Abstract
Background
Ganoderma lucidum is reportedly the best source of traditional natural bioactive constituents. Ganoderma triterpenoids (GTs) have been verified as an alternative adjuvant for treating leukemia, cancer, hepatitis and diabetes. One of the major triterpenoids, Resinacein S, has been found to regulate lipid metabolism and mitochondrial biogenesis. Nonalcoholic fatty liver disease (NAFLD) is a common chronic liver disease that has become a major public health problem. Given the regulatory effects on lipid metabolism of Resinacein S, we sought to explore potential protective effects against NAFLD.
Methods
Resinacein S was extracted and isolated from G. lucidum. And mice were fed with high fat diet with or without Resinacein S to detect hepatic steatosis. According to Network Pharmacology and RNA-seq, we analyzed the hub genes of Resinacein S against NAFLD disease.
Results
Our results can be summarized as follows: (1) The structure of Resinacein S was elucidated using NMR and MS methods. (2) Resinacein S treatment could significantly attenuate high-fat diet (HFD)-induced hepatic steatosis and hepatic lipid accumulation in mouse. (3) GO terms, KEGG pathways and the PPI network of Resinacein S induced Differentially Expressed Genes (DEGs) demonstrated the key target genes of Resinacein S against NAFLD. (4) The hub proteins in PPI network analysis could be used for NAFLD diagnosis and treatment as drug targets.
Conclusion
Resinacein S can significantly change the lipid metabolism in liver cells and yield a protective effect against steatosis and liver injury. Intersected proteins between NAFLD related genes and Resinacein S-induced DEGs, especially the hub protein in PPI network analysis, can be used to characterize targets of Resinacein S against NAFLD.
Keywords: Ganoderma resinaceum, Resinacein S, NAFLD, lipid metabolism, network pharmacology
1. Introduction
Ganoderma lucidum is a medicinal mushroom that can prolong life and promote health and has a long history in traditional Chinese medicine. Given its growing consumption, it has been intensively planted and sold since the 1970s. It is widely thought to be effective in preventing and treating many diseases and has anti-cancer properties (1). G. lucidum is considered the best source of traditional natural bioactive components. It contains various compounds, including polyphenols, polysaccharides, steroids, triterpenes, nucleotides, amino acids, trace elements, and vitamins. Over the past few years, G. lucidum extract has been used as a dietary addition to treating various diseases (2). Among various active components of G. lucidum, polysaccharides (GL-PS) and terpenoids (GL-T) predominantly exert physiological activities. They can inhibit the cell cycle and yield cytotoxicity and anti-metastasis, immunomodulation, antioxidant, antibacterial, anti-inflammatory and other effects (3). Moreover, G. lucidum has been recognized as an alternative adjuvant for treating leukemia, cancer, hepatitis and diabetes (4). Current evidence suggests that Ganoderma triterpenes represent one of the main active ingredients of mushrooms and yield an inhibitory effect on adipogenesis, leading to decreased lipid synthesis and accumulation. Other studies revealed that G. lucidum extracts and ethanol extracts of chigger mites rich in triterpenes contribute to adipogenesis and adipocyte differentiation. Ganoderma resinaceum is generally utilized for treating hepatitis, hyperglycemia, and dysimmunity in China and Nigeria (5).
Previous researchers had separated four new triterpenoids and four identified triterpenoids with anti-obesity effects from G. resinaceum. What’s more, one of the triterpenoids, Resinacein S has been found to induce beige and brown phenotypes, which may be relevant to the activation of AMPK/PGC1α signaling pathway to inhibit and treat obesity and relevant diseases. At the molecular level, Resinacein S treatment could significantly induce the expression of genes and/or proteins related to thermogenesis, fatty acid oxidation and lipolysis (6). Resinacein S, as one of the major triterpenoids from G. resinaceum, provides a therapeutic strategy for lipid metabolic diseases such as NAFLD, but whether Resinacein S treatment could provide protective aspects against NAFLD remains unknown.
Nonalcoholic fatty liver disease (NAFLD) is currently recognized as the most common liver disease in the world, affecting about 25% of adults worldwide. It encompasses steatosis simplex to nonalcoholic steatohepatitis, fibrosis, cirrhosis, and hepatocellular carcinoma. The clinical manifestation of nonalcoholic steatohepatitis (NASH) is a serious form of nonalcoholic fatty liver disease (NAFLD) characterized by the accumulation (steatosis) of triglycerides in liver cells, inflammation, injury and apoptosis, which may lead to cirrhosis and liver cancer in extreme cases (7–9). According to the regional epidemiological model of NAFLD, economy, environment and lifestyle are the key factors of disease progression (10). Hepatic steatosis or fatty liver refers to the increase of lipid accumulation in liver cells caused by increased production or decreased clearance of hepatic triglycerides or fatty acids (11). It has been established that Resinacein S can reduce fat and triglyceride accumulation by inducing the expression of genes and/or proteins related to thermogenesis, fatty acid oxidation and lipolysis at the molecular level. Nevertheless, the protective aspects and molecular mechanisms underlying the ability of Resinacein S to target NAFLD development remain to be illustrated. Therefore, this study aimed to explore the protective effects of Resinacein S against NAFLD.
2. Materials and methods
2.1. General experimental procedures
The NMR spectra were recorded on Bruker AV500 and AVIII600 instruments, with TMS used as an internal benchmark. Optical rotations were measured on an Autopol VI-91058 digital polarimeter. The UV spectra were detected by a UV-2700 spectrophotometer, while IR spectra were measured on a Nicolet iS10 spectrometer. HR-TOF ESI/MS was conducted on an Agilent 6,200 Q-TOF MS system spectrometer. Column chromatography was carried out with silica gel, reversed-phase C18 silica gel and Sephadex LH-20 as packing materials. Fractions were quantitated by thin-layer chromatography after spraying with sulphuric acid and heating. TLC was carried out on silica gel GF254. HPLC was performed on an Agilent 1,100 liquid chromatography system coupled with a diode-array detector and an Agilent Zorbax SB-C18 column (5 μm, 9.4 × 250 mm).
2.2. Fungal material
Fruiting bodies of G. resinaceum were collected from Vientiane, Laos, identified by Peigui Liu and a voucher specimen (accession number: KGR-201606) was deposited at the State Key Laboratory of Phytochemistry and Plant Resources in West China, Kunming Institute of Botany, Chinese Academy of Sciences, China (6).
Extraction and isolation procedures have been described before (6).
2.3. Animal treatments and histological evaluation
Four-week-old male C57BL/6 mice were kept under the standard environment. Mice were fed with high fat diet with or without Resinacein S (intraperitoneal injection, interval for 48 h) at a dose of 10 mg/kg/day for 15 weeks (9). Then mice were sacrificed and the liver were fixed in 4% formaldehyde for HE staining and Oil Red O (G1262, Solarbio, China) staining (abs7049, Absin Bioscience, China). Images were acquired with a light microscope (Zeiss, Germany). Triglycerides (TG) were detected by a commercial kit (E1025-105, Applygen, Beijing, China) according to manufacturer’s instructions. All the experiments were approved by the ethics committee of Shanghai tenth People’s Hospital, Shanghai, China.
2.4. Collection and sorting of nonalcoholic fatty liver disease disease-related genes
Genes related to NAFLD were acquired from DisGeNET,1 GeneCards,2 and OMIM3 databases. The keyword “nonalcoholic fatty liver disease” was entered into the above three databases to search for genes related to NAFLD. Then, the intersection was obtained using the Venny Venn diagram tool (version 2.1.0).4
2.5. Determination of relevant target of Resinacein S
First, the canonical SMILES structure of Resinacein S was obtained through PubChem.5 Later, canonical SMILES information of Resinacein S was input into the SwissTargetPrediction6 database. Finally, the Uniprot ID obtained from the TargetNet database was transformed into Gene ID in UniProt7 database, yielding a total of 124 target genes associated with Resinacein S.
2.6. Protein–protein interactions network construction
The above Resinacein S targets of NAFLD were imported in String,8 with the organization parameter and threshold of combination score being set as Homo sapiens and 0.7 for obtaining protein–protein interactions (PPI). Then, the protein interaction network data from the STRING database was incorporated into Cytoscape 3.9.0 for classification and mapping according to the degree value.
2.7. RNA-seq analysis
TRIzol reagent was utilized for extracting total cellular RNA, which was later subject to spectrophotometry and agarose gel electrophoresis (AGE) with the NanoDrop ND-1000 instrument to analyze RNA integrity. Using a KAPA Stranded RNA-seq Library Prep Kit, this study built an RNA library, while a 2100 Bioanalyzer was employed for library quality assessment. Quantification of the library was performed by qRT-PCR. NCBI Gene Expression Omnibus was utilized to import raw RNA-seq information.
2.8. Functional analysis of identified genes
FastQC v0.11.8 was applied to analyze the raw RNA-seq data. Fragments per kilobase of gene/transcript model per million mapped fragments (FPKM) values of diverse genes and transcripts were determined by the Ballgown package of R v2.10.0 software. Additionally, R was used to generate volcano plots and heatmaps to further analyze the gene expression profiles.
Gene Ontology (GO) enrichment of Differentially Expressed Genes (DEGs) was conducted to identify significantly enriched GO-biological processes (BP), GO-molecular functions (MF) and GO-cellular components (CC). Meanwhile, DEG-related pathways were identified based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (12).
2.9. Statistical analysis
All data were expressed as mean ± standard deviation (SD). Experiments were repeated at least three times. The statistical significance of differences was evaluated using either the Student’s unpaired t-test, One-way analysis of variance (ANOVA), or two-way ANOVA as indicated. A p-value <0.05 was statistically significant.
3. Results
3.1. Structure elucidation of Resinacein S
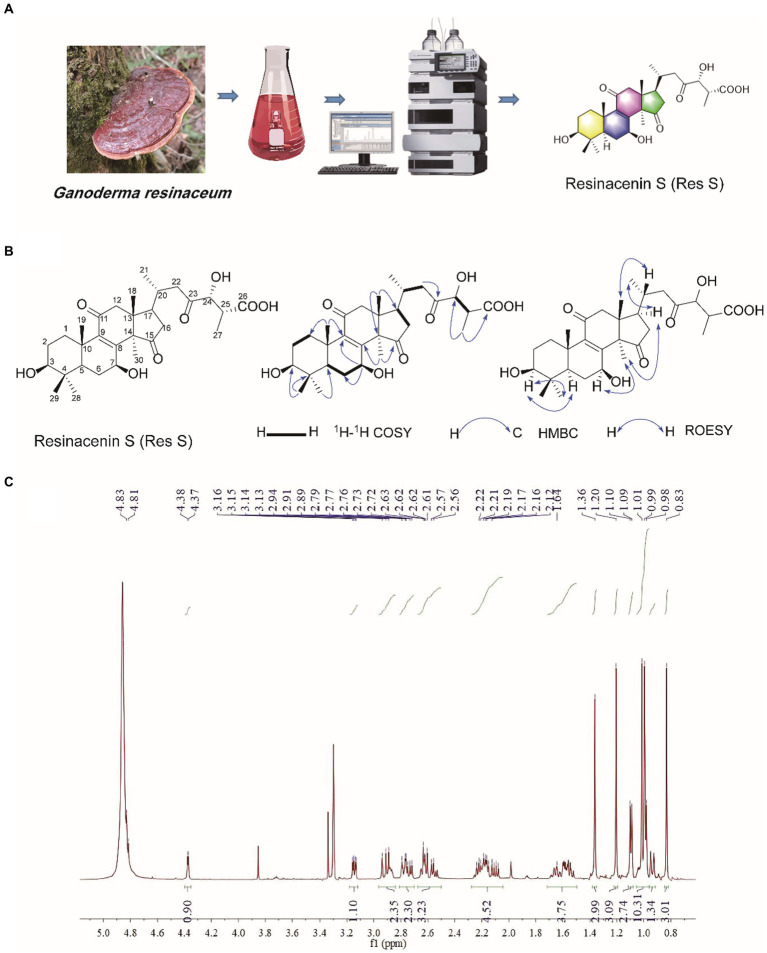
Resinacein S is a natural compound that has only been documented in G. resinaceum. We obtained Resinacein S white powder from the ethanol extracts of G. resinaceum through a series partition and column chromatography as previously described (Figure 1A) (13). Resinacein S was established as C30H44O8 based on the HRESIMS (Supplementary Figure S6), and the X-ray crystallographic structure of Resinacein S was obtained (Figure 1B). The 1H NMR and 13C NMR data of Resinacein S are shown in Table 1. The 1H NMR, 13C NMR and DEPT, HSQC, HMBC, 1H-1H COSY, and ROESY spectra are displayed in Figures 1B,C and Supplementary Figures S1–S6. Resinacein S was elucidated as (6R)-6-((3S,5R,7S,10S,13R,14R,17R)-3,7-dihydroxy-4,4,10,13,14-pentamethyl-11,15-dioxo-,3,4,5,6,7,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl)-3-hydroxy-2-methyl-4-oxo heptanoic acid.
Figure 1.
Resinacein S was obtained from Ganoderma resinaceum (A), and 1H–1H COSY correlations, key HMBC, ROESY correlations for Resinacein S (B), and 1H NMR spectrum (600 MHz) of Resinacein S in CD3OD (C).
Table 1.
1H NMR and 13C NMR Data (δ) for Resinacein S (600 MHz) in CD3OD (δ in ppm, J in Hz).
| Position | Resinacein S | Position | Resinacein S | ||
|---|---|---|---|---|---|
| δC, type | δH, (J in Hz) | δC, type | δH, (J in Hz) | ||
| 1 | 35.9, CH2 | 2.78, overlapped; 1.01, overlapped | 16 | 41.8, CH2 | 2.10, dd (19.3, 9.3); 2.74, dd (19.3, 7.8) |
| 2 | 28.2, CH2 | 1.65, m; 1.56, overlapped | 17 | 46.8, CH | 2.17, dd (18.5, 9.3) |
| 3 | 78.9, CH | 3.14, dd (11.7, 4.5) | 18 | 17.8, CH3 | 0.99, s |
| 4 | 39.7, C | 19 | 18.8, CH3 | 1.20, s | |
| 5 | 50.3, CH | 0.96, d (13.0) | 20 | 32.6, CH | 2.23, overlapped |
| 6 | 28.0, CH2 | 1.56, overlapped; 2.17, overlapped | 21 | 20.2, CH3 | 0.98, d (6.8) |
| 7 | 67.9, CH | 4.83, overlapped | 22 | 46.4, CH2 | 2.55, dd (17.7, 8.5); 2.62, overlapped |
| 8 | 158.9, C | 23 | 212.5, C | ||
| 9 | 144.1, C | 24 | 78.7, CH | 4.37, d (5.0) | |
| 10 | 39.9, C | 25 | 43.3, CH | 2.87, overlapped | |
| 11 | 200.4, C | 26 | 177.7, C | ||
| 12 | 51.5, CH2 | 2.92, d (16.7); 2.62, d (16.7) | 27 | 11.6, CH3 | 1.09, d (7.0) |
| 13 | 46.7, C | 28 | 28.7, CH3 | 1.01, s | |
| 14 | 60.4, C | 29 | 16.2, CH3 | 0.83, s | |
| 15 | 218.3, C | 30 | 24.9, CH3 | 1.36, s | |
3.2. Resinacein S alleviated liver damage and hepatic lipid accumulation
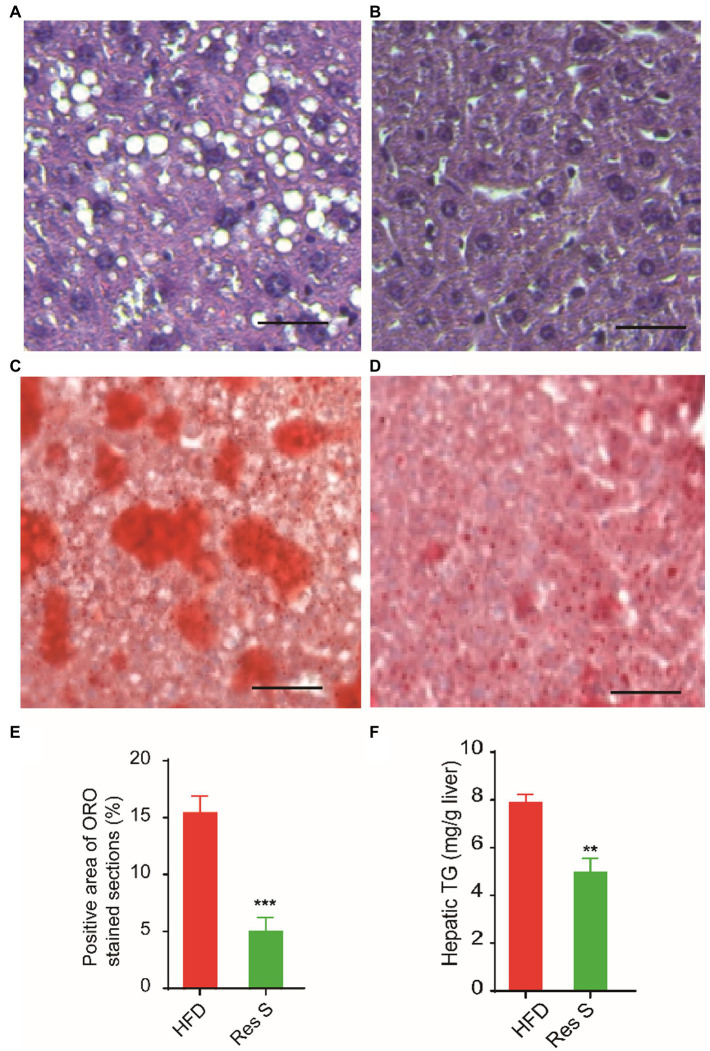
To investigate the benefits of Resinacein S against the HFD-induced extraphysiological process of NAFLD, we employed Resinacein S to treat HFD-diet mice and found that Resinacein S could effectively reduce liver damage and hepatic steatosis in HFD-fed mice (Figures 2A,B). In addition, Oil Red O staining of livers in Resinacein S-treated HFD-fed mice yielded few lipid droplets of small size. Compared to the control, the livers of Resinacein S-treated mice displayed an almost normal phenotype (Figures 2C–E). Consistently, significantly lower TG levels were found in the livers of the Resinacein S-treated group (Figure 2F).
Figure 2.
Resinacein S reduced liver damage and hepatic steatosis in HFD-fed mice. (A,B) H&E staining of livers in HFD-fed mice (A) and Resinacein S treated HFD-fed mice (B). (C–E) Oil Red O staining of livers in HFD-fed mice (C) and Resinacein S treated HFD-fed mice (D). (E) is the quantitative analysis of Oil Red O staining. (F) TG levels in livers of Resinacein S treated or not HFD-fed mice were detected. Values were means ± SD, and for statistical analysis, one-way ANOVA were performed between indicated groups.
3.3. Target recognition results of Resinacein S and nonalcoholic fatty liver disease
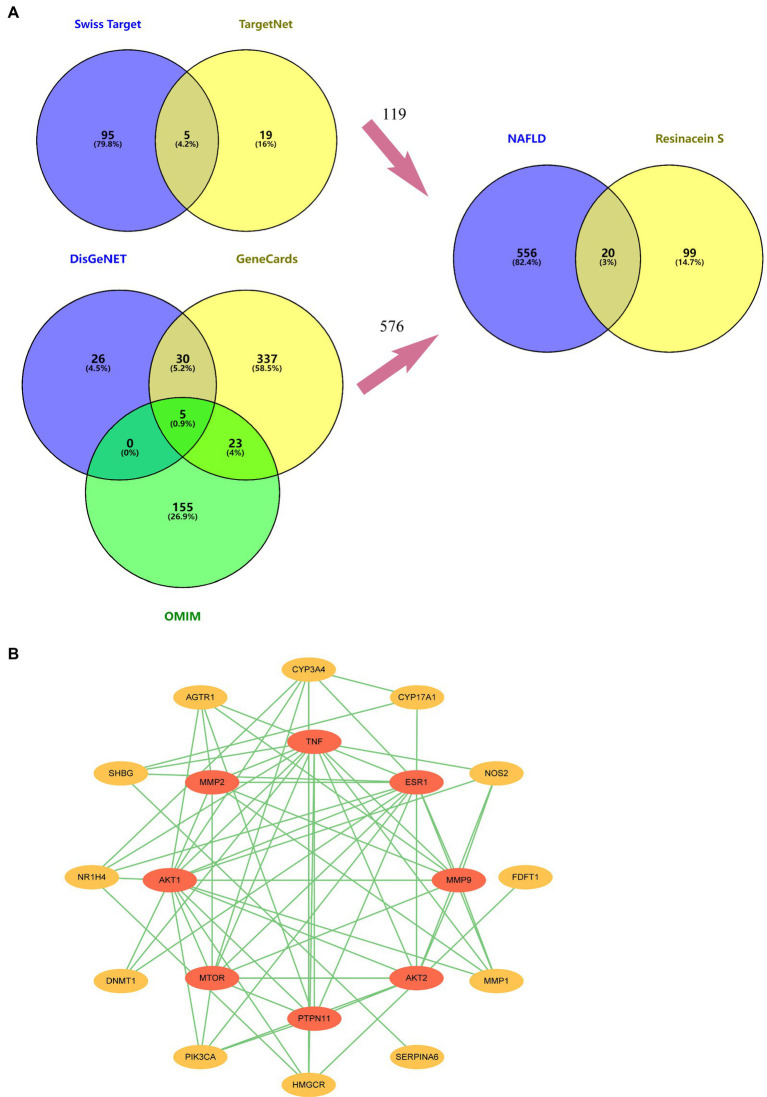
Twenty and 101 gene targets in human related to Resinacein S were obtained from SwissTargetPrediction and TargetNet databases, respectively. Moreover, 61, 395 and 183 gene targets related to NAFLD disease were obtained from DisGeNET, GeneCards and OMIM, respectively. The Venny 2.1.0 online system was used for target recognition (Figure 3A). One hundred nineteen action targets of Resinacein S were acquired, along with 576 NAFLD related genes. Finally, 20 action targets of Resinacein S against NAFLD come to light. Detailed information on the action targets is provided in Table 2.
Figure 3.
(A) 119 drug treatment targets based on the examination and combination of Swiss Target Prediction and TargetNet databases. Five hundred and seventy six genes related to NAFLD based on the examination and combination of DisGeNET, GeneCards and OMIM databases. There are 20 intersection targets between NAFLD and Resinacein S. (B) PPI protein interaction network diagram, in which the innermost circle degree value is 0–6, and the outer circle degree value is 7–16.
Table 2.
Twenty key target genes of Resinacein S related to NAFLD.
| Genes | Protein names | UniProt ID |
|---|---|---|
| NR1H4 | Bile acid receptor | Q96RI1 |
| CYP17A1 | Cytochrome P450 17A1 | P05093 |
| MMP1 | Matrix metalloproteinase 1 | P03956 |
| FDFT1 | Squalene synthetase | P37268 |
| TNF | TNF-alpha | P01375 |
| AGTR1 | Type-1 angiotensin II receptor (by homology) | P30556 |
| AKT1 | Serine/threonine-protein kinase AKT | P31749 |
| AKT2 | Serine/threonine-protein kinase AKT2 | P31751 |
| CYP3A4 | Cytochrome P450 3A4 | P08684 |
| DNMT1 | DNA (cytosine-5)-methyltransferase 1 | P26358 |
| ESR1 | Estrogen receptor alpha | P03372 |
| HMGCR | HMG-CoA reductase | P04035 |
| MMP2 | Matrix metalloproteinase 2 | P08253 |
| MMP9 | Matrix metalloproteinase 9 | P14780 |
| MTOR | Serine/threonine-protein kinase mTOR | P42345 |
| NOS2 | Nitric oxide synthase, inducible (by homology) | P35228 |
| PIK3CA | PI3-kinase p110-alpha subunit | P42336 |
| PTPN11 | Protein-tyrosine phosphatase 2C | Q06124 |
| SERPINA6 | Corticosteroid binding globulin | P08185 |
| SHBG | Testis-specific androgen-binding protein | P04278 |
3.4. Protein–protein interactions network results
The top 20 action targets of Resinacein S for NAFLD were put into the STRING database. Moreover, the PPI network was obtained according to the method described in 2.5. Data from the STRING database was put into Cytoscape 3.9.0 for beautification and classification according to the degree value. The degree values were classified into 0–6 and 7–16, and the protein–protein interaction network was generated (Figure 3B).
3.5. Resinacein S induced gene expression profiles in human liver cells
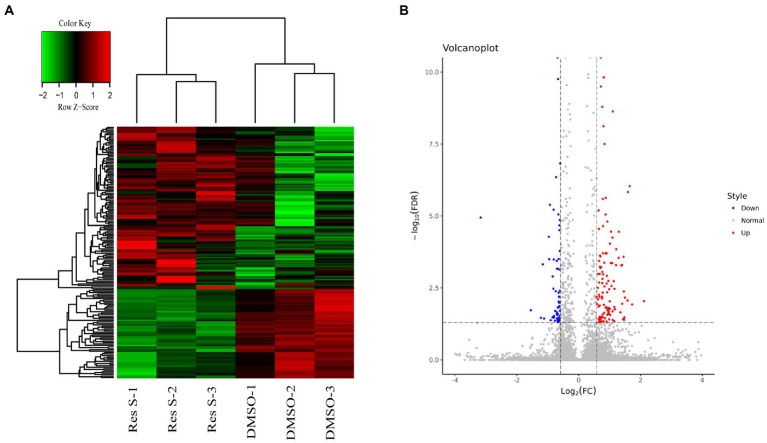
In order to further investigate the hub regulated genes of Resinacein S against NAFLD in human liver cells, we treated the human normal liver cell line L02 with Resinacein S and detected the gene expression profiles by RNA-Seq. According to log2FC > 0.585 or < −0.585 and FDR < 0.05, we obtained 172 DEGs (111 up-regulated and 61 down-regulated) in Resinacein S treated group compared to DMSO group (Figure 4, Supplementary Table 1).
Figure 4.
(A) The heatmap of the DEGs between Resinacein S treated group and DMSO group. (B) The volcano plot of the DEGs between Resinacein S treated group and DMSO group.
3.6. Functional enrichment analysis of Resinacein S induced genes in human liver cells
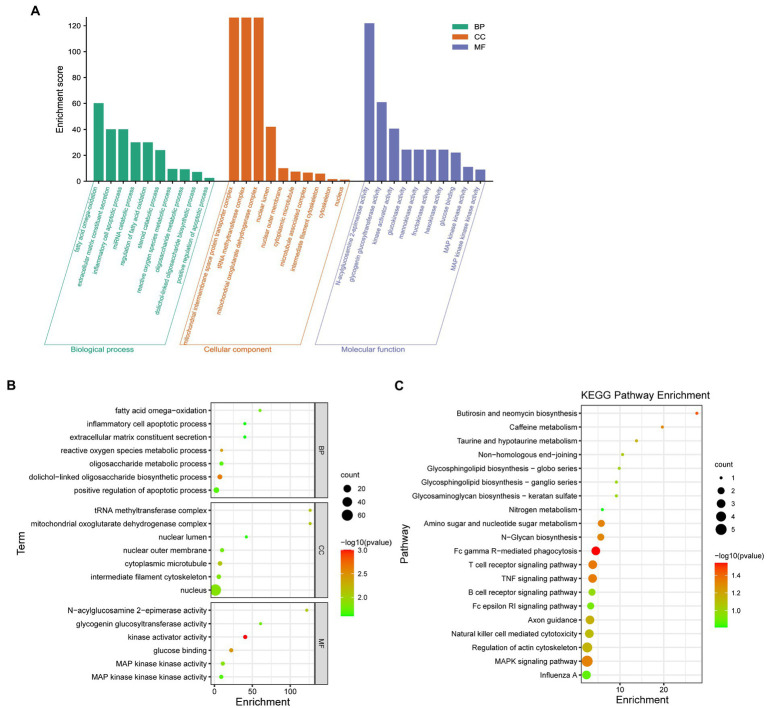
To study the biological function of Resinacein S induced genes in human liver cells, GO annotation was conducted. The significantly enriched GO terms in cellular component (CC), molecular function (MF) and biological process (BP) are shown in Figure 5A and Table 3. Significantly enriched GO terms associated with cellular components included mitochondrial oxoglutarate dehydrogenase complex, tRNA methyltransferase complex, mitochondrial oxoglutarate dehydrogenase complex, nuclear lumen, nuclear outer membrane, cytoplasmic microtubule, microtubule-associated complex, intermediate filament cytoskeleton, cytoskeleton, and nucleus. Significantly enriched GO terms associated with molecular function (MF) included N-acylglucosamine 2-epimerase activity, glycogenin glucosyltransferase activity, kinase activator activity, glucokinase activity, mannokinase activity, fructokinase activity, hexokinase activity, glucose binding, MAP kinase kinase activity, and MAP kinase kinase kinase activity. Significantly enriched GO terms associated with biological process (BP) consisted of fatty acid omega-oxidation, extracellular matrix constituent secretion, inflammatory cell apoptotic process, miRNA catabolic process, regulation of fatty acid oxidation, steroid catabolic process, reactive oxygen species metabolic process, oligosaccharide metabolic process, dolichol-linked oligosaccharide biosynthetic process, and positive regulation of the apoptotic process.
Figure 5.
GO and KEGG pathway enrichment analysis was conducted with Resinacein S induced genes in human liver cells. (A) The categories in GO terms included cellular components (CC), molecular function (MF), and biological process (BP) were analyzed. (B) The top 20 significant GO terms of the overlapped DEGs were chosen based on the order of p value from small to large. (C) KEGG pathway enrichment analysis was performed with Resinacein S-induced genes. The top 20 significant KEGG pathways of the DEGs were chosen based on the order of p value from small to large.
Table 3.
Top 20 significant GO terms of the overlapped DEGs in livers.
| Category | GO ID | Term | List | p value |
|---|---|---|---|---|
| MF | GO:0019209 | Kinase activator activity | 2 | 0.000981 |
| BP | GO:0006488 | Dolichol-linked oligosaccharide biosynthetic process | 4 | 0.002334 |
| MF | GO:0005536 | Glucose binding | 2 | 0.0035 |
| BP | GO:0072593 | Reactive oxygen species metabolic process | 1 | 0.00383 |
| CC | GO:0005881 | Cytoplasmic microtubule | 3 | 0.007664 |
| CC | GO:0009353 | Mitochondrial oxoglutarate dehydrogenase complex | 1 | 0.007916 |
| CC | GO:0043527 | tRNA methyltransferase complex | 1 | 0.007916 |
| MF | GO:0050121 | N-acylglucosamine 2-epimerase activity | 1 | 0.008203 |
| CC | GO:0005634 | Nucleus | 64 | 0.013785 |
| MF | GO:0004708 | MAP kinase kinase activity | 2 | 0.013861 |
| CC | GO:0045111 | Intermediate filament cytoskeleton | 3 | 0.014839 |
| MF | GO:0008466 | Glycogenin glucosyltransferase activity | 1 | 0.016339 |
| BP | GO:0010430 | Fatty acid omega-oxidation | 1 | 0.016551 |
| CC | GO:0005640 | Nuclear outer membrane | 2 | 0.016571 |
| BP | GO:0009311 | Oligosaccharide metabolic process | 2 | 0.019557 |
| BP | GO:0043065 | Positive regulation of apoptotic process | 7 | 0.020125 |
| MF | GO:0004709 | MAP kinase kinase kinase activity | 2 | 0.020507 |
| CC | GO:0031981 | Nuclear lumen | 1 | 0.023562 |
| BP | GO:0070278 | Extracellular matrix constituent secretion | 1 | 0.024724 |
| BP | GO:0006925 | Inflammatory cell apoptotic process | 1 | 0.024724 |
The top 20 GO terms associated with Resinacein S induced genes in human liver cells were selected according to the order of value of p in ascending order (Figure 5B; Table 3). It was found that the DEGs were mainly enriched in the fatty acid omega-oxidation, dolichol-linked oligosaccharide biosynthetic process, glucose binding, inflammatory cell apoptotic process, glycogenin glucosyltransferase activity, and fatty acid omega-oxidation, etc. These functions were closely related to the physiological regulation of liver metabolism, especially the metabolic homeostasis of lipids and cholesterol.
KEGG enrichment analysis was carried out to explore the biological pathways of the Resinacein S-induced DEGs (Figure 5C; Table 4). We found that the DEGs were significantly enriched in KEGG pathways, including T cell receptor signaling pathway, N-Glycan biosynthesis, Amino sugar and nucleotide sugar metabolism, Glycosphingolipid biosynthesis-globo series, MAPK signaling pathway, and TNF signaling pathway, etc.
Table 4.
Top 20 significant enriched pathways of the overlapped DEGs in livers.
| Category | Pathway ID | p value | Genes |
|---|---|---|---|
| Fc gamma R-mediated phagocytosis | PATH:04666 | 0.028775 | VAV3/SCIN/LIMK2 |
| Butirosin and neomycin biosynthesis | PATH:00524 | 0.035794 | HKDC1 |
| T cell receptor signaling pathway | PATH:04660 | 0.044183 | MAP3K8/VAV3/MALT1 |
| TNF signaling pathway | PATH:04668 | 0.045195 | MAP3K8/FAS/MAP2K6 |
| Amino sugar and nucleotide sugar metabolism | PATH:00520 | 0.047149 | RENBP/HKDC1 |
| MAPK signaling pathway | PATH:04010 | 0.047367 | MAP3K8/FAS/MAP2K5 MAP2K6/CACNA1H |
| N-Glycan biosynthesis | PATH:00510 | 0.048932 | ALG9/DOLPP1 |
| Caffeine metabolism | PATH:00232 | 0.049758 | NAT1 |
| Axon guidance | PATH:04360 | 0.069072 | LIMK2/ABLIM3/NTN4 |
| Taurine and hypotaurine metabolism | PATH:00430 | 0.070332 | CSAD |
| Regulation of actin cytoskeleton | PATH:04810 | 0.071639 | VAV3/SCIN/LIMK2 IQGAP2 |
| Natural killer cell mediated cytotoxicity | PATH:04650 | 0.076769 | FAS/VAV3/ULBP3 |
| Non-homologous end-joining | PATH:03450 | 0.09047 | POLM |
| Glycosphingolipid biosynthesis - globo series | PATH:00603 | 0.097087 | ST3GAL1 |
| Glycosaminoglycan biosynthesis - keratan sulfate | PATH:00533 | 0.103657 | ST3GAL1 |
| Glycosphingolipid biosynthesis - ganglio series | PATH:00604 | 0.103657 | ST3GAL1 |
| B cell receptor signaling pathway | PATH:04662 | 0.104971 | VAV3/MALT1 |
| Fc epsilon RI signaling pathway | PATH:04664 | 0.121474 | VAV3/MAP2K6 |
| Influenza A | PATH:05164 | 0.133377 | FAS/RAE1/MAP2K6 |
| Nitrogen metabolism | PATH:00910 | 0.154559 | CA5B |
Overall, these metabolic pathways are closely related to liver metabolic balance, which substantiates the protective effect of Resinacein S against NAFLD.
3.7. The hub proteins of Resinacein S regulated against nonalcoholic fatty liver disease
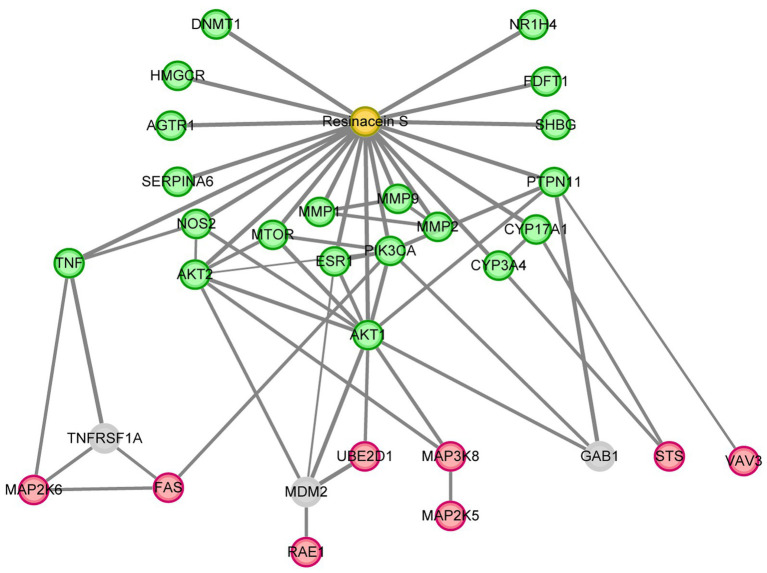
To determine the hub proteins that mediate the protective role of Resinacein S against NAFLD in human liver cells, we analyzed the interaction relationships of 172 DEGs of Resinacein S regulated in human normal liver cell line L02 according to RNA-Seq (Figure 3), and 20 target genes of Resinacein S against NAFLD from public database (Figure 4). The relationships were obtained by the STRING database, including evidence from experiments, databases, and co-expression data, and binding scores greater than 0.7 (high confidence). Therefore, according to the STRING analysis, the Resinacein S-mediated junction proteins against NAFLD were TNF, PIK3CA, AKT1, AKT2, ESR1, CYP3A4, CYP17A1, and PTPN11 (Figure 6). Among them, AKT1 and AKT2 play an important role in the interaction relationships which indicated the regulation of AKT pathway by Resinacein S against NAFLD.
Figure 6.
The hub proteins of Resinacein S regulated against NAFLD. The interaction relationship analysis was performed with the DEGs of Resinacein S regulated in human liver cells and the target genes of Resinacein S against NAFLD from public database. DEGs of Resinacein S regulated in human normal liver cells were shown in red circle, and the 20 target genes of Resinacein S against NAFLD from public database were shown in green. The analysis was generated by Cytoscape 3.9.0.
4. Discussion
NAFLD is widely thought to be the most common chronic liver disease, concomitant with the global obesity epidemic. During the development and progress of NAFLD, liver steatosis is considered a benign state, while NASH exhibits chronic progressive liver damage. Fatty degeneration is related to inflammation and fibrosis and gradually develops into cirrhosis. The pathogenesis of NAFLD is closely related to dysregulated metabolism in obese patients, excess free fatty acid (FFA) accumulation and the excessive secretion of several cellular inflammatory mediators (14). There is an increasing consensus that NAFLD is a heterogeneous disease associated with multiple-hit pathogenesis yielding different phenotypes. Although the clinical presentation of NAFLD can be heterogeneous, insulin resistance plays a leading role in NAFLD (15).
In this study, we focused on the role of the Resinacein S compound extracted from G. resinaceum in treating NAFLD patients. First, we identified the extracted triterpenoid compounds and obtained the molecular structure of Resinacein S through mass spectrometry analysis. By interaction relationship analysis of the DEGs of Resinacein S regulated in human liver cells and the target genes of Resinacein S against NAFLD from public database, we found 8 proteins TNF, PIK3CA, AKT1, AKT2, ESR1, CYP3A4, CYP17A1, and PTPN11 may be the hub proteins of Resinacein S regulated against NAFLD.
According to former studies, Resinacein S treatment can dramatically induce the expression of thermogenesis related genes such as Ucp1 and Pgc1α, fatty acid oxidation related genes such as Pparα and Cpt1α, lipolysis related genes such as Hsl and Atgl, and mitochondriogenesis-related genes. Resinacein S may be involved in activation of AMPK/PGC1α signaling pathway (6). Now we obtained eight Resinacein S targeting genes with potential therapeutic effects against NAFLD through RNA seq analysis and PPI network analysis.
Obesity is related to the increase of circulating Tumor Necrosis Factor (TNF, TNF-α), a pro-inflammatory cytokine that induces the death of liver cells (16). In the process of inflammation, TNF is one of the main pro-inflammatory cytokines, which regulates innate immunity and adaptive immune response (17). Clinical evidence shows that the level of circulating TNF-α in patients with nonalcoholic steatohepatitis (NASH) is highly correlated with the degree of liver fibrosis (18). AKT Serine/Threonine Kinase 1 (AKT1) is a serine/threonine protein kinase, which is called an important downstream target signal pathway of insulin and has anti-apoptosis and peripheral metabolic effects. Studies have shown that the inhibition of ROS production mediated by AKT1 inhibits the fibrosis transformation from NAFLD to liver (19). AKT1 deficiency led to the inhibition of AKT/mTOR/S6K signaling pathway in hepatocytes, which was crucial for the development of hepatic steatosis and provided a new scheme for the development of NAFLD therapy (20). Studies have shown that AKT Serine/Threonine Kinase 2 (AKT2) is essential for lipid synthesis in the liver (21). It was found that hepatocyte-specific Phosphatase and Tensin Homolog (PTEN) deficiency in mice showed liver steatosis due to over-activation of AKT2 (22). As the precursor of NAFLD, deleting AKT2, the downstream target of PTEN signal, can block the development of NASH and reduce the development of liver fibrosis, which also reduces the occurrence of NAFLD from another development process (23). Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA) is an important component of PI3K/AKT/mTOR pathway. PIK3CA mutated cells showed inflated induction of the de novo lipogenesis transcriptional regulator SREBP1 and elevated exogenous FA uptake capacity both of which can lead to lipid-enriched phenotype (24, 25). ESR1 is a nuclear and membrane hormone receptor. Previous studies showed that ESR1 could negatively regulate hepatocyte pyroptosis by directly interacting with gasdermin D (GSDMD). ESR1 deficiency could induce pyroptosis, impaired glucose tolerance, and reduce lipid accumulation in hepatocytes (26–28). Cytochrome P450 Family 3 Subfamily A Member 4 (CYP3A4) is involved in the metabolism of sterols, retinoids and fatty acids. The activity of CYP3A4 is usually reduced in mouse and cell models of NAFLD, also in human. In NAFLD-model mice, the luciferase activity of CYP3A4 from livers was about 38% lower than the normal ones (29, 30). Cytochrome P450 Family 17 Subfamily A Member 1 (CYP17A1) is an important enzyme for Dehydroepiandrosterone (DHEA) synthesis. Former evidences showed that DHEA can modulate oxidative stress, insulin resistance, and fibrosis observed in serious NAFLD (31, 32). Protein Tyrosine Phosphatase Non-Receptor Type 11 (PTPN11) is the first identified oncogenic tyrosine phosphatase which can cooperate with PTEN to maintain the liver homeostasis and function. PTPN11 in hepatocytes could induce early-onset non-alcoholic steatohepatitis (NASH) (33).
Besides, according to our KEGG enrichment analysis, we revealed that the targets of Resinacein S mainly focused on T cell receptor signaling pathway, N-Glycan biosynthesis, Amino sugar and nucleotide sugar metabolism, Glycosphingolipid biosynthesis-globo series, MAPK signaling pathway, and TNF signaling pathway.
In conclusion, we have revealed the structure of Resinacein S and demonstrated that Resinacein S could significantly attenuate high-fat diet induced hepatic steatosis and hepatic lipid accumulation. We have also developed a new gene expression feature related to NAFLD by using Resinacein S-dependent DEGs, especially hub proteins in PPI network analysis, which can assist in diagnosing and treating NAFLD populations as well as drug discovery and development in the near future. Nevertheless, although the extracts of Ganoderma are usually safe and most of the side effects are very mild, the side effect of Resinacein S deserves further studies especially on human. Besides, the molecular mechanisms of Resinacein S against NAFLD and in vivo models for further proving the metabolic phenotypes of Resinacein S need to be further investigated.
5. Conclusion
Overall, Resinacein S yields a protective effect against steatosis and liver injury and can significantly change the gene expression profile in fatty liver cells. Functional enrichment analysis showed significant enrichment in lipid and glucose metabolism. In addition, intersected genes between NAFLD and Resinacein S-induced DEGs, especially the hub protein in PPI network analysis, can be used to characterize NAFLD-related gene expression characteristics for NAFLD diagnosis, treatment and drug development.
Data availability statement
The data presented in the study are deposited in the GEO repository, accession number GSE223990.
Ethics statement
The animal study was reviewed and approved by Animal Care and Use Committee of Shanghai Tenth People’s Hospital of Tongji University.
Author contributions
S-QC, GW, and F-FM conceived the project. The data analysis was done by FM, S-SG, and Y-JH. F-FM and S-SG wrote the drafts of the manuscript. Y-JH checked and revised the manuscript. NZ and J-KF participated in collection of the published datasets. Z-HL, Y-QZ, and L-YY gave many suggestions about the manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
This work was partially supported by grants from the National Natural Science Foundation of China (Nos. 82002480, 8200032).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank NovelBio Bio-Pharm Technology Co., Ltd. (Shanghai, China) for their assistance on RNA-seq data analysis.
Footnotes
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2023.1076569/full#supplementary-material
References
- 1.Unlu A, Nayir E, Kirca O, Ozdogan M. (2016) Ganoderma lucidum (Reishi mushroom) and cancer. J BUON. 21:792–8. [PubMed] [Google Scholar]
- 2.Ahmad R, Riaz M, Khan A, Aljamea A, Algheryafi M, Sewaket D, et al. (2021) Ganoderma lucidum (Reishi) an edible mushroom; a comprehensive and critical review of its nutritional, cosmeceutical, mycochemical, pharmacological, clinical, and toxicological properties. Phytother Res. 35:6030–62. doi: 10.1002/ptr.7215, PMID: [DOI] [PubMed] [Google Scholar]
- 3.Cizmarikova M. (2017) The efficacy and toxicity of using the Lingzhi or Reishi medicinal mushroom, Ganoderma lucidum (Agaricomycetes), and its products in chemotherapy (review). Int J Med Mushrooms. 19:861–77. doi: 10.1615/IntJMedMushrooms.2017024537, PMID: [DOI] [PubMed] [Google Scholar]
- 4.Sanodiya BS, Thakur GS, Baghel RK, Prasad GB, Bisen PS. (2009) Ganoderma lucidum: a potent pharmacological macrofungus. Curr Pharm Biotechnol. 10:717–42. doi: 10.2174/138920109789978757, PMID: [DOI] [PubMed] [Google Scholar]
- 5.Niu XM, Li SH, Xiao WL, Sun HD, Che CT. (2007) Two new Lanostanoids from Ganoderma resinaceum. J Asian Nat Prod Res. 9:659–64. doi: 10.1080/10286020600979910, PMID: [DOI] [PubMed] [Google Scholar]
- 6.Huang Y, Wei G, Peng X, Hu G, Su H, Liu J, et al. (2020) Triterpenoids from functional mushroom Ganoderma resinaceum and the novel role of Resinacein S in enhancing the activity of Brown/beige adipocytes. Food Res Int. 136:109303. doi: 10.1016/j.foodres.2020.109303, PMID: [DOI] [PubMed] [Google Scholar]
- 7.Eslam M, Valenti L, Romeo S. (2018) Genetics and epigenetics of NAFLD and NASH: clinical impact. J Hepatol. 68:268–79. doi: 10.1016/j.jhep.2017.09.003, PMID: [DOI] [PubMed] [Google Scholar]
- 8.Raza S, Rajak S, Upadhyay A, Tewari A, Anthony SR. (2021) Current treatment paradigms and emerging therapies for NAFLD/NASH. Front Biosci. 26:206–37. doi: 10.2741/4892, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao M, Zhang R, Wang Y, Mao Z, Wu J, Guo H, et al. (2022) Corilagin prevents non-alcoholic fatty liver disease via improving lipid metabolism and glucose homeostasis in high fat diet-fed mice. Front Nutr. 9:983450. doi: 10.3389/fnut.2022.983450, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Zhou F, Wang W, Zhang XJ, Ji YX, Zhang P, et al. (2020) Epidemiological features of NAFLD from 1999 to 2018 in China. Hepatology. 71:1851–64. doi: 10.1002/hep.31150, PMID: [DOI] [PubMed] [Google Scholar]
- 11.Manne V, Handa P, Kowdley KV. (2018) Pathophysiology of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. Clin Liver Dis. 22:23–37. doi: 10.1016/j.cld.2017.08.007 [DOI] [PubMed] [Google Scholar]
- 12.Sui Y, Lu K, Fu L. (2021) Prediction and analysis of novel key genes ITGAX, LAPTM5, SERPINE1 in clear cell renal cell carcinoma through bioinformatics analysis. PeerJ. 9:e11272. doi: 10.7717/peerj.11272, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oyetayo OV. (2011) Medicinal uses of mushrooms in Nigeria: towards full and sustainable exploitation. Afr J Tradit Complement Altern Med. 8:267–74. doi: 10.4314/ajtcam.v8i3.65289, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Wang N, Tan HY, Li S, Zhang C, Feng Y. (2021) Gut-liver Axis modulation of Panax Notoginseng Saponins in nonalcoholic fatty liver disease. Hepatol Int. 15:350–65. doi: 10.1007/s12072-021-10138-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goh GB, Pagadala MR, Dasarathy J, Unalp-Arida A, Sargent R, Hawkins C, et al. (2015) Clinical spectrum of non-alcoholic fatty liver disease in diabetic and non-diabetic patients. BBA Clin. 3:141–5. doi: 10.1016/j.bbacli.2014.09.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ezquerro S, Mocha F, Fruhbeck G, Guzman-Ruiz R, Valenti V, Mugueta C, et al. (2019) Ghrelin reduces TNF-alpha-induced human hepatocyte apoptosis, autophagy, and pyroptosis: role in obesity-associated NAFLD. J Clin Endocrinol Metab. 104:21–37. doi: 10.1210/jc.2018-01171, PMID: [DOI] [PubMed] [Google Scholar]
- 17.Jorge ASB, Andrade JMO, Paraiso AF, Jorge GCB, Silveira CM, de Souza LR, et al. (2018) Body mass index and the visceral adipose tissue expression of Il-6 and TNF-alpha are associated with the morphological severity of non-alcoholic fatty liver disease in individuals with class III obesity. Obes Res Clin Pract. 12:1–8. doi: 10.1016/j.orcp.2016.03.009, PMID: [DOI] [PubMed] [Google Scholar]
- 18.Ma R, Zhan Y, Zhang Y, Wu L, Wang X, Guo M. (2022) Schisandrin B ameliorates non-alcoholic liver disease through anti-inflammation activation in diabetic mice. Drug Dev Res. 83:735–44. doi: 10.1002/ddr.21905, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Q, Wei S, Wen J, Zhang W, Jiang Y, Qu C, et al. (2020) Network pharmacology reveals the multiple mechanisms of Xiaochaihu decoction in the treatment of non-alcoholic fatty liver disease. BioData Min. 13:11. doi: 10.1186/s13040-020-00224-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li CH, Tang SC, Wong CH, Wang Y, Jiang JD, Chen Y. (2018) Berberine induces miR-373 expression in hepatocytes to inactivate hepatic steatosis associated AKT-S6 kinase pathway. Eur J Pharmacol. 825:107–18. doi: 10.1016/j.ejphar.2018.02.035, PMID: [DOI] [PubMed] [Google Scholar]
- 21.Pan Q, Qin T, Gao Y, Li S, Li D, Peng M, et al. (2019) Hepatic mTOR-AKT2-Insig2 signaling pathway contributes to the improvement of hepatic steatosis after Roux-en-Y gastric bypass in mice. Biochim Biophys Acta Mol basis Dis. 1865:525–34. doi: 10.1016/j.bbadis.2018.12.014, PMID: [DOI] [PubMed] [Google Scholar]
- 22.Schultze SM, Dietrich M, Hynx D, Geier A, Niessen M, Spinas GA, et al. (2015) Reduced hepatic lipid content in Pten-Haplodeficient mice because of enhanced AKT2/PKBbeta activation in skeletal muscle. Liver Int. 35:1354–66. doi: 10.1111/liv.12600, PMID: [DOI] [PubMed] [Google Scholar]
- 23.He L, Gubbins J, Peng Z, Medina V, Fei F, Asahina K, et al. (2016) Activation of hepatic stellate cell in Pten null liver injury model. Fibrogenesis Tissue Repair. 9:8. doi: 10.1186/s13069-016-0045-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koundouros N, Karali E, Tripp A, Valle A, Inglese P, Perry NJS, et al. (2020) Metabolic fingerprinting links oncogenic Pik3ca with enhanced arachidonic acid-derived eicosanoids. Cells. 181:1596–1611.e27. doi: 10.1016/j.cell.2020.05.053, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricoult SJH, Yecies JL, Ben-Sahra I, Manning BD. (2016) Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene. 35:1250–60. doi: 10.1038/onc.2015.179, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao X, Liu S, Tan L, Ding C, Fan W, Gao Z, et al. (2021) Estrogen receptor alpha regulates metabolic-associated fatty liver disease by targeting NLRP3-GSDMD axis-mediated hepatocyte pyroptosis. J Agric Food Chem. 69:14544–56. doi: 10.1021/acs.jafc.1c05400, PMID: [DOI] [PubMed] [Google Scholar]
- 27.Palmisano BT, Zhu L, Stafford JM. (2017) Role of estrogens in the regulation of liver lipid metabolism. Adv Exp Med Biol. 1043:227–56. doi: 10.1007/978-3-319-70178-3_12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen KL, Madak-Erdogan Z. (2018) Estrogens and female liver health. Steroids. 133:38–43. doi: 10.1016/j.steroids.2017.10.015 [DOI] [PubMed] [Google Scholar]
- 29.Woolsey SJ, Mansell SE, Kim RB, Tirona RG, Beaton MD. (2015) CYP3A activity and expression in nonalcoholic fatty liver disease. Drug Metab Dispos. 43:1484–90. doi: 10.1124/dmd.115.065979, PMID: [DOI] [PubMed] [Google Scholar]
- 30.Woolsey SJ, Beaton MD, Mansell SE, Leon-Ponte M, Yu J, Pin CL, et al. (2016) A fibroblast growth factor 21-Pregnane X receptor pathway downregulates hepatic CYP3A4 in nonalcoholic fatty liver disease. Mol Pharmacol. 90:437–46. doi: 10.1124/mol.116.104687, PMID: [DOI] [PubMed] [Google Scholar]
- 31.Charlton M, Angulo P, Chalasani N, Merriman R, Viker K, Charatcharoenwitthaya P, et al. (2008) Low circulating levels of Dehydroepiandrosterone in histologically advanced nonalcoholic fatty liver disease. Hepatology. 47:484–92. doi: 10.1002/hep.22063, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Wang H, Yao Y, Cao J, Jiang Z, Yan W, et al. (2021) The sex steroid precursor Dehydroepiandrosterone prevents nonalcoholic steatohepatitis by activating the AMPK pathway mediated by GPR30. Redox Biol. 48:102187. doi: 10.1016/j.redox.2021.102187, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo X, Liao R, Hanley KL, Zhu HH, Malo KN, Hernandez C, et al. (2016) Dual Shp2 and Pten deficiencies promote non-alcoholic steatohepatitis and genesis of liver tumor-initiating cells. Cell Rep. 17:2979–93. doi: 10.1016/j.celrep.2016.11.048, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in the study are deposited in the GEO repository, accession number GSE223990.