fig. 4B-C). Histological analyses confirmed the development of NASH with severe steatosis and hepatic inflammation reaching a NAS >4 (Fig. 4E, Table S5) in line with previous reports.56 While treatment with S217879 at 3 and 30 mg/kg/day had no effect on total body weight vs. vehicle (Fig. 4A), it significantly improved liver histology with a dose-dependent reduction in NAS (Fig. 4E, Fig. S2). A reduction in both steatosis and lobular inflammation scores was recorded at the highest dose (Table S5). Interestingly, this reduction in NAS was not followed by a reduction of plasma liver enzymes (Fig. 4B,C). These data are consistent with previous results obtained by Okada and colleagues who reported an impact of constitutive NRF2 activation on liver enzymes only after 13 weeks on MCDD.34 Biochemical analyses confirmed a dose-dependent reduction in liver triglycerides in response to S217879 treatment in MCDD-fed mice (Fig. S3) in agreement with our histological analyses (Fig. 4E, Table S5). As expected, the S217879-mediated reduction in NAS occurred at doses which triggered significant NRF2 target engagement as measured by the dose-dependent increase in liver Nqo1 mRNA levels (Fig. 4F, Fig. S4) in line with the dose-dependent increase in drug exposure (Table S3). It is noteworthy that the MCDD itself triggered the upregulation of antioxidant response as shown previously.57 Gene expression studies confirmed the clear upregulation of the antioxidant response in response to S217879 with significant Gpx2 gene induction (Fig. S4). In addition, S217879 treatment led to significant inhibition of the expression of proinflammatory genes such as Ccl5, Cd68, Il1b and Il6 (Fig. S4). These results are consistent with the anti-inflammatory properties of S217879 described in hPBMCs (Fig. 3). Finally, analyses of genes involved in de novo lipogenesis such as Fasn, Acaca and Scd1 revealed a lack of effect of S217879 on their expression levels (Fig. S4). Sugimoto and colleagues failed to demonstrate a major impact of NRF2 activation on fatty acid metabolism genes in MCDD-fed mice.34 Notably, we observed a small but significant increase in liver weight in response to S217879 treatment (maximum already reached at 3 mg/kg). In a separate experiment, we performed a full dose response analysis to determine the minimal effective dose in this model (see Fig. S5). Indeed, S217879 treatment prevented disease progression in a dose-dependent manner as shown by reduced NAS and induction of hepatic Nqo1 expression (Fig. S5AB). The minimal effective dose was set at 1 mg/kg/day in this model. Taken together, these results suggest that S217879-mediated NRF2 activation may provide hepatoprotective properties.
Fig. 4.
S217879 reduced NASH progression in MCDD mice in a dose-dependent manner.
(A) Body weight evolution throughout the study. (B) Plasma ALT levels at termination. (C) Plasma AST levels at termination. (D) Liver weight at termination. (E) NAFLD activity scores. (F) Liver Nqo1 mRNA levels expressed as fold induction over control (A04-fed group). Data expressed as mean ± SEM (n = 15 per group). Statistical significance was assessed by one-way ANOVA on log-transformed data followed by a Holm’s adjustment for the comparisons of each dose of S217879 vs. MCDD vehicle group. ##p <0.01, ###p <0.001: MCD vs. A04 (control diet). ∗∗∗p <0.001, ∗∗p <0.01 S217879-treated vs. vehicle. n.s., non-significant.
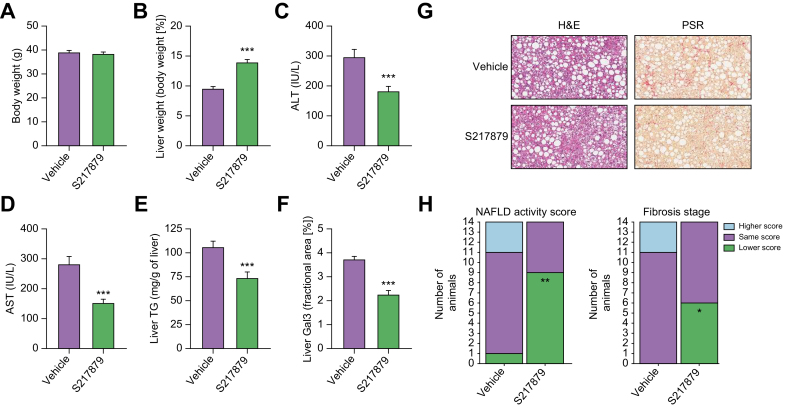
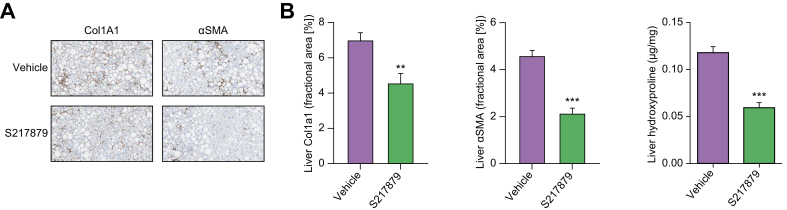
Despite its utility for drug discovery, the MCDD mouse model poorly recapitulates the human NASH etiology with its substantial weight loss and lack of insulin resistance (See55,58 for review). Nevertheless, results from the basic characterization of S217879 responses in the MCDD model (Fig. 4, Figs. S2–5) prompted us to further document the potential of S217879 for the treatment of NASH in the DIO NASH mouse model in a therapeutic setting. This model has been shown to have a good clinical translatability with respect to the histopathological, transcriptional and metabolic aspects of the human disease.59 5–6-week-old C57BL/6JRj mice were fed the DIO NASH diet for 33 weeks prior to initiation of the treatment protocol. Following a liver biopsy for inclusion (see methods), mice were randomized to receive either S217879 (30 mg/kg, PO, QD) or vehicle (HEC 1%) for 8 weeks while being maintained on the DIO NASH diet. S217879 treatment had no impact on food intake (Fig. S6) and body weight (Fig. 5A). It led to a significant reduction of established liver injury as measured by reduced alanine and aspartate aminotransferase (Fig. 5C-D). However, we noticed again a small but significant increase in liver weight in response to treatment (Fig. 5B). Biochemical analyses indicated a reduction in liver triglyceride levels (Fig. 5E). Histological analyses revealed a significant reduction in lobular inflammation resulting in a significant reduction in NAS (Fig. 5G-H, Table S6). These data were confirmed by the decrease of galectin-3 staining (Fig. 5F), a marker of hepatic inflammation.60,61 Interestingly, S217879 treatment led to a marked reduction in liver fibrosis progression as demonstrated by PSR staining (Fig. 5G-H, Table S6). This reduction in liver fibrosis was independently confirmed by a significant reduction in liver hydroxyproline content (vehicle 0.12 ± 0.017, S217879 0.06 ± 0.014 μg/mg liver, p <0.001) and reduced liver collagen 1A1 and αSMA staining (Fig. 6A-B). Taken together, these results indicate that S217879 treatment led to significant improvements of established NASH and decreased liver fibrosis.
Fig. 5.
S217879 reduced NASH progression and liver fibrosis in DIO NASH mice model.
(A) Body weight at the end of the study. (B) Liver weight expressed as percentage of total body weight. (C) Plasma ALT levels. (D) Plasma AST levels. (E) Liver TGs. (F) Liver Gal3-positive area. Data expressed as mean ± SEM (n = 14 per group). (G) H&E and PSR-stained liver sections. (H) Summary of histopathological NAFLD activity (left graph) scoring and fibrosis stage (left graph) of pre- and post-study biopsies. For each group, the number of animals with a higher (worsening), same or lower (improvement) score at post- compared to pre-study is indicated by the height of the bar. (A-F) Significant differences were analyzed using Student’s t test. (H) Significance of number of animals with a lower score vs. vehicle was assessed using Fisher’s exact test followed by correction for multiple comparisons using the Bonferroni method. ∗∗∗p <0.001, ∗∗p <0.01 and ∗p <0.05: S217879-treated vs. vehicle.
Fig. 6.
Markers of liver fibrosis are reduced in response to S217879 treatment in DIO NASH mice.
(A) Col1A1- and αSMA-stained liver sections at termination (n = 14 per group). (B) Quantification of Col1A1- and αSMA-positive areas by morphometry. Liver hydroxyproline levels quantified after extraction and centrifugation using a colorimetric assay. Data expressed as mean ± SEM. Significant differences were analyzed using Student’s t test. ∗∗∗p <0.001 and ∗∗p <0.01: S217879-treated vs. vehicle.
In order to dissect the molecular mechanisms by which S217879-mediated NRF2 activation resulted in reduced NASH and fibrosis progression, RNA-sequencing analysis was performed on liver total RNA at the end of the study. 740 genes were found differentially expressed (361 up and 379 down) between S217879 and vehicle-treated animals (Log2 fold-change >1, false discovery rate-adjusted p value<0.05, Fig. 7A). These transcriptional changes were further explored by gene set-enrichment analysis (Fig. 7B) which confirmed, as expected, that the NRF2-mediated oxidative stress response was ranked among the most affected pathways in response to treatment (“reactive oxygen species pathway”, Fig. 7B) with strong enrichment in NRF2 target genes found upregulated (Fig. 7B–C, Figs. S7–8). The clear and marked upregulation of the overall NRF2 pathway as shown by increased Nqo1, Gstm1, Gclc and Gpx2 expression (Figs. S7–8) demonstrates again that S217879 is a potent NRF2 activator in vivo. More interestingly, key pathways involved in NASH pathophysiology related to inflammation (“inflammatory response”, “TNFA signaling”) and fibrosis (“epithelial mesenchymal transition”) were found significantly suppressed in response to treatment (Fig. 7B-E). In line with the reduction of liver inflammation (Fig. 5), we did record the inhibition of several proinflammatory genes such as Mapk4 (Figs. S7–8). Furthermore, a large number of genes associated with fibrosis and stellate cell activation were downregulated, such as Col1a1, Itgax, Lox and Bmp8b (Figs. S7–8). Again, this is consistent with the significant reduction in liver fibrosis measured in response to treatment. Finally, the expression of several key genes involved in de novo lipogenesis was also significantly downregulated (Fig. S8) which is consistent with the anti-steatotic effects of S217879 in this model (Fig. 5E). Taken together, our RNA-sequencing data confirmed that NRF2 activation leads to the upregulation of the antioxidant response and the coordinated regulation of a wide spectrum of genes involved with disease progression, ultimately leading to reduced NASH and liver fibrosis progression.
Discussion
In this manuscript, we reported the identification and detailed characterization of S217879 as a novel, potent and selective compound activating the NRF2 pathway. This compound selectively binds to KEAP1 leading to the disruption of the KEAP1-NRF2 interaction resulting in NRF2 nuclear translocation and subsequent gene regulation. Moreover, we found that S217879 exhibits an excellent selectivity profile (Table S2) resulting in an excellent safety profile in vitro (LD50 in HepG2 cells >30 μM) in contrast to potent first-generation compounds such as bardoxolone (LD50 in HepG2 cells <1 μM). One key feature of this molecule is its good oral bioavailability which is a major differentiation with respect to first-generation compounds previously identified which displayed good potency and cellular activity but suffered from poor absorption, limited distribution and low metabolic stability (See30 for review). By contrast, we showed that single oral administration of S217879 resulted in marked target engagement as measured by increased Nqo1 mRNA levels in the liver (Fig. S1AB) but also in the kidney (data not shown). Furthermore, we found S217879 to be efficacious in the MCDD mouse model at doses as low as 1 mg/kg/day. These data strongly indicate that S217879 is an excellent compound to probe NRF2 biology in preclinical disease models.
In this study, we have shown using two different preclinical models that selective NRF2 activation is beneficial for the treatment of NASH and liver fibrosis. S217879 treatment led to marked reduction in both steatosis and inflammation, resulting in a lower NAS (Fig. 4, Fig. 5, Fig. 6). Furthermore, S217879 treatment led to a marked reduction in liver fibrosis in DIO NASH mice, as measured by reduced PSR, Col1A1, and αSMA staining as well reduced hydroxyproline content (Fig. 5, Fig. 6). These data are consistent with histological improvements observed in response to first-generation NRF2 activators, such as TBE-31 and omaveloxolone, in high-fat high-fructose diet-fed mice37 and STAM mice,38 respectively. To our knowledge, this is the first report of beneficial properties of a non-covalent NRF2 activator for the treatment NASH and fibrosis. RNA-sequencing analyses indicated that NRF2 activation led to the upregulation of the antioxidant response and the coordinated regulation of a wide spectrum of genes involved with disease progression (Figs. S7–8, Fig. 7). We did observe a pronounced impact on genes belonging to the de novo lipogenesis pathway in line with previous observations.32,62,63 This is in line with the marked reduction in liver triglyceride levels (Fig. 5E). In addition to the expected impact on inflammation (Fig. 5F), one of the most remarkable results obtained in this manuscript is the anti-fibrotic properties of S217879 in vivo as measured at the histological, biochemical and gene expression levels (Fig. 5, Fig. 6, Fig. 7). This is of major interest since long-term follow-up studies revealed that fibrosis is the main driver of mortality in NASH.6,7 NRF2 activation may trigger the reduction of liver fibrosis via multiple mechanisms. First, hepatocyte-specific activation of NRF2 has recently been shown to control fibrogenesis during NASH by regulating, at least in part, the antioxidant stress response and thereby reducing DNA damage and apoptosis.15 Second, NRF2 activation may limit profibrotic macrophage-derived inflammation by negatively interfering with both NF-κB and YAP/NLRP3 pathways.54,64 Finally, NRF2 has been shown to protect hepatic stellate cells from TGF-β-mediated cell activation65 suggesting direct anti-fibrotic properties. Our data are consistent with a model in which S217879-induced NRF2 activation reduces NASH progression and slows down the development of liver fibrosis by targeting both parenchymal and non-parenchymal cells (immune cells and stellate cells mainly) as suggested by cell-specific gain (Keap1 gene deletion) or loss of function (NRF2 gene deletion) studies,15,64 respectively. Additional studies using single cell RNA-sequencing analyses or spatial transcriptomics are required to further document the molecular mechanisms at the cellular level and determine the potential contribution of other relevant cell populations, such as liver sinusoidal endothelial cells66 to S217879-mediated beneficial properties on NASH and fibrosis. Finally, NRF2 activation has been shown to improve gut barrier integrity,67 which may to some extent contribute to the beneficial impact on NASH progression.68
NRF2 activity is reduced during aging69 and considered as an attractive target for a number of chronic diseases including but not limited to autoimmune, respiratory, neurodegenerative, and cardio-metabolic diseases (See30 for review). However, it has also been shown to exert both anti-tumorigenic and pro-tumorigenic actions. It is believed that low NRF2 activity may facilitate the initiation of carcinogenesis, whereas constant high activity may drive cancer progression and resistance to chemotherapy.70 Notably, more than 10% of all hepatocellular carcinomas present a mutation in either KEAP1 or NRF2.71 Interestingly, we found that treatment with S217879 led to small but significant increase in liver weight in both MCDD and DIO NASH models (Fig. 4, Fig. 5B). Similar observations were also made in both rats and mice on chow diet exposed to similar drug levels (data not shown). These results are reminiscent of the phenotype of KEAP1-deficient mice suggesting that constitutive NRF2 activation may trigger hepatomegaly.15,72,73 Notably, constitutive hepatocyte-specific NRF2 activation was shown to significantly reduce NASH-associated hepatocellular carcinoma development in mice with hepatocyte-specific NEMO deficiency.15 Furthermore, He and colleagues reported that NRF2 forced expression leads to hepatomegaly by, at least in part, increasing the expression of several growth factors such as PDGF and EGF in an AKT-dependent manner.74 By contrast, we failed to detect any impact on cell proliferation in vivo and genes related to cell cycle/proliferation (including the gene signature identified by He and colleagues74 [Aurka, Foxm1, Ccnb2, Cdc25b and Cdc25c]) were not significantly modulated upon treatment with S217879 (Fig. 7). Furthermore, in vitro studies performed with S217879 tested up to 10 μM (roughly 500-fold higher than NRF2 cell potency) failed to reveal any impact on cell proliferation using HepG2 cells. Finally, we did not detect any AKT phosphorylation in both hepatocytes and HepG2 cells in response to high concentrations of S217879 (data not shown). Obviously, pharmacological activation of NRF2 with a small molecule in vivo is not equivalent to NRF2 forced expression74 or constitutive NRF2 activation by KEAP1 gene deletion.15,72,73 As a matter of fact, using a different genetic strategy, Kohler and colleagues created a mouse line with constitutive NRF2 activation in hepatocytes. Surprisingly, these mice did not have any increase in liver weight compared to wild-type mice despite a significant increase in NRF2-regulated gene expression.75 Taken together, these independent studies suggest that different levels of NRF2 activation due to either constitutive expression or pharmacological activation may result in different phenotypes. Interestingly, Chan et al. recently reported that NRF2 pharmacological activation may enhance liver regeneration in mice by supporting hepatocellular hypertrophy.76 Preliminary evaluation of S217879’s safety profile using non-GLP (good laboratory practice) dose-ranging studies in both rats and non-human primates did not reveal any major toxicity. Histological analyses confirmed hepatocellular hypertrophy in rodents as measured by reduced nuclei density (data not shown). Additional studies including GLP-toxicology are required to confirm these preliminary findings and determine the long-term safety of S217879-mediated NRF2 activation.
Financial support
This work was funded by Institut de Recherches Servier.
Authors’ contributions
KS and PD coordinated the work. SC, OB, AG, CI, AH, JR and KL performed the in vitro and in vivo studies. VD, NP, KS, PD, CV, SB, VP and LMV contributed to experimental design and data interpretation. CW supervised the medicinal chemistry program (design and synthesis) with the help of AK and DD. VM was responsible for molecular modelling, NM for scale-up syntheses. FM performed bioinformatic analyses. CW and PD drafted the manuscript.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of interest
All the authors are/were employees of Servier.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgments
We would like to thank Michael Feigh and the team at GUBRA (Hørsholm, Denmark) for their excellent support regarding the DIO NASH study.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2022.100651.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Brunt E.M., Wong V.W.S., Nobili V., Day C.P., Sookoian S., Maher J.J., et al. Nonalcoholic fatty liver disease. Nat Rev Dis Prim. 2015:1. doi: 10.1038/nrdp.2015.80. [DOI] [PubMed] [Google Scholar]
- 2.Diehl A.M., Day C. Cause, pathogenesis, and treatment of nonalcoholic steatohepatitis. N Engl J Med. 2017;377:2063–2072. doi: 10.1056/nejmra1503519. [DOI] [PubMed] [Google Scholar]
- 3.Younossi Z.M., Golabi P., de Avila L., Paik J.M., Srishord M., Fukui N., et al. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: a systematic review and meta-analysis. J Hepatol. 2019;71:793–801. doi: 10.1016/j.jhep.2019.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease—meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 5.Loomba R., Friedman S.L., Shulman G.I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell. 2021;184:2537–2564. doi: 10.1016/j.cell.2021.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angulo P., Kleiner D.E., Dam-Larsen S., Adams L.A., Bjornsson E.S., Charatcharoenwitthaya P., et al. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015;149:389–397.e10. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagström H., Nasr P., Ekstedt M., Hammar U., Stål P., Hultcrantz R., et al. Risk for development of severe liver disease in lean patients with nonalcoholic fatty liver disease: a long-term follow-up study. Hepatol Commun. 2018;2:48–57. doi: 10.1002/hep4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romero F.A., Jones C.T., Xu Y., Fenaux M., Halcomb R.L. The race to bash NASH: emerging targets and drug development in a complex liver disease. J Med Chem. 2020 doi: 10.1021/acs.jmedchem.9b01701. [DOI] [PubMed] [Google Scholar]
- 9.Anstee Q.M., Day C.P. The genetics of NAFLD. Nat Rev Gastroenterol Hepatol. 2013;10:645–655. doi: 10.1038/nrgastro.2013.182. [DOI] [PubMed] [Google Scholar]
- 10.Trépo E., Valenti L. Update on NAFLD genetics: from new variants to the clinic. J Hepatol. 2020;72:1196–1209. doi: 10.1016/j.jhep.2020.02.020. [DOI] [PubMed] [Google Scholar]
- 11.Arroyave-Ospina J.C., Wu Z., Geng Y., Moshage H. Role of oxidative stress in the pathogenesis of non-alcoholic fatty liver disease: implications for prevention and therapy. Antioxidants (Basel, Switzerland) 2021;10:1–25. doi: 10.3390/ANTIOX10020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madan K., Bhardwaj P., Thareja S., Gupta S.D., Saraya A. Oxidant stress and antioxidant status among patients with nonalcoholic fatty liver disease (NAFLD) J Clin Gastroenterol. 2006;40:930–935. doi: 10.1097/01.MCG.0000212608.59090.08. [DOI] [PubMed] [Google Scholar]
- 13.Palmieri V.O., Grattagliano I., Portincasa P., Palasciano G. Systemic oxidative alterations are associated with visceral adiposity and liver steatosis in patients with metabolic syndrome. J Nutr. 2006;136:3022–3026. doi: 10.1093/JN/136.12.3022. [DOI] [PubMed] [Google Scholar]
- 14.Podszun M.C., Chung J.Y., Ylaya K., Kleiner D.E., Hewitt S.M., Rotman Y. 4-HNE immunohistochemistry and image analysis for detection of lipid peroxidation in human liver samples using vitamin E treatment in NAFLD as a proof of concept. J Histochem Cytochem. 2020;68:635–643. doi: 10.1369/0022155420946402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohs A., Otto T., Schneider K.M., Peltzer M., Boekschoten M., Holland C.H., et al. Hepatocyte-specific NRF2 activation controls fibrogenesis and carcinogenesis in steatohepatitis. J Hepatol. 2021;74:638–648. doi: 10.1016/J.JHEP.2020.09.037. [DOI] [PubMed] [Google Scholar]
- 16.Masarone M., Rosato V., Dallio M., Gravina A.G., Aglitti A., Loguercio C., et al. Role of oxidative stress in pathophysiology of nonalcoholic fatty liver disease. Oxid Med Cell Longev. 2018;2018 doi: 10.1155/2018/9547613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satapati S., Kucejova B., Duarte J.A.G., Fletcher J.A., Reynolds L., Sunny N.E., et al. Mitochondrial metabolism mediates oxidative stress and inflammation in fatty liver. J Clin Invest. 2015;125:4447–4462. doi: 10.1172/JCI82204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koliaki C., Szendroedi J., Kaul K., Jelenik T., Nowotny P., Jankowiak F., et al. Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metab. 2015;21:739–746. doi: 10.1016/J.CMET.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Karkucinska-Wieckowska A., Simoes I.C.M., Kalinowski P., Lebiedzinska-Arciszewska M., Zieniewicz K., Milkiewicz P., et al. Mitochondria, oxidative stress and nonalcoholic fatty liver disease: a complex relationship. Eur J Clin Invest. 2022:52. doi: 10.1111/ECI.13622. [DOI] [PubMed] [Google Scholar]
- 20.Dodson M., De La Vega M.R., Cholanians A.B., Schmidlin C.J., Chapman E., Zhang D.D. Modulating NRF2 in disease: timing is everything. Annu Rev Pharmacol Toxicol. 2019;59:555–575. doi: 10.1146/ANNUREV-PHARMTOX-010818-021856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong K.I., Katoh Y., Kusunoki H., Itoh K., Tanaka T., Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26:2887–2900. doi: 10.1128/mcb.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong K.I., Padmanabhan B., Kobayashi A., Shang C., Hirotsu Y., Yokoyama S., et al. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D.D., Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D.D., Lo S.-C., Cross J.V., Templeton D.J., Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eggler A.L., Small E., Hannink M., Mesecar A.D. Cul3-mediated Nrf2 ubiquitination and antioxidant response element (ARE) activation are dependent on the partial molar volume at position 151 of Keap1. Biochem J. 2009;422:171–180. doi: 10.1042/BJ20090471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakabayashi N., Dinkova-Kostova A.T., Holtzclaw W.D., Kang-Il M., Kobayashi A., Yamamoto M., et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A. 2004;101:2040–2045. doi: 10.1073/PNAS.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/BBRC.1997.6943. [DOI] [PubMed] [Google Scholar]
- 28.Marini M.G., Chan K., Casula L., Kan Y.W., Cao A., Moi P. hMAF, a small human transcription factor that heterodimerizes specifically with Nrf1 and Nrf2. J Biol Chem. 1997;272:16490–16497. doi: 10.1074/JBC.272.26.16490. [DOI] [PubMed] [Google Scholar]
- 29.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39:199–218. doi: 10.1016/J.TIBS.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat Rev Drug Discov. 2019;18:295–317. doi: 10.1038/S41573-018-0008-X. [DOI] [PubMed] [Google Scholar]
- 31.Tang W., Jiang Y.F., Ponnusamy M., Diallo M. Role of Nrf2 in chronic liver disease. World J Gastroenterol. 2014;20:13079–13087. doi: 10.3748/WJG.V20.I36.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meakin P.J., Chowdhry S., Sharma R.S., Ashford F.B., Walsh S.V., McCrimmon R.J., et al. Susceptibility of Nrf2-null mice to steatohepatitis and cirrhosis upon consumption of a high-fat diet is associated with oxidative stress, perturbation of the unfolded protein response, and disturbance in the expression of metabolic enzymes but not with insulin resistance. Mol Cell Biol. 2014;34:3305–3320. doi: 10.1128/MCB.00677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chowdhry S., Nazmy M.H., Meakin P.J., Dinkova-Kostova A.T., Walsh S.V., Tsujita T., et al. Loss of Nrf2 markedly exacerbates nonalcoholic steatohepatitis. Free Radic Biol Med. 2010;48:357–371. doi: 10.1016/J.FREERADBIOMED.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Sugimoto H., Okada K., Shoda J., Warabi E., Ishige K., Ueda T., et al. Deletion of nuclear factor-E2-related factor-2 leads to rapid onset and progression of nutritional steatohepatitis in mice. Am J Physiol - Gastrointest Liver Physiol. 2010:298. doi: 10.1152/ajpgi.00296.2009. [DOI] [PubMed] [Google Scholar]
- 35.Okada K., Warabi E., Sugimoto H., Horie M., Tokushige K., Ueda T., et al. Nrf2 inhibits hepatic iron accumulation and counteracts oxidative stress-induced liver injury in nutritional steatohepatitis. J Gastroenterol. 2012;47:924–935. doi: 10.1007/S00535-012-0552-9. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka Y., Aleksunes L.M., Yeager R.L., Gyamfi M.A., Esterly N., Guo G.L., et al. NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. J Pharmacol Exp Ther. 2008;325:655–664. doi: 10.1124/jpet.107.135822. [DOI] [PubMed] [Google Scholar]
- 37.Sharma R.S., Harrison D.J., Kisielewski D., Cassidy D.M., McNeilly A.D., Gallagher J.R., et al. Experimental nonalcoholic steatohepatitis and liver fibrosis are ameliorated by pharmacologic activation of Nrf2 (NF-E2 p45-related factor 2) Cell Mol Gastroenterol Hepatol. 2018;5:367–398. doi: 10.1016/j.jcmgh.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reisman S.A., Ferguson D.A., Lee C.Y.I., Proksch J.W. Omaveloxolone and TX63682 are hepatoprotective in the STAM mouse model of nonalcoholic steatohepatitis. J Biochem Mol Toxicol. 2020;2020:34. doi: 10.1002/JBT.22526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azzimato V., Jager J., Chen P., Morgantini C., Levi L., Barreby E., et al. Liver macrophages inhibit the endogenous antioxidant response in obesity-associated insulin resistance. Sci Transl Med. 2020;12 doi: 10.1126/SCITRANSLMED.AAW9709. [DOI] [PubMed] [Google Scholar]
- 40.Bricambert J., Alves-Guerra M.C., Esteves P., Prip-Buus C., Bertrand-Michel J., Guillou H., et al. The histone demethylase Phf2 acts as a molecular checkpoint to prevent NAFLD progression during obesity. Nat Commun. 2018:9. doi: 10.1038/S41467-018-04361-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yore M.M., Kettenbach A.N., Sporn M.B., Gerber S.A., Liby K.T. Proteomic analysis shows synthetic oleanane triterpenoid binds to mTOR. PLoS One. 2011;6 doi: 10.1371/JOURNAL.PONE.0022862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Zeeuw D., Akizawa T., Audhya P., Bakris G.L., Chin M., Christ-Schmidt H., et al. Bardoxolone methyl in type 2 diabetes and stage 4 chronic kidney disease. N Engl J Med. 2013;369:2492–2503. doi: 10.1056/NEJMOA1306033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chin M.P., Wrolstad D., Bakris G.L., Chertow G.M., De Zeeuw D., Goldsberry A., et al. Risk factors for heart failure in patients with type 2 diabetes mellitus and stage 4 chronic kidney disease treated with bardoxolone methyl. J Card Fail. 2014;20:953–958. doi: 10.1016/J.CARDFAIL.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Li X., Zhang D., Hannink M., Beamer L.J. Crystal structure of the Kelch domain of human Keap1. J Biol Chem. 2004;279:54750–54758. doi: 10.1074/JBC.M410073200. [DOI] [PubMed] [Google Scholar]
- 45.Schmoll D., Engel C.K., Glombik H. The Keap1–Nrf2 protein–protein interaction: a suitable target for small molecules. Drug Discov Today Tech. 2017;24:11–17. doi: 10.1016/j.ddtec.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Davies T.G., Wixted W.E., Coyle J.E., Griffiths-Jones C., Hearn K., McMenamin R., et al. Monoacidic inhibitors of the Kelch-like ECH-associated protein 1: nuclear factor erythroid 2-related factor 2 (KEAP1:NRF2) protein-protein interaction with high cell potency identified by fragment-based discovery. J Med Chem. 2016;59:3991–4006. doi: 10.1021/ACS.JMEDCHEM.6B00228. [DOI] [PubMed] [Google Scholar]
- 47.Kristiansen M.N.B., Veidal S.S., Rigbolt K.T.G., Tølbøl K.S., Roth J.D., Jelsing J., et al. Obese diet-induced mouse models of nonalcoholic steatohepatitis-tracking disease by liver biopsy. World J Hepatol. 2016;8:673. doi: 10.4254/WJH.V8.I16.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 49.Morel P.S., Duvivier V., Bertin F., Provost N., Hammoutene A., Hubert E.L., et al. Procollagen C-Proteinase Enhancer-1 (PCPE-1) deficiency in mice reduces liver fibrosis but not NASH progression. PLoS One. 2022;17 doi: 10.1371/JOURNAL.PONE.0263828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tran K.T., Pallesen J.S., Solbak S.M., Narayanan D., Baig A., Zang J., et al. A comparative assessment study of known small-molecule Keap1-Nrf2 protein-protein interaction inhibitors: chemical synthesis, binding properties, and cellular activity. J Med Chem. 2019;62:8028–8052. doi: 10.1021/ACS.JMEDCHEM.9B00723/SUPPL_FILE/JM9B00723_SI_002.CSV. [DOI] [PubMed] [Google Scholar]
- 51.Ma B., Lucas B., Capacci A., Lin E.Y.S., Jones J.H., Dechantsreiter M., et al. Design, synthesis and identification of novel, orally bioavailable non-covalent Nrf2 activators. Bioorg Med Chem Lett. 2020;30:126852. doi: 10.1016/J.BMCL.2019.126852. [DOI] [PubMed] [Google Scholar]
- 52.Mallinson J., Collins I. Macrocycles in new drug discovery. Future Med Chem. 2012;4:1409–1438. doi: 10.4155/FMC.12.93/ASSET/IMAGES/LARGE/FIGURE49.JPEG. [DOI] [PubMed] [Google Scholar]
- 53.Wen Wong D.P., Ng M.Y., Leung J.Y., Boh B.K., Lim E.C., Tan S.H., et al. Regulation of the NRF2 transcription factor by andrographolide and organic extracts from plant endophytes. PLoS One. 2018;13 doi: 10.1371/JOURNAL.PONE.0204853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kobayashi E.H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun. 2016;7 doi: 10.1038/NCOMMS11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santhekadur P.K., Kumar D.P., Sanyal A.J. Preclinical models of non-alcoholic fatty liver disease. J Hepatol. 2018;68:230–237. doi: 10.1016/j.jhep.2017.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Machado M.V., Michelotti G.A., Xie G., De Almeida T.P., Boursier J., Bohnic B., et al. Mouse models of diet-induced nonalcoholic steatohepatitis reproduce the heterogeneity of the human disease. PLoS One. 2015;10 doi: 10.1371/JOURNAL.PONE.0127991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lickteig A.J., Fisher C.D., Augustine L.M., Cherrington N.J. Genes of the antioxidant response undergo upregulation in a rodent model of nonalcoholic steatohepatitis. J Biochem Mol Toxicol. 2007;21:216–220. doi: 10.1002/JBT.20177. [DOI] [PubMed] [Google Scholar]
- 58.Rinella M.E., Green R.M. The methionine-choline deficient dietary model of steatohepatitis does not exhibit insulin resistance. J Hepatol. 2004;40:47–51. doi: 10.1016/J.JHEP.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 59.Hansen H.H., Ægidius H.M., Oró D., Evers S.S., Heebøll S., Eriksen P.L., et al. Human translatability of the GAN diet-induced obese mouse model of non-alcoholic steatohepatitis. BMC Gastroenterol. 2020;20 doi: 10.1186/s12876-020-01356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henderson N.C., Mackinnon A.C., Farnworth S.L., Poirier F., Russo F.P., Iredale J.P., et al. Galectin-3 regulates myofibroblast activation and hepatic fibrosis. Proc Natl Acad Sci U S A. 2006;103:5060–5065. doi: 10.1073/PNAS.0511167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iacobini C., Menini S., Ricci C., Fantauzzi C.B., Scipioni A., Salvi L., et al. Galectin-3 ablation protects mice from diet-induced NASH: a major scavenging role for galectin-3 in liver. J Hepatol. 2011;54:975–983. doi: 10.1016/J.JHEP.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 62.Slocum S.L., Skoko J.J., Wakabayashi N., Aja S., Yamamoto M., Kensler T.W., et al. Keap1/Nrf2 pathway activation leads to a repressed hepatic gluconeogenic and lipogenic program in mice on a high-fat diet. Arch Biochem Biophys. 2016;591:57–65. doi: 10.1016/J.ABB.2015.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knatko E.V., Tatham M.H., Zhang Y., Castro C., Higgins M., Dayalan Naidu S., et al. Downregulation of Keap1 confers features of a fasted metabolic state. IScience. 2020;2020:23. doi: 10.1016/J.ISCI.2020.101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang P., Ni M., Tian Y., Wang H., Qiu J., You W., et al. Myeloid Nrf2 deficiency aggravates non-alcoholic steatohepatitis progression by regulating YAP-mediated NLRP3 inflammasome signaling. IScience. 2021;24 doi: 10.1016/J.ISCI.2021.102427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prestigiacomo V., Suter-Dick L.. Nrf2 protects stellate cells from Smad-dependent cell activation. PLoS One. 2018;13 doi: 10.1371/JOURNAL.PONE.0201044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Furuta K., Guo Q., Hirsova P., Ibrahim S.H. Emerging roles of liver sinusoidal endothelial cells in nonalcoholic steatohepatitis. Biol. 2020;9:395. doi: 10.3390/BIOLOGY9110395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh R., Chandrashekharappa S., Bodduluri S.R., Baby B.V., Hegde B., Kotla N.G., et al. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat Commun. 2019;10 doi: 10.1038/S41467-018-07859-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mouries J., Brescia P., Silvestri A., Spadoni I., Sorribas M., Wiest R., et al. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J Hepatol. 2019;71:1216–1228. doi: 10.1016/J.JHEP.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidlin C.J., Dodson M.B., Madhavan L., Zhang D.D. Redox regulation by NRF2 in aging and disease. Free Radic Biol Med. 2019;134:702–707. doi: 10.1016/J.FREERADBIOMED.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kensler T.W., Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31:90–99. doi: 10.1093/CARCIN/BGP231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schulze K., Imbeaud S., Letouzé E., Alexandrov L.B., Calderaro J., Rebouissou S., et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okawa H., Motohashi H., Kobayashi A., Aburatani H., Kensler T.W., Yamamoto M.. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun. 2006;339:79–88. doi: 10.1016/J.BBRC.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 73.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 74.He F., Antonucci L., Yamachika S., Zhang Z., Taniguchi K., Umemura A., et al. NRF2 activates growth factor genes and downstream AKT signaling to induce mouse and human hepatomegaly. J Hepatol. 2020;72:1182–1195. doi: 10.1016/J.JHEP.2020.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Köhler U.A., Kurinna S., Schwitter D., Marti A., Schäfer M., Hellerbrand C., et al. Activated Nrf2 impairs liver regeneration in mice by activation of genes involved in cell-cycle control and apoptosis. Hepatology. 2014;60:670–678. doi: 10.1002/HEP.26964. [DOI] [PubMed] [Google Scholar]
- 76.Chan B.K.Y., Elmasry M., Forootan S.S., Russomanno G., Bunday T.M., Zhang F., et al. Pharmacological activation of Nrf2 enhances functional liver regeneration. Hepatology. 2021;74:973–986. doi: 10.1002/HEP.31859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.