Version Changes
Revised. Amendments from Version 1
1. We added the full form of NCBI and BLASTn in the section of abstract. 2. At the end of the introduction, we rewrite the importance and objectives of the present research. 3. At the beginning of methods, we clarify the research area, fungi isolation and characterization techniques. 4. At the end of the discussion, we added some relevant research reports and findings with updated references which supports our present research work.
Abstract
Background: Fusarium rot is a newly introduced, devastating disease of citrus fruits. The current investigation was undertaken to characterize the microbes responsible for fruit rot in Citrus reticulata.
Methods: Pathogens were isolated from infected citrus fruits using morphological and molecular approaches. For confirmation of the isolated fungi, polymerase chain reaction (PCR) amplification and internal transcribed spacer gene sequencing techniques were used.
Results: The isolated fungus was grown on potato dextrose agar for three days and it produced clamydospores, hyphae and macroconidia. PCR amplification of isolated fungal DNA gave a 650 bp product. The sequence obtained from isolated fungi had 99.42% similarity with the reference Fusarium concentricum sequence in NCBI GenBank. The obtained sequence was deposited in GenBank (Accession No. MT856371). Two isolates showed virulence capability on fresh guava, sweet orange and tomato fruits, which confirmed species identification and Koch’s postulates. Artificially inoculated fungal species grown on tested fruits showed typical Fusarium species symptoms.
Conclusions: Outcomes of the present study are beneficial for the detection of this detrimental disease in postharvest Citrus reticulata fruits. Further research is needed for the control of this economically important disease. This is the first study of fruit rot in Citrus reticulata caused by Fusarium in Bangladesh.
Keywords: Citrus reticulata, Fruit Rot, Fusarium sp., PCR, ITS rRNA gene
Introduction
Citrus reticulata Blanco, commonly known as mandarin, is an oblate fruit resembling other oranges, belonging to the family of Rutaceae ( Ahmed et al., 2014) and originating from hybridization with Citrus maxima ( Wang et al., 2018). Citrus fruits contain different vitamins, minerals and trace elements. Citrus aurantifolia fruits are usually eaten fresh or used in salads and also used as flavoring in some liqueurs ( Morton, 1987). In traditional medicine, they are also used for the treatment of rheumatoid arthritis and obesity ( Srinivasan et al., 2008).
Fusarium species can cause superficial infections in plants and animals with high mortality in persistently and severely neutropenic patients ( Dignani & Anaissie, 2004). Fusarium species are highly competent at contamination, possessing several mycotoxins ( O’Donnell et al., 2009) and different fruits decay in different storage and postharvest conditions ( Whiteside et al., 1988).
The novel Fusarium fungi were isolated and identified through applying advanced methods on different crops from different countries ( Aktaruzzaman et al., 2018; Al-Najada & Gherbawy, 2015; Geiser et al., 2004; Sun et al., 2018).
Fusarium species are one of the most imperative pathogenic fungi responsible for fruits rots of citrus causing lose their market value ( Ezrari et al., 2022 Numerous Fusarium species have been reported with citrus and other fruits decay in different countries ( Fogliata et al., 2013; Kurt et al., 2020; Mahmud et al., 2021; Paul et al., 2022). Fusarium fruit rot is a very common and destructive problem for mandarin due to harvesting and marketing in Bangladesh ( Ahmed et al., 2014).
To the best of our knowledge, there is no report of Fusarium based mandarin fruit rot in Rajshahi, Bangladesh. Therefore, this study aims to give a clear understanding of the Fusarium species association with mandarin rot in Bangladesh using molecular approaches.
Methods
Fungi isolation from the infected fruits
Research location and samples collection. The research was conducted at Professor Joarder DNA and Chromosome Research Lab., Department of Genetic Engineering and Biotechnology, University of Rajshahi, Bangladesh during 2018 to 2019. The ten different rotten Citrus reticulata fruits ( Figure 1A) were collected from fruit market in Rajshahi, Bangladesh. Out of 10 fruits, three showed symptoms of rot which were used for pathogen isolation.
Figure 1. Naturally infected postharvest mandarin fruit showing symptoms of Fusarium rot and morphological phenotypes (F1).
( A) Infected mandarin fruit; ( B) fungus growth; ( C) clamydospores, hyphae, appressoria; and ( D) conidia under the microscope from seven-day-old culture at 35°C on potato dextrose agar.
Fungi isolation from the infected fruits. Collected fruits were cleaned under running tap water to remove foreign agents and kept in a Biosafety Cabinet (Esco, Singapore). Moreover, the fruits were disinfested with 1% sodium hypochlorite (NaOCl) for 30 seconds, followed by five rinses in autoclave distilled water. Disinfested tissue was excised and plated on potato dextrose agar (PDA) (Hi-Media, India) at 35°C in the dark for three days. The colonies showing typical morphological characteristics including, colony color, pigmentation, growth rate and size of macroconidia of Fusarium species were selected ( Hafizi et al., 2013) and isolated using the single spore technique ( Chowdhury et al., 2019). Isolated colonies were transferred onto a Petri plate with PDA and incubated for seven days at 35°C in dark conditions. Isolates were grouped into two on the basis of morphological color (blackish color in the first group and whitish in the second group). Finally, one isolate from each of the two groups was selected for morphological and molecular analysis.
Morphological characterization of isolates
The selected fungal colony was characterized by macromorphological and micromorphological investigation ( Al-Najada & Gherbawy, 2015). The isolate was sub-cultured in fresh PDA medium and three-day-old cultures were mounted using the lacto-phenol cotton blue (LPCB) staining method ( Sathya et al., 2017). The mounted microscope slide was covered with a cover slip and conidia were observed under a light microscope (LABOMED LX400, USA) at 40X magnification.
Molecular characterization of Fusarium species
Genomic DNA was extracted from 15 gm of mycelia, collected from day three-day-old PDA cultures. DNA was extracted using a MaxMaxwell® 16 LEV Plant DNA Kit (Cat No. AS1420, Promega, USA) and DNA quantity and quality were checked using a NanoDrop 2000 Spectrophotometer (Thermo Scientific, USA).
To amplify the internal transcribed spacer (ITS) gene, primer pairs ITS4 (5 ′-TCCTCCGCTTATTGATATGC-3 ′) and ITS5 (5 ′-GGAAGTAAAAGTCGTAACAAGG-3 ′) were used ( Gardes & Bruns, 1993; White et al., 1990). The PCR reaction was performed using the method described by Hassan et al. (2018) using hot start green master mix (dNTPs, Buffer, MgCl2, Taq Pol) (Cat # M7432, Promega, USA). A total of 25 µl reaction volume containing 2 µl genomic DNA, 2.5 µl 1X PCR buffer, 1.0 µl MgCl 2, 1.5 µl dNTPs, 0.5 µl of each primer, 0.5 µl of Taq polymerase and 16.5 µl of deionized water was used. The PCR was programmed with an initialization step at 95°C for 2 min, followed by 32 cycles of denaturation at 95°C for 30 seconds, primer annealing at 48°C for 30 seconds, and extension at 72°C for 45 seconds and a final extension at 72°C for 10 minutes using a G02 GeneAtlas PCR machine (Astec, Japan). PCR products were run by horizontal electrophoresis (Mini-Gel, CBS Scientific, USA) on 1% agarose (Cat # V3125, Promega, USA) gel with 0.5% ethidium bromide solution (Cat # H5041, Promega, USA) in 1x TAE buffer (Cat # V4251, Promega, USA) using a 1kb DNA ladder (Cat # G5711, Promega, USA) as marker and visualized under alpha imager UV trans-illumination (Mini, Protein Simple, USA). PCR products were purified from the agarose gel, using the Wizard SV Gel and PCR Clean-Up System (Cat # A9281, Promega, USA).
Purified PCR products were sequenced commercially by Sanger sequencing (Apical Scientific, Malaysia). Sequences were used in a search using the NCBI BLAST tool. Sequences were submitted to GenBank and compared with the GenBank database. A phylogenetic tree was constructed using MEGA 10 software using the neighbor joining method ( Kumar et al., 2016; Tamura et al., 2004).
Virulence test
Virulence competency of the isolate was carried out using the method described by Tafinta et al. (2013). The surfaces of mature, fresh guava, lemon and tomato were sterilized using water and 70% ethanol. The fruits were holed using a 2 mm sized cork borer and selected fungal inoculums were aseptically placed in the holes. The inoculated samples and the control were placed in sterile polythene bags and incubated at 35°C for seven days in dark. Isolates from the fruits and colonies from the diseased lesions were sub-cultured in PDA. The isolated fungal stains were identified based on colony features, growth rates and pigmentation. For confirmation, genomic DNA was isolated from subcultured colonies, which were isolated from artificially infected fruits. PCR amplification was performed using same procedure described in our previous article ( Hasan et al., 2020) for virulence potency test through ITS rDNA gene amplification.
The study performed using a completely randomized design (CRD). Data were analyzed using Analysis of Variance (ANOVA) to see the differences between Fusarium species pathogenicity ( Kasiamdari & Sangadah, 2015). The analysis was performed using SAS software version 9.4.
Results
Morphological characterization
Collected samples were incubated on PDA medium following the single spore technique, and after seven days, white colored fungal colonies appeared ( Figure 1B).
Isolated fungus was whitish in color and produced clamydospores, hyphae, appressoria and macroconidia ( Figure 1C–D) at day three on PDA medium. Isolates produced microconidia that were 0-septate, oval, obovoid with a truncate base, elliptical or reniform. Macroconidia were sporodochia and fusiform. Chlamydospores were monophialides.
Molecular characterization
DNA amplification through PCR produced a bright band at approximately 650 bp where a 1kb DNA ladder was used as marker. No band was found in the negative control where water was used instead of template DNA ( Figure 2).
Figure 2. PCR amplification of internal transcribed spacer region yielded ~650 bp product (F1).

1kb DNA marker is used for size determination, M - marker, I - isolate, N - negative control.
The dendrogram tree showed a close relationship with Fusarium concentricum and dissimilarity with Fusarium begoniae ( Figure 3). Therefore, molecular identification confirmed the isolates as Fusarium sp. The sequence of the total isolate was compared to Fusarium sequences in GenBank using BLASTN, which revealed closely related sequences and 99.42% homology with the reference sequence for F. concentricum (Accession No. NR_111886.1).
Figure 3. Phylogenetic tree based on the internal transcribed spacer region of rRNA showing closest relatives of fungal species isolated from citrus fruit samples (F1).
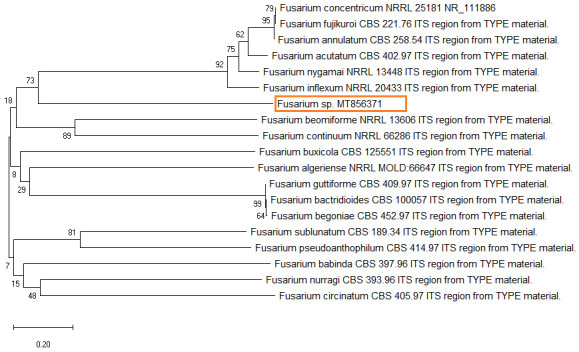
The tree was constructed by neighbor joining method. The scale bar on the rooted tree indicates a 0.20 substitution per nucleotide position.
Virulence test
The virulence test was conducted to characterize the fungus as pathogenic or saprophytic on mature, fresh and healthy guava, lemon and tomato. All fruits showed similar morphological characteristics of Fusarium symptoms ( Figure 4A–C). Isolated ribosomal DNA (rDNA) of fungus from artificially inoculated guava, lemon and tomato showed clear bands of approximately 650bp in length ( Figure 5).
Figure 4. Fusarium rot symptoms in artificially inoculated fruits (F1).
( A) Guava, ( B) lemon and ( C) tomato, the image was taken 10 days after inoculation.
Figure 5. PCR amplification of internal transcribed spacer region yielded ~650 bp product for virulence test.

1kb DNA marker is used for size determination of inoculated fungus in different fruit samples. M - marker, G - guava, C - lemon, T - tomato, N - negative control.
Discussion
Traditionally identification based on colony morphology, conidial morphology and other phenotypic characteristics has been used previously for different fungi of citrus fruits ( Leslie & Summerell, 2006; Tafinta et al., 2013). Further confirmation of the isolated fungi using advanced morphological and molecular approaches is required for characterization and differentiation of closely related Fusarium species ( Geiser et al., 2004). rDNA sequences of Fusarium, isolated from eggplant, lemon and onion (frequencies of occurrence ranging from 40% to 100%) were reported by Al-Najada & Gherbawy (2015).
The present Fusarium sp. responsible for mandarin fruit rot was identified using morpho- molecular approaches. Fusarium appeared as white or blackish-white and showed chlamydospores and macroconidia on PDA after seven days of culture. Huda-Shakirah et al. (2020) found 3–5 long, thin walled, septate macroconidia on a F. concentricum fugal stain under microscopic observation, which supports our present findings. Zhu et al. (2014) also found similar morphological characteristics for Fusarium isolated from dry root rot of citrus, lemon rot, dragon fruit rot, sugarcane wilt, eggplant rot and onion rot (frequencies of occurrence ranging from 40% to 100%) were reported by several researcher ( Al-Najada & Gherbawy, 2015; Ezrari et al., 2022; Fogliata et al., 2013; Paul et al., 2022).
The present Fusarium spp. responsible for mandarin fruit rot was identified using morpho- molecular approaches. To confirm the morphological characterization, mandarin isolate of Fusarium were culture on PDA medium for seven days. Fusarium appeared as white or blackish-white and showed chlamydospores and macroconidia on PDA after seven days of culture. Microconidia were septate, oval and elliptical. Macroconidia were sporodochia and fusiform. Huda-Shakirah et al. (2020) found 3-5µm long, thin walled, septate macroconidia on a F. concentricum fugal stain under microscopic observation, which supports our present findings. Zhu et al. (2014) also found similar morphological characteristics for Fusarium isolated from Eleocharis dulcis. To confirm the individuality of the isolated fungi, the ITS4 and ITS5 were amplified using primers ITS4F/ITS5R. PCR amplification of ITS regions of the isolated fungal strains gave ~ 650 bp products in size and the sequences showed 99.42% similarity with the Fusarium concentricum sequence in the database. Kurt et al. (2020) obtained approximately 538 bp PCR amplicon on Fusarium isolated from mandarin using ITS regions. Huda-Shakirah et al. (2020) reported 99.53% similarity with Fusarium concentricum isolated from Hibiscus sabdariffa. These results are very similar to the present findings. Phylogenetic analysis was done using comparative analysis with different ITS regions of sequences published in NCBI database. Present findings of phylogenetic analysis showed that isolates of Fusarium species were in same clade with robust bootstrap support. Results of PCR products and ITS sequencing confirm the isolated fungus as F. concentricum, which is supported by some other researcher’s findings ( Chowdhury et al., 2019; Ezrari et al., 2022; Hasan et al., 2020; Hyun et al., 2000; Paul et al., 2022). Fusarium species are considered one of the most varied fungal species. It was related with numerous plant hosts and is a thoughtful risk to Citrus reticulata production due to rot. It is also responsible for twig rot, decline dieback, blight and wilt of citrus. Fusarium rot of citrus is fetching a significant worldwide problem. This report for the first time confirmed that Fusarium species are the causative microbe of citrus fruit rot in Bangladesh. This report has significance to develop suitable management practices to control the Fusarium rot diseases of citrus fruits.
Conclusions
Fusarium fruit rot is a big problem for the citrus fruit industry in Bangladesh. In this study, Fusarium species were found to cause mandarin fruit rot. Moreover, pathogenicity was confirmed according to Koch’s postulates using three different types of fresh fruits. Fusarium species fruit rot leads to declines in the Bangladeshi fruit industry as well as fruit markets. Therefore, the current study may help the development of control measures for postharvest mandarin rot.
Funding Statement
This work was supported by the Ministry of Sciences and Technology, Government of the People's Republic of Bangladesh (2019) [39.00.0000.009.06.024.19-12/239BS].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; peer review: 2 approved]
Data availability
Underlying data
Fusarium sp. pure cultured isolate containing small subunit ribosomal RNA, internal transcribed spacer 1 and 5.8S ribosomal RNA on GenBank. Accession number, MT856371: https://www.ncbi.nlm.nih.gov/nuccore/MT856371.1?report=genbank.
Figshare: PCR amplification of ITS region yielded ~650 bp product. https://doi.org/10.6084/m9.figshare.13014209.v1 ( Hasan, 2020a).
This project contains the following underlying data:
-
-
Figure 2.jpg (original, unedited gel image from Figure 2)
Figshare: PCR amplification of ITS region for virulence test. https://doi.org/10.6084/m9.figshare.13008458.v1 ( Hasan, 2020b).
This project contains the following underlying data:
-
-
Gel doc.2.jpg (original, unedited gel image from Figure 5)
Figshare: Molecular identification of Fusarium species causing fruit rot. https://doi.org/10.6084/m9.figshare.12990746.v1 ( Hasan, 2020c).
This project contains the following underlying data:
-
-
Micro.imag.1.jpg (original, unedited microscopy image showing clamydospores, hyphae and appressoria from Figure 1C)
-
-
Micro.imag.2.jpg (original, unedited microscopy image showing conidia from Figure 1D)
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
References
- Ahmed HU, Latif MA, Haq MF, et al. : Pest Risk Analysis (PRA) of Citrus in Bangladesh.SPCB project report, DAE, Farmgate, Dhaka-1215.2014;1–104. Reference Source [Google Scholar]
- Aktaruzzaman M, Afroz T, Lee Y, et al. : Morphological and molecular characterization of Fusarium tricinctum causing postharvest fruit rot of pumpkin in Korea. J General Plant Pathol. 2018;84:407–13. 10.1007/s10327-018-0803-6 [DOI] [Google Scholar]
- Al-Najada AR, Gherbawy YA: Molecular Identification of Spoilage Fungi Isolated from Fruit and Vegetables and Their Control with Chitosan. Food Biotechnol. 2015;29(2):166–184. 10.1080/08905436.2015.1027222 [DOI] [Google Scholar]
- Chowdhury MEK, Jahan MS, Akhtar S, et al. : Characterization of fungal pathogens causing diseases in bitter gourd and establishment of their eco-friendly control measure. Int J Multidis Res Develop. 2019;6(1):109–15. Reference Source [Google Scholar]
- Dignani MC, Anaissie EJ: Human fusariosis. Clin Microbiol Infect. 2004;10(Suppl. 1):67–75. 10.1111/j.1470-9465.2004.00845.x [DOI] [PubMed] [Google Scholar]
- Ezrari S, Radouane N, Tahiria A, et al. : Dry root rot disease, an emerging threat to citrus industry worldwide under climate change: A review. Physiol Mol Plant Pathol. 2022;117:01753. 10.1016/j.pmpp.2021.101753 [DOI] [Google Scholar]
- Fogliata GM, Martínez CV, Acosta ME, et al. : First Report of Fusarium Rot Caused by Fusarium oxysporum on Lemon in Tucumán, Argentina. Plant Dis. 2013;97(7):989. 10.1094/PDIS-01-12-0069-PDN [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD: ITS primers with enhanced specificity for basidiomycetes application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2(2):113–118. 10.1111/j.1365-294x.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Geiser DM, Jiménez-Gasco MM, Kang S, et al. : FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur J Plant Pathol. 2004;110:473–9. 10.1023/B:EJPP.0000032386.75915.a0 [DOI] [Google Scholar]
- Hafizi R, Salleh B, Latiffah Z: Morphological and molecular characterization of Fusarium. solani and F. oxysporum associated with crown disease of oil palm. Braz J Microbiol. 2013;44(3):959–968. 10.1590/s1517-83822013000300047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan Md: PCR amplification of ITS region yielded ~650 bp product. figshare.Figure.2020a. 10.6084/m9.figshare.13014209.v1 [DOI] [Google Scholar]
- Hasan Md: PCR amplification of ITS region for virulence test. figshare.Figure.2020b. 10.6084/m9.figshare.13008458.v1 [DOI] [Google Scholar]
- Hasan Md: Molecular identification of Fusarium species causing fruit rot. figshare.Figure.2020c. 10.6084/m9.figshare.12990746.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan MF, Islam MA, Sikdar B: PCR and Sequencing Base Detection of Gummosis Disease on Citrus aurantifolia Caused by Lasiodiplodia theobromae and Evaluation of Its Antagonisms. J adv microbiol. 2020;20(3):77–90. 10.9734/jamb/2020/v20i330230 [DOI] [Google Scholar]
- Hassan O, Jeon JY, Chang T, et al. : Molecular and morphological characterization of Colletotrichum species in the Colletotrichum gloeosporioides complex associated with persimmon anthracnose in South Korea. Plant Dis. 2018;102(5):1015–24. 10.1094/PDIS-10-17-1564-RE [DOI] [PubMed] [Google Scholar]
- Huda-Shakirah AR, Nur-Salsabila K, Mohd MH: First report of Fusarium concentricum causing fruit blotch on roselle ( Hibiscus sabdariffa). Australas Plant Dis Notes. 2020;15:15. 10.1007/s13314-020-00385-w [DOI] [Google Scholar]
- Hyun JW, Seong CL, Dong HK, et al. : Fusarium Fruit Rot of Citrus in Jeju Island. Mycobiology. 2000;28(3):158–62. 10.1080/12298093.2000.12015743 [DOI] [Google Scholar]
- Kasiamdari RS, Sangadah U: Identification of anthrachnose disease on strawberry fruit ( Fragraria vesca L.) and its control by betel ( Piper betle L.) leaf extract. The 3rd International Conference on Biological Science. 2015;2:458–465. 10.18502/kls.v2i1.192 [DOI] [Google Scholar]
- Kumar S, Stecher G, Tamura K: MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–74. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt S, Uysal A, Soylu EM, et al. : Characterization and pathogenicity of Fusarium solani associated with dry root rot of citrus in the eastern Mediterranean region of Turkey. J General Plant Pathol. 2020;86:326–332. 10.1007/s10327-020-00922-6 [DOI] [Google Scholar]
- Leslie JF, Summerell BA: The Fusarium laboratory manual.Blackwell Publishing, Ames,2006;388. 10.1002/9780470278376 [DOI] [Google Scholar]
- Mahmud NU, Chakraborty M, Paul SK, et al. : First Report of Basal Rot of Dragon Fruit Caused by Fusarium oxysporum in Bangladesh. Plant Dis. 2021;105(1):218. 10.1094/PDIS-01-20-0005-PDN 32748716 [DOI] [Google Scholar]
- Morton JF: Mandarin orange. In: Fruits of Warm Climates.1987;142–145. [Google Scholar]
- O’Donnell K, Sutton DA, Rinaldi MG, et al. : Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. J Clin Microbiol. 2009;47(12):3851–61. 10.1128/JCM.01616-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul SK, Mahmud NU, Gupta DR, et al. : First Report of Fusarium sacchari Causing Sugarcane Wilt in Bangladesh. Plant Dis. 2022;106(1):319. 10.1094/PDIS-04-21-0681-PDN [DOI] [PubMed] [Google Scholar]
- Sathya K, Parthasarathy S, Thiribhuvanamala G, et al. : Morphological and molecular variability of Lasiodiplodia theobromae causing stem end rot of mango in Tamil Nadu, India. Int J Pure App Biosci. 2017;5(6):1024– 31. 10.18782/2320-7051.5892 [DOI] [Google Scholar]
- Srinivasan D, Ramasamy S, Sengottuvelu S: Protective effect of polyherbal formulation on experimentally induced ulcer in rats. Pharmacology Online. 2008;1:331–50. Reference Source [Google Scholar]
- Sun S, Lui Q, Han L, et al. : Identification and Characterization of Fusarium proliferatum, a New Species of Fungi that Cause Fungal Keratitis. Sci Rep. 2018;8(1):4859. 10.1038/s41598-018-23255-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafinta IY, Shehu K, Abdulganiyyu H, et al. : Isolation and Identification of Fungi Associated with the Spoilage of Sweet Orange ( Citrus Sinensis) Fruits In Sokoto State. Nigerian Journal of Basic and Applied Science. 2013;21(3):193–6. 10.4314/njbas.v21i3.4 [DOI] [Google Scholar]
- Tamura K, Nei M, Kumar S: Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101(30):11030–35. 10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, He F, Huang Y, et al. : Genome of wild mandarin and domestication history of mandarin. Mol Plant. 2018;11(8):1024–1037. 10.1016/j.molp.2018.06.001 [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee SJWT, et al. : Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: A guide to methods and applications. 1990;18(1):315–22. 10.1016/b978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Whiteside J, Bennett J, Holtzblatt K: Usability engineering: our experience and evolution.In: M. Helander (Ed.), Handbook of Human-Computer Interaction. New York, North Holland,1988;791–817. 10.1016/B978-0-444-70536-5.50041-5 [DOI] [Google Scholar]
- Zhu Z, Zheng L, Pan L, et al. : Identification and characterization of Fusarium species associated with wilt of Eleocharis dulcis (Chinese water chestnut) in China. Plant Dis. 2014;98(7):977–987. 10.1094/PDIS-08-13-0805-RE [DOI] [PubMed] [Google Scholar]




