Abstract
To study phage-mediated gene transfer in Yersinia, the ability of Yersinia phages to transduce naturally occurring plasmids was investigated. The transduction experiments were performed with a temperate phage isolated from a pathogenic Yersinia enterocolitica strain and phage mixtures isolated from sewage. Small plasmids (4.3 and 5.8 kb) were transduced at a frequency of 10−5 to 10−7/PFU. However, we could not detect the transduction of any indigenous virulence plasmid (ca. 72 kb) in pathogenic Yersinia strains. Transductants obtained by infection with the temperate phage were lysogenic and harbored the phage genome in their chromosomes.
The genus Yersinia contains 11 species, of which Yersinia enterocolitica, Y. pseudotuberculosis, and Y. pestis are known to be pathogenic for humans. The species Y. enterocolitica consists of enteropathogenic and nonpathogenic strains, the latter belonging mostly to biogroup 1A (11, 12). Biogroup 1A strains of Y. enterocolitica are ubiquitous and are most frequently isolated from the environment, e.g., water, soil, and plant surfaces (26, 38); from food (8, 18); or from pigs, which are an important reservoir for food-borne infections of humans with pathogenic Y. enterocolitica O:3 and O:9 strains (19, 20). All pathogenic strains of Y. enterocolitica as well as the pathogenic species Y. pestis and Y. pseudotuberculosis possess a conserved 70-kb plasmid carrying essential virulence genes, e.g., for Yersinia outer proteins (7). Plasmids similar in size to the virulence plasmid are also present in biogroup 1A strains of Y. enterocolitica (31, 39, 41). Only scant information regarding the properties of these plasmids is available. Hybridization studies revealed, in some cases, partial homologies to the virulence plasmid (25). However, the well-known plasmid-encoded virulence genes were not detected on the plasmids of biogroup 1A strains (17). Nevertheless, the occurrence of pathogenic strains and biogroup 1A strains in the same host, the isolation of biogroup 1A strains from clinical samples, and the detected plasmid homologies suggest the possibility of horizontal gene transfer between these strains.
Gene exchange by transduction with temperate phages might be widespread in Y. enterocolitica, as 70 to 85% of isolates of this species, on average, have been reported to be lysogenic (28, 30). In addition, phages lytic for Yersinia strains can be readily isolated from sewage (2, 6, 9). In spite of the narrow host range of many Y. enterocolitica phages, which is useful for the differentiation of Yersinia strains by phage typing (3), some phages have a wide host range and even lyse members of other genera, e.g., Enterobacter, Escherichia, and Klebsiella (37). Moreover, temperate phages of Y. frederiksenii, Y. intermedia, and Y. kristensenii have been reported to have a wider host range than those of Y. enterocolitica and are often able to infect pathogenic strains of Y. enterocolitica (10). However, only 9% of the strains of these nonpathogenic Yersinia species are considered lysogenic (40). Finally, until now, naturally occurring lysogeny has never been reported for Y. pseudotuberculosis and Y. pestis, although infection by phages isolated from the environment has been observed (3).
Apart from host range analysis and some electron microscopic studies (21, 22), only scant information and no molecular data about Yersinia phages are available. In addition, it is unknown what role Yersinia phages might play in gene transfer in natural environments. Therefore, we have isolated a temperate phage from a pathogenic Y. enterocolitica strain and a mixture of Yersinia phages from sewage. Transduction experiments revealed that the temperate phage as well as the Yersinia phages from sewage are able to transduce small plasmids isolated from Y. enterocolitica biogroup 1A strains.
MATERIALS AND METHODS
Bacterial strains.
All Yersinia strains used in this study were obtained from the strain collection of the Institut für Mikrobiologie und Tierseuchen, Freie Universität Berlin, and the strain collection of the Robert Koch-Institut. The nonpathogenic Yersinia strains were originally isolated from environmental sources (freshwater and manure), food, animals (pigs, cattle, chickens, sheep, dogs, and wild animals), and humans. The pathogenic Yersinia strains were mainly isolated from diseased pigs and humans (25). The bacteria were routinely grown in Luria-Bertani (LB) medium at 28°C.
Isolation of bacteriophages.
The temperate bacteriophage PY20 was isolated from biogroup 4 serogroup O:3 strain Y. enterocolitica 29820 by mitomycin C (Sigma Chemical Co., St. Louis, Mo.) induction. Briefly, bacteria were grown in 8 ml of LB medium in the dark. During the early logarithmic growth phase, mitomycin C (1.5 to 2.5 μg ml−1) was added to the culture, and the optical density at 588 nm was measured every hour and additionally after incubation for 24 h. Bacteria were sedimented (5,000 × g, 20 min, 4°C), and the supernatant was passed through a sterile filter (0.2-μm-pore size). The cell lysate was examined for plaque-forming activity by dropping 20 μl of the lysate onto LB soft agar containing growing Yersinia indicator strains. To isolate phages from raw sewage, water samples were obtained from two sewage treatment plants in Brandenburg, Germany. A 90-ml portion of each water sample was centrifuged (5,000 × g, 20 min, 4°C), and the supernatants were passed through 0.45-μm-pore-size filters. Phages were sedimented by ultracentrifugation (112,000 × g, 2 h, 10°C), resuspended in 3 ml of SM buffer (33), and passed through 0.2-μm-pore-size filters. The plaque-forming activity on Yersinia strains was investigated by the spot test described above. Phages were isolated from the areas of lysis on agar plates by removing some agar and resuspending it in 500 μl of SM buffer. Dilutions of these phage cocktails were plated on the respective indicator strains to isolate single plaques. Single phages and phage mixtures were propagated in order to obtain high-titer lysates for DNA extraction and transduction experiments (see below).
Phage propagation, purification, and isolation of phage DNA.
Starting from a single plaque, phages were propagated by the soft-agar overlay method described by Sambrook et al. (33). In order to obtain high-titer lysates, soft agar from 20 agar plates with confluent lysis was harvested. The agar was resuspended in 200 ml of SM buffer by stirring for at least 2 h at room temperature. Bacteria and debris were removed by centrifugation (9,000 × g, 10 min, 4°C) and subsequent filtration (0.2 μm). The phages were concentrated by ultracentrifugation (112,000 × g, 90 min) and purified with cesium chloride step gradients (33). The CsCl was removed by filtration with Centricon 500 filtration units (Amicon, Witten, Germany). The phages were resuspended in 1 ml of SM buffer. In order to isolate phage DNA, phage lysates were mixed with an equal volume of phenol, vortexed for 30 s, incubated for 1 min at room temperature, and vortexed again for 30 s. The subsequent centrifugation and extraction steps with phenol-chloroform and chloroform and precipitation of the phage DNA were performed by standard procedures (33).
Analysis of phage DNA.
Restriction analysis was performed with restriction endonucleases according to the manufacturer’s (MBI Fermentas, St. Leon Roth, Germany) recommendations. DNA fragments were separated on 0.8% agarose gels in TBE buffer (33). Hybridization studies with fluorescein-labelled DNA probes were performed with a Renaissance Kit from NEN Dupont as outlined by the manufacturer.
Electron microscopy.
CsCl-purified phage particles were adsorbed to carbon films and negatively stained with 1% uranyl acetate (pH 4.5) or 2% potassium phosphowolframate (pH 7.2) as described by Steven et al. (36). The samples were examined with a Philips CM 100 electron microscope. The droplet technique described by Lang and Mitani (24) was used for the determination of phage genome size. The 7,250-bp cloning vector m13mp18 (New England BioLabs) was used as an internal standard.
Preparation of plasmid and chromosomal DNAs.
Plasmid DNA of Yersinia strains was isolated by a procedure based on the alkaline lysis method described by Birnboim and Doly (5). Chromosomal DNA was purified from bacterial cells with a MasterPure Genomic DNA Purification Kit (Epicentre Technologies, Madison, Wis.).
Generalized transduction.
The transduction experiments were performed with the following plasmids: p13169neo (virulence plasmid of O:3 strain Y. enterocolitica 13169 [25]) and p29807neo and p49neo (small plasmids originally isolated from Y. enterocolitica biogroup 1A strains) (see Fig. 2). All plasmids contained a neomycin resistance gene inserted by use of the mini-Tn5 transposon derivative pUTKm described by De Lorenzo et al. (13) and Herrero et al. (16). In order to create suitable donor strains for the transduction experiment, both small plasmids were transformed by electroporation into O:3 strain Y. enterocolitica 13169/2, which was cured of its virulence plasmid (15). After infection of the donor strains, high-titer lysates were prepared (see above). Wild-type strain 13169 was used as a recipient for lysates of the donor strains containing the marked small plasmids. The cured derivative 13169/2 was used as a recipient for the lysate of the donor strain containing the marked virulence plasmid. The recipient strains were infected by the isolated phages at a multiplicity of infection of about 1 and plated on LB agar supplemented with 100 μg of neomycin per ml. As controls, the phage lysates as well as the recipient strains were plated on the same medium. Colonies which appeared on the agar plates were investigated by comparison of their plasmid patterns with those of the donor and recipient strains, by DNA hybridization with the respective plasmids and with phage PY20, and by phage induction (see above).
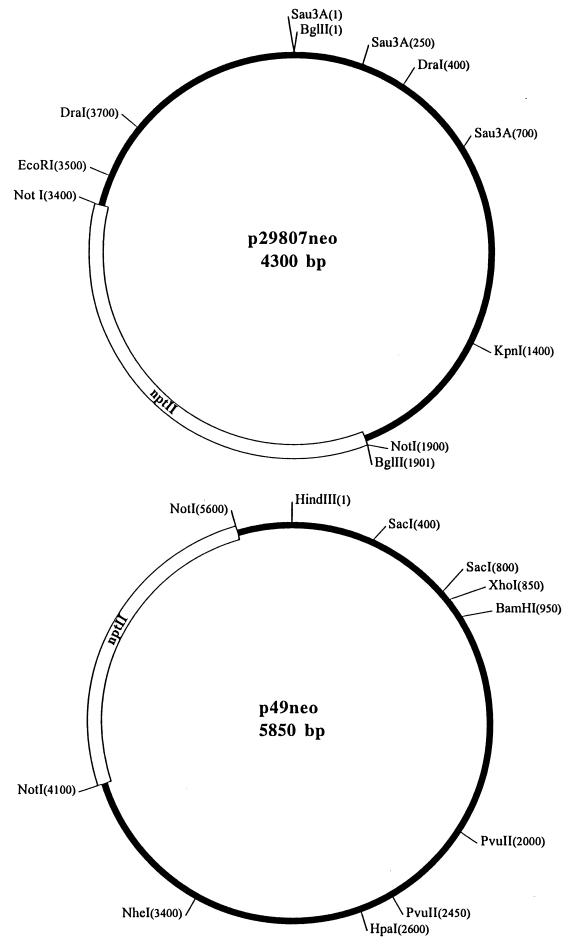
FIG. 2.
Restriction endonuclease cleavage maps of p29807neo and p49neo. nptII, neomycin resistance gene.
RESULTS AND DISCUSSION
Characteristics of the temperate phage PY20.
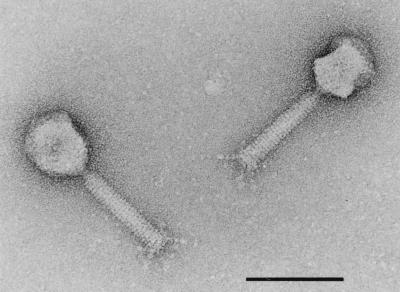
Phage PY20 was induced in a pathogenic Y. enterocolitica strain which had been isolated from a diseased pig. The phage showed typical morphological features of Myoviridae, a phage family exhibiting a long contractile tail with a core (Fig. 1). The morphology of Y. enterocolitica phages belonging to the Myoviridae family has been previously described (21). The average length of phage PY20 particles was 186 nm (n = 4). The average sizes of the elongated heads and tails were 71 by 59 nm and 106 by 19 nm, respectively, in good agreement with the published data (21). The genomes of tailed phages generally consist of double-stranded DNA (1). The genome size of phage PY20 was determined by restriction enzyme analysis and by electron microscopy. After digestion with HpaI, 13 DNA fragments ranging from 0.8 to 7.7 kb were obtained, and a molecular size of approximately 46 kb was estimated. By electron microscopy, a genome size of 51 ± 0.9 kb (n = 8) was determined. Only linear genome structures were detected, indicating that phage PY20 does not possess cohesive ends. This finding was confirmed by treatment of the phage DNA with T4 ligase prior to digestion or heating of the DNA after digestion. No change in the restriction pattern was observed in either case.
FIG. 1.
Electron micrograph of temperate phage PY20 isolated from Y. enterocolitica. Bar, 100 nm.
In order to determine the host range of the phage, 132 Yersinia indicator strains belonging to six species were investigated. Phage PY20 could be exclusively propagated on Y. enterocolitica biogroup 4 serogroup O:3 strains. Of 27 strains belonging to this group, 24 were susceptible, corresponding to a frequency of 88%. Thus, phage PY20 exhibited a narrow host range and might be suitable for identifying Y. enterocolitica serogroup O:3 strains by phage typing. Specific phages for Y. enterocolitica O:3 strains have been described and used to subdivide this predominant pathogenic serogroup in Europe (29).
Characteristics of phages isolated from sewage.
The lytic activity of the phage mixtures on growing Yersinia indicator strains was analyzed by a spot test. Altogether, 62 pathogenic and nonpathogenic Yersinia strains were investigated. The spot test revealed that the phage cocktails were lytic for a wide range of Yersinia strains (Table 1). In most cases, large clear areas of lysis were visible. After resuspension of agar from the areas of lysis and plating of dilutions on the respective indicator strain, many more clear plaques than turbid plaques were obtained. This finding indicated that the sewage samples contained mainly virulent phages. To elucidate if single phages in the sewage samples were able to infect pathogenic and nonpathogenic Yersinia strains, we isolated phages from 14 individual plaques and investigated their host range. Thirteen phages were isolated from plaques on pathogenic Y. enterocolitica strains (O:3 and O:5,27), and 1 phage was isolated from a plaque on a Y. pseudotuberculosis strain. Of the 14 isolated phages, 11 were not active on nonpathogenic Y. enterocolitica biogroup 1A strains. The three remaining phages, which were isolated from plaques on Y. enterocolitica O:5,27 strains, lysed the nonpathogenic biogroup 1A serogroup O:5 strains. One of these phages was also lytic for Y. enterocolitica O:3 strains.
TABLE 1.
Lytic activity of the phage mixtures isolated from raw sewage
| Species | No. of strains tested | No. of strains susceptible to phages in sewage samplea:
|
|
|---|---|---|---|
| 1 | 2 | ||
| Y. enterocolitica | |||
| O:3 | 11 | 9 | 8 |
| O:5,27 | 3 | 3 | 3 |
| O:8 | 1 | — | — |
| O:9 | 4 | 1 | — |
| O:5 biogroup 1A | 13 | 6 | 7 |
| Y. frederiksenii | 10 | 5 | 4 |
| Y. intermedia | 8 | 2 | 3 |
| Y. kristensenii | 7 | 3 | 2 |
| Y. pseudotuberculosis | 5 | — | 1 |
—, no area of lysis detected with any strain tested.
Besides the host range, we compared the HindIII restriction enzyme patterns of the isolated phages. Six of the 14 phages (5 of them isolated from the same sewage sample) were identical in this respect (data not shown). Their genomes were cut by HindIII into nine fragments with an overall molecular size of about 40 kb. Furthermore, since all phages showing this restriction fragment pattern were active on Y. enterocolitica serogroup O:3 strains but not on biogroup 1A serogroup O:5 strains, they might have been identical. This means that one of the investigated sewage samples obviously contained a large number of phages specific for Y. enterocolitica serogroup O:3 strains.
The extended host range of phages lysing serogroup O:5 as well as O:5,27 strains is probably caused by the related O antigens on the bacterial surface, which may be used as phage receptors (23). There are some reports about the host range of Yersinia phages isolated from sewage (2, 9, 37). According to the published data, phages which are active on several serogroups of Y. enterocolitica, several Yersinia species, or even different members of the Enterobacteriaceae are often found in sewage.
Generalized transduction of Yersinia plasmids.
In order to study the capabilities of phage PY20 and of phage mixtures from sewage to transduce Yersinia plasmids, a test system comprising donor and recipient strains as well as marked plasmids was established. In these experiments, we investigated the potential of the phages to transduce the virulence plasmid p13169neo (ca. 72 kb) and two small plasmids isolated from biogroup 1A strains of Y. enterocolitica (p29807neo [4.3 kb] and p49neo [5.8 kb]) (Fig. 2). Until now, we could not detect any phenotypic traits conferred by the small plasmids, although the total DNA sequence of p29807 was determined (accession no. AJ132618; EMBL, Cambridge, United Kingdom).
The transduction experiments revealed that none of the investigated phages was capable of transducing the marked virulence plasmid. With regard to phage PY20, this result could be expected, because the genome of this phage is at least 20 kb smaller than that of the virulence plasmid. Hence, the phage PY20 particles probably were not large enough to encapsidate the whole plasmid. The same might be true for the phages isolated from sewage, as the analysis of HindIII restriction enzyme patterns of selected phages showed that their genomes were smaller than 60 kb. However, it should be noted that certainly only a small portion of the phages in the sewage samples were isolated from single plaques.
It was reported that the transfer of the Yersinia virulence plasmid from Y. pestis to Y. pseudotuberculosis was possible by P1 transduction (42). This finding is not surprising, since phage P1 has a genome of about 100 kb and is well known as a specialized and generalized transducing particle. Moreover, P1 is able to inject its DNA into an extensive range of bacteria (43).
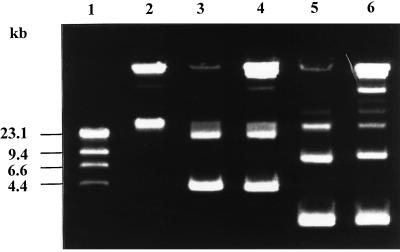
Our experiments showed that in contrast to the virulence plasmid, the smaller Yersinia plasmids could be transduced by either phage PY20 or the phage mixtures isolated from sewage. Phage PY20 transduced both plasmids at a frequency of 10−5 to 10−6 transductants/PFU. The phage mixtures from sewage transduced p29807neo and p49neo at a frequency of 2 × 10−5 and 1 × 10−7 transductants/PFU, respectively. Two days after infection of the recipient strain, neomycin-resistant colonies appeared on agar plates, while no colonies were detected on agar plates on which lysates of only the donor strains or the recipient strain had been incubated. Analysis of the plasmid profiles of the donor strains, the recipient strain, and the transductants (Fig. 3) as well as hybridization studies confirmed that the resistance to neomycin was conferred by acquisition of the marked plasmids.
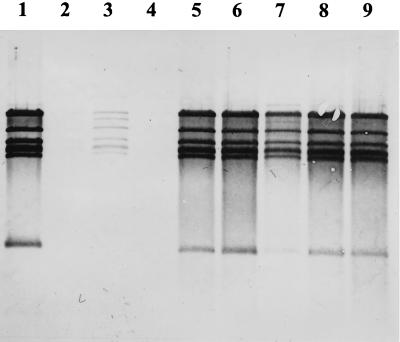
FIG. 3.
Plasmid profiles of the Y. enterocolitica donor strains and recipient strain used for generalized transduction and of two transductants. Lanes: 1, marker DNA (phage lambda DNA digested with HindIII); 2, Y. enterocolitica 13169 (recipient); 3, Y. enterocolitica 13169(p49neo) (donor); 4, transductant containing p49neo; 5, Y. enterocolitica 13169 (p29807neo) (donor); 6, transductant containing p29807neo.
In order to examine if the transductants obtained by infection with the temperate phage PY20 were lysogenic, total DNA was extracted and digested with HindIII. Hybridization experiments with labelled phage DNA clearly showed that the transductants contained the phage PY20 genome (Fig. 4). The hybridization signals obtained with DNA from the transductants were much stronger than those obtained with DNA from Y. enterocolitica 29820, from which phage PY20 originally was isolated. This result indicated that the transductants contained multiple copies of the phage genome. Since generalized transducing particles completely lack DNA originating from the viral vector, coinfection with normal viral particles must have occurred in our experiments. After induction by mitomycin C, transductants released phage PY20 particles which were active on other Y. enterocolitica serogroup O:3 strains. For this reason, transfer of plasmids from the transductants into other strains is possible.
FIG. 4.
Southern hybridization of phage PY20 DNA with total DNA from Yersinia strains. All DNAs were digested with HindIII and separated on an 0.8% agarose gel followed by Southern blotting. Lanes: 1, PY20; 2, empty; 3, Y. enterocolitica 29820 (original host strain of PY20); 4, Y. enterocolitica 13169 (recipient for the transduction experiments); 5 to 9, total DNA of five transductants.
The results reported here indicate that in nature, phage-mediated transfer of small plasmids between Yersinia strains may occur. Our study was initiated by the finding that partial homologies exist between the virulence plasmid of pathogenic Yersinia strains and large plasmids of biogroup 1A strains of Y. enterocolitica (17, 25). For this reason, we investigated if generalized transduction might contribute to transfer of the virulence plasmid. It seems rather unlikely that under natural conditions the whole virulence plasmid, which is known to be nonconjugative, is efficiently transduced into nonpathogenic Yersinia strains. One prerequisite for the transduction is that the phage must be capable of packaging a plasmid of about 70 kb. The phage genomes which we analyzed had molecular sizes of between 40 and 60 kb, a very common size range in phages. Although the virulence plasmid was not transferred in our experiments, smaller plasmids were efficiently exchanged. Based on phage titers of 5 × 106 to 2.5 × 108/ml, as have been found in natural waters (4), and a total phage concentration of 1.6 × 105 to 2.2 × 107 phage particles/ml in sewage (14), a transduction frequency of 10−5 to 10−7/PFU achieved under laboratory conditions might have significance for the transfer of plasmids between Yersinia strains. Generalized transduction of plasmids has already been reported from gram-negative and gram-positive bacterial species (27, 32, 34, 35). It can be assumed that the importance of this mechanism for the spread of nonconjugative plasmids in bacterial populations is greater than previously thought.
ACKNOWLEDGMENT
We thank Eckhard Strauch for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Ackerman H W, DuBow M S, Jarvis A W, Jones L A, Krylov V N, Maniloff J, Rocourt J, Safferman R S, Schneider J, Seldin L, Sozzi T, Stewart P R, Wequin M, Wünsche L. The species concept and its application to tailed phages. Arch Virol. 1992;124:69–82. doi: 10.1007/BF01314626. [DOI] [PubMed] [Google Scholar]
- 2.Baker P M, Farmer J J., III New bacteriophage typing system for Yersinia enterocolitica, Y. kristensenii, Y. frederiksenii, and Y. intermedia: correlation with serotyping, biotyping, and antibiotic susceptibility. J Clin Microbiol. 1982;15:491–502. doi: 10.1128/jcm.15.3.491-502.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergan T, Norris J R. Bacteriophage typing of Yersinia enterocolitica. Methods Microbiol. 1978;12:25–36. [Google Scholar]
- 4.Bergh O, Borsheim K Y, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1522. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bitton G. Fate of bacteriophages in water and wastewater treatment plants. In: Goyal S M, Gerba C P, Bitton G, editors. Phage ecology. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 181–195. [Google Scholar]
- 7.Brubaker B. Factors promoting acute and chronic diseases caused by yersiniae. Clin Microbiol Rev. 1991;4:309–324. doi: 10.1128/cmr.4.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butt H L, Gordon D L, Lee-Archer T, Moritz A, Merrell W H. Relationship between clinical and milk isolates of Yersinia enterocolitica. Pathology. 1991;23:153–157. doi: 10.3109/00313029109060816. [DOI] [PubMed] [Google Scholar]
- 9.Calvo C, Brault J, Alonso J M, Mollaret H H. New waterborne bacteriophages active on Yersinia enterocolitica. Appl Environ Microbiol. 1981;42:35–38. doi: 10.1128/aem.42.1.35-38.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvo C, Brault J. Lysogénie chez Yersinia frederiksenii, Y. kristensenii, et Y. intermedia. Ann Microbiol (Inst Pasteur) 1983;134A:183–188. [PubMed] [Google Scholar]
- 11.Carniel E, Guielvot I, Prentice M. Characterization of large chromosomal high-pathogenicity island in biotype 1B Yersinia enterocolitica. J Bacteriol. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelis G, Laroche Y, Balligand G, Sory M P, Wauters G. Yersinia enterocolitica, a primary model for bacterial invasiveness. Rev Infect Dis. 1987;9:64–87. doi: 10.1093/clinids/9.1.64. [DOI] [PubMed] [Google Scholar]
- 13.De Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewert D L, Paynter M J B. Enumeration of bacteriophages and host bacteria in sewage and the activated sludge treatment process. Appl Environ Microbiol. 1980;39:576–583. doi: 10.1128/aem.39.3.576-583.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heesemann J, Laufs R. Construction of a mobilizable Yersinia enterocolitica virulence plasmid. J Bacteriol. 1983;155:761–767. doi: 10.1128/jb.155.2.761-767.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrero M, De Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann B, Strauch E, Gewinner C, Nattermann H, Appel B. Characterization of plasmid regions of foodborne Yersinia enterocolitica biogroup 1A strains hybridizing to the Yersinia enterocolitica virulence plasmid. Syst Appl Microbiol. 1998;21:201–211. doi: 10.1016/S0723-2020(98)80024-6. [DOI] [PubMed] [Google Scholar]
- 18.Hudson J A, Mott S J, Delacy K M, Edridge A L. Incidence and coincidence of Listeria spp., motile aeromonas and Yersinia enterocolitica on ready-to-eat fleshfoods. Int J Food Microbiol. 1992;16:99–108. doi: 10.1016/0168-1605(92)90002-k. [DOI] [PubMed] [Google Scholar]
- 19.Kapperud G, Nesbakken T, Aleksic S, Mollaret H H. Comparison of restriction endonuclease analysis and phenotypic methods for differentiation of Yersinia enterocolitica isolates. J Clin Microbiol. 1990;28:1125–1131. doi: 10.1128/jcm.28.6.1125-1131.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapperud G. Yersinia enterocolitica in food hygiene. Int J Food Microbiol. 1991;12:53–66. doi: 10.1016/0168-1605(91)90047-s. [DOI] [PubMed] [Google Scholar]
- 21.Kasatiya S, Ackermann H W. Morphology of Yersinia enterocolitica phages. Ann Inst Pasteur Virol. 1986;134E:59–69. [Google Scholar]
- 22.Kawaoka Y, Otsuki K, Tsubokura M. Characteristics of Yersinia enterocolitica bacteriophages. Zentbl Bakteriol Hyg I Abt Orig. 1982;A253:102–109. [PubMed] [Google Scholar]
- 23.Kawaoka Y, Otsuki K, Tsubokura M. Growth temperature-dependent variation in the bacteriophage-inactivating capacity and antigenicity of Yersinia enterocolitica lipopolysaccharide. J Gen Microbiol. 1983;129:2739–2747. doi: 10.1099/00221287-129-9-2739. [DOI] [PubMed] [Google Scholar]
- 24.Lang D, Mitani M. Simplified quantitative electron microscopy of biopolymers. Biopolymers. 1970;9:373–379. doi: 10.1002/bip.1970.360090310. [DOI] [PubMed] [Google Scholar]
- 25.Lewin A, Strauch E, Hertwig S, Hoffmann B, Nattermann H, Appel B. Comparison of plasmids of strains of biogroup 1A with the virulence plasmid of a pathogenic Y. enterocolitica strain. Zentbl Bakteriol. 1996;285:52–63. doi: 10.1016/s0934-8840(96)80022-3. [DOI] [PubMed] [Google Scholar]
- 26.Marranzano M, Agodi A, Gulisano M. Molecular characterization of Yersinia strains isolated from human and environmental sources. Microbiologica. 1993;16:57–62. [PubMed] [Google Scholar]
- 27.Masters M. Generalized transduction. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: American Society for Microbiology; 1996. pp. 2421–2448. [Google Scholar]
- 28.Nicolle P, Mollaret H H, Hamon Y, Vieu J F. Etude lysogénique, bacteriogénique, et lysotypique de l’espéce Yersinia enterocolitica. Ann Inst Pasteur (Paris) 1967;112:86–92. [PubMed] [Google Scholar]
- 29.Nilehn B. Host range, temperature characteristics and serologic relationships among Yersinia phages. Microbiol Immunol. 1973;2:59–67. [Google Scholar]
- 30.Nilehn B, Ericson C. Studies on Yersinia enterocolitica bacteriophages liberated from chloroform treated cultures. Acta Pathol Microbiol Scand. 1969;75:177–187. [PubMed] [Google Scholar]
- 31.Prpic J K, Robins-Browne R M, Davey R B. In vitro assessment of virulence in Yersinia enterocolitica and related species. J Clin Microbiol. 1985;22:105–110. doi: 10.1128/jcm.22.1.105-110.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruhfel R E, Robillard N J, Thorne C B. Interspecies transduction of plasmids among Bacillus anthracis, B. cereus, and B. thuringiensis. J Bacteriol. 1984;157:708–711. doi: 10.1128/jb.157.3.708-711.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Saye D J, Ogunseitan O, Sayler G S, Miller R V. Potential for transduction of plasmids in a natural freshwater environment: effect of plasmid donor concentration and a natural microbial community on transduction in Pseudomonas aeruginosa. Appl Environ Microbiol. 1987;53:987–995. doi: 10.1128/aem.53.5.987-995.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schicklmaier P, Schmieger H. Frequency of generalized transducing phages in natural isolates of the Salmonella typhimurium complex. Appl Environ Microbiol. 1995;61:1637–1640. doi: 10.1128/aem.61.4.1637-1640.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steven A C, Trus B L, Maizel J V, Unser M, Parry D A D, Wall J S, Hainfeld J F, Studier F W. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988;200:351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- 37.Stevenson R M W, Airdrie D W. Isolation of Yersinia ruckeri bacteriophages. Appl Environ Microbiol. 1984;47:1201–1205. doi: 10.1128/aem.47.6.1201-1205.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki A, Hayashidani H, Kaneko K-I, Ogawa M. Isolation of Yersinia from wild animals living in suburbs of Tokyo and Yokohama. Contrib Microbiol Immunol. 1995;13:43–45. [PubMed] [Google Scholar]
- 39.Tanaka M, Nakamura M, Kato Y, Ohmae K, Nogawa H, Ogawa M. Plasmids and lack of pathogenicity of Yersinia enterocolitica isolated from wild-living birds, Japanese serows and environmental specimens. Jpn J Vet Sci. 1987;49:511–513. doi: 10.1292/jvms1939.49.511. [DOI] [PubMed] [Google Scholar]
- 40.Tsubokura M, Otsuki K, Kawaoka Y. Lysogenicity and phage typing of Yersinia enterocolitica isolated in Japan. Jpn J Vet Sci. 1982;44:433–438. doi: 10.1292/jvms1939.44.433. [DOI] [PubMed] [Google Scholar]
- 41.Wachsmuth K, Bradford A K, Birkness K A. Diagnostic value of plasmid analyses and assays for virulence in Yersinia enterocolitica. Diagn Microbiol Infect Dis. 1984;2:219–228. doi: 10.1016/0732-8893(84)90034-8. [DOI] [PubMed] [Google Scholar]
- 42.Wolf-Watz H, Portnoy D A, Bölin I, Falkow S. Transfer of the virulence plasmid of Yersinia pestis to Yersinia pseudotuberculosis. Infect Immun. 1985;48:241–243. doi: 10.1128/iai.48.1.241-243.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yarmolinsky M B, Sternberg N. Bacteriophage P1. In: Calendar R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Press; 1988. pp. 291–438. [Google Scholar]