Abstract
Climate fluctuation mediated abiotic stress consequences loss in crop yields. These stresses have a negative impact on plant growth and development by causing physiological and molecular changes. In this review, we have attempted to outline recent studies (5 years) associated with abiotic stress resistance in plants. We investigated the various factors that contribute to coping with abiotic challenges, such as transcription factors (TFs), microRNAs (miRNAs), epigenetic changes, chemical priming, transgenic breeding, autophagy, and non-coding RNAs. Stress responsive genes are regulated mostly by TFs, and these can be used to enhance stress resistance in plants. Plants express some miRNA during stress imposition that act on stress-related target genes to help them survive. Epigenetic alterations govern gene expression and facilitate stress tolerance. Chemical priming enhances growth in plants by modulating physiological parameters. Transgenic breeding enables identification of genes involved in precise plant responses during stressful situations. In addition to protein coding genes, non-coding RNAs also influence the growth of the plant by causing alterations at gene expression levels. For achieving sustainable agriculture for a rising world population, it is crucial to develop abiotic-resistant crops with anticipated agronomical traits. To achieve this objective, understanding the diverse mechanisms by which plants protect themselves against abiotic stresses is imperative. This review emphasizes on recent progress and future prospects for abiotic stress tolerance and productivity in plants.
Keywords: Abiotic stress, Defense, Environment, Sustainable agriculture, Tolerance
Introduction
The world population is rising without interruption, and food safety is becoming a principal concern worldwide. With the ever-fluctuating universal environmental climates, plant growth is suppressed which threaten the agricultural productivity plant growth is suppressed by the ever-changing global environmental climates, threatening agricultural productivity. These stresses also subsidize the variance in dispersal of plant species among several earth segments (Gong et al. 2020). Plants are sessile organisms, and they have to confront repetitively varying environmental conditions that are unfavorable or stressful for their growth and development. These stressors can be either biotic or abiotic. Biotic stress is demonstrated by pathogen infection and herbivore attack. Abiotic stresses comprise drought, cold, salinity, heat, heavy metals, ultraviolet (UV) radiation, and flooding (Chang et al. 2020a, b, c). Salinity stress is one of the major abiotic stresses and exhibit detrimental effects on plant growth and yield. It is estimated that soil and irrigation-induced salinity has affected approximately 831 Mha of land worldwide, resulting in crop losses totalling more than US$ 27 billion per year. Salinity stress induces osmotic stress and ion toxicity in plants, thereby affecting the plants’ growth primarily because of a reduction in the water potential (Morton et al. 2019). For the cultivation of plants, drought is one of the most fatal abiotic stresses, and its manifestation has been amplified due to changes in evaporation, transpiration, or precipitation Because of global warming, the consumption of fresh water for agriculture will increase in the future (it is currently 70%). Plant production, growth, and geographical dispersal are also restricted by an imperative environmental factor, i.e., temperature. As there is always a variation in environmental conditions, high temperature (heat stress) becomes a threat to sustainable agriculture due to the repetitive rise in global air temperature (Rai et al. 2020). Plant growth, expansion, and survival are also influenced by constant cold stress through interference/demolition of cellular assembly, physiology, and biochemical metabolism (Zeng et al. 2018). Heavy metals (HMs) stress is one of the thoughtful abiotic stresses due to the burning of fossil fuels, mining, smelting, the practice of fertilizers, pesticides, and industrial releases that negatively affect plant productivity (Dubey et al. 2018). Globally abundant plant species are influenced by soil water logging and submergence, which is one of the lethal abiotic stresses. Although rigorous and/or widespread rainfall over a long period of time is the foremost reason for flooding, but it may also be caused by water bodies overflowing onto land. During the flooding, submerged plant portions have an insufficient supply of oxygen because of the lower dispersion of oxygen in water in contrast to air. Plant’s growth is impacted by an oxygen shortage due to an amendment in the levels of ethylene hormone (Pandey and Gautam 2020). Consequences of flooding stress are submergence, waterlogging, hypoxia, and anoxia. Agricultural productivity and consequently human life are affected by flooding due to the destruction of infrastructure and equipment, food scarcity, damage to cattle and seed stocks, and disease spread that leads to poverty and migrations (Fukao et al. 2019). In general, abiotic stress exposures to plants cause overproduction of reactive oxygen species (ROS), leading to the generation of oxidative stress by undesirably affecting the enzyme activities, carbohydrate biogenesis, DNA, proteins, and biochemical activities. Expression of several genes involved in significant processes (such as growth, development, the cell cycle, abiotic and biotic responses, systemic signalling, and programmed cell death) is also stimulated by ROS (Pandey et al. 2017). Different antioxidant enzymes [superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), glutathione reductase (GR), ascorbate peroxide (APX), etc.] play a substantial role in the protection of plants from abiotic stresses by regulating the generation of ROS. Plants protect themselves from various environmental stresses using a variety of mechanisms (Chang et al. 2020a). There are two key ways in which plants respond to abiotic stresses: either by stress circumvention or stress tolerance. Stress tolerance of plants comprises vigorous reversible modification mediated by alteration in gene expression leading to change in transcriptome, proteomics along with metabolomics, while stress avoidance is a physiologically sedentary stage such as mature seeds (Marothia et al. 2021). As we earlier discussed, plants have to confront divergent environmental circumstances in a habitual way that hampers their growth and development. For achieving sustainable agriculture for a rising world population, it is crucial to develop abiotically resistant crops with anticipated agronomical traits. To achieve this objective, understanding the diverse mechanisms by which plants protect themselves against abiotic stresses is imperative. Therefore, in this review, we have attempted to present an outline of the general defense mechanisms (over the past 5 years) in plants during abiotic stresses (Table 1).
Table 1.
Recent studies associated with abiotic stress tolerance in plants
| Stress | Plant species | Defense mechanisms | References |
|---|---|---|---|
| Salinity | Sorghum bicolor | SbWRKY50 regulate ion homoeostasis | Song et al. (2020) |
| Sorghum bicolor | Ion homeostasis | Song et al. (2020) | |
| Arachis hypogaea | miR160-ARF18-mediated response | Tang et al. (2022) | |
| Malus x domestica | miR156/SPL activate MdWRKY100 expression | Ma et al. (2021) | |
| Gossypium hirsutum | lncRNA354 | Zhang et al. (2021) | |
| Annona muricata | Exogenous H2S | da Silva et al. (2019) | |
| Populus | Escalation of selective autophagy and antioxidant levels | Su et al. (2021) | |
| Capsicum annuum | Supplementation of proline, yeast extract, salicylic acid | Abdelaal et al. (2019) | |
| Pistacia vera | Supplementation of H2O2 | Bagheri et al. (2019) | |
| Oryza sativa | Exogenously applied nitric oxide | Adamu et al. (2018) | |
| Arabidopsis thaliana | Salinity-induced R2R3-MYB transcription factor | Yu et al. (2017) | |
| Oryza sativa | Tissue precise cytosine methylation | Kumar et al. (2017) | |
| Solanum tuberosum | Brassinosteroids decreases oxidative stress | Efimova et al. (2018) | |
| Salvia rosmarinus | Salicylic acid governs antioxidant mechanisms and gene expression | El-Esawi et al. (2017) | |
| Arabidopsis thaliana | Polyamine oxidase 5 loss-of-function mutations | Zarza et al. (2017) | |
| Arabidopsis thaliana | Autophagy facilitates speedy protein turnover | Luo et al. (2017) | |
| Arabidopsis thaliana | Exogenous allantoin | Irani and Todd, (2018) | |
| Arabidopsis thaliana | Autophagy | Luo et al. (2017) | |
| Salt and osmotic | Arabidopsis thaliana | Knockdown mutants ANAC069 achieved by T-DNA insertion | He et al. (2017) |
| Malus domestica | Autophagy-related genes overexpression | Wang et al. (2017) | |
| Cold or chilling | Arabidopsis thaliana | Long non-coding RNA SVALKA | Kindgren et al. (2018) |
| Arabidopsis thaliana | COR gene expression modulation | Chang et al. (2020a, b, c) | |
| Solanum lycopersicum | miR 319 target TCP and MYB genes at transcriptional level | Shi et al. (2018) | |
| High temperature | Fortunella crasifolia | Overexpression of FcWRKY40 | Gong et al. (2020) |
| Arabidopsis thaliana | Autophagy participates in degradation of heat shock proteins | Sedaghatmehr et al. (2019) | |
| Brassica rapa | Long non-coding RNAs | Wang et al. (2019) | |
| Actinidia deliciosa | Advances in antioxidant defense | Arnao and Hernández-Ruiz, (2019) | |
| Betula luminifera | Heat responsive miRNAs | Zhang et al. (2021) | |
| Betula luminifera | Heat responsive miRNA | Pan et al. (2017) | |
| Oryza sativa | Salicylic acid and nitric oxide signalling | Rai et al. (2020) | |
| Solanum tuberosum | CRISPR-Cas9 arbitrated genome editing | Klap et al., (2017) | |
| Drought and cold/heat | Zea Mays | WRKY Transcription Factor ZmWRKY106 | Wang et al. (2018) |
| cassava | Long non-coding RNAs | Li et al. (2017a, b) | |
| Drought | Oryza sativa | Exogenous SNP and H2S affect osmoregulation and antioxidant levels | Habib et al. (2020) |
| Arabidopsis thaliana | Overexpression of COST1 via adjustment of autophagy | Bao et al. (2020) | |
| Oryza sativa | Drought responsive miRNAs | Nadarajah and Kumar, (2019) | |
| Brassica juncea | Nitrogen metabolism involved via exposure of nitric oxide and ABA | Sahay et al. (2021) | |
| Dendrobium huoshanense | miRNAs moderate antioxidant level, auxin signalling | Wang et al. (2021) | |
| Malus x domestica | Positive feedback regulatory loop of miR160-Auxin Response Factor | Shen et al. (2022) | |
| Arabidopsis thaliana | Interaction between miR160 and miR165/166 | Yang et al. (2019a, b) | |
| Pyrus betulaefolia | WRKY transcription factor PbrWRKY53 | Liu et al. (2019) | |
| Oryza sativa | NAC-domain transcription factors | Chung et al. (2018) | |
| Arabidopsis thaliana | FtbHLH3 | Yao et al. (2017) | |
| Arabidopsis thaliana | Upregulation of AtbHLH68 gene consequence in ABA homeostasis | Le et al. (2017) | |
| Lycopersicum esculantum | Drought responsive miRNAs | Liu et al. (2018) | |
| Arabidosis thaliana | miRNA 166 cause leaf rolling and stem xylem growth alteration | Zhang et al. (2018a, b) | |
| Malus x domestica | Dynamic epigenomic differences | Xu et al. (2018) | |
| Oryza rufipogon | Amendment in expression levels of 67 new miRNAs | Zhang et al. (2018a, b) | |
| Populus | Variations in the epigenome and transcriptome | Lafon-Placette et al. (2017) | |
| Medicago sativa | Exogenous application of NO and H2S | Antoniou et al. (2020) | |
| Populus trichocarpa | AREB1 mediated histone acetylation | Li et al. (2019) | |
| Medicago sativa | Nitro-oxidative homeostasis and proline metabolism | Antoniou et al. (2017) | |
| Sorghum bicolor | Cerium oxide nanoparticles supplementation reduces oxidative damage | Djanaguiraman et al. (2018) | |
| Arabidopsis thaliana | Exogenous H2S Ion flux mediated Stomatal closure | Jin et al. (2017) | |
| Zea Mays | Next generation breeding methods | Nepolean et al. (2018) | |
| Zea Mays | ARGOS8 variants generated by CRISPR-Cas9 | Shi et al. (2018) | |
| Malus domestica | Overexpression of ATG18a via activation of autophagy | Sun et al. (2018a, b) | |
| Heavy metal | Oryza sativa | Chromium responsive miRNAs | Dubey et al. (2020) |
| Oryza sativa | Transgenerational memory of gene expression | Cong et al. (2019) | |
| Brassica napus | H2S prevent transport of Cd from root to shoot | Yu et al. (2019a, b) | |
| Oryza sativa | Exogenous SNP | Praveen et al. (2020) | |
| Brassica juncea | Exogenous Silicon | Praveen et al. (2020) | |
| Brassica juncea | Reduction of oxidative damage via application of nitric oxide | Praveen et al. (2019) | |
| Triticum astivum | Salicylic acid and thiamine application | El-Esawi et al. (2017) | |
| Cucurbita pepo | Seed priming with H2S stimulate signal memory | Valivand et al. (2019) | |
| Oryza sativa | H2S improve nitrogen metabolism and reduce chloroplast damage | Rizwan et al. (2019) | |
| Brassica juncea | Exogenous Silicon affect phytochemicals and thiol compunds | Pandey et al. (2017) | |
| Oryza sativa | Exogenous H2S Govern ROS production | Chen et al. (2017a, b, c) | |
| Flooding | Arabidopsis thaliana | Autophagy contributes in hypoxia tolerance | Chen et al. (2017a, b, c) |
| Arabidopsis thaliana | Autophagy contributes in hypoxia regulation | Soto-Burgos et al. (2018) | |
| Glycine max | Exogenous H2S | Andrade et al. (2018a, b) | |
| Glycine max | Lessening in ROS generation | Andrade et al. (2018a, b) | |
| Nutrient | Solanum lycopersicum | H2S affect physiological response and transcriptome | Guo et al. (2018) |
| Lycopersicum esculantum | Antioxidant enzyme activation | Liang et al. (2018) | |
| Arabidopsis thaliana | Vacuolar protein degradation via autophagy | Hirota et al. (2018) | |
| Lactuca sativa | Exogenous 5-aminolevulinic acid | Aksakal et al. (2017) | |
| Multiple stress | Arabidopsis thaliana | Autophagy via overexpression of ATG5 and ATG7 genes | Meena et al. (2017) |
| Glycine max | Histone deacetylase alteration | Yang et al. (2018) | |
| Arabidopsis thaliana | Exogenous H2S involved in stress-responsive gene regulation | Pandey and Gautam, (2020) | |
| Solanum tuberosum | New breeding approach | Hameed et al. (2018) | |
| Elaeis guineensis | Modification in WRKY gene expression | Xiao et al. (2017) | |
| Arabidopsis thaliana | Efficient genome editing using CRISPR–Cpf1 | Tang et al. (2017) | |
| Arabidopsis thaliana | Autophagy establishment via tubulin acetylation | Olenieva et al. (2019) |
Understanding the mechanisms regulating abiotic stress resistance in plants is an important question in today’s environmental biology/plant science research. Lots of groups are working in this area, but we still need to explore more defense mechanisms that enable the plants to have better nutritional properties and yield with the minimum toxic effects of abiotic stress. The main objectives of the present work are to examine the role of different factors that are involved in abiotic stress tolerance in plants and to comprehensively review the available information. Owing to diverse abiotic stresses and global climate deterioration, agricultural production worldwide is suffering serious losses. Breeding stress-resilient crops with higher quality and yield against multiple environmental stresses via conventional breeding and the practice of biotechnological tools is presently the most promising approach. Overall, this review provides a systemic glimpse of breeding method, from conventional to the latest innovation in genetic engineering of stress-tolerant crop varieties. This information could be useful for researchers and rice breeders for the generation of abiotic stress-tolerant crop varieties. Although several researchers have investigated it in the past, several factors influencing abiotic stress resistance are not covered in one article. In most cases, a single factor/tool of genome editing is present in a single research article. In our article, we have attempted to present an outline of the general defense mechanisms in plants during abiotic stresses (over the past 5 years). Deciphering physiological, biological, and molecular mechanisms and mining stress-associated genes that govern plant responses against abiotic stresses is one of the prerequisites for developing stress-resistant crop varieties.
Plants are one of the most successful inhabitants on the planet, and they have undoubtedly developed perceptive ways to deal with abiotic stresses. Although plants have a general defense system that allows them to tolerate various types of stresses, there are still numerous flaws that must be addressed before they can be tested in the field. The battle to recognize the multifaceted machinery is enduring, and current new technology advancements for high-throughput phenotyping/genotyping provide us with an innovative gleam of expectations. This review employs cutting-edge science to identify the gaps between lab-based research and how far it is feasible in field trials. This study will unravel how future studies regarding the roles of several TFs under different stresses and the crosstalk among them will facilitate stress-resilient plants. Though many stress-responsive miRNAs have been identified, many more need to be recognized in specific stress and in particular plant species. Limited knowledge is available about miRNAs and their target genes during stressful circumstances. Epigenetic changes are involved in the governance of gene expression as well as their transmission to the next succussing generation. Therefore, these two facets need to be explored equivalently equally in the future. There is little understanding of epigenetic control mechanisms, such as chromatin modifications and the subsequent regulation of transcription. Because these mechanisms are intricately linked to environmental challenges, they will need to be studied further in the future. It is also necessary to investigate inherited stress memory mechanisms. Prior to priming agent approval for agricultural use, the safety of these agents in terms of endurance in soil and water, as well as their impact on ecosystems, should be broadly estimated. To alleviate drought and temperature stress, the application of priming agents needs to be coordinated for exact forecast and an understanding of climate fluctuation. Although the majority of plant autophagy is founded on yeast and mammals, there are still genes that have to be recognized that are conserved in plants. Other novel autophagy in plants exists and should be investigated. Do the genes of one family depict variation in functions? What is the basic molecular mechanism behind selective autophagy? Experiments should be based on these queries for increasing yield and abiotic stress tolerance. ncRNAs interact with factors for the regulation of gene expression, but this area of research is not well explored in plants. Further study is required to unravel their role in functional analysis and the development of abiotic stress-tolerant transgenics to get higher yields and better-quality crops. This information could serve as guidance for researchers and rice breeders for the generation of stress-resilient crops with better yield and quality.
Protection against abiotic stresses
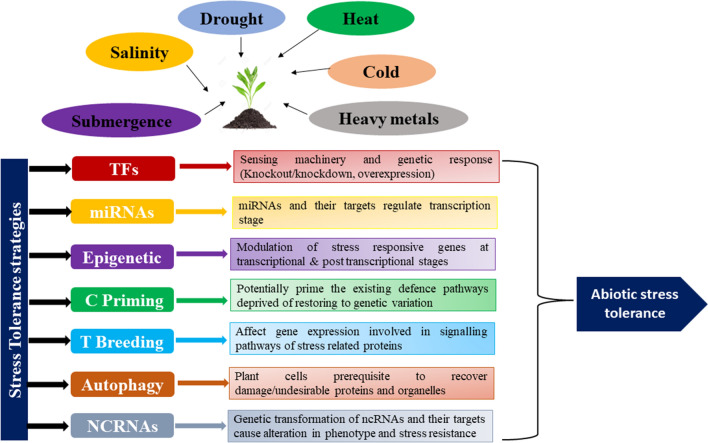
Numerous factors that participate in plant abiotic stress tolerance will be discussed in this section. These factors have a significant role in abiotic stress tolerance in plants and in maintaining the sustainable agriculture, either directly or indirectly (Fig. 1). These factors include transcription factors (such as WRKY, CDF, MYB, bHLH, bZIP), microRNA (miRNA), chemical priming, epigenetic regulation, Ethylene Response Factor (ERF) Family Proteins, autophagy, calcium-dependent protein kinases (CPKs), brassinosteroid, non-coding RNA, helicase, ligase, receptor-like protein kinases (RLKs), NADP, hydrogen sulfide (H2S), etc. To cope with adverse environmental conditions, plants have developed innumerable complex acclimatization approaches at the physiological, biochemical, and cellular levels, such as membrane alterations, antioxidants, cuticular wax, osmolytes synthesis, phytohormones, phenylpropanoid-derived compound production, cell growth and proliferation inhibition, and stomata closure (Khan et al. 2018). Presently, one of the most auspicious strategies to develop a stress-tolerant crop with better yield and quality is transgenic technologies using the stress-associated genes accountable for stress resistance and improved agronomic traits.
Fig. 1.
Diagrammatic illustration of different factors and mode of actions involved in abiotic stress tolerance. Several abiotic stresses, such as drought, cold, salinity, heavy metals, heat, and flooding, severely affect the growth and metabolism of plants. Numerous factors, such as transcription factors (TFs), miRNAs, Epigenetic regulation, chemical (C) priming, transgenic (T) breeding, autophagy, and NC (non-coding) RNAs
Transcription factors (TFs)
Plants combat abiotic stress at the molecular level by linking sensing machinery and genetic response. In general, signal perception, signal transduction, and stress response are the three major stages of the stress signal transduction pathway (Fig. 2). For any abiotic stress’s signalling cascade activation, the first stage is stress signal identification with the help of receptors present on the membrane of the plant cell. Abiotic stress sensors [such as histidine kinases, GPCRs (G protein-coupled receptors), calcium channels, etc.] recognize the signals and transmit them downstream via phytohormones and secondary messengers (ROS, Ca++). These secondary messengers are involved in the activation of a multitude of signalling cascades, such as MAPK (mitogen-activated protein kinase), CIPK (CBL-interacting protein kinase), CBLs (calcineurin-B-like proteins), CDPKs (calcium-dependent protein kinases), PPs such as PP2Cs (protein phosphatase 2Cs), and numerous protein kinases (PKs). A series of phosphorylation/dephosphorylation cascades stimulated primarily by TFs, after information is transferred downstream by PKs and PPs, that eventually end up directly (expression of functional genes participates in cell defenses) or indirectly (expression of regulatory genes contributes to signalling cascades/transcription control of gene expression). Because TFs are involved in the regulation of stress-responsive gene expression via phosphorylation and dephosphorylation, they can be an excellent candidate gene for genetic manipulation of stress resistance traits (Shrestha et al. 2018). Currently, in distinct plant species, plenty of TFs present in divergent families have been recognized depending upon genome-wide analysis, such as WRKY, MYB, bZIP, bHLH, and NAC (Liu et al. 2019). It has been reported that TFs contribute to modulation of abiotic stress tolerance (such as drought, salt, and cold) in model plants and crop plants by knockout/knockdown mutants and overexpression transgenic lines (Khan et al. 2018).
Fig. 2.
Pictorial depiction for regulation of abiotic stress signalling pathways by transcription factors. Signals delivered by abiotic stresses received by receptors located on plant cells. Downriver transmission of these signals facilitated by phytohormones/secondary messengers. Signalling cascades (MAPK, CIPK) are activated by secondary messenger. Phosphorylation or dephosphorylation stimulates the TFs which in turn regulate the stress-responsive genes. By doing the genetic manipulation of stress-responsive genes, abiotic stress resistance can be achieved
WRKY TFs
The most widely recognized TF, WRKY, is found throughout green plants and has a domain (a 60-amino-acid DNA-binding domain) followed by a zinc-finger motif at the C-terminus. WRKY family members have several regulatory mechanisms. In brief, to activate or suppress the transcription of downstream target genes and protein encoded by them, it effectively combined with W-box elements. It also plays a role in increasing transcriptional binding activity by forming protein complexes with other acting elements. WRKY TF self-regulation illuminates the regulatory network of abiotic stress responses (Li et al. 2020b). WRKY proteins play an important role in plant biotic and abiotic responses, because they interact with the W-box (TTGACT/C), a consensus cis-element found in the promoter regions of many abiotic resistance and tolerance genes (Song et al. 2020). The majority of WRKY TFs have a profusion of W-box elements in the self-promoter; self or cross-regulation is accomplished by binding with their own promoters (Jiang et al. 2017). For instance, in Capsicum frutescens, CaWRKY40 is activated by CaWRKY6, enabling the plants to develop better resistance to humidity and high temperatures. GmWRKY27 improved drought and salt stress tolerance in Glycine max by preventing the action of downstream promoter GmNAC29 autonomously as well as collaborating with GmMYB174, resulting in GmNAC29 expression obstruction (Erpen et al. 2017a). In salinity-exposed Arabidopsis, it has been observed that salinity-inducible WRKY71 accelerates flowering to circumvent salt stress and facilitate the prior completion of its life cycle (Yu et al. 2017). Apart from drought, cold, heat, and salinity, WRKY TF is also tangled in other abiotic stresses, such as UV radiation, mechanical damage, and oxidative stress. In transgenic tobacco, overexpression of FcWRKY40 can notably improve tolerance to oxidative stress (Gong et al. 2020). Exposure of Arabidopsis with ROS remarkably upregulated the expression of AtWRKY30, AtWRKY40, AtWRKY75, AtWRKY6, AtWRKY26, and AtWRKY45 (Li et al. 2020c). AtWRKY11, AtWRKY15, AtWRKY22, AtWRKY33, AtWRKY40, AtWRKY53, and AtWRKY6 gene expression levels were upregulated during the mechanical injury (Wu et al. 2017).
MYB TFs
MYB (v-myb avian myeloblastosis viral oncogene homolog) proteins are distinguished via an extremely conserved DNA-binding domain, i.e., the MYB domain, at the N-terminus, and in plants, constitutes the largest TF families (Lloyd et al. 2017). In plants, MYB TF consists of 1–4 DNA-binding repeats. Although the majority of MYB proteins are made up of two repeats from the R2R3-MYB superfamily. The C-terminus is more variable than the N-terminus and plays a regulatory role (Wang et al. 2021a, 2021b). R2R3–MYB proteins have been reported to contribute in to several processes, including plant growth, primary and secondary metabolism, and responses to biotic and abiotic stresses. Among these jobs, MYB proteins have been particularly well delineated as regulators of phenylpropanoid metabolism, like biosynthesis of proanthocyanidins, flavonols, lignin, and anthocyanins in plants. It has also been reported that MYB plays a critical role in abiotic stress resilience in plants. Demerit of MYB is a complicated mechanism for the coregulation of TFs, because it can have opposite functions when it interacts with two different TFs in the same signalling pathway (Deneweth et al. 2022). Transcriptional regulation of abundant physiological and biochemical processes is done by MYB TFs, such as plant growth, secondary metabolism, and cell fate determination (Khan et al. 2018). In several model plants and crop species, MYB proteins contribute in abiotic stress responses (Erpen et al. 2017b; Raza et al. 2019). During cold stress, MYB96 gene can be rapidly persuaded in an Arabidopsis plant and deliver freezing resistance through the interaction of cold and ABA (abscisic acid) signalling pathways (Lv et al. 2017). In Camelina sativa, drought tolerance is endorsed with the overexpression of Arabidopsis MYB96 TF via cuticular wax accretion (Yoon et al. 2020). In salt (NaCl)-exposed Arabidopsis, AtMYB74, which was transcriptionally controlled by the RdDM (RNA-directed DNA methylation) pathway through suppression of 24-nt siRNA buildup, has been intensely increased (Yang and Guo 2018). Enhanced biomass of the plant and reduction in leaf damage are connected with high temperature and drought exposure caused by overexpression of OsMYB55 in Zea mays, reason might be the improvement in expression of stress-related genes (Zandalinas et al. 2018).
bHLH (basic helix–loop–helix) TFs
The bHLH superfamily is named based on its highly conserved alkaline/helix–loop–helix domain. The domain of bHLH consists of around 60 conserved amino acid residues and contains two conserved motifs, particularly a basic region and the helix–loop–helix (HLH region). In the target genes, the basic region contributes to DNA binding to the E-box (usually CANNTG) or G-box (CACGTG) motif. While, the HLH region is made up of two alpha helices that contain hydrophobic residues involved in the dimerization of target genes in signalling cascades (Hao et al. 2021). Members of this superfamily contribute significantly to plant growth, development, light signal transduction, and stress responses (Jang et al. 2022). They also participate in hormone signalling crosstalk, such as salicylic acid (SA), ethylene, jasmonic acid (JA), and brassinosteroid (BR), and are crucial for plant survival. Demerits of the bHLH are due to the complicated crosstalk with other TFs, due to which many aspects have not been discovered (Xue et al. 2022). These TFs disperse globally in eukaryotic organisms, while they create major families in angiosperms (le Hir et al. 2017). bHLH conserved domain comprises two discrete functional regions: the N-terminal region, where DNA will attach, and the C-terminal end, homo or heterodimeric complexes form via protein–protein interactions in the helix–loop–helix region (Li et al. 2020a). The bHLH TFs are involved in numerous biological processes, as confirmed by their functional analysis, like plant growth and development (Yang et al. 2017), photosynthesis (Sun et al. 2018b), flowering (Yao et al. 2018), and the biosynthesis of flavonoids (Niu et al. 2017). bHLH TFs also contribute significantly to abiotic stress resistance in plants. Exogenous ABA application to Arabidopsis plants alters gene expression (AtbHLH68) in organ-specific ways. Further, drought stress resistance was established in transgenic plants due to the upregulation of this gene through ABA homeostasis (le Hir et al. 2017). Salt stress is conferred by OsbHLH068 and its homologous gene AtbHLH112, as testified by their functional characterization, although both genes have contrasting actions in flower phenotype regulation. They provide resistance through root length elongation and lesser H2O2 accretion (Chen et al. 2017a). Upregulation of FtbHLH3 in Arabidopsis contributed to a reduction in drought stress compared to the wild type through the lessening of MDA and stimulation of antioxidant enzymes (Yao et al. 2017).
MicroRNAs (miRNAs)
A class of single-stranded RNA molecules of approximately 21–24 nucleotide in length generated by Dicer enzymes and working on pioneers with intramolecular stem-loop structures are known as miRNA. Mature miRNAs have a phosphate group at the 5' end and a hydroxyl group at the 3' end, and by requiring precise spots in the target mRNA's 3' UTR region, they enable post-transcriptional silencing followed by prompting deprivation or/and translational inhibition (Sun et al. 2019). They have emerged as crucial aspects of transcriptional regulation and complicated numerous processes, including plant growth and biotic or abiotic stresses. The biosynthesis of miRNA is a multistep procedure that takes place mainly in the nucleus. In concise terms, miRNA genes are transcribed into primary miRNA (pri-miRNA) by the enzyme RNA polymerase II. This pri-miRNA is processed to precursor miRNA (pre-miRNA), of around 70–100 nucleotide, with the help of DICER- like (DCL) proteins, and further processed to form miRNA and miRNA duplexes. One strand of the duplex encumbered the RISC (RNA-induced silencing complex), entailing AGO1 (ARGONAUTE) proteins. The RISC complex containing the miRNA recognizes normally cleaved target transcripts; however, central mismatches in the miRNA:miRNA pair predispose to translation inhibition (Ma et al. 2022). Our understanding of miRNAs and their probable targets for plant’s abiotic stresses has improved due to high-throughput genomic techniques and other genomic technologies (Bej and Basak 2014). Arabidopsis thaliana was the first plant in which abiotic stress-regulated miRNAs were perceived (Sunkar et al. 2006). Plant growth and development are undesirably affected by incessant cold stress by obstructing or abolishing cell structure, physiology, and biochemical metabolism (Saleem et al. 2020). In transgenic Agrostis stolonifera (Bent grass), upregulation of osa-miR393a enhances thermotolerance by repressing its targets AsAFB2 and AsTIR1 (Zhao et al. 2019). In S. lycopersicum, overexpression of miR319 (which has a high level of sequence similarity with miR159) can deliver tolerance to heat stress by targeting the TCP and MYB genes and can govern heat-responsive genes at the transcriptional level (Shi et al. 2018). During salinity exposure in peanuts, the expression of miR160 and its ARF target gene is stimulated, while overexpression of miRNA facilitates the ARF 18 pathway, which is intricately involved in protection against generated ROS (Tang et al. 2022). Upregulation of miR156a in apples diminishes salt resistance, while its target gene MdSPL13 enhances salt tolerance (Ma et al. 2021). Drought resistance is mediated in plants by numerous miRNAs via antioxidant hunting, ABA arbitrated governance, and auxin signalling (Wang et al. 2021b). For improvement of root development in apple trees, Mdm-miR160 can be transported from the scion to the rootstock, resulting in drought resistance in the rootstock. Furthermore, targets of miR160 combine with MdHYL1 (HYPONASTIC LEAVES 1) and their promoter for activation of miR160 expression, establishing positive feedback regulation (Shen et al. 2022). In chromium-exposed rice seedlings, diverse miRNAs, such as miR156, miR159, miR160, and miR166, contribute substantially to the protection and detoxification of chromium via ATP-binding cassette transporters, the auxin reaction, and metal ion transference (Dubey et al. 2020). Drought resistance is offered by miR165/166-mediated HDZIP regulation in Arabidopsis via ABA signalling. Drought is induced by a combination of miR160 and miR165/166, which activate variance expression of IAA and ABA signalling, both of which are associated with drought resistance (Yang et al. 2019b). Enhanced cold tolerance is induced in rice plants through overexpression of miR1320. AP2/ERF TF OsERF096 regulates cold resilience by suppressing the jasmonic acid-mediated cold signalling pathway (Sun et al. 2022). Similarly, there was an improvement in cold tolerance in rice plants due to overexpression of Osa-miR156, Osa-miR319, and Osa-miR528 (Huo et al. 2021). In several plant species, miRNAs are associated with abiotic stress resilience through modulation of oxidative damage and alteration of gene expression (Pagano et al. 2021). It was recently reported that advanced tolerance to water scarcity developed during the early stages of tomato plant development as a result of miR169 overexpression. Transgenic tomatoes have demonstrated the ability to hold more water within the cell and require less soil (Rao et al. 2020). In transgenic Arabidopsis, improved tolerance to drought stress developed due to overexpression of GmNFYA5 via a reduction in water loss from leaves and stomatal apertures. In soyabean plant, enhanced and declining drought resilience were observed through overexpression and suppression of GmNFYA5, respectively, with an empty vector (Ma et al. 2020). In the roots of Arabidopsis, miRNA expression and the phytohormone auxin regulate the plant’s growth and development under cold stress. It has been observed that SRL/IAA14 acts as a transcriptional repressor of auxin signalling contributes significantly to cold tolerance through integrating auxin and miRNAs (Aslam et al. 2020). Tomato plants grown in field conditions frequently express synchronized exposure to drought and heat stress. Through the small RNA deep sequencing method, 335 known miRNAs and 430 potential novel miRNAs were identified. It has been reported that there are some unique miRNAs and their target genes that are involved in combined drought and heat stress resistance (Zhou et al. 2020). In cotton plants, during the various crucial phases of anther development, miRNAs are involved in heat resistance. For instance, at the sporogenous cell proliferation stage, miR2949, miR167, and miR160 play significant roles, while at the meiotic phase, miR156 and miR172, the microspore release period, miR156, and at the pollen maturity stage, miR393 and miR3476 do. In heat-sensitive and heat tolerant cotton varieties, these miRNAs act as chief regulators of the heat stress response (Chen et al. 2020).
Non-coding RNAs
Plant growth, progress, and adjustment to abiotic stresses are governed by the functional genes. The functional genes control plant growth, development, and adaptation to abiotic stresses. In the eukaryotic genome, functional proteins are encoded by solitary cells on about 2% of transcribed RNA throughout the process of gene expression (Rai et al. 2019). However, during the advancement of functional genomics and transcriptome sequencing via RNA-sequencing, various non-coding RNAs (ncRNAs) have been recognized that are associated with numerous growth and stress responses (Jha et al. 2020). Functional proteins are not coded by these ncRNAs, and their examples are regulatory ncRNAs, snRNA in the spliceosome, tRNA, and rRNA (Yu et al. 2019a). In plants, major categories of ncRNAs include miRNAs, siRNAs, and long ncRNAs (length greater than 200 nucleotides) (Li et al. 2017a). miRNAs can generate phased small interfering RNAs (phasiRNAs) by targeting the long ncRNAs (lncRNAs), and in contrast, lncRNAs govern the miRNAs expression or can also act as its origin (Meng et al. 2021). Consideration of the ncRNA-regulated growth and controlling network can be useful in increasing plant resistance to abiotic stresses. Current research indicates that genetic transformation of ncRNAs and their target genes alters phenotypes and stress resistance (Meng et al. 2021; Ma et al. 2022). Understanding the molecular mechanisms and numerous supervisory contributions of ncRNAs will facilitate the generation of transgenic plants with better resistance to abiotic stress. In Chinese cabbage, lncRNA provided heat resistance by acting as an eTM (endogenous target mimic) of bra-miR164a and may also be a miR164a target (Wang et al. 2019). Under cold exposure, cold-repressive lincRNA159, which an miRNA164 target mimics, significantly reduced, which activates the expression of miR164, which targets the NAC (NAM, ATAF1/2, and CUC2) genes for conferring cold tolerance (Li et al. 2017b). In Arabidopsis, lncRNAs called DROUGHT-INDUCED LNCRNA (DRIR) positively control salt and drought stress via ABA synchronization. Plants that increase DRIR function upstream of gene transcription in stress/ABA signalling cascades exhibit salt/drought resistance (Qin et al. 2017). Salt stress response in cotton plants is induced by modulating auxin signalling, and lncRNA354 competes with endogenous RNA of miR160b and normalizes GhARF17/18 genes (Zhang et al. 2021). In cotton plants, another LncRNA (XH123) is linked to cold stress resistance. Their mutants are cold susceptible and depict damage in chloroplasts, enhancing ROS levels (Cao et al. 2021). In Arabidopsis, expression of the cryptic antisense CBF1 lncRNA (asCBF1) was arbitrated by transcription of the lncRNA SVALKA, resulting in downregulation of CBF1 and a reduction in freezing resistance (Kindgren et al. 2018). The origin of plant ncRNA is intronic regions of chromosomal DNA, and it governs growth, development gene expression, and biotic and abiotic stress-responsive genes at various levels, such as transcriptional, post-transcriptional, and epigenetic (Ghorbanzadeh et al. 2022). Understanding ncRNA-guided development and stress regulatory networks can deliver new insights to improve plant resilience to environmental stresses. The ncRNA regulatory pathways are multifaceted interaction networks, and the same ncRNA can act as a significant regulator in both plant growth and a variety of abiotic stresses. Genetic transformation of ncRNAs and their target genes can facilitate altering the phenotypes as well as abiotic stress resistance in plants (Bian et al. 2021).
Epigenetic regulation
Modulation of stress-responsive genes at the transcriptional and post-transcriptional stages is mediated by epigenetic mechanisms (EM) through alteration in chromatin gene status (Luo and He 2020). Stress memory is generated in plants with the help of EM and can be genetically transmitted to the progeny of the stressed plants.
Epigenetic modification can be defined as inheritable alteration in gene expression at the meiotic or mitotic levels that are unaffected by the DNA sequence itself. In plants and other organisms, it includes histone modifications, chromatin remodeling, DNA methylation, and histone variations (R M et al. 2020). They can change chromatin architecture, affecting DNA availability, and gene activity, resulting in regulation of several molecular processes, such as gene transcription, replication, recombination, and repair of DNA (Zhang et al. 2018a). These modifications contribute significantly to plant growth and development, including cell differentiation, reproduction, regeneration, flowering, and senescence. They also regulate plant adaptation to various environmental stresses, for instance drought, high salinity, temperature, and heavy metal stress (Chang et al. 2020b). Most of the epigenetic modifications are reversible under the influence of multiple factors, like environmental stresses, diverse developmental stages, and phytohormone signals (Mladenov et al. 2021). The epigenome can be collectively described as an assembly of biochemical variation on DNA sequences and related proteins involved in genome regulation. These epigenetic changes occur not only under stress conditions but also in other circumstances, and they continue over generations for the creation of memory (Pérez-Pizá et al. 2022). A recent study suggested that mild exposure to stress could activate the stress responses of plants, resulting in a faster and stronger response during stress reoccurrence (Villagómez-Aranda et al. 2022). Consequently, to produce stress-tolerant crops, interpretation of epigenetic codes of plant stress reactions could be an important tool (Friedrich et al. 2019). Biological processes, specifically environmental incentives, are affected by vigorous modification of dissimilar epigenetic marks. It has been informed that during modulation of stress responses (such as drought, cold, and heat) in plants, apart from histone methylation, other histone marks like acetylation, ubiquitination, and phosphorylation also contribute significantly (Zhang et al. 2018b). Generally, in plants, there is an insertion of transposable elements (TEs) into the gene body or genome of stress-related genes, and these intragenic epigenetic alterations modulate the gene expression of stress-responsive genes via influencing the alternative splicing of the transcripts (Chang et al. 2020c). After the removal of the inducement causing the stress, epigenetic modification habitually arrives at the prestress stage. Although some reports indicate that this modification may persist even after stress removal and serve as stress memory for plants, assisting them in their adjustment and even evolution over a longer period of time (Ramakrishnan et al. 2021). A variety of epigenetic modifications such as histone modifications, DNA methylation, long non-coding RNAs, small RNAs, histone variants, and other undefined epigenetic alterations are used to protect plants from high temperatures (Pan et al. 2017). In plants, heat stress resistance is also influenced by heat shock proteins (HSPs). After heat exposure, there will be deposition of H3 lysine 9 acetylation (H3K9Ac) and histone H3 lysine 4 trimethylation (H3K4me3) by HSPs like HSP70, HSP22.0, APX2, and HSP18. Heat stress tolerance is also assisted by DNA methylation and histone variation through the RdDM (RNA-directed DNA methylation) pathway (Yang and Guo 2018). During the imposition of cold and salt stress in Arabidopsis, the alleged CHD3-type chromatin remodeling factor (PICKLE) plays a significant role in plant growth and development and provides tolerance to these stresses (Yang et al. 2019a). In ever-changing environmental conditions, swapping from repressed to active status in chromatin regulation contributes significantly to the survival of plants. In Arabidopsis plants, during exposure to cold stress, HOS15-mediated chromatin changes are stimulated by HD2C degradation. As a result, chromatin structure status is switched, assisting CBF enrollment in promoter regions of the COR gene, and providing cold resistance. ABA is overexpressed during stress in the plant system. REPRESSOR OF SILENCING 1 (ROS1)-induced DNA demethylation is required at the promoter for transcriptional activation of NICOTINAMIDASE 3 (NIC3) which is responsive to ABA. It demonstrates that ROS1-induced active DNA demethylation is required for the maintenance of the active state of NIC3 transcription responsive to ABA and confers stress tolerance (Kim et al. 2019). Cytosine methylation was observed in the genomes of drought tolerant and sensitive varieties of apple. Drought stress was linked to changes in methylation at the assembly of genes such as TFs and TEs, as well as DNA methylation modification (Xu et al. 2018). Previous research found that heavy metal-transporting P-type ATPases (HMAs) gene expression changes, as well as numerous low copy cellular genes, showed transgenerational inheritance of the altered expression conditions in heavy metal-treated rice. These heritable changes facilitate heavy metal resistance (Cong et al. 2019). One of the prominent mechanisms during plant cold stress tolerance is the C-repeat binding factor (CBF)-cold responsive gene (COR) pathway, in which TFs such as CBF gene expression are prompted by the cold response and COR gene expression is activated by the binding of these TFs to promoter downstream sites (Chang et al. 2020a, b, c). HEAT SHOCK TRANSCRIPTION FACTOR A1s (HSFA1) are the main transcription factors that contribute to heat stress resistance, and their multistage regulation governs the density of heat stress. Phosphorylation/dephosphorylation, SUMOylation, and protein–protein interactions are the factors that moderately regulate HSFA1s. It is estimated that expression of heat-mediated TFs, which are responsible for activation of heat-responsive genes, is directly controlled by HSFA1s (Ohama et al. 2017). High salinity causes ion toxicity in plants, and the Ca2+Calcineurin B-Like Protein (CBL)-CBL Interacting Protein Kinase (CIPK) component maintains cellular ion homeostasis (Yang and Guo 2018). For salinity stress tolerance in plants, apart from DNA methylation, another definite histone modification such as acetylation by HATs and HDACs is also imperative in fluctuating environmental situations (Kumar et al. 2017). On salt stress-receptive genes, there is a decline in the accumulation of repressive marks such as H3K9me2 and H3K27me3, and high salinity exposure frequently tempts the deposition of active histone marks such as H3K9K14Ac and H3K4me3 (Meena et al. 2017). ABRE sequences are found in the promoter region of drought-responsive NAC TFs, which AREB1 fixes, resulting in the recruitment of the ADA2bGCN5 histone acetylation unit, which activates PtrNAC genes and confers drought tolerance (Li et al. 2019).
Chemical priming
Although several approaches (such as traditional breeding) have been established to improve plant stress tolerance, advance plant productivity, and improve plant survival, these practices have limitations and have resulted in insufficient plant generation with upgraded desire traits in field environments. Plants can also battle environmental stresses by being primed instead of using plant engineering strategies or other tactics. A priming agent caused the activation of stimuli, resulting in direct alteration in the plant at the morphology, physiology, transcriptome, metabolome, and epigenetic levels, resulting in defense mechanism enhancement. Plant pre-exposure with priming agents induces mild stress symptoms (Hasanuzzaman et al. 2020a). Base resilience is provided by priming just after the initial exposure of priming agents. As a result, it is a type of immunological memory that allows plants to remember stressful situations. Therefore, primed plants are better equipped to tolerate abiotic stress as compared to non-primed plants, which will pledge their effect a little later, resulting in fatal costs. Acquired memory is induced in plants through various mechanisms, and one of them is epigenetic modification, which modulates gene expression for a longer period (Pascale et al. 2020). One of the priming agents is sodium nitroprusside, which acts as an NO donor. The key mechanism for the relocation of NO bioactivity is supposed to be the build up of NO portion on the thiol group of cysteine to form S-nitrosothiol (SNO) by the process of S-nitrosylation. This post-translational alteration regulates several actions, such as growth, development, and environmental interfaces. The enzyme S-nitrosoglutathione reductase (GSNOR) obliquely regulates the total cellular S-nitrosylation by decreasing S-nitrosoglutathione (GSNO), which acts as the chief NO donor inside the cell. NO-based S-nitrosylation of GSNOR1 at cysteine-10 residues causes a conformational change and the exposure of the AIM (ATG 8-interacting motif) motif. GSNOR1 protein is selectively degraded via autophagy upon binding ATG8 to AIM, leading to an enhanced NO level that endorses seed germination during hypoxia. A few S-nitrosylated proteins play a significant role in defense and stress responses, phytohormone signalling, and the homeostasis of ROS (Praveen 2022). Progress in plant’s abiotic stress tolerance can also be achieved through chemical priming, as they potentially prime the established defense pathways without resorting to genetic variations (Nguyen et al. 2018). Signalling pathways are stimulated after the exposure of plants to chemical priming mediators, leading to the accretion of resistance signals. Transcriptional and post-transcriptional manifestations are also governed by these chemical mediators, resulting in activation of molecular mechanisms caused by regulatory and functional gene expression (Hasanuzzaman et al. 2020b). Genetic engineering can also express these genes, which lead to stress tolerance through the creation of countless metabolites, proteins, and enzymes (Jacob et al. 2017; Ohama et al. 2017). Collaborative or opponent interfaces caused by exchange among the altered signalling molecules have substantial utility in abiotic stress tolerance in plants. Chemical priming on plants can be accomplished in two ways: by exposing plants to chemical agents prior to the onset of stress, or by exposing entire plants to chemicals after the stress attack (Li et al. 2018). It has previously been stated that the use of chemical compounds as priming mediators was initiated after discovering their potential in improving a variety of abiotic stress tolerance in plants (Irani and Todd 2018). Examples of priming agents are reactive oxygen–nitrogen–sulfur species, hormones, and synthetic compounds, and they facilitate reduction of abiotic stresses (Antoniou et al. 2017; Koo 2018). Many endogenous compounds in plants such as Hydrogen peroxide (H2O2), nitric oxide (NO), and hydrogen sulfide (H2S), have governing roles in the abiotic stress battle (Jin et al. 2017; Praveen et al. 2019). A rise in easiness was detected primarily when plants were pre-exposed to these exogenous compounds without harming the plant growth (Praveen et al. 2020; Liang et al. 2018). In soybean, exogenously supplied H2O2 exposure causes augmented waterlogging tolerance in context with ROS (Andrade et al. 2018a, b). Exogenous NO (SNP as donor) supplementation alleviated arsenic toxicity in rice and drought toxicity in Indian mustard (Praveen and Gupta 2018; Sahay et al. 2021). Earlier studies demonstrated the beneficial role of exogenously supplied hydrogen peroxide (H2O2) in abiotic stress resistance in plant systems, such as providing drought tolerance in G. maxima, Triticum, and quinoa (Andrade et al. 2018b; Iqbal et al. 2018; Habib et al. 2020); convincing toxicity reduction of salinity in basil, soursop, and pistachio (Bagheri et al. 2019; da Silva et al. 2019; Gohari et al. 2020); and reduce oxidative damage in tomato (Liu et al. 2018). H2S donors, namely sodium hydrosulfide (NaHS), function in low quantities in many plant species and are thus highly encouraged to facilitate stress acclimatization and thus stress damage recovery (Sako et al. 2021). Priming of seedlings or seeds with NaHS enables abiotic stress adaptation and depicts improved resistance to chilling in wheat, rye, and avocado (Kolupaev et al. 2019; Joshi et al. 2020); surplus nitrate in tomato (Guo et al. 2018); and heavy metal toxicity reduction in rapeseed, zucchini, and rice (Chen et al. 2017c; Yu et al. 2019b; Rizwan et al. 2019; Valivand et al. 2019). During the environmental stress exposure, metabolic pathways significantly changed, and plants produced precise amino acids, such as 5-aminolevulinic acid (ALA), polyamines, and proline for defense against injury. ALA, which is an imperative biochemical initiator for the biosynthesis of chlorophyll, improves photosynthesis in plant systems under stress imposition (Aksakal et al. 2017). It has recently been investigated whether there is a link between ROS and ALA. Exogenous supplementation of ALA encouraged deposition of H2O2 inside roots, resulting in gene expression of the Na+ transporter and compartmentalization of Na+ in the roots, which therefore diminished its accumulation in leaves in strawberry plants (Wu et al. 2019). Studies have confirmed that the gathering of osmo-protectants such as proline is enhanced as a result of phytohormone supplementation administered leading to stress adaptation in plants (El-Esawi et al. 2017; Min et al. 2018; Fu et al. 2019). Priming with other phytohormones like brassinosteroids delivered stress resistance to salt, cold, and heat (Efimova et al. 2018; Fu et al. 2019). Heat stress is acclimatized by cytokinin in ryegrass (Zhang et al. 2019). Improvement in drought and salt resistance in plants was demonstrated by the application of polyamines (Sequera-Mutiozabal et al. 2017). Because of advances in antioxidant defense, exogenously supplied melatonin has the ability to surge heat tolerance in kiwifruit and chill tolerance in cucumber seeds (Liang et al. 2018; Arnao and Hernández-Ruiz 2019).
Transgenic breeding
To satisfy the food mandate of an unceasingly rising population and for crop upgrading, innovative techniques are required apart from traditional breeding. Therefore, the creation of transgenic lines becomes requisite to meet the food demand as well as to reduce the undesirable impact of abiotic stresses on crop production (Pandey et al. 2017). Transgenic breeding techniques and genetic engineering enable outstanding developments in gene manipulation for the introduction of desired traits into transgenic plants. Transgenic breeding (TB) approaches for the generation of abiotic stress resilience in plants are based on the gene expression involved in regulatory and signalling pathways that regulate genes encoding stress-resistant proteins or enzymes for the synthesis of functional metabolites (Anwar and Kim 2020). For the identification of candidate genes, several molecular techniques have been used, such as QTL mapping, genomic selection, and marker-assisted selection. In Arabidopsis, for the generation of drought stress-tolerant transgenic lines, CRISPR-Cas9 has been used. Sucrose non-ferment 1-related kinase 2 protein kinases (subgroup III SnRK2) genes are manipulated, since they act as positive regulators of the ABA signalling pathway (Huang et al. 2018; Nadarajah and Kumar 2019). Rice root diameter increased under field conditions due to OsNAC5 overexpression, resulting in increased grain yield and drought stress resistance (Shim et al. 2018). Upregulation of cold-regulated C-repeat binding factors (CBFs) and the associated downstream genes in Arabidopsis due to overexpression of miR397 significantly enhanced resilience to chilling and freezing stresses (Abla et al. 2019). Overexpression of MdMIPS1 (Myo-inositol-1-phosphate synthase) in transgenic apples improved salt tolerance through modulation of ion balance, osmosis, and antioxidant levels (Hu et al. 2020). Improvement of salinity and drought stress resistance was observed in rice plants through overexpression of OsMYB6. These rice plants also produced higher levels of proline and antioxidants (Tang et al. 2019). Currently, several plant species, such as rice, Arabidopsis, tomato, etc. along with huge and composite genomes consisting of species, have accessible genome sequences (Hameed et al. 2018; Nepolean et al. 2018). Additionally, the arrival of next-generation sequencing techniques furnishes the opportunity to sequence novel crops, and diverse varieties of a similar crop in comparatively short time and at low cost, which can be utilized in molecular breeding for the generation of innovative markers (Djami-Tchatchou et al. 2017). Presently, deteriorating genetic properties of plants and increased time consumption limit the employment of traditional breeding practices for crop upgrading (Gantait and Mondal, 2018). To achieve novel genome edited crops, there is a requirement for a new technique for crop advancement that is effective, rapid, convenient, and precise, such as CRISPR-Cas9. To cope with the different environmental stresses, researchers are involved in finding the candidate genes with the help of the CRISPR-Cas9 method (Tang et al. 2017; Xiao et al. 2017). To modify the expression level of numerous downstream genes or the stimulation of stress signals, major targets are regulatory genes like phosphatase, kinases, and TFs (Zafar et al. 2020). In Arabidopsis, certain TFs, such as NAC, CUC2, NAM-ATAF1/2, and others, negatively control the expression level of abiotic responsive genes. The specific binding of ANAC069 to the promoter region near the core motif sequence C[A/G]CG[T/G] regulates the expression of multiple stress-receptive genes. Among the consequences are increased susceptibility to salt and osmotic stress due to increased proline biosynthesis and decreased ROS foraging potential. Knockdown mutants ANAC069 achieved by T-DNA insertion demonstrated improved resistance to salt and osmotic stress (He et al. 2016). In maize transgenic plants, upregulation of WRKY TFs (ZmWRKY106) results in a decline in ROS, amplification of antioxidant levels, and the development of heat and drought resistance (Wang et al. 2018). Cis-regulatory sequences enable the binding of precise TFs and thus play a significant role in the amendment of gene expression levels and abiotic stress management (Zafar et al. 2020). Heat stress resistance was accomplished in tomato, followed by an upgrade of fruit setting via targeting an S gene, SlAGAMOUS-LIKE 6 (SIAGL6), using CRISPR-Cas9 genome editing (Klap et al. 2017). CRISPR-Cas9 genome editing has been successfully induced and augmented in a variety of crops, including maize and cotton, by targeting multiple genes in a single organism (Char et al. 2017; Gao et al. 2017). In transgenic maize plants, drought resistance was observed and even established in a field trial via specific gene editing of AGROS8 using CRISPR/Cas9 (Shi et al. 2018). With the help of biotechnological tools, researchers have identified several genes, TFs, and miRNAs accountable for salinity resistance in plants. For instance, in higher plants, transference of Na+ and K+ is synchronized by high-affinity potassium transporter (HKT) genes, which are members of the Trk/Ktr (K+ transporter)/HKT transporter family (Jiang et al. 2018). TaHKT2;1 is the opening plant HKT gene and is present in wheat (Kobayashi et al. 2017) and HKT genes must be elaborated in the elimination of Na + from crops' leaves (Ismail and Horie 2017; Assaha et al. 2017). Ion evenness and salinity stress resistance are primarily delimited by the salt overly sensitive (SOS) signalling pathway (Zelm et al. 2020). In rice plants, OsSOS1 transport activity is regulated by OsCIPK24 (CBL-interacting protein kinases) and OsCBL4 to reduce Na + ion accumulation inside the cell and also to reduce salinity stress detrimental properties (Isayenkov and Maathuis 2019). In crops, numerous molecular procedures, such as genomic and marker-assisted selection, QTL mapping, and CRISPR-Cas9, were used to identify desired genes for drought stress resistance (Nadarajah and Kumar 2019). Water use efficiency was significantly higher in transgenic plants containing the ABA independent transcription factor (DREB1A) than in wild-type plants (Yang et al. 2017). Previously, researchers confirmed that transgenic DREB1A groundnut augments root length density in soil contours to simplify water abstraction occurrence (Chung et al. 2018). Low-temperature stress tolerance is mediated by abundant aspirant genes, which are delimited by C-repeat binding factor/dehydration-responsive element binding (CBF/DREB1) TFs (Shi et al. 2018). CBF1/DREB1b and CBF1/DREB1b overexpression lines amplified the chilling stress adaptions by enhancing the gene expression level of COR and the gathering of proline and sugar in innumerable plants. In Arabidopsis, overexpression of these lines stimulated homologous expression of COR genes at damaging temperatures (Hussain et al. 2018). It has been reported that diverse mitogen-activated protein kinase (MAPK) cataracts are involved in metal stress tolerance (Ghori et al. 2019). Exposure of Cd/Cu to Medicago sativa seedlings caused stimulation of several MAPK genes, such as mitotic activity in Arabidopsis primary and secondary root tips changes, as do auxin and cytokinin accretions mediated by Cu exposure (Jalmi et al. 2018). Under Fe stress, AUX and PIN proteins participated in the shielding of lateral roots efficiently, which is depicted by aux1-7 and pin2 mutants having great resistance to Fe stress relative to that of wild-type plants (Ronzan et al. 2018).
Autophagy
The term autophagy derived from two words: auto-self and phagy-eating. It is a conserved degradation mechanism of cellular constituents. Lytic organelles are the main site for autophagy such as the vacuole in plants and yeast, and the lysosome in animals (Michaeli et al. 2016). Plant cells must recover damaged or undesirable proteins and organelles to adapt during a stressful situation. During non-stressful situations, autophagy arises at basal levels (Inoue et al. 2006). While oxidative stress, heat, drought, saline, osmotic, and endoplasmic reticulum (ER) stress, carbon/nitrogen deprivation, a higher sugar level, and senescence are all factors that influence autophagic flux (Janse van Rensburg et al. 2019). It is confirmed by earlier researchers that during nutrient scarcity and organ senescence, autophagy subsidizes the recovery and recirculation of nutrients (Masclaux-Daubresse et al. 2017). Sugars, fatty acids, and amino acids are generated via autophagic reprocessing, which can act as anabolic substrates or energy producers and are further utilized by the organism. As a result, autophagy is a process that allows plants to grow and develop even when nutrients are scarce (Hirota et al. 2018). Researchers have found that autophagy is present in several crop plants, such as rice, wheat, tomato, and maize (Pei et al. 2014; Wada et al. 2015). In plants with a damaged autophagy mechanism (ATG mutant), symptoms, such as yield reduction, early senescence, and hypersensitivity to carbon and nitrogen deprivation, have been observed (Li et al. 2014; Barros et al. 2021). Overexpression of ATG genes enhanced autophagic flux, endorsed higher yield and late senescence (Minina et al. 2018). Proline catabolism increased during the stress state, and it may help with cell retrieval by enhancing autophagic action. Abiotic stress tolerance is also provided in several plant species by increased levels of amino acid derivatives, such as polyamines and glycine-betaine (Chen and Murata 2011; Zarza et al. 2017). Autophagosome establishment entails two ubiquitin-like systems conjugating Atg12 with Atg5, and Atg8 with lipid phosphatidylethanolamine (PE), respectively. In Arabidopsis, upregulation of ATG5 and ATG7 delivers tolerance to necrotrophic pathogens and oxidative stress and promotes growth, and delays senescence, seed oil content, and seed set (Minina et al. 2018). Several salt exposed atg mutants displayed phenotypic changes as well as enhanced quantities of oxidized proteins as compared to the wild-type (WT) plants (Luo et al. 2017). Furthermore, upregulation of ATG3 homologs in Arabidopsis from Malus domestica improved their tolerance to salt and osmotic stress (Wang et al. 2017). Drought stress resistance was amplified in tomato and apple plants via upregulation of ATG18a in apples when compared with WT plants (Sun et al. 2018a). Autophagy also contributes to submergence tolerance. Hypoxia is generated in plants as a consequence of waterlogging and submergence, which limit gas diffusion inside and outside the plant cells. Autophagosome formation was stimulated during Arabidopsis plant submergence, and atg mutants were found to be more sensitive to submergence than WT plants (Soto-Burgos et al. 2018). Flooding is one of the abiotic stresses that creates a hypoxic state inside the root cells, and the Arabidopsis plant consisting of overexpression of KIN10, able to resist the hypoxia exposure via enhancement of autophagy (Chen et al. 2017b). Autophagy plays a significant role in the alleviation of heat stress in Arabidopsis by degrading the heat shock proteins at an advanced stage of the thermos retrieval stage, which results in the gathering of protein aggregates after a second heat exposure (Sedaghatmehr et al. 2019). Salt resistance in the Arabidopsis plant is facilitated by autophagy via speedy protein turnover (Luo et al. 2017). Autophagy progression in plant cells is enabled by microtubules (MTs). The study investigated that when Arabidopsis plant exposed to different abiotic stimuli like osmotic, salt, and UV rays, then stresses compromised through development of autophagy via tubulin acetylation (Olenieva et al. 2019). In transgenic lines of Populus under salt stress condition, overexpression of PagNBR1 confers stress resistance via escalation of the antioxidant system and selective autophagy (Su et al. 2021). Study investigated that a plant-specific COST1 (constitutively stressed 1) gene confers drought resistance and promotes growth in Arabidopsis via straight synchronization of autophagy (Bao et al. 2020).
Conclusions and future prospective
Globally, crop production is undesirably affected by abiotic stresses, and it has been estimated that these things will rise in the future due to global warming, urbanization, and other fluctuations in the environment. Plants are among the most thriving inhabitants on the planet, and they have undoubtedly developed perceptive ways to deal with abiotic stresses. Although plants have a general defense system that allows them to tolerate various types of stresses, there are still numerous flaws that must be addressed before they can be tested in the field. The battle to recognize the multifaceted machinery is enduring, and the current new technology advancements for high-throughput phenotyping and genotyping provide us with an innovative gleam of expectations. TFs act as major regulators of stress-related genes and can be outstanding candidate genes for crop development. Although work has been done regarding the role of TFs in abiotic stress resistance in plants, much remains to be explored about the various plant species. Only a few TF families, such as WRKY, MYB, bZIP, NAC, and ERF/DREB, have had extensive research into their functions in stress challenges. Current investigations confirmed that validation of TFs is done in both model and non-model plants, but these are limited only to the laboratory. Field trials are mandatory to examine candidate TFs and their role in transgenic plants. Most of the TFs are involved in more than one kind of stress in a fluctuating situation. Therefore, future studies will require the study of several TFs and several stresses to illuminate the crosstalk among them. Significant transcriptional regulators are miRNAs, which affect the growth and development of plants. Though many stress-responsive miRNAs have been identified, many more need to be recognized in specific stress and in particular plant species. Limited knowledge is available about miRNAs and their target genes during stressful circumstances. Epigenetic changes are involved in the governance of gene expression as well as their transmission to the next succussing generation. Therefore, these two facets need to be explored equally in the future. There is little understanding of epigenetic control mechanisms, such as chromatin modifications and the subsequent regulation of transcription. Because these mechanisms are intricately linked to environmental challenges, they will need to be studied further in the future. It is also necessary to investigate inherited stress memory mechanisms. Currently, chemical priming agents are mostly applied to plants to mitigate the toxicity of stress factors and boost their defense mechanisms. Still, prior to their approval for use in agriculture, the security of these priming agents in terms of endurance in soil and water; and their effect on ecosystems should be broadly estimated. To alleviate drought and temperature stress, the application of priming agents needs to be coordinated with an exact forecast and an understanding of climate fluctuation. Although the use of priming agents is a feasible strategy for improving stress resistance in plants, it has yet to be studied comprehensively to enhance crop yield. Transgenic breeding facilitates stress resistance as well as speedy potential multiplication to get an improved cultivar. CRISPR/Cas9 genome editing technology has recently been used in several plant species and transformed agriculture, with the goal of ensuring food demand and nutritional safety for an ever-increasing population. Although the majority of plant autophagy is founded on yeast and mammals, there are still genes that have to be recognized that are conserved in plants. Other novel autophagy in plants exists and should be investigated. Do the genes of one family depict variation in functions? What is the basic molecular mechanism behind selective autophagy? These questions should be the basis for research into increasing yield and abiotic stress tolerance. ncRNAs interact with factors for the regulation of gene expression, but this area of research is not well explored in plants. Further study is required to unravel their role in functional analysis and the development of abiotic stress-tolerant transgenics to get higher yields and better-quality crops. In summary, a thorough examination of plant physiological, biochemical, and molecular mechanisms for abiotic stress tolerance will aid in the management of susceptible crop plants for increased agricultural productivity. With the advancement of genomics, approaches such as genetic engineering and genome editing have contributed to new opportunities for crop improvement by targeting desired traits in crop plants.
Acknowledgements
The authors did not receive support from any organization for the submitted work.
Author contributions
All the authors contributed in some way to the manuscript preparation. AP collected data, contributed new models, analyzed data, and prepared the manuscript. This article was designed by SD. Both SD, SS, and VKS analyzed the final drafts.
Funding
The authors did not receive support from any organization for the submitted work.
Availability of data and materials
Not applicable.
Declarations
Conflict of interest
There is no conflict of interest among authors.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Accession number
Not applicable.
Contributor Information
Afsana Praveen, Email: afsana735@gmail.com, Email: afsana.praveen@niu.edu.in.
Shilpy Singh, Email: shilpy.singh@niu.edu.in.
Varun Kumar Sharma, Email: hod.btmb@niu.edu.in.
References
- Abla M, Sun H, Li Z, et al. Identification of miRNAs and their response to cold stress in astragalus membranaceus. Biomolecules. 2019;182(9):182. doi: 10.3390/BIOM9050182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksakal O, Algur OF, Icoglu Aksakal F, Aysin F. Exogenous 5-aminolevulinic acid alleviates the detrimental effects of UV-B stress on lettuce (Lactuca sativa L) seedlings. Acta Physiol Plant. 2017;39:1–10. doi: 10.1007/S11738-017-2347-3. [DOI] [Google Scholar]
- Andrade CA, de Souza KRD, de Santos MO, et al. Hydrogen peroxide promotes the tolerance of soybeans to waterlogging. Sci Hortic. 2018 doi: 10.1016/j.scienta.2017.12.048. [DOI] [Google Scholar]
- Andrade CA, de Souza KRD, de Santos MO, et al. Hydrogen peroxide promotes the tolerance of soybeans to waterlogging. Sci Hortic. 2018;232:40–45. doi: 10.1016/J.SCIENTA.2017.12.048. [DOI] [Google Scholar]
- Antoniou C, Chatzimichail G, Xenofontos R, et al. Melatonin systemically ameliorates drought stress-induced damage in Medicago sativa plants by modulating nitro-oxidative homeostasis and proline metabolism. J Pineal Res. 2017 doi: 10.1111/jpi.12401. [DOI] [PubMed] [Google Scholar]
- Anwar A, Kim JK. Transgenic breeding approaches for improving abiotic stress tolerance: recent progress and future perspectives. Int J Mol Sci. 2020;21:2695. doi: 10.3390/IJMS21082695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnao MB, Hernández-Ruiz J. Melatonin: a new plant hormone and/or a plant master regulator? Trends Plant Sci. 2019;24:38. doi: 10.1016/j.tplants.2018.10.010. [DOI] [PubMed] [Google Scholar]
- Aslam M, Sugita K, Qin Y, Rahman A. Aux/IAA14 regulates microRNA-mediated cold stress response in arabidopsis roots. Int J Mol Sci. 2020;21:8441. doi: 10.3390/IJMS21228441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaha DVM, Ueda A, Saneoka H, et al. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol. 2017;8:509. doi: 10.3389/FPHYS.2017.00509/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri M, Gholami M, Baninasab B. Hydrogen peroxide-induced salt tolerance in relation to antioxidant systems in pistachio seedlings. Sci Hortic. 2019;243:207–213. doi: 10.1016/J.SCIENTA.2018.08.026. [DOI] [Google Scholar]
- Bao Y, Song WM, Wang P, et al. COST1 regulates autophagy to control plant drought tolerance. Proc Natl Acad Sci USA. 2020;117:7482–7493. doi: 10.1073/PNAS.1918539117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros JAS, Magen S, Lapidot-Cohen T, et al. Autophagy is required for lipid homeostasis during dark-induced senescence. Plant Physiol. 2021;185:1542–1558. doi: 10.1093/PLPHYS/KIAA120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bej S, Basak J. MicroRNAs: the potential biomarkers in plant stress response. Am J Plant Sci. 2014 doi: 10.4236/ajps.2014.55089. [DOI] [Google Scholar]
- Bian X, Yu P, Dong L, et al. Regulatory role of non-coding RNA in ginseng rusty root symptom tissue. Sci Rep. 2021;11:1–23. doi: 10.1038/s41598-021-88709-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Zhao T, Wang L, et al. The lincRNA XH123 is involved in cotton cold-stress regulation. Plant Mol Biol. 2021;106:521–531. doi: 10.1007/S11103-021-01169-1. [DOI] [PubMed] [Google Scholar]
- Chang YN, Zhu C, Jiang J, et al. Epigenetic regulation in plant abiotic stress responses. J Integr Plant Biol. 2020 doi: 10.1111/jipb.12901. [DOI] [PubMed] [Google Scholar]
- Chang YN, Zhu C, Jiang J, et al. Epigenetic regulation in plant abiotic stress responses. J Integr Plant Biol. 2020;62:563–580. doi: 10.1111/JIPB.12901. [DOI] [PubMed] [Google Scholar]
- Char SN, Neelakandan AK, Nahampun H, et al. An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol J. 2017;15:257–268. doi: 10.1111/PBI.12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen THH, Murata N. Glycinebetaine protects plants against abiotic stress: mechanisms and biotechnological applications. Plant Cell Environ. 2011;34:1–20. doi: 10.1111/J.1365-3040.2010.02232.X. [DOI] [PubMed] [Google Scholar]
- Chen C, Li Q, Wang Q, et al. Transcriptional profiling provides new insights into the role of nitric oxide in enhancing Ganoderma oregonense resistance to heat stress. Sci Rep. 2017;7:1–14. doi: 10.1038/s41598-017-15340-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Su ZZ, Huang L, et al. The AMP-activated protein kinase kin10 is involved in the regulation of autophagy in arabidopsis. Front Plant Sci. 2017;8:1201. doi: 10.3389/FPLS.2017.01201/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Chen M, Jiang M. Hydrogen sulfide alleviates mercury toxicity by sequestering it in roots or regulating reactive oxygen species productions in rice seedlings. Plant Physiol Biochem. 2017;111:179–192. doi: 10.1016/J.PLAPHY.2016.11.027. [DOI] [PubMed] [Google Scholar]
- Chen J, Pan A, He S, et al. Different MicroRNA families involved in regulating high temperature stress response during cotton (Gossypium hirsutum L) anther development. Int J Mol Sci . 2020;21:1280. doi: 10.3390/IJMS21041280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung PJ, Jung H, do Choi Y, Kim JK. Genome-wide analyses of direct target genes of four rice NAC-domain transcription factors involved in drought tolerance. BMC Genom. 2018;19:1–17. doi: 10.1186/S12864-017-4367-1/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong W, Miao Y, Xu L, et al. Transgenerational memory of gene expression changes induced by heavy metal stress in rice (Oryza sativa L.) BMC Plant Biol. 2019;19:1–14. doi: 10.1186/S12870-019-1887-7/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva AAR, de Lima GS, de Azevedo CAV, et al (2019) Induction of tolerance to salt stress in soursop seedlings using hydrogen peroxide. Comun Sci 10:484–490. 10.14295/CS.V10I4.3036
- Deneweth J, van de Peer Y, Vermeirssen V. Nearby transposable elements impact plant stress gene regulatory networks: a meta-analysis in A. thaliana and S. lycopersicum. BMC Genom. 2022;23:1–19. doi: 10.1186/S12864-021-08215-8/FIGURES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djami-Tchatchou AT, Sanan-Mishra N, Ntushelo K, Dubery IA. Functional roles of microRNAs in agronomically important plants-potential as targets for crop improvement and protection. Front Plant Sci. 2017 doi: 10.3389/fpls.2017.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey S, Shri M, Gupta A, et al. Toxicity and detoxification of heavy metals during plant growth and metabolism. Environ Chem Lett. 2018;16:1169. doi: 10.1007/s10311-018-0741-8. [DOI] [Google Scholar]
- Dubey S, Saxena S, Chauhan AS, et al. Identification and expression analysis of conserved microRNAs during short and prolonged chromium stress in rice (Oryza sativa) Environ Sci Pollut Res. 2020;27:380–390. doi: 10.1007/S11356-019-06760-0/FIGURES/8. [DOI] [PubMed] [Google Scholar]
- Efimova MV, Khripach VA, Boyko EV, et al. The priming of potato plants induced by brassinosteroids reduces oxidative stress and increases salt tolerance. Doklady Biol Sci. 2018;478(1):33–36. doi: 10.1134/S0012496618010106. [DOI] [PubMed] [Google Scholar]
- El-Esawi MA, Elansary HO, El-Shanhorey NA, et al. Salicylic acid-regulated antioxidant mechanisms and gene expression enhance rosemary performance under saline conditions. Front Physiol. 2017;8:716. doi: 10.3389/FPHYS.2017.00716/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erpen L, Devi HS, Grosser JW, Dutt M. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Culture (PCTOC) 2017;132(1):1–25. doi: 10.1007/S11240-017-1320-6. [DOI] [Google Scholar]
- Erpen L, Devi HS, Grosser JW, Dutt M. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Cult (PCTOC) 2017;132(1):1–25. doi: 10.1007/S11240-017-1320-6. [DOI] [Google Scholar]
- Friedrich T, Faivre L, Bäurle I, Schubert D. Chromatin-based mechanisms of temperature memory in plants. Plant Cell Environ. 2019;42:762. doi: 10.1111/pce.13373. [DOI] [PubMed] [Google Scholar]
- Fu J, Sun P, Luo Y, et al. Brassinosteroids enhance cold tolerance in Elymus nutans via mediating redox homeostasis and proline biosynthesis. Environ Exp Bot. 2019;167:103831. doi: 10.1016/J.ENVEXPBOT.2019.103831. [DOI] [Google Scholar]
- Fukao T, Barrera-Figueroa BE, Juntawong P, Peña-Castro JM. Submergence and waterlogging stress in plants: a review highlighting research opportunities and understudied aspects. Front Plant Sci. 2019 doi: 10.3389/fpls.2019.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Long L, Tian X, et al. Genome editing in cotton with the CRISPR/Cas9 system. Front Plant Sci. 2017;8:1364. doi: 10.3389/FPLS.2017.01364/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbanzadeh Z, Hamid R, Jacob F, et al. Non-coding RNA: chief architects of drought-resilient roots. Rhizosphere. 2022;23:100572. doi: 10.1016/J.RHISPH.2022.100572. [DOI] [Google Scholar]
- Ghori NH, Ghori T, Hayat MQ, et al. Heavy metal stress and responses in plants. Int J Environ Sci Technol. 2019;16(3):1807–1828. doi: 10.1007/S13762-019-02215-8. [DOI] [Google Scholar]
- Gohari G, Alavi Z, Esfandiari E, et al. Interaction between hydrogen peroxide and sodium nitroprusside following chemical priming of Ocimum basilicum L. against salt stress. Physiol Plant. 2020;168:361–373. doi: 10.1111/PPL.13020. [DOI] [PubMed] [Google Scholar]
- Gong Z, Xiong L, Shi H, et al. Plant abiotic stress response and nutrient use efficiency. Sci China Life Sci. 2020;63:635. doi: 10.1007/s11427-020-1683-x. [DOI] [PubMed] [Google Scholar]
- Habib N, Ali Q, Ali S, et al. Use of nitric oxide and hydrogen peroxide for better yield of wheat (Oryza sativa L) under water deficit conditions: growth, osmoregulation, and antioxidative defense mechanism. Plants. 2020;9:285. doi: 10.3390/PLANTS9020285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed A, Zaidi SSEA, Shakir S, Mansoor S. Applications of new breeding technologies for potato improvement. Front Plant Sci. 2018 doi: 10.3389/fpls.2018.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Zong X, Ren P, et al. Basic helix–loop–helix (bHLH) transcription factors regulate a wide range of functions in arabidopsis. Int J Mol Sci. 2021;22:7152. doi: 10.3390/IJMS22137152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasanuzzaman M, Bhuyan MHMB, Zulfiqar F, et al. Reactive oxygen species and antioxidant defense in plants under abiotic stress: revisiting the crucial role of a universal defense regulator. Antioxidants. 2020;9:681. doi: 10.3390/ANTIOX9080681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasanuzzaman M, Bhuyan MHMB, Zulfiqar F, et al. Reactive oxygen species and antioxidant defense in plants under abiotic stress: revisiting the crucial role of a universal defense regulator. Antioxidants. 2020;9:681. doi: 10.3390/antiox9080681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Shi X, Wang Y, et al. Arabidopsis ANAC069 binds to C[A/G]CG[T/G] sequences to negatively regulate salt and osmotic stress tolerance. Plant Mol Biol. 2016;93(4):369–387. doi: 10.1007/S11103-016-0567-3. [DOI] [PubMed] [Google Scholar]
- Hirota T, Izumi M, Wada S, et al. Vacuolar protein degradation via autophagy provides substrates to amino acid catabolic pathways as an adaptive response to sugar starvation in Arabidopsis thaliana. Plant Cell Physiol. 2018;59:1363–1376. doi: 10.1093/PCP/PCY005. [DOI] [PubMed] [Google Scholar]
- Hu L, Zhou K, Liu Y, et al. Overexpression of MdMIPS1 enhances salt tolerance by improving osmosis, ion balance, and antioxidant activity in transgenic apple. Plant Sci. 2020;301:110654. doi: 10.1016/J.PLANTSCI.2020.110654. [DOI] [PubMed] [Google Scholar]
- Huang X, Hou L, Meng J, et al. The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in arabidopsis. Mol Plant. 2018;11:970–982. doi: 10.1016/J.MOLP.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Huo C, Zhang B, Wang R. Research progress on plant noncoding RNAs in response to low-temperature stress. Plant Signal Behav. 2021 doi: 10.1080/15592324.2021.2004035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain HA, Hussain S, Khaliq A, et al. Chilling and drought stresses in crop plants: implications, cross talk, and potential management opportunities. Front Plant Sci. 2018;9:393. doi: 10.3389/FPLS.2018.00393/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Suzuki T, Hattori M, et al. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in arabidopsis root tip cells. Plant Cell Physiol. 2006;47:1641–1652. doi: 10.1093/PCP/PCL031. [DOI] [PubMed] [Google Scholar]
- Iqbal H, Yaning C, Waqas M, et al. Hydrogen peroxide application improves quinoa performance by affecting physiological and biochemical mechanisms under water-deficit conditions. J Agron Crop Sci. 2018;204:541–553. doi: 10.1111/JAC.12284. [DOI] [Google Scholar]
- Irani S, Todd CD. Exogenous allantoin increases Arabidopsis seedlings tolerance to NaCl stress and regulates expression of oxidative stress response genes. J Plant Physiol. 2018 doi: 10.1016/j.jplph.2017.11.011. [DOI] [PubMed] [Google Scholar]
- Isayenkov SV, Maathuis FJM. Plant salinity stress: many unanswered questions remain. Front Plant Sci. 2019;10:80. doi: 10.3389/FPLS.2019.00080/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AM, Horie T. Genomics physiology, and molecular breeding approaches for improving salt tolerance. Annu Rev Plant Biol. 2017 doi: 10.1146/annurev-arplant-042916. [DOI] [PubMed] [Google Scholar]
- Jacob P, Hirt H, Bendahmane A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol J. 2017;15:405–414. doi: 10.1111/pbi.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalmi SK, Bhagat PK, Verma D, et al. Traversing the links between heavy metal stress and plant signaling. Front Plant Sci. 2018;9:12. doi: 10.3389/FPLS.2018.00012/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang YH, Park JR, Kim EG, Kim KM. OsbHLHq11, the basic helix–loop–helix transcription factor, involved in regulation of chlorophyll content in rice. Biol (basel) 2022;11:1000. doi: 10.3390/BIOLOGY11071000/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse van Rensburg HC, van den Ende W, Signorelli S. Autophagy in plants: both a puppet and a puppet master of sugars. Front Plant Sci. 2019;10:14. doi: 10.3389/FPLS.2019.00014/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha UC, Nayyar H, Jha R, et al. Long non-coding RNAs: emerging players regulating plant abiotic stress response and adaptation. BMC Plant Biol. 2020;20(1):1–20. doi: 10.1186/S12870-020-02595-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Ma S, Ye N, et al. WRKY transcription factors in plant responses to stresses. J Integr Plant Biol. 2017;59:86–101. doi: 10.1111/JIPB.12513. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Song G, Shan X, et al. Association analysis and identification of ZmHKT1;5 variation with salt-stress tolerance. Front Plant Sci. 2018 doi: 10.3389/fpls.2018.01485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Wang Z, Ma Q, et al. Hydrogen sulfide mediates ion fluxes inducing stomatal closure in response to drought stress in Arabidopsis thaliana. Plant Soil. 2017 doi: 10.1007/s11104-017-3335-5. [DOI] [Google Scholar]
- Joshi NC, Yadav D, Ratner K, et al. Sodium hydrosulfide priming improves the response of photosynthesis to overnight frost and day high light in avocado (Persea americana Mill, cv. ‘Hass’) Physiol Plant. 2020;168:394–405. doi: 10.1111/PPL.13023. [DOI] [PubMed] [Google Scholar]
- Khan SA, Li MZ, Wang SM, Yin HJ. Revisiting the role of plant transcription factors in the battle against abiotic stress. Int J Mol Sci. 2018;19:1634. doi: 10.3390/IJMS19061634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Lim JY, Shin H, et al. ROS1-dependent DNA demethylation is required for ABA-inducible NIC3 expression. Plant Physiol. 2019;179:1810–1821. doi: 10.1104/PP.18.01471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P, Ard R, Ivanov M, Marquardt S. Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation. Nat Commun. 2018;9(1):1–11. doi: 10.1038/s41467-018-07010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klap C, Yeshayahou E, Bolger AM, et al. Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol J. 2017;15:634–647. doi: 10.1111/PBI.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi NI, Yamaji N, Yamamoto H, et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017 doi: 10.1111/tpj.13595. [DOI] [PubMed] [Google Scholar]
- Kolupaev YE, Horielova EI, Yastreb TO, et al. Stress-protective responses of wheat and rye seedlings whose chilling resistance was induced with a donor of hydrogen sulfide. Russ J Plant Physiol. 2019;66:540–547. doi: 10.1134/S1021443719040058. [DOI] [Google Scholar]
- Koo AJ. Metabolism of the plant hormone jasmonate: a sentinel for tissue damage and master regulator of stress response. Phytochem Rev. 2018 doi: 10.1007/s11101-017-9510-8. [DOI] [Google Scholar]
- Kumar S, Beena AS, Awana M, Singh A. Salt-Induced tissue-specific cytosine methylation downregulates expression of HKT genes in contrasting wheat (Oryza sativa L) genotypes. DNA Cell Biol. 2017 doi: 10.1089/dna.2016.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Hir R, Castelain M, Chakraborti D, et al. AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana. Physiol Plant. 2017;160:312–327. doi: 10.1111/PPL.12549. [DOI] [PubMed] [Google Scholar]
- Li S, Castillo-González C, Yu B, Zhang X. The functions of plant small RNAs in development and in stress responses. Plant J. 2017;90:654–670. doi: 10.1111/TPJ.13444. [DOI] [PubMed] [Google Scholar]
- Li S, Yu X, Lei N, et al. Genome-wide identification and functional prediction of cold and/or drought-responsive lncRNAs in cassava. Sci Rep. 2017;7(1):1–17. doi: 10.1038/srep45981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Yu HM, Meng XF, et al. Ectopic expression of glycosyltransferase UGT76E11 increases flavonoid accumulation and enhances abiotic stress tolerance in Arabidopsis. Plant Biol. 2018 doi: 10.1111/plb.12627. [DOI] [PubMed] [Google Scholar]
- Li S, Lin YCJ, Wang P, et al. The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa. Plant Cell. 2019 doi: 10.1105/tpc.18.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yang Z, Zeng Q, et al. Abnormal expression of bHLH3 disrupts a flavonoid homeostasis network, causing differences in pigment composition among mulberry fruits. Hortic Res. 2020 doi: 10.1038/S41438-020-0302-8/41990011/41438_2020_ARTICLE_302.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Pang S, Lu Z, Jin B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants. 2020;9:1515. doi: 10.3390/PLANTS9111515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Pang S, Lu Z, Jin B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants. 2020;9:1515. doi: 10.3390/PLANTS9111515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Zheng P, Li S, et al. Nitrate reductase-dependent NO production is involved in H2S-induced nitrate stress tolerance in tomato via activation of antioxidant enzymes. Sci Hortic. 2018 doi: 10.1016/j.scienta.2017.10.044. [DOI] [Google Scholar]
- Liu T, Hu X, Zhang J, et al. H2O2 mediates ALA-induced glutathione and ascorbate accumulation in the perception and resistance to oxidative stress in Solanum lycopersicum at low temperatures. BMC Plant Biol. 2018;18:1–10. doi: 10.1186/S12870-018-1254-0/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yang T, Lin Z, et al. A WRKY transcription factor PbrWRKY53 from Pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol J. 2019 doi: 10.1111/pbi.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A, Brockman A, Aguirre L, et al. Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017;58:1431–1441. doi: 10.1093/PCP/PCX075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, He Y. Experiencing winter for spring flowering: a molecular epigenetic perspective on vernalization. J Integr Plant Biol. 2020;62:104–117. doi: 10.1111/JIPB.12896. [DOI] [PubMed] [Google Scholar]
- Luo L, Zhang P, Zhu R, et al. Autophagy is rapidly induced by salt stress and is required for salt tolerance in arabidopsis. Front Plant Sci. 2017;8:1459. doi: 10.3389/FPLS.2017.01459/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y, Yang M, Hu D, et al. The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing β-amylase expression. Plant Physiol. 2017;173:1475–1491. doi: 10.1104/PP.16.01725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XJ, Yu TF, Li XH, et al. Overexpression of GmNFYA5 confers drought tolerance to transgenic Arabidopsis and soybean plants. BMC Plant Biol. 2020;20:1–18. doi: 10.1186/S12870-020-02337-Z/FIGURES/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Xue H, Zhang F, et al. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol J. 2021;19:311–323. doi: 10.1111/PBI.13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhao F, Zhou B. The characters of non-coding RNAs and their biological roles in plant development and abiotic stress response. Int J Mol Sci. 2022;23:4124. doi: 10.3390/IJMS23084124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marothia D, Kaur N, Kumar Pati P (2021) Abiotic stress responses in plants: current knowledge and future prospects. In: Abiotic stress in plants
- Masclaux-Daubresse C, Chen Q, Havé M. Regulation of nutrient recycling via autophagy. Curr Opin Plant Biol. 2017;39:8–17. doi: 10.1016/J.PBI.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Meena KK, Sorty AM, Bitla UM, et al. Abiotic stress responses and microbe-mediated mitigation in plants: the omics strategies. Front Plant Sci. 2017 doi: 10.3389/fpls.2017.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Li A, Yu B, Li S. Interplay between miRNAs and lncRNAs: mode of action and biological roles in plant development and stress adaptation. Comput Struct Biotechnol J. 2021;19:2567–2574. doi: 10.1016/J.CSBJ.2021.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S, Galili G, Genschik P, et al. Autophagy in plants—what’s new on the menu? Trends Plant Sci. 2016;21:134–144. doi: 10.1016/J.TPLANTS.2015.10.008. [DOI] [PubMed] [Google Scholar]
- Min K, Showman L, Perera A, Arora R. Salicylic acid-induced freezing tolerance in spinach (Spinacia oleracea L.) leaves explored through metabolite profiling. Environ Exp Bot. 2018;156:214–227. doi: 10.1016/J.ENVEXPBOT.2018.09.011. [DOI] [Google Scholar]
- Minina EA, Moschou PN, Vetukuri RR, et al. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J Exp Bot. 2018;69:1415–1432. doi: 10.1093/JXB/ERY010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenov V, Fotopoulos V, Kaiserli E, et al. Deciphering the epigenetic alphabet involved in transgenerational stress memory in crops. Int J Mol Sci. 2021;22:7118. doi: 10.3390/IJMS22137118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton MJL, Awlia M, Al-Tamimi N, et al. Salt stress under the scalpel—dissecting the genetics of salt tolerance. Plant J. 2019 doi: 10.1111/tpj.14189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah K, Kumar IS. Drought response in rice: the miRNA story. Int J Mol Sci. 2019;20:3766. doi: 10.3390/IJMS20153766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepolean T, Kaul J, Mukri G, Mittal S. Genomics-enabled next-generation breeding approaches for developing system-specific drought tolerant hybrids in maize. Front Plant Sci. 2018 doi: 10.3389/fpls.2018.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HC, Lin KH, Ho SL, et al. Enhancing the abiotic stress tolerance of plants: from chemical treatment to biotechnological approaches. Physiol Plant. 2018;164:452–466. doi: 10.1111/ppl.12812. [DOI] [PubMed] [Google Scholar]
- Niu X, Guan Y, Chen S, Li H. Genome-wide analysis of basic helix–loop–helix (bHLH) transcription factors in Brachypodium distachyon. BMC Genom. 2017;18(1):1–20. doi: 10.1186/S12864-017-4044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017;22:53–65. doi: 10.1016/j.tplants.2016.08.015. [DOI] [PubMed] [Google Scholar]
- Olenieva V, Lytvyn D, Yemets A, et al. Tubulin acetylation accompanies autophagy development induced by different abiotic stimuli in Arabidopsis thaliana. Cell Biol Int. 2019;43:1056–1064. doi: 10.1002/CBIN.10843. [DOI] [PubMed] [Google Scholar]
- Pagano L, Rossi R, Paesano L, et al. miRNA regulation and stress adaptation in plants. Environ Exp Bot. 2021;184:104369. doi: 10.1016/J.ENVEXPBOT.2020.104369. [DOI] [Google Scholar]
- Pan T, Sun X, Liu Y, et al. Heat stress alters genome-wide profiles of circular RNAs in Arabidopsis. Plant Mol Biol. 2017;96(3):217–229. doi: 10.1007/S11103-017-0684-7. [DOI] [PubMed] [Google Scholar]
- Pandey AK, Gautam A. Stress responsive gene regulation in relation to hydrogen sulfide in plants under abiotic stress. Physiol Plant. 2020 doi: 10.1111/ppl.13064. [DOI] [PubMed] [Google Scholar]
- Pandey C, Augustine R, Panthri M, et al. Arsenic affects the production of glucosinolate, thiol and phytochemical compounds: a comparison of two Brassica cultivars. Plant Physiol Biochem. 2017;111:144–154. doi: 10.1016/J.PLAPHY.2016.11.026. [DOI] [PubMed] [Google Scholar]
- Pascale A, Proietti S, Pantelides IS, Stringlis IA. Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion. Front Plant Sci. 2020;10:1741. doi: 10.3389/FPLS.2019.01741/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei D, Zhang W, Sun H, et al. Identification of autophagy-related genes ATG4 and ATG8 from wheat (Oryza sativa L) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep. 2014;33(10):1697–1710. doi: 10.1007/S00299-014-1648-X. [DOI] [PubMed] [Google Scholar]
- Pérez-Pizá MC, Ibañez VN, Varela A, et al. Non-thermal plasmas affect plant growth and DNA methylation patterns in glycine max. J Plant Growth Regul. 2022;41:2732–2742. doi: 10.1007/S00344-021-10470-8/METRICS. [DOI] [Google Scholar]
- Praveen A. Nitric oxide mediated alleviation of abiotic challenges in plants. Nitric Oxide. 2022;128:37–49. doi: 10.1016/J.NIOX.2022.08.005. [DOI] [PubMed] [Google Scholar]
- Praveen A, Gupta M. Nitric oxide confronts arsenic stimulated oxidative stress and root architecture through distinct gene expression of auxin transporters, nutrient related genes and modulates biochemical responses in Oryza sativa L. Environ Pollut. 2018;240:950–962. doi: 10.1016/J.ENVPOL.2018.04.096. [DOI] [PubMed] [Google Scholar]
- Praveen A, Pandey C, Khan E, et al. Silicon mediated genotoxic alterations in Brassica juncea under arsenic stress: comparative study of biochemical and molecular markers. Pedosphere. 2017 doi: 10.1016/S1002-0160(17)60435-1. [DOI] [Google Scholar]
- Praveen A, Pandey A, Gupta M. Nitric oxide alters nitrogen metabolism and PIN gene expressions by playing protective role in arsenic challenged Brassica juncea L. Ecotoxicol Environ Saf. 2019;176:95–107. doi: 10.1016/J.ECOENV.2019.03.054. [DOI] [PubMed] [Google Scholar]
- Qin T, Zhao H, Cui P, et al. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 2017;175:1321–1336. doi: 10.1104/PP.17.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai MI, Alam M, Lightfoot DA, et al. Classification and experimental identification of plant long non-coding RNAs. Genomics. 2019;111:997–1005. doi: 10.1016/J.YGENO.2018.04.014. [DOI] [PubMed] [Google Scholar]
- Rai KK, Pandey N, Rai SP. Salicylic acid and nitric oxide signaling in plant heat stress. Physiol Plant. 2020 doi: 10.1111/ppl.12958. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan M, Satish L, Kalendar R, et al. The dynamism of transposon methylation for plant development and stress adaptation. Int J Mol Sci. 2021;22:11387. doi: 10.3390/IJMS222111387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Balyan S, Jha S, Mathur S. Novel insights into expansion and functional diversification of MIR169 family in tomato. Planta. 2020;251:1–17. doi: 10.1007/S00425-020-03346-W/METRICS. [DOI] [PubMed] [Google Scholar]
- Raza A, Razzaq A, Mehmood SS, et al. Impact of climate change on crops adaptation and strategies to tackle its outcome: a review. Plants. 2019;8:34. doi: 10.3390/PLANTS8020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizwan M, Mostofa MG, Ahmad MZ, et al. Hydrogen sulfide enhances rice tolerance to nickel through the prevention of chloroplast damage and the improvement of nitrogen metabolism under excessive nickel. Plant Physiol Biochem. 2019;138:100–111. doi: 10.1016/J.PLAPHY.2019.02.023. [DOI] [PubMed] [Google Scholar]
- Ronzan M, Piacentini D, Fattorini L, et al. Cadmium and arsenic affect root development in Oryza sativa L. negatively interacting with auxin. Environ Exp Bot. 2018;151:64–75. doi: 10.1016/J.ENVEXPBOT.2018.04.008. [DOI] [Google Scholar]
- Sahay S, Robledo-Arratia L, Glowacka K, Gupta M (2021) Root NRT, NiR, AMT, GS, GOGAT and GDH expression levels reveal NO and ABA mediated drought tolerance in Brassica juncea L. Sci Rep. 10.1038/s41598-021-86401-0 [DOI] [PMC free article] [PubMed]
- Sako K, Nguyen HM, Seki M. Advances in chemical priming to enhance abiotic stress tolerance in plants. Plant Cell Physiol. 2021;61:1995–2003. doi: 10.1093/PCP/PCAA119. [DOI] [PubMed] [Google Scholar]
- Saleem M, Fariduddin Q, Janda T. Multifaceted role of salicylic acid in combating cold stress in plants: a review. J Plant Growth Regul. 2020;40:464–485. doi: 10.1007/S00344-020-10152-X. [DOI] [Google Scholar]
- Saravana Kumar RM, Wang Y, Zhang X, et al. Redox components: key regulators of epigenetic modifications in plants. Int J Mol Sci. 2020;21:1419. doi: 10.3390/IJMS21041419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedaghatmehr M, Thirumalaikumar VP, Kamranfar I, et al. A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ. 2019;42:1054–1064. doi: 10.1111/PCE.13426. [DOI] [PubMed] [Google Scholar]
- Sequera-Mutiozabal M, Antoniou C, Tiburcio AF, et al. Polyamines: emerging hubs promoting drought and salt stress tolerance in plants. Curr Mol Biol Rep. 2017;3:28–36. doi: 10.1007/S40610-017-0052-Z. [DOI] [Google Scholar]
- Shen X, He J, Ping Y, et al. The positive feedback regulatory loop of miR160-auxin response factor 17-HYPONASTIC LEAVES 1 mediates drought tolerance in apple trees. Plant Physiol. 2022;188:1686–1708. doi: 10.1093/PLPHYS/KIAB565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Ding Y, Yang S. Molecular regulation of cbf signaling in cold acclimation. Trends Plant Sci. 2018;23:623–637. doi: 10.1016/J.TPLANTS.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Shim JS, Oh N, Chung PJ, et al. Overexpression of OsNAC14 improves drought tolerance in rice. Front Plant Sci. 2018;9:310. doi: 10.3389/FPLS.2018.00310/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha A, Khan A, Dey N. Cis–trans engineering: advances and perspectives on customized transcriptional regulation in plants. Mol Plant. 2018;11:886–898. doi: 10.1016/j.molp.2018.05.008. [DOI] [PubMed] [Google Scholar]
- Song Y, Li J, Sui Y, et al. The sweet sorghum SbWRKY50 is negatively involved in salt response by regulating ion homeostasis. Plant Mol Biol. 2020 doi: 10.1007/s11103-020-00966-4. [DOI] [PubMed] [Google Scholar]
- Soto-Burgos J, Zhuang X, Jiang L, Bassham DC. Dynamics of autophagosome formation. Plant Physiol. 2018;176:219–229. doi: 10.1104/PP.17.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Bao Y, Lu Y, et al. Poplar autophagy receptor NBR1 enhances salt stress tolerance by regulating selective autophagy and antioxidant system. Front Plant Sci. 2021;11:2232. doi: 10.3389/FPLS.2020.568411/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wang P, Jia X, et al. Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol J. 2018;16:545–557. doi: 10.1111/PBI.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wang Y, Sui N. Transcriptional regulation of bHLH during plant response to stress. Biochem Biophys Res Commun. 2018;503:397–401. doi: 10.1016/J.BBRC.2018.07.123. [DOI] [PubMed] [Google Scholar]
- Sun X, Lin L, Sui N. Regulation mechanism of microRNA in plant response to abiotic stress and breeding. Mol Biol Rep. 2019;46:1447–1457. doi: 10.1007/s11033-018-4511-2. [DOI] [PubMed] [Google Scholar]
- Sun J, Chen C, Miyamoto N, et al. The scaly-foot snail genome and implications for the origins of biomineralised armour. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Shen Y, Chen Y, et al. Osa-miR1320 targets the ERF transcription factor OsERF096 to regulate cold tolerance via JA-mediated signaling. Plant Physiol. 2022;189:2500–2516. doi: 10.1093/PLPHYS/KIAC208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006 doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Lowder LG, Zhang T, et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants. 2017 doi: 10.1038/nplants.2017.18. [DOI] [PubMed] [Google Scholar]
- Tang Y, Bao X, Zhi Y, et al. Overexpression of a myb family gene, Osmyb6, increases drought and salinity stress tolerance in transgenic rice. Front Plant Sci. 2019;10:168. doi: 10.3389/FPLS.2019.00168/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Du G, Xiang J, et al. Genome-wide identification of auxin response factor (ARF) gene family and the miR160-ARF18-mediated response to salt stress in peanut (Arachis hypogaea L.) Genomics. 2022;114:171–184. doi: 10.1016/J.YGENO.2021.12.015. [DOI] [PubMed] [Google Scholar]
- Valivand M, Amooaghaie R, Ahadi A. Seed priming with H2S and Ca2+ trigger signal memory that induces cross-adaptation against nickel stress in zucchini seedlings. Plant Physiol Biochem. 2019;143:286–298. doi: 10.1016/J.PLAPHY.2019.09.016. [DOI] [PubMed] [Google Scholar]
- Villagómez-Aranda AL, Feregrino-Pérez AA, García-Ortega LF, et al. Activating stress memory: eustressors as potential tools for plant breeding. Plant Cell Rep. 2022;41:1481–1498. doi: 10.1007/S00299-022-02858-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Hayashida Y, Izumi M, et al. Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol. 2015;168:60–73. doi: 10.1104/PP.15.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Sun X, Jia X, Ma F. Apple autophagy-related protein MdATG3s afford tolerance to multiple abiotic stresses. Plant Sci. 2017;256:53–64. doi: 10.1016/J.PLANTSCI.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Wang CT, Ru JN, Liu YW, et al. Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int J Mol Sci. 2018;19:3046. doi: 10.3390/IJMS19103046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Hu J, Gao C, et al. Genome-wide analysis of long non-coding RNAs unveils the regulatory roles in the heat tolerance of Chinese cabbage (Brassica rapa ssp. chinensis) Sci Rep. 2019;9:1–14. doi: 10.1038/s41598-019-41428-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dai J, Chen R, et al. miRNA-based drought regulation in the important medicinal plant Dendrobium huoshanense. J Plant Growth Regul. 2021;41:1099–1108. doi: 10.1007/S00344-021-10366-7. [DOI] [Google Scholar]
- Wang X, Niu Y, Zheng Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int J Mol Sci. 2021;22:6125. doi: 10.3390/IJMS22116125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Chen J, Wang L, Wang S. Genome-wide investigation of WRKY transcription factors involved in terminal drought stress response in common bean. Front Plant Sci. 2017;8:380. doi: 10.3389/FPLS.2017.00380/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WW, He SS, An YY, et al. Hydrogen peroxide as a mediator of 5-aminolevulinic acid-induced Na+ retention in roots for improving salt tolerance of strawberries. Physiol Plant. 2019;167:5–20. doi: 10.1111/PPL.12967. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Zhou L, Lei X, et al. Genome-wide identification of WRKY genes and their expression profiles under different abiotic stresses in Elaeis guineensis. PLoS ONE. 2017 doi: 10.1371/journal.pone.0189224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhou S, Gong X, et al. Single-base methylome analysis reveals dynamic epigenomic differences associated with water deficit in apple. Plant Biotechnol J. 2018;16:672–687. doi: 10.1111/PBI.12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Wei Z, Zhai H, et al. The IbPYL8–IbbHLH66–IbbHLH118 complex mediates the abscisic acid-dependent drought response in sweet potato. New Phytol. 2022;236:2151–2171. doi: 10.1111/NPH.18502. [DOI] [PubMed] [Google Scholar]
- Yang H, Wu JJ, Tang T, et al. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci Rep. 2017;7:1–13. doi: 10.1038/s41598-017-07871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Hong Y, Ren Z, et al. A role for PICKLE in the regulation of cold and salt stress tolerance in arabidopsis. Front Plant Sci. 2019;10:900. doi: 10.3389/FPLS.2019.00900/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Wang Y, Teotia S, et al. The interaction between miR160 and miR165/166 in the control of leaf development and drought tolerance in Arabidopsis. Sci Re. 2019;9:1–13. doi: 10.1038/s41598-019-39397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Guo Y (2018) Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol 217 [DOI] [PubMed]
- Yao PF, Li CL, Zhao XR, et al. Overexpression of a tartary buckwheat gene, FtbHLH3, enhances drought/oxidative stress tolerance in transgenic Arabidopsis. Front Plant Sci. 2017;8:625. doi: 10.3389/FPLS.2017.00625/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao P, Sun Z, Li C, et al. Overexpression of Fagopyrum tataricum FtbHLH2 enhances tolerance to cold stress in transgenic Arabidopsis. Plant Physiol Biochem. 2018;125:85–94. doi: 10.1016/J.PLAPHY.2018.01.028. [DOI] [PubMed] [Google Scholar]
- Yu Y, Ni Z, Chen Q, Qu Y. The wheat salinity-induced R2R3-MYB transcription factor TaSIM confers salt stress tolerance in Arabidopsis thaliana. Biochem Biophys Res Commun. 2017;491:642–648. doi: 10.1016/J.BBRC.2017.07.150. [DOI] [PubMed] [Google Scholar]
- Yu Y, Zhang Y, Chen X, Chen Y. Plant noncoding RNAs: hidden players in development and stress responses. Annu Rev Cell Dev Biol. 2019;35:407. doi: 10.1146/ANNUREV-CELLBIO-100818-125218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Zhou X, Zhu Z, Zhou K. Sodium hydrosulfide mitigates cadmium toxicity by promoting cadmium retention and inhibiting its translocation from roots to shoots in Brassica napus. J Agric Food Chem. 2019;67:433–440. doi: 10.1021/ACS.JAFC.8B04622/SUPPL_FILE/JF8B04622_SI_001.PDF. [DOI] [PubMed] [Google Scholar]
- Zafar SA, Zaidi SSEA, Gaba Y, et al. Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J Exp Bot. 2020;71:470–479. doi: 10.1093/JXB/ERZ476. [DOI] [PubMed] [Google Scholar]
- Zandalinas SI, Mittler R, Balfagón D, et al. Plant adaptations to the combination of drought and high temperatures. Physiol Plant. 2018;162:2–12. doi: 10.1111/PPL.12540. [DOI] [PubMed] [Google Scholar]
- Zarza X, Atanasov KE, Marco F, et al. Polyamine oxidase 5 loss-of-function mutations in Arabidopsis thaliana trigger metabolic and transcriptional reprogramming and promote salt stress tolerance. Plant Cell Environ. 2017;40:527–542. doi: 10.1111/PCE.12714. [DOI] [PubMed] [Google Scholar]
- Zelm EV, Zhang Yanxia, Testerink C (2020) Salt tolerance mechanisms of plants. https://library.wur.nl/WebQuery/wurpubs/565510. Accessed 24 Jun 2022 [DOI] [PubMed]
- Zhang H, Lang Z, Zhu JK. Dynamics and function of DNA methylation in plants. Na Rev Mol Cell Biol. 2018;19:489–506. doi: 10.1038/s41580-018-0016-z. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang H, Srivastava AK, et al. Knockdown of rice MicroRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 2018;176:2082–2094. doi: 10.1104/PP.17.01432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xing J, Lu Q, et al. Transcriptional regulation of chlorophyll-catabolic genes associated with exogenous chemical effects and genotypic variations in heat-induced leaf senescence for perennial ryegrass. Environ Exp Bot. 2019;167:103858. doi: 10.1016/J.ENVEXPBOT.2019.103858. [DOI] [Google Scholar]
- Zhang X, Shen J, Xu Q, et al. Long noncoding RNA lncRNA354 functions as a competing endogenous RNA of miR160b to regulate ARF genes in response to salt stress in upland cotton. Plant Cell Environ. 2021;44:3302–3321. doi: 10.1111/PCE.14133. [DOI] [PubMed] [Google Scholar]
- Zhao J, Yuan S, Zhou M, et al. Transgenic creeping bentgrass overexpressing Osa-miR393a exhibits altered plant development and improved multiple stress tolerance. Plant Biotechnol J. 2019;17:233–251. doi: 10.1111/PBI.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yu X, Ottosen CO, et al. Unique miRNAs and their targets in tomato leaf responding to combined drought and heat stress. BMC Plant Biol. 2020;20:1–10. doi: 10.1186/S12870-020-2313-X/FIGURES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.