Abstract
A structural class of forty glycine transporter type 1 (GlyT1) inhibitors, was examined using molecular modeling techniques. The quantitative structure-activity relationships (QSAR) technology confirmed that human GlyT1 activity is strongly and significantly affected by constitutional, geometrical, physicochemical and topological descriptors. ADME-Tox in-silico pharmacokinetics revealed that L28 and L30 ligands were predicted as non-toxic inhibitors with a good ADME profile and the highest probability to penetrate the central nervous system (CNS). Molecular docking results indicated that the predicted inhibitors block GlyT1, reacting specifically with Phe319, Phe325, Tyr123, Tyr 124, Arg52, Asp475, Ala117, Ala479, Ile116 and Ile483 amino acids of the dopamine transporter (DAT) membrane protein. These results were qualified and strengthened using molecular dynamics (MD) study, which affirmed that the established intermolecular interactions for (L28, L30–DAT protein) complexes remain perfectly stable along 50 ns of MD simulation time. Therefore, they could be strongly recommended as therapeutics in medicine to improve memory performance.
Keywords: QSAR, GlyT1, ADME-Tox, CNS, DAT, Molecular docking, MD simulations
1. Introduction
About 25% of the worldwide population suffers from neurological disorders, according to the World Health Organization (WHO) report, published in 2001 [1]. Many of these neurological diseases are the result of memory dysfunctions [2]. The present work aims to discover successful candidate drugs that can improve memory performance. To achieve this objective, we have processed with the assistance of molecular modeling techniques, a set of forty pyrrolo [3,4-c]pyrazole and azetidine-based analogs, acting as selective glycine transporter type 1 (GlyT1) inhibitors, considering that GluT1 is one of the most essential neurotransmitters that control learning and memory functions [3,4].
Over the last decade, the glycine transporter type 1 (GlyT1) has received enormous interest as it is one of the key constituents of glycine metabolism in the central nervous system (CNS). Whereas, the amino acid glycine is an excitatory and inhibitory neurotransmitter in the vertebrate central nervous system. In the cortex, hippocampus, and thalamus areas, glycine regulates excitatory neurotransmission as a mandatory co-agonist at N-methyl-d-aspartate (NMDA) receptors (glycine B sites). Inversely, it acts as an inhibitory neurotransmitter at strychnine-sensitive glycine receptor ion channels (glycine A sites) mainly present in the brainstem, cerebellum, and spinal cord [3,5]. Hence, GlyT1 inhibitors have been introduced into the clinic as potent treatments for obsessive-compulsive disorder (OCD), negative and cognitive symptoms associated with schizophrenia, addiction, depression, and Parkinson's disease [3,5,6]. Nowadays, computer-assisted drug design (CADD) based on in silico techniques has gained enormous importance and has been widely reported in the literature. In this regard, we examined all 40 GlyT1 inhibitors, through the use of CADD technology, to detect and identify non-toxic, highly effective CNS drug candidates, that will be able to produce highly stable chemical bonds between these candidate inhibitors of glycine transporter and the membrane protein of drosophila melanogaster dopamine transporter (DAT), which would be intended to treat memory impairments [7].
In the first stage, we started this task, using quantitative structure-activity relationships (QSAR) as a technique largely applied in the field of drug discovery, predicting the human GlyT1 activity of 40 pyrrolo [3,4-c]pyrazole and azetidine derivatives, based on two mathematical QSAR models, which were established thanks to the use of multiple linear regression (MLR) and multiple non-linear regression (MNLR) respectively, to model the linear and nonlinear relationships between human GlyT1 activity and different types of molecular descriptors [6,8].
In the second part, we have predicted adsorption, distribution, metabolism, excretion, and toxicity (ADMET) properties for forty selective inhibitors of glycine transporter type 1, with the aim of discovering qualified drugs with a favorable ADMET profile [9,10]. Subsequently, we have performed the molecular docking study for the predicted ligands that traverse the central nervous system, to identify the established intermolecular interactions towards the amino acids from DAT protein encoded as 4M48. pdb [11,12].
Lastly, this work has been concluded using molecular dynamics simulations during 50 ns, to verify the stability of (predicted ligands-protein target) complexes, and monitoring the total free energy variation in Kcal/mol of each complex based on the MM-GBSA approach [6,13,14].
2. Materials and methods
2.1. Experimental database
The current study includes forty pyrrolo [3,4-c]pyrazole and azetidine-based derivatives (Fig. S1) having human GlyT1 activities [3,5], that were subsequently expressed on a logarithmic decimal scale (pIC50 = - log10 IC50), as presented in Table S1.
2.2. Molecular descriptors calculation
To model the quantitative structure-activity relationships (QSAR), various types of molecular descriptors have been calculated as presented in Table S2. With the help of Gaussian 09 software [15], we have calculated the quantum descriptors, to optimize the 3D geometries and ensure molecular stability with minimized energies using the density functional theory (DFT) coupled with B3LYP/6-31 + G (d,p) basis [16,17]. Then, we executed the thermodynamic, topological, and physicochemical descriptors based on the MM2 technique using the ChemBio3D software [18]. Finally, the constitutional descriptors have been processed through the ACD/chemsketch software [19].
2.3. Statistical methods
To minimize the size of the molecular descriptors into a reduced number of factorial axes (or principal components), we have applied the principal component analysis (PCA), as a very important step to identify the most predictive variables that can influence the biological activity of inhibitors [20]. Thereafter, two statistical methods were performed on the training set (N = 32) to develop a predictive QSAR model using XLSTAT 2014 software [21]. The first one is the multiple linear regression (MLR) technique, which is widely used to investigate the linear relationship between the biological activity of pIC50 order and the previously calculated molecular descriptors. The second, is the multiple non-linear regression (MNLR) analysis, which is adapted to examine the nonlinear relationship established between the inhibitory activity and the calculated descriptors [22]. Next, the MNLR and MLR QSAR models were checked with external validation for a test set (N = 8) and further evaluated with internal validation using the leave-one-out (LOO) procedure of the cross-validation (CV) technique. Additionally, the Tropsha and Golbreikh criteria, applicability domain, and the Y-randomization test were also utilized to evaluate the predictive potential and robustness of the developed QSAR models [23,24].
2.4. In silico pharmacokinetics, drug-likeliness, and ADMET depiction exploration
To obtain a novel medication with high efficiency and less experimental investigation time, it's mandatory to evaluate their absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties in the human organism before beginning any clinical trials, verifying the five rules of Lipinski [25], and respecting the Veber, Ghose, Egan and Muegge violations number of drug candidates [[26], [27], [28], [29], [30]]. For this purpose, we have predicted in silico pharmacokinetics and ADMET properties of the candidate's molecules as GlyT1 inhibitors enhancing memory performance, using pkCSM and online SwissADME servers, respectively [31,32].
2.5. Molecular docking simulation
Molecular docking is an accurate, quick, and successful technology in the field of drug discovery [33]. It is largely applied to examine the intermolecular interactions established between the candidate drug and the targeted protein. For this goal, we have extracted the 3D coordinates of the drosophila melanogaster dopamine transporter encoded as 4M48 from the data bank of proteins (pdb) [34], with a resolution equal to 2.96 Å, using the X-ray diffraction method [35]. With the assistance of the Discovery Studio 2021 (BIOVIA) software package [36], we have deleted the water molecules (H2O) and suspended ligands that were bound to the protein, such as nortriptyline (C19H21N), cholesterol (C27H46O), chloride and sodium ions (Na+, Cl-). Then, we added the polar hydrogens, to ensure that the performance of the cavity method is improved [37]. Thereafter, we launched the prepared protein, L28, and L30 ligands in AutoDock 4.2 software [38], going from PDB to PDBQT format. With the assistance of the AUTOGRID algorithm, the grid box dimensions were fixed on (−42.56 Å, −0.46 Å, −55.07 Å), choosing the 3D-structure sizes on (110, 110, 110) with a spacing of 0.375 Å, and running ten genetic algorithms for twenty-five million evals. Lastly, 3D and 2D intermolecular interactions of candidate drug-targeted protein complexes were visualized with the help of Discovery Studio 2021 (BIOVIA) software [36,37].
2.6. Molecular dynamic
To ensure the intermolecular interactions stability of protein-ligand complexes, we have performed the molecular dynamics technique during 50 ns using Desmond software, a package of Schrödinger LLC [39]. The output files of molecular docking were defined as input files of molecular dynamic simulations, which were prepared by Protein Preparation Maestro, which optimizes and minimizes the pre-treated complexes in the System Builder utilizing a solvent model having an orthorhombic box of transferable intermolecular interaction potential 3 points (TIP3P) type, and an OPLS-like force field [40]. In addition, the models were neutralized with the insertion of water molecules and counter ions, where the physiological conditions were reproduced in 0.15 M salts (Na+, Cl-), under a temperature and pressure of 300 K and 1 atm, respectively. After every 10 ps of evaluation, we recorded the trajectories, and we examined the stability of the simulations, studying the root mean square deviation (RMSD) for each complex. Thereafter, we have supervised the root-mean-square fluctuation (RMSF), polar surface area (PSA), solvent accessible surface area (SASA), gyration radius (Rg), and molecular surface area (MolSA) variations [6]. Lastly, the free energies of the protein-candidate drug interactions were tested at 50 ns of simulations time, with the help of MM-GBSA technology [41].
3. Results and discussion
3.1. Principal component analysis
To minimize the dimension of molecular descriptors in a reduced number of principal components, we have focused on the correlation matrix given by the principal component analysis (PCA) technique, as one of the most applied multivariate analyses [42]. We have started this task with a database of 40 molecules and 40 different descriptors as illustrated in Table S2. Then, we reduced this uncontrollable number of descriptors into 25 uncorrelated descriptors, as the strongly correlated descriptors between them (Pearson correlation coefficient>0.9) were removed, so to facilitate the development of QSAR mathematical models.
3.2. Experimental database
The present study is intended to develop a mathematical QSAR model between the structural descriptors of human GlyT1 inhibitors and their biological activities of pIC50 order. For this reason, we have worked on an experimental database of forty compounds and twenty-five uncorrelated descriptors. Then, we based on thousands of randomizations with a ‘stepwise’ option, dividing our statistical database into training and test sets. The first set contains 80% of global data (32 molecules) taken to establish the QSAR model, and the second set includes 20% (8 molecules) selected to validate the established model [6,37,43].
3.3. Multiple linear regression
The first QSAR model was constructed for a training set of thirty-two molecules (N = 32) using multiple linear regression (MLR) as a statistical method capable to evaluate the linear relationship between the molecular descriptors and the inhibitory activity [44]. The process was based on the best values of the correlation, determination, and adjustment coefficients (R, R2, and R2adj). So, the optimal MLR QSAR model was developed in Eq. (1):
| pIC50 = 3.82 + 0.01 × MW – 0.32 × NRB + 0.28 × Log P – 0.22 × TD + 0.045 × PSA | (1) |
The predictive QSAR model introduces the following selected descriptors: molecular weight (MW), partition coefficient (Log P), number of rotatable bonds (NRB), polar surface area (PSA), and topological diameter (TD), as included in Table S3.
The slope of each selected descriptor has a probability inferior to 5%, as indicated by the significance test in Table 1. So, the selected variables have a significant impact on the activity at a confidence interval of 95%. Molecular weight, partition coefficient, and polar surface area have a positive effect on activity, but the number of rotatable bonds and topological diameter influence it negatively, as illustrated in Fig. 1. Thus, the compound can be highly active if it's defined by a small value of TD and NRB and given by higher values of MW, Log P, and PSA. According to Fisher's statistical test, the null hypothesis (H0) is rejected because the analysis of variance (Anova) proves that the critical value: [F (32, 5) = 2.51, p < 0.0001] is much smaller than its calculated value (F = 13.832), as presented in Table 2. Thus, the dependent variable (pIC50) and the independent variables (5 selected descriptors) are defined by a homogeneous variance. In addition, the correlation, determination, and adjustment coefficients are equal to R = 0.85, R2 = 0.73, and R2adj = 0.67, respectively. Which confirms a solid relationship between the inhibitory activity and the molecular descriptors, and a strong predictive performance of the MLR QSAR model, with a minimal root mean square error (RMSE = 0.297). Consequently, we conclude that human GlyT1 activity is strongly and significantly correlated with constitutional (MW), physicochemical (Log P), geometrical (NRB), and topological (PSA and TD) descriptors.
Table 1.
The significant weights of the selected descriptors.
| Source | Value | Standard deviation | t | Pr > |t| | Lower terminal (95%) | Higher terminal (95%) |
|---|---|---|---|---|---|---|
| Constante | 3.823 | 1.307 | 2.926 | 0.007 | 1.137 | 6.509 |
| MW | 0.010 | 0.002 | 5.608 | < 0.0001 | 0.007 | 0.014 |
| NRB | −0.321 | 0.056 | −5.769 | < 0.0001 | −0.436 | −0.207 |
| Log P | 0.280 | 0.095 | 2.950 | 0.007 | 0.085 | 0.474 |
| TD | −0.222 | 0.105 | −2.118 | 0.044 | −0.437 | −0.007 |
| PSA | 0.045 | 0.010 | 4.453 | 0.000 | 0.024 | 0.065 |
Fig. 1.
The impact of molecular descriptors on the biological activity.
Table 2.
Analysis of variance using the fisher test.
| Source | DDL | Total square | Mean square | F | Pr > F |
|---|---|---|---|---|---|
| Model | 5 | 6.086 | 1.217 | 13.832 | <0.0001 |
| Error | 26 | 2.288 | 0.088 | ||
| Adjusted total | 31 | 8.374 |
3.4. Multiple non-linear regression
To model the non-linear relationship between the previously selected descriptors and human GlyT1 activity, we have applied the multiple non-linear regression (MNLR) technique [45], for a training set of thirty-two molecules (N = 32) using the following programmed function Eq. (2) [6]:
| (2) |
Where, Y: is the predicted activity with MNLR QSAR model, a0: is the constant of the mathematical model, Xi: is the selected descriptor, ai and bi: are the slopes of each molecular descriptor for 1 and 2°, respectively [6]. Thus, we have obtained the second QSAR model as presented by Eq. (3):
| pIC50 = - 0.487 + 0.006 × MW - 0.629 × NRB + 0.577 × Log P - 0.477 × TD + 0.232 × PSA + 3.887E-6 × MW^2 + 0.024 × NRB^2–0.055 × Log P^2 + 0.010 × TD^2–0.001 × PSA^2 | (3) |
This predictive model is qualified by a strong correlation coefficient of R = 0.88, a good determination coefficient (R2 = 0.78) and a low root mean square error (RMSE = 0.296).
In conclusion, the MLR and MNLR QSAR models noted in Equations (1), (3) respectively, affirm that human GlyT1 activity is actually affected by five selected descriptors. Where, the weight of active molecule (MW), its lipophilicity in octanol/water solvent (expressed by the partition coefficient: Log P), and the region of its polar surface area (PSA) have all a positive impact. While, the topological diameter (TD) and the number of rotatable bonds (NRP) have a negative effect on GluT1 activity.
3.5. Validation of predictive QSAR models
3.5.1. Applicability domain
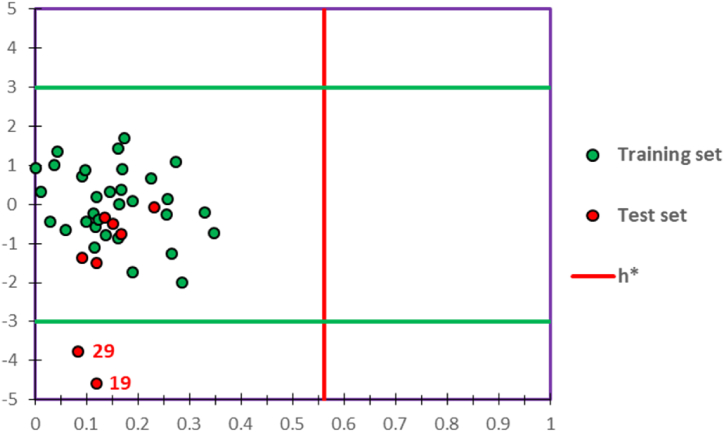
To examine the applicability of quantitative activity-structure relationship models, we have performed the applicability domain (AD) technology. Using IBM SPSS Statistics 22 software. Based on William's diagram shown in Fig. 2. The warning leverage (h* = 0.56) was indicated using Eq. (4) [46]:
| h* = [3 × (p + 1) / n], (p = 5, n = 32) | (4) |
Where, p: is the number of selected descriptors. n: is the number of molecules in the training set, and 3: is the absolute value limit of the standardized residuals interval on the Y-axis [47]. We note that all molecules in the training and test sets have a leverage under the critical leverage value, and they are all placed in the confidence interval of [-3, +3], except two molecules (19 and 29) of the test set, which are considered as outliers because they have a standardized residual of −4.601 and −3.785, respectively. So, the QSAR model was correctly predicted, where the outliers were removed and the 38 remaining molecules were successfully examined in the applicability domain [6,43,48].
Fig. 2.
William's diagram of the predictive QSAR models. (Green and red dots display the molecules of training and test sets, respectively. The red line indicates the warning leverage, and the green lines show the confidence interval of [-3, +3])
3.5.2. External validation
To evaluate the reliability of the QSAR predictive model and to ensure its application, it is strictly necessary to validate it on some novel compounds comprised in the test set using the external validation technique [49]. Based on a training test of thirty-two molecules, we have tested six new compounds from the test set and we have obtained the results of predicted activities using the linear and nonlinear regression techniques, as displayed in Table 3.
Table 3.
The observed and predicted activities of pIC50 order using MLR and MNLR QSAR models.
| Molecules number | Observed pIC50 | Predicted pIC50 (MLR) | Predicted pIC50 (MNLR) |
|---|---|---|---|
| 4a | 7.638 | 8.044 | 7.897 |
| 10a | 7.796 | 7.825 | 7.908 |
| 14a | 8.222 | 8.323 | 8.410 |
| 17a | 7.569 | 8.017 | 7.954 |
| 18a | 7.921 | 8.069 | 8.012 |
| 31a | 6.848 | 7.076 | 7.200 |
Indicates the test set molecules.
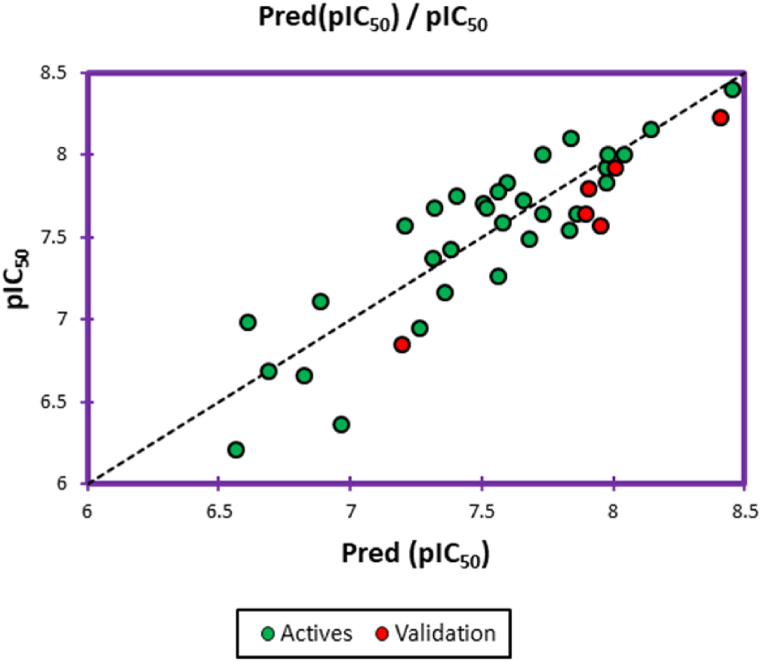
The first QSAR model, which was developed using the multiple linear regression (MLR) technique, is given by an external validation correlation coefficient equal to R2ext = 0.867, as presented in Fig. 3, and the second QSAR model, developed by multiple non-linear regression (MNLR) technique, is defined by an external validation correlation coefficient of R2ext = 0.946, as illustrated in Fig. 4. Thus, MLR and MNLR QSAR models are externally validated because they are characterized by an external validation correlation coefficient greater than 0.6.
Fig. 3.
The correlation among the predicted and observed activities using multiple linear regression. (Green and red dots indicate the molecules in the training and test sets, respectively)
Fig. 4.
The correlation among the predicted and observed activities using multiple nonlinear regression. (Green and red dots indicate the molecules in the training and test sets, respectively)
3.5.3. Internal validation
The leave-one-out procedure (LOO) of the cross-validation technique (CV) was performed to validate internally the developed QSAR models. For this purpose, we tested each molecule once, with the development of a new mathematical model on thirty-one compounds (N-1 = 31) and prediction of the inhibitory activity of this tested molecule [50], as resulted in Table 4, this process was repeated for all training set molecules [46]. Thereafter, we calculated the quadratic coefficient (Q2 cv) of cross-validation using Eq. (5) [43]:
| (5) |
Where, Yobs: signifies the observed activity, Ypred: is the predicted activity, and Ymean: presents the average of the observed activities. Although the obtained value of quadratic coefficient (Q2cv = 0.55) is higher than 0.5, the established QSAR model is robust, reliable, and has a stronger internal predictivity.
Table 4.
The observed and predicted activities using MLR, MNLR and CVLOO techniques.
| Molecules number | Observed pIC50 | Predicted pIC50 (MLR) | Predicted pIC50 (MNLR) | Predicted pIC50 (CVLOO) |
|---|---|---|---|---|
| 1 | 7.824 | 7.551 | 7.601 | 7.541 |
| 2 | 8.097 | 7.802 | 7.844 | 7.780 |
| 3 | 7.638 | 7.836 | 7.866 | 7.856 |
| 5 | 7.921 | 8.097 | 7.976 | 8.128 |
| 6 | 7.770 | 7.559 | 7.563 | 7.528 |
| 7 | 7.638 | 7.709 | 7.732 | 7.721 |
| 8 | 8.000 | 7.743 | 7.734 | 7.704 |
| 9 | 7.420 | 7.329 | 7.387 | 7.325 |
| 11 | 8.000 | 8.067 | 8.042 | 8.105 |
| 12 | 8.155 | 8.119 | 8.148 | 8.105 |
| 13 | 8.000 | 8.081 | 7.983 | 8.114 |
| 15 | 8.398 | 8.376 | 8.458 | 8.370 |
| 16 | 7.538 | 7.775 | 7.833 | 7.824 |
| 20 | 7.108 | 7.056 | 6.887 | 7.047 |
| 21 | 7.161 | 7.298 | 7.361 | 7.307 |
| 22 | 7.745 | 7.351 | 7.406 | 7.318 |
| 23 | 6.678 | 6.813 | 6.691 | 6.833 |
| 24 | 6.979 | 6.712 | 6.616 | 6.645 |
| 25 | 6.947 | 7.205 | 7.266 | 7.267 |
| 26 | 7.678 | 7.258 | 7.322 | 7.156 |
| 27 | 6.356 | 6.878 | 6.970 | 7.027 |
| 28 | 7.699 | 7.376 | 7.511 | 7.233 |
| 30 | 7.260 | 7.592 | 7.562 | 7.650 |
| 32 | 7.481 | 7.602 | 7.684 | 7.625 |
| 33 | 7.678 | 7.485 | 7.522 | 7.418 |
| 34 | 7.721 | 8.098 | 7.660 | 8.259 |
| 35 | 7.569 | 7.073 | 7.208 | 6.944 |
| 36 | 6.201 | 6.797 | 6.569 | 7.075 |
| 37 | 6.658 | 6.882 | 6.825 | 7.019 |
| 38 | 7.367 | 7.257 | 7.316 | 7.230 |
| 39 | 7.585 | 7.493 | 7.580 | 7.473 |
| 40 | 7.824 | 7.823 | 7.973 | 7.823 |
3.5.4. Validation using Y-randomization test
Golbraikh and Tropsha's study argues that the internal predictive accuracy of the quantitative structure-activity relationships (QSAR) model, explained by a large value of the quadratic coefficient of leave one out cross-validation (CVLOO), has a tendency to be overestimated and may be the result of a lucky correlation. For this reason, the Y-Randomization test is necessary [6,51]. With the assistance of java Platform SE binary, we have tested the quality of the established QSAR model running 100 randomizations as resulted in Table S4. The results of the Y-Randomization test indicate that the value of the cR2p criteria is 0.65, which is greater than 0.5. Additionally, the values of correlation, determination, and cross-validation with the leave-one-out procedure coefficients of the original model are so much higher than the resulting values for all one hundred randomizations. As a consequence, the human GlyT1 activities as predicted by the originally designed model are not due to random chance [6].
3.5.5. Statistical criteria of golbreikh and Tropsha's study
The predictive MLR QSAR model given in Equation (1), successfully meets the postulated threshold conditions of Tropsha and Golbraikh's theory, as illustrated in the following Table 5.
Table 5.
Tropsha and Golbraikh conditions to examine the accuracy of generated MLR QSAR model.
| Parameter | Equation | Model score | Threshold | Comment |
|---|---|---|---|---|
| 0.73 | >0.6 | Accepted | ||
| 0.67 | >0.6 | Accepted | ||
| 0.87 | >0.6 | Accepted | ||
| 0.55 | >0.5 | Accepted | ||
| R2rand | Average of the 100 R2rand (i) | 0.16 | < R2 | Accepted |
| ‘LOO’ rand | Average of the 100 ‘LOO’ rand (i) | −0.30 | < Q2cv | Accepted |
| cR2p | cR2p = R | 0.65 | >0.5 | Accepted |
3.6. In silico pharmacokinetics ADMET prediction
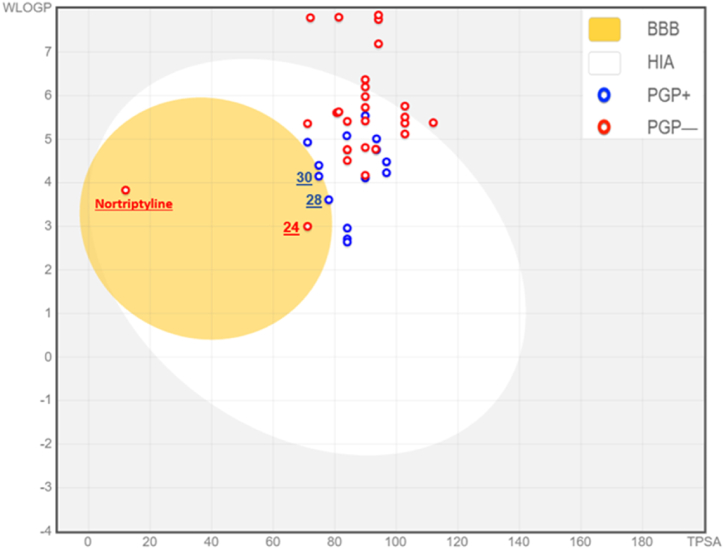
In the beginning, we predicted the human GlyT1 inhibitors using BOILED-Egg as an accurate predictive model, very practical in medicinal chemistry to discover the candidate drugs as presented in Fig. 5. We have noted that 28 and 30 molecules in blue are part of the yellow Egan-egg, so they are predicted as inhibitors with the greatest ability to cross the blood-brain barrier (BBB) and predicted to be effluated from the central nervous system (CNS) by the P-glycoprotein. Thus, 24 molecules and the nortriptyline as a co-crystallized ligand bound to the membrane protein of dopamine transporter (DAT), as colored in red, are also part of yellow Egan-egg, but they are predicted not to be effluated from the central nervous system (CNS) by the P-glycoprotein. In contrast, the compounds that are part of the white Egan-egg, are predicted as molecules passively absorbed by the gastrointestinal tract [52]. At the second stage, we have predicted the nortriptyline as a co-crystallized ligand compared to 28 and 30 inhibitors which are considered as candidate inhibitors able to penetrate the blood-brain barrier (BBB) and evacuated from the central nervous system (CNS) by the P-glycoprotein in the basis of Lipinski, Egan, Veber, Ghose, and Muegge rules, as shown in Table 6. Thus, we have noted that these candidate drugs respect all Lipinski rules like as: MW ≤ 500, 40≤ MR ≤ 130, NRB<10, Log P(octanol/water) < 5, HBA≤10, and HBD<5 [43]. Additionally, they satisfy Veber, Egan, Ghose, and Muegge regulations [53]. In the last step, we studied the pharmacokinetic properties of adsorption, distribution, metabolism, excretion, and toxicity (ADME-Tox) of the candidate molecules. Which were compared to the obtained results for nortriptyline, as resulted in Table 7. We have noticed that L28 and L30 ligands have an excellent human intestinal absorption (HIA superior than 96%), good distribution because they are defined by the human distribution volumes higher than −0.44 Log L/kg, and characterized by a BBB and CNC permeability values greater than −1 Log BB and included in (−2 to −3) Log PS, respectively. So, they are well permeable to the blood-brain barrier (BBB) and the central nervous system (CNS). Moreover, they are all predicted as potent inhibitors of 2C9, 2C19 and 3A4 cytochromes. Then, they present a total clearance of 0.563 and 0.711 Log ml/min/kg respectively, without any toxicity on the human body. Therefore, we have concluded that L28 and L30 ligands are successfully predicted as not toxic inhibitors of 2C9, 2C19, and 3A4 cytochromes, respecting Lipinski, Veber, Egan, Ghose, and Muegge rules, and they are estimated to penetrate the blood-brain barrier (BBB) with the highest probability. So, they are designed as powerful central nervous system (CNS) agents.
Fig. 5.
BOILED-Egg model of nortriptyline and forty pyrrolo [3,4-c]pyrazole and azetidine derivatives. (BBB: blood-brain barrier; HIA: human intestinal absorption; PGP: P-glycoprotein).
Table 6.
Prediction of physico-chemical parameters of L28, L30, and nortriptyline ligands based on Lipinski, Egan, Veber, Ghose, and Muegge regulations.
| Ligands number | Physical and chemical properties |
Lipinski violations | Veber violations | Egan violations | Ghose violations | Muegge violations | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight (g/mol) | Molar refractive index | Rotatable bonds Number | Log P (Octanol/Water) | Hydrogen Bond acceptors |
H-bond donors Number | ||||||
| Threshold | MW ≤ 500 | 40≤ MR ≤ 130 | <10 | <5 | ≤10 | <5 | Yes/No | Yes/No | Yes/No | Yes/No | Yes/No |
| L28 | 448.47 | 124.97 | 4 | 3.35 | 6 | 0 | Yes | Yes | Yes | Yes | Yes |
| L30 | 423.44 | 120.78 | 4 | 3.23 | 5 | 0 | Yes | Yes | Yes | Yes | Yes |
| Nortriptyline | 263.38 | 86.06 | 3 | 3.22 | 1 | 1 | Yes | Yes | Yes | Yes | No |
Table 7.
ADMET-Tox pharmacokinetic properties of L28, L30, and Nortriptyline ligands.
| Absorption |
Distribution |
Metabolism |
Excretion |
Toxicity |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ligands number | Intestinal Absorption (human) |
VDss (human) |
BBB permeability |
CNS permeability |
Substrate |
Inhibitor |
Total Clearance |
AMES toxicity |
|||||
| CYP | |||||||||||||
| 2D6 |
3A4 |
1A2 |
2C19 |
2C9 |
2D6 |
3A4 |
|||||||
| Numeric (% Absorbed) | Numeric (Log L/kg) | Numeric (Log BB) | Numeric (Log PS) | Categorical (Yes/No) | Numeric (Log ml/min/kg) | Categorical (Yes/No) | |||||||
| L28 | 96.007 | 0.234 | −0.818 | −2.839 | No | Yes | No | Yes | Yes | No | Yes | 0.563 | Not toxic |
| L30 | 98.673 | 0.219 | −0.353 | −1.865 | No | Yes | Yes | Yes | Yes | No | Yes | 0.711 | Not toxic |
| Nortriptyline | 97.482 | 1.575 | 0.808 | −1.196 | No | Yes | Yes | No | No | Yes | No | 0.929 | Not toxic |
3.7. Molecular docking
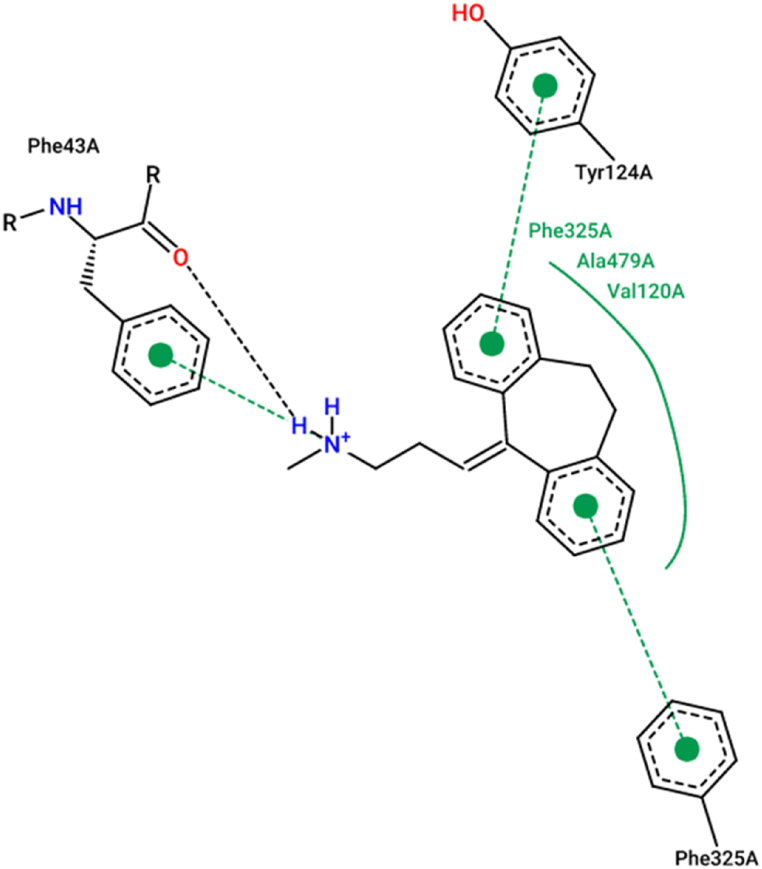
Although L28 and L30 ligands are engineered as potent inhibitors with a favorable ADMET profile, thus we have studied their intermolecular interactions towards the drosophila melanogaster dopamine transporter (DAT) of 4M48. pdb code, as a transmembrane protein able to extract the neurotransmitter dopamine out of the synaptic cleft and transfer it into the cytosol of neighboring cells [6,[54], [55], [56]]. The molecular docking simulations of (L28, L30-protein) complexes were done in the active sites of tricyclic antidepressant nortriptyline (as co-crystallized ligand linked to the targeted protein in his A-chain) which resulted from the ProteinsPlus online server [57], like Tyr124 A, Phe325 A, and Phe43 A amino acids, as presented in Fig. 6.
Fig. 6.
The active sites of co-crystalized ligand bound to 4M48. pdb protein in A-chain.
The molecular docking results presented in Fig. 7, indicate that L28 and L30 ligands have several common intermolecular interactions such as: Two stacked Pi-Pi chemical bonds, established between Phe325 and Phe319 amino acids of the targeted protein and the candidate drugs. A common Pi-Sigma chemical bond was formed between the candidate drugs and Ile116 amino acid with a nuclear distance of 5.91 Å. Three Pi-Alkyl chemical bonds were produced with Ala479, Ile483, and Ala117 amino acids of the responsible protein. A common bond of attractive charge with the nitrogen atom was created with Asp475 amino acid. Additionally, we note that the L28 ligand formed at 6.44 Å a bond of Alkyl type towards Tyr124 amino acid. And created for each one of its two fluorine atoms, a hydrogen bond with Tyr123 amino acid, rendering the L28-protein complex more stable. Also, the L30-protein complex was stabilized via the formation of a hydrogen bond between the candidate drug and the Arg52 amino acid of the targeted protein. Therefore, we conclude that Phe325, Phe319, Ile116, Ala479, Ile483, Ala117, Asp475, Tyr124, Tyr123, and Arg52 amino acids are the responsible sites of human glycine transporter type one (GLUT1) inhibitory activities to improve memory performance.
Fig. 7.
2D (left) and 3D (right) inter-molecular interactions, produced between L28 and L30 ligands and DAT protein of 4M48. pdb code with binding energies of −8.72 kcal/mol and −8.70 kcal/mol, respectively. (The left colored rectangles show all types of intermolecular interactions, produced between the candidate ligands and the target protein, while the right key show the donor and acceptor effects in the studied complexes).
3.8. Docking validation protocol
The efficiency and accuracy of the molecular docking algorithms were examined through the re-docking method, based on the superposition of the co-crystallized ligand on the docked nortriptyline ligand. The overlay results shown in Fig. 8C, reflects a root mean square deviation (RMSD) of 0.552 Å, which is inferior than 2 Å, so the prediction pose is precise and accurate. In addition, 2D and 3D intermolecular interactions, successively presented in Fig. 8A and B, reveal that docked nortriptyline ligand of cyan color, reacts to Tyr124 and Phe43 amino acids, which are the same sites as those experimentally produced. As a result, the docking validation protocol was successfully examined [6,37,58].
Fig. 8.
2D (A) and 3D (B) visualization of intermolecular interactions created between the docked nortriptyline and 4M48. pdb protein with a binding energy of −8.36 kcal/mol, and re-docking pose (c) defined by a RMSD value of 0.552 Å (docked nortriptyline in cyan and co-crystallized nortriptyline colored by the atomic elements). (The key-colored rectangles show all types of intermolecular interactions, produced between the nortriptyline and the target protein)
3.9. Molecular dynamic simulation
To examine the stability of intermolecular interactions, produced between L28 and L30 candidate drugs and the targeted protein encoded as 4M48. pdb, we have performed the molecular dynamics simulations of these two complexes during 50 ns [6,14].
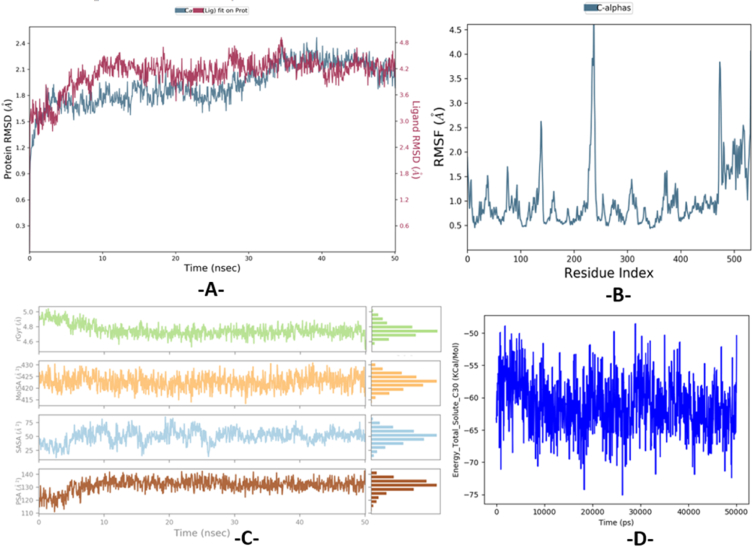
The results of dynamic conformational changes of (the L28-DAT protein) complex shown in Fig. 9A, illustrate that protein root mean square deviation (RMSD) in the left Y-axis. Fluctuates around the mean thermal structure during 50 ns of simulation time, because deviation changes in the range of 1–3 Å are acceptable for small globular proteins. Thus, the simulation was well-balanced. In addition, the evolution of the ligand RMSD in the right Y-axis through the heavy atoms, reveals the ligand stability with respect to the protein targets, when the studied complex was first aligned with the reference protein backbone, as the observed values of ligand RMSDs are nearly the same as the protein RMSDs values. Thus, the ligand had not diffused away of its primary binding site. Moreover, the evolution of the root mean square fluctuation (RMSF) was also controlled to investigate the influence of ligand binding on the internal dynamics of DAT protein over 50 ns, in which C- and N-terminal tails tend to fluctuate more than any other protein part. Similarly, alpha helices and beta strands as secondary structure elements are generally more rigorously rigid than the non-structured protein part, so they fluctuate considerably less than the buckle regions. Just one oscillation of 3.7 Å, has been detected in the buckle region of the residue 390. In contrast, all other fluctuations were inferior to 3 Å, as noted in Fig. 9B, which indicates the absence of a conformational change of the protein caused by its binding towards the ligand, and the stability of the produced complex. The values of gyration radiuses (r-Gyr) have oscillated in a relatively short range of 4.40–5.35 Å along the simulation time, as noticed in Fig. 9C, which reveals a few conformational changes in the compactness of the L28 ligand, so the DAT protein has an acceptable flexibility after being bound to the candidate drug. Also, the evolution of solvent accessible surface area (SASA), was done to evaluate the solvent accessibility of (L28-DAT protein) complex over 50 ns of simulation time, as we have recorded a minimal oscillation, included between 20 and 90 Å2, which demonstrates that the drug structure was approximately unchanged during the simulation process. Moreover, the molecular surface area (MolSA) associated with a Van Der Waals surface was calculated with a probe radius of 1.4 Å, has been fluctuated between 400 and 430 Å2. Furthermore, we have noticed that the polar surface area (PSA) which is mainly influenced by nitrogen and oxygen atoms, changes between 75 and 145 Å2, with some very small maximum and minimum fluctuations. Eventually, the graph of total energy, obtained using the MM-GBSA approach, displays some minimal oscillations around the mean of −58.360388 kcal.mol-1, which are progressively stabilized from 25 ns until the end of the simulation as displayed in Fig. 9D.
Fig. 9.
Evolution of RMSD, RMSF, Rg, MolSA, SASA, PSA and total free energy during 50 ns, for (L28-DAT protein) complex.
Regarding the second complex, we noticed that the DAT protein was also characterized by a root mean square deviation (RMSD) included between 1 and 3 Å, where the observed values of the ligand RMSD oscillate in parallel with those of the protein RMSD, as presented in Fig. 10A. So, the simulation was well equilibrated for the (L30-DAT protein) complex, and the candidate drug had not diffused away from its primary link site. Additionally, the graph of root mean square fluctuations (RMSFs), indicates only two fluctuations greater than 3 Å. The first one was recorded at 4.6 Å for residue 230, and the second one was detected at 4.7 Å for residue 480, as shown in Fig. 10B, indicating that the DAT protein binds to the drug with extremely small conformational changes. Subsequently, we have monitored the evolution of gyration radiuses (r-Gyr), solvent accessible surface area (SASA), molecular surface area (MolSA), and polar surface area (PSA) properties, which are generally subject to very small fluctuations as displayed in Fig. 10C. So, the drug structure remained approximately unchanged during 50 ns of simulation time, and the DAT protein has an agreeable flexibility during its interaction with the L30 candidate drug. Finally, the total energy graph of the (L30-DAT protein) complex, given with the assistance of the MM-GBSA approach, displays slight fluctuations about the mean of −61.193294 kcal.mol-1, as shown in Fig. 10D, which means that the complex remains very stable over the simulation time.
Fig. 10.
Evolution of RMSD, RMSF, Rg, MolSA, SASA, PSA, and total free energy during 50 ns, for (L30-DAT protein) complex.
Consequently, we conclude that intermolecular interactions produced in the (DAT protein-L28, and L30 ligands) complexes are dynamically more stable because their studied properties remained in equilibrium throughout the MD simulation. Moreover, they present similar stability compared to MD calculations of nortriptyline as a positive control molecule crystallized to the DAT protein complex. Where the RMSD values do exceed not 3 Å, and RMSF values display just negligible fluctuations. Furthermore, the variation of free energies obtained with the use of MM-GBSA approach remains stable during the molecular dynamics simulation time, as displayed in Fig. 11.
Fig. 11.
Evolution of RMSD, RMSF, Rg, MolSA, SASA, PSA, and total free energy during 50 ns, for (Nortriptyline -DAT protein) complex.
4. Conclusion
An in silico investigation was performed for forty pyrrolo [3,4-c]pyrazole and azetidine-based derivatives, to discover an effective drug for enhancing human memory performance. In the beginning, two predictive QSAR models were developed using MNLR and MLR techniques and were successfully validated by internal and external validation, Y-randomization test, applicability domain, Tropsha, and Golbreikh criteria, indicating that MW, Log P, NRB, PSA, and TD descriptors have a key function in the human glycine transporter type one (GlyT1) activity. Afterward, a pharmacoinformatic-based approach revealed that L28 and L30 ligands were predicted as non-toxic inhibitors of 2C9, 2C19, and 3A4 cytochromes, with a desirable ADMET profile, meeting the rules of Lipinski, Egan Veber, Muegge, and Ghose. So, they are designed as candidate drugs, most likely to penetrate the central nervous system (CNS). For this reason, they were specially selected for molecular docking simulation, which revealed that they react with Phe319, Phe325, Tyr123, Tyr 124, Arg52, Asp475, Ala117, Ala479, Ile116, and Ile483 amino acids of a membrane protein from the dopamine transporter (DAT) of 4M48. pdb code. Finally, the stability of intermolecular interactions produced between the candidate drugs and the targeted protein was successfully examined through the molecular dynamics technique along 50 ns of simulation time. Therefore, L28 and L30 drugs could be safely and securely applicable as therapeutics in medicine to enhance human memory performance.
Author contribution statement
Mohamed El fadili: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Mohammed Er-rajy: Conceived and designed the experiments; Contributed reagents, and analysis tools.
Hamada Imtara: Analyzed and interpreted the data; Contributed reagents; Wrote the paper.
Omar M. Noman, Ramzi A Mothana: Analyzed and interpreted the data; Wrote the paper.
Sheaf Abdullah, Sara Zerougui: Analyzed and interpreted the data.
Menana Elhallaoui: Conceived and designed the experiments.
Funding statement
This work was supported by King Saud University, Riyadh, Saudi Arabia [RSP2023R119].
Acknowledgment
The authors extend their appreciation to Researchers Supporting Project number (RSP2023R119), King Saud University, Riyadh, Saudi Arabia for funding this work.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interest's statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Supplementary content related to this article has been published online at [URL].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e13706.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Nazir N., Nisar M., Zahoor M., Uddin F., Ullah S., Ullah R., Ansari S.A., Mahmood H.M., Bari A., Alobaid A. Phytochemical analysis, in vitro anticholinesterase, antioxidant activity and in vivo nootropic effect of Ferula ammoniacum (dorema ammoniacum) D. Don. In scopolamine-induced memory impairment in mice. Brain Sci. 2021;11:259. doi: 10.3390/brainsci11020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Couto B., Rojas G., Gelormini-Lezama C., O'Neill S. Encyclopedia of Behavioral Neuroscience. second ed. Elsevier; 2022. Memory, executive function and social cognition in neurological disorders; pp. 140–147. [DOI] [Google Scholar]
- 3.Santora V.J., Almos T.A., Barido R., Basinger J., Bellows C.L., Bookser B.C., Breitenbucher J.G., Broadbent N.J., Cabebe C., Chai C.-K., Chen M., Chow S., Chung D.M., Crickard L., Danks A.M., Freestone G.C., Gitnick D., Gupta V., Hoffmaster C., Hudson A.R., Kaplan A.P., Kennedy M.R., Lee D., Limberis J., Ly K., Mak C.C., Masatsugu B., Morse A.C., Na J., Neul D., Nikpur J., Peters M., Petroski R.E., Renick J., Sebring K., Sevidal S., Tabatabaei A., Wen J., Yan Y., Yoder Z.W., Zook D. Design and synthesis of novel and selective Glycine transporter-1 (GlyT1) inhibitors with memory enhancing properties. J. Med. Chem. 2018;61:6018–6033. doi: 10.1021/acs.jmedchem.8b00372. [DOI] [PubMed] [Google Scholar]
- 4.Eulenburg V., Armsen W., Betz H., Gomeza J. Glycine transporters: essential regulators of neurotransmission. Trends Biochem. Sci. 2005;30:325–333. doi: 10.1016/j.tibs.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Hudson A.R., Santora V.J., Petroski R.E., Almos T.A., Anderson G., Barido R., Basinger J., Bellows C.L., Bookser B.C., Broadbent N.J., Cabebe C., Chai C.-K., Chen M., Chow S., Chung D.M., Heger L., Danks A.M., Freestone G.C., Gitnick D., Gupta V., Hoffmaster C., Kaplan A.P., Kennedy M.R., Lee D., Limberis J., Ly K., Mak C.C., Masatsugu B., Morse A.C., Na J., Neul D., Nikpur J., Renick J., Sebring K., Sevidal S., Tabatabaei A., Wen J., Xia S., Yan Y., Yoder Z.W., Zook D., Peters M., Breitenbucher J.G. Azetidine-based selective glycine transporter-1 (GlyT1) inhibitors with memory enhancing properties. Bioorg. Med. Chem. Lett. 2020;30 doi: 10.1016/j.bmcl.2020.127214. [DOI] [PubMed] [Google Scholar]
- 6.El fadili M., Er-Rajy M., Kara M., Assouguem A., Belhassan A., Alotaibi A., Mrabti N.N., Fidan H., Ullah R., Ercisli S., Zarougui S., Elhallaoui M. QSAR, ADMET in silico pharmacokinetics, molecular docking and molecular dynamics studies of novel bicyclo (aryl methyl) benzamides as potent GlyT1 inhibitors for the treatment of schizophrenia. Pharmaceuticals. 2022;15:670. doi: 10.3390/ph15060670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radan M., Djikic T., Obradovic D., Nikolic K. Application of in vitro PAMPA technique and in silico computational methods for blood-brain barrier permeability prediction of novel CNS drug candidates. Eur. J. Pharmaceut. Sci. 2022;168 doi: 10.1016/j.ejps.2021.106056. [DOI] [PubMed] [Google Scholar]
- 8.Vilar S., Cozza G., Moro S. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008;8:1555–1572. doi: 10.2174/156802608786786624. [DOI] [PubMed] [Google Scholar]
- 9.Ebenezer O., Damoyi N., Jordaan M.A., Shapi M. Unveiling of pyrimidindinones as potential anti-norovirus agents—a pharmacoinformatic-based approach. Molecules. 2022;27:380. doi: 10.3390/molecules27020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian S., Wang J., Li Y., Li D., Xu L., Hou T. The application of in silico drug-likeness predictions in pharmaceutical research. Adv. Drug Deliv. Rev. 2015;86:2–10. doi: 10.1016/j.addr.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Serrano A., Imbernón B., Pérez-Sánchez H., Cecilia J.M., Bueno-Crespo A., Abellán J.L., Qn-Docking An innovative molecular docking methodology based on Q-Networks. Appl. Soft Comput. 2020;96 doi: 10.1016/j.asoc.2020.106678. [DOI] [Google Scholar]
- 12.R.P.D. Bank, RCSB PDB - 4M48: X-ray structure of dopamine transporter elucidates antidepressant mechanism, (n.d.). https://www.rcsb.org/structure/4M48 (accessed July 4, 2022). [DOI] [PMC free article] [PubMed]
- 13.Wang E., Sun H., Wang J., Wang Z., Liu H., Zhang J.Z.H., Hou T. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem. Rev. 2019;119:9478–9508. doi: 10.1021/acs.chemrev.9b00055. [DOI] [PubMed] [Google Scholar]
- 14.de Oliveira T.A., Medaglia L.R., Maia E.H.B., Assis L.C., de Carvalho P.B., da Silva A.M., Taranto A.G. Evaluation of docking machine learning and molecular dynamics methodologies for DNA-ligand systems. Pharmaceuticals. 2022;15:132. doi: 10.3390/ph15020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaussian 09 Citation | Gaussian.com, (n.d.). https://gaussian.com/g09citation/(accessed July 6, 2022).
- 16.Parr R.G. In: Horizons of Quantum Chemistry. Fukui K., Pullman B., editors. Springer Netherlands; Dordrecht: 1980. Density functional theory of atoms and molecules; pp. 5–15. [DOI] [Google Scholar]
- 17.Wang H., Jia Y., Jing G., Wu X. A novel toxicity prediction model for hydrazine compounds based on 1D–3D molecular descriptors. Computational Toxicology. 2021;18 doi: 10.1016/j.comtox.2021.100169. [DOI] [Google Scholar]
- 18.Milne G.W.A. Software review of ChemBioDraw 12.0. J. Chem. Inf. Model. 2010;50:2053. doi: 10.1021/ci100385n. [DOI] [Google Scholar]
- 19.Österberg T., Norinder U. Prediction of drug transport processes using simple parameters and PLS statistics the use of ACD/logP and ACD/ChemSketch descriptors. Eur. J. Pharmaceut. Sci. 2001;12:327–337. doi: 10.1016/S0928-0987(00)00189-5. [DOI] [PubMed] [Google Scholar]
- 20.Guerra Tort C., Aguiar Pulido V., Suárez Ulloa V., Docampo Boedo F., López Gestal J.M., Pereira Loureiro J. 3rd XoveTIC Conference, MDPI. 2020. Electronic Health records exploitation using artificial intelligence techniques; p. 60. [DOI] [Google Scholar]
- 21.Introducing the XLSTAT Free edition, XLSTAT, Your data analysis solution. (n.d.). https://www.xlstat.com/en/news/introducing-the-xlstat-free-edition (accessed March 8, 2022).
- 22.Mostoufi N., Constantinides A. In: Applied Numerical Methods for Chemical Engineers. Mostoufi N., Constantinides A., editors. Academic Press; 2023. Chapter 8 - linear and nonlinear regression analysis; pp. 403–476. [DOI] [Google Scholar]
- 23.Halder A.K., Jha T. Validated predictive QSAR modeling of N-aryl-oxazolidinone-5-carboxamides for anti-HIV protease activity. Bioorg. Med. Chem. Lett. 2010;20:6082–6087. doi: 10.1016/j.bmcl.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 24.Roy K., Kar S., Das R.N. Understanding the Basics of QSAR for Applications in Pharmaceutical Sciences and Risk Assessment. Elsevier; 2015. Validation of QSAR models; pp. 231–289. [DOI] [Google Scholar]
- 25.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- 26.Veber D.F., Johnson S.R., Cheng H.-Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 27.Egan W.J., Merz Kenneth M., Baldwin J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000;43:3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- 28.Egan W.J., Lauri G. Prediction of intestinal permeability. Adv. Drug Deliv. Rev. 2002;54:273–289. doi: 10.1016/S0169-409X(02)00004-2. [DOI] [PubMed] [Google Scholar]
- 29.Yalcin S. Molecular docking, drug likeness, and ADMET analyses of passiflora compounds as P-glycoprotein (P-gp) inhibitor for the treatment of cancer. Current Pharmacology Reports. 2020;6:429–440. doi: 10.1007/s40495-020-00241-6. [DOI] [Google Scholar]
- 30.Muegge I., Heald S.L., Brittelli D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001;44:1841–1846. doi: 10.1021/jm015507e. [DOI] [PubMed] [Google Scholar]
- 31.pkCSM (n.d. http://biosig.unimelb.edu.au/pkcsm/run_example? accessed.
- 32.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7 doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bassani D., Pavan M., Bolcato G., Sturlese M., Moro S. Re-exploring the ability of common docking programs to correctly reproduce the binding modes of non-covalent inhibitors of SARS-CoV-2 protease mpro. Pharmaceuticals. 2022;15:180. doi: 10.3390/ph15020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R.P.D. Bank, RCSB PDB - 4M48: X-ray structure of dopamine transporter elucidates antidepressant mechanism, (n.d.). https://www.rcsb.org/structure/4M48 (accessed August 23, 2022). [DOI] [PMC free article] [PubMed]
- 35.Kouranov A. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkj120. D302–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.BIOVIA Discovery Studio - BIOVIA - Dassault Systèmes®, (n.d.). https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio/(accessed August 23, 2022).
- 37.El fadili M., Er-rajy M., Imtara H., Kara M., Zarougui S., Altwaijry N., Al kamaly O., Al Sfouk A., Elhallaoui M. 3D-QSAR, ADME-tox in silico prediction and molecular docking studies for modeling the analgesic activity against neuropathic pain of novel NR2B-selective NMDA receptor antagonists. Processes. 2022;10:1462. doi: 10.3390/pr10081462. [DOI] [Google Scholar]
- 38.Norgan A.P., Coffman P.K., Kocher J.-P.A., Katzmann D.J., Sosa C.P. Multilevel parallelization of AutoDock 4.2. J. Cheminf. 2011;3:12. doi: 10.1186/1758-2946-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desmond | Schrodinger, (n.d.). https://www.schrodinger.com/products/desmond (accessed April 28, 2022).
- 40.Kaminski G.A., Friesner R.A., Tirado-Rives J., Jorgensen W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B. 2001;105:6474–6487. doi: 10.1021/jp003919d. [DOI] [Google Scholar]
- 41.Wang E., Sun H., Wang J., Wang Z., Liu H., Zhang J.Z.H., Hou T. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem. Rev. 2019;119:9478–9508. doi: 10.1021/acs.chemrev.9b00055. [DOI] [PubMed] [Google Scholar]
- 42.Bastianoni A., Guastaldi E., Barbagli A., Bernardinetti S., Zirulia A., Brancale M., Colonna T. Multivariate analysis applied to aquifer hydrogeochemical evaluation: a case study in the coastal significant subterranean water body between “cecina river and san vincenzo”. Tuscany (Italy), Applied Sciences. 2021;11:7595. doi: 10.3390/app11167595. [DOI] [Google Scholar]
- 43.Er-rajy M., El Fadili M., Hadni H., Mrabti N.N., Zarougui S., Elhallaoui M. 2D-QSAR modeling, drug-likeness studies, ADMET prediction, and molecular docking for anti-lung cancer activity of 3-substituted-5-(phenylamino) indolone derivatives. Struct. Chem. 2022;33:973–986. doi: 10.1007/s11224-022-01913-3. [DOI] [Google Scholar]
- 44.Uyanık G.K., Güler N. A study on multiple linear regression analysis. Procedia - Social and Behavioral Sciences. 2013;106:234–240. doi: 10.1016/j.sbspro.2013.12.027. [DOI] [Google Scholar]
- 45.Kravić N., Savosina J., Agafonova-Moroz M., Babain V., Legin A., Kirsanov D. Nonlinear multivariate regression algorithms for improving precision of multisensor potentiometry in analysis of spent nuclear fuel reprocessing solutions. Chemosensors. 2022;10:90. doi: 10.3390/chemosensors10030090. [DOI] [Google Scholar]
- 46.Kasmi R., Hadaji E., Chedadi O., El Aissouq A., Bouachrine M., Ouammou A. 2D-QSAR and docking study of a series of coumarin derivatives as inhibitors of CDK (anticancer activity) with an application of the molecular docking method. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e04514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pourbasheer E., Riahi S., Ganjali M.R., Norouzi P. Quantitative structure–activity relationship (QSAR) study of interleukin-1 receptor associated kinase 4 (IRAK-4) inhibitor activity by the genetic algorithm and multiple linear regression (GA-MLR) method. J. Enzym. Inhib. Med. Chem. 2010;25:844–853. doi: 10.3109/14756361003757893. [DOI] [PubMed] [Google Scholar]
- 48.Er-rajy M., El fadili M., Mrabti N.N., Zarougui S., Elhallaoui M. QSAR, Molecular docking, ADMET properties in silico studies for a series of 7-propanamide benzoxaboroles as potent anti-cancer Agents. Chin. J. Anal. Chem. 2022 doi: 10.1016/j.cjac.2022.100163. [DOI] [Google Scholar]
- 49.Rezende K.B. de C., da Cunha A.J.L.A., Amim Junior J., Bornia R.G. External validation of the Fetal Medicine Foundation algorithm for the prediction of preeclampsia in a Brazilian population. Pregnancy Hypertension. 2019;17:64–68. doi: 10.1016/j.preghy.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 50.Rafało M. ICT Express; 2021. Cross Validation Methods: Analysis Based on Diagnostics of Thyroid Cancer Metastasis. S2405959521000552. [DOI] [Google Scholar]
- 51.Golbraikh A., Tropsha A. Beware of q2. J. Mol. Graph. Model. 2002;20:269–276. doi: 10.1016/S1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- 52.Daina A., Zoete V. A BOILED-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem. 2016;11:1117–1121. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia C.-Y., Li J.-Y., Hao G.-F., Yang G.-F. A drug-likeness toolbox facilitates ADMET study in drug discovery. Drug Discov. Today. 2020;25:248–258. doi: 10.1016/j.drudis.2019.10.014. [DOI] [PubMed] [Google Scholar]
- 54.Adamski A., Kruszka D., Dutkiewicz Z., Kubicki M., Gorczyński A., Patroniak V. Novel family of fused tricyclic [1,4]diazepines: design, synthesis, crystal structures and molecular docking studies. Tetrahedron. 2017;73:3377–3386. doi: 10.1016/j.tet.2017.05.015. [DOI] [Google Scholar]
- 55.Penmatsa A., Wang K.H., Gouaux E. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature. 2013;503:85–90. doi: 10.1038/nature12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belhassan A., Zaki H., Benlyas M., Lakhlifi T., Bouachrine M. Study of novel triazolo-benzodiazepine analogues as antidepressants targeting by molecular docking and ADMET properties prediction. Heliyon. 2019;5 doi: 10.1016/j.heliyon.2019.e02446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zentrum für Bioinformatik: Universität Hamburg - Proteins Plus Server, (n.d.). https://proteins.plus/(accessed August 29, 2022).
- 58.Abdullahi M., Shallangwa G.A., Uzairu A. In silico QSAR and molecular docking simulation of some novel aryl sulfonamide derivatives as inhibitors of H5N1 influenza A virus subtype. Beni-Suef Univ J Basic Appl Sci. 2020;9:2. doi: 10.1186/s43088-019-0023-y. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.