Abstract
Previously, members of our group reported the isolation and characterization of mutacin II from Streptococcus mutans T8 and the genetic analyses of the mutacin II biosynthesis genes (J. Novak, P. W. Caufield, and E. J. Miller, J. Bacteriol. 176:4316–4320, 1994; F. Qi, P. Chen, and P. W. Caufield, Appl. Environ. Microbiol. 65:652–658, 1999; P. Chen, F. Qi, J. Novak, and P. W. Caufield, Appl. Environ. Microbiol. 65:1356–1360, 1999). In this study, we cloned and sequenced the mutacin III biosynthesis gene locus from a group III strain of S. mutans, UA787. DNA sequence analysis revealed eight open reading frames, which we designated mutR, -A, -A′, -B, -C, -D, -P, and -T. MutR bears strong homology with MutR of mutacin II, while MutA, -B, -C, -D, -P, and -T are counterparts of proteins in the lantibiotic epidermin group. MutA′ has 60% amino acid identity with MutA and therefore appears to be a duplicate of MutA. Insertional inactivation demonstrated that mutA is an essential gene for mutacin III production, while mutA′ is not required. Mutacin III was purified to homogeneity by using reverse-phase high-pressure liquid chromatography. N-terminal peptide sequencing of the purified mutacin III determined mutA to be the structural gene for prepromutacin III. The molecular mass of the purified peptide was measured by laser disorption mass spectrophotometry and found to be 2,266.43 Da, consistent with our supposition that mutacin III has posttranslational modifications similar to those of the lantibiotic epidermin.
Several species of bacteria inhabit the human oral cavity; among them, Streptococcus mutans is considered a major agent responsible for dental caries (31). Previous studies showed a certain percentage of clinical isolates of S. mutans producing antimicrobial substances called mutacins (4, 15). Mutacins are active against closely related species as well as a surprisingly wide spectrum of other gram-positive bacteria (35). The ability of S. mutans to produce mutacins, combined with its lactic acid production, may contribute to the pathogenesis of these bacteria (25). Production of mutacins by S. mutans and other oral streptococci may also play a protective role for the host against pathogens such as group A streptococci and Streptococcus pneumoniae. In this respect, mutacins may serve as antimicrobial agents in the future.
Previously, members of our group divided mutacin-producing S. mutans strains into three groups, I, II, and III, based on the existence of a resident plasmid and the antagonistic activity of the strains towards each other (4). Groups I and II harbor a 5.6-kb plasmid but differ in the restriction patterns of their plasmids (4), while group III strains are plasmid free. The three groups have overlapping but different antimicrobial spectra (37a). Mutacin II was isolated from the group II strain T8 and partially characterized (7, 34). It belongs to a family of peptide antibiotics called lantibiotics, which are ribosomally synthesized and posttranslationally modified, containing lanthionines, β-methylanthionines, and didehydro amino acid residues (20, 39).
The biosynthesis of lantibiotics includes translation of the structural gene to produce a prepropeptide, which consists of an N-terminal leader peptide and a C-terminal propeptide. Modification of the propeptide includes dehydration of serine and threonine residues to form 2,3-didehydro amino acids and then addition of the thiol groups from cysteine residues to the double bonds to form lanthionines or β-methyllanthionines (40). The prepeptide is transported across the cell membrane, processed, and then secreted into the outside medium. This series of posttranslational modifications is carried out by specific sets of enzymes encoded by the lantibiotic biosynthesis operon (16). Also included in the biosynthesis gene cluster is a set of immunity genes, which functions in protecting the producer cells from being killed by the antibiotics they produce (41). These characteristics of lantibiotics make them good candidates for protein engineering to improve their properties and for genetic manipulations to increase their production levels under industrial conditions.
Expression systems for study of structure-function and modification of lantibiotics have been constructed in the nisin, Pep5, and epidermin systems (27). Recently, members of our group reported construction of a mutacin II expression system for gene replacement studies of S. mutans (5). While this system can be successfully used for structure-function studies, studies on improving the properties and increasing the production levels of mutacin II become complicated because of the involvement of multiple genes and factors in the modification, processing, and regulation of mutacin II production (37). To understand the molecular mechanism of mutacin modification, processing, and control of expression, we continued to characterize other mutacins and their biosynthesis genes. By comparing the similarities and differences, we hoped to gain insights into the intricate network of mutacin production. Our initial attempt to isolate mutacin I and mutacin III was limited due to the low production levels of these mutacins in liquid culture. After cloning and sequencing of the mutacin II biosynthesis locus (6, 49), DNA probes were made with the mutacin II biosynthesis genes and hybridized with chromosomal DNA isolated from all three mutacin-producing groups, in the hope that sequences conserved among the three mutacins could be found. Unfortunately, the mutacin II probes hybridized only with chromosomal DNA isolated from group II strains, not with that isolated from group I or group III strains, suggesting a fundamental difference between mutacin II and other mutacins. In this communication, we report the cloning and sequencing of the mutacin III biosynthesis genes by using information from the conserved sequence derived from several other lantibiotics, and we also report the isolation and purification of mutacin III from a modified semisolid culture medium.
MATERIALS AND METHODS
Bacterial strains and media.
The group III S. mutans strain UA787 was isolated from a caries-active white female patient in the late 1980s. Streptococcus sanguis NY101 was used as the indicator for mutacin activity assays. Methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus faecium (VRE), penicillin-resistant S. pneumoniae (PRSP), and group A streptococcus (GA) were clinical isolates from the University of Alabama, Birmingham, Hospital. For propagation, UA787 and NY101 were grown on TSBY plates containing 3% each of Trypticase soy broth (BBL Becton Dickinson, Cockeysville, Md.) and yeast extract, with 1.6% agar. The other strains were stored as frozen cultures in Todd-Hewitt (TH) broth (Difco Laboratories, Detroit, Mich.) and grown in TH broth when needed.
Cloning and sequencing of the mutacin III biosynthesis genes.
For cloning the mutacin III structural gene, a pair of primers was designed. The forward primer, III-F1 (5′-AGTTTCAATAGTTACTGTTGC-3′), was based on the conserved amino acid sequence, S-F-N-S-Y-C-C, of mutacin 1140 (18), mutacin NY266 (33), epidermin (42), and gallidermin (43) and the codon preference in S. mutans (49). The reverse primer, III-R1 (5′-GCCAAACGGAGTTGATCTCGT-3′), was based on the conserved amino acid sequence, T-R-S-T-P-F-G, of LanB (18), SpaB (14, 24), NisB (10, 26), and EpiB (42). PCR amplification was performed with chromosomal DNA of UA787 as the template and with Elongase (Gibco BRL, Gaithersburg, Md.) under the following conditions: 94°C for 1 min, 60°C for 1 min, and 68°C for 3 min for 25 cycles. The PCR amplicon (III-F1–III-R1) was cloned into pGEM-T Easy vector (Promega Corp., Madison, Wis.) and sequenced. For cloning the upstream and downstream regions of the structural genes, we used a circular PCR technique. Briefly, two primers (up1 and dn1), one going upstream from the 5′ portion of the DNA fragment and one going downstream from the 3′ portion of the DNA fragment, were designed based on the sequence of the III-F1–III-R1 PCR amplicon. The chromosomal DNA of UA787 was digested to completion with a panel of restriction enzymes and self-ligated. The ligation mixtures were used as templates in PCRs with the up1 and dn1 primers. The PCR products were then cloned and sequenced. The upstream and downstream sequences could be distinguished at the unique restriction site where the chromosomal DNA was initially cut.
Insertional inactivation.
To construct a vector to facilitate a double-crossover insertional inactivation of mutA and mutA′, a 1.5-kb DNA fragment encompassing regions upstream of mutA and downstream of mutA′ of the mutacin III operon was generated by PCR and cloned into pGEM-T Easy to generate pGEM787. Two divergent primers were then designed from the coding region of the structural gene to be inactivated, one of which had a ClaI restriction site incorporated at its 5′ end. The two primers were phosphorylated and used to copy pGEM787 by inverse PCR. The PCR product was treated with DpnI restriction enzyme to destroy the parental plasmid, precipitated with ethanol, ligated, and transformed into Escherichia coli DH5α to obtain pGEM787A or pGEM787A′. The kanamycin resistance cassette was a derivative of the aphIII gene (46), from which the transcription terminator was deleted to prevent polar effects and in which a ClaI site was incorporated at the 5′ and 3′ ends to facilitate cloning. The aphIII gene was amplified by PCR and then cut with ClaI and cloned into pGEM787A and pGEM787A′ to create pGEM787AKm and pGEM787A′Km, respectively. The plasmids were cut with EcoRI to release the mutAKm or mutA′Km insert and transformed into competent UA787 by the standard procedure (44). Recombinants were selected on TH agar plates with 400 μg of kanamycin per ml and tested for mutacin production, with NY101 as the indicator.
Isolation and purification of mutacin III.
Isolation of mutacin III was performed based on methods described previously (18, 34), with some modifications. Briefly, UA787 was grown for 72 h on TH agarose plates containing 0.3% agarose supplemented with trace elements (5 mg of FeSO4 · 7H2O per liter, 200 mg of K2HPO4 per liter, 1 g of KH2PO4 per liter, 0.7 g of MgSO4 · 7H2O per liter, and 5 mg of MnSO4 per liter). The plates were then frozen at −70°C and thawed quickly in a 60°C water bath. The culture was transferred into a centrifuge tube and spun for 30 min at 20,000 × g. The supernatant was passed through a 0.45-μm-pore-size membrane and extracted with an equal volume of chloroform. The emulsion at the chloroform-aqueous interface was collected by centrifugation. The pellet was dried under a stream of air and washed once with double-distilled H2O. The water-insoluble material (crude extract) was dissolved in 0.25% trifluoroacetic acid (TFA). Both water-soluble and -insoluble fractions were tested for antimicrobial activity after a serial dilution with phosphate-buffered saline. One arbitrary unit of activity was defined as the highest dilution that exhibited a clear zone of inhibition of growth of the indicator strain, NY101. For purification, the crude extract of mutacin III was applied to a Source 15RPC column and eluted with a fragmented gradient of buffer A (0.1% TFA) and buffer B (0.085% TFA in 80% methanol) with the AKTA purifier and the UNICORN control system (Amersham Pharmacia Biotech, Piscataway, N.J.). The active fractions (fraction 1) were pooled and dried in a lyophilizer. The pellet was redissolved in 0.25% TFA and subjected to a second round of purification with the same column and protocol. The single active peak fraction was collected, dried in a lyophilizer, and used for sequence analysis and matrix-assisted laser disorption mass spectrometry (MALD-MS).
Determination of MICs of purified mutacin III.
High-pressure liquid chromatography (HPLC)-purified mutacin III was dissolved in 0.11% TFA at a concentration of 25 mg/ml. The solution was first diluted to 2.5 mg/ml with phosphate-buffered saline; then a series of 1:2 dilutions was performed with this starting material. As a control, the lantibiotic nisin (2.5%, balanced with sodium chloride and denatured milk solid; Sigma Biochemical Inc., St. Louis, Mo.) was suspended in 0.11% TFA to obtain a concentration of 2.5 mg/ml (pure nisin), and the solution was diluted in the same manner as mutacin III. Ten microliters from each dilution was added to each well in a 96-well culture plate. The indicator strain, S. sanguis NY101, and clinical isolates of MRSA, VRE, PRSP, and GA were grown in TH broth overnight under anaerobic conditions, except for MRSA, which was grown aerobically. The overnight culture was diluted in TH broth to obtain 105 CFU/ml, and 90 μl of this cell suspension was added to the 10 μl of lantibiotic solution in each well of the 96-well plate. The plate was then covered and incubated at 37°C anaerobically overnight (the plate with MRSA was incubated under aerobic conditions). The highest dilution of mutacin III or nisin that exhibited complete inhibition of cell growth was used to calculate the MIC.
Nucleotide sequence accession number.
The sequence of the mutacin III genes has been deposited in GenBank with the accession no. AF154675.
RESULTS
Cloning and sequencing of the mutacin III biosynthesis genes.
Our previous attempts to clone mutacin I and mutacin III biosynthesis genes with mutacin II gene probes were not successful due to the low degree of similarity between mutacin II and other mutacins. When mutacins 1140 and NY266 demonstrated remarkable similarity to epidermin and gallidermin (18, 33), we reasoned that the group I or group III mutacin might be a member of this family of lantibiotics. To test this notion, we designed two primers, one based on the sequence conserved among mutacin 1140, mutacin NY266, epidermin, and gallidermin and the other based on the sequence conserved among LanB, SpaB, NisB, and EpiB (see Materials and Methods). With these two primers, we screened a panel of S. mutans isolates, which produced either group I, group II, or group III mutacins, by PCR amplification for the presence of these genes on their chromosomes. Two DNA fragments of ∼700 and 450 bp were amplified from all isolates of group I and group III but not from those of group II (data not shown). We chose the PCR amplicons from the group III strain UA787 for sequence analysis (details of the cloning and sequencing of the group I mutacin will be published elsewhere).
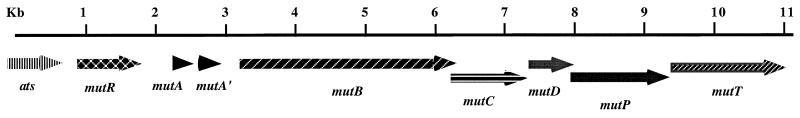
The 700- and 450-bp PCR amplicons of UA787 were cloned and sequenced. Sequence analysis revealed that the 450-bp fragment was colinear with the 700-bp fragment from the 3′ end. It turned out that the 5′ primer (III-F1) bound to two conserved regions within two partially duplicated genes (mutA and mutA′ [see Fig. 1 and 2]). Based on the sequence of the 700-bp fragment, upward and downward primers were designed to clone the upstream and downstream genes by chromosome walking by using a circular PCR technique (see Materials and Methods). A total of >11 kb of DNA was thus cloned and sequenced. Sequence analysis revealed nine open reading frames in the order of ats-mutR-mutA-mutA′-mutB-mutC-mutD-mutP-mutT (Fig. 1), which were followed by the immunity genes mutF and -E and possibly -G (data not shown). All genes were transcribed in the same direction. The ats gene encodes alanyl tRNA synthetase and therefore was presumed to define the upstream border of the mutacin III gene locus.
FIG. 1.
Mutacin III biosynthesis genes. The orientation of the genes and their relative distances are shown. The ats gene encodes alanyl tRNA synthetase and is therefore presumed not to be part of the mutacin III biosynthesis gene operon. MutA is the prepropeptide of mutacin III, and MutA′ has no known function. MutB, -C, and -D are probably involved in posttranslational modification of mutacin III, and MutP and -T are possibly responsible for processing and transport of the prepeptide mutacin III.
FIG. 2.
Comparison of some of the mutacin III gene products with other lantibiotic proteins. (A) Amino acid sequences of the MutRs of mutacin II and mutacin III. The middle row shows the identical amino acids and the conserved changes. (B) Alignment of MutA′ with OrfY of mutacin 1140. The N-terminal 13 aa in OrfY may be a result of improper translation from an upstream illegitimate ATG start codon. Note the identical leader peptide sequence between the two proteins. (C) Peptide sequences of MutA and MutA′. The C-terminal amino acid sequence, S-F-N-S-Y-C-C, identical for the two peptides, was the one upon which the PCR primer III-F1 was based (see Materials and Methods).
Inspection of the upstream region of mutR revealed a ribosomal binding site with the sequence GGAG which was 6 bp upstream of the initiation codon, TTG. A rho-independent transcription terminator-like sequence followed the stop codon for MutR. The intergenic region between mutR and mutA (590 bp) consisted of promoter-like sequences, AT-rich elements, and direct repeats, which would likely play regulatory roles in promoter activity. A stem-loop structure which could act as a transcription attenuator for differential gene expression existed in the intergenic region between mutA and mutA′. A promoter-like sequence may be present upstream of mutB, but no terminator-like sequence was apparent in the intergenic region between mutA′ and mutB. The reading frames of mutB and mutC overlapped by 11 bp, suggesting a cotranscription for the two genes. The MutD initiation codon (ATG) began 25 bp after the stop codon for MutC. The reading frame of mutP overlapped with that of mutD by 18 bp and was followed closely by mutT. This arrangement suggests that the mutBCDPT genes are likely cotranscribed. Downstream of the mutAA′BCDPT operon is a separate transcriptional unit consisting of the mutFE and possibly -G genes, which are probably involved in immunity to mutacin III (data not shown).
Similarity of the mutacin III gene products with other proteins.
The first gene in the mutacin III locus, mutR, encoded a protein of 284 amino acids (aa), which exhibited strong homology (62% identity and 79% similarity) with MutR encoded in the mutacin II operon (37) (Fig. 2A). MutR of mutacin II is a transcription activator required for expression of the mutAMTFEG genes (37). Like mutR of mutacin III, mutR of mutacin II is located upstream of the structural gene for prepromutacin II. The fact that the two genes have similarities in sequence as well as genomic organizations suggests that they may assume the same function. The second gene, mutA, and the 5′ half of mutA′ encoded peptides with sequences identical to those of LanA and the N-terminal half of OrfY of mutacin 1140 (18), respectively. The C-terminal half of MutA′, however, was markedly different from that of OrfY (Fig. 2B). In addition, mutA and mutA′ also had significant homology, with 60% identity and 73% similarity at the amino acid level and 89% identity at the nucleotide level (Fig. 2C). The higher degree of similarity of the two genes at the nucleotide level suggests that they may have arisen from a single gene ancestor by gene duplication events. The fourth gene, mutB, encoded a protein of 990 aa, which showed significant similarity to LanB proteins in the lantibiotic nisin group (10, 14, 24, 26, 42). The fifth gene, mutC, encoded a protein of 424 aa, which bore strong similarity to EpiC (42) and weaker similarity to SpaC (8), NisC (10), EciC (17), and PepC (accession no. S58361), the LanC proteins in the nisin group. The LanB and LanC proteins are assumed or inferred to catalyze the posttranslational dehydration of serine and threonine residues and formation of the thioether bridges in the prepeptide (28, 32, 45). The sixth gene in the operon, mutD, encoded a protein of 188 aa, which resembled EpiD (42). EpiD is a flavoprotein shown to catalyze the C-terminal oxidative decarboxylation of the lantibiotic precursor peptide EpiA (29, 30). MutP was 447 aa in size and had strong similarity to NisP (47), EpiP (42), and other serine proteases. MutT contained 541 aa and had strong similarity to the multidrug resistance protein LmrA in Lactococcus lactis (48), PepT of the lantibiotic Pep5 (32), and other ATP-binding-cassette transporters. The LanP and LanT proteins were shown to be required for secretion and processing of the prepeptide lantibiotic (11, 38, 47). Taken together, these findings indicate that mutacin III is a lantibiotic peptide which likely shares structural features with the lantibiotics in the nisin and epidermin groups.
Insertional inactivations of mutA and mutA′ and their effects on mutacin III production.
The finding that there were two tandem genes (mutA and mutA′) encoding two highly homologous peptides suggested, among other possibilities, that (i) both peptides were required for mutacin III activity or (ii) one peptide may serve as a peptide pheromone (3, 9, 22) for activation of transcription of the mutacin III biosynthesis genes. To determine the function of the two genes in the production of mutacin III, we disrupted the two genes separately by inserting a kanamycin-resistant gene cassette within the gene. The disrupted genes were then recombined back into the chromosome to replace the wild-type copy. The two resulting mutants, UA787AKm and UA787A′Km, were assayed for mutacin production by a deferred-antagonism test (4, 35) with S. sanguis NY101 as the indicator. As shown in Fig. 3, inactivation of mutA abolished mutacin production, while inactivation of mutA′ did not exert a noticeable effect. This result suggested that mutA may be the only structural gene encoding mutacin III.
FIG. 3.
Effects of mutA and mutA′ mutations on mutacin III production. Cells from an overnight culture plate were stabbed onto a TH agar plate and incubated at 37°C for 24 h. The plate was heated at 80°C for 1 h to kill the producing bacteria, and then an overnight culture of the indicator strain, NY101, was overlaid on top of the plate. The plate was inspected after an overnight incubation at 37°C.
Purification and characterization of mutacin III.
To confirm that there was only one component in mutacin III activity and that mutA was indeed the structural gene for mutacin III, we isolated mutacin III from the semisolid culture as described in Materials and Methods. The TH agarose plate with trace elements and 0.3% agarose was used because it supported a relatively high level of mutacin production yet had a low level of peptide contaminants in the supernatant. The production level of mutacin III on such a plate, after chloroform precipitation, was calculated to be ∼8,000 arbitrary units of activity per liter of bacterial culture, with NY101 as the indicator.
Pure mutacin III was obtained by reverse-phase HPLC after two rounds of purification. Fractions (1 ml) were collected during the first purification and tested for antimicrobial activity, with NY101 as the indicator. Only fractions 8, 9, and 10 (Fig. 4A) showed activity against the indicator. These fractions were then collected and subjected to a second purification (Fig. 4B). Two fractions (1 and 2) from the second purification were collected and tested for purity and antimicrobial activity (Fig. 4C). Only Fraction 5 showed antimicrobial activity, suggesting that mutacin III may have only one component. To rule out the possibility that a minor component was coeluted with the major component of mutacin III, the material(s) from fraction 5 (Fig. 4C) was analyzed by MALD-MS. As shown in Fig. 4D, a single peak with a molecular mass of 2,266.49 Da was revealed, and no minor peaks were detected. Taken together, these results strongly suggest that mutacin III had only one component and that mutA may be the structural gene for prepromutacin III.
FIG. 4.
Purification and MALD-MS analyses of mutacin III. (A) Elution profile of the first-round purification of crude extract of mutacin III by reverse-phase HPLC. Fractions (1 ml) were collected during the course of elution and tested for antimicrobial activity. Only fractions 8, 9, and 10 were active against the indicator. (B) Elution profile of the second-round purification with pooled fractions 8, 9, and 10 from the first pass as starting material. (C) Determinations of the purity and antimicrobial activity of the material collected from the second-round purification. (D) MALD-MS analysis of the purified mutacin III. The calculated molecular mass is given at the top of each peak. The major peak (2,266.49) was mutacin III; the next peak (2,281.68) was assumed to be the oxidized form of mutacin III. The third peak (2,287.99) was mutacin III plus sodium, which was incorporated during the analysis, and the fourth peak was the oxidized mutacin III plus sodium. AU, arbitrary units; % B, percentage of solution B (see Materials and Methods).
Peptide sequence analyses of the purified mutacin III.
The purified mutacin III was subjected to sequence analysis by Edman degradation. The sequencing reaction revealed a sequence, F1-K2-blank-W4, and was blocked at the fifth position. This result suggested that the third residue may be involved in thioether bridge formation and that the fifth residue may be a dehydrated amino acid, which was known to block protein sequencing reactions in other lantibiotics (33, 34). The N-terminal sequence of mutacin III corresponded to the deduced prepropeptide sequence of MutA, F42-K43-S44-W45 (Fig. 5), suggesting that mutA was the structural gene for mutacin III and that the leader peptide was cleaved at the R41-F42 junction. Comparison of the amino acid sequence of mutacin III with those of mutacin NY266, epidermin, and gallidermin revealed striking similarities (Fig. 5). In fact, the sequence was nearly identical from C7 to C22 (the C-terminal residue), except for a conserved change, R13→K. In addition, S3, which was shown to form a thioether bridge with C7 in epidermin (1), was also conserved in mutacin III. This result suggests that this group of lantibiotics may form similar bridging patterns, thus suggesting similar secondary structures.
FIG. 5.
Comparison of mutacin III with other lantibiotics in the epidermin group. (A) Sequences of the leader peptide of mutacin III and those of mutacin 1140, epidermin, and gallidermin. The FNLD motif is underlined. (B) Alignment of the mature mutacin III peptide with mutacin 1140, mutacin NY266, epidermin, and gallidermin. Filled boxes represent the identical amino acids, and the open boxes denote the conserved changes. Brackets indicate the pairs of amino acid residues involved in thioether bridge formation in epidermin.
MICs of mutacin III.
The MICs of mutacin III, along with those of the control lantibiotic nisin, for a panel of antibiotic-resistant pathogens are shown in Table 1. Mutacin III was more active than nisin against the pathogens that we tested, especially the drug-resistant pathogens, such as MRSA and VRE. These results suggest that mutacin III has potential as an alternative agent against drug-resistant pathogens.
TABLE 1.
MICs of mutacin III and the control
| Lantibiotic | MIC (μg/ml) for:
|
||||
|---|---|---|---|---|---|
| MRSA | VRE | PRSP | GA | NY101 | |
| Mutacin III | 3.12 | 6.24 | 0.78 | 3.12 | 6.24 |
| Nisin | 62.5 | 62.5 | 3.9 | 62.5 | 62.5 |
DISCUSSION
In this study, we cloned and sequenced the mutacin III biosynthesis genes by PCR using the information obtained from the amino acid sequence conserved among LanA and LanB proteins of several lantibiotics and the codon preference of S. mutans. Sequence analyses revealed two highly homologous, tandem open reading frames, mutA and mutA′, encoding 63 and 64 aa, respectively. The N-terminal parts of the proteins constituting the leader peptides were identical in size (41 aa) and had 28 identical amino acids (68%), while the C-terminal parts (the mature peptide) differed in size by 1 aa and had 9 identical amino acids (∼41%), mainly at the C terminus (Fig. 2C). At the nucleotide level, the two genes had 89% identity, indicating that they likely arose from a gene duplication event during evolution of the producer strain. Interestingly, a similar gene duplication also occurred in the lantibiotic streptococcin A-M49 structural gene (19). However, the respective arrangements between the two duplicated genes of mutacin III and streptococcin A-M49 are different. In mutacin III, a stem-loop structure, which is reminiscent of the transcription attenuator between mutA and mutM in the mutacin II operon, exists between mutA and mutA′, (37). This arrangement suggests that the transcript level of mutA′ will be much lower than the transcript level of mutA because of transcription termination following mutA. In the case of streptococcin A-M49, no stem-loop structure exists between the two duplicated genes, suggesting a coordinated transcription. This difference in gene arrangement may suggest different roles played by the two duplicated genes.
As mentioned above, the arrangement of mutA and mutA′ suggests that MutA′ may not be the second component in mutacin III as is the case with some of the nonlantibiotic bacteriocins (2, 9). Supporting this notion, disruption of mutA′ by antibiotic resistance cassette insertion showed no effect on mutacin III production (Fig. 3). Further support came from analyses of the crude mutacin extract by reverse-phase HPLC. During the first round of purification, every fraction of the eluate was tested for antimicrobial activity, and only fractions 8, 9, and 10, which constituted a single peak, showed activity against the indicator strain (Fig. 4A). When these fractions were further purified to homogeneity and analyzed by MALD-MS, a single peak, which had a molecular weight consistent with that predicted from the mature peptide of MutA, was revealed (Fig. 4D). Furthermore, N-terminal sequencing of the purified mutacin III peptide revealed the sequence F-K-blank-W, which corresponded to the internal sequence, F42-K43-S44-W45, of the predicted prepeptide MutA, confirming that mutA is indeed the structural gene for mutacin III. This question remains: what is the function of mutA′? The results obtained from insertional inactivation of mutA′ also ruled out the possibility of MutA′ being the inducer peptide for mutIII operon expression, because its inactivation would have resulted in diminished expression of mutacin III. The function of mutA′ remains undetermined at present.
Comparison of the DNA sequence of mutA and mutA′ of mutacin III with that of lanA and orfY of mutacin 1140 (18) revealed nearly identical sequences, except for a 5-bp insertion in the middle of orfY. However, at the amino acid level, MutA′ and OrfY are dramatically different. Whether this difference reflects the evolutionary distance between the strains producing these two mutacins or is merely a sequencing error in mutacin 1140 is not known. It is important to note, however, that unlike mutA′, inactivation of orfY in mutacin 1140 by Tn917 insertion abolished mutacin production (18). While a polar effect by Tn917 insertion may be accountable for the result, a true difference in function may also exist between the two peptides.
The deduced amino acid sequence of the mature peptide of mutacin III showed extensive similarities to the peptides in the epidermin group (Fig. 5) (39). In fact, it has the same size (22 aa) as epidermin and 17 aa identical (except for a K→R change) to those of epidermin; the most conserved regions are the C-terminal half and the amino acid residues that are involved in thioether bridge formation. These features suggest that mutacin III may assume the same bridging pattern and C-terminal oxidative decarboxylation as the other members of the epidermin group do (39). Peptide sequencing and MALD-MS analyses of the purified mutacin III and DNA sequence analysis of the mutacin III operon support this notion. In the peptide sequencing analysis, Edman degradation was blocked at position 5 and a blank cycle was obtained at position 3. Based on observations with other lantibiotics (12, 13, 21, 33, 34), Edman cleavage of a residue forming a lanthionine or β-methylanthionine would result in a blank cycle, but subsequent reactions would continue; however, sequencing would be blocked completely at an α,β-unsaturated amino acid residue. Thus, we assume that S3 would be involved in thioether bridge formation, probably with C7, while S5 would remain a dehydroalanine. In the DNA sequence analyses, we found mutD, the counterpart of epiD, whose gene product carries out the C-terminal oxidative decarboxylation reaction for the propeptide of epidermin (29, 30). As further support for the above argument, the molecular mass of mutacin III as determined by MALD-MS is 2,266.47 Da. This value is in good agreement with the calculated molecular mass of unmodified mutacin III of 2,417 Da minus six molecules of water (by dehydration of the six residues of threonine and serine) and one carboxy residue, HCOOH (by decarboxylation of the C-terminal cysteine).
It is noteworthy that the proposed secondary structure of mutacin 1140 was significantly different at the N-terminal half than the structure of epidermin (1) and the proposed structure of mutacin NY266 (33). In the proposed structure of mutacin 1140, S3 remains a dehydroalanine, while S5 is involved in thioether bridge formation with C7 (18). This difference may result from different modifications, as is discussed below.
Jung divided the lantibiotics into two groups, type A (linear) and type B (globular), based on their secondary structures (20). Sahl and Bierbaum (39) further divided each group into subgroups based on the primary sequence of the peptide. Subgroup AI comprises nisin- and Pep5-like lantibiotics, which have an FNLD motif in the leader peptide and are modified by the LanB and LanC enzymes, transported by LanT, and processed by LanP. Within this subgroup, epidermin and gallidermin have an extra gene, lanD, whose gene product catalyzes the oxidation and decarboxylation of the C-terminal cysteine (30). The AII subgroup comprises the lacticin 481-like peptides, which have a double glycine-type leader peptide, are modified by a single enzyme, LanM, and are transported and processed by LanT. The peptide sequence and genomic organization of mutacin III place it in subgroup AI with the epidermin-like lantibiotics. However, the leader peptide of mutacin III is dramatically different from those of the other lantibiotics in this group in that it lacks a conserved FNLD motif (Fig. 5). More interestingly, the mutacin III operon contains a mutR gene, which does not resemble other regulatory genes in this group, such as epiQ (36), nisR (47), and spaR (23), but is similar to the transcription regulator gene mutR of the mutacin II operon (47). Mutacin II is a member of subgroup AII. These findings suggest that the mutacin biosynthesis genes may have been acquired by horizontal gene transfer from other species, while the regulatory gene may have evolved within the species.
Mutacin III and mutacin 1140 have identical structural genes. However, the modifying enzyme genes of the two lantibiotics are dramatically different. MutB is 990 aa in size, comparable with NisB (993 aa), EpiB (990 aa), and SpaB (1,030 aa), while LanB of mutacin 1140 contains only 184 aa (18). It would be interesting to know whether this small LanB protein performs the same function as the other LanB proteins or whether it requires an accessory factor for function. It will be more interesting to compare the secondary structures of mutacin III and mutacin 1140, which would result from different modifications by the two different enzymes. We anticipate that elucidation of the secondary structures of both mutacins would help us gain insights into the mechanism of propeptide modification of lantibiotics. In fact, we also found two active reverse-phase HPLC peaks for mutacin I in one producer strain (UA140) but not in the other (CH43), although the two strains have identical structural genes (37a). These observations suggest that the secondary structure of the mature lantibiotic peptide is determined not only by the sequence of the structural gene but also by the modifying enzymes and possibly by other factors in the producing strain.
Mutacin III is active against a panel of antibiotic-resistant pathogens, among them, MRSA, VRE, and PRSP. PRSP showed even more sensitivity to mutacin III on the plate than it did in liquid medium (data not shown). We anticipate that as resistance to conventional antibiotics surges lantibiotics such as mutacin III will prove to be valuable alternative therapies against these emerging pathogens.
ACKNOWLEDGMENTS
We thank W. H. Benjamin for providing the pathogenic strains, K. Morrison for assistance in the N-terminal sequencing of mutacin III, and M. Kirk for assistance with MALD-MS.
This work was supported in part by NIH grant RO1 DE09082.
REFERENCES
- 1.Allgaier H, Jung G, Werner R G, Schneider U, Zahner H. Epidermin: sequencing of a heterodetic tetracyclic 21-peptide amide antibiotic. Eur J Biochem. 1986;160:9–22. doi: 10.1111/j.1432-1033.1986.tb09933.x. [DOI] [PubMed] [Google Scholar]
- 2.Anderssen E L, Diep D B, Nes I F, Eijsink V G H, Nissen-Meyer J. Antagonistic activity of Lactobacillus plantarum C11: two new two-peptide bacteriocins, plantaricins EF and JK, and the induction factor plantaricin A. Appl Environ Microbiol. 1998;64:2269–2272. doi: 10.1128/aem.64.6.2269-2272.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brurberg M B, Nes I F, Eijsink V G H. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol Microbiol. 1997;26:347–360. doi: 10.1046/j.1365-2958.1997.5821951.x. [DOI] [PubMed] [Google Scholar]
- 4.Caufield P W, Childers N K, Allen D N, Hansen J B. Distinct bacteriocin groups correlate with different groups of Streptococcus mutans plasmids. Infect Immun. 1985;48:51–56. doi: 10.1128/iai.48.1.51-56.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen P, Novak J, Kirk M, Barnes S, Qi F, Caufield P W. Structure-activity study of the lantibiotic mutacin II from Streptococcus mutans T8 by a gene replacement strategy. Appl Environ Microbiol. 1998;64:2335–2340. doi: 10.1128/aem.64.7.2335-2340.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen P, Qi F, Novak J, Caufield P W. The specific genes for lantibiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc. Appl Environ Microbiol. 1999;65:1356–1360. doi: 10.1128/aem.65.3.1356-1360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chikindas M L, Novák J, Driessen A J M, Konings W N, Schilling K M, Caufield P W. Mutacin II, a bactericidal lantibiotic from Streptococcus mutans. Antimicrob Agents Chemother. 1995;39:2656–2660. doi: 10.1128/aac.39.12.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung Y J, Steen M T, Hansen J N. The subtilin gene of Bacillus subtilis ATCC 6633 is encoded in an operon that contains a homolog of the hemolysin B transport protein. J Bacteriol. 1992;174:1417–1422. doi: 10.1128/jb.174.4.1417-1422.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diep D B, Havarstein L S, Nes I F. A bacteriocin-like peptide induces bacteriocin synthesis in Lactobacillus plantarum C11. Mol Microbiol. 1995;18:631–639. doi: 10.1111/j.1365-2958.1995.mmi_18040631.x. [DOI] [PubMed] [Google Scholar]
- 10.Engelke G, Gutowski-Eckel Z, Hammelmann M, Entian K-D. Biosynthesis of the lantibiotic nisin: genomic organization and membrane localization of the NisB protein. Appl Environ Microbiol. 1992;58:3730–3743. doi: 10.1128/aem.58.11.3730-3743.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geißler S, Götz F, Kupke T. Serine protease EpiP from Staphylococcus epidermidis catalyzes the processing of the epidermin precursor peptide. J Bacteriol. 1996;178:284–288. doi: 10.1128/jb.178.1.284-288.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross E, Kiltz H H. The number and nature of α,β-unsaturated amino acids in subtilin. Biochem Biophys Res Commun. 1973;50:559–565. doi: 10.1016/0006-291x(73)90876-0. [DOI] [PubMed] [Google Scholar]
- 13.Gross E, Morell J L. The number and nature of α,β-unsaturated amino acids in nisin. FEBS Lett. 1968;2:61–64. doi: 10.1016/0014-5793(68)80101-2. [DOI] [PubMed] [Google Scholar]
- 14.Gutowski-Eckel Z, Klein C, Siegers K, Bohm K, Hammelmann M, Entian K-D. Growth phase-dependent regulation and membrane localization of SpaB, a protein involved in biosynthesis of the lantibiotic subtilin. Appl Environ Microbiol. 1994;60:1–11. doi: 10.1128/aem.60.1.1-11.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamada S, Ooshima T. Production and properties of bacteriocins (mutacins) from Streptococcus mutans. Arch Oral Biol. 1975;20:641–648. doi: 10.1016/0003-9969(75)90131-4. [DOI] [PubMed] [Google Scholar]
- 16.Hansen J N. Antibiotics synthesized by posttranslational modifications. Annu Rev Microbiol. 1993;47:535–564. doi: 10.1146/annurev.mi.47.100193.002535. [DOI] [PubMed] [Google Scholar]
- 17.Heidrich C, Pag U, Josten M, Metzger J, Jack R W, Bierbaum G, Jung G, Sahl H-G. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl Environ Microbiol. 1998;64:3140–3146. doi: 10.1128/aem.64.9.3140-3146.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hillman J D, Novák J, Sagura E, Gutierrez J A, Brooks T A, Crowley P J, Hess M, Azizi A, Leung K-P, Cvitkovitch D, Bleiweis A S. Genetic and biochemical analysis of mutacin 1140, a lantibiotic from Streptococcus mutans. Infect Immun. 1998;66:2743–2749. doi: 10.1128/iai.66.6.2743-2749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hynes W L, Friend V L, Ferretti J J. Duplication of the lantibiotic structural gene in M-type 49 group A streptococcus strains producing streptococcin A-M49. Appl Environ Microbiol. 1994;60:4207–4209. doi: 10.1128/aem.60.11.4207-4209.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung G. Lantibiotics: a survey. In: Jung G, Sahl H G, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: ESCOM Science Publishers; 1991. pp. 1–34. [Google Scholar]
- 21.Kellner R, Jung G, Sahl H-G. Structure elucidation of the tricyclic lantibiotic Pep5 containing eight positively charged amino acids. In: Jung G, Sahl H G, editors. Nisin and novel lantibiotics. Leiden, The Netherlands: ESCOM Science Publishers; 1991. pp. 141–158. [Google Scholar]
- 22.Kleerebezem M, Quadri L E N, Kuipers O P, De Vos W M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 23.Klein C, Kaletta C, Entian K-D. Biosynthesis of the lantibiotic subtilin is regulated by a histidine kinase/response regulator system. Appl Environ Microbiol. 1993;59:296–303. doi: 10.1128/aem.59.1.296-303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein C, Kaletta C, Schnell N, Entian K-D. Analysis of genes involved in biosynthesis of the lantibiotic subtilin. Appl Environ Microbiol. 1992;58:132–142. doi: 10.1128/aem.58.1.132-142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleinberg I. Dental caries. In: Nolte W A, editor. Oral microbiology. St. Louis, Mo: The C. V. Mosby Company; 1982. pp. 605–624. [Google Scholar]
- 26.Kuipers O P, Beerthuyzen M M, Siezen R J, De Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactobacillus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 27.Kuipers O P, Bierbaum G, Ottenwalder B, Dodd H M, Horn N, Metzger J, Kupke T, Gnau V, Bongers R, van den Bogaard P, et al. Protein engineering of lantibiotics. Antonie Leeuwenhoek. 1996;69:161–170. doi: 10.1007/BF00399421. [DOI] [PubMed] [Google Scholar]
- 28.Kupke T, Götz F. Expression, purification, and characterization of EpiC, an enzyme involved in the biosynthesis of the lantibiotic epidermin, and sequence analysis of Staphylococcus epidermidis epiC mutants. J Bacteriol. 1996;178:1335–1340. doi: 10.1128/jb.178.5.1335-1340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kupke T, Kempter C, Gnau V, Jung G, Götz F. Mass spectroscopic analysis of a novel enzymatic reaction: oxidative decarboxylation of the lantibiotic precursor peptide EpiA catalyzed by the flavoprotein EpiD. J Biol Chem. 1994;269:5653–5659. [PubMed] [Google Scholar]
- 30.Kupke T, Kempter C, Jung G, Götz F. Oxidative decarboxylation of peptides catalyzed by flavoprotein EpiD: determination of substrate specificity using peptide libraries and neutral loss mass spectrometry. J Biol Chem. 1995;270:11282–11289. doi: 10.1074/jbc.270.19.11282. [DOI] [PubMed] [Google Scholar]
- 31.Loesche W J. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer C, Bierbaum G, Heidrich C, Reis M, Suling J, Iglesias-Wind M I, Kemper C, Molitor E, Sahl H G. Nucleotide sequence of the lantibiotic Pep5 biosynthetic gene cluster and functional analysis of PepP and PepC. Evidence for a role of PepC in thioether formation. Eur J Biochem. 1995;232:478–489. doi: 10.1111/j.1432-1033.1995.tb20834.x. [DOI] [PubMed] [Google Scholar]
- 33.Mota-Meira M, Lacroix C, LaPointe G, Lavoie M C. Purification and structure of mutacin B-Ny266: a new lantibiotic produced by Streptococcus mutans. FEBS Lett. 1997;410:275–279. doi: 10.1016/s0014-5793(97)00425-0. [DOI] [PubMed] [Google Scholar]
- 34.Novák J, Caufield P W, Miller E J. Isolation and biochemical characterization of a novel lantibiotic mutacin from Streptococcus mutans. J Bacteriol. 1994;176:4316–4320. doi: 10.1128/jb.176.14.4316-4320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parrot M, Caufield P W, Lavoie M C. Preliminary characterization of four bacteriocins from Streptococcus mutans. Can J Microbiol. 1990;36:123–130. doi: 10.1139/m90-022. [DOI] [PubMed] [Google Scholar]
- 36.Peschel A, Augustin J, Kupke T, Stevanovic S, Götz F. Regulation of epidermin biosynthetic genes by EpiQ. Mol Microbiol. 1993;9:31–39. doi: 10.1111/j.1365-2958.1993.tb01666.x. [DOI] [PubMed] [Google Scholar]
- 37.Qi F, Chen P, Caufield P W. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl Environ Microbiol. 1999;65:652–658. doi: 10.1128/aem.65.2.652-658.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Qi, F., et al. Unpublished data.
- 38.Qiao M, Saris P E J. Evidence for a role of NisT in transport of the lantibiotic nisin produced by Lactococcus lactis N8. FEMS Microbiol Lett. 1996;144:89–93. doi: 10.1111/j.1574-6968.1996.tb08513.x. [DOI] [PubMed] [Google Scholar]
- 39.Sahl H-G, Bierbaum G. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from Gram-positive bacteria. Annu Rev Microbiol. 1998;52:41–79. doi: 10.1146/annurev.micro.52.1.41. [DOI] [PubMed] [Google Scholar]
- 40.Sahl H-G, Jack R W, Bierbaum G. Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. Eur J Biochem. 1995;230:827–853. doi: 10.1111/j.1432-1033.1995.tb20627.x. [DOI] [PubMed] [Google Scholar]
- 41.Saris P, Immonen T, Reis M, Sahl H. Immunity to lantibiotics. Antonie Leewenhoek. 1996;69:151–159. doi: 10.1007/BF00399420. [DOI] [PubMed] [Google Scholar]
- 42.Schnell N, Engelke G, Augustin J, Rosenstein R, Ungermann V, Götz F, Entian K-D. Analysis of genes involved in the biosynthesis of lantibiotic epidermin. Eur J Biochem. 1992;204:57–68. doi: 10.1111/j.1432-1033.1992.tb16605.x. [DOI] [PubMed] [Google Scholar]
- 43.Schnell N, Entian K D, Götz F, Horner T, Kellner R, Jung G. Structural gene isolation and prepeptide sequence of gallidermin, a new lanthionine containing antibiotic. FEMS Microbiol Lett. 1989;49:263–267. doi: 10.1016/0378-1097(89)90050-5. [DOI] [PubMed] [Google Scholar]
- 44.Shah G R, Caufield P W. Enhanced transformation of Streptococcus mutans by modifications in culture conditions. Anal Biochem. 1993;214:343–346. doi: 10.1006/abio.1993.1503. [DOI] [PubMed] [Google Scholar]
- 45.Siegers K, Heinzmann S, Entian K-D. Biosynthesis of lantibiotic nisin. J Biol Chem. 1996;271:12294–12301. doi: 10.1074/jbc.271.21.12294. [DOI] [PubMed] [Google Scholar]
- 46.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-amino glycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- 47.van der Meer J R, Polman J, Beerthuyzen M M, Siezen R J, Kuipers O P, De Vos W M. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol. 1993;175:2578–2588. doi: 10.1128/jb.175.9.2578-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Veen H W, Venema K, Bolhuis H, Oussenko I, Kok J, Poolman B, Driessen A J, Konings W N. Multidrug resistance mediated by a bacterial homolog of the human multidrug transporter MDR1. Proc Natl Acad Sci USA. 1996;93:10668–10672. doi: 10.1073/pnas.93.20.10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodruff W A, Novak J, Caufield P W. Characterization of mutA and mutM genes involved in the biosynthesis of the lantibiotic mutacin II in Streptococcus mutans. Gene. 1998;206:37–43. doi: 10.1016/s0378-1119(97)00578-7. [DOI] [PubMed] [Google Scholar]