Abstract
Gene editing-based therapeutic strategies grant the power to override cell machinery and alter faulty genes contributing to disease development like cancer. Nowadays, the principal tool for gene editing is the clustered regularly interspaced short palindromic repeats-associated nuclease 9 (CRISPR/Cas9) system. In order to bring this gene-editing system from the bench to the bedside, a significant hurdle remains, and that is the delivery of CRISPR/Cas to various target cells in vivo and in vitro. The CRISPR-Cas system can be delivered into mammalian cells using various strategies; among all, we have reviewed recent research around two natural gene delivery systems that have been proven to be compatible with human cells. Herein, we have discussed the advantages and limitations of viral vectors, and extracellular vesicles (EVs) in delivering the CRISPR/Cas system for cancer therapy purposes.
Subject terms: Genetic vectors, Cancer therapy
Introduction
Cancer is one of the top fatal diseases in the world, which causes numerous complications in many aspects. Despite all the advances in diagnostics and therapeutics, it remains too challenging to be effectively treated [1, 2]. Various therapeutic methods have been developed, including small molecules [3], and gene therapies [4], to fight cancer by targeting oncogenic signaling pathways or modulating the immune system, which in some cases has led to complete remission. Nevertheless, novel therapeutic approaches are still needed to cure different kinds of cancer.
The advances achieved by prokaryote-derived gene alteration systems have greatly aided the understanding of diseases mechanisms and finding new treatment strategies. In this regard, the Clustered regularly interspaced short palindromic repeats-associated nuclease 9 (CRISPR/Cas9)-based approaches provide a novel strategy toward gene editing for clinical utilization [5]. Hence, knowing the history of the CRISPR/Cas system and its features can help comprehend this revolutionary technology.
Initially, In 1987, when Ishino and colleagues attempted to discover an alkaline-phosphatase-isozyme convertase in Escherichia coli, they encountered repetitive sequences interspersed with spacers belonging to the CRISPR/Cas system [6]. The next encounter with such sequences was in Haloferax mediterranei, where the repeats contained 35 bp spacer sequences [7]. During the identification phase of the CRISPR/Cas, these spacer regions caught the attention of scientists. Afterward, finding similar enigmatic orders of repeats and spacer in archaea and bacteria emphasized the value of their biological importance [8]. In 2002, the repetitive sequences found in the DNA of Haloferax mediterranei, near the region related to the DNA repair system were defined as Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs). These genes accompanied by CRISPR-associated (Cas) genes were initially thought to be involved in the DNA repair system [9]. Investigations in 2005 could connect the link between the origin of the spacers and bacteriophages [9, 10]. The Koonin et al. study followed it in 2006, where they found the connection related to the activity of CRISPR and Cas genes as an entirety, which showcased a degree of resemblance between CRISPR-Cas system and prokaryotic RNA interference (RNAi) participating in the immune system [11]. CRISPR has been used for many purposes and in 2020 Emmanuelle Charpentier and Jennifer Doudna received the Nobel prize for introducing the CRISPR/Cas9 system as a gene editing tool for precise manipulation human genome.
CRISPR–Cas9-targeted fragmentation of DNA can be used as a means to pinpoint the changes in cancer-specific sequences. Moreover, in recent years, the development of chimeric antigen receptors (CAR) has made a milestone in cancer therapy. Although CAR T-cell therapy has shown some encouraging results, there are still some limitations that must be addressed, and the CRISPR/Cas system has shown promise for improving CAR T-cell-based cancer immunotherapy [12].
Moreover, in gene therapy, oncogenic viruses have been a significant concern for many years, and CRISPR appears to offer a solution to preventing them from causing cancer [13]. CRISPR-Cas system can be delivered into mammalian cells using various strategies such as viral vectors, extracellular vesicles (EVs), physical methods, and nanocomplexes [14]. In this review, we discussed the advantages and limitations of viral vectors and EVs for delivering the CRISPR/Cas system for cancer diagnosis and treatment.
The CRISPR/Cas systems
Recent advancements in prokaryote-isolated editing systems have paved the way for a better understanding of tumorigenesis mechanisms. CRISPR/Cas-based methods offer an ingenious path toward utilizing gene therapy and immunotherapy for cancer treatment. The CRISPR/Cas systems used in cancer treatment approaches are mostly based on the Cas nucleases (Cas9, Cas12a, and Cas13a) and their orthologs [15].
CRISPR/Cas9 systems are generally utilized as genetic engineering tools in most species. Cas9 is a crRNA-guided endonuclease that contains two domains called RuvC and HNH, participating in cleaving double-strand DNA (dsDNA) [16]. Furthermore, CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA) are required to form the RNP complex (Cas9–crRNA–tracrRNA ribonucleoprotein). The most frequent kind of Cas9 is Streptococcus pyogenes Cas9 (SpCas9) which is able to target DNA via protospacer adjacent motif (PAM) recognition (Fig. 1) [17]. Following Cas9-mediated DNA cleavage, the gene editing effect occurs via either the non-homologous end-joining (NHEJ) or homology-directed repair (HDR) pathway [18]. Staphylococcus aureus Cas9 (SaCas9), a variant of Cas9, has a distinctive PAM recognition capability, which can target the 5′-NNGRRT PAMs [19]. Recently, CasX, a variant of Cas9 that is both smaller and more efficient at gene editing, was discovered and is regarded as the smallest of them all [20]. Moreover, by modifying CRISPR/Cas9 into a nuclease-deficient system (called dCas9) and fusing an additional effector domain, this system can be repurposed for a variety of purposes. Specifically, CRISPR/dCas9 systems fused with the KRAB domain (CRISPR/dCas9-KRAB), a transcriptional repressor domain, are used to interfere with target gene transcription [21]. The CRISPR interference (CRISPRi) system created in this way can be used for numerous purposes, such as cancer diagnosis and treatment.
Fig. 1. Overview on different mechanisms of action carried out by CRISPR/Cas9 systems.
One of the significant advantages of the CRISPR/Cas system is that it can be modified readily to carry out various functions. Cas9 can induce DSB and edit genes with high accuracy or with the help of modified Cas9 enzymes like dCas9; this system can be used as a transcription activator (CRISPRa) or transcription repressor (CRISPRi).
The breakthroughs achieved by the CRISPR/Cas9 system have prompted sweeping exploration to uncover novel methods for extending applications. Another endonuclease with unique features for the CRISPR system is Cas12a (so-called Cpf1) [22]. In contrast to Cas9, which creates blunt ends in DNA, Cas12a is able to make the staggered ends in a distinctive cleavage pattern, promoting the DNA integration in a stringent position. Of note, the Cas12a enzymes do not require tracrRNA and process the pre-crRNA on their own. In order to cover a wide range of targeting locus, variants of Cas12a were developed to target different PAMs (5′-TATV, 5′-VTTV, 5′-TTTT, 5′-TATV, 5′-TTCN) [23, 24]. Moreover, it has been shown that CRISPR/Cas12a system potentially detects viral DNA in samples, though it hinges on the non-specific cutting of single-stranded DNA (ssDNA) [25]. This particular CRISPR system seems to have the potential to provide an opportunity for multiplex genome editing; therefore, its utilization might be much broader in cancer therapy.
Cas13a also termed C2c2, is a novel form of RNA-guided endonuclease with the ability to target RNA [26]. Upon recognizing and attaching to RNA, Cas13a can collaterally cut untargeted RNAs. As of yet, the Cas13a function in eukaryotes has not been discovered, and its mechanism of action is not fully understood [27]. CRISPR systems able to target RNAs have been used in clinical research for the detection of RNA viruses and tumor-derived circulating RNA [28, 29]. Altering different types of RNAs such as messenger RNA (mRNA), microRNA (miR), long non-coding RNA (lncRNA), and circular RNA (circRNA) by gene-editing methods, especially CRISPR, offers splendid potential in cancer therapy [30]. CRISPR/Cas is originally a system for defending bacteria against invaders, which is now being used for gene editing in mammalian cells and is able to provide immunity that transcends bacterial boundaries [31, 32].
CRISPR/Cas systems are categorized into two classes based on how many Cas endonucleases participate in their machinery. Class I operates using several Cas proteins, whereas class II requires a single Cas enzyme [16, 33]. As compared to the class I CRISPR-Cas system, elements (Cas protein, crRNA, and tracrRNA) involved in class II can be generated more readily; therefore, it would be a more appealing option for gene editing [16, 34–36]. Cas9 is a crRNA-guided endonuclease that contains two domains, HNH and RuvC, which cut complementary and non-complementary DNA strands, respectively [16, 31]. To facilitate mammalian gene manipulation, the crRNA and tracrRNA elements of CRISPR/Cas9 have been replaced with single-guide RNA (sgRNA) [37]. SgRNA’s 20 nucleotides at the 5’-terminus bind the target gene, while its 3’-duplex enables interaction with Cas9, ensuring accurate and guided gene editing [16]. The Cas9 should recognize the PAM right next to the target DNA prior to Cas9-induced double-strand break (DSB). Afterward, Cas9 undergoes a conformational change that enables it to execute a DSB three to four nucleotides upstream of the PAM [16, 35, 38]. Following that, a conformational transition occurs to the Cas9 letting it execute a DSB three to four nucleotides upstream of the PAM [16, 35, 38]. Upon Cas9-induced DSB, DNA repair pathways, such as NHEJ and HDR, become activated.
Since NEHJ, as the leading DSB repair pathway, ligates DNA break ends regardless of homologous templates, there is a risk of error in the ligation process due to arbitrary nucleotide insertions and deletions (indels), which could lead to frameshifts and nonsense mutations in genes [39]. In addition to gene alteration, NHEJ can result in deletions when it executes DNA repair with two distinct sgRNAs. These effects of NHEJ can be beneficial in the treatment of diseases indicated by an overproduction of a particular protein.
The HDR pathway repairs DNA with greater precision because a piece of DNA template homologous to the targeted part of chromosomal DNA is required for gene insertion. This allows transgenes to be integrated with more specificity after DSBs [40] HDR-based genome editing can be beneficial for diseases resulting from gene deletion, such as X-linked retinitis pigmentosa [41], hemophilia A/B [42, 43], and phenylketonuria [44]. However, NHEJ is preferred over HDR in mammalian cells for several reasons: NHEJ is active during the entire cell cycle, whereas HDR is only active in the S/G2 phase and NHEJ is faster than HDR. The use of NHEJ exceeds that of HDR, particularly in terminally differentiated cells, such as neurons, cardiac myocytes, and mature muscle cells [45, 46].
Since these repair pathways provide a simple approach to edit genes, they have become the most commonly used strategies in cancer research, even though there are alternatives such as homology-independent targeted integration (HITI), homology-mediated end joining (HMEJ), and microhomology-mediated end joining (MMEJ) [47–50].
Viral vectors and extracellular vesicles used in CRISPR-Cas9 delivery
CRISPR-Cas system can be delivered into mammalian cells using various strategies such as viral vectors and extracellular vesicles (Table 1). In this section, we will discuss these vectors in more detail. However, other approaches, such as physical methods and nanocomplexes, have not been addressed in this review. For further information on these methods, readers can refer to the cited papers discussing them [51–54].
Table 1.
Overview of recent studies that used exosomes and viral vectors for CRISPR/Cas delivery in cancer cells.
| Study | Vector | Type of cancer | Approach | Outcome |
|---|---|---|---|---|
| A New Tool for CRISPR-Cas13a-Based Cancer Gene Therapy [55] | AAVs | HCC | Decoy minimal promoter (DMP)-controlled CRISPR-Cas13a system used to knock down several oncogenes (TERT, EZH2, and RelA) | Significant inhibition of HCC cell growth |
| CRISPR-Cas9 disruption of PD-1 enhances activity of universal EGFRvIII CAR T cells in a preclinical model of human glioblastoma [56] | AAV6 | Glioblastoma (GBM) | Allogeneic EGFRvIII CAR T-cell deficient in PD-1, TCR and B2M generated by CRISPR-Cas9 | Enhanced antitumor efficacy in preclinical models of GBM. |
| Targeting HPV16 DNA using CRISPR/Cas inhibits anal cancer growth in vivo [57] | AAVs | Anal cancer | Cleaving the HPV16 E6 or E7 genes in primary human anal cancer cells utilizing CRISPR/Cas9 | Significant and selective tumor suppression |
| CRISPR/Cas9-mediated cervical cancer treatment targeting human papillomavirus E6 [58] | AAVs | Cervical cancer | Causing multiple mutations in targeted HPV E6 gene in cervical cancer cells by CRISPR/Cas9 | Increased expression of tumor suppressors, tumor growth inhibition, and increased apoptosis in the cancer cells |
| Disruption of PD-1 Enhanced the Anti-tumor Activity of Chimeric Antigen Receptor T Cells Against Hepatocellular Carcinoma [59] | Lentivirus | Hepatocellular Carcinoma (HCC) | The PD-1 disruption in the second-generation GPC3-targeted CAR T cells by CRISPR/Cas9. | Promoted Anti-tumor activity of the CAR T cells against HCC |
| CRISPR knock out of programmed cell death protein 1 enhances anti-tumor activity of cytotoxic T lymphocytes [60] | Lentivirus | Multiple Myeloma (MM) | Impairing PD-1/PD-L1 pathway in CTLs with CRISPR-Cas9 system. | CTLs repressed MM tumor growth and prolonged survival. |
| Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer [61] | Lentivirus | Refractory non-small-cell lung cancer (NSCLC) | CRISPR-Cas9-mediated knock down of PD-1 of T cells in patients with NSCLC | Results showcased the clinical safety and feasibility of CRISPR-Cas9 gene-edited T cells in NSCLC patients. |
| CRISPR-engineered T cells in patients with refractory cancer [62] | Lentivirus | Refractory cancer | NY-ESO-1 TCR-expressing engineered cells deprived of PD-1 (PDCD1), TCRα (TRAC), and TCRβ (TRBC) | Enhanced anti-tumor immunity and feasibility multiplex CRISPR-Cas9 editing at clinical scale |
| Activation of concurrent apoptosis and necroptosis by SMAC mimetics for the treatment of refractory and relapsed ALL [63] | Lentivirus | Refractory acute lymphoblastic leukemia (ALL) | Disruption of receptor-interacting protein kinase 1 (RIP1) by using CRISPR and pharmacologic interference (SMAC mimetics). | Hampering cancer cells evading apoptosis, and activating concurrent apoptosis and necroptosis |
| Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing [64] | Lentivirus | Prostate cancer | CRISPR/Cas9-based gene knockout of essential spliceosome and RNA binding protein (RBP) genes. | A RBP gene aclled HNRNPL is capable of increasing prostate cancer growth. |
| Genome-wide CRISPR/Cas9 library screen identifies PCMT1 as a critical driver of ovarian cancer metastasis [65] | Lentivirus | Ovarian cancer | PCMT1 (protein-L-isoaspartate (D-aspartate) O-methyltransferase) knockdown by CRISPR/Cas9 | PCMT1 increases in vivo metastasis formation and its serum levels may serve as a potential metastatic marker. |
| Genome-wide CRISPR screen reveals SGOL1 as a druggable target of sorafenib-treated hepatocellular carcinoma [66] | Lentivirus | HCC | In combination with NGS, the genome-wide CRISPR screen was used to determine loss-of-function mutations bestowing sorafenib resistance upon HCC cells. | SGOL1 found to be a druggable target that its inhibition may reduce drug resistance against sorafenib treatment. |
| Genome-Wide CRISPR-Cas9 Screen Identifies MicroRNAs That Regulate Myeloid Leukemia Cell Growth [67] | Lentivirus | Acute Myeloid Leukemia (AML) | miRNA loss-of-function screening was exerted by CRISPR-Cas9 technology. | Disruption of miR-150 (targeting p53) and miR-155 are therapeutic targets in AML. |
| Genome-wide CRISPR screen identifies LGALS2 as an oxidative stress-responsive gene with an inhibitory function on colon tumor growth [68] | Lentivirus | Colon cancer | The CRISPR-based screening alongside NGS evaluated the genetic factors involved in the regulation of oxidative stress. | It is reported that Glycan-binding protein Galectin 2 (Gal2) overexpression reduces the human colon tumor growth. |
| Genome-wide CRISPR-Cas9 screen identified KLF11 as a druggable suppressor for sarcoma cancer stem cells [69] | Lentivirus | Osteosarcoma | The genome-wide CRISPR screening of cancer stem cells (CSCs) of Osteosarcoma identified the regulator of osteosarcoma. | Results showed that Low KLF11 correlates with osteosarcoma’s poor prognosis and inadequate chemotherapy response. |
| Genome-Scale CRISPR-Cas9 Transcriptional Activation Screening in Metformin Resistance Related Gene of Prostate Cancer [70] | Lentivirus | Prostate cancer | CRISPR-based screening of metformin resistance in prostate cancer to find genes involved in metformin insensitivity. | Activation of ECE1, ABCA12, BPY2, EEF1A1, RAD9A, and NIPSNAP1 associated with in vitro resistance to metformin. |
| Genome-wide CRISPR-Cas9 knockout library screening identified PTPMT1 in cardiolipin synthesis is crucial to survival in hypoxia in liver cancer [71] | Lentivirus | HCC | Genome-wide CRISPR-Cas9 screening showcased therapeutic factors responsible for hypoxic survival in HCC. | Knockout of PTPMT1 provokes ROS and apoptosis in hypoxic HCC cells. |
| Identifying novel therapeutic targets in gastric cancer using genome-wide CRISPR-Cas9 screening [72] | Lentivirus | Gastric cancer | The genome-scale CRISPR-Cas9 knock-out library of gastric cancer cells | Among 184 novel genes involved in gastric cancer, methyltransferase 1 (METTL1) inhibition was the most validated approach for cancer-targeted therapy. |
| In vivo CRISPR/Cas9 targeting of fusion oncogenes for selective elimination of cancer cells [73] | Adenovirus | PDX (patient-derived xenograft) cancer models | CRISPR/Cas9-mediated targeting of two introns of the translocated genes, results in | Disruption of the fusion oncogene in cancer cells, followed by a selective and efficient activity for cancer cell elimination. |
| Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing [74] | Lentivirus and Adenovirus | Pancreatic cancer | CRISPR/Cas9-mediated genomic manipulation of pancreatic cancer cells to develop transgenic mouse lines, allowing titratable initiation of pancreatic tumors | This method paves the way for the investigation of molecular alterations, driving each step of pancreatic cancer development. |
| Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting [75] | Exosome | Ovarian cancer | Tumor-derived exosomes loaded with cas9 and PARP-1 sgRNA expression plasmids via electroporation | CRISPR/Cas9-induced inhibition of PARP-1 resulted in ovarian cancer cell apoptosis and increased sensitivity to chemotherapeutic agent (cisplatin). |
| Exosome-mediated delivery of CRISPR/Cas9 for targeting of oncogenic KrasG12D in pancreatic cancer [76] | Exosome | Pancreatic cancer | Exosomes loaded with CRISPR/Cas9 capable of targeting the mutant KrasG12D oncogenic | Suppressed proliferation and hampered tumor growth in syngeneic subcutaneous and orthotopic models of pancreatic cancer. |
| Tropism-facilitated delivery of CRISPR/Cas9 system with chimeric antigen receptor-extracellular vesicles against B-cell malignancies [77] | EVs | B cell malignancies | The CRISPR/Cas9 system aiming at the MYC oncogene, was loaded into selective EVs having anti-CD19-CAR on their surface. | The induced CRISPR/Cas9-mediated loss-of-function mutations of the MYC gene in CD19 + cells exhibited the significant potential of this approach. |
| Efficient RNA drug delivery using red blood cell extracellular vesicles [78] | RBC extracellular vesicles (RBCEVs) | AML M5 | Electroporation of HA-tagged Cas9 mRNA and gRNA of human mir-125b-2 into RBCEVs, and used them to treat MOLM13 cells. | Exosomes successfully transfected both human cells and xenograft mice, with no notable cytotoxicity. |
| Exosome–Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs [79] | Exosome | Mesenchymal stem cells (MSCs) | Hybrid exosomes-liposomes capable of carrying large cargoes such as CRISPR/Cas9 System | Hybrid exosomes efficiently delivered CRISPR/dCas9 to inhibit the expression of mRunx2 and hCTNNB1 in MSCs |
| In vitro and in vivo RNA inhibition by CD9-HuR functionalized exosomes encapsulated with miRNA or CRISPR/dCas9 [80] | Exosome | Recipient cells | Since Hur is an RNA binding protein, CD9-HuR exosomes could efficiently encapsulate the miR-155 or CRISPR/dCas9 | Increased RNA cargo loading into engineered exosomes |
| Activation of Necroptosis by Engineered Self Tumor-Derived Exosomes Loaded with CRISPR/Cas9 [55] | Exosome | GBM, Thyroid cancer, lung adenocarcinoma | Engineered exosomes for TNFR activation and impairment of IAP 1/2 and Caspase 8 expression loaded with CRISPR/CAS9 | Activation of TNFR and subsequently inactivation of IAP 1/2 and Caspase results in blocked cell survival and Necroptosis activation. |
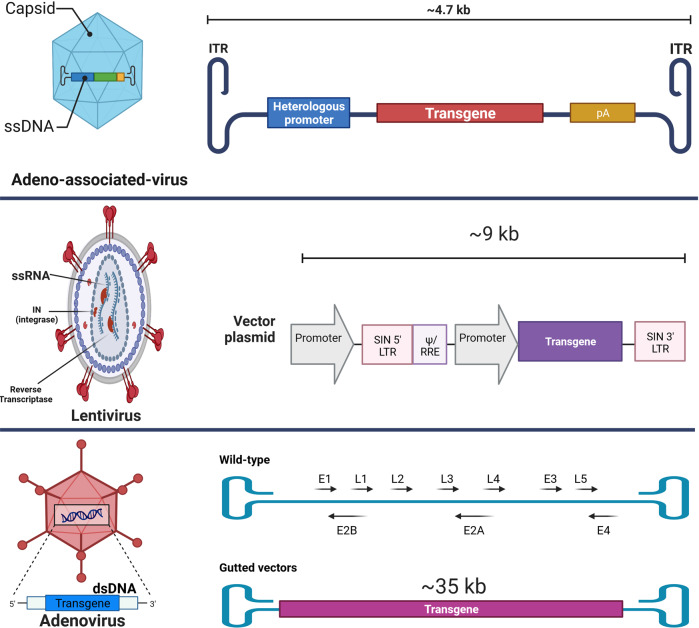
Viral vectors
Viruses are genuine vehicles for gene delivery. Mammalian gene therapy has been made possible with recombinant and pseudotyped viral vectors. The most frequently used viral vectors for CRISPR/Cas delivery are adeno-associated viruses (AAVs) [55], adenoviral vectors (AdVs) [56], and lentiviral vectors (LVs) [57] (Fig. 2), which are being used in clinical trials.
Fig. 2. Commonly used viral vectors for gene delivery.
AAVs are single-stranded DNA viruses with no envelope and small size that are not pathogenic to humans. The gutted Adenoviral vectors are double-strand DNA viruses devoid of most genes from their wild-type, although still capable of transducing a broad spectrum of both dividing and non-dividing cells with a capacity of carrying genetic cargo up to 35 kb. Lentiviral vectors, primarily derived from HIV-1, are capable of integrating their transgene (~9 kb) into the human genome and are suitable for long-term gene expression.
Adeno-associated viruses (AAVs)
AAVs are small, non-enveloped single-stranded DNA viruses that are not pathogenic to humans. These members of the Parvoviridae family have attracted the attention of researchers as gene delivery systems (Fig. 3) [58]. Despite the fact that around 80% of the general population is sero-positive for these viruses, no connection between AAVs and human diseases has been reported. Various characteristics of AAVs, including relatively low immunogenicity, cytotoxicity, and chromosomal integration probability, make them quintessential delivery systems for CRISPR/Cas, particularly in vivo [59, 60]. Furthermore, different types of AAVs are available for gene delivery into a variety of cells, such as lung, heart, neuron, and muscle cells, thus making them excellent tropism vectors for tissue-specific applications [61].
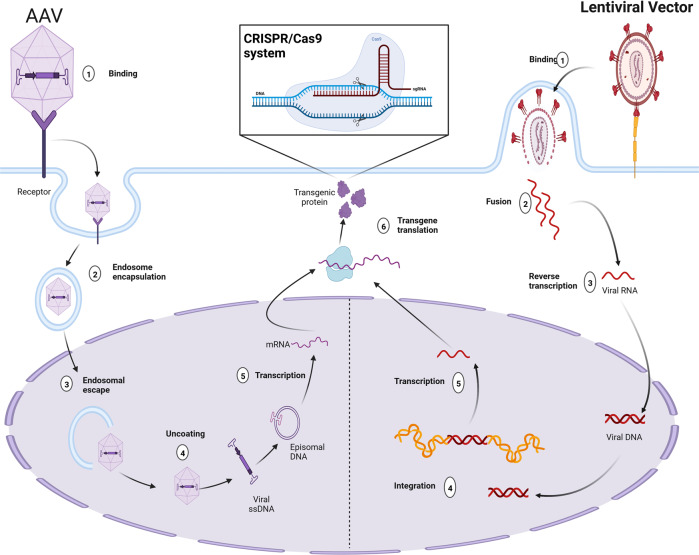
Fig. 3. Viral Vectors encoding CRISPR/Cas.
Mechanism of action of two types of viral vectors for delivery of CRISPR/Cas systems: integrative (lentiviral vectors) and non-integrative (Adenoviral and Adeno-associated viral (AAV) vectors).
To some degree, AAVs still have the ability to integrate their genes into the host genome, to overcome this limitation recombinant AAV has been developed [62]. AAVs’ gene called Rep is responsible for the production of Rep proteins (Rep78, Rep68, Rep52, Rep40), involved in the packaging of the viral genome, replication, gene expression, and integration of the genetic material [63]. For accurate site-specific integration, AAVs require Rep proteins, particularly Rep78 and 68, although in recombinant forms of AAVs the Rep gene has been removed [64]. Overall, these recombinant AAVs have proven to be effective gene delivery systems; for instance, delivering CRISPR/Cas9 to mice bearing a mutation in the low-density lipoprotein receptor (LDLR) gene exhibited therapeutic effects [65]. Thus far, many AAV-based gene therapies have achieved FDA approval, for example, in Pompe disease and inherited retinal disease (IRD) retinal pigment epithelium (RPE)65-LCA (LCA2), which indicates the effectiveness of AAVs vectors [66]. Furthermore, efficient delivery of the CRISPR/Cas9 system by AAVs has paved the way for disease modelings, such as neurodegenerative abnormalities, muscular dystrophy, liver diseases, and sickle cell disease [67–71].
Although AAVs have impressive achievement records, their limited packaging capacity and genome length limit their ability to harbor genetic payloads that exceed 5 kb. In this case, carrying large cargoes like SpCas9 protein remains a considerable limitation [72]. A number of methods have been designed by scientists to circumvent the barricade of the AAVs’ limited packaging capacity. One of which is to transduce cells with AAVs only carrying sgRNA, knowing that these cells have already been induced to express the Cas9 protein. Another strategy is to co-transduce the cells using two AAVs tagged distinctively, one carrying sgRNA and another the Cas9 protein. Nevertheless, there are concerns about this approach since it demands a high viral dose and accurate tracking of the vectors in order to confirm co-transduction [73, 74].
There are some genes that are even larger than the capacity of dual AVV vectors (9 kb); for example, the CDH23 (Usher syndrome) and DMD (muscular dystrophy) genes are about 10 kb and 11.1 kb, respectively. In order to deliver these kinds of genes into host cells, scientists have designed a triple AAV system [75, 76].
Since a novel approach called CRISPR/Cas-mediated base and prime editing uses dead or Cas9 nickase (nCas9) enzymes, it can modify the CRISPR-Cas system so that it is possible to use single-vector delivery and overcome challenges pertaining to limited viral-vector packaging capacity [77]. Further characteristics of this system include not inducing DSB, does not need a DNA donor template, and has great capability for editing non-dividing cells [78]. AAVs have proven to be potential systems for delivering CRISPR DNA base-editing tools [58]; for instance, a study reported that in vivo delivery of the CRISPR/Cas-based cytidine-base editor by dual AAVs could treat amyotrophic lateral sclerosis (ALS) in an animal model. It has been shown that the AAV-delivered prime editor is an effective tool for correcting pathogenic alleles and cancer modeling in adult mice, because it has a significant lower off-target effect than the CRISPR/Cas-based base editor [78, 79]. Although AAV-base and prime editing systems improve some drawbacks related to AAV-mediated CRISPR/Cas9 delivery, the limitations, including viral-induced immune response, vector persistency, and off-target activity, are still present to some extent [77].
The packaging capacity limitation of AAVs can be managed by utilizing other classes of Cas protein like Cas12a or different forms of Cas9 having smaller sizes, such as SaCas9 and St1-Cas9 [55, 80–82]. Moreover, breaking large transgenes into two separate cuts and packaging each half into two sets of AAVs can extend AAVs’ delivery capacity. This approach is carried out by a process called intein-mediated trans-splicing, which to some degree is similar to mRNA splicing [83, 84]. This method has been used to deliver base and prime editing systems, which were found to be quite promising [85, 86]. Another challenging issue for in vivo delivery is the pre-existing immunogenicity against the bacterial Cas9 protein and AAV capsid. However, the AAV capsid can be improved by altering its antigens, creating a chimeric AAV capsid to decrease antibody response and evade the immune system [87, 88].
The characteristics of AAVs, which include low immunogenicity and cytotoxicity, as well as a slim chance of chromosomal integration, make them the ideal delivery vessels for CRISPR/Cas system. Alongside these, having various types and diverse delivery approaches, like dual and triple AAV vectors, covers their limitations compared to other viral vectors and brings them to the forefront of gene delivery.
Adenoviral vectors
The AdV is a double-strand DNA virus that can transduce a broad spectrum of both dividing and non-dividing cells. These vectors can carry a genetic cargo up to 37 kb, and following transduction, they create an episomal DNA adjacent to the host DNA instead of integrating into the genome. Noteworthy that the episomal gene expression of AdVs removes the off-target effects, a limitation in CRISPR/Cas-based gene editing [89, 90].
There are different generations of AdVs; in the first one, the E1 gene was deleted, although using this generation could provoke acute and chronic immune responses [88]. Furthermore, in the second generation, the E2 and E4 genes of AdV were removed to diminish the immune response. Of note, the capacity of second-generation AdVs (~14 kb) for incorporating transgene is substantially superior to the first-generation (~8 kb) [91, 92]. The helper-dependent vectors (gutted vectors), which represent the third generation of AdV vectors, do not contain viral genes that allow cloning up to 35 kb; therefore, they move beyond the size limitation barrier for delivering the CRISPR/Cas9 system. It is worth mentioning that, like second-generation AdVs, this generation does not trigger chronic immune responses [93, 94].
The recombinant AdV5 has shown potential for in vivo CRISPR/Cas9-mediated knock-in of the human alpha-1-antitrypsin gene in mice, and it has also been demonstrated that gene expression continued for more than 200 days [95]. In a recent study, AdV-delivered CRISPR/Cas12a to human hepatocytes indicated its potential toward gene editing of human cells, although the provoked immune system against the vector and Cas protein was reported [96]. Furthermore, AdV structural proteins (Hexon, penton, and fiber) are quite manipulatable, which is advantageous for creating AdVs with tissue-specific tropism. Moreover, these properties, along with the fact that AdVs are safe for clinical trials, can be manufactured at large scales, and are cost-effectively manufactured, make them indispensable for CRISPR/Cas delivery [56, 97]. These beneficial features could be verified by the fact that AdVs became the top choice for creating mRNA vaccines for Coronavirus disease 2019 (COVID-19). This betokens the idea that AdVs can be produced as a delivery system on a global scale [98].
All in all, in CRISPR/Cas-based gene editing, AdVs’ episomal gene expression eliminates off-target effects. Also, its huge packaging capacity allows for loading large genetic payloads, which is a concern in other viral vectors.
Lentiviral vectors
The HIV-1-derived LVs are single-stranded RNA (ssRNA) viruses primarily used for integrating the desired transgene into dividing and non-dividing cells (Fig. 3). Their delivery capacity for genetic cargo is around 9 kb, encompassed by a lipid-enriched capsid. There are four generations of LVs, where the third and fourth generations are mostly safe for clinical applications [99].
The effectiveness of CRISPR/Cas-based gene editing greatly depends on the delivery of its components into the host cell. As LVs are a great armamentarium for gene delivery, and the CRISPR/Cas system was proven to be functional in human cells, LVs expressing the whole system were chosen [100]. Like shRNAs, LVs carrying CRISPR/Cas9 systems have also been designed initially in a library model with numerous sgRNAs [101]. A study showed that an LV-based genome-scale CRISPR-Cas9 knockout (GeCKO) library that targets more than 18,000 human genes is quite valuable for negative and positive selection screening, which are generally utilized for in vitro assays to determine disturbances in cells, especially cancer cells affected by various stimuli [100]. In particular, this library has been used to specify genes critical for the viability of cancer cells and pluripotent stem cells. It also allowed for screening of loss of function in genes that cause resistance to vemurafenib in melanoma cells [100]. Further studies have also developed LV-based CRISPR/Cas libraries which have resulted in the identification of novel tumor-suppressor genes involved in myeloid leukemia (Nf1, Ezh2, Dnmt3a, Tet2, and Runx1), and fetal hemoglobin reinduction (BCL11A) [102, 103].
With time, the capabilities of the LV-delivered CRISPR/Cas9 system are expanding into the fields of targeted therapy for HIV-1 and HBV infections as well as the treatment of genetically defective diseases, such as cystic fibrosis and neurodegenerative diseases [104]. Additionally, other methods, including base and prime editing CRISPR systems and epigenetic modifiers, have also been paired with LVs [105]. Since LVs integrate the encoding CRISPR/Cas transgene into the host genome, and due to their long-lasting gene expression, the possibility of off-target effects could be raised as a concern. To address this risk, integration-deficient lentiviral vectors (IDLV) were developed to provide an impermanent CRISPR/Cas expression [106].
In unresectable hepatocellular carcinoma, HIF-1α expression is capable of worsening the prognosis of the disease and hampering the overall improvement of patients. A recent study knocked out the HIF-1α in mice using an LV-delivered CRISPR/Cas9 system where it drastically diminished the HIF-1α expression in the tumor tissues three days post-injection, showcasing valuable antitumor effects [107]. Chromosomal translocations creating fusion oncogenes are frequent in certain cancers and are substantial tumorigenesis factors [108, 109]. For example, a tyrosine kinase produced by BCR-ABL rearrangement can develop chronic myeloid leukemia (CML) [110]. Tyrosine kinase inhibitors (TKIs) such as Imatinib can significantly inhibit the product of BCR-ABL, though there are reports of drug resistance against these TKIs [111]. Accordingly, Martinez-Lage and colleagues used an LV-based CRISPR/Cas9 system to induce deletion in introns of the BCR-ABL rearranged gene, which resulted in a significant deletion of the BCR transactivation domain as well as frameshift mutation in the DNA-binding domain of ABL [112].
LV has always been an intriguing viral vector for delivering genes, particularly for clinical purposes. This is because the transgene can be integrated into the host genome and expressed for a long time. Furthermore, the desired gene can be passed along to new cells following cell division, thus ensuring that CRISPR/Cas functions are expressed for a long time to come.
Extracellular vesicles (EVs)
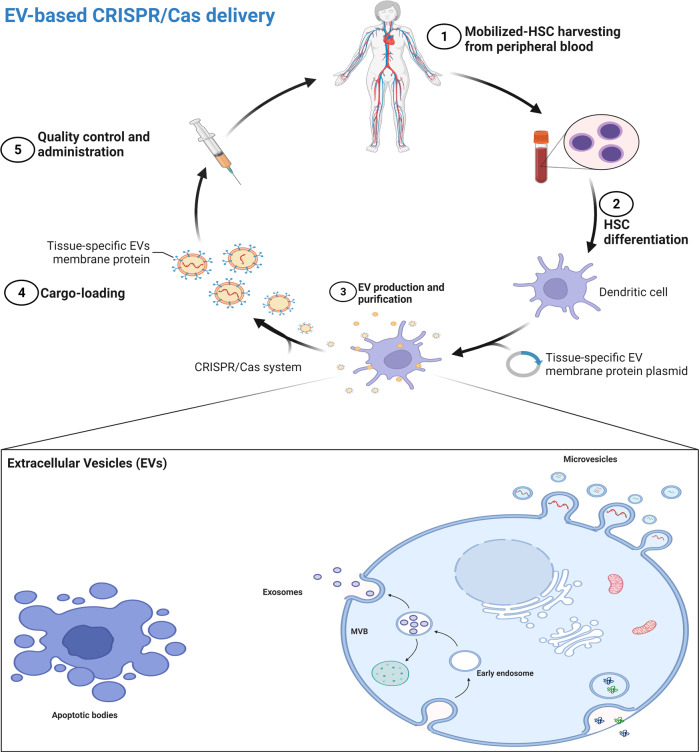
Extracellular vesicles are nano-scaled non-viral delivery vessels that can be used for various purposes, one of which is to be used as a targeted delivery system. EVs are lipid-coated particles produced by different cells with the innate objective of cell–cell transportation of cargoes such as genetic material, and proteins [113]. EVs can be grouped into three main categories: microvesicles (MVs), exosomes, and apoptotic bodies. They are different in packaging capacity, biogenesis, function, and releasing mechanism. Among them, exosomes having 30–150 nm in diameter are genuine options for selective delivery of proteins, and genetic components, including CRISPR/Cas system (Fig. 4) [113–115].
Fig. 4. EV-based gene delivery.
There are different types of EVs, including microvesicles, exosomes, and apoptotic bodies, that can be collected from human cells. These EVs can carry proteins, DNA, and different types of RNA from one cell to another. These exosomes can be engineered to target a specific tissue while carrying our desired cargo like CRISPR/Cas system. This approach allows for autologous tissue-specific gene editing.
Exosomes have drawn much attention for being used in cancer diagnosis and therapy. Exosomes secreted from tumor cells could be an excellent option for tumor-targeted therapy since they are similar to their source and, therefore, more likely to be taken up by the cells. Cancer therapy with a cell-specific tropism approach can be facilitated by controlling this feature and loading desired cargoes into tumor-derived exosomes. Accordingly, a study indicated that loading tumor-derived exosomes with a cancer therapeutic agent called Doxil and injecting it systemically into the tissue of origin can result in promoted tumor suppression compared to the application of the drug alone [116]. In an intriguing strategy, biocompatible porous silicon nanoparticles (PSiNPs) harboring doxorubicin called DOX@E-PSiNPs were introduced to isolated tumor cells (Fig. 5). Afterward, tumor cell-released exosomes sheltering DOX@E-PSiNPs were injected systematically into mice, which were uptaken by both bulk cancer cells and cancer stem cells (CSCs), resulting in considerable tumor suppression [117]. The promising potential of exosomes for CRISPR/Cas delivery in tumor cells can be extrapolated from their ability to deliver selective and efficient delivery systems.
Fig. 5. Tumor-derived exosomes.
Transfecting tumor cells with DOX@E-PSiNPs, CRISPR/Cas system, and viral vector plasmids while inducing the tumor cells to produce exosomes can provide tumor-derived exosomes encapsulating our desired cargo. Since these exosomes originated from the tumor, they can be harvested and injected systematically back into tumor cells for therapeutic purposes.
According to the potency of exosomes, a study used tumor-derived exosomes for in vivo targeted delivery of CRISPR/Cas9 into SKOV3 xenograft mice cells with ovarian cancer. They compared epithelial cell-derived exosomes and cancer-derived exosomes to deliver a CRISPR/Cas9 system capable of suppressing the expression of poly (ADP-ribose) polymerase-1 (PARP-1). The results showed significant apoptosis in ovarian cancer cells and a synergic enhancement of chemosensitivity to cisplatin. The combination of both therapeutic approaches resulted in 57% inhibition of cancer proliferation, almost twice the effect of exosomes- or cisplatin-sole treatment [118]. One serious concern about using exosome-mediated gene delivery, especially in the case of delivering the CRISPR/Cas system, is the potential for off-target negative impacts on peripheral and distant tissues.
In addition to the packaging capabilities of exosomes, engineering them to be targeted delivery vehicles is an essential goal. Alvarez-Erviti and colleagues produced brain-targeting exosomes carrying short interfering RNA (siRNA) from engineered dendritic cells capable of expressing Lamp2b, an exosomal membrane protein, fused with nervous system-specific rabies viral glycoprotein (RVG). The results of in vivo delivery of these exosomes in mice showcased strong therapeutic potential and no non-specific uptake by other tissues [119]. Another study used a similar approach to deliver miRNA in the cartilage as a treatment for osteoarthritis, which exhibited a significant potential in targeting hard-penetrating tissues [120, 121].
Necroptosis is a form of necrosis in response to a pathogen or inflammation, in which cells undergo non-programmed cell death. Of note, in this pathway caspase 8 and IAP1/2, which are involved in cell survival and apoptosis, should be suppressed. In a recent study, a particular kind of tumor-derived exosomes was designed to have TNF receptor (TNFR) ligands on their surface. These exosomes were carrying CRISPR/Cas9 systems which are capable of inhibiting caspase 8 and IAP1/2; therefore, following exosome-mediated activation of TNFR signaling and CRISPR/Cas9-mediated inactivation of caspase 8 and IAP1/2, tumor cells underwent necroptosis (Fig. 6). The advantage of necroptosis over apoptosis is that the former also provokes T-cells to eliminate the remaining cancer cells [122].
Fig. 6. Necroptosis induction using exosomes.
Engineered exosomes with TFNα on their surface, carrying two vectors of the CRISPR/Cas system capable of inhibiting cIAPs and caspase 8. In this approach, the exosome-TNFα ligand provokes TNFR signaling pathways, which can activate the cIAPs, propelling the cell toward survival. CRISPR/Cas9-mediated inhibition of cIAPs alongside active caspase 8 results in apoptosis of cancer cells. Moreover, dual inhibiting of cIAPs and caspase 8 alongside TNFR signaling lead to necroptosis. The advantage of necroptosis over apoptosis is that the former also provokes T-cells to eliminate the remaining cancer cells.
Unlike viruses, exosomes can be modified in a matter of size. Hybrid exosomes, a combination of cell-derived exosomes and synthetic liposomes, have been developed to carry large cargoes such as CRISPR/Cas system [123]. Not only do hybrid exosomes have greater packaging capacity but also, due to the positive charge of the liposomes, they interact more efficiently with negatively charged RNA and DNA to uptake them by membrane fusion. This approach can remove the need for loading genetic material into the exosomes by mechanical methods such as electroporation [123]. Noteworthy that this alteration to exosomes had no effects on the efficiency of their cell-type tropism and uptake [124]. This method has been used by Lin and colleagues to efficiently deliver CRISPR/Cas9 expression vectors to mesenchymal stem cells (MSCs), which are hard to transfect [125].
Exosomes can also be optimized with DNA aptamer, a short synthetic oligonucleotide, in order to use exosomes as targeted delivery systems. DNA aptamers are cost-effective, readily available, and non-provocative for the immune system; thus making them a suitable alternative to antibodies or other probes [126, 127]. A recent study used cholesterol-anchored valency-controlled tetrahedral DNA nanostructures (TDNs) conjugated with DNA aptamer on the surface of exosomes to selectively deliver the CRISPR/Cas system inhibiting the WNT10B gene into hepatocellular carcinoma cells in vitro, ex vivo, and in vivo. The promising results of targeted gene suppression in hepatocellular carcinoma cells by this method highlight the outstanding potential of EVs for being used as labeled and targeted delivery systems for CRISPR/Cas system [128].
The risk of nucleated cell-derived EVs-mediated horizontal gene transfer could be viewed as a limitation for EVs, though RBC-derived EVs bypass this safety-related issue. Since RBCs are non-nucleated, O blood group RBCs can be used as a universal source for the harvesting of EVs without DNA. The RBC-EVs have been used in vivo and in vitro for delivering CRISPR/Cas9, demonstrating high transfection efficiency and without detectable cytotoxicity [129]. Further, Pham and colleagues reported that RBC-EVs could be targeted for selective delivery of cargoes. Accordingly, they conjugated the epidermal growth factor receptor (EGFR)-targeting peptide to paclitaxel-carrying EVs by Sortase A and OaAEP1 ligase. The results indicated that EVs could efficiently deliver the chemotherapeutic agent to EGFR-positive lung cancer cells, causing significant apoptosis and shrinking the tumor [129]. It is plausible to extrapolate the considerable potential of targeted RBC-EVs for the delivery of CRISPR/Cas9 systems to specific cancer cells.
EVs, especially tumor-derived EVs have been shown to be extremely effective in bypassing tumor defense mechanisms in CRISPR/Cas-based cancer therapy. They come in different sizes and can contain different genetic materials. These qualities, as well as many others, including easy manipulation, reliable gene delivery, and high safety, make EVs formidable cancer therapy weapons.
Vexosomes: exosome-enveloped viral vectors
Exosomes-enveloped viral vector or vexosome is a novel gene delivery approach that confers exosome features such as a broad spectrum of cell tropism, almost non-immunogenic, and scalability, to viral vectors for enhancing gene therapy [130]. The production of vexosomes requires infecting the packaging cells with plasmids encoding the viral genome. The cells’ cytoplasm becomes enriched with the produced viral genome and proteins, which are recognized by the cell’s receptors in the plasma membrane, endosomes, and phagosomes. Therefore, EVs encapsulate the viral components upon their secretion from the cells. Saari and co-workers used this method to produce capsid-free EV-based vectors carrying oncolytic AAV components (AAV/EVs) into cancer cells [131]. Vexosomes have not been used for delivery of CRISPR/Cas system, though their potential in combining the characteristics of viral vectors and EVs is promising for efficient gene editing.
AAV/EVs can be modified to deliver genes more selectively by including targeting peptides on their surfaces. Multiple studies have reported benefits from using targeted AAV/EVs in vivo and in vitro; for example, Wood et al. reported that these vectors can pass the blood-brain barrier and effectively transduce neural cells [132]. Different serotypes of AAV, including AAV1, AAV6, and AAV9, have been used for targeted delivery of transgenes into cochlear and vestibular hair cells, neurons, and oligodendrocytes, respectively [133–136]. It indicates the compatibility of this approach for gene delivery into various cell types.
The EVs encapsulating the AAVs can be used as a trojan horse to circumvent the pre-existing immunity against the viral vectors. In this context, a recent study systematically injected AAV9/EVs into mice with pre-existing immunity to AAV9, and the results demonstrated successful evading from the immune system [137]. Furthermore, this approach has shown that it can reduce the number of injections required for efficient delivery of vectors, reducing the risk of AAV-induced cytotoxicity [119, 138–140]. Despite the benefits of this delivery method, it has not yet been explored as a delivery method for the CRISPR/Cas system.
Combining the power and potential of EVs and viral vectors promises a great future in the field of gene delivery, especially for CRISPR/Cas delivery. Due to the fact that this approach is new, it will be more interesting for researchers to explore its full potential as a cancer therapy.
Clinical translation for cancer diagnostics and therapies
The CRISPR/Cas technology provides a quintessential gene-editing application [80, 141], though in the case of clinical cancer therapy it has not been a major player. That said, the potential of CRISPR-based genome editing is promising for cancer diagnostics and therapies in the near future (Table 2).
Table 2.
Clinical trials on CRISPR/Cas-mediated cancer therapy (https://www.clinicaltrials.gov).
| Study title | Status | Conditions | Interventions | Gender | Age | Phases | Clinical trial identifier |
|---|---|---|---|---|---|---|---|
| Study of CRISPR-Cas9 Mediated PD-1 and TCR Gene-knocked Out Mesothelin-directed CAR-T Cells in Patients with Mesothelin Positive Multiple Solid Tumors. | Recruiting | Solid Tumor | Biological: anti-mesothelin CAR-T cells | All | 18–70 | Phase 1 | NCT03545815 |
| A Study of Metastatic Gastrointestinal Cancers Treated with Tumor Infiltrating Lymphocytes in Which the Gene Encoding the Intracellular Immune Checkpoint CISH Is Inhibited Using CRISPR Genetic Engineering | Recruiting | Gastrointestinal Cancer | Drug: Cyclophosphamide|Drug: Fludarabine| Biological: Tumor-Infiltrating Lymphocytes (TIL)|Drug: Aldesleukin | All | 18–70 | Phase1 and Phase 2 | NCT04426669 |
| A Safety and Efficacy Study of TALEN and CRISPR/Cas9 in the Treatment of HPV-related Cervical Intraepithelial Neoplasia | Unknown status | Human Papillomavirus-Related Malignant Neoplasm | Biological: TALEN|Biological: CRISPR/Cas9 | Female | 18–50 | Phase 1 | NCT03057912 |
| Study of PD-1 Gene-knocked Out Mesothelin-directed CAR-T Cells with the Conditioning of PC in Mesothelin Positive Multiple Solid Tumors | Unknown status | Solid Tumor | Biological: Mesothelin-directed CAR-T cells | All | 18–70 | Phase 1 | NCT03747965 |
| A Safety and Efficacy Study Evaluating CTX110 in Subjects With Relapsed or Refractory B-Cell Malignancies (CARBON) | Recruiting | B-cell Malignancy|Non-Hodgkin Lymphoma|B-cell Lymphoma|Adult B Cell ALL | Biological: CTX110 | All | 18< | Phase 1 | NCT04035434 |
| PD-1 Knockout Engineered T Cells for Advanced Esophageal Cancer | Completed | Esophageal Cancer | Other: PD-1 Knockout T Cells | All | 18–80 | Not Applicable | NCT03081715 |
| PD-1 Knockout Engineered T Cells for Metastatic Non-small Cell Lung Cancer | Completed | Metastatic Non-small Cell Lung Cancer | Drug: Cyclophosphamide|Other: PD-1 Knockout T Cells | All | 18–70 | Phase 1 | NCT02793856 |
| Stem Cells in NF1 Patients With Tumors of the Central Nervous System | Suspended | Neurofibromatosis Type 1|Tumors of the Central Nervous System | Diagnostic Test: Collection of Stem Cells | All | Child, Adult, Older Adult | NCT03332030 | |
| A Safety and Efficacy Study Evaluating CTX130 in Subjects With Relapsed or Refractory T or B Cell Malignancies (COBALT-LYM) | Recruiting | T Cell Lymphoma | Biological: CTX130 | All | 18< | Phase 1 | NCT04502446 |
| First-time-in-human (FTIH) Study of GSK3145095 Alone and in Combination With Other Anticancer Agents in Adults With Advanced Solid Tumors | Terminated | Neoplasms, Pancreatic | Drug: GSK3145095|Drug: Pembrolizumab | All | 18< | Phase 2 | NCT03681951 |
| Lavage of the Uterine Cavity for Diagnosis of Ovarian Cancer | Recruiting | Ovarian Cancer | Other: Biospecimen Collection|Other: Laboratory Biomarker Analysis|Device: Lavage|Other: Pap Smear | Female | 18< | Not Applicable | NCT03606486 |
| TGF²R-KO CAR-EGFR T Cells in Previously Treated Advanced EGFR-positive Solid Tumors | Recruiting | Solid Tumor| EGFR Overexpression | Biological: TGF²R-KO CAR-EGFR T Cells | All | 18–75 | Phase 1 | NCT04976218 |
| PD-1 Knockout Engineered T Cells for Castration Resistant Prostate Cancer | Withdrawn | Hormone Refractory Prostate Cancer | Biological: PD-1 Knockout T Cells|Drug: Cyclophosphamide|Drug: IL-2 | Male | 45–85 | NCT02867345 | |
| PD-1 Knockout Engineered T Cells for Muscle-invasive Bladder Cancer | Withdrawn | Invasive Bladder Cancer Stage IV | Biological: PD-1 Knockout T Cells|Drug: Cyclophosphamide|Drug: IL-2 | All | 18–75 | Phase 1 | NCT02863913 |
| CRISPR (HPK1) Edited CD19-specific CAR-T Cells (XYF19 CAR-T Cells) for CD19 + Leukemia or Lymphoma. | Recruiting | Leukemia Lymphocytic Acute (ALL) in Relapse|Leukemia Lymphocytic Acute (All) Refractory|Lymphoma, B-Cell|CD19 Positive | Genetic: XYF19 CAR-T cell|Drug: Cyclophosphamide|Drug: Fludarabine | All | 18–55 | Phase 1 | NCT04037566 |
| Safety and Efficacy of CT125A Cells for Treatment of Relapsed/Refractory CD5 + Hematopoietic Malignancies | Not yet recruiting | CD5+ Relapsed/Refractory Hematopoietic Malignancies|Chronic Lymphocytic Leukemia (CLL)|Mantle Cell Lymphoma (MCL)|Diffuse Large B-cell Lymphoma (DLBCL)|Follicular Lymphoma (FL)|Peripheral T-cell Lymphomas (PTCL) | Biological: CT125A cells|Drug: Cyclophosphamide, fludarabine | All | 18–70 | Early Phase 1 | NCT04767308 |
| A Safety and Efficacy Study Evaluating CTX120 in Subjects With Relapsed or Refractory Multiple Myeloma | Recruiting | Multiple Myeloma | Biological: CTX120 | All | 18< | Phase 1 | NCT04244656 |
| Study of Base Edited CAR7 T Cells to Treat T Cell Malignancies (TvT CAR7) | Recruiting | Relapsed/Refractory T-cell Acute Lymphoid Leukemia | Biological: Cryopreserved BE CAR7 T cells (BE752TBCCLCAR7PBL) | All | 6–16 | Phase 1 | NCT05397184 |
| PD-1 Knockout EBV-CTLs for Advanced Stage Epstein-Barr Virus (EBV) Associated Malignancies | Unknown status | Stage IV Gastric Carcinoma|Stage IV Nasopharyngeal Carcinoma|T-Cell Lymphoma Stage IV|Stage IV Adult Hodgkin Lymphoma|Stage IV Diffuse Large B-Cell Lymphoma | Drug: Fludarabine|Drug: Cyclophosphamide|Drug: Interleukin-2 | All | 18–75 | Phase 1|Phase 2 | NCT03044743 |
| Study Investigating NTLA-5001 in Subjects With Acute Myeloid Leukemia | Recruiting | Acute Myeloid Leukemia | Genetic: Arm 1: NTLA-5001|Genetic: Arm 2: NTLA-5001 | All | 18< | Phase 1|Phase 2 | NCT05066165 |
| A Study Evaluating UCART019 in Patients With Relapsed or Refractory CD19+ Leukemia and Lymphoma | Unknown status | B Cell Leukemia|B Cell Lymphoma | Biological: UCART019 | All | 12–75 | Phase 1|Phase 2 | NCT03166878 |
| A Long-term Follow-up Study of Patients Who Received VOR33 | Recruiting | Leukemia, Myeloid, Acute | Genetic: VOR33 | All | Child, Adult, Older Adult | NCT05309733 | |
| NY-ESO-1-redirected CRISPR (TCRendo and PD1) Edited T Cells (NYCE T Cells) | Terminated | Multiple Myeloma |Melanoma| Synovial Sarcoma |Myxoid/Round Cell Liposarcoma | Biological: NY-ESO-1 redirected autologous T cells with CRISPR edited endogenous TCR and PD-1| Drug: Cyclophosphamide|Drug: Fludarabine|Device: NY-ESO-1 expression testing | All | 18< | Phase 1 | NCT03399448 |
| A Safety and Efficacy Study Evaluating CTX130 in Subjects With Relapsed or Refractory Renal Cell Carcinoma (COBALT-RCC) | Recruiting | Renal Cell Carcinoma | Biological: CTX130 | All | 18< | Phase 1 | NCT04438083 |
| CRISPR-Edited Allogeneic Anti-CD19 CAR-T Cell Therapy for Relapsed/Refractory B Cell Non-Hodgkin Lymphoma | Recruiting | Lymphoma | Genetic: CB-010|Drug: Cyclophosphamide|Drug: Fludarabine | All | 18< | Phase 1 | NCT04637763 |
| Programmed Allogeneic CRISPR-edited T Cells Engineered to Express Anti-CD19 Chimeric Antigen Receptor (PACE CART19) in Patients With Relapsed Or Refractory CD19 + Leukemia and Lymphoma | Withdrawn | Acute Lymphoblastic Leukemia |Chronic Lymphocytic Leukemia| Non-Hodgkin Lymphoma | Biological: PACE CART19 | All | 18< | Phase 1 | NCT05037669 |
| TT52CAR19 Therapy for B-cell Acute Lymphoblastic Leukemia (B-ALL) | Recruiting | B Acute Lymphoblastic Leukemia | Drug: PBLTT52CAR19 | All | 6 Months to 18 Years | Phase 1 | NCT04557436 |
| A Feasibility and Safety Study of Universal Dual Specificity CD19 and CD20 or CD22 CAR-T Cell Immunotherapy for Relapsed or Refractory Leukemia and Lymphoma | Unknown status | B Cell Leukemia Cell Lymphoma | Biological: Universal Dual Specificity CD19 and CD20 or CD22 CAR-T Cells | All | 12–70 | Phase 1|Phase 2 | NCT03398967 |
| TACE Combined With PD-1 Knockout Engineered T Cell in Advanced Hepatocellular Carcinoma. | Unknown status | Advanced Hepatocellular Carcinoma | Procedure: Transcatheter arterial chemoembolization Biological: PD-1 knockout engineered T cells | All | 18–70 | Phase 1 | NCT04417764 |
| PD-1 Knockout Engineered T Cells for Metastatic Renal Cell Carcinoma. | Withdrawn | Metastatic Renal Cell Carcinoma | Biological: PD-1 Knockout T Cells Drug: Cyclophosphamide Drug: IL-2 | All | 18–75 | Phase 1 | NCT02867332 |
CRISPR–Cas9-targeted fragmentation of DNA can be used as a means to pinpoint the changes in cancer-specific sequence. For example, CRISPR/Cas9 can detect microsatellite sequences, called short tandem repeats (STRs), which can serve as cancer markers. Most STR assays depend on PCR amplicons, which are limited to a few. In comparison, CRISPR/Cas9-mediated STR-sequencing is capable of accurately and sensitively analyzing more than 2000 STRs in parallel [142, 143]. There are regions in the human genome housing complex megabase-sized fragments, containing biologically essential genes. Owing to their complexity and variations, these regions are not thoroughly uncovered, although Baker and colleagues used a potential approach called CISMR (CRISPR-mediated isolation of specific megabase-sized regions of the genome), for targeted sequencing of these fragments [144]. Moreover, if CRISPR-mediated fragmentation of genomic DNA is combined with duplex sequencing, correcting sequencing errors, it can result in ultra-accurate sequencing with low DNA input, termed CRISPR-DS [145]. As compared to duplex sequencing, CRISPR-DS had a superior capability in detecting TP53 mutations in the peritoneal fluid of ovarian cancer patients using 10 to 100-fold less DNA than the latter [145]. In light of CRISPR-DS’ diagnostic potential, it has now reached the stage of clinical trials [NCT03606486]. Other highly accurate enzymatic nucleic acid detection systems have also been developed for cancer-related mutations, called DETECTR and SHERLOCK, which use Cas12a and Cas13, respectively [25, 28, 146, 147].
Additional to cancer diagnosis, CRISPR has made its way to be applied for clinical cancer treatment, which demonstrates the astonishing potential of a recently developed gene-editing tool. Initially, the CRISPR/Cas9 system was used for treating non-small-cell lung carcinoma (NSCLC) at West China Hospital, Sichuan University, in 2016 [148–150]. In that study, patients’ T-cells were genetically engineered not to express the T-cell activation inhibitor, PD-1. Accordingly, this approach has also been used for cancer of other tissues, including bladder, prostate, renal, and esophageal [150]. It is noteworthy that some of these clinical trials have withdrawn their studies [151].
The development of CAR T cell receptors has made significant strides in cancer treatment, with Kymriah and Yescarta (CAR T-cell against CD19) being approved by the FDA for the treatment of B-cell leukemia and lymphoma [152]. Despite all the encouraging results from autologous CAR T-cell therapy, there are still some limitations that need to be tackled. For example, in some cases, such as infants, there may not be enough T-cells to generate CAR-T cells and perform autologous transplantation. The CRISPR/Cas system can be integrated into CAR T-cell engineering to produce universal CAR T-cells from healthy donors [153].
Viral vectors integrate CAR genes into T-cells in a random manner, not in a site-specific manner, which might adversely affect the genome. Eyquem et al. used a CRISPR/Cas9-based site-specific integration of the CD19-specific CAR gene to the T-cell receptor α constant (TRAC) locus. These engineered CAR T-cells outperformed the previous generations regarding safety, precision, and effectiveness in targeting acute lymphoblastic leukemia cells [154].
CAR T-cell-induced cytokine storms and neuroinflammation are not favorable outcomes and are thought to be the effects of GM-CSF. Thus, to address these limitations, lenzilumab has been used to suppress the GM-CSF, which not only reduces the side effects of CAR-T cell therapy but also increases its proliferation, and improves the control of leukemia. According to that, Sterner and colleagues manufactured GM-CSF knocked-out CAR-T cells with CRISPR/Cas9 system, which showcased significant in vivo anti-tumor activity while keeping the side effects at their lowest [155].
Targeting CD33, a myeloid marker in acute myeloid leukemia (AML) by CAR T-cells leads to induced toxicity due to the destruction of normal myeloid cells. An intricate approach, with CRISPR/Cas9-mediated knock-out of the CD33 gene from normal stem cells, followed by autologous transplantation in the rhesus monkey, paved the way for CD33-specific CAR T-cell-mediated elimination of leukemic cells, without affecting normal hematopoietic stem cells [156]. Additionally, Multiplex CRISPR-Cas9 system has been used for simultaneous knock-out of multiple genes in T-cells, including T cell receptor (TCR) chains, and PD-1. This could create cancer-specific T-cells (NY-ESO-1) having features such as minor mismatch pairing to the target, and enhanced durability, as well as offering risk-free graft-versus-host disease (GVHD) [157]. All these efforts have made it clear that CRISPR/Cas9 system is a great armamentarium in improving immunotherapy against cancer.
Given that CRISPR/Cas system can be introduced to human cells, it might be a potent tool to protect us against carcinogenic viruses [158]. Some viruses are capable of initiating cancer in humans, such as hepatitis B and C viruses (HBV and HCV), human papillomavirus (HPV), and Epstein-Barr virus (EBV), which can cause hepatocellular carcinoma (HCC) [159], cervical cancer [160] and Burkitt lymphoma [161], respectively. Given that CRISPR/Cas system can be introduced to human cells, it might be a potent tool to protect us against carcinogenic viruses [162].
The role of HBV in HCC is tightly pertained to the activity of covalent closed-loop DNA (cccDNA) of the HBV in hepatocytes [163]. A number of studies have shown that CRISPR/Cas9-induced mutations in HBV cccDNA can reduce its levels, impairing virus replication [164–166]. Moreover, with the help of CRISRP/Cas system it was demonstrated that a lncRNA PCNAP1 contributes in HBV replication and promotes hepatocarcinogenesis by modulating miR-154/PCNA/HBV cccDNA signaling [167]. In addition, HCV also contributes to HCC development. In a recent study, Francisella novicida (FnCas9), a type of Cas9 that can target both DNA and RNA viruses, was used to inhibit the ssRNA of HCV inside HCC cells [168]. The potential of FnCas9 seems to be promising for targeting different types of oncogenic viruses.
The persistence of HPV infection can lead to the development of cervical cancer, and it is responsible for the death of nearly 200,000 patients each year [168]. Among HPV functional proteins, E6 and E7 have been associated with the carcinogenic properties of HPV [169]. These two proteins are capable of sabotaging the major tumor suppressors in cells, including p53 and Rb. Based on that, researchers suppressed the E6 and E7 genes of HPV-16 and HPV-18 using the CRISPR/Cas9 system, which then restored the expression of p53 and Rb, leading to apoptosis in the cancer cells [170, 171].
EBV, another oncogenic virus mostly related to Burkitt lymphoma, has been targeted by CRISPR/Cas9. Wang et al. stated that targeting EBV genes, such as EBNA-1, LMP-1, or EBNA-3C by CRISPR/Cas9 in Burkitt’s lymphoma cells derived from a patient with latent EBV infection resulted in a drastic reduction in cell proliferation and a decrease in viral load [172]. This approach has also been supported by other studies [172], and since EBV causes several types of malignancies, CRISPR/Cas9-based targeted therapies can be effective in preventing these diseases.
Conclusion
Though the CRISPR system was only recently developed, it has already made significant advances in cancer diagnosis and treatment. Researchers can inhibit or induce gene expression in vitro, in vivo, or ex vivo using CRISPR/Cas. Although CRISPR has some challenges ahead, it has attracted much attention in recent years due to its potential for precise cancer therapy and immunotherapy.
Against the wide spectrum utility of CRISPR in cancer diagnosis and therapy, there are still limitations and concerns that should be addressed in the future. One of these is the probability of CRISPR/Cas-induced DSBs, causing unwanted large deletions in the genome, and in the worst case scenario, it can lead to chromothripsis, which can disrupt tumor suppressors and cell regulatory systems [172, 173]. Another issue is the off-target effects of the CRISPR/Cas system, although in this review some solutions have been mentioned to avoid them. There has been concern about off-target effects, which might lead to CRISPR-induced cancer growth, but the good news is there is no evidence that this is the case. Noteworthy is that these effects can be minimized by using precise protocols and standards [173–175]. As widely used Cas9 comes from bacteria, pre-existing immunity to it can be considered a limitation of the CRISPR/Cas system [176–178]. Modification of Cas enzymes, and creating variants with different antigenic properties may help in this case [179].
Delivering the CRISPR/Cas system to target cells either in vivo or in vitro has its own challenges. To address them different delivery systems have been used, including physical methods, viral vectors, extracellular vesicles, nanocomplexes, etc. Among all, viral vectors and EVs are non-synthetic and natural systems that are destined to deliver their genetic cargoes. In recent years these two genuine systems have been combined together creating a novel vector called vexosomes. Overall, advances in delivery systems promise a bright future for efficient CRISPR/Cas-based cancer therapy.
Acknowledgements
The authors wish to thank the Department of Hematology and Blood banking at the Iran University of Medical Science for supporting this study. Also, we would like to thank the Parscoders Team for their help designing the figures.
Author contributions
MS conceived, edited, and revised the manuscript; SEA, MS, MAZ, MDF, FS, and MO wrote the manuscript. SEA, MS, and MDF designed the figures. FS, MAZ, and SEA prepared tables. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cao W, Chen H-D, Yu Y-W, Li N, Chen W-Q. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J. 2021;134:783–91. doi: 10.1097/CM9.0000000000001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorgalaleh A, Bahraini M, Ahmadi SE. Personalized anesthesia in hematology. In: Dabbagh A, editor. Personalized medicine in anesthesia, pain and perioperative medicine. Cham: Springer International Publishing; 2021. p. 231–74.
- 3.Zahavi D, Weiner L. Monoclonal antibodies in cancer therapy. Antibodies. 2020;9:34. doi: 10.3390/antib9030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belete TM. The current status of gene therapy for the treatment of cancer. Biologics: Targets Ther. 2021;15:67. doi: 10.2147/BTT.S302095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Gent M, Gack MU. Viral anti-CRISPR tactics: no success without sacrifice. Immunity. 2018;49:391–3. doi: 10.1016/j.immuni.2018.08.023. [DOI] [PubMed] [Google Scholar]
- 6.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–33. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mojica FJ, Juez G, Rodríguez-Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol Microbiol. 1993;9:613–21. doi: 10.1111/j.1365-2958.1993.tb01721.x. [DOI] [PubMed] [Google Scholar]
- 8.Jore MM, Brouns SJ, van der Oost J. RNA in defense: CRISPRs protect prokaryotes against mobile genetic elements. Cold Spring Harb Perspect Biol. 2012;4:a003657. doi: 10.1101/cshperspect.a003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EVA. DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–96. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–63. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 11.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C, Mei H, Hu Y. Applications and explorations of CRISPR/Cas9 in CAR T-cell therapy. Brief Funct Genom. 2020;19:175–82. doi: 10.1093/bfgp/elz042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balon K, Sheriff A, Jacków J, Łaczmański Ł. Targeting cancer with CRISPR/Cas9-based therapy. Int J Mol Sci. 2022;23:573. doi: 10.3390/ijms23010573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song X, Liu C, Wang N, Huang H, He S, Gong C, et al. Delivery of CRISPR/Cas systems for cancer gene therapy and immunotherapy. Adv drug Deliv Rev. 2021;168:158–80. doi: 10.1016/j.addr.2020.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–49. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–6. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ran F, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–91. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, et al. New CRISPR–Cas systems from uncultivated microbes. Nature. 2017;542:237–41. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–51. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–71. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, et al. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol. 2019;37:276–82. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao L, Cox DB, Yan WX, Manteiga JC, Schneider MW, Yamano T, et al. Engineered Cpf1 variants with altered PAM specificities. Nat Biotechnol. 2017;35:789–92. doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–9. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, et al. RNA targeting with CRISPR–Cas13. Nature. 2017;550:280–4. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–42. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dincer C, Bruch R, Costa‐Rama E, Fernández‐Abedul MT, Merkoçi A, Manz A, et al. Disposable sensors in diagnostics, food, and environmental monitoring. Adv Mater. 2019;31:1806739. doi: 10.1002/adma.201806739. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Li S, Wang J, Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–43. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 32.Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat Rev Microbiol. 2014;12:479–92. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–7. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–7. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end joining pathway. Annu Rev Biochem. 2010;79:181. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J-P, Li X-L, Li G-H, Chen W, Arakaki C, Botimer GD, et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017;18:1–18. doi: 10.1186/s13059-017-1164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu S, Du J, Chen N, Jia R, Zhang J, Liu X, et al. In vivo CRISPR/Cas9-mediated genome editing mitigates photoreceptor degeneration in a mouse model of X-linked retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2020;61:31. doi: 10.1167/iovs.61.4.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen H, Shi M, Gilam A, Zheng Q, Zhang Y, Afrikanova I, et al. Hemophilia A ameliorated in mice by CRISPR-based in vivo genome editing of human Factor VIII. Sci Rep. 2019;9:1–15. doi: 10.1038/s41598-019-53198-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao J, Bergmann T, Zhang W, Schiwon M, Ehrke-Schulz E, Ehrhardt A. Viral vector-based delivery of CRISPR/Cas9 and donor DNA for homology-directed repair in an in vitro model for canine hemophilia B. Mol Ther - Nucleic Acids. 2019;14:364–76. doi: 10.1016/j.omtn.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richards DY, Winn SR, Dudley S, Nygaard S, Mighell TL, Grompe M, et al. AAV-mediated CRISPR/Cas9 gene editing in murine phenylketonuria. Mol Ther-Methods Clin Dev. 2020;17:234–45. doi: 10.1016/j.omtm.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Behr M, Zhou J, Xu B, Zhang H. In vivo delivery of CRISPR-Cas9 therapeutics: progress and challenges. Acta Pharm Sin B. 2021;11:2150–71. doi: 10.1016/j.apsb.2021.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valikhani M, Rahimian E, Ahmadi SE, Chegeni R, Safa M. Involvement of classic and alternative non-homologous end joining pathways in hematologic malignancies: targeting strategies for treatment. Exp Hematol Oncol. 2021;10:51. doi: 10.1186/s40164-021-00242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–38. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakuma T, Nakade S, Sakane Y, Suzuki K-IT, Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc. 2016;11:118–33. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- 49.Yao X, Wang X, Hu X, Liu Z, Liu J, Zhou H, et al. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 2017;27:801–14. doi: 10.1038/cr.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–9. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H-X, Li M, Lee CM, Chakraborty S, Kim H-W, Bao G, et al. CRISPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery. Chem Rev. 2017;117:9874–906. doi: 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- 52.Glass Z, Lee M, Li Y, Xu Q. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 2018;36:173–85. doi: 10.1016/j.tibtech.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wan T, Ping Y. Delivery of genome-editing biomacromolecules for treatment of lung genetic disorders. Adv drug Deliv Rev. 2021;168:196–216. doi: 10.1016/j.addr.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 54.Yin H, Kauffman KJ, Anderson DG. Delivery technologies for genome editing. Nat Rev Drug Discov. 2017;16:387–99. doi: 10.1038/nrd.2016.280. [DOI] [PubMed] [Google Scholar]
- 55.Wang D, Zhang F, Gao G. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell. 2020;181:136–50. doi: 10.1016/j.cell.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boucher P, Cui X, Curiel DT. Adenoviral vectors for in vivo delivery of CRISPR-Cas gene editors. J Control Release. 2020;327:788–800. doi: 10.1016/j.jconrel.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong W, Kantor B. Lentiviral vectors for delivery of gene-editing systems based on CRISPR/Cas: current state and perspectives. Viruses. 2021;13:1288. doi: 10.3390/v13071288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naso MF, Tomkowicz B, Perry WL, Strohl WR. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs. 2017;31:317–34. doi: 10.1007/s40259-017-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asmamaw M, Zawdie B. Mechanism and applications of CRISPR/Cas-9-mediated genome editing. Biologics: Targets Ther. 2021;15:353. doi: 10.2147/BTT.S326422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verdera HC, Kuranda K, Mingozzi F. AAV vector immunogenicity in humans: a long journey to successful gene transfer. Mol Ther. 2020;28:723–46. doi: 10.1016/j.ymthe.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson RC, Gilbert LA. The promise and challenge of in vivo delivery for genome therapeutics. ACS Chem Biol. 2018;13:376–82. doi: 10.1021/acschembio.7b00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flotte TR. Gene Therapy Progress and Prospects: recombinant adeno-associated virus (rAAV) vectors. Gene Ther. 2004;11:805–10. doi: 10.1038/sj.gt.3302233. [DOI] [PubMed] [Google Scholar]
- 63.Vogel R, Seyffert M, Pereira Bde A. Fraefel C. Viral and cellular components of AAV2 replication compartments. Open Virol J. 2013;7:98–120. doi: 10.2174/1874357901307010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conlon TJ, Flotte TR. Recombinant adeno-associated virus vectors for gene therapy. Expert Opin Biol Ther. 2004;4:1093–101. doi: 10.1517/14712598.4.7.1093. [DOI] [PubMed] [Google Scholar]
- 65.Zhao H, Li Y, He L, Pu W, Yu W, Li Y, et al. In vivo AAV-CRISPR/Cas9-mediated gene editing ameliorates atherosclerosis in familial hypercholesterolemia. Circulation. 2020;141:67–79. doi: 10.1161/CIRCULATIONAHA.119.042476. [DOI] [PubMed] [Google Scholar]
- 66.Mendell JR, Al-Zaidy SA, Rodino-Klapac LR, Goodspeed K, Gray SJ, Kay CN, et al. Current clinical applications of in vivo gene therapy with AAVs. Mol Ther. 2021;29:464–88. doi: 10.1016/j.ymthe.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fuentes CM, Schaffer DV. Adeno-associated virus-mediated delivery of CRISPR-Cas9 for genome editing in the central nervous system. Curr Opin Biomed Eng. 2018;7:33–41. doi: 10.1016/j.cobme.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–3. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–7. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yin S, Ma L, Shao T, Zhang M, Guan Y, Wang L, et al. Enhanced genome editing to ameliorate a genetic metabolic liver disease through co-delivery of adeno-associated virus receptor. Sci China Life Sci. 2022;65:718–30. doi: 10.1007/s11427-020-1744-6. [DOI] [PubMed] [Google Scholar]
- 71.Park SH, Bao G. CRISPR/Cas9 gene editing for curing sickle cell disease. Transfus Apher Sci. 2021;60:103060. doi: 10.1016/j.transci.2021.103060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le Rhun A, Escalera-Maurer A, Bratovič M, Charpentier E. CRISPR-Cas in Streptococcus pyogenes. RNA Biol. 2019;16:380–9. doi: 10.1080/15476286.2019.1582974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hayashi H, Kubo Y, Izumida M, Matsuyama T. Efficient viral delivery of Cas9 into human safe harbor. Sci Rep. 2020;10:21474. doi: 10.1038/s41598-020-78450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fang H, Bygrave AM, Roth RH, Johnson RC, Huganir RL. An optimized CRISPR/Cas9 approach for precise genome editing in neurons. Elife. 2021;10:e65202.. doi: 10.7554/eLife.65202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maddalena A, Tornabene P, Tiberi P, Minopoli R, Manfredi A, Mutarelli M, et al. Triple vectors expand AAV transfer capacity in the retina. Mol Ther. 2018;26:524–41. doi: 10.1016/j.ymthe.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koo T, Popplewell L, Athanasopoulos T, Dickson G. Triple trans-splicing adeno-associated virus vectors capable of transferring the coding sequence for full-length dystrophin protein into dystrophic mice. Hum Gene Ther. 2014;25:98–108. doi: 10.1089/hum.2013.164. [DOI] [PubMed] [Google Scholar]
- 77.Kantor A, McClements ME, MacLaren RE. CRISPR-Cas9 DNA base-editing and prime-editing. Int. J. Mol. Sci. 2020;21:6240.. doi: 10.3390/ijms21176240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38:824–44. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 79.Liu P, Liang S-Q, Zheng C, Mintzer E, Zhao YG, Ponnienselvan K, et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat Commun. 2021;12:2121. doi: 10.1038/s41467-021-22295-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Uddin F, Rudin CM, Sen T. CRISPR gene therapy: applications, limitations, and implications for the future. Front Oncol. 2020;10:1387. doi: 10.3389/fonc.2020.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim H, Kim ST, Ryu J, Kang BC, Kim JS, Kim SG. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun. 2017;8:14406. doi: 10.1038/ncomms14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alok A, Sandhya D, Jogam P, Rodrigues V, Bhati KK, Sharma H, et al. The rise of the CRISPR/Cpf1 system for efficient genome editing in plants. Front Plant Sci. 2020;11:264. doi: 10.3389/fpls.2020.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chamberlain K, Riyad JM, Weber T. Expressing transgenes that exceed the packaging capacity of adeno-associated virus capsids. Hum Gene Ther Methods. 2016;27:1–12. doi: 10.1089/hgtb.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang H, Wang L, Zhong B, Dai Z. Protein splicing of inteins: a powerful tool in synthetic biology. Front Bioeng Biotechnol. 2022;10:810180. doi: 10.3389/fbioe.2022.810180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Villiger L, Grisch-Chan HM, Lindsay H, Ringnalda F, Pogliano CB, Allegri G, et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med. 2018;24:1519–25. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 86.Lim JM, Kim HH. Basic principles and clinical applications of CRISPR-based genome editing. Yonsei Med J. 2022;63:105–13. doi: 10.3349/ymj.2022.63.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ibraheim R, Tai PWL, Mir A, Javeed N, Wang J, Rodríguez TC, et al. Self-inactivating, all-in-one AAV vectors for precision Cas9 genome editing via homology-directed repair in vivo. Nat Commun. 2021;12:6267. doi: 10.1038/s41467-021-26518-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barnes C, Scheideler O, Schaffer D. Engineering the AAV capsid to evade immune responses. Curr Opin Biotechnol. 2019;60:99–103. doi: 10.1016/j.copbio.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tatsis N, Ertl HC. Adenoviruses as vaccine vectors. Mol Ther. 2004;10:616–29. doi: 10.1016/j.ymthe.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Danthinne X, Imperiale MJ. Production of first generation adenovirus vectors: a review. Gene Ther. 2000;7:1707–14. doi: 10.1038/sj.gt.3301301. [DOI] [PubMed] [Google Scholar]
- 91.Armentano D, Sookdeo CC, Hehir KM, Gregory RJ, St George JA, Prince GA, et al. Characterization of an adenovirus gene transfer vector containing an E4 deletion. Hum Gene Ther. 1995;6:1343–53. doi: 10.1089/hum.1995.6.10-1343. [DOI] [PubMed] [Google Scholar]
- 92.Engelhardt JF, Ye X, Doranz B, Wilson JM. Ablation of E2A in recombinant adenoviruses improves transgene persistence and decreases inflammatory response in mouse liver. Proc Natl Acad Sci USA. 1994;91:6196–200. doi: 10.1073/pnas.91.13.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parks RJ, Chen L, Anton M, Sankar U, Rudnicki MA, Graham FL. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc Natl Acad Sci USA. 1996;93:13565–70. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Palmer D, Ng P. Improved system for helper-dependent adenoviral vector production. Mol Ther. 2003;8:846–52. doi: 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 95.Stephens CJ, Kashentseva E, Everett W, Kaliberova L, Curiel DT. Targeted in vivo knock-in of human alpha-1-antitrypsin cDNA using adenoviral delivery of CRISPR/Cas9. Gene Ther. 2018;25:139–56. doi: 10.1038/s41434-018-0003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tsukamoto T, Sakai E, Iizuka S, Taracena-Gándara M, Sakurai F, Mizuguchi H. Generation of the adenovirus vector-mediated CRISPR/Cpf1 system and the application for primary human hepatocytes prepared from humanized mice with chimeric liver. Biol Pharm Bull. 2018;41:1089–95. doi: 10.1248/bpb.b18-00222. [DOI] [PubMed] [Google Scholar]
- 97.Srivastava A, Mallela KMG, Deorkar N, Brophy G. Manufacturing challenges and rational formulation development for AAV viral vectors. J Pharm Sci. 2021;110:2609–24. doi: 10.1016/j.xphs.2021.03.024. [DOI] [PubMed] [Google Scholar]
- 98.Jacob-Dolan C, Barouch DH. COVID-19 vaccines: adenoviral vectors. Annu Rev Med. 2022;73:41–54. doi: 10.1146/annurev-med-012621-102252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gouvarchin Ghaleh HE, Bolandian M, Dorostkar R, Jafari A, Pour MF. Concise review on optimized methods in production and transduction of lentiviral vectors in order to facilitate immunotherapy and gene therapy. Biomed. Pharmacother. 2020;128:110276. doi: 10.1016/j.biopha.2020.110276. [DOI] [PubMed] [Google Scholar]
- 100.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST, et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol. 2018;36:536–9. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 102.Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32:941–6. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–7. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Choi JG, Dang Y, Abraham S, Ma H, Zhang J, Guo H, et al. Lentivirus pre-packed with Cas9 protein for safer gene editing. Gene Ther. 2016;23:627–33. doi: 10.1038/gt.2016.27. [DOI] [PubMed] [Google Scholar]
- 105.Joglekar AV, Hollis RP, Kuftinec G, Senadheera S, Chan R, Kohn DB. Integrase-defective lentiviral vectors as a delivery platform for targeted modification of adenosine deaminase locus. Mol Ther. 2013;21:1705–17. doi: 10.1038/mt.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ortinski PI, O’Donovan B, Dong X, Kantor B. Integrase-deficient lentiviral vector as an all-in-one platform for highly efficient CRISPR/Cas9-mediated gene editing. Mol Ther Methods Clin Dev. 2017;5:153–64. doi: 10.1016/j.omtm.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu Q, Fan D, Adah D, Wu Z, Liu R, Yan QT, et al. CRISPR/Cas9‑mediated hypoxia inducible factor‑1α knockout enhances the antitumor effect of transarterial embolization in hepatocellular carcinoma. Oncol Rep. 2018;40:2547–57. doi: 10.3892/or.2018.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ahmadi SE, Rahimi S, Zarandi B, Chegeni R, Safa M. MYC: a multipurpose oncogene with prognostic and therapeutic implications in blood malignancies. J Hematol Oncol. 2021;14:121. doi: 10.1186/s13045-021-01111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Paridar M, Zibara K, Ahmadi SE, Khosravi A, Soleymani M, Azizi E, et al. Clinico-Hematological and cytogenetic spectrum of adult myelodysplastic syndrome: The first retrospective cross-sectional study in Iranian patients. Mol Cytogenet. 2021;14:24. doi: 10.1186/s13039-021-00548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ghalesardi OK, Khosravi A, Azizi E, Ahmadi SE, Hajifathali A, Bonakchi H, et al. The prognostic importance of BCR-ABL transcripts in Chronic Myeloid Leukemia: a systematic review and meta-analysis. Leuk Res. 2021;101:106512. doi: 10.1016/j.leukres.2021.106512. [DOI] [PubMed] [Google Scholar]
- 111.Volpe G, Panuzzo C, Ulisciani S, Cilloni D. Imatinib resistance in CML. Cancer Lett. 2009;274:1–9. doi: 10.1016/j.canlet.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 112.Martinez-Lage M, Torres-Ruiz R, Puig-Serra P, Moreno-Gaona P, Martin MC, Moya FJ, et al. In vivo CRISPR/Cas9 targeting of fusion oncogenes for selective elimination of cancer cells. Nat Commun. 2020;11:5060. doi: 10.1038/s41467-020-18875-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gee P, Lung MSY, Okuzaki Y, Sasakawa N, Iguchi T, Makita Y, et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat Commun. 2020;11:1334. doi: 10.1038/s41467-020-14957-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Duan L, Xu L, Xu X, Qin Z, Zhou X, Xiao Y, et al. Exosome-mediated delivery of gene vectors for gene therapy. Nanoscale. 2021;13:1387–97. doi: 10.1039/D0NR07622H. [DOI] [PubMed] [Google Scholar]
- 116.Qiao L, Hu S, Huang K, Su T, Li Z, Vandergriff A, et al. Tumor cell-derived exosomes home to their cells of origin and can be used as Trojan horses to deliver cancer drugs. Theranostics. 2020;10:3474–87. doi: 10.7150/thno.39434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yong T, Zhang X, Bie N, Zhang H, Zhang X, Li F, et al. Tumor exosome-based nanoparticles are efficient drug carriers for chemotherapy. Nat Commun. 2019;10:3838. doi: 10.1038/s41467-019-11718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim SM, Yang Y, Oh SJ, Hong Y, Seo M, Jang M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J Control Release. 2017;266:8–16. doi: 10.1016/j.jconrel.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 119.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–5. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 120.Liang Y, Duan L, Lu J, Xia J. Engineering exosomes for targeted drug delivery. Theranostics. 2021;11:3183–95. doi: 10.7150/thno.52570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liang Y, Xu X, Li X, Xiong J, Li B, Duan L, et al. Chondrocyte-targeted microRNA delivery by engineered exosomes toward a cell-free osteoarthritis therapy. ACS Appl Mater Interfaces. 2020;12:36938–47. doi: 10.1021/acsami.0c10458. [DOI] [PubMed] [Google Scholar]
- 122.Gulei D, Berindan-Neagoe I. Activation of necroptosis by engineered self tumor-derived exosomes loaded with CRISPR/Cas9. Mol Ther Nucleic Acids. 2019;17:448–51. doi: 10.1016/j.omtn.2019.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lamichhane TN, Raiker RS, Jay SM, Exogenous DNA. Loading into extracellular vesicles via electroporation is size-dependent and enables limited gene delivery. Mol Pharm. 2015;12:3650–7. doi: 10.1021/acs.molpharmaceut.5b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Emam SE, Abu Lila AS, Elsadek NE, Ando H, Shimizu T, Okuhira K, et al. Cancer cell-type tropism is one of crucial determinants for the efficient systemic delivery of cancer cell-derived exosomes to tumor tissues. Eur J Pharm Biopharm. 2019;145:27–34. doi: 10.1016/j.ejpb.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 125.Lin Y, Wu J, Gu W, Huang Y, Tong Z, Huang L, et al. Exosome-liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Adv Sci. 2018;5:1700611. doi: 10.1002/advs.201700611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sun H, Zu Y. Aptamers and their applications in nanomedicine. Small. 2015;11:2352–64. doi: 10.1002/smll.201403073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li J, Fan C, Pei H, Shi J, Huang Q. Smart drug delivery nanocarriers with self-assembled DNA nanostructures. Adv Mater. 2013;25:4386–96. doi: 10.1002/adma.201300875. [DOI] [PubMed] [Google Scholar]
- 128.Zhuang J, Tan J, Wu C, Zhang J, Liu T, Fan C, et al. Extracellular vesicles engineered with valency-controlled DNA nanostructures deliver CRISPR/Cas9 system for gene therapy. Nucleic Acids Res. 2020;48:8870–82. doi: 10.1093/nar/gkaa683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Usman WM, Pham TC, Kwok YY, Vu LT, Ma V, Peng B, et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat Commun. 2018;9:2359. doi: 10.1038/s41467-018-04791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Firquet S, Beaujard S, Lobert PE, Sané F, Caloone D, Izard D, et al. Survival of enveloped and non-enveloped viruses on inanimate surfaces. Microbes Environ. 2015;30:140–4. doi: 10.1264/jsme2.ME14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Saari H, Turunen T, Lõhmus A, Turunen M, Jalasvuori M, Butcher SJ, et al. Extracellular vesicles provide a capsid-free vector for oncolytic adenoviral DNA delivery. J Extracell Vesicles. 2020;9:1747206. doi: 10.1080/20013078.2020.1747206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wood MJ, O’Loughlin AJ, Samira L. Exosomes and the blood-brain barrier: implications for neurological diseases. Ther Deliv. 2011;2:1095–9. doi: 10.4155/tde.11.83. [DOI] [PubMed] [Google Scholar]
- 133.Hudry E, Andres-Mateos E, Lerner EP, Volak A, Cohen O, Hyman BT, et al. Efficient gene transfer to the central nervous system by single-stranded Anc80L65. Mol Ther Methods Clin Dev. 2018;10:197–209. doi: 10.1016/j.omtm.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Frank SA. Evolution in health and medicine Sackler colloquium: somatic evolutionary genomics: mutations during development cause highly variable genetic mosaicism with risk of cancer and neurodegeneration. Proc Natl Acad Sci USA. 2010;107(Suppl 1):1725–30. doi: 10.1073/pnas.0909343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.György B, Sage C, Indzhykulian AA, Scheffer DI, Brisson AR, Tan S, et al. Rescue of hearing by gene delivery to inner-ear hair cells using exosome-associated AAV. Mol Ther. 2017;25:379–91. doi: 10.1016/j.ymthe.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Orefice NS, Souchet B, Braudeau J, Alves S, Piguet F, Collaud F, et al. Real-time monitoring of exosome enveloped-AAV spreading by endomicroscopy approach: a new tool for gene delivery in the brain. Mol Ther Methods Clin Dev. 2019;14:237–51. doi: 10.1016/j.omtm.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Meliani A, Boisgerault F, Fitzpatrick Z, Marmier S, Leborgne C, Collaud F, et al. Enhanced liver gene transfer and evasion of preexisting humoral immunity with exosome-enveloped AAV vectors. Blood Adv. 2017;1:2019–31. doi: 10.1182/bloodadvances.2017010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 139.Zhang D, Lee H, Jin Y. Delivery of functional small rnas via extracellular vesicles in vitro and in vivo. Methods Mol Biol. 2020;2115:107–17. doi: 10.1007/978-1-0716-0290-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhao L, Gu C, Gan Y, Shao L, Chen H, Zhu H. Exosome-mediated siRNA delivery to suppress postoperative breast cancer metastasis. J Control Release. 2020;318:1–15. doi: 10.1016/j.jconrel.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 141.Konishi CT, Long C. Progress and challenges in CRISPR-mediated therapeutic genome editing for monogenic diseases. J Biomed Res. 2020;35:148–62. doi: 10.7555/JBR.34.20200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li K, Luo H, Huang L, Luo H, Zhu X. Microsatellite instability: a review of what the oncologist should know. Cancer Cell Int. 2020;20:16. doi: 10.1186/s12935-019-1091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shin G, Grimes SM, Lee H, Lau BT, Xia LC, Ji HP. CRISPR-Cas9-targeted fragmentation and selective sequencing enable massively parallel microsatellite analysis. Nat Commun. 2017;8:14291. doi: 10.1038/ncomms14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bennett-Baker PE, Mueller JL. CRISPR-mediated isolation of specific megabase segments of genomic DNA. Nucleic Acids Res. 2017;45:e165. doi: 10.1093/nar/gkx749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nachmanson D, Lian S, Schmidt EK, Hipp MJ, Baker KT, Zhang Y, et al. Targeted genome fragmentation with CRISPR/Cas9 enables fast and efficient enrichment of small genomic regions and ultra-accurate sequencing with low DNA input (CRISPR-DS) Genome Res. 2018;28:1589–99. doi: 10.1101/gr.235291.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–44. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature. 2016;539:479. doi: 10.1038/nature.2016.20988. [DOI] [PubMed] [Google Scholar]
- 149.Yang X. Applications of CRISPR-Cas9 mediated genome engineering. Mil Med Res. 2015;2:11. doi: 10.1186/s40779-015-0038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.You L, Tong R, Li M, Liu Y, Xue J, Lu Y. Advancements and obstacles of CRISPR-Cas9 technology in translational research. Mol Ther Methods Clin Dev. 2019;13:359–70. doi: 10.1016/j.omtm.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhan T, Rindtorff N, Betge J, Ebert MP, Boutros M. CRISPR/Cas9 for cancer research and therapy. Semin Cancer Biol. 2019;55:106–19. doi: 10.1016/j.semcancer.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 152.Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl J Med. 2017;377:2531–44. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lin H, Cheng J, Mu W, Zhou J, Zhu L. Advances in universal CAR-T cell therapy. Front Immunol. 2021;12:744823.. doi: 10.3389/fimmu.2021.744823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–7. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sterner RM, Sakemura R, Cox MJ, Yang N, Khadka RH, Forsman CL, et al. GM-CSF inhibition reduces cytokine release syndrome and neuroinflammation but enhances CAR-T cell function in xenografts. Blood. 2019;133:697–709. doi: 10.1182/blood-2018-10-881722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kim MY, Yu KR, Kenderian SS, Ruella M, Chen S, Shin TH, et al. Genetic inactivation of CD33 in hematopoietic stem cells to enable CAR T cell immunotherapy for acute myeloid leukemia. Cell. 2018;173:1439–1453.e19. doi: 10.1016/j.cell.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Stadtmauer EA, Fraietta JA, Davis MM, Cohen AD, Weber KL, Lancaster E, et al. CRISPR-engineered T cells in patients with refractory cancer. Science (New York, N.Y.) 2020;367:eaba7365.. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Saayman S, Ali SA, Morris KV, Weinberg MS. The therapeutic application of CRISPR/Cas9 technologies for HIV. Expert Opin Biol Ther. 2015;15:819–30. doi: 10.1517/14712598.2015.1036736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Arzumanyan A, Reis HMGPV, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123–35. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- 160.Wang X, Huang X, Zhang Y. Involvement of human papillomaviruses in cervical cancer. Front Microbiol. 2018;9:2896.. doi: 10.3389/fmicb.2018.02896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kanda T, Yajima M, Ikuta K. Epstein-Barr virus strain variation and cancer. Cancer Sci. 2019;110:1132–9. doi: 10.1111/cas.13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gilani U, Shaukat M, Rasheed A, Shahid M, Tasneem F, Arshad M, et al. The implication of CRISPR/Cas9 genome editing technology in combating human oncoviruses. J Med Virol. 2019;91:1–13. doi: 10.1002/jmv.25292. [DOI] [PubMed] [Google Scholar]
- 163.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–8. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ramanan V, Shlomai A, Cox DB, Schwartz RE, Michailidis E, Bhatta A, et al. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Seeger C, Sohn JA, Targeting Hepatitis B. Virus with CRISPR/Cas9. Mol Ther Nucleic Acids. 2014;3:e216. doi: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Seeger C, Sohn JA. Complete spectrum of CRISPR/Cas9-induced mutations on HBV cccDNA. Mol Ther. 2016;24:1258–66. doi: 10.1038/mt.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Feng J, Yang G, Liu Y, Gao Y, Zhao M, Bu Y, et al. LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics. 2019;9:5227–45. doi: 10.7150/thno.34273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Price AA, Sampson TR, Ratner HK, Grakoui A, Weiss DS. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc Natl Acad Sci USA. 2015;112:6164–9. doi: 10.1073/pnas.1422340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mirabello L, Yeager M, Yu K, Clifford GM, Xiao Y, Zhu B, et al. HPV16 E7 genetic conservation is critical to carcinogenesis. Cell. 2017;170:1164–1174.e6. doi: 10.1016/j.cell.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kennedy EM, Kornepati AV, Goldstein M, Bogerd HP, Poling BC, Whisnant AW, et al. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol. 2014;88:11965–72. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhen S, Hua L, Takahashi Y, Narita S, Liu YH, Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem Biophys Res Commun. 2014;450:1422–6. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 172.Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci USA. 2014;111:13157–62. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Leibowitz ML, Papathanasiou S, Doerfler PA, Blaine LJ, Sun L, Yao Y, et al. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing. Nat Genet. 2021;53:895–905. doi: 10.1038/s41588-021-00838-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med. 2018;24:1216–24. doi: 10.1038/s41591-018-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ikeda A, Fujii W, Sugiura K, Naito K. High-fidelity endonuclease variant HypaCas9 facilitates accurate allele-specific gene modification in mouse zygotes. Commun Biol. 2019;2:371. doi: 10.1038/s42003-019-0627-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25:249–54. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wagner DL, Amini L, Wendering DJ, Burkhardt LM, Akyüz L, Reinke P, et al. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat Med. 2019;25:242–8. doi: 10.1038/s41591-018-0204-6. [DOI] [PubMed] [Google Scholar]
- 178.Simhadri VL, McGill J, McMahon S, Wang J, Jiang H, Sauna ZE. Prevalence of pre-existing antibodies to CRISPR-associated nuclease Cas9 in the USA population. Mol Ther Methods Clin Dev. 2018;10:105–12. doi: 10.1016/j.omtm.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ferdosi SR, Ewaisha R, Moghadam F, Krishna S, Park JG, Ebrahimkhani MR, et al. Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nat Commun. 2019;10:1842. doi: 10.1038/s41467-019-09693-x. [DOI] [PMC free article] [PubMed] [Google Scholar]